Abstract
The single-strand overhang present at all known telomeres plays a critical role in mediating both the capping and telomerase regulation functions of telomeres. The telomere end-binding proteins, Cdc13 in S. cerevisiae, Pot1 in higher eukaryotes, and TEBP in the ciliated protozoa O. nova, exhibit sequence-specific binding to their respective single-strand overhangs. S. cerevisiae telomeres are composed of a heterogeneous mixture of GT-rich telomeric sequence, unlike in higher eukaryotes which have a simple repeat that is maintained with high fidelity. In yeast, the telomeric overhang is recognized by the essential protein Cdc13, which coordinates end-capping and telomerase activities at the telomere. The Cdc13-DNA-binding domain (Cdc13-DBD) binds these telomere sequences with high affinity (3 pM) and sequence specificity. To better understand the basis for this remarkable recognition, we have investigated the binding of the Cdc13-DBD to a series of altered DNA substrates. Although an 11-mer of GT-rich sequence is required for full binding affinity, only 3 of these 11 bases are recognized with high specificity. This specificity differs from that observed in the other known telomere end-binding proteins, but is well suited to the specific role of Cdc13 at yeast telomeres. These studies expand our understanding of telomere recognition by the Cdc13-DBD and of the unique molecular recognition properties of ssDNA binding.
Recognition of single-strand DNA (ssDNA) is essential for many fundamental cellular activities, including recombination, repair, replication, transcription, translation, and telomere maintenance. Structural and biochemical studies of the proteins that mediate ssDNA-binding activity have provided key insights into the mechanism of recognition. Generally, small domains containing mixed α/β topologies, such as the OB-fold and KH domain, are used for ssDNA recognition, and binding is achieved through base and backbone interactions that are completely distinct from those used in double-strand DNA recognition (1–5). In many biological contexts DNA needs to be uniformly recognized, thus it is appropriate for this recognition to be non-sequence specific. However, in a few cases, including telomere maintenance, transcription, and viral packaging, it is imperative that recognition be exquisitely sequence specific (3, 6–8).
Telomeres, the DNA, RNA and protein complexes at the ends of chromosomes, play important roles in many essential cellular functions. They regulate the proliferative lifetime of the cell and protect chromosomes from degradation or end-to-end fusion, as well as participate in meiotic segregation and chromatic silencing (reviewed in (9–13)). Telomeres are critical to the maintenance of the genome and normal development. Thus, telomere malfunction can have dramatic adverse impacts on human health, leading to cancer, aging disorders and increased human mortality (14–19). Therefore, it is essential to understand the mechanisms through which telomeres are properly maintained.
In all organisms studied thus far, telomeres terminate in a highly regulated single-strand overhang comprised of the GT-rich strand. This end must be sequestered from cellular factors to prevent chromosomal degradation, as well as to discriminate the end structure from damaged DNA (20–22). A family of single-strand DNA-binding proteins with binding specificity for the GT sequences at telomeres has been identified whose functions include protection of the overhang from degradation and regulation of telomerase activity (8, 23–26). Members of this family include the Pot1 (protection of telomeres) proteins from S. pombe to human (24), the TEBPs (telomere end-binding proteins) from ciliates (27, 28), and the Cdc13 proteins from budding yeasts (8).
Cdc13 is strictly required for proper S. cerevisiae telomere function (8, 29, 30) and mediates the activity of two primary telomere functions: end protection and telomerase regulation. Deletion of Cdc13 is lethal due to loss of the essential end protection function. Loss of Cdc13 activity leads to dramatic resection of the C-rich strand of the chromosome (up to 10,000 bases) (31). This DNA damage activates the Rad9 DNA damage pathway, resulting in cell-cycle arrest at G2/M (31, 32). In addition to end protection, Cdc13 is both a positive and negative regulator of telomere length (23, 33). Loss of the ability of Cdc13 to interact with telomerase leads to progressive erosion of telomeres, resulting in a delayed lethal phenotype (34, 35). Negative regulation of telomere length is less well understood, but is believed to involve the interplay of additional end capping proteins (33). Extensive biochemical and genetic data indicate that Cdc13 serves as the primary anchor point for assembly of the functional complexes at the telomere through its specific interaction with the G-rich tail of telomeres. This ssDNA-binding activity is performed by a centrally located domain within the full-length protein that spans residues 497–694 (Cdc13-DBD) (36).
High fidelity recognition of S. cerevisiae telomeres presents a biochemical challenge due to the intrinsic heterogeneity of the sequence. Rather than consisting of a simple repeat that is maintained with high integrity, yeast telomeres are a mixture of sequential guanines interspersed within runs of GTs. Sequence patterns can be summarized with the consensus sequence {(TG)0–6TGGGTGTG(G)n}(39). This heterogeneity can be understood by examining the features of the unusually large telomerase RNA templating sequence, (here 3’–5’) ACACACACCCACACCAC (41). The variability observed of yeast telomeres can be completely explained by a combination of abortive reverse transcription of this template and the ability of the template to bind substrate in more than one register (39). The Cdc13-DBD must recognize these variable sequences and also remain specific for telomeric DNA to ensure proper telomere maintenance and to prevent aberrant telomerase activity at other sites, such as double-strand breaks.
Cdc13-DBD binds single-strand telomeric DNA with unusually high affinity, with a dissociation constant of 3 pM at low ionic strength (36). The NMR structure of the Cdc13-DBD/DNA complex revealed that the Cdc13-DBD adopts an oligosaccharide/oligonucleotide binding motif (OB-fold), the topology present in both the Pot1 (42, 43) and TEBP (44) proteins. An extensive binding interface is formed between Cdc13-DBD and the minimal telomeric DNA (GTGTGGGTGTG, Tel-11) (45–47). Specific sites in the protein that contribute to affinity for DNA were identified by a complete alanine scan of the protein/DNA interface (48). Analysis of the mutations that abolish DNA binding in vivo revealed that most are lethal, highlighting the requirement for appropriate Cdc13 DNA-binding activity for cellular viability (R. B. Cervantes, L. Ricks, J. N. Roberts, DSW, and V. Lundblad unpublished data). Here, we report the in vitro binding affinity of Cdc13-DBD to a series of DNA ligands containing several natural and unnatural base substitutions to probe the molecular requirements for the unusual specificity necessary for biological function. Our results indicate that while a surprisingly small set of bases confers sequence-specific binding, this minimal set is enough to ensure the biologically required telomere localization. The specificity exhibited by the Cdc13-DBD is distinct from that observed in other telomere end-binding proteins, illustrating the diversity exhibited by the OB-fold in performing this specialized function.
EXPERIMENTAL PROCEDURES (MATERIALS AND METHODS)
Cdc13-DBD protein
The minimal DNA-binding domain of the telomere end-binding protein from S. cerevisiae, Cdc13 spans residues 497–694 of the full-length protein (36). The C-terminally His-tagged Cdc13-DBD was expressed in E. coli and purified using a nickel-charged HiTrap Chelating HP column (GE Healthcare/Amersham Biosciences) as previously described (36). This His-tagged protein binds to the single-strand DNA ligand Tel-11 (dGTGTGGGTGTG) with affinity indistinguishable from the non-His-tagged version. Protein concentrations were determined by absorbance at 280 nm (ε=14320 M−1cm−1).
DNA ligands
DNA ligands were purchased at µg scale (Operon) and purified by reversed-phase HPLC (36). DNA concentrations were determined by absorbance at 260 nm using standard extinction coefficients calculated from the sequence. Binding reactions contained 32P-labeled single-strand DNA ligands (Tables 1 and 2) at concentrations (30 to 50 pM) at least 100-fold below the measured dissociation constants.
Table 1.
Thermodynamic and affinity changes for Cdc13-DBD binding to Tel-11 containing a degenerate library substitution at each site in the DNA.
| Sample | Sequence | ΔΔG (kcal/mol) ensemble |
Average relative Kd |
|---|---|---|---|
| WT | GTGTGGGTGTG | 1.0 | |
| 1 | 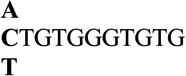 |
2.5 ± 0.3 | 95 |
| 2 | 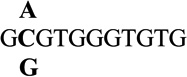 |
1.0 ± 0.1 | 5.9 |
| 3 | 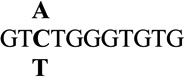 |
3.4 ± 0.1 | 450 |
| 4 | 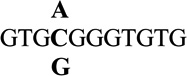 |
3.2 ± 0.3 | 360 |
| 5 | 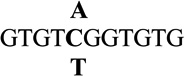 |
−0.6 ± 0.1 | 0.4 |
| 6 | 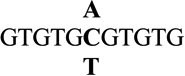 |
0.1 ± 0.1 | 1.1 |
| 7 | 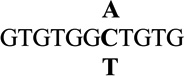 |
0.9 ± 0.1 | 5.0 |
| 8 | 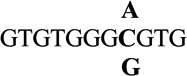 |
−0.5 ± 0.2 | 0.4 |
| 9 | 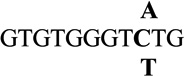 |
0.8 ± 0.2 | 4.1 |
| 10 | 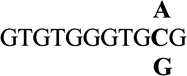 |
0.2 ± 0.2 | 1.4 |
| 11 | 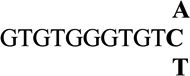 |
−0.1 ± 0.02 | 0.9 |
Table 2.
Thermodynamic and affinity changes for Cdc13-DBD binding to Tel-11 containing single substitutions.
| Sample | Sequence | Average ΔΔG (kcal/mol) |
Average relative Kd |
|---|---|---|---|
| WT | GTGTGGGTGTG | 0 | 1 |
| A1 | ATGTGGGTGTG | 2.9 ± 0.1 | 200 |
| C1 | CTGTGGGTGTG | 3.3 ± 0.1 | 370 |
| T1 | TTGTGGGTGTG | 2.8 ± 0.1 | 170 |
| S1 | STGTGGGTGTG | 2.8 ± 0.2 | 160 |
| I1 | ITGTGGGTGTG | 0.7 ± 0.1 | 3.8 |
| A3 | GTATGGGTGTG | 3.3 ± 0.1 | 410 |
| C3 | GTCTGGGTGTG | 4.3 ± 0.1 | >2300 |
| T3 | GTTTGGGTGTG | 3.5 ± 0.4 | >540 |
| S3 | GTSTGGGTGTG | 3.2 ± 0.4 | 340 |
| I3 | GTITGGGTGTG | 0.2 ± 0.1 | 1.4 |
| A4 | GTGAGGGTGTG | 3.9 ± 0.1 | >1100 |
| C4 | GTGCGGGTGTG | 3.7 ± 0.1 | >830 |
| G4 | GTGGGGGTGTG | 2.8 ± 0.1 | 170 |
| U4 | GTGUGGGTGTG | 0.4 ± 0.1 | 2.2 |
Nitrocellulose filter binding studies
Equilibrium binding reactions were performed with a fixed concentration of DNA ligand by varying the concentration of protein over a broad range in binding buffer (5 mM HEPES pH 7.8, 600 mM LiCl, 2.5 mM MgCl2, 0.1 mM Na2EDTA, 2 mM DTT, and 0.1 mg/ml BSA) (36, 48). This buffer is identical to one used in our previous studies with the exception of the monovalent salt (36, 48). In this study, we substituted 600 mM LiCl for 750 mM KCl because LiCl has been shown to reduce the propensity of G-quadruplex formation (49, 50). Although these structures have not been observed in our gel electrophoresis assays with the canonical ligand, Tel-11 GTGTGGGTGTG, we wanted to minimize the possibility of G-quadruplex formation for some of the G-rich mutant ligands in this study. The binding constant for Cdc13-DBD with Tel-11 in 600 mM LiCl (2.7 nM) was comparable to the binding constant in 750 mM KCl (1 nM). While the possibility exists that substitutions in the Tel-11 DNA cause structural changes in the Cdc13-DBD, preliminary NMR data shows that global structure of Cdc13-DBD is not significantly altered when several different DNA variants are bound (M. P. Latham and DSW unpublished data).
The 96-well MultiScreen MAHA N4550 nitrocellulose filter plates (Millipore) were prepared for binding by pre-wetting for 1 minute with 80 µl of the binding buffer lacking DTT and BSA. Samples containing protein and DNA were incubated an hour on ice and were then loaded onto the filter paper in the wells and incubated for 1 minute before filtering with a MultiScreen Resist Vacuum manifold. The wells were washed twice with 200 µl of buffer without DTT and BSA, air dried and then exposed to a Phosphorimager screen. ImageQuant (Molecular Dynamics) was used to quantify radioactivity on the filters. Data was fit to a standard two-state binding model, y = C1(x/(x+Kd)) + C2, where y is the magnitude of radioactivity bound to the protein, x is the protein concentration, C2 is the baseline or background counts, C1 is the total change in counts (value at the plateau minus the background counts) and Kd is the apparent equilibrium constant. The reported ΔΔG values of the ensemble in the degenerate experiment and in the single substitution experiment are an average of experiments repeated under the same conditions. ΔΔG values for the single substitution 9 experiment were determined using an average value for the wild-type Kd. In the case of the degenerate library experiment, the ΔΔG values were calculated using a value for the wild-type Kd determined for each plate to correct for a systematic difference in sample binding from plate to plate.
RESULTS
Identification of “hot spots” for Cdc13 specificity within telomere sequence
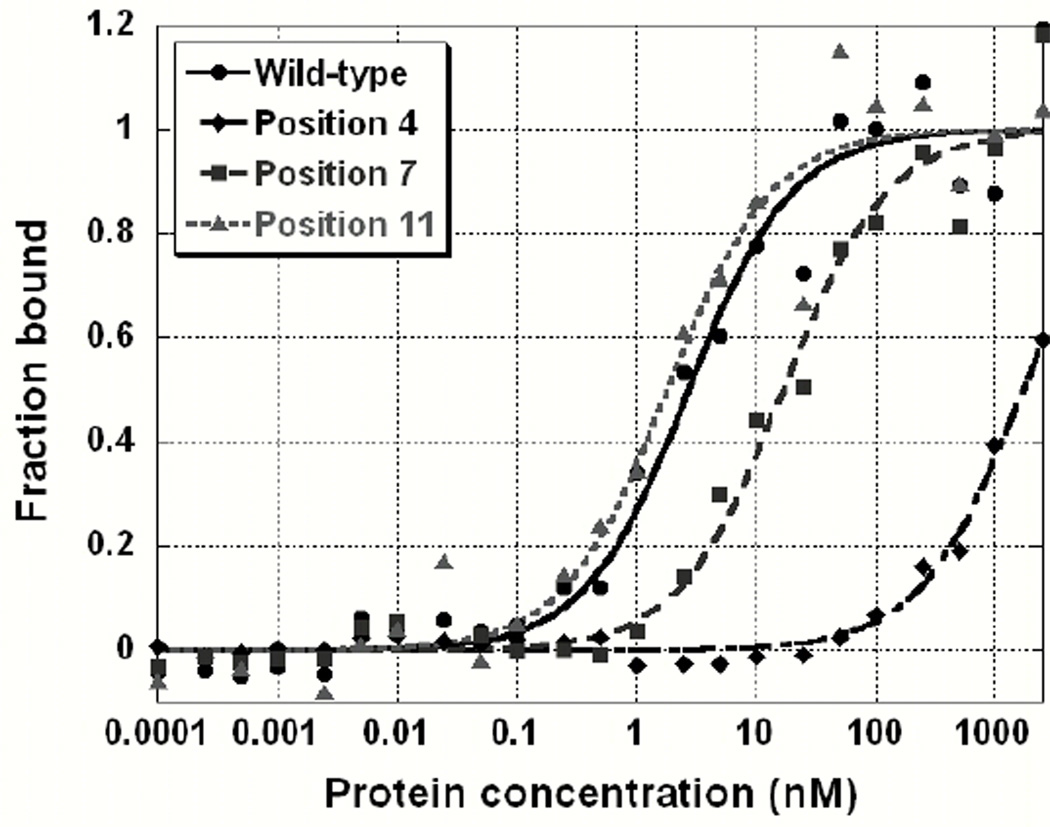
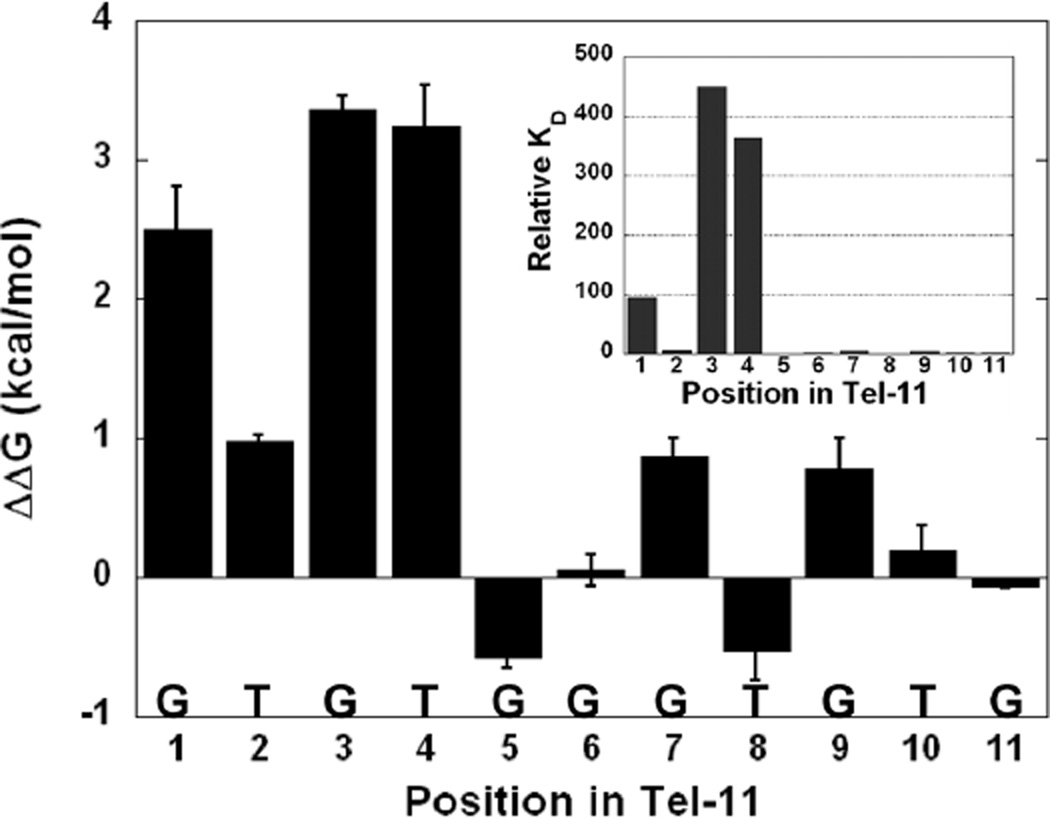
In order to efficiently identify the bases in Tel-11 critical for recognition, we substituted an approximately equimolar mixture of the 3 other non-canonical natural bases for the wild-type base at a given position in the 11-mer (Table 1). Specificity was assessed by comparing of the apparent binding affinities for the cognate and randomized sequences. Surprisingly, the effects of substitution varied dramatically throughout Tel-11. The relative binding affinities spanned a wide range, from slight changes to a 450-fold reduction in affinity. The effects partitioned into three classes: large (>100-fold), small (4–100-fold), and negligible (<2-fold) (Table 1). Figure 1 shows representative binding data from these binding experiments using one example from each class compared to the wild-type Tel-11. Large reductions in affinity were observed for substitutions to bases in positions 1, 3 and 4 at ~100-fold, ~450-fold, and ~400-fold, respectively. Replacing bases at positions 2, 7, and 9 has a small effect (~4–6-fold) on affinity while substitution of any of the remaining positions (5, 6, 8, 10, and 11) results in minor effects on measured binding. Figure 2 highlights the distinction between the bases at 5’ end of Tel-11 required for specificity and the bases at the 3’ end that are not individually required for specificity (although they are required for high affinity binding, vide infra).
Figure 1.
Binding curves from the degenerate library experiment illustrating the range of measured binding affinities. Data are fit to a two-state model as described in Materials and Methods. Wild-type binding data is shown in filled circles. Data obtained from replacement of the canonical base with a library of alternate natural bases for positions 4, 7 and 11 are shown by diamonds, squares, and triangles, respectively.
Figure 2.
Graphical representation of the ensemble average ΔΔG (kcal/mol) values for substitution of individual bases with a library of other natural bases. The inset graph shows the relative binding constants at each position.
Role of the 3’ end of Tel-11
None of the bases at the 3’ end of Tel-11 are individually required for binding specificity since substitution at each position with the pool of natural bases has only small effects on affinity. However, the 3’ bases are known to be important for maintaining affinity since binding to shorter ligands results in decreased binding (data not shown and (45)). Loss of the terminal base results in a 20-fold reduction in binding and no binding is observed for an 8-mer ligand as assessed by analogous filter binding experiments (data not shown). In addition, changing every site that is not strongly specifically recognized to its complementary base (creating the GAGTCCCACAC substrate) results in a complete loss of binding. This indicates that, while a wholesale substitution of the 3’ region is not tolerated by Cdc13-DBD, this region maintains a preference for GT-rich sequences. The striking difference in the magnitude of affinity effects depending on position in Tel-11 led to further study of atomic determinants for specificity at the 5’ end of the DNA ligand.
Single-substitution experiments delineate the chemical moieties required for specific recognition at the 5’ end
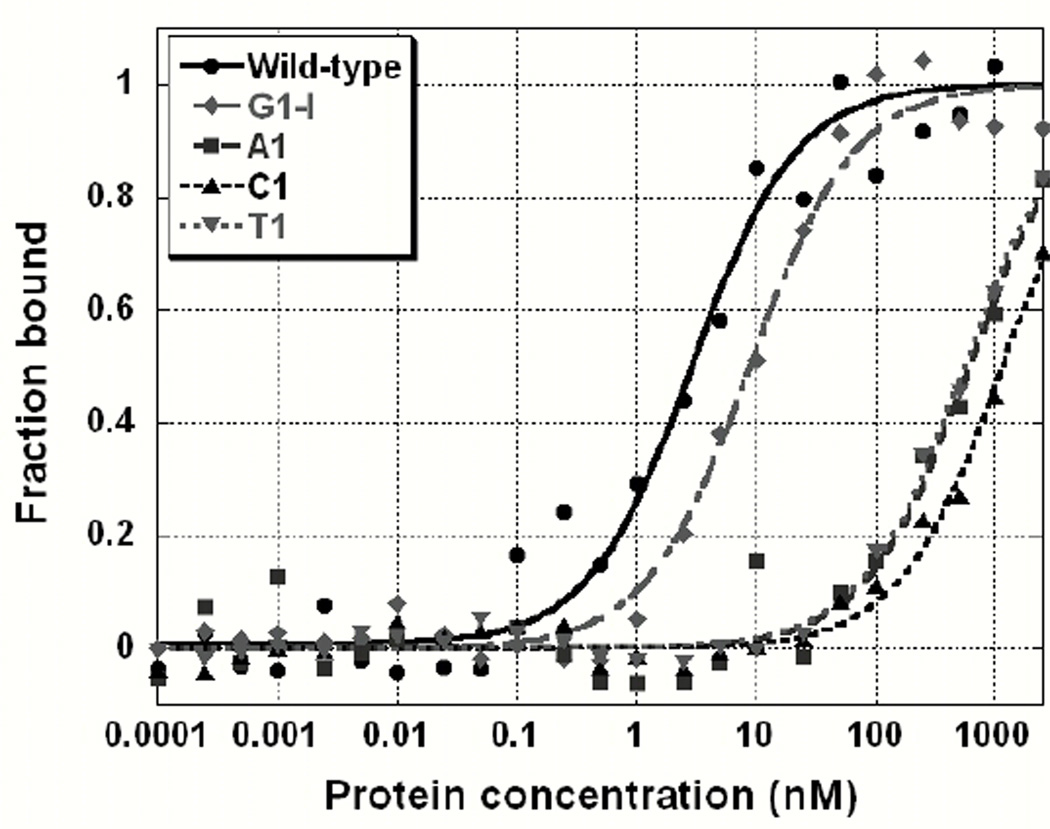
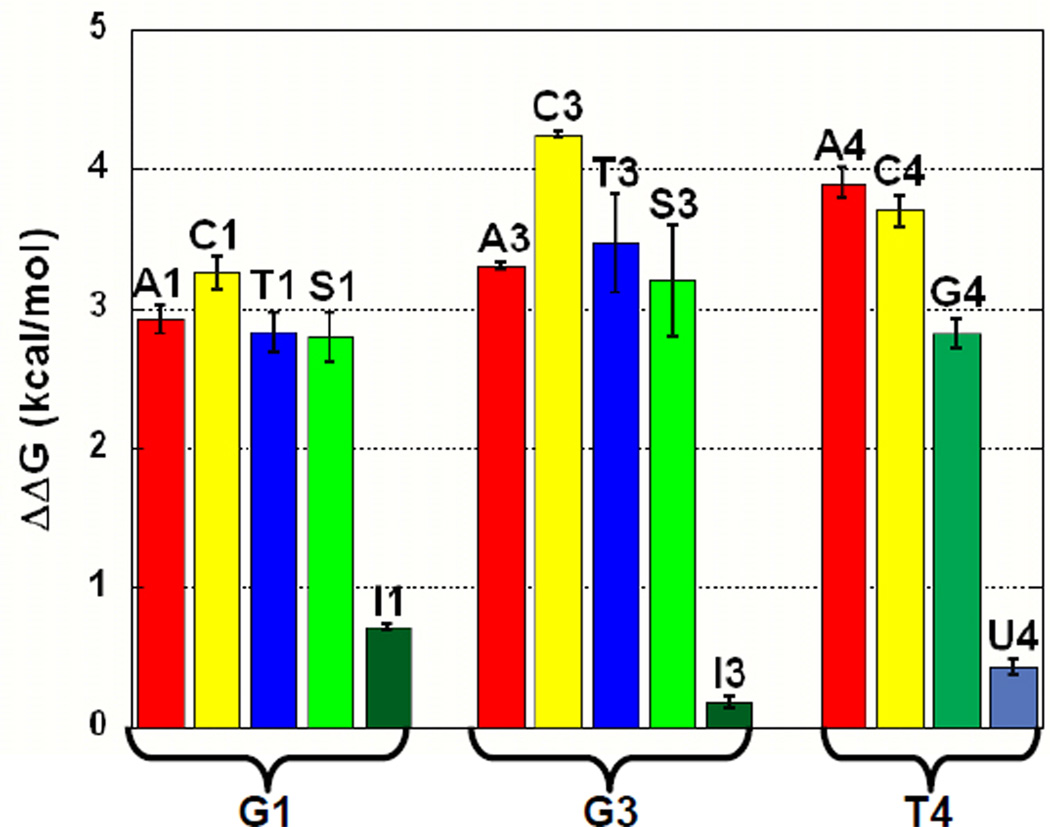
The chemical moieties required for specific binding at the 5’ end of the DNA were identified using a series of singly substituted ligands (see Materials and Methods, Table 2). Bases with the largest affinity reductions in the library experiment (positions 1, 3, and 4) were individually substituted with the three alternative natural bases and several non-natural variants. The natural base substitutions test for the importance of base size and hydrogen bond donor/acceptor patterns while the non-natural variants test specific chemical moieties. For example, inosine is identical to guanine but lacks the extra-cyclic amine at the 2 carbon. Inosine was substituted at positions 1 and 3 to test the thermodynamic importance of the extra-cyclic amine on binding by guanine. The variant thio-G, in which the 6-carbonyl oxygen is replaced with sulfur, was tested to determine the importance of the oxygen as a hydrogen-bonding acceptor. Thio-G reports on hydrogen bonding because the hydrogen bond accepting potential of the sp2 hybridized sulfur is much weaker than that of oxygen (51). Deoxy-uracil was substituted at position 4 to test the importance of the ring methyl of thymine on binding. Qualitatively, the aggregate binding data from this single-substitution experiment mirrored the results from the library experiment. Substitution of any other natural base at positions 1, 3, and 4 resulted in a marked decrease in binding affinity. The differences in the relative affinities between the substituted ligands and for the base variants provide insights into the base features required for specific recognition by Cdc13-DBD.
G1 substitutions
Guanine has several possible specificity determinants, including the size of the purine base and specific interactions with hydrogen bond acceptors/donors. Recognition of the base could occur through the Watson-Crick (WC) interaction face, which includes the extra cyclic amine, the 1-imino and the 6-carbonyl oxygen, or the Hoogstein face, which includes the N7 and the 6-carbonyl oxygen. Figure 3 illustrates the large range of binding affinities observed when G1 is substituted with natural and non-natural ligands. Replacing G1 with inosine results in a small reduction in affinity (~3-fold) indicating that the extra cyclic amine is not required for recognition or affinity. In contrast, the 6-oxygen is very important for recognition, since substitution of G1 with thio-G results in ~150-fold reduction in affinity. Substitution with adenine represents a complete change in the WC acceptors and donors and leads to a ~200-fold decrease in affinity. This loss of binding is approximately equal to swapping the 6-carbonyl oxygen for sulfur (thio-G), indicating that the 1-imino donor contributes minimally to binding. Also, this result suggests that the Hoogstein hydrogen bond acceptor (N7) cannot compensate for the loss of the 6-oxygen. We substituted thymine for G1 to test the importance of the size and/or shape of the purine base on binding, while preserving a WC-face carbonyl oxygen hydrogen bond acceptor. A decrease in affinity of 160-fold indicates that size/shape of the purine base is important for binding. This conclusion is supported by the cytosine substitution, which causes a reduction in affinity (~370-fold) that is more than twice the effect of the thymine or thio-G substitutions and nearly double the adenine substitution. These data indicate that both the purine base geometry and the 6-oxygen are critical for recognition and binding of the guanine at position 1.
Figure 3.
Binding curves for site-specific substitutions at G1. Replacement of G1 with other natural bases has a large effect on binding affinity, while inosine has a modest effect. Data are fit to a two-state model as described in Materials and Methods. Wild-type data is shown in circles, inosine with diamonds, A, C and T are shown in squares, traditional triangles, and upside down triangles, respectively.
G3 substitutions
Guanine at position 3 is absolutely essential for recognition of Tel-11 by Cdc13-DBD as no other natural base is tolerated at this position. The trends in affinity changes upon substitution closely parallel those observed for substitution at position 1. Substitution with adenine, thymine, and thio-G have large effects on affinity (410-, 540-, and 340-fold decreases, respectively), while substitution with inosine results in essentially no change in affinity (~1.3-fold decrease). Also, replacing G3 with a cytosine leads to the greatest reduction in apparent Kd of any substitution in the study (>2000-fold decrease). Therefore, both the purine base geometry and the 6-oxygen are critical for recognition and affinity. Interestingly, thymine is less well tolerated here compared to position 1, suggesting that the geometry of the purine base is more important for specificity and affinity at position 3. Nucleotide G3 is the most important base for recognition and specificity, since position 3 has a much lower tolerance for altering any of these determinants than any other site in Tel-11.
T4 substitutions
The thymine at position 4 is highly specifically recognized by the Cdc13-DBD. The thermodynamic effects of substitution at position 4 are second in magnitude only to those at G3 (Table 2), revealing that the GT pair at position 3 is a stringent specificity determinant for Cdc13. Substitution of T4 with cytosine results in a >830-fold reduction in binding affinity, suggesting that this pyrimidine ligand cannot be accommodated in the binding pocket to satisfy either steric or hydrogen-bonding requirements. Substitution of T4 with deoxy-uracil produces a very small change (~2.2-fold decrease), indicating that the ring methyl is not important for specificity. Purine bases are not well tolerated at this position, with binding by adenine decreased by >1100 fold and guanine somewhat better tolerated, exhibiting a 170-fold decrease in affinity. Thus, base size is clearly an important discriminant at this position. The pronounced lack of tolerance for substitution at this site suggests that the alternate bases cannot reorient to find favorable steric or hydrogen-bonding interactions, consistent with limited conformational malleability at this site. In contrast to the pronounced effects of substitution at T4, modification from T at position 2 to any other base is well tolerated and results in modest (< 30-fold) decreases in affinity (data not shown), consistent with the data from the pooled oligonucleotide experiment.
DISCUSSION
A Cdc13-DBD specificity motif within Tel-11
A subset of bases at the 5’ end of Tel-11, G1, G3 and T4, are specifically recognized by the Cdc13-DBD. Substitution of these bases to either a mixture of alternate bases or to each of the other natural bases results in sizable decreases in binding free energy (see Figures 1–4). In contrast, individual substitution of any of the bases at the 3’ end (5–11) to a library of the other natural bases results in only modest changes in binding (Table 1, Figure 2). Thus, the G1X2G3T4 motif is the specificity element for Cdc13. Recognition of the two guanines requires a hydrogen bond acceptor, the 6-carbonyl oxygen, and favors the purine base geometry. The hydrogen-bonding pattern on T4 is also required for recognition with base size or shape being less important. In contrast, all of the bases in Tel-11 are required for high affinity binding as Cdc13-DBD binds weakly to shorter ligands, suggesting that non-specific backbone and/or stacking interactions are dominant in the 3’ end of the Tel-11 ligand.
Figure 4.
Graphical representation of the energetic impact of single substitutions on binding to Cdc13-DBD. The alternative bases substituted for each of the first four bases in Tel-11 are shown color coded by base type, adenine in red, cytosine in yellow, thymine in blue, and guanine in green. Analogs of guanine are in alternate shades of green, thio-G (S) in light green and dark green for inosine (I). The analog of thymine, uracil (U), is shown in light blue.
Coincidence of the protein and DNA determinants of binding
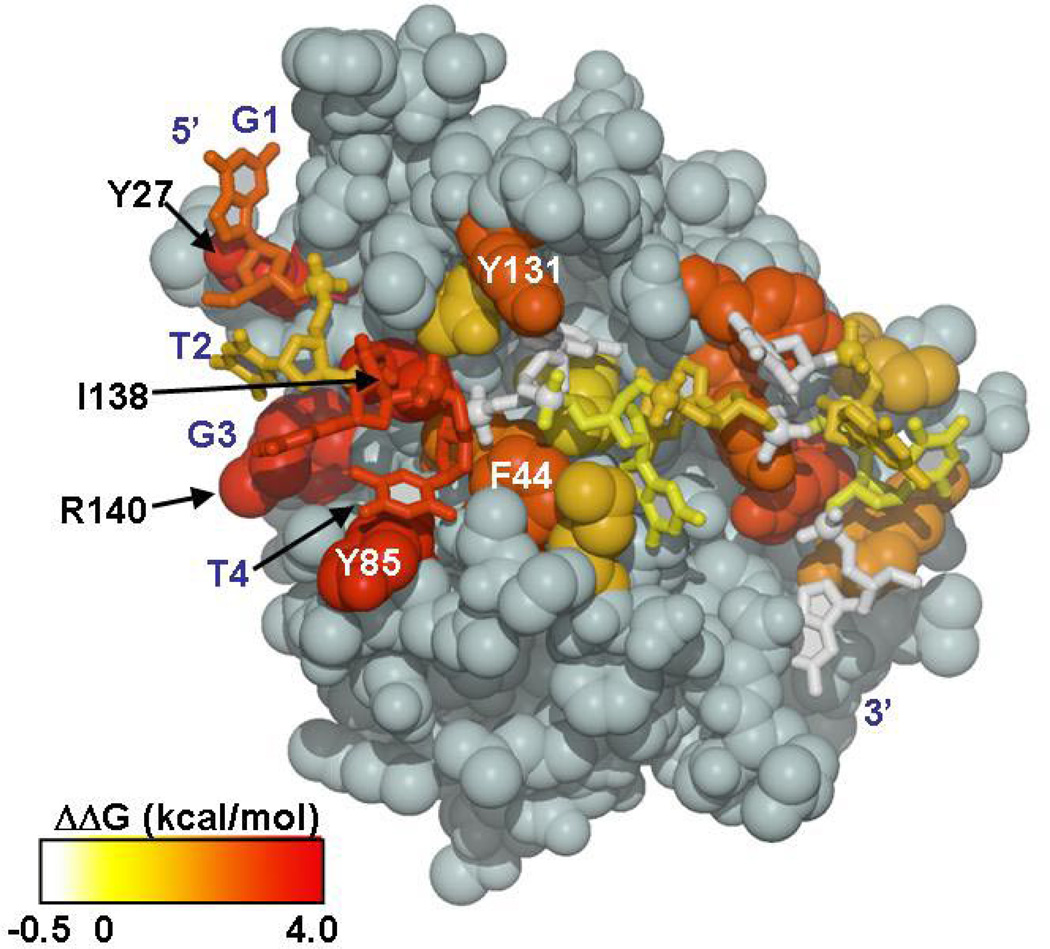
The specificity determining region of Tel-11, the 5’ G1X2G3T4 interaction site, overlaps precisely with the ‘hotspot’ of affinity on the protein surface identified by the complete alanine-scanning mutagenesis of the residues at the interface (48). The spatial coincidence of the thermodynamically important bases in Tel-11 and residues in Cdc13-DBD is striking when the average ΔΔG value resulting from mutation of both the DNA and protein are simultaneously mapped on the structure of the complex (Figure 5). Therefore, the direct contacts between the important protein residues and the critical DNA bases confer specificity as well as affinity.
Figure 5.
The energetic contributions of each DNA base and individual protein residues on binding mapped onto Cdc13-DBD/Tel-11 structure. The magnitude of the change in binding free energy is illustrated using color ramp from white to red, for −0.5 – 4.0 kcal/mol. Values for ΔΔG are from the ensemble average values obtained in the degenerate library experiment and from the alanine mutagenesis study by Anderson et al. 2003 (48). Figure prepared using MOLSCRIPT (63).
Analysis of protein determinants of specificity
The structure of the Cdc13-DBD/Tel-11 complex identified the contact partners but not the precise geometry of the protein/DNA contacts. With the knowledge of the specificity-determining features of the DNA, the structure can be re-examined to predict the protein elements that confer DNA-binding specificity. The two features of the G1 base that confer specificity are the shape of the purine base and the oxygen at the 6-carbon. The orientation of this base with respect to the protein surface is not well defined in the structure due to fraying near the end, therefore we cannot unambiguously identify the protein residues involved in recognition. There are, however, several candidates that may participate in a hydrogen bond interaction with G1. A nearby residue, Y27, has one of the largest decreases in binding affinity (650-fold) when mutated to alanine and this mutation is lethal to the cell (R. B. Cervantes, L. Ricks, J. N. Roberts, DSW, and V. Lundblad unpublished data) (48). The side-chain hydroxyl of Y27 is close to the phosphate backbone of G1 and does not appear to participate in a direct H-bonding interaction with the base. One possibility is a conformational rearrangement of Y27 to place the aromatic ring in line to form a hydrogen bond (H-bond) contact with G1. Alternate H-bonding partners for G1 include lysine 5, which would require the side-chain to rotate toward G1, or lysine 26, where G1 would have to flip the solvent-exposed 6-oxygen inward toward the protein. There are no obvious structural constraints on the size or shape of the base, but perhaps the requirement for a purine at this position is due to proper alignment of the 6-oxygen for hydrogen bonding interactions or for stacking interactions with Y27.
The important recognition elements of G3 are the 6-oxygen and the geometry of the purine base. The requirement for an oxygen atom at position 6 suggests that a hydrogen bonding interaction between the protein and the DNA is important for the specific recognition of this critical base. The side chain of R140 likely provides the hydrogen bond donor. In the structure, R140 is poised to interact directly with the base of G3. Mutation of this conserved residue to alanine decreases affinity by 540-fold and is lethal in vivo (R. B. Cervantes, L. Ricks, J. N. Roberts, DSW, and V. Lundblad unpublished data) (48). Similarly, a substitution of adenine for G3, which results in a loss of a hydrogen bond acceptor at position 6, decreases affinity by 380-fold. Furthermore, mutating R140 to lysine restores binding and cellular viability, implying a requirement for a hydrogen bond donor at this position. Another candidate hydrogen bonding residue is K127 since mutation to alanine is lethal to cells and reduces binding affinity (R. B. Cervantes, L. Ricks, J. N. Roberts, DSW, and V. Lundblad unpublished data). However, K127 is closer to T2 in the structure. Since T2 is not strongly recognized by Cdc13, the role of K127 may be to form a hydrogen bonding interaction with oxygen 4 in order to correctly orient T2 for a stacking interaction with G3.
The specific recognition of T4 appears to be achieved by a combination of space complementarity and hydrogen-bonding interactions. Examination of the structure reveals a hydrophobic pocket at this site formed by Y85, F44 and I83. Tyrosine 85, one of the most important residues for binding DNA (a 700-fold reduction in binding is observed in Y85A) is involved in a stacking interaction with T4 and forms one side of a buried pocket that accommodates T4. Additionally, the side-chain hydroxyl of Y85 is in position to hydrogen bond to the oxygen on the 4 carbon, which may direct specificity for a T at this position. I138 and F44 also play a role in maintaining affinity (replacement to alanine results in 710-fold and 170-fold reduction in affinity, respectively) perhaps by sterically constraining the pocket to accommodate the smaller pyrimidine base.
Recognition trends in the telomere end-binding protein family
Specific recognition of telomeric ssDNA by the telomere end-binding proteins from different organisms, Cdc13 from budding yeast, TEBP from ciliated protozoa, and Pot1 from fission yeast and humans, follow similar but not identical trends (42–44, 46, 47). All members of this family specifically recognize a subset of the bases in their canonical ligand. The cognate sequence for the SpPot1 N-terminal DBD is GGTTAC. Substitution of any of the first five bases with the complementary base results in large decreases in affinity, ranging from 200-fold (G1) to greater than 1000-fold (G2-A5) (7). This specificity has been attributed to the unusual non-sequential G-T hydrogen bonding interactions that occur within the compact DNA ligand (42). The full-length S. pombe Pot1 protein binds a larger substrate containing at least two GGTTAC repeats, with relaxed specificity for the second repeat relative to the first (52). Similarly, hPot1, which binds the sequence TTAGGGTTAG, is most sensitive to substitutions to the five underlined bases (43). The 5’ end of the ligand is specifically recognized and the final 3’ end guanine is sequestered within the protein. Thus, the Cdc13-DBD specificity requirements mirror those observed for the Pot1 proteins, whereby only a subset of the canonical ligand, G1X2G3T4, is specifically recognized by Cdc13-DBD. In contrast, TEBPαβ is distinct from the other end-binding proteins in that only two bases in the minimal 12-mer (GGGGTTTTGGGG) produce large affinity changes upon substitution (44, 53, 54). These affinity changes range from 10 to 80-fold, several orders of magnitude smaller than the changes observed in Pot1 or Cdc13-DBD binding to non-cognate sequences.
Structures of the TEBPαβ complex with non-cognate sequences reveal the basis for the observed tolerance for base substitution. When additional bases are either deleted or inserted into the sequence, the ligand reorients such that the sequence register retains some cognate character. The result of this “nucleotide shuffling” is a relatively small change in binding energy, despite large structural rearrangements (54). This type of structural malleability may explain the relatively small effects of individual substitutions in the 3’ end of the Tel-11 ligand but the apparent requirement for GT-rich sequence in Cdc13 binding. When all of the 3’ bases are substituted with their complement, however, no binding by Cdc13-DBD is observed. However, at any given position in the 3’ end, substitution with a pool of alternate bases produces little change in binding, in contrast to the strict sequence requirements at the 5’ end of the ligand. The biochemical implication that the binding site at the 5’ end is fixed, while the binding site at the 3’ end is more malleable, can be understood in the context of the structure. The 5’ end of the cognate DNA interacts with the β-barrel and β1–β2 and β4–β5 loops, the canonical ligand binding site used by almost all OB-fold proteins (1). However, the 3’ end of the DNA interacts with the unusually large ordered loop (between β2–β3), a feature present only in Cdc13-DBD and, interestingly, the second OB-fold of hPot1(43, 46). This loop serves to provide a higher affinity binding site and may be important for the recognition of heterogeneous sequences.
While Cdc13, hPot1, SpPot1, and TEBPαβ all employ OB-folds to recognize ssDNA, the chemical nature of the protein/DNA interface is surprisingly diverse among the members of this family. Cdc13 and SpPot1 each possess a single OB-fold that alone exhibits high affinity and specific binding to DNA, while hPot1 and TEBPαβ require two or more of these protein modules for interaction with telomeric DNA in vitro. Interestingly, only hPot1 and TEBPαβ are currently known to recognize the extreme 3’ terminus by sequestering the final base within the protein structure. Cdc13 and SpPot1 may also have a role in end recognition as both proteins have been proposed to contain at least one more OB-fold (52, 55, 56). Similar to Cdc13-DBD, the DNA recognition interface of TEBPαβ contains a large number of aromatic and hydrophobic residues; 19 aromatic and hydrophobic residues cluster at the protein/DNA interface in TEBPαβ and 12 for Cdc13-DBD (44, 46, 53, 57). In TEBPαβ, the aromatic residues participate in a contiguous extended stacking structure with guanine bases, which is further stabilized by extensive hydrophobic packing. For Cdc13, nearly all, nine out of ten tested, of the interfacial aromatic or hydrophobic residues are crucial for high affinity binding, however the stacking interactions are localized (47, 48). The ten aromatic and hydrophobic interfacial residues in the hPot1 complex flank the base stacking pairs and also do not form an extended stacked system (43). In contrast to the other end-binding proteins, there are only five aromatic and hydrophobic residues at the SpPot1 interface. These residues form stabilizing stacking interactions similar to those observed in hPot that flank the highly compact DNA structure (42).
One unusual feature in Cdc13 is a dearth of charged or polar residues at the protein/DNA interface that can participate in specific hydrogen bonding interactions. In contrast, SpPot1 and hPot1 are rich in these types of residues which form specific hydrogen bonding interactions between the DNA and protein. In SpPot1, there are thirteen sequence specific hydrogen bonding interactions that form a network arising from positively charged and polar side chains in the protein to the Watson-Crick donor/acceptors of the first four bases, GGTT (42). Similarly, hPot1 has 22 sequence specific hydrogen bonding interactions over two OB-folds to nine of the ten bases recognized (43). TEBPαβ is less hydrophilic with nine hydrogen bonding interactions from charged and polar residues to specific moieties on the WC face of the bases within the 12-mer DNA ligand (44). There are only seven charged residues at the interface between Cdc13-DBD and the Tel-11 DNA and only one polar amino acid. Two of these residues, R140, which is predicted to hydrogen bond to G3, and K127, are the only charged or polar residues that greatly reduce binding when mutated to alanine. Several other lysines along the interface have only modest reductions (< 8 fold) in affinity when altered to alanine suggesting that they are not important for hydrogen bonding interactions. Tyrosine is well represented across the interface between Cdc13-DBD and Tel-11 and can contribute a hydrogen bonding-donor through the side chain hydroxyl. G1 may be specifically recognized by Y27 through a hydrogen bonding interaction to the 6-carbonyl oxygen. The absence of traditional hydrogen-bonding partners may assist in the recognition of heterogeneous yeast telomeres.
Recognition of heterogeneous yeast telomere sequences
Cdc13 binds specifically to heterogeneous yeast telomeric sequence with high affinity. While an 11-mer of DNA sequence is necessary for high affinity binding, we have shown that the determinants of specificity reside in the first four bases with the ideal binding site G1X2G3T4. Examination of the consensus sequence representing yeast telomeres, [(TG)0–6TGGGTGTG(G)], reveals that despite the heterogeneity, yeast telomeres always contain a consensus Cdc13 recognition site (that is, a G1X2G3T4 sequence followed by GT-rich sequence). Inspection of several typical yeast telomere sequences (40), shown in Figure 6, reveals that there are many potential binding sites for the Cdc13-DBD contained within the telomere sequence. Thus, the specificity exhibited by the Cdc13-DBD is therefore ideally suited for Cdc13 to fulfill its essential role at the yeast telomere, as a consensus binding sequence is invariably present. The unusual specificity exhibited by Cdc13 also provides an alternate explanation for the intolerance of budding yeast to changes in the templating region of telomerase RNA (58, 59). Complete mutation of a region of the TLC1 template indicates that yeast tolerate only conservative A/C rich sequence in the telomerase RNA even though the reverse transcriptase is still enzymatically active. While some of this tolerance could be attributed to loss of the double-strand binding protein Rap1 at telomeres, the patterns of viability are also consistent with the binding preferences exhibited by Cdc13 (58, 59). In addition, the Cdc13-DBD binding specificity reported here explains why a short stretch of GT repeats is sufficient for the Cdc13-dependent recruitment of telomerase to double-strand breaks (60–62), leading to telomere-mediated healing of the DNA break.
Figure 6.
Potential Cdc13 binding sites mapped on representative yeast telomere sequences (40). Multiple Cdc13 binding sites are in every heterogeneous telomere sequence.
Acknowledgements
We thank Vicki Lundblad, Rachel Cervantes, and Leslie Ricks for providing data prior to publication and Douglas Theobald and Fiona Jucker for insightful discussions and thoughtful reading of the manuscript.
Acknowledgement of financial aid: NIH GM59414, the Arnold and Mabel Beckman Foundation, and the Keck Foundation
Abbreviations used
- BSA
bovine serum albumin
- DBD
DNA-binding domain
- DTT
1,4 dithiothreitol
- EDTA
ethylenediamine tetraacetic acid
- OB
oligonucleotide/oligosaccharide binding
- Pot
protection of telomeres
- ssDNA
single-strand DNA
- TEBP
telomere end-binding protein
- thio-G
6-thioguanosine
- WC
Watson-Crick
REFERENCES
- 1.Theobald DL, Mitton-Fry RM, Wuttke DS. Nucleic acid recognition by OB-fold proteins. Annu. Rev. Biophys. Biomol. Struct. 2003;32:115–133. doi: 10.1146/annurev.biophys.32.110601.142506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braddock DT, Baber JL, Levens D, Clore GM. Molecular basis of sequence-specific single-stranded DNA recognition by KH domains: solution structure of a complex between hnRNP K KH3 and single-stranded DNA. EMBO J. 2002;21:3476–3485. doi: 10.1093/emboj/cdf352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braddock DT, Louis JM, Baber JL, Levens D, Clore GM. Structure and dynamics of KH domains from FBP bound to single-stranded DNA. Nature. 2002;415:1051–1056. doi: 10.1038/4151051a. [DOI] [PubMed] [Google Scholar]
- 4.Datta S, Larkin C, Shildbach JF. Structural insights into single-stranded DNA binding and cleavage by F factor TraI. Structure. 2003;11:1369–1379. doi: 10.1016/j.str.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 5.Desveaux D, Allard J, Brisson N, Sygusch J. A new family of plant transcription factors displays a novel ssDNA-binding surface. Nat. Struct. Biol. 2002;9:512–517. doi: 10.1038/nsb814. [DOI] [PubMed] [Google Scholar]
- 6.Stern JC, Anderson BJ, Owens TJ, Shildbach JF. Energetics of the sequence-specific binding of single-stranded DNA by the F factor relaxase domain. J. Biol. Chem. 2004;279:29155–29159. doi: 10.1074/jbc.M402965200. [DOI] [PubMed] [Google Scholar]
- 7.Lei M, Baumann P, Cech TR. Cooperative binding of single-stranded telomeric DNA by the Pot1 protein of Schizosaccharomyces pombe . Biochemistry. 2002;41:14560–14568. doi: 10.1021/bi026674z. [DOI] [PubMed] [Google Scholar]
- 8.Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- 9.Cong YS, Wright WE, Shay JW. Human telomerase and its regulation. Microbiol. Mol. Biol. Rev. 2002;66:407–425. doi: 10.1128/MMBR.66.3.407-425.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cech TR. Beginning to understand the end of the chromosome. Cell. 2004;116:273–279. doi: 10.1016/s0092-8674(04)00038-8. [DOI] [PubMed] [Google Scholar]
- 11.Ferreira MG, Miller KM, Cooper JA. Indecent exposure: when telomeres become uncapped. Mol. Cell. 2004;13:7–18. doi: 10.1016/s1097-2765(03)00531-8. [DOI] [PubMed] [Google Scholar]
- 12.Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu. Rev. Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- 13.Blackburn EH. Switching and signaling at the telomere. Cell. 2001;106:661–673. doi: 10.1016/s0092-8674(01)00492-5. [DOI] [PubMed] [Google Scholar]
- 14.Shiloh Y, Kastan MB. ATM: genome stability, neuronal development, and cancer cross paths. Adv. Cancer Res. 2001;83:209–254. doi: 10.1016/s0065-230x(01)83007-4. [DOI] [PubMed] [Google Scholar]
- 15.Wong K-K, Maser RS, Bachoo RM, Menon J, Carrasco RD, Gu Y, Alt FW, DePinho RA. Telomere dysfunction and ATM deficiency comprises organ homeostasis and accelerates aging. Nature. 2003;421:643–648. doi: 10.1038/nature01385. [DOI] [PubMed] [Google Scholar]
- 16.Maser RS, DePinho RA. Connecting chromosomes, crisis, and cancer. Science. 2002;297:565–569. doi: 10.1126/science.297.5581.565. [DOI] [PubMed] [Google Scholar]
- 17.Cawthon RM, Smith KR, O'Brien E, Sivatchenko A, Kerber RA. Association between telomere length in blood and mortality in people aged 60 years or older. Lancet. 2003;361:393–395. doi: 10.1016/S0140-6736(03)12384-7. [DOI] [PubMed] [Google Scholar]
- 18.Harley CB. Telomerase is not an oncogene. Oncogene. 2002;21:494–502. doi: 10.1038/sj.onc.1205076. [DOI] [PubMed] [Google Scholar]
- 19.Collins K, Mitchell JR. Telomerase in the human organism. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- 20.Cervantes RB, Lundblad V. Mechanisms of chromosome-end protection. Curr. Opin. Cell Biol. 2002;14:351–356. doi: 10.1016/s0955-0674(02)00325-3. [DOI] [PubMed] [Google Scholar]
- 21.Harrington L. Those dam-aged telomeres. Curr. Opin. Genetics and Devel. 2004;14:22–28. doi: 10.1016/j.gde.2003.12.007. [DOI] [PubMed] [Google Scholar]
- 22.Lydall D. Hiding at the ends of yeast chromosomes: telomeres, nucleases and checkpoint pathways. J. Cell Sci. 2003;116:4057–4065. doi: 10.1242/jcs.00765. [DOI] [PubMed] [Google Scholar]
- 23.Evans SK, Lundblad V. Positive and negative regulation of telomerase access to the telomere. J. Cell. Sci. 2000;113(Pt 19):3357–3364. doi: 10.1242/jcs.113.19.3357. [DOI] [PubMed] [Google Scholar]
- 24.Baumann P, Cech TR. Pot1, the putative telomere end-binding protein in fission yeast and humans. Science. 2001;292:1171–1175. doi: 10.1126/science.1060036. [DOI] [PubMed] [Google Scholar]
- 25.Loayza D, de Lange T. POT1 as a terminal transducer of TRF1 telomere length control. Nature. 2003;423:1013–1018. doi: 10.1038/nature01688. [DOI] [PubMed] [Google Scholar]
- 26.Colgin LM, Baran K, Baumann P, Cech TR, Reddel RR. Human POT1 facilitates telomere elongation by telomerase. Curr. Biol. 2003;13:942–946. doi: 10.1016/s0960-9822(03)00339-7. [DOI] [PubMed] [Google Scholar]
- 27.Gottschling DE, Zakian VA. Telomere proteins: specific recognition and protection of the natural termini of Oxytricha macronuclear DNA. Cell. 1986;47:195–205. doi: 10.1016/0092-8674(86)90442-3. [DOI] [PubMed] [Google Scholar]
- 28.Price CM, Cech TR. Telomeric DNA-protein interactions of Oxytricha macronuclear DNA. Genes & Dev. 1987;1:783–793. doi: 10.1101/gad.1.8.783. [DOI] [PubMed] [Google Scholar]
- 29.Lustig AJ. Cdc13 subcomplexes regulate multiple telomere functions. Nat. Struct. Biol. 2001;8:297–299. doi: 10.1038/86157. [DOI] [PubMed] [Google Scholar]
- 30.Lin J-J, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo . Proc. Natl. Acad. Sci. USA. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Booth C, Griffith E, Brady G, Lydall D. Quantitative amplification of single-stranded DNA (QAOS) demonstrates that cdc13-1 mutants generate ssDNA in a telomere to centromere direction. Nucl. Acids Res. 2001;29:4414–4422. doi: 10.1093/nar/29.21.4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chandra A, Hughes TR, Nugent CI, Lundblad V. Cdc13 both positively and negatively regulates telomere replication. Genes & Dev. 2001;15:404–414. doi: 10.1101/gad.861001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lendvay TS, Morris DK, Sah J, Balasubramanian B, Lundblad V. Senescence mutants of Saccharomyces cerevisiae with a defect in telomere replication identify three additional EST genes. Genetics. 1996;144:1399–1412. doi: 10.1093/genetics/144.4.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- 36.Anderson EM, Halsey WA, Wuttke DS. Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucl. Acids Res. 2002;30:4305–4313. doi: 10.1093/nar/gkf554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szostak JW, Blackburn EH. Cloning yeast telomeres on linear plasmid. Cell. 1982;29:245–255. doi: 10.1016/0092-8674(82)90109-x. [DOI] [PubMed] [Google Scholar]
- 38.Förstemann K, Hoss M, Lingner J. Telomerase-dependent repeat divergence at the 3' end of yeast telomeres. Nucl. Acids Res. 2000;28:2690–2694. doi: 10.1093/nar/28.14.2690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Förstemann K, Lingner J. Molecular basis for telomere repeat divergence in budding yeast. Mol. Cell. Biol. 2001;21:7277–7286. doi: 10.1128/MCB.21.21.7277-7286.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teixeira MT, Arneric M, Sperisen P, Lingner J. Telomere length homeostasis is achieved via a switch between telomerase-extendible and -nonextendible states. Cell. 2004;117:323–335. doi: 10.1016/s0092-8674(04)00334-4. [DOI] [PubMed] [Google Scholar]
- 41.Singer MS, Gottschling DE. TLC1: Template RNA component of Saccharomyces cerevisiae telomerase. Science. 1994;266:404–409. doi: 10.1126/science.7545955. [DOI] [PubMed] [Google Scholar]
- 42.Lei M, Podell ER, Baumann P, Cech TR. DNA self-recognition in the crystal structure of the Pot1 (Protection of Telomeres)-ssDNA complex. Nature. 2003;426:198–204. doi: 10.1038/nature02092. [DOI] [PubMed] [Google Scholar]
- 43.Lei M, Podell E, Cech TR. Structure of human Pot1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat. Struct. and Mol. Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 44.Horvath MP, Schweiker VL, Bevilacqua JM, Ruggles JA, Schultz SC. Crystal structure of the Oxytricha nova telomere end binding protein complexed with single strand DNA. Cell. 1998;95:963–974. doi: 10.1016/s0092-8674(00)81720-1. [DOI] [PubMed] [Google Scholar]
- 45.Hughes TR, Weilbaecher RG, Walterscheid M, Lundblad V. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc. Natl. Acad. Sci. USA. 2000;97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 47.Mitton-Fry RM, Anderson EM, Glustrom LW, Theobald DL, Wuttke DS. Structural basis for telomeric single-stranded DNA recognition by yeast Cdc13. J. Mol. Biol. 2004;338:241–255. doi: 10.1016/j.jmb.2004.01.063. [DOI] [PubMed] [Google Scholar]
- 48.Anderson EM, Halsey WA, Wuttke DS. Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry. 2003;42:3751–3758. doi: 10.1021/bi027047c. [DOI] [PubMed] [Google Scholar]
- 49.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 50.Meyer M, Suhnel J. Interaction of cyclic cytosine-, guanine-, thymine-, uracil-, and mixed guanine-cytosine base tetrads with K+, Na+, and Li+ ions - a density functional study. J. Biomol. Struct. and Dynamics. 2003;20:507–517. doi: 10.1080/07391102.2003.10506868. [DOI] [PubMed] [Google Scholar]
- 51.Allen FH, Bird CM, Rowland RS, Raithby PR. Resonance-induced hydrogen bonding at sulfur acceptors in R 1 R 2C=S and R 1CS2 − systems. Acta Cryst. 1997;B53:680–695. [Google Scholar]
- 52.Trujillo KM, Bunch JT. Extended DNA binding site in Pot1 broadens sequence specificity to allow recognition of hetergeneous fission yeast telomeres. J. Biol. Chem. 2005;280:9119–9128. doi: 10.1074/jbc.M414511200. [DOI] [PubMed] [Google Scholar]
- 53.Classen S, Ruggles JA, Schultz SC. Crystal structure of the N-terminal domain of Oxytricha nova telomere end-binding protein α subunit both uncomplexed and complexed with telomeric ssDNA. J. Mol. Biol. 2001;314:1113–1125. doi: 10.1006/jmbi.2000.5191. [DOI] [PubMed] [Google Scholar]
- 54.Theobald DL, Schultz SC. Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J. 2003;22:4314–4324. doi: 10.1093/emboj/cdg415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Theobald DL, Cervantes RB, Lundblad V, Wuttke DS. Homology among telomeric end-protection proteins. Structure. 2003;11:1049–1050. doi: 10.1016/s0969-2126(03)00183-7. [DOI] [PubMed] [Google Scholar]
- 56.Theobald DL, Wuttke DS. Prediction of multiple tandem OB-folds in telomere end-binding proteins Pot1 and Cdc13. Structure. 2004;12:1877–1879. doi: 10.1016/j.str.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 57.Classen S, Lyons D, Cech TR, Schultz SC. Sequence-specific and 3'-end selective single-strand DNA binding by the Oxytricha nova telomere end binding protein α subunit. Biochemistry. 2003;42:9269–9277. doi: 10.1021/bi0273718. [DOI] [PubMed] [Google Scholar]
- 58.Förstemann K, Zaug AJ, Cech TR, Lingner J. Yeast telomerase is specialized for C/A-rich RNA templates. Nucl. Acids Res. 2003;31:1646–1655. doi: 10.1093/nar/gkg261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lin J, Smith DL, Blackburn EH. Mutant telomere sequences lead to impaired chromosome separation and a unique checkpoint response. Mol. Bio. Cell. 2004;15:1623–1634. doi: 10.1091/mbc.E03-10-0740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Diede SJ, Gottschling DE. Exonuclease activity is required for sequence addition and Cdc13p loading at a de novo telomere. Curr Biol. 2001;11:1336–1340. doi: 10.1016/s0960-9822(01)00400-6. [DOI] [PubMed] [Google Scholar]
- 61.Stellwagon AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes & Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bianchi A, Negrini S. Delivery of the yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol. Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 63.Kraulis PJ. MOLSCRIPT: a program for display and analysis of macromolecular structure. J Appl Cryst. 1991;24:946–950. [Google Scholar]