Abstract
A large fraction of the genes from sequenced organisms are of unknown function. This limits biological insight, and for pathogenic microorganisms hampers the development of new approaches to battle infections. There is thus a great need for novel strategies that link genotypes to phenotypes for microorganisms. We describe a high-throughput strategy based on the method Tn-seq that can be applied to any genetically manipulatable microorganism. By screening 17 in vitro and two in vivo (carriage and infection) conditions for the pathogen Streptococcus pneumoniae, we create a resource consisting of >1800 interactions that is rich in new genotype–phenotype relationships. We describe genes that are involved in differential carbon source utilization in the host, as well as genes that are involved both in virulence and in resistance against specific in vitro stresses, thereby revealing selection pressures that the pathogen experiences in vivo. We reveal the secondary response to an antibiotic, including a dual role efflux pump also involved in resistance to pH stress. Through genetic-interaction mapping and gene-expression analysis we define the mechanism of attenuation and the regulatory relationship between a two-component system and a core biosynthetic pathway specific to microorganisms. Thus, we have generated a resource that provides detailed insight into the biology and virulence of S. pneumoniae and provided a road map for similar discovery in other microorganisms.
An important goal in biology is to understand the relationship between genotype and phenotype. With respect to pathogenic microorganisms, this goal is especially relevant because the lack of understanding about the function of a significant part of the pan-genome (Medini et al. 2008) is hampering the design of novel strategies to battle infectious diseases. Developing high-throughput approaches for non-model organisms that can match genotypes to phenotypes under in vitro and in vivo (infection) conditions is therefore crucial.
A reverse genetics approach based on genome-wide ordered arrays of single gene knockouts (Tong et al. 2001; Schuldiner et al 2005) has been applied to several model organisms (Giaever et al. 2004; Lee et al. 2005; Baba et al. 2006; St Onge et al. 2007; de Berardinis et al. 2008; Liu et al. 2008; Kim et al. 2010; Noble et al. 2010). By determining their growth rate or fitness under defined conditions, genotype–phenotype patterns are obtained. A limitation of this approach is that genome-wide knockout libraries are only available for a handful of organisms. Even for model organisms, experiments often remain restricted to a small number of strains, because constructing new knockout arrays is extremely laborious. In order to make both model and non-model organisms accessible to high-throughput phenotypic profiling and genetic interaction mapping, we recently developed the method Tn-seq with which it is possible to determine each gene's contribution to fitness in a single experiment (van Opijnen et al. 2009).
Here, we report a strategy using Tn-seq to generate detailed genotype–phenotype maps of a microorganism. We apply this strategy to the pathogen Streptococcus pneumoniae, a gram-positive bacterial species and commensal of the human nasopharynx. Dissemination of S. pneumoniae from the nasopharynx frequently leads to otitis media or less often to invasive diseases including pneumonia, meningitis, and bacteremia. Antibiotic resistance is on the rise and each year over a million people succumb to invasive infection with S. pneumoniae, making it one of the most important bacteria clinically (Tuomanen et al. 2004; Harboe et al. 2009; Liñares et al. 2010). Here we measured the fitness of mutant libraries in 17 different in vitro conditions and in two in vivo environments in mice, yielding numerous phenotypes that allowed us to study conditional gene essentiality, discover leads for gene function and antibiotic action, and match defined in vitro stress conditions with in vivo colonization and disease states. Besides creating a resource that provides insight into the biology and virulence of this pathogen, we have drawn up a detailed roadmap that can be used to navigate similar discovery in other microorganisms.
Results and Discussion
Genotype–phenotype profiling generates a fine-scale gene-condition interaction map and suggests that the nasopharynx is the primary adaptive niche
S. pneumoniae can be found in different niches within the host (e.g., nasopharynx, inner-ear, lung, bloodstream, and brain) and is exposed to a wide range of largely unknown conditions. Six independent transposon insertion libraries were evaluated in 17 different in vitro growth conditions, which were chosen to represent selective pressures the bacterium may encounter in vivo (Hava et al. 2003; Kadioglu et al. 2008).
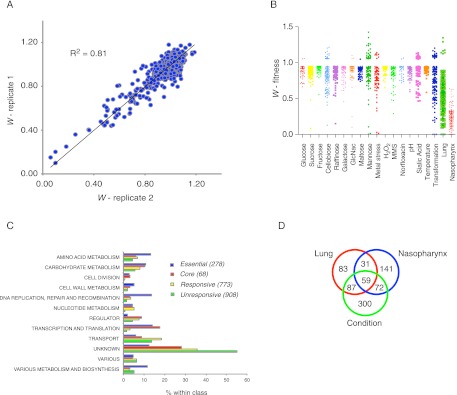
S. pneumoniae has a significant part of its genome dedicated to growth on host carbohydrates (Tettelin 2001), including genes that cleave terminal sialic acid, galactose, and N-acetylglucosamine (GlcNac) residues from host glycans (King et al. 2006). Although S. pneumoniae can grow on these carbon sources in vitro, it is unknown which of these are utilized in which host tissue, or whether it can exploit other carbohydrates for growth (Tettelin 2001). Therefore, in addition to these three carbohydrates, we screened the monosaccharides glucose, fructose, mannose; the disaccharides sucrose, maltose, cellobiose; and the trisaccharide raffinose as the main carbon and energy source. The seven other tested conditions consisted of stresses including hydrogen peroxide, which is produced by naturally catalase-negative S. pneumoniae and by host phagocytic cells; low-temperature and acid pH; reduced divalent cation concentration (referred to as metal stress); exposure to antibiotics; DNA damage; and the induction of natural competence followed by DNA transformation. Six biological replicates were evaluated under each test condition and fitness was calculated for each insertion mutant in the population by Tn-seq. For each condition, reproducibility was determined by comparing fitness values between different libraries, which in each case was high (R2 = 0.63–0.85) (Fig. 1A).
Figure 1.
Condition-specific phenotypic profiling with Tn-seq generates a robust and novel data set. (A) Reproducibility of Tn-seq data was high (R2 = 0.63–0.85). Shown is a representative correlation between two libraries. (B) Significant average Tn-seq fitness scores for 17 in vitro conditions, lung and nasopharynx (also see Supplemental Table S1). (C) Classification of 2027 genes into four classes, which are divided into 12 functional categories. (D) Overlap of genes that respond to nasopharynx and/or lung and at least one in vitro condition.
Mouse models were used to sample the nasopharynx and the lung, which represent carriage and the invasive disease state pneumonia, respectively. Because we take into account the number of generations of the population during a given Tn-seq experiment, fitness equates with growth rate, and thus allows for comparisons between experiments and conditions. While the expansion can be easily determined for in vitro growth experiments, this is not so under in vivo conditions, because the bacterial load in a tissue at any particular time is a function of prior growth, death, and clearance. Indeed, the doubling times of S. pneumoniae in the nasopharynx and lung are unknown. In order to determine these values, we transformed S. pneumoniae with a temperature-sensitive plasmid that cannot replicate at physiological temperatures in the mouse lung and nasopharynx, and thus the rate of loss of the plasmid from the population was used as a proxy for in vivo growth rates. Exponential growth curves were fit on the data and the doubling time was calculated for each niche, resulting in an average of 161 min in the nasopharynx and 108 min in the lung (Supplemental Fig. S1A,B).
For each in vitro and in vivo condition it was determined which genes had a significant response. Although the distribution and average fitness effects were similar across the in vitro environments, there was a clear difference between the in vivo conditions; the nasopharynx responsive genes having a stronger defect on average than the lung responsive genes. Comparison of the 90 overlapping genes that responded both in the lung and nasopharynx revealed that the average fitness defect is significantly larger in the nasopharynx (Wnasopharynx = 0.12, Wlung = 0.52, paired t-test P < 0.0001; Supplemental Fig. S2A). In combination with the slower growth rate of the bacterium in the nasopharynx, this suggests that the nasopharynx is a harsher environment than the lung, possibly due to the presence of more rigorous and variable selection pressures. The lung in that sense may have had a smaller impact on the evolution of S. pneumoniae, which makes sense when we consider that colonization is much more prevalent than pneumonia.
Based on their contribution to fitness, genes were grouped into four classes. (1) Essential: required for growth; (2) Core: fitness defect in ≥13 conditions; (3) Responsive: phenotype in at least one condition; and (4) Unresponsive: no response in any of the sampled conditions. In total, 278 of 2027 annotated genes were found to be essential, 272 of which were previously suggested as essential in TIGR4 or in other strains (Supplemental Fig. S2B; Supplemental Table S1). Thirty-eight genes previously reported as essential are now classified as core genes (Supplemental Table S1). Additionally, several genes that are assumed to be pseudogenes have a phenotype in our data set, and one gene is marked as essential (SP0193). This suggests that these genes may be functional, which for instance has been shown for RadA (SP0023) (Burghout et al. 2007).
Genes in each of the four classes were divided into 12 functional categories (Fig. 1C; Supplemental Table S1). Five of these categories (cell division; cell wall metabolism; DNA replication, repair and recombination; nucleotide metabolism; and transcription and translation) are predominantly made up of essential, core, and responsive genes, emphasizing their importance for bacterial growth. In contrast, the category transport has, after the category unknown, the highest proportion of unresponsive genes, suggesting that there is redundancy among transporters. In addition to the notion that transporters are often able to transport different compounds (Lewinson et al. 2006; Fluman and Bibi 2009), we were not exhaustive in the number of sampled conditions and we anticipate that additional test conditions will reveal more phenotypes. Interestingly, ∼50% of all the unresponsive genes are <500 bp, in contrast to ∼16% of the genes in the other three classes, with most of these genes being annotated as hypothetical (Supplemental Table S1). As a consequence, their function remains elusive and their overall contribution unclear. Moreover, a large fraction (>30%) within the unknown category contains responsive genes (279) and core genes (19), indicating that the data set is rich in novel genotype–phenotype relationships.
Recently, it was shown for Escherichia coli that both essential and responsive genes were preferentially located on the leading strand of the genome, while unresponsive genes had a higher probability of being located on the lagging strand (Nichols et al. 2011). In S. pneumoniae, genes from every category, including unresponsive, are preferentially located on the leading strand (Supplemental Fig. S3; Supplemental Table S2).
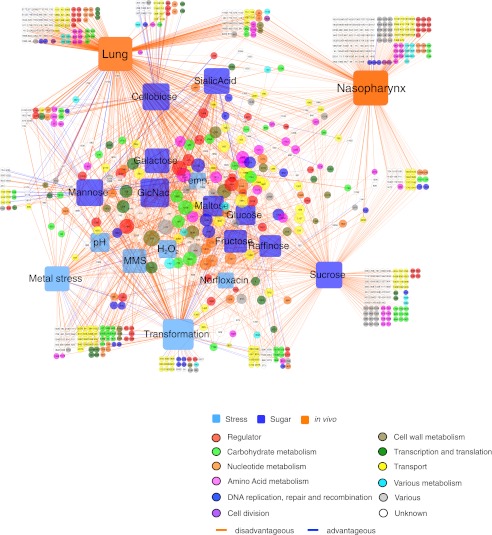
Analysis of responsive genes between the in vitro and in vivo conditions revealed an extensive amount of overlap: 218 genes critical for fitness in the lung and/or nasopharynx were also important in one or more specific in vitro conditions (Fig. 1D). Thus, our experimental strategy yielded a genotype–phenotype data set that can serve as a valuable resource for linking specific metabolic pathways and stress responses to pathogenicity. In total, 1828 significant genotype–phenotype relationships were identified, which could be visualized in a single gene-condition interaction network (Fig. 2). Six percent of the scored interactions resulted in enhancement of bacterial fitness upon gene disruption, while 94% of interactions indicate a fitness defect (Supplemental Table S1). This network provides functional information for 48% of the annotated, nonessential genes of S. pneumoniae. Each condition contains unique responsive genes, which are located away from the center of the network, while the central part of the network is characterized by genes that respond to multiple conditions. Among genes that were sensitive to multiple test conditions are several regulators, indicating their involvement in orchestrating an appropriate general response.
Figure 2.
A gene-condition interaction network. A total of 1828 significant genotype–phenotype interactions were scored and visualized in a gene-condition interaction network. Conditions are represented as rounded squares and color-coded as carbon source, stress, or in vivo niche. Genes are color-coded according to their functional category. Interactions between a gene and a condition (nodes) are indicated by a line (edge); blue for a positive interaction (mutant fitness > wild-type), orange for a negative interaction (mutant fitness < wild-type). For both conditions and genes, the size of the node increases with the number of interactions, while the thickness of the line increases with the fitness effect.
The lung and nasopharynx data were tested for the enrichment of specific gene sets that are active in the same pathway or that have a related biological function (Subramanian et al. 2005). With extensive overlap between in vivo niches, 17 gene sets were negatively enriched in the lung, while 16 were negatively enriched in the nasopharynx (overlapping gene sets are made up of pathways such as fatty acid biosynthesis, cell division, and DNA replication; P ≤ 0.01, FDR < 25%; Supplemental Tables S3, S4). Additionally, gene sets made up of different transporter types (e.g., Phosphotransferase Systems or ABC-transporters) were positively enriched in both niches and thus, in general, are dispensable for survival (Supplemental Tables S5, S6), again indicating redundancy between transporters.
We also determined whether there was enrichment in the in vivo niches for any of the in vitro-specific responsive genes. This analysis showed that sucrose and temperature-responsive genes were significantly enriched in the nasopharynx, while cellobiose, GlcNac, transformation, and H2O2-responsive genes were enriched in the lung (Supplemental Table S7). Direct comparisons between the nasopharynx and the lung showed that, in particular, sucrose and temperature-responsive genes were significantly different between these niches (Supplemental Fig. S4), suggesting that these are factors that differentiate the nasopharynx from the lung.
Although the gene-set enrichment analyses above are valuable, we hypothesized that they overlook more subtle differences between in vivo niches. Below we show that by exploring subnetworks from the complete data set we are able to identify leads toward gene function discovery, new functional and regulatory relationships, and selection pressures that the pathogen encounters and responds to in the host nasopharynx and lung.
Subnetworks illustrate in vivo niche-specific virulence subpathways
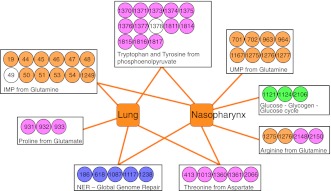
By focusing on specific parts instead of entire pathways, we identified one subpathway that is important in the lung only, three that are important in the nasopharynx only, and four that are important in both (Fig. 3). For instance, the finding that synthesis of proline is needed in the lung but not in the nasopharynx and vice versa for arginine suggests that these amino acids are present at sufficient amounts in one host niche but not the other. We identified three genes (SP1121/SP1124/SP2106) involved in glucose/glycogen cycling that are necessary in the nasopharynx but not lung, which suggests that excess carbon is stored as glycogen and cycled back to glucose to sustain colonization. Additionally, glutamine is required in the nasopharynx for the generation of an essential precursor for pyrimidine synthesis, uridine monophosphate (UMP; also see below), while in the lung UMP is acquired in a different way, possibly from the extracellular environment or via pyrR-mediated synthesis using uracil (SP1278; WpyrR = 0.48). The specific importance of these distinct subpathways indicates that there are distinct in vivo environmental differences.
Figure 3.
Subnetworks illustrate in vivo niche-specific pathways. A pathway-specific subnetwork indicating the importance and specificity of each pathway for the nasopharynx and lung niches is shown. For the tryptophan/tyrosine and IMP producing pathways, we identified two neighboring genes of unknown function (SP0049 and SP1378) with similar fitness defects as their direct neighbors on the chromosome (Supplemental Table S1), and thus we expect that they are equally involved in the respective pathways.
Additional grouping of genes into subnetworks according to the in vitro environment in which they have the most striking response revealed associations that provide insight into gene function and further suggests that each niche differs in stresses such as DNA damage, pH, metal stress, and hydrogen peroxide level, in addition to the availability of specific carbon and energy sources that push the bacterium to utilize both specific as well as overlapping (carbon utilization) genes and pathways in each niche (Supplemental Fig. S5A–E).
Validation of in vitro genotype–phenotype relationships leads to new gene functions
Each tested condition yielded new relationships and individual genes required for survival in the presence of a specific carbon source or under pressure of a defined stress. To evaluate the accuracy of the fitness data, we first examined the in vitro data set and compared it with established functions and genotype–phenotype relationships. For different carbon sources, several previously implicated genes could be confirmed. For example, sucrose-6P hydrolase (SP1724/scrH) is required to generate Glucose-6P from Sucrose-6P and is here conditionally essential (W1724 = 0.08) in the sucrose environment. Likewise, N-acetylglucosamine-6P deacetylase (SP2056), which forms GlcN-6P from GlcNac-6P, is important in the presence of sialic acid (W2056 = 0.47), while conditionally essential (W2056 = 0.1) with GlcNac.
Also, stress conditions yielded expected genes. For example, addition of exogenous hydrogen peroxide revealed the importance of pyruvate oxidase (SP0730/spxB), a gene involved in both producing and conferring resistance to hydrogen peroxide (W0730 = 0.9). Growth in the presence of the DNA-damaging agent MMS, revealed the importance of the base excision repair gene DNA-3-methyladenine glycosylase I (SP0180; W0180 = 0.29). For transformation, we found nine out of 17 previously identified genes that are directly involved in transformation, such as the TCS comDE (SP2235, SP2236) and ciaRH (SP0798, SP0799). Furthermore, we identified 113 other genes involved in transformation. This large number of genes reflects the complex nature of transformation, which includes quorum sensing, competence induction, DNA uptake, recombination and DNA repair, and other stress responses. The overlap of 49 genes between transformation and genes identified under DNA damaging conditions, hydrogen peroxide, and pH stress confirms the involvement of diverse stress responses (Supplemental Fig. S2C).
For each in vitro environment we performed enrichment analyses to determine whether genes belonging to different gene lists were preferentially responsive in a specific in vitro environment. Indeed, we found that, for instance, sucrose genes belonging to the sucrose pathway gene-set were enriched in the sucrose environment (P < 0.0001), fructose genes in the fructose environment (P = 0.0001), and galactose-genes in the galactose environment (P = 0.0001, Supplemental Table S8).
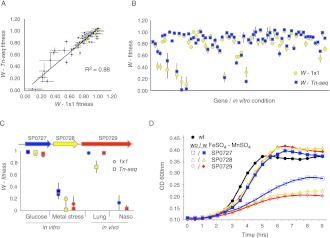
In a quantitative test to validate genotype–phenotype relationships we compared Tn-seq fitness with 1 × 1 deletion-mutant versus wild-type competition assays. In total, 75 in vitro competitions were performed under different growth conditions, which correlated strongly with Tn-seq fitness (R2 = 0.88) (Fig. 4A), and none of the comparisons were significantly different (Fig. 4B; Supplemental Table S9). We confirmed multiple specific genotype–phenotype relationships, thereby demonstrating that the data set consists of high-confidence gene-condition interactions. These data were used to infer new roles for genes; for instance, we identified a new role for the response regulator from TCS 6 (SP2193) in transformation. Dextran glucosidase S (SP1883) is a hydrolase that is important in all tested carbon-source environments except for sucrose, which requires the specific hydrolase scrH (SP1724). Moreover, SP1883 was especially important in the presence of the disaccharide cellobiose for which no hydrolase had previously been identified (Supplemental Table S9). Roles for several hypothetical genes were also revealed and confirmed, such as SP0160, which demonstrates a strong defect in the metal stress environment (Supplemental Fig. S6A). The hypothetical gene SP0181, which is located downstream from the ruvA (SP0179) homologous recombination machinery component and the DNA-3 methyladenine glycosylase I (SP0180) base excision repair gene, is sensitive to MMS, and thus, like its neighbors, is likely involved in DNA repair (Supplemental Fig. S6B). Similarly, spoJ (SP2240) is sensitive to MMS-induced DNA damage (Supplemental Fig. S6B) and contains a ParB-like domain, which has been found in genes involved in processes such as DNA partitioning and nick-closing (Moscoso et al. 1997; Lin and Grossman 1998).
Figure 4.
Tn-seq fitness strongly correlates with traditional 1 × 1 competition experiments, and overlapping in vitro condition-specific fitness with in vivo disease state-specific fitness provides leads toward in vivo selection pressures. (A) Correlation between Tn-seq fitness and 1 × 1 fitness, where a single deletion mutant is competed against the wild type (n = 75, shown are average ± SEM). Each competition was done at least four times, while each Tn-seq fitness value was calculated from at least three independent insertions. (B) No significant differences were found between Tn-seq fitness and 25 deletion mutants that were competed 1 × 1 in several different conditions against the wild type. (C) A strong overlap was found between the three genes SP0727–0729, their sensitivity for metal stress (depleted divalent cations), and their attenuation in the nasopharynx. According to both Tn-seq fitness and 1 × 1 fitness, none of the genes exhibit growth defects in glucose, while growth is severely attenuated in the presence of bipyridyl (metal stress; one-sample t-test with Bonferroni correction; P < 0.0001). In addition, according to Tn-seq fitness for SP0729 and 1 × 1 fitness for SP0727, the operon is important for growth in the nasopharynx (P < 0.001), while Tn-seq fitness for SP0728 and SP0729 and 1 × 1 fitness for SP0727 show there is no defect in the lung. Tn-seq fitness could not be determined in the lung for SP0727 and for SP0727 and SP0728 in the nasopharynx due to a lack of insertions. (D) The growth defect of the three genes SP0727–0729 under metal stress (open symbols) is compensated for by adding 0.2 mM FeSO4 and MnSO4 (closed symbols). These data suggest that S. pneumoniae experiences metal imbalance of divalent cations in the nasopharynx, but not in the lung.
Overlapping virulence and in vitro condition-specific genes reveal selective pressures in the host
We hypothesized that linking gene function with virulence phenotypes can reveal selective pressures in the host and identify target genes and pathways within the pathogen for therapeutic intervention. Because of the biological variation inherent within live mammalian hosts, it was important to validate the accuracy of in vivo Tn-seq fitness. Our Tn-seq data set confirmed 72% (31 of 43) of previously published in vivo virulence phenotypes (Supplemental Table S1). Phenotypes we could not confirm were due to strain differences, highly variable outcomes in the published data, and genes such as nanA, bgaA, and strH, which can be compensated in trans by other clones. We selected eight genes having an in vivo defect in colonization and/or lung infection and a specific in vitro growth defect, and confirmed 14 in vivo virulence phenotypes by competitions (Supplemental Table S9). For instance, we confirmed fitness of the amiACDEF oligopeptide ABC transporter (SP1887/1891) in five different carbon sources as well as the importance of the operon in the lung and the nasopharynx (Supplemental Table S9). Two hypothetical genes (SP0826 and SP1043) and the operon SP1340–1342, consisting of a hypothetical gene (SP1340) and an ABC transporter (SP1341/SP1342), were confirmed for their defect in the presence of sucrose and their attenuation in the nasopharynx, but not the lung (Supplemental Fig. S6C,D). Interestingly, a homology search links the transporter to a macrolide multidrug efflux pump, indicating that it may have a double function; however, we found no association between the pump and the macrolides erythromycin or azithromycin (data not shown), leaving this possible antibiotic efflux function unresolved. These validation examples show how associations can be made between defects found in vitro and in vivo, thereby generating hypotheses regarding the kinds of selective pressures the bacterium experiences in vivo.
Besides the association we found through the enrichment analysis between sucrose and the nasopharynx, which we further confirmed through individual competitions (Supplemental Fig. S5A; Supplemental Table S9), a particularly strong association was identified for a three-gene operon between the in vitro metal stress condition and colonization of the nasopharynx. This in vitro condition, created by adding the divalent cation-chelator bipyridyl to the growth media, identified 68 responsive genes (Supplemental Table S1), among them the operon SP0727–0729, whose genes are annotated as a transcriptional repressor (SP0727), a hypothetical gene (SP0728), and a cation transporter (SP0729) (boxed in the subnetwork in Supplemental Fig. S5D). Confirmation of Tn-seq fitness with 1 × 1 competitions showed that the operon is indeed sensitive to the lowering of divalent cations in the growth medium (Fig. 4C). It was shown recently that this operon could be transcriptionally induced by, and is sensitive to, toxic levels of copper sulfate (Shafeeq et al. 2011), a phenotype which we confirmed (data not shown). Since S. pneumoniae is unlikely to encounter such nonphysiological levels of copper, our findings suggest that, instead, the operon is used to counter imbalances in divalent cation concentrations. In support of this we showed that by adding a slight molar excess of both iron sulfate and manganese sulfate over bipyridyl to the growth media, it was possible to counter the effect of bipyridyl, thereby restoring the balance in divalent cation concentration and rescuing the defect of each of the gene knockouts (Fig. 4D). In addition, we confirmed that the operon is essential for colonization of the nasopharynx, but not for lung infection (Fig. 4C). Although we cannot rule out additional roles for these genes and we did not find overall enrichment in the meta-analysis for metal stress-responsive genes in the nasopharynx, this finding nevertheless suggests that the bacterium may have to deal with some form of imbalance in divalent cations such as Fe2+, Cu2+, and Mn2+ in the nasopharynx, and that this operon plays an important role under this stress condition.
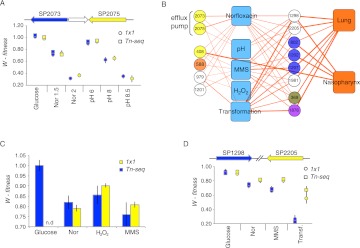
The identification of a dual-role efflux pump and the secondary DNA-damage response of Norfloxacin
The increase in antibiotic resistance in S. pneumoniae (Zhanel et al. 1999; Liñares et al. 2010) and in other bacterial pathogens is an expanding problem. In order to identify genes involved in low-level antibiotic resistance or sensitivity, we screened in the presence of Norfloxacin, a member of the fluoroquinolone class of antibiotics that represent a first line of treatment for lung infection. Norfloxacin inhibits DNA gyrase and topoisomerase IV, which are required for DNA replication and genome segregation. Two identified and confirmed genes (SP2073/SP2075) (Fig. 5A) form a previously reported ABC-transporter, or efflux pump, shown to mediate resistance to multiple drugs and chemicals including Norfloxacin (Robertson et al. 2005). We found that in the absence of this efflux pump, S. pneumoniae is also exquisitely sensitive to alkaline pH (Fig. 5A). Although there are transporters that function as multidrug:cation/proton antiporters (Cheng et al. 1996; Lewinson et al. 2004), there are no direct examples of ABC-transporters that mediate both multidrug resistance and pH homeostasis. Notably, in the absence of the antibiotic, the SP2073/SP2075 efflux pump mediates pH homeostasis, which shows that the two functions are separable, and to our knowledge this is the first dual role transporter identified for S. pneumoniae. Linking antibiotic efflux with a basic physiological function like pH homeostasis may serve the purpose to retain an antibiotic resistance determinant in the absence of the antibiotic.
Figure 5.
The identification of a dual-role efflux pump and the secondary DNA-damage response to Norfloxacin. (A) Tn-seq and 1 × 1 fitness data demonstrate that both genes in the efflux pump SP2073/SP2075 are involved in Norfloxacin resistance in a dose-dependent manner (concentrations 1.5 and 2 μg/mL are indicated by Nor 1.5 and Nor 2). Efflux pump mutants are also sensitive to pH stress, with a small but significant defect at pH 6 (P < 0.001) and an increasing defect at pHs greater than 8 (P < 0.001). Thus, the efflux pump appears to have a dual function—contributing to antibiotic resistance and to pH homeostasis. (B) A subnetwork of Norfloxacin responsive genes (color-coding same as in Fig. 2) indicates that 12 genes contribute to Norfloxacin resistance and also have interactions with the DNA-damaging stress conditions MMS and H2O2. Those genes that interact with MMS and H2O2 also often interact with transformation and have a defect in vivo in the lung and/or the nasopharynx. (C) Confirmation of the significant interactions of recN (SP1202) with Norfloxacin, H2O2, and MMS by 1 × 1 competitions (n.d., 1 × 1 fitness of the single deletion mutant in glucose was not determined). (D) Confirmation of the significant interactions of the DHH family genes SP1298 and SP2205 with Norfloxacin, MMS, and in DNA transformation by 1 × 1 competitions.
Amongst other genes involved in resistance to Norfloxacin we identified 12 that were also sensitive to the DNA-damaging agents, hydrogen peroxide, and/or MMS (Fig. 5B). Three of those genes are annotated as being involved in DNA replication, repair, and recombination, of which we confirmed recN (SP1202) (Fig. 5C; Supplemental Table S9). Five of the 12 genes have unknown functions, but are likely involved in DNA repair and recombination. These include SP1201, located immediately downstream from recN; SP1981, a hypothetical gene and member of the RmuC family that include genes that both protect against nuclease activity and are themselves involved in DNA cleavage; and two genes, SP1298 and SP2205, (Fig. 5D, Supplemental Table S9) that belong to the DHH phosphatase family that includes the exonuclease recJ (SP0611), which degrades single-stranded DNA and is involved in homologous recombination and DNA repair pathways. Consistent with their role in DNA repair and recombination, SP1298 and SP2205 also contribute to DNA transformation. Finally, nine of these 12 genes are important for colonization of the nasopharynx and/or lung infection (including SP1298 and SP2205) (Cron et al. 2011), indicating that DNA damage represents another in vivo stress experienced by S. pneumoniae.
These data demonstrate that Norfloxacin creates an internal bacterial environment in which DNA repair and recombination genes become important for bacterial survival, possibly, as has been shown in E. coli, by creating reactive oxygen species (Kohanski et al. 2010a). Furthermore, this shows that a network approach focused on interactions between antibiotics and the genome provides valuable insight into both the function of an antibiotic as well as the factors that may promote the emergence of resistance (Yeh et al. 2006; Kohanski et al. 2010b).
Genetic interaction mapping reveals a part of the pyrimidine synthesis pathway as a regulatory module controlled by a two-component system response regulator
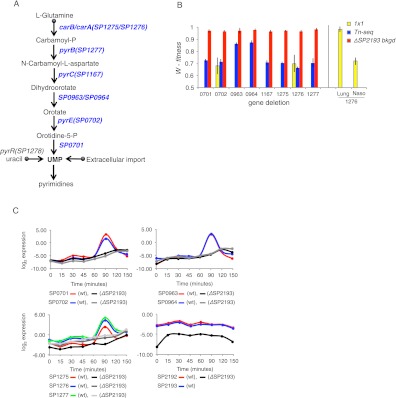
The gene-condition network in Figure 2 does not readily reveal more complex regulatory relationships that are responsible for orchestrating bacterial responses under the various test conditions. Nevertheless, the set of single gene-condition fitness values on which the network is built can be used for genetic interaction mapping, which in theory should be able to reveal such regulatory higher-order relationships. By focusing our analysis on smaller parts of pathways (Fig. 3) we revealed that some of the genes involved in de novo pyrimidine synthesis, specifically the generation of uridine monophosphate (UMP) from glutamine (Fig. 6A; Kanehisa et al. 2008), have a fitness defect during growth in most of the carbon sources tested and in the nasopharynx. This arm of the pyrimidine biosynthetic pathway is specific to bacteria, and although its genes and enzymatic steps are relatively well-characterized, there is very little known about its regulation. Uncovering regulatory relationships can lead toward the stimuli, sensor, and of course regulator, that trigger the bacterial response, and it is therefore possible to identify ways to manipulate this response and potentially new avenues for therapeutic intervention.
Figure 6.
Genetic interaction mapping in combination with gene-expression analysis reveals temporally constrained regulation of the pyrimidine pathway. (A) A schematic of the pyrimidine pathway that produces UMP from L-glutamine by utilizing eight genes that mediate seven enzymatic reactions. (B) Tn-seq fitness for all eight genes from the pyrimidine pathway and 1 × 1 fitness to confirm the phenotypes for genes SP0702 and SP1276. Also, 1 × 1 fitness in the lung and nasopharynx is shown for SP1276 and the observed Tn-seq fitness for all double mutants between SP2193 and the eight genes from the pyrimidine pathway. (C) Quantitative reverse transcription–polymerase chain reaction experiments were done for the first seven of the genes in the de novo pyrimidine biosynthetic pathway, the response regulator SP2193, and the cognate sensor kinase SP2192. A strong induction is observed after 90 min of exposure to glucose for all seven genes in a wild-type background, while the induction is absent in a Δ2193 background.
For all eight genes involved we found very similar fitness defects in glucose (Fig. 6B), and we confirmed phenotypes for the genes involved in the first step (carA/SP1276) and the sixth step (pyrE/SP0702) of this pathway (Fig. 6B). To investigate the regulation of these genes we used genetic interaction mapping to screen for higher-order relationships between carA and the rest of the genome. Six transposon insertion libraries were created in the ΔcarA background, and fitness of each double mutant in the library was measured using Tn-seq. The observed double-mutant fitness was then compared with the fitness expected from the multiplicative model, which was calculated from the initial Tn-seq data by WcarA× Wj, where j is every other gene in the genome. Analyzing the data for fitness values that deviated from the multiplicative model yielded a genetic interaction between carA and the response regulator SP2193 from TCS 6, with an expected fitness of 0.66 ± 0.04 (WcarA × W2193 = 0.66 × 1.00), but an observed fitness of 0.97 ± 0.04 (SEM; P < 0.0002). Deletion of the response regulator thus appeared to suppress the carA fitness defect. To confirm this interaction, we did the reciprocal genetic interaction experiment in the ΔSP2193 background. This experiment revealed that the defect of each of the eight genes in the pathway could be suppressed by ΔSP2193 (Fig. 6B; Supplemental Fig. S7A). To further confirm these genetic interactions, we constructed a double knockout of carA and SP2193. In order to verify that the sensor kinase SP2192 of TCS 6 is not involved, we also constructed a single gene knockout of SP2192 and a double knockout with carA. As shown by the growth curves in glucose (Supplemental Fig. S7B), only ΔSP2193 was able to suppress the carA mutation.
From these data, we hypothesized that SP2193 is a transcriptional activator of the genes in the pyrimidine pathway. To test this we did temporal qRT–PCR experiments on the pathway and the TCS gene transcripts. In the wild-type background the pathway genes were activated in a spike-like manner at ∼90 min after exposure to glucose, while there was no activation when SP2193 was deleted (Fig. 6C). These data suggest that when a gene within the pathway is mutated, the pathway remains active and negatively affects fitness of the bacterium, possibly by accumulation of toxic intermediate products and/or wasteful energy expenditure.
In the event of the disruption of, or failure to express the arm of the pathway from glutamine to UMP, pyrimidines can still be synthesized through production of UMP from uracil via pyrR (SP1278). In addition, it is possible to acquire UMP from the extracellular environment (Fig. 6A). Tn-seq data demonstrates that pyrR is indeed used to produce UMP in the lung (WpyrR = 0.44), thereby explaining why the longer route is not absolutely required, and suggests possible environmental differences between the nasopharynx and lung. This is further confirmed by the requirement of SP2193 in the nasopharynx (WSP2193 = 0.47) but not lung (WSP2193 = 0.95). Downstream from UMP is where both host niches overlap in their requirements such that the paths that lead toward uridine-di/tri-phosphate converge and are required to establish both colonization in the nasopharynx and infection of the lung.
By means of generating genetic interaction profiles, we were successful in discovering a functional regulatory relationship between a core pathway and its transcriptional regulator. This suggests that by perturbing the signal that triggers SP2193 activity in the nasopharynx, or alternatively, by preventing these conserved bacteria-specific enzymatic reactions that lead up to production of UMP, colonization by S. pneumoniae can be inhibited.
A roadmap toward developing new strategies to battle infectious diseases
The development of multidrug resistance by several major human pathogens has highlighted the need for new vaccines, new antibiotics, and strategies to limit the evolution of resistance. This quest can be aided by knowledge of a pathogen's response to antibiotic pressure as well as to host selective pressures during disease and nondisease (carrier) states. In this report we present an experimental strategy that takes a significant step in that direction by utilizing Tn-seq to generate a detailed gene-condition network in response to various defined in vitro and host-specific stresses. With modest resources it will be possible to rapidly generate similar networks for any culturable microorganism for which insertional mutagenesis is available, thus creating a wealth of information that can keep pace with the rapidly accumulating genome sequence data and enable comparisons across different strains and species.
Methods
Bacterial strains, growth, and media
The in vitro experiments were done using an acapsular derivative of S. pneumoniae strain TIGR4 (NCBI Reference Sequence: NC_003028.3), while in vivo experiments were done with the original encapsulated strain. Single gene knockouts were constructed by replacing the coding sequence with a Cm or Spec resistance cassette as described previously (Iyer et al. 2005; van Opijnen and Camilli 2010). Except for specific growth and selection experiments, S. pneumoniae was grown statically in Todd Hewitt broth supplemented with yeast extract (THY) and 5 μL/mL Oxyrase (Oxyrase, Inc) or on Sheep's blood agar plates at 37°C in a 5% CO2 atmosphere. Where appropriate, cultures and blood plates contained 4 μg/mL chloramphenicol (Cm), 200 μg/mL Spectinomycin (Spec), or 1 μg/mL Erythromycin (Erm).
In vivo bacterial doubling time
The temperature-sensitive plasmid pGh9 (Maguin et al. 1996), which does not replicate above 30°C and confers resistance to erythromycin, was transformed into S. pneumoniae. In vitro control experiments indicated that multiple copies of the plasmid were initially present but were reduced to ≤1 per bacterium by culturing the population for seven generations at 37°C. Subsequent growth experiments confirmed the temperature-sensitive nature of the plasmid and a rate of loss in accordance with an in vitro doubling time of 33 min. The strain was used in lung infection and nasopharynx colonization experiments (15 and 16 mice, respectively). Mice were euthanized at different time points post-infection and bacterial loads were determined by plating dilutions on blood agar plates supplemented with and without erythromycin (Supplemental Fig. S1A,B). The growth rate of the population was determined by summing the growth rate of the whole population (titer on plates lacking erythromycin, k-erm) with the absolute value of the rate at which the plasmid disappears from the population (titer on plates plus erythromycin, k+erm). Exponential growth functions were fit to both populations harvested from the lung (k+erm = −0.41, k-erm = −0.025) and the nasopahrynx (k+erm = −0.28, k−erm = −0.022), resulting in a growth rate in the lung of klung = 0.385 which translates to a doubling time of 108 min, and a growth rate of knasopharynx = 0.258, translating to a doubling time of 161 min.
Transposon library construction and selection experiments
Library construction was done as described (van Opijnen et al. 2009; van Opijnen and Camilli 2010). Note that the magellan6 minitransposon we designed lacks transcriptional terminators, therefore allowing for read-through transcription, which explains why no relevant polar effects were observed by examining fitness of downstream genes (Supplemental Table S1). Additionally, the minitransposon contains stop codons in all three frames in either orientation when inserted into a coding sequence. In vitro selection experiments were done with six independently generated libraries each with a size of ∼8000 transposon insertion mutants covering 88% of nonessential genes. Growth conditions where the carbon source was varied consisted of semi-defined minimal media (SDMM) at pH 7.3 supplemented with 10 mM of one of the following carbon sources: glucose, fructose, mannose, galactose, N-acetylglucosamine (GlcNac), sialic acid, sucrose, maltose, cellobiose, or raffinose. Stress conditions consisted of SDMM with 10 mM glucose at pH 7.3 and one of the following stresses: Metal stress, 0.5 mM of 2,2′-Bipyridyl (Sigma-Aldrich); DNA damage, Methyl methanesulfonate 0.015% (MMS, Fluka); hydrogen peroxide exposure, H2O2 4.5 mM (Sigma-Aldrich); acidic pH stress, pH6; temperature stress, growth at 30°C; antibiotic exposure, norfloxacin 1.5 μg/mL (Sigma-Aldrich); and DNA transformation.
Nasopharynx colonization experiments were done in 17 mice with eight independently generated libraries each with a size of ∼4000 mutants, while lung infection experiments were done in 20 mice with six libraries each with a size of ∼30,000 mutants. Because of differences in the bacterial load, 105–106 colony forming units (cfu) for nasopharynx and 107–108 cfu for lung, smaller libraries were used for the nasopharynx in order to minimize the stochastic loss of mutants. Mice were euthanized after 24 h for lung infection, followed by removal and homogenization of the lungs, and 48 h for nasopharynx colonization, followed by flushing of the nasopharynx with 500 μL of PBS.
Sample preparation, sequencing, and fitness
Sample preparation, Illumina sequencing, and fitness calculations using Tn-seq were done as described (van Opijnen et al. 2006, 2009; van Opijnen and Camilli 2010). In vivo fitness was corrected for stochastic loss of mutants due to bottleneck effects, which was calculated for each mouse by determining the proportion of insertion mutants that were lost from the neutral gene set. Subsequently, for each gene the same proportion of insertions is removed from the set of insertions that were lost. On average, 74% of insertions disappeared due to a bottleneck from the nasopharynx population, and 31% from the lung population. The resulting fitness Wi for each gene represents the growth rate per generation. To determine whether Wi significantly differed from wild type, three requirements had to be fulfilled: (1) Wi had to be calculated from at least three data points, (2) Wi had to deviate by >5%, (thus, Wi = <0.95 or >1.05), and (3) Wi had to be significantly different in a one sample t-test with Bonferroni correction for multiple testing. Due to the higher degree of noise in the in vivo data we set more stringent cut offs: Wi in the lung had to deviate by >10%, while in the nasopharynx Wi had to deviate by at least 35%.
Competition assays and single strain growth
Competitions were done at least four times as described previously (van Opijnen et al. 2009). Single strain growth assays were done at least four times using a BioTek Synergy HT plate reader (BioTek Instruments).
Expression analysis
RNA was isolated from cultures at different times using the Qiagen RNAeasy kit (Qiagen). RNA was treated with the TURBO-DNAfree kit (Ambion), after which cDNA was generated with the iScript complete kit (BioRad) from 1 μg of RNA with random hexamers. Quantitative PCR was done using a Stratagene Mv3005P. Each sample was measured in both technical and biological triplicates, and controls lacking reverse transcriptase were included. All samples were normalized against the 50S ribosomal genes SP2204 and SP0973.
Data access
Sequence data can be found at the NCBI Sequence Read Archive (SRA) (http://www.ncbi.nlm.nih.gov/sra) under accession number SRA053099.
Acknowledgments
We thank M. Shainheit and A. Dalia for discussions and detailed comments on the manuscript, A. Boorsma for help in compiling data, and A. Gruss for the plasmid pGh9. T.v.O is supported by a fellowship from the Charles H. Hood Foundation. A.C. is a Howard Hughes investigator. T.v.O and A.C. designed research and wrote the paper. T.v.O performed and analyzed research.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.137430.112.
References
- Baba T, Ara T, Hasegawa M, Takai Y, Okumura Y, Baba M, Datsenko KA, Tomita M, Wanner BL, Mori H 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol Syst Biol 2 doi: 10.1038/msb4100050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghout P, Bootsma HJ, Kloosterman TG, Bijlsma JJE, de Jongh CE, Kuipers OP, Hermans PWM 2007. Search for genes essential for pneumococcal transformation: The RADA DNA repair protein plays a role in genomic recombination of donor DNA. J Bacteriol 189: 6540–6550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng J, Hicks DB, Krulwich TA 1996. The purified Bacillus subtilis tetracycline efflux protein TetA(L) reconstitutes both tetracycline–cobalt/H+ and Na+(K+)/H+ exchange. Proc Natl Acad Sci 93: 14446–14451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cron LE, Stol K, Burghout P, Van Selm S, Simonetti ER, Bootsma HJ, Hermans PWM 2011. Two DHH subfamily 1 proteins contribute to pneumococcal virulence and confer protection against pneumococcal disease. Infect Immun 79: 3697–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Berardinis V, Vallenet D, Castelli V, Besnard M, Pinet A, Cruaud C, Samair S, Lechaplais C, Gyapay G, Richez C, et al. 2008. A complete collection of single-gene deletion mutants of Acinetobacter baylyi ADP1. Mol Syst Biol 4 doi: 10.1038/msb.2008.1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluman N, Bibi E 2009. Bacterial multidrug transport through the lens of the major facilitator superfamily. Biochim Biophys Acta 1794: 738–747 [DOI] [PubMed] [Google Scholar]
- Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW 2004. Chemogenomic profiling: Identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci 101: 793–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harboe ZB, Thomsen RW, Riis A, Valentiner-Branth P, Christensen JJ, Lambertsen L, Krogfelt KA, Konradsen HB, Benfield TL, Klugman KP 2009. Pneumococcal serotypes and mortality following invasive pneumococcal disease: A population-based cohort study. PLoS Med 6: e1000081 doi: 10.1371/journal.pmed.1000081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hava D, Lemieux J, Camilli A 2003. From nose to lung: The regulation behind Streptococcus pneumoniae virulence factors. Mol Microbiol 50: 1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer R, Baliga NS, Camilli A 2005. Catabolite control protein A (CcpA) contributes to virulence and regulation of sugar metabolism in Streptococcus pneumoniae. J Bacteriol 187: 8340–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadioglu A, Weiser J, Paton J, Andrew P 2008. The role of Streptococcus pneumoniae virulence factors in host respiratory colonization and disease. Nat Rev Microbiol 6: 288–301 [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D-U, Hayles J, Kim D, Wood V, Park H-O, Won M, Yoo H-S, Duhig T, Nam M, Palmer G, et al. 2010. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol 28: 617–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King SJ, Hippe KR, Weiser J 2006. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol Microbiol 59: 961–974 [DOI] [PubMed] [Google Scholar]
- Kohanski MA, Depristo MA, Collins JJ 2010a. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell 37: 311–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohanski MA, Dwyer DJ, Collins JJ 2010b. How antibiotics kill bacteria: From targets to networks. Nat Rev Microbiol 8: 423–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, St.Onge R, Proctor M, Flaherty P, Jordan M, Arkin A, Davis R, Nislow C, Giaever G 2005. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet 1: e24 doi: 10.1371/jourmal.pgen.0010024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinson O, Padan E, Bibi E 2004. Alkalitolerance: A biological function for a multidrug transporter in pH homeostasis. Proc Natl Acad Sci 101: 14073–14078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewinson O, Adler J, Sigal N, Bibi E 2006. Promiscuity in multidrug recognition and transport: The bacterial MFS Mdr transporters. Mol Microbiol 61: 277–284 [DOI] [PubMed] [Google Scholar]
- Lin DC-H, Grossman AD 1998. Identification and characterization of a bacterial chromosome partitioning site. Cell 92: 675–685 [DOI] [PubMed] [Google Scholar]
- Liñares J, Ardanuy C, Pallares R, Fenoll A 2010. Changes in antimicrobial resistance, serotypes and genotypes in Streptococcus pneumoniae over a 30-year period. Clin Microbiol Infect 16: 402–410 [DOI] [PubMed] [Google Scholar]
- Liu OW, Chun CD, Chow ED, Chen C, Madhani HD, Noble SM 2008. Systematic genetic analysis of virulence in the human fungal pathogen Cryptococcus neoformans. Cell 135: 174–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maguin E, Prévost H, Ehrlich SD, Gruss A 1996. Efficient insertional mutagenesis in lactococci and other gram-positive bacteria. J Bacteriol 178: 931–935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medini D, Serruto D, Parkhill J, Relman DA, Donati C, Moxon R, Falkow S, Rappuoli R 2008. Microbiology in the post-genomic era. Nat Rev Microbiol 6: 419–430 [DOI] [PubMed] [Google Scholar]
- Moscoso M, Eritja R, Espinosa M 1997. Initiation of replication of plasmid pMV158: Mechanisms of DNA strand-transfer reactions mediated by the initiator RepB protein1. J Mol Biol 268: 840–856 [DOI] [PubMed] [Google Scholar]
- Nichols RJ, Sen S, Choo YJ, Beltrao P, Zietek M, Chaba R, Lee S, Kazmierczak KM, Lee KJ, Wong A, et al. 2011. Phenotypic landscape of a bacterial cell. Cell 144: 143–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble SM, French S, Kohn LA, Chen V, Johnson AD 2010. Systematic screens of a Candida albicans homozygous deletion library decouple morphogenetic switching and pathogenicity. Nat Genet 42: 590–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson GT, Doyle TB, Lynch AS 2005. Use of an efflux-deficient Streptococcus pneumoniae strain panel to identify ABC-class multidrug transporters involved in intrinsic resistance to antimicrobial agents. Antimicrob Agents Chemother 49: 4781–4783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuldiner M, Collins S, Thompson N, Denic V, Bhamidipati A, Punna T, Ihmels J, Andrews B, Boone C, Greenblatt J 2005. Exploration of the function and organization of the yeast early secretory pathway through an epistatic miniarray profile. Cell 123: 507–519 [DOI] [PubMed] [Google Scholar]
- Shafeeq S, Yesilkaya H, Kloosterman TG, Narayanan G, Andrew PW, Kuipers OP, Morrissey JA 2011. The cop operon is required for copper homeostasis and contributes to virulence in Streptococcus pneumoniae. Mol Microbiol 81: 1255–1270 [DOI] [PubMed] [Google Scholar]
- St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis R, Nislow C, Roth F, Giaever G 2007. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat Genet 39: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S 2005. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102: 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293: 498–506 [DOI] [PubMed] [Google Scholar]
- Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Page N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, et al. 2001. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science 294: 2364–2368 [DOI] [PubMed] [Google Scholar]
- Tuomanen L, Mitchel TJ, Morrison DA, Spratt BG 2004. The Pneumococcus. ASM Press, Washington, DC [Google Scholar]
- van Opijnen T, Camilli A 2010. Genome-wide fitness and genetic interactions determined by Tn-seq, a high-throughput massively parallel sequencing method for microorganisms. Curr Protoc Microbiol 1E.3.1–1E.3.16. doi: 10.1002/9780471729259.mc01e03s19 [Google Scholar]
- van Opijnen T, Boerlijst MC, Berkhout B 2006. Effects of random mutations in the human immunodeficiency virus Type 1 transcriptional promoter on viral fitness in different host cell environments. J Virol 80: 6678–6685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Opijnen T, Bodi KL, Camilli A 2009. Tn-seq: High-throughput parallel sequencing for fitness and genetic interaction studies in microorganisms. Nat Methods 6: 767–772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeh P, Tschumi AI, Kishony R 2006. Functional classification of drugs by properties of their pairwise interactions. Nat Genet 38: 489–494 [DOI] [PubMed] [Google Scholar]
- Zhanel G, Karlowsky J, Palatnick L, Vercaigne L, Low DE, Hoban DJ 1999. Prevalence of antimicrobial resistance in respiratory tract isolates of Streptococcus pneumoniae: Results of a Canadian national surveillance study. Antimicrob Agents Chemother 43: 2504–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]