Abstract
The mechanism by which myristoylated proteins are targeted to specific subcellular membrane compartments is poorly understood. Two novel acyl-binding proteins, UNC119A and UNC119B, have been shown recently to function as chaperones/co-factors in the transport of myristoylated G protein α-subunits and src-type tyrosine kinases. UNC119 polypeptides feature an immunoglobulin-like β-sandwich fold that forms a hydrophobic pocket capable of binding lauroyl (C12) and myristoyl (C14) side chains. UNC119A in rod photoreceptors facilitates the transfer of transducin α subunits (Tα) from inner segment to outer segment membranes by forming an intermediate diffusible UNC119-Tα complex. Similar complexes are formed in other sensory neurons, as the G proteins ODR-3 and GPA-13 in C. elegans unc-119 mutants traffic inappropriately. UNC119B knockdown in IMCD3 cells prevents trafficking of myristoylated nephrocystin-3 (NPHP3), a protein associated with nephronophthisis, to cilia. Further, UNC119A was shown to transpot myristoylated src-type tyrosine kinases to cell membranes and to affect T-cell receptor (TCR) and interleukin-5 receptor (IL-5R) activities. These interactions establish UNC119 polypeptides as novel lipid-binding chaperones with specificity for a diverse subset of myristoylated proteins.
Keywords: Uncoordinated 119 (UNC 119), acyl binding proteins, UNC119 crystal structure, C. elegans olfactory neurons, rod photoreceptors, G protein α-subunits, src-type tyrosine kinases, nephrocystin-3, T-cell receptor
1. Introduction
Gα subunits carrying the myristoylation consensus sequence (Farazi et al., 2001) are presumed to be acylated cotranslationally, and combine with Gβγ subunits following the prenylation of Gγ. While there is general agreement that Gαβγ heterotrimer formation is essential for targeting (Marrari et al., 2007), it remains unclear how the complex traffics to the plasma membrane in non-polarized cells, or to cilia of polarized sensory neurons. Trafficking of Gα subunits to cilia mediated by the acyl-binding proteins, UNC119A and UNC119B, has been proposed recently for mouse rod photoreceptors and C. elegans olfactory neurons. Here we discuss the structure and function of the two lipid-binding proteins and the effects of their deletion or knockdown on trafficking/localization of the respective binding partners.
2. UNC119 supergene family
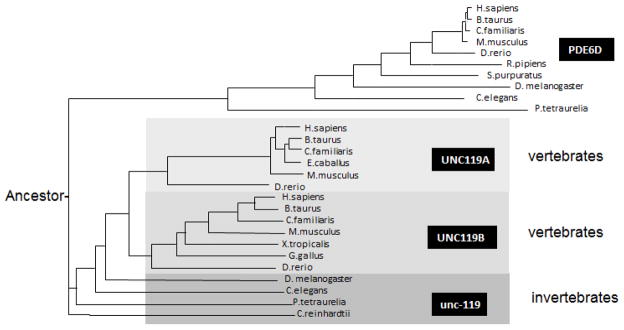
UNC119 polypeptides derive from a supergene family encoding proteins whose function has been maintained through metazoan evolution. Members of this family include prenyl-binding proteins encoded by the PDE6D gene, and two UNC119 paralogs generated by duplication of an ancestral gene (Fig. 1). Polypeptides of this supergene family share an immunoglobulin-like β-sandwich structure for lipid-binding and participate in trafficking of lipidated proteins.
Figure 1.
Phylogram of PDE6D (PrBP/δ) and UNC119 paralogs, generated by ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Four main branches are PDE6D, UNC119A, UNC119B and the invertebrate Unc-119. Only sequences from selected species are shown (from hundreds of genbank hits). PDE6D and UNC119 homologs are present throughout the animal kingdom (unc119 even in plants, Chlamydomonas reinhardtii).
Mammalian UNC119 encodes a widely-expressed 27 kDa polypeptide (Higashide et al., 1996). UNC119 homologs have been identified in plants (POC7 in C. reinhardtii, (Keller et al., 2009)) and all animals, including unicellular protozoa such as Paramecium and Tetrahymena. Subcellularly, UNC119 homologs were identified in the basal body proteome of C. reinhardtii (Keller et al., 2009), the flagellar rootlet of Naegleria (Chung et al., 2007), neurons of C. elegans (Maduro and Pilgrim, 1995) and in the mouse photoreceptor sensory cilium complex (Liu et al., 2007).
3. The multifunctional UNC119A/RG4 protein
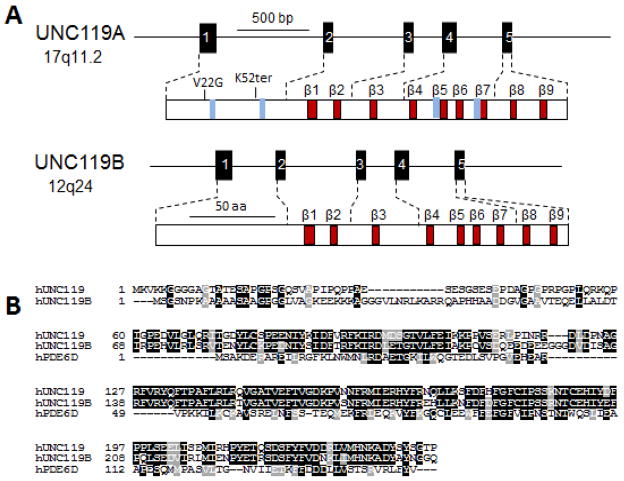
The C. elegans unc-119 gene was first discovered due to a spontaneous mutation resulting in defects in chemosensation, locomotion and feeding behavior (Maduro and Pilgrim, 1995). Predominant expression of an unc-119/LacZ fusion construct in C. elegans neurons suggested that the unc-119 mutant phenotype was caused by a nervous system defect. A human UNC119A ortholog termed HRG4 (Human Retina-specific Gene 4) was identified shortly thereafter by subtractive cloning. Retina specificity referred to abundant expression in the retina as observed by northern blotting. A closely related paralog, UNC119B, was identified later in zebrafish by library screening with degenerate primers (Manning et al., 2004). Ubiquitously-expressed human UNC119A and UNC119B are comprised of 240 and 251 amino acids, respectively. UNC119A mRNA exhibits strong expression in retina with the protein localized to photoreceptor synapses and inner segments (Higashide et al., 1998). The human UNC119 and UNC119B genes (Fig. 2) have an identical gene structure consisting of 5 exons (Higashide and Inana, 1999). UNC119A produces a splice variant in which the 3′ most intron is retained, yielding a distinct C-terminal end (Swanson et al., 1998) lacking the β-sandwich structure. Both paralogs exhibit significant sequence similarity with PrBP/δ, particularly at the PrBP/δ C-terminal region, which forms the β-sandwich fold into which lipid side chains may insert.
Figure 2.
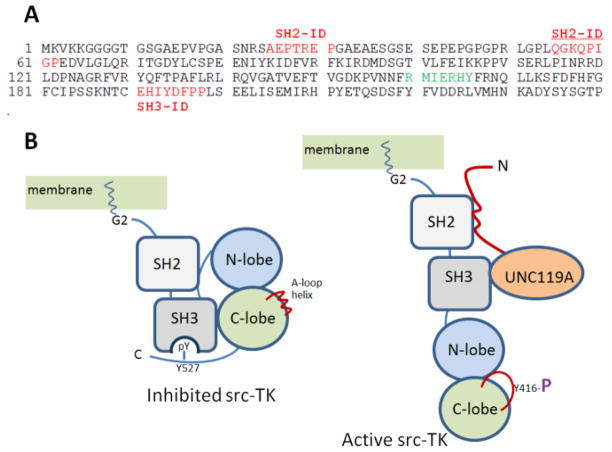
Unc119 genes and proteins. A, Structure of Unc119 genes and gene products. Mutations associated with disease are noted in the UNC119A protein. Blue bars denote approximate positions of SH2- and SH3-interacting domains. B, Sequence alignment of UNC119 paralogs and PrBP/δ. Conserved residues are shown white-on-black background, conservative substitutions white-on-grey background.
A heterozygous stop codon in the UNC119 gene (K57ter) was linked to cone dystrophy in a single patient (Kobayashi et al., 2000) and this phenotype was replicated in a transgenic mouse model expressing the murine Unc119 gene with an identical stop codon (Kobayashi et al., 2000). Further, a UNC119(G22V) dominant negative mutation has been suggested to cause idiopathic CD4 lymphopenia (ICL), an immunodeficiency disorder associated with reduced T-cell stimulation (Gorska and Alam, 2011).
4. UNC119A-interacting partners
Yeast two-hybrid screening and in-vitro pulldown assays reveal that UNC119A interacts with a large number of both related and unrelated polypeptides, including acylated Gα-subunits, receptor-associated src-type tyrosine kinases, non-receptor protein kinases, small Arf-like GTPases (ARL proteins), the GTPase dynamin and the synaptic ribbon-associated protein RIBEYE (Table 1). Pulldowns and isothermal titration calorimetry showed a tight interaction between UNC119A and acylated Gα peptides. Interaction with myristoylated G proteins was rather specific as several myristoylated Ca2+-binding proteins (e.g., recoverin, GCAP1) failed to interact with UNC119A (Zhang et al., 2011b). These proteins have either a Ca2+-dependent myristoyl switch that obscures the acyl chain intramolecularly (recoverin) (Zozulya and Stryer, 1992), or interact with another protein (GCAP1 binding to guanylate cyclase) (Baehr and Palczewski, 2009), thus preventing interaction with UNC119.
Table 1.
UNC119 interacts with a large number of diverse proteins. Interactants include acylated G protein α-subunits, src-type tyrosine kinases, the large GTPase dynamin, small Arf-like GTPases (Arl2 and Arl3), the Ca2+-binding protein CaBP4 and RIBEYE. Column 1, interacting proteins; proteins known to insert an acyl side chain into the β-sandwich fold of UNC119 are bold-faced and underlined. Red, proteins known to play a role in photoreceptors; green, G protein a-subunits in C. elegans olfactory neurons; black, all others. Column 2, function. Arrow directions indicate enzymatic activation (up) or inhibition (down). Column 3, N-terminal sequence; N-termini known to be N-acylated at G2 are shaded black. Column 4, pathways or cells in which interactants play a role. Column 5, references.
| Molecule | function | N-term | pathway/cells | reference |
|---|---|---|---|---|
| Gnat1 | Gα subunit |

|
photoreceptor | (Zhang et al., 2011a) |
| Gnat2 | Gα subunit | photoreceptor | (Zhang et al., 2011a) | |
| Rab11 | GTPase | MGTRDDEY | vesicular transport | (Gorska et al., 2009) |
| Arl2 | GDF | MGLLTILK | vesicular transport | (Kobayashi et al., 2003) |
| Arl3 | GDF | MGLLSILR | vesicular transport | (Veltel et al., 2008b) |
| RP2 | GAP | MGCCFFSK | vesicular transport | (Veltel et al., 2008a) |
| CaBP4 | Ca2+ binding | MTTEQARQ | synapse | (Haeseleer, 2008) |
| RIBEYE | Scaffold protein | MPVPSRHI | synaptic ribbon | (Alpadi et al., 2008) |
| ODR-3 | Gα subunit |

|
C. elegans olfaction | (Zhang et al., 2011a) |
| Gpa-13 | Gα subunit | C. elegans olfaction | (Zhang et al., 2011a) | |
| NPHP3 | unknown | IMCD3 cilia | (Wright et al., 2011) | |
| CYS1 | unknown | MGSGSSR | IMCD3 cilia | (Wright et al., 2011) |
| C5orf30 | unknown | MEVDINGD | IMCD3 cilia | (Wright et al., 2011) |
| Lyn | srcTK↑ |

|
eosinophils | (Cen et al., 2003) |
| Hck | srcTK↑ | eosinophils | (Cen et al., 2003) | |
| Lck | srcTK↑ | T-cells | (Gorska et al., 2004) | |
| Fyn | srcTK↑ | T-cells | (Gorska et al., 2004) | |
| Abl | pTK↓ | (Vepachedu et al., 2009) | ||
| Arg | pTK↓ | (Vepachedu et al., 2009) | ||
| Dynamin | GTPase | MGNRGMEE | endocytosis | (Gorska et al., 2006) |
Of the 17 known mammalian G-protein α-subunits, nine contain glycine as residue 2 (G2) and only seven of these contain a myristoylation consensus sequence. Both bovine rod and cone transducin α-subunits (GNAT1 and GNAT2) have been shown experimentally to interact with UNC119A. GNAT3 (gustducin, expressed in the gustatory system) is closely related in sequence and is therefore predicted to interact as well. ODR-3, a G protein present in C. elegans olfactory sensory neurons has also been shown experimentally to interact directly with UNC119A. Isothermal titration calorimetry experiments indicate that ODR-3 interacts with human UNC119A one order of magnitude weaker (Zhang et al., 2011b) than GNAT1, owing to the differences between the N-terminal 10 residues of ODR-3 and GNAT1 (Table 1).
5. UNC119A crystal structure
The crystal structure at 1.95Å resolution (Zhang et al., 2011b) reveals that UNC119A adopts an immunoglobulin-like β-sandwich fold that forms a hydrophobic cavity into which the lipid may be inserted. The co-crystal structure of UNC119A in complex with a lauroylated Tα peptide at 2.0 Å shows that the binding pocket easily accommodates a lauroyl (C12) moiety (Fig. 3). The cavity is lined predominantly by hydrophobic residues (mostly Phe and Tyr) that mediate the interaction with the lauroylated T peptide primarily via Van der Waals forces, consistent with properties of a lipid-binding site. The lauroyl group and the first six residues of the Tα peptide are buried deeply in the hydrophobic pocket of UNC119A whereas peptide residues 7–10 make only peripheral contact with UNC119A and are therefore poorly ordered in the structure. In general, the surrounding UNC119A residues form a surface that is highly complementary to the shape of the ligand, thereby rationalizing the high degree of conservation among the N-terminal residues of Tα and the contacting residues of UNC119A. These interactions provide specific recognition of N-terminal groups that define the location of the peptide-acyl junction within the UNC119 cavity, thereby limiting the acyl chain length that can be accommodated.
Figure 3.
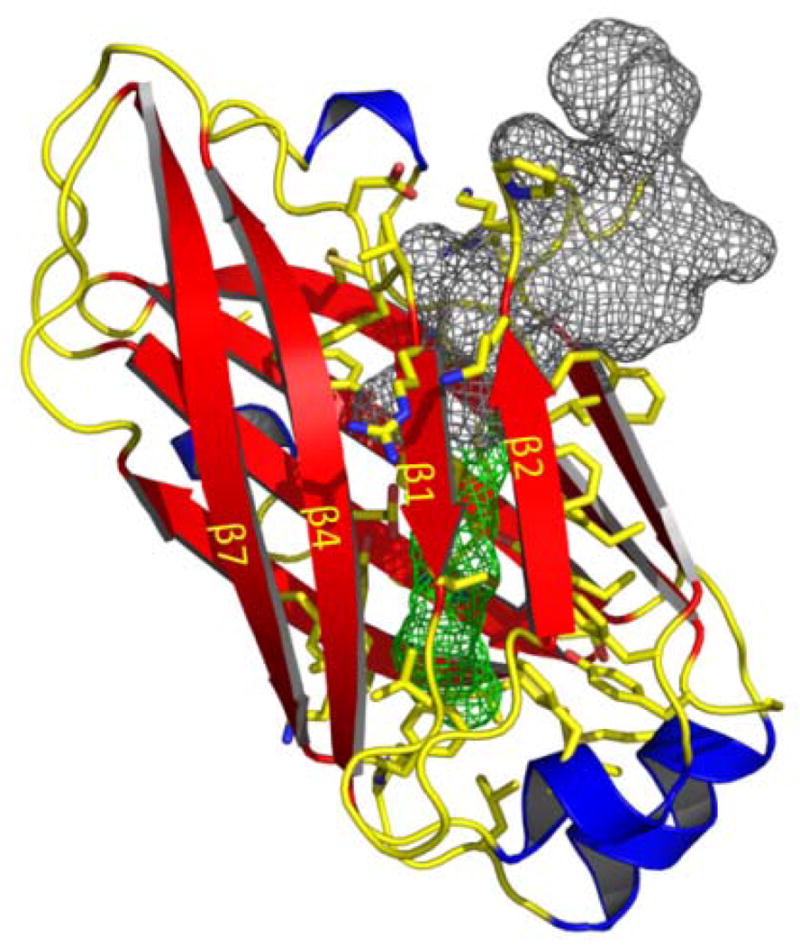
Structure of UNC119 and lipid-binding cavity. Ribbon representation of human UNC119 structure (residues 57–237). Nine β-strands (β1-β9, red) form two β-sheets that splay apart at one end to create an entrance to the β-sandwich cavity. Lauroyl-GAGASAEEKH is depicted as a space-filling mesh with its lipid (green) and amino acids (grey).
Crystal structures of PrBP/δ, encoded by the PDE6D gene, and RhoGDI with cognate lipids inserted into the hydrophobic pocket have been determined with high resolution and structural similarities have been discussed in detail (Hanzal-Bayer et al., 2002; Zhang et al., 2011b; Chandra et al., 2012; Ismail et al., 2011). Remarkably, the sites through which the lipid chains enter the pocket distinguish UNC119A from RhoGDI and PrBP/δ: the entrance to the hydrophobic pocket in UNC119A lies between strands β2 and β3, whereas the entrances of RhoGDI and PrBP/δ are located on the opposite side of the β-sandwich (Zhang et al., 2011b).
6. UNC119A and trafficking in rod photoreceptors
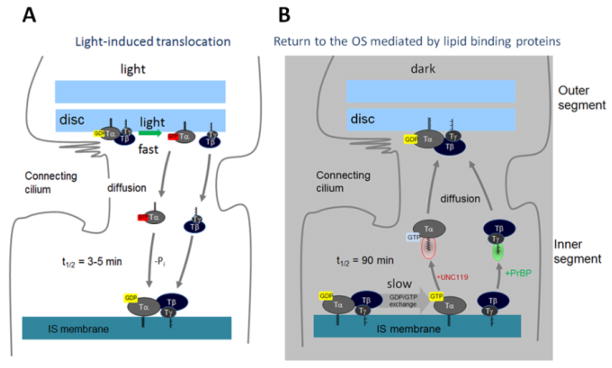
The Unc119−/− mouse shows no obvious retinal degeneration early in life (up to 6 months postnatally) (Ishiba et al., 2007), but degeneration accelerates by 17 months and is complete at 20 months. The slow progression of retinal degeneration may, in part, be attributed to the presence of UNC119B in photoreceptors (Zhang et al., 2011b). In predegenerate Unc119−/− retina, Tα partially localized to the inner segment and outer nuclear layer suggesting defects in trafficking. Under intense light conditions, TαGTP and Tβγ are known to translocate to the inner segments by diffusion in minutes and in darkness return to the outer segments very slowly over the course of hours (Fig. 4) (Sokolov et al., 2002). After three hours of dark-adaptation return of Tα was nearly complete in wild-type retinas, but failed in the retinas of Unc119−/− mice providing evidence that UNC119A participates in targeting transducin-α to the outer segment via interaction with its acylated N-terminus (Zhang et al., 2011b).
Figure 4.
Depiction of transducin translocation to the inner segment (IS) induced by constant light, and return to the outer segment (OS) in dark. A, Following GTP/GDP exchange, Tα-GTP and Tβγ diffuse separately to the inner segment. UNC119 is not required in this phase. As GTP hydrolyzes, the subunits recombine and associate with IS membranes. B, In dark, GTP/GDP exchange is slow in the absence of a GEF, and may be rate-limiting in extraction of Tα in the presence of UNC119A. Following exchange, TαGTP-UNC119 and Tβγ-PrBP/δ solubilize and return by diffusion to the OS discs. Most likely, ARL3-GTP acts as a GDF to discharge Tα-GDP to membranes.
The delayed return to the outer segment is likely the result of an enzymatically switchable sink in the inner segment, and relatively low amount of UNC119 compared to transducin (Zhang et al., 2011b; Gopalakrishna et al., 2011). Upon arrival at the inner segment, transducin subunits are presumed to recombine following GTP hydrolysis and dock to inner segment membranes. In the absence of a GEF (rhodopsin), GTP/GDP exchange is presumed to be very slow (rate constant 10−4/sec) (Cowan et al., 2000). Both the GTP requirement for Tα extraction from membranes by UNC119A and the slow rate constant suggest that this event may be the rate-limiting step for return to the outer segment. Upon solubilization of the complex, UNC119A/Tα-GTP likely diffuses passively through the inner segment and connecting cilium, depositing Tα directly onto outer segment discs. Separate trafficking of Tα by diffusion to the OS is supported by the formation of a stable Tα-UNC119 complex, the competition of UNC119 with Tβγ for binding to Tα regardless of the state of the nucleotide, and by the effect of PrBP/δ deletion on Tβγ trafficking (retention in the inner segments). Detection of UNC119A in the mouse photoreceptor sensory cilium complex (Liu et al., 2007) suggests a possible role for UNC119A in discharging of cargo (Tα). This observation is strengthened further by the association of UNC119A with ARL3 and RP2 (retinitis pigmentosa protein 2) which localize to mouse and human cilia (Liu et al., 2007; Veltel et al., 2008b). It is intriguing to speculate that ARL3-GTP may serve as a GDF (GDI displacement factor) that discharges Tα from UNC119A, similarly as shown for discharging myristoylated NPHP3 from UNC119B in IMCD3 cells (Wright et al., 2011) (see Fig. 5, next paragraph).
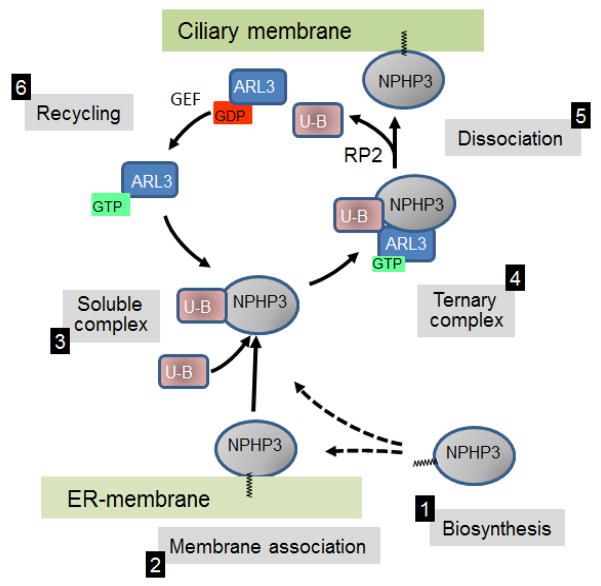
Figure 5.
Targeting of myr-NPHP3 to cilia (adapted from Wright et al). Following biosynthesis and co-translational acylation (1), NPHP5 forms a soluble complex with UNC119B (U–B) (3), either directly or indirectly (broken arrows) after membrane association (2). The UNC119B/NPHP3 complex combines with the GDI-displacement factor (GDF) ARL3-GTP forming a ternary complex (4). GTP hydrolysis mediated by the GAP RP2 discharges NPHP3 to the ciliary membrane (5). GDP/GTP exchange catalyzed by a guanine nucleotide exchange factor (GEF) recycles ARL3-GTP and UNC119B (6).
7. UNC119B targets NPHP3 to cilia
Very little is known about the function of UNC119B. In mouse, Unc119B ESTs have been identified in retina, brain, cerebellum, kidney, pancreas and mammary gland. Recently, Wright, et al. (Wright et al., 2011) showed that knockdown of UNC119B prevented proper ciliary targeting of nephrocystin-3 (NPHP3) in transfected IMCD3 cells. UNC119A could not substitute for the UNC119B deficiency, suggesting that these two closely related proteins have different functions (Wright et al., 2011). NPHP3 is a myristoylated protein which is mutated in 1% of all nephronophthisis cases. A hypomorphic mutation of Nphp3 causes cystic kidney disease in the naturally occurring pcy mouse (Olbrich et al., 2003) while an Nphp3 knockout is embryonic lethal. Targeting of NPHP3 to primary cilia is controlled by N-terminal myristoylation as a NPHP3(G2A) mutant did not target to cilia (Nakata et al., 2012). Ciliary targeting experiments in IMCD3 cells further established that RP2, which functions as an ARL3 GTPase activator (GAP), is required for unloading cargo (Wright et al., 2011) (Fig. 5). The ability of UNC119B to control ciliary targeting suggests that null alleles of human UNC119B may cause of ciliopathies such as Senior Løken or Joubert syndromes, both of which are associated with nephronophthisis and retinal degeneration.
Knockdown of unc119a in zebrafish resulted in a MO-mediated dose-dependent curved body axis, Kupffer’s vesicle (KV) defects and reduced visual function (Wright et al., 2011), while knockdown of zebrafish unc119b yielded only a mild KV defect with no other obvious cilia-related phenotypes (Wright et al., 2011). A “curly tail down” phenotype accompanied by axon pathfinding defects was observed during embryonic development (Manning et al., 2004). These experimental findings suggest that UNC119A and UNC119B have distinct functions. However, as shown for UNC119A (Maduro et al., 2000), human UNC119B was able to rescue the C. elegans unc-119 mutant phenotype (Wright et al., 2011). Thus, functions of UNC119A and UNC119B are likely in part overlapping.
8. G protein mistrafficking in unc-119 mutant C. elegans olfactory neurons
C. elegans relies on its 11 pairs of amphid neurons to detect odorants and soluble molecules (Perkins et al., 1986; Ward et al., 1975). These sensory neurons are polarized ciliated cells with an overall morphology similar to vertebrate photoreceptors. The neuron dendritic tips possess specialized cilia which host a variety of odorant GPCRs as well as downstream components, e.g., G proteins and ion channels. C. elegans uses AWA and AWC neurons to sense volatile substances (Bargmann, 2006). Several G proteins participate in chemosensory signal transduction including ODR-3 and GPA-13. ODR-3 is the most important of these G proteins in AWA and AWC sensory neurons (Roayaie et al., 1998). AWA cilia are filamentous and extensively branched, whereas AWC cilia form a large membranous sheet that encircles the tip of the nose. GPA-13 appears exclusively in cilia of ADF, ASH and AWC chemosensory neurons (Lans et al., 2004). In unc-119 mutants, ODR-3 protein is mislocalized to the cell bodies and GPA-13 was found to be down-regulated. Consequences of unc-119 deletion in the worm are mislocalization and mistargeting of the G proteins ODR-3 and GPA-13 expressed in AWA, AWB, and AWC as well as in ASH and ADF amphid cells. These data suggest that Unc-119 participates in regulating the transport of ODR-3 and GPA-13 from the cell bodies to the cilia of C. elegans olfactory neurons, a function similar to that in mouse photoreceptors. The unc-119 mutant phenotype in C. elegans is more severe, likely because the worm genome harbors only one unc-119 gene and lacks the redundancy present in mouse (Fig. 1).
9. UNC119A and src-type Tyrosine Kinases
UNC119A is known to interact with the α-chain of the interleukin-5 receptor (IL-5R), which is associated with the src-type tyrosine kinases Lyn and Hck (Cen et al., 2003). IL-5 is one of the primary regulators of eosinophil differentiation and function (Gorska et al., 2006). UNC119A induces the catalytic activity of these kinases through interaction with domains SH2 and SH3 (Src homology domains 2 and 3), which recognize the phosphorylated state of critical tyrosine residues. Two SH2-interacting domains (SH2-ID) on UNC119A are located near the N-terminus and the SH3-ID is found in the C-terminal β-sandwich domain (Cen et al., 2003) (Fig. 6). The inactive state of scr-type kinases is stabilized by intramolecular interactions between the SH2/SH3 domains and the kinase domains. Binding of the activator, UNC119A, to these domains induces autophosphorylation, stimulating kinase activity. Eosinophils (a leukocyte subgroup) transduced with UNC119A both activated Lyn and prolonged eosinophil survival in the absence of IL-5 stimulation. Additionally, antibody-mediated inhibition of UNC119A decreased eosinophil survival (Cen et al., 2003). Lyn and Hck carry myristoylation consensus sequences at their N-termini (G2…S6) (Table 1) and are known to be acylated depending on which splice variant is transcribed (Silverman et al., 1993). However, it is unknown whether the Lyn and Hck acyl chains interact with UNC119A as described for Gα subunits.
Figure 6.
Model of src-TK activation by UNC119A. A, UNC119 sequence with locations of SH2- and SH3-interacting domains (ID, red) (Cen et al., 2003). Two SH2-IDs are located in the N-terminal region, one SH3-ID is located on β7. The tyrosine phosphorylation site (green) on β5 is shown. B, Depiction of inhibited Src-type tyrosine kinase (left) and its activation by Unc119A (right) (adapted from (Xu et al., 1999). In the inhibited form, Y527 is phosphorylated and interacts with SH2; the loop forms an inhibitory helix. In the active form, kinase domains are displaced by UNC119’s SH2- and SH3-IDs, allowing phosphorylation of Y416 and activation.
10. UNC119A regulates trafficking of Lck to the T-cell receptor (TCR)
As indicated above, UNC119A overexpression in eosinophils stimulated the activities of Lck and Fyn, tyrosine kinases associated with the T-cell receptor (Gorska et al., 2004). Like Lyn and Hck, Lck and Fyn are src-type tyrosine kinases, carry G2 at the N-terminus and are myristoylated (Silverman et al., 1993) (Table 1). UNC119A-deficient cells exhibited reduced tyrosine phosphorylation and the TCR was unable to stimulate Lck and Fyn tyrosine kinases, or downstream pathways. However, reconstitution of these depleted cells with UNC119A was able to fully rescue the phenotype (Gorska et al., 2004). Furthermore, this UNC119A-regulated pathway is essential for immunological synapse formation and T-cell activation (Gorska et al., 2009).
The T-cell receptor lacks enzymatic activity and relies on Lck to initiate signaling. UNC119A not only activates Lck, but also appears to regulate trafficking of Lck on vesicles to the plasma membrane (Gorska et al., 2009), likely through an interaction with the myristoyl moiety on Lck. Transport of Lck to the membrane depends on both UNC119A and Rab11, which strongly colocalize in Jurkat T-cells. UNC119A was also able to control the GTPase activity of Rab11, most likely by interfering with its GAP. In UNC119A-knockdown cells, Lck failed to transport to the plasma membrane and accumulated in perinuclear membranes (Gorska et al., 2009). In contrast, Fyn was not affected by UNC119 knockdown. Data also indicate that UNC119A orchestrates the recruitment of the actin-based motor protein, myosin 5B, and the organization of multiprotein complexes on endosomes (Gorska et al., 2009).
11. UNC119A regulates endocytosis and partially protects against Shigella infection
Dynamin-2 is a GTPase that has been shown to be critical for endocytosis and interact with the IL-5R α-subunit (Gorska et al., 2006). Dynamin also interacts with IL-5 receptor-associated tyrosine kinases, Lyn and JAK2, in eosinophils. Data indicate that UNC119A inhibits the GTPase activity of dynamin, which is critical for endocytosis, implying a role for UNC119A in clathrin- and caveolae-based endocytosis as well as macro pinocytosis (Karim et al., 2010). Knockdown of UNC119A in fibroblasts increases the uptake of transferrin, albumin, viruses and ligand-coated beads, whereas overexpression inhibits uptake of these entities (Karim et al., 2010). Furthermore, UNC119A inhibits signaling pathways used by Shigella bacteria to enter the cell and therefore provides partial but significant protection from Shigella infection, which causes Shigellosis in primates but not in other mammals, typically resulting in dysentery. UNC119A induction in-vivo has been shown to boost host defense against infections (Vepachedu et al., 2009).
12. G22V missense mutation in UNC119A causes idiopathic CD4 lymphocytopenia (ICL)
ICL is a rare heterogeneous syndrome in which the body has too few CD4 T-lymphocytes, in the absence of HIV infection. Patients with ICL have weakened immune systems and are susceptible to a variety of infections and autoimmune diseases. Recently, Gorska, et al. identified a G22V missense mutation in the human UNC119A gene of a single patient (Gorska and Alam, 2011) with reduced TCR responses and mislocalization of Lck. The patient presented with a history of recurrent sinusitis/otitis media, frequent episodes of shingles, a widespread fungal nail infection, fungal dermatitis and oral herpetic lesions. The G22V mutation, located adjacent to the first SH2-interacting domain near the N-terminus of UNC119A (Fig. 6), was absent in 60 healthy controls. Mechanistically, mutant UNC119A is unable to interact with Lck, an interaction necessary for both the localization of Lck to the TCR and also the activation of Lck. As a consequence, T-cell proliferation is blocked impairing the normal immune response (Gorska and Alam, 2011).
13. Summary
UNC119 paralogs are a novel class of polypeptides involved in targeting a subset of acylated polypeptides to primary cilia. These include acylated Gα subunits and nephrocystin-3 expressed in species as varied ashuman. mouse and worm. This interaction is based on the insertion of the acyl chain into a hydrophobic pocket formed by a immunoglobulin-like β-sandwich fold. UNC119 may interact with myristoylated src-type tyrosine kinases by binding to SH2 and SH3- interacting domains, a pathway essential for T-cell activation and formation of an immunological synapse. Furthermore, UNC119 forms complexes with the GTPase ARL3-GTP and its GAP, the retinitis pigmentosa protein RP2. Interactions with multiple partners and modes assign UNC119 polypeptides a central role in trafficking a subgroup of acylated proteins to their subcellular destinations.
Highlights.
UNC119 polypeptides are multifunctional proteins and part of a supergene family
UNC119A interacts with a subset of acylated G protein α-subunits
UNC119A affects trafficking of transducin in rod photoreceptors
UNC119A interacts with src-type tyrosine kinases through SH2 and SH3 interacting domains
UNC119B traffics NPHP3 to cilia in IMCD3 cells
Acknowledgments
This work was supported by National Institute of Health grants EY08123, EY019298 (WB), EY014800–039003 (NEI core grant), by a Center Grant of the Foundation Fighting Blindness, Inc., to the University of Utah, and unrestricted grants to the Departments of Ophthalmology, University of Utah from Research to Prevent Blindness (RPB; New York). WB is a recipient of a Research to Prevent Blindness Senior Investigator Award.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- Alpadi K, Magupalli VG, Kappel S, Koblitz L, Schwarz K, Seigel GM, Sung CH, Schmitz F. RIBEYE recruits Munc119, a mammalian ortholog of the Caenorhabditis elegans protein unc119, to synaptic ribbons of photoreceptor synapses. J Biol Chem. 2008;283:26461–26467. doi: 10.1074/jbc.M801625200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baehr W, Palczewski K. Focus on molecules: guanylate cyclase-activating proteins (GCAPs) Exp Eye Res. 2009;89:2–3. doi: 10.1016/j.exer.2008.12.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bargmann CI. WormBook. 2006. Chemosensation in C. elegans; pp. 1–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen O, Gorska MM, Stafford SJ, Sur S, Alam R. Identification of UNC119 as a novel activator of SRC-type tyrosine kinases. J Biol Chem. 2003;278:8837–8845. doi: 10.1074/jbc.M208261200. [DOI] [PubMed] [Google Scholar]
- Chandra A, Grecco HE, Pisupati V, Perera D, Cassidy L, Skoulidis F, Ismail SA, Hedberg C, Hanzal-Bayer M, Venkitaraman AR, et al. The GDI-like solubilizing factor PDEdelta sustains the spatial organization and signalling of Ras family proteins. Nat Cell Biol. 2012;14:148–158. doi: 10.1038/ncb2394. [DOI] [PubMed] [Google Scholar]
- Chung S, Kang S, Paik S, Lee J. NgUNC-119, Naegleria homologue of UNC-119, localizes to the flagellar rootlet. Gene. 2007;389:45–51. doi: 10.1016/j.gene.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Cowan CW, Wensel TG, Arshavsky VY. Enzymology of GTPase acceleration in phototransduction. Methods Enzymol. 2000;315:524–538. doi: 10.1016/s0076-6879(00)15865-3. [DOI] [PubMed] [Google Scholar]
- Farazi TA, Waksman G, Gordon JI. The biology and enzymology of protein N-myristoylation. J Biol Chem. 2001;276:39501–39504. doi: 10.1074/jbc.R100042200. [DOI] [PubMed] [Google Scholar]
- Gopalakrishna KN, Doddapuneni K, Boyd KK, Masuho I, Martemyanov KA, Artemyev NO. Interaction of transducin with uncoordinated 119 protein (UNC119): implications for the model of transducin trafficking in rod photoreceptors. J Biol Chem. 2011;286:28954–28962. doi: 10.1074/jbc.M111.268821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorska MM, Alam R. A mutation in the human Uncoordinated 119 gene impairs TCR signaling and is associated with CD4 lymphopenia. Blood. 2011 doi: 10.1182/blood-2011-04-350686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorska MM, Cen O, Liang Q, Stafford SJ, Alam R. Differential regulation of interleukin 5-stimulated signaling pathways by dynamin. J Biol Chem. 2006;281:14429–14439. doi: 10.1074/jbc.M512718200. [DOI] [PubMed] [Google Scholar]
- Gorska MM, Liang Q, Karim Z, Alam R. Uncoordinated 119 protein controls trafficking of Lck via the Rab11 endosome and is critical for immunological synapse formation. J Immunol. 2009;183:1675–1684. doi: 10.4049/jimmunol.0900792. [DOI] [PubMed] [Google Scholar]
- Gorska MM, Stafford SJ, Cen O, Sur S, Alam R. Unc119, a novel activator of Lck/Fyn, is essential for T cell activation. J Exp Med. 2004;199:369–379. doi: 10.1084/jem.20030589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haeseleer F. Interaction and Colocalization of CaBP4 and Unc119 (MRG4) in Photoreceptors. Invest Ophthalmol Vis Sci. 2008;49:2366–2375. doi: 10.1167/iovs.07-1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanzal-Bayer M, Renault L, Roversi P, Wittinghofer A, Hillig RC. The complex of Arl2-GTP and PDE delta: from structure to function. EMBO J. 2002;21:2095–2106. doi: 10.1093/emboj/21.9.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higashide T, Inana G. Characterization of the gene for HRG4 (UNC119), a novel photoreceptor synaptic protein homologous to unc-119. Genomics. 1999;57:446–450. doi: 10.1006/geno.1999.5791. [DOI] [PubMed] [Google Scholar]
- Higashide T, McLaren MJ, Inana G. Localization of HRG4, a photoreceptor protein homologous to Unc-119, in ribbon synapse. Invest Ophthalmol Vis Sci. 1998;39:690–698. [PubMed] [Google Scholar]
- Higashide T, Murakami A, McLaren MJ, Inana G. Cloning of the cDNA for a novel photoreceptor protein. J Biol Chem. 1996;271:1797–1804. doi: 10.1074/jbc.271.3.1797. [DOI] [PubMed] [Google Scholar]
- Ishiba Y, Higashide T, Mori N, Kobayashi A, Kubota S, McLaren MJ, Satoh H, Wong F, Inana G. Targeted inactivation of synaptic HRG4 (UNC119) causes dysfunction in the distal photoreceptor and slow retinal degeneration, revealing a new function. Exp Eye Research. 2007;84:473–485. doi: 10.1016/j.exer.2006.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ismail SA, Chen YX, Rusinova A, Chandra A, Bierbaum M, Gremer L, Triola G, Waldmann H, Bastiaens PI, Wittinghofer A. Arl2-GTP and Arl3-GTP regulate a GDI-like transport system for farnesylated cargo. Nat Chem Biol. 2011;7:942–949. doi: 10.1038/nchembio.686. [DOI] [PubMed] [Google Scholar]
- Karim Z, Vepachedu R, Gorska M, Alam R. UNC119 inhibits dynamin and dynamin-dependent endocytic processes. Cell Signal. 2010;22:128–137. doi: 10.1016/j.cellsig.2009.09.022. [DOI] [PubMed] [Google Scholar]
- Keller LC, Geimer S, Romijn E, Yates J, III, Zamora I, Marshall WF. Molecular architecture of the centriole proteome: the conserved WD40 domain protein POC1 is required for centriole duplication and length control. Mol Biol Cell. 2009;20:1150–1166. doi: 10.1091/mbc.E08-06-0619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi A, Higashide T, Hamasaki D, Kubota S, Sakuma H, An W, Fujimaki T, McLaren MJ, Weleber RG, Inana G. HRG4 (UNC119) mutation found in cone-rod dystrophy causes retinal degeneration in a transgenic model. Invest Ophthalmol Vis Sci. 2000;41:3268–3277. [PubMed] [Google Scholar]
- Kobayashi A, Kubota S, Mori N, McLaren MJ, Inana G. Photoreceptor synaptic protein HRG4 (UNC119) interacts with ARL2 via a putative conserved domain. FEBS Lett. 2003;534:26–32. doi: 10.1016/s0014-5793(02)03766-3. [DOI] [PubMed] [Google Scholar]
- Lans H, Rademakers S, Jansen G. A network of stimulatory and inhibitory Galpha-subunits regulates olfaction in Caenorhabditis elegans. Genetics. 2004;167:1677–1687. doi: 10.1534/genetics.103.024786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Jr, Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro M, Pilgrim D. Identification and cloning of unc-119, a gene expressed in the Caenorhabditis elegans nervous system. Genetics. 1995;141:977–988. doi: 10.1093/genetics/141.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maduro MF, Gordon M, Jacobs R, Pilgrim DB. The UNC-119 family of neural proteins is functionally conserved between humans, Drosophila and C. elegans. J Neurogenet. 2000;13:191–212. doi: 10.3109/01677060009084494. [DOI] [PubMed] [Google Scholar]
- Manning AG, Crawford BD, Waskiewicz AJ, Pilgrim DB. unc-119 homolog required for normal development of the zebrafish nervous system. Genesis. 2004;40:223–230. doi: 10.1002/gene.20089. [DOI] [PubMed] [Google Scholar]
- Marrari Y, Crouthamel M, Irannejad R, Wedegaertner PB. Assembly and trafficking of heterotrimeric G proteins. Biochemistry. 2007;46:7665–7677. doi: 10.1021/bi700338m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K, Shiba D, Kobayashi D, Yokoyama T. Targeting of Nphp3 to the primary cilia is controlled by an N-terminal myristoylation site and coiled-coil domains. Cytoskeleton (Hoboken) 2012;69:221–234. doi: 10.1002/cm.21014. [DOI] [PubMed] [Google Scholar]
- Olbrich H, Fliegauf M, Hoefele J, Kispert A, Otto E, Volz A, Wolf MT, Sasmaz G, Trauer U, Reinhardt R, et al. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat Genet. 2003;34:455–459. doi: 10.1038/ng1216. [DOI] [PubMed] [Google Scholar]
- Perkins LA, Hedgecock EM, Thomson JN, Culotti JG. Mutant sensory cilia in the nematode Caenorhabditis elegans. Dev Biol. 1986;117:456–487. doi: 10.1016/0012-1606(86)90314-3. [DOI] [PubMed] [Google Scholar]
- Roayaie K, Crump JG, Sagasti A, Bargmann CI. The G alpha protein ODR-3 mediates olfactory and nociceptive function and controls cilium morphogenesis in C. elegans olfactory neurons. Neuron. 1998;20:55–67. doi: 10.1016/s0896-6273(00)80434-1. [DOI] [PubMed] [Google Scholar]
- Silverman L, Sudol M, Resh MD. Members of the src family of nonreceptor tyrosine kinases share a common mechanism for membrane binding. Cell Growth Differ. 1993;4:475–482. [PubMed] [Google Scholar]
- Sokolov M, Lyubarsky AL, Strissel KJ, Savchenko AB, Govardovskii VI, Pugh EN, Jr, Arshavsky VY. Massive light-driven translocation of transducin between the two major compartments of rod cells: a novel mechanism of light adaptation. Neuron. 2002;34:95–106. doi: 10.1016/s0896-6273(02)00636-0. [DOI] [PubMed] [Google Scholar]
- Swanson DA, Chang JT, Campochiaro PA, Zack DJ, Valle D. Mammalian orthologs of C. elegans unc-119 highly expressed in photoreceptors. Invest Ophthalmol Vis Sci. 1998;39:2085–2094. [PubMed] [Google Scholar]
- Veltel S, Gasper R, Eisenacher E, Wittinghofer A. The retinitis pigmentosa 2 gene product is a GTPase-activating protein for Arf-like 3. Nat Struct Mol Biol. 2008a;15:373–380. doi: 10.1038/nsmb.1396. [DOI] [PubMed] [Google Scholar]
- Veltel S, Kravchenko A, Ismail S, Wittinghofer A. Specificity of Arl2/Arl3 signaling is mediated by a ternary Arl3-effector-GAP complex. FEBS Lett. 2008b;582:2501–2507. doi: 10.1016/j.febslet.2008.05.053. [DOI] [PubMed] [Google Scholar]
- Vepachedu R, Karim Z, Patel O, Goplen N, Alam R. Unc119 protects from Shigella infection by inhibiting the Abl family kinases. PLoS ONE. 2009;4:e5211. doi: 10.1371/journal.pone.0005211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward S, Thomson N, White JG, Brenner S. Electron microscopical reconstruction of the anterior sensory anatomy of the nematode Caenorhabditis elegans.?2UU. J Comp Neurol. 1975;160:313–337. doi: 10.1002/cne.901600305. [DOI] [PubMed] [Google Scholar]
- Wright KJ, Baye LM, Olivier-Mason A, Mukhopadhyay S, Sang L, Kwong M, Wang W, Pretorius PR, Sheffield VC, Sengupta P, et al. An ARL3-UNC119-RP2 GTPase cycle targets myristoylated NPHP3 to the primary cilium. Genes Dev. 2011;25:2347–2360. doi: 10.1101/gad.173443.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Constantine R, Vorobiev S, Chen Y, Seetharaman J, Huang YJ, Xiao R, Montelione GT, Gerstner CD, Davis MW, et al. UNC119 is required for G protein trafficking in sensory neurons. Nat Neurosci. 2011a;14:874–880. doi: 10.1038/nn.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Constantine R, Vorobiev V, Chen Y, Seetharaman J, Huang YJ, Xie G, Montelione GT, Gerstner CD, Davis MW, et al. UNC119 regulates G protein trafficking in sensory neurons. Nature Neuroscience. 2011b;14:874–880. doi: 10.1038/nn.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zozulya S, Stryer L. Calcium-myristoyl protein switch. PNAS. 1992;89:11569–11573. doi: 10.1073/pnas.89.23.11569. [DOI] [PMC free article] [PubMed] [Google Scholar]