Abstract
Intestinal commensal bacteria induce protective and regulatory responses that maintain host-microbial mutualism. However, the contribution of tissue-resident commensals to immunity and inflammation at other barrier sites has not been addressed. We found that in mice, the skin microbiota have an autonomous role in controlling the local inflammatory milieu and tuning resident T lymphocyte function. Protective immunity to a cutaneous pathogen was found to be critically dependent on the skin microbiota but not the gut microbiota. Furthermore, skin commensals tuned the function of local T cells in a manner dependent on signaling downstream of the interleukin-1 receptor. These findings underscore the importance of the microbiota as a distinctive feature of tissue compartmentalization, and provide insight into mechanisms of immune system regulation by resident commensal niches in health and disease.
Mammals and their microbiota have formed an evolutionary partnership that is critical for metabolism, tissue development, and host defense (1–3). In particular, the gut flora has been implicated in intestinal immune tissue development and function, as well as in promoting systemic inflammation in the context of autoimmunity and infection (1, 4–8). Despite our growing understanding of the consequences of this host-microbe alliance for intestinal immune function, the degree to which the gut flora contributes to immunity at distal sites remains unclear.
The skin represents the primary interface between the host and the environment. Microbial profiling has revealed the presence of highly diverse commensal communities along distinct topographical skin sites (9, 10). Moreover, cutaneous inflammatory disorders such as psoriasis, atopic dermatitis, and rosacea have been associated with dysbiosis in the cutaneous microbiota (11, 12). Indeed, microbial products from skin commensals are known to exert immunoregulatory effects (13).
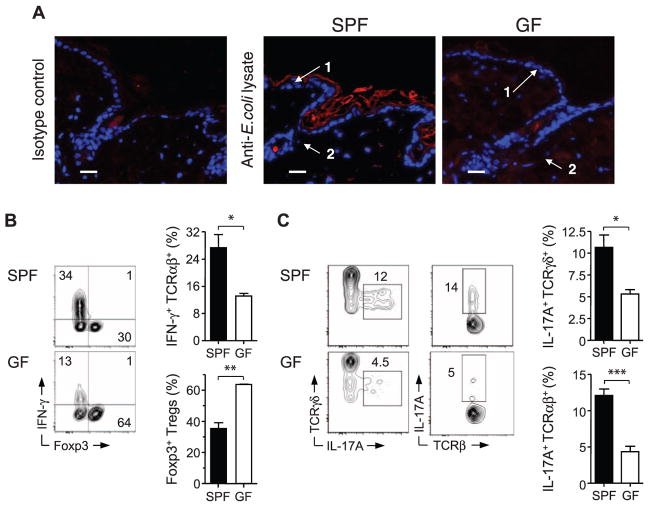
In humans, commensals reside in defined niches such as hair follicles and sebaceous glands (14). Microbes or microbial products densely coat the outer skin layer, hair follicles, and sebaceous glands in mice raised under specific pathogen-free (SPF) conditions (Fig. 1A). As previously described (15), the skin tissue of SPF mice contains a high frequency of Foxp3+ regulatory T cells (Tregs) (Fig. 1B and fig. S1, A and B). This compartment is also home to T cell receptor (TCR) αβ+ T cells with the potential to produce substantial amounts of the cytokines interferon-γ (IFN-γ) or interleukin-17A (IL-17A), the latter produced by CD4+ and CD4− CD8− T cells (Fig. 1, B and C, and fig. S1, A to C), as well as TCRγδlow dermal T cells (Fig. 1C and fig. S1, A and B) (16). Microbe-derived products were undetectable in the skin of germ-free (GF) mice born and raised in aseptic conditions (Fig. 1A). In the intestine, the balance between effector and regulatory T lymphocytes is tightly controlled by commensal signals (17–19). Similarly, we observed a significant reduction in IFN-γ and IL-17A production by αβ T cells, and IL-17A by γδlow T cells, in skin tissue of GF mice relative to SPF mice (Fig. 1, B and C, and fig. S1D). Concomitantly, the frequency and absolute numbers of cutaneous Foxp3+ Tregs were increased in the absence of commensals (Fig. 1B and fig. S1D). The influence of commensals on T cell subset frequencies did not extend to draining lymph nodes (fig. S2A).
Fig. 1.
Commensal microbiota control the balance of effector and regulatory T cells in the skin tissue. (A) Immunofluorescence labeling of bacterial products in interfollicular keratinocytes (1) and hair follicles (2) from skin tissue of SPF and GF mice. Representative images show naïve skin stained with anti–E. coli lysate antibody (red) or isotype control and Hoechst (blue); scale bars, 25 μm. (B) Representative flow cytometric plots and summarized bar graphs of IFN-γ and Foxp3 expression by live CD45+ TCRβ+ cells extracted from skin tissue of SPF and GF mice after stimulation with phorbol myristate acetate (PMA) and ionomycin. Graphs show means ± SEM of three or four mice (*P < 0.05, **P < 0.005). Results are representative of three experiments. (C) Representative flow cytometric plots and summarized bar graphs of IL-17A expression in live CD45+ TCRγδ+ or CD45+ TCRαβ+ cells from skin tissue of SPF and GF mice after stimulation with PMA and ionomycin. Graphs show means ± SEM of three or four mice (*P < 0.05, ***P < 0.0005). Results are representative of three experiments.
Defects in effector responses, previously observed in the gut of GF mice, have in part been attributed to impaired development of tissue and associated lymphoid structures (5). Surveys of skin homing receptors on peripheral T cells and frequencies of cutaneous γδ or αβ T cells, eosinophils, mast cells, and dendritic cell subsets revealed no differences between GF and SPF mice (fig. S2, B to F). In line with previous analyses (20, 21), the cellularity and architecture of skin-draining lymph nodes were comparable between SPF and GF mice (fig. S2, C and G). Thus, effector T cell functional potential in the skin is critically dependent on signals from the commensal microbiota and independent of developmental defects.
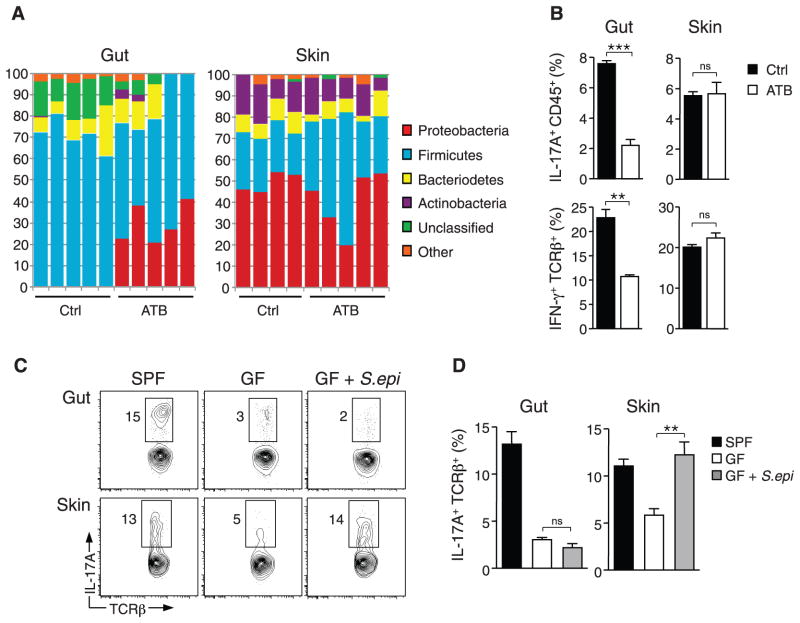
Differences in dermal T cell effector profiles in GF mice may result from a lack of signals from the gut flora, which have been implicated in the control of both local and systemic immune responses (4, 7, 8, 22). In particular, mice harboring segmented filamentous bacteria (SFB) in their gastrointestinal (GI) tract display increased intestinal levels of CD4+ T cell–derived IL-17A and IFN-γ relative to mice devoid of such bacteria (23, 24). Consistent with previous reports, mono-association of GF mice with SFB reconstituted IL-17A and IFN-γ to levels observed in the GI tract of SPF mice (fig. S3, A and B). In contrast, the presence of SFB did not restore effector cytokine production or alter Treg frequencies in the skin (fig. S3, A to C). To further address this point, we used a well-established oral antibiotic regimen known to decrease the density of intestinal flora (17, 19, 25). After oral antibiotic treatment, the overall density and composition of gut microbiota, but not skin microbiota, were profoundly altered (17) (Fig. 2A and fig. S3, D and E). Alterations of gut flora had no effect on the capacity of T cells to produce inflammatory cytokines (Fig. 2B).
Fig. 2.
Distinct commensal niches control T cell cytokine production in the gut and skin. (A) Taxonomic classifications at the phylum level for 16S ribosomal RNA gene sequence data clustered at 97% identity from skin tissue and fecal pellet of control mice and mice treated with oral antibiotic cocktail (ATB) for 4 weeks. Each column represents an individual mouse. (B) Assessment of IFN-γ production in live CD45+ TCRβ+ cells and IL-17A production in live CD45+ cells from skin and intestine of mice treated with oral antibiotic cocktail or water (Ctrl) for 4 weeks. Graphs show means ± SEM of four mice (**P < 0.005, ***P < 0.0005; ns, not significant). Results are representative of two or three experiments. (C and D) Flow cytometric analysis of IL-17A production in live CD45+ TCRβ+ cells from the gut and skin of SPF mice, GF mice, and GF mice monoassociated with S. epidermidis (GF + S.epi) for 2 to 3 weeks. Graphs show means ± SEM of three to five mice (**P < 0.005). Results are representative of two experiments.
We next monoassociated GF mice with the skin commensal Staphylococcus epidermidis (9). Colonization with this single commensal organism was sufficient to rescue IL-17A production in the skin but not the gut (Fig. 2, C and D). Furthermore, treatment with oral vancomycin to prevent S. epidermidis from colonizing the gut had no effect on this bacterium’s ability to rescue cutaneous IL-17A levels (fig. S3F). Together, our results show that resident bacteria are necessary to drive effector T cell function in the skin, and that fluctuations in the gut microbiota have no direct effect on cutaneous immune homeostasis.
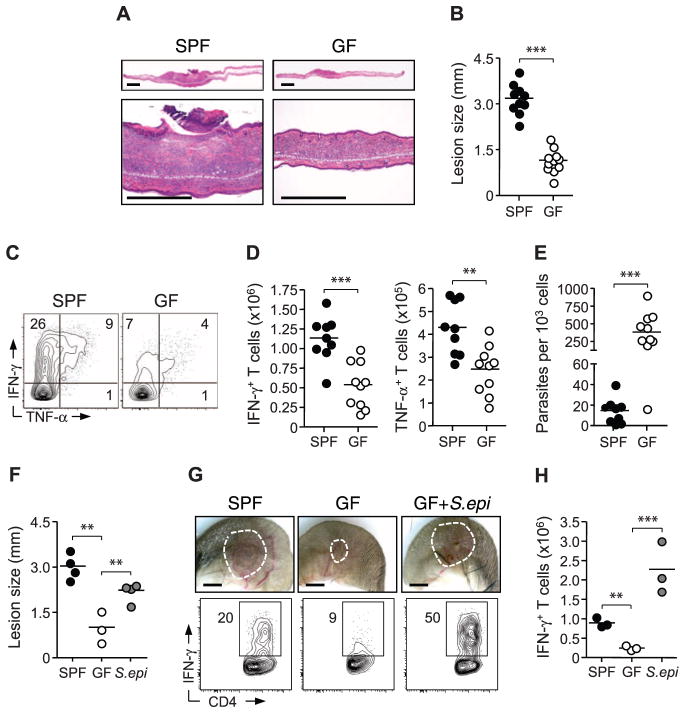
To evaluate the functional consequences of skin commensals on local immunity, we used a model of dermal infection induced by the protozoan parasite Leishmania major (26). Protective immunity to L. major is dependent on T cell–derived IFN-γ (26). Previous work demonstrated impaired parasite control in the absence of commensals upon intramuscular parasite delivery (27). Relative to SPF mice, GF mice infected intradermally with L. major manifested smaller lesions with reduced edema and necrosis (Fig. 3, A and B). Effector responses were severely impaired in GF mice, as evidenced by a reduction in Leishmania-specific IFN-γ and production of the pro-inflammatory cytokine tumor necrosis factor–α (TNF-α) by cutaneous T cells (Fig. 3, C to E, and fig. S4A). Altered immunity and parasite control were not associated with increased Treg frequencies or IL-10 responses (fig. S4, B and C).
Fig. 3.
Cutaneous commensals drive immunity and promote pathology in L. major infection. (A) Histopathological comparison of ear pinnae skin lesions from L. major–infected SPF and GF mice. Scale bars, 500 μm. (B) Assessment of lesion size in SPF and GF mice. Each data point represents an individual mouse (***P < 0.0005). (C and D) Flow cytometric analysis of Leishmania antigen-specific IFN-γ and TNF-α production by TCRβ+ CD4+ dermal cells from L. major–infected SPF and GF mice. Each data point represents an individual mouse (**P < 0.005, ***P < 0.0005). Results are representative of three experiments. (E) Number of L. major parasites per 1000 nucleated cells from dermal lesions of infected SPF and GF mice. Each data point represents an individual mouse (***P < 0.0005). (F) Assessment of lesion size in SPF mice, GF mice, and GF mice monoassociated with S. epidermidis (S.epi). Each data point represents an individual mouse (**P < 0.005). Results are representative of two experiments. (G and H) Representative images of L. major skin lesions and analysis of IFN-γ production by live TCRβ+ CD4+ cells from SPF mice, GF mice, and GF mice mono-associated with S. epidermidis. Each data point represents an individual mouse (***P < 0.0005). Results are representative of two experiments.
To corroborate a role for skin commensals in promoting immunity to L. major, we mono-associated GF mice with S. epidermidis at the time of infection. Colonization of GF skin with this commensal was sufficient to rescue protective immunity in these animals (Fig. 3, F to H, and fig. S4D). Monoassociation of GF mice with S. epidermidis also restored pathology with increased necrosis; this finding supports the idea that pathological consequences of this skin infection depend on the presence of commensals rather than the parasite (Fig. 3, F and G). Further implicating cutaneous commensals in the direct control of skin immunity and inflammation, oral vancomycin treatment of GF animals associated with S. epidermidis had no influence on immunity and pathology during L. major infection (fig. S4, E and F).
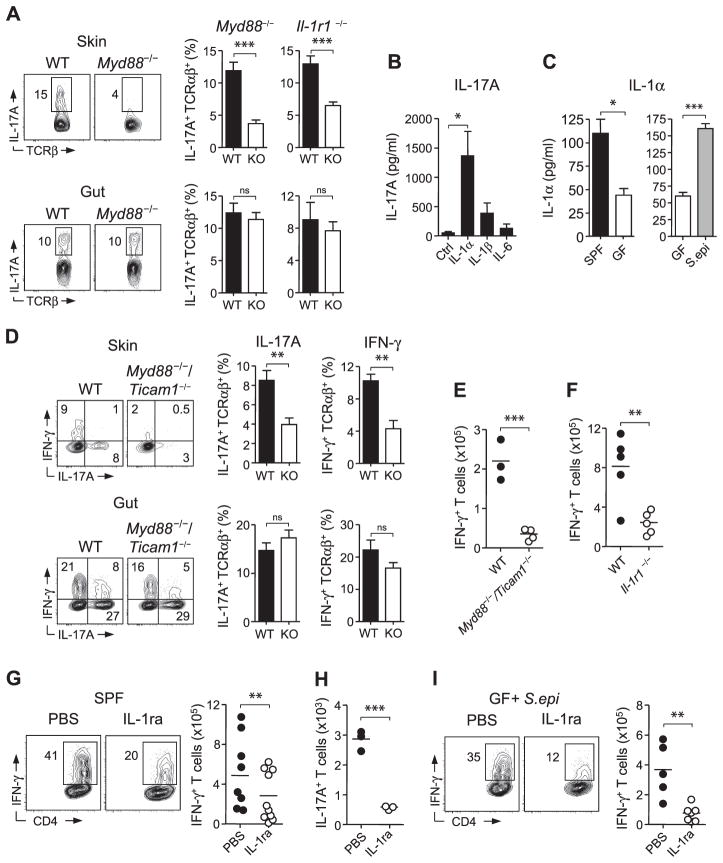
The dominant cytokine signals that drive effector cytokine production by T cells in the skin have not yet been elucidated. By screening mice that were deficient in factors known to drive IL-17A production, we determined that IL-1R1 and its downstream signaling complex MyD88, but not IL-23R or IL-6, played a dominant role in controlling the production of IL-17A, but not IFN-γ, by cutaneous T cells (Fig. 4A and fig. S5, A to C). Toll-like receptor 2 (TLR2), which also relies on MyD88-dependent signaling, is required to sense by-products from S. epidermidis during skin inflammation (13). However, mice deficient in TLR2 as well as TLR3, 5, and 9 did not display a reduction in T cells or cytokine production relative to control mice under steady-state conditions (fig. S5D). Hence, our data support the idea that the defect in IL-17A observed in MyD88-deficient mice is primarily a consequence of altered IL-1R1–mediated signaling. Consistent with a partitioning of the dominant signals controlling the skin and gut environment, MyD88 or IL-1R1 deficiency had no impact on T cell potential to produce IL-17A in the intestine (Fig. 4A).
Fig. 4.
Skin-resident commensals modulate dermal T cells in a manner dependent on IL-1 and MyD88. (A) Flow cytometric analysis of IL-17A production by live CD45+ TCRβ+ cells in skin and intestine of age-matched Myd88−/− and Il1r1−/− mice. WT, wild type; graphs show means ± SEM of three or four mice (***P < 0.0005). Results are representative of two or three experiments. (B) IL-17A production from purified skin CD45+ TCRβ+ T cells cultured in vitro in the presence of anti-CD3 and either IL-1α, IL-1β, or IL-6. Graphs represent the mean of three experimental groups ± SEM (*P < 0.05). Results are representative of three experiments. (C) Spontaneous release (± SEM) of IL-1α from skin-derived cells of SPF mice, GF mice, and GF mice monoassociated with S. epidermidis (S.epi) as measured by enzyme-linked immunosorbent assay (*P < 0.05, ***P < 0.0005). (D) Comparative assessment of IFN-γ and IL-17A production from WT and Myd88−/−/Ticam1−/− TCRβ+ cells from mixed bone marrow chimeric mice. Bar graphs show frequency (± SEM) of cytokine production by WT and knockout TCRβ+ cells. Results are representative of two experiments in the skin and one experiment in the gut (**P < 0.005). (E and F) Analysis of L. major–specific IFN-γ production by TCRB+ CD4+ T cells from WT and Myd88−/−/Ticam1−/− or Il1r1−/− mice. Results are representative of two experiments. (G) Flow cytometric assessment of L. major–specific IFN-γ production from TCRβ+ CD4+ T cells from the skin of SPF animals treated with either IL-1ra or phosphate-buffered saline (PBS). Results are a compilation of two experiments. (H) Number of CD45+ TCRβ+ IL-17A+ T cells from the skin of GF mice monoassociated with S. epidermidis treated with either IL-1ra or PBS. Results are representative of one experiment. (I) L. major–specific IFN-γ produced by TCRβ+ CD4+ T cells from the skin of GF mice monoassociated with S. epidermidis and treated with either IL-1ra or PBS. Results are representative of two experiments. For (E) to (I), each data point represents an individual mouse (**P < 0.005, ***P < 0.0005).
Both αβ and γδ effector T cells are known to express IL-1R1 and to respond directly to IL-1 (28–31). We purified skin lymphocytes and stimulated them via their TCR in the presence of IL-1α, IL-1β, or IL-6. Under these conditions, IL-1α and IL-1β, but not IL-6, potently increased the capacity of T cells to release IL-17A (Fig. 4B and fig. S5E). Thus, T cells that reside at dermal sites can be functionally tuned by the local cytokine milieu, and in particular by IL-1.
We next explored the possibility that IL-1 signaling may be diminished in the absence of commensals. Indeed, IL-1α production by cutaneous cells was significantly reduced in GF relative to SPF mice, and monoassociation of GF mice with S. epidermidis restored the production of this cytokine (Fig. 4C). Additionally, keratinocytes from GF mice displayed increased levels of the IL-1 receptor antagonist (IL-1ra) mRNA relative to SPF mice, indicating that commensals control various aspects of functional IL-1 signaling (fig. S5F). Complementing these observations, the addition of S. epidermidis to GF mice significantly reduced IL-1ra from cutaneous cells (fig. S5G). Therefore, resident commensals are required for optimal IL-1 signaling in the skin, which in turn promotes local effector responses.
To ascertain the importance of the MyD88 pathway in hematopoietic cells, we generated mixed bone marrow chimeras using MyD88/TRIF knockout mice that phenocopied MyD88 knockout animals in their cutaneous IL-17A deficiency. (fig. S5, A, H, and I). Irradiation required for generating chimeras induces inflammation and homeostatic proliferation of T cells. Under these inflammatory conditions, MyD88 signaling in hematopoietic cells was required for the production of both IFN-γ and IL-17A in the skin, but not in the gut (Fig. 4D). Furthermore, mice deficient in MyD88 or IL1R1 displayed impaired effector responses and parasite control during L. major infection, as did mice treated with IL1ra (Fig. 4, E to G, and fig. S5J).
These results support the idea that defects in T cell function at steady state or during inflammation result from an impaired dialogue with skin commensals. To functionally link commensally driven IL-1 signaling to T cell immunity in the skin, we treated GF mice monoassociated with S. epidermidis with IL-1ra at steady state and during L. major infection. Neutralizing IL-1 activity in these animals hindered this bacterium’s ability to rescue IL-17A at steady state and to promote L. major–specific IFN-γ during infection (Fig. 4, H and I). Thus, via their capacity to control IL-1 signaling, skin commensals promote effector T cell responses according to local inflammatory cues. On the basis of the pleiotropic role of IL-1, this effect may result from direct IL-1 signaling in T cells and/or modulation of various innate inflammatory cells such as neutrophils (32).
Our results indicate that resident commensals are necessary for optimal skin immune fitness. Specifically, we find that cutaneous commensals exert their effect by augmenting IL-1 signaling and amplifying responses in accordance with the local inflammatory milieu. The IL-1 pathway is an evolutionarily conserved arm of the innate immune system that may have arisen as an early mediator of host skin-commensal cross talk. This pathway is also linked to a multitude of chronic inflammatory disorders such as arthritis and asthma. Moreover, IL-1 has been implicated in the etiology and pathology of psoriasis and other cutaneous disorders (33). Thus, via their capacity to promote IL-1 signaling and consequently effector T cell function, skin commensals are likely important drivers and amplifiers of skin pathologies. Understanding the role of the skin microbiota in maintaining tissue function is not only of primary importance for human health, but will also lead to the development of more rational tissue-specific adjuvants and vaccine approaches.
Supplementary Material
Acknowledgments
Supported by the Division of Intramural Research of the National Institute of Allergy and Infectious Diseases (NIAID) and by the National Human Genome Research Institute, the National Cancer Institute, NIH grant F30 DK094708 (S.S.), and the Human Frontier Science Program (C.W.). We thank the NIAID gnotobiotic facility staff, in particular C. Acevedo and D. Trageser-Cesler; K. Holmes and the NIAID sorting facility; K. Beacht, B. Kelsall, T. Tamachi, C. Brown, J. Benchley, A. MacDonald, L. Mijares, K. Shenderov, and A. Sher; and Yakult Central Institute for Microbiological Research for sharing SFB via a material transfer agreement. DNA sequence data are available via GenBank (accession no. SRS345653). The data reported in this paper are tabulated in the main paper and in the supplementary materials.
Footnotes
References and Notes
- 1.Lee YK, Mazmanian SK. Science. 2010;330:1768. doi: 10.1126/science.1195568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huttenhower C, et al. Human Microbiome Project Consortium. Nature. 2012;486:207. [Google Scholar]
- 3.Methé BA, et al. Human Microbiome Project Consortium. Nature. 2012;486:215. doi: 10.1038/nature11209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ichinohe T, et al. Proc Natl Acad Sci USA. 2011;108:5354. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cebra JJ. Am J Clin Nutr. 1999;69:1046S. doi: 10.1093/ajcn/69.5.1046s. [DOI] [PubMed] [Google Scholar]
- 6.Clarke TB, et al. Nat Med. 2010;16:228. doi: 10.1038/nm.2087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee YK, Menezes JS, Umesaki Y, Mazmanian SK. Proc Natl Acad Sci USA. 2011;108(suppl 1):4615. doi: 10.1073/pnas.1000082107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu HJ, et al. Immunity. 2010;32:815. doi: 10.1016/j.immuni.2010.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grice EA, et al. Science. 2009;324:1190. doi: 10.1126/science.1171700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costello EK, et al. Science. 2009;326:1694. doi: 10.1126/science.1177486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallo RL, Nakatsuji T. J Invest Dermatol. 2011;131:1974. doi: 10.1038/jid.2011.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kong HH, et al. NISC Comparative Sequence Program. Genome Res. 2012;22:850. [Google Scholar]
- 13.Lai Y, et al. Nat Med. 2009;15:1377. doi: 10.1038/nm.2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Grice EA, Segre JA. Nat Rev Microbiol. 2011;9:244. doi: 10.1038/nrmicro2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Belkaid Y, Piccirillo CA, Mendez S, Shevach EM, Sacks DL. Nature. 2002;420:502. doi: 10.1038/nature01152. [DOI] [PubMed] [Google Scholar]
- 16.Sumaria N, et al. J Exp Med. 2011;208:505. doi: 10.1084/jem.20101824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hill DA, et al. Mucosal Immunol. 2010;3:148. doi: 10.1038/mi.2009.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ivanov II, et al. Cell. 2009;139:485. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall JA, et al. Immunity. 2008;29:637. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson NS, et al. Immunol Cell Biol. 2008;86:200. doi: 10.1038/sj.icb.7100125. [DOI] [PubMed] [Google Scholar]
- 21.Guilliams M, et al. Blood. 2010;115:1958. doi: 10.1182/blood-2009-09-245274. [DOI] [PubMed] [Google Scholar]
- 22.Wen L, et al. Nature. 2008;455:1109. doi: 10.1038/nature07336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ivanov II, et al. Cell Host Microbe. 2008;4:337. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Microbiol Immunol. 1995;39:555. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 25.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Cell. 2004;118:229. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 26.Belkaid Y, et al. J Immunol. 2000;165:969. doi: 10.4049/jimmunol.165.2.969. [DOI] [PubMed] [Google Scholar]
- 27.de Oliveira MR, et al. Rev Inst Med Trop Sao Paulo. 1999;41:87. doi: 10.1590/s0036-46651999000200005. [DOI] [PubMed] [Google Scholar]
- 28.Sutton CE, et al. Immunity. 2009;31:331. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 29.Guo L, et al. Proc Natl Acad Sci USA. 2009;106:13463. doi: 10.1073/pnas.0906988106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shaw MH, Kamada N, Kim YG, Núñez G. J Exp Med. 2012;209:251. doi: 10.1084/jem.20111703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu W, Troutman TD, Edukulla R, Pasare C. Immunity. 2011;35:1010. doi: 10.1016/j.immuni.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller LS, et al. Immunity. 2006;24:79. doi: 10.1016/j.immuni.2005.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Sims JE, Smith DE. Nat Rev Immunol. 2010;10:89. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.