Abstract
India is known for its vast human diversity, consisting of more than four and a half thousand anthropologically well-defined populations. Each population differs in terms of language, culture, physical features and, most importantly, genetic architecture. The size of populations varies from a few hundred to millions. Based on the social structure, Indians are classified into various caste, tribe and religious groups. These social classifications are very rigid and have remained undisturbed by emerging urbanisation and cultural changes. The variable social customs, strict endogamy marriage practices, long-term isolation and evolutionary forces have added immensely to the diversification of the Indian populations. These factors have also led to these populations acquiring a set of Indian-specific genetic variations responsible for various diseases in India. Interestingly, most of these variations are absent outside the Indian subcontinent. Thus, this review is focused on the peopling of India, the caste system, marriage practice and the resulting health and forensic implications.
Keywords: Admixture, caste, Indians, mtDNA, tribe, Y-chromosome
Review
Introduction
India is well-known for its human and geographical diversities. It has a variety of landscapes ranging from desert to evergreen forest, fertile plains to southern dry plateaus, shallow swamps to deep bays, and lowlands to the high Himalayas. They all give refuge to diverse populations of humans, plants, animals and microbes. India also has extensive river systems that feed the fertile plains and provide adequate food. It is bordered by Nepal, China and Bhutan in the north, Burma and Bangladesh in the east, and Pakistan in the west. There are 4,635 anthropologically well-defined human populations; 532 are tribes, including 72 ancestral tribes with the hunter and gatherer lifestyle [1]. The size of these populations ranges from a few hundred to several millions. The total population count of India has reached 1.21 billion [2].
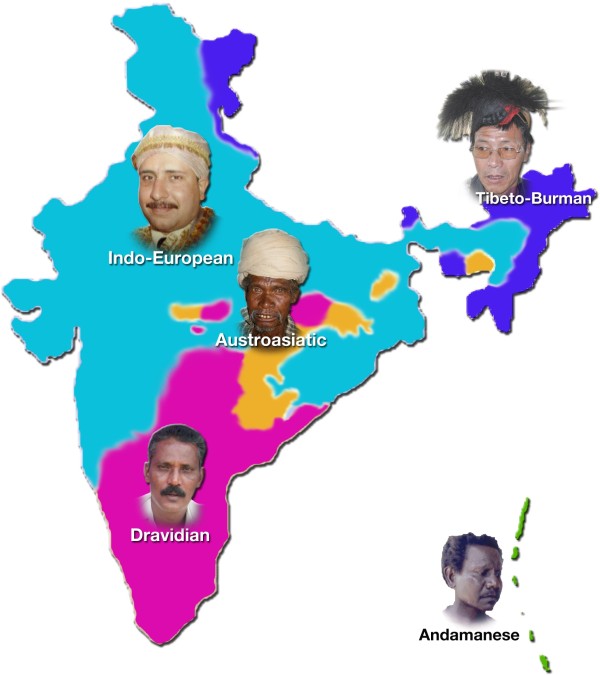
Linguistically, Indians are classified into four major language families; Indo-European, Dravidian, Austroasiatic and Tibeto-Burman (Figure 1). Indo-European is the most widely spoken language family in India, particularly in northern, central and western India. Dravidian speakers are mainly confined to southern parts of the country. Austroasiatic speakers are dispersed mostly in the central and eastern parts, while the Tibeto-Burman speakers are concentrated in and around the foothills of the Himalayas and north east states (Figure 1). In addition to these major language families, there are a few isolated languages, such as Andamanese [3,4], spoken by the enigmatic tribal populations of Andaman and Nicobar Islands, and Nihali spoken in the pocket of the Western Ghats.
Figure 1.
Geographical distribution of different language families in India.
There are various evidences supporting the peopling of India by the early modern humans. It is well established that the modern human originated in Africa about 200,000 years before present (YBP) [5,6]. They started migrating out-of-Africa between 55,000 and 85,000 YBP. There are several thoughts regarding the cause and timing of this migration. One compelling view being put forward is based on geological finding. It states that there was a mega drought in East Africa between 135,000 and 75,000 YBP, when the water volume of Lake Malawi was reduced by at least 95% [7]. The timing of this mega drought corresponds with the timing of the exodus of anatomically modern humans out-of-Africa along the southern coastal route [4,8]. The firm establishment of the southern coastal route of modern human migration reveals India as a major corridor for early human migration. The anthropological, historical, linguistic and genetic evidence for early peopling of India is found imprinted all over the country.
Recently, archaeological evidence supporting the early peopling of India was discovered in the Kurnool district of Andhra Pradesh, one of the southern Indian states [9,10]. This study shows that the country was inhabited by modern humans before and after the Toba eruption around 74,000 YBP. The evidence is in the form of stone tools. The stone tools of this study most likely resemble contemporaneous Homo sapiens technologies in Africa. Further, a partial cranium recovered from Narmada Basin was dated back to around 300,000 to 250,000 YBP [11,12]. Over the past two decades, several independent studies have been carried out in various Indian populations with ancient and modern DNA using haploid and diploid markers. Almost all the studies found signs of early settlement by the first group of modern human venturing out-of-Africa and very recent gene flow from west and east Eurasia [4,8,13-34].
There has been tremendous interest among historians, archaeologists, anthropologists, linguists and geneticists to understand the unique structure of Indian populations and their affinities with the rest of the world. Most importantly, researchers working on various diseases often find that disease-causing genetic variations are different in Indian populations. During the last two decades, many exciting observations have been made regarding Indian people by several investigators; however, these findings have remained scattered. Thus, we have made an attempt to present an overview of the peopling of India, the caste system, endogamous marriage practice, and the resulting health and forensic implications.
Evidence of the first out-of-Africa human in the Indian subcontinent
An interesting finding about the Indian population is the evidence of the early settlement in Andaman and Nicobar Islands by a group of modern humans, whose ancestors made the first journey out-of-Africa. The Andaman and Nicobar Islands are located in the Bay of Bengal (Figure 1). The study of the isolated people of these islands provides important clues to the evolution and dispersal of early modern humans. There live two groups of tribes in the Andaman and Nicobar Islands: those who share physical features with African pygmies and other similarly featured Asian people, such as short stature, dark skin, peppercorn hair, and scant body hair; and those who share physical features with the Chinese, Malays and Burmese. Linguistically, the people of Andaman Islands can be divided into Little Andaman and the Great Andaman groups. The Little Andaman group is divided into three branches, the Onge, Sentinelese and Jarawa. The Great Andaman group is divided into the Northern and Southern groups [3].
There are fewer archaeological and genetic records on the origin of Andaman Islanders. Our group [35] and Endicott and his group [15] undertook parallel studies on the people of Andaman and Nicobar Islands. The latter group dealt with the ancient remains of these populations using mitochondrial DNA (mtDNA), while we studied modern DNA using mtDNA and Y-chromosomal markers [4,35]. Surprisingly, both of the studies established Andamanese affinity with Asian populations. Subsequently, we performed a high resolution study using complete mtDNA of the Onge and Greater Andamanese. We found unique mutations in their mtDNA. Hence, we have assigned two novel haplogroups, M31 and M32, and estimated the age of these haplogroups to be about 65,000 years. This suggests an ancient origin of the people of Andaman and Nicobar Islands and long-term isolation [4]. Even though researchers have come forward with different time estimates on the peopling of Andaman and Nicobar Islands [36], the early peopling of this region cannot be ruled out [36,37]. Recently, our study using 560,123 autosomal SNPs suggested a unique genetic identity of the Andamanese and ancient isolation from mainland south Asian populations. However, they shared genetic affinities with ancient south Indians (ASI) [30].
The caste system in India
India is a land of social stratifications, such as castes, tribes and religious groups. Although the precise date of origin for the caste system in India is unclear, the written evidence about the organisation of the caste system exists in the Rig Veda, which was written between 1700 and 1100 BC [38]. Caste is a social hierarchy based on occupation. There are four broad categories of castes in Hindu society: Brahmin, Kshatriya, Vaishya and Sudra. Each caste is known to perform a specific duty. Brahmins perform rituals and are in charge of teaching society; Kshatriyas are rulers and warriors, and are involved in ruling and defending the territories; Vaishyas are cultivators and businessmen; Sudras rank last in society and are labourers by profession. Each caste is further subdivided into smaller units generally known as subcastes, which in turn are further divided into multiple exogamous clans known as Gotras. The caste system governs all social, religious and economic activities of the people. The long-term social boundaries and endogamy practice among all social groups has given birth to diverse, population-specific social traditions and the development of distinct linguistic dialects [39]. The divergent endogamous cultural and social structures are helpful in understanding genetic variation among the populations and their ancestry [39].
There are several genetic studies on the caste systems in India based on mtDNA and Y-chromosome markers [16,19,33,34,40-44]. These studies reveal that the origin of the caste system is mainly rooted in male-mediated Indo-Aryan migration that pushed indigenous Dravidian speaking populations towards southern India and Sri Lanka, and suggest that the Indo-Aryans established themselves as the upper caste [39]. Further, it has been shown that the caste populations are closer to Europeans and Central Asians and differ significantly from tribal populations [34,39,43,45]. These studies succeeded in drawing a trend to investigate the integrity of the caste system. Most of the researchers started by including the Indo-Aryan invasion concept in their studies, assuming that it was a universally accepted and proven fact [33,46-50].
Emergence of caste system in India and its amalgamation with the waves of migrations
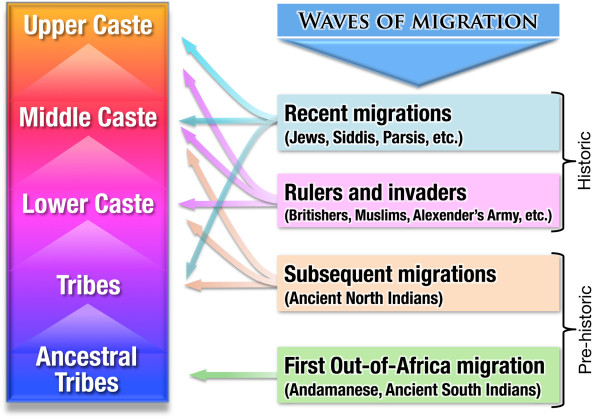
A noteworthy view can be put forward on the development and maintenance of the caste system on the basis of genetic observations (Figure 2) [33,51,52]. The ancestral tribes might have given birth to various subtribes over time. Some of the subtribes might have migrated and gradually established themselves as lower caste groups through better knowledge of procuring necessary resources. Further, the empowerments of some of the lower caste group might have helped them to establish themselves as middle caste groups. Increased mastery over technological and economic measures among some of the middle caste group might have facilitated attainment of the upper caste level (Figure 2), thus giving rise to a complete caste system. Hence, a person’s profession became the symbol of the caste to which he or she belonged. As time passed, the caste system might have fortified itself in society by dividing populations into several endogamous pockets, and has undoubtedly played an essential role in shaping present day Indian genetic architecture. Furthermore, India has witnessed several waves of immigrations. The immigrants were absorbed into different hierarchal levels of the caste system (Figure 2).
Figure 2.
Schematic showing the emergence of the caste system in India and its amalgamation with the waves of migrations.
No support for the Aryan invasion
Even though there is a continued debate on the Aryan migration into India, detectable gene flow from west Eurasia has been shown by many studies [13,16,23,24,30-32,44,51,53]. Interestingly, we have detected gene flow from the west prior to the Aryan invasion [30,32]. There is now universal agreement that various Indian populations share a common late Pleistocene maternal and paternal ancestry, along with detectable east and west Eurasian ancestries [31,54]. Using hundreds of thousands of autosomal markers, we illustrated that the Indian populations have two major distinct ancestry components; one restricted to southern India, the second one restricted to the northern region of India [30,32]. It is noteworthy that both of the ancestry components show higher haplotypic diversity than those predominant in west Eurasia [32]. This rejects the idea of an Aryan invasion/migration and suggests an ancient demographic history and/or higher long-term larger effective population size in India than in west Eurasia.
Genetic structure of ancient India
Indigenous population
During the past two decades, we have witnessed remarkable advancements in technology. We have advanced from low resolution genetic markers to high throughput whole genome sequencing. Despite these advancements, studies using high density markers were lacking in the Indian scenario. Therefore, we made an attempt to extensively study various Indian populations using Affymetrix (SNP 6.0) array [30].
We predicted that the present day Indian populations might have arisen from a relatively small group of ancestors with limited gene flow and long term isolation. We also predicted that there might have been two ancestral groups in prehistoric India: an ancestral North Indian (ANI) population distantly related to those in the Middle East, Central Asia and Europe (30% to 70%), and an ASI population not related to groups outside India. We estimated that the ASI populations showed 39% to 71% ANI ancestry. Thus, the extant Indian populations are admixtures of both ANI and ASI. Interestingly, indigenous Andaman Islanders are the only ASI-related groups without ANI ancestry [30].
Recent migrants
In addition to the large number of indigenous populations, India has experienced immigration of several populations in the past, further adding to the complexities of Indian population structure. Among the recent migrants, we have extensively studied Siddi, Muslim and Jewish populations inhabiting India.
The Siddis are mainly found in three Indian states - Gujarat, Karnataka and Andhra Pradesh -and have typical African features such as dark skin, curly hair, broad nose, and so on. Historically, it is known that they were brought to India by Portuguese traders between the 17th and 19th centuries and sold to the Nawabs and the Sultans of India to serve as soldiers and slaves. Earlier, we showed the presence of a Y-chromosome Alu insertion; an African-specific marker, in 40% of Siddis [55]. The mitochondrial DNA hypervariable sequence also confirmed the Siddis’ affinity with Africans. However, there was no high resolution study pertaining to the origin, affinity and genetic structure of the Indian Siddis [56-58]. In an attempt to reveal these issues together, we screened the Siddi populations from the Junagarh district of Gujarat and Uttara Kannad district of Karnataka using mtDNA and Y-chromosomal and high density autosomal markers. Along with Siddis, six populations with geographical proximity to Siddis have also been studied to capture a scenario of gene flow. Our analysis revealed that the Siddi population has a combined ancestry (that is, 70% African and 30% Indian and European) [24]. We further estimated that the Siddis have admixed with the neighbouring Indian populations for about 200 years ago (eight generations). Our genetic finding coincides with the historical record of the arrival of Siddi people in India [24].
Y-chromosome results revealed the presence of African-specific haplogroups. Further, extensive investigation on Y-STRs revealed that the Siddis are the direct descendants of the Bantu-speakers of sub-Saharan Africa. We observed an Indian-specific gene pool in Siddis but the Siddi-specific gene pool was not observed in neighbouring Indian populations. This firmly suggests the unidirectional gene flow from the Indian population to the Siddis, confirming the rigidity of the Indian social structure [24]. We also predicted that the approximate number of males Siddis who reached India between the 17th and 19th centuries was about 1,500.
Another well-documented recent migration is that of the Muslim population. The Arab Muslims established their first kingdom in India by conquering Sindh in 711 AD [59,60]. During the 13th century, a Turkic kingdom was established in Delhi and in the 16th century the famous Mughal Empire appeared in India. The Muslim immigrations were especially male-mediated in the form of invaders from Iran and Arabia. The majority of the present day Indian Muslims are believed to be the descendants of converts from local Hindu (caste and tribal) populations. Thus, we undertook an extensive study among Indian Muslims from diverse geographical regions to trace their genetic structure, origin and affinity using Y-chromosome and mtDNA markers. Even though we observed their genetic affinity with indigenous Indian non-Muslim populations, a small frequency of the Middle Eastern ancestry was also noticed. Therefore, the genetic analysis of Indian Muslims shows the spread of Muslims in India was mainly mediated by cultural adaptation linked with minor gene flow from the Middle East [61,62].
Discrepancy between linguistic affiliation and genetic signature
Each linguistic group in India has a strong genetic affinity between its members. Therefore, any recent change in language could be reflected in the genome. When people move from one place to another, as per human tendency, they try to adapt themselves to the new place. Over time, some of the migrants adopt the local language for better communication and, in turn, for better living. Thus, language shift is a phenomenon where a new language is adopted by a population with virtually no influence on their genetic make-up. There are several examples of language shift around the world [63-65]. In India, we have documented two case studies, where linguistic affiliation and gene pool reveal different scenarios.
In India, the first evidence of inconsistency between language and gene pool was noticed by us among the Hindi (Indo-European)-speaking Mushar population [52]. mtDNA and Y-chromosomal datasets of this population were compared with those of the neighbouring Indo-European- and Austroasiatic-speaking populations of similar traditional social status. mtDNA analysis revealed the presence of both Austroasiatic and Indo-European haplogroups. However, the Austroasiatic-specific haplogroups were observed in higher frequencies. A similar scenario was noticed with Y-chromosomal marker data. Further, principal component analysis revealed a closer genetic affinity to the Austroasiatic (Mundari) populations than to the neighbouring Hindi-speaking populations. This showed that the Mushar population was originally an Austroasiatic group (genetically) but linguistically, it is an Indo-European population [52]. The rapid loss of original tongue was facilitated by the schooling of Mushar children in the Indo-European language-based schools [52].
The second evidence of discrepancy was observed among the Bharia tribe of the central Indian state of Madhya Pradesh. We carried out an extensive characterization of genetic ancestry of Bharia, a Dravidian speaking group, and its Indo-European-speaking neighbours (Bhil and Sahariya) along with other Austroasiatic-speaking groups. mtDNA analysis showed the presence of lineages that are commonly found among the Austroasiatic groups [54,66]. In addition, Y-chromosomal biallelic markers also revealed a high frequency of the Austroasiatic-specific haplogroup in Bharia. Further, principal component analysis showed a strong genetic affinity of Bharia with the Austroasiatic (Munda) group [23]. The gene flow from the Munda group was also confirmed by analysis of the Y-STRs haplotype sharing. The autosomal markers also established the Austroasiatic gene pool among Bharia. Hence, in a landscape like India, the linguistic label of a population does not unequivocally follow the genetic footprints [23].
The origin of Austroasiatic speakers
There are 104 million Austroasiatic speakers around the globe. There exist two major branches of the Austroasiatic language in India. The Munda branch is mostly spoken by eastern, northeastern, and central Indians, and the Khasi-Aslian branch is located in Meghalaya and (Andaman) Nicobar Islands [67]. There are disputes on the geographic origin of the Austroasiatic language family [27,68-71]. The first hypothesis places the origin of Austroasiatic speakers in Southeast Asia with a later dispersal to South Asia during the Neolithic period [72]. The second hypothesis advocates pre-Neolithic origins and dispersal of this language family from South Asia [68]. The main lacuna in previous studies was the absence of extensive sampling from both Southeast Asia and India, and a smaller number of markers used. Therefore, to test the two alternative models, we performed combined analysis of uniparentally inherited markers and one million common SNP loci from the nuclear genome [27]. Our result showed higher diversity of haplogroup M95-O2a with an older coalescent time of 17,000 to 28,000 years ago in Southeast Asia samples than in their Indian counterparts, supporting the first of the two hypotheses. A principal component analysis and structure-like analysis on autosomal loci showed a more complex population history of Austroasiatic speakers in India. Thus, we propose that the Austroasiatic speakers in India dispersed from Southeast Asia, followed by extensive sex-specific admixture with local Indian populations [27].
Endogamy as a barrier to the general perception about patrilocality and matrilocality
The presence of both patrilocal and matrilocal societies further adds to the complexities of the Indian social structure. The societies in which a woman moves to her husband’s home after marriage are called patrilocal societies, and the societies in which a man moves to his wife’s home is a matrilocal society. Thus, these social customs are expected to have an impact on the patterns of genetic variation among the population of a given region. It is indeed observed that there is localization of male-specific Y-chromosomal variants and wide dispersal of female-specific mtDNA variants in patrilocal societies and vice versa in matrilocal societies [73-75]. However, Indian societies are strictly endogamous and have very rigid social customs. Therefore, marriage always takes places within a population, and these different patterns of genetic variations among patrilocal and matrilocal Indian populations are not the expected Indian scenario. We, along with other researchers [76], have analysed 10 patrilocal and five matrilocal Indian populations. We collectively suggest that there is an insignificant difference between the patrilocal and matrilocal societies. Hence, the patterns of genetic variation in humans are not always universal, but depend on local cultural practice [76].
Unique genetic variations in Indian populations and their implications
Implications in health and disease
Several boundaries acting at different levels within the Indian populations have added to the human diversity in the country. Indian populations have been shaped by a long-term genetic isolation of different populations that predates the caste system in India. The allele frequency differences between the groups in India are larger than those in Europe, reflecting strong founder effects whose signatures have been preserved for thousands of years due to firm social customs and strict endogamy. We therefore predict that there is an excess of population and region-specific diseases in India. This prediction is well correlated with several population- or region-specific diseases and medical conditions, for example, Madras motor neuron disease [77,78], Handigodu disease [79,80], and pseudocholinesterase deficiency among Vysyas [81,82]. Further, our study on high density markers in 142 samples from 30 ethnic groups in India predicted the positive selection of MSTN and DOK5 genes, which have potential implications in lipid metabolism and the aetiology of type 2 diabetes in Indians [32].
Our previous study revealed that the deletion of 25 bp in the myosin binding protein-C3 gene (MYBPC3) is associated with inheritable cardiomyopathies in India due to skipping of wild type exon 33 [83]. We also demonstrated that this mutation accounts for 45% of cardiac deaths in India from sudden heart attack. This mutation is widely distributed (4.5%) across India, but was not found in the tribes of Andaman and Nicobar Islands and northeastern populations of India, who have different ancestry. We have screened this mutation in the worldwide population, representing 26 countries, and found this mutation only in the populations of India, Pakistan, Sri Lanka, Indonesia and Malaysia (South and Southeast Asian countries). We further estimated that this mutation probably originated in India about 33,000 years ago.
We have also observed the implications of the admixture of ethnically diverse populations, who have undergone selection processes through environmental influence. The best example is the Siddis, the Indo-African population, whose admixture with the neighbouring Indian populations has remarkable medical and social implications. The A-variant of the G6PD gene that gives protection against malaria is commonly found in Africa. Even though the Siddis have African origin, the A-variant was found only in 10% of the Siddi individuals. This is primarily because of their admixture with the neighbouring Indian populations, which in turn has diluted the frequency of the A-variant. This makes the present day Siddis more susceptible to malaria than their African ancestors.
The differences in disease rates between populations are largely due to the frequency differences in disease-causing genetic variants. It is said that these genetic variants occur frequently on chromosome segments inherited from the ancestral population with the higher disease-variant frequency [84]. Thus, efficient mapping of disease-causing variants are improved if the divergence between the ancestral populations is large. A large difference in the prevalence of the disease between the ancestral and derived populations adds to the successfully mapping of disease-causing variants, which is more often observed in the case of the admixed populations [85]. A study on the Indian populations spanning African descendants has shown the importance of admixture in mapping the diseases [85]. A significant enrichment of processes related to ion-channel activity and cadherin genes was found. Selection in ion channel genes among populations of African ancestry can be explained by the fact that this population resides in an extremely saline region of the country [85]. This finding was similar to results regarding African Americans, where the family of genes related to ion channels, cadherins, and calmodulins, had disproportionate influence on hypertension and associated complications [86].
Forensic implication
The structure of the Indian population also has implications in forensic genetics. Highly polymorphic STR markers provided by Applied Biosystems, Foster City, CA, USA, and Promega, Madison, WI, USA are being extensively used for medicolegal purposes. The abilities of these markers to differentiate individuals are quite efficient. However, the strict endogamy practices and small population size of some Indian populations has presented a potential challenge to the efficiency of these methods. For example, three Y-STRs loci were found to be monomorphic for Onges, and one for Nicobarese people [35]. There may be more cases in different endogamous Indian populations. Therefore, it is difficult to distinguish individuals using only commercially available marker sets. Thus, further refinement of commonly used STRs is necessary to differentiate each individual to its best possible resolution.
The amelogenin gene, which is present on both X and Y chromosomes differs in size. This fact has been widely used in forensic casework and prenatal diagnosis to identify sex; however, regarding the same marker among the diverse Indian populations, we have observed a deletion of Y-chromosome-specific amelogenin in 1.85% of Indian males [87]. These individuals would have been considered female through routine analysis, thus creating a massive misinterpretation of fact. Therefore, considering the false consequences of the result obtained by only using the amelogenin marker, we suggest the use of additional Y-chromosome markers for unambiguous gender identification.
In a mass disaster or crime investigation, genetic markers play a major role in victim identification. We have shown earlier that every Indian population forms a separate cluster [30]. Therefore, by using a larger number of SNPs (for example, Affymetrix 6.0 array), it is possible to distinguish every population involved. The Y-chromosomal and mitochondrial markers used in classifying the individuals into haplogroups can also be used to elucidate additional facts.
Conclusions
Based on the extensive genetic studies carried out on different Indian populations, we infer that each of them is a genetically distinct ethnic population in part due to high levels of endogamy. High resolution genetic studies revealed the in situ origin of several deep-rooting mtDNA lineages in India, suggesting that Indian populations are genetically unique and harbour the second highest genetic diversity after Africans. The genomic complexity brought on by endogamous practice for thousands of years, language shifts and sex-specific admixture are highly challenging to study and need further, extensive genetic characterization. The complex genomic architecture of Indian populations adds difficulty in understanding diseases and implementing preventive measures. The accumulation of various mutations due to endogamy leads to recessive diseases in the Indian population, which further increases the total disease burden of the country. Several questions pertaining to the origin of the caste system, the arrival and spread of the major language families and finding disease-associated genetic variants require effective approaches combining various disciplines such as archaeology, historical linguistics, genetics, bioinformatics and pharmacology. Even though studies with high density markers have added much to the knowledge about the Indian populations, whole genome approaches are expected to answer many of the existing questions.
Abbreviations
ANI: ancestral North Indian; ASI: ancestral South Indian; bp: base pairs; mtDNA: mitochondrial DNA; SNP: single nucleotide polymorphism; STR: short tandem repeat; YBP: years before present.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RT and KT wrote the manuscript and performed graphical work. Both the authors read and approved the final manuscript.
Contributor Information
Rakesh Tamang, Email: tamang@ccmb.res.in.
Kumarasamy Thangaraj, Email: thangs@ccmb.res.in.
Acknowledgements
We thank all our national and international collaborators for their continuous cooperation and efficient team work. Appreciation is due for valuable contributions from the students of various universities across the country. We also thank Dr. Gyaneshwer Chaubey for his valuable suggestions, and Dr. B N Sarkar for providing census on tribe/caste ratio. KT is supported by the Council of Scientific and Industrial Research (CSIR), India. The financial support of Department of Biotechnology (DBT), India and Department of Science and Technology (DST), India are gratefully acknowledged.
References
- Census of India. 2001. http://censusindia.gov.in/.
- Census of India. 2011. http://censusindia.gov.in/.
- Abbi A. Endangered languages of the Andaman Islands. München: Lincom Europa; 2006. [Google Scholar]
- Thangaraj K, Chaubey G, Kivisild T, Reddy AG, Singh VK, Rasalkar AA, Singh L. Reconstructing the origin of Andaman Islanders. Science. 2005;308:996. doi: 10.1126/science.1109987. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza LL. The DNA revolution in population genetics. Trends Genet. 1998;14:60–65. doi: 10.1016/S0168-9525(97)01327-9. [DOI] [PubMed] [Google Scholar]
- Yotova V, Lefebvre JF, Kohany O, Jurka J, Michalski R, Modiano D, Utermann G, Williams SM, Labuda D. Tracing genetic history of modern humans using X-chromosome lineages. Hum Genet. 2007;122:431–443. doi: 10.1007/s00439-007-0413-4. [DOI] [PubMed] [Google Scholar]
- Scholz CA, Johnson TC, Cohen AS, King JW, Peck JA, Overpeck JT, Talbot MR, Brown ET, Kalindekafe L, Amoako PY, Lyons RP, Shanahan TM, Castañeda IS, Heil CW, Forman SL, McHargue LR, Beuning KR, Gomez J, Pierson J. East African megadroughts between 135 and 75 thousand years ago and bearing on early-modern human origins. Proc Natl Acad Sci U S A. 2007;104:16416–16421. doi: 10.1073/pnas.0703874104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macaulay V, Hill C, Achilli A, Rengo C, Clarke D, Meehan W, Blackburn J, Semino O, Scozzari R, Cruciani F, Taha A, Shaari NK, Raja JM, Ismail P, Zainuddin Z, Goodwin W, Bulbeck D, Bandelt HJ, Oppenheimer S, Torroni A, Richards M. Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science. 2005;308:1034–1036. doi: 10.1126/science.1109792. [DOI] [PubMed] [Google Scholar]
- Clarkson C, Petraglia M, Korisettar R, Haslam M, Boivin N, Crowther A, Ditchfield P, Fuller D, Miracle P, Harris C, Connell K, James H, Koshy J. The oldest and longest enduring microlithic sequence in India: 35 000 years of modern human occupation and change at the Jwalapuram Locality 9 rockshelter. Antiquity. 2009;83(320):326–348. [Google Scholar]
- Petraglia M, Korisettar R, Boivin N, Clarkson C, Ditchfield P, Jones S, Koshy J, Lahr MM, Oppenheimer C, Pyle D, Roberts R, Schwenninger JL, Arnold L, White K. Middle Paleolithic assemblages from the Indian subcontinent before and after the Toba super-eruption. Science. 2007;317:114–116. doi: 10.1126/science.1141564. [DOI] [PubMed] [Google Scholar]
- Patnaik R, Chauhan P. India at the cross-roads of human evolution. J Biosci. 2009;34:729. doi: 10.1007/s12038-009-0056-9. [DOI] [PubMed] [Google Scholar]
- Patnaik R, Chauhan PR, Rao MR, Blackwell BA, Skinner AR, Sahni A, Chauhan MS, Khan HS. New geochronological, paleoclimatological, and archaeological data from the Narmada Valley hominin locality, central India. J Hum Evol. 2009;56:114–133. doi: 10.1016/j.jhevol.2008.08.023. [DOI] [PubMed] [Google Scholar]
- Kivisild T, Rootsi S, Metspalu M, Mastana S, Kaldma K, Parik J, Metspalu E, Adojaan M, Tolk HV, Stepanov V, Gölge M, Usanga E, Papiha SS, Cinnioğlu C, King R, Cavalli-Sforza L, Underhill PA, Villems R. The genetic heritage of the earliest settlers persists both in Indian tribal and caste populations. Am J Hum Genet. 2003;72:313–332. doi: 10.1086/346068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott P, Metspalu M, Stringer C, Macaulay V, Cooper A, Sanchez JJ. Multiplexed SNP typing of ancient DNA clarifies the origin of Andaman mtDNA haplogroups amongst South Asian tribal populations. PLoS One. 2006;1:e81. doi: 10.1371/journal.pone.0000081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endicott P, Gilbert MT, Stringer C, Lalueza-Fox C, Willerslev E, Hansen AJ, Cooper A. The genetic origins of the Andaman Islanders. Am J Hum Genet. 2003;72:178–184. doi: 10.1086/345487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S, Zhivotovsky LA, King R, Mehdi SQ, Edmonds CA, Chow CE, Lin AA, Mitra M, Sil SK, Ramesh A, Usha Rani MV, Thakur CM, Cavalli-Sforza LL, Majumder PP, Underhill PA. Polarity and temporality of high-resolution y-chromosome distributions in India identify both indigenous and exogenous expansions and reveal minor genetic influence of Central Asian pastoralists. Am J Hum Genet. 2006;78:202–221. doi: 10.1086/499411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Underhill PA, Myres NM, Rootsi S, Metspalu M, Zhivotovsky LA, King RJ, Lin AA, Chow CE, Semino O, Battaglia V, Kutuev I, Järve M, Chaubey G, Ayub Q, Mohyuddin A, Mehdi SQ, Sengupta S, Rogaev EI, Khusnutdinova EK, Pshenichnov A, Balanovsky O, Balanovska E, Jeran N, Augustin DH, Baldovic M, Herrera RJ, Thangaraj K, Singh V, Singh L, Majumder P. et al. Separating the post-Glacial coancestry of European and Asian Y chromosomes within haplogroup R1a. Eur J Hum Genet. 2010;18:479–484. doi: 10.1038/ejhg.2009.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder PP. The human genetic history of South Asia. Curr Biol. 2010;20:R184–R187. doi: 10.1016/j.cub.2009.11.053. [DOI] [PubMed] [Google Scholar]
- Basu A, Mukherjee N, Roy S, Sengupta S, Banerjee S, Chakraborty M, Dey B, Roy M, Roy B, Bhattacharyya NP, Roychoudhury S, Majumder PP. Ethnic India: a genomic view, with special reference to peopling and structure. Genome Res. 2003;13:2277–2290. doi: 10.1101/gr.1413403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabarti CS, Roy M, Sengupta NK, Lalthantluanga R, Majumder PP. Genetic relationships among some tribal groups inhabiting the north-eastern, eastern and sub-Himalayan regions of India. Ann Hum Genet. 2002;66:361–368. doi: 10.1046/j.1469-1809.2002.00132.x. [DOI] [PubMed] [Google Scholar]
- Mukherjee N, Nebel A, Oppenheim A, Majumder PP. High-resolution analysis of Y-chromosomal polymorphisms reveals signatures of population movements from Central Asia and West Asia into India. J Genet. 2001;80:125–135. doi: 10.1007/BF02717908. [DOI] [PubMed] [Google Scholar]
- Roychoudhury S, Roy S, Basu A, Banerjee R, Vishwanathan H, Usha Rani MV, Sil SK, Mitra M, Majumder PP. Genomic structures and population histories of linguistically distinct tribal groups of India. Hum Genet. 2001;109:339–350. doi: 10.1007/s004390100577. [DOI] [PubMed] [Google Scholar]
- Sharma G, Tamang R, Chaudhary R, Singh VK, Shah AM, Anugula S, Rani DS, Reddy AG, Eaaswarkhanth M, Chaubey G, Singh L, Thangaraj K. Genetic affinities of the central Indian tribal populations. PLoS One. 2012;7:e32546. doi: 10.1371/journal.pone.0032546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah AM, Tamang R, Moorjani P, Rani DS, Govindaraj P, Kulkarni G, Bhattacharya T, Mustak MS, Bhaskar LV, Reddy AG, Gadhvi D, Gai PB, Chaubey G, Patterson N, Reich D, Tyler-Smith C, Singh L, Thangaraj K. Indian Siddis: African descendants with Indian admixture. Am J Hum Genet. 2011;89:154–161. doi: 10.1016/j.ajhg.2011.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraj K, Naidu BP, Crivellaro F, Tamang R, Upadhyay S, Sharma VK, Reddy AG, Walimbe SR, Chaubey G, Kivisild T, Singh L. The influence of natural barriers in shaping the genetic structure of Maharashtra populations. PLoS One. 2010;5:e15283. doi: 10.1371/journal.pone.0015283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraj K, Chaubey G, Singh VK, Vanniarajan A, Thanseem I, Reddy AG, Singh L. In situ origin of deep rooting lineages of mitochondrial Macrohaplogroup ‘M’ in India. BMC Genomics. 2006;7:151. doi: 10.1186/1471-2164-7-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaubey G, Metspalu M, Choi Y, Magi R, Romero IG, Soares P, van Oven M, Behar DM, Rootsi S, Hudjashov G, Mallick CB, Karmin M, Nelis M, Parik J, Reddy AG, Metspalu E, van Driem G, Xue Y, Tyler-Smith C, Thangaraj K, Singh L, Remm M, Richards MB, Lahr MM, Kayser M, Villems R, Kivisild T. Population genetic structure in Indian Austroasiatic speakers: the role of landscape barriers and sex-specific admixture. Mol Biol Evol. 2011;28:1013–1024. doi: 10.1093/molbev/msq288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayden T, Cadenas AM, Regueiro M, Singh NB, Zhivotovsky LA, Underhill PA, Cavalli-Sforza LL, Herrera RJ. The Himalayas as a directional barrier to gene flow. Am J Hum Genet. 2007;80:884–894. doi: 10.1086/516757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar DM, Yunusbayev B, Metspalu M, Metspalu E, Rosset S, Parik J, Rootsi S, Chaubey G, Kutuev I, Yudkovsky G, Khusnutdinova EK, Balanovsky O, Semino O, Pereira L, Comas D, Gurwitz D, Bonne-Tamir B, Parfitt T, Hammer MF, Skorecki K, Villems R. The genome-wide structure of the Jewish people. Nature. 2010;466:238–242. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009;461:489–494. doi: 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metspalu M, Kivisild T, Metspalu E, Parik J, Hudjashov G, Kaldma K, Serk P, Karmin M, Behar DM, Gilbert MT, Endicott P, Mastana S, Papiha SS, Skorecki K, Torroni A, Villems R. Most of the extant mtDNA boundaries in south and southwest Asia were likely shaped during the initial settlement of Eurasia by anatomically modern humans. BMC Genet. 2004;5:26. doi: 10.1186/1471-2156-5-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metspalu M, Romero IG, Yunusbayev B, Chaubey G, Mallick CB, Hudjashov G, Nelis M, Magi R, Metspalu E, Remm M, Pitchappan R, Singh L, Thangaraj K, Villems R, Kivisild T. Shared and unique components of human population structure and genome-wide signals of positive selection in South Asia. Am J Hum Genet. 2011;89:731–744. doi: 10.1016/j.ajhg.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanseem I, Thangaraj K, Chaubey G, Singh VK, Bhaskar LV, Reddy BM, Reddy AG, Singh L. Genetic affinities among the lower castes and tribal groups of India: inference from Y chromosome and mitochondrial DNA. BMC Genet. 2006;7:42. doi: 10.1186/1471-2156-7-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad M, Kivisild T, Watkins WS, Dixon ME, Ricker CE, Rao BB, Naidu JM, Prasad BV, Reddy PG, Rasanayagam A, Papiha SS, Villems R, Redd AJ, Hammer MF, Nguyen SV, Carroll ML, Batzer MA, Jorde LB. Genetic evidence on the origins of Indian caste populations. Genome Res. 2001;11:994–1004. doi: 10.1101/gr.GR-1733RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraj K, Singh L, Reddy AG, Rao VR, Sehgal SC, Underhill PA, Pierson M, Frame IG, Hagelberg E. Genetic affinities of the Andaman Islanders, a vanishing human population. Curr Biol. 2003;13:86–93. doi: 10.1016/S0960-9822(02)01336-2. [DOI] [PubMed] [Google Scholar]
- Barik SS, Sahani R, Prasad BV, Endicott P, Metspalu M, Sarkar BN, Bhattacharya S, Annapoorna PC, Sreenath J, Sun D, Sanchez JJ, Ho SY, Chandrasekar A, Rao VR. Detailed mtDNA genotypes permit a reassessment of the settlement and population structure of the Andaman Islands. Am J Phys Anthropol. 2008;136:19–27. doi: 10.1002/ajpa.20773. [DOI] [PubMed] [Google Scholar]
- Wang HW, Mitra B, Chaudhuri TK, Palanichamy MG, Kong QP, Zhang YP. Mitochondrial DNA evidence supports northeast Indian origin of the aboriginal Andamanese in the Late Paleolithic. J Genet Genomics. 2011;38:117–122. doi: 10.1016/j.jgg.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Rao RP, Yadav N, Vahia MN, Joglekar H, Adhikari R, Mahadevan I. Entropic evidence for linguistic structure in the Indus script. Science. 2009;324:1165. doi: 10.1126/science.1170391. [DOI] [PubMed] [Google Scholar]
- Chaubey G. The demographic history of India: a perspective based on genetic evidence. University of Tartu and Estonian Biocentre; 2010. pp. 45–49. (PhD thesis). [Google Scholar]
- Wells RS, Yuldasheva N, Ruzibakiev R, Underhill PA, Evseeva I, Blue-Smith J, Jin L, Su B, Pitchappan R, Shanmugalakshmi S, Balakrishnan K, Read M, Pearson NM, Zerjal T, Webster MT, Zholoshvili I, Jamarjashvili E, Gambarov S, Nikbin B, Dostiev A, Aknazarov O, Zalloua P, Tsoy I, Kitaev M, Mirrakhimov M, Chariev A, Bodmer WF. The Eurasian heartland: a continental perspective on Y-chromosome diversity. Proc Natl Acad Sci U S A. 2001;98:10244–10249. doi: 10.1073/pnas.171305098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana-Murci L, Krausz C, Zerjal T, Sayar SH, Hammer MF, Mehdi SQ, Ayub Q, Qamar R, Mohyuddin A, Radhakrishna U, Jobling MA, Tyler-Smith C, McElreavey K. Y-chromosome lineages trace diffusion of people and languages in southwestern Asia. Am J Hum Genet. 2001;68:537–542. doi: 10.1086/318200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Deepa E, Vishwanathan H, Stoneking M. Genetic evidence for the demic diffusion of agriculture to India. Science. 2004;304:1125. doi: 10.1126/science.1095819. [DOI] [PubMed] [Google Scholar]
- Cordaux R, Aunger R, Bentley G, Nasidze I, Sirajuddin SM, Stoneking M. Independent origins of Indian caste and tribal paternal lineages. Curr Biol. 2004;14:231–235. doi: 10.1016/j.cub.2004.01.024. [DOI] [PubMed] [Google Scholar]
- Sahoo S, Singh A, Himabindu G, Banerjee J, Sitalaximi T, Gaikwad S, Trivedi R, Endicott P, Kivisild T, Metspalu M, Villems R, Kashyap VK. A prehistory of Indian Y chromosomes: evaluating demic diffusion scenarios. Proc Natl Acad Sci U S A. 2006;103:843–848. doi: 10.1073/pnas.0507714103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamshad MJ, Watkins WS, Dixon ME, Jorde LB, Rao BB, Naidu JM, Prasad BV, Rasanayagam A, Hammer MF. Female gene flow stratifies Hindu castes. Nature. 1998;395:651–652. doi: 10.1038/27103. [DOI] [PubMed] [Google Scholar]
- Quintana-Murci L, Chaix R, Wells RS, Behar DM, Sayar H, Scozzari R, Rengo C, Al-Zahery N, Semino O, Santachiara-Benerecetti AS, Coppa A, Ayub Q, Mohyuddin A, Tyler-Smith C, Qasim Mehdi S, Torroni A, McElreavey K. Where west meets east: the complex mtDNA landscape of the southwest and Central Asian corridor. Am J Hum Genet. 2004;74:827–845. doi: 10.1086/383236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S, Saha A, Rai E, Bhat A, Bamezai R. Human mtDNA hypervariable regions, HVR I and II, hint at deep common maternal founder and subsequent maternal gene flow in Indian population groups. J Hum Genet. 2005;50:497–506. doi: 10.1007/s10038-005-0284-2. [DOI] [PubMed] [Google Scholar]
- Zerjal T, Pandya A, Thangaraj K, Ling EY, Kearley J, Bertoneri S, Paracchini S, Singh L, Tyler-Smith C. Y-chromosomal insights into the genetic impact of the caste system in India. Hum Genet. 2007;121:137–144. doi: 10.1007/s00439-006-0282-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayub Q, Tyler-Smith C. Genetic variation in South Asia: assessing the influences of geography, language and ethnicity for understanding history and disease risk. Brief Funct Genomic Proteomic. 2009;8:395–404. doi: 10.1093/bfgp/elp015. [DOI] [PubMed] [Google Scholar]
- Chakravarti A. Human genetics: tracing India’s invisible threads. Nature. 2009;461:487–488. doi: 10.1038/461487a. [DOI] [PubMed] [Google Scholar]
- Chaubey G, Metspalu M, Kivisild T, Villems R. Peopling of South Asia: investigating the caste-tribe continuum in India. Bioessays. 2007;29:91–100. doi: 10.1002/bies.20525. [DOI] [PubMed] [Google Scholar]
- Chaubey G, Metspalu M, Karmin M, Thangaraj K, Rootsi S, Parik J, Solnik A, Selvi-Rani D, Singh V, Reddy A, Metspalu E, Singh L, Kivisild T, Villems R. Language shift by indigenous population: a model genetic study in South Asia. Int J Hum Genet. 2008;8:41–50. [Google Scholar]
- Kivisild T, Bamshad MJ, Kaldma K, Metspalu M, Metspalu E, Reidla M, Laos S, Parik J, Watkins WS, Dixon ME, Papiha SS, Mastana SS, Mir MR, Ferak V, Villems R. Deep common ancestry of Indian and western-Eurasian mitochondrial DNA lineages. Curr Biol. 1999;9:1331–1334. doi: 10.1016/S0960-9822(00)80057-3. [DOI] [PubMed] [Google Scholar]
- Chaubey G, Karmin M, Metspalu E, Metspalu M, Selvi-Rani D, Singh VK, Parik J, Solnik A, Naidu BP, Kumar A, Adarsh N, Mallick CB, Trivedi B, Prakash S, Reddy R, Shukla P, Bhagat S, Verma S, Vasnik S, Khan I, Barwa A, Sahoo D, Sharma A, Rashid M, Chandra V, Reddy AG, Torroni A, Foley RA, Thangaraj K, Singh L, Kivisild T. et al. Phylogeography of mtDNA haplogroup R7 in the Indian peninsula. BMC Evol Biol. 2008;8:227. doi: 10.1186/1471-2148-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraj K, Ramana GV, Singh L. Y-chromosome and mitochondrial DNA polymorphisms in Indian populations. Electrophoresis. 1999;20:1743–1747. doi: 10.1002/(SICI)1522-2683(19990101)20:8<1743::AID-ELPS1743>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Gauniyal M, Chahal SM, Kshatriya GK. Genetic affinities of the Siddis of South India: an emigrant population of East Africa. Hum Biol. 2008;80:251–270. doi: 10.3378/1534-6617-80.3.251. [DOI] [PubMed] [Google Scholar]
- Gauniyal M, Aggarwal A, Kshatriya GK. Genomic structure of the immigrant Siddis of East Africa to southern India: a study of 20 autosomal DNA markers. Biochem Genet. 2011;49:427–442. doi: 10.1007/s10528-011-9419-7. [DOI] [PubMed] [Google Scholar]
- Ramana GV, Su B, Jin L, Singh L, Wang N, Underhill P, Chakraborty R. Y-chromosome SNP haplotypes suggest evidence of gene flow among caste, tribe, and the migrant Siddi populations of Andhra Pradesh, South India. Eur J Hum Genet. 2001;9:695–700. doi: 10.1038/sj.ejhg.5200708. [DOI] [PubMed] [Google Scholar]
- Schimmel A. Islam in India and Pakistan. Leiden: Brill; 1982. [Google Scholar]
- Robb P. A History of India. Houndmills: Palgrave; 2002. [Google Scholar]
- Eaaswarkhanth M, Dubey B, Meganathan PR, Ravesh Z, Khan FA, Singh L, Thangaraj K, Haque I. Diverse genetic origin of Indian Muslims: evidence from autosomal STR loci. J Hum Genet. 2009;54:340–348. doi: 10.1038/jhg.2009.38. [DOI] [PubMed] [Google Scholar]
- Eaaswarkhanth M, Haque I, Ravesh Z, Romero IG, Meganathan PR, Dubey B, Khan FA, Chaubey G, Kivisild T, Tyler-Smith C, Singh L, Thangaraj K. Traces of sub-Saharan and Middle Eastern lineages in Indian Muslim populations. Eur J Hum Genet. 2010;18:354–363. doi: 10.1038/ejhg.2009.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, Reed FA, Friedlaender FR, Ehret C, Ranciaro A, Froment A, Hirbo JB, Awomoyi AA, Bodo JM, Doumbo O, Ibrahim M, Juma AT, Kotze MJ, Lema G, Moore JH, Mortensen H, Nyambo TB, Omar SA, Powell K, Pretorius GS, Smith MW, Thera MA, Wambebe C, Weber JL, Williams SM. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veltman CJ. Language Shift in the United States. Berlin: Mouton; 1983. [Google Scholar]
- Clyne M, Kipp S. Language maintenance and language shift: community languages in Australia, 1996. People Place. 1997;5:19–27. [PubMed] [Google Scholar]
- Thangaraj K, Nandan A, Sharma V, Sharma VK, Eaaswarkhanth M, Patra PK, Singh S, Rekha S, Dua M, Verma N, Reddy AG, Singh L. Deep rooting in-situ expansion of mtDNA Haplogroup R8 in South Asia. PLoS One. 2009;4:e6545. doi: 10.1371/journal.pone.0006545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diffloth G. More on Dvaravati Old Mon. Salaya: Mahidol University; 2009. Fourth International Conference on Austroasiatic Linguistics; 2009 October 29–30. [Google Scholar]
- Fuller D. In: Non-human genetics, agricultural origins and historical linguistics in South Asia. Petraglia MD, Allchin B, editor. Springer; 2007. The Evolution and History of Human Populations in South Asia; pp. 393–443. Vert Paleobiol Paleoanthropol Part III. [Google Scholar]
- van Driem G. Languages of the Himalayas: an ethnolinguistic handbook of the greater Himalayan region containing an introduction to the symbiotic theory of language. Leiden: Brill; 2001. [Google Scholar]
- Reddy BM, Langstieh BT, Kumar V, Nagaraja T, Reddy AN, Meka A, Reddy AG, Thangaraj K, Singh L. Austro-Asiatic tribes of Northeast India provide hitherto missing genetic link between South and Southeast Asia. PLoS One. 2007;2:e1141. doi: 10.1371/journal.pone.0001141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V, Reddy AN, Babu JP, Rao TN, Langstieh BT, Thangaraj K, Reddy AG, Singh L, Reddy BM. Y-chromosome evidence suggests a common paternal heritage of Austro-Asiatic populations. BMC Evol Biol. 2007;7:47. doi: 10.1186/1471-2148-7-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higham CJ. Languages and Farming Dispersals: Austroasiatic Languages and Rice Cultivation. Cambridge: The McDonald Institute for Archaeological Research; 2003. [Google Scholar]
- Salem AH, Badr FM, Gaballah MF, Paabo S. The genetics of traditional living: Y-chromosomal and mitochondrial lineages in the Sinai Peninsula. Am J Hum Genet. 1996;59:741–743. [PMC free article] [PubMed] [Google Scholar]
- Seielstad MT, Minch E, Cavalli-Sforza LL. Genetic evidence for a higher female migration rate in humans. Nat Genet. 1998;20:278–280. doi: 10.1038/3088. [DOI] [PubMed] [Google Scholar]
- Oota H, Kitano T, Jin F, Yuasa I, Wang L, Ueda S, Saitou N, Stoneking M. Extreme mtDNA homogeneity in continental Asian populations. Am J Phys Anthropol. 2002;118:146–153. doi: 10.1002/ajpa.10056. [DOI] [PubMed] [Google Scholar]
- Kumar V, Langstieh BT, Madhavi KV, Naidu VM, Singh HP, Biswas S, Thangaraj K, Singh L, Reddy BM. Global patterns in human mitochondrial DNA and Y-chromosome variation caused by spatial instability of the local cultural processes. PLoS Genet. 2006;2:e53. doi: 10.1371/journal.pgen.0020053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalini A, Yamini BK, Gayatri N, Thennarasu K, Gope R. Familial Madras motor neuron disease (FMMND): study of 15 families from southern India. J Neurol Sci. 2006;250:140–146. doi: 10.1016/j.jns.2006.08.010. [DOI] [PubMed] [Google Scholar]
- Nalini A, Thennarasu K, Yamini BK, Shivashankar D, Krishna N. Madras motor neuron disease (MMND): clinical description and survival pattern of 116 patients from Southern India seen over 36 years (1971–2007) J Neurol Sci. 2008;269:65–73. doi: 10.1016/j.jns.2007.12.026. [DOI] [PubMed] [Google Scholar]
- Agarwal SS, Phadke SR, Phadke RV, Das SK, Singh GK, Sharma JP, Teotia SP, Saxena BN. Handigodu disease: a radiological study. A new variety of spondyloepi(meta)physeal dysplasia of the autosomal dominant type. Skeletal Radiol. 1994;23:611–619. [PubMed] [Google Scholar]
- Badadani M, Shetty KT, Babu SV, Agarwal SS. Metabolic status of magnesium and ceruloplasmin in Handigodu Joint Disease: a variety of spondylo epi (meta) physeal dysplasia. Clin Chim Acta. 2008;395:170–171. doi: 10.1016/j.cca.2008.05.017. [DOI] [PubMed] [Google Scholar]
- Rao PR, Gopalam KB. High incidence of the silent allele at cholinesterase locus I in Vysyas of Andhra Pradesh (S. India) Hum Genet. 1979;52:139–141. doi: 10.1007/BF00284608. [DOI] [PubMed] [Google Scholar]
- Pandit JJ, Gopa S, Arora J. A hypothesis to explain the high prevalence of pseudo-cholinesterase deficiency in specific population groups. Eur J Anaesthesiol. 2011;28:550–552. doi: 10.1097/EJA.0b013e3283457cfb. [DOI] [PubMed] [Google Scholar]
- Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, Rai TS, Khullar M, Soares P, Bahl A, Tharkan JM, Vaideeswar P, Rathinavel A, Narasimhan C, Ayapati DR, Ayub Q, Mehdi SQ, Oppenheimer S, Richards MB, Price AL, Patterson N, Reich D, Singh L, Tyler-Smith C, Thangaraj K. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat Genet. 2009;41:187–191. doi: 10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler CA, Nelson GW, Smith MW. Admixture mapping comes of age. Annu Rev Genomics Hum Genet. 2010;11:65–89. doi: 10.1146/annurev-genom-082509-141523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narang A, Jha P, Rawat V, Mukhopadhyay A, Dash D, Basu A, Mukerji M. Recent admixture in an Indian population of African ancestry. Am J Hum Genet. 2011;89:111–120. doi: 10.1016/j.ajhg.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adeyemo A, Gerry N, Chen G, Herbert A, Doumatey A, Huang H, Zhou J, Lashley K, Chen Y, Christman M, Rotimi C. A genome-wide association study of hypertension and blood pressure in African Americans. PLoS Genet. 2009;5:e1000564. doi: 10.1371/journal.pgen.1000564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thangaraj K, Reddy AG, Singh L. Is the amelogenin gene reliable for gender identification in forensic casework and prenatal diagnosis? Int J Legal Med. 2002;116:121–123. doi: 10.1007/s00414-001-0262-y. [DOI] [PubMed] [Google Scholar]