SUMMARY
Widespread changes in gene expression drive tumorigenesis, yet our knowledge of how aberrant epigenomic and transcriptome profiles arise in cancer cells is poorly understood. Here, we demonstrate that metabolic transformation plays an important role. Butyrate is the primary energy source of normal colonocytes and is metabolized to acetyl-CoA, which was shown to be important not only for energetics but also for HAT activity. Due to the Warburg effect, cancerous colonocytes rely on glucose as their primary energy source so butyrate accumulated and functioned as an HDAC inhibitor. Although both mechanisms increased histone acetylation, different target genes were upregulated. Consequently, butyrate stimulated the proliferation of normal colonocytes and cancerous colonocytes when the Warburg effect was prevented from occurring, whereas it inhibited the proliferation of cancerous colonocytes undergoing the Warburg effect. These findings link a common metabolite to epigenetic mechanisms that are differentially utilized by normal and cancerous cells because of their inherent metabolic differences.
Keywords: Warburg effect, colorectal cancer, butyrate paradox, ATP citrate lyase, acetyl-CoA, HAT, HDAC, metaboloepigenetics
INTRODUCTION
It is generally accepted that diet and energy metabolism influence gene expression, yet our knowledge of how this occurs is extremely limited. Understanding the mechanisms that link cellular energy metabolism and regulation of gene expression is of fundamental importance and could potentially be exploited for the treatment of various disease states such as cancer. Due to the Warburg effect, cancer cells primarily undergo aerobic glycolysis instead of oxidative metabolism (Vander Heiden et al., 2009). This metabolic shift alters the production and utilization of numerous metabolites including acetyl-CoA. In addition to being a crucial metabolite in several metabolic pathways, acetyl-CoA is an essential co-factor for histone acetyl transferases (HATs) that epigenetically regulate gene expression (Donohoe and Bultman, 2012; Wellen et al., 2009). The importance of epigenetic perturbations in driving tumorigenesis is well documented. Chromosomal rearrangements that deregulate HATs are a common cause of leukemias (Yang, 2004), and promoter CpG hypermethylation (coupled with histone hypoacetylation) silences tumor-suppressor genes in leukemias and solid tumors (Esteller, 2008). At a genome-wide level, cancer cells exhibit aberrant profiles of histone acetylation and other epigenomic marks that result in altered patterns of gene expression and genomic instability (Baylin and Jones, 2011).
Butyrate is a short-chain fatty acid produced by bacterial fermentation of dietary fiber in the colon (Hamer et al., 2008; Scheppach and Weiler, 2004). Present at high levels (mM) in the lumen, butyrate is the primary energy source for colonocytes (Fleming et al., 1991; Roediger, 1982) and also functions as a histone deacetylase (HDAC) inhibitor [reviewed in (Davie, 2003). Rodent and human studies have shown that fiber and butyrate ameliorate inflammation associated with colitis and may prevent colorectal cancer (Hamer et al., 2008; Kim and Milner, 2007; Pierre et al., 1997; Scheppach et al., 1992; Sengupta et al., 2006). Other studies have demonstrated that butyrate inhibits the growth of colorectal cancer cell lines and other tumor-derived cell lines (Hamer et al., 2008; Scheppach and Weiler, 2004). This inhibitory effect on cancer cell growth, although complicated, has been attributed to butyrate functioning as an HDAC inhibitor, which alters the expression of many genes with diverse functions, some of which include cell proliferation, apoptosis, and differentiation (Archer et al., 1998; Chopin et al., 2002; Velcich et al., 1995). In contrast to colorectal cancer cells, butyrate does not inhibit cell growth when it is delivered to the normal colonic epithelium in rodents or when it is added to noncancerous colonocytes in vitro. Instead, butyrate has either no significant effect or the opposite effect of stimulating cell growth under these conditions [reviewed in (Lupton, 2004)]. The fact that butyrate can have opposing effects on the growth of normal versus cancerous colonocytes has been referred to as the butyrate paradox and is poorly understood.
We reasoned that the dual functions of butyrate in energetics and epigenetics combined with its differential effects in normal and cancerous cells make it an ideal model system to investigate mechanistic links between cellular energy metabolism and regulation of gene expression. Specifically, we hypothesized that the butyrate paradox can be explained by the Warburg effect. Thus, butyrate may stimulate the growth of normal colonocytes by functioning as an oxidative energy source, since it is a fatty acid metabolized by β-oxidation followed by the tricarboxylic acid (TCA) cycle, whereas butyrate may inhibit the growth of cancerous colonocytes because it is metabolized inefficiently due to the Warburg effect, accumulates in the nucleus, and functions as an HDAC inhibitor to upregulate the expression of downstream target genes. While performing experiments that support this hypothesis, we also discovered that the role of butyrate in histone acetylation is more complex than previously realized. In addition to functioning as an HDAC inhibitor, butyrate increases histone acetylation by being metabolized to acetyl-CoA and stimulating HAT activity. Furthermore, the metabolic state of the cell influences intranuclear butyrate and acetyl-CoA levels and determines whether butyrate functions to inhibit HDACs or stimulate HATs to epigenetically regulate the expression of different target genes.
RESULTS
Prevention of the Warburg Effect in Cancer Cells
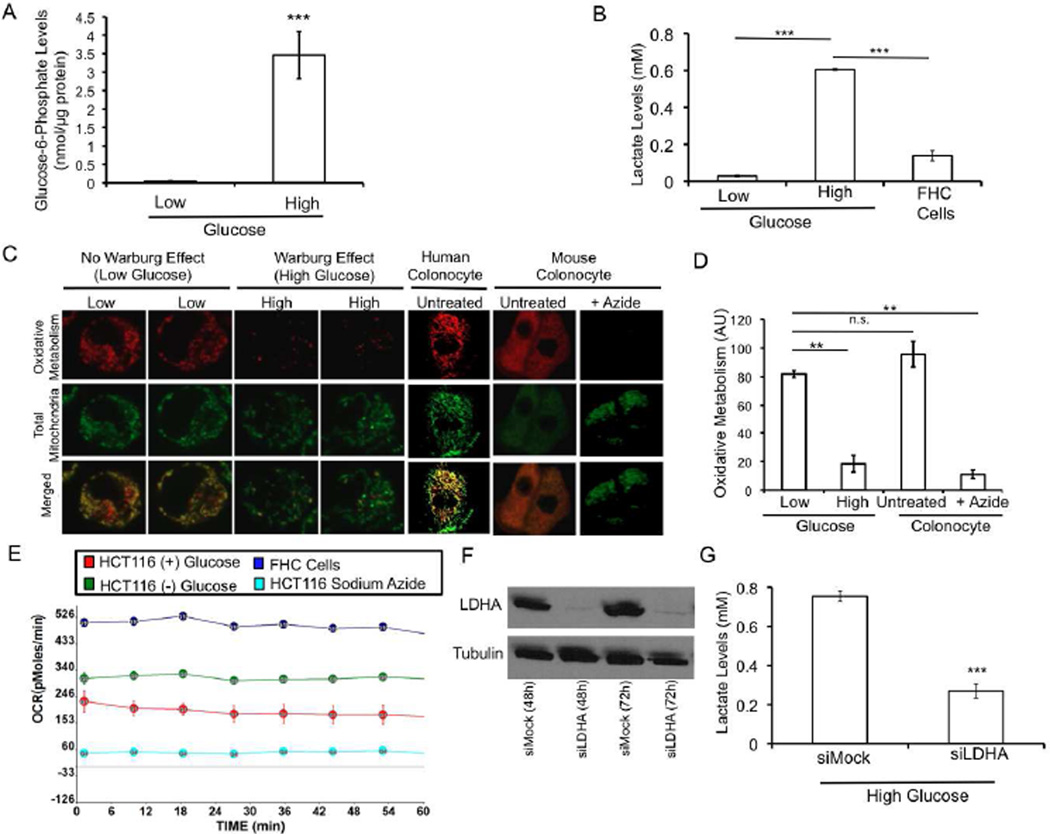
To test whether the Warburg effect would account for the differential effects of butyrate on cell proliferation and apoptosis, it was first necessary to prevent the Warburg effect from occurring in cancer cells within the context of a tightly controlled experimental system. Therefore, we grew HCT116 colon carcinoma cells in Dulbecco’s Modified Eagle Medium (DMEM) formulated without glucose but supplemented with 10% fetal bovine serum (FBS). The glucose concentration was ~0.5 mM because of the contribution of glucose from the FBS (Ramakrishnan et al., 2011). Under these conditions (herein referred to as low glucose), glucose levels were too low to support aerobic glycolysis since we detected negligible levels of glucose-6-phosphate (the first intermediate of glycolysis) and lactate (an end product of glycolysis) (Figures 1A and B). These low lactate levels were similar to noncancerous fetal human colonic (FHC) epithelial cells that do not undergo the Warburg effect regardless of glucose availability (Figure 1B). As a control, we grew HCT116 cells in DMEM formulated with glucose (25 mM) and supplemented with 10% FBS (herein referred to as the high-glucose condition). With the glucose substrate not limiting, aerobic glycolysis was elevated as judged by increased levels of glucose-6-phosphate and lactate (Figures 1A and B).
Figure 1. Prevention of the Warburg Effect in Colon Carcinoma Cells.
(A and B) Glucose-6-phosphate (A) and lactate (B) levels in HCT116 colon cancer cells grown in low or high glucose conditions and in noncancerous fetal human colonocyte (FHC) epithelial cells. Results are from 3 independent experiments and are presented as mean ± SE with significant differences indicated (***p < 0.001).
(C) Confocal microscopic images of colonocytes showing MitoTracker Red as a measure of oxidative metabolism (top row), MitoTracker Green as a measure of total mitochondria (middle row), and both MitoTracker probes merged (bottom row). The first four columns correspond to HCT116 cells grown in low or high glucose as indicated. The final three columns are two positive controls consisting of noncancerous human and mouse colonocytes followed by a negative control where the mouse colonocytes were treated with sodium azide to block oxidative metabolism.
(D) Quantification of oxidative metabolism. Results are based on 20 cells per condition from 3 independent experiments and represent the mean ± SE with significant differences indicated (**p<0.01; n.s., not significant).
(E) Oxygen consumption rates (OCR) for HCT116 cells grown in low or high glucose conditions and in FHC epithelial cells over a 1 hr period.
(F) Western blot analysis of lactate dehydrogenase A (LDHA) levels in HCT116 cells at two timepoints (48h and 72h) following RNAi (siLDHA) and in control cells (siMock). Tubulin (lower panel) serves as a loading control.
(G) Lactate levels in siLDHA and siMock HCT116 cells. Results are from 3 independent experiments and are presented as mean ± SE with significant differences indicated (***p < 0.001).
We hypothesized that cells grown in low glucose to prevent the Warburg effect would have elevated oxidative metabolism compared to their counterparts grown in high glucose. To test whether this is the case, we performed experiments where HCT116 colon carcinoma cells were incubated with two mitotracker probes simultaneously. One probe fluoresced red upon being oxidized and provided a measure of oxidative metabolism (Paxinou et al., 2001), while the other probe fluoresced green regardless of oxidative metabolism and therefore served as an internal control. Oxidative metabolism was elevated 4-fold in cells grown in low glucose compared to cells grown in high glucose (Figures 1C and D). We found that HCT116 colon carcinoma cells grown in low glucose underwent oxidative metabolism at levels comparable to noncancerous colonocytes from humans and mice (Figures 1C and D). We verified these findings by performing extracellular flux experiments. These experiments demonstrated that the rate of oxygen consumption in HCT116 cells grown in low glucose and in noncancerous FHC cells was 2- to 3-fold greater than HCT116 cells undergoing the Warburg effect (Figure 1E).
To prevent the Warburg effect from occurring in HCT116 cells using an independent methodology, we used RNAi to knockdown lactate dehydrogenase A (LDHA). Western blot analysis demonstrated a robust depletion of LDHA (Figure 1F), and lactate production was diminished by 64% in siLDHA cells compared to siMock cells (Figure 1G). This knockdown is also known to increase oxygen consumption (Le et al., 2010). Taken together, these results define a controlled cellular system by which the Warburg effect can be manipulated to analyze its role in cell proliferation and apoptosis.
Butyrate Increases or Decreases Cell Proliferation Depending on the Warburg Effect
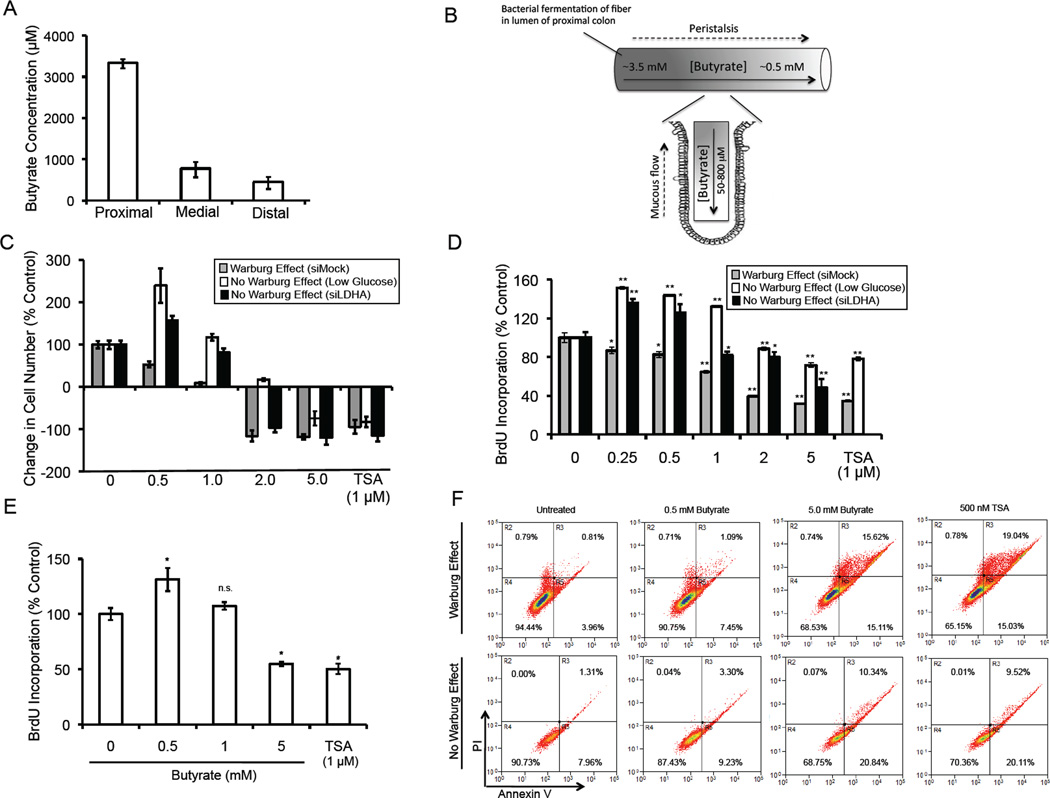
To define a physiological range of butyrate doses, we used liquid chromatography tandem mass spectrometry (LC-MS/MS) to measure butyrate in the lumen of mouse colons. We detected 3.5 mM, 0.8 mM, and 0.5 mM concentration of butyrate in the proximal, medial, and distal segments, respectively (Figure 2A). We also calculated low levels of butyrate (50–800 µM) in the colonic crypts (data not shown). This is consistent with a model (Figure 2B) predicting two butyrate gradients in the colon in vivo (Csordas, 1996; Sengupta et al., 2006).
Figure 2. Butyrate-Induced Changes in Cell Proliferation and Apoptosis.
(A) Butyrate levels in the lumen of proximal, medial, and distal segments of mouse colons.
(B) A model showing two butyrate gradients in the mammalian colon. The proximal-to-distal luminal gradient arises because most bacterial fermentation occurs in the proximal colon and butyrate that is not absorbed by proximal colonocytes moves distally with luminal contents due to peristalsis. The luminal-to-crypt gradient arises because of peristalsis and the upward flow of mucous produced by goblet cells as it is sloughed away from the epithelium into the lumen.
(C and D) Changes in HCT116 cell number (C) and BrdU incorporation (D) in response to 3 days of butyrate or TSA treatment relative to untreated control cells. Cells were grown in high glucose to facilitate the Warburg effect or either low glucose or siLDHA conditions (but in high glucose) to prevent the Warburg effect from occurring. In panel A, the histograms at the far left represent the growth of untreated control cells over the 3-day period (set at 100%), and the other histograms with SE bars represent the corresponding changes in cell growth relative to these controls. The histograms at the right that show negative values in panel A have decreased numbers of cells compared to the start of the 3-day culture period. Results are from 3 independent experiments.
(E) Changes in BrdU incorporation in response to 3 days of butyrate in FHC epithelial cells grown in high glucose. Results are presented as mean ± SE from 3 independent experiments with significant differences indicated (*p < 0.05).
(F) Flow cytometry displaying annexin V (x-axis) and propidium iodide (y-axis) levels in HCT116 cells grown in the presence (top row) or absence (bottom row) of the Warburg effect as indicated at the left. Cells were untreated or treated with either butyrate or TSA (from left to right, as indicated at the top). The percentage of cells positive or negative for annexin V and PI are indicated within the quadrants. Results are representative of 3 independent experiments.
To analyze cell proliferation in the presence of the Warburg effect at different physiological doses of butyrate, we grew HCT116 cells in high glucose. Butyrate inhibited cell growth in a dose-dependent manner compared to untreated controls (Figure 2C). The 0.5-mM dose showed modest growth but to a lesser extent (53%) than the untreated controls. The 2- and 5-mM doses of butyrate led to negative cell growth with fewer cells after 3 days than at the start of the culture period (Figure 2C). Under these conditions, butyrate also decreased cell proliferation in a dose-dependent manner based on BrdU incorporation assays (Figure 2D). On the other hand, when we grew HCT116 cells while preventing the Warburg effect by either low glucose conditions or siLDHA, butyrate had the opposite effect at lower doses (0.5 and 1.0 mM), where it stimulated cell growth compared to untreated controls, although it did inhibit cell growth at higher doses (2.0 and 5.0 mM) (Figure 2C). A similar effect was observed for cell proliferation as butyrate increased BrdU incorporation at low doses but decreased BrdU incorporation at high doses (Figure 2D). These results demonstrate that lower doses of butyrate have a differential effect on cell proliferation depending on the Warburg effect, while higher doses of butyrate inhibit proliferation regardless of the Warburg effect. The higher doses of butyrate had an inhibitory effect on cell growth and BrdU incorporation similar to trichostatin A (TSA), which is a structurally distinct HDAC inhibitor (Figures 2C and D). As expected, noncancerous FHC epithelial cells, which do not undergo the Warburg effect, responded to butyrate with a dose-response profile very similar to colorectal cancer cells when the Warburg effect was prevented from occurring (Figure 2E).
We repeated the above experiments with other colorectal cancer cell lines, including HT-29 cells, and butyrate had dose-response profiles similar to HCT116 cells with lower doses having opposing effects on cell proliferation in the presence versus absence of the Warburg effect (Figure S1A). We also performed experiments showing that lower doses of butyrate stimulate HCT116 cell proliferation in DMEM supplemented with low concentrations (0.25 mM and 0.5 mM) of glucose (as opposed to no glucose) in addition to 10% FBS (Figure S1B). Low glucose conditions may affect processes other than glycolysis such as AMPK activation. To test whether activation of AMPK in HCT116 cells maintained in high glucose mimics low glucose conditions in their proliferative response to butyrate, we performed experiments with the AMP analog AICAR. Although AICAR activated AMPK in our experiments (Figure S1C), it did not affect BrdU incorporation when HCT116 cells were grown in butyrate at several doses (Figure S1D). Thus, AMPK activation in low glucose where the Warburg effect has been prevented from occurring does not explain the effects of butyrate on cell proliferation. These results demonstrate that the Warburg effect influences the role of butyrate as a stimulator or inhibitor of cell growth and proliferation.
We also evaluated whether the Warburg effect is required for the induction of apoptosis and cell death by butyrate by measuring cellular annexin V and intracellular propidium iodide (PI) levels using flow cytometry. Untreated HCT116 cells grown in the presence of the Warburg effect had relatively low levels of apoptosis (4.8% were annexin V positive) and cell death (1.6% stained with PI) as expected (Figure 2F, upper-left panel). Both 0.5- and 5-mM butyrate increased the percentage of cells positive for annexin V (8.5% and 30.7%, respectively) and PI (1.8% and 16.4%, respectively) (Figure 2F, upper panels). The magnitude of these effects at 5 mM was similar to that observed following TSA treatment (Figure 2F, upper-right panel). This cell death is compatible with the negative cell growth induced by 5-mM butyrate and TSA in Figure 2C. Both doses of butyrate induced apoptosis and cell death to a similar extent in HCT116 cells when they were grown in low glucose to prevent the Warburg effect from occurring (Figure 2F, bottom panels). Therefore, butyrate exerts a differential effect on cell growth, which is dependent on the Warburg effect, and this is correlated with a differential effect on cell proliferation but not on apoptosis. For this reason, we decided to investigate the relationship between butyrate and cell proliferation in more detail.
Cell Proliferation is Regulated by Energy Metabolism and Epigenetic Mechanisms
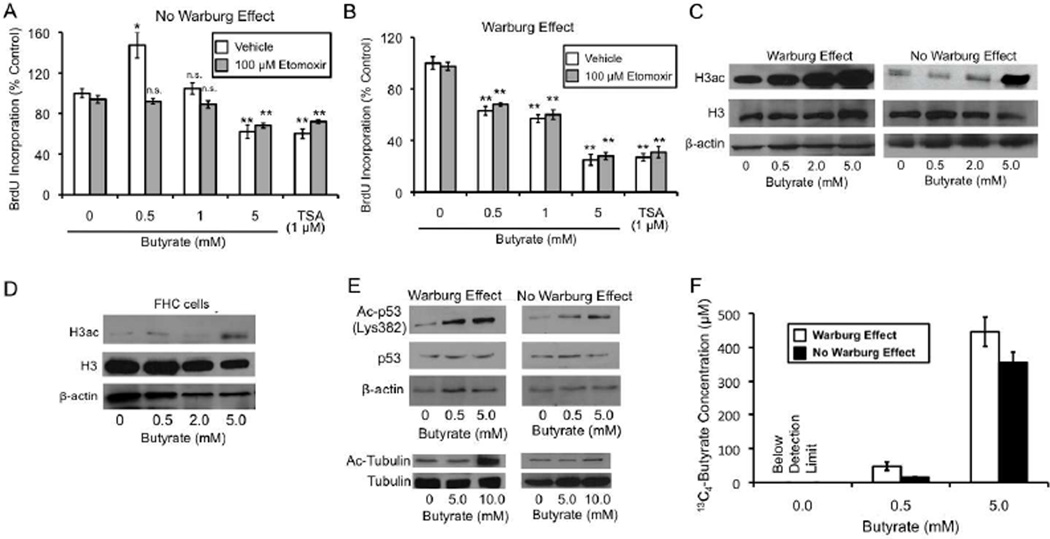
We hypothesized that butyrate promotes the proliferation of normal colonocytes by functioning as an oxidative energy source, while it inhibits the proliferation of cancerous colonocytes because it is metabolized at relatively low levels and functions as an epigenetic factor that inhibits HDACs. To test this hypothesis, we manipulated oxidative metabolism and HDAC activity in HCT116 cells. We used the pharmacological inhibitor etomoxir to inhibit β-oxidation. To confirm that etomoxir was efficacious in our experimental system, we demonstrated that it prevented butyrate from stimulating oxidative metabolism using the same mitotracker probes as described above (Figure S2A). Next, we evaluated whether etomoxir could alter the differential effects of butyrate on cell proliferation. When the Warburg effect was prevented from occurring in HCT116 cells so their metabolism resembled noncancerous cells, 0.5-mM butyrate increased BrdU incorporation in the absence of etomoxir (vehicle only) (Figure 3A), as it did previously (Figure 2D), but not in the presence of etomoxir (Figure 3A). Etomoxir completely blocked the mitogenic effect (Figure 3A). As a control, etomoxir did not affect BrdU incorporation when butyrate was not added (Figures 3A), which indicates that etomoxir was not having toxic effects. To provide further support for the idea that butyrate is metabolized as an oxidative energy source to stimulate cell proliferation, we used other fatty acids that are oxidative substrates but do not inhibit HDACs. Similar to butyrate, these fatty acids (oleate and decanoate) stimulated BrdU incorportion in HCT116 cells under the same conditions (Figure S2B).
Figure 3. The Warburg Effect Influences Energetic and Epigenetic Mechanisms that Regulate Cell Proliferation and Histone Acetylation.
(A and B) Changes in BrdU incorporation in HCT116 cells in response to butyrate or TSA treatment relative to untreated control cells in the absence (A) or presence (B) of the Warburg effect. The effect of etomoxir and vehicle on BrdU incorporation was evaluated under each condition. Results in each panel are from 3 independent experiments and are presented as mean ± SE with significant differences indicated (*p < 0.05; **p < 0.01 compared to no-butyrate control; n.s., not significant).
(C) Western blot analysis showing global H3ac levels (top panels) in HCT116 cells in the presence (left panel) or absence (right panel) of the Warburg effect. Cells were either untreated or treated with butyrate as indicated at the bottom. Total H3 (middle panels) and β-actin (bottom panels) served as loading controls.
(D) Western blot analysis of H3ac (top panel), total H3 (middle panel), and β-actin loading control (bottom panel) in fetal FHC epithelial cells that were untreated or treated with butyrate as indicated.
(E) Western blot analysis of acetyl-p53 (Ac-p53) and acetyl-tubulin (Ac-Tubulin) in HCT116 cells in response to butyrate in the presence or absence of the Warburg effect. p53, β-actin, and tubulin were analyzed as loading controls. The p53 experiments were performed on HCT116 cells following exposure to ionizing radiation.
(F) 13C4-butyrate concentration within nuclei isolated from HCT116 cells that were untreated (0.0 at bottom) or treated with 13C4-butyrate (at 0.5- or 5-mM as indicated at the bottom). Cells were grown in the presence or absence of the Warburg effect as indicated. Measurements were based on analysis of LC-MS spectra from three independent experiments and are presented as mean ± SE. The limit of detection for 13C4-butyrate was 0.5 µM.
In contrast to Figure 3A, when we repeated the etomoxir experiments using HCT116 cells in the presence of the Warburg effect to represent the metabolic state of cancer cells, etomoxir had no effect on the ability of butyrate to inhibit BrdU incorporation (Figure 3B). This result is compatible with butyrate functioning as an HDAC inhibitor rather than an energy source under these conditions. To support this interpretation, we treated HCT116 cells with TSA because it is a potent HDAC inhibitor but is not an energy source. TSA inhibited BrdU incorporation, and this was unaffected by etomoxir (Figure 3B). In fact, TSA mimicked the effects of 5-mM butyrate (Figure 3B).
A Metabolic Link Between Histone Acetylation and Cell Proliferation
To test whether there is a relationship between butyrate-induced changes in cell proliferation and histone acetylation, we analyzed the effect of butyrate on global histone acetylation levels in HCT116 cells in the presence or absence of the Warburg effect. We expected butyrate to increase global histone acetylation levels in the presence of the Warburg effect because it is a well-known HDAC inhibitor. We confirmed that this is the case as butyrate increased pan-histone 3 acetylation (H3ac) levels in a dose-dependent manner based on western blot analyses (Figure 3C, left panel). Therefore, as the dose of butyrate increased, it had a progressively stronger effect on both histone acetylation (Figure 3C, left panel) and the inhibition of cell proliferation (Figure 2D). On the other hand, if the Warburg effect were prevented from occurring, then one would expect butyrate to be metabolized at relatively high levels and fewer molecules should exist, especially at lower doses, to inhibit HDAC activity. The dose-response profile suggested that this is true as well. A low dose (0.5 mM) of butyrate had no effect on H3ac (Figure 3C, right panel), and this same dose did not decrease, but increased, cell proliferation (Figure 2D). Higher doses of butyrate (2 and 5 mM) did increase H3ac (Figure 3C, right panel), albeit to a lesser extent than in the presence of the Warburg effect (Figure 3C, left panel), and also decreased cell proliferation (Figure 2D). This result is compatible with the higher doses of butyrate exceeding the rate which can be metabolized [0.5 mM has been shown to be at 74% of the maximum metabolic capacity based on conversion to CO2, and saturation is reached at 1–2 mM (Andriamihaja et al., 2009)], resulting in the accumulation of butyrate in the nucleus where it functions as an HDAC inhibitor to suppress cell proliferation. As expected, noncancerous FHC epithelial cells responded to butyrate similarly to HCT116 cells in the absence of the Warburg effect. 0.5 mM butyrate had no effect on H3ac (Figure 3D) and increased cell proliferation (Figure 2E). 5 mM was the only dose that induced H3ac in FHC cells (Figure 3D), and it also decreased cell proliferation (Figure 2E). To demonstrate that butyrate also acetylates non-histone substrates, we analyzed tubulin and p53 in HCT116 cells. Similar to H3, butyrate increased tubulin and p53 acetylation, and this effect was more pronounced in the presence of the Warburg effect (Figure 3E).
The objective of the next series of experiments was to evaluate butyrate metabolism and accumulation in the presence versus absence of the Warburg effect because this is the proposed mechanistic basis for the differential effects on histone acetylation and cell proliferation. We added isotopically labeled 13C4-butyrate to HCT116 cells, isolated nuclei, and measured nuclear 13C4-butyrate by LC-MS/MS. Prerequisite control experiments demonstrated that 13C4-butyrate has identical effects on H3ac and cell proliferation as unlabelled butyrate (data not shown). In the presence of the Warburg effect, we observed a dose-dependent effect as the 0.5- and 5-mM doses of 13C4-butyrate resulted in 13C4-butyrate being detected in nuclear extracts at concentrations of 39.5 µM and 477 mM, respectively (Figure 3F). When the Warburg effect was prevented from occurring, the intranuclear 13C4-butyrate concentrations dropped to 14.5 µM and 377.5 µM, respectively (Figure 3F). These lower intranuclear concentrations are compatible with the increased metabolism of butyrate as an oxidative energy source when the Warburg effect is prevented. We also measured 13C4-butyrate levels in the media after the 3-day culture period and did not observe a significant difference (Figure S2C). This result suggests that the lower intranuclear 13C4-butyrate concentrations in cells lacking the Warburg effect cannot be attributed to diminished uptake of 13C4-butyrate. These results demonstrate that the energetics of the cell influences the abundance of butyrate in the nucleus.
Butyrate Induces Histone Acetylation by Stimulating HATs as well as Inhibiting HDACs
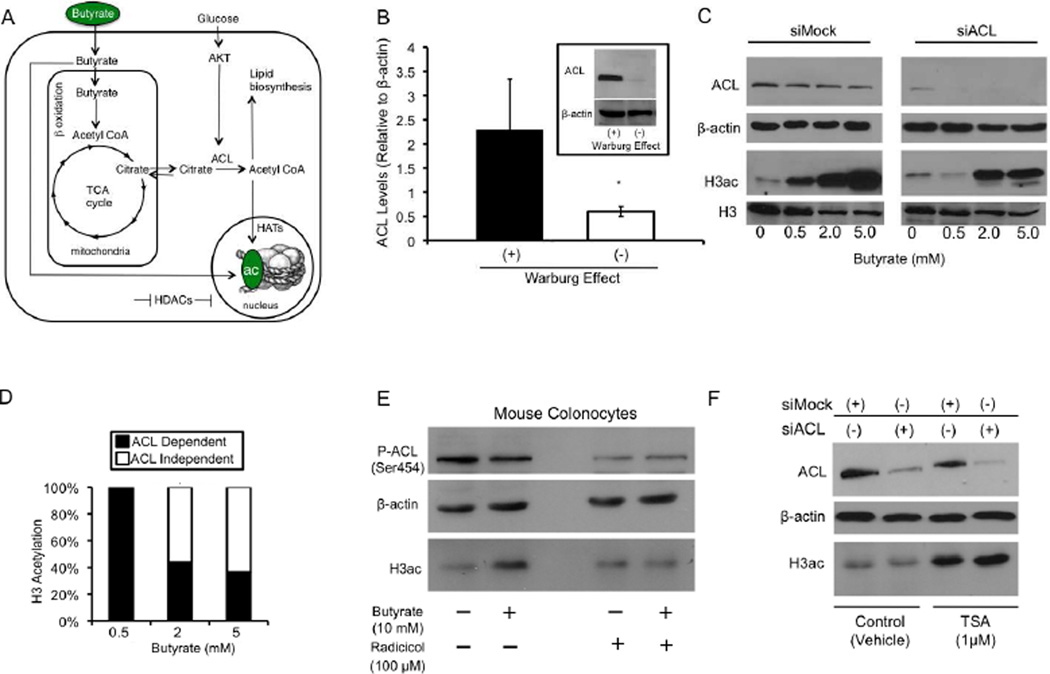
The results of our butyrate measurements raised an important question about the epigenetic mechanism by which butyrate regulates histone acetylation and the contribution of the Warburg effect in this epigenetic regulation. The concentration of butyrate required to inhibit HDAC activity by 50%, referred to as the IC50, is 100–350 mM (Buggy et al., 2000; Sekhavat et al., 2007). This concentration range is consistent with butyrate functioning as a potent HDAC inhibitor at the 5-mM dose but not at the 0.5-mM dose where the concentration is only 0.04–0.40 of the IC50 and inhibits HDAC activity by ≤ 10–20%. This finding suggested that butyrate (especially at low doses such as 0.5 mM) might induce histone acetylation by an alternative mechanism that is distinct from its role as an HDAC inhibitor. One possible model to explain this mechanism involves metabolism of butyrate to acetyl-CoA, which in turn would stimulate HAT activity by functioning as a HAT co-factor and acetyl-group donor (Figure 4A). In this model, butyrate undergoes β-oxidation to acetyl-CoA inside of the mitochondria, followed by the first step of the TCA cycle to yield citrate. Citrate is then transported out of the mitochondria via the citrate shuttle and converted by the enzyme ATP citrate lyase (ACL) to acetyl-CoA and oxaloacetate (Figure 4A). Although it has been known for decades that ACL produces acetyl-CoA in the cytosol for lipid biosynthesis, it was recently shown that ACL is abundant in the nucleus and regulates histone acetylation (Wellen et al., 2009). This model also includes glucose-induced signal transduction that upregulates ACL activity [via phosphorylation of ACL by AKT (Berwick et al., 2002)], which makes it susceptible to being regulated by the Warburg effect (Figure 4A). We confirmed and extended this idea by showing that preventing the Warburg effect from occurring in HCT116 cells resulted in total ACL protein being downregulated to negligible levels (Figure 4B).
Figure 4. Butyrate Increases Histone Acetylation in an ACL-Dependent Manner.
(A) A model for two butyrate-induced histone acetylation mechanisms. In addition to acting as an HDAC inhibitor (depicted by the horizontal line at the bottom), butyrate can act as an acetyl-CoA donor and stimulate HAT activity (depicted by the vertical line pointing downward at the right). See text for a detailed description.
(B) Total ACL protein levels in HCT116 cells grown in the presence (+) or absence (−) of the Warburg effect for 3 days. ACL levels were normalized to β-actin and are presented as mean relative units ± SE (*p < 0.05) based on 3 independent experiments.
(C) Western blot analysis for ACL (top panels), β-actin loading control, H3ac, and total H3 (bottom panels) in siMock (left) and siACL (right) HCT116 cells that were untreated or treated with butyrate as indicated at the bottom.
(D) Quantification of the ACL dependence versus independence of butyrate-induced H3ac from western blots. H3ac levels were normalized using β-actin. Each butyrate treatment had the untreated value subtracted, and the siACL/siMock ratio was computed. Results are from 3 independent experiments.
(E) Western blot analysis of ACL phosphorylated at Ser454 (top panel), β-actin loading control (middle panel), and H3ac (bottom panel) in primary colonocytes freshly isolated from C57BL/6 mice. Colonocytes were treated with butyrate and/or radicicol (as indicated at the bottom) as they were being harvested over a 60-minute period.
(F) Western blot analysis for ACL (top panel), β-actin loading control (middle panel), and H3ac (bottom panel) in siMock and siACL HCT116 cells that were treated with vehicle or TSA as indicated at the bottom.
To test whether butyrate can induce histone acetylation in an ACL-dependent manner, we added butyrate to HCT116 cells grown in the presence of the Warburg effect but with ACL levels depleted by RNAi. As shown in Figure 4C, butyrate increased H3ac in a dose-dependent manner in control cells (siMock) but had a different dose-response profile in ACL-depleted (siACL) cells. A 0.5-mM dose of butyrate had no effect on H3ac in siACL-treated cells (Figure 4C). Higher doses of butyrate (2 and 5 mM) did induce H3ac in siACL but to a lesser extent than siMock. Quantification of results from three independent experiments indicated that 0.5-mM butyrate induced H3ac in a completely ACL-dependent manner (Figure 4D). At higher doses, butyrate continued to have an ACL-dependent effect, but the majority of the histone acetylation shifted to being ACL independent (Figure 4D). It is noteworthy that butyrate had a similar dose-response profile in siACL cells (Figure 4C, right panel) as when the Warburg effect was prevented from occurring (Figure 3C, right panel). This similarity suggests that ACL is an important part of the Warburg effect, presumably because of its role in the biosynthesis of phospholipids/plasma membranes of rapidly dividing cancer cells, but acetylation of histones and other proteins (including metabolic enzymes) may also be important.
The dose-response profile of siACL cells also resembled noncancerous FHC epithelial cells that do undergo the Warburg effect as only 5 mM induced H3ac (Figure 3D). To follow up on this finding and test whether the ACL-dependent mechanism exists in normal, uncultured colonocytes, we added butyrate and/or the ACL inhibitor radicicol (Ki et al., 2000) to mouse colonocytes as they were being isolated over a 1-hr period. Compared to untreated colonocytes, butyrate increased H3ac (Figure 4E, compare first two lanes) but not in the presence of radicicol (Figure 4E, compare second and fourth lanes). Radicicol did diminish the levels of ACL in its active, phosphophorylated form (Figure 4E). These results demonstrate that the ACL-dependent mechanism functions in normal colonocytes to regulate histone acetylation. The short incubation period was probably not sufficient for butyrate to accumulate and may have precluded detectable levels of HDAC inhibition. To demonstrate that the ACL-dependent mechanism does not involve HDAC inhibition, we treated HCT116 cells with TSA. TSA increased H3ac to the same extent under siMock and siACL conditions, which confirms that HDAC inhibition is an ACL-independent mechanism (Figure 4F).
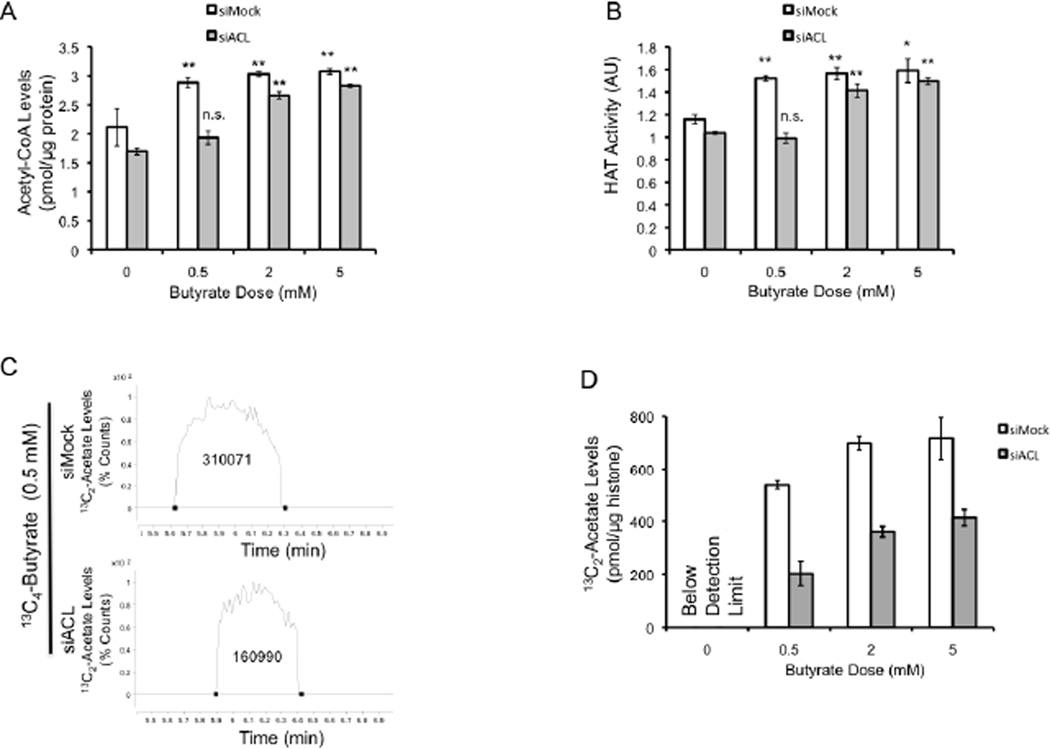
Based on our model (Figure 4A), butyrate should function as an acetyl-CoA donor and stimulate HAT activity in an ACL-dependent manner. Accordingly, biochemical experiments performed in the presence of the Warburg effect showed that butyrate increased both acetyl-CoA and HAT activity in siMock cells at all doses (Figures 5A and B), and that ACL depletion negated these effects at the 0.5-mM dose, where the ACL mechanism predominates (as shown in Figure 4D) but not at the 2- and 5-mM doses where butyrate primarily acts as an HDAC inhibitor (Figures 5A and B) [presumably because the oxidative metabolic capacity of butyrate is exceeded (Andriamihaja et al., 2009)]. To test whether butyrate elevated HAT activity through inducing the direct acetylation of HATs, we analyzed p300/CBP acetylation over several butyrate doses. Butyrate did not change the acetylation status of p300/CBP (Figure S3). Taken together, these data suggest that butyrate stimulates HAT activity by increasing the abundance of acetyl-CoA co-factor (via ACL-dependent metabolism) rather than increasing HAT catalytic activity independent of co-factor availability.
Figure 5. Butyrate is a Carbon Donor for Acetyl-CoA and Histone Acetylation.
(A and B) Nuclear acetyl-CoA levels (A) and HAT activity (B) in siMock and siACL (as indicated at the top) HCT116 cells. Cells were untreated or treated with butyrate as indicated at the bottom. Results are from 3 independent experiments and are presented as mean ± SE with significant differences indicated (*p < 0.05; **p < 0.01 compared to no-butyrate control; n.s., not significant).
(C) LC-MS chromatograms of hydrolyzed histones from HCT116 cells treated with 0.5 mM of uniformly labeled 13C4-butyrate for 3 days. The 13C2-acetate peak is larger in siMock cells (top spectrum) than siACL cells (bottom spectrum) as indicated by the numbers, which correspond to the area under the peaks.
(D) Quantification of 13C2-acetate levels from hydrolyzed histones isolated from untreated or 13C4-butyrate-treated siMock and siACL HCT116 cells. The results are normalized to histone levels (as indicated on the y axis) and are presented as the mean ± SE from 3 independent experiments.
To rigorously confirm the ACL/acetyl-CoA/HAT pathway as the mechanism, we performed metabolic flux experiments where we added uniformly-labeled butyrate (13C4-butyrate) to HCT116 cells undergoing the Warburg effect, purified histones, hydrolyzed their posttranslational modifications, and quantitatively measured 13C2-acetyl groups (13C2-acetate) using LC/MS-MS. All 13C4-butyrate treatment groups had detectable amounts of 13C2-acetate, whereas untreated negative control samples did not as expected (Figures 5C and D). These results demonstrate that carbons derived from butyrate contribute acetyl groups on histones. In siMock cells, 13C4-butyrate contributed carbons to the acetyl groups, but the dose-response profile was not linear between 0.5, 2, and 5 mM (Figure 5D). The observed plateau between 2 and 5 mM was expected because these doses exceed the concentration of butyrate that can be metabolized efficiently and result in butyrate accumulation (as shown in Figure 3F) and HDAC inhibition. 13C2-acetate levels were significantly diminished in siACL cells compared to siMock cells by 62%, 51%, and 58% in the 0.5, 2, and 5 mM doses, respectively (Figure 5D). These flux experiments unequivocally demonstrate that butyrate contributes to histone acetylation by being metabolized to acetyl-CoA and increasing HAT activity although they do not provide insight into how this mechanism influences gene expression.
Target Genes are Upregulated by HAT and HDAC Inhibition Mechanisms in a Dose-Dependent Manner
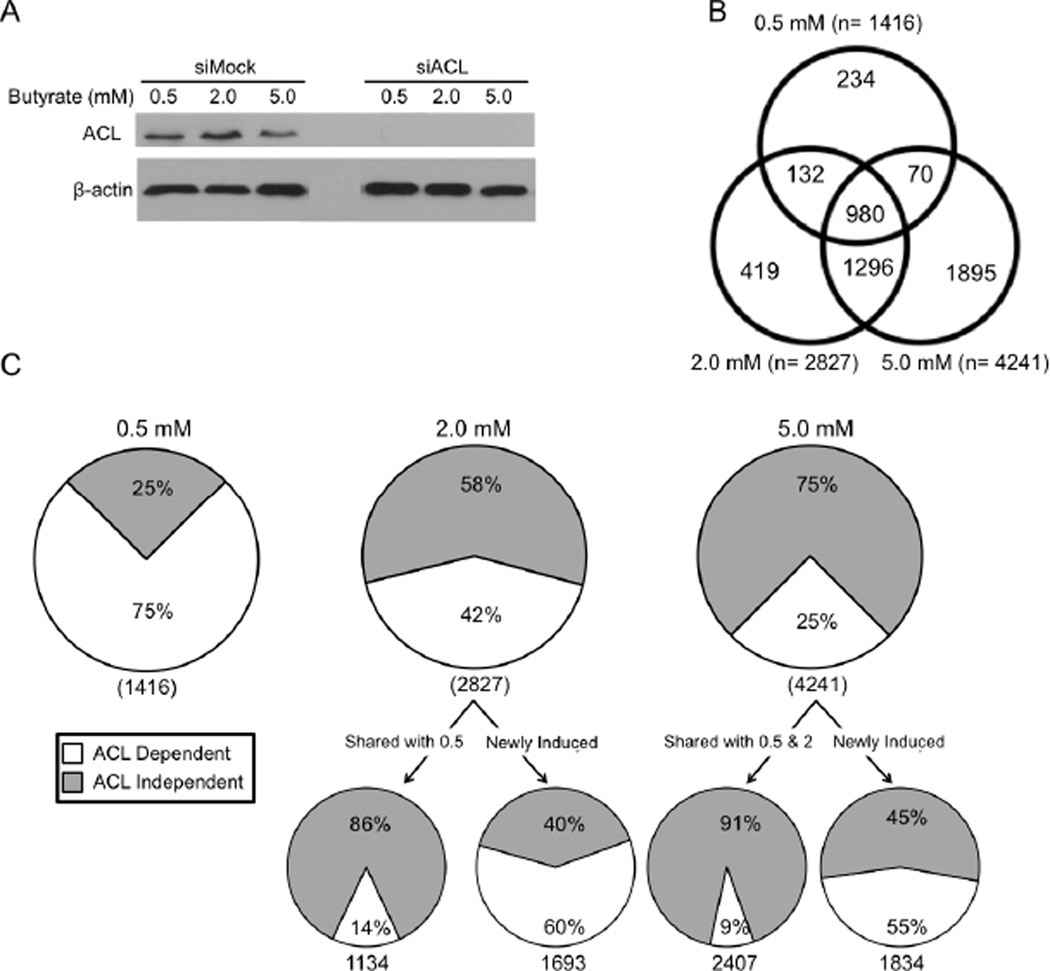
To understand the relative contribution of the HAT and HDAC inhibition mechanisms on gene expression, we performed a series of transcriptome profiling experiments. We analyzed siMock and siACL HCT116 cells undergoing the Warburg effect that were either untreated or treated with 0.5-, 2-, or 5-mM butyrate. The entire data set and statistical values are included as Supplemental Table S1, and a dendogram produced by hierarchical clustering of these data are included as Figure S4A. In the absence of butyrate, the abundance of only 38 mRNAs was significantly different between siMock and siACL samples. This result indicates that the knockdown of ACL, as shown by western blot analysis (Figure 6A), had a minimal effect on gene expression in untreated cells. Consistent with butyrate stimulating H3ac in a dose-dependent manner (Figure 3C), it increased mRNA levels in a dose-dependent manner. Compared to untreated cells, butyrate upregulated 1416 genes at 0.5 mM, 2827 genes at 2 mM, and 4241 genes at 5 mM in siMock cells (Figure 6B). The vast majority of genes that were induced at lower doses stayed upregulated at higher doses. For example, 84% of the genes induced by 0.5 mM were also upregulated at 2 mM and/or 5 mM (Figure 6B); 69% were upregulated at all three doses (Figure 6B). Because of this high degree of overlap, only 17% (234/1416) and 15% (419/2827) of genes were unique to the 0.5-mM and 2-mM doses, respectively. The percentage of unique genes was higher at the 5-mM dose (1895/4241 = 45%), but this is presumably due to it being the highest dose and genes first induced at 5 mM did not have an opportunity to be shown to be upregulated at an even higher dose.
Figure 6. Butyrate Regulates Gene Expression Using ACL-Dependent and -Independent Mechanisms.
(A) Western blot analyses of HCT116 cells used for transcriptome profiling. ACL (top panel) and β-actin loading control (bottom panel) levels are shown in siMock and siACL cells in response to different butyrate treatments.
(B) Venn diagram showing the overlap between genes upregulated by butyrate at doses of 0.5, 2, and 5 mM.
(C) Pie charts showing the percentage of genes upregulated by 0.5-mM (left), 2-mM (middle), and 5-mM (right) butyrate in an ACL-dependent (white) and -independent (gray) manner. Beneath the 2 mM and 5 mM pie charts are two additional pie charts. The chart shown on the left corresponds to genes that were also upregulated at the previous dose (shared). The chart shown on the right corresponds to genes that were not upregulated at the previous dose (newly induced).
By comparing the siMock and siACL gene lists, we identified two classes of genes. ACL-dependent genes were upregulated by butyrate in siMock samples but not in siACL samples, whereas ACL-independent genes were upregulated in both siMock and siACL samples. 75% of the 1416 genes induced by 0.5-mM butyrate were ACL dependent while 25% were ACL independent (Figure 6C). This finding is consistent with our results presented above showing that 0.5-mM butyrate was metabolized and converted by ACL to acetyl-CoA and relatively little accumulated to act as an HDAC inhibitor. For the HCT116 samples treated with either 2- or 5-mM butyrate, the number of ACL-dependent genes was overtaken by ACL-independent genes. For example, the percentage of ACL-dependent genes dropped from 75% at 0.5 mM to 42% and 25% (for 2 and 5 mM, respectively), and there was a corresponding increase in the percentage of ACL-independent genes from 25% to 58% and 75%, respectively (Figure 6C).
1134/1416 (80%) of the genes induced at 0.5 mM were also upregulated at 2 mM. However, there was a marked change in the regulatory mechanism at 2 mM with these genes shifting away from ACL dependence (14%) toward ACL independence (86%) (Figure 6C). ACL-dependent genes at 0.5 mM tended to shift to the ACL-independent category at 2 mM (649/808 = 80%), while ACL-independent genes at 0.5 mM stayed ACL independent at 2 mM (324/326 = 99%). This result is consistent with butyrate accumulating at the higher dose and primarily functioning as an HDAC inhibitor such that target genes are upregulated even when ACL is depleted. 1693 genes at 2 mM were not shared with 0.5 mM, and these newly-induced genes showed a profile that was distinct from the shared genes. The majority of these newly-induced genes were ACL dependent (60%) rather than ACL independent (40%) (Figure 6C).
The same trend that was observed from 0.5 to 2 mM also occurred at the 2 to 5 mM transition. Overall, 42% of the 2827 genes upregulated at 2 mM were ACL dependent compared to 58% that were independent (Figure 6C). 2407/2827 (85%) of these genes were also upregulated at 5 mM. Once again, there was a marked change in the mechanism with these shared genes shifting away from ACL dependence (9%) to ACL independence (91%) at 5 mM (Figure 6C). Nearly identical to the first transition, ACL-dependent genes at 2 mM tended to switch to the ACL-independent category at 5 mM (682/860 = 79%), while ACL-independent genes at 2 mM stayed independent at 5 mM (1446/1473 = 98%). Also similar to the first transition, the newly-induced genes were much more likely to be ACL dependent compared to the shared genes. Specifically, 55% of the 1834 genes newly induced at 5 mM were ACL dependent and 45% were ACL independent (Figure 6C). Taken together, these results support the idea that newly-induced genes are upregulated in an ACL-dependent manner and then switch to ACL independence at higher doses (Figure S3B). There were very few exceptions with a total of only 32 genes switching in the opposite direction (from ACL independent to ACL dependent) in aggregate (at both transitions combined) (Figure S4B).
Butyrate increased the number of upregulated genes in a dose-dependent manner, as described above, but there was a surprisingly weak correlation between butyrate dose and mRNA abundance for many genes. Butyrate increased mRNA levels by an average (mean) of 2.5 fold at 0.5 mM but had only a marginally stronger effect at 2 mM (2.8 fold) and 5 mM (3.0 fold). For genes that stayed ACL dependent at the 0.5–2 mM or 2–5 mM transitions, mRNA levels increased by only 4% at the higher dose, which was not significant. However, the mRNA abundance for each gene increased by an average of 16% as the mechanism shifted from ACL dependent to ACL independent. This result is consistent with two mechanisms being engaged and acting additively. However, the ACL/acetyl-CoA/HAT activity mechanism accounts for the majority of the upregulation since the magnitude of change was only 4% and 16%. Overall, these results indicate that the ACL-dependent/acetyl-CoA/HAT activity mechanism acts first to induce gene expression, and the ACL-independent/HDAC inhibition mechanism subsequently works to maintain the upregulated state and has only a modest effect on further increasing mRNA levels.
To interrogate the biological significance of the ACL-dependent and -independent mechanisms, we used Ingenuity Pathway Analysis (IPA) to identify different Gene Ontology (GO) or Kyoto Encyclopedia of Genes and Genomes (KEGG) categories that were enriched. The ACL-dependent mechanism accounted for 75% of the upregulated genes at 0.5 mM (Figure 6C), and there was a significant enrichment for genes associated with cell proliferation but not cell death under these conditions (Figure S4C). Conversely, the ACL-independent mechanism accounted for 75% of the upregulated genes at 5 mM (Figure 6C), and these genes exhibited a reciprocal pattern with enrichment for cell death but not cell proliferation (Figure S4C).
For the 0.5 mM ACL-dependent genes upregulated by >2-fold, we found that 2% were associated with cell proliferation as compared to only 0.1% of the genes in the 5.0 mM ACL-independent group (Figure S5A). Consistent with the 0.5 mM ACL-dependent genes being enriched for cell proliferation functions, ACL knockdown protected HCT116 cells from diminished BrdU incorporation at 0.5 mM but not at 2 or 5 mM (Figure S5B). This result demonstrates that ACL-dependent genes, in the presence of the Warburg effect, inhibit cell proliferation and may have tumor-suppressor properties. The tumor-suppressor gene, TES, whose expression is suppressed in cancer cells (Drusco et al., 2005; Ma et al., 2010; Sarti et al., 2005; Tatarelli et al., 2000), was the most highly upregulated gene (221-fold) induced by 0.5 mM butyrate (Figure S5A) and was ACL-dependent. On the other hand, we found that 3% of the 5 mM ACL-independent genes upregulated by >2-fold were associated with apoptosis as compared to only 0.2% of the genes in the 0.5 mM ACL-dependent group (Figure S5C).
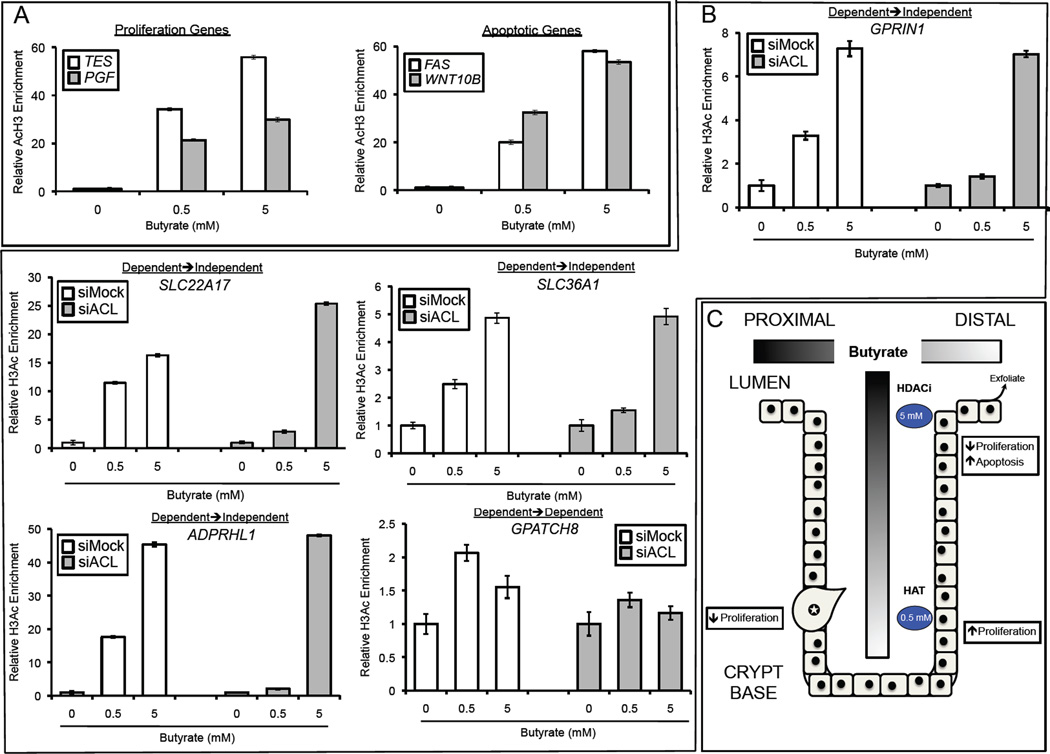
ACL-Dependent and -Independent Histone Acetylation of Target Genes
To test whether ACL-dependent and -independent expression changes are associated with corresponding changes in histone acetylation, we performed quantitative ChIP assays to characterize H3ac enrichment at the proximal promoters of the 4 most highly upregulated genes involved in cell proliferation and apoptosis. In a dose-dependent manner, butyrate induced H3ac of the proliferation genes (TES and PGF) and the apoptotic genes (FAS and WNT10B) in HCT116 cells (Figure 7A). To address whether the mechanism of H3ac switched from ACL dependence at low doses to ACL independence at higher doses, we performed additional ChIP assays. For these experiments, HCT116 treatment groups were nearly identical to the transcriptome profiling experiments with siMock and siACL cells being untreated or treated with butyrate (0.5 or 5 mM). The expression of 4 of the target genes (SLC22A17, SLC36A1, GPRIN1, ADPRHL1) switched from being ACL dependent at 0.5 mM to ACL independent at 5 mM (Table S1). qPCR of the ChIPs showed that H3ac enrichment in each promoter also switched from ACL dependent at 0.5 mM to ACL independent at 5 mM (Figure 7B). The expression of the other gene (GPATCH8) was ACL dependent at both 0.5 and 5 mM (Table S1), and H3ac enrichment showed a similar profile (Figure 7B).
Figure 7. Butyrate Induces Histone Acetylation by Dual Mechanisms.
(A, B) Quantitative ChIP assays showing H3ac enrichment at the promoters of genes in HCT116 cells that were untreated or treated with butyrate as indicated. In panel A, the most-highly induced cell proliferation (left) and apoptotic (right) genes are shown. In panel B, the assays were performed under siMock and siACL conditions. The first 4 panels correspond to genes (SLC22A17, SLC36A1, GPRIN1, ADPRHL1) whose expression switched from ACL dependence at 0.5 mM to ACL independence at 5 mM. The final gene (GPATCH8) was expressed in an ACL-dependent manner at both doses. qPCR results for each ChIP were normalized to input, and the values of the untreated samples (0-mM butyrate) were set at 1.0. The 0.5- and 5-mM butyrate-treated samples are presented as histograms that show H3ac enrichment relative to the untreated controls. Each histogram represents the mean ± SE for 3 independent experiments.
(C) A model depicting how butyrate might regulate histone acetylation and cellular turnover in the colonic epithelium. Butyrate is produced by microbiota in the proximal lumen, and mucous flow and peristalsis result in butyrate concentration gradients shown as two shaded bars. Near the crypt base, where butyrate concentrations are relatively low (0.5 mM), most of the butyrate molecules are metabolized and contribute to histone acetylation by the acetyl-CoA/HAT mechanism. This is associated with the proliferation of mitotically-active colonocytes. Near the lumen, butyrate concentrations are relatively high (5 mM) and exceed the concentration (1–2 mM) that can be metabolized efficiently. As a result, butyrate accumulates in the nucleus and acts as an HDAC inhibitor (HDACi). These colonocytes are not mitotically active, undergo apoptosis, and exfoliate into the lumen. Due to the Warburg effect, cancerous colonocytes (labeled with a star) metabolize relatively little butyrate regardless of their position within the epithelium. Consequently, the HDAC inhibition mechanism predominates, and butyrate is associated with decreased proliferation and increased apoptosis.
DISCUSSION
In the work presented here, we characterize two histone acetylation mechanisms regulated by butyrate that are utilized differently in cancer cells because of their unique metabolism. These mechanisms are defined as ACL dependent (acetyl-CoA/HAT) and ACL independent (HDAC inhibition). The acetyl-CoA/HAT mechanism involves metabolism of butyrate in the mitochondria followed by the subsequent ACL-catalyzed production of acetyl-CoA (Figure 4A). Acetyl-CoA then serves as an essential co-factor for the acetylation of histones and other substrates. This pathway is compatible with previous flux experiments that followed 14C-butyrate metabolism in colon carcinoma cells and reported that the 14C-labeled carbon was converted to CO2, histones, and lipids (Andriamihaja et al., 2009; Leschelle et al., 2000). These experiments did not use mass spectrometry to identify compounds and could not follow the specific fate of 14C-labeled carbons within histones, but our flux experiments indicate that the butyrate-derived carbons are incorporated as the acetyl group moieties of histones when they are acetylated and therefore provide mechanistic insight as to how butyrate can epigenetically regulate gene expression. We demonstrate that the HDAC inhibition (ACL independent) mechanism is primarily utilized by cancer cells because they rely on glucose and aerobic glycolysis to a greater extent than oxidative metabolism. In this situation, cancerous colonocytes metabolize relatively little butyrate (since it is an oxidative energy source) so it accumulates at greater levels inside of nuclei (and is therefore a candidate oncometabolite), and there is a corresponding increase in HDAC inhibition. Our experiments preventing the Warburg effect from occurring in cancerous colonocytes demonstrate that the Warburg effect is a cause, rather than a consequence, of this mechanistic shift in histone acetylation. This is a significant conceptual advance because it represents a fundamental mechanism whereby metabolic transformation (i.e., cancer cell metabolism) can drive the divergence of epigenomic and transcriptome profiles in cancer cells away from their cell of origin and contribute to tumorigenesis. This idea is supported by recent reports showing that the oncometabolite 2-hydroxyglutarate, which accumulates in tumors as a result of isocitrate dehydrogenase (IDH1/2) mutations, inhibits JmjC histone demethylases and DNA methyltransferses to alter the transcription of genes necessary for differentiation (Lu et al., 2012; Turcan et al., 2012; Xu et al., 2011).
Previous studies have used relatively high doses of butyrate (~5 mM) where it has been interpreted to function as an HDAC inhibitor [and possibly as a product inhibitor of deacetylation (Corfe, 2012)]. The acetyl-CoA/HAT mechanism may have been overlooked until now because few studies have used lower doses of butyrate (0.5 mM), where this mechanism is most prevalent, and butyrate-induced histone acetylation has generally been assumed to arise because of HDAC inhibition without the mechanism being analyzed (Corfe, 2012). A 5-mM dose of butyrate in vitro is physiologically relevant because it is routinely found at this concentration in the lumen of the colon (Cummings et al., 1987; Hamer et al., 2008; Scheppach and Weiler, 2004). However, at this or any other dose, it is likely that fewer butyrate molecules reach colonocytes in vivo than in vitro because the colonic epithelium is covered by a thick (~100 mm) layer of mucous. Furthermore, mucous is constantly being produced by goblet cells and sloughed away from the epithelium as it flows up from the crypt bases into the lumen and then down the lumen due to peristalsis (Forstner and Forstner, 1994). Consequently, butyrate concentration gradients are expected to form along the lumen-to-crypt axis and the luminal proximal-distal axis (Csordas, 1996; Sengupta et al., 2006) (Figures 2A and B). Therefore, colonocytes at the base of crypts receive an estimated 50–800 µM dose equivalent. The results from this study showing that butyrate stimulates histone acetylation in a dose-dependent manner by two distinct mechanisms suggests that colonocytes near the base of crypts, which are exposed to relatively small amounts of butyrate, exclusively utilize the acetyl-CoA/HAT mechanism, while luminal colonocytes that are chronically exposed to higher levels of butyrate (which surpass oxidative capacity resulting in accumulation) primarily utilize the HDAC-inhibition mechanism (Figure 7C). Although both mechanisms increase histone acetylation, our transcriptome profiling results indicate that they upregulate different targets with the former enriched for cell proliferation genes and the latter being enriched for apoptotic genes. These changes in gene expression are consistent with the lower doses of butyrate stimulating cell proliferation, while higher doses inhibit proliferation and increase apoptosis. These findings lead to a model where butyrate facilitates the normal turnover of the colonic epithelium by promoting colonocyte proliferation in the bottom half of each crypt while increasing apoptosis in those cells that exfoliate into the lumen (Figure 7C). This model is supported by previous experiments demonstrating that stimulation of proliferation by butyrate occurs at low doses and only at the base of the crypt (Velazquez et al., 1997).
The data presented in this study supports previous hypotheses proposing that butyrate metabolism is impaired in cancer cells (Jass, 1985; Singh et al., 1997) and provides a mechanism (the Warburg effect) for why this is the case. It also supports and expands upon previous studies (Comalada et al., 2006; Jass, 1985; Singh et al., 1997) by explaining why butyrate has opposing effects on cell growth: it inhibits cancer cell proliferation as an HDAC inhibitor but stimulates the proliferation of noncancerous cells (and cancerous cells when the Warburg effect is blocked) by being oxidized as an energy source. However, when ACL levels were depleted by RNAi, the stimulatory effect was attenuated from 240% to 160% at 0.5 mM compared to untreated cells (Figure S5). This result is surprising because it suggests that butyrate exerts its stimulatory effect not only by serving as a carbon source for β-oxidation and the TCA cycle but also by increasing acetyl-CoA production for lipid biosynthesis and/or acetylation of lysine residues. This effect may not involve histone acetylation [0.5 mM did not increase H3ac when the Warburg effect was blocked (Figure 3C)] but could be mediated by the acetylation of non-histone proteins including metabolic enzymes, which are often regulated by acetylation status (Guan and Xiong; Ma et al., 2010; Zhao et al.).
Butyrate is an attractive candidate for chemotherapy or chemoprevention because it selectively inhibits tumor growth and has minimal adverse effects in clinical trials (Pouillart, 1998). However, the efficacy of butyrate as a chemotherapeutic agent has been limited by its rapid uptake and metabolism by normal cells [resulting in a half-life of 6 minutes and peak blood levels below 0.05 mM (Miller et al., 1987)] before reaching tumors (Pouillart, 1998). More stable butyrate derivatives such as tributyrin have also not been successful on a consistent basis (Pouillart, 1998). A fiber-rich diet might be more successful for chemoprevention because it delivers mM levels of butyrate (via the microbiota) to the correct place (the colon) before the onset of tumorigenesis or at an early stage. Evidence for this idea comes from recent human studies demonstrating lower levels of butyrate-producing bacteria among the gut microbiota of colorectal cancer patients compared to healthy participants (Balamurugan et al., 2008; Wang et al., 2012), and studies showing an inverse correlation between fecal butyrate levels and tumor size in colorectal cancer (Boutron-Ruault et al., 2005; Monleon et al., 2009). Taken together, these findings suggest that human studies should be conducted to investigate the combined interaction between fiber/butyrate and gut microbiota, in the prevention of colorectal cancer.
EXPERIMENTAL PROCEDURES
Cell culture and Transfections
HCT116 and HT-29 cells were obtained from ATCC (Manassas, VI) and grown in DMEM formulated without glucose (Invitrogen, Carlsbad, CA) and supplemented with glucose to achieve a 25 mM concentration and 10% FBS. In experiments that prevented the Warburg effect, cells were grown in the same DMEM/FBS media but without any glucose supplementation. FHC cells were obtained from ATCC (CRL-1831) and grown in accordance to previously determined conditions (Siddiqui and Chopra, 1984) with the exception that DMEM minus glucose was used instead of high glucose DMEM as a 1:1 mixture with F12 medium. RNAi transfections were performed as described (Wellen et al., 2009), and siRNA pools for human ACL (Thermo Scientific/Dharmacon #L-004915-00), human LDHA (Thermo Scientific/Dharmacon #L-008201-00), and siMock non-targeting control (Dharmacon #D-001810-01-20) were used at a 20 nM final concentration.
Biochemical Assays
Histone acetyltransferase (K332-100), acetyl CoA (K317-100), glucose-6-phosphate (K657-100), and lactate (K607-100) assays were performed as specified by manufacturer (Biovision, Mountain View, CA) instructions. Perchloric acid extraction (K808-200) of cells was used for acetyl CoA, glucose-6-phosphate, and lactate assays.
Confocal Fluorescence Microscopy
MitoTracker Red CM-H2XRos and MitoTracker Green FM (Invitrogen) were added to cells at a final concentration of 500 nM. Cells were viewed with an Olympus FV500 laser-scanning confocal microscope equipped with three confocal channels, excitation lines 458, 488, 514, 543, an 633. All fluorescent micrographs were acquired using Olympus Fluoview software with identical settings. Image J (Bethesda, MD) was used to analyze the integrated density per cell for both mitotracker dyes. All micrographs were taken with identical exposures. A total of 3 independent biological replicates were analyzed per experimental group.
Cell Proliferation and Apoptosis Assays
Cell proliferation assays were performed using the BrdU Cell Proliferation kit (2750) from Millipore/Chemicon (Tecmecula, CA). Apoptosis assays were performed using Annexin V (K101-100) from Biovision. Cells were incubated with Annexin V and propidium iodide after a 24-hr treatment with butyrate. Cells were then analyzed for Annexin V and PI incorporation by flow cytometry using standard procedures.
Western Blotting
Experimental details can be found in supplemental methods. Antibodies that were used included phospho-ACL (Cell Signaling, Beverly, MA), ACL (4332; Cell Signaling), pan-acetyl H3 (06-599; Millipore/Upstate), β-actin (ab8226; Abcam), total Histone H3 (05-928; Upstate), LDHA (3582; Cell Signaling), acetyl-tubulin (T6793; Sigma), acetyl-p53 (2525; Cell Signaling), p53 (9282; Cell Signaling), α-tubulin (T6199; Sigma), acetyl-CBP (4771; Cell Signaling), CBP (7389; Cell Signaling), P-AMPK (2535; Cell Signaling), AMPK (2603; Cell Signaling).
Flux Experiments
Cells were treated with 13C4-butyrate (#488380; Sigma-Aldrich, St. Louis, MO). After the treatment period, nuclei were isolated using a kit (#266-100; Biovision), an internal 13C1-butyrate standard was added, and nuclei were resuspended. Details can be found in supplemental methods.
A Seahorse XF24 Analyzer (Seahorse Bioscience) was used to measure oxygen consumption rates in HCT116 and FHC cells. Experiments were performed in accordance to manufacturer instructions.
Gene Expression
Treated and untreated HCT116 cells were flash-frozen (3 biological replicates each). Total RNA was prepared using Trizol reagent (Invitrogen), and integrity was assessed using the RNA 6000 Nano-LabChip kit followed by analysis using a Bioanalyzer (Agilent, Palo Alto, CA). Hybridizations to the HumanWG-6 v2.0 BeadChip arrays (Illumina, San Diego, CA), washing, and scanning were performed at the University of Tennessee at Memphis Core Facility. Data was analyzed using R/limma (Smyth, 2004).
ChIP Assays
Experimental details can be found in supplemental methods. A pan-acetyl H3 (#06-599; Millipore) was used in ChIP experiments. Each ChIP was normalized to input. For each treatment group, data are expressed relative to untreated controls. The primers used are included in supplemental methods. No PCR product was detected in no template controls or in mock ChIP samples immunopreciptated with normal rabbit serum (IgG).
Statistics
For biochemical assays, BrdU incorportation assays, and mitotracker experiments, differences between experimental groups were determined by ANOVA followed by a Tukey post-hoc test. All data are expressed as mean ± SE.
Supplementary Material
ACKNOWLEDGEMENTS
We thank V. Madden, S. Ray, and B. Bagnell at the UNC Microscopy Services Laboratory for advice with confocal microscopy. We would also like to thank the Flow Cytometry and Biomarker Mass Spectrometry Core Facilities for making flow cytometry and LC-MS instruments available to us. This work was supported by the National Institutes of Health (CA125237 to S.J.B.), The American Institute of Cancer Research (S.J.B), and The Prevent Cancer Foundation (D.R.D. and S.J.B.). The Biomarker Mass Spectrometry Facility is supported by NIH grant P30-ES10126.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS
Microarray data have been submitted to the GEO database.
SUPPLEMENTAL INFORMATION
Supplemental information includes Supplemental Experimental Procedures, five figures, and one table.
The authors declare no conflicts of interest.
REFERENCES
- Andriamihaja M, Chaumontet C, Tome D, Blachier F. Butyrate metabolism in human colon carcinoma cells: implications concerning its growth-inhibitory effect. Journal of cellular physiology. 2009;218:58–65. doi: 10.1002/jcp.21556. [DOI] [PubMed] [Google Scholar]
- Archer SY, Meng S, Shei A, Hodin RA. p21(WAF1) is required for butyrate-mediated growth inhibition of human colon cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:6791–6796. doi: 10.1073/pnas.95.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balamurugan R, Rajendiran E, George S, Samuel GV, Ramakrishna BS. Real-time polymerase chain reaction quantification of specific butyrate-producing bacteria, Desulfovibrio and Enterococcus faecalis in the feces of patients with colorectal cancer. Journal of gastroenterology and hepatology. 2008;23:1298–1303. doi: 10.1111/j.1440-1746.2008.05490.x. [DOI] [PubMed] [Google Scholar]
- Baylin SB, Jones PA. A decade of exploring the cancer epigenome - biological and translational implications. Nature reviews. Cancer. 2011;11:726–734. doi: 10.1038/nrc3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berwick DC, Hers I, Heesom KJ, Moule SK, Tavare JM. The identification of ATP-citrate lyase as a protein kinase B (Akt) substrate in primary adipocytes. The Journal of biological chemistry. 2002;277:33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- Boutron-Ruault MC, Marteau P, Lavergne-Slove A, Myara A, Gerhardt MF, Franchisseur C, Bornet F. Effects of a 3-mo consumption of short-chain fructo-oligosaccharides on parameters of colorectal carcinogenesis in patients with or without small or large colorectal adenomas. Nutrition and cancer. 2005;53:160–168. doi: 10.1207/s15327914nc5302_5. [DOI] [PubMed] [Google Scholar]
- Buggy JJ, Sideris ML, Mak P, Lorimer DD, McIntosh B, Clark JM. Cloning and characterization of a novel human histone deacetylase, HDAC8. The Biochemical journal. 2000;350(Pt 1):199–205. [PMC free article] [PubMed] [Google Scholar]
- Chopin V, Toillon RA, Jouy N, Le Bourhis X. Sodium butyrate induces P53-independent, Fas-mediated apoptosis in MCF-7 human breast cancer cells. British journal of pharmacology. 2002;135:79–86. doi: 10.1038/sj.bjp.0704456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comalada M, Bailon E, de Haro O, Lara-Villoslada F, Xaus J, Zarzuelo A, Galvez J. The effects of short-chain fatty acids on colon epithelial proliferation and survival depend on the cellular phenotype. Journal of cancer research and clinical oncology. 2006;132:487–497. doi: 10.1007/s00432-006-0092-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corfe BM. Hypothesis: butyrate is not an HDAC inhibitor, but a product inhibitor of deacetylation. Molecular bioSystems. 2012;8:1609–1612. doi: 10.1039/c2mb25028d. [DOI] [PubMed] [Google Scholar]
- Csordas A. Butyrate, aspirin and colorectal cancer. Eur J Cancer Prev. 1996;5:221–231. doi: 10.1097/00008469-199608000-00002. [DOI] [PubMed] [Google Scholar]
- Cummings JH, Pomare EW, Branch WJ, Naylor CP, Macfarlane GT. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut. 1987;28:1221–1227. doi: 10.1136/gut.28.10.1221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davie JR. Inhibition of histone deacetylase activity by butyrate. The Journal of nutrition. 2003;133:2485S–2493S. doi: 10.1093/jn/133.7.2485S. [DOI] [PubMed] [Google Scholar]
- Donohoe DR, Bultman SJ. Metaboloepigenetics: Interrelationships between energy metabolism and epigenetic control of gene expression. Journal of cellular physiology. 2012 doi: 10.1002/jcp.24054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drusco A, Zanesi N, Roldo C, Trapasso F, Farber JL, Fong LY, Croce CM. Knockout mice reveal a tumor suppressor function for Testin. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:10947–10951. doi: 10.1073/pnas.0504934102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteller M. Epigenetics in cancer. The New England journal of medicine. 2008;358:1148–1159. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- Fleming SE, Fitch MD, DeVries S, Liu ML, Kight C. Nutrient utilization by cells isolated from rat jejunum, cecum and colon. The Journal of nutrition. 1991;121:869–878. doi: 10.1093/jn/121.6.869. [DOI] [PubMed] [Google Scholar]
- Forstner JF, Forstner GG. Gastrointestinal mucus. In: Johnson LR, editor. Physiology of the Gastrointestinal Tract. New York: Raven; 1994. pp. 1255–1283. [Google Scholar]
- Guan KL, Xiong Y. Regulation of intermediary metabolism by protein acetylation. Trends Biochem Sci. 36:108–116. doi: 10.1016/j.tibs.2010.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer RJ. Review article: the role of butyrate on colonic function. Alimentary pharmacology & therapeutics. 2008;27:104–119. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- Jass JR. Diet, butyric acid and differentiation of gastrointestinal tract tumours. Medical hypotheses. 1985;18:113–118. doi: 10.1016/0306-9877(85)90043-x. [DOI] [PubMed] [Google Scholar]
- Ki SW, Ishigami K, Kitahara T, Kasahara K, Yoshida M, Horinouchi S. Radicicol binds and inhibits mammalian ATP citrate lyase. The Journal of biological chemistry. 2000;275:39231–39236. doi: 10.1074/jbc.M006192200. [DOI] [PubMed] [Google Scholar]
- Kim YS, Milner JA. Dietary modulation of colon cancer risk. The Journal of nutrition. 2007;137:2576S–2579S. doi: 10.1093/jn/137.11.2576S. [DOI] [PubMed] [Google Scholar]
- Le A, Cooper CR, Gouw AM, Dinavahi R, Maitra A, Deck LM, Royer RE, Vander Jagt DL, Semenza GL, Dang CV. Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2037–2042. doi: 10.1073/pnas.0914433107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leschelle X, Delpal S, Goubern M, Blottiere HM, Blachier F. Butyrate metabolism upstream and downstream acetyl-CoA synthesis and growth control of human colon carcinoma cells. European journal of biochemistry / FEBS. 2000;267:6435–6442. doi: 10.1046/j.1432-1327.2000.01731.x. [DOI] [PubMed] [Google Scholar]
- Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupton JR. Microbial degradation products influence colon cancer risk: the butyrate controversy. The Journal of nutrition. 2004;134:479–482. doi: 10.1093/jn/134.2.479. [DOI] [PubMed] [Google Scholar]
- Ma H, Weng D, Chen Y, Huang W, Pan K, Wang H, Sun J, Wang Q, Zhou Z, Wang H, et al. Extensive analysis of D7S486 in primary gastric cancer supports TESTIN as a candidate tumor suppressor gene. Molecular cancer. 2010;9:190. doi: 10.1186/1476-4598-9-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AA, Kurschel E, Osieka R, Schmidt CG. Clinical pharmacology of sodium butyrate in patients with acute leukemia. Eur J Cancer Clin Oncol. 1987;23:1283–1287. doi: 10.1016/0277-5379(87)90109-x. [DOI] [PubMed] [Google Scholar]
- Monleon D, Morales JM, Barrasa A, Lopez JA, Vazquez C, Celda B. Metabolite profiling of fecal water extracts from human colorectal cancer. NMR in biomedicine. 2009;22:342–348. doi: 10.1002/nbm.1345. [DOI] [PubMed] [Google Scholar]
- Paxinou E, Weisse M, Chen Q, Souza JM, Hertkorn C, Selak M, Daikhin E, Yudkoff M, Sowa G, Sessa WC, et al. Dynamic regulation of metabolism and respiration by endogenously produced nitric oxide protects against oxidative stress. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:11575–11580. doi: 10.1073/pnas.201293198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre F, Perrin P, Champ M, Bornet F, Meflah K, Menanteau J. Short-chain fructo-oligosaccharides reduce the occurrence of colon tumors and develop gut-associated lymphoid tissue in Min mice. Cancer Res. 1997;57:225–228. [PubMed] [Google Scholar]
- Pouillart PR. Role of butyric acid and its derivatives in the treatment of colorectal cancer and hemoglobinopathies. Life sciences. 1998;63:1739–1760. doi: 10.1016/s0024-3205(98)00279-3. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan P, Kahn DA, Baltimore D. Anti-apoptotic effect of hyperglycemia can allow survival of potentially autoreactive T cells. Cell death and differentiation. 2011;18:690–699. doi: 10.1038/cdd.2010.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roediger WE. Utilization of nutrients by isolated epithelial cells of the rat colon. Gastroenterology. 1982;83:424–429. [PubMed] [Google Scholar]
- Sarti M, Sevignani C, Calin GA, Aqeilan R, Shimizu M, Pentimalli F, Picchio MC, Godwin A, Rosenberg A, Drusco A, et al. Adenoviral transduction of TESTIN gene into breast and uterine cancer cell lines promotes apoptosis and tumor reduction in vivo. Clinical cancer research : an official journal of the American Association for Cancer Research. 2005;11:806–813. [PubMed] [Google Scholar]
- Scheppach W, Sommer H, Kirchner T, Paganelli GM, Bartram P, Christl S, Richter F, Dusel G, Kasper H. Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology. 1992;103:51–56. doi: 10.1016/0016-5085(92)91094-k. [DOI] [PubMed] [Google Scholar]
- Scheppach W, Weiler F. The butyrate story: old wine in new bottles? Current opinion in clinical nutrition and metabolic care. 2004;7:563–567. doi: 10.1097/00075197-200409000-00009. [DOI] [PubMed] [Google Scholar]
- Sekhavat A, Sun JM, Davie JR. Competitive inhibition of histone deacetylase activity by trichostatin A and butyrate. Biochemistry and cell biology = Biochimie et biologie cellulaire. 2007;85:751–758. doi: 10.1139/o07-145. [DOI] [PubMed] [Google Scholar]
- Sengupta S, Muir JG, Gibson PR. Does butyrate protect from colorectal cancer? Journal of gastroenterology and hepatology. 2006;21:209–218. doi: 10.1111/j.1440-1746.2006.04213.x. [DOI] [PubMed] [Google Scholar]
- Siddiqui KM, Chopra DP. Primary and long term epithelial cell cultures from human fetal normal colonic mucosa. In vitro. 1984;20:859–868. doi: 10.1007/BF02619632. [DOI] [PubMed] [Google Scholar]
- Singh B, Halestrap AP, Paraskeva C. Butyrate can act as a stimulator of growth or inducer of apoptosis in human colonic epithelial cell lines depending on the presence of alternative energy sources. Carcinogenesis. 1997;18:1265–1270. doi: 10.1093/carcin/18.6.1265. [DOI] [PubMed] [Google Scholar]
- Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3 doi: 10.2202/1544-6115.1027. Article3. [DOI] [PubMed] [Google Scholar]
- Tatarelli C, Linnenbach A, Mimori K, Croce CM. Characterization of the human TESTIN gene localized in the FRA7G region at 7q31.2. Genomics. 2000;68:1–12. doi: 10.1006/geno.2000.6272. [DOI] [PubMed] [Google Scholar]
- Turcan S, Rohle D, Goenka A, Walsh LA, Fang F, Yilmaz E, Campos C, Fabius AW, Lu C, Ward PS, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg effect: the metabolic requirements of cell proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velazquez OC, Seto RW, Bain AM, Fisher J, Rombeau JL. Deoxycholate inhibits in vivo butyrate-mediated BrDU labeling of the colonic crypt. The Journal of surgical research. 1997;69:344–348. doi: 10.1006/jsre.1997.5075. [DOI] [PubMed] [Google Scholar]
- Velcich A, Palumbo L, Jarry A, Laboisse C, Racevskis J, Augenlicht L. Patterns of expression of lineage-specific markers during the in vitro-induced differentiation of HT29 colon carcinoma cells. Cell growth & differentiation : the molecular biology journal of the American Association for Cancer Research. 1995;6:749–757. [PubMed] [Google Scholar]
- Wang T, Cai G, Qiu Y, Fei N, Zhang M, Pang X, Jia W, Cai S, Zhao L. Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. The ISME journal. 2012;6:320–329. doi: 10.1038/ismej.2011.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellen KE, Hatzivassiliou G, Sachdeva UM, Bui TV, Cross JR, Thompson CB. ATP-citrate lyase links cellular metabolism to histone acetylation. Science. 2009;324:1076–1080. doi: 10.1126/science.1164097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic acids research. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.