Summary
During oncogenesis, cells acquire multiple genetic alterations that confer essential tumor-specific traits, including immortalization, escape from anti-mitogenic signaling, neovascularization, invasiveness, and metastatic potential. In most instances, these alterations are thought to arise incrementally over years if not decades. However, recent progress in sequencing cancer genomes has begun to challenge this paradigm, as a radically different phenomenon, termed chromothripsis, has been suggested to cause complex intra- and inter-chromosomal rearrangements on short timescales. In this article, we review established pathways crucial for genome integrity and discuss how their dysfunction could precipitate widespread chromosome breakage and rearrangement in the course of malignancy.
Decades of research have established that cancer is, at its core, a genetic disease (Vogelstein and Kinzler, 2004). However, unlike classical Mendelian disorders that are monogenically transmitted via the germline, somatic alterations in multiple unlinked loci (i.e., proto-oncogenes and tumor suppressor genes) are generally required to produce most types of cancer, whether sporadic or familial (Hanahan and Weinberg, 2011). The coincidence of these events vastly exceeds their expected frequency based on the locus-specific rates of single-nucleotide changes (~10−6 to 10−7 per division; (Loeb et al., 2008)) and genomic rearrangements (~10−4 to 10−5 per division; (Lupski, 2007)) in normal cells. Therefore, additional forms of genomic instability fuel the evolution of most cancers. Even with this support, some pre-malignant lesions (e.g., adenomatous polyps, Barrett’s esophagus, or cervical dysplasia) may still take years to acquire the final mutation(s) needed for progression into active disease (Fearon, 2011; Reid et al., 2010; Woodman et al., 2007). Therefore, multi-step carcinogenesis requires genomic instability, either due to environmental genotoxic stressors (e.g., radiation or DNA-damagingcompounds) or diminished fidelity of endogenous DNA replication, repair, and chromosome segregation pathways, to explain the spectrum and extent of mutation in most tumors.
Nucleotide and whole-chromosome instability
A small but important group of cancers are linked to elevated rates of simple sequence changes (i.e., base substitutions, insertions, or deletions affecting one or few contiguous nucleotides; Figure 1, left panel). For example, patients with xeroderma pigmentosum lack a functional nucleotide excision repair (NER) pathway and hence are unable to reverse DNA lesions induced by ultraviolet light, resulting in widespread mutagenesis and cancer development in sunlight-exposed regions of the epidermis (Lehmann et al., 2011). Similarly, inherited or somatic mutations in the mismatch repair (MMR) machinery lead to error-prone replication and recombination of short DNA repeats known as microsatellites, resulting in their dynamic expansion and contraction across the genome, and account for 15% of colon cancers (Umar et al., 2004). However, most solid tumors are associated with a different form of genetic or genomic instability, whereby whole chromosomes are gained or lost through mitotic missegregation (Schvartzman et al., 2010; Thompson et al., 2010) (Figure 1, right panel). This tendency, often described as chromosome instability (CIN), not only results in numerical deviation from the normal 2N karyotype (i.e., aneuploidy), but also facilitates loss of heterozygosity, a key mechanism for tumor suppressor gene inactivation (Baker et al., 2009). A number of mechanisms for CIN have been proposed, including dysregulation of the spindle checkpoint, which restrains mitosis until all sister chromatid pairs have been captured by microtubules from opposite spindle poles (Musacchio and Salmon, 2007), centrosome overduplication and multipolar spindle assembly (Bettencourt-Dias et al., 2011), deficiencies in sister chromatid cohesion (Solomon et al., 2011), and formation of improper kinetochore-microtubule attachments that are not monitored by the spindle checkpoint (i.e., merotelism; (Bakhoum et al., 2009a; Bakhoum et al., 2009b)). These pathways and their relevance to tumorigenesis have been reviewed at length elsewhere (Holland and Cleveland, 2012; Schvartzman et al., 2010; Thompson et al., 2010).
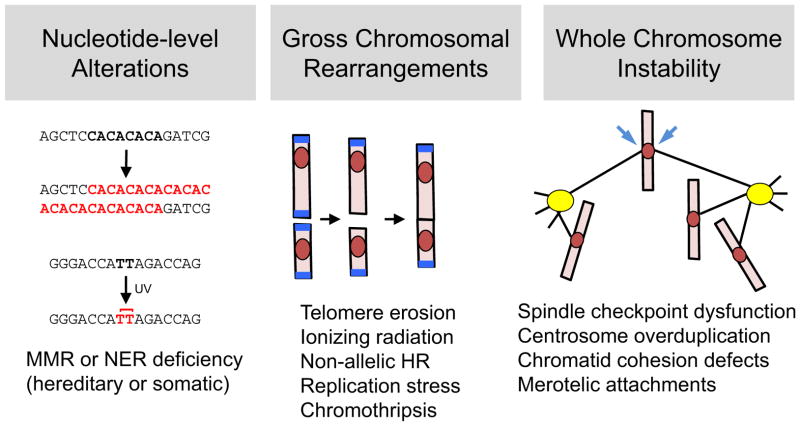
Figure 1. The spectrum of genetic instability in malignancy.
Genomic alterations in tumors can be subdivided into three main groups. At the smallest scale, subtle sequence changes may affect one or a few adjacent nucleotides (left). Examples include deficiencies in mismatch repair (MMR) and nucleotide excision repair (NER) systems, which result in unstable microsatellite repeats (top) and retention of UV-induced photoproducts, such as thymine dimers (bottom). At an intermediate scale (middle), gross chromosomal rearrangements – including deletions, amplifications, inversions, and translocations – are a ubiquitous feature of most cancer genomes. GCRs can arise from multiple mechanisms, including telomere erosion and end-to-end fusion (depicted here), non-allelic homologous recombination, replication stress, and chromothripsis. At the largest scale, whole chromosome instability (right) causes not only aneuploidy but also loss of heterozygosity (LOH), which is crucial for unmasking recessive mutations in tumor suppressor genes. This form of instability can result from errors in virtually any aspect of mitosis, including dysregulation of the spindle assembly checkpoint; biogenesis of supernumerary centrosomes and multipolar spindles; untimely dissolution of sister chromatid cohesion; and formation of merotelic kinetochore-microtubule attachments (arrows).
Inter- and intra-chromosomal rearrangements
In addition to whole-chromosome gains and losses, most cancer genomes contain evidence of gross chromosomal rearrangements (GCRs) (Figure 1, middle panel). This class of genomic alterations has played a major role in the history of cancer genetics, beginning with the discovery in 1960 of a recurrent t(9;22) translocation (the ‘Philadelphia chromosome’) in patients with chronic myelogenous leukemia, resulting in expression of the oncogenic BCR-ABL kinase fusion (reviewed in (Mitelman et al., 2007)). Similarly, homozygous deletions affecting the long arm of chromosome 13 (13q14) were crucial in identifying the RB locus as the causative tumor suppressor in retinoblastoma (Berger et al., 2011), while high-copy amplifications of 17q21–22 were likewise important in delineating HER2/neu as a major oncogene and therapeutic target in breast and ovarian cancer (Baselga and Swain, 2009).
Fundamentally, all GCRs stem from erroneous repair of double-stranded DNA breaks (DSBs). Although often considered in the context of exogenous sources of damage like ionizing radiation (IR) or genotoxic drugs, the vast majority of DSBs are caused by reactive oxygen species generated through normal cellular metabolism, as well as by DNA replication and recombination pathways themselves (Jackson and Bartek, 2009). In specialized compartments (e.g., the adaptive immune systems of jawed vertebrates and the germlines of most sexually reproducing species) programmed DSBs are intentionally created to induce genetic diversity (Bassing and Alt, 2004; Sasaki et al., 2010). Because even a single unrepaired DSB would lead to irreversible loss of genetic material, multiple repair systems of varying fidelity exist. However, this redundancy comes at the price of collateral lesions caused by “off-target” DSB repair.
Modes of DSB repair
Broadly speaking, DSB repair mechanisms can be classified according to their use of non-homologous end joining (NHEJ) versus homologous recombination (HR) in the pairing and ligation of exposed DNA ends (reviewed in Ciccia and Elledge, 2010). To a first approximation this choice is dictated by cell-cycle position, as HR requires a pre-existing sister chromatid and thus becomes available only during S and G2 phase (Heyer et al., 2010). These pathways also differ in their repair signatures. NHEJ ligates DSBs in an error-prone fashion, with the repair junctions displaying little or no homology and frequently marked by small deletions, insertions, or duplications (Weinstock et al., 2006). In contrast, HR repairs DSBs in a potentially error-free manner, via controlled single-strand DNA resection to create free 3′ ends that can recruit the Rad51 recombinase and search for homology within the unbroken sister chromatid (San Filippo et al., 2008). The presence of dispersed high-copy repeats in complex genomes (e.g., Alu elements) means that non-allelic HR is also possible, resulting in mutagenic repair outcomes (e.g., deletions, inversions, and reciprocal translocations; Moynahan and Jasin, 2010). Fortunately, the frequency of such illegitimate recombination events is considerably reduced by the divergent evolution of these elements at the sequence level (Elliott and Jasin, 2001).
Gross chromosomal rearrangements and replication stress
To a significant degree our understanding of GCR-relevant pathways has been shaped by genetic studies in Saccharomyces cerevisiae, a model system in which these rare events can be positively selected and quantified over a wide dynamic range, in both wildtype and mutant genetic backgrounds (Kolodner et al., 2002). Importantly, the rearrangements detected in these assays (interstitial or terminal deletions, reciprocal or non-reciprocal translocations, and telomere addition) parallel those observed in cancer genomes (Chen et al., 1998; Kolodner et al., 2002; Myung et al., 2001). This approach exposed a major role for the intra-S phase checkpoint (a surveillance system that alleviates replication stress by stabilizing stalled forks, inhibiting new origin firing, and delaying cell cycle progression) in GCR suppression (Myung et al., 2001). While the precise contribution of each branch remains to be elucidated, mutations that compromise the intra-S phase checkpoint in mammals are clearly linked to chromosome aberrations and cancer predisposition (Bartek et al., 2004), suggesting that replication-dependent DSBs are a major source of GCRs during malignancy. Consistently, it has been reported that overexpression of commonly activated oncogenes (e.g., H-RasV12) or S-phase regulators normally held in check by Rb and other pocket proteins (e.g., Cdc6 and cyclin E) can induce both replication stress and S phase-specific DSBs in primary cells (Bartkova et al., 2006; Di Micco et al., 2006), possibly through precocious origin firing and accelerated depletion of nucleotide pools (Bester et al., 2011).
Telomere erosion and chromosome breakage-fusion-bridge cycles
GCRs can also arise through non-replicative mechanisms, such as the classic breakage-fusion-bridge (BFB) cycle first proposed by Barbara McClintock (McClintock, 1941). Under this scenario, progressive telomere shortening depletes the shelterin complex from chromosome ends, resulting in their detection as (virtual) DSBs and ligation via NHEJ (Palm and de Lange, 2008). Subsequent attempts to segregate the fused and now-dicentric chromosome give rise to anaphase bridges that can only be resolved through a new round of chromosome breakage, thereby starting the cycle anew (O’Sullivan and Karlseder, 2010). Iteration of this process leads to the non-reciprocal translocations and cancer predisposition seen in telomerase-deficient mice (Artandi et al., 2000; Chin et al., 1999). Interestingly, the frequency and complexity of these alterations seems to be substantially greater than in telomerase-proficient mouse cancer models, similar to the situation in human tumors (O’Hagan et al., 2002) and consistent with the notion that telomere attrition is a significant early event in human (but not murine) carcinogenesis.
Cancer genomics and chromothripsis
Historically, the extent of GCRs within tumors was inferred from cytogenetic methods – such as spectral karyotyping (SKY) and comparative genomic hybridization (CGH)) – that survey chromosome integrity with megabase resolution. When applied to a curated set of sixty cancer-derived cell lines, these techniques identified ~16 structurally aberrant chromosomes per tumor (Roschke et al., 2003). However, recent advances in sequencing and assembling cancer genomes suggested that these estimates might be off by one or more orders of magnitude (Korbel et al., 2007; Stephens et al., 2009). This discrepancy is most clearly evident in the small fraction of solid tumors that exhibit hundreds of GCR breakpoints that are clustered into discrete subchromosomal territories (Stephens et al., 2011) (see Figure 2 for illustrative examples). As discussed below, statistical considerations have led to the proposal that this unprecedented pattern of chromosome rearrangement (now termed chromothripsis, after the Greek word for “shattering into pieces”) may occur as a single catastrophic event. Chromothripsis has been invoked to explain similar clusters of GCRs across a wide variety of tumor types (Kloosterman et al., 2011; Lapuk et al., 2012; Magrangeas et al., 2011; Molenaar et al., 2012; Rausch et al., 2012) (Table 1).
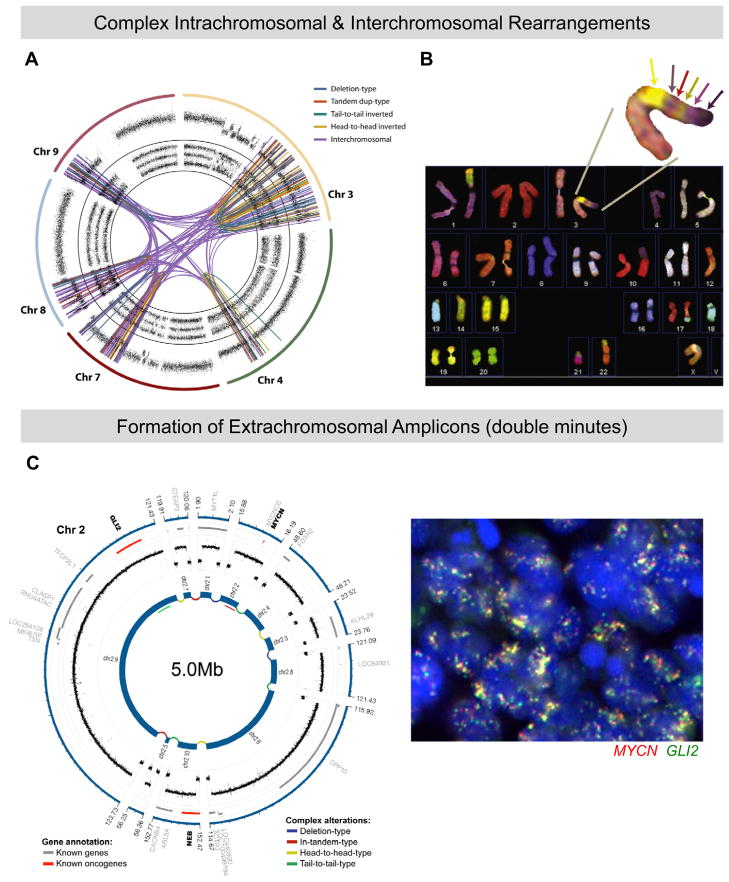
Figure 2. Manifestations of chromothripsis in human tumors.
(A) Sequencing of a chordoma (notochord-derived tumor) reveals 147 linkages between chromosomes 3q, 4q, 7q, 8p, and 9p, as well as various intrachromosomal rearrangements. Copy number profiles and allelic ratios are displayed in outer and inner rings of the circos plot. Adapted from Stephens et al (2011). (B) Spectral karyotyping of pancreatic adenocarcinoma. Enlargement shows a derivative chromosome containing at least six distinct regions from heterologous chromosomes, including 22, 1, 4, 10, and 14. Note that formation of this derivative has resulted in multiple monosomies, indicating widespread allelic and copy-number losses. Adapted from Stephens et al. (2011). (C) Chromothripsis in a Sonic hedgehog (Shh)-type medulloblastoma arising in a patient with Li-Fraumeni syndrome. The circos plot on the left shows a double minute generated through fragmentation of chromosome 2. Two major mediators of Shh signaling, MYCN and GLI2, are contained in this 5-megabase interval. On the right is shown a fluorescence in situ hybridization (FISH) experiment confirming ubiquitous amplification of both MYCN (red) and GLI2 (green) in the primary tumor. Adapted from Rausch et al. (2012).
Table 1.
Chromothripsis in human malignancies
| Tumor type | Chromothripsis incidence | Recurrently affected loci (examples) | Reference |
|---|---|---|---|
| Cancer-derived cell lines (multiple sites) | 2.4% (n = 746) | MYC, CDKN2A, AVEN, IDH2, PML, RYR3, TCF12, MLLT3 | (Stephens et al., 2011) |
| Chronic lymphocytic leukemia | 10% (n = 10) | CDKN2A, miR-15a/16-1 | (Stephens et al., 2011) |
| Osteosarcoma | 33% (n = 9) | (Stephens et al., 2011) | |
| Chordoma | 18% (n = 11) | CDKN2A, FBXW7, WRN, ARID1A | (Stephens et al., 2011) |
| Neuroblastoma | 11% (n = 87) | MYCN, CDK4, C-MYC, FAN1, FANCM, ATRX, ODZ3, PTPRD, CSMD1, TIAM1 | (Molenaar et al., 2012) |
| Medulloblastoma (all types) | 13% (n = 98) | MYCN, GLI2 | (Rausch et al., 2012) |
| Sonic Hedgehog subtype medulloblastoma (SHH-MB) with point-mutated TP53 | 100% (n = 10) | BOC, ADAM2A, NEK11, CDK6, NAMPT, IGF1R, MYCN, GLI2 | (Rausch et al., 2012) |
| SHH-MB with hemizygous TP53 loss | 33% (n = 3) | (Rausch et al., 2012) | |
| SHH-MB with wildtype TP53 | 0% (n = 22) | (Rausch et al., 2012) | |
| WNT-MB | 0% (n = 11) | (Rausch et al., 2012) | |
| Multiple myeloma | 1.3% (n = 764) | (Magrangeas et al., 2011) | |
| Colorectal carcinoma | not indicated | NOTCH2, EXO1, MLL3 | (Kloosterman et al., 2011) |
Information on the incidence, organ/tissue distribution, and target loci affected by chromothripsis was extracted from the indicated studies and summarized herein.
In general, genomic segments impacted by chromothripsis tend to display partial or complete loss of genetic information (i.e., hemizygous or homozygous deletion), rather than a balanced mix of gains and losses. Based on in silico modeling, it has been argued that this pattern is a poor fit with models in which GCRs are induced sequentially over time, rather than in a single massively parallel event (Stephens et al., 2011). On the other hand, because the underlying rates and mechanisms of GCR induction are unknown, serial models in which losses and gains are generated in unequal proportions (or alternatively associated with differences in cellular fitness during clonal expansion) cannot be excluded at this point.
What induces the massive, localized, and perhaps synchronous wave of DSBs during chromothripsis? Various triggers have been suggested, including ionizing radiation, telomere erosion and bridge-breakage-fusion cycles, abortive apoptosis, premature chromosome compaction, and replication stress (Figure 3). Here we discuss these mechanisms and how they may account for the signature patterns of chromosome rearrangements in chromothripsis-positive tumors.
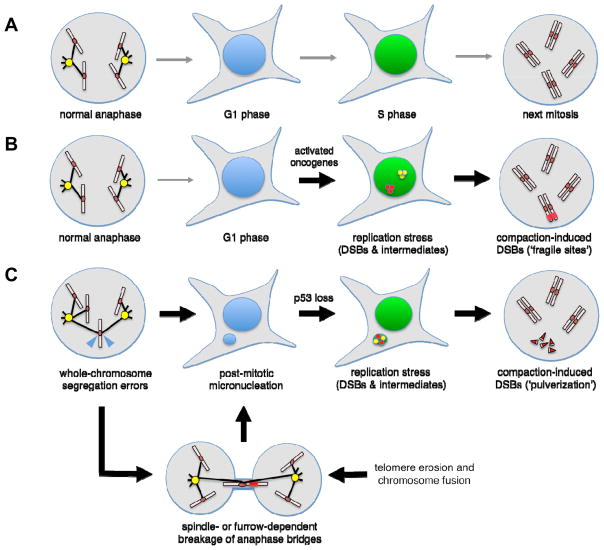
Figure 3. Replication stress and mitotic errors may synergize to induce high levels of genomic instability and precipitate chromothripsis.
(A) Untransformed cells pass through mitosis normally, resulting in correct partitioning of sister genomes in daughter cell nuclei. Once the restriction point is passed, orderly activation of replication origins leads to faithful S-phase progression and genome duplication with minimal loads of incidental DSBs. (B) Under the influence of activated oncogenes, G1/S-phase regulation is corrupted, leading to inappropriate firing of replication origins, depletion of nucleotide precursors, and elevated rates of fork stalling and collapse. Such replication stress may be particularly severe in hard-to-duplicate regions of the genome that harbor low densities of replication origins (i.e., ‘fragile sites’) and thus demand high fork processivity. (C) Replication stress can be induced by antecedent mitotic errors. Whole-chromosome segregation errors frequently result in the formation post-mitotic micronuclei and also trigger a p53-dependent arrest in late G1 phase (Uetake and Sluder, 2010). However, if p53 is mutated or experimentally inactivated, micronucleated cells can proceed into S-phase, resulting in delayed and breakage-prone DNA synthesis within the micronucleus. In addition, persistent replication intermediates within the micronucleus can be further destabilized by transit into mitosis, resulting in haphazard chromatin compaction and ‘pulverization’. A similar fate can also befall lagging anaphase chromatids generated through other mechanisms (for example, telomere erosion and end-to-end fusion, resulting in a dicentric chromosome).
Ionizing radiation and telomere erosion
The initial observation that tens to hundreds of DSBs were clustered into relatively small (megabase-sized) chromosomal regions led Campbell and colleagues to propose a model in which a stray pulse of IR travels through a small subcompartment of the interphase nucleus (Misteli, 2007), or perhaps more plausibly, a condensed mitotic chromosome, thereby generating a spatially restricted zone of DNA damage (Stephens et al., 2011). While difficult to exclude, this model is somewhat constrained by the fact that few natural sources of IR are both strong enough and sufficiently focused to deliver 2 to 4 Gy (the energy required to create 50 to 100 DSBs) into a sub-picoliter target volume while simultaneously avoiding the rest of the genome. (In comparison, the average person receives a cumulative dose of ~2.5 mGy/year from environmental sources of IR.) Further refinement of this model (for example, under conditions of therapeutic or accidental exposure to high-dose IR) is therefore warranted.
A second suggestion relates to the BFB cycles previously discussed that are precipitated by telomere erosion. Specifically, excess stretching of the bridging chromosome by the anaphase spindle and/or physical entrapment by the cytokinetic furrow might not result in a single DSB, but rather a series of closely spaced breaks (Stephens et al., 2011). On the one hand, this idea is consistent with the fact that lagging mitotic chromosomes trapped within the furrow can indeed undergo breakage and rejoining, resulting in non-reciprocal translocations (Janssen et al., 2011), and also supported by examples of complex rearrangements involving telomeric or subtelomeric regions (Stephens et al., 2011). However, most chromothripsis-associated rearrangements neither affect chromosome ends nor implicate a dicentric fusion as an intermediate, and thus are difficult to explain through this particular mechanism.
Abortive apoptosis
Endonucleolytic DNA cleavage is a key aspect of programmed cell death (Taylor et al., 2008). Consequently, it has been proposed that chromothripsis arises from rare instances where apoptosis was transiently initiated and then aborted (Tubio and Estivill, 2011). While an intriguing hypothesis, how cells might escape not only complete DNA fragmentation but also endonuclease-independent cell death pathways remains to be clarified. One possibility is that cleavage is restricted to regions of high chromatin accessibility or influenced by the activity of viruses that inhibit apoptosis (Tubio and Estivill, 2011). Importantly, this hypothesis makes the striking and testable prediction that mice lacking key apoptotic nucleases (e.g., caspase-activated DNAse and endonuclease G (Samejima and Earnshaw, 2005)) will be prevented from inducing chromothripsis-dependent tumors in vivo.
Replication stress and mitotic errors: a synergistic combination?
In our view, replication stress must be considered a prime suspect among potential inducers of chromothripsis (Figure 3). As noted above, commonly activated oncogenes and targets of the Rb-E2F circuit are known to induce premature fork termination, S-phase checkpoint signaling, and DSBs in otherwise naïve human cells, at least in part due to the accelerated consumption of nucleotide precursors. Likewise, the resulting tendency of replication forks to stall or collapse at so-called “fragile sites,” which are especially hard to duplicate fully because of their low origin density (Letessier et al., 2011), may explain the apparent clustering of DSBs within a subchromosomal region (Figure 3B).
In addition to causing immediate fork collapse and breakage during S phase, replication stress can induce genomic instability on a delayed basis. This possibility arises because certain types of replication intermediates (specifically those which do not expose long tracts of single-stranded DNA) activate the DNA damage response inefficiently if at all, thus allowing cells to enter M phase in their presence (Chan et al., 2009; Lukas et al., 2011). Ensuing attempts to compact these unresolved intermediates into mitotic chromosomes can then induce DNA damage de novo (Lukas et al., 2011), and thus may be a source of DSBs during chromothripsis ((Maher and Wilson, 2012) (Figure 3B). Such haphazard chromatin compaction is reminiscent of the now-classic findings of Rao and Johnson, who found that fusing S-phase and mitotic cells caused the nuclei of the former to enter mitosis prematurely, resulting in unscheduled chromatin condensation and fragmentation (Rao and Johnson, 1970). Moreover, recent work indicates that mitotic errors can play a primary role in fomenting replication stress. In particular, intact chromosomes that missegregated during anaphase were found to form extranuclear bodies (micronuclei) that import key replication factors inefficiently, resulting in sluggish and DSB-prone DNA synthesis, as well as aberrant chromatin compaction and fragmentation in the next mitosis (Crasta et al., 2012) (Figure 3C). Whether whole-chromosome aneuploidy stimulates chromothripsis in vivo remains to be addressed, but in principle ought to be evident in mouse models of CIN that generate aneuploidy at high rates (Schvartzman et al., 2010). Collectively, these results suggest ways in which replication stress and mitotic errors can synergize over one or a few cell divisions to establish the high degree of genomic instability needed for chromothripsis.
Chromothripsis and p53
Interestingly, complex chromosome rearrangements resembling chromothripsis have also been found in the tumors of patients affected by Li-Fraumeni syndrome (LFS), a cancer predisposition syndrome caused by germline mutations in the TP53 (p53) tumor suppressor (Li and Fraumeni, 1969; Rausch et al., 2012). The effect of p53 inactivation seems to be especially pronounced in medulloblastomas (tumors of the cerebellum and brain stem) driven by excess Sonic Hedgehog activity: essentially all such tumors undergo chromothripsis if they arise in LFS patients, but rarely if ever do so in patients with wildtype TP53 alleles (Table 1). In principle, p53 inactivation might support chromothripsis by preventing cells with high levels of DSBs from being eliminated via p53-mediated cell cycle arrest, apoptosis, or senesence (Bartek et al., 2004). Alternatively, loss of p53-dependent control over S-phase entry may conspire with latent sources of replication stress (e.g., activated oncogenes or post-mitotic micronuclei) to trigger DSBs directly (Bester et al., 2011; Crasta et al., 2012) (Figure 3C).
Possible mechanisms of chromosome reassembly and resynthesis
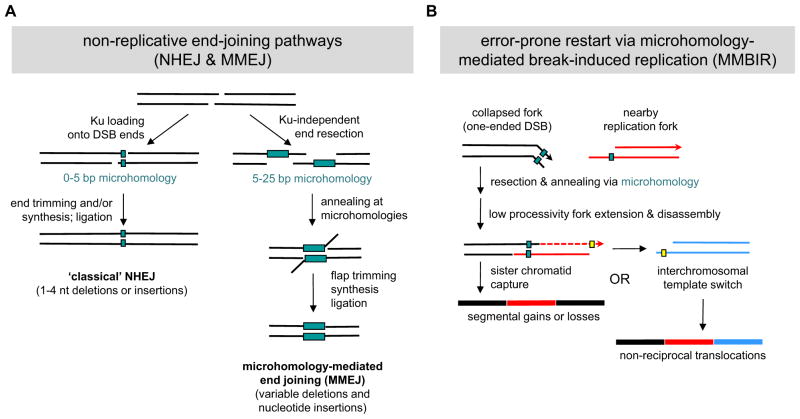
Ultimately, the breaks created in the first stage of chromothripsis must be healed by one of several DNA repair systems, each of which has a characteristic signature. In the case of cancer-associated chromothripsis, most breakpoint junctions exhibit tracts of microhomology, as well as variably sized deletions and insertions of nucleotides (Rausch et al., 2012; Stephens et al., 2011). Together these features suggest the predominant use of non-replicative repair pathways (Figure 4, upper panel), specifically NHEJ and microhomology-mediated end joining (MMEJ). The former uses the Ku70–Ku80 heterodimer and DNA-dependent protein kinase (DNA-PK) to recognize, process, and ligate DSB ends via short (0–5 bp) stretches of microhomology, often with the insertion or removal of a few nucleotides (Figure 5A). In contrast, MMEJ uses Ku- and DNA-PK-independent strand resection to expose somewhat longer (5–25 bp) microhomology tracts. Subsequent annealing and flap trimming produce the final mutagenic repair products, which are skewed towards deletions and translocations (Lieber, 2010) (Figure 5A).
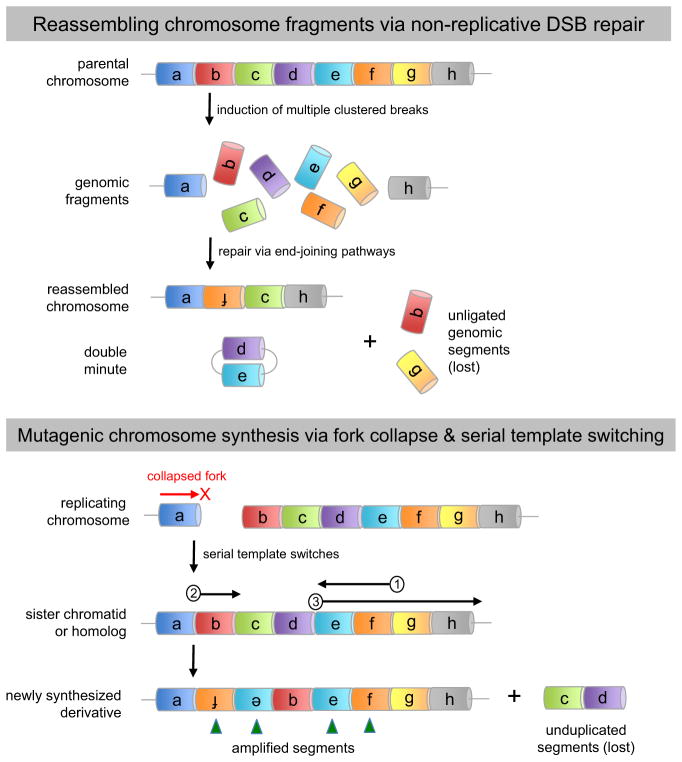
Figure 4. Error-prone modes of DNA repair may underlie chromothripsis.
(A) Non-replicative end joining. Intense but localized induction of DSBs may liberate genomic fragments that are recognized, processed, and ligated by one of several end-joining pathways that are intrinsically error-prone (see figure for mechanistic details). Fragments not ligated to the founding chromosome ‘stub’ either self-ligate to form extrachromosomal arrays (double-minutes) or are lost from the genome entirely. (B) Mutagenic fork restart. Under conditions where HR-dependent pathways are compromised or limiting, a collapsed replication fork may be restarted via microhomology-mediated template switching. This error-prone mode of lesion bypass can generate the full spectrum of structural alterations, including deletions, amplifications, inversions, and non-reciprocal translocations (see figure for mechanistic details).
Figure 5. Mechanisms and signatures of mutagenic chromosome reassembly.
(A) Non-replicative end-joining pathways. Under NHEJ, DSB ends are first recognized and protected from resection by the Ku70–Ku80 heterodimer. Subsequent interactions between DSB ends occur via short (0–5 bp) patches of microhomology. One to four nucleotides are then added or removed to create a ligatable end, resulting in an imprecise junction or “information scar” at the breakpoint. In contrast, MMEJ uses Ku-independent strand resection to expose longer (5–25 bp) microhomology tracts. After annealing of these tracts, the resulting 3′ flaps are trimmed off prior to fill-in synthesis and ligation. Consequently, the final repair products generated by MMEJ typically contain variably sized deletions as well as nucleotide insertions. (B) MMBIR is initiated when a replication fork collapses, resulting in a single-ended DSB (black lines; for clarity the nicked or gapped duplex representing the opposite side of the collapsed fork has been omitted). Thereafter the 5′ end of the broken arm is resected to reveal a free 3′ ssDNA tail, which then uses short-tract microhomology (2–5 nucleotides) to anneal with other exposed ssDNAs, such as the lagging strand of an upstream replication fork (red lines). Elongation of this primer-template junction occurs with low processivity, resulting in eventual fork disassembly, release of the extension products, and further cycles of template switching. While a switch back to the original sister chromatid can re-establish processive DNA replication (resulting in a segmental duplication or higher-order amplification), interchromosomal switching can also occur, resulting in a complex non-reciprocal translocation.
While NHEJ and MMEJ can account for the net loss of genetic information during chromothripsis, they cannot easily explain how other genomic regions become amplified at the same time (Magrangeas et al., 2011; Rausch et al., 2012; Stephens et al., 2011). Instead, recent analyses of germline chromothripsis in patients with congenital neurodevelopmental disorders (Kloosterman et al., 2012; Liu et al., 2011) suggest that replication-based repair pathways can support both copy number gains and losses (Chen et al., 2012; Hastings et al., 2009; Liu et al., 2012) (Figure 4, bottom panel). Indeed, such pathways have recently been implicated in the amplification of subtelomeric regions (Yatsenko et al., 2012). These pathways, variously referred to as microhomology-mediated break-induced replication (MMBIR) or replication fork stalling and template switching (FoSTeS), begin by converting the one-sided DSB associated with a collapsed fork into a free 3′ end (Figure 5B). Through microhomology-dependent annealing between this 3′ end and exposed single strand DNA (ssDNA) regions at nearby replication forks, the resumption of DNA synthesis becomes possible. However, because such forks are only weakly processive, most rounds of DNA synthesis are punctuated by fork disassembly and release of the extension products, thereby licensing further cycles of template switching. Depending on whether the switch involves sequences on the same chromosome, its homolog, or a different chromosome, complex deletions, amplifications, and non-reciprocal translocations can be generated (Figure 5B). In addition, more elaborate template switches involving pairs of replication forks (i.e., replication bubbles) have been proposed to explain certain complex rearrangements in human breast cancer cell lines (Howarth et al., 2011) and may also contribute to chromothripsis.
What drives cells to use these DNA repair pathways, given their low fidelity? One possibility is that other mutations within the tumor reduce the prospects for error-free repair, even under conditions where a sister chromatid is present. In the case of familial breast and ovarian tumors, attenuated HR-dependent repair can be explained by germline alterations in BRCA1 and BRCA2, which regulate DSB metabolism at multiple levels (Roy et al., 2012). Alternatively, high loads of replication-dependent DSBs may overwhelm the HR machinery even if it is present at full strength. For example, recent work suggests that as few as 20 DSBs can saturate one key aspect of the DNA damage response (chromatin ubiquitylation) that is important for shaping repair outcomes (Gudjonsson et al., 2012). Thus, DSBs in excess of this threshold may lack an error-free option for repair. As the breakpoints generated by MMBIR and MMEJ/NHEJ are somewhat overlapping due to their use of microhomology, further work will be required to clarify whether the two pathways are indeed mutually exclusive during germline versus somatic chromothripsis, or instead active to differing degrees in both tissue contexts. In part, this will require functional studies that look at the dynamics of (and requirements for) DSB induction and resolution in chromothripsis-prone animal and cell culture models.
Concluding perspective: The costs and benefits of chromothripsis
Similar to aneuploidy (Siegel and Amon, 2012), chromothripsis deranges the structure and dosage of multiple genes at one stroke, through the induction of extensive rearrangements involving one or several subchromosomal regions (Figure 2). While such random changes may occasionally enhance growth by disrupting haploinsufficient tumor suppressor genes (Berger and Pandolfi, 2011; Solimini et al., 2012; Xue et al., 2012) or activating proto-oncogenes, most would be expected to result in deleterious consequences (e.g., derangement of essential or dosage-sensitive genes) and thus be prone to elimination during clonal outgrowth. Moreover, the DSBs required to produce both beneficial and detrimental rearrangements are themselves liable to inhibit proliferation via activation of the DNA damage response (Jackson and Bartek, 2009). Collectively, these adverse selection pressures may explain why chromothripsis is relatively rare (or at least rarely results in clonal fixation) in most forms of cancer. On the other hand, under circumstances where a sufficiently large population of cells can initiate and survive chromosome breakage, the probability of recovering clones with one or more beneficial ‘driver’ alterations and a minimal load of deleterious ‘passenger’ mutations might be close to 100%. Clearly, further experimental work and evolutionary modeling is needed to illuminate both the costs and benefits of chromothripsis, especially as compared with more incremental and chronic forms of genetic instability (Figure 1). While potentially offering a shortcut to the acquisition of multiple cancer-causing mutations, chromothripsis may also make tumors vulnerable to therapeutic strategies that specifically target regions of genomic loss (Nijhawan et al., 2012) and hence sharply escalate the negative costs associated with ‘passenger’ rearrangements. One attractive venue for exploring this paradigm is afforded by Sonic hedgehog-dependent medulloblastoma, which undergoes chromothripsis uniformly in the absence of p53 (Rausch et al., 2012). If such personalized genomics-based treatments can indeed be realized, then the recent discovery of chromothripsis in human tumors might someday be considered a lucky break.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature. 2000;406:641–645. doi: 10.1038/35020592. [DOI] [PubMed] [Google Scholar]
- Baker DJ, Jin F, Jeganathan KB, van Deursen JM. Whole chromosome instability caused by Bub1 insufficiency drives tumorigenesis through tumor suppressor gene loss of heterozygosity. Cancer Cell. 2009;16:475–486. doi: 10.1016/j.ccr.2009.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009a;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakhoum SF, Thompson SL, Manning AL, Compton DA. Genome stability is ensured by temporal control of kinetochore-microtubule dynamics. Nat Cell Biol. 2009b;11:27–35. doi: 10.1038/ncb1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Lukas C, Lukas J. Checking on DNA damage in S phase. Nature reviews Molecular cell biology. 2004;5:792–804. doi: 10.1038/nrm1493. [DOI] [PubMed] [Google Scholar]
- Bartkova J, Rezaei N, Liontos M, Karakaidos P, Kletsas D, Issaeva N, Vassiliou LV, Kolettas E, Niforou K, Zoumpourlis VC, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444:633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- Baselga J, Swain SM. Novel anticancer targets: revisiting ERBB2 and discovering ERBB3. Nat Rev Cancer. 2009;9:463–475. doi: 10.1038/nrc2656. [DOI] [PubMed] [Google Scholar]
- Bassing CH, Alt FW. The cellular response to general and programmed DNA double strand breaks. DNA Repair (Amst) 2004;3:781–796. doi: 10.1016/j.dnarep.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Berger AH, Knudson AG, Pandolfi PP. A continuum model for tumour suppression. Nature. 2011;476:163–169. doi: 10.1038/nature10275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger AH, Pandolfi PP. Haplo-insufficiency: a driving force in cancer. J Pathol. 2011;223:137–146. doi: 10.1002/path.2800. [DOI] [PubMed] [Google Scholar]
- Bester AC, Roniger M, Oren YS, Im MM, Sarni D, Chaoat M, Bensimon A, Zamir G, Shewach DS, Kerem B. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011;145:435–446. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends in genetics : TIG. 2011;27:307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KL, Palmai-Pallag T, Ying S, Hickson ID. Replication stress induces sister-chromatid bridging at fragile site loci in mitosis. Nat Cell Biol. 2009;11:753–760. doi: 10.1038/ncb1882. [DOI] [PubMed] [Google Scholar]
- Chen C, Umezu K, Kolodner RD. Chromosomal rearrangements occur in S. cerevisiae rfa1 mutator mutants due to mutagenic lesions processed by double-strand-break repair. Mol Cell. 1998;2:9–22. doi: 10.1016/s1097-2765(00)80109-4. [DOI] [PubMed] [Google Scholar]
- Chen JM, Ferec C, Cooper DN. Transient hypermutability, chromothripsis and replication-based mechanisms in the generation of concurrent clustered mutations. Mutat Res. 2012;750:52–59. doi: 10.1016/j.mrrev.2011.10.002. [DOI] [PubMed] [Google Scholar]
- Chin L, Artandi SE, Shen Q, Tam A, Lee SL, Gottlieb GJ, Greider CW, DePinho RA. p53 deficiency rescues the adverse effects of telomere loss and cooperates with telomere dysfunction to accelerate carcinogenesis. Cell. 1999;97:527–538. doi: 10.1016/s0092-8674(00)80762-x. [DOI] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crasta K, Ganem NJ, Dagher R, Lantermann AB, Ivanova EV, Pan Y, Nezi L, Protopopov A, Chowdhury D, Pellman D. DNA breaks and chromosome pulverization from errors in mitosis. Nature. 2012;482:53–58. doi: 10.1038/nature10802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Micco R, Fumagalli M, Cicalese A, Piccinin S, Gasparini P, Luise C, Schurra C, Garre M, Nuciforo PG, Bensimon A, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444:638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- Elliott B, Jasin M. Repair of double-strand breaks by homologous recombination in mismatch repair-defective mammalian cells. Mol Cell Biol. 2001;21:2671–2682. doi: 10.1128/MCB.21.8.2671-2682.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- Gudjonsson T, Altmeyer M, Savic V, Toledo L, Dinant C, Grofte M, Bartkova J, Poulsen M, Oka Y, Bekker-Jensen S, et al. TRIP12 and UBR5 Suppress Spreading of Chromatin Ubiquitylation at Damaged Chromosomes. Cell. 2012;150:697–709. doi: 10.1016/j.cell.2012.06.039. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Hastings PJ, Lupski JR, Rosenberg SM, Ira G. Mechanisms of change in gene copy number. Nat Rev Genet. 2009;10:551–564. doi: 10.1038/nrg2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–139. doi: 10.1146/annurev-genet-051710-150955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland AJ, Cleveland DW. Losing balance: the origin and impact of aneuploidy in cancer. EMBO Rep. 2012;13:501–514. doi: 10.1038/embor.2012.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howarth KD, Pole JC, Beavis JC, Batty EM, Newman S, Bignell GR, Edwards PA. Large duplications at reciprocal translocation breakpoints that might be the counterpart of large deletions and could arise from stalled replication bubbles. Genome research. 2011;21:525–534. doi: 10.1101/gr.114116.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461:1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333:1895–1898. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- Kloosterman WP, Hoogstraat M, Paling O, Tavakoli-Yaraki M, Renkens I, Vermaat JS, van Roosmalen MJ, van Lieshout S, Nijman IJ, Roessingh W, et al. Chromothripsis is a common mechanism driving genomic rearrangements in primary and metastatic colorectal cancer. Genome Biol. 2011;12:R103. doi: 10.1186/gb-2011-12-10-r103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloosterman WP, Tavakoli-Yaraki M, van Roosmalen MJ, van Binsbergen E, Renkens I, Duran K, Ballarati L, Vergult S, Giardino D, Hansson K, et al. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 2012;1:648–655. doi: 10.1016/j.celrep.2012.05.009. [DOI] [PubMed] [Google Scholar]
- Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapuk AV, Wu C, Wyatt AW, McPherson A, McConeghy BJ, Brahmbhatt S, Mo F, Zoubeidi A, Anderson S, Bell RH, et al. From sequence to molecular pathology, and a mechanism driving the neuroendocrine phenotype in prostate cancer. J Pathol. 2012;227:286–297. doi: 10.1002/path.4047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann AR, McGibbon D, Stefanini M. Xeroderma pigmentosum. Orphanet J Rare Dis. 2011;6:70. doi: 10.1186/1750-1172-6-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letessier A, Millot GA, Koundrioukoff S, Lachages AM, Vogt N, Hansen RS, Malfoy B, Brison O, Debatisse M. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature. 2011;470:120–123. doi: 10.1038/nature09745. [DOI] [PubMed] [Google Scholar]
- Li FP, Fraumeni JF., Jr Soft-tissue sarcomas, breast cancer, and other neoplasms. A familial syndrome? Ann Intern Med. 1969;71:747–752. doi: 10.7326/0003-4819-71-4-747. [DOI] [PubMed] [Google Scholar]
- Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 2010;79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Carvalho CM, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev. 2012;22:211–220. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu P, Erez A, Nagamani SC, Dhar SU, Kolodziejska KE, Dharmadhikari AV, Cooper ML, Wiszniewska J, Zhang F, Withers MA, et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell. 2011;146:889–903. doi: 10.1016/j.cell.2011.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer research. 2008;68:3551–3557. doi: 10.1158/0008-5472.CAN-07-5835. discussion 3557. [DOI] [PubMed] [Google Scholar]
- Lukas C, Savic V, Bekker-Jensen S, Doil C, Neumann B, Pedersen RS, Grofte M, Chan KL, Hickson ID, Bartek J, et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol. 2011;13:243–253. doi: 10.1038/ncb2201. [DOI] [PubMed] [Google Scholar]
- Lupski JR. Genomic rearrangements and sporadic disease. Nat Genet. 2007;39:S43–47. doi: 10.1038/ng2084. [DOI] [PubMed] [Google Scholar]
- Magrangeas F, Avet-Loiseau H, Munshi NC, Minvielle S. Chromothripsis identifies a rare and aggressive entity among newly diagnosed multiple myeloma patients. Blood. 2011;118:675–678. doi: 10.1182/blood-2011-03-344069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maher CA, Wilson RK. Chromothripsis and human disease: piecing together the shattering process. Cell. 2012;148:29–32. doi: 10.1016/j.cell.2012.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClintock B. The Stability of Broken Ends of Chromosomes in Zea Mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misteli T. Beyond the sequence: cellular organization of genome function. Cell. 2007;128:787–800. doi: 10.1016/j.cell.2007.01.028. [DOI] [PubMed] [Google Scholar]
- Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat Rev Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- Molenaar JJ, Koster J, Zwijnenburg DA, van Sluis P, Valentijn LJ, van der Ploeg I, Hamdi M, van Nes J, Westerman BA, van Arkel J, et al. Sequencing of neuroblastoma identifies chromothripsis and defects in neuritogenesis genes. Nature. 2012;483:589–593. doi: 10.1038/nature10910. [DOI] [PubMed] [Google Scholar]
- Moynahan ME, Jasin M. Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nature reviews Molecular cell biology. 2010;11:196–207. doi: 10.1038/nrm2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED. The spindle-assembly checkpoint in space and time. Nature reviews Molecular cell biology. 2007;8:379–393. doi: 10.1038/nrm2163. [DOI] [PubMed] [Google Scholar]
- Myung K, Datta A, Kolodner RD. Suppression of spontaneous chromosomal rearrangements by S phase checkpoint functions in Saccharomyces cerevisiae. Cell. 2001;104:397–408. doi: 10.1016/s0092-8674(01)00227-6. [DOI] [PubMed] [Google Scholar]
- Nijhawan D, Zack TI, Ren Y, Strickland MR, Lamothe R, Schumacher SE, Tsherniak A, Besche HC, Rosenbluh J, Shehata S, et al. Cancer vulnerabilities unveiled by genomic loss. Cell. 2012;150:842–854. doi: 10.1016/j.cell.2012.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Hagan RC, Chang S, Maser RS, Mohan R, Artandi SE, Chin L, DePinho RA. Telomere dysfunction provokes regional amplification and deletion in cancer genomes. Cancer Cell. 2002;2:149–155. doi: 10.1016/s1535-6108(02)00094-6. [DOI] [PubMed] [Google Scholar]
- O’Sullivan RJ, Karlseder J. Telomeres: protecting chromosomes against genome instability. Nature reviews Molecular cell biology. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:301–334. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- Rao PN, Johnson RT. Mammalian cell fusion: studies on the regulation of DNA synthesis and mitosis. Nature. 1970;225:159–164. doi: 10.1038/225159a0. [DOI] [PubMed] [Google Scholar]
- Rausch T, Jones DT, Zapatka M, Stutz AM, Zichner T, Weischenfeldt J, Jager N, Remke M, Shih D, Northcott PA, et al. Genome sequencing of pediatric medulloblastoma links catastrophic DNA rearrangements with TP53 mutations. Cell. 2012;148:59–71. doi: 10.1016/j.cell.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid BJ, Li X, Galipeau PC, Vaughan TL. Barrett’s oesophagus and oesophageal adenocarcinoma: time for a new synthesis. Nat Rev Cancer. 2010;10:87–101. doi: 10.1038/nrc2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roschke AV, Tonon G, Gehlhaus KS, McTyre N, Bussey KJ, Lababidi S, Scudiero DA, Weinstein JN, Kirsch IR. Karyotypic complexity of the NCI–60 drug-screening panel. Cancer Res. 2003;63:8634–8647. [PubMed] [Google Scholar]
- Roy R, Chun J, Powell SN. BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer. 2012;12:68–78. doi: 10.1038/nrc3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samejima K, Earnshaw WC. Trashing the genome: the role of nucleases during apoptosis. Nature reviews Molecular cell biology. 2005;6:677–688. doi: 10.1038/nrm1715. [DOI] [PubMed] [Google Scholar]
- San Filippo J, Sung P, Klein H. Mechanism of eukaryotic homologous recombination. Annu Rev Biochem. 2008;77:229–257. doi: 10.1146/annurev.biochem.77.061306.125255. [DOI] [PubMed] [Google Scholar]
- Sasaki M, Lange J, Keeney S. Genome destabilization by homologous recombination in the germ line. Nat Rev Mol Cell Biol. 2010;11:182–195. doi: 10.1038/nrm2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman JM, Sotillo R, Benezra R. Mitotic chromosomal instability and cancer: mouse modelling of the human disease. Nat Rev Cancer. 2010;10:102–115. doi: 10.1038/nrc2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel JJ, Amon A. New Insights into the Troubles of Aneuploidy. Annual review of cell and developmental biology. 2012 doi: 10.1146/annurev-cellbio-101011-155807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solimini NL, Xu Q, Mermel CH, Liang AC, Schlabach MR, Luo J, Burrows AE, Anselmo AN, Bredemeyer AL, Li MZ, et al. Recurrent hemizygous deletions in cancers may optimize proliferative potential. Science. 2012;337:104–109. doi: 10.1126/science.1219580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon DA, Kim T, Diaz-Martinez LA, Fair J, Elkahloun AG, Harris BT, Toretsky JA, Rosenberg SA, Shukla N, Ladanyi M, et al. Mutational inactivation of STAG2 causes aneuploidy in human cancer. Science. 2011;333:1039–1043. doi: 10.1126/science.1203619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu B, Yang F, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Cullen SP, Martin SJ. Apoptosis: controlled demolition at the cellular level. Nat Rev Mol Cell Biol. 2008;9:231–241. doi: 10.1038/nrm2312. [DOI] [PubMed] [Google Scholar]
- Thompson SL, Bakhoum SF, Compton DA. Mechanisms of chromosomal instability. Curr Biol. 2010;20:R285–295. doi: 10.1016/j.cub.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tubio JM, Estivill X. Cancer: When catastrophe strikes a cell. Nature. 2011;470:476–477. doi: 10.1038/470476a. [DOI] [PubMed] [Google Scholar]
- Uetake Y, Sluder G. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr Biol. 2010;20:1666–1671. doi: 10.1016/j.cub.2010.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Ruschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, et al. Revised Bethesda Guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96:261–268. doi: 10.1093/jnci/djh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- Weinstock DM, Richardson CA, Elliott B, Jasin M. Modeling oncogenic translocations: distinct roles for double-strand break repair pathways in translocation formation in mammalian cells. DNA Repair (Amst) 2006;5:1065–1074. doi: 10.1016/j.dnarep.2006.05.028. [DOI] [PubMed] [Google Scholar]
- Woodman CB, Collins SI, Young LS. The natural history of cervical HPV infection: unresolved issues. Nat Rev Cancer. 2007;7:11–22. doi: 10.1038/nrc2050. [DOI] [PubMed] [Google Scholar]
- Xue W, Kitzing T, Roessler S, Zuber J, Krasnitz A, Schultz N, Revill K, Weissmueller S, Rappaport AR, Simon J, et al. A cluster of cooperating tumor-suppressor gene candidates in chromosomal deletions. Proc Natl Acad Sci U S A. 2012;109:8212–8217. doi: 10.1073/pnas.1206062109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsenko SA, Hixson P, Roney EK, Scott DA, Schaaf CP, Ng YT, Palmer R, Fisher RB, Patel A, Cheung SW, et al. Human subtelomeric copy number gains suggest a DNA replication mechanism for formation: beyond breakage-fusion-bridge for telomere stabilization. Human genetics. 2012 doi: 10.1007/s00439-012-1216-9. [DOI] [PMC free article] [PubMed] [Google Scholar]