Abstract
Background
Mesenchymal stem cells (MSCs) are multipotent stem cells with capacity to differentiate into several mesenchymal lineages. This quality makes MSCs good candidates for use in cell therapy. MSCs can be isolated from a variety of tissues including bone marrow and adipose tissue, which are the most common sources of these cells. However, MSCs can also be isolated from peripheral blood. Sheep has been proposed as an ideal model for biomedical studies including those of orthopaedics and transmissible spongiform encephalopathies (TSEs). The aim of this work was to advance these studies by investigating the possibility of MSC isolation from ovine peripheral blood (oPB-MSCs) and by subsequently characterizing there in vitro properties.
Results
Plastic-adherent fibroblast-like cells were obtained from the mononuclear fraction of blood samples. These cells were analysed for their proliferative and differentiation potential into adipocytes, osteoblasts and chondrocytes, as well as for the gene expression of cell surface markers. The isolated cells expressed transcripts for markers CD29, CD73 and CD90, but failed to express the haematopoietic marker CD45 and expressed only low levels of CD105. The expression of CD34 was variable. The differentiation potential of this cell population was evaluated using specific differentiation media. Although the ability of the cultures derived from different animals to differentiate into adipocytes, osteoblasts and chondrocytes was heterogeneous, we confirmed this feature using specific staining and analysing the gene expression of differentiation markers. Finally, we tested the ability of oPB-MSCs to transdifferentiate into neuronal-like cells. Morphological changes were observed after 24-hour culture in neurogenic media, and the transcript levels of the neurogenic markers increased during the prolonged induction period. Moreover, oPB-MSCs expressed the cellular prion protein gene (PRNP), which was up-regulated during neurogenesis.
Conclusions
This study describes for the first time the isolation and characterization of oPB-MSCs. Albeit some variability was observed between animals, these cells retained their capacity to differentiate into mesenchymal lineages and to transdifferentiate into neuron-like cells in vitro. Therefore, oPB-MSCs could serve as a valuable tool for biomedical research in fields including orthopaedics or prion diseases.
Keywords: Sheep, Mesenchymal stem cell, Peripheral blood, Neurogenesis
Background
Mesenchymal stem cells (MSCs) are morphologically fibroblast-like cells that are characterized by their ability to both self-renew and differentiate into tissues of mesodermal origin (osteoblasts, adipocytes, chondrocytes and myocytes) [1]. However, MSCs can also give rise to other cell types such as astrocytes and neurons [2,3]. This indicates cellular pluripotency and suggests that MSCs are responsible for the normal turnover and maintenance of adult mesenchymal tissues [4].
Sheep is an ideal model for bone tissue engineering [5] and has been proposed as an animal model for a wide range of applications in biomedical research, such as for the studies of respiratory diseases [6], cardiomyopathies [7,8], neurological disorders [9] and prion diseases [10,11].
Although MSCs are generally obtained from the bone marrow [12], they can also be isolated from other sources such as adipose tissue, umbilical cord blood and foetal tissues [13,14]. The isolation of MSCs from peripheral blood (PB-MSCs) has been reported for a variety of mammals including guinea pigs, rabbits, dogs, mice, rats, horses and humans [15-19]. Because blood harvesting is a less invasive procedure to obtain stem cells, this method would represent a significant advantage for patients and, therefore, would be an ideal candidate technique to obtain PB-MSCs for future clinical applications. Moreover, monitoring the presence and the proportional quantity of MSCs in the peripheral blood could possibly help in the understanding of the patients’ reaction to a disease.
The isolation procedure of ovine PB-MSCs (oPB-MSCs) would facilitate the sampling of these progenitor cells for use in a wide variety of applications, including fundamental and applied studies of orthopaedics or prion diseases. Here, we present the first study describing the isolation and characterization of oPB-MSCs. The osteogenic, chondrogenic and adipogenic differentiation potential of oPB-MSCs was analysed in vitro and monitored by specific staining and molecular differentiation markers. We also demonstrate the capacity of these cells to differentiate into neuron-like cells and the expression of the gene coding for the prion protein (PRNP) in both regular and differentiated cells.
Results
Isolation and characterization of peripheral blood derived fibroblast-like cells
Isolation and expansion of peripheral blood derived fibroblast-like cells
Plastic-adherent fibroblast-like cells were observed within the first days of culture of the nucleated cell fraction of peripheral blood obtained from total six sheep. Although the volume of blood collected was similar for all animals (approximately 25 mL), the number of peripheral blood nucleated cells (PBNC) obtained was variable, ranging from 0.594 106 to 1.9 106 PBNC/mL, with mean 1.36 106 ± 682646. After the isolation process, a mean of 281400 ± 178051 adherent cells were obtained from each individual, varying between 2.7 and 9.3 adherent cells for every 1000 PBNC (mean: 5.85 ± 2.7).
Cells were expanded until the second passage and then frozen. The proliferation capacity of the adherent cells was measured during the first two passages. An average of 12.6 days was necessary to complete the first passage. Mean cell doubling during the first passage was 2.29 ± 0.887 and the doubling time was 5.99 ± 1.86 days. Time required to complete the second passage was shortened to 7.33 days, cell doubling decreased to 1.84 ± 0.975 and the doubling time was 4.88 ± 2.68 days.
After thawing, the cells from passage 2 were expanded for two more passages to obtain sufficient amount of cells for the differentiation assays. The cells were then characterized by analysing the expression of cell surface markers and the tri-lineage differentiation potential into adipocytes, osteoblasts and chondrocytes.
Expression of mesenchymal cell surface markers
To initiate the characterization of oPB-MSCs, the expression of six cell surface markers specific for mesenchymal and haematopoietic cells were first analysed at the transcript level by quantitative real time PCR (RT-qPCR). All analysed cultures expressed CD29 (integrin β1), CD73 (ecto-5’-nucleotidase) and CD90 (Thy-1), whereas the expression of CD34 (CD34 molecule) was detected in five out of six of these cultures. The amplification of the hematopoietic marker CD45 (protein tyrosine phosphatase, receptor type, C) was not detected and CD105 (endoglin) was only weakly amplified at threshold cycles above 35.
Adipogenic potential
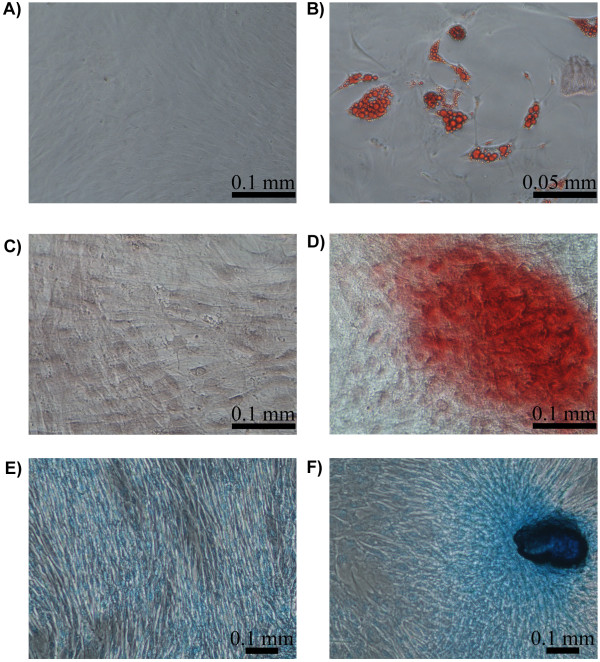
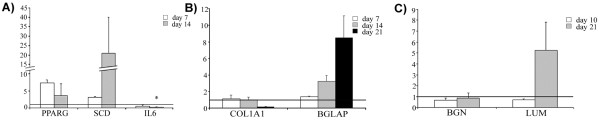
Cells cultured under adipogenic conditions presented cytoplasmic lipid droplets under light microscope, although the size of the droplets was variable depending on the donor animal. To confirm that the contents of the droplets were lipids, the cultures were stained with oil red O (Figure 1A and B). The expression of adipogenic markers was analysed on days 7 and 14 of post-induction. The expression profiles of PPARG (peroxisome proliferator-activated receptor gamma), SCD (stearoyl-CoA desaturase) and IL6 (interleukin 6) are shown in Figure 2. During the induction of differentiation, the PPARG and SCD mRNA expression levels increased to 7.3- and 20.8-fold, respectively. However, these changes were not statistically significant due to the high variability observed between animals. A significant downregulation of IL6 (−31-fold, P < 0.05) was detected after two weeks of culture (Figure 2A).
Figure 1.
Staining for adipogenic, osteogenic and chondrogenic differentiation of PB-MSCs. Oil red O staining of cells cultured for 15 days in basal (A) and adipogenic differentiation medium (B). Alizarin red staining of cells cultured for 21 days in basal (C) and osteogenic differentiation medium (D). Alcian blue staining of cells cultured for 21 days in basal (E) and chondrogenic medium (F).
Figure 2.
Quantitative real time PCR analysis. Expression of the adipogenic (A), osteogenic (B) and chondrogenic (C) markers at different times of the differentiation process relative to the levels observed in control cultures (values set to 1, horizontal line). Data are shown as mean ± standard errors. Statistically significant differences between differentiated and control cells were determined by Student t test (*P < 0.05).
Osteogenic potential
The ability of oPB-MSCs to differentiate into osteoblasts was demonstrated using alizarin red staining (Figure 1C and D). Nodule-like aggregations stained in red appeared in the osteogenic media on the 21st day of culture exclusively, indicating that these cultures were mineralized at a relatively late stage. However, the cells from different animals displayed variable osteogenic potential. The expression of osteogenic markers was evaluated in the cultures that displayed positive staining (n = 2). The expression levels of COL1A1 (collagen, type 1, α 1) were not altered during the first 2 weeks in osteogenic media. However, a strong downregulation of COL1A1 was observed at 3 weeks of culture. In contrast, the expression levels of BGLAP (bone gamma-carboxyglutamate (gla) protein, or osteocalcin) increased drastically throughout the culture period (Figure 2B).
Chondrogenic potential
The chondrogenic potential was evaluated in monolayer cultures. Ovine PB-MSCs formed nodule-like aggregations in both control and induced conditions. However, the oPB-MSCs in chondrogenic media displayed a stronger staining with alcian blue (Figure 1F). Although the chondrogenic marker expression analysis did not reveal variations in the gene expression levels of the BGN (biglycan), LUM (lumican) was found to be upregulated on the 21st day of culture (Figure 2C).
Neuronal differentiation of oPB-MSCs
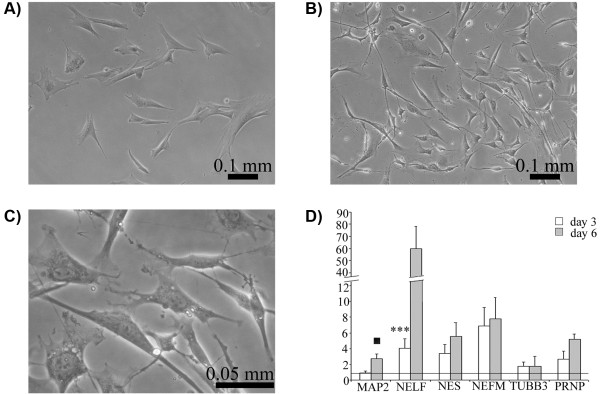
The ability of the isolated cells to transdifferentiate into neuronal cells was evaluated in vitro. The cells cultured under neurogenic conditions displayed distinctly altered morphology after the first 24 hours of induction. Differentiated cells were sharply defined, retracted towards the nucleus displaying phase-bright bodies, and some neurite-like processes (thin, long, and often branched) became apparent (Figure 3B,C). Neuronal differentiation was also demonstrated using RT-qPCR analysis. Control cells displayed none or very low levels of NELF (nasal embryonic LHRH factor) expression on 3 and 6 days of culture, while low expression levels of the remaining markers (MAP2 [microtubule-associated protein 2], NES [nestin], NEFM [neurofilament, medium polypeptide], TUBB3 [tubulin, beta 3]) were observed. The expression of these markers increased in neurogenic conditions, with a peak of expression on day 6 post-induction. Statistically significant changes were found for NELF on day 3 of culture (5.85 fold induction, P < 0.001) and an over-expression tendency was observed for MAP2 on day 6 (2.4 fold induction, P < 0.1). Moreover, oPB-MSCs expressed transcripts of the prion protein (PRNP), which increased up to 5 times during the neurogenic period (Figure 3D).
Figure 3.
Neurogenic differentiation of PB-MSCs. Phase contrast micrograph of ovine PB-MSCs at passage 3 cultured on basal and neurogenic medium for 3 days. Control cells (A) showed a fibroblast-like shape whereas differentiated cells (B) displayed neuronal-like features such as phase-bright bodies, long multipolar extensions and branching ends (C: higher magnification). Increase of the expression levels of neurogenic markers after 3 (white bars) and 6 (grey bars) days of induction relative to the levels observed in cells cultured on basal medium (values set to 1, horizontal line) as assessed by real time PCR. Data are shown as mean ± standard errors. Statistically significant differences between differentiated and control cells were determined by Student t test (▪ P < 0.10, *** P < 0.001).
Discussion
Despite the importance of ovine as a large animal model for many conditions (i.e., orthopaedic injuries or Transmissible Spongiform Encephalopathies) the characterisation of ovine MSCs (oMSCs) is still limited. During the last decade, there has been an important effort within the scientific community to focus on the characterisation of MSCs obtained from different species, including the sheep. However, most research on MSCs has been performed on cells derived from bone marrow and, to a lesser degree, adipose tissue. The osteogenic and chondrogenic differentiation potential of MSCs in vitro[20-22] and in vivo[23,24] is currently relatively well understood. Their phenotype for mesenchymal surface cell markers has also been analysed [25], and their proliferative potential has been shown to be heterogeneous [26]. Although the existence of MSCs in peripheral blood has been demonstrated in many species [17,18], this work represents the first report describing the isolation of these cells from sheep circulation.
The minimal criteria to define human MSCs proposed by the Mesenchymal and Tissue Stem Cell Committee of the International Society for Cellular Therapy are: (1) plastic-adhesion when maintained in standard culture conditions; (2) expression of CD105, CD73 and CD90, and lack expression of the haematopoietic markers CD45, CD34, CD14 or CD11b, CD79alpha or CD19 and HLA-DR surface molecules and; (3) ability to differentiate to osteoblasts, adipocytes and chondroblasts in vitro[27]. In our study, plastic-adherent cells with a fibroblast-like morphology were obtained from all experimental sheep and were further analysed to determine the expression of mesenchymal markers and their ability to differentiate into adipocytes, osteoblasts and chondrocytes.
In other domestic species [28] the proportion of MSCs in the peripheral blood is low, which is in agreement with the few colonies of MSCs detected in our original oPB-MSCs cultures. Although the proliferation ability of oPB-MSCs was very different between individuals, the doubling time was generally longer than in other species, such as horse [29]. This difference may be due to the higher percentage of FBS used in the isolation of equine PB-MSCs (30%) and also due to the addition of dexamethasone to the growth media, which has been demonstrated to favour the expansion of MSCs [30]. The variability observed in this work is in accordance with the high heterogeneity in the proliferative potential of oMSCs obtained from bone marrow (oBM-MSCs) [26].
The absence of a well-defined immunophenotype for PB-MSCs renders the comparison of studies difficult. Moreover, most of the cell surface markers utilized to sort subpopulations of human MSC by flow cytometry have not been validated in sheep [21]. Gene expression-based technologies may be useful for the identification of possible molecules described as MSC markers [31,32]. In our study, RT-qPCR was performed to quantify the mRNA expression levels of six cell surface antigens considered as either positive (CD29, CD73, CD90 and CD105) or negative (CD34 and CD45) MSC markers in humans.
In accordance with the immunophenotype described for human PB-MSCs [33-35], our expression analysis revealed significant amplification of the typical MSC markers, CD29, CD73 and CD90, and a weak signal for CD105. In contrast, the haematopoietic marker CD45 was not expressed. To our knowledge, there are no published data concerning the gene expression of cell surface markers in oMSCs obtained from other tissues. However, we have observed amplification of CD29, CD73 and CD90 in oBM-MSCs, as well as the lack of CD34 and CD45 expression (unpublished work from our group). Using flow cytometry, the presence of CD29 and CD105 has also been detected in oBM-MSCs [21,36]. Additionally, oMSCs isolated from adipose tissue (oAT-MSCs) display high expression of CD90 and low immunoreactivity for CD105 [25]. The immunophenotypes of oBM-MSCs [36] and oAT-MSCs [25] are negative for the haematopoietic CD34 marker. However, this marker is expressed at low levels in human PB-MSCs [37] and in equine MSCs derived from adipose tissue [38] as demonstrated by RT-qPCR. We detected CD34 expression in 5 out of 6 cultures, which may indicate individual variability. Finally, the cells analysed were negative for the haematopoietic marker CD45, as are human MSCs [16]. We have previously found a good correlation between MSC marker gene expression and the immunophenotype detected by flow cytometry in equine MSCs [39]. Although flow cytometry analysis is necessary to validate the immunophenotype of the isolated cells, the gene expression profile observed in this work strongly suggests that the peripheral blood derived fibroblast-like cells obtained as described would fulfill the requirements to be considered as MSCs.
Ovine BM-MSCs can be differentiated into adipocytes, showing lipid droplets in their cytoplasm and the induction of adipogenic markers [20,36]. Similarly, adipogenic differentiation has been achieved here in all peripheral blood derived cell cultures, although great variability in the size of lipid droplets was observed. The expression of two adipogenic markers was evaluated in the cultures using RT-qPCR. PPARG is considered the master regulator of adipogenesis [40,41] and is up-regulated in MSCs under adipogenic conditions [42]. SCD is expressed uniquely in adipocytes and catalyzes the rate-limiting step in the synthesis of poly-unsaturated fatty acids, thereby exhibiting a pivotal role in adipocyte metabolism [43]. Inter-individual variability was also noticeable in the expression of these adipogenic markers, which explains the lack of statistically significant differences in PPARG and SCD expression results despite the strong overexpression observed throughout the culture period. We also determined the expression of IL-6, which maintains the proliferative and undifferentiated state of bone marrow-derived MSCs [44] and is down-regulated during lineage-specific differentiation [45]. In accordance with these reports, a significant decrease was detected in the expression of IL6 in the differentiated cultures. Therefore, using specific staining and gene expression profiles of adipogenic markers, we have confirmed the adipogenic potential of oPB-MSCs.
Similar to the adipogenic analysis, a great individual variation was also observed in the osteogenic potential. Osteogenic mineralization was confirmed on the last day of culture in osteogenic conditions (21 days) by staining calcium deposits with alizarin red. The induction period necessary for visualization of matrix mineralisation in oMSCs varies among different studies. The period reported for oBM-MSC mineralisation ranges from 21 days [36] to 5 weeks [21], while 4 weeks are required to differentiate periodontal oMSCs [46]. The weak alizarin red staining observed in some of our experiments could be due to the relatively short period of induction.
Although COL1A1 is considered an early marker of osteoprogenitor cells [47], we observed either no changes or a strong down-regulation on the 21st day of culture. Besides displaying a rapid mineralisation, oBM-MSCs cultured under osteogenic conditions express increased or declined levels of COL1A1 depending on the differentiation moment [21]. Other authors have reported either no significant increase in COL1A1 mRNA expression levels after osteogenic differentiation in human [48], porcine [49] and equine [39] MSCs, or a down-regulation of this marker in human PB-MSCs during osteogenesis [50]. Therefore, COL1A1 may not be suitable for monitoring osteogenesis in oPB-MSCs. In contrast, BGLAP was upregulated during the differentiation process and was maximally expressed on the last day of culture (day 21), coinciding with the positive alizarin red staining. This is in accordance with the role of BGLAP as a late marker of developing osteoblasts [51].
The sheep has been used as a large animal model for the studies of chondrogenesis both in vivo[52] and in vitro[22]. The chondrogenic potential of oMSCs has been evaluated mainly in micromass cultures of cells derived from bone marrow [20,21,36]. Chondrogenesis was evaluated in our study using a bidimensional culture with a high cell concentration seeding, according to the protocol described by Jäger et al. [53] for chondrogenic differentiation of ovine umbilical cord blood-derived MSCs. Chondrogenic nodules were observed in both control and chondrogenic media, although the staining was stronger in the induced cultures. The confirmation with molecular markers was not straightforward as the expression of the two components of the extracellular matrix BGN and LUM changed in opposite directions during chondrogenic differentiation. In accordance to our results, the lack of strong BGN overexpression has been reported for chondrogenic induced micropellets of oBM-MSCs [36]. However, further analysis is necessary to fully confirm the ability of oPB-MSCs to differentiate into chondrocytes.
During the last decade, many reports have described the in vitro neural transdifferentiation of MSCs derived from a range of species [2,54,55] but, to our knowledge, this has never been investigated in oMSCs. Neurogenic capacity of PB-MSCs would offer exciting possibilities for autologous therapeutic treatments for a variety of neurological disorders. As ovine is a natural model for prion diseases, the transdifferentiation of MSCs into neural cells could provide an excellent in vitro model for the study of these pathologies. Here, we described alterations in the morphology and expression profiles of neurogenic markers (MAP2, NEFM, NELF, NES and TUBB3) that are consistent with neural differentiation. In addition, we detected up-regulation of PRNP, which could also be involved in the morphological changes as the cellular prion protein seems to be necessary for neuritogenesis [56]. The variable success in the ability to transdifferentiate MSCs to a neural phenotype could be influenced by the inter-donor variability of expression of neural-related markers in MSCs prior to differentiation [57]. Nevertheless, our study shows that oPB-MSCs retain the ability to transdifferentiate. Finally, although murine bone marrow stromal cells express the prion protein [58], this has not been previously shown in species susceptible to prion diseases. In the present work, we have demonstrated the expression of PRNP in oPB-MSCs and its overexpression during neuronal differentiation.
Conclusions
In this study we describe, for the first time, the isolation of mesenchymal stem cells from ovine peripheral blood. These cells express mesenchymal markers and retain the ability to differentiate into adipocytes and osteoblasts. Although oPB-MSCs seem to differentiate into chondrocytes, further studies are necessary to confirm the suitability of these cells for chondrogenesis studies. Finally, these cells can transdifferentiate into neuron-like cells and express PRNP.
Methods
Animals and MSC isolation
Peripheral blood (25 mL) was obtained from a total of 6 sheep aged 1.5 to 6 years. The animals belonged to the Rasa Aragonesa breed and came from regional flocks. The procedure for blood collection from commercial farm animals was performed according to the recommendations of the Joint Working Group on Refinement [59]. The ethics committee of the University of Zaragoza approved the study (PI38/10). The blood was collected in 5 mL tubes with sodium heparin. Immediately after, blood was diluted in 1 volume of PBS and layered over Lymphoprep (Atom) in a 1:1 proportion. The mononuclear fraction was harvested after a density gradient centrifugation step of 20 min at 1600 g. Mononuclear cells were rinsed twice in the same volume of PBS by centrifugation for 5 min at 1600 g. The cells were resuspended, counted and plated at 106 cells/cm2 in 6-well plates with basal medium consisting of low glucose Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) supplemented with 20% foetal bovine serum (FBS), 1% L-glutamine (Sigma-Aldrich) and 1% streptomycin/penicillin (Sigma-Aldrich).
Non-adherent cells were removed washing the mononuclear cells twice with PBS after 24, 48 and 72 h of incubation at 37°C and 5% CO2 and were maintained in growth medium until reaching approximately 80% confluence. The cells were then treated with trypsin (Sigma Aldrich) and plated either in T75 or T175 flasks (Becton Dickinson) at 5000 cells/cm2 in basal medium with 10% FBS. The cells were trypsinised until the second passage (P2) and then cryopreserved in FBS with 10% DMSO.
The yield of adherent cells during these two passages was used to characterize the self renewal capacity of the cells isolated towards the estimation of the cell doubling (CD) and the doubling time (DT) parameters. These values were calculated using the formula: CD = ln (Nf/Ni)/ ln2; DT: time (days)/ CD, Nf being the final number of cells in the culture, and Ni the initial number.
Approximately 106 cells from passage two were thawed at 37°C and plated in a T75 flask. Cells were grown for two more passages prior to being used for the differentiation analyses.
Adipogenic differentiation
The cells obtained from the 6 sheep were seeded at 5000 cells/cm2 in 24-well plates with a previously described adipogenic medium [39]. Four replicates were seeded for each sheep, two were cultured with growth (control) medium and the other two with the adipogenic medium. The medium was changed every 3 days, and the differentiation was maintained for 14 days. To analyse the adipogenic differentiation, cells were fixed in 10% formalin (Sigma-Aldrich) for 15 min, and lipid droplets formed inside the cells were stained with 0.3% oil red O (Sigma-Aldrich). The expression of the adipogenic markers PPARG, SCD and IL6 was analysed at days 7 and 14 of culture using RT-qPCR.
Osteogenic differentiation
Cells from the 6 sheep were plated at 2 x 104 cells/cm2 in 24-well plates and cultured under osteogenic conditions (two replicates) or with growth medium (two replicates) for 21 days as previously described [39]. To assess their osteogenic potential, cells at days 7, 14 and 21 were fixed in 70% ethanol for 1 h and stained with 2% Alizarin Red S (Sigma Aldrich) for 10 min. The transcript expression of the osteogenic markers COL1A1 and BGLAP was evaluated by RT-qPCR at days 7, 14 and 21 of culture.
Chondrogenic differentiation
For chondrogenic differentiation in monolayer cultures (n = 5), 105 cells/cm2 were seeded in 24-well plates with the chondrogenic media described by Jäger et al. [53] (two replicates) or with growth medium (two replicates). The culture was maintained for 21 days with the media being changed twice per week. To determine chondrogenic differentiation, the cultures were stained with alcian blue dye (Sigma-Aldrich). Briefly, cells were washed with PBS, fixed with 70% ethanol for 1 h at room temperature, washed three times with distilled water, stained with alcian blue stain diluted in methanol at a 1:1 proportion and washed with water until the excess staining was removed. BGN and LUM transcripts were quantified at days 10 and 21 of chondrogenic induction using RT-qPCR.
Neuronal differentiation
The neurogenic potential of the isolated cells was tested in two cell lines. The cells were seeded at 2500 cells/cm2 in 24-well plates with the neurogenic medium (Thermo Scientific) (two replicates) or under growth medium (two replicates) and maintained for 6 days, changing the media every 3 days. Differentiation was monitored by both light microscope and analysis of the mRNA expression levels of neurogenic markers (MAP2, NELF, NES, NEFM and TUBB3) and PRNP by RT-qPCR at days 3 and 6.
Real Time quantitative PCR
The potential of cultured cells to differentiate into adipocytes, osteoblasts, chondrocytes and nervous cells was monitored via analysis of the expression levels of differentiation markers (Table 1) using RT-qPCR. The same methodology was used to evaluate the expression levels of cell surface markers for mesenchymal (CD29, CD73, CD90 and CD105) and haematopoietic (CD34 and CD45) stem cells in undifferentiated cells. The primers for RT-qPCR were designed using Primer Express 2.0 software (Applied Biosystems).
Table 1.
Cell surface, adipogenic, osteogenic, chondrogenic and neurogenic markers analysed by RT-qPCR
|
Genes |
Accession number |
Primer sequences |
Amplicon size (bp) |
||
|---|---|---|---|---|---|
| Forward (5’ → 3’) | Reverse (5’ → 3’) | ||||
|
Cell Surface Markers |
|
|
|
||
|
CD29 |
AF349461 |
GTGCCCGAGCCTTCAATAAAG |
CCCGATTTTCAACCTTGGTAATG |
87 |
|
|
CD34 |
AB021662 |
TGGGCATCGAGGACATCTCT |
GATCAAGATGGCCAGCAGGAT |
107 |
|
|
CD45 |
NM_001206523 |
CCTGGACACCACCTCAAAGCT |
TCCGTCCTGGGTTTTATCCTG |
101 |
|
|
CD73 |
BC114093 |
TGGTCCAGGCCTATGCTTTTG |
GGGATGCTGCTGTTGAGAAGAA |
115 |
|
|
CD90 |
BC104530 |
CAGAATACAGCTCCCGAACCAA |
CACGTGTAGATCCCCTCATCCTT |
96 |
|
|
CD105 |
NM_001076397 |
CGGACAGTGACCGTGAAGTTG |
TGTTGTGGTTGGCCTCGATTA |
115 |
|
|
Differentiation Markers |
|
|
|
||
|
PPARG |
NM_001100921 |
GCCCTGGCAAAGCATTTGTA |
TGTCTGTCGTCTTTCCCGTCA |
94 |
|
|
SCD1 |
AJ001048 |
CCCAGCTGTCAGAGAAAAGG |
GATGAAGCACAACAGCAGGA |
115 |
|
|
IL6 |
FJ409227.1 |
CAGCAAGGAGACACTGGCAG |
TGATCAAGCAAATCGCCTGAT |
101 |
|
|
COL1A1 |
AF129287 |
CCTGCGTACAGAACGGCCT |
ACAGCACGTTGCCGTTGTC |
93 |
|
|
BGLAP |
DQ418490 |
CCCAGGAGGGAGGTGTGTG |
CTAGACCGGGCCGTAGAAGC |
99 |
|
|
BGN |
NM_001009201.1 |
AACATGAACTGCATTGAGATGGG |
GCGAAGGTAGTTGAGCTTCAGG |
93 |
|
|
LUM |
NM_173934.1 |
AAGCAATTGAAGAAGCTGCACA |
TTAGTGAGCTGCAGGTCCACC |
92 |
|
|
NES |
194665083 |
CAAATCGCCCAGGTCCTG |
GCCTCTAGGAGGGTCCTGTATGT |
95 |
|
|
NEFM |
194669578 |
GCTCGTCATCTGCGAGAATACC |
CACCCTCCAGGAGTTTCCTGTA |
91 |
|
|
NELF |
27806522 |
CGCTATGCAGGACACAATCAAC |
GGGTCTCCTCACCTTCCAAGA |
161 |
|
|
TUBB3 |
116004470 |
GACCTCGAGCCTGGAACCAT |
GCCCCACTCTGACCAAAGATG |
92 |
|
|
MAP2 |
194664873 |
TGTCCCAGTGGAGGAAGGTTT |
TCTTGTCTAGTGGCTCGGCTG |
95 |
|
|
PRNP |
BC119821 |
CGCAGAAGCAGGACTTCTGAA |
TGGATTTGTGTCTCTGGGAAGA |
86 |
|
|
Housekeeping genes |
|||||
|
G6PDH2 |
AJ507200 |
TGACCTATGGCAACCGATACAA |
CCGCAAAAGACATCCAGGAT |
76 |
|
| HPRT3 | EF078978 | AGGTGTTTATTCCTCATGGAGTAATTATG | GGCCTCCCATCTCCTTCATC | 79 | |
GenBank accession numbers of the sequences used for primer design. Primer sequences (F: Forward and R: Reverse) and the length of the amplicon in base pairs (bp).
1 Primers described in Dervishi et al. [60].
2 Primers described in García-Crespo et al. 2005 [61].
3 Primers described in Lyahyai et al. 2010 [62].
RNA extraction and cDNA synthesis were performed on both differentiated and control oPB-MSC cultures using the cells-to-cDNA kit (Ambion). The isolated cDNA was diluted 1:5 in water for further analysis. Amplification experiments were performed in triplicate using Fast SYBR Green Master Mix reagent (Life Technologies) and the StepOne™ Real Time System (Life Technologies). The levels of gene expression were determined using the comparative Ct method. A normalization factor (NF) calculated as the geometric mean of the quantity of two housekeeping genes (GAPDH and HPRT) was used to normalize the expression levels for each gene. Variations in gene expression between differentiated and control oPB-MSCs were evaluated with the Student’s t test. Statistical significance was defined as P < 0.05.
Abbreviations
BGLAP: Bone Gamma-Carboxyglutamate (Gla) Protein; BGN: Biglycan; CD105: Endoglin; CD29: Integrin beta 1; CD34: CD34 molecule; CD45: Protein Tyrosine Phosphatase Receptor Type C; CD73: Ecto-5’-nucleotidase; CD90: Thy-1 cell surface antigen; COL1A1: Collagen type I alpha 1; FBS: Foetal Bovine Serum IL6, interleukin 6 (interferon, beta 2); LUM: Lumican; MAP2: Microtubule-Associated Protein 2; MSCs: Mesenchymal Stem Cells; NEFM: Neurofilament, Medium Polypeptide; NELF: Nasal Embryonic LHRH Factor; NES: Nestin; oAT-MSCs: ovine Adipose Tissue-Derived Mesenchymal Stem Cells; oBM-MSCs: ovine Bone Marrow-Derived Mesenchymal Stem Cells; oPB-MSCs: ovine Peripheral Blood-derived Mesenchymal Stem Cells; oMSCs: ovine Mesenchymal Stem Cells; PPARG: Peroxisome Proliferator-Activated Receptor Gamma; PRNP: Prion Protein; RT-qPCR: quantitative Real Time PCR; SCD: Stearoyl-CoA Desaturase (delta-9-desaturase); TUBB3: Tubulin Beta 3.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JL and DRM carried out the isolation and expansion of the cells, differentiation assays, gene expression analyses, statistical analysis and participated in drafting the manuscript. BR, AS and ARR helped in the culture and differentiation assays and manuscript drafting. RB performed the sample collections from the animals and participated in cell isolation. PZ and CR helped to draft the manuscript. IMB conceived the study, participated in its design and draft the manuscript. All authors read and approved the final manuscript.
Contributor Information
Jaber Lyahyai, Email: jaber.lyahyai@gmail.com.
Diego R Mediano, Email: mediano_87@hotmail.com.
Beatriz Ranera, Email: branera@unizar.es.
Arianne Sanz, Email: arianne@unizar.es.
Ana Rosa Remacha, Email: aremacha@unizar.es.
Rosa Bolea, Email: rbolea@unizar.es.
Pilar Zaragoza, Email: pilarzar@unizar.es.
Clementina Rodellar, Email: rodellar@unizar.es.
Inmaculada Martín-Burriel, Email: minma@unizar.es.
Acknowledgements
We thank Belén Marín from the CIEETE for her assistance in blood sampling. This work was performed as part of the AGL2008-02428/GAN (MICINN/FEDER) project and was partially financed by the Gobierno de Aragón/Fondo Social Europeo (Grupo de Excelencia LAGENBIO) and Instituto Aragonés de Ciencias de la Salud (I + CS). B. Ranera was supported by a doctoral grant from DGA.
References
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, Moorman MA, Simonetti DW, Craig S, Marshak DR. Multilineage potential of adult human mesenchymal stem cells. Science. 1999;284:143–147. doi: 10.1126/science.284.5411.143. [DOI] [PubMed] [Google Scholar]
- Bossolasco P, Cova L, Calzarossa C, Rimoldi SG, Borsotti C, Deliliers GL, Silani V, Soligo D, Polli E. Neuro-glial differentiation of human bone marrow stem cells in vitro. Exp Neurol. 2005;193:312–325. doi: 10.1016/j.expneurol.2004.12.013. [DOI] [PubMed] [Google Scholar]
- Gimble J, Guilak F. Adipose-derived adult stem cells: isolation, characterization, and differentiation potential. Cytotherapy. 2003;5:362–369. doi: 10.1080/14653240310003026. [DOI] [PubMed] [Google Scholar]
- Caplan AI. Review: mesenchymal stem cells: cell-based reconstructive therapy in orthopedics. Tissue Eng. 2005;11:1198–1211. doi: 10.1089/ten.2005.11.1198. [DOI] [PubMed] [Google Scholar]
- Guo X, Wang C, Duan C, Descamps M, Zhao Q, Dong L, Lu S, Anselme K, Lu J, Song YQ. Repair of osteochondral defects with autologous chondrocytes seeded onto bioceramic scaffold in sheep. Tissue Eng. 2004;10:1830–1840. doi: 10.1089/ten.2004.10.1830. [DOI] [PubMed] [Google Scholar]
- Scheerlinck JP, Snibson KJ, Bowles VM, Sutton P. Biomedical applications of sheep models: from asthma to vaccines. Trends Biotechnol. 2008;26:259–266. doi: 10.1016/j.tibtech.2008.02.002. [DOI] [PubMed] [Google Scholar]
- Psaltis PJ, Carbone A, Nelson AJ, Lau DH, Jantzen T, Manavis J, Williams K, Itescu S, Sanders P, Gronthos S, Zannettino AC, Worthley SG. Reparative effects of allogeneic mesenchymal precursor cells delivered transendocardially in experimental nonischemic cardiomyopathy. JACC Cardiovasc Interv. 2010;3:974–983. doi: 10.1016/j.jcin.2010.05.016. [DOI] [PubMed] [Google Scholar]
- Sill B, Roy N, Hammer PE, Triedman JK, Sigg DC, Kelly MF, Nedder A, Dunning PS, Cowan DB. Development of an ovine model of pediatric complete heart block. J Surg Res. 2011;166:e103–e108. doi: 10.1016/j.jss.2010.11.878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fauza DO, Jennings RW, Teng YD, Snyder EY. Neural stem cell delivery to the spinal cord in an ovine model of fetal surgery for spina bifida. Surgery. 2008;144:367–373. doi: 10.1016/j.surg.2008.05.009. [DOI] [PubMed] [Google Scholar]
- Hunter N. Scrapie and experimental BSE in sheep. Br Med Bull. 2003;66:171–183. doi: 10.1093/bmb/66.1.171. [DOI] [PubMed] [Google Scholar]
- Lyahyai J, Bolea R, Serrano C, Monleon E, Moreno C, Osta R, Zaragoza P, Badiola JJ, Martin-Burriel I. Correlation between Bax overexpression and prion deposition in medulla oblongata from natural scrapie without evidence of apoptosis. Acta Neuropathol. 2006;112:451–460. doi: 10.1007/s00401-006-0094-4. [DOI] [PubMed] [Google Scholar]
- Le Blanc K, Pittenger M. Mesenchymal stem cells: progress toward promise. Cytotherapy. 2005;7:36–45. doi: 10.1080/14653240510018118. [DOI] [PubMed] [Google Scholar]
- Krampera M, Marconi S, Pasini A, Galie M, Rigotti G, Mosna F, Tinelli M, Lovato L, Anghileri E, Andreini A, Pizzolo G, Sbarbati A, Bonetti B. Induction of neural-like differentiation in human mesenchymal stem cells derived from bone marrow, fat, spleen and thymus. Bone. 2007;40:382–390. doi: 10.1016/j.bone.2006.09.006. [DOI] [PubMed] [Google Scholar]
- Fadel L, Viana BR, Feitosa ML, Ercolin AC, Roballo KC, Casals JB, Pieri NC, Meirelles FV, Martins Ddos S, Miglino MA, Ambrosio CE. Protocols for obtainment and isolation of two mesenchymal stem cell sources in sheep. Acta Cir Bras. 2011;26:267–273. doi: 10.1590/S0102-86502011000400004. [DOI] [PubMed] [Google Scholar]
- He Q, Wan C, Li G. Concise review: multipotent mesenchymal stromal cells in blood. Stem Cells. 2007;25:69–77. doi: 10.1634/stemcells.2006-0335. [DOI] [PubMed] [Google Scholar]
- Valenti MT, Dalle Carbonare L, Donatelli L, Bertoldo F, Zanatta M, Lo Cascio V. Gene expression analysis in osteoblastic differentiation from peripheral blood mesenchymal stem cells. Bone. 2008;43:1084–1092. doi: 10.1016/j.bone.2008.07.252. [DOI] [PubMed] [Google Scholar]
- Koerner J, Nesic D, Romero JD, Brehm W, Mainil-Varlet P, Grogan SP. Equine peripheral blood-derived progenitors in comparison to bone marrow-derived mesenchymal stem cells. Stem Cells. 2006;24:1613–1619. doi: 10.1634/stemcells.2005-0264. [DOI] [PubMed] [Google Scholar]
- Zvaifler NJ, Marinova-Mutafchieva L, Adams G, Edwards CJ, Moss J, Burger JA, Maini RN. Mesenchymal precursor cells in the blood of normal individuals. Arthritis Res. 2000;2:477–488. doi: 10.1186/ar130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwana M, Okazaki Y, Kodama H, Izumi K, Yasuoka H, Ogawa Y, Kawakami Y, Ikeda Y. Human circulating CD14+ monocytes as a source of progenitors that exhibit mesenchymal cell differentiation. J Leukoc Biol. 2003;74:833–845. doi: 10.1189/jlb.0403170. [DOI] [PubMed] [Google Scholar]
- Rentsch C, Hess R, Rentsch B, Hofmann A, Manthey S, Scharnweber D, Biewener A, Zwipp H. Ovine bone marrow mesenchymal stem cells: isolation and characterization of the cells and their osteogenic differentiation potential on embroidered and surface-modified polycaprolactone-co-lactide scaffolds. In Vitro Cell Dev Biol Anim. 2010;46:624–634. doi: 10.1007/s11626-010-9316-0. [DOI] [PubMed] [Google Scholar]
- McCarty RC, Gronthos S, Zannettino AC, Foster BK, Xian CJ. Characterisation and developmental potential of ovine bone marrow derived mesenchymal stem cells. J Cell Physiol. 2009;219:324–333. doi: 10.1002/jcp.21670. [DOI] [PubMed] [Google Scholar]
- Zscharnack M, Poesel C, Galle J, Bader A. Low oxygen expansion improves subsequent chondrogenesis of ovine bone-marrow-derived mesenchymal stem cells in collagen type I hydrogel. Cells Tissues Organs. 2009;190:81–93. doi: 10.1159/000178024. [DOI] [PubMed] [Google Scholar]
- Niemeyer P, Fechner K, Milz S, Richter W, Suedkamp NP, Mehlhorn AT, Pearce S, Kasten P. Comparison of mesenchymal stem cells from bone marrow and adipose tissue for bone regeneration in a critical size defect of the sheep tibia and the influence of platelet-rich plasma. Biomaterials. 2010;31:3572–3579. doi: 10.1016/j.biomaterials.2010.01.085. [DOI] [PubMed] [Google Scholar]
- Kunisaki SM, Fuchs JR, Steigman SA, Fauza DO. A comparative analysis of cartilage engineered from different perinatal mesenchymal progenitor cells. Tissue Eng. 2007;13:2633–2644. doi: 10.1089/ten.2006.0407. [DOI] [PubMed] [Google Scholar]
- Martinez-Lorenzo MJ, Royo-Canas M, Alegre-Aguaron E, Desportes P, Castiella T, Garcia-Alvarez F, Larrad L. Phenotype and chondrogenic differentiation of mesenchymal cells from adipose tissue of different species. J Orthop Res. 2009;27:1499–1507. doi: 10.1002/jor.20898. [DOI] [PubMed] [Google Scholar]
- Rhodes NP, Srivastava JK, Smith RF, Longinotti C. Heterogeneity in proliferative potential of ovine mesenchymal stem cell colonies. J Mater Sci Mater Med. 2004;15:397–402. doi: 10.1023/b:jmsm.0000021109.21807.f0. [DOI] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, Deans R, Keating A, Prockop D, Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- Wang X, Moutsoglou D. Osteogenic and adipogenic differentiation potential of an immortalized fibroblast-like cell line derived from porcine peripheral blood. In Vitro Cell Dev Biol Anim. 2009;45:584–591. doi: 10.1007/s11626-009-9231-4. [DOI] [PubMed] [Google Scholar]
- Spaas JH, Schauwer CD, Cornillie P, Meyer E, Soom AV, Van de Walle GR. Culture and characterisation of equine peripheral blood mesenchymal stromal cells. Vet J. 2012. in press. [DOI] [PubMed]
- Wang H, Pang B, Li Y, Zhu D, Pang T, Liu Y. Dexamethasone has variable effects on mesenchymal stromal cells. Cytotherapy. 2012;14:423–430. doi: 10.3109/14653249.2011.652735. [DOI] [PubMed] [Google Scholar]
- Radcliffe CH, Flaminio MJ, Fortier LA. Temporal analysis of equine bone marrow aspirate during establishment of putative mesenchymal progenitor cell populations. Stem Cells Dev. 2010;19:269–282. doi: 10.1089/scd.2009.0091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rallapalli S, Bishi DK, Verma RS, Cherian KM, Guhathakurta S. A multiplex PCR technique to characterize human bone marrow derived mesenchymal stem cells. Biotechnol Lett. 2009;31:1843–1850. doi: 10.1007/s10529-009-0106-2. [DOI] [PubMed] [Google Scholar]
- Chong PP, Selvaratnam L, Abbas AA, Kamarul T. Human peripheral blood derived mesenchymal stem cells demonstrate similar characteristics and chondrogenic differentiation potential to bone marrow derived mesenchymal stem cells. J Orthop Res. 2012;30:634–642. doi: 10.1002/jor.21556. [DOI] [PubMed] [Google Scholar]
- Kassis I, Zangi L, Rivkin R, Levdansky L, Samuel S, Marx G, Gorodetsky R. Isolation of mesenchymal stem cells from G-CSF-mobilized human peripheral blood using fibrin microbeads. Bone Marrow Transplant. 2006;37:967–976. doi: 10.1038/sj.bmt.1705358. [DOI] [PubMed] [Google Scholar]
- Tondreau T, Meuleman N, Delforge A, Dejeneffe M, Leroy R, Massy M, Mortier C, Bron D, Lagneaux L. Mesenchymal stem cells derived from CD133-positive cells in mobilized peripheral blood and cord blood: proliferation, Oct4 expression, and plasticity. Stem Cells. 2005;23:1105–1112. doi: 10.1634/stemcells.2004-0330. [DOI] [PubMed] [Google Scholar]
- Mrugala D, Bony C, Neves N, Caillot L, Fabre S, Moukoko D, Jorgensen C, Noel D. Phenotypic and functional characterisation of ovine mesenchymal stem cells: application to a cartilage defect model. Ann Rheum Dis. 2008;67:288–295. doi: 10.1136/ard.2007.076620. [DOI] [PubMed] [Google Scholar]
- Bian ZY, Li G, Gan YK, Hao YQ, Xu WT, Tang TT. Increased number of mesenchymal stem cell-like cells in peripheral blood of patients with bone sarcomas. Arch Med Res. 2009;40:163–168. doi: 10.1016/j.arcmed.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Ranera B, Lyahyai J, Romero A, Vazquez FJ, Remacha AR, Bernal ML, Zaragoza P, Rodellar C, Martin-Burriel I. Immunophenotype and gene expression profiles of cell surface markers of mesenchymal stem cells derived from equine bone marrow and adipose tissue. Vet Immunol Immunopathol. 2011;144:147–154. doi: 10.1016/j.vetimm.2011.06.033. [DOI] [PubMed] [Google Scholar]
- Ranera B, Ordovás L, Lyahyai J, Bernal ML, Fernandes F, Romero A, Vázquez FJ, Osta R, Cons C, Varona L, Zaragoza P, Martín-Burriel I, Rodellar C. Comparative study of equine bone marrow- and adipose tissue-derived mesenchymal stem cells. Equine Vet J. 2012;44:33–42. doi: 10.1111/j.2042-3306.2010.00353.x. [DOI] [PubMed] [Google Scholar]
- Seo JB, Moon HM, Kim WS, Lee YS, Jeong HW, Yoo EJ, Ham J, Kang H, Park MG, Steffensen KR, Stulnig TM, Gustafsson JA, Park SD, Kim JB. Activated liver X receptors stimulate adipocyte differentiation through induction of peroxisome proliferator-activated receptor gamma expression. Mol Cell Biol. 2004;24:3430–3444. doi: 10.1128/MCB.24.8.3430-3444.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aguilar V, Annicotte JS, Escote X, Vendrell J, Langin D, Fajas L. Cyclin G2 regulates adipogenesis through PPAR gamma coactivation. Endocrinology. 2010;151:5247–5254. doi: 10.1210/en.2010-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menssen A, Haupl T, Sittinger M, Delorme B, Charbord P, Ringe J. Differential gene expression profiling of human bone marrow-derived mesenchymal stem cells during adipogenic development. BMC Genomics. 2011;12:461. doi: 10.1186/1471-2164-12-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YC, Ntambi JM. Regulation of stearoyl-CoA desaturase genes: role in cellular metabolism and preadipocyte differentiation. Biochem Biophys Res Commun. 1999;266:1–4. doi: 10.1006/bbrc.1999.1704. [DOI] [PubMed] [Google Scholar]
- Pricola KL, Kuhn NZ, Haleem-Smith H, Song Y, Tuan RS. Interleukin-6 maintains bone marrow-derived mesenchymal stem cell stemness by an ERK1/2-dependent mechanism. J Cell Biochem. 2009;108:577–588. doi: 10.1002/jcb.22289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song L, Webb NE, Song Y, Tuan RS. Identification and functional analysis of candidate genes regulating mesenchymal stem cell self-renewal and multipotency. Stem Cells. 2006;24:1707–1718. doi: 10.1634/stemcells.2005-0604. [DOI] [PubMed] [Google Scholar]
- Gronthos S, Mrozik K, Shi S, Bartold PM. Ovine periodontal ligament stem cells: isolation, characterization, and differentiation potential. Calcif Tissue Int. 2006;79:310–317. doi: 10.1007/s00223-006-0040-4. [DOI] [PubMed] [Google Scholar]
- Jikko A, Harris SE, Chen D, Mendrick DL, Damsky CH. Collagen integrin receptors regulate early osteoblast differentiation induced by BMP-2. J Bone Miner Res. 1999;14:1075–1083. doi: 10.1359/jbmr.1999.14.7.1075. [DOI] [PubMed] [Google Scholar]
- Liu F, Akiyama Y, Tai S, Maruyama K, Kawaguchi Y, Muramatsu K, Yamaguchi K. Changes in the expression of CD106, osteogenic genes, and transcription factors involved in the osteogenic differentiation of human bone marrow mesenchymal stem cells. J Bone Miner Metab. 2008;26:312–320. doi: 10.1007/s00774-007-0842-0. [DOI] [PubMed] [Google Scholar]
- Zou L, Zou X, Chen L, Li H, Mygind T, Kassem M, Bunger C. Multilineage differentiation of porcine bone marrow stromal cells associated with specific gene expression pattern. J Orthop Res. 2008;26:56–64. doi: 10.1002/jor.20467. [DOI] [PubMed] [Google Scholar]
- Sollazzo V, Palmieri A, Scapoli L, Martinelli M, Girardi A, Pellati A, Scarano A, Perrotti V, Spinelli G, Carinci F. Allogro® acts on stem cells derived from peripheral blood. The Internet Journal of Dental Science. 2009;8 [Google Scholar]
- Aubin JE. Bone stem cells. J Cell Biochem Suppl. 1998;30-31:73–82. [PubMed] [Google Scholar]
- Zscharnack M, Hepp P, Richter R, Aigner T, Schulz R, Somerson J, Josten C, Bader A, Marquass B. Repair of chronic osteochondral defects using predifferentiated mesenchymal stem cells in an ovine model. Am J Sports Med. 2010;38:1857–1869. doi: 10.1177/0363546510365296. [DOI] [PubMed] [Google Scholar]
- Jager M, Bachmann R, Scharfstadt A, Krauspe R. Ovine cord blood accommodates multipotent mesenchymal progenitor cells. In Vivo. 2006;20:205–214. [PubMed] [Google Scholar]
- Woodbury D, Schwarz EJ, Prockop DJ, Black IB. Adult rat and human bone marrow stromal cells differentiate into neurons. J Neurosci Res. 2000;61:364–370. doi: 10.1002/1097-4547(20000815)61:4<364::AID-JNR2>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Jori FP, Napolitano MA, Melone MA, Cipollaro M, Cascino A, Altucci L, Peluso G, Giordano A, Galderisi U. Molecular pathways involved in neural in vitro differentiation of marrow stromal stem cells. J Cell Biochem. 2005;94:645–655. doi: 10.1002/jcb.20315. [DOI] [PubMed] [Google Scholar]
- Loubet D, Dakowski C, Pietri M, Pradines E, Bernard S, Callebert J, Ardila-Osorio H, Mouillet-Richard S, Launay JM, Kellermann O, Schneider B. Neuritogenesis: the prion protein controls beta1 integrin signaling activity. FASEB J. 2012;26:678–690. doi: 10.1096/fj.11-185579. [DOI] [PubMed] [Google Scholar]
- Montzka K, Lassonczyk N, Tschoke B, Neuss S, Fuhrmann T, Franzen R, Smeets R, Brook GA, Woltje M. Neural differentiation potential of human bone marrow-derived mesenchymal stromal cells: misleading marker gene expression. BMC Neurosci. 2009;10:16. doi: 10.1186/1471-2202-10-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takakura Y, Yamaguchi N, Nakagaki T, Satoh K, Kira J, Nishida N. Bone marrow stroma cells are susceptible to prion infection. Biochem Biophys Res Commun. 2008;377:957–961. doi: 10.1016/j.bbrc.2008.10.099. [DOI] [PubMed] [Google Scholar]
- Joint Working Group on Refinement. Removal of blood from laboratory mammals and birds. First report of the BVA/FRAME/RSPCA/UFAW Joint Working Group on Refinement. Lab Anim. 1993;27:1–22. doi: 10.1258/002367793781082412. [DOI] [PubMed] [Google Scholar]
- Dervishi E, Serrano C, Joy M, Serrano M, Rodellar C, Calvo JH. Effect of the feeding system on the fatty acid composition, expression of the Delta9-desaturase, Peroxisome Proliferator-Activated Receptor Alpha, Gamma, and Sterol Regulatory Element Binding Protein 1 genes in the semitendinous muscle of light lambs of the Rasa Aragonesa breed. BMC Vet Res. 2010;6:40. doi: 10.1186/1746-6148-6-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Crespo D, Juste RA, Hurtado A. Selection of ovine housekeeping genes for normalisation by real-time RT-PCR; analysis of PrP gene expression and genetic susceptibility to scrapie. BMC Vet Res. 2005;1:3. doi: 10.1186/1746-6148-1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyahyai J, Serrano C, Ranera B, Badiola JJ, Zaragoza P, Martin-Burriel I. Effect of scrapie on the stability of housekeeping genes. Anim Biotechnol. 2010;21:1–13. doi: 10.1080/10495390903323851. [DOI] [PubMed] [Google Scholar]