Abstract
Packing of about two meters of the human genome DNA into chromatin occupying a several micron-sized cell nucleus requires a high degree of compaction in a manner that allows the information encoded on DNA to remain easily accessible. This packing is mediated by repeated coiling of DNA double helix around histones to form nucleosome arrays that are further folded into higher-order structures. Relatively straight DNA linkers separate the nucleosomes and the spacing between consecutive nucleosome varies between different cells and between different chromosomal loci. In a recent work1 our group used a biochemically defined in vitro reconstituted system to explore how do various DNA linkers mediate nucleosome array packing into higher-order chromatin structures. For long nucleosome linkers (about 60 bp) we observed a more open chromatin structure and no effect of small linker length alterations (±2−4 bp) on chromatin folding. In striking contrast, for shorter linkers (20−32 bp) we found more compact packing with strong periodical dependence upon the linker DNA lengths. Our data together with high-resolution nucleosome position mapping provide evidence for the natural nucleosome repeats to support a chromatin architecture that, by default, restricts spontaneous folding of nucleosome arrays into compact chromatin fibers. We suggest that incomplete folding of the nucleosome arrays may promote global inter-array interactions that lead to chromatin condensation in metaphase chromosomes and heterochromatin.
Keywords: chromatin, chromatin higher-order structure, nucleosome, sedimentation, electron microscopy
Introduction
In eukaryotic cells, the DNA is organized into the nucleosome arrays, and at the basic level of compaction, this involves wrapping of 145–147 bp of DNA around an octamer of core histone proteins to form the nucleosome core particle. The nucleosome cores are connected by 10–70 bp of linker DNA forming the nucleosome arrays that are further compacted in higher-order chromatin structures. The nucleosome core structure has been resolved by X-ray crystallography revealing the DNA conformation and the protein core of the nucleosome with outstanding precision.2 In contrast to the nucleosome core, the conformation of linker DNA is variable and is not yet completely resolved in compact chromatin.3,4 In addition, the linker DNA length varies widely among different organisms and tissues from the shortest (7 bp) linkers found in fission yeast5 to the longest linker DNA in echinoderm sperm (~100 bp).6 Even within one cell type, transcriptionally active genes may have linker DNA about 40 bp shorter than repressed or noncoding sequences.7 This extraordinary size and conformational variability of the linker DNA makes it extremely challenging to figure out which molecular mechanism(s) are engaged to pack nucleosome arrays into compact higher-order structures of interphase nuclei and metaphase chromosomes in a way that still renders the DNA accessible to all key genetic processes such as transcription, replication, and repair. Several modeling studies have previously suggested that linker DNA length and internucleosomal rotational variations could be important determinants in chromatin higher order folding8-10 and underlie irregular organization of the natural chromatin fibers.11,12 Most recent in situ studies revealing a surprisingly open and irregular conformation of nucleosome arrays in the condensed states (e.g., see refs. 13 and 14) make it even more exciting to explore the mechanism(s) that mediate packing of highly irregular nucleosome arrays into condensed chromatin and metaphase chromosomes.
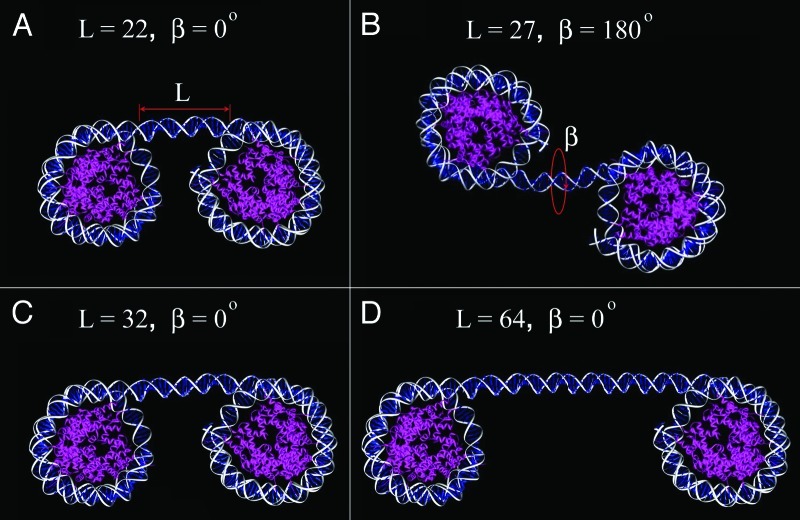
Previous modeling work prompted us to expect two distinct effects of the DNA linker lengths on the chromatin structure. We assumed that changes within the natural range of linker DNA length, L (see ref. 11), measured in base pairs would determine the distance between consecutive nucleosomes (Fig. 1) and establish the diameter of the nucleosome fiber especially for zigzag or cross-linker structures observed in vitro15 and in situ16 where the nucleosome linkers are mostly extended. Note that the variability within the few bp range would not considerably change the distance between nucleosomes but due to the helical nature of DNA, should strongly affect the internucleosomal rotation angle β (see ref. 11) between the nucleosome disks (Fig. 1, cf. structures A and B) and might interfere with the association between nucleosome surfaces that contributes to tight packing of chromatin. We focused on nucleosome arrays with linker lengths varying between 22 and 64 bp which cover most of the vertebrate linker DNA range. To model natural linker DNA variations, we constructed nucleosome arrays using Widom’s clone 601 nucleosome core positioning sequence17 that has been shown to form a nucleosme core containing 145 bp of DNA by X-ray crystallography18,19 and a set of variable linkers that we designed in the lab. We then examined how these changes affected the formation of chromatin higher-order structures using a combination of solution structural studies and EM visualization of single folded particles.
Figure 1. Linker DNA length variations change spacing and orientation between nucleosomes. Molecular models of dinucleosomes with linker lengths L = 22 (A), 27 (B), 32 (C), and 64 (D) bp were constructed based on the clone 601 nucleosome core X-ray crystal structure PDB 3MVD18 after removing RCC protein from the file. For adding linker DNA, we used a DNA fragment from PDB 1ZBB. For modeling details see reference 1.
Increasing Linker DNA Length Inhibits Nucleosome Array Compaction
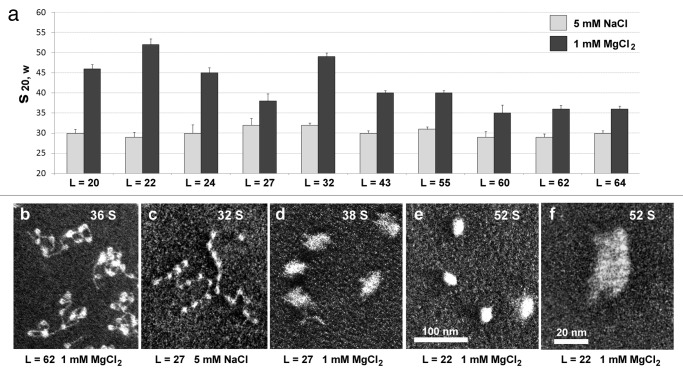
To experimentally address structural changes resulting from the increased nucleosome spacing, we constructed a set of 12-mer arrays differing by linker length and examined their folding in the presence of millimolar concentration of MgCl2 as divalent cations were previously shown to promote complete folding of nucleosomes arrays.20 We assayed chromatin compaction by analytical centrifugation expecting an increase of sedimentation coefficient s20º,w from 30 S to more than 50 S for compact 12-mer arrays. First, we observed that for the linker lengths close to an integer number of DNA double helix turns (10.5 bp/turn): L = 22, 32, 43, 64, there was a clear negative correlation of linker DNA length with chromatin compaction. Arrays with shorter nucleosome linkers (L = 22 and 32 corresponding to ~2 and ~3 turns of DNA) could fold almost completely with increasing salt concentrations while nucleosome arrays with linkers of L = 43 (~4 turns) and 64 (~6 turns could not completely fold at physiological salt conditions and required linker histone for maximal compactness (Fig. 2A). It was reassuring that our sedimentation data obtained with a set of systematically altered linkers were consistent with several independent experiments showing similar folded conformation of repeats with short linkers20-23 contrasting with unfolded conformations of arrays with long linkers.21,22,24
Figure 2. Linker DNA length variations affect chromatin folding in vitro. (a) Histograms showing peaks of sedimentation coefficients s20º,w distribution (average of three independent experiments) for 12-mer oligonucleosome core arrays with varying linker lengths L as indicated. Sedimentation data are taken from experiments conducted at 5 mM NaCl and 1 mM MgCl2.1 (b–f) Electron micrographs (positive uranyl acetate staining, dark-field imaging) of 12-mer nucleosome core arrays. Linker lengths, L (base pairs) and ionic conditions during glutaraldehyde fixation are indicated. S values shown on the panels are taken from parallel sedimentation experiments conducted under the same conditions. Electron microscopy samples were prepared and imaged as described.1 Magnification: panels b–e, 42,000; panel f, 150,000.
From comparison with native chromatin, where increase in DNA linker length often accompanies chromatin compaction6,25 this result may seem surprising. However, differentiated cell with increased nucleosome repeats also tend to accumulate linker histones that are sufficient to alleviate the folding difference between the short and long repeats in vitro. A positive correlation between linker histone levels and the linker DNA length has been discussed in detail elsewhere.26-28
The nucleosomes at the upstream portions of the active genes are most uniformly spaced and there is an evidence that the nucleosome spacing in active chromatin is mediated by transcription.26,29 Earlier hybridization mapping30 as well as most recent genome-wide mapping of nucleosome positions7 reported a strikingly shorter nucleosome repeat of transcribed genes as compared with repressed or noncoding sequences in the same cells. In addition, transcribed chromatin has been shown to display a compact higher-order structure exceeding compaction expected for the 30 nm fiber31,32 despite evidence for decreased association of active chromatin with linker histone.33,34 Linker histone depletion at the active genes may be mediated by core histone variants disfavoring H1 binding such as H3.3 and H2A.Z35,36 and/or by linker histone modification regulating its affinity to chromatin.37,38 It is also possible that the arrays with shorter nucleosome linkers have intrinsically lower affinity for linker histone. Since total linker histone levels in most cells with active chromatin are below saturation (1 molecule per nucleosome) it will be important to examine whether nucleosome arrays with shorter DNA linkers and/or special core histones display lower linker histone affinity especially under conditions of competition for linker histone.
The ability of the shorter DNA linkers to promote chromatin compaction in the absence of linker histone (Fig. 2) suggests that the chromatin with shorter repeats (22–32 bp) may fold independently of linker histone in vivo and, after being subjected to dynamic stretching induced by passage of RNA polymerase or chromatin remodelers, could restore its conformation spontaneously. In addition, packing of nucleosomes in such structures would depend on their interface interaction and may be limited from folding in the most compact fibers (such as shown on Fig. 2F and Fig. 3C) by histone H4 acetylation at lysine 1623 or by including histone variants altering the nucleosome surface.39 It thus looks likely that decreasing the linker DNA length in the active chromatin makes the underlying higher order structure to be more dependent on complex regulation mediated by core histone variants and their modifications than in case of transcriptionally inactive chromatin with longer DNA linkers.
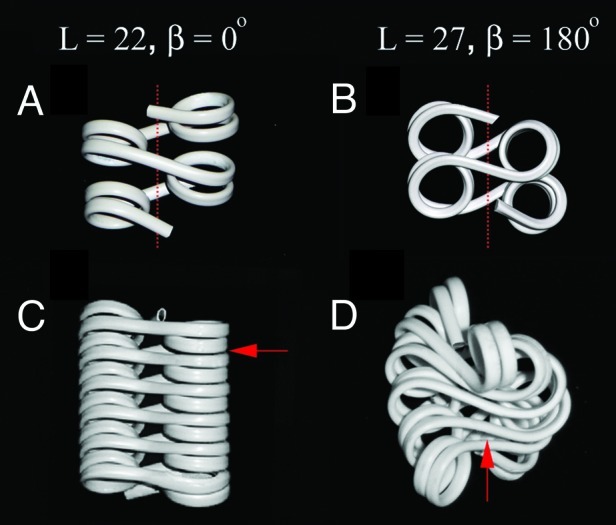
Figure 3. A 5 bp difference between short linkers is predicted to have a strong effect on chromatin fiber architecture. Flexible wire models1 for DNA geometry in tetranucleosomes with linker DNA length L = 22 (A) and 27 (B) and in compact 12-mer nucleosome fibers with linker DNA length L = 22 (C) and 27 (D). The arrows show the points of contacts that limit the longitudinal compaction of the fiber. The dashed lines show the 30 nm fiber axis.
Short But Not Long Linkers Impose Periodical Rotational Alterations on Chromatin Folding
In spite of the fact that native nucleosomal arrays have linker DNA length variations of ± 2 to ± 4 bp8,40 and structural modeling suggests a significant dependence of chromatin folding on internucleosomal rotational variations11,12,41 most nucleosome reconstitutes used in in vitro experiments elsewhere employed regular linker DNA lengths. To explore the importance of local variations in the rotational setting mimicking native chromatin, we constructed nucleosome arrays with 62 bp linkers varying by either ± 2 or ± 4 bp. Based on earlier modeling of nucleosomes within similar linker length variations11 we expected to have different orientations, reflecting local changes in the internucleosomal rotational angle β. Despite the model predictions, however, we observed no significant difference in the folding of the regular and variable arrays. Furthermore, when we tested a set of nucleosome arrays with regular repeats differing by two base pairs from the base 62 bp repeat (60, 62 and 64 bp), we did not observe any difference in chromatin folding by sedimentation (Fig. 2A).
So, why is chromatin fiber compaction independent of the internucleosomal rotations for the ~60 bp arrays? Based on nucleosome X-ray crystal studies, it was suggested that nucleosome core DNA may be stretched or contracted to absorb up to 4 bp variations in linker DNA length2,18 and thus alleviate its effect on chromatin folding. It is, however, not clear whether such transitions do happen in solution. If this was the case, then we would expect that the nucleosome core could absorb the length difference in short linkers as well as in the longer ones. However, we observed that in a striking contrast to the longer arrays (55–64 bp), folding of the arrays with shorter linkers (18–30 bp) was significantly affected by small linker length variations. The most pronounced changes observed between maximally folded nucleosomes with L = 22 linkers and minimally folded nucleosome arrays with L = 27 bp (Fig. 2A). Our results showing a strong dependence of chromatin compaction for shorter nucleosome linker length are clearly inconsistent with linker DNA length variations absorbed by the nucleosome core but may be better explained by a recent theoretical work42 suggesting that longer linkers readily adopt multiple fiber conformations with only small changes in torsional energy while shorter linkers have a limited conformational flexibility in determining nucleosome packing of arrays and are much more sensitive to rotational changes between the nucleosomes.
Electron microscopy experiments (Fig. 2D–F) and modeling provided further explanation to the effect of nucleosome linker lengths on chromatin folding observed by sedimentation. The arrays with L = 22 (167 bp nucleosome repeat) have a clear advantage of being the only type of nucleosome array solved by X-ray crystallography.43 In this crystal structure, the linker DNA is slightly twisted and the nucleosome disks are rotated for about 90° to allow the nucleosome packing. The actual twist of linker DNA and the exact number of DNA bp associated with histone octamers in solution is not known. However, a simple space-filling model that recapitulates the DNA linker lengths shown in Figure 1 and allows some variation in the DNA twisting and the linker DNA path similar to those in the tetranucleosome structure43 allows one to easily fold a nucleosome array with L = 22 in a conformation promoting stacking of the nucleosome disks and close contacts between the nucleosome surfaces (Fig. 3A and C). In this structure, the internucleosomal rotations and the chromatin fiber size and shape are consistent with EM images of the 61-nucleosome repeats,21 our high-magnification EM images of the 12-nucleosome repeat (Fig. 2F), as well as chromatin fiber model based on the tetranucleosome crystal structure of the same linker length.43 Thus, for the nucleosome array with 22 bp linkers there seems to be a universal agreement between different structural studies on a two-start zigzag fiber folded by close stacking interactions between nucleosome surfaces.
A similar model but with 27 bp linkers, in a remarkable agreement with sedimentation and EM, shows increasing spatial clashes between linker DNA upon compaction (Fig. 3B and D) that prevent the nucleosome disks from coming into close contacts and folding as tightly as with 22 bp linkers. Previously, our computation modeling suggested that intersections of linker DNA stems in the middle of the fiber limited complete folding and that bending of some DNA linkers by linker histone and Mg2+ was needed to bring the fiber into full compaction.22 Our new data showing that linker histone alleviates the folding difference between the two arrays and promotes the complete folding of the 172 bp array1 provides additional strong argument for linker DNA conformation being a major factor limiting complete compaction of the nucleosome arrays.
Natural Nucleosome Repeats Are Likely to Restrict Nucleosome Array Folding
The striking effect of a few bp difference of the linker length on chromatin folding in vitro appeals for careful measurement of DNA spacing between native nucleosomes in order to predict the possible higher-order structure of the associated chromatin. Changes in average spacing between the nucleosomes such as those between active and repressed genes an/or during terminal differentiation are well documented. Most previous nucleosome repeat data have been obtained in experiments that describe average nucleosome spacing in the genome but do not account for precise rotational settings between neighboring nucleosomes which may considerably differ from the average. Yeast chromatin, for example, is known to have an average nucleosome repeat of 167 bp that is equivalent to 20–22 bp of linker DNA close to an integer number of DNA turns (2) in the linker. However, earlier biochemical experiments by Dennis Lohr who used micrococcal nuclease and DNase I to map overlapping phases of nuclease digestion periodicity showed, surprisingly, that the DNase I pattern matches a non-integer (N + 0.5) number of DNA turns, such as 0.5, 1.5 and or 2.5.44,45
Recent developments in parallel sequencing technology allowed researchers to account for millions of nucleosomal DNA sequences genome-wide and map them to eukaryotic genomes. Remarkably, sequencing and statistical analysis of yeast genome dinucleosomes by late Jonathan Widom and his colleagues showed that indeed, yeast DNA linkers lengths are quantized with peaks close to N + 0.5 DNA turns (e.g., 5, 15, 25 bp) and the linkers with integer number of DNA turns such as the 20 bp linker size, average in yeast chromatin, corresponding to a minimum in the size distribution.46 Note that in this analysis the nucleosome core DNA length is fixed at 147 bp so that the total nucleosome repeat (147 + 20) is equal to 145 + 22 in our structural model (Fig. 1A). A most recent study from the same laboratory readdressed nucleosome positioning in yeast with chemical crosslinking approach that eliminates nuclease digestion artifacts and confirmed the predominant linker lengths to contain N + 0.5 turns of DNA.47 Together with our studies, nucleosome position mapping suggests that the yeast genome developed a nucleosome spacing mechanism to support an open zigzag chromatin structure similar to the one of the 172 bp repeat (Fig. 2B and D). This is also consistent with the open chromatin structure in S. cerevisiae nuclei with just about 2 nucleosomes per 11 nm observed by chromatin interaction (3C) mapping in situ.48
It thus appears that vast majority of nucleosome repeats found in vivo in contrast to the two regular repeats, 167 and 177 bp, extensively studied in vitro, support a relatively open conformation of the chromatin fiber and cannot be completely folded without linker histone or some other architectural factor. It has been noticed before that linker histone levels correlate positively with the linker DNA length27 apparently because more linker histone is needed to condense chromatin with longer linkers. In cells with linker histone knockouts, the linker length decreases49 reflecting some mechanism that allows assembling nucleosomes at higher density and compensating for linker histone losses. Remarkably, our results showing that linker histone can also fold short linkers with non-integer linker DNA lengths provide an explanation for neuronal chromatin that has anomalously high levels of linker histone and a 162 bp nucleosome repeat unfavorable for folding.50
Native chromatin is known to be highly dynamic. Its dynamic transitions may include spontaneous dissociation of the outer DNA segments from nucleosomes, transient association of the linker histone and architectural proteins like heterochromatin protein HP1 with chromatin, and including histone variants that leads to nucleosome cores with less than 147 bp of DNA (reviewed in refs. 3 and 4). In addition, native DNA has a dramatically less affinity to histones that clone 60117 and may facilitate repositioning of the nucleosome cores to accommodate to local energetically favorable structures. Our new observation that precise linker length controls folding of short nucleosome repeats similar to those found in fission and budding yeast5,46 and cortical neurons50 in vivo necessitates developing new experimental approaches to capture transient structural intermediates of chromatin folding in vitro and in vivo and to relate them to cell-specific chromatin models that take in consideration linker lengths, levels of linker histone and other specific architectural factors, as well as genomic maps and bioinformatic analyses of specific nucleosome positions in situ.
It has been shown before that elimination of linker DNA in situ makes the nuclear chromatin significantly more condensed.51 Rendering chromatin higher-order folding in an open state by varying nucleosome linkers provides additional mechanisms for regulating its dynamic transitions both locally and globally. Paradoxically, an open conformation of a nucleosome array may be essential for its packing in most condensed chromatin structures. For example, in budding yeast where heterochromatin occupies only a small fraction of the genome, special heterochromatin architectural proteins such as Sir352 organize heterochromatin structure locally and promote its transcriptional silencing while linker histone (whose level is very low in yeast nuclei) inhibits transcriptional silencing.53 Recent X-ray crystal structure study showed that Sir3 recognizes acidic patch at the nucleosome disk surface54 so that complete fiber folding and nucleosome disk stacking such as shown in Figure 3C could perturb heterochromatin formation by Sir3 by interfering with its binding to the nucleosome surface. The fact that the acidic patch is recognized by heterochromatin protein HP1 when enhanced by acidic residues of histone variant H2A.Z55 is also consistent with the necessity of opening the nucleosome surface for heterochromatin formation. Finally, as it becomes apparent that nucleosome arrays are not folded into 30 nm fibers in mitotic chromosomes56 and heterochromatin13 it looks more likely that global chromatin folding in mitosis and heterochromatin is mediated by lateral interactions between the nucleosome arrays4,57 rather than by longitudinal compaction of chromatin fibers. Future studies should clarify whether natural variations of the nucleosome linkers could regulate interactions between nucleosomes in vivo and segregate individual chromatin fibers from the compact chromatin mass cemented by lateral inter-array interactions.
Acknowledgments
I thank S. Correll and M. Schubert who contributed to work published in EMBO Journal. This work was supported by NSF grant MCB-1021681 to S.A.G.
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/22168
References
- 1.Correll SJ, Schubert MH, Grigoryev SA. Short nucleosome repeats impose rotational modulations on chromatin fibre folding. EMBO J. 2012;31:2416–26. doi: 10.1038/emboj.2012.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richmond TJ, Davey CA. The structure of DNA in the nucleosome core. Nature. 2003;423:145–50. doi: 10.1038/nature01595. [DOI] [PubMed] [Google Scholar]
- 3.Schlick T, Hayes J, Grigoryev S. Toward convergence of experimental studies and theoretical modeling of the chromatin fiber. J Biol Chem. 2012;287:5183–91. doi: 10.1074/jbc.R111.305763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luger K, Dechassa ML, Tremethick DJ. New insights into nucleosome and chromatin structure: an ordered state or a disordered affair? Nat Rev Mol Cell Biol. 2012;13:436–47. doi: 10.1038/nrm3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lantermann AB, Straub T, Strålfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat Struct Mol Biol. 2010;17:251–7. doi: 10.1038/nsmb.1741. [DOI] [PubMed] [Google Scholar]
- 6.Athey BD, Smith MF, Rankert DA, Williams SP, Langmore JP. The diameters of frozen-hydrated chromatin fibers increase with DNA linker length: evidence in support of variable diameter models for chromatin. J Cell Biol. 1990;111:795–806. doi: 10.1083/jcb.111.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A. Determinants of nucleosome organization in primary human cells. Nature. 2011;474:516–20. doi: 10.1038/nature10002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Widom J. A relationship between the helical twist of DNA and the ordered positioning of nucleosomes in all eukaryotic cells. Proc Natl Acad Sci U S A. 1992;89:1095–9. doi: 10.1073/pnas.89.3.1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wedemann G, Langowski J. Computer simulation of the 30-nanometer chromatin fiber. Biophys J. 2002;82:2847–59. doi: 10.1016/S0006-3495(02)75627-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stehr R, Kepper N, Rippe K, Wedemann G. The effect of internucleosomal interaction on folding of the chromatin fiber. Biophys J. 2008;95:3677–91. doi: 10.1529/biophysj.107.120543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Woodcock CL, Grigoryev SA, Horowitz RA, Whitaker N. A chromatin folding model that incorporates linker variability generates fibers resembling the native structures. Proc Natl Acad Sci U S A. 1993;90:9021–5. doi: 10.1073/pnas.90.19.9021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Leuba SH, Yang G, Robert C, Samori B, van Holde K, Zlatanova J, et al. Three-dimensional structure of extended chromatin fibers as revealed by tapping-mode scanning force microscopy. Proc Natl Acad Sci U S A. 1994;91:11621–5. doi: 10.1073/pnas.91.24.11621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fussner E, Djuric U, Strauss M, Hotta A, Perez-Iratxeta C, Lanner F, et al. Constitutive heterochromatin reorganization during somatic cell reprogramming. EMBO J. 2011;30:1778–89. doi: 10.1038/emboj.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Joti Y, Hikima T, Nishino Y, Kamda F, Hihara S, Takata H, et al. Chromosomes without a 30-nm chromatin fiber. Nucleus. 2012;3:3. doi: 10.4161/nucl.21222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Williams SP, Langmore JP. Small angle x-ray scattering of chromatin. Radius and mass per unit length depend on linker length. Biophys J. 1991;59:606–18. doi: 10.1016/S0006-3495(91)82276-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horowitz RA, Agard DA, Sedat JW, Woodcock CL. The three-dimensional architecture of chromatin in situ: electron tomography reveals fibers composed of a continuously variable zig-zag nucleosomal ribbon. J Cell Biol. 1994;125:1–10. doi: 10.1083/jcb.125.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lowary PT, Widom J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J Mol Biol. 1998;276:19–42. doi: 10.1006/jmbi.1997.1494. [DOI] [PubMed] [Google Scholar]
- 18.Makde RD, England JR, Yennawar HP, Tan S. Structure of RCC1 chromatin factor bound to the nucleosome core particle. Nature. 2010;467:562–6. doi: 10.1038/nature09321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vasudevan D, Chua EY, Davey CA. Crystal structures of nucleosome core particles containing the ‘601’ strong positioning sequence. J Mol Biol. 2010;403:1–10. doi: 10.1016/j.jmb.2010.08.039. [DOI] [PubMed] [Google Scholar]
- 20.Dorigo B, Schalch T, Kulangara A, Duda S, Schroeder RR, Richmond TJ. Nucleosome arrays reveal the two-start organization of the chromatin fiber. Science. 2004;306:1571–3. doi: 10.1126/science.1103124. [DOI] [PubMed] [Google Scholar]
- 21.Routh A, Sandin S, Rhodes D. Nucleosome repeat length and linker histone stoichiometry determine chromatin fiber structure. Proc Natl Acad Sci U S A. 2008;105:8872–7. doi: 10.1073/pnas.0802336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grigoryev SA, Arya G, Correll S, Woodcock CL, Schlick T. Evidence for heteromorphic chromatin fibers from analysis of nucleosome interactions. Proc Natl Acad Sci U S A. 2009;106:13317–22. doi: 10.1073/pnas.0903280106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–7. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 24.Carruthers LM, Bednar J, Woodcock CL, Hansen JC. Linker histones stabilize the intrinsic salt-dependent folding of nucleosomal arrays: mechanistic ramifications for higher-order chromatin folding. Biochemistry. 1998;37:14776–87. doi: 10.1021/bi981684e. [DOI] [PubMed] [Google Scholar]
- 25.Weintraub H. The nucleosome repeat length increases during erythropoiesis in the chick. Nucleic Acids Res. 1978;5:1179–88. doi: 10.1093/nar/5.4.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szerlong HJ, Hansen JC. Nucleosome distribution and linker DNA: connecting nuclear function to dynamic chromatin structure. Biochem Cell Biol. 2011;89:24–34. doi: 10.1139/O10-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Woodcock CL, Skoultchi AI, Fan Y. Role of linker histone in chromatin structure and function: H1 stoichiometry and nucleosome repeat length. Chromosome Res. 2006;14:17–25. doi: 10.1007/s10577-005-1024-3. [DOI] [PubMed] [Google Scholar]
- 28.Perišić O, Collepardo-Guevara R, Schlick T. Modeling studies of chromatin fiber structure as a function of DNA linker length. J Mol Biol. 2010;403:777–802. doi: 10.1016/j.jmb.2010.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weiner A, Hughes A, Yassour M, Rando OJ, Friedman N. High-resolution nucleosome mapping reveals transcription-dependent promoter packaging. Genome Res. 2010;20:90–100. doi: 10.1101/gr.098509.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sun YL, Xu YZ, Bellard M, Chambon P. Digestion of the chicken beta-globin gene chromatin with micrococcal nuclease reveals the presence of an altered nucleosomal array characterized by an atypical ladder of DNA fragments. EMBO J. 1986;5:293–300. doi: 10.1002/j.1460-2075.1986.tb04212.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu Y, Kireev I, Plutz M, Ashourian N, Belmont AS. Large-scale chromatin structure of inducible genes: transcription on a condensed, linear template. J Cell Biol. 2009;185:87–100. doi: 10.1083/jcb.200809196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Georgel PT, Fletcher TM, Hager GL, Hansen JC. Formation of higher-order secondary and tertiary chromatin structures by genomic mouse mammary tumor virus promoters. Genes Dev. 2003;17:1617–29. doi: 10.1101/gad.1097603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kamakaka RT, Thomas JO. Chromatin structure of transcriptionally competent and repressed genes. EMBO J. 1990;9:3997–4006. doi: 10.1002/j.1460-2075.1990.tb07621.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Misteli T, Gunjan A, Hock R, Bustin M, Brown DT. Dynamic binding of histone H1 to chromatin in living cells. Nature. 2000;408:877–81. doi: 10.1038/35048610. [DOI] [PubMed] [Google Scholar]
- 35.Braunschweig U, Hogan GJ, Pagie L, van Steensel B. Histone H1 binding is inhibited by histone variant H3.3. EMBO J. 2009;28:3635–45. doi: 10.1038/emboj.2009.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thakar A, Gupta P, Ishibashi T, Finn R, Silva-Moreno B, Uchiyama S, et al. H2A.Z and H3.3 histone variants affect nucleosome structure: biochemical and biophysical studies. Biochemistry. 2009;48:10852–7. doi: 10.1021/bi901129e. [DOI] [PubMed] [Google Scholar]
- 37.Bleher R, Martin R. Nucleo-cytoplasmic translocation of histone H1 during the HeLa cell cycle. Chromosoma. 1999;108:308–16. doi: 10.1007/s004120050382. [DOI] [PubMed] [Google Scholar]
- 38.Trojer P, Zhang J, Yonezawa M, Schmidt A, Zheng H, Jenuwein T, et al. Dynamic Histone H1 Isotype 4 Methylation and Demethylation by Histone Lysine Methyltransferase G9a/KMT1C and the Jumonji Domain-containing JMJD2/KDM4 Proteins. J Biol Chem. 2009;284:8395–405. doi: 10.1074/jbc.M807818200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou J, Fan JY, Rangasamy D, Tremethick DJ. The nucleosome surface regulates chromatin compaction and couples it with transcriptional repression. Nat Struct Mol Biol. 2007;14:1070–6. doi: 10.1038/nsmb1323. [DOI] [PubMed] [Google Scholar]
- 40.Strauss F, Prunell A. Nucleosome spacing in rat liver chromatin. A study with exonuclease III. Nucleic Acids Res. 1982;10:2275–93. doi: 10.1093/nar/10.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stehr R, Schöpflin R, Ettig R, Kepper N, Rippe K, Wedemann G. Exploring the conformational space of chromatin fibers and their stability by numerical dynamic phase diagrams. Biophys J. 2010;98:1028–37. doi: 10.1016/j.bpj.2009.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Scipioni A, Turchetti G, Morosetti S, De Santis P. Geometrical, conformational and topological restraints in regular nucleosome compaction in chromatin. Biophys Chem. 2010;148:56–67. doi: 10.1016/j.bpc.2010.02.010. [DOI] [PubMed] [Google Scholar]
- 43.Schalch T, Duda S, Sargent DF, Richmond TJ. X-ray structure of a tetranucleosome and its implications for the chromatin fibre. Nature. 2005;436:138–41. doi: 10.1038/nature03686. [DOI] [PubMed] [Google Scholar]
- 44.Lohr D, Van Holde KE. Organization of spacer DNA in chromatin. Proc Natl Acad Sci U S A. 1979;76:6326–30. doi: 10.1073/pnas.76.12.6326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lohr DE. Detailed analysis of the nucleosomal organization of transcribed DNA in yeast chromatin. Biochemistry. 1981;20:5966–72. doi: 10.1021/bi00524a007. [DOI] [PubMed] [Google Scholar]
- 46.Wang JP, Fondufe-Mittendorf Y, Xi L, Tsai GF, Segal E, Widom J. Preferentially quantized linker DNA lengths in Saccharomyces cerevisiae. PLoS Comput Biol. 2008;4:e1000175. doi: 10.1371/journal.pcbi.1000175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brogaard K, Xi L, Wang JP, Widom J. A map of nucleosome positions in yeast at base-pair resolution. Nature. 2012;486:496–501. doi: 10.1038/nature11142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dekker J. Mapping in vivo chromatin interactions in yeast suggests an extended chromatin fiber with regional variation in compaction. J Biol Chem. 2008;283:34532–40. doi: 10.1074/jbc.M806479200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan Y, Nikitina T, Zhao J, Fleury TJ, Bhattacharyya R, Bouhassira EE, et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell. 2005;123:1199–212. doi: 10.1016/j.cell.2005.10.028. [DOI] [PubMed] [Google Scholar]
- 50.Pearson EC, Bates DL, Prospero TD, Thomas JO. Neuronal nuclei and glial nuclei from mammalian cerebral cortex. Nucleosome repeat lengths, DNA contents and H1 contents. Eur J Biochem. 1984;144:353–60. doi: 10.1111/j.1432-1033.1984.tb08471.x. [DOI] [PubMed] [Google Scholar]
- 51.Green GR, Ferlita RR, Walkenhorst WF, Poccia DL. Linker DNA destabilizes condensed chromatin. Biochem Cell Biol. 2001;79:349–63. doi: 10.1139/o01-115. [DOI] [PubMed] [Google Scholar]
- 52.McBryant SJ, Krause C, Woodcock CL, Hansen JC. The silent information regulator 3 protein, SIR3p, binds to chromatin fibers and assembles a hypercondensed chromatin architecture in the presence of salt. Mol Cell Biol. 2008;28:3563–72. doi: 10.1128/MCB.01389-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yu Q, Kuzmiak H, Zou Y, Olsen L, Defossez PA, Bi X. Saccharomyces cerevisiae linker histone Hho1p functionally interacts with core histone H4 and negatively regulates the establishment of transcriptionally silent chromatin. J Biol Chem. 2009;284:740–50. doi: 10.1074/jbc.M806274200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Armache KJ, Garlick JD, Canzio D, Narlikar GJ, Kingston RE. Structural basis of silencing: Sir3 BAH domain in complex with a nucleosome at 3.0 Å resolution. Science. 2011;334:977–82. doi: 10.1126/science.1210915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fan JY, Rangasamy D, Luger K, Tremethick DJ. H2A.Z alters the nucleosome surface to promote HP1alpha-mediated chromatin fiber folding. Mol Cell. 2004;16:655–61. doi: 10.1016/j.molcel.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 56.Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–53. doi: 10.1038/emboj.2012.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Grigoryev SA. Keeping fingers crossed: heterochromatin spreading through interdigitation of nucleosome arrays. FEBS Lett. 2004;564:4–8. doi: 10.1016/S0014-5793(04)00258-3. [DOI] [PubMed] [Google Scholar]