Abstract
The connection between genotype and phenotype was assessed by determining the adhesion phenotype for the same mutation in two closely related yeast strains, S288c and Sigma, using two identical deletion libraries. Previous studies, all in Sigma, had shown that the adhesion phenotype was controlled by the filamentation mitogen-activated kinase (fMAPK) pathway, which activates a set of transcription factors required for the transcription of the structural gene FLO11. Unexpectedly, the fMAPK pathway is not required for FLO11 transcription in S288c despite the fact that the fMAPK genes are present and active in other pathways. Using transformation and a sensitized reporter, it was possible to isolate RPI1, one of the modifiers that permits the bypass of the fMAPK pathway in S288c. RPI1 encodes a transcription factor with allelic differences between the two strains: The RPI1 allele from S288c but not the one from Sigma can confer fMAPK pathway-independent transcription of FLO11. Biochemical analysis reveals differences in phosphorylation between the alleles. At the nucleotide level the two alleles differ in the number of tandem repeats in the ORF. A comparison of genomes between the two strains shows that many genes differ in size due to variation in repeat length.
Keywords: budding yeast, natural variation, repeat length polymorphisms, agar adhesion
RECENT advances in DNA sequencing have identified many nucleotide polymorphisms in the human genome, but it has been challenging to associate this genetic variation to specific phenotypic differences among individuals for complex traits (Jakobsdottir et al. 2009; Manolio et al. 2009; Dickson et al. 2010). This difficulty has been variously attributed to both genetic and nongenetic factors (Hartman et al. 2001; Carlborg and Haley 2004; Korbel et al. 2007; Dickson et al. 2010). Among the genetic factors are many genes contributing a small effect to the final phenotype (QTL) and complex (epistatic) gene interactions. The baker’s yeast Saccharomyces cerevisiae, with its compact and easily manipulated genome, offers the potential for identifying the relevant polymorphisms and, more importantly, identifying the molecular basis for the phenotypic differences.
Sequence studies comparing S. cerevisiae to other yeast species that diverged by 20 million years advanced our understanding of yeast evolution, but did not address how small genetic differences affect phenotypes (Kellis et al. 2003). Other studies have examined large numbers of both feral and laboratory S. cerevisiae strains, but have focused on population structure and evolutionary origins of the strains rather than the problem of connecting genotype to phenotype (Liti et al. 2009; Schacherer et al. 2009).
More recently, insights into the genotype-to-phenotype problem have been gained from linkage studies using modern genotyping techniques. Several examples can be seen in the cross of the wild vineyard strain RM11 to the standard laboratory strain S288c. A number of traits have been examined using this cross, including gene expression, cell morphology, resistance to DNA-damaging agents, and telomere length (Brem et al. 2002; Gatbonton et al. 2006; Nogami et al. 2007; Demogines et al. 2008). The genetic complexity for most of these traits is high, with many of them influenced by more than three loci. By examining large pools of progeny, recent techniques have further increased the ability to map relevant loci; however, it is still challenging to determine the exact alleles responsible and to understand how those alleles affect the phenotype (Ehrenreich et al. 2010; Connelly and Akey 2012).
Recent studies developed a model system that enables a comprehensive assessment of phenotypic differences for the same mutation in the two genetic backgrounds S288c and Σ1278b (Sigma) (Dowell et al. 2010). The two strains have very similar genomic sequences: Their divergence of ∼0.3% is similar to that between unrelated humans. To assess functional differences between these two strains, ∼5100 genes were deleted in Sigma for comparison with the same set of deletions in S288c (Winzeler et al. 1999; Dowell et al. 2010). The analysis identified strain-specific essential genes. The basis for the strain specificity was likely a complex set of background modifiers.
Here we compare these deletion libraries for the genes that control the key morphogenetic trait of adhesion/filamentation. In Sigma, adhesion requires the filamentation mitogen-activated kinase (fMAPK) pathway, but our library comparison showed that S288c can adhere in the absence of the fMAPK pathway. Although fMAPK-independent adhesion is a complex genetic trait, we devised a transformation protocol that enabled the isolation of RPI1, one of the modifiers responsible for the bypass of the fMAPK pathway. RPI1 is a transcription factor that is polymorphic between S288c and Sigma; the RPI1 allele from S288c (RPI1S288c) confers fMAPK pathway independence by activating FLO11 transcription, whereas the RPI1 allele from Sigma (RPI1Sigma) cannot. RPI1S288c confers fMAPK pathway independence in either genetic background. Moreover, there is a biochemical difference between the alleles; RPI1S288c, but not RPI1Sigma is hyperphosphorylated in both S288c and Sigma. The two forms of RPI1 differ in the number of tandem repeats in the ORF. A comparison of the S288c and Sigma genomes shows that many other genes with intragenic tandem repeats are highly polymorphic with respect to repeat size, a polymorphism that has been associated with phenotypic changes (Verstrepen et al. 2005).
Materials and Methods
Strains, media, microbiological techniques, and growth conditions
Yeast strains used in this study are derived from S288c and Σ1278b. Standard yeast media were prepared and genetic manipulation techniques were carried out as described in Guthrie and Fink (2002). The list of strains used in this study can be found in Supporting Information, Table S5. Adhesion assays were carried out by densely patching strains onto YPD or SC plates. These were grown overnight at 30° and then replica plated onto YPD or SC plates. The replica plates were grown at 30° for 3 days and then washed. The S288c strain expresses FLO1, which leads to flocculation that can influence agar adhesion phenotypes. To compare agar adhesion between S288c and Sigma, which does not express FLO1, the washes were performed by partially filling the petri dishes with 10 mM EDTA (which disrupts FLO1-dependent aggregates) and gentle shaking at ∼75 rpm on an orbital shaker. To visualize the difference between the strains, the media used for both the adhesion and the transcription assays were optimized for intrinsic growth differences between S288c and Sigma (e.g., flocculation and mother–daughter cell separation). However, the controls intrinsic to each experiment always permitted a comparison between strains grown under the same media conditions. To induce pseudohyphal growth, single cells were microdissected and grown on SLAD media (Gimeno et al. 1992).
The S288c library was constructed using previously published methods (Voynov et al. 2006). Each of the 4705 deletion strains in the standard S288c flo8 library was transformed with a CEN/ARS plasmid carrying the Sigma FLO8 gene under the control of its own promoter. The 4633 FLO8 deletion strains successfully recovered from these transformations formed the S288c deletion library. Screening the S288c library and the comparable Sigma deletion library for adhesion uncovered 599 deletions with decreased adhesion (Ahs−) (Table S1, Table S2, and Table S3). Only 46 deletions affected adhesion the same way in both strains (Table S3).
For quantitative (q)PCR and chromatin immunoprecipitation (ChIP), cells were grown overnight in liquid media as noted, diluted to OD600 = 0.25, and grown to OD600 = 4–4.5. For protein preparations, cells were grown as for qPCR in synthetic complete media.
Yeast strains carrying gene deletions were constructed by PCR amplification of kanamycin-resistance gene cassettes from the yeast deletion library (Winzeler et al. 2000) with ∼200 bases of flanking sequence. The list of oligos used in this study can be found in Supporting Information, Table S6. Correct integrants were identified by PCR, with the exception of tec1Δ, which was additionally checked by Southern blot using standard techniques (Brown 2001). FLO11 promoter swaps were carried out by first deleting the FLO11 promoter with the URA3 cassette. The reciprocal swap was carried out by PCR amplifying the sequences from each strain and using the PCR products to transform the opposite strain from which the sequence was amplified. The same procedure was performed for the RPI1 swaps but with only the ORF sequences. 3× FLAG-tagged constructs were created by amplifying the URA3 cassette from PRS306, using a primer (BCP534) that contained the 3× FLAG epitope. This construct was then subjected to another round of PCR to add 50 bp of flanking homology to the RPI1 C terminus. The resulting PCR product was used for transformation. The haploid MATa deletion collection was transformed with plasmid pHL1, using previously published protocols (Liu et al. 1996; Voynov et al. 2006).
GFP measurements
Cultures for GFP measurements were grown overnight in liquid YPD in 96-well plates and then pelleted and resuspended in water. Samples were transferred to Corning 96-well black clear-bottom plates and OD600 and GFP fluorescence were measured in a Tecan Safire2 plate reader. For backcrosses, high-fluorescing progeny were backcrossed to the low-fluorescing Sigma tec1Δ for three generations.
tec1Δ bypass screen
The CLN2 PEST sequence was added to the end of the HIS3 gene to target the protein product to the proteasome. Without this modification, a Sigma FLO11pr-HIS3, tec1Δ strain produces enough His3p protein from the FLO11 promoter to be His+, even in relatively high concentrations of the His3p competitive inhibitor 3-aminotriazole. The HIS3-PEST construct was created by Infusion PCR cloning (Clontech) the PEST sequence from CLN2 immediately upstream of the HIS3 stop codon in PRS315. The CLN2 PEST sequence was amplified using primers BCP316 and BCP317 and PRS315 was linearized by PCR using primers BCP320 and BCP321. To create the FLO11pr-HIS3-PEST strain, the HIS3-PEST construct was PCR amplified with primers BCP249 and BCP324. These primers have homology to replace the endogenous FLO11 ORF with the HIS3-PEST ORF, and the PCR product was transformed into yBC172. Transformants were selected on −HIS media and then correct transformants were screened for by PCR. TEC1 was deleted in FLO11pr-HIS3-PEST transformants by PCR transformation.
The FLO11pr-HIS3-PEST, tec1Δ strain was transformed with an S288c CEN/ARS genomic library (Rose et al. 1987). Transformants were first selected for 24 hr on −URA plates and then replica plated onto −URA, −HIS plates plus 5 mM 3-amino-1,2,4-triazole.
We obtained ∼300 His+ transformants from >15,000 total transformants, and we examined whether the His+ phenotype was dependent upon the plasmid by selecting for strains that had lost the plasmid on 5-FOA. After 5-FOA selection, these strains were examined, by dilution series, on −HIS plates.
Fifty-four strains required the library plasmid to be His+, and the plasmid from these strains was isolated and the ends of the insert were sequenced. Potential bypass strains were identified by examining the overlapping regions among the inserts.
qPCR
Total RNA was obtained by standard acid phenol extraction from 2 ml of culture. The QIAGEN (Valencia, CA) QuantiTect Reverse Transcription Kit was used to remove residual genomic DNA and reverse transcribe the RNA templates to generate cDNAs. Aliquots of cDNA were used in real-time PCR analyses with reagent from Applied Biosystems (Foster City, CA) and the ABI7500 real-time PCR system.
Chromatin IP
Protocols have been described in Lee et al. (2006). Briefly, IPs were performed with Dynal Protein G magnetic beads preincubated with antibodies against FLAG epitope (Sigma M2). To examine enrichment, SYBR Green qPCR (Applied Biosystems) was performed on IP and whole cell extract, using gene-specific primers.
Protein manipulations
Total protein was extracted using standard TCA precipitation with slight modifications (Graham 2001). Namely, after TCA precipitation the acetone wash was omitted and instead the cells were washed once with 1 M Tris, pH 8. For phosphatase assays, 5 μl of total protein was treated with 2 μl λ-phosphatase (New England Biolabs, Beverly, MA) for 2 hr at 30° and the reaction was stopped by adding 6× Laemmli loading buffer to 1× concentration and boiling for 10 min. Samples were run out on a 10% TGX gel [Bio-Rad (Hercules, CA) 456-1036S]. The phosphorylation of RPI1 causes it to run as a diffuse smear and the amount of signal is distributed across this entire range. To visualize phosphorylated RPI1 alongside phosphatase-treated RPI1, up to five times the amount of phosphorylated RPI1 was loaded. Blotting against FLAG was performed using HRP-conjugated anti-FLAG M2 antibody (Sigma A8592).
Bioinformatics
Gene ontology term enrichment was performed using the AMIGO term enrichment tool version 1.8 (http://amigo.geneontology.org/cgi-bin/amigo/term_enrichment).
To find intragenic repeats, the EMBOSS program ETANDEM (Rice et al. 2000) was used to screen the sequences of all S. cerevisiae (S288c version 2010 downloaded from the Saccharomyces Genome Database in April 2011) and the ∑1278b strain (Sigma downloaded from http://mcdb.colorado.edu/labs1/dowelllab/pubs/DowellRyan/ in October 2010) for repeat units of length 3–500 bp. For each ORF, we compared the length in the two strains. We screened 6685 ORFs in S288c and 6450 ORFs in Sigma. A total of 6439 ORFs were common to both strains. Of these 6439 ORFs, 5928 were identical in length. Of the remaining 511 ORFs, 127 ORFs differed in total length by at least 6 bp and showed a length difference in the repeat region of at least 6 bp. We eliminated an additional 11 ORFs because of large truncations in either the 5′ or the 3′ region of the ORF, accounting for the length differences between strains. All but 9 of the length differences in the 116 ORFs were a multiple of 3. These discrepancies could be due to sequencing errors. The length of the ORF was longer in Sigma for 60 ORFs (43 ORFs with base pair differences of 6–33, and 17 ORFs with base pair differences of ≥36). A total of 56 ORFs were longer in S288c (43 ORFS with base pair differences of 6–33, and 13 ORFs with base pair differences of ≥36).
Repeat length PCRs
Primers flanking the repeat region were designed using PRIMER3 (Rozen and Skaletsky 2000). PCR products were visualized on 10% polyacrylamide gels.
Results
Creation of an S288c FLO8 deletion library
Systematic deletion library comparison of S288c and Sigma for the adhesion phenotype required the creation of a new S288c FLO8 library because the progenitor to the standard S288c deletion library carries a flo8 mutation that prevents adhesion to agar. When S288c flo8 is transformed to Flo8+, it adheres in a FLO11-dependent fashion (Liu et al. 1996). We next assayed the entire library for the adhesion phenotype (Adh+ or Adh−) and identified deletions in the S288c library with the Adh− phenotype.
The fMAPK pathway is required for adhesion and FLO11 transcription in Sigma but not in S288c
Comparison of the loss of adhesion mutants in the Sigma and S288c deletion libraries revealed that many genes have strain-specific roles in adhesion (Table S1, Table S2, and Table S3). The strain specificity of the Ahs− phenotypes is not attributable to an integrated FLO8 in the Sigma library, but to a plasmid-borne FLO8 in the S288c library. The Ahs− phenotype was the same in 28/30 deletions tested from the S288c deletion library whether FLO8 was plasmid borne or integrated at the resident FLO8 locus (replacing the flo8 allele). All strains pursued further had the FLO8 gene integrated at its native locus in S288c.
The comparison of S288c and Sigma adhesion mutants revealed that the fMAPK pathway is required for adhesion in Sigma but it is not required for adhesion in S288c (Figure 1A). Strains carrying deletions in kinase genes—STE7, STE11, and KSS1—and the transcription factor genes—STE12 and TEC1—have a clear adhesion defect in Sigma but adhere well in S288c (Figure 1A). qPCR measurements revealed that wild-type S288c and S288c tec1Δ both show strong expression of FLO11, whereas Sigma tec1Δ has a 50-fold decrease in FLO11 RNA levels relative to the wild-type control (Figure 1C). The distinct requirement for the fMAPK pathway in Sigma but not in S288c suggests that adhesion is controlled differently in the two strains.
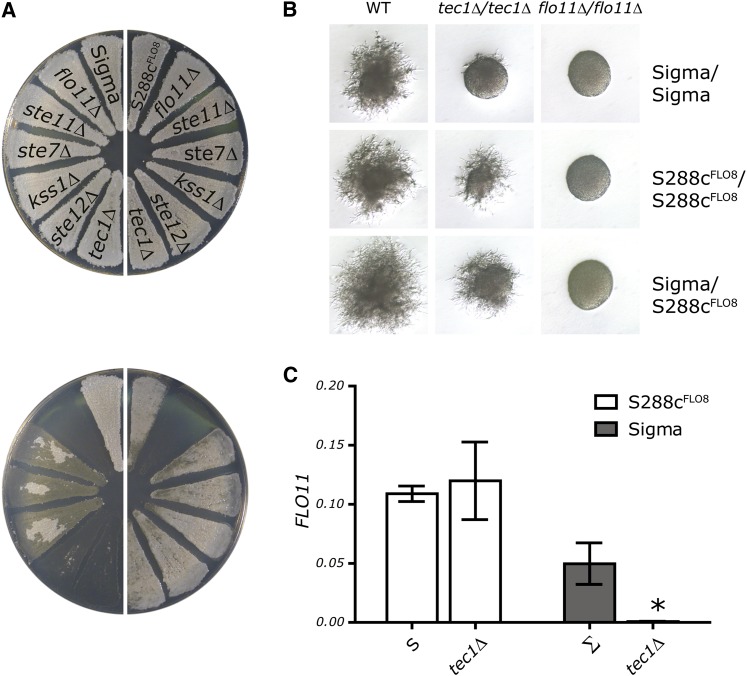
Figure 1 .
The fMAPK pathway is not required for FLO11 expression in S288c. (A) Adhesion assays performed on S288c strains (right half of the plate) or Sigma strains (left half of the plate). The same plate is shown before (top) and after (bottom) washing. (B) Pseudohyphal growth on SLAD media for diploid Sigma, S288c, or Sigma/S288c hybrids. (C) qPCR assay of FLO11 transcript levels was performed on Sigma and S288c strains that were WT or tec1Δ. Mean FLO11 levels normalized to ACT1 levels are presented ± SD. *P < 0.01 compared to WT.
The fMAPk pathway in Sigma activates FLO11 transcription for haploid adhesion and diploid filamentation (Liu et al. 1993; Roberts and Fink 1994; Lo and Dranginis 1998). To determine whether the fMAPK pathway is dispensable for diploid filamentation in S288c, we constructed diploid S288c strains. Filamentation in the S288c tec1Δ/tec1Δ strain is indistinguishable from that in wild type, whereas the Sigma tec1Δ/tec1Δ strain has a filamentation defect (Figure 1B). A hybrid S288c/Sigma tec1Δ/tec1Δ strain is able to filament, showing that the ability of S288c to bypass an fMAPK defect for filamentation is dominant. Homozygous diploid S288c flo11Δ/flo11Δ and Sigma flo11Δ/flo11Δ strains failed to form filaments. Thus, FLO11 function is required for adherence and filamentation in both S288c and Sigma even though the requirement for the fMAPK pathway is restricted to Sigma.
Differences in the FLO11 promoter sequence do not account for S288c fMAPK-independent FLO11 expression
Reciprocal promoter swap strains were used to determine whether the sequence differences between the S288c and Sigma FLO11 promoters (FLO11prS288c and FLO11prSigma, respectively) could account for the fMAPK independence of S288c. S288c FLO11prSigma adhered like a wild-type S288c as did S288c FLO11prSigmatec1Δ, showing that FLO11prS288c is not necessary for fMAPK-independent adhesion of S288c cells (Figure 2). FLO11 RNA levels in the S288c FLO11prSigma strain were consistent with the adhesion phenotypes; specifically, in S288c there was no significant difference in FLO11 RNA levels, regardless of the promoter or the presence of a tec1Δ (Figure S1A).
Figure 2 .
S288c with FLO11prSigma::FLO11 is fMAPK independent. Agar adhesion assays were performed on S288c strains (right half of the plate) or Sigma strains (left half of the plate) in the FLO11 promoter swap experiment (see text). The same plate is shown before (left) and after (right) washing. Strains with their endogenous FLO11 promoter are labeled with their relevant genotype. Strains carrying a swapped FLO11 promoter are labeled numerically: (1) S288c FLO11prSigma::FLO11; (2) S288c FLO11prSigma::FLO11, tec1Δ; (3) Sigma FLO11prS288c::FLO11, tec1Δ; and (4) Sigma FLO11prS288c::FLO11.
The FLO11prS288c does not promote FLO11 transcription as efficiently in Sigma as it does in S288c. This difference is reflected both in the adhesion assay and in the qPCR measurement of FLO11 RNA levels (Figure 2 and Figure S1B). Nevertheless, the FLO11prS288c in Sigma is TEC1 dependent for both adhesion and FLO11 transcription, whereas it is TEC1 independent in S288c. These results imply that the sequence differences in the promoters are not responsible for the fMAPK independence of S288c.
The strain difference in FLO11 regulation is genetically complex
Crosses between the adherent S288c tec1Δ strain and the nonadherent Sigma tec1Δ strain did not yield a simple segregation pattern for adherence:nonadherence. Analysis of 24 complete meiotic tetrads produced novel phenotypes (24/96 progeny were clearly adherent, 56/96 were nonadherent, and 16/96 displayed various partially adherent phenotypes) (Figure S2). Backcrosses of the F1 adherent progeny to the Sigma tec1Δ strain continued to yield non-Mendelian segregations and novel adherent phenotypes.
We considered the possibility that the failure to isolate modifiers by backcrosses was due to the lack of robustness of the adhesion assay. Moreover, agar adhesion can be affected by both transcriptional and posttranscriptional regulation of FLO11 (Voynov et al. 2006; Wolf et al. 2010). In addition, FLO11 manifests epigenetic switching between on and off states (Halme et al. 2004; Bumgarner et al. 2009). To quantitatively assess the FLO11 phenotype we used a FLO11pr::GFP construct to monitor the segregation of FLO11 transcription in S288c tec1Δ × Sigma tec1Δ crosses. These crosses directly examined the variation affecting FLO11 transcription, yet the segregation of GFP fluorescence was still complex in both the F1 generation and subsequent backcrosses (Figure S3).
Tetrad analysis of crosses between the adherent wild-type S288c and Sigma strains provided further insight into the cause of the anomalous segregation patterns. Since both wild-type strains were adherent, we expected the F1 progeny would all be adherent. However, many of the F1 progeny were nonadherent (Figure S4). These data suggest that polymorphisms between wild-type Sigma and S288c combine in the progeny to suppress FLO11 expression. This situation considerably complicates using either conventional tetrad genetic analysis or bulk segregation analysis to find alleles that bypass the fMAPK pathway. Isolation and analysis of any of the many polymorphisms contributing to fMAPK independence required another approach.
Transformation permits the isolation of a modifier from S288c conferring fMAPK-independent expression of FLO11
To overcome the challenges of mapping polymorphisms for fMAPK-independent adhesion, we developed a transformation protocol to select for plasmids carrying S288c genes that bypass the fMAPK pathway. The selection required replacement of the FLO11 ORF with a HIS3-PEST construct in the Sigma tec1Δ strain. This PEST modification enabled the visualization of slight differences in FLO11 expression when selecting for His+ transformants. The Sigma FLO11pr-HIS3-PEST, tec1Δ strain is His− whereas the S288c FLO11pr-HIS3-PEST, tec1Δ strain is His+. Modifiers from S288c that could bypass the requirement for the fMAPK pathway in Sigma were obtained by transforming the Sigma FLO11pr-HIS3-PEST, tec1Δ strain (His−) with a S288c CEN/ARS genomic library (Rose et al. 1987) and selecting for His+ transformants.
Sequence analysis of the plasmids capable of conferring the His+ phenotype to the Sigma FLO11pr-HIS3-PEST, tec1Δ strain identified several genes (including TEC1 itself). A gene with a relevant S288c polymorphism should have a sequence difference from its Sigma allele and the ability to confer the His+ phenotype (bypass the tec1Δ defect) when integrated in the chromosome in a single copy. RPI1S288c was the only gene obtained that fulfilled these criteria. When RPI1S288c replaced RPI1Sigma in the chromosome, the Sigma FLO11pr-HIS3-PEST, tec1Δ strain was His+. Moreover, RPI1S288c and RPI1Sigma differ in numerous SNPs and stretches of intragenic repeats that differ in length (Figure 3, Figure S5, and Figure S6).
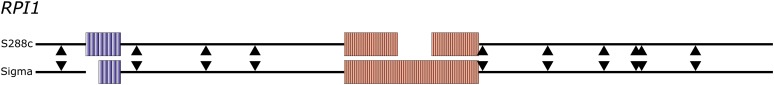
Figure 3 .
RPI1 alleles vary in the number of intragenic repeats. The S288c and Sigma alleles of RPI1 have intragenic repeats, but the repeat lengths differ between the two strains. The schematic illustrates the alignment of the two alleles. The boxes represent individual repeat elements and arrowheads represent locations of SNPs. Open areas represent the shortened repeat length in that allele.
RPI1S288c but not RPI1Sigma is a bypass suppressor of the fMAPK pathway
Consistent with the hypothesis that RPI1S288c has an allele-specific role in FLO11 expression, deletion of RPI1S288c in S288c results in a strong adhesion defect and decreased FLO11 RNA, whereas deletion of RPI1Sigma in Sigma does not (Figure 4, A–C). To further characterize the allele specificity of RPI1, we swapped RPI1 alleles between the strains. S288c RPI1Sigma displayed an adherence phenotype and FLO11 RNA levels that were not significantly different from an rpi1Δ, suggesting that RPI1Sigma is not functional in FLO11 regulation (Figure 4, A and B). Deletion of TEC1 in S288c RPI1Sigma does not further decrease adhesion or FLO11 levels. Reciprocally, the Sigma RPI1S288c strain had FLO11 mRNA levels that were comparable to wild type, and when TEC1 is deleted, Sigma RPI1S288c tec1Δ had more FLO11 RNA than the Sigma RPI1Sigma tec1Δ, but less than wild type (Figure 4C). These results show that the RPI1S288c allele promotes FLO11 expression and can partially bypass the tec1Δ; however, the RPI1Sigma allele is unable to bypass tec1Δ.
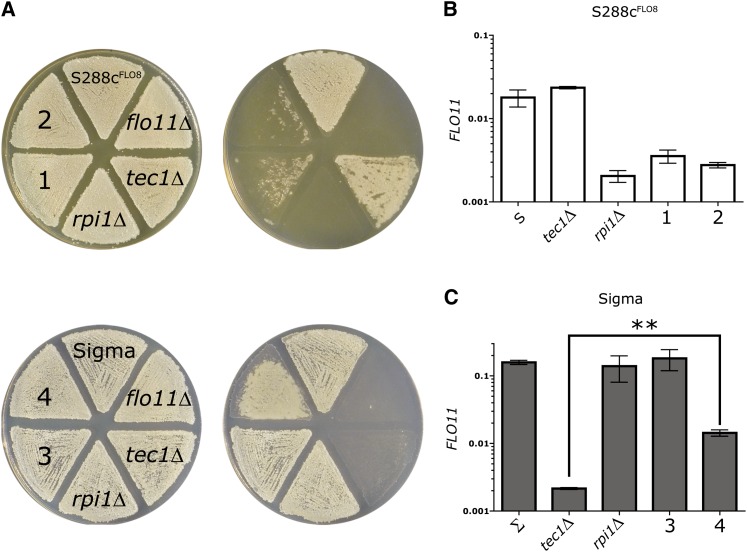
Figure 4 .
RPI1S288c can partially bypass the fMAPK pathway for agar adhesion and FLO11 expression. (A) Agar adhesion of S288c and Sigma strains carrying reciprocal allele swaps of RPI1. The top row shows adhesion assays performed on S288c strains grown on YPD and the bottom row shows adhesion assays performed on Sigma strains grown on synthetic media (see Materials and Methods). The same plates are shown before and after washing. (B and C) qPCR assay of FLO11 transcript levels performed on (B) S288c strains grown in synthetic media and (C) Sigma strains grown on YPD. Mean FLO11 levels normalized to ACT1 levels are presented ± SD. **P < 0.01. Strains with their endogenous RPI1 allele are labeled with their relevant genotype. Strains carrying a swapped RPI1 allele are labeled numerically: (1) S288c RPI1Sigma; (2) S288c RPI1Sigma, tec1Δ; (3) Sigma RPI1S288c; and (4) Sigma RPI1S288c, tec1Δ.
Rpi1p interaction with the FLO11 promoter is Rpi1p allele specific
To determine whether the difference in fMAPK-independent FLO11 expression is a consequence of differences in the ability of Rpi1pSigma and Rpi1pS288c to interact with the FLO11 promoter, we performed ChIP and tested for enrichment of the FLO11 promoter. Rpi1pS288c interacts with the FLO11 promoter with a peak around −1300 bp (Figure 5A), the site bound by other positive activators of FLO11 such as Tec1p, and Flo8p (Zeitlinger et al. 2003; Borneman et al. 2006). Immunoprecipitation of the Rpi1pS288c allele enriches for the FLO11 promoter regardless of the strain background. In contrast to Rpi1pS288c, immunoprecipitation of Rpi1pSigma results in strain-background–specific enrichment for this same region of the FLO11 promoter. When Rpi1pSigma is immunoprecipitated from a Sigma strain, it enriches for the FLO11 promoter; when it is immunoprecipitated from an S288c strain, it does not.
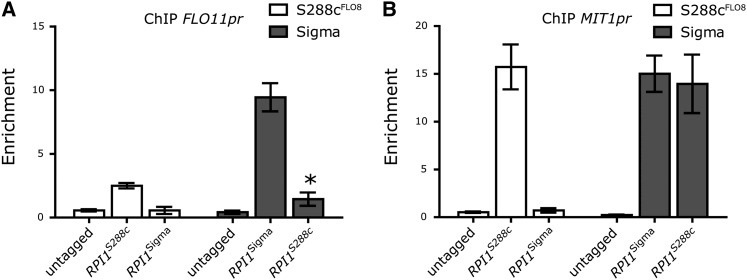
Figure 5 .
RPI1S288c shows strain-independent localization to the MIT1 and FLO11 promoters. (A and B) Localization of Rpi1p using FLAG-tagged alleles in Sigma and S288c assayed by ChIP followed by qPCR for enrichment at (A) −1.3 kb in the FLO11 promoter and (B) −1 kb in the MIT1 promoter. Data were normalized to ACT1 and are expressed as the mean fold enrichment ± SD. *P < 0.01 compared to untagged.
This difference between Rpi1pS288c and Rpi1pSigma promoter binding is also observed at the promoter of MIT1, previously identified as a target of Rpi1p and a “master regulator” of FLO11 transcription (Zeitlinger et al. 2003; Cain et al. 2011; Wang et al. 2011). However, Wang et al. and Cain et al. provided only strain-specific analyses of MIT1 and RPI1 function: The Mit1pSigma protein was shown to bind to the FLO11 promoter in Sigma, and Rpi1pS288c has been reported to localize to the promoter of MIT1S288c in S288c. Our ChIP data show that Rpi1pS288c localizes to the MIT1 promoter, regardless of strain background, but Rpi1pSigma localizes to the MIT1 promoter only in the Sigma background (Figure 5B). Furthermore, Rpi1pS288c requires a functional MIT1 to suppress a defect in the fMAPK pathway in both S288c and Sigma. Rpi1pSigma can interact with both the FLO11 and the MIT1 promoters in Sigma, but not in S288c. Thus, Rpi1pSigma must be structurally different from Rpi1pS288c and require additional factors to function.
The Rpi1p protein is differentially phosphorylated in the two strains
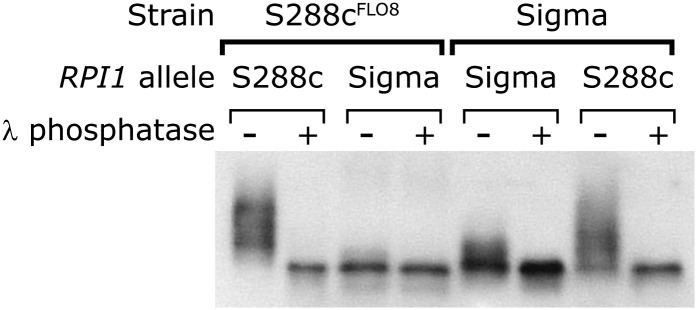
Analysis of the Rpi1p protein showed that Rpi1pS288c is structurally different from Rpi1pSigma. Figure 6 shows that 3× FLAG-tagged Rpi1pS288c extracted from S288c and visualized on Western blots runs as a diffuse species different from the Rpi1pSigma band from Sigma. When Rpi1pS288c is expressed in Sigma, it again runs as a diffuse higher molecular weight species, but when Rpi1pSigma is expressed in S288c, it runs as a single band (Figure 6).
Figure 6 .
The Rpi1pS288c protein is hyperphosphorylated. Shown is Western blot analysis of Rpi1p phosphorylation state in strains expressing either 3× flag-tagged RPI1S288c or RPI1Sigma. Samples were treated with either buffer or λ-phosphatase.
To determine whether the difference between the isoforms of Rpi1p is due to phosphorylation, protein extracts were treated with λ-phosphatase. The broad Rpi1pS288c band collapsed to a single band. This change in migration pattern occurs regardless of the strain background that expresses Rpi1pS288c. Treatment of Rpi1pSigma with phosphatase changed its migration only if the protein was obtained from a Sigma strain. These experiments show that Rpi1pSigma has strain-specific phosphorylation and likely has a different phosphorylation pattern from that of Rpi1pS288c. This altered phosphorylation pattern of Rpi1pSigma may account for its inability to activate FLO11 transcription in either strain.
The RPI1 polymorphism is not restricted to laboratory strains
The striking difference in the control of FLO11 transcription between these two strains could be attributed to their long-term culture in the laboratory. Indeed, all S288c strains have a nonsense mutation in FLO8 and many have a mutation in the KSS1 gene as well, both affecting FLO11 expression (Elion et al. 1991; Liu et al. 1996). However, an assessment RPI1 sequences shows that the S288c-like polymorphisms are widespread and found in both feral and laboratory strains (Figure S6). Thus, the expansion and contraction of RPI1 appears to be a common avenue for diversity both in the laboratory and in the wild.
Intragenic tandem repeats are highly polymorphic within a species
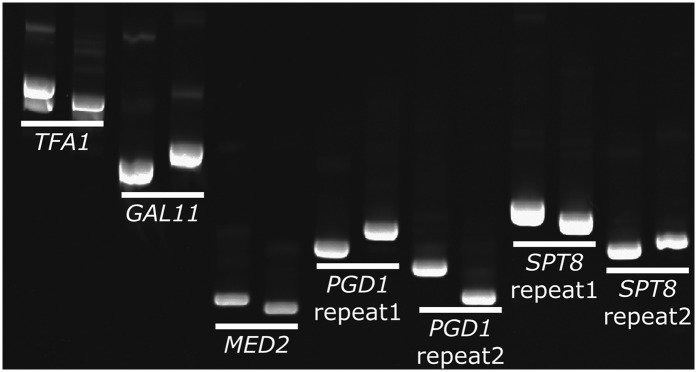
The difference in repeat length between the RPI1 alleles of S288c and Sigma led us to ask how many other genes differ in this way. Previous studies focused on cell surface proteins and have found profound phenotypic consequences for changes in the size of an internal repeat region (MacDonald et al. 1993; Verstrepen et al. 2005; Levdansky et al. 2007; Fidalgo et al. 2008; Tan et al. 2010; Sheets and St. Geme 2011), but it is difficult to perform genome-wide examinations of repeat length changes because few organisms have multiple genomes of sufficiently high quality to compare repeat regions. With the release of the Sigma genome, this comparison can be done because both the S288c and the Sigma genomes are of a high enough quality to ask, like in RPI1, how many genes differ in size due to repeat length changes? By computationally comparing the size of every ORF between S288c and Sigma, we identified 107 genes that differ in length due to in-frame expansions or contractions of intragenic repeat sequences (Table S4). The set of genes with intragenic repeat length differences includes genes involved in diverse biological processes, including transcription, chromatin modification, and signal transduction. To ensure that these differences are not due to sequencing errors, 24 of these length differences were verified by PCR (Figure 7 and Figure S7). Twenty-two of 24 genes show the predicted size difference, confirming the size differences predicted from the genome sequences’ reflected length differences in the repeats.
Figure 7 .
Many S288c genes differ from Sigma genes due to changes in intragenic tandem repeats. Twenty-four of the 107 genes predicted to differ between S288c and Sigma in the length of internal repeats were examined by PCR. Twenty-two of these genes had the predicted size difference. Five genes are shown and the results for the other genes are shown in Figure S7. PGD1 and SPT8 have two repeat regions that both change in size. For each pair the left sample is the S288c product and the right sample is the Sigma product.
Discussion
Individuals within a species may signal gene expression through different pathways
Our analysis of comparable deletion libraries in two interfertile strains of S. cerevisiae (Sigma and S288c) with nearly identical genomes (Dowell et al. 2010) allowed us to ask the question: Do the same signal transduction pathways control development in both strains? Previous mutational analyses identified the fMAPK pathway as required for adhesion and FLO11 transcription in Sigma (Roberts and Fink 1994; Cook et al. 1996; Lorenz and Heitman 1998). A recent comprehensive genome-wide analysis of the Sigma deletion library for adhesion, filamentation, and biofilm formation again uncovered the fMAPK genes (Ryan et al. 2012). Therefore, the finding that S288c does not require the fMAPK pathway was unanticipated. This functional difference is not a consequence of gene duplication but rather involves distinct genes encoding two separate pathways, each capable of eliciting the same phenotype. The two strains differ by polymorphisms in the transcription factor RPI1; the RPI1S288c allele is active and suppresses the loss of function of the fMAPK pathway; the RPI1Sigma allele is inactive and incapable of suppressing of a defect in the fMAPK pathway. These RPI1 polymorphisms must alter phosphorylation sites, change the conformation to prevent access to the sites, or prevent interaction with a kinase.
The discovery of RPI1S288c as a bypass suppressor of the fMAPK pathway provides insight into the mechanism by which allelic polymorphisms can buffer the effect of mutations and rewire a signaling pathway. Although previous studies have identified many QTL in intraspecies crosses of S. cerevisiae, many of these polymorphisms have not been connected to differences in function. As with the adhesion phenotype, each of the polymorphisms may have only a modest effect on the phenotype, making it difficult to isolate and assess the mechanism of action. We were able to tune the conditions so that we could use transformation to select for modifiers such as RPI1 that only partially restore FLO11 expression.
The presence of RPI1S288c in S288c means that loss of function of any member of the fMAPK pathway will fail to manifest an adhesion phenotype because FLO11 can now be activated by RPI1S288c. Even MSB2, the protein believed to be the sensor for the fMAPK pathway, is not needed for S288c adhesion (Table S2). The activation of FLO11 by RPI1S288c raises the question: What is upstream of RPI1 in S288c? Our genome-wide screen of the S288c library for strains with adhesion defects identified a number of potential candidates that do not have adhesion/filamentation defects in Sigma. In the future a systematic analysis of these is likely to identify those genes required for RPI1 activation.
The evolution of circuit diversification begins within a species
Comparing species that evolved from a common ancestor before and after the whole-genome duplication (WGD) (Kellis et al. 2004; Wapinski et al. 2007) has elucidated the gradual rewiring of transcription circuits in the fungal lineage. For example, yeast species post-WGD have two proteins controlling the ribosomal protein stress response, a positive (IFH1) and a negative (CRF1) regulator, whereas organisms that did not undergo the WGD have a single ancestral protein with both positive and negative activities (Wapinski et al. 2010). Post-WGD, the duplicate genes specialized with one losing a positive function and the other a negative one, while both retained “stress response control.”
The plasticity of these regulatory networks is most dramatically seen in the comparison of the regulatory circuit that regulates mating type in the human fungal pathogen Candida albicans with that of S. cerevisiae. The ensemble of genes controlling mating is largely conserved in the two organisms; however, the a-specific genes in Candida are under positive control by the a2 protein and in S. cerevisiae they are under negative control by the α2 protein. This transition from positive to negative regulation of the a-specific genes involved slight changes over evolutionary time in both the cis-acting elements in the promoters of the a-specific genes and the trans-acting regulatory proteins a2 and α2 (Tsong et al. 2006).
These variations in regulatory control observed in different species, which evolved over evolutionary time, must have arisen from variations that occurred within a single species and subsequently became fixed as sexual isolation took place. As we have shown, such variation in the circuitry of key signaling pathways exists among contemporary members of the same species. This apparent redundancy in FLO11 activation raises the question: Why are the two pathways retained? Despite the overlapping functions of the fMAPK pathway and RPI1, the organization of these genes into complex networks likely imposes constraints on the loss of one or the other of these activation pathways. The elements of the fMAPK pathway that have been conserved in both S288c and Sigma (Ste20p, Ste11p, Ste7p, and Ste12p) are under strong positive selection because they have cross-pathway functions in additional signal transduction pathways (mating, osmotic sensing). Since RPI1 regulates the cell wall under different conditions, it is also likely to function in conjunction with many pathways (Sobering et al. 2002; Puria et al. 2009; Wang et al. 2011). The finding that RPI1 localizes not only to the FLO11 promoter but also the MIT1 promoter (Wang et al. 2011), itself a transcriptional activator of FLO11 and many other genes (Cain et al. 2011), is consistent with the idea that RPI1 is also constrained by its participation in many regulatory networks.
RPI1S288c and RPI1Sigma differ by intragenic tandem repeat expansions
Although the two RPI1 alleles differ by several nucleotide changes, the most striking difference is the alteration in the size of a repeat region present in the coding sequence of the gene. These repeat polymorphisms in RPI1 are present in wild isolates of yeast as well as in many laboratory strains (Figure S6). Some wild isolates have the RPI1S288c length repeat and others have the RPI1Sigma length.
Repeats within a coding sequence create enormous flexibility for the evolution of diversity within a species. Because repeats can expand and contract at high frequencies, they permit a species to adapt to changing environments without becoming irreversibly committed to a phenotype (Rando and Verstrepen 2007). Although SNPs remain the major type of variation between S288c and Sigma, >100 genes differ in size due to repeat length differences. These data suggest that in a cross between S288c and Sigma these size polymorphisms could generate as many as 2100 genotypes in a cross. Phenotypic effects from even a tiny fraction of this variation, would provide ample grist for evolution’s mill.
Supplementary Material
Acknowledgments
We thank William Timberlake for critical reading of this manuscript. This work was supported by National Institutes of Health grant GM035010. C.B. was supported by the Natural Sciences and Engineering Research Council of Canada and Howard Hughes Medical Institute.
Footnotes
Communicating editor: C. D. Jones
Literature Cited
- Borneman A. R., Leigh-Bell J. A., Yu H., Bertone P., Gerstein M., et al. , 2006. Target hub proteins serve as master regulators of development in yeast. Genes Dev. 20: 435–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brem R. B., Yvert G., Clinton R., Kruglyak L., 2002. Genetic dissection of transcriptional regulation in budding yeast. Science 296: 752–755 [DOI] [PubMed] [Google Scholar]
- Brown T., 2001. Southern Blotting in Current Protocols in Molecular Biology. John Wiley & Sons, New York. [DOI] [PubMed] [Google Scholar]
- Bumgarner S. L., Dowell R. D., Grisafi P., Gifford D. K., Fink G. R., 2009. Toggle involving cis-interfering noncoding RNAs controls variegated gene expression in yeast. Proc. Natl. Acad. Sci. USA 106: 18321–18326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cain C. W., Lohse M. B., Homann O. R., Johnson A. D., 2011. A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics 190: 511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlborg O., Haley C. S., 2004. Epistasis: Too often neglected in complex trait studies? Nat. Rev. Genet. 5: 618–625 [DOI] [PubMed] [Google Scholar]
- Connelly C. F., Akey J. M., 2012. On the prospects of whole-genome association mapping in Saccharomyces cerevisiae. Genetics 191: 1345–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook J. G., Bardwell L., Kron S. J., Thorner J., 1996. Two novel targets of the MAP kinase Kss1 are negative regulators of invasive growth in the yeast Saccharomyces cerevisiae. Genes Dev. 10: 2831–2848 [DOI] [PubMed] [Google Scholar]
- Demogines A., Smith E., Kruglyak L., Alani E., 2008. Identification and dissection of a complex DNA repair sensitivity phenotype in Baker’s yeast. PLoS Genet. 4: e1000123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson S. P., Wang K., Krantz I., Hakonarson H., Goldstein D. B., 2010. Rare variants create synthetic genome-wide associations. PLoS Biol. 8: e1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowell R. D., Ryan O., Jansen A., Cheung D., Agarwala S., et al. , 2010. Genotype to phenotype: a complex problem. Science 328: 469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich I. M., Torabi N., Jia Y., Kent J., Martis S., et al. , 2010. Dissection of genetically complex traits with extremely large pools of yeast segregants. Nature 464: 1039–1042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elion E. A., Brill J. A., Fink G. R., 1991. FUS3 represses CLN1 and CLN2 and in concert with KSS1 promotes signal transduction. Proc. Natl. Acad. Sci. USA 88: 9392–9396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fidalgo M., Barrales R. R., Jimenez J., 2008. Coding repeat instability in the FLO11 gene of Saccharomyces yeasts. Yeast 25: 879–889 [DOI] [PubMed] [Google Scholar]
- Gatbonton T., Imbesi M., Nelson M., Akey J. M., Ruderfer D. M., et al. , 2006. Telomere length as a quantitative trait: genome-wide survey and genetic mapping of telomere length-control genes in yeast. PLoS Genet. 2: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimeno C. J., Ljungdahl P. O., Styles C. A., Fink G. R., 1992. Unipolar cell divisions in the yeast S. cerevisiae lead to filamentous growth: regulation by starvation and RAS. Cell 68: 1077–1090 [DOI] [PubMed] [Google Scholar]
- Graham, T. R., 2001 Metabolic labeling and immunoprecipitation of yeast proteins, pp. 7.6.1–7.6.9 in Current Protocols in Cell Biology, edited by J. S. Bonifacino, M. Dasso, J. B. Harford, J. Lippincott-Schwartz, and K. M. Yamada. John Wiley & Sons, New York. [DOI] [PubMed] [Google Scholar]
- Guthrie, C., and G. Fink, 2002 Guide to Yeast Genetics and Molecular and Cellular Biology Academic Press, San Diego, CA.
- Halme A., Bumgarner S., Styles C., Fink G. R., 2004. Genetic and epigenetic regulation of the FLO gene family generates cell-surface variation in yeast. Cell 116: 405–415 [DOI] [PubMed] [Google Scholar]
- Hartman J. L., IV, Garvik B., Hartwell L., 2001. Principles for the buffering of genetic variation. Science 291: 1001–1004 [DOI] [PubMed] [Google Scholar]
- Jakobsdottir J., Gorin M. B., Conley Y. P., Ferrell R. E., Weeks D. E., 2009. Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 5: e1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S., 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423: 241–254 [DOI] [PubMed] [Google Scholar]
- Kellis M., Birren B. W., Lander E. S., 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624 [DOI] [PubMed] [Google Scholar]
- Korbel J. O., Urban A. E., Affourtit J. P., Godwin B., Grubert F., et al. , 2007. Paired-end mapping reveals extensive structural variation in the human genome. Science 318: 420–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T. I., Johnstone S. E., Young R. A., 2006. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat. Protoc. 1: 729–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levdansky E., Romano J., Shadkchan Y., Sharon H., Verstrepen K. J., et al. , 2007. Coding tandem repeats generate diversity in Aspergillus fumigatus genes. Eukaryot. Cell 6: 1380–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liti G., Carter D. M., Moses A. M., Warringer J., Parts L., et al. , 2009. Population genomics of domestic and wild yeasts. Nature 458: 337–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Styles C. A., Fink G. R., 1993. Elements of the yeast pheromone response pathway required for filamentous growth of diploids. Science 262: 1741–1744 [DOI] [PubMed] [Google Scholar]
- Liu H., Styles C. A., Fink G. R., 1996. Saccharomyces cerevisiae S288C has a mutation in FLO8, a gene required for filamentous growth. Genetics 144: 967–978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W. S., Dranginis A. M., 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9: 161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M. C., Heitman J., 1998. Regulators of pseudohyphal differentiation in Saccharomyces cerevisiae identified through multicopy suppressor analysis in ammonium permease mutant strains. Genetics 150: 1443–1457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald M. E., Ambrose C. M., Duyao M. P., Myers R. H., Lin C., et al. , 1993. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. Cell 72: 971–983 [DOI] [PubMed] [Google Scholar]
- Manolio T. A., Collins F. S., Cox N. J., Goldstein D. B., Hindorff L. A., et al. , 2009. Finding the missing heritability of complex diseases. Nature 461: 747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogami S., Ohya Y., Yvert G., 2007. Genetic complexity and quantitative trait loci mapping of yeast morphological traits. PLoS Genet. 3: e31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puria R., Mannan M. A., Chopra-Dewasthaly R., Ganesan K., 2009. Critical role of RPI1 in the stress tolerance of yeast during ethanolic fermentation. FEMS Yeast Res. 9: 1161–1171 [DOI] [PubMed] [Google Scholar]
- Rando O. J., Verstrepen K. J., 2007. Timescales of genetic and epigenetic inheritance. Cell 128: 655–668 [DOI] [PubMed] [Google Scholar]
- Rice P., Longden I., Bleasby A., 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16: 276–277 [DOI] [PubMed] [Google Scholar]
- Roberts R. L., Fink G. R., 1994. Elements of a single MAP kinase cascade in Saccharomyces cerevisiae mediate two developmental programs in the same cell type: mating and invasive growth. Genes Dev. 8: 2974–2985 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Novick P., Thomas J. H., Botstein D., Fink G. R., 1987. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene 60: 237–243 [DOI] [PubMed] [Google Scholar]
- Rozen S., Skaletsky H., 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Ryan O., Shapiro R. S., Kurat C. F., Mayhew D., Baryshnikova A., et al. , 2012. Global gene deletion analysis exploring yeast filamentous growth. Science 337: 1353–1356. [DOI] [PubMed] [Google Scholar]
- Schacherer J., Shapiro J. A., Ruderfer D. M., Kruglyak L., 2009. Comprehensive polymorphism survey elucidates population structure of Saccharomyces cerevisiae. Nature 458: 342–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheets A. J., St. Geme J. W., III 2011. Adhesive activity of the haemophilus cryptic genospecies cha autotransporter is modulated by variation in tandem peptide repeats. J. Bacteriol. 193: 329–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobering A. K., Jung U. S., Lee K. S., Levin D. E., 2002. Yeast Rpi1 is a putative transcriptional regulator that contributes to preparation for stationary phase. Eukaryot. Cell 1: 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J. C., Tan A., Checkley L., Honsa C. M., Ferdig M. T., 2010. Variable numbers of tandem repeats in Plasmodium falciparum genes. J. Mol. Evol. 71: 268–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsong A. E., Tuch B. B., Li H., Johnson A. D., 2006. Evolution of alternative transcriptional circuits with identical logic. Nature 443: 415–420 [DOI] [PubMed] [Google Scholar]
- Verstrepen K. J., Jansen A., Lewitter F., Fink G. R., 2005. Intragenic tandem repeats generate functional variability. Nat. Genet. 37: 986–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voynov V., Verstrepen K. J., Jansen A., Runner V. M., Buratowski S., et al. , 2006. Genes with internal repeats require the THO complex for transcription. Proc. Natl. Acad. Sci. USA 103: 14423–14428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Mayhew D., Chen X., Johnston M., Mitra R. D., 2011. Calling cards enable multiplexed identification of the genomic targets of DNA-binding proteins. Genome Res. 21: 748–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wapinski I., Pfeffer A., Friedman N., Regev A., 2007. Natural history and evolutionary principles of gene duplication in fungi. Nature 449: 54–61 [DOI] [PubMed] [Google Scholar]
- Wapinski I., Pfiffner J., French C., Socha A., Thompson D. A., et al. , 2010. Gene duplication and the evolution of ribosomal protein gene regulation in yeast. Proc. Natl. Acad. Sci. USA 107: 5505–5510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler E. A., Shoemaker D. D., Astromoff A., Liang H., Anderson K., et al. , 1999. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285: 901–906 [DOI] [PubMed] [Google Scholar]
- Winzeler E. A., Liang H., Shoemaker D. D., Davis R. W., 2000. Functional analysis of the yeast genome by precise deletion and parallel phenotypic characterization. Novartis Found. Symp. 229: 105–109, discussion 109–111 [DOI] [PubMed] [Google Scholar]
- Wolf J. J., Dowell R. D., Mahony S., Rabani M., Gifford D. K., et al. , 2010. Feed-forward regulation of a cell fate determinant by an RNA-binding protein generates asymmetry in yeast. Genetics 185: 513–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlinger J., Simon I., Harbison C. T., Hannett N. M., Volkert T. L., et al. , 2003. Program-specific distribution of a transcription factor dependent on partner transcription factor and MAPK signaling. Cell 113: 395–404 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.