Abstract
Background
Companion diagnostic tests can depend on accurate measurement of protein expression in tissues. Preanalytic variables, especially cold ischemic time (time from tissue removal to fixation in formalin) can affect the measurement and may cause false-negative results. We examined 23 proteins, including four commonly used breast cancer biomarker proteins, to quantify their sensitivity to cold ischemia in breast cancer tissues.
Methods
A series of 93 breast cancer specimens with known time-to-fixation represented in a tissue microarray and a second series of 25 matched pairs of core needle biopsies and breast cancer resections were used to evaluate changes in antigenicity as a function of cold ischemic time. Estrogen receptor (ER), progesterone receptor (PgR), HER2 or Ki67, and 19 other antigens were tested. Each antigen was measured using the AQUA method of quantitative immunofluorescence on at least one series. All statistical tests were two-sided.
Results
We found no evidence for loss of antigenicity with time-to-fixation for ER, PgR, HER2, or Ki67 in a 4-hour time window. However, with a bootstrapping analysis, we observed a trend toward loss for ER and PgR, a statistically significant loss of antigenicity for phosphorylated tyrosine (P = .0048), and trends toward loss for other proteins. There was evidence of increased antigenicity in acetylated lysine, AKAP13 (P = .009), and HIF1A (P = .046), which are proteins known to be expressed in conditions of hypoxia. The loss of antigenicity for phosphorylated tyrosine and increase in expression of AKAP13, and HIF1A were confirmed in the biopsy/resection series.
Conclusions
Key breast cancer biomarkers show no evidence of loss of antigenicity, although this dataset assesses the relatively short time beyond the 1-hour limit in recent guidelines. Other proteins show changes in antigenicity in both directions. Future studies that extend the time range and normalize for heterogeneity will provide more comprehensive information on preanalytic variation due to cold ischemic time.
An accurate assessment of tissue biomarkers is increasingly important as efforts are made towards individualized molecular targeted therapy of cancer patients. Therefore, it is necessary to determine the exact amount of protein in the tissue specimens not only to better understand the signaling pathways involved in cancer progression but also in order to be able to characterize the biology of a tumor and plan treatment according to the expression of the biomarkers (1). For example, breast cancer therapy is based on the expression level of estrogen receptor (ER), progesterone receptor (PgR), and HER2, as well as the expression of the proliferation marker Ki67 (2–8). However, the accuracy of these assessments, which is dependent on a range of variables related to tissue handling before the expression analysis is conducted (preanalytic variables), has recently been brought into question (10–12).
The field of biospecimen science has recognized the impact of biospecimen handling and preanalytical variables on the expression of biomarkers (9–12) and the need for standardization in biobanking and standard operating procedures (13). Preanalytical procedures affecting tissue quality are not generally standardized and have been historically poorly controlled. Anecdotes about specimens sitting unfixed in a refrigerator during the weekend are not uncommon, and the effects have been shown in analysis of ER status in breast cancer as a function of the day of the week (14). It has been shown that warm and cold ischemic time (ie, time from tissue removal to fixation in warm or cold buffer) during and after surgery affect gene and protein expression patterns in the tissue (15–17). Many other preanalytic variables have also been identified, including the size of the tissue, type of fixative used, time of fixation, temperatures during fixation processes, types of tissue processing and paraffin embedding, variations in antigen retrieval and staining protocols, as well as the use of different antibody clones. Although this is an incomplete list, it illustrates the complexity of the problem of preanalytic variation.
To address the issue of preanalytic variation in the clinical setting, physician’s societies published guidelines and white papers (14,18–20). These papers provide protocols and parameters for analysis to control these variables and minimize their impact on the biospecimen, including definition of maximum time before formalin fixation and an optimal range of time for specimen fixation. Although the authors of the guidelines attempted to have each parameter supported by scientific evidence, quantitative data are sparse or nonexistent for many variables. Loss of immunoreactivity for ER and PgR has been reported after time-to-formalin fixation was delayed for 1–2 hours (21), though the results are controversial and others report no changes for ER, PgR, HER2, and Ki67 antigenicity (10,22,23). There is more uniform evidence for loss of antigenicity of phosphorylated proteins during routine fixation of surgical resection specimens (10,22,24). However, most of these studies have used nonquantitative methods to assess expression as a function of time-to-formalin fixation.
Here we report a systematic, prospectively designed effort to characterize the effects of cold ischemic time. We examined the levels of 23 proteins in breast cancer, which included ER, PgR, HER2, and Ki67, as well as a series of other markers commonly used in research settings. We first validated the reagents for each biomarker and then assessed the antigenicity levels using quantitative immunofluorescence on two complementary series where cold ischemic time was defined.
Patients and Methods
Study Cohorts
We included breast cancer patients from two different cohorts to assess possible changes in protein antigenicity in the tissue specimens according to delay to formalin fixation. The tissue microarray (TMA) series served as a continuous cohort. In case of a suggestion of change in antigenicity of a given protein, further evaluation was performed on the conventional whole slide series of matched pairs of core needle biopsies (CNBs) and surgical resections (although time-to-formalin fixation was not available for this series). All tissue was used after approval from the Rochester Institutional Review Board or from the Yale Human Investigation Committee protocol #8219, which approved the patient consent forms or in some cases waiver of consent.
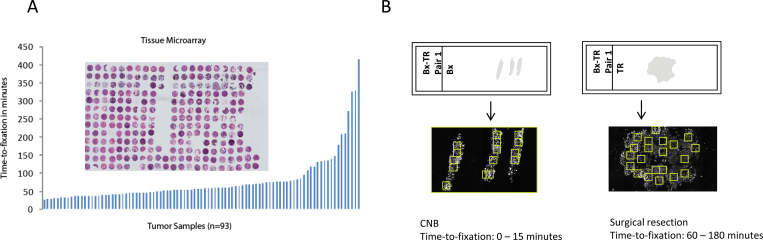
TMA Series. Formalin-fixed, paraffin-embedded (FFPE) tissues of 93 breast cancer patients, who had surgery at the University of Rochester, School of Medicine, (Rochester, NY), were collected. Time from surgical resection to immersion of the specimen in formalin was recorded and ranged from 25 to 415 minutes. A TMA was constructed, consisting of these 93 breast cancer specimens, some cell lines and controls, all represented in twofold redundancy (two histospots per patient specimen) (Figure 1, A). Each marker was analyzed using all data (time up to 415 minutes).
Figure 1.
Series used for evaluation of preanalytical variables on protein expression in patient tissues. Two different series of formalin-fixed, paraffin embedded tissues were used for evaluation of expression of 23 proteins. A) The first series is represented on a tissue microarray, consisting of tissue specimens from 93 breast cancer patients with recorded time from surgical removal of the tissue to immersion of the specimen into formalin, ranging from 25 to 415 minutes. B) The second series consists of matched pairs of biopsy (Bx) and tumor resection (TR) specimen. The time-to-formalin fixation of biopsies ranges from 0 to 15 minutes, whereas the routinely fixed surgical resection specimens usually suffered a processing delay of 60–180 minutes. CNB = core needle biopsy.
Conventional Series. FFPE tissues from approximately 500 breast cancer patients, who had a diagnostic biopsy followed by surgical resection at the Yale University hospital between 1995 and 2005, were obtained from the archives of the Pathology Department of Yale University (New Haven, CT). Only patient tissues with matched specimens consisting of a core needle biopsy specimen and a surgical resection specimen were collected. Out of this collection, 25 matched pairs were analyzed for markers of interest. Time to immersion of the specimen into formalin is generally less than 3 minutes for the CNB, so fixation is limited by the diffusion of formalin into tissues (1mm per hour). Time-to-formalin fixation of the surgical resection specimen was not recorded, but routine specimen handling generally resulted in delayed time-to-formalin fixation of surgical specimens of at least 60 minutes (Figure 1, B).
Cell Lines and Cell Culture
The cell lines T47D, BT474, SKBR3, MB231, MB468, CHO, A431, HT29, A59-195, and A82-68-B were purchased from ATCC (Manassas, VA), cultured in our lab, and used to create control cell line cell blocks for TMA standardization. Culture conditions and construction of cell pellets for TMAs have been described previously (25). No authentication was done on the cell lines because they were used as controls rather than as model systems.
TMA Construction
The TMA of breast cancer specimens with recorded time-to-formalin fixation was constructed as described previously (25). Representative tumor areas from FFPE breast tissue were placed in a recipient block using 0.6mm core size. Breast cancer tissues as well as cell lines were used as internal controls.
Antibodies
The clinical standard biomarkers, ER, PgR, HER2, and Ki67, were each tested with two antibodies, commonly used in the clinical pathology laboratory setting; for ER: monoclonal mouse antihuman ERalpha 1D5 (DAKO, Carpinteria, CA) and monoclonal rabbit antihuman ERalpha SP1 (Thermo Scientific); for PgR: mouse antihuman PgR monoclonal mouse antihuman 636 (DAKO) and mouse antihuman PgR A/B C89F7 (Cell Signaling Technology, Danvers, MA); for HER2: monoclonal mouse antihuman CB11 (Biocare Medical, Concord, CA) and rabbit polyclonal c-erbB2-oncoprotein, also known as Herceptest (DAKO); for Ki67: monoclonal mouse antihuman MIB-1 (DAKO) and monoclonal rabbit antihuman SP6 (Lab Vision). Other antibodies were selected for changes that might occur during time periods before fixation. This included antibodies for proteins sensitive to hypoxia and post-translational modifications or others that are commonly used in a research lab setting. Details of all antibodies selected for study are shown in Table 1.
Table 1.
Biomarkers used in the study*
| Antibody | ||||
|---|---|---|---|---|
| Name (symbol) | Clone/isotype | Clone/isotype | Catalog No. | Vendor |
| Biomarkers for companion diagnostic tests | ||||
| Estrogen receptor-alpha (ERα) | Mouse | 1D5/IgG1kappa | M7047 | DAKO |
| Rabbit | SP1/IgG | RM-9101 | Thermo Scientific | |
| Progesterone receptor (PgR) | Mouse | PgR636/IgG1kappa | M3569 | DAKO |
| Rabbit | PgRA/B (C89F7) | 3153 | Cell Signaling Technology | |
| HER2, also known as ERBB2 | Mouse | CB11/IgG1 | CM 076 AA | Biocare Medical |
| Rabbit | polyclonal | A0485 | DAKO | |
| Ki67 | Mouse | MIB-1/IgG1kappa | M7240 | DAKO |
| Rabbit | SP6/IgG | 9106-S0 | Lab Vision | |
| Markers of cold ischaemia | ||||
| Beta-actin (ACTB) | Rabbit | 13E5/IgG | 13E5/IgG | Cell Signaling Technology |
| Beta-tubulin (TUBB) | Rabbit | pF3/IgG | 2128 | Cell Signaling Technology |
| Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) | Rabbit | 14C10/IgG | 2118 | Cell Signaling Technology |
| Histone 4 (HIST4H4) | Mouse | L64C1 | 2935 | Cell Signaling Technology |
| Histone 3 (HIST3H3) | Mouse | 96C10/IgG1, kappa | 3680 | Cell Signaling Technology |
| Lamin A/C (LMNA) | Rabbit | polyclonal | 2032 | Cell Signaling Technology |
| Lactate dehydrogenase A (LDHA) | Rabbit | IgG, C4B5 | 3582 | Cell Signaling Technology |
| Cytokeratin (KRT X) | Mouse | AE1/AE3/IgG1 | M3515 | DAKO |
| Rabbit | polyclonal | ZO622 | DAKO | |
| Markers of hypoxia | ||||
| Cyclin D1 (CCND1) | Rabbit | IgG/SP4 | RM-9104 | Thermo Fisher Fremont |
| Cyclin B1 (CCNB1) | Mouse | GNS-11/IgG2 | 554178 | BD Biosciences |
| A kinase (PRKA) anchor protein 13 (AKAP13) | Mouse | IgG2a/ZX-18 | sc-81902 | Santa Cruz Biotechnology |
| Cell division cycle 42 (CDC42) | Mouse | IgG3/B-8 | sc-8401 | Santa Cruz Biotechnology |
| Cleaved caspase3 (CASP3) | Rabbit | polyclonal | 9661 | Cell Signaling Technology |
| Hypoxia inducible factor 1-alpha (HIF1A) | Rabbit | polyclonal | NB 100–449 | Novus Biological |
| Hypoxia inducible factor 2-alpha (HIF2A) | Mouse | ep190b/IgG1 | ab8365 | abcam |
| Markers of phosphorylated proteins | ||||
| Phosphorylated tyrosine (4G10) | Mouse | IgG2b | 05-1050 | Millipore |
| Markers of post-translational modification | ||||
| SMT3 suppressor of mif two 3 homolog 1 (SUMO1) | Rabbit | Y299/IgG | ab32058 | abcam |
| Acetylated lysine | Rabbit | polyclonal, purified | 9441 | Cell Signaling Technology |
| Neural precursor cell expressed, developmentally down-regulated 8 (NEDD8) | Rabbit | IgG, 19E3 | 2754 | Cell Signaling Technology |
* Antibodies were validated and used for quantitative immunofluorescence assays as described in the Patients and Methods section.
Conventional Whole Tissue Sections
The TMA and conventional whole tissue section slides were deparaffinized with xylene and rehydrated with ethanol. Antigen retrieval was performed using citrate buffer (pH = 6) at a temperature of 97ºC for 20 minutes. Endogenous peroxidase activity was blocked with 2.5% hydroxyl peroxide in methanol, and slides were preincubated with 0.3% bovine serum albumin in 0.1mol/L of Tris-buffered saline for 30 minutes at room temperature. This was followed by incubation of the slides with the primary antibody and cytokeratin. For standard markers such as ER, PgR, HER2, and Ki67, which are routinely used for companion diagnostic tests, the slides were incubated with primary antibodies for 1 hour at room temperature. Markers commonly used in research settings were incubated for 2 hours at room temperature. Mouse/rabbit EnVision reagent (DAKO, neat) and Alexa 546 conjugated goat antirabbit/mouse secondary antibody (Molecular Probes, Eugene, OR) (1:100 dilution) were used as secondary antibodies followed by Cy5-tyramide (Perker Elmer, Life Science, MA). 4′6-Diamidino-2-phenylindol (DAPI) staining was used to identify cell nuclei. All staining was performed using the Lab Vision Autostainer 720 (Thermo Scientific).
Quantitative Immunofluorescence Using AQUA
The automated quantitative analysis (AQUA) system is a method to objectively and accurately measure protein expression within defined tumor areas and subcellular compartments, as described previously (25). Briefly, after immunofluorescent staining of the TMAs or conventional series, a set of monochromatic, high-resolution images was captured using the PM2000 image workstation (HistoRx, Branford, CT) and analyzed using the AQUA software (AQUA analysis). For each histospot represented on a TMA and each field of view (FOV) on a whole tissue section, three images were collected, one for each wavelength matching the DAPI, Alexa 546, and Cy5 fluorophores. Cell nuclei were visualized by the signal from DAPI stain; cytokeratin was visualized with Alexa 546 fluorophore; and the protein of interest was visualized with the Cy5 dye.
The image collected by visualizing cytokeratin with Alexa 546 dye was manipulated to fill holes, which created a region of interest for subsequent analysis or a “mask.” The intensity of Cy5 fluorescence used to visualize each target is then summed and divided by the area to create an AQUA score for each target (25). CNBs and whole tissue sections were included if we were able to capture at least eight FOVs where the tumor area represented at least 4% of the total area of the tissue specimen.
Validation of Antibodies for Protein Expression Analysis
To show specificity and reproducibility, all antibodies were validated using an antibody validation protocol described previously (26). Antibodies were titered on test arrays and the optimal titer was chosen according to visual assessment or the use of an expression range graph, which allows objective assessment of the optimal dynamic range as well as signal-to-noise ratio of a given protein of interest. Specificity was evaluated by immunoblot analysis for every protein-specific antibody to confirm the recognition of a single band at the correct molecular weight. These assays were followed by immunostaining and AQUA analysis of the cell line TMA. When protein expression levels of a given antibody as measured by AQUA score on the TMA correlated with the immunoblot result, the antibody was considered validated for specificity. Reproducibility of the antibody was assessed by AQUA analysis of serial sections of test arrays stained under the same conditions on different days. Sufficient antibody reproducibility was defined by Pearson’s correlation coefficient (R 2) values of 0.75 and greater. Antibodies for protein modifications were generally more heterogeneous and showed lower R 2 values. Only antibodies that were validated by this protocol were used to further investigate the effect of preanalytical variables on protein expression (Table 1).
Validation of Antibodies for Phosphorylated Proteins
To further validate the specificity of antibodies targeting phosphorylated proteins, small randomly selected breast cancer TMAs (test TMAs) were preincubated with Lambda protein phosphatase (New England Biolabs, Danvers, MA) according to the manufacturer’s instructions for 2 hours at 37ºC, followed by regular immunostaining as described above. Test TMAs without phosphatase treatment were simultaneously incubated with the antibody of interest and served as positive controls. When the phosphatase-treated slides showed a reduction in AQUA score levels at or below the signal-to-noise threshold, then the phosphorylated antibodies were considered validated and used for further studies.
Statistical Analysis
Pearson’s correlation coefficient was used to assess the correlation of AQUA scores for a given marker between corresponding histospots on the TMAs and to assess reproducibility of an assay. On the TMA series, two histospots were measured and averaged with the exemption of ER and PgR where, according to current diagnostic standards, the maximum AQUA score of the two measured histospots was used. The average sample intensities were then log2-transformed and standardized; the time-to-fixation was also log2-transformed.
For the conventional series of matched pairs of CNBs and surgical resections, a minimum of eight FOVs with 4% tumor mask or more was analyzed for every protein, and the scores were averaged. The difference between protein expression of CNBs or surgical specimens was calculated using the paired Student t-test. All P-values were based on two-sided tests, and all values less than .05 were considered statistically significant. Statistical analysis was performed using Statview software and the R package, available on the internet.
Results
Antigenicity of Biomarkers as a Function of Time-to-Formalin Fixation
In order to investigate the possible influence of preanalytical variables on protein assessment, we analyzed 23 proteins on the TMA series with recorded time-to-formalin fixation (Table 1). The markers that are commonly used in routine breast cancer testing are ER, PgR, HER2, and Ki67. Each of these proteins was assessed with two different antibodies, which are routinely used in clinical settings. For each marker, we performed least squares univariate linear regressions and computed the slope and intercept. Because of missing values, none of the markers had 93 pairs of intensity and time-to-fixation measurements. Results for the slopes are shown in Table 2. A slope of 0.25 indicates that for every time doubling the signal intensity increases by 25%. In order to compute meaningful 95% confidence intervals (CIs) for the regression estimates, we analyzed the residuals of each regression independently using the Wilk–Shapiro test. For several regressions, our analyses suggested that the residuals were not normally distributed. Therefore, we used a bootstrapping technique with 5000 replications in order to derive meaningful confidence intervals. For each marker, we constructed a new sample of M pairs of intensity and time-to-fixation measurements by resampling with replacement just from the pairs of measurements of nonmissing histospot measurements, where M is the number of nonmissing specimen histospot measurements of a given marker; we then determined both the slope and the intercept estimates using a least squares linear regression method and stored the predicted values for the entire time-to-fixation domain. For each time-to-fixation in the range between 0.1 and 8 hours, we computed the 95% confidence interval of the estimated log2-transformed standardized intensity. To examine the sample size effect on the 95% confidence intervals, we also estimated the 95% confidence intervals for a sample of 10 times the number of nonmissing pairs of intensity and time-to-fixation (10 times the actual number of nonmissing specimens in our study). We divided the markers into five categories. A marker was labeled as “increase” when the 0.025 quantile of the slopes obtained by bootstrapping for n = M was greater than zero. Similarly, a marker was labeled as “decrease” when the 0.975 quantile of the slopes obtained by bootstrapping for n = M was less than zero. A marker was labeled as “no change” when the 95% confidence intervals for the slope with both n = M and n = 10 × M included the zero slope. A marker was labeled as “trend up” or “trend down” when the 95% confidence interval for the slope with n = M included the zero slope or when the 95% confidence interval for the slope with n = 10 × M did not include it. In this situation, it is possible that a larger sample size might have enabled detection of a monotonic relationship between the measured intensities and the time-to-fixation.
Table 2.
Summary of the proteins tested on the TMA series and the conventional series of biopsies and resections*
| TMA series | Conventional series | |||||
|---|---|---|---|---|---|---|
| Protein tested, name (symbol) | Sample size | Inferred slope | 95% CI, same sample size | 95% CI, 10x sample size | Category of the TMA analysis | Change on biopsy–resection cohort |
| Estrogen receptor alpha (ESR1) | 87 | −0.091 | −0.324 to 0.171 | −0.163 to −0.013 | trend down | none |
| Progesterone receptor (PgR) | 51 | −0.112 | −0.486 to 0.215 | −0.219 to −0.011 | trend down | none |
| HER2 | 71 | −0.018 | −0.283 to 0.244 | −0.097 to 0.061 | no change | none |
| Ki67 (MKI67) | 81 | −0.052 | −0.124 to 0.026 | −0.124 to 0.026 | no change | none |
| Cytokeratin (KRT X) | 82 | 0.122 | −0.148 to 0.409 | 0.036 to 0.212 | trend up | none |
| Beta-actin (ACTB) | 78 | −0.153 | −0.446 to 0.113 | −0.238 to −0.071 | trend down | not evaluated |
| Beta-tubulin (TUBB) | 81 | −0.033 | −0.249 to 0.173 | −0.098 to 0.031 | no change | not evaluated |
| Glyceraldehyde-3-phosphatede hydrogenase (GAPDH) | 85 | 0.009 | −0.16 to 0.204 | −0.046 to 0.066 | no change | not evaluated |
| Lamin A/C (LMNA) | 83 | 0.111 | −0.191 to 0.363 | 0.029 to 0.194 | trend up | not evaluated |
| Lactate dehydrogenase A (LDHA) | 84 | 0.039 | −0.215 to 0.256 | −0.032 to 0.107 | no change | not evaluated |
| Cyclin D1 (CCND1) | 76 | −0.052 | −0.289 to 0.183 | −0.123 to 0.017 | no change | not evaluated |
| Cyclin B1 (CCNB1) | 64 | 0.151 | −0.172 to 0.512 | 0.046 to 0.261 | trend up | not evaluated |
| Histone 3 (HIST3H3) | 78 | 0.093 | −0.104 to 0.317 | 0.029 to 0.158 | trend up | not evaluated |
| Histone 4 (HIST4H4) | 77 | −0.391 | −0.742 to −0.044 | −0.504 to −0.277 | decrease | none |
| SMT3 suppressor of mif two 3 homolog 1 (SUMO1) | 84 | 0.014 | −0.24 to 0.245 | −0.06 to 0.084 | no change | not evaluated |
| Cell division cycle 42 (CDC42) | 93 | 0.026 | −0.113 to 0.192 | −0.021 to 0.075 | no change | not evaluated |
| Cleaved caspase3 (CASP3) | 78 | 0.183 | −0.053 to 0.427 | 0.112 to 0.259 | trend up | not evaluated |
| HIF-2-alpha (EPAS1) | 43 | 0.061 | −0.289 to 0.413 | −0.046 to 0.168 | no change | not evaluated |
| HIF-1-alpha (HIF1A) | 77 | 0.151 | −0.088 to 0.38 | 0.082 to 0.221 | trend up | increase, P = .046 |
| A kinase (PRKA) anchor protein 13 (AKAP13) | 66 | 0.274 | −0.016 to 0.576 | 0.185 to 0.368 | trend up | increase, P = .009 |
| Acetylated lysine | 78 | 0.255 | 0.056 to 0.457 | 0.195 to 0.315 | increase | not evaluated |
| Neural precursor cell expressed, developmentally down-regulated 8 (NEDD8) | 77 | 0.139 | −0.082 to 0.397 | 0.067 to 0.212 | trend up | not evaluated |
| Phosphorylated tyrosine (4G10) | 79 | −0.376 | −0.578 to −0.172 | −0.441 to −0.311 | decrease | decrease, P = .0048 |
* TMA = tissue microarray; CI = confidence interval.
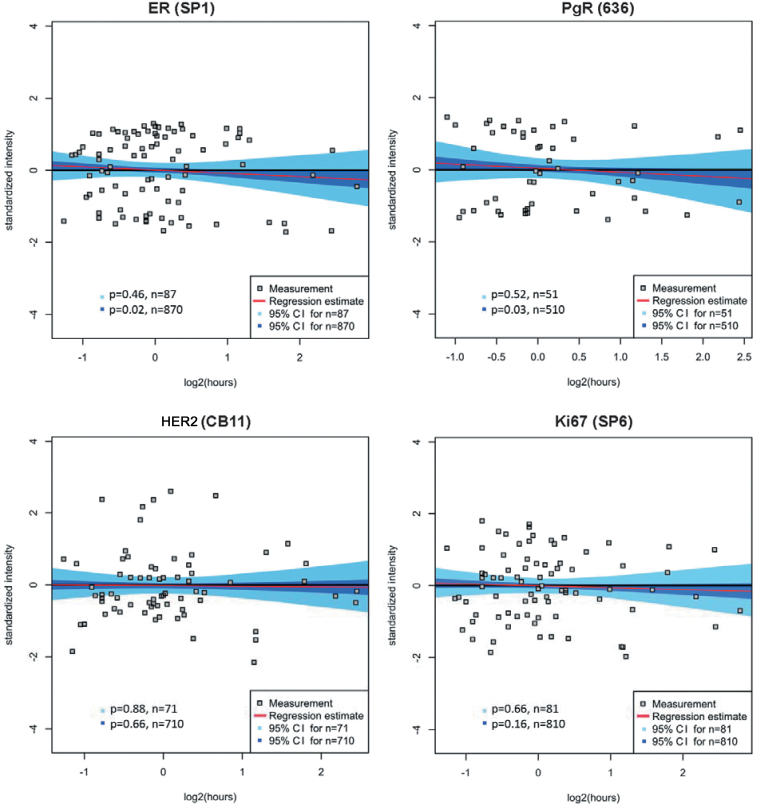
Within the ranges of cold ischemic time tested (25–415 minutes), ER, PgR, HER2, and Ki67 showed a non-statistically significant change in level of antigen (Figure 2 and Table 2); seven other proteins shown in Table 2 did not show any change in antigen level according to delayed formalin fixation. For these proteins, the 95% confidence interval for both n = M and n = 10 × M included the zero slope. For ER and PgR (Figure 2), a trend was seen towards decreased antigen level with further delay in fixation time. However, the trend was not statistically significant without a larger sample size, because the 95% confidence intervals for ER (n = 87) and PgR (n = 51) contained the zero slopes, whereas the 95% confidence intervals for ER (n = 870) and PgR (n = 510) did not. Ki67 was analyzed using continuous AQUA scoring instead of accounting for percentage of Ki67-positive nuclei within a field of interest. Neither Ki67 nor HER2 showed a decrease or even a trend toward a decrease in this time period.
Figure 2.
Regression models for time-to-formalin fixation and category determination for commonly used biomarkers. The commonly used biomarker, ER, PgR, HER2, and Ki67, antibodies were tested on the tissue microarray series and their change of antigenicity as a function of increasing time-to-formalin fixation was assessed using the regression model and category determination. Measured log2-transformed and standardized intensities are shown as a function of log2-transformed time-to-fixation where each grey square represents one tumor sample. The least squares linear regression estimate is shown in red, together with the 95% confidence interval for n = M (light blue area) and for n = 10 × M (blue area) by bootstrapping, where M is the number of nonmissing cases of a given marker. The zero slope line is shown in black.
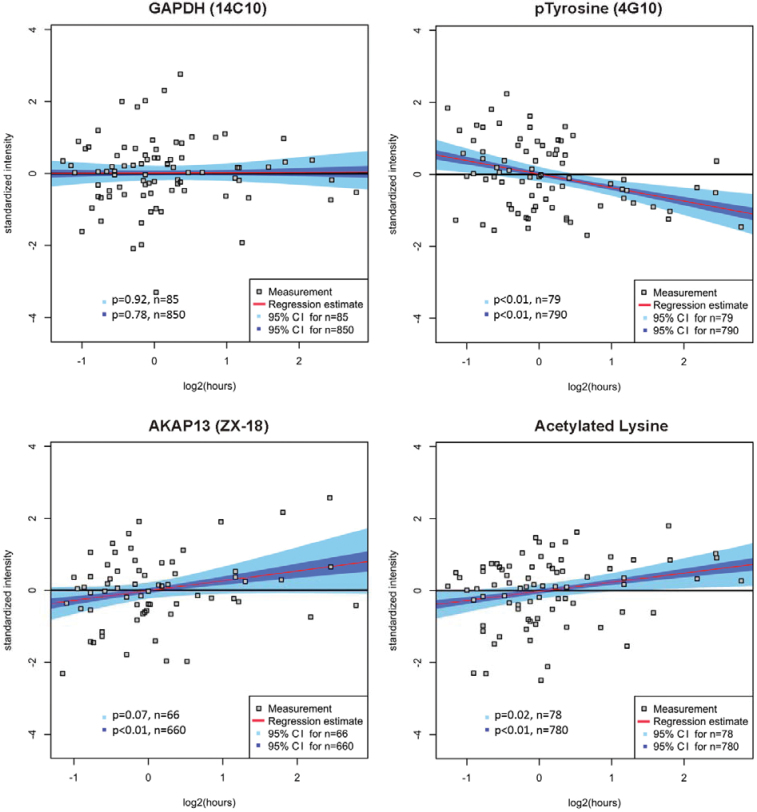
Nineteen other proteins commonly used in research settings were assessed in a similar manner (Table 1). Of these, seven proteins did not show any loss of level of antigen as a function of delayed time-to-formalin fixation (Table 2). Of the other proteins assessed, two (HIST4H4 and phospho-tyrosine [4G10]) showed statistically significant decrease in antigenicity (both P < .01) and one showed an increase (P = .02). The remainder did not show a statistically significant change in antigenicity, but trends were noted including eight proteins showing trend up and one protein showing trend down. The trends were only visualized after bootstrapping and require future validation. Examples of statistically significant changes and trends are shown in Figure 3. These data are illustrated and summarized in Table 2.
Figure 3.
Regression models for time-to-formalin fixation and category determination illustrating examples from Table 2. Four examples illustrating no change, increase, and decrease with time-to-formalin fixation are shown. GAPDH is an example for a protein with stable antigenicity (the 95% confidence intervals for n = 85 and n = 850 including the zero slope). In contrast, phosphorylated tyrosine is an example of decreased epitope antigenicity, indicated by the fact that every slope in the 95% confidence intervals for n = 79 is smaller than zero, and examples for increase of protein expression with every slope in the 95% confidence intervals for n = 79 being greater than zero. Finally, acetylated lysine is shown as an example of statistically significantly increased antigenicity, whereas the analysis of AKAP13 reveals a trend toward increase but is only seen by bootstrapping analysis. The results of all markers tested are summarized in Table 2.
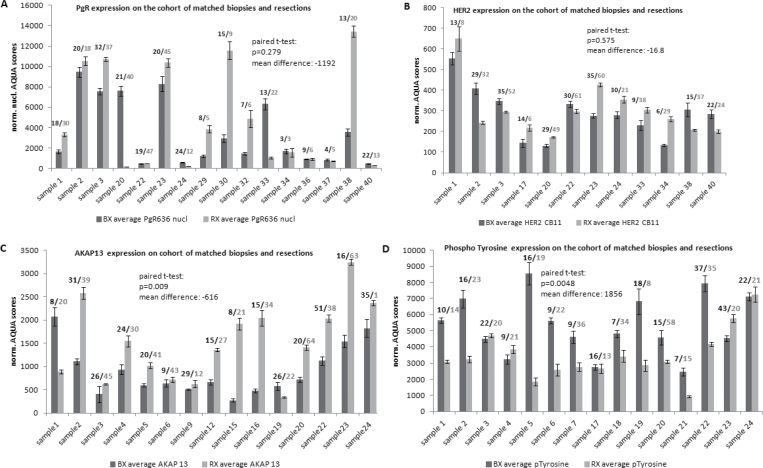
The standard clinical biomarkers, ER, PgR, HER2, and Ki67, were also assessed on the conventional series of matched pairs of biopsies and resections. Though the exact time-to-fixation has not been recorded for these surgical specimens, we estimated that this time ranged between 60 and 180 minutes in our routine clinical setting. This cohort allowed us to analyze several fields of view for both the biopsy and the surgical resection specimens in a manner that mimics the evaluation of actual patient specimens (as compared to the TMAs discussed above). No statistically significant loss of protein antigenicity was seen for ER, PgR, HER2, and Ki67. Examples are shown for PgR and HER2 (Figure 4, A and B).
Figure 4.
Examples of change of protein expression according to time-to-formalin fixation assessed on the conventional series of matched pairs of biopsies and resections. The numbers above the error bars describe the field of views captured for each slide; the means are shown; and the error bars represent the standard deviation. The change from delayed fixation time was assessed with the paired t-test. Panels A and B show examples of standard-of-care proteins. A) PgR. B) HER2. C) Expression levels of AKAP13, a marker of hypoxia. D) Antigenicity levels of phosphorylated tyrosine as detected by the 4G10 antibody, where increased time-to-fixation was associated with loss of antigenicity.
Expression of Proteins Associated With Hypoxia as a Function of Time-to-Formalin Fixation
Expression of hypoxia markers may increase with increasing time-to-formalin fixation, because the tissue still undergoes biological processes even after ligation of vessels or removal from the body. We assessed the expression of known hypoxia markers in the TMA series and found increased expression of HIF1A, AKAP13, CCNB1, cleaved CASP3, NEDD8, KRT-X, LMNA, HIST3H3, and acetylated lysine with increased time-to-formalin fixation when analyzed with bootstraping of data. However, only the increase in expression of acetylated lysine was statistically significant (P = .02). To determine the predictive value of this analysis, a subset of proteins that trended upward (AKAP13 and HIF1A) were selected for validation on the conventional series. This analysis confirmed the initial result but now showing a statistically significant increase of protein expression (P = .009 and .046, respectively) within the resection specimens where time-to-fixation was longer (Figure 4 and Table 2).
Expression of Phosphorylated Proteins as a Function of Time-to-Formalin Fixation
We and others have previously reported loss of antigenicity of phosphorylated epitopes with increased time-to-formalin fixation (10,30). We measured the level of phospho-tyrosine using the monoclonal anti-phospho-tyrosine antibody (4G10) to confirm and generalize findings from previous studies. Assessment of both the TMA series and the series of biopsies and resections showed a statistically significant decrease in epitope with increased time-to-fixation (P < 0.01 and P = 0.0048, respectively). Nearly every specimen in the conventional series showed less antigenicity in the resection specimen than the biopsy (Figures 3 and 4 and Table 2).
Discussion
Here we report a standardized approach to assess the influence of delayed formalin fixation on the antigenicity of breast biomarkers analyzed on two different series—a TMA series as well as a second series of biopsies and resections that more accurately represent tissues seen in routine patient care. The results of our quantitative protein analysis on these series suggest that ER, PgR, HER2, and Ki67 antigenicity as well as seven other proteins are not affected by delayed time-to-formalin fixation within the standard recommended time from surgical removal of the specimen (60 minutes). The analysis of ER and PgR on the TMA series shows a trend toward decreased epitope with increased time-to-fixation, but this was only seen with bootstrapping analysis. Two proteins associated with hypoxia showed a statistically significant increase in levels with increasing time-to-fixation. In contrast, the phospho-tyrosine epitope showed statistically significant degradation with increasing time-to-fixation.
The observations in this study agree with some previous work, but not all, depending on the time points examined. This trend was not seen on the conventional series of biopsies and resections measured in a previous small series from our lab (30). Because time-to-fixation is not recorded for these cases, it is likely that degradation of protein, or epitopes on proteins, occurs in time periods beyond 2 hours. The change between the core biopsy and resection specimen may be highly dependent on which specimens were selected and the time delay associated with each case. Our findings are consistent with those reported in a recent study by Yildiz-Aktaz et al. (37), where loss of protein epitopes is initially seen at around 3–4 hours but is not substantial until 24 or 48 hours.
Perhaps the most unique finding of this work is that some proteins that are expressed in response to hypoxia show increased expression with increased time-to-fixation. Although this result would be expected based on mechanistic and cell line model data, we are unaware of any similar data on real human tumors. Given the relationship between expression and time before fixation, scientists who study these proteins should be aware that results may be unexpectedly high if care is not taken to fix tissue rapidly. This observation may also be of value in the construction of a tissue quality index. One can envision usage to help quantify the degree of cold ischemic artifact within a given tissue specimen.
Although our study reports quantitative assessment of the effect of delayed time-to-formalin fixation on proteins in breast cancer, it is subject to two major limitations: 1) the limited time range tested and 2) the problem of heterogeneity of expression. The TMA series consists of 93 patients with recorded time-to-formalin fixation between 25 and 415 minutes. Furthermore, the conventional series of breast cancer specimens does not have information on the exact time of delay to formalin fixation but ranges between 60 and 180 minutes. Because there are many anecdotal reports of tissue being stored for 4 hours or even much longer before being cut and fixed, future quantitative studies are required to assess detrimental effects beyond 2 hours. This may also explain the discrepancy between this study and other studies in the literature related to time-to-fixation. In fact, in a previous study by our group using older core vs resection specimens with unknown times-to-fixation and a relatively small sample size (10 cases), we reported a statistically significant decrease in ER levels with time-to-fixation (30). We believe these studies are not discrepant in that the current study is focused on earlier time periods to address current practices. However, the lack of agreement between these two studies emphasizes the need for future efforts to extend the time window.
The second key limitation of this, or any study in anatomic pathology, is tumor heterogeneity. All tumors are known to be heterogeneous, and this problem has historically raised issues related to tumor sampling (31–33). In fact, the standard of care in anatomic pathology, which is one 5-µ section per 1cm of tumor, means that we sample only 0.05% of the tumor. Statistically significant differences of ER expression in different samples of one tumor have been reported previously (34–36) as well as for PgR, HER2, Ki67, and a few other markers used in research settings. The heterogeneity is a problem both within cases and within a population. For example, the range of each of the markers at early time points is broad (see Figures 2 and 3), illustrating the dynamic range of the population. So a low value of a given biomarker may be interpreted either as the natural expression level of that particular protein in that particular tumor, or it may be interpreted as evidence of loss of antigenicity due to preanalytic variation. This heterogeneity is further complicated by heterogeneity within a tumor specimen. A TMA spot of 0.6mm in diameter might not necessarily reflect the protein status of the entire tumor. Similarly, delayed time to cutting and immersion of tissue in formalin might affect different areas of a given tumor at different rates. Therefore, differences in protein antigenicity interpreted as a function of delayed time-to-fixation or cold ischemic time here are only based on statistically significant trends seen in whole populations. Some proteins, like Histone 4 in this study, are an example of an inconclusive result. This protein seemed to lose protein antigenicity as a factor of delayed time-to-formalin fixation as assessed on the TMA series. However, this result could not be confirmed on the conventional series of matched pairs of biopsies and resections. Thus, heterogeneity limits the strength of the conclusions of this work.
Although these limitations generally decrease the strength of the result for any individual protein, the converse is that when a protein is found to be increased or decreased, it is likely to be quite robust and unlikely to represent a false-positive result. For example, we report a decrease to complete loss of antigenicity of phosphorylated proteins as a function of delayed tissue fixation. These results confirm concerns about the accuracy of assessment of phosphorylated proteins in breast cancer tissue and their loss due to delayed processing and fixation, as reported by others (10,22,30). They also illustrate that the effects of cold ischemic time are epitope-specific. The lack of effect seen in some of the proteins tested is also likely to be robust. That is, certain antigens, like those on nearly every “housekeeping” protein we have tested, appear stable even at the extremes of our assays.
These limitations apply to most of the studies in the literature and are probably responsible for the reported incongruous results in biospecimen science papers. For example, Pinhel et al. described the loss of antigenicity for phospho-AKT and phospho-ERK1/2, whereas ER, PgR, HER2, and Ki67 did not show any statistically significant change in antigen levels (10). Bai et al. described the loss of ER antigenicity in addition to decreased levels of phosphorylated epitopes (30), Khoury et al. found that both ER and PgR antigenicity were diminished after 2 hours of delay to fixation (21), and Yildiz-Aktaz et al. reported loss of immunoreactivity for ER and PgR after 3–4 hours of fixation delay (37). None of these works comprehensively addresses either the full range of times-to-fixation or the issue of heterogeneity. However, agreement is found addressing the problem of accurate assessment of phosphorylated proteins where it is thought that phosphatase activity leads to dephosphorylation of epitopes during delayed fixation time.
In summary, the key finding of this study is that there is no evidence for loss of four critical breast cancer proteins as a function of time-to-fixation when tested within a range equal to twice the current standard recommended by the American Society of Clinical Oncology/College of American Pathologists (ASCO/CAP) guidelines. Although it is difficult to prove the negative, these quantitative results were confirmed in both experimental (ie, TMA) and clinical specimen studies. We believe it represents the first quantitative assessment of these markers in that time window, providing scientific support for the guidelines. To further address the exact time course of degradation of epitopes, broader new studies are required. We have begun a prospective effort to investigate these and other markers and their change as a function of cold ischemic time on normal breast tissue where we can extend the time-to- fixation without jeopardizing patient safety and where heterogeneity is more limited than that seen in malignancy. We are hopeful that these and other studies will ultimately help define the acceptable parameters for time-to-fixation for critical biomarkers needed for companion diagnostic tests.
Funding
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health (contract number HHSN261200800001E to DLR).
Notes
D. L. Rimm is a stockholder in and consultant to HistoRx Inc, the exclusive licensee to the Yale University–owned AQUA technology. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the US Government.
The authors are responsible for the study design, data collection, data analysis, interpretation, writing of the report, and the decision to submit the report for publication.
References
- 1. Dowsett M. Preoperative models to evaluate endocrine strategies for breast cancer. Clin Cancer Res 2003. 9(1, pt 2):502S–510S [PubMed] [Google Scholar]
- 2. Caldarella A, et al. Female breast cancer status according to ER, PR and HER2 expression: a population based analysis. Pathol Oncol Res. 2011. 17(3): 753–758 [DOI] [PubMed] [Google Scholar]
- 3. Kim EK, et al. Prognostic significance of young age (<35 years) by subtype based on ER, PR, and HER2 status in breast cancer: a nationwide registry-based study. World J Surg. 2011. 35(6):1244–1253 [DOI] [PubMed] [Google Scholar]
- 4. Lips EH, et al. Neoadjuvant chemotherapy in ER+ HER2- breast cancer: response prediction based on immunohistochemical and molecular characteristics. Breast Cancer Res Treat. 2012. 131(3):827–836 [DOI] [PubMed] [Google Scholar]
- 5. Shao MM, Liu J, Vong JS, et al. A subset of breast cancer predisposes to brain metastasis. Med Mol Morphol 2011. 44(1):15–20 [DOI] [PubMed] [Google Scholar]
- 6. Moon YW, Park S, Sohn JH, et al. Clinical significance of progesterone receptor and HER2 status in estrogen receptor-positive, operable breast cancer with adjuvant tamoxifen. J Cancer Res Clin Oncol 2011. 137(7):1123–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Anderson H, et al. Relationship between estrogen receptor, progesterone receptor, HER-2 and Ki67 expression and efficacy of aromatase inhibitors in advanced breast cancer. Ann Oncol. 2011. 22(8):1770–1776 [DOI] [PubMed] [Google Scholar]
- 8. Tanei T, Shimomura A, Shimazu K, et al. Prognostic significance of Ki67 index after neoadjuvant chemotherapy in breast cancer. Eur J Surg Oncol 2011. 37(2):155–161 [DOI] [PubMed] [Google Scholar]
- 9. Moore HM, Compton CC, Alper J, Vaught JB. International approaches to advancing biospecimen science. Cancer Epidemiol Biomarkers Prev 2011. 20(5):729–732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pinhel IF, Macneill FA, Hills MJ, et al. Extreme loss of immunoreactive p-Akt and p-Erk1/2 during routine fixation of primary breast cancer. Breast Cancer Res 2010. 12(5):R76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Betsou F, Barnes R, Burke T, et al. ; International Society for Biological and Environmental Repositories (ISBER) Working Group on Biospecimen Science Human biospecimen research: experimental protocol and quality control tools. Cancer Epidemiol Biomarkers Prev 2009. 18(4):1017–1025 [DOI] [PubMed] [Google Scholar]
- 12.De Cecco L, Musella V, Veneroni S, et al. Impact of biospecimens handling on biomarker research in breast cancer. BMC Cancer. 2009;9:409. doi: 10.1186/1471-2407-9-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lim MD, Dickherber A, Compton CC. Before you analyze a human specimen, think quality, variability, and bias. Anal Chem 2011. 83(1):8–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hammond ME, Hayes DF, Dowsett M, et al. ; American Society of Clinical Oncology; College of American Pathologists American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer (unabridged version). Arch Pathol Lab Med 2010. 134(7):e48–e72 [DOI] [PubMed] [Google Scholar]
- 15. Juhl H. Preanalytical aspects: a neglected issue. Scand J Clin Lab Invest Suppl 2010. 242 63–65 [DOI] [PubMed] [Google Scholar]
- 16. Spruessel A, Steimann G, Jung M, et al. Tissue ischemia time affects gene and protein expression patterns within minutes following surgical tumor excision. BioTechniques 2004. 36(6):1030–1037. [DOI] [PubMed] [Google Scholar]
- 17. Hatzis C, Sun H, Yao H, et al. Effects of tissue handling on RNA integrity and microarray measurements from resected breast cancers. J Natl Cancer Inst 2011. 103(24):1871–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yaziji H, Taylor CR, Goldstein NS, et al. ; Members of the Standardization Ad-Hoc Consensus Committee Consensus recommendations on estrogen receptor testing in breast cancer by immunohistochemistry. Appl Immunohistochem Mol Morphol 2008. 16(6):513–520 [DOI] [PubMed] [Google Scholar]
- 19. Carlson RW, Allred DC, Anderson BO, et al. ; NCCN Breast Cancer Clinical Practice Guidelines Panel Breast cancer. Clinical practice guidelines in oncology. J Natl Compr Canc Netw 2009. 7(2):122–192 [DOI] [PubMed] [Google Scholar]
- 20. Wolff AC, Hammond ME, Schwartz JN, et al. ; American Society of Clinical Oncology; College of American Pathologists American Society of Clinical Oncology/College of American Pathologists guideline recommendations for human epidermal growth factor receptor 2 testing in breast cancer. J Clin Oncol 2007. 25(1):118–145 [DOI] [PubMed] [Google Scholar]
- 21. Khoury T, Sait S, Hwang H, et al. Delay to formalin fixation effect on breast biomarkers. Mod Pathol 2009. 22(11):1457–1467 [DOI] [PubMed] [Google Scholar]
- 22. Mueller C, Edmiston KH, Carpenter C, et al. One-step preservation of phosphoproteins and tissue morphology at room temperature for diagnostic and research specimens. PLoS ONE 2011. 6(8):e23780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Apple S, Pucci R, Lowe AC, Shintaku I, Shapourifar-Tehrani S, Moatamed N. The effect of delay in fixation, different fixatives, and duration of fixation in estrogen and progesterone receptor results in breast carcinoma. Am J Clin Pathol 2011. 135(4):592–598 [DOI] [PubMed] [Google Scholar]
- 24. Baker AF, Dragovich T, Ihle NT, Williams R, Fenoglio-Preiser C, Powis G. Stability of phosphoprotein as a biological marker of tumor signaling. Clin Cancer Res 2005. 11(12):4338–4340 [DOI] [PubMed] [Google Scholar]
- 25. Camp RL, Chung GG, Rimm DL. Automated subcellular localization and quantification of protein expression in tissue microarrays. Nat Med. 2002. 8(11):1323–1327 [DOI] [PubMed] [Google Scholar]
- 26. Bordeaux J, Welsh A, Agarwal S, et al. Antibody validation. BioTechniques 2010. 48(3):197–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Espina V, Edmiston KH, Heiby M, et al. A portrait of tissue phosphoprotein stability in the clinical tissue procurement process. Mol Cell Proteomics 2008. 7(10):1998–2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Araujo RP, Liotta LA, Petricoin EF. Proteins, drug targets and the mechanisms they control: the simple truth about complex networks. Nat Rev Drug Discov 2007. 6(11):871–880 [DOI] [PubMed] [Google Scholar]
- 29. Petricoin EF, III, Bichsel VE, Calvert VS, et al. Mapping molecular networks using proteomics: a vision for patient-tailored combination therapy. J Clin Oncol 2005. 23(15):3614–3621 [DOI] [PubMed] [Google Scholar]
- 30. Bai Y, Tolles J, Cheng H, et al. Quantitative assessment shows loss of antigenic epitopes as a function of pre-analytic variables. Lab Invest 2011. 91(8):1253–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Davis BW, Zava DT, Locher GW, Goldhirsch A, Hartmann WH. Receptor heterogeneity of human breast cancer as measured by multiple intratumoral assays of estrogen and progesterone receptor. Eur J Cancer Clin Oncol 1984. 20(3):375–382 [DOI] [PubMed] [Google Scholar]
- 32. Douglas-Jones AG, Collett N, Morgan JM, Jasani B. Comparison of core oestrogen receptor (ER) assay with excised tumour: intratumoral distribution of ER in breast carcinoma. J Clin Pathol 2001. 54(12):951–955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sapino A, Marchiò C, Senetta R, et al. Routine assessment of prognostic factors in breast cancer using a multicore tissue microarray procedure. Virchows Arch 2006. 449(3):288–296 [DOI] [PubMed] [Google Scholar]
- 34. Chung GG, Zerkowski MP, Ghosh S, Camp RL, Rimm DL. Quantitative analysis of estrogen receptor heterogeneity in breast cancer. Lab Invest 2007. 87(7):662–669 [DOI] [PubMed] [Google Scholar]
- 35. Tolles J, Bai Y, Baquero M, Harris LN, Rimm DL, Molinaro AM. Optimal tumor sampling for immunostaining of biomarkers in breast carcinoma. Breast Cancer Res 2011. 13(3):R51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nassar A, Radhakrishnan A, Cabrero IA, Cotsonis GA, Cohen C. Intratumoral heterogeneity of immunohistochemical marker expression in breast carcinoma: a tissue microarray-based study. Appl Immunohistochem Mol Morphol 2010. 18(5):433–441 [DOI] [PubMed] [Google Scholar]
- 37. Yildiz-Aktas IZ, Dabbs DJ, Bhargava R. The effect of cold ischemic time on the immunohistochemical evaluation of estrogen receptor, progesterone receptor, and HER2 expression in invasive breast carcinoma. Mod Pathol. 2012. 25(8):1098–1105 [DOI] [PubMed] [Google Scholar]