Abstract
Isocitrate dehydrogenase 1 (IDH1) gene aberrations have recently been reported in acute myeloid leukemia (AML). To evaluate the prognostic significance of IDH1 mutations in AML, we performed a meta-analysis. Fifteen studies covering a total of 8121 subjects were included in this analysis. The frequency of IDH1 R132 mutations were 4.4–9.3% for AML patients and 10.9–16.0% for cytogenetically normal (CN)-AML patients. The IDH1 mutations were associated with NPM1 mutations in 6 studies and normal cytogenetics in 5 studies. AML patients with IDH1 mutations had inferior overall survival compared to patients without the mutations (hazard ratio 1.17, 95% CI: 1.02–1.36). Additionally, in CN-AML patients, IDH1 mutations were associated with a lower complete remission rate (risk ratio 1.30, 95% CI: 1.04–1.63). Although the available literature is limited to observational studies, these results may justify the risk-adapted therapeutic strategies for AML according to the IDH1 status.
Keywords: Acute myeloid leukemia, IDH1, mutation, prognosis, meta-analysis
Introduction
Acute myeloid leukemia (AML) is a group of heterogeneous diseases with respect to its underlying cellular and molecular biology, acquired genetic profiles, and associated clinical responses to treatment [1,2]. To date, cytogenetic aberrations provide the most important prognostic information of this heterogeneous disease [3,4]. Furthermore, the molecular genetic alterations have been reported with prognostic significance. The increasing number of genetic alterations discovered in AML has additionally contributed to our understanding of mechanisms of leukemogenesis, to an improvement of individual risk assessment, and eventually to the development of risk stratification and molecularly based therapies [5].
Among these genetic alterations, recurrent somatic mutations in nicotinamide adenine dinucleotide phosphate (NADPH)-dependent isocitrate dehydrogenase 1 (IDH1) gene affecting codon R132 [6] are special in that the gene is involved in metabolism [7,8], rather than signaling pathways or transcription factors. IDH1, a citric acid cycle enzyme encoded by the IDH1 gene, converts isocitrate to α-ketoglutarate in an NADP+-dependent manner and is supposed to control redox status in cells [9,10]. Mutations of IDH1 were found to cause dominant-negative inhibition of normal enzymatic function and gain the neomorphic enzyme activity and, ultimately, catalyze the NADPH-dependent reduction of α-ketoglutarate to 2-hydroxyglutarate (2-HG) [8,11]. It is thought that consumption of NADPH and production of 2-HG could contribute to leukemogenesis [8,11,12], which might be due to the damage of DNA via the elevated levels of reactive oxygen species [9,11] and/or the induction of DNA hypermethylation via the disruption of TET2 function [13]. IDH1 mutations have been reported in 4.4% to 9.6% of patients with AML [6,14-18]. However, the prognostic implications of IDH1 mutations are less clear and wildly variable among different institutions [14,16-20]. A recent meta-analysis conducted by Zhou et al. including 11 studies suggested subtle but significant inferior eventfree survival (EFS) and possible adverse overall survival (OS) for AML patients with IDH1 mutations [21]. However, only the studies dealing with non-promyelocytic AML were considered eligible for inclusion in the Zhou study, while some current studies focus on cytogenetically normal (CN)-AML or all AML subtypes. Therefore, in order to gain a full insight into the prognostic value of IDH1 mutations in patients with AML, we conducted an updated meta-analysis, including all available clinical evidences to date.
Materials and methods
Selection of studies
Studies were eligible for inclusion in the meta-analysis if they met all of the following criteria. (1) published up to October 2012 as original articles, (2) dealt only with untreated AML patients, (3) offered survival information based on the IDH1 status: IDH1 mutations and wild type, and (4) described survival information (overall survival (OS)) and/or response to induction therapy (complete remission (CR)). Studies were excluded if they focused exclusively on children or on acute promyelocytic leukemia.
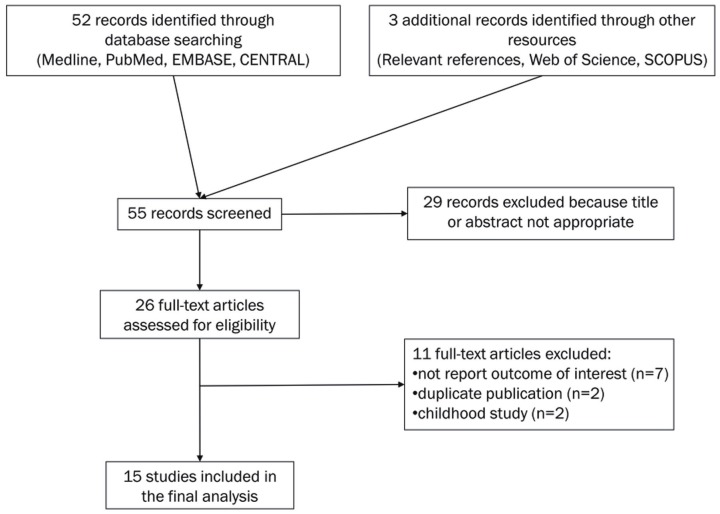
A computerized literature search of Medline, PubMed, EMBASE and The Cochrane Central Register of Controlled Trials (CENTRAL) was conducted by using the free text search term AML and isocitrate dehydrogenase and survival, with the publication period limited to before October 2012. The search was restricted to human studies with no language limitations. The initial database search yielded 52 citations. Three studies were found through other resources (Relevant references, Web of Science and SCOPUS). Abstracts of the 55 papers were reviewed, resulting in 29 of them being excluded, and leaving 26 as candidate articles. Of these, 15 studies satisfied eligibility criteria and were included in the meta-analysis (Table 1). The reasons for excluding 11 articles are shown in Figure 1 [22-32].
Table 1.
List of studies included in the meta-analysis
| Author [ref.] | Publication year | Region | N | Age (years) | Tumor types | IDH1 R132 mutation (%) |
|---|---|---|---|---|---|---|
| Ravandi [43] | 2012 | USA | 170 | 53 (17-73) | AML | 7.1% |
| Patel [44] | 2012 | USA | 657 | 48 (17-60) | AML | 7% |
| Shen [41] | 2011 | China | 605 | 43 (18-86) | AMLa | 9.3% |
| Zhang [42] | 2011 | China | 365 | 39 (15-74) | AML | 6.3% |
| Schnittger [39] | 2010 | Germany | 1414 | 66 (17-93) | AML | 6.6% |
| Abbas [17] | 2010 | Netherlands | 893 | 46 (15-77) | AML | 6.2% |
| Paschka [20] | 2010 | Germany | 805 | NA (16-60) | AML | 7.2% |
| Ho [15] | 2010 | USA | 274 | 63 (18-88) | AML | 4.4% |
| Chou [14] | 2010 | Taiwan | 493 | 53 (18-90) | AML | 5.5% |
| Ward [11] | 2010 | USA | 78 | 61 (6-91) | AML | 7.7% |
| Green [16] | 2010 | England | 1333 | 43 (15-68) | AML | 8.0% |
| Mardisb [6] | 2009 | USA | 188 | 47 (16-81) | AML | 8.5% |
| Boissel [18] | 2010 | France | 213 | 48 (17-70) | CN-AML | 16.0% |
| Marcucci [19] | 2010 | USA | 358 | 61 (19-83) | CN-AML | 13.1% |
| Wagner [40] | 2010 | Germany | 275 | 47 (17-60) | CN-AML | 10.9% |
Ref, reference; AML, acute myeloid leukemia; CN-AML, cytogenetically normal AML; —, not applicable; NA, not assessed.
Only including AML without prognostic cytogenetic markers;
Including 30 AML patients (16%) who underwent transplantation.
Figure 1.
Flow diagram showing the process of identifying and selecting relevant studies.
Data extraction and quality assessment
To avoid bias in the data abstraction process, the two reviewers (J.-H.F and Y.-M.T) independently abstracted the data from the articles and subsequently compared the results. All data were checked for internal consistency, and disagreements were resolved by discussion. Characteristics abstracted from the articles included the name of the first author, year of publication, location of the study, number of subjects, mean or median values of age and initial white blood cell (WBC) counts, the incidence of IDH1 mutations, incidence of NPM1 mutations, incidence of FLT3-ITD and percentage of cases with normal cytogenetics, and outcomes including hematologic complete remission (CR) rate and hazard ratio (HR) for OS according to the IDH1 status based on multivariate analysis. When the data required for the analysis could not be abstracted, attempts were made to contact the investigators who conducted the studies.
The quality of evidence and the strength of recommendations were evaluated by GRADE profiler (version 3.2) [33]. Any discrepancies in quality assessments were resolved by consensus amongst authors. The overall quality of the evidences was graded as moderate.
Quantitative data synthesis
HR was used to assess the survival effect of IDH1 mutations compared with wild type. The natural logarithm of a crude HR and its variance within the study was calculated by using the abstracted survival probabilities at each time point with the methods proposed by Parmar et al. [34] and described elsewhere [35]. HR was calculated to show how many times higher the probability of the survival failure was for patients with IDH1 mutations than for those with wild type, as an HR higher than unity indicates that IDH1 mutations yield a worse survival rate than wild type.
Risk ratio (RR) was calculated to describe the probability of response failure to induction treatment based on IDH1 mutation status. RR greater than one indicates that the patients with IDH1 mutations are associated with a worse outcome as compared to those without the mutations.
A Der-Simonian Laird random method was used to calculate summary HRs or RRs and their 95% confidence intervals (CI). Begg’s funnel plots [36] and Egger’s test [37] were used to detect possible publication bias. We also calculated the between-study variation (τ2) from the Q statistic [38]. All statistical analyses were conducted with Stata ver. 12 software (College Station, TX, USA). We defined a P-value of less than 0.05 as a statistically significant test result for a summary HR or RR.
Results
Study characteristics
As shown in Table 1, 15 studies with a total of 8121 subjects were included in the meta-analysis. Six studies originated from Europe [16-18,20,39,40], three from Asia [14,41,42] and six from the United States [6,11,15,19,43,44]. The frequency of IDH1 R132 mutations varied between 4.4-9.3% for AML patients and 10.9-16.0% for CN-AML patients.
IDH1 mutations were associated with a higher frequency of NPM1 mutations in six studies [14,16-18,39,43]. No significant correlation was reported between IDH1 mutations and FLT-ITD although one study showed that IDH1 mutations were associated with a lower frequency of FLT-ITD [19]. The frequency of normal cytogenetics was higher among IDH1 mutant patients in 5 studies (Table 2) [6,14,16,17,39]. We find no evidence of publication bias for either CR or OS.
Table 2.
Diagnostic characteristics according to the IDH1 status in the patients with AML and CN-AML
| Author [ref.] | IDH1 status | N | Age (years) | Initial WBC count (109/L) | NPM1 mutation (%) | FLT3-ITD (%) | Normal cytogenetics (%) |
|---|---|---|---|---|---|---|---|
| Ravandi [43] | IDH1 wild type | 158 | 53 (17-72) | 4.9 (0.3-161.5) | 24%* | 20% | 59% |
| IDH1 Mutation | 12 | 53 (36-73) | 8.8 (0.6-50.7) | 67% | 33% | 92% | |
| Shen [41] | IDH1 wild type | 585 | 38±19* | 7.8 (0.3-453) | NR | NR | NR |
| IDH1 Mutation | 34 | 48±18 | 10.1 (0.6-255) | NR | NR | NR | |
| Zhang [42] | IDH1 wild type | 342 | 39 (15-74) | 38.3 (0.5-443) | NR | NR | 36.1% |
| IDH1 Mutation | 23 | 44 (16-67) | 28.0 (1.0-127) | NR | NR | 33.3% | |
| Schnittger [39] | IDH1 wild type | 1321 | 66 (17-93) | 8.6 (0.4-600) | 25%* | 18% | 45.9%* |
| IDH1 Mutation | 93 | 67 (22-86) | 5.0 (0.3-255) | 47% | 19% | 72.0% | |
| Abbas [17] | IDH1 wild type | 743 | 45 (15-77) | 46 (0-510) | 26%* | 23.82% | 38.1%* |
| IDH1 Mutation | 55 | 50 (20-71) | 48 (1-400) | 64% | 27.27% | 71.0% | |
| Ho [15] | IDH1 wild type | 252 | 63 (18-88) | 29.1 (0.7-298) | NR | 34% | 41% |
| IDH1 Mutation | 12 | 61 (34-81) | 59.2 (1.2-98.2) | NR | 50% | 60% | |
| Chou [14] | IDH1 wild type | 466 | 38.41% ≥60 years | NA | 19%* | 23% | 46.0%* |
| IDH1 Mutation | 27 | 44.44% ≥60 years | NA | 56% | 37% | 76.9% | |
| Ward [11] | IDH1 wild type | 60 | 58 (6-86) | NR | 7% | NA | 86.3% |
| IDH1 Mutation | 6 | 70 (51-91) | NR | 17% | NA | 100% | |
| Green [16] | IDH1 wild type | 1226 | 42 (15-68) | 22.9 (0.4-480) | 36%* | 26% | 47%* |
| IDH1 Mutation | 107 | 49 (16-67) | 22.5 (0.4-502) | 65% | 25% | 74% | |
| Mardis [6] | IDH1 wild type | 172 | 46.3±15.8 | NR | 21% | 21% | 39%* |
| IDH1 Mutation | 16 | 48.9±15.4 | NR | 44% | 25% | 81% | |
| Boissel [18] | IDH1 wild type | 179 | 48 (17-70) | 12 (0.5-250) | 37%* | 20% | — |
| IDH1 Mutation | 34 | 54 (19-70) | 20 (0.8-120) | 62% | 18% | — | |
| Marcucci [19] | IDH1 wild type | 240 | 60 (19-81) | 28.4 (0.9-450) | 60% | 38%* | — |
| IDH1 Mutation | 49 | 62 (21-82) | 24.6 (0.9-152) | 71% | 20% | — | |
| Wagner [40] | IDH1 wild type | 245 | 47 (17-60) | 23.2 (0.5-328.2) | 55% | 32% | — |
| IDH1 Mutation | 30 | 50 (33-60) | 21.1 (0.65-192.0) | 57% | 13% | — |
WBC, white blood cell; NR, not reported; NA, not assessed; —, not applicable.
Statistically significant difference (P < 0.05).
Treatment outcomes
Table 3 and Table 4 show CR rate and HR for OS among AML and CN-AML patients with IDH1 mutations compared to patients without the mutations in individual studies.
Table 3.
IDH1 mutations and outcomes in acute myeloid leukemia
| Author [ref.] | IDH1 status | N | CR (%) | P-value | HR for OS | 95% CI for OS |
|---|---|---|---|---|---|---|
| Patel [44] | IDH1 wild type | 372 | NR | 1.00 | Reference | |
| IDH1 Mutation | 23 | NR | 0.86 | 0.52-1.43 | ||
| Zhang [42] | IDH1 wild type | 194 | 65.5% | NS | NR | NR |
| IDH1 Mutation | 15 | 66.70% | NR | NR | ||
| Choua [31] | IDH1 wild type | 287 | 73.60% | NR | NR | NR |
| IDH1 Mutation | 22 | 86.44% | NR | NR | ||
| Schnittger [39] | IDH1 wild type | 717 | NR | 1.00 | Reference | |
| IDH1 Mutation | 52 | NR | 1.36 | 0.93-1.97 | ||
| Abbas [17] | IDH wild type | 694 | NR | 1.00 | Reference | |
| IDH1 Mutation | 49 | NR | 1.09 | 0.76-1.58 | ||
| Paschka [20] | IDH wild type | 607 | NR | 1.00 | Reference | |
| IDH1 Mutation | NA | NR | 1.42 | 1.03-1.99 | ||
| Ho [15] | IDH1 wild type | 262 | 51% | 0.14 | 1.00 | Reference |
| IDH1 Mutation | 12 | 75% | 0.90 | 0.48-1.71 | ||
| Chou [14] | IDH1 wild type | 466 | NR | 1.00 | Reference | |
| IDH1 Mutation | 27 | NR | 0.82 | 0.38-1.78 | ||
| Ward [11] | IDH1 wild type | 72 | NR | 1.00 | Reference | |
| IDH1 Mutation | 6 | NR | 1.60 | 0.57-4.45 | ||
| Green [16] | IDH1 wild type | 1226 | 83% | NR | 1.00 | Reference |
| IDH1 Mutation | 107 | 81% | 1.06 | 0.79-1.40 | ||
| Mardis [6] | IDH1 wild type | 172 | NR | 1.00 | Reference | |
| IDH1 Mutation | 16 | NR | 1.72 | 0.98-3.01 |
CR, complete remission; HR, hazard ratio; 95% CI, 95% confidence interval; OS, overall survival; NS, not significant; NA, not assessed; NR, not reported.
For the Abbas and Paschka studies, AML patients younger than 60 years were used for survival analysis.
Table 4.
IDH1 mutations and outcomes in cytogenetically normal acute myeloid leukemia
| Author [ref.] | IDH1 status | N | CR (%) | P-value | HR for OS | 95% CI for OS |
|---|---|---|---|---|---|---|
| Ravandi [43] | IDH1 wild type | 93 | NR | 1.00 | Reference | |
| IDH1 Mutation | 11 | NR | 0.59 | 0.24-1.44 | ||
| Shen [41] | IDH1 wild type | 506 | 61.30% | 0.223 | NR | NR |
| IDH1 Mutation | 52 | 51.90% | NR | NR | ||
| Abbas [17] | IDH wild type | 268 | NR | 1.00 | Reference | |
| IDH1 Mutation | 35 | NR | 1.19 | 0.64-2.22 | ||
| Green [16] | IDH wild type | 468 | NR | 1.00 | Reference | |
| IDH1 Mutation | 60 | NR | 1.06 | 0.75-1.49 | ||
| Boissel [18] | IDH1 wild type | 179 | 86% | 0.19 | 1.00 | Reference |
| IDH1 Mutation | 34 | 76% | 0.95 | 0.58-1.54 | ||
| Marcucci [19] | IDH wild type | 240 | 75% | 0.86 | 1.00 | Reference |
| IDH1 Mutation | 49 | 73% | 1.20 | 0.83-1.73 | ||
| Wagner [40] | IDH1 wild type | 245 | 80% | 0.097 | 1.00 | Reference |
| IDH1 Mutation | 30 | 67% | 1.19 | 0.72-1.96 | ||
| Mardis [6] | IDH1 wild type | 67 | NR | 1.00 | Reference | |
| IDH1 Mutation | 13 | NR | 1.18 | 0.61-2.27 |
CR, complete remission; HR, hazard ratio; 95% CI, 95% confidence interval; OS, overall survival; NR, not reported.
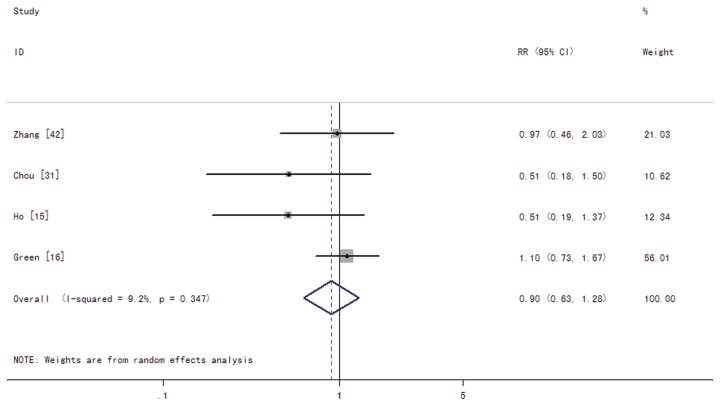
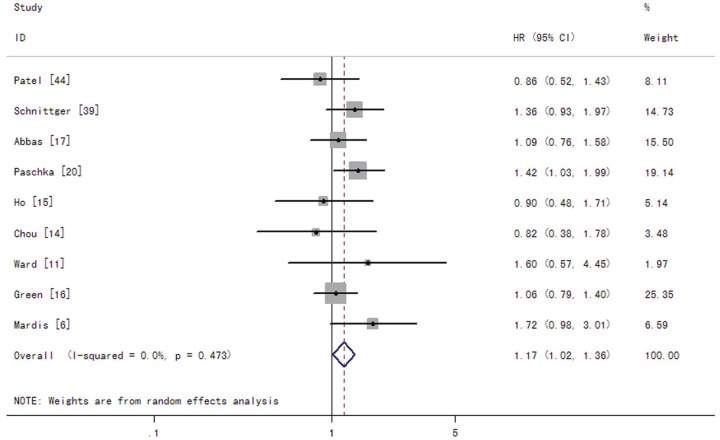
In patients with AML, the summary RRs for CR in the IDH1 mutant group were 0.90 (95% CI: 0.63-1.28 with a P-value of 0.559) (Figure 2). The summary HRs for OS were 1.17 (95% CI: 1.02-1.36 with a P-value of 0.029) for patients with the IDH1 mutations compared to those without the mutations (Figure 3). The test for heterogeneity, which evaluates variation in study outcomes between studies in a metaanalysis, showed no significant heterogeneity among studies included in OS analysis (Q = 7.61, df = 8, P = 0.473, T2 = 0).
Figure 2.
Forest plots of the risk ratios (RRs) and 95% confidence intervals for complete remission in AML patients. The size of the blocks or diamonds represents the weight for the random-effect model in the meta-analysis. A RR higher than one would indicate that the presence of IDH1 mutations is associated with a lower CR rate.
Figure 3.
Forest plots of the hazard ratios (HRs) and 95% confidence intervals for overall survival in AML patients. The size of the blocks or diamonds represents the weight for the random-effect model in the meta-analysis. A HR higher than one indicates that the presence of IDH1 mutations is associated with a worse prognosis.
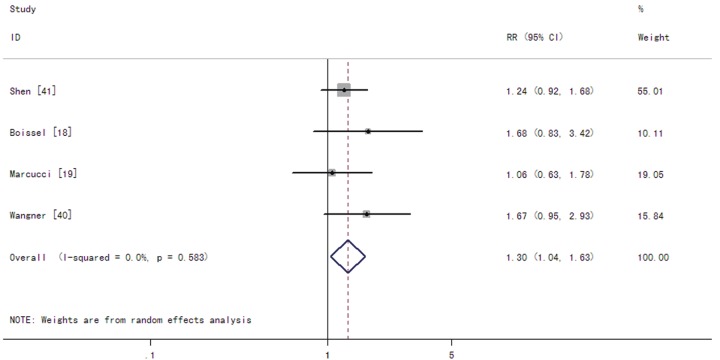
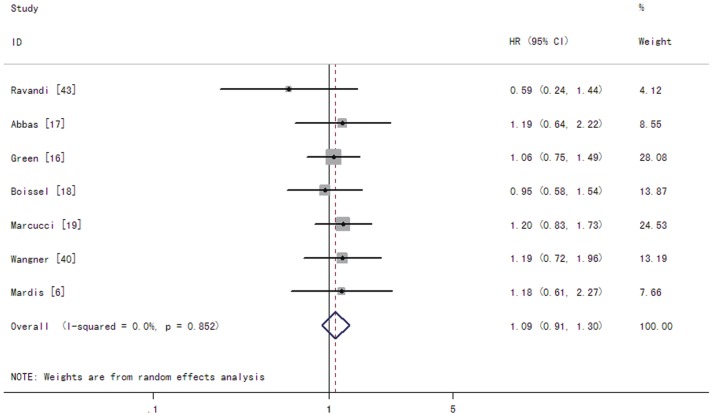
Among CN-AML patients, the summary RRs for CR of IDH1 mutations were 1.30 (95% CI: 1.04-1.63 with a P-value of 0.021) (Figure 4). The summary HRs for OS were 1.09 (95% CI: 0.91-1.30 with a P-value of 0.373) in patients with the IDH1 mutations compared to those without the mutations (Figure 5). The test for heterogeneity for OS between studies showed no evidence of heterogeneity related to IDH1 status (Q = 2.65, df = 6, P = 0.852, T2 = 0).
Figure 4.
Forest plots of the risk ratios (RRs) and 95% confidence intervals for complete remission in CN-AML patients. The size of the blocks or diamonds represents the weight for the random-effect model in the meta-analysis. A RR higher than one would indicate that the presence of IDH1 mutations is associated with a lower CR rate.
Figure 5.
Forest plots of the hazard ratios (HRs) and 95% confidence intervals for overall survival in CN-AML patients. The size of the blocks or diamonds represents the weight for the random-effect model in the meta-analysis. A HR higher than one indicates that the presence of IDH1 mutations is associated with a worse prognosis.
Furthermore, we conducted a sensitivity test during the process of meta-analysis. Exclusion of any single study did not change the overall results in any way.
Discussion
While the prognostic implication of IDH1 mutation status in patients with gliomas is well described [45,46], its importance in AML remains a matter of discussion. There have been several studies investigating the prognostic significance of IDH1 mutation status in AML patients, some of which demonstrated the negative prognostic effect of IDH1 mutations [16-20,39,42], whereas others found no clinical outcome difference between patients with and without IDH1 mutations [14,15,40,41]. The aim of the present meta-analysis was to clarify the prognostic significance of IDH1 mutation status in AML patients. Our study is a recent update on the prior meta-analysis by Zhou et al. [21] with the largest sample size and power. Also, it includes studies focusing on CN-AML and all AML subtypes, which reflects a real-world scenario. Meta-analysis is a useful statistical method for integrating results from independent studies for a specified outcome. Combining the relevant studies increases statistical power, and makes it possible to detect effects that may be missed by individual studies.
The meta-analysis reported here suggests that IDH1 mutations are associated with a higher frequency of NPM1 mutations and normal cytogenetics and with poor OS in AML patients. Interestingly, the presence of IDH1 mutations did not impact CR rates in AML patients, suggesting that the poor survival is not likely due to death during induction or induction failure. Another interesting observation of our study is that, unlike the situation in AML patients, IDH1 mutations were found to be associated with a lower CR rate in CN-AML patients, thereby supporting the notion that the clinical importance of molecular aberrations may vary according to distinct biologic and/or therapeutic contexts in which they are evaluated.
Notably, when the prognostic significance of IDH1 mutations were analyzed in CN-AML patients, the OS difference was weakened to become insignificant. This finding suggests that the issue surrounding IDH1 mutations is far more complicated than the simple presence or absence of the mutations, and needs to be put in the context of other collaborating factors. Several recurrent transcription factor aberrations such as AML1/ETO, PML/RARα and CBFβ/MYH11 were recently showed co-existence with IDH1 mutations in AML [42]. The coexistence implicates that IDH1 mutations may cooperate with these fusion genes in leukemogenesis and impact the outcome of AML with abnormal cytogenetics, which may be one of the reasons why the poor prognostic effect of IDH1 mutations is not evident in CN-AML, but only in genetically heterogeneous series of AML.
Our study has several limitations. The first problem is that the analyses were based on observational studies rather than prospective controlled studies or randomized trials. Secondly, we used abstracted data, while an individual patient data-based meta-analysis would have provided a more robust estimate of the association. The results reported here should therefore be interpreted carefully by clinical physicians. Thirdly, as is often the case with meta-analysis, there was some heterogeneity among studies in terms of diagnostic characteristics such as WBC count at the time of diagnosis as well as other confounding factors such as differences in treatment and distinct cytogenetic categories, which were not examined in our analysis. Finally, publication bias is also possible and we do not have information about studies that were not reported or published.
Although these limitations need to be borne in mind, our meta-analysis showed that IDH1 mutations have an unfavorable impact on OS for AML. Additionally, in CN-AML patients, IDH1 mutations can predict a decreased CR rate. These findings may make it advisable to distinguish AML with IDH1 mutations from AML without mutations and justify the risk-adapted therapeutic strategy for AML based on the IDH1 status. However, these conclusions should be verified in prospective clinical trials with a large number of patients. Furthermore, comprehensive functional studies are needed to understand the biologic role of the mutations in leukemogenesis.
Acknowledgements
We thank the library of Zhejiang University for entering database and acquiring full texts. This study was supported in part by grants from the National Natural Science Foundation of China (No: 30170391, No: 30971283, No: 81170502), the Natural Science Foundation of Zhejiang Province (No: Z205166) and the Zhejiang Provincial Fund of Science and Technology Bureau (No: 2007C23007).
Conflict of interest statement
The authors declare no conflict of interest.
References
- 1.Marcucci G, Haferlach T, Dohner H. Molecular genetics of adult acute myeloid leukemia: prognostic and therapeutic implications. J. Clin. Oncol. 2011;29:475–486. doi: 10.1200/JCO.2010.30.2554. [DOI] [PubMed] [Google Scholar]
- 2.Burnett A, Wetzler M, Lowenberg B. Therapeutic advances in acute myeloid leukemia. J. Clin. Oncol. 2011;29:487–494. doi: 10.1200/JCO.2010.30.1820. [DOI] [PubMed] [Google Scholar]
- 3.Mrozek K, Heerema NA, Bloomfield CD. Cytogenetics in acute leukemia. Blood Rev. 2004;18:115–136. doi: 10.1016/S0268-960X(03)00040-7. [DOI] [PubMed] [Google Scholar]
- 4.Grimwade D. The clinical significance of cytogenetic abnormalities in acute myeloid leukaemia. Best Pract Res Clin Haematol. 2001;14:497–529. doi: 10.1053/beha.2001.0152. [DOI] [PubMed] [Google Scholar]
- 5.Dohner H, Estey EH, Amadori S, Appelbaum FR, Buchner T, Burnett AK, Dombret H, Fenaux P, Grimwade D, Larson RA, Lo-Coco F, Naoe T, Niederwieser D, Ossenkoppele GJ, Sanz MA, Sierra J, Tallman MS, Lowenberg B, Bloomfield CD. Diagnosis and management of acute myeloid leukemia in adults: recommendations from an international expert panel, on behalf of the European LeukemiaNet. Blood. 2010;115:453–474. doi: 10.1182/blood-2009-07-235358. [DOI] [PubMed] [Google Scholar]
- 6.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ. Recurring mutations found by sequencing an acute myeloid leukemia genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gross S, Cairns RA, Minden MD, Driggers EM, Bittinger MA, Jang HG, Sasaki M, Jin S, Schenkein DP, Su SM, Dang L, Fantin VR, Mak TW. Cancer-associated metabolite 2-hydroxyglutarate accumulates in acute myelogenous leukemia with isocitrate dehydrogenase 1 and 2 mutations. J Exp Med. 2010;207:339–344. doi: 10.1084/jem.20092506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, Ding J, Lei Q, Guan KL, Xiong Y. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reitman ZJ, Yan H. Isocitrate dehydrogenase 1 and 2 mutations in cancer: alterations at a crossroads of cellular metabolism. J Natl Cancer Inst. 2010;102:932–941. doi: 10.1093/jnci/djq187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward PS, Patel J, Wise DR, Abdel-Wahab O, Bennett BD, Coller HA, Cross JR, Fantin VR, Hedvat CV, Perl AE, Rabinowitz JD, Carroll M, Su SM, Sharp KA, Levine RL, Thompson CB. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdel-Wahab O, Levine RL. Metabolism and the leukemic stem cell. J Exp Med. 2010;207:677–680. doi: 10.1084/jem.20100523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Figueroa ME, Abdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJ, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chou WC, Hou HA, Chen CY, Tang JL, Yao M, Tsay W, Ko BS, Wu SJ, Huang SY, Hsu SC, Chen YC, Huang YN, Chang YC, Lee FY, Liu MC, Liu CW, Tseng MH, Huang CF, Tien HF. Distinct clinical and biologic characteristics in adult acute myeloid leukemia bearing the isocitrate dehydrogenase 1 mutation. Blood. 2010;115:2749–2754. doi: 10.1182/blood-2009-11-253070. [DOI] [PubMed] [Google Scholar]
- 15.Ho PA, Alonzo TA, Kopecky KJ, Miller KL, Kuhn J, Zeng R, Gerbing RB, Raimondi SC, Hirsch BA, Oehler V, Hurwitz CA, Franklin JL, Gamis AS, Petersdorf SH, Anderson JE, Reaman GH, Baker LH, Willman CL, Bernstein ID, Radich JP, Appelbaum FR, Stirewalt DL, Meshinchi S. Molecular alterations of the IDH1 gene in AML: a Children’s Oncology Group and Southwest Oncology Group study. Leukemia. 2010;24:909–913. doi: 10.1038/leu.2010.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green CL, Evans CM, Hills RK, Burnett AK, Linch DC, Gale RE. The prognostic significance of IDH1 mutations in younger adult patients with acute myeloid leukemia is dependent on FLT3/ITD status. Blood. 2010;116:2779–2782. doi: 10.1182/blood-2010-02-270926. [DOI] [PubMed] [Google Scholar]
- 17.Abbas S, Lugthart S, Kavelaars FG, Schelen A, Koenders JE, Zeilemaker A, van Putten WJ, Rijneveld AW, Lowenberg B, Valk PJ. Acquired mutations in the genes encoding IDH1 and IDH2 both are recurrent aberrations in acute myeloid leukemia: prevalence and prognostic value. Blood. 2010;116:2122–2126. doi: 10.1182/blood-2009-11-250878. [DOI] [PubMed] [Google Scholar]
- 18.Boissel N, Nibourel O, Renneville A, Gardin C, Reman O, Contentin N, Bordessoule D, Pautas C, de Revel T, Quesnel B, Huchette P, Philippe N, Geffroy S, Terre C, Thomas X, Castaigne S, Dombret H, Preudhomme C. Prognostic impact of isocitrate dehydrogenase enzyme isoforms 1 and 2 mutations in acute myeloid leukemia: a study by the Acute Leukemia French Association group. J. Clin. Oncol. 2010;28:3717–3723. doi: 10.1200/JCO.2010.28.2285. [DOI] [PubMed] [Google Scholar]
- 19.Marcucci G, Maharry K, Wu YZ, Radmacher MD, Mrozek K, Margeson D, Holland KB, Whitman SP, Becker H, Schwind S, Metzeler KH, Powell BL, Carter TH, Kolitz JE, Wetzler M, Carroll AJ, Baer MR, Caligiuri MA, Larson RA, Bloomfield CD. IDH1 and IDH2 gene mutations identify novel molecular subsets within de novo cytogenetically normal acute myeloid leukemia: a Cancer and Leukemia Group B study. J. Clin. Oncol. 2010;28:2348–2355. doi: 10.1200/JCO.2009.27.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paschka P, Schlenk RF, Gaidzik VI, Habdank M, Kronke J, Bullinger L, Spath D, Kayser S, Zucknick M, Gotze K, Horst HA, Germing U, Dohner H, Dohner K. IDH1 and IDH2 mutations are frequent genetic alterations in acute myeloid leukemia and confer adverse prognosis in cytogenetically normal acute myeloid leukemia with NPM1 mutation without FLT3 internal tandem duplication. J. Clin. Oncol. 2010;28:3636–3643. doi: 10.1200/JCO.2010.28.3762. [DOI] [PubMed] [Google Scholar]
- 21.Zhou KG, Jiang LJ, Shang Z, Wang J, Huang L, Zhou JF. Potential application of IDH1 and IDH2 mutations as prognostic indicators in non-promyelocytic acute myeloid leukemia: a meta-analysis. Leuk Lymphoma. 2012;53:2423–9. doi: 10.3109/10428194.2012.695359. [DOI] [PubMed] [Google Scholar]
- 22.Damm F, Thol F, Hollink I, Zimmermann M, Reinhardt K, van den Heuvel-Eibrink MM, Zwaan CM, de Haas V, Creutzig U, Klusmann JH, Krauter J, Heuser M, Ganser A, Reinhardt D, Thiede C. Prevalence and prognostic value of IDH1 and IDH2 mutations in childhood AML: a study of the AML-BFM and DCOG study groups. Leukemia. 2011;25:1704–1710. doi: 10.1038/leu.2011.142. [DOI] [PubMed] [Google Scholar]
- 23.Ho PA, Kutny MA, Alonzo TA, Gerbing RB, Joaquin J, Raimondi SC, Gamis AS, Meshinchi S. Leukemic mutations in the methylation-associated genes DNMT3A and IDH2 are rare events in pediatric AML: a report from the Children’s Oncology Group. Pediatr Blood Cancer. 2011;57:204–209. doi: 10.1002/pbc.23179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Green CL, Evans CM, Zhao L, Hills RK, Burnett AK, Linch DC, Gale RE. The prognostic significance of IDH2 mutations in AML depends on the location of the mutation. Blood. 2011;118:409–412. doi: 10.1182/blood-2010-12-322479. [DOI] [PubMed] [Google Scholar]
- 25.Lin J, Yao DM, Qian J, Chen Q, Qian W, Li Y, Yang J, Wang CZ, Chai HY, Qian Z, Xiao GF, Xu WR. IDH1 and IDH2 mutation analysis in Chinese patients with acute myeloid leukemia and myelodysplastic syndrome. Ann Hematol. 2012;91:519–525. doi: 10.1007/s00277-011-1352-7. [DOI] [PubMed] [Google Scholar]
- 26.Thol F, Damm F, Wagner K, Gohring G, Schlegelberger B, Hoelzer D, Lubbert M, Heit W, Kanz L, Schlimok G, Raghavachar A, Fiedler W, Kirchner H, Heil G, Heuser M, Krauter J, Ganser A. Prognostic impact of IDH2 mutations in cytogenetically normal acute myeloid leukemia. Blood. 2010;116:614–616. doi: 10.1182/blood-2010-03-272146. [DOI] [PubMed] [Google Scholar]
- 27.Ho PA, Kopecky KJ, Alonzo TA, Gerbing RB, Miller KL, Kuhn J, Zeng R, Ries RE, Raimondi SC, Hirsch BA, Oehler V, Hurwitz CA, Franklin JL, Gamis AS, Petersdorf SH, Anderson JE, Godwin JE, Reaman GH, Willman CL, Bernstein ID, Radich JP, Appelbaum FR, Stirewalt DL, Meshinchi S. Prognostic implications of the IDH1 synonymous SNP rs11554137 in pediatric and adult AML: a report from the Children’s Oncology Group and SWOG. Blood. 2011;118:4561–4566. doi: 10.1182/blood-2011-04-348888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chotirat S, Thongnoppakhun W, Promsuwicha O, Boonthimat C, Auewarakul CU. Molecular alterations of isocitrate dehydrogenase 1 and 2 (IDH1 and IDH2) metabolic genes and additional genetic mutations in newly diagnosed acute myeloid leukemia patients. J Hematol Oncol. 2012;5:5. doi: 10.1186/1756-8722-5-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Byers R, Hornick JL, Tholouli E, Kutok J, Rodig SJ. Detection of IDH1 R132H mutation in acute myeloid leukemia by mutation-specific immunohistochemistry. Appl Immunohistochem Mol Morphol. 2012;20:37–40. doi: 10.1097/PAI.0b013e31822c132e. [DOI] [PubMed] [Google Scholar]
- 30.Rockova V, Abbas S, Wouters BJ, Erpelinck CA, Beverloo HB, Delwel R, van Putten WL, Lowenberg B, Valk PJ. Risk stratification of intermediate-risk acute myeloid leukemia: integrative analysis of a multitude of gene mutation and gene expression markers. Blood. 2011;118:1069–1076. doi: 10.1182/blood-2011-02-334748. [DOI] [PubMed] [Google Scholar]
- 31.Chou WC, Lei WC, Ko BS, Hou HA, Chen CY, Tang JL, Yao M, Tsay W, Wu SJ, Huang SY, Hsu SC, Chen YC, Chang YC, Kuo KT, Lee FY, Liu MC, Liu CW, Tseng MH, Huang CF, Tien HF. The prognostic impact and stability of Isocitrate dehydrogenase 2 mutation in adult patients with acute myeloid leukemia. Leukemia. 2011;25:246–253. doi: 10.1038/leu.2010.267. [DOI] [PubMed] [Google Scholar]
- 32.Nomdedeu J, Hoyos M, Carricondo M, Esteve J, Bussaglia E, Estivill C, Ribera JM, Duarte R, Salamero O, Gallardo D, Pedro C, Aventin A, Brunet S, Sierra J. Adverse impact of IDH1 and IDH2 mutations in primary AML: experience of the Spanish CETLAM group. Leuk Res. 2012;36:990–997. doi: 10.1016/j.leukres.2012.03.019. [DOI] [PubMed] [Google Scholar]
- 33.Guyatt GH, Oxman AD, Vist GE, Kunz R, Falck- Ytter Y, Alonso-Coello P, Schunemann HJ. GRADE: an emerging consensus on rating quality of evidence and strength of recommendations. BMJ. 2008;336:924–926. doi: 10.1136/bmj.39489.470347.AD. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parmar MK, Torri V, Stewart L. Extracting summary statistics to perform meta-analyses of the published literature for survival endpoints. Stat Med. 1998;17:2815–2834. doi: 10.1002/(sici)1097-0258(19981230)17:24<2815::aid-sim110>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 35.Hotta K, Matsuo K, Ueoka H, Kiura K, Tabata M, Tanimoto M. Meta-analysis of randomized clinical trials comparing Cisplatin to Carboplatin in patients with advanced non-small-cell lung cancer. J. Clin. Oncol. 2004;22:3852–3859. doi: 10.1200/JCO.2004.02.109. [DOI] [PubMed] [Google Scholar]
- 36.Begg CB, Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- 37.Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DerSimonian R, Laird N. Meta-analysis in clinical trials. Control Clin Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 39.Schnittger S, Haferlach C, Ulke M, Alpermann T, Kern W, Haferlach T. IDH1 mutations are detected in 6.6% of 1414 AML patients and are associated with intermediate risk karyotype and unfavorable prognosis in adults younger than 60 years and unmutated NPM1 status. Blood. 2010;116:5486–5496. doi: 10.1182/blood-2010-02-267955. [DOI] [PubMed] [Google Scholar]
- 40.Wagner K, Damm F, Gohring G, Gorlich K, Heuser M, Schafer I, Ottmann O, Lubbert M, Heit W, Kanz L, Schlimok G, Raghavachar AA, Fiedler W, Kirchner HH, Brugger W, Zucknick M, Schlegelberger B, Heil G, Ganser A, Krauter J. Impact of IDH1 R132 mutations and an IDH1 single nucleotide polymorphism in cytogenetically normal acute myeloid leukemia: SNP rs11554137 is an adverse prognostic factor. J. Clin. Oncol. 2010;28:2356–2364. doi: 10.1200/JCO.2009.27.6899. [DOI] [PubMed] [Google Scholar]
- 41.Shen Y, Zhu YM, Fan X, Shi JY, Wang QR, Yan XJ, Gu ZH, Wang YY, Chen B, Jiang CL, Yan H, Chen FF, Chen HM, Chen Z, Jin J, Chen SJ. Gene mutation patterns and their prognostic impact in a cohort of 1185 patients with acute myeloid leukemia. Blood. 2011;118:5593–5603. doi: 10.1182/blood-2011-03-343988. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Wei H, Wang M, Huai L, Mi Y, Lin D, Liu B, Li W, Zhou C, Rao Q, Wang J. Some novel features of IDH1-mutated acute myeloid leukemia revealed in Chinese patients. Leuk Res. 2011;35:1301–1306. doi: 10.1016/j.leukres.2011.01.019. [DOI] [PubMed] [Google Scholar]
- 43.Ravandi F, Patel K, Luthra R, Faderl S, Konopleva M, Kadia T, Brandt M, Pierce S, Kornblau S, Andreeff M, Wang X, Garcia-Manero G, Cortes J, Kantarjian H. Prognostic significance of alterations in IDH enzyme isoforms in patients with AML treated with high-dose cytarabine and idarubicin. Cancer. 2012;118:2665–2673. doi: 10.1002/cncr.26580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J, Van Vlierberghe P, Dolgalev I, Thomas S, Aminova O, Huberman K, Cheng J, Viale A, Socci ND, Heguy A, Cherry A, Vance G, Higgins RR, Ketterling RP, Gallagher RE, Litzow M, van den Brink MR, Lazarus HM, Rowe JM, Luger S, Ferrando A, Paietta E, Tallman MS, Melnick A, Abdel-Wahab O, Levine RL. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med. 2012;366:1079–1089. doi: 10.1056/NEJMoa1112304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weller M, Felsberg J, Hartmann C, Berger H, Steinbach JP, Schramm J, Westphal M, Schackert G, Simon M, Tonn JC, Heese O, Krex D, Nikkhah G, Pietsch T, Wiestler O, Reifenberger G, von Deimling A, Loeffler M. Molecular predictors of progression-free and overall survival in patients with newly diagnosed glioblastoma: a prospective translational study of the German Glioma Network. J. Clin. Oncol. 2009;27:5743–5750. doi: 10.1200/JCO.2009.23.0805. [DOI] [PubMed] [Google Scholar]