Abstract
Small nucleolar RNAs (snoRNAs) and small Cajal body-specific RNAs (scaRNAs) are non-coding RNAs involved in the maturation of other RNA molecules and generally located in the introns of host genes. It is now emerging that altered sno/scaRNAs expression may have a pathological role in cancer. This study elucidates the patterns of sno/scaRNAs expression in multiple myeloma (MM) by profiling purified malignant plasma cells from 55 MMs, 8 secondary plasma cell leukemias (sPCLs) and 4 normal controls. Overall, a global sno/scaRNAs downregulation was found in MMs and, even more, in sPCLs compared with normal plasma cells. Whereas SCARNA22 resulted the only sno/scaRNA characterizing the translocation/cyclin D4 (TC4) MM, TC2 group displayed a distinct sno/scaRNA signature overexpressing members of SNORD115 and SNORD116 families located in a region finely regulated by an imprinting center at 15q11, which, however, resulted overall hypomethylated in MMs independently of the SNORD115 and SNORD116 expression levels. Finally, integrative analyses with available gene expression and genome-wide data revealed the occurrence of significant sno/scaRNAs/host genes co-expression and the putative influence of allelic imbalances on specific snoRNAs expression. Our data extend the current view of sno/scaRNAs deregulation in cancer and add novel information to the bio-molecular complexity of plasma cell dyscrasias.
Keywords: ncRNA, snoRNA, multiple myeloma.
Introduction
Multiple myeloma (MM) is a fatal B-cell malignancy characterized by a heterogeneous clinical course and a remarkable genomic instability that encompasses a large array of structural and numerical chromosomal aberrations.1 So far, many efforts have been undertaken to define the different molecular types of MM, with the aim to dissect the clinical heterogeneity of the disease, identify patients at higher risk of progression or relapse and provide insights for the development of more effective therapeutic regimens. Furthermore, global gene expression profiling of purified myeloma cells identified peculiar tumor-associated transcriptional profiles, which are able to discriminate between normal and tumor phenotypes, or specifically associated with distinct MM molecular subtypes with different prognosis.2, 3
In recent years, the increasing discovery of non-coding RNAs (ncRNAs) have changed the landscape of cancer biology. ncRNAs are functional transcripts that do not code for proteins, however, they have a major role in regulating almost every level of gene expression. In addition to highly abundant and functionally important transfer and ribosomal RNAs (tRNA and rRNA), ncRNAs include a large variety of molecules such as microRNAs (miRNAs), short interfering RNAs, piwi-associated RNAs, small nucleolar RNAs (snoRNAs), small Cajal body-specific RNAs (scaRNAs), small nuclear RNAs (snRNAs) and long ncRNAs that are still only partially understood.4 Among these, miRNAs have been extensively studied in carcinogenesis.4 In the context of MM, we and other authors have recently shown that deregulated patterns of miRNA's expression are associated with distinct clinical and molecular entities of disease, giving evidence of their pathogenetic role.5, 6
snoRNAs are molecules of 60–300 nucleotides in length that associate with specific proteins, highly conserved among eukaryotes, to form small nucleolar ribonucleoparticles. Two main groups of snoRNAs have been described: the box H/ACA snoRNAs (SNORAs), which bind conserved core box H/ACA snoRNP proteins, such as DKC1, GAR1, NHP2 and NOP10, mainly acting as guide RNAs in the site-specific pseudouridylation of target rRNA; and the box C/D snoRNAs (SNORDs), which bind conserved core box C/D snoRNP proteins, such as fibrillarin, NOP56, NOP5/NOP58 and NHP2L1, involved in 2′-o-ribose methylation. The peculiar nucleolar localization of snoRNAs provides a direct connection with their classical function: in fact, most snoRNAs function as guide RNAs for the post-transcriptional modification of ribosomal RNAs and some spliceosomal RNAs, with a few others involved in nucleolytic processing of the original rRNA transcript. These post-transcriptional modifications are important for the production of efficient and accurate ribosomes. Unlike snoRNAs, scaRNAs accumulate within the Cajal bodies (conserved subnuclear organelles that are present in the nucleoplasm); moreover, scaRNA molecules, such as U92, ACA47, ACA11, U109 and ACA57, can possess both box C/D and box H/ACA sequence motifs, thus guiding both the methylation and pseudouridylation of snRNAs as reported for U85.7 sno/scaRNAs can also target other RNAs, including snRNAs and mRNAs. However, a subgroup of so-called ‘orphan' sno/scaRNAs exists that lacks any apparent complementarities to cellular RNAs and their function remains to be clarified. Notably, in vertebrates, most sno/scaRNAs reside in the introns of protein-coding host genes and are processed out of the excised introns.8
Experimental evidence has recently shown that sno/scaRNAs may have malfunctioning roles in human cancers, suggesting that they could be actively involved in tumorigenesis.9 However, data concerning the involvement of sno/scaRNAs in cancer, and hematological malignancies in particular, are extremely limited. Recently, a global downregulation of snoRNAs in acute leukemia cells when compared with non-neoplastic counterparts has been described along with the significant ectopic expression of SNORD112-114 in acute promyelocytic leukemia.10 Lopez-Corral et al.11 reported the overexpression of SNORD25, SNORD27, SNORD30, and SNORD31 in smoldering MM patients showing a rapid progression to symptomatic disease. Very recently, Chu et al.12 reported the upregulation of SCARNA22 in MMs harboring the t(4;14); notably, this scaRNA is localized within intron 18 of the WHSC1/MMSET gene involved in the translocation and constitutively expressed in these patients. Interestingly, these authors provided experimental evidence that SCARNA22 can suppress oxidative stress both in vitro and in vivo, facilitate cell proliferation and protect cells from the effects of chemotherapy, suggesting an oncogenic role for this type of ncRNA in MM.
In the present study, we investigated the sno/scaRNA expression profiling in a molecularly characterized cohort of MM and secondary plasma cell leukemia (sPCL) patients. Furthermore, sno/scaRNA profiles have been correlated with the expression levels of the respective host-genes and the allelic imbalances involving their genomic location.
Materials and methods
Samples
Bone marrow specimens from 55 newly diagnosed untreated MM and 8 sPCL patients, all described in our previous reports,13, 14, 15 were obtained during standard diagnostic procedures after informed consent in accordance with the Institutional guidelines. The diagnosis of MM was made according to previously described criteria.16 Plasma cells (PCs) were purified from bone marrow samples using CD138 immunomagnetic microbeads (MidiMACS system, Miltenyi Biotec, Auburn, CA) as previously described.2, 17 PCs from tonsils of four healthy donors were also purified and included in the study. The purity of the positively selected PCs was assessed by means of morphology and flow cytometry and was ⩾90% in all the cases. The 55 MM used for microarray analysis were representative of the major molecular characteristics of the disease. All of the samples were characterized for the presence of the most frequent chromosomal translocations and the ploidy status based on fluorescence in situ hybridization (FISH) evaluation criteria.18 MM samples were stratified in five groups according to the translocation/cyclin D (TC) classification.19, 20 Specifically, 12 samples were in the TC1 group, characterized by the t(11;14) or t(6;14) translocations, with the consequent overexpression of either cyclin D1 (CCND1) or CCND3 and a non-hyperdiploid (HD) status; 13 in the TC2, showing low-to-moderate levels of the CCND1 gene in the absence of any primary immunoglobulin H (IGH) translocation and HD status; 12 in the TC3, including tumors that do not fall into any of the other groups, most of which express CCND2; 12 in the TC4, showing high CCND2 levels and the presence of the t(4;14) translocation; and 6 in the TC5, expressing the highest levels of CCND2 in association with either the t(14;16) or t(14;20) translocations. Deletions of chromosome 17p13, 13q14 and gain of chromosome 1q were also evaluated by FISH. Clinical and biomolecular features concerning all the investigated patients are reported in Supplementary Table S1.
Gene expression profiling
The MM-sPCL panel and tonsillar-purified PCs from four normal controls were analyzed on the GeneChip Human Gene 1.0 ST array (Affymetrix, Santa Clara, CA). The raw intensity expression values were processed by Robust Multi-array Average procedure,21 with the re-annotated Chip Definition Files from BrainArray libraries version 15.0.022 available at http://brainarray.mbni.med.umich.edu.
Supervised analyses were carried out using the Significant Analysis of Microarrays software version 4.00 (excel front-end publicly available at http://www-stat.stanford.edu/~tibs/SAM/index.html).23 The cutoff point for statistical significance (at a q-value 0) was determined by tuning the Δ parameter on the false discovery rate and controlling the q-value of the selected probes. Hierarchical agglomerative clustering of the most significant probesets found was performed adopting Euclidean and average as distance and linkage methods, respectively. DNA-Chip Analyzer software (dChip)24 was used to perform clustering and to graphically represent it. Gene expression profiling data have been deposited in the NCBI Gene Expression Omnibus database (GEO; http//www.ncbi.nlm.nih.gov/geo).
Quantitative real-time PCR (QRT-PCR)
For QRT-PCR, 500 ng of the total RNA was used for reverse transcription using random primer mix and reagents from INVITROGEN (Life Technologies, Foster City, CA). Real-time PCR was performed in triplicate using TaqMan Non-coding RNA Assays together with the TaqMan Fast Universal PCR Master Mix on an Applied Biosystems 7900 Sequence Detection System (Life Technologies). All of the RNA samples were normalized on the basis of the UBC housekeeping gene. The threshold cycle (CT) was defined as the fractional cycle number at which the fluorescence passes the fixed threshold. snoRNA expression was relatively quantified using the 2−ΔCt method (Applied Biosystems User Bulletin No. 2) and expressed as the relative quantity of target snoRNA normalized to the UBC housekeeping gene.
Genome-wide DNA profiling
Twenty-five of the 63 MM/sPCL samples profiled for sno/scaRNA expression were used for genome-wide DNA analysis using Affymetrix GeneChip Human Mapping 50K XbaI microarrays as previously reported by us.13
Methylation analysis at 15q11 by pyrosequencing
DNA methylation was quantitated using bisulfite-PCR and Pyrosequencing assays. In brief, the samples were bisulfite treated, and PCR amplified by means of the PyroMark Q96 CpG Prader-Willi kit (Qiagen, Dusseldorf, Germany), as recommended by the manufacturer. Pyrosequencing was performed using the PyroMark 96 (Qiagen). Methylation quantification was performed using the provided software. The degree of methylation was expressed as %5-methylated cytosines over the sum of methylated and unmethylated cytosines.
Statistical analysis
Conventional statistical procedures were applied using standard packages for R software (Kendall τ correlations and Wilcoxon rank-sum tests). Multiple-test correction has been applied in order to measure the false discovery rate that incurred when calling the test significant based on P-value (q-value <0.05 as threshold).
Results
Global sno/scaRNA expression profiling in MM patients
The expression profiles of sno/scaRNA genes have been investigated in a panel of 55 MMs, 8 sPCLs and 4 normal tonsil PCs. MM patients were fully representative of the different molecular groups according to the TC classification (see Material and Methods). In addition, the HD status, del(13), del(17) and 1q gain were investigated by FISH in almost all the samples (Figure 1 and Supplementary Table S1 for further details).
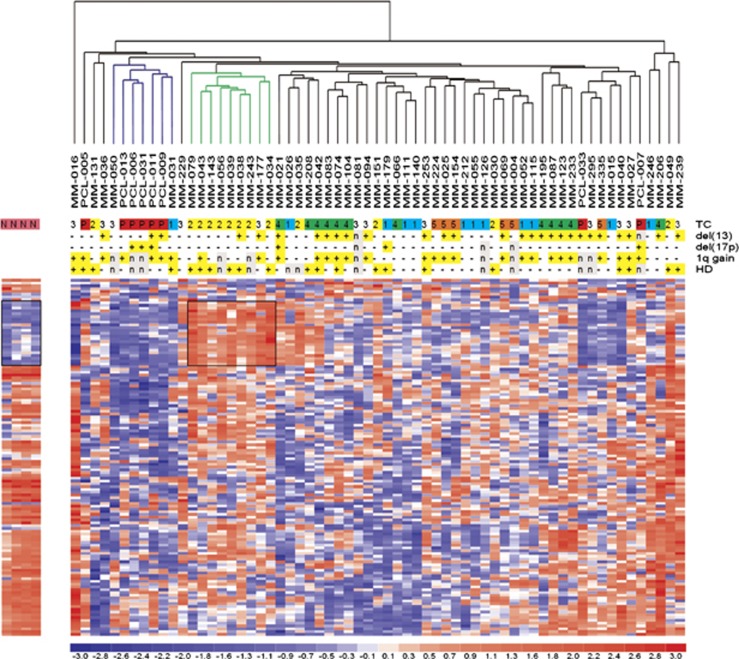
Figure 1.
Unsupervised analysis of sno/scaRNA expression profiles. Hierarchical clustering of the samples using the 128 most variable sno/scaRNAs (patients in columns, snoRNAs in rows). sno/scaRNA expression levels in CD138-positive cells from tonsils are shown on the left. The color scale bar represents the relative sno/scaRNA expression changes normalized by the standard deviation. The patients' molecular characteristics are shown above the matrix; n indicates unavailable information. Specific characteristics are enriched in colored sub-branches (see text). Black boxes identify SNORD115 and -116 clusters at 15q11.
Sno/scaRNA expression profiles were analyzed using Human Gene 1.0 ST arrays. Oligonucleotide probes on Gene Chips were re-mapped according to the latest human genome and transcriptome information (see materials and methods) identifying 215 human snoRNA (76 SNORA and 139 SNORD transcripts) and 17 human scaRNA genes. To determine whether the natural grouping of global sno/scaRNA expression profiling could be associated with specific molecular groups, we performed an unsupervised analysis using conventional hierarchical agglomerative clustering of the 115 snoRNAs and 13 scaRNAs whose average change in expression levels varied at least 1.5-fold from the mean across the data set (Figure 1). The most striking finding was that 8 of 13 TC2 patients clustered together (Figure 1, green cluster, P<0.0001), as did 5 of 8 sPCL cases (blue cluster, P=0.0006), whereas the TC1, TC3, TC4 and TC5 samples were scattered along the dendrogram.
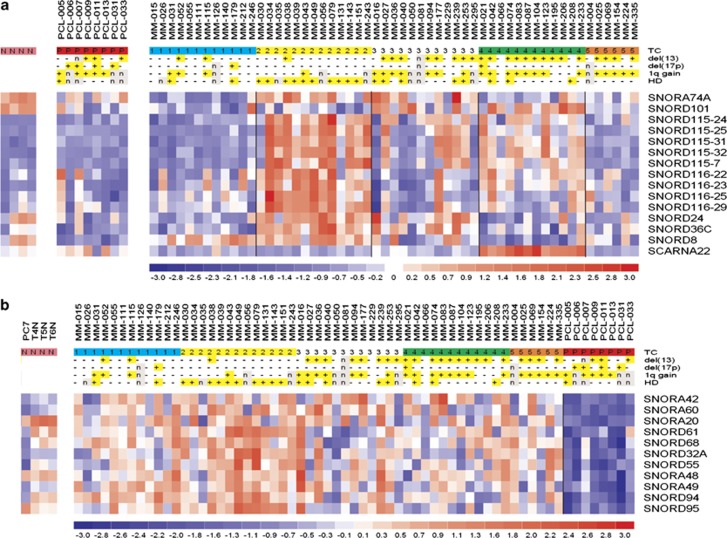
Next, multiclass analysis of MM samples involving all the five TC groups, identified a set of 14 snoRNAs and 1 scaRNA, showing highly significant differential expression between the groups (at q-value=0). As shown in Figure 2a, all identified ncRNAs, with the exception of SNORD101 and SCARNA22, were found to be upregulated in the TC2 group and partially expressed in some TC4 and TC3 patients, whereas in TC1 and TC5, they were downmodulated (Supplementary Table S2). In addition, SCARNA22 was selectively expressed in TC4 samples. Notably, SNOR115-7, -24, -25, -31, -32 and SNORD116-22, -23, -25, -29 are multiple transcripts of SNORD115 and SNORD116 families that belong to a cluster located at 15q11. Similarly, supervised analysis comparing HD versus not-HD cases (five HD patients harboring concurrently IGH translocation were excluded from the analysis) revealed seven SNORA (SNORA70D, -40, -74A, -64, -23, -22 and -68) and five SNORD (SNORD24, -36B, -63, -36C and -D95) transcripts, all but one (SNORA70D) upregulated in the HD setting (Supplementary Table S3). Notably, SNORA74A, SNORD24 and SNORD36C were also found upregulated in TC2 patients by the multiclass analysis (see above), in all likelihood because of their representativeness within HD cases. Furthermore, we investigated sno/scaRNA expression in the context of other recurrent chromosomal alterations in MM, such as del(13) and 1q gain, but no significantly differentially expressed sno/scaRNAs between positive and negative patients were recognized. Finally, the comparison of MM versus sPCL samples identified 11 differentially expressed snoRNAs, all downregulated in sPCL patients (Figure 2b and Supplementary Table S4).
Figure 2.
Identification of sno/scaRNA signatures characterizing distinct MM genetic subgroups. (a) Heatmap of the differentially expressed sno/scaRNAs in MM patients stratified into the five TC groups; sno/scaRNA expression levels in sPCLs and CD138-positive cells from tonsils are shown on the left. (b) Supervised analyses identifying the snoRNAs that are differentially expressed in MM and sPCL patients; sno/scaRNA expression levels in CD138-positive cells from tonsils are shown on the left.
As a final point, the profiling of normal tonsils PCs provided us some insight into the sno/scaRNA expression pattern in a putative normal PC counterpart. In particular, compared with normal PCs, it appears that both MM and, even more, sPCL samples exhibited lower expression levels of the most variables sno/scaRNA genes in our data set (Figure 1), with the exception of snoRNAs located at 15q11 region, which are overexpressed in the TC2 group (see black boxes in Figures 1 and 2a).
QRT-PCR validation of differentially expressed snoRNAs
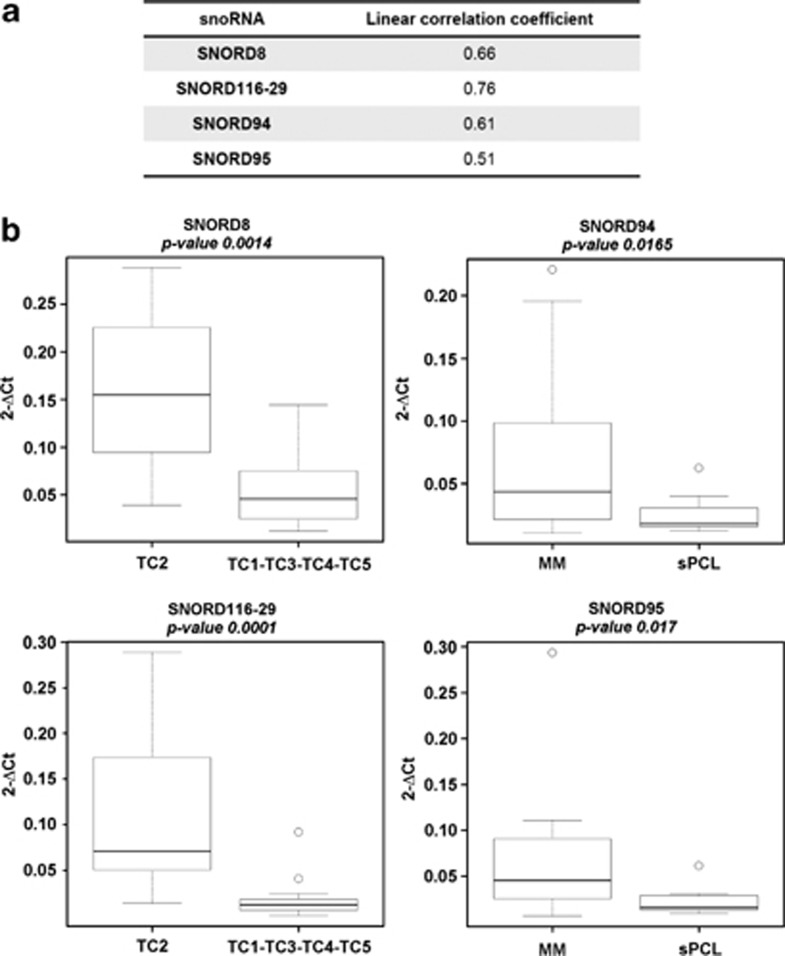
To validate the microarray data, the levels of SNORD8, SNORD116-29, SNORD94 and SNORD95 were quantified in 33 samples by QRT-PCR. As reported in Figure 3a, linear correlation analysis indicated a good correspondence between the two techniques. QRT-PCR results also confirmed the statistically significant and specific overexpression of SNORD116-29 and SNORD8 in the TC2 group and the downregulation of SNORD94 and SNORD95 in sPCL samples (Figure 3b).
Figure 3.
QRT-PCR validation of snoRNA expression. For each snoRNA, the Pearson's correlation coefficient was calculated between gene expression profiling data and QRT-PCR results expressed as 2−ΔCt (a). Box plots of snoRNAs quantified in QRT-PCR in 33 MM-PCL cases, whose expression significantly correlated to group membership on the basis of Wilcoxon rank-sum tests (P-values are shown above each panel). (b) The expression levels are represented as 2−ΔCt.
Correlation of sno/scaRNAs expression levels with those of their corresponding host genes
The vast majority of human sno/scaRNAs are encoded in introns of so-called host genes, in the sense orientation, and are produced by exonucleolytic processing of the debranched intron after splicing.25 To assess whether deregulated sno/scaRNAs were coordinately expressed with their host mRNAs, we investigated the 160 sno/scaRNA-host gene couples for which the host gene expression levels are available on Human Gene 1.0 ST arrays. Non parametric Kendall τ correlation test identified 72 snoRNAs and 7 scaRNAs whose expression levels significantly correlated with those of their corresponding host gene (Supplementary Table S5). Concerning sno/scaRNAs resulting from the multiclass and two-class analyses (Figure 2), the expression levels of SNORD36C, -63, -95, -101, SNORA23, -40, -70D and SCARNA22 were significantly associated with that of their host transcripts encoding for the ribosomal protein L7A (RPL7A, q-value=2E-2), the heat shock protein 9 (HSPA9, q-value=5E-4), the receptor for activated protein Kinase C (RACK1, q-value=4E-4), the ribosomal protein S12 (RPS12, q-value=1E-3), the cytoplasmic shuttle protein IPO7 (q-value=9E-3), the TATA box-binding factor 1D (TAF1D, q-value=4.3E-13), the AKT protein phosphatase PHLPPL (q-value=2.9E-11) and MMSET (q-value=1.29E-09), respectively (Table 1).
Table 1. Correlation of the expression level of sno/scaRNAs (resulting from supervised analyses) and corresponding host genes.
| snoRNA | Alias | Cytoband | HOST GENE | Kendall q-value | Target RNA |
|---|---|---|---|---|---|
| SNORA23 | ACA23 | 11p15 | IPO7 | 9.0E-03 | 28S rRNA U3737 and 28S rRNA U4331 |
| SNORA40 | ACA40 | 11q21 | TAF1D | 4.3E-13 | 28S rRNA U4546 and 18 S rRNA U1174 |
| SNORA49 | ACA49 | 12q24 | EP400 | 6.0E-01 | Unknown |
| SNORD8 | mgU6-53 | 14q11 | CHD8 | 3.4E-01 | U6 snRNA A53 |
| SNORD115-24 | HBII-52-24 | 15q11 | SNURF-SNRPN | 5.0E-01 | Serotonin receptor 5HT-2C mRNA |
| SNORD115-25 | HBII-52-25 | 15q11 | SNURF-SNRPN | 1.2E-01 | Serotonin receptor 5HT-2C mRNA |
| SNORD115-31 | HBII-52-31 | 15q11 | SNURF-SNRPN | 3.9E-01 | Serotonin receptor 5HT-2C mRNA |
| SNORD115-32 | HBII-52-32 | 15q11 | SNURF-SNRPN | 3.8E-01 | Serotonin receptor 5HT-2C mRNA |
| SNORD115-7 | HBII-52-7 | 15q11 | SNURF-SNRPN | 1.6E-01 | Serotonin receptor 5HT-2C mRNA |
| SNORD116-22 | HBII-85-22 | 15q11 | SNURF-SNRPN | 9.0E-01 | Unknown |
| SNORD116-23 | HBII-85-23 | 15q11 | SNURF-SNRPN | 6.1E-01 | Unknown |
| SNORD116-25 | HBII-85-25 | 15q11 | SNURF-SNRPN | 4.4E-01 | Unknown |
| SNORD116-29 | HBII-85-29 | 15q11 | SNURF-SNRPN | 5.6E-01 | Unknown |
| SNORA64 | U64 | 16p13 | RPS2 | Aa | 28S rRNA U4975 |
| SNORA70D | U70D | 16q22 | PHLPPL | 3.0E-11 | 18S rRNA U1692 |
| SNORD68 | HBII-202 | 16q24 | RPL13 | 3.0E-01 | 18S rRNA U428 and 28S rRNA A2388 |
| SNORA48 | ACA48 | 17p13 | EIF4A1 | Aa | 28S rRNA U3797 |
| SNORA68 | U68 | 19p13 | RPL18A | Aa | 28S rRNA U4393 |
| SNORD32A | U32A | 19q13 | RPL13A | Aa | 18S rRNA G1328 and 28S rRNA A1511 |
| SNORD55 | U55 | 1p34 | RPS8 | Aa | 28S rRNA C2791 |
| SNORA42 | ACA42 | 1q22 | KIAA0907 | 9.3E-01 | 8S rRNA U572 and 18S rRNA U109 |
| SNORA60 | ACA60 | 20q11 | SNHG11 | 8.3E-01 | 18S rRNA U1004 |
| SNORD94 | U94 | 2p11 | PTCD3 | 8.3E-01 | U6 snRNA C62 |
| SNORA74A | U19 | 5q31 | MATR | 1.3E-01 | 28S rRNA U3741 and 28S rRNA U3743 and U3 snRNA U8 |
| SNORD63 | U63 | 5q31 | HSPA9B | 5.7E-04 | 28S rRNA A4541 |
| SNORD95 | U95 | 5q35 | RACK1 | 4.2E-04 | 28S rRNA A2802 and 28S rRNA C2811 |
| SNORA20 | ACA20 | 6p25 | TCP1 | 9.5E-02 | 18S rRNA U651 |
| SNORD101 | U101 | 6q23 | RPS12 | 1.4E-03 | Unknown |
| SNORA22 | ACA22 | 7q11 | CCT6P1 | Aa | 28S rRNA U4966 and 28S rRNA U4975 |
| SNORD24 | U24 | 9q34 | RPL7A | 3.6E-01 | 28S rRNA C2338 and 28S rRNA C2352 |
| SNORD36B | U36B | 9q34 | RPL7A | 5.8E-01 | 18S rRNA A668 |
| SNORD36C | U36C | 9q34 | RPL7A | 2.9E-02 | 28S rRNA A3703 |
| SNORD61 | U61 | Xq26 | RBMX | 4.2E-01 | 18S rRNA U1442 |
| SCARNA22 | ACA11 | 4p16 | MMSET | 1.3E-09 | Unknown |
Abbreviations: scaRNA, small Cajal body-specific RNA; snoRNA, small nucleolar RNA.
Probe not present onto Human Gene 1.0 ST arrays; snoRNA significantly correlating with corresponding host gene is typed in bold.
Integrative analysis of sno/scaRNA expression profiles with genome-wide copy number alterations and occurrence of loss of heterozygositys
Next, we investigated the influence on sno/scaRNA expression of the allelic imbalances detected by single nucleotide polymorphism array analysis available in 25 of 63 cases.13 By referring to snoRNA-LBME-database25 based on the UCSC Genome Browser Database annotations (genome assembly NCBI36/hg18), we inferred the DNA copy number of each sno/scaRNA locus according to our previous report.13 The integrative analysis identified 16 snoRNAs whose expression was significantly associated with their matching copy number (Kendall q-value <0.05; Table 2, Supplementary Figure S1). Among these, we found SNORD116-23, -25, -29 overexpressed in TC2 patients, SNORA68 upregulated in HD samples, SNORD95 downregulated in sPCL but upregulated in HD MM and SNORD32A downregulated in sPCL. Notably, SNOR116-11, -23, -25, -26, -28, and -29 and SNORD115-8 belong to the SNORD115 and SNORD116 cluster at 15q11; SNORA68, SNORD105, SNORD41 and SNORD32A are located on chromosome 19; SNORD15B is positioned at 11q13, whereas SNORD95 is mapped at 5q35. These chromosome regions are gained in HD patients and exhibit transcriptional imbalances, as previously reported by us.18 Moreover, a gene-dosage effect might also be associated with the expression of SNORD81, -75 and -44, all of them located on chromosome 1q25, a region known to be involved in gain/amplification in MM.26
Table 2. Correlation of snoRNA expression and CN alteration.
| snoRNA | Alias | Cytoband | START LOC | END LOC | Kendall q-value |
|---|---|---|---|---|---|
| SNORD95 | U95 | 5q35.3 | 180602920 | 180602982 | 5.7E-3 |
| SNORA68 | U68 | 19p13.11 | 17834397 | 17834529 | 1.8E-2 |
| SNORD105 | U105 | 19p13.2 | 10079327 | 10079411 | 1.8E-2 |
| SNORD116-25 | HBII-85-25 | 15q11.2 | 22893902 | 22893995 | 1.8E-2 |
| SNORD116-26 | HBII-85-26 | 15q11.2 | 22895738 | 22895835 | 1.8E-2 |
| SNORD116-28 | HBII-85-28 | 15q11.2 | 22900881 | 22900973 | 1.8E-2 |
| SNORD32A | U32A | 19q13.33 | 54685037 | 54685113 | 1.8E-2 |
| SNORD115-8 | HBII-52-8 | 15q11.2 | 22980546 | 22980627 | 2.4E-2 |
| SNORD116-29 | HBII-85-29 | 15q11.2 | 22902760 | 22902844 | 2.8E-2 |
| SNORD81 | U81 | 1q25.1 | 172099907 | 172099983 | 2.9E-2 |
| SNORD116-11 | HBII-85-11 | 15q11.2 | 22872168 | 22872261 | 3.3E-2 |
| SNORD41 | U41 | 19p13.13 | 12678263 | 12678332 | 3.9E-2 |
| SNORD116-23 | HBII-85-23 | 15q11.2 | 22888025 | 22888118 | 4.8E-2 |
| SNORD15B | U15B | 11q13.4 | 74793113 | 74793258 | 4.8E-2 |
| SNORD75 | U75 | 1q25.1 | 172102640 | 172102699 | 4.8E-2 |
| SNORD44 | U44 | 1q25.1 | 172101727 | 172101789 | 4.9E-2 |
Abbreviations: CN, copy number; snoRNA, small nucleolar RNAs.
snoRNAs resulting from multiclass or two-class analyses and significantly correlating with corresponding CN are typed in bold.
Conversely, no significant correlation was identified between sno/scaRNAs expression and the occurrence of loss of heterozygosity in the same panel of cases using Wilcoxon rank-sum test as previously described13 (data not shown).
Upregulation of SNORD115 and SNORD116 does not correlate with altered methylation status of the imprinting center at 15q11
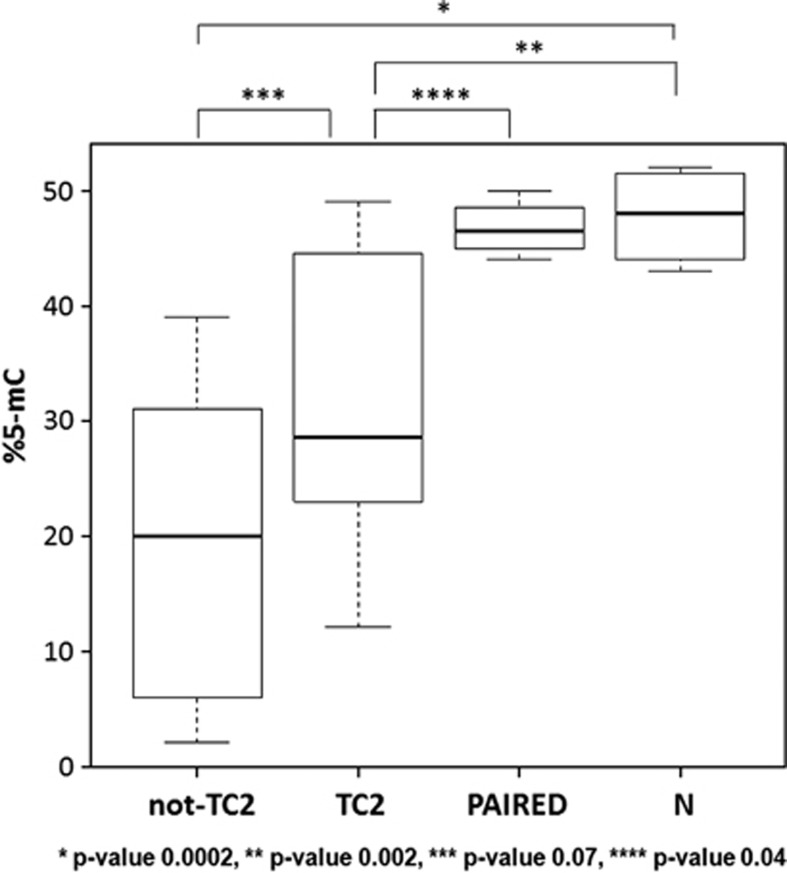
SNORD115 and SNORD116 are located at 15q11, in a region known to be affected by minimal deletions involved in the Prader–Willi syndrome (PWS).27 SNORD115 and SNORD116 are present in highly similar multiple copies (47 and 24 copies, respectively) that are tandemly arranged within the introns of the SNURF–SNRPN transcript, which spans more than 460 kb and contains at least 148 exons. The expression of SNURF–SNRPN, as well as that of several genes in this region, is finely regulated by a bipartite imprinting center (IC) that silences most maternal genes of the PWS critical region: owing to this imprinting, the SNURF–SNRPN pre-mRNA is expressed only from the paternal allele.27 To test whether the IC methylation status may contribute to the modulated expression of SNORD115 and SNORD116 in MM, we performed a pyrosequencing analysis of 30 MM patients (12 TC2 samples and 18 MM of other TC groups), 4 DNA from purified peripheral T cells of paired TC2 samples and the 4 normal controls. As shown in Figure 4, all myeloma samples displayed a significant IC hypomethylation compared with normal controls.
Figure 4.
Methylation status of IC at 15q11. Box plots of methylation values quantified as %5-methylated cytosines (%5-mC) with pyrosequencing analysis in 12 TC2 samples and 18 MM of other TC groups, 4 paired controls (purified peripheral T cells) of TC2 samples and 4 normal controls. Imprinted normal locus presents 50% methylation, with respect to which both TC2 and not-TC2 MM are significantly hypomethylated (Wilcoxon rank-sum tests' P-values are reported).
Discussion
snoRNAs and scaRNAs have been found to have crucial roles in rRNA, tRNA, snRNA as well as in mRNA biogenesis.28 More recently, experimental evidence indicated that sno/scaRNAs may also have important roles in human cancers.9 To the best of our knowledge, this is the first report describing the global expression profiles of sno/scaRNAs in MM. Our results identify specific patterns of sno/scaRNAs associated with distinct molecular types of MM and provide novel insights into our understanding the complex biological scenario of the disease.
Our study indicated a general pattern of downregulation of sno/scaRNAs expression in MM and sPCL patients compared with a non-neoplastic counterpart (Figure 1), a finding in agreement with the recent evidence of snoRNAs global downregulation reported in acute myeloblastic and lymphoblastic leukemias.10 This finding is particularly striking in sPCL; in such a context, the downregulation of SNORD32A must be noted (Figure 2b), for which a non-canonical role in the mediation of oxidative and endoplasmic reticulum stress-induced response pathways has been recently reported in vitro and in mouse models.29 In this perspective, one can speculate that SNORD32A downregulation could be involved in the regulation of oxidative stress during oncogenesis and might imply tumoral PCs resistance to endoplasmic reticulum stress-induced response, as it was recently demonstrated for SCARNA22 in myeloma cell lines.12 Surprisingly, sPCLs showed the downregulation of SNORA42, which is located at chromosome 1q22, a region frequently amplified in plasma cell dyscrasias. SNORA42 has been described to act as a putative oncogene in non-small cell lung cancer cell lines being overexpressed in association with the gain of 1q region and positively correlated to poor survival.30 The different patterns found in our study may suggest that some snoRNAs could have different roles depending on the cellular context, as largely demonstrated for miRNAs.
Despite the general pattern of downregulation observed in myeloma cells, HD MMs upregulate specific snoRNAs. Considering the canonical function of snoRNAs, their expression in HD tumors may be not surprising, as this molecular group is characterized by a global upregulation of the translational machinery, including genes involved in protein biosynthesis, such as ribosomal proteins, mitochondrial ribonucleoproteins or proteins acting in rRNA processing.18 Furthermore, SNORD36C, -63, -95 and SNORA40 appear to be upregulated in HD samples in correlation with their matched host genes RPL7A, HSPA9B, RACK1 and TAF1D, respectively (Table 1), whose biological functions are related to the above-mentioned processes as well.31, 32, 33 Notably, RACK1 is the major component of translating ribosomes and it has been reported to provide a physical and functional link between protein kinase C signaling and ribosome activation.31 Interestingly, it has been recently described as a pro-apoptotic gene in human breast and colon cancer.34 Finally, the chromosomal regions where these snoRNAs are located (see Table 1) have been previously reported by us to be transcriptionally active or specifically downregulated in HD patients.18 Based on these findings, it could be suggested that snoRNAs expression in HD samples may have a role in the overall upregulation of the translational machinery.
Interestingly, MM patients included in the TC2 group showed a quite different signature, consisting of 14 specific snoRNAs (Figure 2a). In particular, these patients expressed nine members of the SNORD115 and SNORD116 snoRNA families that are specifically expressed in the brain in mouse, while only SNORD115 is strictly brain-specific in humans.35 Our study provide evidences that, similarly to what was described for SNORD112, -113 and -114 in leukemia,10 not all the SNORD115 or SNORD116 variants have similar expression patterns in MM, a finding that could be consistent with the different putative targets predicted for these SNORDs.36 Furthermore, it should be considered that the expression of some variants of SNORD115 or SNORD116 appeared to be significantly correlated with the DNA copy number (Table 2). In addition, as suggested by the hypomethylation of the IC at 15q11, other mechanisms may regulate their expression in specific MM groups. With regard to this finding, the hypomethylation of the IC could be related to the general pattern of global de-methylation described in tumors and, in particular, in MM, as recently reported by us and others.37, 38
Notably, the only scaRNA differentially expressed among the molecular MM groups is SCARNA22, which is specifically upregulated in patients with the t(4;14). As specified above, SCARNA22 is located within intron 18 of the MMSET gene, which is constitutively activated by the translocation. Therefore, as demonstrated by the correlation analysis (Supplementary Table S5), SCARNA22 is overexpressed in association with its host gene, as also found in sPCLs and human MM cell lines harboring t(4;14) (Supplementary Figure S2). This is in line with very recent evidence of the upregulation of this scaRNA in t(4;14) patients, suggesting a key role for this molecule in the pathogenesis of the TC4 group of MM.12
Concerning the significance of the sno/scaRNA expression in MM patients, it should be considered that these molecules may have also a role in the regulation of alternative splicing mRNAs. This has been suggested for murine SNORD115 that has been found to be processed to a shorter form in order to hybridize to complementary target sequences on the HTR2C pre-mRNA for the correct alternative splicing of the serotonin receptor mRNA.39 Furthermore, a role in the 3′ processing of selected snoRNAs has been very recently proposed for SCARNA22.12 In line with this evidence, a similar function could be awaited for SNORD101 and the ‘orphan' SNORD116 variants, as predicted by the recent available snoTARGET open source.36
In the attempt to elucidate the role of snoRNAs in cancer, it should be considered that they may be involved in mRNA stability. Indeed, recent studies have expanded the range of regulatory non-coding RNAs, based on the evidence of human miRNA precursors resembling SNORA or SNORD genes or snoRNAs showing miRNA-like functions.40, 41, 42, 43 Although miRNAs and snoRNAs are currently believed to be generated by different cellular pathways and to function in different cellular compartments, at least some of these molecules seem to have dual functions; that is, the same transcript can give rise both to a snoRNA and—probably through subsequent processing steps—to a miRNA.44
In conclusion, our data extend the current view of sno/scaRNA deregulation in cancer. In the context of MM, altered sno/scaRNA expression could be consistent with the role that the activation of the translational machinery may have in the pathogenesis of a large fraction of MM. Indeed, the recent discovery of inactivating mutations in genes involved in RNA processing, protein translation and the unfolded protein response in MM patients has already led to the suggestion that dysregulation of such processes may represent oncogenic mechanisms in MM.45, 46 Overall, sno/scaRNAs deregulation in plasma cell dyscrasias adds a further level of complexity to the heterogeneity of these forms and may contribute to discover new biomarkers and altered molecular pathways associated with the disease.
Acknowledgments
This work was supported by: Associazione Italiana Ricerca sul Cancro (AIRC) Grant IG4569 to AN; AIRC 5xmille Grant 9980, 2010–15 to AN, MF and PT; Ministero Italiano dell'Istruzione, Università e Ricerca (MIUR) (2009PKMYA2) to AN; and Fondazione Matarelli, Milano. ML is supported by a fellowship from the Fondazione Italiana Ricerca sul Cancro (FIRC).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Blood Cancer Journal website (http://www.nature.com/bcj)
Supplementary Material
References
- Fonseca R, Barlogie B, Bataille R, Bastard C, Bergsagel PL, Chesi M, et al. Genetics and cytogenetics of multiple myeloma: a workshop report. Cancer Res. 2004;64:1546–1558. doi: 10.1158/0008-5472.can-03-2876. [DOI] [PubMed] [Google Scholar]
- Mattioli M, Agnelli L, Fabris S, Baldini L, Morabito F, Bicciato S, et al. Gene expression profiling of plasma cell dyscrasias reveals molecular patterns associated with distinct IGH translocations in multiple myeloma. Oncogene. 2005;24:2461–2473. doi: 10.1038/sj.onc.1208447. [DOI] [PubMed] [Google Scholar]
- Shaughnessy JD, Zhan F, Burington BE, Huang Y, Colla S, Hanamura I, et al. A validated gene expression model of high-risk multiple myeloma is defined by deregulated expression of genes mapping to chromosome 1. Blood. 2007;109:2276–2284. doi: 10.1182/blood-2006-07-038430. [DOI] [PubMed] [Google Scholar]
- Galasso M, Elena SM, Volinia S. Non-coding RNAs: a key to future personalized molecular therapy. Genome Med. 2010;2:12. doi: 10.1186/gm133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionetti M, Agnelli L, Lombardi L, Neri A. MicroRNAs in the pathobiology of multiple myeloma. Curr Cancer Drug Targets. 2012;12:823–837. doi: 10.2174/156800912802429274. [DOI] [PubMed] [Google Scholar]
- Pichiorri F, De LL, Aqeilan RI. MicroRNAs: new players in multiple myeloma. Front Genet. 2011;2:22. doi: 10.3389/fgene.2011.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jady BE, Kiss T. A small nucleolar guide RNA functions both in 2'-O-ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J. 2001;20:541–551. doi: 10.1093/emboj/20.3.541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss T. Small nucleolar RNAs: an abundant group of noncoding RNAs with diverse cellular functions. Cell. 2002;109:145–148. doi: 10.1016/s0092-8674(02)00718-3. [DOI] [PubMed] [Google Scholar]
- Williams GT, Farzaneh F. Are snoRNAs and snoRNA host genes new players in cancer. Nat Rev Cancer. 2012;12:84–88. doi: 10.1038/nrc3195. [DOI] [PubMed] [Google Scholar]
- Valleron W, Laprevotte E, Gautier EF, Quelen C, Demur C, Delabesse E, et al. Specific small nucleolar RNA expression profiles in acute leukemia. Leukemia. 2012;26:2052–2060. doi: 10.1038/leu.2012.111. [DOI] [PubMed] [Google Scholar]
- Lopez-Corral L, Mateos MV, Corchete LA, Sarasquete ME, de la RJ, de AF, et al. Genomic analysis of high risk smoldering multiple myeloma. Haematologica. 2012;97:1439–1443. doi: 10.3324/haematol.2011.060780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu L, Su MY, Maggi LB, Lu L, Mullins C, Crosby S, et al. Multiple myeloma-associated chromosomal translocation activates orphan snoRNA ACA11 to suppress oxidative stress. J Clin Invest. 2012;122:2793–2806. doi: 10.1172/JCI63051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agnelli L, Mosca L, Fabris S, Lionetti M, Andronache A, Kwee I, et al. A SNP microarray and FISH-based procedure to detect allelic imbalances in multiple myeloma: an integrated genomics approach reveals a wide gene dosage effect. Genes Chromosomes Cancer. 2009;48:603–614. doi: 10.1002/gcc.20668. [DOI] [PubMed] [Google Scholar]
- Lionetti M, Biasiolo M, Agnelli L, Todoerti K, Mosca L, Fabris S, et al. Identification of microRNA expression patterns and definition of a microRNA/mRNA regulatory network in distinct molecular groups of multiple myeloma. Blood. 2009;114:e20–e26. doi: 10.1182/blood-2009-08-237495. [DOI] [PubMed] [Google Scholar]
- Ronchetti D, Lionetti M, Mosca L, Agnelli L, Andronache A, Fabris S, et al. An integrative genomic approach reveals coordinated expression of intronic miR-335, miR-342, and miR-561 with deregulated host genes in multiple myeloma. BMC Med Genomics. 2008;1:37. doi: 10.1186/1755-8794-1-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- IMWG Criteria for the classification of monoclonal gammopathies, multiple myeloma and related disorders: a report of the International Myeloma Working Group. Br J Haematol. 2003;121:749–757. [PubMed] [Google Scholar]
- Fabris S, Agnelli L, Mattioli M, Baldini L, Ronchetti D, Morabito F, et al. Characterization of oncogene dysregulation in multiple myeloma by combined FISH and DNA microarray analyses. Genes Chromosomes Cancer. 2005;42:117–127. doi: 10.1002/gcc.20123. [DOI] [PubMed] [Google Scholar]
- Agnelli L, Fabris S, Bicciato S, Basso D, Baldini L, Morabito F, et al. Upregulation of translational machinery and distinct genetic subgroups characterise hyperdiploidy in multiple myeloma. Br J Haematol. 2007;136:565–573. doi: 10.1111/j.1365-2141.2006.06467.x. [DOI] [PubMed] [Google Scholar]
- Agnelli L, Bicciato S, Mattioli M, Fabris S, Intini D, Verdelli D, et al. Molecular classification of multiple myeloma: a distinct transcriptional profile characterizes patients expressing CCND1 and negative for 14q32 translocations. J Clin Oncol. 2005;23:7296–7306. doi: 10.1200/JCO.2005.01.3870. [DOI] [PubMed] [Google Scholar]
- Hideshima T, Bergsagel PL, Kuehl WM, Anderson KC. Advances in biology of multiple myeloma: clinical applications. Blood. 2004;104:607–618. doi: 10.1182/blood-2004-01-0037. [DOI] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Li C, Ellis B, Wong WH.Feature extraction and normalization algorithms for high-density oligonucleotide gene expression array data J Cell Biochem Suppl 2001. Suppl 37120–125. [DOI] [PubMed]
- Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabris S, Ronchetti D, Agnelli L, Baldini L, Morabito F, Bicciato S, et al. Transcriptional features of multiple myeloma patients with chromosome 1q gain. Leukemia. 2007;21:1113–1116. doi: 10.1038/sj.leu.2404616. [DOI] [PubMed] [Google Scholar]
- Runte M, Huttenhofer A, Gross S, Kiefmann M, Horsthemke B, Buiting K. The IC-SNURF-SNRPN transcript serves as a host for multiple small nucleolar RNA species and as an antisense RNA for UBE3A. Hum Mol Genet. 2001;10:2687–2700. doi: 10.1093/hmg/10.23.2687. [DOI] [PubMed] [Google Scholar]
- Xie J, Zhang M, Zhou T, Hua X, Tang L, Wu W. Sno/scaRNAbase: a curated database for small nucleolar RNAs and cajal body-specific RNAs. Nucleic Acids Res. 2007;35:D183–D187. doi: 10.1093/nar/gkl873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel CI, Holley CL, Scruggs BS, Sidhu R, Brookheart RT, Listenberger LL, et al. Small nucleolar RNAs U32a, U33, and U35a are critical mediators of metabolic stress. Cell Metab. 2011;14:33–44. doi: 10.1016/j.cmet.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei YP, Liao JP, Shen J, Yu L, Liu BL, Liu L, et al. Small nucleolar RNA 42 acts as an oncogene in lung tumorigenesis. Oncogene. 2012;31:2794–2804. doi: 10.1038/onc.2011.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceci M, Gaviraghi C, Gorrini C, Sala LA, Offenhauser N, Marchisio PC, et al. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature. 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- Chen TH, Kambal A, Krysiak K, Walshauser MA, Raju G, Tibbitts JF, et al. Knockdown of Hspa9, a del(5q31.2) gene, results in a decrease in hematopoietic progenitors in mice. Blood. 2011;117:1530–1539. doi: 10.1182/blood-2010-06-293167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorski JJ, Pathak S, Panov K, Kasciukovic T, Panova T, Russell J, et al. A novel TBP-associated factor of SL1 functions in RNA polymerase I transcription. EMBO J. 2007;26:1560–1568. doi: 10.1038/sj.emboj.7601601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Reefy S, Osman H, Jiang W, Mokbel K. Evidence for a pro-apoptotic function of RACK1 in human breast cancer. Oncogene. 2010;29:5651. doi: 10.1038/onc.2010.291. [DOI] [PubMed] [Google Scholar]
- Bortolin-Cavaille ML, Cavaille J. The SNORD115 (H/MBII-52) and SNORD116 (H/MBII-85) gene clusters at the imprinted Prader-Willi locus generate canonical box C/D snoRNAs. Nucleic Acids Res. 2012;40:6800–6807. doi: 10.1093/nar/gks321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazeley PS, Shepelev V, Talebizadeh Z, Butler MG, Fedorova L, Filatov V, et al. snoTARGET shows that human orphan snoRNA targets locate close to alternative splice junctions. Gene. 2008;408:172–179. doi: 10.1016/j.gene.2007.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollati V, Fabris S, Pegoraro V, Ronchetti D, Mosca L, Deliliers GL, et al. Differential repetitive DNA methylation in multiple myeloma molecular subgroups. Carcinogenesis. 2009;30:1330–1335. doi: 10.1093/carcin/bgp149. [DOI] [PubMed] [Google Scholar]
- Walker BA, Wardell CP, Chiecchio L, Smith EM, Boyd KD, Neri A, et al. Aberrant global methylation patterns affect the molecular pathogenesis and prognosis of multiple myeloma. Blood. 2011;117:553–562. doi: 10.1182/blood-2010-04-279539. [DOI] [PubMed] [Google Scholar]
- Kishore S, Khanna A, Zhang Z, Hui J, Balwierz PJ, Stefan M, et al. The snoRNA MBII-52 (SNORD 115) is processed into smaller RNAs and regulates alternative splicing. Hum Mol Genet. 2010;19:1153–1164. doi: 10.1093/hmg/ddp585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brameier M, Herwig A, Reinhardt R, Walter L, Gruber J. Human box C/D snoRNAs with miRNA like functions: expanding the range of regulatory RNAs. Nucleic Acids Res. 2011;39:675–686. doi: 10.1093/nar/gkq776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ono M, Scott MS, Yamada K, Avolio F, Barton GJ, Lamond AI. Identification of human miRNA precursors that resemble box C/D snoRNAs. Nucleic Acids Res. 2011;39:3879–3891. doi: 10.1093/nar/gkq1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Avolio F, Ono M, Lamond AI, Barton GJ. Human miRNA precursors with box H/ACA snoRNA features. PLoS Comput Biol. 2009;5:e1000507. doi: 10.1371/journal.pcbi.1000507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Ono M, Yamada K, Endo A, Barton GJ, Lamond AI. Human box C/D snoRNA processing conservation across multiple cell types. Nucleic Acids Res. 2012;40:3676–3688. doi: 10.1093/nar/gkr1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Ono M. From snoRNA to miRNA: dual function regulatory non-coding RNAs. Biochimie. 2011;93:1987–1992. doi: 10.1016/j.biochi.2011.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, et al. Initial genome sequencing and analysis of multiple myeloma. Nature. 2011;471:467–472. doi: 10.1038/nature09837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan GJ, Walker BA, Davies FE. The genetic architecture of multiple myeloma. Nat Rev Cancer. 2012;12:335–348. doi: 10.1038/nrc3257. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.