Abstract
A number of studies show that mitochondrial DNA (mtDNA) depletion and attendant activation of retrograde signaling induces tumor progression. We have reported previously that activation of a novel nuclear factor-Kappa B pathway is critical for the propagation of mitochondrial retrograde signaling, which induces both phenotypic and morphological changes in C2C12 myoblasts and A549 lung carcinoma cells. In this study, we investigated the role of stress-induced nuclear factor-Kappa B in tumor progression in xenotransplanted mice. We used a retroviral system for the inducible expression of small interfering RNA against IkBα and IkBβ mRNAs. Expression of small interfering RNA against IkBβ markedly impaired tumor growth and invasive ability of mtDNA-depleted C2C12 myoblasts and also thwarted anchorage-independent growth of the cells. Knockdown of IkBα mRNA, however, did not have any modulatory effect in this cell system. Moreover, expression of small interfering RNA against IkBβ reduced the expression of marker genes for retrograde signaling and tumor growth in xenografts of mtDNA-depleted cells. Our findings demonstrate that IkBβ is a master regulator of mitochondrial retrograde signaling pathway and that the retrograde signaling plays a role in tumor growth in vivo. In this regard, IkBβ supports the tumorigenic potential of mtDNA-depleted C2C12 cells.
Introduction
Studies over the past two decades have established that mitochondrial DNA (mtDNA) depletion or mutations in both unicellular eukaryotes or metazoan cells alters nuclear gene expression (1–4). This unique mitochondria-generated signaling pathway has been alternatively termed retrograde signaling, mitochondrial respiratory stress or mitochondria-to-nucleus stress signaling (1,5,6). Retrograde signaling was first reported in the yeast, Saccharomyces cerevisiae (7), where it was shown to involve activation of the bHLH factor Rtg, and results in alterations in both nuclear gene expression and cellular metabolism (6). In mammalian cells, retrograde signaling is induced by multiple stimuli, including mtDNA depletion or mutation, nuclear mutations that cause perturbations in the mitochondrial electron transport chain complexes and stress due to the mitochondrial unfolded protein response (6). A number of different Ca2+ responsive PKCs, AMPK, protein phosphatases, transcription factors (CREB, C/EBP family C/EBPδ and CHOP, NFAT, and nuclear factor-Kappa B [NF-κB]) are activated under these various stress-inducing conditions (6). Although some studies also suggest that mitochondrial oxidative stress-induced HIF factors may be involved in retrograde signaling (1,5,6,8–13), we have shown that mtDNA depletion- or mitochondrial electron transport chain complex inhibitor-induced retrograde signaling in C2C12 skeletal myoblasts and A549 lung cancer cells is predominantly mediated by a Ca2+-activated calcineurin pathway and is not accompanied by HIF activation (1,14).
Mitochondrial DNA depletion, and/or, mutations are associated with a wide spectrum of human cancers (15,16). Under experimental conditions, mtDNA depletion (50–70% of normal levels) induces tumor growth and phenotypic transformation of immortalized skeletal muscle C2C12 myoblasts, lung, breast and prostate cancer cells in vitro (8,17,18), and promotes tumor progression in a xenograft mouse model of colorectal cancer cells (19). Notably, mtDNA depletion in C2C12 cells also induces the expression of tumor cell markers including transforming growth factor-β (TGF-β), insulin-like growth factor 1 receptor (IGF1R), Akt/PI3-K, Glut 4, cathepsin L, as well as other genes involved in cell metabolism and proliferation (8). Finally, we have demonstrated that mtDNA depletion in C2C12 cells induced morphological changes that may contribute to transformation, including appearance of pseudopodia-like structures, increased glucose uptake, activation of the PI3-K/Akt pathway, increased resistance to apoptotic stimuli and increased invasive properties in vitro (20). Results emerging from our own and others’ studies show that retrograde signaling drives phenotypic transformation and tumor progression.
To further understand the molecular pathways that contribute to retrograde-signaling-induced phenotypic transformation, we used transient transfection with promoter–reporter constructs, coupled with mutational analysis, DNA–protein binding and ChIP analysis. Notably, this analysis revealed that stress response genes, including cathepsin L, RyR1, Glut4 and Akt, contain functionally important cis-DNA motifs for binding to CREB, C/EBPδ, NFAT and NF-κB factors immediately upstream of transcription start sites (11,20,21). We also showed that hnRNPA2, an RNA-binding protein involved in RNA maturation and trafficking, with potential roles in cancer promotion, binds to enhanceosomes of stress target genes by protein–protein interaction through DNA bound proteins listed above. Finally, gene expression profiling by cDNA array analysis showed that nearly 120 genes with diverse functions including cell surface receptors, cell cycle control, mitochondrial OXPHOS function, cell metabolism and oncogenesis were upregulated in mtDNA-depleted cells (1,21). Such global changes in nuclear gene expression and growth potential following mtDNA depletion are supported by studies in other mitochondrial stress-induced tumor cell models (2–4).
The NF-κB pathway activated in response to mitochondrial retrograde signaling is distinctly different from the more extensively studied canonical and non-canonical NF-κB pathways that participate in multiple physiological and pathological processes (22). Specifically, NF-κB activation by retrograde signaling involves the dephosphorylation of the IkBβ–cRel/p50 complex by calcineurin, a Ca2+-dependent phosphatase. The subsequent release of the cRel:p50 heteromeric factor then activates the transcription of target genes that are distinctly different from the classical NF-κB pathways (5,6). To further assess the role of this novel NF-κB pathway in propagating stress signaling, we carried out a microarray analysis of mRNA from cells stably expressing small interfering RNA (siRNA) against IkBβ. Results showed that IkBβ silencing, but not IkBα silencing, retarded the expression of almost all of the 120 genes overexpressed in response to mtDNA depletion (6). In this article, we have tested whether siRNA-mediated silencing of IkBβ in mtDNA-depleted C2C12 cells can suppress tumor progression in vivo in a mouse xenograft model. Notably, inducible knockdown of IkBβ, but not that of IkBα, suppressed tumor growth and metastasis in subcutaneously transplanted nude mice. Our results provide evidence that IkBβ is a master regulator of mitochondrial stress-induced retrograde signaling and tumor progression and thus describe a novel role in the oncogenic process.
Materials and methods
Expression of siRNA using lentiviral vectors
We used the siRNA sequences against mouse IkBα (5ʹ-AGGCCAGCGTCTGACATTA-3ʹ and GGCCAGCGTCTGACA TTAT-3ʹ), IkBβ (5ʹ-GACTGGAGGCTACAACTAG-3ʹ and 5ʹ-CAGA GATGAGGGCGATGAA-3ʹ) and scrambled siRNA described in our previous study (1). The target DNAs were subcloned into inducible lentivirus vector pLVPT-tTRKRAB obtained from Didier Trono (23). We also subcloned click beetle luciferase cDNA obtained from Günter J.Hämmerling (24) into the FUGW lentivirus vector obtained from David Baltimore and Carlos Lois (25). Lentiviruses were produced by transfection of triple plasmids (VSV-G, Gag-pol and target gene vector) in 293T cells via calcium precipitation method. Lentiviral particles were harvested at 48h post-transfection and C2C12 cells were transduced with viral particles in the presence of 6 µg/ml polybrene. Single colonies were picked and screened for inducible knockdown of IkBα, IkBβ or scrambled sequence by western blot analysis. The expression of click beetle luciferase was detected by bioluminescence signal detected as described (24).
Cell lines and culture conditions
Murine C2C12 skeletal myoblasts (ATCC CRL1772) were grown in Dulbecco’s modified Eagle’s medium (Invitrogen, 11965) supplemented with 10% fetal bovine serum and 0.1% gentamycin as described (1). Although mtDNA depletion to 50–70% of normal cell levels generally promotes cell growth, depletion to near ρo level is detrimental (8,26). Therefore, we partially depleted mtDNA by treatment with 100ng/ml ethidium bromide as described (1). Selected clones with 80–85% reduced mtDNA content were grown in the presence of 1mM sodium pyruvate and 50 µg/ml uridine. The mtDNA contents were routinely monitored by real-time PCR as described (1).
Cell fractionation, immunoblot analysis and immunostaining of paraffin-embedded tumor tissue
Subcellular fractions from cultured cells and tumor tissues were prepared in the presence of protease and phosphatase inhibitors and 30–50 µg of protein was resolved on 10 or 12% SDS-polyacrylamide gels for immunoblot analysis as described (1). Blots were developed using Super Signal West Femto maximum sensitivity substrate (Pierce, 34095). Antibodies against IkBα (sc-371) and IkBβ (sc-945) from Santa Crutz Biotech were used for immunoblot and immunostaining of paraffin-embedded tumor tissue. Ki-67 proliferation antigen antibody (Thermo Scientific) and fluorescence-labeled secondary antibody Alexa Fluor® 488 Dye (Invitrogen) were used for immunostaining and fluorescence images were pseudo colored.
Invasion assay and anchorage-independent assay
Invasion assays were performed as described by Albini et al. (27) with some modification. Fifty micrograms of Matrigel (BD Biosciences) was diluted in 100 µl of ice cold growth medium. The mixture was coated on the surface of a cell culture insert (BD Biosciences) and left to dry in the cell culture hood before rehydration with cell culture medium for 2h prior to the assay. Cells (10 000 cells) were seeded and allowed to invade the Matrigel barrier and membrane. The invaded cells were fixed, stained and counted essentially as described (27). In anchorage-independent assays, cells (500 000 cells) were plated on the top of a double layer agar plate consisting of a top layer of 0.35% soft agar in bacterial medium and a bottom layer of 0.5% agar in complete bacterial medium. After ~8 weeks, colonies were stained and counted as described by Scholl et al. (28).
Mouse xenograft tumor model
A suspension of 100 000 cells in 100 µl of phosphate buffered saline (PBS) was injected subcutaneously into the flanks of nude mice (NCRNU-M; Taconic) as described previously (28,29). After 2 months, tumor was detected by bioluminescence imaging using a Xenogen IVIS-200 imaging system as described (30). Mice were euthanized by CO2 asphyxiation and tumors were removed and weighed. Animal experiments were performed in accordance with the University of Pennsylvania Animal Care and Use Committee guidelines under an approved protocol.
Statistical analysis
Data are presented as mean ± SEM based on four separate experiments. Student’s t-test was applied in comparison of means of four independent groups with and without tetracycline treatment. A value of P < 0.05 was considered statistically significant and P < 0.001 was considered highly significant.
Results
Establishment of stable cell lines expressing luciferase and inducible siRNA
As shown in Supplementary Figure 1, available at Carcinogenesis Online, the sequence-specific siRNAs markedly suppressed the levels of IkBα and IkBβ proteins, respectively, whereas the scrambled RNA construct had no effect. In this study, we used a doxycycline (Dox)- (an analog of tetracycline) inducible lentiviral system for the expression of siRNA against IkBβ and IkBα. We also introduced click beetle luciferase through lentiviral transduction (24) to enable the monitoring of tumor cell growth and metastasis through bioluminescence imaging. Although several inducible siRNA expression systems are available (23), we selected this inducible system for expressing siRNA because of its low basal expression and high amplitude of induction.
To generate the double stable cell lines, we first transduced mtDNA-depleted cells with lentivirus expressing click beetle luciferase cDNA, driven by a CMV promoter. Clones of stable cells were picked, amplified and transduced with lentiviral preparations encoding the inducible siRNAs (scrambled, IkBα or IkBβ siRNAs). The inducible siRNA vector contains green fluorescent protein (GFP) for selection in fluorescence-activated cell sorting following induction with Dox. Initially, we enriched lentivirus transduced cells by fluorescence-activated cell sorting by isolating cells that expressed GFP upon addition of Dox. Following expansion, Dox-induced GFP expression was confirmed and the ability of individual cell clones to silence IkBα or IkBβ following Dox treatment for 2 days was assessed. In addition, clones were monitored for their ability to express click beetle luciferase.
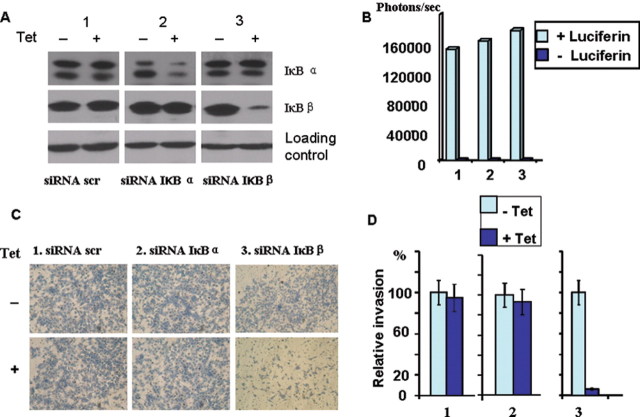
Western blot analysis in Figure 1A shows that the IkBα and IkBβ levels were markedly reduced following treatment of siRNA-expressing cells with Dox. There was no obvious change in the protein level of IkBα and IkBβ in the cells expressing scrambled vector with or without Dox treatment. Although staining with the anti-IkBα antibody revealed two bands, their coordinate downregulation by IkBα suggests that the faster migrating band may be a proteolytic product of the full length protein. In addition, all three cell lines yielded similar levels of bioluminescence following addition of d-luciferin in an in vitro bioluminescence quantitation assay (24) (Figure 1B) indicating that nearly comparable levels of luciferase genes are transduced in all three cell lines.
Fig. 1.
Characterization of the mtDNA-depleted C2C12 cell lines expressing Dox-inducible siRNAs and stable CBG 99 luciferase. Transduction of mtDNA-depleted cells with viral vectors was as described in Materials and methods. The three cell lines stably expressing CBG 99 luciferase and inducible expression of scrambled siRNA, siRNA against IκBα and siRNA against IκBβ are marked as 1, 2 and 3, respectively. (A) Western blot analysis of cell extracts (30 µg total protein) from stable cell lines treated with or without Dox (0.4mg/ml) with antibodies against IκBα and IκB (1:1000 dilution). The panel marked as loading control was developed with antibody to glyceraldehyde 3-phosphate dehydrogenase. (B) The level of luciferase gene expression in stable cell lines. The three stable cell lines were grown in 96-well plates and after 24h of culturing, cell medium was removed, cells were washed with PBS and incubated with d-luciferin (300 µg/ml in PBS) for 15min before bioluminescence signal detection in a Hidex Microwin 200 plate reader. (C) In vitro invasion of the three stable cell lines through the Matrigel membrane system. Each of the three siRNA expressing cell lines (10 000 cells) were treated with or without Dox, seeded on top of the Matrigel chamber and allowed to grow/invade for 24h. Cells that invaded the membrane were stained with eosin–hematoxylin solution and photographed for quantitation. (D) Matrigel invasion assays from Figure C were quantified. Error bars represent the mean ± SEM of triplicate experiments.
Knockdown of IkBβ but not IkBα reduces invasiveness and impairs anchorage-independent growth of mtDNA-depleted cells
The in vitro Matrigel invasion in Figure 1D shows that Dox-mediated induction of siRNA against IkBβ markedly reduced the number of invasive cells as compared with untreated cells. Dox-mediated induction of scrambled siRNA or siRNA against IkBα had no significant effect on the number of invading cells.
The anchorage-independent growth assay, also called soft agar assay, is an excellent indicator of tumorigenic potential of cells (28,29).We carried out an anchorage-independent growth assay with the three isogenic stable cell lines in the presence or absence of Dox. Notably, Dox can pass through soft agar and penetrate cell membranes to induce siRNA. In this study, Dox was replenished every third day to ensure its optimal level for induction of siRNA expression. Figure 2A shows the colony forming ability of control and mtDNA-depleted cells; control cells formed small microcolonies that are not readily visible to the naked eye. The mtDNA-depleted cells, on the other hand, formed large visible colonies characteristic of anchorage-independent growth of tumor cells. Figure 2B shows the growth patterns of cells expressing scrambled siRNA, siRNA against IkBβ and IkBα. The mtDNA-depleted cells expressing either scrambled siRNA or siRNA against IkBα showed similar level of growth and number of colonies per unit area both in the presence and absence of Dox. In contrast, addition of Dox to cells expressing siRNA against IkBβ reduced the number of colonies by <80%, suggesting the selective and specific involvement of this factor in cancer progression. These results suggest the role of IkBβ in the propagation of mitochondrial retrograde signaling and phenotypic transformation.
Fig. 2.
Anchorage-independent growth of control C2C12 cells and mtDNA-depleted cells expressing different siRNA constructs. Cells suspensions (500 000 cells) was plated on soft agar plates and allowed to grow for ~8 weeks. Colonies formed in the 0.35% agar layer were stained with 0.005% crystal violet, counted and photographed. In panel A normal C2C12 cell and mtDNA-depleted C2C12 cells were compared. In panel B, mtDNA-depleted cells expressing scrambled (1), siRNA against IkBα (2) and siRNA against IkBβ (3) induced with and without Dox were grown on soft agar and compared for colony growth. Panel C represents quantitative analysis of gel patterns in B. The bars represent mean ± SEM of three different plates in each case.
Inducible knockdown of IkBβ retards tumor growth in the mouse xenograft model
We next tested the effects of Dox-induced expression of siRNA against IkBα and IkBβ using the three isogenic stable mtDNA-depleted cells described above in xenografts of immunocompromised nude mice. Cell lines were transplanted subcutaneously on the back of nude mice (four mice in each group). The control group received normal sterile water and the experimental group was administered Dox (1mg/ml) in drinking water. As shown from the size of the tumor and the area of the bioluminescence by in vivo imaging, all three cell lines yielded large and similarly sized tumors in mice that were provided with water alone (Figure 3A). Addition of Dox had no effect on the growth of mtDNA-depleted cells expressing either control siRNA or siRNA against IkBα. In sharp contrast, Dox treatment of mice expressing siRNA against IkBβ exhibited a marked reduction in tumor size. Tumor dissected from representative mice belonging to each group and statistical variations in tumor mass within each group of four animals are presented in Figure 3B and 3C. These results recapitulate the in vivo imaging data presented in Figure 3A, as IkBβ mRNA knockdown effectively reduced the tumor size by >80%. In addition, the results are consistent with data from the in vitro invasion and anchorage-independent assays (Figures 1 and 2). As shown in Supplementary Figure 2, available at Carcinogenesis Online, the inhibitory effects of IkBβ siRNA expression can be reversed by Dox withdrawal, with tumors reappearing within 3 weeks. Finally, hematoxylin and eosin staining of tumor sections indicate that the morphology and growth patterns of cells expressing IkBβ siRNA are quite distinct (Supplementary Figure 3F is available at Carcinogenesis Online) from other tumor samples (A–E), in that, cells are much larger and more elongated and exhibit a more distinctive osteoid-type of matrix pattern consistent with a slow growing tumor type.
Fig. 3.

Effects of IkBα and IkBβ mRNA knockdown on tumor growth in xenografted mice. A suspension of 100 000 cells in 100 µl PBS (50% matrigel) was injected subcutaneously into the flanks of each nude mouse. The mice were provided either normal water or water containing 1mg/ml Dox. A minimum of three mice were used in each group. After 2 months, tumors were detected by bioluminescence imaging of mice anesthetized with isoflurane. (A) Top panel shows the photographs of representative members of each group. The bottom panel shows the bioluminescence image of tumors in each group. (B) The mice from Figure A were euthanized, and the tumors were extracted and weighed. The top panel shows the photographs of tumors extracted from representative mice from each group. The bottom panel shows the mean weight ± SEM of tumors from three mice within each group. The weight of tumors expressing scrambled siRNA from mice that were not treated with Dox was used as 100% in calculating these values.
We used the tail vein injection method for testing the invasive property of mtDNA-depleted C2C12 cells under in vivo conditions. As seen in vivo imaging of mice (Figure 4A) and visualization of extracted lung tissue (Figure 4B), mtDNA-depleted cells expressing IkBβ siRNA injected through tail vein migrated and colonized in the lung in the absence of Dox. However, Dox-induced expression of IkBβ siRNA markedly retarded the invasive behavior of mtDNA-depleted cells and also appearance of nodular growth in the lung. These results show that IkBβ-dependent mitochondrial stress signaling is essential for both tumor growth and metastasis of mtDNA-depleted C2C12 cells.
Fig. 4.
(A) Effects of IkBβ mRNA knockdown on tumor metastasis by mtDNA-depleted C2C12 cells. A suspension of 10 000 cells in 100 µl PBS was injected into the tail vein of each nude mouse and the mice were either provided with normal water (–) or water containing 1mg/ml of Dox (+). After 4 months, the tumors were detected by bioluminescence imaging through a Xenogen IVIS-200 imager. Mice administered Dox containing water showed markedly reduced bioluminescence, suggesting minimal invasion in the lung, compared with their counterpart administered water alone. (B) Tumors were extracted from euthanized mice and photographed. The results show a markedly enlarged lung with tumor growth and tumor-associated lesions.
Levels of expression of mitochondrial stress responsive genes in tumor tissues
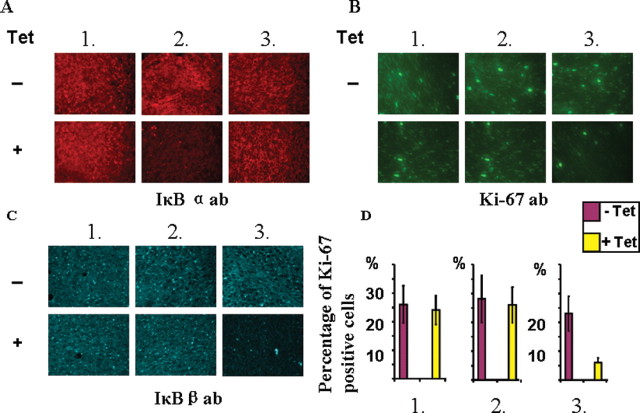
The effectiveness of siRNA-dependent downregulation of IkBα and IkBβ protein levels in tumor tissues from xenografted mice was ascertained by immunostaining of tissue sections. In support of a specific effect of siRNA, immunostaining patterns in Figure 5A and 5B show that the intensity of staining with antibodies to IkBα and IkBβ were reduced in tumors expressing the respective siRNAs. Dox-induced expression of scrambled RNA had no effect on the extent of immunostaining. Although not shown, the extent of staining with actin antibody showed no difference in all six siRNA expressing and non-expressing cells.
Fig. 5.
Immunohistochemical analysis of tumor sections for the detection of IkBα, IkBβ and tumor invasive marker Ki-67 expressing different siRNA constructs. Paraffin-embedded tumor tissues from the six different groups shown in Figure 4 were sectioned and 5 μm sections were stained with primary antibodies against IkBα (Figure A), antibody to Ki-67 (Figure B) and antibody to IkBβ (Figure C). The sections were co-stained with Alexa Fluor 488 dye conjugated secondary antibody in A, B and C. Figure D shows the relative percentage of Ki-67 positive cells in each group calculated from three separate experiments represented in Figure B. Results show mean ± SEM of three separate experiments.
Immunostaining of tumor sections with Ki-67 antibody has been used as a cell proliferation marker in many tumorigenesis studies. Immunohistograms in Figure 5D show that the number of cells staining with Ki-67 antibody was markedly less in tumors expressing siRNA against IkBβ than that in tumors without siRNA expression. There was no significant change in the number of cells staining with Ki-67 antibody in tumors with and without induction of siRNA against IkBα. Consistent with the results of in vivo tumor growth in Figure 3, Dox-induced expression of siRNA against IkBβ markedly reduced the number of Ki-67 antibody positive cells in tumor tissue (the average cell counts from at least five different fields, Figure 5D).
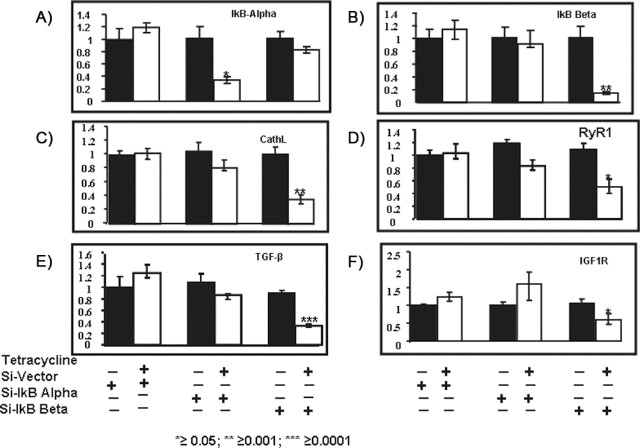
We next assessed if siRNA for either IkBα or IkBβ affected the expression of genes induced by mitochondrial stress signaling in tumors derived from mtDNA-depleted cells. As expected, real-time PCR data revealed that IkBα and IkBβ mRNA levels were specifically downregulated in tumor tissues by their respective siRNA following Dox treatment (Figure 6A and 6B) and levels are near those in control tumor tissue in mice given water alone. Notably, real-time PCR analyses reveal that the levels of cathepsin L, RyR1, IGF1R, and TGF-β mRNAs, important marker genes for the mitochondrial stress signaling pathway, are reduced selectively in tumors expressing siRNA to IkBβ mRNA. In agreement with our published results (1), results in Figure 6F also suggest a negative modulatory effect of IkBα on IGF1R gene expression. Finally, consistent with our previous data with mtDNA-depleted C2C12 cells in culture, ChIP analysis data in Figure 7 show that the occupancy of cathepsin L promoter sites by C/EBPδ, cRel and p50 were markedly lower in tumor tissues expressing IkBβ specific siRNA. Together, our results conclusively show that IkBβ plays an important role in the propagation of mitochondrial stress response and strongly supports a role for IkBβ in the tumor promoting effects of the stress signaling.
Fig. 6.
Levels of expression of marker genes for mitochondrial stress signaling in tumor tissues from mtDNA-depleted cells expressing different siRNA constructs. The steady state levels of mRNAs for IkBα, IkBβ, cathepsin L, TGF-β RyR1 and IGF1R were quantified by real-time PCR as described in the Materials and methods. About 25ng archived template DNA was used in each case. The level of β-actin mRNA was used as an internal control in each series. The results represent mean ± SEM of three separate experiments.
Fig. 7.
Relative occupancy of mitochondrial stress response region of the cathepsin L promoter by transcription factors and a model for stress-induced NF-κB activation. (A) ChIP analysis of the mouse cathepsin L promoter region (–273 to –53) from mtDNA-depleted/scrambled siRNA and mtDNA-depleted/IkBβ-silenced cells was carried out as described in the Materials and methods. Immunoprecipitation of chromatin fragments was carried out with antibodies to C/EBPδ, p50 and cRel proteins as indicated. Real-time PCR analysis was carried out using 10% input DNA for normalization. Values for factor binding relative to the control depleted/scrambled siRNA cell values are presented. Preimmune serum was used as a negative antibody control. Mean ± SEM values were calculated from four independent experiments. Statistical analysis was carried out using analysis of variance. (B) A proposed models for the mitochondrial stress-induced activation of NF-κB and its role in transcription activation of nuclear gene targets resulting in tumor growth. (C) The model shows how siRNA-mediated knockdown of IkBβ causes disruption of stress signaling and retards tumor growth.
Discussion
Epidemiological studies show that >50% of human liver and gastric tumors contain significantly lower levels of mtDNA than do normal tissues (15,31). Moreover, a number of studies have shown that mtDNA depletion induces proliferative and metastasizing phenotypes in renal, gastric, lung, breast, colorectal cancer and hepatocellular carcinomas (31–36). It is also known that a large fraction of human tumors harbor mtDNA mutations. A causal relationship between mtDNA damage and tumorigenesis was demonstrated in prostate cancer cells (37,38) where mutations in mtDNA encoded ATPase6 or ND5 induced tumorigenic property of cells (37,39). In previous studies, we have shown that partial depletion of mtDNA (~70–80%) induced phenotypic transformation of C2C12 myocytes and A549 cells and markedly altered their nuclear gene expression profiles. We also showed that these changes in nuclear gene expression profiles occurred through activation of calcineurin and a number of Ca2+ responsive transcription factors, including C/EBPδ, NFAT, CREB and NF-κB. Here, we further investigate the role of IkBβ-dependent mitochondrial stress signaling in the tumor growth and metastasis of C2C12 cells in vivo.
A metabolic switch to increased utilization of glucose and glutamine are hallmarks of proliferating cancer cells (40). As pointed out in many recent studies (41,42), glucose and glutamine serve as a carbon source for both energy production and anaplurotic biosynthetic processes. They also generate most of their energy through glycolysis and excrete high levels of lactic acid. Thus, although tumor cells are not most efficient in terms of energy production, they nevertheless possess an efficient system for the synthesis of ‘biomass’ (nucleotides, amino acids, fatty acids) that are required for cell growth and proliferation. Working with C2C12 myocytes and A549 lung carcinoma cells, we demonstrated previously that mitochondrial respiratory stress initiated by mtDNA depletion induced IGF1R pathway, which in turn induced PI3-K and Akt and upregulation of Glut4 expression (11,20,21). Activation of Akt and PI3-K pathways were also shown in other cancer cells in response to chemically induced or mutant polγ-induced mtDNA depletion in other cancer cells (43–45). In C2C12 cells, these changes were accompanied by increased glucose uptake, increased glycolysis, resistance to apoptotic stimuli and rapid cell proliferation (11,20,21,46). The mtDNA-depleted cells used in our studies exhibit significant, but low levels of ATP (~30%), partially disrupted Δψm but ability to import proteins and nearly intact tricarboxylic acid cycle activity. These cells therefore contain enzyme systems and pathways to fully support metabolic needs of proliferating tumor cells and promote tumorigenesis. Our present and published results show that intact mitochondrial stress signaling is important for supporting these metabolic changes (11,21).
NF-κB transcription factors are known to regulate a multitude of physiological and pathophysiological processes including the immune response, inflammation, musculoskeletal disorders, atherosclerosis, apoptosis and cancer. In cancer, NF-κB pathways regulate multiple steps of cell proliferation, survival and metastasis (47). At least two major pathways, canonical and non-canonical pathways have been described, both requiring the action of IkB kinases for phosphorylation and subsequent degradation of inhibitory proteins (IkBβ, IkBα or p100) that allow for translocation of active NF-κB complexes to the nucleus. In a recent study implicating a distinct functional role to IkBβ, Rao et al. (48) reported that hypophosphorylated IkBβ bound to p65:cRel dimer promotes the chronic phase of tumor necrosis factor-α production by maintaining prolonged mRNA expression. In another study, Wright et al. showed that IkBβ-dependent NF-κB plays an essential role in preventing oxidant-induced cell death (49). Studies from our own laboratory revealed a distinctive role for IkBβ in the propagation of mitochondrial stress signaling. We have demonstrated that mitochondrial stress-induced NF-κB activation occurs through a IkB kinases-independent process (both IKKα and IKKβ) in which activation and release of cRel:p50 dimers from IkBβ occurs in calcineurin-dependent manner. Thus, IkBβ plays a permissive role in the propagation of mitochondrial stress signaling since depletion of this protein diminishes mitochondrial stress signaling. In this regard, NF-κB pathway we described is distinctly different from that described by Rao et al. (48), but probably similar to that reported by Wright et al. (49). A model showing the mechanism of stress-induced NF-κB activation in mtDNA-depleted cells resulting in the activation of nuclear target genes is presented in Figure 7B. Figure 7B shows the mechanism by which the stress signaling is abrogated in cells depleted of IkBβ by siRNA expression that also retards tumor growth.
In this study we show that IkBβ knockdown inhibits colony formation in soft agar, as well as tumor growth and metastasis in a mouse xenograft model. IkBββ knockdown also reversed the expression of a number of mitochondrial stress markers such as IGF1R, TGF-β, RyR1 and cathepsin L. In addition to inhibition of tumor growth, IkBβ knockdown also altered the morphology of tumor tissue. These results confirm and extend our mitochondrial stress-dependent transcription model and show that IkBβ is a master regulator of stress signaling (see Figure 7B and 7C). This is probably the first study showing the distinct role of IkBβ in potentiating oncogenic potential of cells subjected to mitochondrial stress. Although not shown, transcription activation and attendant effects on tumor progression in C2C12 cells is independent of HIF activation.
Previously we showed that transcription activation of mitochondrial stress response genes involves the binding of the signature factors, C/EBPδ, NF-κB (cRel:p50), CREB and NFAT to canonical factor binding sites located within 200–600 bases immediately upstream of transcription activation sites of the stress-activated genes cathepsin L, IGF1R, Glut4 and RyR1 (12,21). The transcriptional complex is stabilized by recruitment of hnRNPA2 as a transcriptional co-activator. Results of the ChIP analysis in Figure 7 show that IkBβ knockdown markedly reduced the occupancy of cathepsin L promoter by cRel, p50 and C/EBPδ. These results provide further confirmation on a distinct transcriptional reprogramming and patterns of gene expression in response to mitochondrial stress. It should be noted that the mitochondrial stress response genes are distinctly different from those targeted by the canonical and non-canonical NF-κB pathways. This distinction allows us to target the mitochondrial stress pathway while leaving the physiological canonical/non-canonical pathways intact. Our results also suggest that IkBβ and its dependent NF-κB pathway may be good therapeutic targets for controlling cancer progression in some tumor systems.
Supplementary material
Supplementary Figures 1–3 can be found at http://carcin.oxfordjournals.org/
Funding
National Institutes of Health (CA-22762).
Conflict of Interest Statement: None declared.
Acknowledgements
We thank Drs David Baltimore and Carlos Lois for the FUGW lentivirus vector, Dr Didier Trono for the inducible lentivirus vector pLVPT-tTRKRAB and Dr Günter J. Hämmerling for click beetle luciferase cDNA. We thank Dr Leslie King for editorial help. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked advertisement in accordance with 18U.S.C. Section 1734 solely to indicate this fact.
Glossary
Abbreviations:
- Dox
doxycycline
- GFP
green fluorescent protein
- IGF1R
insulin-like growth factor 1 receptor
- mtDNA
mitochondrial DNA
- PBS
phosphate-buffered saline
- siRNA
small interfering RNA
- TGF-β
transforming growth factor-β
References
- 1. Biswas G., et al. (2008). A distinctive physiological role for IkappaBbeta in the propagation of mitochondrial respiratory stress signaling J. Biol. Chem 283 12586–12594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crimi M., et al. (2005). Skeletal muscle gene expression profiling in mitochondrial disorders FASEB J 19 866–868 [DOI] [PubMed] [Google Scholar]
- 3.Delsite R., et al. Nuclear genes involved in mitochondria-to-nucleus communication in breast cancer cells. Mol. Cancer. (2002);1:6. doi: 10.1186/1476-4598-1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jahangir Tafrechi R.S., et al. (2005). Distinct nuclear gene expression profiles in cells with mtDNA depletion and homoplasmic A3243G mutation Mutat. Res 578 43–52 [DOI] [PubMed] [Google Scholar]
- 5. Biswas G., et al. (2003). Mitochondria to nucleus stress signaling: a distinctive mechanism of NFkappaB/Rel activation through calcineurin-mediated inactivation of IkappaBbeta J. Cell Biol 161 507–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Butow R.A., et al. (2004). Mitochondrial signaling: the retrograde response Mol. Cell 14 1–15 [DOI] [PubMed] [Google Scholar]
- 7. Liu Z., et al. (2006). Mitochondrial retrograde signaling Annu. Rev. Genet 40 159–185 [DOI] [PubMed] [Google Scholar]
- 8. Amuthan G., et al. (2001). Mitochondria-to-nucleus stress signaling induces phenotypic changes, tumor progression and cell invasion EMBO J 20 1910–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arnould T., et al. (2002). CREB activation induced by mitochondrial dysfunction is a new signaling pathway that impairs cell proliferation EMBO J 21 53–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Biswas G., et al. (1999). Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk EMBO J 18 522–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guha M., et al. (2007). Activation of a novel calcineurin-mediated insulin-like growth factor-1 receptor pathway, altered metabolism, and tumor cell invasion in cells subjected to mitochondrial respiratory stress J. Biol. Chem 282 14536–14546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Guha M., et al. (2009). Heterogeneous nuclear ribonucleoprotein A2 is a common transcriptional coactivator in the nuclear transcription response to mitochondrial respiratory stress Mol. Biol. Cell 20 4107–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ryan M.T., et al. (2007). Mitochondrial-nuclear communications Annu. Rev. Biochem 76 701–722 [DOI] [PubMed] [Google Scholar]
- 14. Amuthan G., et al. (2002). Mitochondrial stress-induced calcium signaling, phenotypic changes and invasive behavior in human lung carcinoma A549 cells Oncogene 21 7839–7849 [DOI] [PubMed] [Google Scholar]
- 15. Lee H.C., et al. (2010). Somatic mutations of mitochondrial DNA in aging and cancer progression Ageing Res. Rev 9 (suppl 1) S47–S58 [DOI] [PubMed] [Google Scholar]
- 16. Modica-Napolitano J.S., et al. (2002). Mitochondria as targets for detection and treatment of cancer Expert Rev. Mol. Med 4 1–19 [DOI] [PubMed] [Google Scholar]
- 17. Naito A., et al. (2008). Progressive tumor features accompany epithelial-mesenchymal transition induced in mitochondrial DNA-depleted cells Cancer Sci 99 1584–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Singh K.K., et al. (2009). Mutations in mitochondrial DNA polymerase-gamma promote breast tumorigenesis J. Hum. Genet 54 516–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Guo J., et al. (2011). Frequent truncating mutation of TFAM induces mitochondrial DNA depletion and apoptotic resistance in microsatellite-unstable colorectal cancer Cancer Res 71 2978–2987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Guha M., et al. (2010). Activation of Akt is essential for the propagation of mitochondrial respiratory stress signaling and activation of the transcriptional coactivator heterogeneous ribonucleoprotein A2 Mol. Biol. Cell 21 3578–3589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Guha M., et al. (2010). Role of calcineurin, hnRNPA2 and Akt in mitochondrial respiratory stress-mediated transcription activation of nuclear gene targets Biochim. Biophys. Acta 1797 1055–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ghosh S., et al. (2002). Missing pieces in the NF-kappaB puzzle Cell 109 (suppl) S81–S96 [DOI] [PubMed] [Google Scholar]
- 23. Wiznerowicz M., et al. (2006). Tuning silence: conditional systems for RNA interference Nat. Methods 3 682–688 [DOI] [PubMed] [Google Scholar]
- 24.Miloud T., et al. Quantitative comparison of click beetle and firefly luciferases for in vivo bioluminescence imaging. J. Biomed. Opt. (2007);12:054018. doi: 10.1117/1.2800386. [DOI] [PubMed] [Google Scholar]
- 25. Lois C., et al. (2002). Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors Science 295 868–872 [DOI] [PubMed] [Google Scholar]
- 26. Jazayeri M., et al. (2003). Inducible expression of a dominant negative DNA polymerase-gamma depletes mitochondrial DNA and produces a rho0 phenotype J. Biol. Chem 278 9823–9830 [DOI] [PubMed] [Google Scholar]
- 27. Albini A., et al. (1987). A rapid in vitro assay for quantitating the invasive potential of tumor cells Cancer Res 47 3239–3245 [PubMed] [Google Scholar]
- 28. Scholl C., et al. (2009). Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells Cell 137 821–834 [DOI] [PubMed] [Google Scholar]
- 29. Luo J., et al. (2009). A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene Cell 137 835–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Naik S., et al. (2007). Real-time imaging of beta-catenin dynamics in cells and living mice Proc. Natl. Acad. Sci. U.S.A 104 17465–17470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wu C.W., et al. (2005). Mitochondrial DNA mutations and mitochondrial DNA depletion in gastric cancer Genes. Chromosomes Cancer 44 19–28 [DOI] [PubMed] [Google Scholar]
- 32. Lee H.C., et al. (2005). Mitochondrial genome instability and mtDNA depletion in human cancers Ann. N. Y. Acad. Sci 1042 109–122 [DOI] [PubMed] [Google Scholar]
- 33. Mambo E., et al. (2005). Tumor-specific changes in mtDNA content in human cancer Int. J. Cancer 116 920–924 [DOI] [PubMed] [Google Scholar]
- 34. Selvanayagam P., et al. (1996). Detection of mitochondrial genome depletion by a novel cDNA in renal cell carcinoma Lab. Invest 74 592–599 [PubMed] [Google Scholar]
- 35. Xing J., et al. (2008). Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma J. Natl. Cancer Inst 100 1104–1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Yu M., et al. (2007). Reduced mitochondrial DNA copy number is correlated with tumor progression and prognosis in Chinese breast cancer patients IUBMB Life 59 450–457 [DOI] [PubMed] [Google Scholar]
- 37. Petros J.A., et al. (2005). mtDNA mutations increase tumorigenicity in prostate cancer Proc. Natl. Acad. Sci. U.S.A 102 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shidara Y., et al. (2005). Positive contribution of pathogenic mutations in the mitochondrial genome to the promotion of cancer by prevention from apoptosis Cancer Res 65 1655–1663 [DOI] [PubMed] [Google Scholar]
- 39. Park J.S., et al. (2009). A heteroplasmic, not homoplasmic, mitochondrial DNA mutation promotes tumorigenesis via alteration in reactive oxygen species generation and apoptosis Hum. Mol. Genet 18 1578–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Warburg O. (1956). On the origin of cancer cells Science 123 309–314 [DOI] [PubMed] [Google Scholar]
- 41. Baggetto L.G. (1992). Deviant energetic metabolism of glycolytic cancer cells Biochimie 74 959–974 [DOI] [PubMed] [Google Scholar]
- 42. DeBerardinis R.J., et al. (2008). The biology of cancer: metabolic reprogramming fuels cell growth and proliferation Cell Metab 7 11–20 [DOI] [PubMed] [Google Scholar]
- 43. Moro L., et al. (2009). Mitochondrial DNA depletion in prostate epithelial cells promotes anoikis resistance and invasion through activation of PI3K/Akt2 Cell Death Differ 16 571–583 [DOI] [PubMed] [Google Scholar]
- 44. Pelicano H., et al. (2006). Mitochondrial respiration defects in cancer cells cause activation of Akt survival pathway through a redox-mediated mechanism J. Cell Biol 175 913–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Singh K.K. (2004). Mitochondrial dysfunction is a common phenotype in aging and cancer Ann. N. Y. Acad. Sci 1019 260–264 [DOI] [PubMed] [Google Scholar]
- 46. Biswas G., et al. (2005). Mechanism of mitochondrial stress-induced resistance to apoptosis in mitochondrial DNA-depleted C2C12 myocytes Cell Death Differ 12 266–278 [DOI] [PubMed] [Google Scholar]
- 47. Grivennikov S.I., et al. (2010). Immunity, inflammation, and cancer Cell 140 883–899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Rao P., et al. (2010). IkappaBbeta acts to inhibit and activate gene expression during the inflammatory response Nature 466 1115–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wright C.J., et al. (2012). Nuclear factor-kB (NF-kB) inhibitory protein IkBβ determines apoptotic cell death following exposure to oxidative stress J. Biol. Chem 287: 6230–6239 [DOI] [PMC free article] [PubMed] [Google Scholar]