The Hsp70–Hsp110 chaperone complex antagonizes Cin8 plus-end motility and prevents premature spindle elongation in S phase.
Abstract
Systematic affinity purification combined with mass spectrometry analysis of N- and C-tagged cytoplasmic Hsp70/Hsp110 chaperones was used to identify new roles of Hsp70/Hsp110 in the cell. This allowed the mapping of a chaperone–protein network consisting of 1,227 unique interactions between the 9 chaperones and 473 proteins and highlighted roles for Hsp70/Hsp110 in 14 broad biological processes. Using this information, we uncovered an essential role for Hsp110 in spindle assembly and, more specifically, in modulating the activity of the widely conserved kinesin-5 motor Cin8. The role of Hsp110 Sse1 as a nucleotide exchange factor for the Hsp70 chaperones Ssa1/Ssa2 was found to be required for maintaining the proper distribution of kinesin-5 motors within the spindle, which was subsequently required for bipolar spindle assembly in S phase. These data suggest a model whereby the Hsp70–Hsp110 chaperone complex antagonizes Cin8 plus-end motility and prevents premature spindle elongation in S phase.
Introduction
The Hsp70 family of chaperones is a multifunctional group of highly related stress proteins with diverse cellular roles. Saccharomyces cerevisiae (baker’s yeast) has 14 different Hsp70s, with 9 of these localized to the cytoplasm/nucleus (Kampinga and Craig, 2010): Ssa1, Ssa2, Ssa3, Ssa4, Ssb1, Ssb2, Ssz1, Sse1, and Sse2. The overall structure of these proteins is highly conserved and typically consists of an N-terminal ATPase domain, a central substrate-binding domain (SBD), and a C-terminal variable domain (Fig. S1 A).
Ssa1, Ssa2, Ssa3, and Ssa4 encode four closely related proteins that define the Ssa subclass of Hsp70 chaperones in yeast (Fig. S1 B). The majority of Ssa proteins are found in the cytoplasm, although both Ssa1 and Ssa2 have also been detected in the nucleus and are associated with the cell wall (López-Ribot and Chaffin, 1996; van den Bosch and Lowndes, 2004). Ssb1 and Ssb2 define the Ssb subclass of yeast Hsp70 chaperones (Fig. S1 B). Both Ssb1 and Ssb2 have been found to be localized to the ribosome as part of the ribosome-associated complex (Pfund et al., 1998), which also includes Ssb1 or Ssb2, Ssz1, and Zuo1 (an Hsp40). For Ssz1, neither the ATPase nor the peptide-binding activities appear to be necessary for its function (Huang et al., 2005). It has been suggested that Ssz1 does not function in binding unfolded polypeptides but rather acts to enhance the ATPase-stimulating activity of Zuo1 on Ssb1 and Ssb2 (Huang et al., 2005). Sse1 and Sse2 define the Hsp110 subclass of the Hsp70 chaperones in yeast. They have an Hsp110 insertion sequence within the SBD (Fig. S1 A). The Sse/Hsp110 subclass is only found in eukaryotic cells. These chaperones are not thought to actively assist in protein folding; rather, they are thought to bind unfolded polypeptides and to hold them in a folding-competent state (Easton et al., 2000). More recently, based on biochemical and structural data, Sse1 and Sse2 have been proposed to act as nucleotide exchange factors (NEFs) for the Ssa and Ssb chaperones (Polier et al., 2008; Schuermann et al., 2008). Nevertheless, the actual cellular function of the Sse/Hsp110 proteins has remained elusive.
In an effort to reveal novel functions of the Hsp70s/Hsp110s in the cell, we performed a systematic pull-down of all the nine cytoplasmic Hsp70/Hsp110 proteins in yeast using strains in which these proteins were endogenously N- or C-terminally tandem affinity purification (TAP) tagged. This allowed us to identify novel pathways and complexes affected by these chaperones. In-depth analysis of the protein–protein interaction map showed that many of Hsp70/Hsp110 chaperones copurified with proteins involved in spindle organization. We were able to demonstrate that the nucleotide exchange activity of Sse1 on Ssa1/Ssa2 is required for proper spindle assembly. More specifically, the Hsp70/Hsp110 chaperone system was found to antagonize kinesin-5 motor Cin8 plus-end motility to prevent premature spindle elongation in S phase.
Results
Overview of the Hsp70/Hsp110 interactors
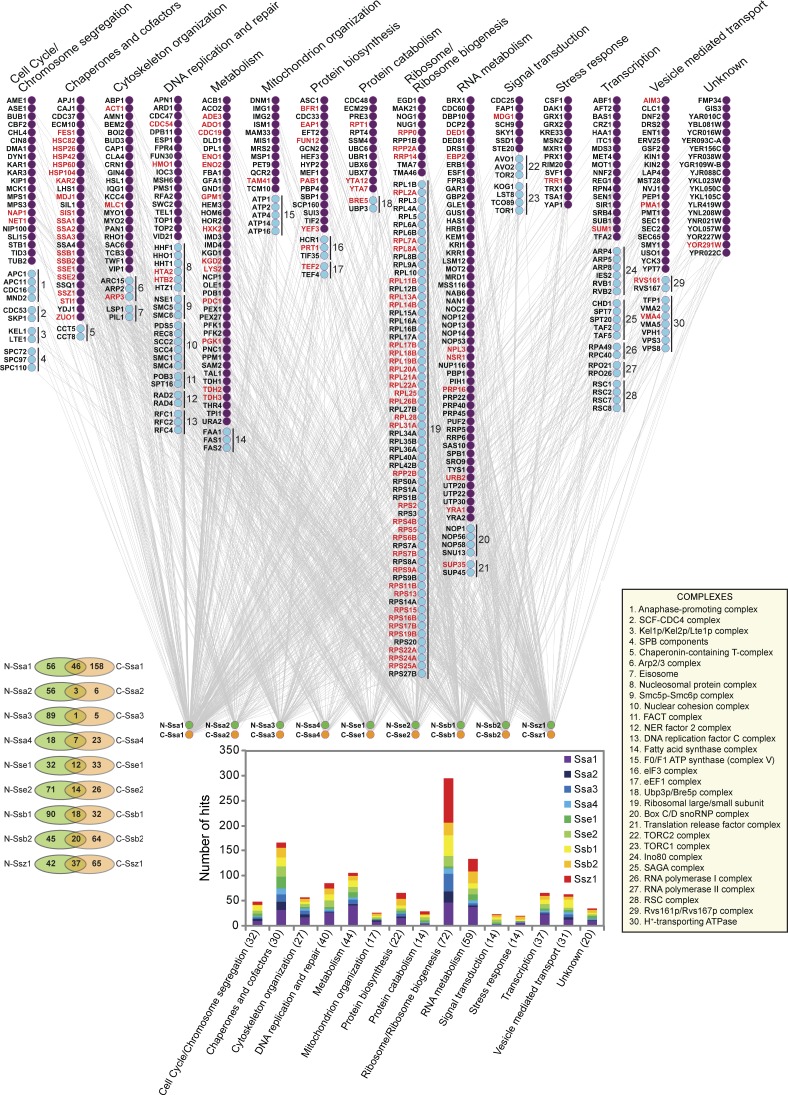
Our previous analysis of the global physical interaction network of the yeast chaperones was based on using yeast strains in which all the chaperones were C-terminally TAP tagged (Gong et al., 2009). The analysis provided insights into the general principles that govern chaperone–chaperone and chaperone–substrate interactions. In this study, we performed a more detailed physical interaction mapping of the cytoplasmic Hsp70/Hsp110 chaperones. To this end, the nine cytoplasmic Hsp70/Hsp110 chaperones were endogenously TAP tagged at the N and C termini, and the tag was used to pull down the chaperones; protein interactors were then identified using mass spectrometry (see Materials and methods). Western blot analysis of cells grown at 30°C shows that Ssa1 and Ssa2 are most abundant followed by Ssb1, Ssb2, Sse1, and Ssa4 (Fig. S1 C), which is consistent with the published literature, indicating that the tags did not affect the expression levels of the chaperones. Mass spectrometry analysis allowed us to obtain a physical interaction map consisting of 1,227 interactions between the 9 chaperones and 473 proteins (Fig. 1 and Table S2) and highlighted roles for Hsp70/Hsp110 chaperones in 14 broad biological processes and 30 well-represented protein complexes (Fig. 1). 591 of these interactions were observed in previous published work (Table S3) including our own large-scale proteomic study (Gong et al., 2009). Out of the 473 interacting proteins, only 26 can be classified as transmembrane proteins (see Materials and methods); hence, most of the interactors are soluble proteins under the conditions of our experiments, as would be expected for these chaperones.
Figure 1.
The TAP tag–based Hsp70/Hsp110 interaction network. The main figure shows the hits obtained from the TAP tag pull-down of the nine Hsp70s/Hsp110s. The hits are grouped according to Gene Ontology categories (also shown in the bottom graph; The Gene Ontology Consortium, 2000) and are organized into complexes as defined by Pu et al. (2009; bottom right inset). The overlap in the hits between N- and C-tagged chaperones is shown in the bottom left. Data are from experiments completed once. Complexes identified are shown in the top right inset. FACT, facilitates chromatin transcription; NER, nucleotide excision repair; snoRNP, small nucleolar ribonucleoprotein; SCF, Skp, Cullin, F-box–containing complex.
657 out of the 1,227 interactions were detected using N-tagged chaperones (Fig. 1 and Table S2). A significantly smaller proportion of the interactions, 570, was detected using C-tagged chaperones (z test for proportions: P < 0.001). 158 hits overlapped between the N- and C-tagged chaperones (Fig. 1 [left inset] and Table S4). 61% (96 out of 158) of these are supported by other published experiments (Table S3). Proteins interacting with both N- and C-terminally TAP-tagged chaperones have a significantly greater proportion supported by published literature (Table S3) than all captured proteins in our dataset (z test for proportions: P < 0.05). Ssa1 and Ssb1, Ssa1 and Ssb2, and Ssa1 and Ssz1 have the largest number of overlapping interactors (Fig. S2 A), suggesting cooperativity between Ssa1 and each of Ssb1, Ssb2, and Ssz1 chaperones. Chaperone–chaperone TAP tag interactions were also detected between the different chaperones using both N- and C-TAP interactions (Fig. S2 B). However, N-tagged chaperones were generally more efficient in binding other chaperones (Fig. S2 B) or other proteins (Table S2) than their C-tagged variants, possibly highlighting the importance of an unblocked C terminus for proper chaperone–chaperone and chaperone–substrate interactions. For example, the C terminus in all Ssa chaperones ends with the highly conserved EEVD residues, which mediate interaction with cochaperones and cofactors (Scheufler et al., 2000; Wegele et al., 2003).
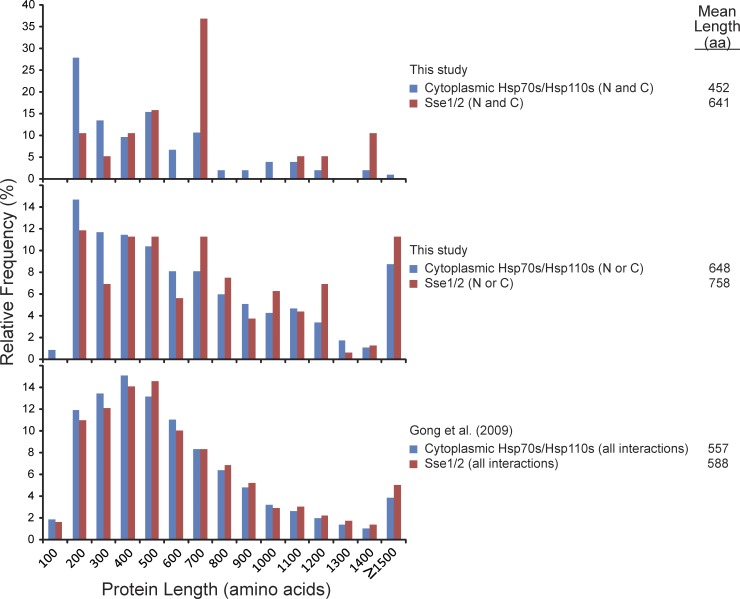
In analyzing the physical interaction data, we noted that the interactors of Sse1/2 were, on average, larger proteins than those of the total cytoplasmic Hsp70s. This observation held true whether we used the total hits obtained from the N- or C-tagged chaperones or only the hits that overlapped between N- and C-tagged chaperones (Fig. 2). Furthermore, the trend was also observed in the data from our large-scale proteomic study (Fig. 2; Gong et al., 2009). This intriguing finding prompted us to further analyze specific Sse hits.
Figure 2.
Size distribution of chaperone hits. The size distribution of the hits obtained in this work for the cytoplasmic Hsp70s/Hsp110s compared with those obtained for the Hsp110s (Sse1/2) alone is shown as a bar graph. (N and C) refers to the hits that overlapped between N- and C-TAP tag pull-down experiments, whereas (N or C) refers to the hits obtained in either N- or C-TAP tag pull-down experiments. The size distribution of the hits obtained in our previous study (Gong et al., 2009) is also shown. Proteins interacting with Sse1/2 were found to be longer than proteins interacting with the cytoplasmic Hsp70/Hsp110 chaperones (Mann–Whitney test, P < 0.04). Data are from experiments completed once.
Role of Hsp110/Sse1 in spindle assembly
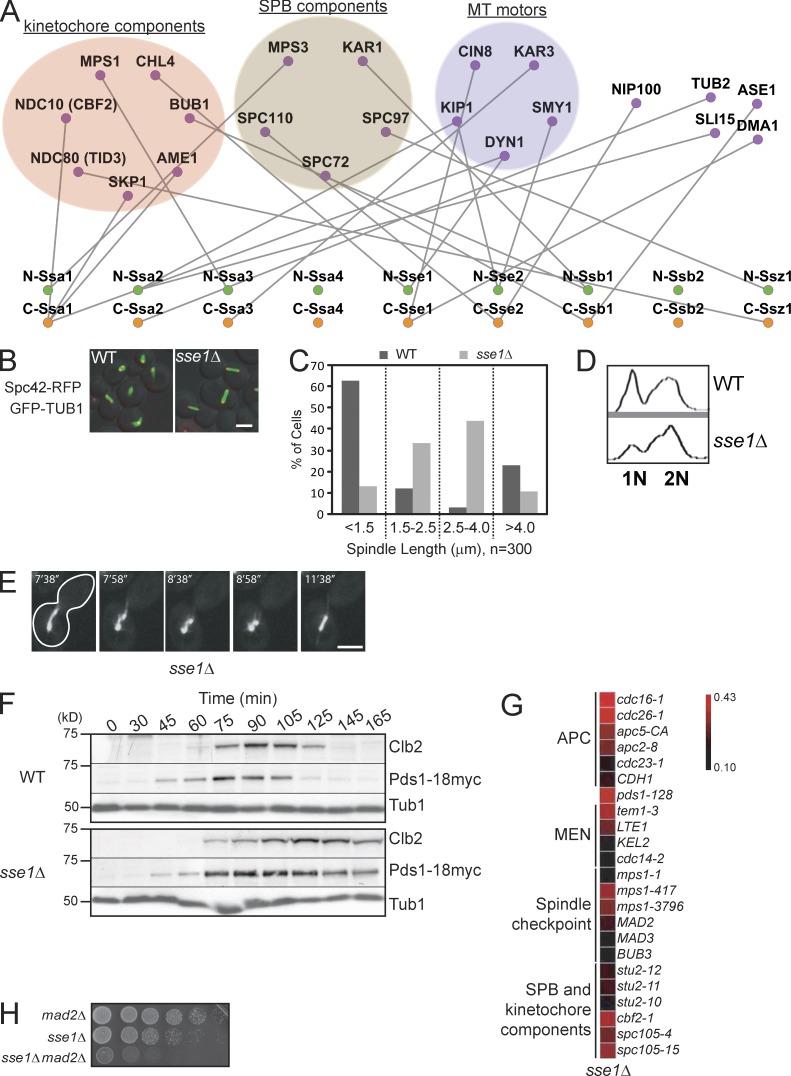
One observation made from the comprehensive analysis of the obtained network (Fig. 1) is that Hsp70/Hsp110 copurified with different components of the spindle pole body (SPB; Kar1, Mps3, Spc72, Spc97, and Spc110), kinetochore (Ame1, Bub1, Chl4, Mps1, Ndc10, Ndc80, and Skp1), and microtubule (MT) motors (Cin8, Dyn1, Kar3, Kip1, and Smy1) as well as the large subunit of the dynactin complex (Nip100), proteins with spindle midzone organization roles (Ase1 and Sli15), and the establishment of mitotic spindle orientation (Dma1) in addition to tubulin (Tub2; Fig. 3 A), strongly suggesting a role for these chaperones in spindle organization/assembly and MT-based movement. This is consistent with previous observations indicating that the Hsp70/Hsp110 chaperones might be components of the SPB and kinetochore in yeast and mammalian cells (Lechner and Carbon, 1991; Oka et al., 1998; Wigge et al., 1998). Several of these interacting proteins are particularly large, e.g., Cin8 (1,000 residues), Dyn1 (4,092 residues), and Kip1 (1,111 residues). However, the molecular mechanism of Hsp70/Hsp110 function in MT organization has not been characterized.
Figure 3.
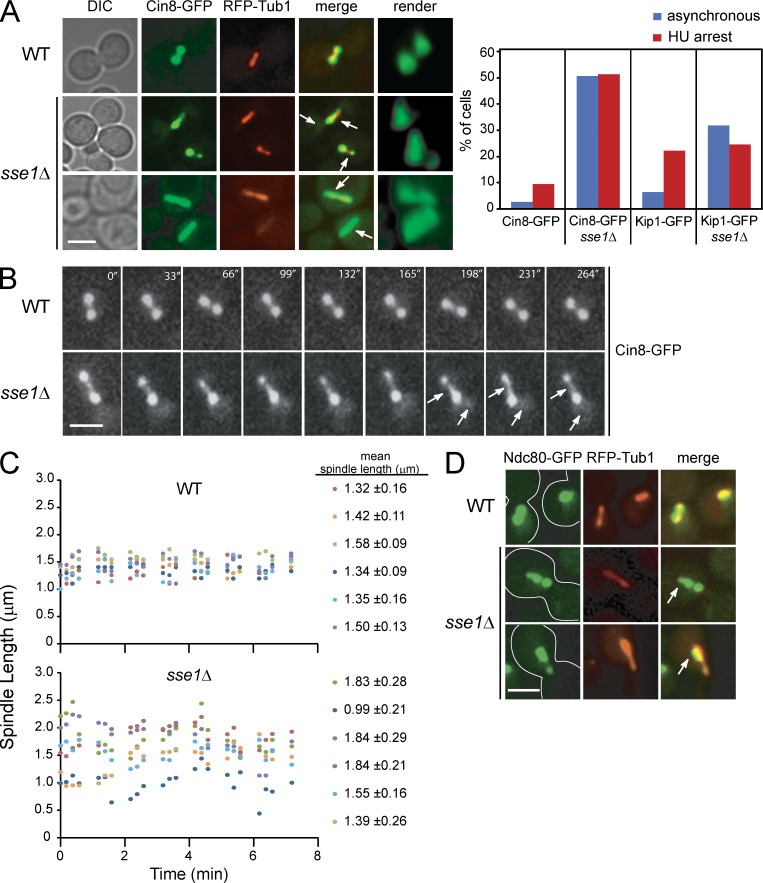
The role of Hsp70/Hsp110 in spindle organization. (A) A subnetwork of the Hsp70/Hsp110 protein interactions (also refer to Fig. 1 and Table S2) highlighting TAP tag–based physical interactions between N- and C-tagged Hsp70/Hsp110 and components of SPB, kinetochore, and MT motors. (B) The fluorescence images show spindle morphology examined by confocal microscopy in logarithmically growing WT and sse1Δ strains at 30°C containing an SPB marker, endogenous Spc42-RFP (red), and harboring the pGFP-TUB1 (green) plasmid. Bar, 5 µm. (C) Bar graph showing the distribution of spindle lengths observed in WT and sse1Δ cells. The data shown are from a single representative experiment out of three repeats. (D) FACS profiles of WT and sse1Δ cells. 3 ml of early log–phase culture was used. (E) The spindle of an sse1Δ cell was visualized using plasmid-borne GFP-Tub1 and examined by time-lapse confocal microscopy. The cell outline is traced by the white line. Bar, 5 µm. (F) Logarithmically growing cultures of WT and sse1Δ expressing Pds1-18myc were synchronized in G1 with α-factor (time = 0 min). α-factor was then removed, and cells were grown at 26°C in YPD. Samples were taken at the indicated time points, and total cell lysates were analyzed by Western blot analysis using antibodies directed against cMyc, Clb2, and tubulin. Molecular mass markers are shown on the left of the gels. (G) A synthetic genetic array subnetwork highlighting genetic interactions between sse1Δ and genes involved in chromosome segregation and cell cycle progression. APC and MEN refer to anaphase-promoting complex and mitotic exit network, respectively. The intensity of the red color correlates with the strength of the genetic interaction (refer to Costanzo et al. [2010]). (H) 10× serial dilutions of log-phase cells of the indicated genotypes spotted onto YPD and incubated at 26°C for 2 d are shown.
To determine the physiological consequences underlying the physical interactions we observed, the morphology of the mitotic spindle was examined in strains deleted of each of the nine different Hsp70/Hsp110 chaperones. The spindle was visualized using confocal microscopy by expressing GFP-Tub1 from a plasmid in ∼150 to ∼300 cells that were grown in appropriate synthetic media to early log phase. Only cells deleted of SSE1 exhibited significant cell cycle transition defects (Fig. S3 A). Furthermore, an increased number of sse1Δ cells was found to have spindle length between 1.5 and 4 µm (Figs. 3 [B and C] and S3 B). Consistent with these results, FACS analysis demonstrated that the majority of these sse1Δ cells accumulated with G2 DNA content (Fig. 3 D). In addition, using time-lapse microscopy, the spindle stability was found to be dramatically decreased in ∼5% of the sse1Δ cells, as several rounds of spindle collapse were observed, and the cells were unable to elongate spindles to >4 µm (Fig. 3 E and Video 1).
To further investigate the cell cycle delay exhibited by sse1Δ mutant cells, we monitored levels of Pds1, an inhibitor of the anaphase activator Esp1 (Peters, 2002) and mitotic cyclin Clb2 during the cell cycle progression. Upon release from α-factor–induced G1 arrest, levels of Pds1 increased at a similar rate in both wild-type (WT) and sse1Δ cells, suggesting that the progression to S phase is not affected by SSE1 deletion (Fig. 3 F). However, the levels of Pds1 remained elevated for a significantly longer time in sse1Δ compared with WT cells. Similarly to Pds1, Clb2 degradation was also significantly delayed. This indicates that SSE1 deletion results in a delay in the metaphase-to-anaphase transition (Fig. 3 F). This result is further supported by previous published experiments showing a genetic interaction between PDS1 and SSE1 (Fig. 3 G; Sarin et al., 2004; Costanzo et al., 2010) and indicates that the proper timing of anaphase initiation becomes critical in the absence of Sse1. Consistently, deletion of SSE1 when combined with gene mutants encoding different kinetochore, SPB, and anaphase-promoting complex components produced synthetic fitness defects (Fig. 3 G). We also confirmed the reported genetic interaction between SSE1 and the spindle assembly checkpoint MAD2 (Fig. 3 H; Daniel et al., 2006), indicating that spindle assembly is compromised in the sse1Δ mutant. These data strongly suggest a role for Sse1 in cell cycle progression.
Deletion of SSE1 results in spindle elongation in S phase
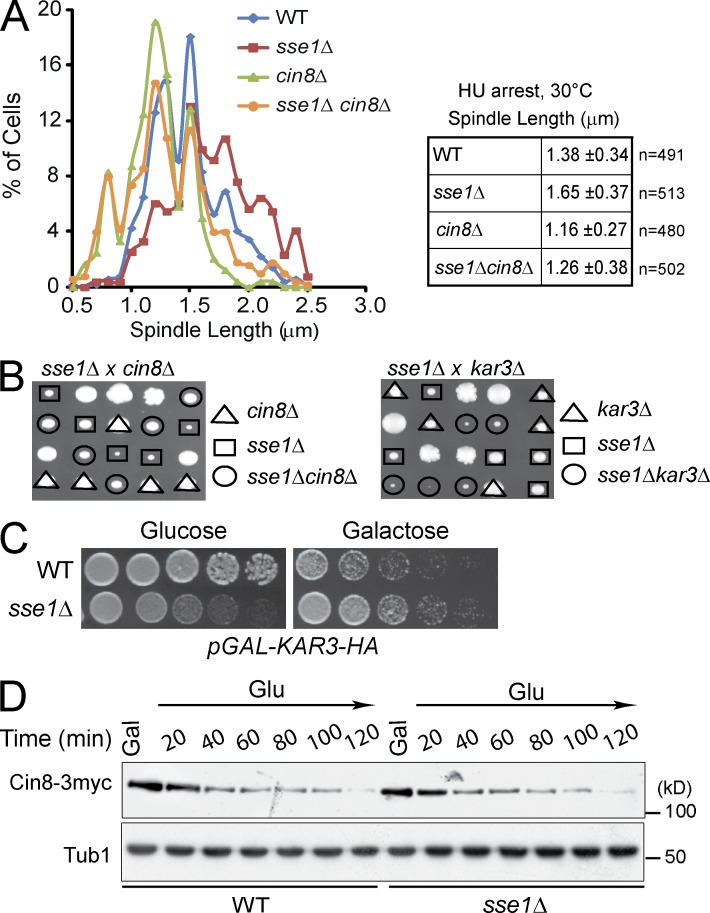
To more directly investigate the role of Sse1 in spindle organization, the consequences of SSE1 deletion on spindle assembly in S phase were investigated. When DNA replication was stalled using the DNA synthesis inhibitor hydroxyurea (HU), both WT and sse1Δ cells arrested in S phase with a large bud, an undivided nucleus positioned at the mother bud neck, and a short bipolar spindle (Allen et al., 1994). Spindle length was measured in HU-arrested cells containing Spc42-RFP, an SPB marker, or by expressing GFP-TUB1 from a plasmid to visualize the spindle (Saunders et al., 1997b). The data indicate that the mean spindle length in sse1Δ cells is longer compared with that in WT cells (Fig. 4 A). More specifically, only 32% of the spindles in WT cells was ≥1.5 µm, whereas 62% of the spindles in sse1Δ was ≥1.5 µm (Fig. 4 A). Importantly, sse1Δ cells are not HU sensitive, and there is no genetic interaction between DNA damage checkpoint genes and sse1Δ mutation (Sarin et al., 2004; Costanzo et al., 2010). Spindle assembly in S phase is orchestrated by the kinesin-5 motors Cin8 and Kip1, with Cin8 playing a major role (Hildebrandt and Hoyt, 2000). Therefore, we tested whether the increased elongation of the spindle in sse1Δ cells also depends on the kinesin-5 motors’ activity. Interestingly, the spindle length of the double knockout strain, sse1Δcin8Δ, was shorter than that of the WT or sse1Δ strains but similar to that of cin8Δ strain (Fig. 4 A). Hence, the spindles in sse1Δ cells undergo Cin8-dependent premature elongation in S phase.
Figure 4.
Deletion of Sse1 promotes Cin8-dependent spindle elongation in S phase. (A) Logarithmically growing cultures of WT, sse1Δ, cin8Δ, and sse1Δcin8Δ and cells expressing a plasmid-borne copy of GFP-Tub1 and/or containing Spc42-RFP were synchronized in S phase with 100 mM HU, and spindle length was measured by confocal microscopy. The data shown are from a single representative experiment out of three repeats. (B) The dissection of sse1Δcin8Δ and sse1Δkar3Δ diploid strains demonstrating the presence of alleviating genetic interaction between sse1Δ and cin8Δ and aggravating genetic interaction between sse1Δ and kar3Δ is shown. (C) 10× serial dilutions of log-phase WT and sse1Δ cells harboring pGAL-KAR3-HA were spotted onto glucose or galactose medium and incubated at 26°C for 2 d. (D) WT and sse1Δ cells expressing Cin8-3myc from a CEN plasmid were grown in yeast extract peptone + raffinose at 26°C and then arrested using α-factor. Cells were then released from arrest into yeast extract peptone + raffinose + galactose (Gal) medium containing 100 mM HU to induce the expression of Cin8-3myc expression and arrest cells in S phase. After 120 min, cells were transferred to glucose (Glu) medium containing 100 mM HU and 1 µg/ml cycloheximide to turn off the GAL promoter and prevent further translation, and the level of Cin8 was monitored by Western blot analysis using antibodies directed against cMyc. Molecular mass markers are shown on the right of the gels.
The assembled preanaphase bipolar spindle in yeast is a metastable structure. A force generated by plus-end–directed kinesin-5 motors Cin8 and, to a lesser extent, Kip1 pushes the spindle poles apart, and that force is counterbalanced by a force that pulls them inward, generated by the minus-end–directed kinesin-14 motor Kar3 (Saunders and Hoyt, 1992; Hoyt et al., 1993; Saunders et al., 1997b). Hence, the length of the yeast mitotic spindle is a product of the balancing forces of the different types of kinesin motors. To understand the nature of lengthening of the spindle in sse1Δ in S phase, we tested how the deletion of these kinesin motors affects the growth of sse1Δ cells. Combining sse1Δ and cin8Δ deletions resulted in alleviating genetic interactions, whereas combining sse1Δ and kar3Δ exhibited aggravating genetic interactions. (Fig. 4 B). Furthermore, overexpression of Kar3 was more toxic in WT compared with sse1Δ cells (Fig. 4 C). The simplest explanation for these results is that SSE1 deletion results in the up-regulation or modulation of Cin8 activity, and, consequently, more Kar3 is required in sse1Δ cells to counterbalance the over active Cin8.
It was previously shown that Cin8 level is cell cycle regulated and peaks in S phase (Hildebrandt and Hoyt, 2001). To rule out the possibility that the stability of Cin8 is affected upon SSE1 deletion, WT and sse1Δ cells carrying GAL1 promoter–driven CIN8-3myc were arrested in G1 by α-factor and then released into galactose medium containing HU (Krishnan et al., 2004). After 120 min, cells were transferred to glucose medium containing HU and cycloheximide to turn off the GAL promoter and prevent further translation, and the fate of Cin8-3myc was monitored. In both WT and sse1Δ cells, Cin8-3myc levels decreased at similar rates, indicating that the stability of Cin8 is not affected in the sse1Δ strain (Fig. 4 D).
Sse1 is required for proper distribution of Cin8 and Kip1 within the spindle
Functional analysis of kinesin motors revealed that they are spatially regulated during the cell cycle (Tytell and Sorger, 2006; Gardner et al., 2008; Khmelinskii et al., 2009). To determine whether Sse1 affects the localization of Cin8, Kip1, and Kar3, the cellular localization of the GFP-tagged motors was examined in sse1Δ cells. Cin8-GFP and Kip1-GFP fluorescence had a bilobed pattern in large budded, HU-arrested WT cells (Figs. 5 A and S4 A), which is consistent with the predominant localization of Cin8 and Kip1 motors to kinetochore MTs (kMTs; Tytell and Sorger, 2006). Interestingly, we found that under the same conditions, Cin8-GFP was asymmetrically distributed in 50% of sse1Δ cells within the bilobed spindle and was partially mislocalized to the nucleoplasm (Fig. 5 A). Similar results, albeit less drastic, were obtained for Kip1-GFP (Fig. S4 A). In addition, Cin8-GFP fluorescence concentrated along the spindle in 15% of the sse1Δ cells (Fig. 5 A). Consistently, live-cell imaging showed frequent redistribution of Cin8-GFP to the spindle midzone and to the nucleoplasm in S-phase–arrested sse1Δ cells (Fig. 5 B). Elevated midzone levels of Cin8-GFP correlated with increased distribution of spindle length, as shown in Fig. 5 C, and, consequently, the mean SD of the spindle length of six sse1Δ cells measured over 8 min was twice as high as that of WT (0.24 vs. 0.12 µm). In contrast, no significant difference between WT and sse1Δ cells was observed for the localization of Kar3-GFP (colocalizes with the SPB; Fig. S4 B) or the spindle midzone–organizing protein Ase1-GFP (Fig. S4 C). Based on these results, we propose that elongated spindles in sse1Δ cells could be the result of the abnormal redistribution of the spindle motors Cin8 and Kip1 to the midzone area, which, subsequently, results in increased motor sliding between oppositely oriented interpolar MTs (iMTs).
Figure 5.
Localization of GFP-tagged kinesin-5 motors in WT and sse1Δ cells. (A) WT and sse1Δ cells expressing RFP-Tub1 and Cin8-GFP were synchronized in S phase with HU for 2.5 h at 26°C. Representative images (extended focus and 3D render) of Cin8-GFP (green) and RFP-Tub1 (red) obtained using fluorescence confocal microscopy are shown. Arrows point to the presence of Cin8 in the midzone and nucleoplasm. Bar, 5 µm. The bar graph shows the percentage of cells of the indicated genotype that have asymmetric distribution of Cin8 in either asynchronous or HU-arrested cultures. About 200 cells were observed. The data shown are from a single representative experiment out of three repeats. DIC, differential interference contrast. (B) WT and sse1Δ cells expressing Cin8-GFP were incubated with HU for 2.5 h, and time-lapse microscopy was performed. The images show that deletion of SSE1 results in redistribution of Cin8 localization and, hence, in varied spindle length. The arrows show the presence of Cin8 in the midzone and nucleoplasm. Bar, 5 µm. (C) Spindle length is plotted for six HU-arrested WT or sse1Δ cells as a function of time. The mean spindle length measured for each cell over the indicated time period is given on the right. (D) WT and sse1Δ cells expressing Ndc80-GFP (green) and RFP-Tub1 (red) were synchronized in S phase with HU for 2.5 h at 26°C. Images were obtained using fluorescence confocal microscopy. Arrows point to the tighter clustering of Ndc80-GFP near the SPB. Bar, 5 µm.
Cin8 and Kip1 have been shown to cross-link kMT and control kinetochore position (Tytell and Sorger, 2006; Gardner et al., 2008); therefore, changes in motor localization pattern could also affect kinetochore position. Kinetochores are multiprotein structures that assemble on centromeric DNA and that mediate MT–chromosome interaction. We assayed the localization of the structural kinetochore protein Ndc80-GFP and a kinetochore-associating XMAP215 orthologue, Stu2-3GFP, which was shown to promote spindle integrity when DNA replication is stalled (Ma et al., 2007). Interestingly, we observed tighter clustering of Ndc80-GFP near each SPB (Fig. 5 D) and Stu2-3GFP within each half spindle (Fig. S4 D) in the majority of sse1Δ cells, indicating premature shortening of the kMT. Interestingly, it has been reported that Cin8 overexpression produces a similar phenotype (Gardner et al., 2008). Furthermore, in 30% of sse1Δ cells, Ndc80-GFP fluorescence was asymmetrically distributed within the spindle, indicating an unregulated premature chromosome segregation (Fig. 5 D).
Sse1 does not affect kinetochore–MT attachment
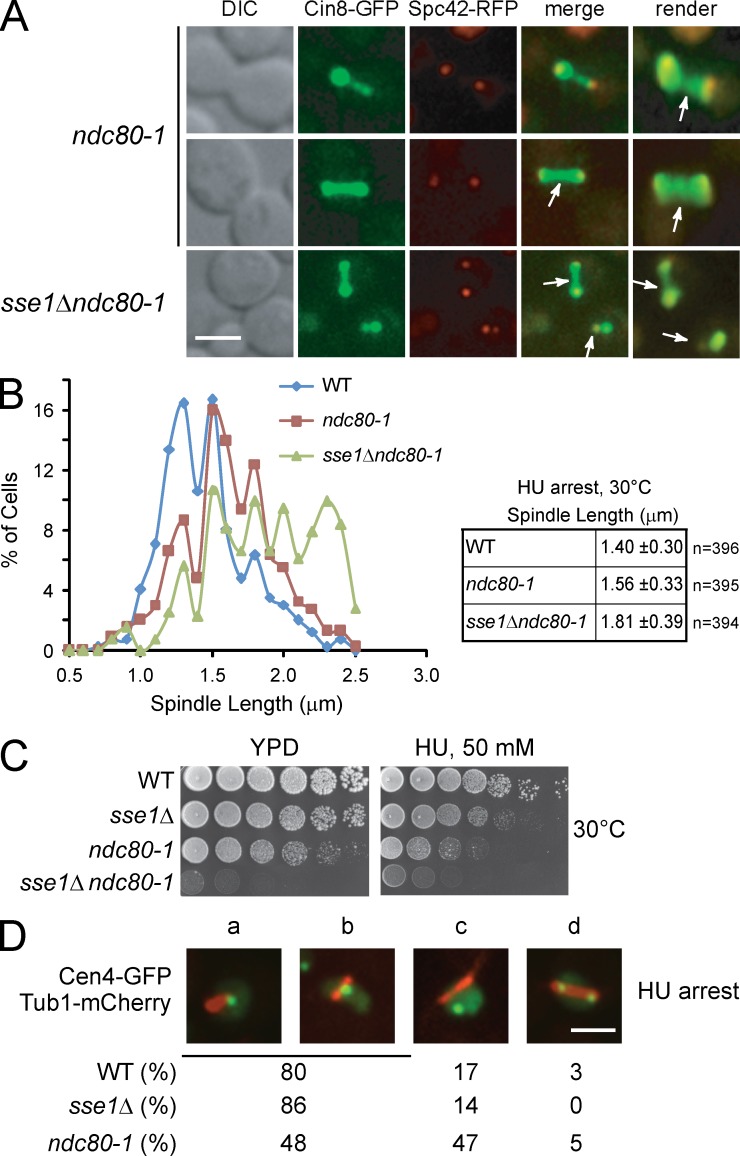
The data so far could be interpreted as either indicating that Sse1 has a direct effect on kinetochore–MT attachment, which is a key factor in spindle length control (Hildebrandt and Hoyt, 2000), or that Sse1 regulates the redistribution of kinesin-5 motors from the kMT to the spindle midzone, which then causes increased motor sliding between oppositely oriented iMTs. The conserved kinetochore-associated Ndc80 complex plays an essential role in forming stable kinetochore–MT attachments (He et al., 2001; Janke et al., 2002). Hence, Cin8-GFP distribution in the temperature-sensitive kinetochore ndc80-1 mutant strain expressing endogenous Spc42-RFP was assessed in S phase. We found that the pattern of Cin8-GFP distribution in ndc80-1 cells at the semipermissive temperature is very similar to that in sse1Δ cells (Fig. 6 A). In both strains, the majority of cells showed Cin8-GFP asymmetrically distributed along the spindle in S-phase–arrested cells (Fig. 6 A). Furthermore, in S-phase–arrested cells, the spindle length in the ndc80-1 mutant was longer than that in WT and similar to that in sse1Δ mutants (Fig. 6 B; also refer to Fig. 4 A). The length of the spindle was further increased in the sse1Δndc80-1 double mutant (Fig. 6 B), suggesting that sse1Δ and ndc80-1 mutations affect parallel pathways in the spindle length control. Consistently, we observed genetic interaction between sse1Δ and ndc80-1 mutants at 30°C (Fig. 6 C). Importantly, the observed genetic interaction did not result in increased HU sensitivity of either single or double mutants (Fig. 6 C), indicating that increased spindle length in these mutants cannot be explained as resulting from transition into mitosis.
Figure 6.
Cin8-GFP localization pattern in kinetochore mutant ndc80-1 mirrors its localization in the sse1Δ strain. (A) Logarithmically growing cultures of ndc80-1 and sse1Δndc80-1 expressing Cin8-GFP and the SPB marker Spc42-RFP were synchronized in S phase with HU for 2.5 h at 30°C. Images were then obtained using fluorescence confocal microscopy. Arrows point to Cin8 accumulation in the midzone area and the nucleoplasm. DIC, differential interference contrast. Bar, 5 µm. (B) Spindle length in HU-arrested ndc80-1 and sse1Δndc80-1 cells grown at 30°C was measured using Spc42-RFP fluorescence by confocal microscopy. The data shown are from a single representative experiment out of three repeats. (C) 10× serial dilutions of log-phase cells of the indicated genotypes were spotted onto YPD and incubated at 30°C for 2 d. (D) The localization of Cen4-GFP and Tub1-mCherry in HU-arrested WT sse1Δ and ndc80-1 cells is shown. A single GFP dot near one end of the spindle might represent cases in which chromosome IV is attached to one SPB, as not all centromeres are replicated in the presence of HU (a). Cen4-GFP was also observed in the middle of the spindle (b), displaced from the spindle, which might represent detached chromosomes (c), and duplicated on both ends of the spindle (d). The percentages were obtained based on observing 100 cells. Bar, 5 µm.
One interpretation of the observations made in Fig. 6 (A–C) is that Sse1 is required for kinetochore–MT interactions in S phase. To investigate this possibility, the localization of GFP-marked centromere of chromosome IV (Cen4-GFP) and mCherry-tagged Tub1 (Khmelinskii et al., 2007; Liu et al., 2008) was determined in WT, sse1Δ, and ndc80-1 cells. We did not observe significant differences in chromosome–MT association between WT and sse1Δ, indicating that Sse1 does not affect centromere–MT interactions (Fig. 6 D). In contrast, in the positive control ndc80-1 mutant, we observed that the number of detached kinetochores increased by ∼2.8 times. Thus, we conclude that kinetochore–MT attachment is not perturbed in sse1Δ cells.
Collectively, these results suggest that the prematurely elongated spindles in S-phase–arrested sse1Δ cells are likely the result of the redistribution of kinesin-5 motors from the kMT to the spindle midzone, which then results in increased motor sliding between oppositely oriented iMTs and leads to spindle lengthening.
The NEF activity of Sse1 on the Hsp70 chaperones Ssa1 and Ssa2 is required for proper spindle assembly in S phase
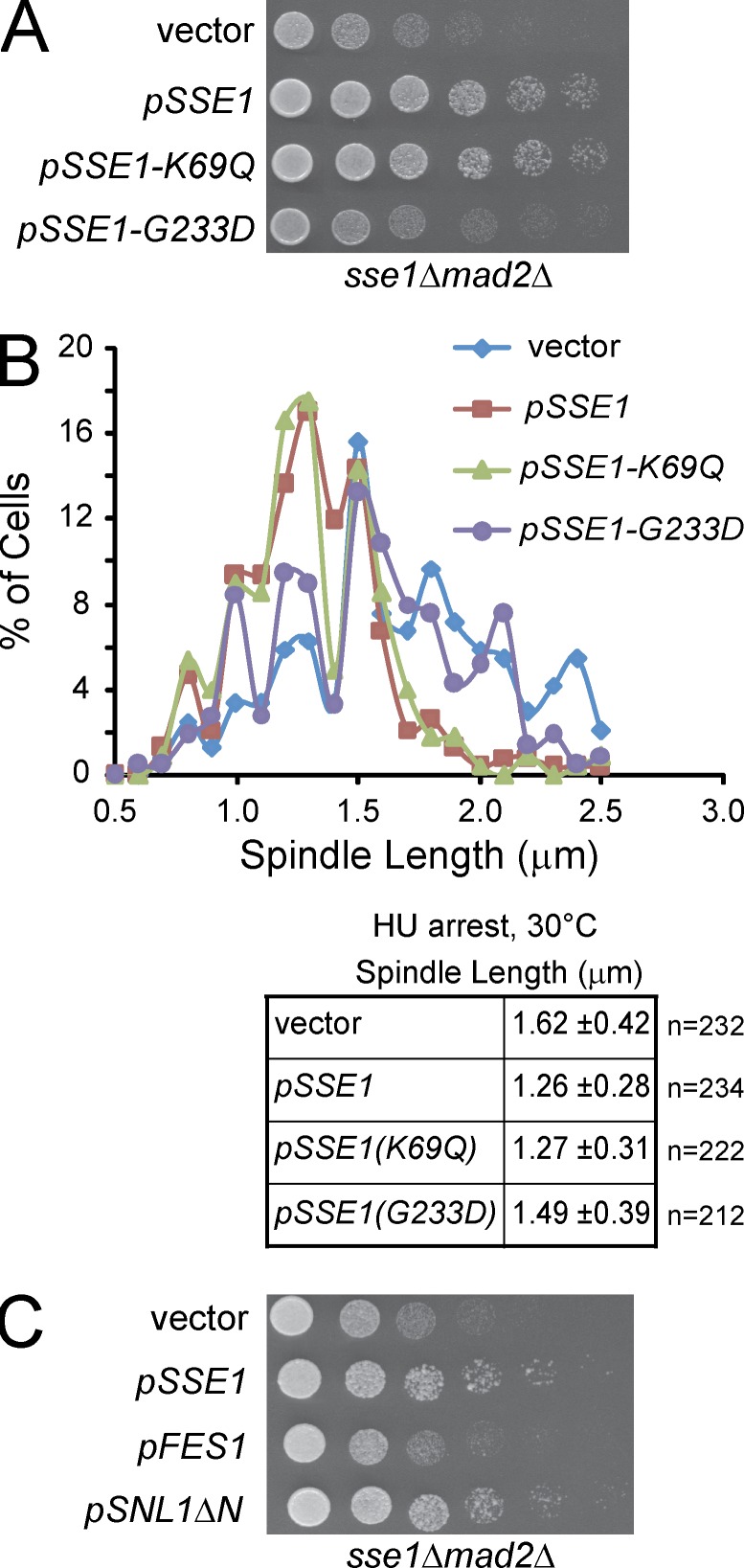
To establish whether Sse1 is working as a chaperone or as an NEF for the Hsp70s (Polier et al., 2008; Schuermann et al., 2008) during spindle assembly, we used SSE1 alleles harboring point mutations that either abolish ATP hydrolysis (pSSE1-K69Q) or ATP binding (pSSE1-G233D; Shaner et al., 2004) and assayed them for their ability to complement sse1Δmad2Δ slow-growth phenotype (Fig. 3 H). It is established that ATP binding but not hydrolysis is required for the interaction of Sse1 with the Hsp70 chaperones and, hence, for its NEF activity on these chaperones (Shaner et al., 2005; Andréasson et al., 2008). The ATPase-deficient mutant Sse1(K69Q) restored growth of the sse1Δmad2Δ strain to WT levels, whereas the ATP-binding–deficient mutant Sse1(G233D) was not able to enhance the growth rate of this strain (Fig. 7 A). Consistently, the expression of WT Sse1 and ATPase-deficient mutant Sse1(K69Q) restored the spindle length in the sse1Δ strain to WT levels for S-phase cells, whereas the ATP-binding–deficient mutant Sse1(G233D) had a spindle length similar to that of vector control (Fig. 7 B). This indicates that Sse1 is not acting as a classical ATP-dependent chaperone during spindle assembly but rather as an NEF for the Hsp70 chaperones.
Figure 7.
Nucleotide exchange activity of Sse1 is required for proper spindle assembly. (A) 10× serial dilutions of log-phase sse1Δmad2Δ cells harboring either empty vector, pSSE1, pSSE1-K69Q, or pSSE1-G233D plasmid were spotted onto YPD and incubated at 30°C for 2 d. (B) Logarithmically growing sse1Δ cells expressing Spc42-RFP as an SPB marker and harboring either empty vector, pSSE1, pSSE1-K69Q, or pSSE1-G233D plasmid were arrested in S phase with 100 mM HU for 2.5 h at 30°C. Spindle length was measured using Spc42-RFP fluorescence. The data shown are from a single representative experiment out of three repeats. (C) 10× serial dilutions of log-phase sse1Δmad2Δ cells harboring either empty vector, pSSE1, pFES1, or pSNL1ΔN plasmid were spotted onto YPD and incubated at 30°C for 2 d.
Three other proteins are known to act as NEFs for the Hsp70s in yeast, namely Fes1, Snl1, and Sil1 (Young, 2010). Fes1 is cytoplasmic (Kabani et al., 2002), and Snl1 has an N-terminal transmembrane segment of ∼20 amino acids that localizes the protein to the nuclear and ER membranes with the remaining bulk of the protein localized to the cytoplasm (Ho et al., 1998), whereas Sil1 is in the lumen of the ER (Tyson and Stirling, 2000). To determine whether other Hsp70 NEFs also affect spindle organization, Fes1 and the soluble catalytic domain of Snl1 (Snl1ΔN) were assayed for their ability to complement the sse1Δmad2Δ slow-growth phenotype. As shown in Fig. 7 C, only the overexpression of Snl1ΔN was able to rescue sse1Δmad2Δ growth phenotype, although it was previously reported that the overexpression of both Fes1 or Snl1ΔN can complement sse1Δ-related phenotypes (Sondermann et al., 2002; Sadlish et al., 2008). As native full-length Snl1 is membrane associated (Ho et al., 1998), the data suggest that only Sse1, through its NEF activity, has a specific role in spindle length control and not the other NEFs.
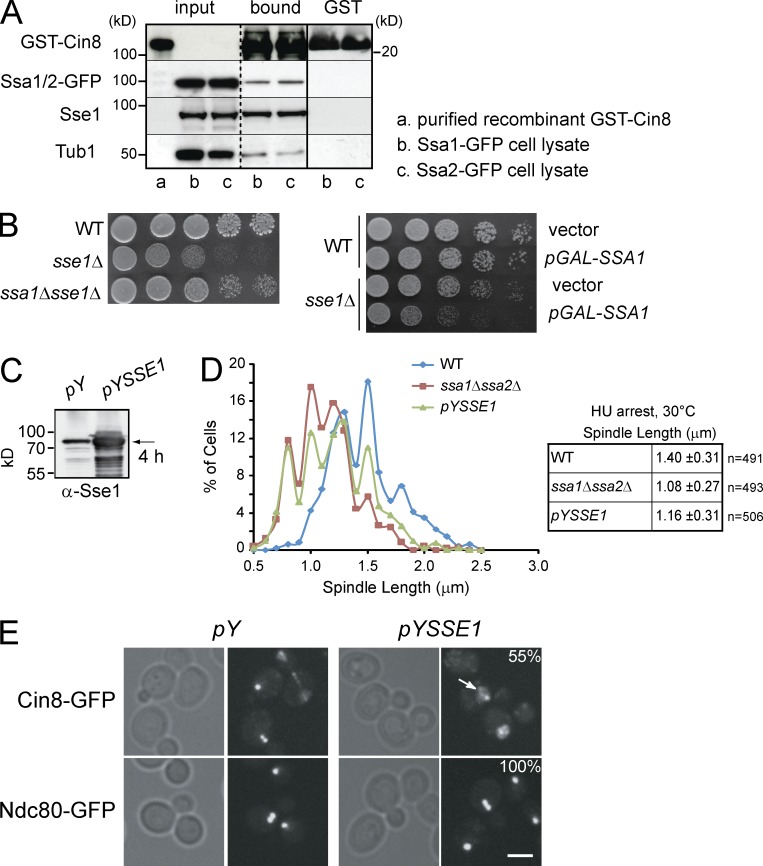
Next, we wanted to determine which Hsp70 chaperone is acting with Sse1 to affect Cin8 activity in S phase. Out of the nine cytoplasmic Hsp70 chaperones, only Ssa1-4 have a significant nuclear localization in addition to their cytoplasmic localization (unpublished data). Furthermore, as shown in Fig. S1 C, the expression levels of the Ssa chaperones vary significantly under nonstress conditions. Typically, Ssa3 and Ssa4 are of low abundance and are only induced during stress, whereas Ssa1 and Ssa2 are highly expressed under normal growth conditions. Hence, Ssa1 and Ssa2 are the most likely Hsp70 chaperones to be involved in spindle assembly in S phase. Experimentally, recombinant GST-Cin8 was incubated with yeast cell extracts obtained from two different strains each producing Ssa1-GFP or Ssa2-GFP expressed from their native loci. GST-Cin8 was found to directly bind Sse1-Ssa1/2 (Fig. 8 A). The ability of GST-Cin8 to bind Tub1 indicates that the recombinant protein is functional. Similar experiments were performed with the other Hsp70 chaperones, but no significant binding to Cin8 was observed. Hence, the effect of Sse1 on Cin8 could be mediated by the effect of Sse1 on Ssa1/Ssa2.
Figure 8.
The effect of Sse1 on spindle assembly is mediated through Ssa1/Ssa2. (A) Recombinant GST-Cin8 or GST alone was incubated with yeast cytosol obtained from strains expressing endogenous Ssa1-GFP or Ssa2-GFP (see Materials and methods). After extensive washing, proteins bound to GST-Cin8 and GST-alone were eluted with SDS sample buffer, separated by SDS-PAGE, and analyzed by Western blot using antibodies directed against GFP, Sse1, Tub1, and GST. Molecular mass markers are shown on both sides of the gels. The broken vertical line indicates that intervening lanes of the gel have been spliced out. The solid vertical line indicates that the samples on the left of the line are from a different gel than those on the right. (B) 10× serial dilutions of log-phase cells of the indicated genotypes were spotted onto YPD and incubated at 30°C for 2 d. The right image shows WT and sse1Δ cells harboring either empty vector or pGAL-SSA1 plasmid spotted onto selective plates containing galactose to induce expression from GAL promoter. Tub1 is used as a control. (C) Western blot analysis using anti-Sse1 antibodies of WT cells harboring empty vector or pYSSE1 expressing Sse1 under the control of inducible CUP1 promoter. Sse1 (indicated by the arrow) was induced with 0.5 mM copper sulfate for 4 h. (D) Spindle length in HU-arrested cells at 30°C measured using GFP-Tub1 fluorescence. The data shown are from a single representative experiment out of three repeats. (E) The localization of Cin8 and Ndc80 in WT cells containing empty vector (pY) or overexpressing Sse1 (pYSSE1). Cells were grown to early log phase, and Sse1 overexpression was induced for 4 h by the addition of copper sulfate to a final concentration of 0.5 mM. Arrow points to the presence of Cin8-GFP in the nucleoplasm. 55% of the cells overexpressing Sse1 (n = 60) showed significant mislocalization of Cin8-GFP, as shown. Bar, 5 µm.
It is reasonable to assume that appropriate levels of Sse1 are needed to ensure that the ATP-dependent functional cycle of the Ssa1/2 chaperones proceeds at an optimal rate. The interplay between Sse1 and Ssa1/2 chaperones is highlighted by the observation that ssa1Δsse1Δ cells grow better than sse1Δ single knockouts and that the overexpression of Ssa1 is more toxic in sse1Δ compared to WT (Fig. 8 B). This is consistent with the conclusion that polypeptide release from the Hsp70 chaperones is reduced in the absence of Sse1, which then affects proper protein refolding activity of these chaperones (Yam et al., 2005).
Overexpression of Sse1 is expected to perturb this functional cycle; indeed, it is already known that high levels of Sse1 inhibit cell growth (Shaner et al., 2004). Overexpression of Sse1 from a copper sulfate–inducible promoter (pYSSE1) resulted in threefold increase in Sse1 levels (Fig. 8 C). In these cells, the length of the spindle in S phase was significantly shorter than that in WT cells (Fig. 8 D). As the ADP-bound state of Ssa1/2 is the high-affinity substrate-binding state and as Sse1 promotes the release of ADP from these chaperones (Yam et al., 2005), high levels of Sse1 might prevent Ssa1/2 from properly interacting with their substrates, such as Cin8. Hence, in the cell, the overexpression of Sse1 might be functionally similar to a deletion of Ssa1/2. Consistent with this argument, although no effect on the spindle length was observed for either SSA1 or SSA2 single deletion (not depicted) highlighting the functional redundancy between these highly homologous chaperones (97% identity; Fig. S1 B), however, the spindle length for the double mutant ssa1Δssa2Δ was significantly shorter than that for WT and similar to that of the Sse1-overexpressing strain (Fig. 8 D). Hence, the effect of overexpressing Sse1 on the spindle length in S-phase cells is similar to that of deleting SSA1 and SSA2 and opposite to what we observed in sse1Δ cells, which have longer spindles (Figs. 3 C and 4 A).
Image analysis of cells expressing endogenous Cin8-GFP or Ndc80-GFP (aforementioned kinetochore marker) demonstrated that the overexpression of Sse1 results in significant mislocalization of Cin8 from spindle MTs (in 55% of cells, n = 60) but not Ndc80 from kinetochores (n = 50; Fig. 8 E). This is consistent with the conclusion that Sse1 does not affect kinetochore–MT attachments but rather affects the distribution of Cin8 within the spindle MT. The displacement of Cin8 from the spindle could explain the shortening of the spindle upon Sse1 overexpression.
In summary, the data presented in this section show that Sse1 NEF activity and the ATP-dependent functional cycle of the Sse1–Ssa1/2 chaperone complex modulate Cin8 distribution within spindle MTs and, consequently, proper spindle assembly.
Sse1–Ssa1/2 chaperone system partially suppresses Cin8 plus-end–directed motility
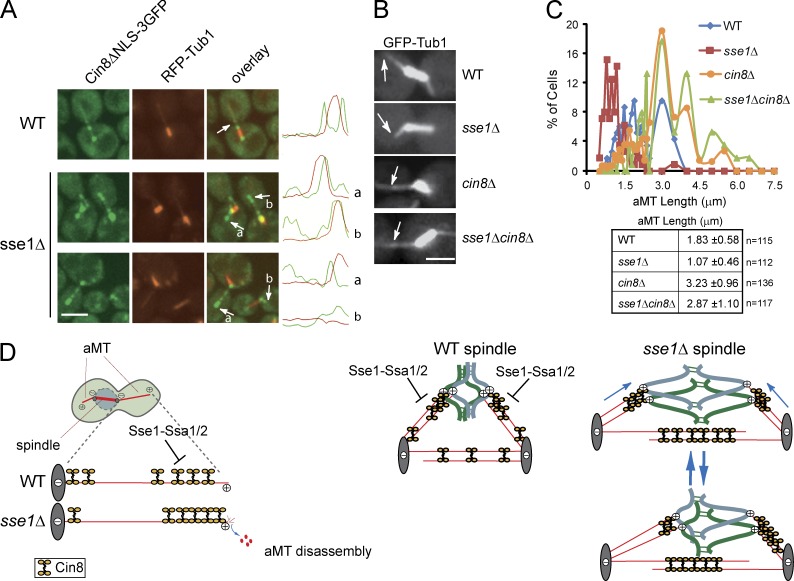
The aforementioned data suggest that Sse1 regulates the activity of Cin8 within the spindle by affecting the distribution of the motor between kMTs and iMTs at the spindle midzone. Hence, our next step was to examine Cin8 movement on individual MTs with the aim of understanding the molecular basis by which Sse1–Ssa1/2 affects Cin8. As individual spindle MTs are tightly packed and cannot be easily visualized by fluorescence microscopy inside the yeast nucleus, we targeted GFP-labeled Cin8 to the astral (cytoplasmic) MTs (aMTs) using Cin8 lacking the NLS, Cin8ΔNLS-3GFP (Hildebrandt and Hoyt, 2001; Roostalu et al., 2011). Individual aMT can be easily visualized in the yeast cytoplasm, which makes individual motor–aMT interactions readily observable.
Consistent with previous observations (Gardner et al., 2008; Roostalu et al., 2011), Cin8ΔNLS-3GFP was found to accumulate near the minus end of aMTs that are organized by the SPB (Fig. 9 A). Surprisingly, in sse1Δ background, we observed predominant clustering of Cin8ΔNLS-3GFP near aMT plus ends (Fig. 9 A).
Figure 9.
The effect of Sse1 on Cin8 oligomerization. (A) Representative images of Cin8ΔNLS-3GFP (green) and RFP-Tub1 (red) expressed in WT and sse1Δ cells. Arrows indicate the localization of Cin8ΔNLS-3GFP to aMT plus ends. The curves on the right show the intensity of the GFP and RFP signals along the spindle. Bar, 5 µm. (B and C) Logarithmically growing cultures of WT, sse1Δ, cin8Δ, and sse1Δcin8Δ cells expressing a plasmid-borne copy of GFP-Tub1 were synchronized in S phase with 100 mM HU for 2.5 h at 26°C. aMT length was measured using GFP-Tub1 and/or Spc42-RFP fluorescence by confocal microscopy. Arrows in B point to aMT in the cytoplasm. Bar, 5 µm. The data shown in C are from a single representative experiment out of three repeats. (D) A model for the regulation of Cin8 motility on MT by Sse1-Ssa1/2. Refer to the text for further details.
To further confirm these observations, we evaluated the physiological consequence of increased accumulation of Cin8 motors at aMTs plus ends in the sse1Δ mutant. Localization of Cin8 to aMT was shown to promote aMT shortening (Gardner et al., 2008). Therefore, we examined the effect of SSE1 deletion by measuring the length of individual aMTs in large budded cells using GFP-Tub1 fluorescence (Gupta et al., 2002; Gardner et al., 2008). Indeed, we found that aMTs were significantly shorter in sse1Δ compared with WT cells (Fig. 9, B and C). Importantly, shortening of aMT, as with lengthening of the spindle, was Cin8 dependent, as we observed increased aMT length in the sse1Δcin8Δ strain to the same extent as observed in cin8Δ (Fig. 9, B and C). Together, this indicates that deletion of SSE1 results in hyperactive Cin8ΔNLS and leads to shortening of aMT. It should be noted that Cin8ΔNLS is, hence, not aggregated in sse1Δ cells but rather hyperactivated, as it leads to shortening of aMTs.
Gardner et al. (2008) proposed that Cin8 distribution on aMTs mirrors its distribution on kMTs. Therefore, it is reasonable to assume that a similar effect of Sse1 on full-length Cin8 occurs at spindle MTs in the nucleus. Hence, the simplest model that explains our results is that Sse1–Ssa1/2 chaperone system interferes with Cin8 plus-end–directed movement. In S phase, for example, Sse1–Ssa1/2-dependent partial inhibition of plus-end–directed motility of Cin8 motor would ensure that the cell has enough time to complete DNA synthesis. On the other hand, SSE1 deletion would perturb normal chaperone function and, hence, would induce redistribution of Cin8 to the spindle midzone, where it would stimulate antiparallel iMT sliding, resulting in premature shortening of kMTs and induction of chromosome congression (Fig. 9 D).
Discussion
Using an unbiased proteomic approach allowed us to identify a yet-uncharacterized role of the Sse1–Ssa1/2 chaperone system in spindle assembly by regulating kinesin-5 motors. The presented experiments indicate that proper distribution of Cin8 on the MTs requires a functional Sse1–Ssa1/2 chaperone complex in which Sse1 serves as an NEF for Ssa1/2. The proper targeting of Cin8 to Ssa1/2 seems to be dependent on Sse1.
Fig. 9 D shows a model of the regulation of Cin8 motility on MTs by Sse1–Ssa1/2 based on our experiments. The left illustration shows the effect of the chaperones on Cin8 motility on aMTs. The chaperone system antagonizes plus-end–directed Cin8 movement. On the other hand, SSE1 deletion results in an increased plus-end–directed motility of Cin8 motors, which leads to accumulation of Cin8 near aMT plus ends and causes aMT shortening. The middle and right illustrations show that Cin8 is recruited to both kMTs and iMTs within the spindle in S phase. The Sse1–Ssa1/2 chaperones antagonize Cin8 plus-end–directed movement by binding to Cin8 (and maybe tubulin). This favors more accumulation of Cin8 near minus ends of kMTs before anaphase. SSE1 deletion compromises the proper function of Ssa1/Ssa2 chaperones, leading to an increased plus-end–directed motility of Cin8. As Cin8 motors move toward the plus end of kMTs, this results in the uncontrolled kMT disassembly. At the same time, this induces Cin8 redistribution to iMTs at the spindle midzone. Increased binding of Cin8 at the midzone stimulates sliding of iMTs and causes premature spindle elongation in S-phase–arrested cells. This would also explain the observed shifts of Cin8-GFP observed in sse1Δ spindles (Fig. 5, A and B).
Increased Cin8 activity in the sse1Δ spindles would also put larger stresses on properly oriented sister kinetochores, especially later in mitosis, potentially leading to chromosome detachment and cell cycle delays/segregation errors (Fig. 3, D, F, and H). The sudden release of high tension after kinetochore detachment could also explain the increased variability in spindle length in sse1Δ spindles (Fig. 5, B and C). The variability in spindle length upon Cin8 overexpression has been previously noted (Saunders et al., 1997a; Gardner et al., 2008). The idea that detachment is occasionally occurring in sse1Δ spindles is supported by the observation that the ndc80-1 mutant at the semipermissive temperature, where occasional detachment is expected, phenocopies sse1Δ spindles (Fig. 6 A). The release of tension in the ndc80-1 mutant would then lead to longer spindles, as observed (Fig. 6 B). Creating a double mutant of sse1Δndc80-1 would create even more attachment defects and even greater tension on the fewer remaining proper attachments, so that spindle will be even longer as observed (Fig. 6 B; also see genetic interactions of Fig. 6 C). Because the detachments are likely occasional, they would not yield significantly different Cen4 marker separation than observed in WT, especially if the marker fails to separate in normal mitosis (Fig. 6 D). Although the ability of Cin8 to switch directionality has recently been demonstrated (Gerson-Gurwitz et al., 2011; Roostalu et al., 2011), however, whether this is physiologically relevant remains to be established.
It would be interesting to consider the physiological role of such regulation of the spindle by chaperones. As heat shock proteins, including Ssa1/2 and Sse1, are induced under stress (Albanèse et al., 2006), it is plausible to suggest that the plus-end–directed motility of Cin8 would be inhibited under such unfavorable growth conditions. This could ultimately result in shortening of the spindle as a result of decreased motor concentration at the midzone (i.e., MT plus ends). Indeed, when the spindle length was measured in HU-treated WT cells at 37°C, we observed spindle shortening compared with normal growth conditions (Fig. S5 A). Furthermore, in agreement with our model, the spindle shortening induced by heat shock was not evident upon deletion of SSE1 (Fig. S5 B). This suggests that the Hsp110–Hsp70 system is being used by the cell to actively adopt favorable spindle length based on environmental cues. This identified cellular role of Hsp110 in spindle length control might explain the recent observation that colorectal cancer cells showing microsatellite instability are sensitized to chemotherapy, as they express a mutant Hsp110 lacking the SBD, which compromises the normal cellular function and antiapoptotic activity of WT Hsp110 in a dominant-negative manner (Dorard et al., 2011).
Materials and methods
Yeast strains and plasmids
Yeast strains used in this study were derived from BY4741 (MATα) or BY4743 (MATa/α; Brachmann et al., 1998) and are listed in Table S1.
Construction of tagged strains
All C-terminally–tagged yeast strains are from the TAP fusion library (Krogan et al., 2006) and are derivatives of a single parental strain: BY4741 MATa, ura3Δ0, leu2Δ0, his3Δ1, met15Δ0. N-terminal tagging was performed as described in Puig et al. (2001). In brief, PCR fragments containing TAP cassette were generated from (N-TAP) plasmid pBS1761. Yeast strain AE146 (W303 background; his3-11, leu2-3, trp1-1, ura3-1, ade2-1, can1-100) was transformed with PCR fragments, and then transformants were selected on Trp− plates with 2% galactose. Correct targeted integration of the TAP cassette was verified by site-specific PCR and Western blotting. To put N-TAP–tagged proteins under the control of their native endogenous promoter, the GAL promoter and TRP selection marker sequences were subsequently removed by recombination. Cells were transformed with a URA3-marked plasmid expressing Cre recombinase, and colonies that grew on Ura− plates with 2% galactose were picked. These colonies were then grown in rich medium and tested for the loss of both the TRP and URA markers. Finally, colonies lacking TRP and URA markers were rechecked for the presence of TAP tag by Western blotting.
Proteomic analysis
TAP-tagged proteins were purified as previously described (Krogan et al., 2002). Cells were collected from 6 liters of cultures grown to late log phase in yeast extract peptone dextrose. Cell pellets (7–10 g) were frozen in liquid nitrogen and lysed by grinding with dry ice in a coffee grinder (model 203-70; Krups). An equal volume of YEB (250 mM KCl, 100 mM Hepes-KOH, pH 7.9, 1 mM EDTA, and 2.5 mM DTT) was added, and, after centrifugation in a rotor (70 Ti; Beckman Coulter) at 4°C for 2 h at 34,000 rpm, the supernatant was dialyzed against IPP buffer (10 mM Tris-HCl, pH 7.9, 0.1% Triton X-100, 0.5 mM DTT, 0.2 mM EDTA, and 20% glycerol containing 100, 125, 150, or 200 mM NaCl). After dialysis, the extract was again centrifuged in a rotor (70 Ti) at 4°C for 30 min at 34,000 rpm, and the supernatant was mixed for 3 h with 200 µl IgG-Sepharose (GE Healthcare) equilibrated with IPP buffer. After binding, the IgG-Sepharose was washed with 1 ml of IPP buffer followed by 400 µl of tobacco etch virus (TEV) protease cleavage buffer (50 mM Tris-HCl, pH 7.9, 1 mM DTT, and 0.1% Triton X-100 containing 200, 150, 125, or 100 mM NaCl). The beads were then incubated overnight at 4°C with 100 U of TEV protease (Life Technologies) in 200 µl of TEV cleavage buffer. The eluate was combined with a 200-µl wash with TEV cleavage buffer. To this sample was added 200 µl of CaM-binding buffer (10 mM Tris-HCl, pH 7.9, 10 mM β-mercaptoethanol, 2 mM CaCl2, and 0.1% Triton X-100 containing 100, 125, 150, or 200 mM NaCl) and 200 µl of CaM beads (GE Healthcare) equilibrated with the same buffer. After binding for 1–2 h at 4°C, the CaM beads were washed with 200 µl of CaM-binding buffer and 200 µl of CaM wash buffer (10 mM Tris-HCl, pH 7.9, 10 mM β-mercaptoethanol, 0.1 mM CaCl2, and 0.1% Triton X-100 containing 100 mM NaCl). The purified protein complexes were eluted from the CaM beads with 5× 100 µl of CaM elution buffer (10 mM Tris-HCl, pH 7.9, 10 mM β-mercaptoethanol, 3 mM EGTA, and 100 mM NH4HCO3), and the eluted proteins were lyophilized.
Purified proteins were separated on 10% SDS-PAGE gels and visualized by silver staining. Visible bands were cut and identified by trypsin digestion and peptide mass fingerprinting using matrix-assisted laser desorption/ionization–time of flight mass spectrometry. In parallel, another aliquot of each purified protein preparation was digested in solution, and the peptides were separated and sequenced by data-dependent liquid chromatography tandem mass spectrometry.
Plasmids and strains
The following plasmids contain inserts synthesized using the Expand High Fidelity PCR system (Roche). pGEX-6P-1-CIN8 was constructed by amplifying CIN8 ORF with BamH1 and Not1 linkers and cloned into corresponding sites of pGEX-6P-1 vector (GE Healthcare). pYSSE1 was constructed by amplifying the SSE1 ORF with linkers containing cut sites for BamH1 and Pst1 and then cloned into corresponding sites in pYEX-BX vector (Takara Bio Inc.). pAG425GAL-SSA1 plasmid was constructed using SSA1 ORF obtained from the Yeast FLEXGene collection (Hu et al., 2007). The ORF was first subcloned into Gateway(R) donor vector pDONR201 and then subcloned into pAG425GAL expression vector following a published protocol (Alberti et al., 2007). pGFP-TUB1 and pRFP-TUB1 were obtained from C. Boone (University of Toronto, Toronto, Canada). Integrating plasmid pPDS1-18myc (Shirayama et al., 1998) and pGAL-CIN8-3myc (Krishnan et al., 2004) were obtained from U. Surana (Institute of Molecular and Cell Biology, Singapore). pRS425-GPD-FES1 and pRS425-GPD-SNL1ΔN were gifts from B. Bukau and A. Mogk (Heidelberg University, Heidelberg, Germany). p415TEF, p415TEFSSE1, p415TEFSSE1-K69Q, and p415TEFSSE1-G233D were gifts from K. Morano (The University of Texas, Austin, TX). pGAL-KAR3-HA was obtained from B. Andrews (University of Toronto, Toronto, Canada; Sopko et al., 2006). The strains TM139, TM195, TM202, and TM204 (Table S1) were obtained from S. Biggins (Fred Hutchinson Cancer Research Center, Seattle, WA), T. Surrey (Cancer Research UK, London, England, UK), T. Huffaker (Cornell University, Ithaca, NY), and Y. Wang (Florida State University, Tallahassee, FL), respectively.
Procedures for cell cycle arrest
Yeast strains were typically grown at 26°C unless otherwise indicated in yeast peptone dextrose (YPD) or in synthetic media supplemented with appropriate nutrients and 2% glucose. To induce cell cycle arrest, cells were typically grown in YPD to early log phase and treated with 5 µg/ml α-factor (Sigma-Aldrich) for 2 h to arrest in G1 phase or 100 mM HU (Sigma-Aldrich) for 2.5 h to arrest in S phase. Cell cycle arrest was confirmed visually by microscopy or by FACS analysis.
Induction of SSE1 and SSA1 overexpression
For the induction of GAL1-controlled expression of SSA1, cells were grown to mid-logarithmic phase in synthetic media–Leu media containing 2% glucose, washed extensively with water, transferred to synthetic media–Leu media containing 2% galactose, and then incubated for 3 h. For the induction of SSE1 in strains containing the pCUP1-SSE1 plasmid, cells were grown to mid-logarithmic phase in selection medium and then induced for 4 h by the addition of copper sulfate to a final concentration of 0.5 mM.
Western blot analysis
Cells derived from 2-ml cultures grown in appropriate media were harvested by centrifugation, washed with water, and sonicated for 20–30 s in SDS-PAGE sample buffer. Samples were then incubated at 75°C for 10 min, and proteins were separated by SDS-PAGE. Proteins were transferred to nitrocellulose membranes, which were then blocked with 5% skim milk and 0.1% Tween 20 in PBS. TAP-tagged Hsp70 chaperones were detected using polyclonal anti-TAP antibodies. Specific polyclonal anti-cMyc and anti-GFP antibodies and mouse monoclonal anti-GST and antitubulin (H-300) antibodies were purchased from Santa Cruz Biotechnology, Inc. Anti-Sse1 rabbit pAbs were obtained from J. Brodsky (University of Pittsburgh, Pittsburgh, PA). Bound primary antibodies were detected using HRP-conjugated appropriate secondary antibodies and the ECL system (GE Healthcare).
Flow cytometry and fluorescence imaging
DNA distribution by flow cytometry was determined as described in Makhnevych et al. (2009). In brief, strains were grown to early log phase, fixed in 70% ethanol, and treated with 10 mg/ml RNaseA in 50 mM Tris-HCl, pH 8.0, for 3 h at 37°C. After resuspension in 50 mM Tris-HCl, pH 7.5, 2 mg/ml proteinase K was added, and cells were further incubated for 60 min at 50°C. DNA was stained with 1 mM SYTOX green (Invitrogen) in 50 mM Tris-HCl, pH 7.5, sonicated at low intensity, and scanned in a guava easyCyte FACS (EMD Millipore) using FlowJo software (Flow Jo, LLC).
Fluorescence imaging was performed using early log–phase cell cultures (OD600 ∼0.35). Cells were pelleted, and a 1.5-µl suspension was spotted onto a glass slide for image analysis. Images were captured using a spinning-disk confocal system (WaveFX; Quorum Technologies Inc.) with an ultra-cooled 512 back-tinned EM charge-coupled device camera or a microscope (E-600FN; Nikon) with an ORCA II camera (Hamamatsu Photonics). Magnification used was 63×. GFP was excited using a 488-nm laser, and its emission was collected using a 505-nm long-pass filter. The z-axis images were converted into a single composite image using the brightest pixel at every position in each of the image planes. This maximum pixel projection technique produced a 2D representation of the GFP fusion proteins within the cell from the 3D dataset. Spindle length was measured using either GFP-Tub1–labeled spindle MTs or the distance between the SPB marker Spc42-RFP. aMT lengths were assessed by measuring the lengths of GFP-Tub1–labeled aMTs, in which both the plus and the minus ends were clearly visible within one focal plane.
Binding assays
Escherichia coli cells transformed with pGST-CIN8 or pGST alone were grown to midlog phase and induced with 0.5 mM IPTG for 4 h. Cells were lysed, and GST fusions were purified on glutathione-Sepharose beads according to the manufacturer’s instructions (GE Healthcare) using 40 mM Hepes-KOH, pH 7.4, 150 mM KCl, 5 mM MgCl2, and 5% vol/vol glycerol as a lysis buffer and 40 mM Hepes-KOH, pH 7.4, 500 mM KCl, 5 mM MgCl2, and 0.1% Tween 20 as a wash buffer supplemented with protease inhibitors (Roche). 5 ml of yeast cultures, each endogenously expressing Ssa1-GFP or Ssa2-GFP (Huh et al., 2003), was grown to saturation. Cells were harvested by centrifugation and resuspended in lysis buffer supplemented with protease inhibitors followed by an addition of glass beads. The samples were then agitated using a vortex mixer in four rounds of 1.5 min each followed by 1.5 min on ice. The lysates were then cleared by centrifugation at 14,000 rpm at 4°C. Ssa chaperone lysates containing 1 mg of protein were incubated with ∼10 µg GST-Cin8 or GST alone preloaded onto glutathione-Sepharose beads for 45 min at 4°C. Beads were collected by centrifugation, and, after extensive washing, bound proteins were eluted with SDS-PAGE sample buffer. Eluted proteins were separated on SDS-PAGE gels and transferred to nitrocellulose for immunodetection.
Online supplemental material
Fig. S1 shows the cytoplasmic yeast Hsp70s/Hsp110s. Fig. S2 shows the Hsp70/Hsp110 chaperone–chaperone interaction network. Fig. S3 shows spindle morphology in different chaperone knockout strains. Fig. S4 shows the effect of SSE1 deletion on Kip1, Kar3, Ase1, and Stu2 localization. Fig. S5 shows the effect of heat shock on spindle length. Video 1 shows time-lapse microscopy of the spindle. Table S1 shows strains used in this study. Table S2 shows interactions identified in this study. Table S3 shows interactions previously reported in published literature. Table S4 shows hits obtained using both N- and C-TAP–tagged chaperones. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201111105/DC1.
Supplementary Material
Acknowledgments
This work was supported by a grant from the Canadian Institutes of Health Research (MOP-81256) to A. Emili and W.A. Houry and Genome Canada through the Ontario Institute of Genomics to J.F. Greenblatt and A. Emili.
Footnotes
Abbreviations used in this paper:
- aMT
- astral MT
- HU
- hydroxyurea
- iMT
- interpolar MT
- kMT
- kinetochore MT
- MT
- microtubule
- NEF
- nucleotide exchange factor
- SBD
- substrate-binding domain
- SPB
- spindle pole body
- TAP
- tandem affinity purification
- TEV
- tobacco etch virus
- WT
- wild type
- YPD
- yeast peptone dextrose
References
- Albanèse V., Yam A.Y., Baughman J., Parnot C., Frydman J. 2006. Systems analyses reveal two chaperone networks with distinct functions in eukaryotic cells. Cell. 124:75–88 10.1016/j.cell.2005.11.039 [DOI] [PubMed] [Google Scholar]
- Alberti S., Gitler A.D., Lindquist S. 2007. A suite of Gateway cloning vectors for high-throughput genetic analysis in Saccharomyces cerevisiae. Yeast. 24:913–919 10.1002/yea.1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen J.B., Zhou Z., Siede W., Friedberg E.C., Elledge S.J. 1994. The SAD1/RAD53 protein kinase controls multiple checkpoints and DNA damage-induced transcription in yeast. Genes Dev. 8:2401–2415 10.1101/gad.8.20.2401 [DOI] [PubMed] [Google Scholar]
- Andréasson C., Fiaux J., Rampelt H., Mayer M.P., Bukau B. 2008. Hsp110 is a nucleotide-activated exchange factor for Hsp70. J. Biol. Chem. 283:8877–8884 10.1074/jbc.M710063200 [DOI] [PubMed] [Google Scholar]
- Brachmann C.B., Davies A., Cost G.J., Caputo E., Li J., Hieter P., Boeke J.D. 1998. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 14:115–132 [DOI] [PubMed] [Google Scholar]
- Costanzo M., Baryshnikova A., Bellay J., Kim Y., Spear E.D., Sevier C.S., Ding H., Koh J.L., Toufighi K., Mostafavi S., et al. 2010. The genetic landscape of a cell. Science. 327:425–431 10.1126/science.1180823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel J.A., Keyes B.E., Ng Y.P., Freeman C.O., Burke D.J. 2006. Diverse functions of spindle assembly checkpoint genes in Saccharomyces cerevisiae. Genetics. 172:53–65 10.1534/genetics.105.046441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorard C., de Thonel A., Collura A., Marisa L., Svrcek M., Lagrange A., Jego G., Wanherdrick K., Joly A.L., Buhard O., et al. 2011. Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat. Med. 17:1283–1289 10.1038/nm.2457 [DOI] [PubMed] [Google Scholar]
- Easton D.P., Kaneko Y., Subjeck J.R. 2000. The hsp110 and Grp1 70 stress proteins: Newly recognized relatives of the Hsp70s. Cell Stress Chaperones. 5:276–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner M.K., Bouck D.C., Paliulis L.V., Meehl J.B., O’Toole E.T., Haase J., Soubry A., Joglekar A.P., Winey M., Salmon E.D., et al. 2008. Chromosome congression by Kinesin-5 motor-mediated disassembly of longer kinetochore microtubules. Cell. 135:894–906 10.1016/j.cell.2008.09.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerson-Gurwitz A., Thiede C., Movshovich N., Fridman V., Podolskaya M., Danieli T., Lakämper S., Klopfenstein D.R., Schmidt C.F., Gheber L. 2011. Directionality of individual kinesin-5 Cin8 motors is modulated by loop 8, ionic strength and microtubule geometry. EMBO J. 30:4942–4954 10.1038/emboj.2011.403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y., Kakihara Y., Krogan N., Greenblatt J., Emili A., Zhang Z., Houry W.A. 2009. An atlas of chaperone-protein interactions in Saccharomyces cerevisiae: Implications to protein folding pathways in the cell. Mol. Syst. Biol. 5:275 10.1038/msb.2009.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta M.L., Jr, Bode C.J., Thrower D.A., Pearson C.G., Suprenant K.A., Bloom K.S., Himes R.H. 2002. beta-Tubulin C354 mutations that severely decrease microtubule dynamics do not prevent nuclear migration in yeast. Mol. Biol. Cell. 13:2919–2932 10.1091/mbc.E02-01-0003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X., Rines D.R., Espelin C.W., Sorger P.K. 2001. Molecular analysis of kinetochore-microtubule attachment in budding yeast. Cell. 106:195–206 10.1016/S0092-8674(01)00438-X [DOI] [PubMed] [Google Scholar]
- Hildebrandt E.R., Hoyt M.A. 2000. Mitotic motors in Saccharomyces cerevisiae. Biochim. Biophys. Acta. 1496:99–116 10.1016/S0167-4889(00)00012-4 [DOI] [PubMed] [Google Scholar]
- Hildebrandt E.R., Hoyt M.A. 2001. Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence. Mol. Biol. Cell. 12:3402–3416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho A.K., Raczniak G.A., Ives E.B., Wente S.R. 1998. The integral membrane protein snl1p is genetically linked to yeast nuclear pore complex function. Mol. Biol. Cell. 9:355–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M.A., He L., Totis L., Saunders W.S. 1993. Loss of function of Saccharomyces cerevisiae kinesin-related CIN8 and KIP1 is suppressed by KAR3 motor domain mutations. Genetics. 135:35–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Rolfs A., Bhullar B., Murthy T.V., Zhu C., Berger M.F., Camargo A.A., Kelley F., McCarron S., Jepson D., et al. 2007. Approaching a complete repository of sequence-verified protein-encoding clones for Saccharomyces cerevisiae. Genome Res. 17:536–543 10.1101/gr.6037607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P., Gautschi M., Walter W., Rospert S., Craig E.A. 2005. The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat. Struct. Mol. Biol. 12:497–504 10.1038/nsmb942 [DOI] [PubMed] [Google Scholar]
- Huh W.K., Falvo J.V., Gerke L.C., Carroll A.S., Howson R.W., Weissman J.S., O’Shea E.K. 2003. Global analysis of protein localization in budding yeast. Nature. 425:686–691 10.1038/nature02026 [DOI] [PubMed] [Google Scholar]
- Janke C., Ortíz J., Tanaka T.U., Lechner J., Schiebel E. 2002. Four new subunits of the Dam1-Duo1 complex reveal novel functions in sister kinetochore biorientation. EMBO J. 21:181–193 10.1093/emboj/21.1.181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabani M., Beckerich J.M., Brodsky J.L. 2002. Nucleotide exchange factor for the yeast Hsp70 molecular chaperone Ssa1p. Mol. Cell. Biol. 22:4677–4689 10.1128/MCB.22.13.4677-4689.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga H.H., Craig E.A. 2010. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat. Rev. Mol. Cell Biol. 11:579–592 10.1038/nrm2941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Lawrence C., Roostalu J., Schiebel E. 2007. Cdc14-regulated midzone assembly controls anaphase B. J. Cell Biol. 177:981–993 10.1083/jcb.200702145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khmelinskii A., Roostalu J., Roque H., Antony C., Schiebel E. 2009. Phosphorylation-dependent protein interactions at the spindle midzone mediate cell cycle regulation of spindle elongation. Dev. Cell. 17:244–256 10.1016/j.devcel.2009.06.011 [DOI] [PubMed] [Google Scholar]
- Krishnan V., Nirantar S., Crasta K., Cheng A.Y., Surana U. 2004. DNA replication checkpoint prevents precocious chromosome segregation by regulating spindle behavior. Mol. Cell. 16:687–700 10.1016/j.molcel.2004.11.001 [DOI] [PubMed] [Google Scholar]
- Krogan N.J., Kim M., Ahn S.H., Zhong G., Kobor M.S., Cagney G., Emili A., Shilatifard A., Buratowski S., Greenblatt J.F. 2002. RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol. Cell. Biol. 22:6979–6992 10.1128/MCB.22.20.6979-6992.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N.J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A.P., et al. 2006. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 440:637–643 10.1038/nature04670 [DOI] [PubMed] [Google Scholar]
- Lechner J., Carbon J. 1991. A 240 kd multisubunit protein complex, CBF3, is a major component of the budding yeast centromere. Cell. 64:717–725 10.1016/0092-8674(91)90501-O [DOI] [PubMed] [Google Scholar]
- Liu H., Liang F., Jin F., Wang Y. 2008. The coordination of centromere replication, spindle formation, and kinetochore-microtubule interaction in budding yeast. PLoS Genet. 4:e1000262 10.1371/journal.pgen.1000262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Ribot J.L., Chaffin W.L. 1996. Members of the Hsp70 family of proteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 178:4724–4726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L., McQueen J., Cuschieri L., Vogel J., Measday V. 2007. Spc24 and Stu2 promote spindle integrity when DNA replication is stalled. Mol. Biol. Cell. 18:2805–2816 10.1091/mbc.E06-09-0882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makhnevych T., Sydorskyy Y., Xin X., Srikumar T., Vizeacoumar F.J., Jeram S.M., Li Z., Bahr S., Andrews B.J., Boone C., Raught B. 2009. Global map of SUMO function revealed by protein-protein interaction and genetic networks. Mol. Cell. 33:124–135 10.1016/j.molcel.2008.12.025 [DOI] [PubMed] [Google Scholar]
- Oka M., Nakai M., Endo T., Lim C.R., Kimata Y., Kohno K. 1998. Loss of Hsp70-Hsp40 chaperone activity causes abnormal nuclear distribution and aberrant microtubule formation in M-phase of Saccharomyces cerevisiae. J. Biol. Chem. 273:29727–29737 10.1074/jbc.273.45.29727 [DOI] [PubMed] [Google Scholar]
- Peters J.M. 2002. The anaphase-promoting complex: Proteolysis in mitosis and beyond. Mol. Cell. 9:931–943 10.1016/S1097-2765(02)00540-3 [DOI] [PubMed] [Google Scholar]
- Pfund C., Lopez-Hoyo N., Ziegelhoffer T., Schilke B.A., Lopez-Buesa P., Walter W.A., Wiedmann M., Craig E.A. 1998. The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J. 17:3981–3989 10.1093/emboj/17.14.3981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polier S., Dragovic Z., Hartl F.U., Bracher A. 2008. Structural basis for the cooperation of Hsp70 and Hsp110 chaperones in protein folding. Cell. 133:1068–1079 10.1016/j.cell.2008.05.022 [DOI] [PubMed] [Google Scholar]
- Pu S., Wong J., Turner B., Cho E., Wodak S.J. 2009. Up-to-date catalogues of yeast protein complexes. Nucleic Acids Res. 37:825–831 10.1093/nar/gkn1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puig O., Caspary F., Rigaut G., Rutz B., Bouveret E., Bragado-Nilsson E., Wilm M., Séraphin B. 2001. The tandem affinity purification (TAP) method: A general procedure of protein complex purification. Methods. 24:218–229 10.1006/meth.2001.1183 [DOI] [PubMed] [Google Scholar]
- Roostalu J., Hentrich C., Bieling P., Telley I.A., Schiebel E., Surrey T. 2011. Directional switching of the kinesin Cin8 through motor coupling. Science. 332:94–99 10.1126/science.1199945 [DOI] [PubMed] [Google Scholar]
- Sadlish H., Rampelt H., Shorter J., Wegrzyn R.D., Andréasson C., Lindquist S., Bukau B. 2008. Hsp110 chaperones regulate prion formation and propagation in S. cerevisiae by two discrete activities. PLoS ONE. 3:e1763 10.1371/journal.pone.0001763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarin S., Ross K.E., Boucher L., Green Y., Tyers M., Cohen-Fix O. 2004. Uncovering novel cell cycle players through the inactivation of securin in budding yeast. Genetics. 168:1763–1771 10.1534/genetics.104.029033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W., Hornack D., Lengyel V., Deng C.C. 1997a. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J. Cell Biol. 137:417–431 10.1083/jcb.137.2.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W., Lengyel V., Hoyt M.A. 1997b. Mitotic spindle function in Saccharomyces cerevisiae requires a balance between different types of kinesin-related motors. Mol. Biol. Cell. 8:1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders W.S., Hoyt M.A. 1992. Kinesin-related proteins required for structural integrity of the mitotic spindle. Cell. 70:451–458 10.1016/0092-8674(92)90169-D [DOI] [PubMed] [Google Scholar]
- Scheufler C., Brinker A., Bourenkov G., Pegoraro S., Moroder L., Bartunik H., Hartl F.U., Moarefi I. 2000. Structure of TPR domain-peptide complexes: Critical elements in the assembly of the Hsp70-Hsp90 multichaperone machine. Cell. 101:199–210 10.1016/S0092-8674(00)80830-2 [DOI] [PubMed] [Google Scholar]
- Schuermann J.P., Jiang J., Cuellar J., Llorca O., Wang L., Gimenez L.E., Jin S., Taylor A.B., Demeler B., Morano K.A., et al. 2008. Structure of the Hsp110:Hsc70 nucleotide exchange machine. Mol. Cell. 31:232–243 10.1016/j.molcel.2008.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaner L., Trott A., Goeckeler J.L., Brodsky J.L., Morano K.A. 2004. The function of the yeast molecular chaperone Sse1 is mechanistically distinct from the closely related hsp70 family. J. Biol. Chem. 279:21992–22001 10.1074/jbc.M313739200 [DOI] [PubMed] [Google Scholar]
- Shaner L., Wegele H., Buchner J., Morano K.A. 2005. The yeast Hsp110 Sse1 functionally interacts with the Hsp70 chaperones Ssa and Ssb. J. Biol. Chem. 280:41262–41269 10.1074/jbc.M503614200 [DOI] [PubMed] [Google Scholar]
- Shirayama M., Zachariae W., Ciosk R., Nasmyth K. 1998. The Polo-like kinase Cdc5p and the WD-repeat protein Cdc20p/fizzy are regulators and substrates of the anaphase promoting complex in Saccharomyces cerevisiae. EMBO J. 17:1336–1349 10.1093/emboj/17.5.1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sondermann H., Ho A.K., Listenberger L.L., Siegers K., Moarefi I., Wente S.R., Hartl F.U., Young J.C. 2002. Prediction of novel Bag-1 homologs based on structure/function analysis identifies Snl1p as an Hsp70 co-chaperone in Saccharomyces cerevisiae. J. Biol. Chem. 277:33220–33227 10.1074/jbc.M204624200 [DOI] [PubMed] [Google Scholar]
- Sopko R., Huang D., Preston N., Chua G., Papp B., Kafadar K., Snyder M., Oliver S.G., Cyert M., Hughes T.R., et al. 2006. Mapping pathways and phenotypes by systematic gene overexpression. Mol. Cell. 21:319–330 10.1016/j.molcel.2005.12.011 [DOI] [PubMed] [Google Scholar]
- The Gene Ontology Consortium 2000. Gene ontology: Tool for the unification of biology. Nat. Genet. 25:25–29 10.1038/75556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyson J.R., Stirling C.J. 2000. LHS1 and SIL1 provide a lumenal function that is essential for protein translocation into the endoplasmic reticulum. EMBO J. 19:6440–6452 10.1093/emboj/19.23.6440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tytell J.D., Sorger P.K. 2006. Analysis of kinesin motor function at budding yeast kinetochores. J. Cell Biol. 172:861–874 10.1083/jcb.200509101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Bosch M., Lowndes N.F. 2004. Remodelling the Rad9 checkpoint complex: preparing Rad53 for action. Cell Cycle. 3:119–122 [PubMed] [Google Scholar]
- Wegele H., Haslbeck M., Reinstein J., Buchner J. 2003. Sti1 is a novel activator of the Ssa proteins. J. Biol. Chem. 278:25970–25976 10.1074/jbc.M301548200 [DOI] [PubMed] [Google Scholar]
- Wigge P.A., Jensen O.N., Holmes S., Souès S., Mann M., Kilmartin J.V. 1998. Analysis of the Saccharomyces spindle pole by matrix-assisted laser desorption/ionization (MALDI) mass spectrometry. J. Cell Biol. 141:967–977 10.1083/jcb.141.4.967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yam A.Y., Albanèse V., Lin H.T., Frydman J. 2005. Hsp110 cooperates with different cytosolic HSP70 systems in a pathway for de novo folding. J. Biol. Chem. 280:41252–41261 10.1074/jbc.M503615200 [DOI] [PubMed] [Google Scholar]
- Young J.C. 2010. Mechanisms of the Hsp70 chaperone system. Biochem. Cell Biol. 88:291–300 10.1139/O09-175 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.