Abstract
The human asparaginase-like protein 1 (hASRGL1) catalyzes the hydrolysis of l-asparagine and isoaspartyl-dipeptides. As an N-terminal nucleophile (Ntn) hydrolase superfamily member, the active form of hASRGL1 is generated by an intramolecular cleavage step with Thr168 as the catalytic residue. However, in vitro, autoprocessing is incomplete (~50 %), fettering the biophysical characterization of hASRGL1. We circumvented this obstacle by constructing a circularly permuted hASRGL1 that uncoupled the autoprocessing reaction, allowing us to kinetically and structurally characterize this enzyme and the precursor-like, hASRGL1-Thr168Ala variant. Crystallographic and biochemical evidence suggest an activation mechanism where a torsional restraint on the Thr168 side-chain helps drive the intramolecular processing reaction. Cleavage and formation of the active site releases the torsional restriction on Thr168, which is facilitated by a small conserved Gly-rich loop near the active site that allows the conformational changes necessary for activation.
Introduction
The human Asparaginase–like Protein 1 (hASRGL1) is an N-terminal nucleophile (Ntn) hydrolase, and a mammalian member of the plant-type asparaginase family catalyzing the hydrolysis of l-asparagine and isoaspartyl-dipeptides (1). Human ASRGL1 has been found to be up-regulated in certain ovarian, colon, and breast carcinomas and its expression correlated with enhanced growth rates in metastatic colon cancer cells (2). Additionally, there is an interest in engineering hASRGL1 for use as a therapeutic agent for the systemic depletion of serum l-asparagine, in the treatment of acute lymphoblastic leukemia (3).
The formation of isoaspartyl peptide bonds is a common source of non-enzymatic protein damage under physiological conditions, as it introduces a kink in the protein backbone that can disrupt normal folding leading to altered susceptibility to proteolysis or loss of function (4). Böhme et al. have shown that isoaspartate containing peptides have a huge impact on substrate recognition and turnover across several classes of proteases, and in some cases even act as protease inhibitors (5). Therefore, hASRGL1 likely plays an important physiological role in the clearing of isoaspartate containing peptides and the return of amino acids back into the metabolic pool. The hASRGL1 protein is first translated as an inactive precursor that then undergoes an autocatalytic intramolecular activation step with the hydroxyl side-chain of Thr168 acting as the catalytic nucleophile (6). This intramolecular processing of enzymes within the Ntn hydrolyase superfamily is believed to share a common first step; namely an acyl shift (N → O or N → S) from nucleophiles Thr, Ser or Cys that results in the formation of a (thio)ester intermediate. This intermediate is subsequently hydrolyzed to expose the N-terminal residue of the nascent β-subunit, creating the active form of the enzyme (7, 8).
However, biochemical and structural studies of hASRGL1 have been hampered by incomplete intramolecular processing, resulting in all cases with a mixture of active and inactive protein (1). In fact partial processing is frequently reported in the Ntn hydrolase family, hindering the precise biochemical characterization of these enzymes. One strategy for the generation of fully processed enzyme is co-expression of the α and β subunits to circumvent incomplete intramolecular processing (9-11). However, individual subunits need to be expressed separately and allowed to associate in vitro which can be a challenge because of low solubility of one or both of the subunits. To uncouple the autoprocessing requirements of hASRGL1, we engineered a circularly permuted form of the enzyme (cp-hASRGL1) that is expressed in the fully processed form. The crystal structures of cp-hASRGL1 and the processing-inactive hASRGL1-Thr168Ala variant were solved at 3.3 and 2.3 Å resolution respectively, and constitute the first structures of a mammalian plant-type asparaginase. Phylogenetic, mutational, and kinetic analyses revealed a strictly conserved His-Gly-Gly motif near the active site which is critical for both autoprocessing and for substrate turnover. We also observed a dramatic change in the thermodynamic stability between unprocessed hASRGL1 and hASRGL1-Thr168Ala indicative of a strain-based activation mechanism.
RESULTS AND DISCUSSION
Construction and characterization of cp-hASRGL1
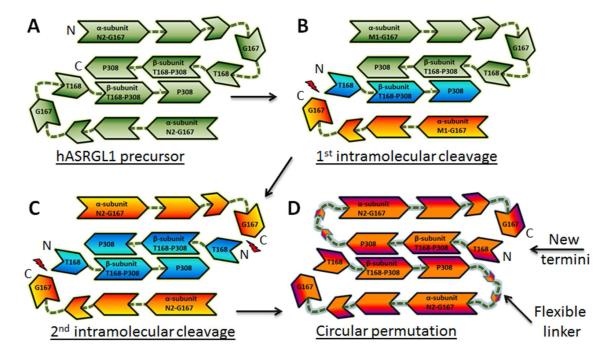
The hASRGL1 precursor is robustly expressed ( >30 mg/L) as a stable dimer in E. coli strain (BL21(DE3)) as confirmed in size exclusion chromatography (1). Within each monomer, intramolecular processing occurs at the peptide bond between Gly167 and Thr168 to activate the protein. In vitro, however, only 40-50 % of the enzyme undergoes autoprocessing as judged by SDS-PAGE gel densitometry under any of the conditions tested (1). The resulting heterogeneity of recombinant hASRGL1, which is present as a mixture of active and inactive protein, is a complication for both structural and kinetic characterization of the enzyme. To solve these problems, we constructed a “circularly permuted” form of the enzyme. In circularly permuted proteins, the authentic N- and C- termini are joined by a flexible linker and new N- and C-termini are generated at a desired position (12). The circularly permuted version of hASRGL1 (cp-hASRGL1) was constructed that physically links the β and α domains between Pro308 and Asn2 with a 12 amino acid linker using small and hydrophilic residues (Pro308-GAGSGAGSGAGG-Asn2) (Figure 1). Met1 was omitted from the linker as the initiating residue is commonly removed by methionine amino peptidase and we wished to avoid a hydrophobic residue within the linker. The cp-hASRGL1 is a homodimer with the catalytic Thr168 residue now being the 2nd amino acid following the initiator l-Met residue. Intracellular processing in E.coli by the native methionine amino peptidase (13) results in the formation of homogenous cp-hASRGL1 having the nucleophilic Thr168 as its first residue (for the sake of clarity, we have kept the numbering scheme for the circular permutant the same as the wild-type enzyme). The cp-hASRGL1 was expressed and purified in bacteria at a high yield (>50 mg/L) in soluble form and was shown to display a kcat/KM of 3.2 ± 0.5 ×103 M−1s−1 for l-Asn hydrolysis, a similar value to that of the parental hASRGL1 (kcat/KM = 2.0 ± 0.4 ×103M−1s−1 (1): the amount of active hASRGL1 is assessed by correcting for the amount of unprocessed enzyme by gel densitometry which likely introduces an error in estimating concentration). Thus, the linker between Pro308 and Asn2 did not adversely affect the catalytic properties of the enzyme.
Figure 1.
A) hASRGL1 is expressed as a full-length precursor which forms a homodimer. B & C) Autocatalytic cleavage between Gly167 and Thr168 produces the α and β subunits and results in the active heterotetrameric form of the enzyme. Intramolecular processing does not proceed to completion in vitro, resulting in a mixture of A, B, and C. D) Circular permutation of hASRGL1 was accomplished by addition of a linker between Pro308 of the β subunit and Asn2 of the α subunit. The active site nucleophile Thr168 becomes the 2nd amino acid following the N-terminal Met residue which is efficiently cleaved during expression in the bacterial cytoplasm by methionine amino peptidase.
To assess the effect of the identity of the nucleophile at position 168 on enzyme kinetics, we expressed and purified cp-hASRGL1-Thr168Cys and cp-hASRGL1-Thr168Ser. Both variants could be expressed at a high yield comparable to that for cp-hASRGL1. ESI-MS analysis of the purified proteins showed that cp-hASRGL1, cp-hASRGL1-Thr168Cys and cp-hASRGL1-Thr168Ser were > 95 % processed, i.e. the initiator Met residue had been cleaved (Table S1). cp-hASRGL1 displays a high catalytic activity with the easily detectable l-asparaginase substrate analog, l-aspartic acid-β-hydroxamate (AHA) (14) ( kcat/KM of 20 ± 3 mM−1 s−1 at pH 7.4 and 25 °C), whereas we could not detect any activity with the cp-hASRGL1-Thr168Cys variant. The cp-hASRGL1-Thr168Ser variant was active, albeit with more than a thousand fold reduction in kcat/KM (Table 1). When these mutations were introduced into the wild-type hASRGL1 protein neither autoprocessing nor substrate hydrolysis could be observed (data not shown) indicating that while Thr168 can be substituted with Ser in the hydrolysis of AHA, albeit inefficiently, it is indispensable for autocatalytic processing.
Table 1.
Kinetics of cp-hASRGL1 and variants
| Variant | kcat (s−1) | KM (mM) | kcat/KM (mM-1 s−1) |
|---|---|---|---|
| cp-hASRGL-1 | 1.80 ± 0.05 | 0.09 ± 0.01 | 20 ± 3 |
| cp-hASRGL-1-Thr168Ser | 0.016 ± 0.001 | 1.1 ± 0.1 | 0.015 ± 0.003 |
| cp-hASRGL-1-Thr168Cys | nd | nd | nd |
All reactions were conducted at 25 °C and pH 7.4, using AHA as a substrate (nd = not within the detection limit)
We then assessed the ionization status of the cp-hASRGL1 active site. The cp-hASRGL1 kcat dependence upon pH, which reflects ionizations important to the enzyme-substrate complex (ES), was well fit to a single ionization model (Supporting Figure S1 A) with an ascending limb pKa of 6.0 ± 0.1. (It should be noted that there is not much data defining the lower bounds of this curve and is thus more of an estimate.) This is similar to the values reported for Ntn hydrolases glutaryl 7-aminocephalosporanic acid acylase (Pseudomonas), and glycosylasparaginase (Spodoptera) (15, 16) and is most likely dictated by the pKa of the Thr168 primary amine when substrate is bound. The fits of the pH dependence of kcat/KM resulted in a narrow bell shaped curve (Supporting Figure S1 B). Because the fitted values are less than 3.5 pH units from each other, we applied Segel’s method (17) to calculate corrected pKa values for each limb of the kcat/KM profiles with an ascending limb pKa1 of 7.3 ± 0.2 and a descending limb pKa2 of 8.0 ± 0.2. The pH dependence of kcat/KM, which reflects ionizations important to the free enzyme and free substrate (E + S), can be described by the de-protonation of the primary amine (pKa1) of Thr168 which is required for activity and is not far from the predicted value of 7.62 for an N-terminal Thr residue (18). Similarly, the descending pKa2 value can be ascribed to de-protonation of the primary amine of the substrate AHA molecule, which closely matches the predicted pKa value of 8.05 (18), and results in reduced substrate binding affinity (Table S2).
Structural studies of the precursor-like hASRGL1-Thr168Ala and the fully activated cp-hASRGL1
Crystals obtained from recombinant hASRGL1, which as discussed above is incompletely processed and therefore consist of a mixture of the two forms of the enzyme, did not yield high-resolution diffraction patterns. Therefore, we determined the structures of cp-hASRGL1 and the hASRGL1-Thr168Ala variant (Table 2). Both proteins fold as intertwined central layers of β-strands surrounded by α-helices, displaying the classic αββα fold conserved amongst the Ntn hydrolyase family members, and similar to other plant-type asparaginases, we observed bound ions that we have modeled as a Na+ ions based on their octahedral geometry and coordination spheres with Na+ …O distances ranging from 2.2 to 2.7 Å (Figure 2A) (19).
Table 2.
Crystallographic Data and Refinement Statistics:
| Data statistics | hASRGL1-Thr168Ala | cp-hASRGL1 |
| Source | Advanced Light Source 5.0.3 | Advanced Light Source 5.0.2 |
| Wavelength (Å) | 0.9765 | 1.00 |
| Temperature of Measurements (K) | 100 | 100 |
| Resolution (Å) | 79-2.3 (2.34-2.30)a | 50-3.3 (3.36-3.30) |
| Space group | C2221 | P6522 |
| Unit Cell (Å, °) | a= 111.7, b= 111.4, c= 119.5, | a = 108.6, b= 108.6, c= 275.1, |
| Molecule per asymmetric unit | 2 | 2 |
| Number of unique reflections | 33467 (1462) | 15355 (1229) |
| Redundancy | 2.9 (2.3) | 3.9 (3.8) |
| Completeness (%) | 95.6 (87.8) | 97.6 (95.6) |
| I/σ(I) | 12.0(1.7) | 8.9 (1.8) |
| Rint (%) | 9.2 (41.5) | 10.5 (26.1) |
| Refinement statistics | ||
| Refinement | ||
| Resolution limit (Å) | 47.62-2.30 | 47.50-3.30 |
| No. reflections (test) | 31963 (1605) | 14817 (745) |
| Rwork/Rfree (%)b | 16.8/ 19.3 | 21.5/24.9 |
| No. atoms | 4578 | 4304 |
| protein | 4331 | 4302 |
| Na+ | 2 | 2 |
| TAMc | 22 | N/A |
| SO4 | 10 | N/A |
| Water | 213 | N/A |
| B-factors (Å2) | ||
| protein | 38 | 127.7 |
| Na+ | 33.8 | 37.4 |
| TAMc | 61.1 | N/A |
| SO4 | 38.6 | N/A |
| Water | 42.1 | N/A |
| R.m.s deviations | ||
| Bond lengths (Å) | 0.010 | 0.010 |
| Bond angles (°) | 1.14 | 1.24 |
| Ramachandran plot (%) | ||
| Most favored regions | 91.8 | 92.6 |
| Additional allowed | ||
| regions | 7.8 | 7 |
| Generously allowed | ||
| regions | 0 | 0 |
| Disallowed regionsd | 0.4 | 0.4 |
Highest resolution shell is shown in parentheses.
Rfree is calculated with 5 % of the data randomly omitted from refinement.
TAM is the abbreviation of Tris(hydroxymethyl)aminomethane which is used as pH buffer in crystallization.
Asp119 in both structures exhibit unusual φ/ Ψ angles after iterative refinements, though electron density clearly indicates its orientation in the hASRGL1 structure.
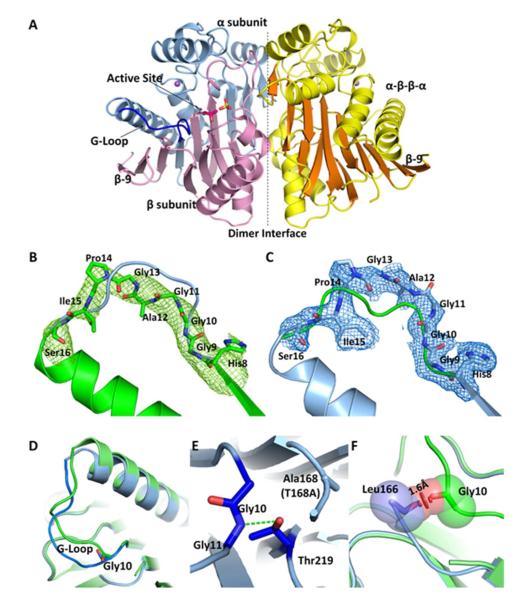
Figure 2.
A) The overall structure of hASRGL1 displays a conserved Ntn αββα fold with α-helices colored yellow and β-sheets colored orange. The dashed line indicates the interface between the dimers. The wild-type enzyme autocatalytically cleaves between Thr168 and Gly167 into α (light blue) and β (light pink) subunits. A sulfate group (yellow sticks) binds in each substrate binding pocket in proximity to the active site nucleophilic residue Thr168 (Thr168Ala) (hot pink sticks). A conserved Gly-rich loop (blue) is close to the active site and plays an important role in protein activation and catalysis. Plant-type asparaginases coordinate a sodium cation (purple sphere) in each monomer. B) The Fo-Fc omit map of cp-hASRGL1 Gly-rich loop (green mesh). The final refined cp-hASRGL1 (green) is well fitted in the cp-hASRGL1 Gly-rich loop omit map representing a dramatic orientation difference with the final refined hASRGL1-Thr168Ala Gly-rich loop model (blue cartoon). C) The Fo-Fc omit map of hASRGL1-Thr168Ala Gly-rich loop (blue mesh). It also shows changed orientation from final refined cp-hASRGL1 model (green cartoon). These two omit maps doubly confirm that the Gly-rich loops of cp-hASRGL1 and hASRGL1-Thr168Ala have different orientations, and that this difference is not due to model bias. □ level of omit map is set as 1.8 Å. D) Superposition of Gly-rich loops of hASRGL1-Thr168Ala (light blue) and of cp-hASRGL1 (green). The carbonyl group of Gly10 points in the opposite direction as shown by sticks. E) The hASRGL1-Thr168Ala amide group of Gly11 forms a hydrogen bond (green dashed line) with the proposed general base residue Thr219. F) The superimposed structure of hASRGL1-Thr168Ala (light blue) and cp-hASRGL1 (green), indicates that the residues preceding Thr168 (Leu166 and Gly167) of hASRGL1 would clash with the Gly-rich loop if the protein were to assume the open conformation observed in cp-hASRGL1. The red disk represents a significant Van der Waals overlay.
A homodimer is modeled in each asymmetric unit in both cases. The hydrophobic dimer interface accounts for 30 % of the protein surface (3149 Å2 of dimerization area) in both structures calculated by Areaimol (20) underscoring its functional importance (Figure 2A). The active site cleft is located at a groove adjacent to the dimer interface and divides the α and β subunits of the protein, with the nucleophilic Thr168 residue positioned within the cleft (Figure 2A). In both hASRGL1 structures of precursor or fully processed forms, the active site residues preceding the nucleophilic Thr168 (Lys155-Leu166) are disordered due to high flexibility. Interestingly, the same regions are also lacking electron density in the other two plant-type asparaginase as well (PDB codes: 2ZAL, residues Thr162-Gly178 and 2GEZ, residues Arg157-Gly192 (21, 22) ). The uniformity of the disordered nature of the active site loop in all plant-type asparaginase structures suggests an intrinsic flexibility in the proximity of the active site. It should be noted that cp-hASRGL1 model exhibits a high B-factor (Table 2), consistent with its low resolution in diffraction and high sensitivity for cryo-soaking and temperature change.
The overall structure of hASRGL1 is highly homologous to its plant and bacterial counterparts. By using PyMOL alignment program, we observed an R.M.S deviation of 0.77 Å in Cα compared to the Lupinus luteus isoaspartyl peptidase (UniProtKB Q9ZSD6, PDB code: 2GEZ) and an R.M.S. deviation of 0.82 Å compared to E. coli isoaspartyl peptidase (UniProtKB P37595, PDB code: 2ZAL (21, 22)), indicating they share nearly identical core structures. In particular, the active site near the catalytic nucleophile Thr168 is invariably located at the center of the sandwich formed by anti-parallel β-strands (Figure 2A), strongly suggesting a conservation of enzymatic mechanism. Consistent with the structural homology to other isoaspartyl peptidases, the oxyanion hole stabilizing the autoprocessing reaction is likely the Asn62 side chain (Asn67 in E.coli isoaspartyl peptidase) and the oxyanion hole stabilizing the tetrahedral intermediate during substrate hydrolysis would be the amide N-H of Gly220 and the side chain oxygen of Thr221 (Gly230 and Thr231 in E.coli isoaspartyl peptidase) (19). A sulfate ion (from the purification buffer) is also observed in the hASRGL1-Thr168Ala structure coordinated to Arg196 which is the residue responsible for coordinating the α-carboxyl moiety of substrates within the superfamily (19). In previous reports of plant-type asparaginase structures, a chloride has been found bound to the same place (19, 22). The coordination distances of four oxygen atom with surrounded residues are within 2.6-3.3 Å. The density of sulfate ion shows a clear tetrahedral shape supporting our hypothesis (Supporting Figure S2). The hASRGL1-Thr168Ala structure was superimposed upon that of the fully activated cp-hASRGL1. Even though the two proteins crystallize in different space groups (hASRGL1-Thr168Ala crystallizes in C2221 space group while cp-hASRGL1 in P6522), their domain orientation is the same and their Cα chain exhibits an average R.M.S. deviation of only 0.2 Å. However, a very significant difference was observed in the conformation of a loop formed by residues 8-16 close to the active site (Figure 2 B, C & D) displaying an average R.M.S. deviation of 3.5 Å for main chain residues (Cα, amide and carbonyl carbon). It is highly unlikely that this is a crystallographic artifact as this loop does not participate in crystal packing contacts, nor is the change of conformation caused by the introduction of linker residues since residues 1-9 superimpose perfectly with the hASRGL1-Thr168Ala structure. There is also no apparent structure refinement bias. We deleted region His8-Ser16 in each model and generated omit maps for hASRGL1-T168A and cp-hASRGL1 respectively; the positive density indicates a clear unbiased orientation change of Gly-rich loop (Figure 2B and 2C). Interestingly, this loop contains a His8-Gly9-Gly10 (HGG) motif that is ~100 % conserved amongst plant-type asparaginases across all phylogenies, while in contrast the remainder of the loop is not stringently conserved (Supporting Figure S3). In the structure of the inactive hASRGL1-Thr168Ala protein, the Gly10 carbonyl group faces away from the Thr168Ala residue, positioning the amide group of Gly11 for hydrogen-bonding with the hydroxyl side chain of Thr219 (Figure 2E). This Thr219 (residue Thr230 in E. coli isoaspartyl peptidase) is highly conserved through all plant-type asparaginases and has been proposed to act as a general base in activating the hydroxyl group of the nucleophilic Thr residue (19). Hydrogen bonding locks the loop in a closed conformation (Figure 2D) in the inactive form of the enzyme. In contrast, in the catalytically active cp-hASRGL1, the enzyme carbonyl group of Gly10 flips 180 degrees with the loop changing its direction (Figure 2D). Had the protein been unprocessed, steric forces would prevent the loop from adopting an open conformation as the carbonyl group of Leu166 would be only 1.6 Å away from the Gly10 residue (Figure 2F). However, after processing, the Gly-rich loop at residues 11-16 is able to adopt an open conformation which may be important for facilitating substrate binding and hydrolysis (a hypothesis we plan to elaborate upon in a future report).
In order to understand if the residues within the Gly-rich loop are important for autoprocessing and/or catalysis, we introduced individual substitutions of Gly9Ala, Gly10Ala or Gly11Ala into both the wild-type and the circularly permuted forms of the enzyme. We then assessed intramolecular processing as a function of time by SDS-PAGE gel densitometry as well as the steady-state kinetics for the hydrolysis of substrate analog AHA (Table 3). Consistent with the strict conservation of Gly9-Gly10 (Supporting Figure S3), Gly9Ala and Gly10Ala substitutions decreased the rate of intramolecular processing by 6 and 30-fold respectively compared to the wild-type enzyme. In contrast, the Gly11Ala substitution, which is observed in ~90 % of available sequences of isoaspartyl-dipeptidases and is in the middle part of the flexible loop, had essentially no effect on the autoprocessing rate, nor on substrate turnover. The Gly9Ala and Gly10Ala mutations in cp-hASRGL1 greatly impacted AHA hydrolysis resulting in respective 14- and 50-fold reductions in kcat/KM. The kinetic results are consistent with the structural observation that residue Gly10 is important for gating the conformational freedom of the Gly-rich loop.
Table 3.
Intramolecular processing and substrate hydrolysis rates.
| Subtrate (AHA) hydrolysis1 | |||||
|---|---|---|---|---|---|
| hASRGL1 Variants | Int. Mol processing rate hr−1 |
Obs. kcat s−1 |
Corrected for processing kcat s−1 |
KM mM |
kcat/KM mM−1 s−1 |
| WT-hASRGL1 | 0.034 ± 0.004 | 6.6 ± 0.2 | 14 | 0.23 ±0.02 | 60 ± 6 |
| hASRGL1-Gly9Ala* | 0.006 ± 0.001 | 0.0126 ± 0.0001 | 0.1 | 0.28 ±0.01 | 0.4 ± 0.02 |
| hASRGL1-Gly10Ala* | 0.0012 ± 0.0001 |
0.0053 ± 0.0001 | 0.3 | 0.19 ± 0.02 | 2 ± 0.2 |
| hASRGL1-Gly11Ala | 0.024 ± 0.003 | 4.4 ± 0.1 | 14 | 0.39 ± 0.03 | 35 ± 3 |
| cp-hASRGL1 | N/A | 25 ± 0.7 | N/A | 0.25 ± 0.02 | 100 ± 11 |
| cp-hASRGL1-Gly9Ala | N/A | 1.2 ± 0.1 | N/A | 0.17 ± 0.06 | 7 ± 3 |
| cp-hASRGL1-Gly10Ala | N/A | 0.6 ± 0.1 | N/A | 0.3 ± 0.07 | 2 ± 0.6 |
| cp-hASRGL1-Gly11Ala | N/A | 29 ± 0.7 | N/A | 0.3 ± 0.02 | 94 ± 8 |
p < 0.01, N/A = not applicable,
37 °C at pH 7.4.
Constraint-driven intramolecular processing
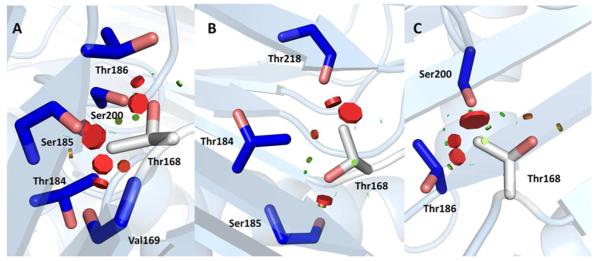
The nucleophilic residue Thr168 is located on the center strand of a β-sheet at the base of a deep and narrow active site funnel. Prior to backbone hydrolysis between Gly167 and Thr168, residues preceding the scissile bond (Leu166–Gly167) are observed to extend the central β-strand in the hASRGL1-Thr168Ala structure. When the Thr168 side chain is modeled into the precursor-like structure, it would appear to be sterically restrained from acting as a nucleophile where it is > 4 Å from the Gly167-Thr168 scissile bond. The β-hydroxyl group of the modeled Thr168 side chain forms favorable hydrogen bond interactions in all three of the most frequently observed rotamers (p, m, or t form) but the γ-methyl group has steric clashes in all conformations (Figure 3 A, B, C) (See Table S3 for a list of interactions). Molecular modeling of torsion angles of the Thr168 side chain shows that the active site Thr168 is restrained within the β-strand in any orientation. Therefore, the active site must accommodate some unfavorable interactions from the Thr168 γ-methyl group, suggestive of a constrained local conformation in the wild-type precursor. Intramolecular cleavage between Gly167 and Thr168 likely relieves the steric restraints upon Thr168 which becomes the first residue in the β-subunit. In the active site of cp-hASRGL1, we observed that the carbonyl group of the activated Thr168 consistently formed a hydrogen bond with hydroxyl group of Thr219. The side chain of Thr168 is flipped 180 degrees compared with Ala168 (Thr168Ala) in the hASRGL1-Thr168Ala structure. This orientation is further stabilized by hydrogen bonds with amide group and carbonyl group of Asn62. Cleavage of the peptide bond between Gly167 and Thr168 releases the side chain of Thr168 from a constrained space and extends the hydroxyl group of Thr168 into the active site. The nascent Thr168 amino group is within 2.7 Å of its side chain hydroxyl group, which is consistent with an intramolecular hydrogen bond as found in other plant-type asparaginases (6) (Supporting Figure S4).
Figure 3.
Modeled threonine rotamer conformations (A-C) in hASRGL1-Thr168Ala structure. Thr168 is represented by white sticks and the surrounding residues forming a close constrained space are represented by blue sticks. The visible disks and their colors indicate pairwise overlap of atomic van der Waals radii (A qualitative representation of clash). The green lines or disks are shown when atoms are almost in contact or slightly overlap, and the red disks represent significant van der Waals overlays which are proportional to disk size.
To investigate whether the active sites in hASRGL1 and variants are under strain, we determined the apparent melting temperature of hASRGL1-Thr168Ala and hASRGL1-Thr168Ser proteins, as well as freshly purified unprocessed wild-type hASRGL1 using differential scanning fluorometry (23). The unprocessed wild-type hASRGL1 (less than 1 % processed form present based on densitometric analysis of SDS-PAGE gels) exhibited a Tm of 61.0 ± 0.1 C° while the hASRGL1-Thr168Ala variant with a methyl side chain substitution exhibited a Tm = 71.0 ± 0.1 °C, a 10 °C increase. Likewise the hASRGL1-Thr168Ser variant, which differs from wild-type by the removal of the γ-methyl group, exhibits a Tm = 71.0 ± 0.1°C (Supporting Figure S5). This data strongly suggests that the γ-methyl group of Thr168 imparts a large steric strain upon the unprocessed enzyme that drives the Thr168 hydroxyl group into a catalytically relevant position suitable for attack on the Gly167-Thr168 scissile bond. The concept of a strain-propelled mechanism of autoprocessing of Ntn hydrolases has been proposed for E.coli isoaspartyl dipeptidase (19), glycosylasparaginase (24), and pantetheine hydrolase (25). More quantitatively, Härd et.al. have exhaustively examined the human Muc1 SEA (sea urchin sperm protein, enterokinase, and agrin) domain for the contribution of conformational strain to autoproteolysis and reported that torsional strain at the scissile bond lowered the energy barrier to cleavage by 7 kcal/mol (26, 27). Our data suggests that torsional strain within the active site may be a unifying feature amongst the Ntn hydrolase family and that in hASRGL1 the strain is provided by steric clashes of the Thr168 γ-methyl group.
Given the evidence for conformational strain in the active site described above, we also investigated whether the identity of Gly167 was crucial to the cleavage of the Gly167-Thr168 scissile bond. The residue preceding the nucleophilic (P−1) Thr residue in isoaspartyl dipeptidases is frequently a Gly residue, although other residues such as l-Asp and l-Ser are also present at the P−1 position. We constructed two variants, hASRGL1-Gly167Asp and hASRGL1-Gly167Ala, and measured their level of autoprocessing and steady-state kinetics for AHA hydrolysis. Both hASRGL1-Gly167Ala and hASRGL1-Gly167Asp were capable of intramolecular processing, although the Gly167Asp substitution resulted in a reduced level of final autoprocessing relative to wild-type enzyme (~5-10 %). After adjusting for the level of processed enzymes, the kinetics for AHA hydrolysis were essentially the same as for the wild-type enzyme. This suggests that for hASRGL1 the identity of this residue is not critical; only a small residue is necessary at the P−1 position and the extra rotational freedom provided by a Gly residue is not required.
SUMMARY
In this work, we used circular permutation to engineer a variant of hASRGL1 to decouple autoprocessing from substrate hydrolysis. This strategy allowed us to determine the structure of the first mammalian plant-type asparaginase in both a precursor and fully-activated form. With autoprocessing of the Gly167-Thr168 scissile bond no longer required, we were able to observe the kinetic effect of mutations in or around the active site that were previously unmeasurable, such as Thr168 substitutions with Ser. The hASRGL1 enzyme is critically dependent on the Thr168 γ-methyl group for orientation of the active site nucleophile as the Thr168Ser substitution results in > 1,000-fold reduction in kcat/KM for AHA hydrolysis. As implicated from structural and thermodynamic experiments, Thr168 γ-methyl is also required to produce a strained local conformation in the active site that drives the proper position and orientation of the Thr168 β-hydroxy group for attack on the Gly167-Thr168 scissile bond for cleavage. The strictly conserved Gly9-Gly10 residues are important for allowing a small loop near the active site (residues 8-16) sufficient flexibility to accommodate the C-terminal peptide from the α subunit and permit proper orientation of the Thr168 side chain relative to the Gly167-Thr168 scissile bond. Substitution of either Gly9 or Gly10 by Ala results in a large decrease in precursor autoprocessing and in the substrate hydrolysis activity of cp-hASRGL1. After intramolecular cleavage, the C-terminal peptide from the α subunit leaves the reaction center, releasing the torsional strain at the Thr168 γ-methyl group and resulting in the fully formed active site. The now exposed N-terminal amine of Thr168 acts as general base (amine pKa ~7.3) in deprotonating and activating its own β-hydroxy group for nucleophilic attack. Interestingly substrate binding appears to further enhance this effect by lowering the pKa to 6.0 for the Michaelis complex.
MATERIALS AND METHODS
All oligonucleotides were from Integrated DNA Technologies. Phusion DNA Polymerase and dNTPs were purchased from New England Biolabs. Crystal Screening Kits were from Hampton Research. O-phthalaldehyde (OPA) was from Agilent Technologies. All other reagents were from Sigma Aldrich unless otherwise noted. (The oligonucleotides used are listed in Supporting Table S4).
Molecular Biology Methods
The construction of hASRGL1 and hASRGL1-Thr168Ala expression vectors was described previously (1). The hASRGL1-Gly167Ala and hASRGL1-Gly167Asp variants were constructed by overlap extension PCR using a codon optimized hASRGL1 gene (1) as the template. The additional hASRGL1 point mutants Thr168Cys, Thr168Ser, Gly9Ala, Gly10Ala, and Gly11Ala were constructed using the QuikChange Site Directed Mutagenesis Kit (Stratagene). Circularly permuted hASRGL1 (cp-hASRGL1) was constructed by overlap extension PCR with oligonucleotides coding for a 12 amino acid linker between the β and α domains, the catalytic Thr168 residue as the 2nd amino acid following the initiator Met codon and finally a C-terminal 6×His tag. The linker length needed was designed by measuring the distance of the N- and C-termini (~26 Å) from the structure of the E.coli plant-type asparaginase (PDB: 2ZAL). Using an estimated length of 3.5 Å per amino acid resulted in 8 amino acids required to span the distance; to which we added four extra residues to accommodate omitting the Met1 residue and to allow for extra degrees of freedom. The linker was comprised of Gly, Ala, and Ser residues in an effort to create a flexible hydrophilic linker.
Two other circularly permuted variants were also constructed replacing the catalytic Thr residue with Cys or Ser. All genes were cloned into pET28a vector and sequenced to ensure no undesired mutations were introduced.
Expression and Purification
E. coli BL21(DE3) cells expressing hASRGL1 or variants as described, were cultured overnight at 37 °C in 2×YT medium supplemented with 30 μg/mL kanamycin and used to inoculate fresh medium (1:100 dilution). When the absorbance at 600 nm (A600) reached 0.5–0.7, the cells were cooled to 16 °C and allowed to equilibrate for 20 min, at which point the culture was supplemented with IPTG to a final concentration of 500 μM to induce protein expression. After incubation for ~16 hr at 16 °C, the cells were harvested by centrifugation at 5000 g for 15 min. The cell pellet was resuspended lysis buffer [50 mM tris-HCl, 100 mM NaCl, 10 mM β-mercaptoethanol, 10 mM imidazole, pH 8], and lysed by sonication on ice (Misonix sonicator 4000, amplitude 90 %) for 5 cycles (30 s each cycle with 1 s on/off pulses; 30 s pause between cycles). Following sonication, cell debris was removed by centrifugation at 15,000 g for 30 min. The resulting supernatant was loaded onto a pre-equilibrated Ni2+-NTA affinity column, and washed with 25 mM imidazole lysis buffer. The protein was finally eluted with 250 mM imidazole elution buffer and dialyzed at 4 °C for 16 hr either in buffer A [50 mM HEPES, 100 mM NaCl, 10 mM β-mercaptoethanol, pH 7.5] for kinetic analyses, or in buffer B [20 mM HEPES, 50 mM NaCl, 10 mM β-mercaptoethanol, 200 mM ammonium sulfate, pH 7.5] for hASRGL1 Thr168Ala, or in buffer C [50 mM Tris-HCl, 50 mM NaCl, 10 mM β-mercaptoethanol, pH 8.5] for cp-hASRGL1. The cp-hASRGL1 variant was then loaded onto a monoQ ion exchange chromatography column using 10 ml sepharose (GE Healthcare). For crystallization setup, both of the proteins had a final polishing step by size exclusion chromatography with a GE HiLoad™ 16/60 Superdex™ 200 column in buffer B for hASRGL1-Thr168Ala or in Buffer D [20 mM HEPES, 50 mM NaCl, 10 mM β-mercaptoethanol pH 7.5] for cp-hASRGL1. Expression and purification of site-directed mutants of hASRGL1 was conducted using a similar protocol as described above. Fresh protein samples were always used for all experiments.
Crystallization
Following purification, hASRGL1 variant proteins were concentrated to ~10 mg/mL with Vivaspin concentrator (GE Healthcare) and set up for sparse-matrix screening using a Crystal Phoenix liquid handling system (Art Robbins Instruments). Diffracting crystals were obtained by mixing 1 μL hASRGL1-Thr168Ala (10 mg/mL) with 1 μL crystallization solution (19-30 % PEG4000 (w/v), 0.1 M sodium acetate, pH 4.6), followed by incubation at 25°C. Within 3-4 days, thin needle-shape crystals were formed through vapor diffusion in sitting drops. The cp-hASRGL1 variant was first pre-incubated on ice with 20mM glycine (in buffer D) for three hours before crystallization setup. Crystals were found after 3–4 days in sitting drops. The crystals used for structural determination were from 17 % PEG3350 (w/v), 0.15 M potassium citrate at 4°C. Hexahedron cone shaped crystals were observed. Crystals were then transferred into stabilization solution (the respective crystallization condition with 25 % glycerol (v/v) for hASRGL1-Thr168Ala, or 20 % glycerol (v/v) for cp-hASRGL1). Following a brief equilibration period, the crystal was captured in a nylon loop and vitrified in liquid nitrogen for data collection.
Diffraction Data Collection and Structure Determination
X-Ray diffraction data were collected from the synchrotron radiation at beamlines 5.0.2 and 5.0.3 of the Advanced Light Source (Berkeley, CA) with 3 × 3 CCD array detectors (ADSC Q315R). Data were processed and scaled using the HKL2000 software suite (28). Data collection statistics are summarized in Table 2.
The structure of hASRGL1-Thr168Ala was determined using molecular replacement with the E. coli isoaspartyl peptidase (PDB code: 2ZAL) as the search model by the program Phaser from the CCP4 package suite (29). Structures were refined with autoBUSTER (30) along with iterative model building in COOT (31, 32). Non-crystallographic symmetry (autoNCS) restraints and translation, libration, and screw-motion (TLS) parameters were applied in autoBUSTER refinements. The finalized coordinates of hASRGL1 Thr168Ala were used as the model for cp-hASRGL1 molecular replacement by Phaser, and its geometry was applied as target restraint in addition to autoNCS restraints for cp-hASRGL1 refinement (33, 34). In autoBUSTER refinement 5 % test set (reflections) was excluded for Rfree cross-validation (35). The final structures were evaluated by PROCHECK (36) and MolProbity (37). The MolProbity scores of the structures reflect good stereochemistry. The cp-hASRGL1, as indicated by MolProbity score of 1.76, ranks 100th percentile among structures with comparable resolution, and an all atom clash score of 8.24 corresponding to 97th percentile among similar resolution structure. The evaluated hASRGL1-Thr168Ala structure gave a MolProbity score of 1.52 (99th percentile), and an all atom clash score of 5.87 (99th percentile). Refinement statistics are summarized in Table 2. Molecular figures were generated using PyMOL (38).
Tm measurement using Differential Scanning Fluorometry
Purified hASRGL1 variants in buffer A were diluted to ~40 μM in 96-well low-profile PCR plates (ABgene, Thermo Scientific) and pre-incubated on ice for 30 minutes. 10X SYPRO® Orange (Molecular Probes) was added into each well and mixed prior to centrifuging the plates at 500 rpm for one minute and placing them in an RT-PCR machine (LightCycler 480, Roche). The plates were sealed with adhesive and heat seals (ABgene, Thermo Scientific) to prevent evaporation. The protein melting experiments were carried out with a continuous temperature acquisition mode using 10 acquisitions per 1 °C in each cycle from 20 °C to 95 °C. The melting curves of the hASRGL1 variants were monophasic and Tm values were derived using Boltzmann equation (23).
AHA assay
Reactions of hASRGL1 or variants (0.5-1 μM total enzyme) with AHA (concentrations from 0–10X KM) were carried out at 37 °C in 50 mM HEPES, 100 mM NaCl, pH 7.4, in a total volume of 100 μL, and were then quenched with 5 μL 12 % (w/v) trichloroacetic acid. Subsequently, 50 μL color reagent (2 % 8-hydroxyquinoline in ethanol/1 M Na2CO3, 1:3 by volume) was added to each reaction well and the plate was covered, heated at 100 °C for 4 min, and allowed to cool at 4 °C for 10 min. The formation of hydroxylamine product was determined by measuring the absorbance at 705 nm (Synergy HT Fluorescent Plate reader, BioTek) and product concentrations were evaluated using a hydroxylamine standard curve generated under the same conditions. All reactions were repeated three times and the observed rates were fitted to the Michaelis-Menten equation using the program Kaleidagraph (Synergy).
pH Rate Dependence
The steady-state kinetics of cp-hASRGL1 catalyzed hydrolysis of AHA was assessed across a broad range of pH values, in triplicate at 25 °C. The following buffers were used: MES (pH 5.9-6.5), HEPES (pH 7-7.4), Tris (pH 8-8.5), and Capso (pH 9-9.5), all at a 100 mM concentration. After calculating the steady state parameters at each condition, kcat and kcat/KM were plotted against pH and fit to the Henderson-Hasselbalch equation reflecting either one or two ionization events. Using software from ChemAxon (18), the pKa values for the primary amines of substrate AHA and peptidyl Thr168 residue were calculated to assess their ionization values.
Intramolecular Processing Characterization
The hASRGL1 Gly9Ala, Gly10Ala, or Gly11Ala variants at 1 mg/mL in buffer A were incubated at 37 °C. At t = 0, 2, 4, 6, 8, 12, 24, and 48 hr, an aliquot of each enzyme was collected, and loaded onto an SDS-PAGE gel. Following Coomassie staining (Pierce), bands corresponding to the precursor and processed enzyme were quantified by densitometry (Odyssey, Li-core, NE). The rates of intramolecular processing for each variant were determined by non-linear fitting of fraction of processed enzyme versus time using KaleidaGraph (Synergy).
Supplementary Material
Acknowledgements
This work was supported by National Institutes of Health Grants 1 R01 CA154754 and R01 CA139059 and by the Cancer Prevention and Research Initiative of Texas (CPRIT) grant RP120314. We thank the staff of the Advanced Light Source (ALS), Berkeley, California for their assistance during data collection at beam lines 5.0.2 and 5.0.3. The authors declare no competing financial interests.
Footnotes
Accession Codes: hASRGL1-Thr168Ala and cp-hASRGL1 have been deposited in Protein Data Bank as 3tkj and 4et0, respectively.
Supporting Information Available: This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Cantor JR, Stone EM, Chantranupong L, Georgiou G. The human asparaginase-like protein 1 hASRGL1 is an Ntn hydrolase with beta-aspartyl peptidase activity. Biochemistry. 2009;48:11026–11031. doi: 10.1021/bi901397h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weidle UH, Evtimova V, Alberti S, Guerra E, Fersis N, Kaul S. Cell growth stimulation by CRASH, an asparaginase-like protein overexpressed in human tumors and metastatic breast cancers. Anticancer Res. 2009;29:951–963. [PubMed] [Google Scholar]
- 3.Dinndorf PA, Gootenberg J, Cohen MH, Keegan P, Pazdur R. FDA drug approval summary: pegaspargase (oncaspar) for the first-line treatment of children with acute lymphoblastic leukemia (ALL) Oncologist. 2007;12:991–998. doi: 10.1634/theoncologist.12-8-991. [DOI] [PubMed] [Google Scholar]
- 4.Aswad DW, Paranandi MV, Schurter BT. Isoaspartate in peptides and proteins: formation, significance, and analysis. J Pharm Biomed Anal. 2000;21:1129–1136. doi: 10.1016/s0731-7085(99)00230-7. [DOI] [PubMed] [Google Scholar]
- 5.Böhme L, Bär JW, Hoffmann T, Manhart S, Ludwig HH, Rosche F, Demuth HU. Isoaspartate residues dramatically influence substrate recognition and turnover by proteases. Biological chemistry. 2008;389:1043–1053. doi: 10.1515/BC.2008.123. [DOI] [PubMed] [Google Scholar]
- 6.Michalska K, Jaskolski M. Structural aspects of L-asparaginases, their friends and relations. Acta Biochim Pol. 2006;53:627–640. [PubMed] [Google Scholar]
- 7.Brannigan JA, Dodson G, Duggleby HJ, Moody PC, Smith JL, Tomchick DR, Murzin AG. A protein catalytic framework with an N-terminal nucleophile is capable of self-activation. Nature. 1995;378:416–419. doi: 10.1038/378416a0. [DOI] [PubMed] [Google Scholar]
- 8.Xu Q, Buckley D, Guan C, Guo HC. Structural insights into the mechanism of intramolecular proteolysis. Cell. 1999;98:651–661. doi: 10.1016/s0092-8674(00)80052-5. [DOI] [PubMed] [Google Scholar]
- 9.Li Y, Chen J, Jiang W, Mao X, Zhao G, Wang E. In vivo post-translational processing and subunit reconstitution of cephalosporin acylase from Pseudomonas sp. 130. European Journal of Biochemistry. 1999;262:713–719. doi: 10.1046/j.1432-1327.1999.00417.x. [DOI] [PubMed] [Google Scholar]
- 10.Burtscher H, Schumacher G. Reconstitution in vivo of penicillin G acylase activity from separately expressed subunits. European Journal of Biochemistry. 1992;205:77–83. doi: 10.1111/j.1432-1033.1992.tb16753.x. [DOI] [PubMed] [Google Scholar]
- 11.Boanca G, Sand A, Barycki JJ. Uncoupling the enzymatic and autoprocessing activities of Helicobacter pylori -glutamyltranspeptidase. Journal of Biological Chemistry. 2006;281:19029. doi: 10.1074/jbc.M603381200. [DOI] [PubMed] [Google Scholar]
- 12.Yu Y, Lutz S. Circular permutation: a different way to engineer enzyme structure and function. Trends in biotechnology. 2011;29:18–25. doi: 10.1016/j.tibtech.2010.10.004. [DOI] [PubMed] [Google Scholar]
- 13.Frottin F, Martinez A, Peynot P, Mitra S, Holz RC, Giglione C, Meinnel T. The proteomics of N-terminal methionine cleavage. Molecular & Cellular Proteomics. 2006;5:2336. doi: 10.1074/mcp.M600225-MCP200. [DOI] [PubMed] [Google Scholar]
- 14.Frohwein YZ, Friedman M, Reizer J, Grossowicz N. Sensitive and rapid assay for L-asparaginase. Nature. 1971;230:158–159. doi: 10.1038/newbio230158a0. [DOI] [PubMed] [Google Scholar]
- 15.Lee YS, Kim HW, Park SS. The role of alpha-amino group of the N-terminal serine of beta subunit for enzyme catalysis and autoproteolytic activation of glutaryl 7-aminocephalosporanic acid acylase. Journal of Biological Chemistry. 2000;275:39200. doi: 10.1074/jbc.M002504200. [DOI] [PubMed] [Google Scholar]
- 16.Liu Y, Dunn GS, Aronson NN., Jr Purification, biochemistry and molecular cloning of an insect glycosylasparaginase from Spodoptera frugiperda. Glycobiology. 1996;6:527–536. doi: 10.1093/glycob/6.5.527. [DOI] [PubMed] [Google Scholar]
- 17.Segel IH. Enzyme kinetics : behavior and analysis of rapid equilibrium and steady state enzyme systems. Wiley; New York: 1975. [Google Scholar]
- 18.MarvinSketch ChemAxon. 5.5.0.1 edn 2011.
- 19.Michalska K, Hernandez-Santoyo A, Jaskolski M. The mechanism of autocatalytic activation of plant-type L-asparaginases. Journal of Biological Chemistry. 2008;283:13388–13397. doi: 10.1074/jbc.M800746200. [DOI] [PubMed] [Google Scholar]
- 20.Potterton E, Briggs P, Turkenburg M, Dodson E. A graphical user interface to the CCP4 program suite. Acta Crystallogr D Biol Crystallogr. 2003;59:1131–1137. doi: 10.1107/s0907444903008126. [DOI] [PubMed] [Google Scholar]
- 21.Michalska K, Brzezinski K, Jaskolski M. Crystal structure of isoaspartyl aminopeptidase in complex with L-aspartate. Journal of Biological Chemistry. 2005;280:28484–28491. doi: 10.1074/jbc.M504501200. [DOI] [PubMed] [Google Scholar]
- 22.Michalska K, Bujacz G, Jaskolski M. Crystal structure of plant asparaginase. Journal of molecular biology. 2006;360:105–116. doi: 10.1016/j.jmb.2006.04.066. [DOI] [PubMed] [Google Scholar]
- 23.Niesen FH, Berglund H, Vedadi M. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nature Protocols. 2007;2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Guo HC. Crystallographic snapshot of glycosylasparaginase precursor poised for autoprocessing. Journal of molecular biology. 2010;403:120–130. doi: 10.1016/j.jmb.2010.08.038. [DOI] [PubMed] [Google Scholar]
- 25.Buller AR, Freeman MF, Wright NT, Schildbach JF, Townsend CA. Insights into cis-autoproteolysis reveal a reactive state formed through conformational rearrangement. Proceedings of the National Academy of Sciences. 2012;109:2308–2313. doi: 10.1073/pnas.1113633109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johansson DGA, Macao B, Sandberg A, Härd T. SEA domain autoproteolysis accelerated by conformational strain: mechanistic aspects. Journal of molecular biology. 2008;377:1130–1143. doi: 10.1016/j.jmb.2008.01.050. [DOI] [PubMed] [Google Scholar]
- 27.Sandberg A, Johansson DGA, Macao B, Härd T. SEA domain autoproteolysis accelerated by conformational strain: energetic aspects. Journal of molecular biology. 2008;377:1117–1129. doi: 10.1016/j.jmb.2008.01.051. [DOI] [PubMed] [Google Scholar]
- 28.Otwinowski Z, M. W. Processing of X-ray Diffraction Data Collected in Oscillation Mode. Vol. 276. Academic Press; New York: 1997. [DOI] [PubMed] [Google Scholar]
- 29.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 30.Bricogne G, B. E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, Sharff A, Smart OS, Vonrhein C, Womack TO. BUSTER. version 2.10 Global Phasing Ltd.; Cambridge, United Kingdom: 2011. [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallographica Section D. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 33.Painter J, Merritt EA. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr D Biol Crystallogr. 2006;62:439–450. doi: 10.1107/S0907444906005270. [DOI] [PubMed] [Google Scholar]
- 34.Smart OS, Womack TO, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. Exploiting structure similarity in refinement: automated NCS and targetstructure restraints in BUSTER. Acta Crystallogr D Biol Crystallogr. 2012;68:368–380. doi: 10.1107/S0907444911056058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brunger AT. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–475. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski RA, Macarthur MW, Moss DS, Thornton JM. Procheck - a Program to Check the Stereochemical Quality of Protein Structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 37.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallographica Section D: Biological Crystallography. 2009;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schrodinger, LLC . The PyMOL Molecular Graphics System. Version 1.3r1 2010. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.