Abstract
Transcriptional regulation is a complex process that requires the integrated action of many multi-protein complexes. The way in which a living cell coordinates the action of these complexes in time and space is still poorly understood. Recent work has shown that nuclear pores, well known for their role in 3′ processing and export of transcripts, also participate in the control of transcriptional initiation. We have recently begun to explore how nuclear pores interface with the well-described machinery that regulates initiation. This work led to the discovery that specific nucleoporins are required for binding of the repressor protein Mig1 to its site in target promoters. Nuclear pores are therefore involved in repressing, as well as activating, transcription. Here we discuss in detail the main models explaining our result and consider what each implies about the roles that nuclear pores play in the regulation of gene expression.
Keywords: AMPK, NPC, Nup120, Nup133, RNA polymerase II, acute myeloid leukemia, glucose, glucose metabolism, glucose-regulated gene expression, nuclear pore complex, transcriptional regulation
Introduction
Cells respond to environmental cues to grow and differentiate by turning discrete sets of genes on and off. Although no one model appears to apply to all genes,1,2 work done over the past 40 y has given us a general understanding of how a eukaryotic cell accomplishes this on/off switch.2-9 The process is complex, requiring the interaction of nucleosomes, chromatin remodelers, chromatin modifiers, sequence-specific repressors and activators, co-activators, basal transcription factors, positive and negative regulators of elongation, termination factors and of course the DNA itself. Control of transcriptional initiation, though historically appreciated as important, is only one way in which information in the genome is managed. Once the signal to express a certain gene has been received and the necessary factors are all associated with the DNA, the polymerase must escape from the promoter and transition to productive elongation, travel down the body of the gene and stop correctly once the termination signal has been reached. At the same time, the mRNA must be capped, spliced and polyadenylated, then exported to the cytoplasm for translation when finished. Each step in this process is highly interdependent and subject to multiple levels of regulation.2,10-12 Developing a precise understanding of the mechanisms that enforce these multiple levels of regulation and the ways in which they interact with each other is now the central challenge in the field of gene expression.
Recently, it has been shown that several different subunits of the nuclear pore, a 60 MDa complex best known for its role in nucleocytoplasmic transport, also play a role in upstream transcriptional regulation. Nuclear pore proteins (nucleoporins) have been found in physical association with both active and repressed regions of the budding yeast, fruit fly, rat and human genomes.13-21 Importantly, a subset of nucleoporins interact with DNA in the nuclear interior, independently of the presence of RNA polymerase II. This finding strongly suggests that proteins of the nuclear pore play a role in transcription that is independent of mRNA transport.19,20 Consistent with such a role, experiments in both flies and rats have shown that knock-down or overexpression of nucleoporins reciprocally alters mRNA levels of the genes with which they interact.14,15,19,20,22 However, almost nothing is known about the nature of the nuclear pore-genome interaction, or the mechanics of how nucleoporins contribute to transcriptional regulation.
Recently, though, we have identified a specific mechanism through which nuclear pore proteins can influence transcription. Deletion of either of two particular subunits of the nuclear pore, NUP120 or NUP133, results in a loss of repression of SUC2, one of the canonical model genes in the Saccharomyces cerevisiae system on which our current understanding of transcriptional regulation has been built. This loss of repression is not attributable to defects in the nucleocytoplasmic shuttling of Mig1, the DNA-binding factor primarily responsible for inhibition of SUC2 transcription. Instead, nuclear Mig1 must physically associate with intact nuclear pore complexes (NPCs) in order to bind DNA and repress transcription. In other words, the ability of the repressor to find its site in the DNA is dependent on its interaction with NPCs; both interaction with NPCs and DNA binding are lost if either NUP120 or NUP133 is deleted. Viewed in the context of earlier work, this result suggests that nuclear pore proteins function as transcriptional regulators by exerting a global influence on chromatin biology, nuclear organization, or both.
Different Biological Functions for Nuclear Pores
Nuclear pores were first identified as electron-dense structures visible in microscopic analyses of the nuclear membrane performed during the 1950s.23-28 Over the next 25 y, their universality in eukaryotic cells, as well as a consensus regarding their overall shape and size, became well established. By 1975 two distinct but mutually compatible functions of nuclear pores had been proposed: nucleocytoplasmic transport and chromatin organization.29 NPCs are now known to be the sole conduit through which macromolecules are exchanged between the nucleus and cytoplasm (reviewed in refs. 30 and 31). The second postulated function, which was predicated on numerous observations of the physical proximity of chromatin to nuclear pores,32-39 has received less attention. Recent work, though, has reintroduced the idea that this physical juxtaposition may indicate a functional link.
ChIP,13-15,40 ChIP-chip16-19 and chromatin cleavage20,41 analyses have shown that subunits of the nuclear pore, or nucleoporins, interact with both active and inactive regions of the genomes of budding yeast, fruit flies, rats and humans (Table 1). Several lines of evidence suggest that these interactions serve a purpose other than facilitating the export of newly synthesized transcripts. First and simplest, some nucleoporins preferentially associate with regions of the genome that are not transcribed.14,18 The next argument for a transport-independent function stems from the observation that some nucleoporins localize to both the nuclear periphery and the nuclear interior,42-46 also called the nucleoplasm; nucleoplasmic pools of these nucleoporins have been shown to interact with loci that are also found in the nuclear interior and to influence steady-state levels of mRNA transcribed from these genes.14,19,20 Finally, work done in fruit flies has directly demonstrated that the association between nucleoporins and active genes is stable to treatment with RNase A and thus not tethered by RNA molecules in the process of being exported.14
Table 1. Nuclear pore proteins in physical interaction with genomic loci.
| Nucleoporin | Yeast homolog | References | |
|---|---|---|---|
| |
Saccharomyces cerevisiae |
|
|
| Active |
Nup60 |
|
16 |
| Nup116 |
|
16 |
|
| Nic96 |
|
16 |
|
| Mlp1 |
|
16 |
|
| Mlp2 |
|
16 |
|
| Active and repressed |
Nup2 |
|
16, 17, 41 |
| Nup145C |
|
13, 16 |
|
| Nup53 |
|
13 |
|
| Nup133 |
|
13 |
|
| Pom152 |
|
13 |
|
| |
|
|
|
| |
Drosophila melanogaster |
|
|
| Active |
mTOR |
Mlp1 |
19, 22 |
| Nup153 |
Nup60 |
19 |
|
| Nup50 |
none |
20 |
|
| Nup62 |
Nsp1 |
20 |
|
| Nup98 |
Nup145N |
14, 20 |
|
| Sec13 |
Sec13 |
14 |
|
| Repressed |
Nup88 |
Nup82 |
14 |
| |
|
|
|
| |
Rattus norvegicus |
|
|
| Active and repressed |
Nup155 |
Nup170 |
15 |
| |
|
|
|
| |
Homo sapiens |
|
|
| Active and repressed | Nup93 | Nic96 | 18 |
Since RNA-only models are ruled out, the way in which nucleoporins interact with the genome is unclear. With one possible exception,47 nucleoporins do not contain recognizable domains for binding DNA or chromatin directly. It therefore seems likely that they recognize specific loci through another protein or proteins, but the identity of these is currently unknown. However, examining the function of nucleoporin-DNA interactions may provide a hint about mechanism of interaction. In budding yeast, nucleoporins interact specifically with intergenic regions, places where the transcriptional machinery and the DNA first come into contact; this contact occurs independently of active transcription.13,41 Two Drosophila nucleoporins, Nup98 and Sec13, have been shown to make contact with their target loci prior to RNA polymerase.14 Consistent with these data, the yeast nucleoporin Nup2 has been shown to contact the promoter of the GAL1,10 locus prior to TBP.41 Sequence specific transcription factors, chromatin remodelers and chromatin modifying complexes are therefore the most logical candidates for proteins that serve as intermediates between nucleoporins and DNA, since only they arrive at the promoter prior to TBP.
Regulation of SUC2 Transcription is Linked to Nuclear Pores
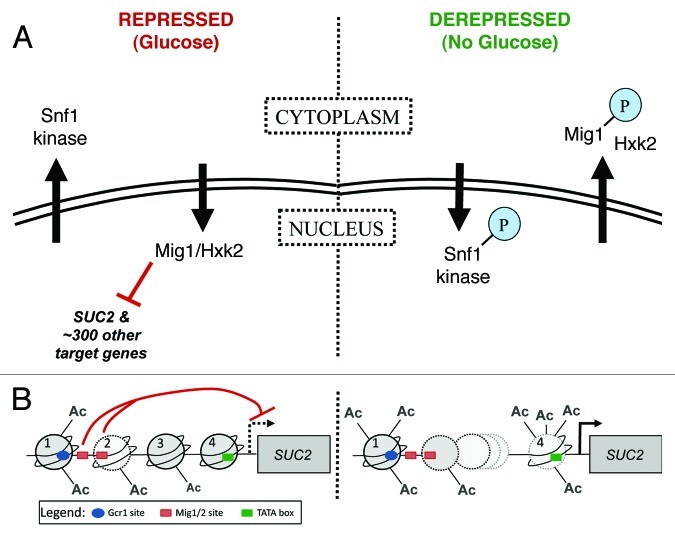
Our understanding of glucose-regulated gene expression in the model eukaryote Saccharomyces cerevisiae has been established largely through study of the SUC2 promoter. Work completed over the past 25 y has thoroughly identified the set of proteins that control its activity (Fig. 1), although the way in which they work together is still not fully understood. Two factors central to this regulatory system are the Snf1 kinase and the DNA-binding repressor protein Mig1. Together they control transcription of not only SUC2, but approximately 300 other genes.48
Figure 1. Regulation of glucose-repressed gene expression in Saccharomyces cerevisiae. Regulation of SUC2 expression is well characterized, and the proteins that carry out this regulation have been studied extensively. The SUC2 system therefore provides a good model for understanding how other genes controlled by the same factors are regulated. (A) A kinase-transcription factor pair work together to control expression of SUC2. Left panel. In the presence of glucose (repressed), the C2H2 zinc finger protein Mig1 binds DNA and inhibits transcription of SUC2, plus approximately 300 other genes. Complete inhibition also requires the hexokinase Hxk2. Snf1 kinase, a structural and functional homolog of mammalian AMPK, is found in the cytoplasm. Right panel. When glucose is depleted or withdrawn (derepressed), Snf1 is phosphorylated by upstream kinases, enters the nucleus and phosphorylates Mig1, which is then exported together with Hxk2. For detailed review, see reference 48. (B) Chromatin structure of the SUC2 promoter. Left panel. In the presence of glucose (repressed), four nucleosomes (numbered 1 through 4) cover the SUC2 promoter; a gradient of histone H3 acetylation decreases from 5′ to 3′.55 Two GC-rich sequences are required for repression of transcription (red t-bars) from the TSS (dashed black arrow).88 These sites are interchangeably bound in vivo by Mig1 and its homolog Mig2; function of the former, but not the latter, is regulated by the Snf1 kinase.48 The average location of the first Mig1/2 site (left-most red rectangle) is between nucleosomes 1 and 2. The second Mig1/2 site (right-most red rectangle) is close to the end of the DNA covered by nucleosome 289-91; nucleosome and repressor may compete for occupancy of this site. The TATA box (green rectangle) is covered by nucleosome 4.89-93 Right panel. Full induction of transcription requires the Swi/Snf chromatin remodeling complex.57,90,91,94-98 Swi/Snf associates more stably with promoter nucleosomes that have been acetylated by SAGA and NuA4 complexes57; in the absence of glucose, acetylation of H3 and H4 tails increases for all nucleosomes. The DNA formerly covered by two nucleosomes (nucleosomes 2 and 3) is more frequently covered by only one. Nucleosome 4, covering the TATA box, is hyperacetlyated and becomes unstable.55 Initiation occurs at the TSS (black arrow). Additionally required for full induction of SUC2 transcription are the nucleosome remodeling protein Spt699,100 and the transcriptional activator Gcr1 (blue oval, Gcr1 binding site).90,101-104 Nuclear pore proteins also interact with the SUC2 promoter13; it is important to learn how they collaborate with the well-characterized transcription factors described here.
Recent work has shown that the promoters of three Mig1 targets—SUC2, HXK1 and GAL1, 1013,41—are also bound by nuclear pore proteins; furthermore, these genes very likely spend some time interacting with intact NPCs, since GFP tagging shows they periodically visit the nuclear periphery when repressed and localize there for prolonged intervals when active.13,49,50 We therefore wondered if the Snf1 and Mig1 proteins themselves might also interact with NPCs. We found that Snf1 co-fractionates with intact NPCs only in the absence of glucose, when SUC2, HXK1 and GAL1,10 are actively transcribed and stably associated with nuclear pores. In contrast, Mig1 co-fractionates with NPCs only in the presence of glucose, when these three target genes are repressed and appear to interact with nuclear pores transiently.13 However, Snf1 and Mig1 undergo nucleocytoplasmic transport in response to changes in the availability of glucose and must associate with the nuclear pores for this purpose. Both proteins cross the pore when glucose appears as well as when it disappears; therefore it is not straightforward to explain the differential physical association we observe by invoking a transport-only function, but we still considered it a possibility. Accordingly, we looked for a way to test whether the association of these regulators with nuclear pores was only a reflection of their transport. It has long been known that deletion of HXK2, which encodes the predominant form of hexokinase in budding yeast, causes defects in Mig1-dependent glucose repression.51-53 We found that in an hxk2∆ mutant, Mig1 is properly localized to the nucleus in response to the appearance of glucose; however, despite being localized to the nucleus in this mutant, Mig1 no longer co-fractionates with nuclear pores.13 In other words, our analysis of an hxk2∆ mutant shows that repressor function of Mig1 requires its physical association with NPCs in a way that is independent of the protein’s nucleocytoplasmic transport.
Specific subunits of the NPC Mediate Binding of the Mig1 Repressor to Target Promoters
We next asked whether mutation of nuclear pores themselves might cause defects in the transcriptional regulation of SUC2. Of particular interest were cells deleted for either NUP120 or NUP133, in which we observed a loss of repression of SUC2 approximately equal to that seen on deleting MIG1 itself.54 Since a reduction in export of SUC2 mRNA would not be expected to lead to an increase in levels of the translated protein product, we reasoned that these nucleoporins likely impact regulation of SUC2 expression at the level of transcription. Visualization of Mig1-GFP shows that import of Mig1 into the nucleus does not require Nup120 or Nup133.54 Given our above-described data on Hxk2, though, we wondered if Nup120 and Nup133 might be required for the interaction of Mig1 with NPCs. We found that this is true; although Mig1 is correctly transported into and out of the nucleus in the absence of these nucleoporins, it is no longer able to co-fractionate with nuclear pores. In the absence of Nup120 or Nup133, nuclear Mig1 is also unable to bind its site in DNA.54 This striking result provides a clear explanation of how these nucleoporins influence SUC2 repression.
How Nuclear Pores Might Influence Gene Expression: Models and Possible Mechanisms
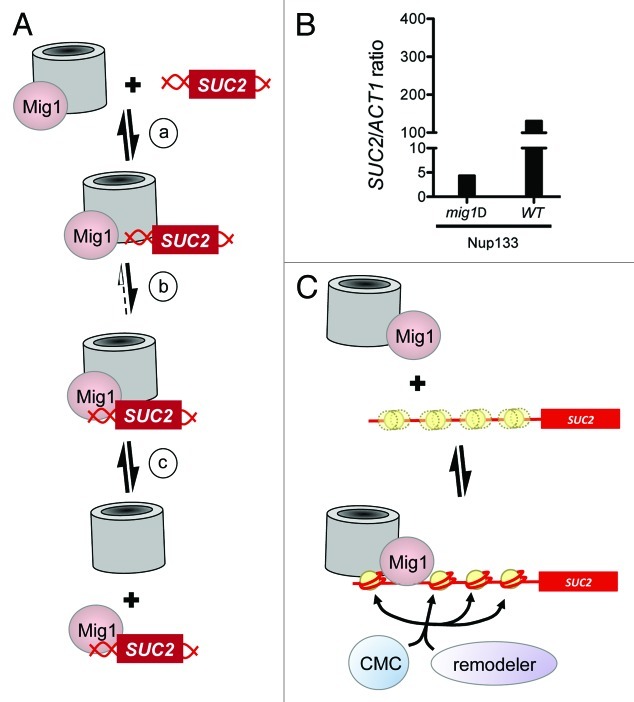
Why does MIg1 require Nup120 and Nup133 to bind DNA? We have shown that Mig1 co-purifies with intact NPCs in glucose-grown cells, in which the repressor binds its target loci.13,54 We have also shown that both Nup133 and Pom152 interact with the SUC2 promoter in glucose-grown cells.54 Nup133 is a structural component of the NPC, and Pom152 is an integral nuclear membrane protein. We therefore interpret this result to mean that in the presence of glucose, the repressed SUC2 promoter makes transient contact with intact NPCs embedded in the nuclear membrane. This interpretation is consistent with the observation that the SUC2 locus can be seen to periodically “visit” the nuclear periphery in glucose grown cells.13 An obvious possibility, therefore, is that during these visits, the NPC facilitates binding of the repressor protein to the DNA by simultaneously interacting with both. Bringing together a gene and its transcriptional regulator in a particular location, at the nuclear periphery, might not only facilitate DNA binding, but also reduce the time it takes that transcription factor to search the genome for its site.
Drawing the simplest possible analogy to cooperative DNA binding by two transcription factors (Fig. 2A), we can infer two significant corollaries to this model. First, SUC2 is required to contact the NPC independently of Mig1. As noted above, this second, Mig1-independent contact may be direct, but more likely occurs through some other, currently unidentified factor. Second, deletion of any one of the components shown in Figure 2A has the potential to reduce the affinity with which the others interact. Consistent with these requirements, we observe that in glucose-grown mig1Δ cells, Nup133 still binds the SUC2 promoter, although the strength of the interaction appears to be reduced (Fig. 2B).
Figure 2. How nucleoporins might influence access to a specific site in DNA. (A) Model for NPC facilitated Mig1 binding to DNA. (a) In the presence of glucose, Mig1 associates with NPCs; the SUC2 locus contacts NPCs transiently. (b) Increased local concentration of both Mig1 and its consensus site facilitates DNA binding by the repressor. (c) The repressor-bound promoter can dissociate from the NPC. (B) Interaction of Nup133 with the SUC2 promoter is reduced in mig1Δ cells. TAP-tagged Nup133 was immunoprecipitated from mig1Δ or wild type cells grown in media containing glucose as the carbon source, then fixed with formaldehyde as described previously.13 Crosslinks were reversed and PCR was used to amplify the promoters of SUC2 and ACT1 (negative control) from recovered material. Amplified target is expressed as a ratio of SUC2/ACT1, normalized to the amount of product amplified whole cell extracts (input). Adding increasing amounts of input DNA shows that amplification of the product is linear. A representative experiment is shown; similar results were obtained for Nup145C (not shown). (C) A model for collaborative nucleosome positioning. Top panel. Mig1 associates with nuclear pores as in (A) above. SUC2 is not associated with nuclear pores, and the nucleosomes across its promoter are poorly positioned (three yellow circles, one distinct and two indistinct, represent the average position of one nucleosome, over time and across a population). Bottom panel. One or more subunits of the nuclear pore associate with the SUC2 locus, serving as a marker or barrier against which chromatin remodelers can position a single nucleosome, thus seeding an ordered array. Alternatively, nuclear pore proteins may direct the activity of chromatin modifiers such as SAGA and NuA4, which in turn direct the activity of remodelers such as Swi/Snf. Models shown in A and C are not mutually exclusive; for example, nuclear pores contribute to nucleosome positioning at step (b) of model A.
Although interesting, this finding is far from conclusive, and further work must be done to test the accuracy of the idea. If it is correct, classical biochemical theory predicts that adding either individual nucleoporin(s) or intact nuclear pores to in vitro DNA-binding reactions should increase the affinity of Mig1 for its site. However, if in vivo nuclear context is important—perhaps for reducing search time by compressing three dimensions into two – then in vitro analyses such as this may not be informative. In which case, quantitative measurement of in vivo DNA binding at different nuclear locations, either by fluorescent resonance energy transfer (FRET) or bimolecular fluorescence complementation (BiFC), may provide the best test of this model. It will also be critical to definitively address the question of where in the yeast nucleus promoter-nucleoporin interactions truly occur. Circumstantial evidence—the periodic localization of the DNA to the nuclear periphery, together with the relative immobility of the nucleoporins we observe to interact with it—strongly suggests that intact, membrane-embedded NPCs influence Mig1 DNA binding. However, at this time, we cannot rule out a role for soluble nucleoplasmic pools of nucleoporins. Determining whether an interaction occurs in one or the other of two specific locations—the nuclear periphery or the nucleoplasm—may seem straightforward at first. In reality, the need for precise localization in a small nucleus, together with the likelihood that no more than a very few molecules of nucleoporin are localized to the nuclear interior, means that a combination of super-resolution microscopy and single molecule detection techniques will be necessary to answer this question in a satisfactory way.
A second model, not mutually exclusive with the one described above, is that NPCs collaborate with chromatin modifying complexes and chromatin remodelers to fine-tune the post-translational modification and/or position of nucleosomes over many Mig1 target promoters (Fig. 2C). In repressed wild type cells, nucleosomes 1 and 2 of the SUC2 promoter are at least 2- to 5-fold more acetylated than nucleosome 455; 1 and 2 flank the first Mig1 binding site, 2 covers the second Mig1 binding site and 4 covers the TATA box (Fig. 1B). The effect of histone acetylation on Mig1 binding has not been studied directly, but modeling of nucleosome positioning data suggests that these are less stably associated with the DNA in the absence of glucose.56 A change in the targeting or activity of histone acetyltransferases and/or in acetylation-dependent Swi/Snf remodeling, might occur in nup120Δ and nup133Δ mutants, thus altering the position or occupancy of promoter nucleosomes so that binding sites for Mig1 become obscured. In this model, Mig1 cannot repress transcription because it is unable to outcompete these nucleosomes.
Some data exist to support a model in which nucleoporins influence transcriptional regulation by co-operating with chromatin modifying complexes, particularly those that control histone acetylation. This is significant, since acetylation state is believed to be an important determinant of the affinity between nucleosomes and DNA. In Drosophila, the histone acetyltransferase MOF has been found in complex with both Nup153 and the NPC associated protein mTOR.22 Nup153 has also been found to preferentially interact with genomic loci where H4K16Ac-containing nucleosomes are enriched.19 In Saccharomyces, the MOF homolog Esa1, catalytic subunit of NuA4, constitutively associates with the SUC2 promoter.57 Deletion of ESA1 is synthetically defective with deletion of NUP120, suggesting the products of these genes share some common function58; this interesting possibility has not yet been explored. Yeast Gcn5, the catalytic subunit of SAGA, has been shown to associate directly with Mlp1 and 2, homologs of Drosophila mTOR.59 Most recently, the Rattus norvegicus nucleoporin Nup155 has been shown to interact with the histone deacetylase HDAC4 and to regulate the association of specific genomic loci with that nucleoporin.15
An intriguing but currently even less-well elaborated connection may exist between nuclear pores and chromatin remodeling complexes. The RSC complex is essential both for the localization of NPCs to the nuclear envelope and for normal nuclear morphology, leading to the suggestion that RSC might be required for establishing contacts between chromatin and the NPC.60 SUC2 is not known to be a target of RSC, but the promoters of other Mig1-regulated genes, including HXK1 and FBP1, are bound at a low level by RSC in glucose-grown cells.61 Again, further work must be done to determine whether changes in the position or acetylation state of promoter nucleosomes are responsible for the failure of Mig1 to bind DNA in nup120Δ and nup133Δ cells.
Relevance to Leukemia: Nucleoporins as Transcription Factors in Human Cells
Genes encoding components of the NPC are the target of chromosomal translocations in several different types of cancer62-66; possibly the best studied of these translocations are found in hematological malignancies, including myelodysplastic syndrome (MDS), acute myeloid leukemia (AML), chronic myeloid leukemia (CML) and T-cell acute lymphocytic leukemia (T-ALL).67-69 Different mechanisms have been proposed to explain how nucleoporin fusions contribute to the development of disease. One hypothesis is that interaction of nucleoporin chimeras with endogenous nucleoporins or RNA export factors may contribute to their leukemogenic properties.70-72 Another is that nucleoporins are involved in spindle formation and this function is disrupted in oncogenic fusions, leading to aneuploidy.69 However, in many cases the translocation partner of the nucleoporin is a transcription factor. NUP98-HOXA9, probably the best-characterized of these leukemogenic nucleoporin fusion proteins,73-77 has long been known to have the ability to bind to DNA and interact with components of the basal transcription machinery, leading to aberrant gene expression.14,20,78-82 The recent work described above strongly suggests that nucleoporins play a role in normal transcriptional regulation; it seems likely, then, that conscription of this function is a primary mechanism of oncogenesis.69 This conclusion is supported by studies of the fusion proteins themselves.
Of particular relevance to our work on Mig1 is a recent study linking NUP98 with the transcription factor AES. AES is a homolog of the Mig1-interacting protein Tup1; both AES and Tup1 are Groucho family proteins that act as co-repressors of transcription. Importantly, AES interacts with both wild type NUP98 and NUP98-HOXA9, consistent with the idea of a conserved, nucleoporin-dependent mechanism of transcriptional regulation that is hijacked in leukemogenesis. Together, AES and NUP98-HOXA9 cooperate to regulate transcription, cell transformation and the impairment of hematopoietic differentiation.79,80
Functional and physical links have also been described between NUP98 fusions and chromatin modifiers. The FG repeats in the NUP98 portion of a NUP98-HOXA9 fusion physically interact with CREB-binding protein (CBP)/p300; transcriptional activation of a reporter gene by NUP98-HOXA9 is dependent on the histone acetyltransferase activity of CBP/p300.81 Chromosomal translocations also fuse NUP98 to the lysine methyltransferases NSD1 and MLL, to the lysine demethylase JARID1A and to the poorly-characterized protein PHF23, which recognizes methylated H3K4.69 NUP98-NSD1 promotes the methylation of histone H3-Lys36 (H3K36) at loci encoding several proto-oncogenes, including HoxA7, HoxA9, HoxA10 and Meis1; enforced activation of these transcription factors then blocks differentiation.83 NUP98-JARID1A and NUP98-PHF23 fusion proteins inhibit demethylation of histone H3-Lys 4 (H3K4me3) at loci encoding lineage-specific transcription factors, again blocking differentiation.84,85 It is not clear how the deregulation of lysine methylation by these NUP98 chimeras relates to nucleoporin-mediated regulation of gene expression described above. However, in Drosophila cells, the methylation state of H3K36 is linked to levels of H4K16 acetylation,86,87 and nucleoporins preferentially bind loci enriched for H4K16Ac.19,20
Conclusions and Future Directions
These observations bring up an important question: to what extent is transcriptional regulation by nucleoporins conserved in healthy and cancerous cells? The work summarized above suggests that it is, to at least some extent. Where this is true, with what partner proteins do they work, and how do these differ at different stages of transcript production? How can their contribution be integrated into existing models of gene regulation? Addressing these questions should help us to understand how multiple steps of transcript synthesis are coupled, bringing us closer to a full understanding of how gene expression is regulated in a nuclear context.
Acknowledgments
We thank George Santangelo for important theoretical contributions; Dan Larson and Vytas Bankaitis for comments on the manuscript and Patrick Varga-Weisz for comments on the manuscript, as well as for a generous amount of time spent in helpful discussions of the text. This work was supported by National Institutes of Health grant GM083309 to K.A.W.
Glossary
Abbreviations:
- NPC
nuclear pore complex
- TBP
TATA binding protein
- GFP
green fluorescent protein
- TSS
transcription start site
- HDAC
histone deacetylase
Footnotes
Previously published online: www.landesbioscience.com/journals/nucleus/article/22427
References
- 1.Cosma MP. Ordered recruitment: gene-specific mechanism of transcription activation. Mol Cell. 2002;10:227–36. doi: 10.1016/S1097-2765(02)00604-4. [DOI] [PubMed] [Google Scholar]
- 2.Orphanides G, Reinberg D. A unified theory of gene expression. Cell. 2002;108:439–51. doi: 10.1016/S0092-8674(02)00655-4. [DOI] [PubMed] [Google Scholar]
- 3.Krishnamurthy S, Hampsey M. Eukaryotic transcription initiation. Curr Biol. 2009;19:R153–6. doi: 10.1016/j.cub.2008.11.052. [DOI] [PubMed] [Google Scholar]
- 4.Weake VM, Workman JL. Inducible gene expression: diverse regulatory mechanisms. Nat Rev Genet. 2010;11:426–37. doi: 10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 5.Hahn S, Young ET. Transcriptional regulation in Saccharomyces cerevisiae: transcription factor regulation and function, mechanisms of initiation, and roles of activators and coactivators. Genetics. 2011;189:705–36. doi: 10.1534/genetics.111.127019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Malik S, Roeder RG. The metazoan Mediator co-activator complex as an integrative hub for transcriptional regulation. Nat Rev Genet. 2010;11:761–72. doi: 10.1038/nrg2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Saunders A, Core LJ, Lis JT. Breaking barriers to transcription elongation. Nat Rev Mol Cell Biol. 2006;7:557–67. doi: 10.1038/nrm1981. [DOI] [PubMed] [Google Scholar]
- 8.Bryant GO, Ptashne M. Independent recruitment in vivo by Gal4 of two complexes required for transcription. Mol Cell. 2003;11:1301–9. doi: 10.1016/S1097-2765(03)00144-8. [DOI] [PubMed] [Google Scholar]
- 9.Carey MF, Peterson CL, Smale ST. Transcriptional Regulation in Eukaryotes: Concepts, Strategies, & Techniques. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press, 2009. [Google Scholar]
- 10.Maniatis T, Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. doi: 10.1038/416499a. [DOI] [PubMed] [Google Scholar]
- 11.Pandit S, Wang D, Fu XD. Functional integration of transcriptional and RNA processing machineries. Curr Opin Cell Biol. 2008;20:260–5. doi: 10.1016/j.ceb.2008.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Proudfoot NJ, Furger A, Dye MJ. Integrating mRNA processing with transcription. Cell. 2002;108:501–12. doi: 10.1016/S0092-8674(02)00617-7. [DOI] [PubMed] [Google Scholar]
- 13.Sarma NJ, Haley TM, Barbara KE, Buford TD, Willis KA, Santangelo GM. Glucose-responsive regulators of gene expression in Saccharomyces cerevisiae function at the nuclear periphery via a reverse recruitment mechanism. Genetics. 2007;175:1127–35. doi: 10.1534/genetics.106.068932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–83. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kehat I, Accornero F, Aronow BJ, Molkentin JD. Modulation of chromatin position and gene expression by HDAC4 interaction with nucleoporins. J Cell Biol. 2011;193:21–9. doi: 10.1083/jcb.201101046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casolari JM, Brown CR, Komili S, West J, Hieronymus H, Silver PA. Genome-wide localization of the nuclear transport machinery couples transcriptional status and nuclear organization. Cell. 2004;117:427–39. doi: 10.1016/S0092-8674(04)00448-9. [DOI] [PubMed] [Google Scholar]
- 17.Dilworth DJ, Tackett AJ, Rogers RS, Yi EC, Christmas RH, Smith JJ, et al. The mobile nucleoporin Nup2p and chromatin-bound Prp20p function in endogenous NPC-mediated transcriptional control. J Cell Biol. 2005;171:955–65. doi: 10.1083/jcb.200509061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brown CR, Kennedy CJ, Delmar VA, Forbes DJ, Silver PA. Global histone acetylation induces functional genomic reorganization at mammalian nuclear pore complexes. Genes Dev. 2008;22:627–39. doi: 10.1101/gad.1632708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaquerizas JM, Suyama R, Kind J, Miura K, Luscombe NM, Akhtar A. Nuclear pore proteins nup153 and megator define transcriptionally active regions in the Drosophila genome. PLoS Genet. 2010;6:e1000846. doi: 10.1371/journal.pgen.1000846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–71. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 21.Finlan LE, Sproul D, Thomson I, Boyle S, Kerr E, Perry P, et al. Recruitment to the nuclear periphery can alter expression of genes in human cells. PLoS Genet. 2008;4:e1000039. doi: 10.1371/journal.pgen.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, et al. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–23. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 23.Watson ML. Further observations on the nuclear envelope of the animal cell. J Biophys Biochem Cytol. 1959;6:147–56. doi: 10.1083/jcb.6.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Callan HG, Randall JT, Tomlin SG. An electron microscope study of the nuclear membrane. Nature. 1949;163:280. doi: 10.1038/163280a0. [DOI] [PubMed] [Google Scholar]
- 25.Callan HG, Tomlin SG. Experimental studies on amphibian oocyte nuclei. I. Investigation of the structure of the nuclear membrane by means of the electron microscope. Proc R Soc Lond B Biol Sci. 1950;137:367–78. doi: 10.1098/rspb.1950.0047. [DOI] [PubMed] [Google Scholar]
- 26.Gall JG. Observations on the nuclear membrane with the electron microscope. Exp Cell Res. 1954;7:197–200. doi: 10.1016/0014-4827(54)90054-3. [DOI] [PubMed] [Google Scholar]
- 27.Anderson E, Beams HW. Evidence from electron micrographs for the passage of material through pores of the nuclear membrane. J Biophys Biochem Cytol. 1956;2(Suppl):439–44. doi: 10.1083/jcb.2.4.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beams HW, Tahmisian TN, Anderson E, Devine RL. The structure of nuclear membrane in larval gonads of Heliothis obsoleta. Proc Soc Exp Biol Med. 1956;91:473–5. doi: 10.3181/00379727-91-22296. [DOI] [PubMed] [Google Scholar]
- 29.Aaronson RP, Blobel G. Isolation of nuclear pore complexes in association with a lamina. Proc Natl Acad Sci U S A. 1975;72:1007–11. doi: 10.1073/pnas.72.3.1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Grünwald D, Singer RH, Rout M. Nuclear export dynamics of RNA-protein complexes. Nature. 2011;475:333–41. doi: 10.1038/nature10318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoelz A, Debler EW, Blobel G. The structure of the nuclear pore complex. Annu Rev Biochem. 2011;80:613–43. doi: 10.1146/annurev-biochem-060109-151030. [DOI] [PubMed] [Google Scholar]
- 32.DuPraw E. Cell and Molecular Biology. New York: Academic Press, 1958:424. [Google Scholar]
- 33.Dupraw EJ. Cell and Molecular Biology. NY: Academic Press, 1954. [Google Scholar]
- 34.Comings DE, Okada TA. Association of nuclear membrane fragments with metaphase and anaphase chromosomes as observed by whole mount electron microscopy. Exp Cell Res. 1970;63:62–8. doi: 10.1016/0014-4827(70)90331-9. [DOI] [PubMed] [Google Scholar]
- 35.Kirschner RH, Rusli M, Martin TE. Characterization of the nuclear envelope, pore complexes, and dense lamina of mouse liver nuclei by high resolution scanning electron microscopy. J Cell Biol. 1977;72:118–32. doi: 10.1083/jcb.72.1.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chan JK, Park PC, De Boni U. Association of DNAse sensitive chromatin domains with the nuclear periphery in 3T3 cells in vitro. Biochem Cell Biol. 2000;78:67–78. doi: 10.1139/o99-074. [DOI] [PubMed] [Google Scholar]
- 37.Hutchison N, Weintraub H. Localization of DNAase I-sensitive sequences to specific regions of interphase nuclei. Cell. 1985;43:471–82. doi: 10.1016/0092-8674(85)90177-1. [DOI] [PubMed] [Google Scholar]
- 38.Markovics J, Glass L, Maul GG. Pore patterns on nuclear membranes. Exp Cell Res. 1974;85:443–51. doi: 10.1016/0014-4827(74)90148-7. [DOI] [PubMed] [Google Scholar]
- 39.Arlucea J, Andrade R, Alonso R, Aréchaga J. The nuclear basket of the nuclear pore complex is part of a higher-order filamentous network that is related to chromatin. J Struct Biol. 1998;124:51–8. doi: 10.1006/jsbi.1998.4054. [DOI] [PubMed] [Google Scholar]
- 40.Light WH, Brickner DG, Brand VR, Brickner JH. Interaction of a DNA zip code with the nuclear pore complex promotes H2A.Z incorporation and INO1 transcriptional memory. Mol Cell. 2010;40:112–25. doi: 10.1016/j.molcel.2010.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmid M, Arib G, Laemmli C, Nishikawa J, Durussel T, Laemmli UK. Nup-PI: the nucleopore-promoter interaction of genes in yeast. Mol Cell. 2006;21:379–91. doi: 10.1016/j.molcel.2005.12.012. [DOI] [PubMed] [Google Scholar]
- 42.Griffis ER, Altan N, Lippincott-Schwartz J, Powers MA. Nup98 is a mobile nucleoporin with transcription-dependent dynamics. Mol Biol Cell. 2002;13:1282–97. doi: 10.1091/mbc.01-11-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Griffis ER, Craige B, Dimaano C, Ullman KS, Powers MA. Distinct functional domains within nucleoporins Nup153 and Nup98 mediate transcription-dependent mobility. Mol Biol Cell. 2004;15:1991–2002. doi: 10.1091/mbc.E03-10-0743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, et al. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol. 2001;153:1465–78. doi: 10.1083/jcb.153.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–95. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rabut G, Doye V, Ellenberg J. Mapping the dynamic organization of the nuclear pore complex inside single living cells. Nat Cell Biol. 2004;6:1114–21. doi: 10.1038/ncb1184. [DOI] [PubMed] [Google Scholar]
- 47.Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci U S A. 2006;103:17801–6. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Santangelo GM. Glucose signaling in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 2006;70:253–82. doi: 10.1128/MMBR.70.1.253-282.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cabal GG, Genovesio A, Rodriguez-Navarro S, Zimmer C, Gadal O, Lesne A, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441:770–3. doi: 10.1038/nature04752. [DOI] [PubMed] [Google Scholar]
- 50.Taddei A, Van Houwe G, Hediger F, Kalck V, Cubizolles F, Schober H, et al. Nuclear pore association confers optimal expression levels for an inducible yeast gene. Nature. 2006;441:774–8. doi: 10.1038/nature04845. [DOI] [PubMed] [Google Scholar]
- 51.Entian KD. Genetic and biochemical evidence for hexokinase PII as a key enzyme involved in carbon catabolite repression in yeast. Mol Gen Genet. 1980;178:633–7. doi: 10.1007/BF00337871. [DOI] [PubMed] [Google Scholar]
- 52.Neigeborn L, Carlson M. Genes affecting the regulation of SUC2 gene expression by glucose repression in Saccharomyces cerevisiae. Genetics. 1984;108:845–58. doi: 10.1093/genetics/108.4.845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Neigeborn L, Carlson M. Mutations causing constitutive invertase synthesis in yeast: genetic interactions with snf mutations. Genetics. 1987;115:247–53. doi: 10.1093/genetics/115.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarma NJ, Buford TD, Haley T, Barbara-Haley K, Santangelo GM, Willis KA. The nuclear pore complex mediates binding of the Mig1 repressor to target promoters. PLoS One. 2011;6:e27117. doi: 10.1371/journal.pone.0027117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Boukaba A, Georgieva EI, Myers FA, Thorne AW, López-Rodas G, Crane-Robinson C, et al. A short-range gradient of histone H3 acetylation and Tup1p redistribution at the promoter of the Saccharomyces cerevisiae SUC2 gene. J Biol Chem. 2004;279:7678–84. doi: 10.1074/jbc.M310849200. [DOI] [PubMed] [Google Scholar]
- 56.Zawadzki KA, Morozov AV, Broach JR. Chromatin-dependent transcription factor accessibility rather than nucleosome remodeling predominates during global transcriptional restructuring in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:3503–13. doi: 10.1091/mbc.E09-02-0111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Geng F, Laurent BC. Roles of SWI/SNF and HATs throughout the dynamic transcription of a yeast glucose-repressible gene. EMBO J. 2004;23:127–37. doi: 10.1038/sj.emboj.7600035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell L, Lambert JP, Gerdes M, Al-Madhoun AS, Skerjanc IS, Figeys D, et al. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol Cell Biol. 2008;28:2244–56. doi: 10.1128/MCB.01653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Luthra R, Kerr SC, Harreman MT, Apponi LH, Fasken MB, Ramineni S, et al. Actively transcribed GAL genes can be physically linked to the nuclear pore by the SAGA chromatin modifying complex. J Biol Chem. 2007;282:3042–9. doi: 10.1074/jbc.M608741200. [DOI] [PubMed] [Google Scholar]
- 60.Titus LC, Dawson TR, Rexer DJ, Ryan KJ, Wente SR. Members of the RSC chromatin-remodeling complex are required for maintaining proper nuclear envelope structure and pore complex localization. Mol Biol Cell. 2010;21:1072–87. doi: 10.1091/mbc.E09-07-0615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ng HH, Robert F, Young RA, Struhl K. Genome-wide location and regulated recruitment of the RSC nucleosome-remodeling complex. Genes Dev. 2002;16:806–19. doi: 10.1101/gad.978902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gould VE, Orucevic A, Zentgraf H, Gattuso P, Martinez N, Alonso A. Nup88 (karyoporin) in human malignant neoplasms and dysplasias: correlations of immunostaining of tissue sections, cytologic smears, and immunoblot analysis. Hum Pathol. 2002;33:536–44. doi: 10.1053/hupa.2002.124785. [DOI] [PubMed] [Google Scholar]
- 63.Graux C, Cools J, Melotte C, Quentmeier H, Ferrando A, Levine R, et al. Fusion of NUP214 to ABL1 on amplified episomes in T-cell acute lymphoblastic leukemia. Nat Genet. 2004;36:1084–9. doi: 10.1038/ng1425. [DOI] [PubMed] [Google Scholar]
- 64.Soman NR, Wogan GN, Rhim JS. TPR-MET oncogenic rearrangement: detection by polymerase chain reaction amplification of the transcript and expression in human tumor cell lines. Proc Natl Acad Sci U S A. 1990;87:738–42. doi: 10.1073/pnas.87.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ma Z, Hill DA, Collins MH, Morris SW, Sumegi J, Zhou M, et al. Fusion of ALK to the Ran-binding protein 2 (RANBP2) gene in inflammatory myofibroblastic tumor. Genes Chromosomes Cancer. 2003;37:98–105. doi: 10.1002/gcc.10177. [DOI] [PubMed] [Google Scholar]
- 66.Köhler A, Hurt E. Gene regulation by nucleoporins and links to cancer. Mol Cell. 2010;38:6–15. doi: 10.1016/j.molcel.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 67.Xu S, Powers MA. Nuclear pore proteins and cancer. Semin Cell Dev Biol. 2009;20:620–30. doi: 10.1016/j.semcdb.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lam DH, Aplan PD. NUP98 gene fusions in hematologic malignancies. Leukemia. 2001;15:1689–95. doi: 10.1038/sj.leu.2402269. [DOI] [PubMed] [Google Scholar]
- 69.Gough SM, Slape CI, Aplan PD. NUP98 gene fusions and hematopoietic malignancies: common themes and new biologic insights. Blood. 2011;118:6247–57. doi: 10.1182/blood-2011-07-328880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Funasaka T, Nakano H, Wu Y, Hashizume C, Gu L, Nakamura T, et al. RNA export factor RAE1 contributes to NUP98-HOXA9-mediated leukemogenesis. Cell Cycle. 2011;10:1456–67. doi: 10.4161/cc.10.9.15494. [DOI] [PubMed] [Google Scholar]
- 71.Ren Y, Seo HS, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc Natl Acad Sci U S A. 2010;107:10406–11. doi: 10.1073/pnas.1005389107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Takeda A, Sarma NJ, Abdul-Nabi AM, Yaseen NR. Inhibition of CRM1-mediated nuclear export of transcription factors by leukemogenic NUP98 fusion proteins. J Biol Chem. 2010;285:16248–57. doi: 10.1074/jbc.M109.048785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Nakamura T, Largaespada DA, Lee MP, Johnson LA, Ohyashiki K, Toyama K, et al. Fusion of the nucleoporin gene NUP98 to HOXA9 by the chromosome translocation t(7;11)(p15;p15) in human myeloid leukaemia. Nat Genet. 1996;12:154–8. doi: 10.1038/ng0296-154. [DOI] [PubMed] [Google Scholar]
- 74.Raza-Egilmez SZ, Jani-Sait SN, Grossi M, Higgins MJ, Shows TB, Aplan PD. NUP98-HOXD13 gene fusion in therapy-related acute myelogenous leukemia. Cancer Res. 1998;58:4269–73. [PubMed] [Google Scholar]
- 75.Nakamura T, Yamazaki Y, Hatano Y, Miura I. NUP98 is fused to PMX1 homeobox gene in human acute myelogenous leukemia with chromosome translocation t(1;11)(q23;p15) Blood. 1999;94:741–7. [PubMed] [Google Scholar]
- 76.Panagopoulos I, Isaksson M, Billström R, Strömbeck B, Mitelman F, Johansson B. Fusion of the NUP98 gene and the homeobox gene HOXC13 in acute myeloid leukemia with t(11;12)(p15;q13) Genes Chromosomes Cancer. 2003;36:107–12. doi: 10.1002/gcc.10139. [DOI] [PubMed] [Google Scholar]
- 77.Jankovic D, Gorello P, Liu T, Ehret S, La Starza R, Desjobert C, et al. Leukemogenic mechanisms and targets of a NUP98/HHEX fusion in acute myeloid leukemia. Blood. 2008;111:5672–82. doi: 10.1182/blood-2007-09-108175. [DOI] [PubMed] [Google Scholar]
- 78.Ghannam G, Takeda A, Camarata T, Moore MA, Viale A, Yaseen NR. The oncogene Nup98-HOXA9 induces gene transcription in myeloid cells. J Biol Chem. 2004;279:866–75. doi: 10.1074/jbc.M307280200. [DOI] [PubMed] [Google Scholar]
- 79.Yassin ER, Sarma NJ, Abdul-Nabi AM, Dombrowski J, Han Y, Takeda A, et al. Dissection of the transformation of primary human hematopoietic cells by the oncogene NUP98-HOXA9. PLoS One. 2009;4:e6719. doi: 10.1371/journal.pone.0006719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sarma NJ, Yaseen NR. Amino-terminal enhancer of split (AES) interacts with the oncoprotein NUP98-HOXA9 and enhances its transforming ability. J Biol Chem. 2011;286:38989–9001. doi: 10.1074/jbc.M111.297952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–76. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Menon BB, Sarma NJ, Pasula S, Deminoff SJ, Willis KA, Barbara KE, et al. Reverse recruitment: the Nup84 nuclear pore subcomplex mediates Rap1/Gcr1/Gcr2 transcriptional activation. Proc Natl Acad Sci U S A. 2005;102:5749–54. doi: 10.1073/pnas.0501768102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol. 2007;9:804–12. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]
- 84.Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, et al. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–51. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Reader JC, Meekins JS, Gojo I, Ning Y. A novel NUP98-PHF23 fusion resulting from a cryptic translocation t(11;17)(p15;p13) in acute myeloid leukemia. Leukemia. 2007;21:842–4. doi: 10.1038/sj.leu.2404579. [DOI] [PubMed] [Google Scholar]
- 86.Wu SF, Zhang H, Cairns BR. Genes for embryo development are packaged in blocks of multivalent chromatin in zebrafish sperm. Genome Res. 2011;21:578–89. doi: 10.1101/gr.113167.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Bell O, Wirbelauer C, Hild M, Scharf AN, Schwaiger M, MacAlpine DM, et al. Localized H3K36 methylation states define histone H4K16 acetylation during transcriptional elongation in Drosophila. EMBO J. 2007;26:4974–84. doi: 10.1038/sj.emboj.7601926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lundin M, Nehlin JO, Ronne H. Importance of a flanking AT-rich region in target site recognition by the GC box-binding zinc finger protein MIG1. Mol Cell Biol. 1994;14:1979–85. doi: 10.1128/mcb.14.3.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pérez-Ortín JE, Estruch F, Matallana E, Franco L. Fine analysis of the chromatin structure of the yeast SUC2 gene and of its changes upon derepression. Comparison between the chromosomal and plasmid-inserted genes. Nucleic Acids Res. 1987;15:6937–56. doi: 10.1093/nar/15.17.6937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wu L, Winston F. Evidence that Snf-Swi controls chromatin structure over both the TATA and UAS regions of the SUC2 promoter in Saccharomyces cerevisiae. Nucleic Acids Res. 1997;25:4230–4. doi: 10.1093/nar/25.21.4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gavin IM, Simpson RT. Interplay of yeast global transcriptional regulators Ssn6p-Tup1p and Swi-Snf and their effect on chromatin structure. EMBO J. 1997;16:6263–71. doi: 10.1093/emboj/16.20.6263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Hirschhorn JN, Bortvin AL, Ricupero-Hovasse SL, Winston F. A new class of histone H2A mutations in Saccharomyces cerevisiae causes specific transcriptional defects in vivo. Mol Cell Biol. 1995;15:1999–2009. doi: 10.1128/mcb.15.4.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hirschhorn JN, Brown SA, Clark CD, Winston F. Evidence that SNF2/SWI2 and SNF5 activate transcription in yeast by altering chromatin structure. Genes Dev. 1992;6(12A):2288–98. doi: 10.1101/gad.6.12a.2288. [DOI] [PubMed] [Google Scholar]
- 94.Schwabish MA, Struhl K. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol. 2007;27:6987–95. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dror V, Winston F. The Swi/Snf chromatin remodeling complex is required for ribosomal DNA and telomeric silencing in Saccharomyces cerevisiae. Mol Cell Biol. 2004;24:8227–35. doi: 10.1128/MCB.24.18.8227-8235.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fleming AB, Pennings S. Tup1-Ssn6 and Swi-Snf remodelling activities influence long-range chromatin organization upstream of the yeast SUC2 gene. Nucleic Acids Res. 2007;35:5520–31. doi: 10.1093/nar/gkm573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pollard KJ, Peterson CL. Role for ADA/GCN5 products in antagonizing chromatin-mediated transcriptional repression. Mol Cell Biol. 1997;17:6212–22. doi: 10.1128/mcb.17.11.6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Sudarsanam P, Cao Y, Wu L, Laurent BC, Winston F. The nucleosome remodeling complex, Snf/Swi, is required for the maintenance of transcription in vivo and is partially redundant with the histone acetyltransferase, Gcn5. EMBO J. 1999;18:3101–6. doi: 10.1093/emboj/18.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Neigeborn L, Celenza JL, Carlson M. SSN20 is an essential gene with mutant alleles that suppress defects in SUC2 transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:672–8. doi: 10.1128/mcb.7.2.672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Adkins MW, Tyler JK. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol Cell. 2006;21:405–16. doi: 10.1016/j.molcel.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 101.Türkel S, Turgut T, López MC, Uemura H, Baker HV. Mutations in GCR1 affect SUC2 gene expression in Saccharomyces cerevisiae. Mol Genet Genomics. 2003;268:825–31. doi: 10.1007/s00438-003-0808-4. [DOI] [PubMed] [Google Scholar]
- 102.Sasaki H, Uemura H. Influence of low glycolytic activities in gcr1 and gcr2 mutants on the expression of other metabolic pathway genes in Saccharomyces cerevisiae. Yeast. 2005;22:111–27. doi: 10.1002/yea.1198. [DOI] [PubMed] [Google Scholar]
- 103.Barbara KE, Haley TM, Willis KA, Santangelo GM. The transcription factor Gcr1 stimulates cell growth by participating in nutrient-responsive gene expression on a global level. Mol Genet Genomics. 2007;277:171–88. doi: 10.1007/s00438-006-0182-0. [DOI] [PubMed] [Google Scholar]
- 104.López MC, Baker HV. Understanding the growth phenotype of the yeast gcr1 mutant in terms of global genomic expression patterns. J Bacteriol. 2000;182:4970–8. doi: 10.1128/JB.182.17.4970-4978.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]