Abstract
In contrast to molecular rates for neutral mitochondrial sequences, rates for constrained sites (including nonsynonymous sites, D-loop, and RNA) in the mitochondrial genome are known to vary with the time frame used for their estimation. Here, we examined this issue for the nuclear genomes using single-nucleotide polymorphisms (SNPs) from six complete human genomes of individuals belonging to different populations. We observed a strong time-dependent distribution of nonsynonymous SNPs (nSNPs) in highly constrained genes. Typically, the proportion of young nSNPs specific to a single population was found to be up to three times higher than that of the ancient nSNPs shared between diverse human populations. In contrast, this trend disappeared, and a uniform distribution of young and old nSNPs was observed in genes under relaxed selective constraints. This suggests that because mutations in constrained genes are highly deleterious, they are removed over time, resulting in a relative overabundance of young nSNPs. In contrast, mutations in genes under relaxed constraints are nearly neutral, which leads to similar proportions of young and old SNPs. These results could be useful to researchers aiming to select appropriate genes or genomic regions for estimating evolutionary rates and species or population divergence times.
Keywords: rates of evolution, natural selection, time dependency, deleterious polymorphisms, population genetic theory
Introduction
The rate of molecular evolution is a fundamental parameter in genetics and evolutionary biology. Although rates that are estimated using different timescales and methods are expected to be similar, recently studies suggested otherwise. For example, studies based on pedigree analyses have recorded higher rates of evolution compared with those estimated using phylogenetics methods (Parsons et al. 1997; Howell et al. 2003; Denver et al. 2004; Haag-Liautard et al. 2007; Millar et al. 2008). Comparative genomic studies have shown that molecular rates estimated using short timescales, for instance, those based on intraspecific data, are much higher than those estimated using interspecies data (Garcia-Moreno 2004; Ho et al. 2005, 2007; Burridge et al. 2008; Subramanian and Lambert 2011). A number of reasons have been suggested for this discrepancy. However, most of these involve biases or errors in estimation such as calibration errors, saturation effects on nucleotide positions, and phylogenetic and demographic model misspecification (Emerson 2007; Bandelt 2008; Debruyne and Poinar 2009; Henn et al. 2009; Ho et al. 2011). The higher intraspecific rates have also been attributed to artifacts such as sequencing errors, postmortem damage (in the case of ancient DNA), and ascertainment bias (reviewed by Ho et al. [2011]).
The major biological factor that produces a time-dependent pattern of molecular rates appears to be purifying selection (Endicott and Ho 2008; Subramanian et al. 2009; Ho et al. 2011; Subramanian and Lambert 2011). At short timescales, slightly deleterious polymorphisms will segregate in populations, and this standing variation results in a higher diversity. This is evident from a number of recent studies that have shown an overabundance of low-frequency nonsynonymous single-nucleotide polymorphism (nSNPs) in human protein-coding genes (Li et al. 2010; Nelson et al. 2012; Subramanian 2012; Tennessen et al. 2012). Thus, an elevated molecular rate is observed for the data from within a population or for closely related populations (Kimura 1983). In contrast, deleterious polymorphisms are removed over time, which results in reduced divergence between species or distantly related populations.
Note that throughout this article, the rate of evolution refers to the observed or estimated divergence divided by calibration time. It does not denote a mutation rate, and hence, we do not intend to suggest that mutation rate varies with time.
Time-dependent rates have been typically reported for the D-loop region of the mitochondrial genome (Parsons et al. 1997; Lambert et al. 2002; Howell et al. 2003; Ho et al. 2005; Hay et al. 2008; Henn et al. 2009). Because the D-loop is hypervariable, the saturation effects on nucleotide sites confound the effects of purifying selection in this region. Hence, later studies (Endicott and Ho 2008; Subramanian et al. 2009; Subramanian and Lambert 2011) used coding genes of mitochondria to examine the time-dependency effect, because these are less prone to estimation errors. These studies showed that rates of molecular evolution at synonymous sites of protein-coding genes are similar across different timescales. In contrast, these studies found a strong time-dependent rate of evolution at nonsynonymous sites. Because synonymous sites are free from selection and nonsynonymous positions are under selective constraints, the observed pattern clearly points to the effects of purifying selection as predicted theoretically (Kimura 1983). This is also evident from the time-dependent decline in the ratio of divergences/diversities at nonsynonymous and synonymous sites (dN/dS or pN/pS) for protein-coding genes of human mitochondrial genes (Subramanian 2009) and nuclear genes of virus (Holmes 2003) and bacteria (Rocha et al. 2006).
Studies on the nuclear genomes of vertebrates did not reveal a clear-cut pattern of time dependency. Although earlier studies suggested a time-dependent pattern in microsatellites (Zhivotovsky et al. 2004, 2006) of the human nuclear genome, later studies based on next-generation sequencing did not identify such a pattern (Xue et al. 2009; Sun et al. 2012). This was further confirmed by a number of genome-wide studies, which showed that the evolutionary rates estimated using complete nuclear genomes of human pedigrees were similar to those estimated using human–chimpanzee species comparison (Conrad et al. 2010; Roach et al. 2010; Kong et al. 2012). However, as discussed earlier, time-dependent variation is expected only for constrained genomic regions. Therefore, in this study, we examined protein-coding genes in the human nuclear genome. It is well known that different nuclear genes are under different levels of selective constraint, depending on the relative importance of their functions. Hence, it would be interesting to examine the time-dependent pattern of evolutionary rates in genes, under various magnitudes of selection pressure. Therefore, we grouped nuclear genes of the human genome into four categories based on the intensities of selection pressure on them. We examined the pattern of time dependency using synonymous SNPs (sSNPs) and nSNPs from the complete genomes of six humans belonging to different populations.
Materials and Methods
DNA Sequence and Polymorphism Data
Protein-coding sequence alignments for humans (build 36) and chimpanzee (build 2) were obtained (for 16,750 known genes) from the University of California–Santa Cruz (UCSC) genome bioinformatics (http://genome.ucsc.edu/). Polymorphism data from six complete genomes belonging to a Khoisan (Schuster et al. 2010), two Yorubans (Bentley et al. 2008; Schuster et al. 2010), a European (Levy et al. 2007), a Chinese (Wang et al. 2008), and a Korean person (Kim et al. 2009) were obtained from UCSC genome bioinformatics and PSU Bioinformatics (http://main.genome-browser.bx.psu.edu/) data repositories. sSNPs and nSNPs of these genomes were determined using the chromosomal coordinates and gene boundary information. The number of sSNPs and nSNPs from each genome is given in table 1. The reference human genome was used to determine the ancestral state of each SNP. However, using the chimpanzee genome to orient the direction of SNP produced similar results (data not shown).
Table 1.
Number of SNPs and Estimates of Ratios for each Branch of the Human Tree
| Branch on the Tree | Nonsynonymous SNPs (A) | Synonymous SNPs (S) | A/S | pN/pS (SE) |
|---|---|---|---|---|
| dN/dS = 0–0.1 | ||||
| SAECK | 423 | 2,309 | 0.18 | 0.067 (0.0035) |
| AECK | 137 | 534 | 0.26 | 0.093 (0.0089) |
| ECK | 122 | 371 | 0.33 | 0.120 (0.0125) |
| CK | 558 | 1,049 | 0.53 | 0.194 (0.0101) |
| dN/dS = 0.1–0.2 | ||||
| SAECK | 721 | 1,552 | 0.46 | 0.169 (0.0076) |
| AECK | 168 | 327 | 0.51 | 0.187 (0.0177) |
| ECK | 169 | 235 | 0.72 | 0.262 (0.0264) |
| CK | 571 | 702 | 0.81 | 0.296 (0.0167) |
| dN/dS = 0.2–0.6 | ||||
| SAECK | 2,406 | 2,695 | 0.89 | 0.325 (0.0091) |
| AECK | 626 | 636 | 0.98 | 0.358 (0.0202) |
| ECK | 461 | 434 | 1.06 | 0.387 (0.0259) |
| CK | 1,571 | 1,188 | 1.32 | 0.481 (0.0185) |
| dN/dS = 0.6–1.0 | ||||
| SAECK | 875 | 571 | 1.53 | 0.558 (0.0300) |
| AECK | 244 | 165 | 1.48 | 0.538 (0.0542) |
| ECK | 160 | 96 | 1.67 | 0.607 (0.0783) |
| CK | 506 | 286 | 1.77 | 0.644 (0.0476) |
Divergence and Diversity Estimation
The numbers of synonymous and nonsynonymous positions and substitutions in each gene were calculated using the codeml program of the software PAML (Yang 2007). Pairwise evolutionary distances at synonymous and nonsynonymous sites for the human–chimpanzee pair were estimated using the Jukes–Cantor method. sSNPs and nSNPs were grouped based on the pattern of sharing, as described in the results (Subramanian 2012). To estimate the proportion of differences (pS or pN), the number of sSNPs or nSNPs belonging to each group was divided by the total number of synonymous or nonsynonymous positions in the genome. The binomial variance was used to compute the standard error.
Results and Discussion
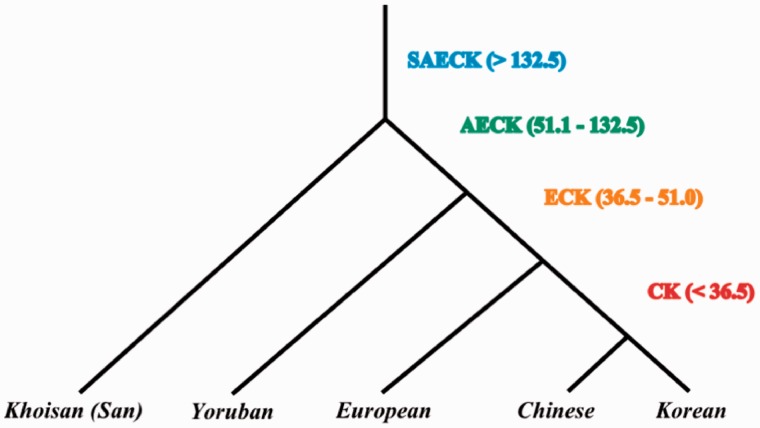
To examine nucleotide diversity at various temporal or evolutionary depths, we obtained SNP data from five complete human genomes belonging to a Chinese, a Korean, a European, a Yoruban (West African), and a Khoisan, an ancient African lineage. The phylogenetic relationship between the genomes of these humans (Tishkoff et al. 2009) is shown in figure 1. sSNPs and nSNPs were grouped based on the pattern of sharing between these genomes. If an SNP is shared between the Khoisan and any other genome, it was considered to be the oldest (see SAECK in fig. 1). If an SNP is shared between European (or an Asian) and Yoruban genomes and not shared by the Khoisan, it was considered to be common to Yorubans (or Africans) and non-Africans (AECK). Similarly, if an SNP is shared between a European and an Asian but not shared by Yoruban or Khoisan, it was considered to be ancestral to Eurasians (ECK). Finally, if an SNP is present in the Chinese or Korean genomes and not shared with any other genome, it was considered to be specific to Asians (CK). These grouping assumed that convergent or parallel mutations are rare. A recent study estimated the population divergence times of 4.5 (1–8), 36.5 (26–47), 51.0 (38–64), and 132.5 (108–157) Kyr for Chinese–Korean, Asian–European, African–Eurasian, and Khoisan–other humans, respectively (Gronau et al. 2011). Hence, these times provide relative ages for the SNPs as <36.5, 36.5–51.0, 51.1–132.5, and >132.5 for CK, ECK, AECK, and SAECK, respectively.
Fig. 1.—
Population phylogeny of the human genomes. The sharing pattern of SNPs used in this study is illustrated. Abbreviation denotes SNPs specific to Asians (CK), those shared between Europeans and Asians (ECK), Yorubans and Eurasians (AECK), and Khoisan and other genomes (SAECK). The relative age of SNPs (Kyr) obtained using the population divergence times estimated by a previous study (Gronau et al. 2011) are given in parenthesis.
We then grouped human genes based on the level of selective constraint on them. To quantify this, we used dN/dS ratio estimated from the human–chimp divergence at nonsynonymous- (dN) and synonymous sites (dS). Because synonymous sites are free from selection, this ratio reveals the extent of selective constraint on amino acids. For each group of genes, we estimated the ratio (pN/pS) of nSNPs (pN) to sSNPs (pS) per site using the polymorphisms shared between different branches of the human population tree (table 1). First, we examined the distribution of SNPs in the highly constrained human genes. This revealed a clear negative relationship between the age of SNPs (based on the extent of sharing) and pN/pS ratios (fig. 2A). For example, the pN/pS ratio of the Asian-specific SNPs (CK) was 2.9 times higher than that estimated using the SNPs shared with the ancient Khoisan genome (SAECK). This is a perfect example for time dependency, as the latter (>132.5 Kyr) is more than three times older than the former (<36.5 Kyr).
Fig. 2.—
The ratio of nonsynonymous- to synonymous SNPs per site (pN/pS) estimated for each of the branches shown in figure 1. The results shown are using the genes with dN/dS ratio (estimated for the human–chimp orthologous pair) of (A) <0.1, (B) 0.1–0.2, (C) 0.2–0.6, and (D) >0.6. Error bars are the standard error of the mean. In (D), the differences in pN/pS ratios between any two categories were not statistically significant (Z test, P > 0.12).
Interestingly, the distribution of pN/pS of genes gradually changes with decreasing selection pressures (fig. 2A–D). The distribution observed for highly constrained genes was strongly right skewed (fig. 2A). In contrast, it is largely uniform for the genes under relaxed selective constraints (fig. 2D) where the difference between pN/pS ratios of young and old SNPs disappeared (Z test, P > 0.12).
In addition to using dN/dS ratios, we also grouped human genes based on a different measure namely the Genome Evolutionary Rate Profiling (GERP) score described previously (Cooper et al. 2005). A GERP score is determined using multiple sequence alignments of orthologous genes and the level of selective constraints on each position. The score is quantified using the deficit of substitutions compared with a neutral site. We estimated the average GERP score for each gene and grouped genes based on their mean GERP scores. The results based on average GERP score (supplementary fig. S1, Supplementary Material online) were very similar to those showed in figure 2. For instance, constrained genes with a mean GERP score > 3.5 showed a clear-cut pattern of time dependency, which was absent for genes under relaxed selective constraints (average GERP score < 0.5).
Theoretical studies suggested that deleterious mutations contribute to the diversity of a population, but they are prevented from reaching higher frequencies and are purged over time (Kimura 1983). The results of this study strongly support this prediction. Because mutations in constrained genes are highly deleterious in nature, they do not typically spread through populations. As they are removed over time, we observe a very strong time-dependent effect on these genes (fig. 2A). In contrast, mutations on genes under relaxed selective constraints are typically only mildly harmful to humans and, therefore, segregate in human population for a long time. Particularly mutations in the most rapidly evolving genes with a dN/dS > 0.6 (fig. 2D) are almost neutral with negligible effects on the fitness of humans. This results in the similarity of pN/pS ratios across various timescales.
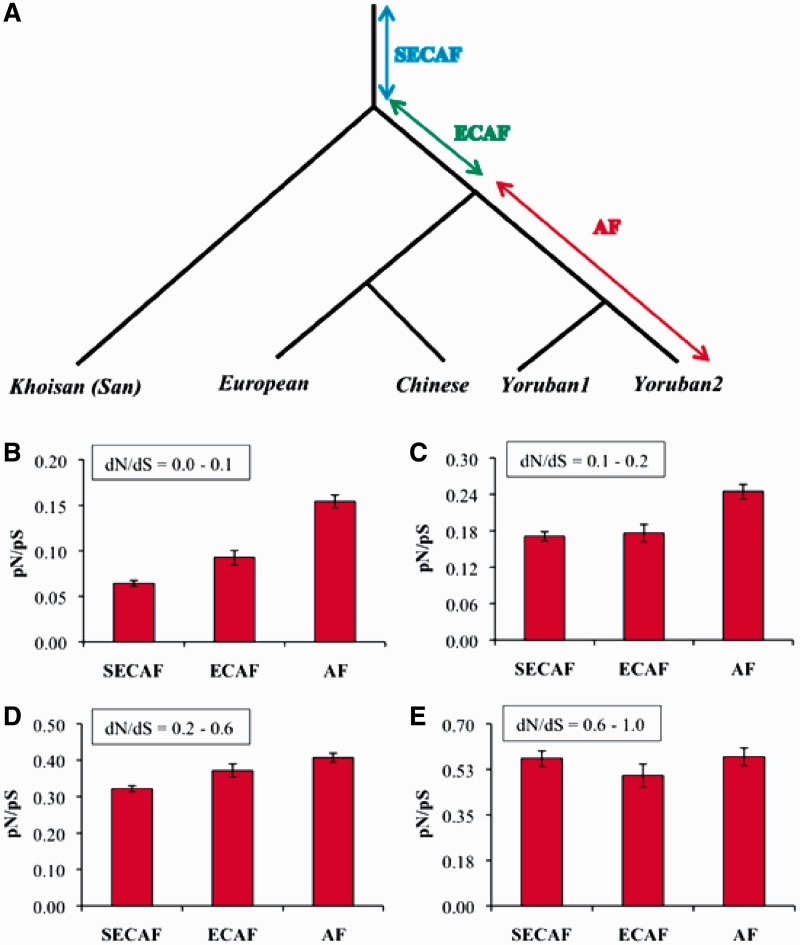
Previous studies suggested a population bottleneck in European and Asian populations (Marth et al. 2004; Li and Durbin 2011), and thus, a much smaller effective population size is expected for non-African populations than Yoruban and Khoisan populations. Hence, this bottleneck effect might have drifted some deleterious SNPs to higher frequencies, and this might have some influence on the pattern observed for constrained genes (fig. 2A). To address this issue, we examined the pattern of nSNPs in the African lineage, which is not bottlenecked in the past (Gronau et al. 2011; Li and Durbin 2011). For this purpose, we included another Yoruban genome and examined pN/pS ratios for the branches of the human population tree constructed using the genomes from two Yorubans, a European, a Chinese, and a Khoisan (fig. 3). For constrained human genes (dN/dS < 0.1), we found a significantly higher pN/pS ratio for Yoruban-specific SNPs (0.15) than that estimated using the SNPs shared between Yorubans and Eurasians (0.09), and this is higher than that computed for those shared between the ancient Khoisan and other genomes (0.06) (fig. 3B). In contrast, for genes under weak purifying selection (dN/dS > 0.6), pN/pS ratios of old and young SNPs were similar (P = 0.44) (fig. 3E). Figure 3B clearly supports a time-dependent pattern of deleterious SNPs rather than any bottleneck effect in the African lineage. Furthermore, a previous study suggested that the common ancestral population of all humans (SECAF) was smaller than the population ancestral to Yorubans (or Africans) and Eurasians (ECAF), which was in turn smaller than the population of Yorubans (AF) (Gronau et al. 2011). Hence, it seems likely that population expansion occurred throughout the African lineage. If population size is assumed to modulate the pN/pS ratios, then a pattern opposite to that shown in figure 3 is expected: high pN/pS for SECAF and low for AF. Therefore, population size effect might not explain our results shown in figures 2 and 3.
Fig. 3.—
(A) Phylogeny of human genomes belonging to different populations. Abbreviations are SNPs specific to Yorubans (AF), those shared between Yorubans and Eurasians (ECAF), and Khoisan and other genomes (SECAF). pN/pS ratios were estimated for three branches at different depths of the tree. Human genes were grouped into four categories mentioned in figure 2. Error bars denote the standard error of the mean. In (E), the differences in pN/pS ratios between any two categories were not statistically significant (Z test, P > 0.20).
This study demonstrated how purifying selection on genes influences the pattern of time dependency of molecular rates. Previous studies on human mitochondrial genes also suggested enrichment of nonsynonymous polymorphisms in the tips of the human population tree, compared with the internal nodes (Kivisild et al. 2006; Subramanian 2009; Pereira et al. 2011). Furthermore, Pereira et al. (2011) showed a predominance of pathogenic mutations in the younger branches of the human tree. Previous studies based on pedigree analysis of D-loop regions reported much higher rates compared with phylogenetic studies, which suggests the presence of deleterious mutations (Parsons et al. 1997; Howell et al. 2003; Millar et al. 2008). In contrast, pedigree analyses based on complete nuclear genomes (Conrad et al. 2010; Roach et al. 2010; Kong et al. 2012; Sun et al. 2012) of humans showed that the rates obtained from these analyses were not significantly different from the interspecies rate estimated using pseudogene data from human and chimp comparison (Nachman and Crowell 2000). Because the majority (>95%) of the sites of human genome are under neutral evolution, the similarity of the pedigree-based rate with the interspecific rate appears to be due to the predominance of neutral mutations.
In this study, we showed the variations in the rate of evolution observed between different timescales could largely be attributed to purifying selection. However, some of the variations could be due to other biological factors such as changes in population sizes. For instance, if the population size (Ne) of a species has been declining over time, this might mimic the pattern of time dependency as pN/pS ratios could be modulated by Ne. However, time dependency has been observed in a large number of species from virus to vertebrates, which suggests that this could be a universal phenomenon. This seems a reasonable view because we do not expect a population decline in the large number of species examined to date. Although multiple substitutions, homoplasy, or back mutations are of biological origin, they might be missed particularly in mutational hotspots because of the lack of methods to identify them. Therefore, long-term divergences are subjected to a higher rate of underestimation than short-term distances, and this might also produce a time-dependent effect. However, we used nuclear genomes of human for which the evolutionary rate is very low (<1 per site per billion years [Conrad et al. 2010; Roach et al. 2010; Kong et al. 2012; Sun et al. 2012]), and therefore multiple, back, or parallel substitutions are unlikely to influence our results as our comparisons involve only closely related species and populations.
Our results suggest that although the observed rates of molecular evolution vary for different timescales, this variation is limited to genes or genomic regions under selection. Importantly, we showed that the extent of variation is determined by the magnitude of selection on these regions. Therefore, to estimate molecular evolutionary rates or divergence times between species/populations, it is advisable to use the genes and genomic regions that are under minimal selective constraints.
Supplementary Material
Supplementary figure S1 is available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgment
This work was supported by the Australian Research Council and Griffith University.
Literature Cited
- Bandelt HJ. Clock debate: when times are a-changin': time dependency of molecular rate estimates: tempest in a teacup. Heredity. 2008;100:1–2. doi: 10.1038/sj.hdy.6801054. [DOI] [PubMed] [Google Scholar]
- Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burridge CP, Craw D, Fletcher D, Waters JM. Geological dates and molecular rates: fish DNA sheds light on time dependency. Mol Biol Evol. 2008;25:624–633. doi: 10.1093/molbev/msm271. [DOI] [PubMed] [Google Scholar]
- Conrad DF, et al. Variation in genome-wide mutation rates within and between human families. Nat Genet. 2010;43:712–714. doi: 10.1038/ng.862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper GM, et al. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15:901–913. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debruyne R, Poinar HN. Time dependency of molecular rates in ancient DNA data sets, a sampling artifact? Syst Biol. 2009;58:348–360. doi: 10.1093/sysbio/syp028. [DOI] [PubMed] [Google Scholar]
- Denver DR, Morris K, Lynch M, Thomas WK. High mutation rate and predominance of insertions in the Caenorhabditis elegans nuclear genome. Nature. 2004;430:679–682. doi: 10.1038/nature02697. [DOI] [PubMed] [Google Scholar]
- Emerson BC. Alarm bells for the molecular clock? No support for Ho et al.'s model of time-dependent molecular rate estimates. Syst Biol. 2007;56:337–345. doi: 10.1080/10635150701258795. [DOI] [PubMed] [Google Scholar]
- Endicott P, Ho SY. A Bayesian evaluation of human mitochondrial substitution rates. Am J Hum Genet. 2008;82:895–902. doi: 10.1016/j.ajhg.2008.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Moreno J. Is there a universal mtDNA clock for birds? J Avian Biol. 2004;35:465–468. [Google Scholar]
- Gronau I, et al. Bayesian inference of ancient human demography from individual genome sequences. Nat Genet. 2011;43:1031–1034. doi: 10.1038/ng.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag-Liautard C, et al. Direct estimation of per nucleotide and genomic deleterious mutation rates in Drosophila. Nature. 2007;445:82–85. doi: 10.1038/nature05388. [DOI] [PubMed] [Google Scholar]
- Hay JM, et al. Rapid molecular evolution in a living fossil. Trends Genet. 2008;24:106–109. doi: 10.1016/j.tig.2007.12.002. [DOI] [PubMed] [Google Scholar]
- Henn BM, Gignoux CR, Feldman MW, Mountain JL. Characterizing the time dependency of human mitochondrial DNA mutation rate estimates. Mol Biol Evol. 2009;26:217–230. doi: 10.1093/molbev/msn244. [DOI] [PubMed] [Google Scholar]
- Ho SY, Kolokotronis SO, Allaby RG. Elevated substitution rates estimated from ancient DNA sequences. Biol Lett. 2007;3:702–705. doi: 10.1098/rsbl.2007.0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho SY, et al. Time-dependent rates of molecular evolution. Mol Ecol. 2011;20:3087–3101. doi: 10.1111/j.1365-294X.2011.05178.x. [DOI] [PubMed] [Google Scholar]
- Ho SYW, Phillips MJ, Cooper A, Drummond AJ. Time dependency of molecular rate estimates and systematic overestimation of recent divergence times. Mol Biol Evol. 2005;22:1561–1568. doi: 10.1093/molbev/msi145. [DOI] [PubMed] [Google Scholar]
- Holmes EC. Patterns of intra- and interhost nonsynonymous variation reveal strong purifying selection in dengue virus. J Virol. 2003;77:11296–11298. doi: 10.1128/JVI.77.20.11296-11298.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N, et al. The pedigree rate of sequence divergence in the human mitochondrial genome: there is a difference between phylogenetic and pedigree rates. Am J Hum Genet. 2003;72:659–670. doi: 10.1086/368264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, et al. A highly annotated whole-genome sequence of a Korean individual. Nature. 2009;460:1011–1015. doi: 10.1038/nature08211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. Cambridge (UK): Cambridge University Press; 1983. [Google Scholar]
- Kivisild T, et al. The role of selection in the evolution of human mitochondrial genomes. Genetics. 2006;172:373–387. doi: 10.1534/genetics.105.043901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, et al. Rate of de novo mutations and the importance of father's age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert DM, et al. Rates of evolution in ancient DNA from Adelie penguins. Science. 2002;295:2270–2273. doi: 10.1126/science.1068105. [DOI] [PubMed] [Google Scholar]
- Levy S, et al. The diploid genome sequence of an individual human. PLoS Biol. 2007;5:e254. doi: 10.1371/journal.pbio.0050254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, et al. Resequencing of 200 human exomes identifies an excess of low-frequency non-synonymous coding variants. Nat Genet. 2010;42:969–972. doi: 10.1038/ng.680. [DOI] [PubMed] [Google Scholar]
- Marth GT, Czabarka E, Murvai J, Sherry ST. The allele frequency spectrum in genome-wide human variation data reveals signals of differential demographic history in three large world populations. Genetics. 2004;166:351–372. doi: 10.1534/genetics.166.1.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar CD, et al. Mutation and evolutionary rates in Adelie penguins from the Antarctic. PLoS Genet. 2008;4:e1000209. doi: 10.1371/journal.pgen.1000209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics. 2000;156:297–304. doi: 10.1093/genetics/156.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MR, et al. An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science. 2012;337:100–104. doi: 10.1126/science.1217876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons TJ, et al. A high observed substitution rate in the human mitochondrial DNA control region. Nat Genet. 1997;15:363–368. doi: 10.1038/ng0497-363. [DOI] [PubMed] [Google Scholar]
- Pereira L, et al. Comparing phylogeny and the predicted pathogenicity of protein variations reveals equal purifying selection across the global human mtDNA diversity. Am J Hum Genet. 2011;88:433–439. doi: 10.1016/j.ajhg.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, et al. Analysis of genetic inheritance in a family quartet by whole-genome sequencing. Science. 2010;328:636–639. doi: 10.1126/science.1186802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocha EPC, et al. Comparisons of dN/dS are time dependent for closely related bacterial genomes. J Theor Biol. 2006;239:226–235. doi: 10.1016/j.jtbi.2005.08.037. [DOI] [PubMed] [Google Scholar]
- Schuster SC, et al. Complete Khoisan and Bantu genomes from southern Africa. Nature. 2010;463:943–947. doi: 10.1038/nature08795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S. Temporal trails of natural selection in human mitogenomes. Mol Biol Evol. 2009;26:715–717. doi: 10.1093/molbev/msp005. [DOI] [PubMed] [Google Scholar]
- Subramanian S. The abundance of deleterious polymorphisms in humans. Genetics. 2012;190:1579–1583. doi: 10.1534/genetics.111.137893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian S, et al. High mitogenomic evolutionary rates and time dependency. Trends Genet. 2009;25:482–486. doi: 10.1016/j.tig.2009.09.005. [DOI] [PubMed] [Google Scholar]
- Subramanian S, Lambert DM. Time dependency of molecular evolutionary rates? Yes and no. Genome Biol Evol. 2011;3:1324–1328. doi: 10.1093/gbe/evr108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun JX, et al. A direct characterization of human mutation based on microsatellites. Nat Genet. 2012;44:1161–1165. doi: 10.1038/ng.2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennessen JA, et al. Evolution and functional impact of rare coding variation from deep sequencing of human exomes. Science. 2012;337:64–69. doi: 10.1126/science.1219240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tishkoff SA, et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–1044. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, et al. The diploid genome sequence of an Asian individual. Nature. 2008;456:60–65. doi: 10.1038/nature07484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, et al. Human Y chromosome base-substitution mutation rate measured by direct sequencing in a deep-rooting pedigree. Curr Biol. 2009;19:1453–1457. doi: 10.1016/j.cub.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang ZH. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Zhivotovsky LA, et al. The effective mutation rate at Y chromosome short tandem repeats, with application to human population-divergence time. Am J Hum Genet. 2004;74:50–61. doi: 10.1086/380911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhivotovsky LA, Underhill PA, Feldman MW. Difference between evolutionarily effective and germ line mutation rate due to stochastically varying haplogroup size. Mol Biol Evol. 2006;23:2268–2270. doi: 10.1093/molbev/msl105. [DOI] [PubMed] [Google Scholar]