Abstract
Age-related macular degeneration (AMD) is a common cause of visual impairment in individuals over 55 years of age worldwide. The varying clinical phenotypes of AMD result from contributions of genetic, epigenetic and non-genetic (environmental) factors. Genetic studies of AMD have come of age as a direct result of tremendous gains from human genome project, genomewide association studies and identification of numerous susceptibility loci. These findings have implicated immune response, high-density lipoprotein cholesterol metabolism, extracellular matrix, and angiogenesis signaling pathways in disease pathophysiology. Here, we address how the wealth of genetic findings in AMD is expected to impact the practice of medicine, providing opportunities for improved risk assessment, molecular diagnosis, preventive and therapeutic intervention. We propose that the potential of using genetic variants for monitoring treatment response (pharmacogenetics) may usher a new era of personalized medicine in the clinical management of AMD.
Keywords: Retinal neurodegeneration, Complex disease, Genomewide association, Risk factors, Genetic diagnosis, Pharmacogenetics, Clinical management
Introduction
Age related macular degeneration (AMD) (MIM; 603075) is a complex, multifactorial disease, characterized by progressive degeneration of photoreceptors and retinal pigment epithelium (RPE) with or without choroidal neovascularization. AMD is the leading cause of irreversible vision loss in elderly individuals, affecting nearly 0.2% of the population between 55 to 64 years of age and 13% of those above 85 years of age in developed countries1. In United States alone, more than 1.75 million individuals are affected with AMD, and this number is likely to increase by 50% in 20202. Although the etiology of AMD remains largely unknown, numerous studies have established age, smoking and genetic predisposition as the key and cardiovascular risk factors (such as hypertension and hyperlipidemia) as inconsistent contributors to disease manifestation. Rather than providing a detailed review of AMD genetic studies (see3–6), we will focus on how genetic findings may be translated to clinical management and therapy.
The human retina undergoes structural and/or functional changes as part of the natural course of aging. Appearance of ophthalmoscopically visible focal yellow depositions, called drusen, between the RPE and Bruch’s membrane is a common observation in aging retina. Drusen are classified as hard or soft, based on the diameter of the lesion and the appearance of their margin. Hard or small drusen are <63 µm in diameter and have discrete margins, whereas soft or large drusen are greater in size (>125 µm) with indistinct edges. In the natural course of the disease, these drusen disappear and the effects on the RPE are manifested in hypo- or hyper-pigmentary changes. Drusen are considered hallmark of AMD, but only individuals with drusen that are larger in size and greater in area and those with hypo- or hyper-pigmentary changes in the RPE will have increased risk of developing advanced AMD. Detachment of the RPE and new blood vessel growth between Bruch's membrane and the retina are lesions of advanced neovascular AMD, and outer retinal atrophy is indicative of geographic atrophy (GA) in AMD. These advanced stages can lead to loss of central vision.
AMD patients have broad severity range of drusen characteristics and pigmentary abnormalities. The Age-Related Eye Disease Study (AREDS) research group developed a simplified 5-step severity scale based on drusen size and pigmentary changes to define risk categories for advanced AMD7. Using this scale, 5-year risk of progression to advanced AMD in at least one eye increases from less than 0.5% in the first step to about 50% in the last step, depending on the presence of the drusen and pigmentary changes of the RPE.
Susceptibility to AMD
Epidemiological studies have provided significant information on the prevalence, incidence, natural history and risk factors of AMD. Fundus features such as large, soft drusen and pigmentary abnormalities are associated with an increased risk of AMD progression8. Among non-genetic components, aging and smoking are among the most consistent risk factors. Racial/ethnic differences affect the prevalence of AMD with higher risk of neovascular AMD in Caucasians compared to Hispanic and black population8. Large Multi-Ethnic Study of Atherosclerosis (MESA) reported lower AMD prevalence in blacks than in whites with overall prevalence varying from 2.4% in blacks, 4.2% in Hispanics, 4.6% in Chinese compared to 5.4% in whites9. Increased risk of AMD in females and an association with hypertension or cardiovascular disease has been indicated in some studies8. Dietary factors (such as intake of nutritional antioxidants and omega-3 fatty acids) and abdominal adiposity have also been correlated to risk of AMD progression10.
Genetic Studies
Familial aggregation, twin studies, and segregation analyses have revealed substantial genetic contributions to AMD pathogenesis5, 6. High concordance of AMD among monozygotic twins compared to dizygotic twin pairs suggested genetic component of the disease. Segregation analysis indicated a higher risk of manifesting the symptoms of AMD among first-degree relatives of patients compared to the general population. These investigations suggested the role of genetic factors and/or shared environmental exposures in disease onset and encouraged researchers to explore genetic component of AMD through candidate gene, linkage and association studies (see Table 1 – Glossary for details of genetic terms and methodology).
Table 1.
Glossary of Genetic Terms
| Genotype: The genetic makeup of an organism, including the entire set of genes in a cell, tissue, organism, or an individual. |
| Phenotype: Outward, expressed physical manifestation of traits in an organism. |
| Candidate gene study: A study of the genes of known biological function, implicated directly or indirectly in modulating the cellular processes associated with traits under investigation. In a candidate gene study, the allele frequency is compared between cases and controls to find the disease-causing mutation or association. This hypothesis-driven but biased approach does not allow investigation of genes with unknown function and is not likely to identify novel genes/pathway involved in the disease. |
| Mendelian disease: Also called a monogenic disease that is a direct consequence of mutations in a single gene and follows Mendelian inheritance pattern (dominant, recessive, or X-linked). |
| Linkage: Genetic linkage is defined as the tendency of two loci (genetic units, genes) that are close to each other on the same chromosome and are inherited together. Linkage is measured by the percentage recombination between loci, with unlinked genes showing 50% recombination. |
| LOD score LOD stands for “Logarithm (base 10) of Odds” and is a statistical test often used in linkage analysis. The LOD score compares the likelihood of obtaining the test data if the two loci are indeed linked, to the likelihood of observing the same data purely by chance. Traditionally, LOD score of more than or equal to 3 (1000:1 odds) is taken as an evidence of linkage. |
| Phenocopy: When a phenotype closely resembles genetically determined trait, but does not carry the genetic variation causing the phenotype. Usually caused by environmentally induced, nonhereditary variation in an organism. |
| Low penetrance: When genetic variation is present, but phenotype is not seen. |
| Complex disease: The disease with genetic component(s) yet lacking a simple inheritance pattern. Instead, a complex disease inheritance may be influenced by contributions of multiple genes, their interactions with each other and/or with the environment. |
| Haplotype: A combination of alleles at different DNA markers that tend to be inherited together. The haplotype may include one locus, several loci, or an entire chromosome depending on the crossover events. |
| Linkage disequilibrium (LD): LD is used to define the non-random association of alleles at adjacent loci. In other words, if two nearby markers are in LD, they will appear on the same haplotype more often than if they were unlinked. |
| Genomewide association study (GWAS): A method of identifying disease-associated variants across the whole genome. A large number of common variants (100,000 to 5,000,000) are tested for significant allele-frequency differences between individuals with the phenotype of interest (“cases”) and a set of unrelated control individuals. Associated allele is not causative, but is a associated marker. The causal mutation could be another common variant, a rare variant, and/or a structural variant, which is in strong LD with the associated allele or in rare cases can be associated allele itself. |
| Genomewide significance: GWAS involves performing multiple individual statistical tests for each SNP, and because of the large number of SNPs a traditional p-value cutoffs of 0.05 can give false positive results (association by chance). Thus, p-value is revised for multiple-hypothesis testing using bonferroni correction, and genomewide significance p-value is derived by dividing p-value cutoffs to total number of SNPs tested in the association study (e.g., p = 0.05 / 500,000 = 10−7). This represents the conservative threshold for declaring a significant association in a genomewide study. |
| Heritability: The proportion of the variance in a phenotype among individuals that is attributable to differences in genotype. |
| Odds ratio (OR): A measure of relative risk that is defined as the odds of exposure to the susceptible genetic variant in cases compared to the odds of exposure in controls. An odds ratio of 1 implies that the event is equally likely in both groups. An odds ratio greater than one implies that the event is more likely in the cases and thus considered harmful. Whereas OR of less than one implies that the variant is protective in cases. |
| Risk prediction: Predicting one’s risk for developing a particular disease, based on known genetic and environment factor(s). |
| Pharmacogenetics and Pharmacogenomics: These terms are used interchangeably in literature. Pertains to the study of effect(s) of a genetic variant in a single gene on response to drug treatment, or may refer to a broad and systematic examination of effects of all inherited variants, their products, inter-individual variation(s), intra-individual variation(s) on the treatment outcome. |
| Exome: Collection of all protein-coding exons in genome, comprising ~ 1% of the genome. |
| Epigenetics: Is study of inherited phenotype that is not attributable to alternation in DNA sequences. The examples are DNA methylation and histone modifications. |
| Transcriptome: Set of all RNA molecules, including mRNA, rRNA, tRNA, and other non-coding RNAs, produced in a cell or population of cells. |
| Copy number variations (CNV): A class of DNA sequence variants (including deletions and duplications) that lead to a departure from the expected diploid representation of DNA sequence. In Ophthalmology, the abbreviation of this genetic term – CNV – can be mistaken for choroidal neovascularization. |
Candidate genes
As Mendelian forms of macular degenerative diseases exhibit similar clinical characteristics, the underlying genes have been investigated as candidates for conferring genetic risk to AMD. Though Stargardt disease gene ABCA411 and ApoE12, 13 have been implicated in AMD, variants in the genes for other macular diseases such as Sorsby fundus dystrophy (STGD3), Best disease (VMD, VMD2), Doyne honeycomb retinal dystrophy (EFEMP1), Stargardt-like macular dystrophy (STGD3, ELVOL4), cone dystrophy (CORDX1, RPGR), malattia leventinese (MLVT, EFEMPI), adult vitelliform dystrophy (AVMD, RDS) were not found to be associated with AMD susceptibility5, 14. These findings led researchers to shift their efforts towards whole genome linkage and association studies to explore the genetic basis of AMD.
Lessons from linkage analysis
Traditional methods of linkage mapping have been successful in discovering genetic defects in large families with multiple affected members. Linkage analysis examines for region(s) in the genome where a marker or set of markers are inherited with the disease phenotype. Thus, all affected but not the unaffected members in a family harbor the linked marker(s) or region of the genome (haplotype). Statistically, the linkage is determined through logarithm of odds (LOD) score calculation, and a score of ≥3 is traditionally taken as an evidence of linkage. This approach is highly successful for Mendelian disorders where penetrance is high and inheritance pattern can be inferred. In rare instances when large multi-generational families were available, linkage study can be extended to complex disorders like AMD though the data analysis may be complicated because of the possibility of independent involvement of different genetic defects. The first genetic locus for AMD was localized in a single large pedigree to chromosome 1q, and a co-segregating HMCN-1 variant was later identified15. As the late onset of clinical manifestations in AMD poses substantial challenges in the collection of large pedigrees, many subsequent studies focused on linkage analysis in large sets of smaller families and in sib/relative pairs, leading to the identification of AMD susceptibility loci at many chromosomal regions including 1q31, 9p13, 10q26, and 17q25 (Table 2)3, 5, 6.
Table 2.
AMD susceptibility genes/variants, identified as of May 2012
| Gene | Chr location |
DNA marker | Disease phenotype |
Genetic method |
Pathway | Validation |
|---|---|---|---|---|---|---|
| ABCA4 | 1p22 | rs1800553, rs1800555 |
AMD/ Stargardt disease |
Candidate gene |
Clearance of all-trans retinal aldehyde (atRAL) from photoreceptors |
Confirmed |
| APOE | 19q13 | rs429358, rs7412 |
AMD | Candidate gene/GWAS |
Transport and metabolism of lipoproteins |
Confirmed |
|
ARMS2/ HTRA1 |
10q26 | rs10490924 (ARMS2), rs11200638 (HTRA1) |
AMD | Linkage/ GWAS |
Unknown | Confirmed |
| C2/CFB | 6p21 | rs9332739 (C2), rs4151667(CFB) |
AMD | Candidate gene/ GWAS |
Complement pathway | Confirmed |
| C3 | 19p13 | rs2230199 | AMD | Candidate gene/ GWAS |
Complement pathway | Confirmed |
| CETP | 16q21 | rs3764261 | AMD | GWAS | Transport and metabolism of lipoproteins |
Confirmed |
| CFH | 1q32 | rs1061170, rs10737680 |
AMD | Linkage/G WAS/candid ate gene |
Complement pathway | Confirmed |
|
CFHR1/ CFHR3 |
1q31-q32 | 84K bp deletion |
AMD | Candidate gene |
Possibly complement pathway |
Tentative |
| CFI | 4q25 | rs2285714 | AMD | Candidate gene/ GWAS |
Complement pathway | Confirmed |
| COL8A1 | 3q12 | rs13095226 | AMD | GWAS | Extracellular/collagen matrix pathway |
Confirmed |
|
EFEMP1/ FBLN3 |
2p16 | R345W | AMD/ Doyne honeycomb retinal dystrophy |
Candidate gene |
Extracellular matrix pathway |
Tentative |
| FBLN5 | 14q32 | 7 variants | AMD | Candidate gene |
Extracellular matrix pathway |
Confirmed |
|
FBLN6/ HMCN1 |
1q24-q25 | Q5345R | AMD | Linkage | Extracellular matrix pathway |
Tentative |
|
FRK/ COL10A1 |
6q21-q22.3 | rs1999930 | AMD | GWAS | Extracellular/collagen matrix pathway |
Confirmed |
| LIPC | 15q22 | rs493258, rs10468017 | AMD | GWAS | Transport and metabolism of lipoproteins |
Confirmed |
|
REST-C4orf14- POLR2B- IGFBP7 |
4q12 | rs1713985 | AMD | GWAS | Unknown | Tentative |
| TIMP3 | 22q12 | rs9621532 | AMD/ Sorsby fundus dystrophy |
GWAS | Degradation of the extracellular matrix |
Confirmed |
| TNFRSF10A | 8p21 | rs13278062 | AMD | GWAS | Unknown | Tentative |
| VEGFA | 6p12 | rs4711751 | AMD | Candidate gene/ GWAS |
Angiogenesis | Confirmed |
| IER3/DDR1 | 6p21 | rs3130783 | AMD | GWAS | Unknown | Tentative |
| SLC16A8 | 22q12 | rs8135665 | AMD | GWAS | Oxidative stress | Tentative |
| TGFBR1 | 9q22 | rs334353 | AMD | GWAS | Angiogenesis | Tentative |
| RAD51B | 14q23 | rs8017304 | AMD | GWAS | DNA repair | Tentative |
| B3GALTL | 13q12 | rs9542236 | AMD | GWAS | Unknown | Tentative |
Confirmed: When identified in two or more studies; Tentative: When reported in only one study and pending independent replication(s); Suggestive: When the p value failed to reach genomewide significance; Chr – chromosome; AMD- Age-related macular degeneration; GWAS- Genome-wide association study
Genomewide Association Studies
Like most common and complex disorders, AMD is caused by risk alleles of moderate effect with substantial influence of epigenetic and environmental factors. Thus, exclusive sharing of risk alleles among affected individuals is much less evident. Genomewide association studies (GWAS), in which a dense genomewide set of SNPs (300,000 to few million haplotype-tagging SNPs) is tested for disease association, offer an alternative approach for genetic dissection of AMD16. GWAS is based on the assumption that if a risk allele exists, it will be either genotyped directly or will be in strong linkage disequilibrium (LD) with one of the SNPs used in the study. In its simplest form, GWAS comprehensively tests for common genetic variants across the genome and compares their allele frequency in a large set of cases to controls from the same population. If the frequency difference is statistically significant, a variant is considered as associated. The Y402H variant within the complement factor H (CFH) gene was the first significantly associated AMD risk allele that was identified by GWAS and using candidate genes in linked chromosomal regions3, 5, 6. This was followed by studies of other candidate genes within the complement pathway. The complement genes have now emerged as major contributors to AMD susceptibility, with C3 on 19p13, CFB/C2 on 6p21, and CFI on 4q253, 6. Two tightly linked genes at 10q26 (age-related maculopathy susceptibility 2 [ARMS2] or LOC387715 and high-temperature requirement factor H [HTRA1]) have also emerged as highly significantly associated loci in AMD patients3, 5, 6. In recent years, several GWAS studies with large number of cases and controls have replicated the finding of CFH and ARMS2/HTRA1 locus as well as have identified additional risk loci (LIPC, CETP, TIMP3), unraveling novel pathways (Table 2)16–18.
Current status of AMD genetics
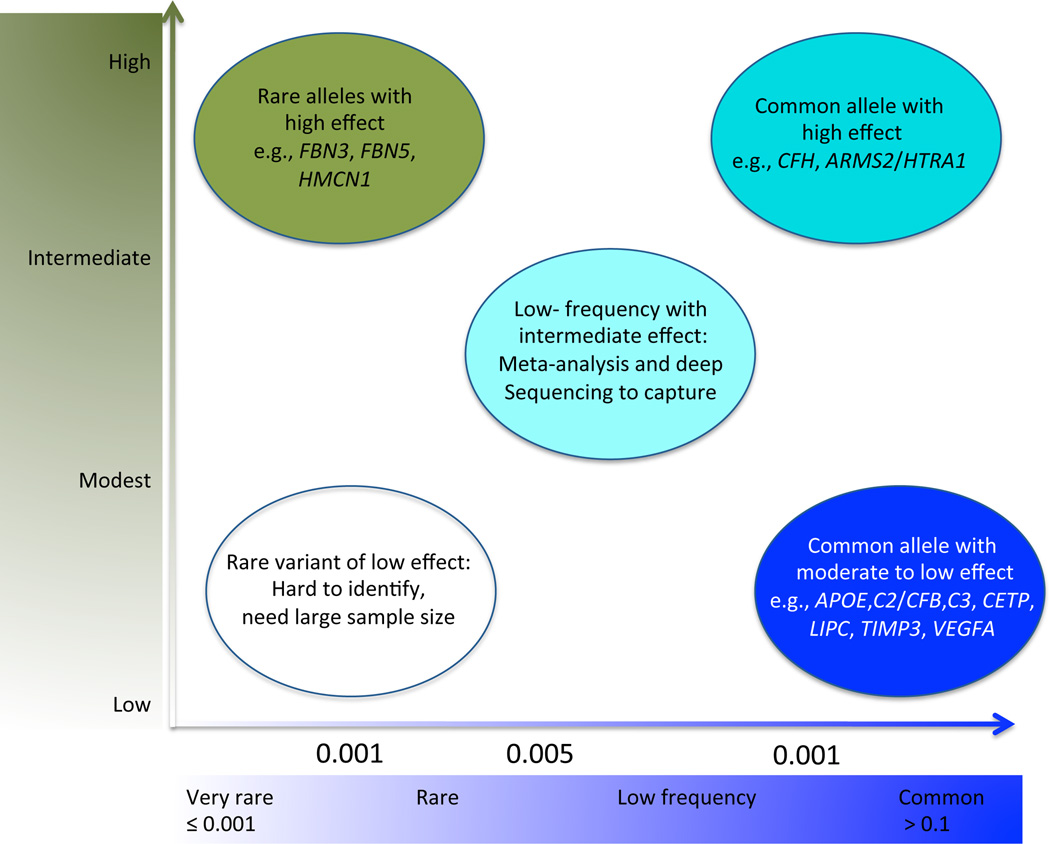
Remarkable progress has been made in delineating the genetics of AMD, with over 50% of the heritability explained by the genetic loci so far16– 18 (Table 2). Among complex disorders, AMD represents a unique case in which a large proportion of genetic basis can be explained by relatively small number of variants (Figure 1, modified from ref.19). Unaccounted heritability can be attributed to additional loci with smaller effects on risk; such variants are often missed in GWAS owing to the sample size and adjustment for multiple testing. Hence, combined analysis of the data from multiple GWAS (meta-analysis) is an important step towards the systematic identification of low-frequency moderate risk loci. A recent international collaborative effort on meta-analysis of AMD-GWAS from 18 centers, involving over 17,000 AMD cases and 60,000 matched controls of European and Asian ancestry, has provided the scale and power necessary to identify additional susceptibility loci. This meta-analysis has revealed and validated 19 AMD loci, including seven novel loci that reach genomewide significance near the genes COL8A1/FILIP1L, IER3/DDR1, SLC16A8, TGFBR1, RAD51B, MIR548A2, and B3GALTL17.
Figure 1.
Genetic architecture of age-related macular degeneration (AMD). Adapted from Manolio et al., 2009
AMD susceptibility loci are now providing the framework to elucidate disease etiology; however, these results must be interpreted with caution since associated allele(s) are not responsible for causing the disease. These variant alleles are present in the control population as well, yet their frequency is significantly altered in AMD patients. Furthermore, the AMD-associated genes might interact with other genes or non-genetic risk factors to produce the clinical phenotypes. The associated marker may reflect the cumulative effect of multiple low-frequency rare causal alleles, and re-sequencing of larger genomic regions is warranted to identify the rare or common pathogenic variant(s) at these loci. More recently, next-generation sequencing technology has provided an opportunity to discover patient-specific variants using whole genome and whole exome sequencing methods20. Functional characterization of genes would also be needed to gain insights into their relevance for AMD causation, especially where genetic studies can’t pinpoint the susceptibility gene (e.g., ARMS2 or HTRA1 at 10q26). Such studies are in progress and would reveal valuable insights in AMD pathogenesis and suggest new avenues for treatment.
Aging, a major risk factor of AMD, is commonly associated with the accumulation of mutations primarily in the non-coding regions of the genome. Repetitive and transposable elements are now recognized as important regulators of development and disease21, 22; hence, recent reports of increased Alu RNA in the RPE of patients with GA23 deserve serious consideration as such changes might lead to aberrant immune activation24.
Clinical implications
The discovery of multiple AMD associated loci has provided an opportunity for exploring risk models for clinical and public health practices in post-GWAS era (Figure 2). Incorporation of patient’s genotype can improve prediction accuracy as genotype is not dependent on external factors and is constant over the patient’s lifetime. This is particularly true for a late onset disease like AMD, where signs of the disease do not appear until the fifth or sixth decade of life. Early detection of genetic risks may translate to more efficacious preventive regimens, and interventions. Genetic tests are already being offered based on the relative risk associated with the most common AMD variants. However, obtaining a robust estimate of the size of the risk effects associated with these variants is challenging, and even the strongly associated allele can be a poor classifier for risk prediction25. It should also be noted that we do not yet have good preventive therapies even if we can identify those at risk.
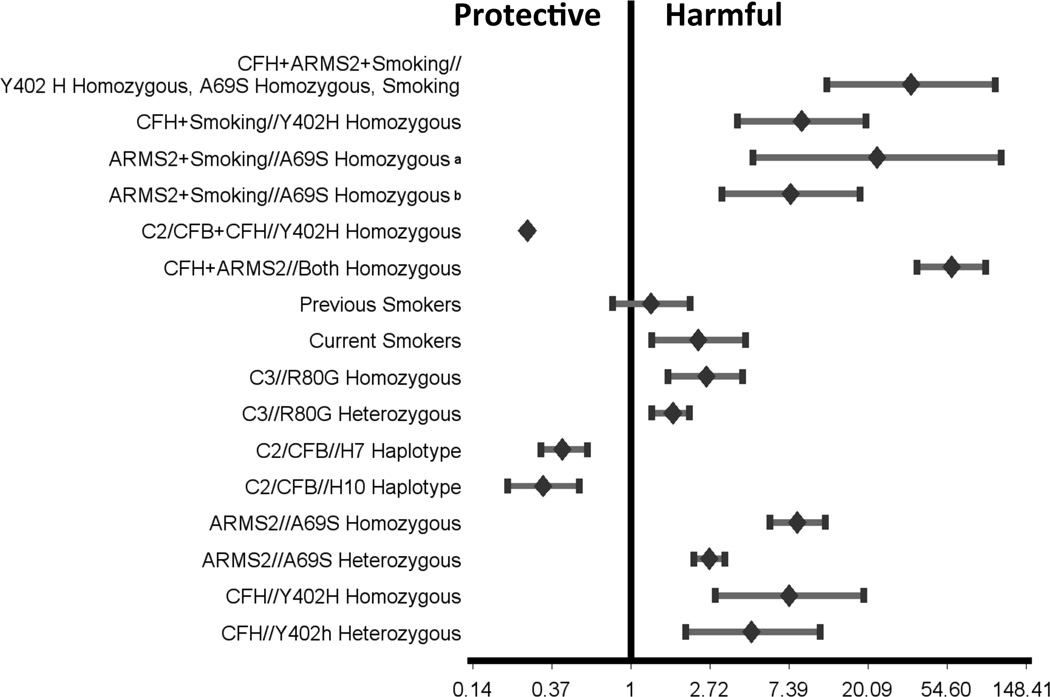
Figure 2.
Individual and combined age-related macular degeneration (AMD) risk estimation conferred by CFH, ARMS2, C3, C2/CFB and smoking. The odds ratio (OR) for each gene or in combination is taken from the first published report because of extensive variations in different studies. Inclusion of additional susceptibility alleles would further assist in diagnosis and disease management, as illustrated in Chen et al., 2010.
Risk prediction based on genotypes and family history
Genotyping individuals at associated loci will provide predictive accuracy of disease risk proportional to the heritability and prevalence of disease. CFH and ARMS2/HTRA1 genes have emerged as the most significantly associated with AMD risk and/or protection in most ethnic populations (Table 2). CFHY402H heterozygotes allele conferred 4.6-fold increased risk for AMD and homozygotes 7.4-fold increased risk, as compared to the homozygous non-risk genotype26. Individuals heterozygous for ARMS2 A69S allele confer ~2.7-fold increased risk of AMD compared to wild type homozygous allele, whereas a 8.2-fold increased risk is associated with the homozygous risk allele27. A 50-fold increase in the risk of AMD in subjects homozygous for both risk allele indicated log-additive effects of CFHY402H and LOC387715 A69S in their interaction28. Together, these two loci might confer approximately 50% of the genetic risks of AMD. Additive accumulation of risk was also observed among five common variants at three loci (CFH, LOC387715 and C2/CFB), where specific genotype combination could identify individuals whose lifetime risk of AMD varied from less than 1% to more than 50%29. However, the use of just three variants in the CFH, ARMS2/HTRA1 and C2/CFB has limited diagnostic value because of the relatively low sensitivity and specificity of the combined test panel25.
Risk prediction based on gene-environment interaction
As non-genetic risk factors contribute substantially to AMD, researchers have realized the need to combine genetic variants with demographic, environmental and macular characteristics of patients. However, quantification of environmental changes remains a challenge. Multivariate regression models that include genetic risk variants, clinical features of AMD and environmental risk factors should be useful in estimating a risk for AMD and its progression from intermediate to advanced forms. An ordered subset analyses with apolipoprotein E alleles, smoking history, and age at onset as stratifying covariates hypothesized that the effect of smoking on the risk of ARMD is accentuated by the locus at the 10q26 region (ARMS2)30. Another study evaluated the association of Y402H (CFH) and A69S (ARMS2/LOC387715) genotypes and multiple environmental factors simultaneously, including body mass index (BMI) and smoking with progression from early or intermediate stages to advanced stages of AMD in 1466 participants of the Age-Related Eye Disease Study31. In this study, the common variants in these two genes were independently related and the risk of AMD progression increased approximately 7-fold in the presence of both homozygous risk genotypes compared to the homozygous non-risk genotypes. Additionally, attributable risks for AMD progression increased to 81% when smoking and BMI were added to the model31. Another study calculated the lifetime risk estimation model based on genetic and environmental factors by multiplying variant-specific odds ratios from disparate sources and multiplying them together32. A predictive model based on multivariate unconditional logistic regression analysis using variants in CFH, ARMS2/HTRA1, C2/ CFB and C3 genes plus environmental risk factors, including age, sex, smoking, and BMI, has been reported for early detection and risk prediction in AMD33. More recently, another predictive model for advanced AMD comprising of demographic and environmental, phenotypic and genetic risk factors was derived from longitudinal data derived from 2846 participants in the AREDS34. However, the predictive value of common genetic variants for AMD and other complex disorders has had limited success, as the value of rare variants has not been appreciated yet25, 35. Next-generation sequencing technologies have now enhanced our ability to identify rare variant with larger effect disease susceptibility at the known as well as unknown AMD loci. Newer models should therefore incorporate a more comprehensive set of rare and common susceptibility variants with environmental factors, such as aging and diet, to access appreciable effect of genetic and non-genetic factors on AMD risk prediction.
Risk prediction based on cellular pathways
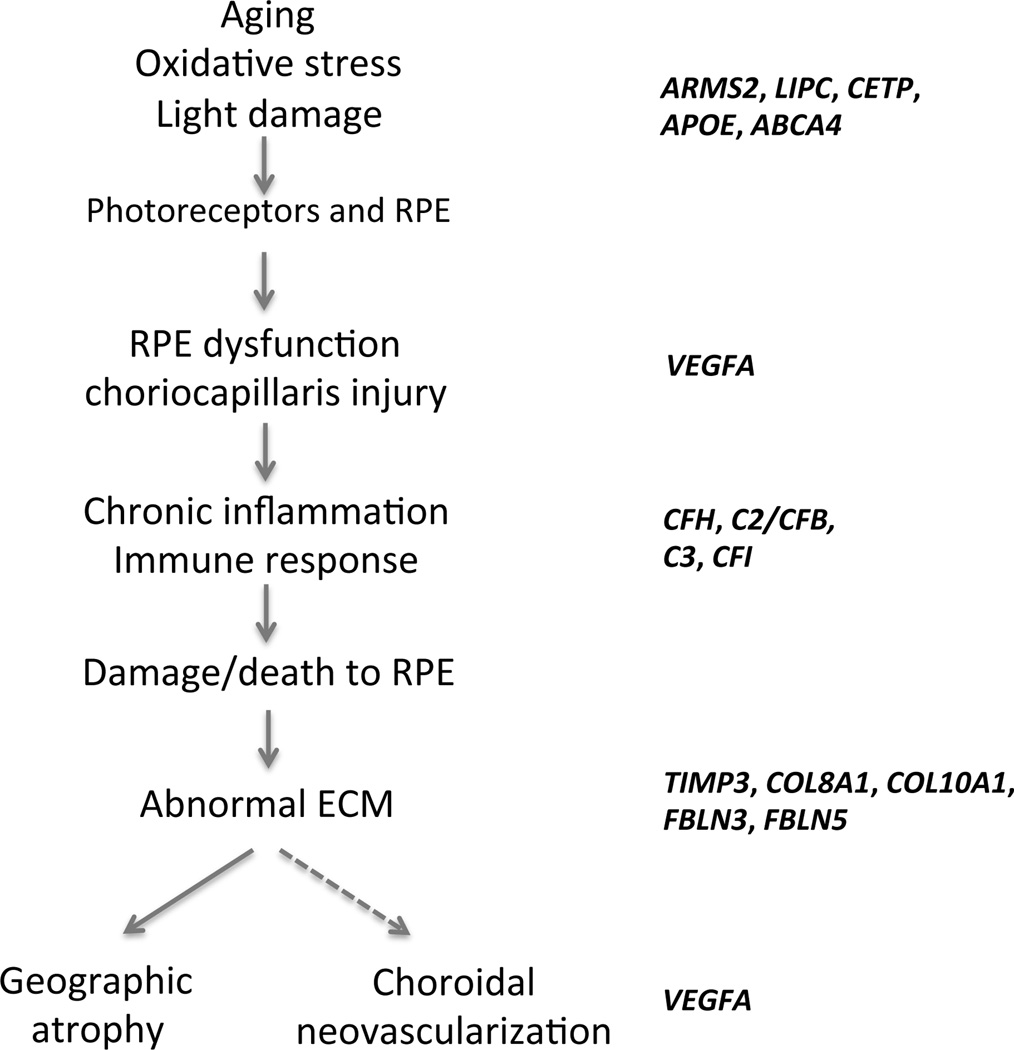
Testing few susceptibility variants cannot yield perfect prediction as the risk may change upon inclusion of loci with small effects. However, risk prediction can be improved considerably if we are able to find out whether different genetic loci interact with each other through few common pathways. A significant genetic association of AMD has been established for genes in complement pathway (CFH, C2/CFB, C3 and CFI), high-density lipoprotein pathway (APOE, LIPC and CETP), extracellular/collagen matrix pathway (TIMP3, COL8A1, COL10A1 and TIMP3) and angiogenesis signaling pathway (VEGFA and TGFBR1), each having largely individual and independent effect(s) on AMD risk16–18 (Table 2). A simplified pathway (modified from ref.36, 37) for AMD pathogenesis based on our current understanding is presented in Figure 3. Pathway-based approach considers multiple variants in interacting or related genes and provides functional relationships within a genetic network, yielding biological significance of disease-associated loci. Additionally, the knowledge of pathways can assist in identifying novel therapeutic targets for intervention. Attempts of risk prediction or drug target discovery based on a single gene/pathway have not produced any meaningful outcome as of yet. With the identification of new loci involved in regulation of lipid metabolism, extracellular matrix remodeling and angiogenesis, we can now implement a better pathway-based study design for improved risk prediction and therapeutic intervention. One limitation, however, is the incompleteness of curated biological pathways and specificity of genetic interactions. Correlation of associated genotypes to whole genome expression data (particularly from control and AMD retina) should be also helpful for risk analysis. Eventually, a systems-based integrated analysis of cellular networks incorporating genetic, epigenetic and expression maps with proteomics, imaging and functional data (e.g., mitochondrial oxidation and stress response) would be needed to create accurate models for AMD risk assessment and clinical management.
Figure 3.
Different stages of age-related macular degeneration. RPE: retinal pigment epithelium, ECM: extracellular matrix. Adapted from Zarbin et al., 2004 and Kanda et al., 2006.
Pharmacogenomics and AMD
Pharmacogenomics focuses on testing for how individual's genetic makeup affects the response to drug therapy, both in terms of efficacy and adverse drug reactions. Unlike pharmacogenetics, which mainly evaluates the effect of single genes on drug responses, pharmacogenomics investigates effect of several genes on a particular trait. A better prognosis of outcome before drug treatment promises reduced side effects and quality-of-life benefits to patients. At present, anti-vascular endothelial growth factor (VEGF) (bevacizumab and ranibizumab) therapy represents the most commonly used treatment in the management of Choroidal neovascularization (CNV), while photodynamic therapy with verteporfin (PDT-V) is used less frequently. No effective treatment is available for GA.
Anti VEGFA therapy
VEGF is a secreted endothelial-specific mitogen, which acts as a key regulator of angiogenesis and vascular permeability. Increased expression of VEGF in CNV is reported in several studies. The hypothesis of retarding pathological angiogenesis through VEGF inhibition has led to current anti-VEGF therapy. Bevacizumab (Avastin) and its derivative ranibizumab (Lucentis) are humanized monoclonal antibodies, which neutralize all active forms of VEGF and thus decrease abnormal vascular permeability and neovascular vessel growth. Comparison of age-related macular degeneration treatment trial (CATT) has shown a correlation between lesion activity and visual acuity that may have implications for treatment of eyes undergoing anti-VEGF therapy for neovascular AMD (ARVO abstract 2893). The use of these drugs has revolutionized the treatment of neovascular AMD. Though highly effective, individual variation in drug response and occasional adverse drug reactions constitute therapeutic challenges. These inter-individual differences in drug response are often attributed to genetic variations and have encouraged several groups to conduct pharmacogenetic testing with a hope of offering personalized medicine for AMD patients. In a study to investigate the association between drug response and CFH Y402H variant, 53% of AMD patients with TT and TC genotype demonstrated improved visual acuity with intra-vitreal bevacizumab treatment, whereas only 10% of the patients with CC genotype showed better visual acuity38. Pharmacogenetic relationship between the CFH Y402H variant and therapeutic response to ranibizumab also showed analogous relationships where individuals homozygous for the variant CFH genotype (CC) were more likely to require reinjection than those with TT or TC genotype; however, the CFH genotype was not significantly associated with post-treatment visual acuity at either 6 or 9 months, after adjusting for pretreatment acuity39. Another study reported more favorable visual acuity outcome after 6 months of intravitreal ranibizumab therapy with the higher AMD risk genotypes at CFH, VEGF and ARMS2/HTRA1 loci40. A prospective study has revealed a correlation between the ARMS2 genotype (homozygous 69S variant) and poor response to ranibizumab therapy41. However, further studies including larger cohorts and more comprehensive genotyping are required before stronger arguments can be made for or against such pharmacogenetic associations42.
Photodynamic therapy (PDT)
The use of PDT for the treatment of neovascular AMD is much more limited but may be important in the treatment of a phenotype of neovascular AMD, known as polypoidal choroidopathy. CNV regression by PDT implicates immune response and/or complement pathway, encouraging researchers to test whether variants in CFH or C-reactive protein (CRP) can affect the treatment outcome; however, the data is inconclusive and further investigations with variants at multiple loci are required to clarify genetic predisposition to treatment outcome after PDT43–45.
Recent pharmacogenetic studies in AMD have concentrated on strongly associated genes (CFH and ARMS2) and have small sample size, limiting the statistical power to detect associations. Notably, drug metabolism and variations in drug response result from interaction of variants at multiple loci. Such studies will therefore require collaborative multi-center efforts with larger patient cohorts, multiple genetic variants, and consistent study design incorporating environmental variables.
Disease Management
Prevention and/or slowing the disease progression remain high priorities in clinical management of AMD. The possible role of oxidative damage in aging-associated diseases led to the exploration of micronutrients with antioxidants properties for treatment/prevention of AMD. A 11-center randomized controlled clinical trial, AREDS, has established that a combination of nutritional supplements consisting of β-carotene (15 mg), vitamins C (500 mg) and E (400 IU), along with zinc (80mg) and copper (2 mg) slowed the progression of AMD in individuals at high risk of developing advanced AMD46. Based upon observational data from AREDS and other epidemiologic studies, AREDS2 is currently evaluating the role of lutein/zeaxanthin and/or omega-3 long-chain polyunsaturated fatty acids, docosahexaenoic acid (DHA) and its precursor eicosapentaenoic acid (EPA) for delaying the progression to advanced AMD47. While AREDS formulation reduced the risk of advanced AMD and prevented the progression to CNV, it was not beneficial in geographic atrophy. Another randomized trial of supplements of B vitamins (B6, B12, and folic acid) was designed to evaluate their impact on cardiovascular disease through lowering homocysteine levels48. While no beneficial effect was observed on the primary outcome of cardiovascular disease, there was a reduction in AMD. As the diagnosis of AMD was based upon validated self-reports, this study did not result in recommendations for AMD and additional trials will be required to evaluate the role of B vitamins for the treatment of AMD48.
Pharmacogenetic interaction has been investigated in individuals with intermediate or unilateral advanced AMD carrying different CFH and ARMS2 genotypes to evaluate treatment response to AREDS-type nutritional supplementation. A greater reduction in AMD progression (68%) in individuals with the low-risk CFH TT genotype compared to those with the high-risk CC genotype (11%) suggested limited benefits of supplementary diets in individuals with strong genetic predisposition49. In another study, pharmacological association was examined between AMD risk variants in 13 genes with AREDS nutritional supplements50. C2/CFB and C3 variants were independently related to progression from early/intermediate to advanced AMD, with no gene–gene or pharmacogenetic interactions. Additional trials in larger cohorts are required to detect potential associations between genotypes and response to drugs or nutritional supplementation.
Summary
With the recent rapid progression in the research, the future holds great promise for patients affected with AMD and for scientists and clinicians involved in such studies. Genetics has provided a basic framework by identifying AMD risk variants and implicated important pathways. However, current associated variants do not explain causality, resulting in a gap in our understanding of how these variations contribute to disease pathogenesis. The integration of genomic, functional and disease model based approaches will be necessary to dissect the pathophysiology of AMD. Additional genetic information can reveal an individual’s overall risk profile, and recent advent of next-generation sequencing offers the possibility of exponential discovery. Whole-genome and whole-exome sequencing projects are already in progress for systematic identification of rare high-risk alleles at AMD susceptibility loci. Whole genome sequence information should also be advantageous in building haplotype-based predictive model(s) for AMD, by facilitating detection of associations driven by cis-interactions among nearby SNPs. As a large number of associated signals are located in the non-coding region of the genes, the loci are predicted to confer AMD susceptibility through quantitative changes in gene expression51, 52. Transcription factor binding sites, promoter, splicing, mRNA stability are some of the relevant mechanisms that can alter expression of associated genes. High-throughput transcriptome sequencing in the retina, preferably macula, offers a starting point for such an analysis. Epigenetic modifications provide another level of gene regulation. An increasing interest in genomewide profiling of epigenetic modifications in normal and AMD retina should reveal new insights in the near future.
The need to elucidate the disease mechanism(s) is imperative, and the major challenges are ahead in incorporating gene-gene interactions, gene copy-number variations, epigenetics, environmental interactions, treatment effects, and clinical covariates. A better understanding of AMD genetics and disease mechanisms will offer potentially new treatment paradigms, improved prediction of disease risk and severity, possibilities for prevention, and eventually personalized therapies and management of this devastating blinding disease.
Acknowledgments
We apologize to our colleagues whose work, especially on original findings of linked, associated and candidate genes/variants for AMD and other dystrophies could not be cited because of the reference limitations. We thank Dr. Wai Wong for comments and suggestions.
Financial Disclosure: Our research is supported by the intramural program of the National Eye Institute, NIH. A.S. is co-inventor on a University of Michigan patent covering the use of genetic markers for diagnosis of AMD.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith W, Assink J, Klein R, et al. Risk factors for age-related macular degeneration: Pooled findings from three continents. Ophthalmology. 2001;108:697–704. doi: 10.1016/s0161-6420(00)00580-7. [DOI] [PubMed] [Google Scholar]
- 2.Friedman DS, O'Colmain BJ, Munoz B, et al. Prevalence of age-related macular degeneration in the United States. Arch Ophthalmol. 2004;122:564–572. doi: 10.1001/archopht.122.4.564. [DOI] [PubMed] [Google Scholar]
- 3.Deangelis MM, Silveira AC, Carr EA, Kim IK. Genetics of age-related macular degeneration: current concepts, future directions. Semin Ophthalmol. 2011;26:77–93. doi: 10.3109/08820538.2011.577129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gorin MB. A clinician's view of the molecular genetics of age-related maculopathy. Arch Ophthalmol. 2007;125:21–29. doi: 10.1001/archopht.125.1.21. [DOI] [PubMed] [Google Scholar]
- 5.Haddad S, Chen CA, Santangelo SL, Seddon JM. The genetics of age-related macular degeneration: a review of progress to date. Surv Ophthalmol. 2006;51:316–363. doi: 10.1016/j.survophthal.2006.05.001. [DOI] [PubMed] [Google Scholar]
- 6.Swaroop A, Chew EY, Rickman CB, Abecasis GR. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu Rev Genomics Hum Genet. 2009;10:19–43. doi: 10.1146/annurev.genom.9.081307.164350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferris FL, Davis MD, Clemons TE, et al. A simplified severity scale for age-related macular degeneration: AREDS Report No. 18. Arch Ophthalmol. 2005;123:1570–1574. doi: 10.1001/archopht.123.11.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klein R, Peto T, Bird A, Vannewkirk MR. The epidemiology of age-related macular degeneration. Am J Ophthalmol. 2004;137:486–495. doi: 10.1016/j.ajo.2003.11.069. [DOI] [PubMed] [Google Scholar]
- 9.Klein R, Klein BE, Knudtson MD, et al. Prevalence of age-related macular degeneration in 4 racial/ethnic groups in the multi-ethnic study of atherosclerosis. Ophthalmology. 2006;113:373–380. doi: 10.1016/j.ophtha.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 10.Krishnadev N, Meleth AD, Chew EY. Nutritional supplements for age-related macular degeneration. Curr Opin Ophthalmol. 2010;21:184–189. doi: 10.1097/ICU.0b013e32833866ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Allikmets R. Further evidence for an association of ABCR alleles with age-related macular degeneration. The International ABCR Screening Consortium. Am J Hum Genet. 2000;67:487–491. doi: 10.1086/303018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klaver CC, Kliffen M, van Duijn CM, et al. Genetic association of apolipoprotein E with age-related macular degeneration. Am J Hum Genet. 1998;63:200–206. doi: 10.1086/301901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmidt S, Klaver C, Saunders A, et al. A pooled case-control study of the apolipoprotein E (APOE) gene in age-related maculopathy. Ophthalmic Genet. 2002;23:209–223. doi: 10.1076/opge.23.4.209.13883. [DOI] [PubMed] [Google Scholar]
- 14.Stone EM, Sheffield VC, Hageman GS. Molecular genetics of age-related macular degeneration. Hum Mol Genet. 2001;10:2285–2292. doi: 10.1093/hmg/10.20.2285. [DOI] [PubMed] [Google Scholar]
- 15.Schultz DW, Klein ML, Humpert AJ, et al. Analysis of the ARMD1 locus: evidence that a mutation in HEMICENTIN-1 is associated with age-related macular degeneration in a large family. Hum Mol Genet. 2003;12:3315–3323. doi: 10.1093/hmg/ddg348. [DOI] [PubMed] [Google Scholar]
- 16.Chen W, Stambolian D, Edwards AO, et al. Genetic variants near TIMP3 and high-density lipoprotein-associated loci influence susceptibility to age-related macular degeneration. Proc Natl Acad Sci U S A. 2010;107:7401–7406. doi: 10.1073/pnas.0912702107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schu MC, Chen W, Fritsche LG, et al. Meta-analysis Of Genome-wide Association Studies Identifies 19 Loci Associated With AMD Risk Investigative ophthalmology & visual science. 2012:53. (ARVO E-Abstract 2259) [Google Scholar]
- 18.Yu Y, Bhangale TR, Fagerness J, et al. Common variants near FRK/COL10A1 and VEGFA are associated with advanced age-related macular degeneration. Hum Mol Genet. 2011;20:3699–3709. doi: 10.1093/hmg/ddr270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nat Rev Genet. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 21.Baillie JK, Barnett MW, Upton KR, et al. Somatic retrotransposition alters the genetic landscape of the human brain. Nature. 2011;479:534–537. doi: 10.1038/nature10531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin L, Chang HY. Uncovering the role of genomic "dark matter" in human disease. J Clin Invest. 2012;122:1589–1595. doi: 10.1172/JCI60020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaneko H, Dridi S, Tarallo V, et al. DICER1 deficit induces Alu RNA toxicity in age-related macular degeneration. Nature. 2011;471:325–330. doi: 10.1038/nature09830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tarallo V, Hirano Y, Gelfand BD, et al. DICER1 Loss and Alu RNA Induce Age-Related Macular Degeneration via the NLRP3 Inflammasome and MyD88. Cell. 2012;149:847–859. doi: 10.1016/j.cell.2012.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jakobsdottir J, Gorin MB, Conley YP, et al. Interpretation of genetic association studies: markers with replicated highly significant odds ratios may be poor classifiers. PLoS Genet. 2009;5:e1000337. doi: 10.1371/journal.pgen.1000337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klein RJ, Zeiss C, Chew EY, et al. Complement factor H polymorphism in age-related macular degeneration. Science. 2005;308:385–389. doi: 10.1126/science.1109557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rivera A, Fisher SA, Fritsche LG, et al. Hypothetical LOC387715 is a second major susceptibility gene for age-related macular degeneration, contributing independently of complement factor H to disease risk. Hum Mol Genet. 2005;14:3227–3236. doi: 10.1093/hmg/ddi353. [DOI] [PubMed] [Google Scholar]
- 28.Schaumberg DA, Hankinson SE, Guo Q, et al. A prospective study of 2 major age-related macular degeneration susceptibility alleles and interactions with modifiable risk factors. Arch Ophthalmol. 2007;125:55–62. doi: 10.1001/archopht.125.1.55. [DOI] [PubMed] [Google Scholar]
- 29.Maller J, George S, Purcell S, et al. Common variation in three genes, including a noncoding variant in CFH, strongly influences risk of age-related macular degeneration. Nat Genet. 2006;38:1055–1059. doi: 10.1038/ng1873. [DOI] [PubMed] [Google Scholar]
- 30.Weeks DE, Conley YP, Tsai HJ, et al. Age-related maculopathy: a genomewide scan with continued evidence of susceptibility loci within the 1q31, 10q26, and 17q25 regions. Am J Hum Genet. 2004;75:174–189. doi: 10.1086/422476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Seddon JM, Francis PJ, George S, et al. Association of CFH Y402H and LOC387715 A69S with progression of age-related macular degeneration. JAMA. 2007;297:1793–1800. doi: 10.1001/jama.297.16.1793. [DOI] [PubMed] [Google Scholar]
- 32.Zanke B, Hawken S, Carter R, Chow D. A genetic approach to stratification of risk for age-related macular degeneration. Can J Ophthalmol. 2010;45:22–27. doi: 10.3129/i09-209. [DOI] [PubMed] [Google Scholar]
- 33.Chen Y, Zeng J, Zhao C, et al. Assessing susceptibility to age-related macular degeneration with genetic markers and environmental factors. Arch Ophthalmol. 2011;129:344–351. doi: 10.1001/archophthalmol.2011.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klein ML, Francis PJ, Ferris FL, 3rd, et al. Risk Assessment Model for Development of Advanced Age-Related Macular Degeneration. Arch Ophthalmol. 2011 doi: 10.1001/archophthalmol.2011.216. [DOI] [PubMed] [Google Scholar]
- 35.Kraft P, Hunter DJ. Genetic risk prediction--are we there yet? N Engl J Med. 2009;360:1701–1703. doi: 10.1056/NEJMp0810107. [DOI] [PubMed] [Google Scholar]
- 36.Zarbin MA. Current concepts in the pathogenesis of age-related macular degeneration. Arch Ophthalmol. 2004;122:598–614. doi: 10.1001/archopht.122.4.598. [DOI] [PubMed] [Google Scholar]
- 37.Kanda A, Abecasis G, Swaroop A. Inflammation in the pathogenesis of age-related macular degeneration. Br J Ophthalmol. 2008;92:448–450. doi: 10.1136/bjo.2007.131581. [DOI] [PubMed] [Google Scholar]
- 38.Brantley MA, Jr, Fang AM, King JM, et al. Association of complement factor H and LOC387715 genotypes with response of exudative age-related macular degeneration to intravitreal bevacizumab. Ophthalmology. 2007;114:2168–2173. doi: 10.1016/j.ophtha.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 39.Lee AY, Raya AK, Kymes SM, et al. Pharmacogenetics of complement factor H (Y402H) and treatment of exudative age-related macular degeneration with ranibizumab. Br J Ophthalmol. 2009;93:610–613. doi: 10.1136/bjo.2008.150995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McKibbin M, Ali M, Bansal S, et al. CFH, VEGF and HTRA1 promoter genotype may influence the response to intravitreal ranibizumab therapy for neovascular age-related macular degeneration. Br J Ophthalmol. 2012;96:208–212. doi: 10.1136/bjo.2010.193680. [DOI] [PubMed] [Google Scholar]
- 41.Teper SJ, Nowinska A, Pilat J, et al. Involvement of genetic factors in the response to a variable-dosing ranibizumab treatment regimen for age-related macular degeneration. Mol Vis. 2010;16:2598–2604. [PMC free article] [PubMed] [Google Scholar]
- 42.Orlin A, Hadley D, Chang W, et al. Association between High-Risk Disease Loci and Response to Anti-Vascular Endothelial Growth Factor Treatment for Wet Age-Related Macular Degeneration. Retina. 2012;32:4–9. doi: 10.1097/IAE.0b013e31822a2c7c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Feng X, Xiao J, Longville B, et al. Complement factor H Y402H and C-reactive protein polymorphism and photodynamic therapy response in age-related macular degeneration. Ophthalmology. 2009;116:1908–12. doi: 10.1016/j.ophtha.2009.03.011. e1. [DOI] [PubMed] [Google Scholar]
- 44.Goverdhan SV, Hannan S, Newsom RB, et al. An analysis of the CFH Y402H genotype in AMD patients and controls from the UK, response to PDT treatment. Eye. 2008;22:849–854. doi: 10.1038/sj.eye.6702830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seitsonen SP, Jarvela IE, Meri S, et al. The effect of complement factor H Y402H polymorphism on the outcome of photodynamic therapy in age-related macular degeneration. Eur J Ophthalmol. 2007;17:943–949. doi: 10.1177/112067210701700612. [DOI] [PubMed] [Google Scholar]
- 46.A randomized, placebo-controlled, clinical trial of high-dose supplementation with vitamins C and E, beta carotene, and zinc for age-related macular degeneration and vision loss: AREDS report no. 8. Arch Ophthalmol. 2001;119:1417–1436. doi: 10.1001/archopht.119.10.1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.SanGiovanni JP, Chew EY, Agron E, et al. The relationship of dietary omega-3 long-chain polyunsaturated fatty acid intake with incident age-related macular degeneration: AREDS report no. 23. Arch Ophthalmol. 2008;126:1274–1279. doi: 10.1001/archopht.126.9.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Christen WG, Glynn RJ, Chew EY, et al. Folic acid, pyridoxine, and cyanocobalamin combination treatment and age-related macular degeneration in women: the Women's Antioxidant and Folic Acid Cardiovascular Study. Arch Intern Med. 2009;169:335–341. doi: 10.1001/archinternmed.2008.574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Klein ML, Francis PJ, Rosner B, et al. CFH and LOC387715/ARMS2 genotypes and treatment with antioxidants and zinc for age-related macular degeneration. Ophthalmology. 2008;115:1019–1025. doi: 10.1016/j.ophtha.2008.01.036. [DOI] [PubMed] [Google Scholar]
- 50.Francis PJ, Hamon SC, Ott J, et al. Polymorphisms in C2, CFB and C3 are associated with progression to advanced age related macular degeneration associated with visual loss. J Med Genet. 2009;46:300–307. doi: 10.1136/jmg.2008.062737. [DOI] [PubMed] [Google Scholar]
- 51.Silveira AC, Morrison MA, Ji F, et al. Convergence of linkage, gene expression and association data demonstrates the influence of the RAR-related orphan receptor alpha (RORA) gene on neovascular AMD: a systems biology based approach. Vision Res. 2010;50:698–715. doi: 10.1016/j.visres.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jun G, Nicolaou M, Morrison MA, et al. Influence of ROBO1 and RORA on risk of age-related macular degeneration reveals genetically distinct phenotypes in disease pathophysiology. PLoS One. 2011;6:e25775. doi: 10.1371/journal.pone.0025775. [DOI] [PMC free article] [PubMed] [Google Scholar]