Abstract
Advances in DNA sequencing technologies, as well as refined bioinformatics methods for interpretation of complex datasets, have provided the opportunity to comprehensively assess gene expression in tumours and their surrounding microenvironment. More recently, these advances have highlighted the interplay between the immune effector mechanisms and breast cancer cell biology, emphasizing the long-recognized link between immunity and cancer. Studying immune-associated genes has not only resulted in further stratification within the broad pathological types of breast cancers, but also provided further biological insights into the complex heterogeneity within breast cancer subgroups. On the basis that anti-cancer therapies can modify the host-tumour interaction, investigators have focused their attention on the predictive value of immune parameters as markers of therapeutic anti-tumour response. We discuss the current status of immune signatures in breast cancer and some of the fundamental limitations that need to be overcome to move these discoveries into clinic.
Keywords: Genomic biomarkers, Immune stromal interface, Breast cancer
Abstract
Fortschritte in der DNA-Sequenzierungstechnologie sowie verfeinerte Bioinformatik-Methoden zur Interpretation komplexer Datensätze haben die umfassende Bestimmung der Genexpression in Tumoren und der sie umgebende Mikroumwelt möglich gemacht. In jüngster Vergangenheit haben diese Fortschritte das Zusammenspiel zwischen Immuneffektormechanismen und der Mammakarzinom-Zell-biologie hervorgehoben und damit die seit langem akzeptierte Verbindung zwischen Immunität und Krebs betont. Die Erforschung von Genen des Immunsystems hat nicht nur die Stratifizierung der vielfältigen pathologischen Formen des Mammakarzinoms erweitert, sondern auch neue zelluläre Einblicke in die komplexe Heterogenität des Mammakarzinoms gegeben. In Anlehnung an die Tatsache, dass antitumoröse Therapien die Wirt-Tumor-Interaktion modifizieren können, haben Forscher den Fokus ihrer Aufmerksamkeit auf den prädiktiven Wert von Immunparametern als Marker des Ansprechens auf die Krebstherapie gerichtet. Wir diskutieren den aktuellen Stand auf dem Gebiet der Immunsignaturen beim Mammakarzinom sowie einige der fundamentalen Limitationen, die es zu überwinden gilt, um diese Entdeckungen in die Klinik zu überführen.
Introduction
A decade from the first draft of the human genome sequence [1, 2], our increasing understanding of the genetic aberrations that drive human malignancies has provided an impetus towards achieving more personalized cancer care. Genome sequence analysis (genomics) is beginning to reveal how DNA sequences vary from individual to individual: epidemiological genome-wide association studies have identified a number of single nucleotide polymorphisms (SNPs) related to disease susceptibility and breast cancer survival [3, 4, 5], and within the tumour a catalogue of sequence polymorphisms and chromosomal aberrations exhibit variations between individuals [6, 7]. Additionally, recent advances in mRNA and microRNA gene expression profiling (transcriptomics) in tumours have helped identify prognostic and predictive biomarkers in many types of human malignancies [8, 9, 10]. No cancer type has seen as much attention to the layers of its genomic background as breast cancer, and early work by Perou et al. [11] showed that transcriptional data generated using microarrays could stratify patients into distinct molecular ‘intrinsic subtypes’ relating to tumour biology and behaviour. 5 groups were identified and named Luminal A, Luminal B, Basal-like, Normal-like and the HER2-enriched subgroups. This classification has since seen further adaptation and evolved to include a 6th subgroup based on the low expression level of tight junction (claudin) genes – the claudin-low group [12, 13, 14]. These seminal publications resulted in an explosion of interest in the field of cancer genomics with countless publications attempting to unravel the complexities of both the inter-patient and intra-tumour heterogeneity of breast tumours at DNA copy number, sequence and transcriptional levels [15, 16, 17].
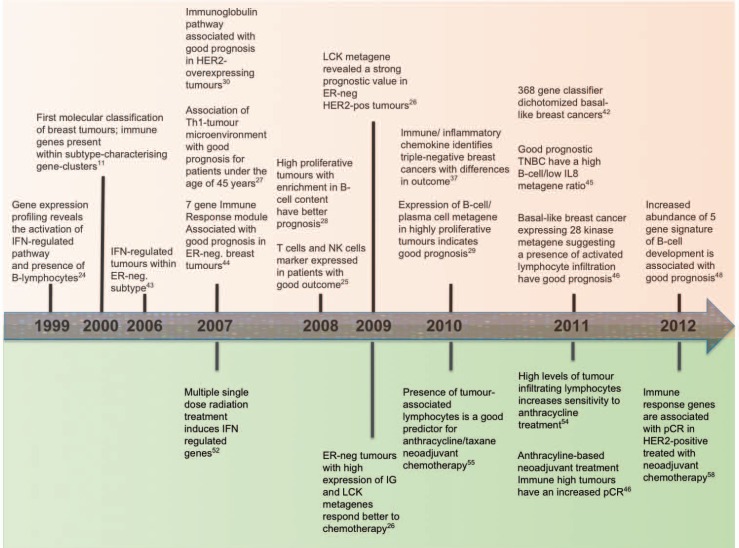
In parallel, recent years have seen a growing appreciation of the concept of ‘cancer immunoediting’ describing the integration of the immune system's dual but opposing impacts: host-protection and tumour promotion [18]. Cancer immunoediting consists of 3 successive steps whereby immune cells in the tumour microenvironment are thought to interact intimately and actively with the transformed cells [18, 19, 20]. In the ‘elimination’ phase, various components of the immune response work together to destroy developing tumours long before they become clinically apparent. During the ‘equilibrium’ phase, a balance is established between the tumour and the immune system, shaping each other reciprocally. Finally, the immune system contributes to the selection of tumour variants that enter the ‘escape’ phase, in which their outgrowth is no longer blocked by immunity resulting in clinically apparent disease [18, 19, 20, 21, 22]. The cells playing a key role in this process have been identified in both the innate (e.g. natural killer cells, natural killer T-cells, macrophages and dendritic cells) and the adaptive (e.g. CD4+ T helper type 1 (TH1) and CD8+ T-cells) immune ‘arms’. While the exact interplay between these components remains to be fully elucidated, the extensive transcriptomics information of breast cancers has provided novel insights into the interaction between the immune and breast cancer cell biology. Figure 1 gives a schematic illustration of the development of immunological gene expression signatures, and highlights that the relevance of early transcriptomics discoveries for identification of immune predictive and prognostic biomarkers is only now being fully appreciated. There remain some fundamental limitations, however, that need to be overcome to move these exciting discoveries from the bench to the bedside.
Fig. 1.
Prognostics.
Immune Signatures in Breast Cancers
The presence of a prognostic impact of lymphocyte infiltration in breast cancer has long been debated and remains controversial. Several lines of evidence support the hypothesis that lymphocytic infiltration is a marker of host anti-tumour immune response. Even the first applications of microarray analysis to whole tumour sections revealed variations in the expression of several genes associated with immune cells; including for example, the interferon-regulated genes [23, 24], B-lymphocyte markers [24], as well as T-lymphocyte-associated genes [23]. Laser-captured microdissection of stromal components identified a good-outcome signature of 26 genes enriched for elements of the TH1 immune response [25]. Rody et al. [26] identified 7 clusters of immune system-related metagenes by large-scale microarray analysis and demonstrated an association with different immunological cell types. A strong positive prognostic value for the T-cell surrogate marker (lymphocyte-specific kinase (LCK) metagene) was observed among all oestrogen receptor (ER)-negative tumours and amongst ER-positive tumours with HER2 overexpression, whereas an IgG metagene as a marker for B-cells had no significant prognostic value. Another earlier study reported that increased expression of TH1-associated genes was protective in breast cancer patients, but only in patients under 45 years of age [27]. The importance of subtle variations in the expression levels of immune cell-associated genes within breast tumours is evident. Schmidt et al. [28] demonstrated that the IgG metagene outperformed the T-cell metagene as a favourable prognostic factor in highly proliferating specimens, while a further study reported that among ER-negative and ER-positive highly proliferative cancers, a subset of tumours with high expression of a B-cell/plasma cell metagene carries a favourable prognosis [29]. In HER2-overexpressing breast cancers, the expression of genes associated with the immunoglobulin pathway correlated with tumour-infiltrating lymphocytes and predicted favourable clinical outcome among highly proliferating tumours [30]. These immunoglobulin genes are likely to be co-regulated, and therefore the prominence of an immunoglobulin pathway does not necessarily imply that the lymphocytic infiltrate is composed mostly of a particular immune cell subset such as B cells. Interestingly, in ovarian cancer, the causal nature of relationship between the strong association of T-cell infiltrate with good patient outcome has been debated: it may be due to the T-cell infiltrate effectively eliminating tumour cells, or might reflect indolent tumour cell biology characterized by slower growth, thus increasing the opportunity for immune cell infiltration into the tumour microenvironment [31]. However, this is challenged by the observation that highly proliferating tumours are more likely to be associated with higher T-cell infiltrates [32], possibly due to mitotically active cancers exhibiting high genomic instability [33]. It can therefore be hypothesized that a higher mutational rate is likely to generate neo-antigens with higher immunogenicity [34]. Similarly, other studies have reported that p53 abnormalities and increased immune cell infiltrates are significantly more common in high-grade serous ovarian cancers with germline and somatic mutations in BRCA1 or BRCA2, compared with tumours lacking BRCA abnormalities [35, 36]. In ER-negative breast cancers, a clear anti-correlation between proliferation genes and an immune signature of 14 genes involved in proinflammatory cytokine/chemokine signalling has been observed [37]; on the other hand, the observed correlation between tumour growth and immune microenvironment [28] raises the possibility that a slow growing tumour is associated with a different tumour-host interaction (e.g. a humoral response to the tumour and its antigens) than a fast growing one. Studies report the enhancement of metastatic potency of breast cancer cell lines enriched with stromal cells [38], lending support to the concept of a symbiotic relationship between tumours and its microenvironment. In contrast, highly proliferative tumour cells may produce different signals which will in turn influence the microenvironment down a different path. Thus, the proliferation rate of a tumour [39] might be a surrogate indicator of the tumour's stromal environment as well as the intrinsic properties within the tumour cell that influence that environment. Unravelling the complexity of this association would be valuable information for targeted therapeutic strategies with potential to target both tumour cells and their interface with an immunologically active and cytokine-rich stroma and might shed further light on the signals influencing tumour growth and its microenvironment.
There has been an increasing focus on the potential for immune-related features for further stratification of breast cancers, alongside proliferation and associated gene expression signatures (genomic grade index [40], 70-gene signature [41] and 76-gene signature [42]) which appear to be successful in particular for stratifying ER-positive tumours where cell proliferation is a key element for outcome prediction. Using a test-and-validation strategy on a comprehensive collection of breast tumours, Hu et al. [43] were the first to report that interferon (IFN)-γ and regulators of STAT1-based immunity may have an anti-tumour effect. IFNs are a family of structurally related cytokines and possess a wide range of immune properties including antiviral activity, promotion of antigen presentation as well as inhibition of cell growth and proliferation. This study provided the first suggestion that the expression of multiple immune-related genes had an influence on the outcome of breast cancers of non-ER-positive type. It was shortly followed by a publication interrogating 3 publicly available microarray data sets, comprising 186 adjuvant therapy-naïve ER-negative breast cancers, and thereby identifying 7 immune-responsive genes capable of specifying tumours with reduced risk for distant metastasis [44]. Interestingly, a correlation between the so-called immune response (IR+) module and the previously determined IFN-regulated cluster was observed. The small overlap of genes (including STAT1) and the difference in their clinico-pathological features (IFN-expressing tumours encompassed more cases with positive lymph node metastasis) strengthened the IR+ module as a key gene signature for subtype classification. Focusing on basal-like breast cancers – the majority of which are ER-negative tumours – revealed a positive relation to outcome with the presence of either a module encompassing 28 kinases or a gene signature derived from medullary breast cancers [45, 46]. Medullary breast carcinomas are highly proliferative breast cancers showing increased lymphocytic infiltration and having an overall good prognosis. Genes involved in the IL15 and IL12 pathway appear to be the main players in the medullary breast cancer signature and indicated a better prognosis [45]. In agreement with this immunological pathway was the expression pattern of the kinome-gene module in basal-like breast cancers, which also pointed to an activation of TH1-biased lymphocytic infiltration in good prognostic cancers [46]. Characterisation of a comprehensive transcriptomic dataset of triple-negative breast tumours, by definition also ER-negative and with significant clinically and biologically overlap to the basal-like subtype, could further dissect their tumour-host interaction. A specific relationship between individual components of an immune response such as the ratio of a high B-cell content to a low IL8 expression seem to infer a positive prognosis for triple-negative breast cancers [47]. A positive association of lymphocytic infiltration and outcome in ER-negative breast tumours has recurrently and robustly been observed and triggered expectation for the development of new therapies based on immune response manipulation for breast cancer subtypes.
However, the fine print within the bulk of observations should not be overlooked, and novel insights into tumour immunology need to be cautiously evaluated. Recently, Ascierto et al. [48] identified among 299 immune function genes, a 5-gene signature (IGKC, GBP1, STATI, 1GLL5 and OCLN) involved in B-cell development with a high predictive accuracy for relapse-free survival of 85%. At the same time, genes involved in primary immunodeficiency signalling, T-cell apoptosis, CTLA4 signalling and production of nitric oxide and reactive oxygen species were also up-regulated in the tumour specimens of patients who were subsequently free of relapse. The authors offer an explanation for this paradoxically concurrent expression of immune effector and suppressor genes whereby tumour-derived factors (e.g. GM-CSF, VEGF and MCP-1) facilitate the expression of immune suppressor genes as well as acting as chemo-attractants for immune cells. Surgical intervention may then disturb this carefully balanced system between immune suppressor and effector genes, leading to the different expression ratio between these 2 sides. It is also possible that the immune activation of cancer cells initiates a positive feedback loop whereby the cancer cells not only invite immune cells to the tumour microenvironment but they are also more sensitive to pre-inflammatory factors secreted by immune cells.
Nonetheless, recent genomic studies have evidently indicated that the expression of genes related to immune response provide important prognostic information in ER-negative, HER2-overexpressing or highly proliferating ER-positive breast cancers. The value of molecular signatures such as the Food and Drug Administration (FDA)-approved 70-gene MammaPrint® (Agendia, Amsterdam, The Netherlands) prognostic panel and Oncotype DX® (Genomic Health, Redwood City, CA, USA) in defining the role of chemotherapy in an intermediate prognostic risk group defined by a 21-gene panel, are currently tested in the context of the prospective randomized phase III trials, MINDACT and TAILORx, respectively. However, since this first generation of gene signatures has largely been focused on hormone receptor-positive disease and only partly includes immune-related genes [49, 50], the integration of these elements into prognostic and predictive models for further breast cancer subgroups is the next step to assess validity and optimize efficacy.
Predictive Immune Biomarkers in Breast Cancer
The development of predictive immune signatures to help guide the use of anti-tumour therapy is still in its infancy. However, on the basis that anti-cancer therapies can modify the host-tumour interaction, cancer genomic experts have focused their attention on the predictive value of immune parameters as markers of therapeutic anti-tumour response. Gianni et al. [51] have shown that immune-related genes, such as CD3, are linked to response to chemotherapy in a cohort of 89 breast cancers, of which 11 had a pathological complete response (pCR). More recently, Sabatier et al. [46] investigated the link between an immune cell-derived 28-kinase metagene and response to anthracycline-based neoadjuvant chemotherapy for basal-like breast cancers. ‘Immune-High’ patients experienced more pCR (59%) than ‘Immune-Low’ patients (43%), and although this was not significant (p = 0.29), similar trends have been observed with modest predictive value for neoadjuvant chemotherapies: high expression of both IgG and LCK metagenes in ER-negative breast carcinomas [26], and a high B-cell/low IL8 ratio for triple-negative breast cancers [47]. Similarly, gene expression profiling of breast tumour cell lines and mouse models exposed to single-dose (10 Gy) versus fractionated (2 Gy × 5) radiation have revealed that only the fractionated regimen induced an interferon-related gene signature, including STAT1 [52]. Taken together, these model systems illustrate that chemotherapeutic agents may restore the immunological equilibrium not only due to the ‘debulking’ of the tumour mass but also due to direct or indirect effects on the immune system. In fact, anthracycline-based chemotherapies have been shown to induce a vigorous infiltration of anticancer immune effectors in mice [53]. A recent clustering analysis of the neoadjuvant (EORTC) cohort defined an 8-gene lymphocyte mRNA expression signature (including CD19, CD3D, CD48, GZMB, LCK, MS4A1, PRF1 and SELL) to examine the association between tumour-infiltrating lymphocytes (TIL) and short-term response to neoadjuvant chemotherapy in ER-negative tumours (n = 113) [54]. TIL-enriched tumours significantly predicted anthracycline sensitivity with an odds ratio of 6.3 for HER2-positive and triple-negative tumour phenotypes. Additionally, Denkert et al. [55] reported a significant relationship between TIL (identified by a combination of H and E assessment, and expression analysis of several TIL genes by polymerase chain reaction) and pathologic response to neoadjuvant anthracycline/taxane therapy in a large group of 1,058 patients (one fourth of whom were ER-negative).
Several studies have suggested possible mechanisms of tumour-immune interaction in response to chemotherapy. Appropriate preclinical models have shown that 2 receptors present on dendritic cells, namely TLR4 (a toll-like receptor) and P2RX7 (a purinergic receptor), are essential for their cross-talk with a dying cell. They recognise 2 soluble molecules released from the dying tumour cells, HMGB1 and ATP, respectively. In the absence of TLR4 or P2RX7, the immune system fails to mount an antitumor immune response after chemotherapy [56, 57]. A loss-of-function polymorphism of the TLR4 is an independent predictive biomarker for response to anthracycline chemotherapy in breast cancer patients [56]. Similarly, anthracycline-treated individuals with breast cancer carrying a loss-of-function allele of P2RX7 developed metastatic disease more rapidly than individuals bearing the normal allele [57]. A comprehensive analysis of publicly available gene expression studies evaluating anthracycline with or without taxane-based neoadjuvant chemotherapy has reported that high immune module scores were associated with increased probability of achieving pCR in all breast cancer subtypes with varying degree of significance [58]. Although the data with regards to predictive immune response gene sets is still very sparse, the above data provide preliminary validation of the concept that selective immune defects can influence the efficacy of anticancer chemotherapies. Hence studies exploring the possibility of predicting therapeutic outcome by assessing dynamic variables such as changes in the frequency, composition, activation status and repertoire of TIL, the expression of immune-relevant metagenes (in repeated lymph node biopsies), or the generation of tumour-specific antibodies (in patient sera) after chemotherapy need to be encouraged.
Discussion
While multigene prognostic and predictive gene signatures were once expected to replace clinicopathological parameters for therapy decision-making, a complete transition has not yet taken place. This is partly due to inherent problems of technical robustness and experimental as well as analytical standardization [14]. Likewise, the inherent complexity of immune gene signatures (fig. 2 A), heterogeneous assay protocols between laboratories, and the use of different statistical strategies are proving to be rate-limiting steps for the development of immune-related biomarkers for clinical application. The high data variability and poor reproducibility complicate meta-analyses comparing results across laboratories, however, efforts are being made internationally to minimize these obstacles. For example, 2 large immunological consortia (the Cancer Immunotherapy Consortium (CIC) in the US and the Association of Cancer Immunoguiding Program (CIP) in Europe) have recently addressed the issue of immune assay harmonization across laboratories with the objective of accelerating immune biomarker identification and drug development [59].
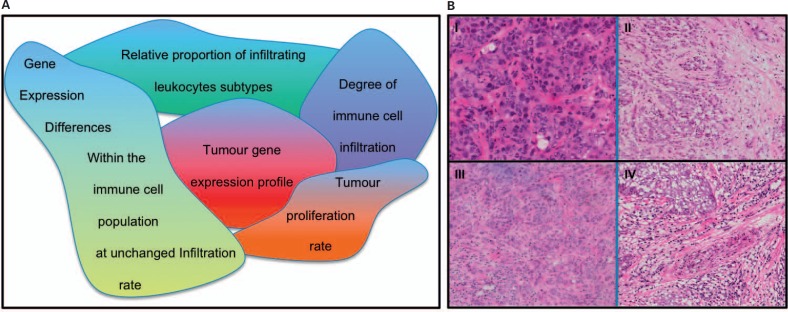
Fig. 2.
A Inherent complexities of interpreting immune gene signatures. B Triple-negative tumours show a different degree of lymphocytic contents: I minimal non-malignant (stromal and lymphocytic) enrichment; II small presence (< 10%); III moderate presence (10–30%), and IV strong enrichment (> 30%) (images courtesy of Patrycja Gazinska).
More specifically, Rody et al. [26] have argued that the level of lymphocytic infiltration has a greater effect on the tumour gene expression profile than the differences in a mixture of distinct types of immune cells. Although immune gene signatures appear to be highly sensitive to the composition of tumour cell content, it may be difficult to detect differences in immune cell composition in a background of differential infiltration levels between tumour samples. Enrichment of samples to > 70% malignant epithelium has become standard for expression profiling experiments and should help to minimize this obstacle for many tumour samples. In triple-negative breast cancer specimens, however, where the lymphocytic cells are intertwined with cancerous cells (fig. 2 B), this is not always feasible.
The dynamic plasticity of the tumour microenvironment itself adds further complexity to the task of translating immune cancer genomics into cancer therapies. Possible alteration of immune milieu within the tumour due to surgical or chemotherapeutic interventions means that the composition of the profiled tumour sample might not resemble the true immune microenvironment. Recent evidence indicates that T-cells alter their end-stage commitment in response to microenvi-ronmental cues, for example shifting from a TH17 commitment to a TH1-, TH2- or T-reg-specific pattern [60], with shift patterns ultimately determining the functional role of these immune cells. Similarly, molecular variations between tumour samples may be occurring at the protein and protein-protein interaction level, for example mediated by post-translational modifications, thus hampering the identification of transcriptomic-based immune gene signatures as robust fingerprints for prediction of survival or treatment response.
Lastly, although gene expression profiling of the primary tumour and its microenvironment are likely to help drive various tumour immunology hypotheses, one important consideration in the development of cancer immunotherapy is tissue source for utilization into biomarker assays. Applying microarray-based expression profiling on peripheral blood mononuclear cell (PBMC) from patient blood samples may represent a useful tool in the identification of immune signatures associated with cancer immunotherapy. Although little is known about PBMC gene expression immune signatures in cancers, several studies in the autoimmune literature have demonstrated utility of PBMC expression arrays to help identify predictors of response to therapy and prognosis [61, 62, 63]. More importantly, validation of immune signatures on a large scale is much more feasible on blood-based technologies, and PBMC-based biomarkers of immune response in patients receiving cancer immunotherapy [64] need to be further explored as means to guide therapy and prognosis.
Conclusion
In conclusion, recent genomic approaches provide an opportunity to evaluate the tumour microenvironment (including stromal, endothelial and immune cells) and offer the possibility of identifying cytokines and signalling molecules that are important for limiting pro-tumourigenic responses and enhancing anti-tumour immune responses. Whilst the majority of the ER-positive breast cancer prognostic signatures are associated with proliferation signals, genes related to immune response appear to provide important prognostic information in other breast cancer subtypes (e.g. ER-negative, HER2-overexpressing or highly proliferating ER-positive breast cancers). Moreover, accumulating evidence indicates that chemotherapy can stimulate anticancer immune responses, and immune-related genes are linked to chemotherapy response and patient outcome. Large-scale studies exploring the composition, intra- and peritumoural distribution, architecture, and functional articulation of the immune infiltrate along with its context are needed to fully comprehend the immune readouts in breast cancer. As massive amounts of biological and immunological data are generated, technological and advances in the biostatistical analysis of genomics represent a remarkable opportunity to fine-tune breast cancer classification, prognosis and treatment prediction.
Disclosure Statement
The authors declare no conflict of interest.
Acknowledgement
We would like to acknowledge the funding to Dr. Sheeba Irshad, Dr. Anita Grigoriadis and Professor Tutt from the Breakthrough Breast Cancer and the Sarah Greene Tribute Funds. Dr. Katherine Lawler is funded by the Comprehensive Cancer Imaging Centre (CCIC) and Professor Ng from the Dimbleby Cancer Care and the Breakthrough Breast Cancer Funds.
References
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, Funke R, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, Gocayne JD, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 3.Campa D, Kaaks R, Le Marchand L, Haiman CA, Travis RC, Berg CD, Buring JE, Chanock SJ, Diver WR, Dostal L, Fournier A, et al. Interactions between genetic variants and breast cancer risk factors in the breast and prostate cancer cohort consortium. J Natl Cancer Inst. 2011;103:1252–1263. doi: 10.1093/jnci/djr265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Closas M, Hall P, Nevanlinna H, Pooley K, Morrison J, Richesson DA, Bojesen SE, Nordest-gaard BG, Axelsson CK, Arias JI, Milne RL, et al. Heterogeneity of breast cancer associations with five susceptibility loci by clinical and pathological characteristics. PLoS Genet. 2008;4:e1000054. doi: 10.1371/journal.pgen.1000054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peng R, Wang S, Shi Y, Liu D, Teng X, Qin T, Zeng Y, Yuan Z. Patients 35 years old or younger with operable breast cancer are more at risk for relapse and survival: a retrospective matched case-control study. Breast. 2011;20:568–573. doi: 10.1016/j.breast.2011.07.012. [DOI] [PubMed] [Google Scholar]
- 6.Kwei KA, Kung Y, Salari K, Holcomb IN, Pollack JR. Genomic instability in breast cancer: pathogenesis and clinical implications. Mol Oncol. 2010;4:255–266. doi: 10.1016/j.molonc.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, Greenman CD, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Iorio MV, Croce CM. MicroRNA involvement in human cancer. Carcinogenesis. 2012 doi: 10.1093/carcin/bgs140. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nambiar PR, Gupta RR, Misra V. An ‘omics’ based survey of human colon cancer. Mutat Res. 2010;693:3–18. doi: 10.1016/j.mrfmmm.2010.07.008. [DOI] [PubMed] [Google Scholar]
- 10.Tran B, Dancey JE, Kamel-Reid S, McPherson JD, Bedard PL, Brown AM, Zhang T, Shaw P, Onetto N, Stein L, Hudson TJ, et al. Cancer genomics: technology, discovery, and translation. J Clin Oncol. 2012;30:647–660. doi: 10.1200/JCO.2011.39.2316. [DOI] [PubMed] [Google Scholar]
- 11.Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, Fluge O, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 12.Prat A, Parker JS, Karginova O, Fan C, Livasy C, Herschkowitz JI, He X, Perou CM. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 2010;12:R68. doi: 10.1186/bcr2635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800. doi: 10.1056/NEJMra0801289. [DOI] [PubMed] [Google Scholar]
- 14.Weigelt B, Mackay A, A'Hern R, Natrajan R, Tan DS, Dowsett M, Ashworth A, Reis-Filho JS. Breast cancer molecular profiling with single sample predictors: a retrospective analysis. Lancet Oncol. 2010;11:339–349. doi: 10.1016/S1470-2045(10)70008-5. [DOI] [PubMed] [Google Scholar]
- 15.Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, et al. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–352. doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, Martinez P, Matthews N, Stewart A, Tarpey P, Varela I, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366:883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shah SP, Roth A, Goya R, Oloumi A, Ha G, Zhao Y, Turashvili G, Ding J, Tse K, Haffari G, Bashashati A, et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature. 2012;486:395–399. doi: 10.1038/nature10933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn GP, Bruce AT, Ikeda H, Old LJ, Schreiber RD. Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol. 2002;3:991–998. doi: 10.1038/ni1102-991. [DOI] [PubMed] [Google Scholar]
- 19.Koebel CM, Vermi W, Swann JB, Zerafa N, Rodig SJ, Old LJ, Smyth MJ, Schreiber RD. Adaptive immunity maintains occult cancer in an equilibrium state. Nature. 2007;450:903–907. doi: 10.1038/nature06309. [DOI] [PubMed] [Google Scholar]
- 20.Mantovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- 21.Kraman M, Bambrough PJ, Arnold JN, Roberts EW, Magiera L, Jones JO, Gopinathan A, Tuveson DA, Fearon DT. Suppression of antitumor immunity by stromal cells expressing fibroblast activation protein-alpha. Science. 2010;330:827–830. doi: 10.1126/science.1195300. [DOI] [PubMed] [Google Scholar]
- 22.Teng MW, Swann JB, Koebel CM, Schreiber RD, Smyth MJ. Immune-mediated dormancy: an equilibrium with cancer. J Leukoc Biol. 2008;84:988–993. doi: 10.1189/jlb.1107774. [DOI] [PubMed] [Google Scholar]
- 23.Huang E, Cheng SH, Dressman H, Pittman J, Tsou MH, Horng CF, Bild A, Iversen ES, Liao M, Chen CM, West M, et al. Gene expression predictors of breast cancer outcomes. Lancet. 2003;361:1590–1596. doi: 10.1016/S0140-6736(03)13308-9. [DOI] [PubMed] [Google Scholar]
- 24.Perou CM, Jeffrey SS, van de Rijn M, Rees CA, Eisen MB, Ross DT, Pergamenschikov A, Williams CF, Zhu SX, Lee JC, Lashkari D, et al. Distinctive gene expression patterns in human mammary epithelial cells and breast cancers. Proc Natl Acad Sci U S A. 1999;96:9212–9217. doi: 10.1073/pnas.96.16.9212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, et al. Stromal gene expression predicts clinical outcome in breast cancer. Nat Med. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]
- 26.Rody A, Holtrich U, Pusztai L, Liedtke C, Gaetje R, Ruckhaeberle E, Solbach C, Hanker L, Ahr A, Metzler D, Engels K, et al. T-cell metagene predicts a favorable prognosis in estrogen receptor-negative and HER2-positive breast cancers. Breast Cancer Res. 2009;11:R15. doi: 10.1186/bcr2234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hsu SD, Anders CK, Acharya CR, Zhang Y, Wang Y, Foekens JA, Blackwell KL, Drake CG, Morse MA, Febbo PG. Immune signatures hold prognostic import across solids tumors. J Clin Oncol. 2007;25:21041. [Google Scholar]
- 28.Schmidt M, Bohm D, von Torne C, Steiner E, Puhl A, Pilch H, Lehr HA, Hengstler JG, Kolbl H, Gehrmann M. The humoral immune system has a key prognostic impact in node-negative breast cancer. Cancer Res. 2008;68:5405–5413. doi: 10.1158/0008-5472.CAN-07-5206. [DOI] [PubMed] [Google Scholar]
- 29.Bianchini G, Qi Y, Alvarez RH, Iwamoto T, Coutant C, Ibrahim NK, Valero V, Cristofanilli M, Green MC, Radvanyi L, Hatzis C, et al. Molecular anatomy of breast cancer stroma and its prognostic value in estrogen receptor-positive and -negative cancers. J Clin Oncol. 2010;28:4316–4323. doi: 10.1200/JCO.2009.27.2419. [DOI] [PubMed] [Google Scholar]
- 30.Alexe G, Dalgin GS, Scanfeld D, Tamayo P, Mesirov JP, DeLisi C, Harris L, Barnard N, Martel M, Levine AJ, Ganesan S, et al. High expression of lymphocyte-associated genes in node-negative HER2+ breast cancers correlates with lower recurrence rates. Cancer Res. 2007;67:10669–10676. doi: 10.1158/0008-5472.CAN-07-0539. [DOI] [PubMed] [Google Scholar]
- 31.Kandalaft LE, Coukos G. The microenvironment of ovarian cancer: lessons on immune mediated tumor rejection or tolerance. In: Wang E, Marincola FM, editors. Signatures of Rejection. 1st ed. New York, NY: Springer; 2010. [Google Scholar]
- 32.Adams SF, Levine DA, Cadungog MG, Hammond R, Facciabene A, Olvera N, Rubin SC, Boyd J, Gimotty PA, Coukos G. Intraepithelial t cells and tumor proliferation: impact on the benefit from surgical cytoreduction in advanced serous ovarian cancer. Cancer. 2009;115:2891–2902. doi: 10.1002/cncr.24317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Belgen H, Einhorn N, Sjovall K, Roschkes A, Ghadimis BM, McShane LM, Nilsson B, Shah K, Ried T, Auer G. Prognostic significance of cell cycle proteins and genomic instability in borderline, early and advanced stage ovarian carcinomas. Int J Gynecol Cancer. 2000;10:477–487. doi: 10.1046/j.1525-1438.2000.00077.x. [DOI] [PubMed] [Google Scholar]
- 34.Duesberg P, Stindl R, Hehlmann R. Explaining the high mutation rates of cancer cells to drug and multidrug resistance by chromosome reassortments that are catalyzed by aneuploidy. Proc Natl Acad Sci U S A. 2000;97:14295–14300. doi: 10.1073/pnas.97.26.14295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McAlpine JN, Porter H, Kobel M, Nelson BH, Prentice LM, Kalloger SE, Senz J, Milne K, Ding J, Shah SP, Huntsman DG, et al. BRCA1 and BRCA2 mutations correlate with tp53 abnormalities and presence of immune cell infiltrates in ovarian high-grade serous carcinoma. Mod Pathol. 2012;25:740–750. doi: 10.1038/modpathol.2011.211. [DOI] [PubMed] [Google Scholar]
- 36.Shah CA, Allison KH, Garcia RL, Gray HJ, Goff BA, Swisher EM. Intratumoral t cells, tumor-associated macrophages, and regulatory t cells: association with p53 mutations, circulating tumor DNA and survival in women with ovarian cancer. Gynecol Oncol. 2008;109:215–219. doi: 10.1016/j.ygyno.2008.01.010. [DOI] [PubMed] [Google Scholar]
- 37.Yau C, Esserman L, Moore DH, Waldman F, Sninsky J, Benz CC. A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer. Breast Cancer Res. 2010;12:R85. doi: 10.1186/bcr2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 39.Beresford MJ, Wilson GD, Makris A. Measuring proliferation in breast cancer: practicalities and applications. Breast Cancer Res. 2006;8:216. doi: 10.1186/bcr1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sotiriou C, Wirapati P, Loi S, Harris A, Fox S, Smeds J, Nordgren H, Farmer P, Praz V, Haibe-Kains B, Desmedt C, et al. Gene expression profiling in breast cancer: understanding the molecular basis of histologic grade to improve prognosis. J Natl Cancer Inst. 2006;98:262–272. doi: 10.1093/jnci/djj052. [DOI] [PubMed] [Google Scholar]
- 41.Van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 42.Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, et al. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–679. doi: 10.1016/S0140-6736(05)17947-1. [DOI] [PubMed] [Google Scholar]
- 43.Hu Z, Fan C, Oh DS, Marron JS, He X, Qaqish BF, Livasy C, Carey LA, Reynolds E, Dressler L, Nobel A, et al. The molecular portraits of breast tumors are conserved across microarray platforms. BMC Genomics. 2006;7:96. doi: 10.1186/1471-2164-7-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Teschendorff AE, Miremadi A, Pinder SE, Ellis IO, Caldas C. An immune response gene expression module identifies a good prognosis subtype in estrogen receptor negative breast cancer. Genome Biol. 2007;8:R157. doi: 10.1186/gb-2007-8-8-r157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sabatier R, Finetti P, Cervera N, Lambaudie E, Esterni B, Mamessier E, Tallet A, Chabannon C, Extra JM, Jacquemier J, Viens P, et al. A gene expression signature identifies two prognostic subgroups of basal breast cancer. Breast Cancer Res Treat. 2011;126:407–420. doi: 10.1007/s10549-010-0897-9. [DOI] [PubMed] [Google Scholar]
- 46.Sabatier R, Finetti P, Mamessier E, Raynaud S, Cervera N, Lambaudie E, Jacquemier J, Viens P, Birnbaum D, Bertucci F. Kinome expression profiling and prognosis of basal breast cancers. Mol Cancer. 2011;10:86. doi: 10.1186/1476-4598-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rody A, Karn T, Liedtke C, Pusztai L, Ruckhaeberle E, Hanker L, Gaetje R, Solbach C, Ahr A, Metzler D, Schmidt M, et al. A clinically relevant gene signature in triple negative and basal-like breast cancer. Breast Cancer Res. 2011;13:R97. doi: 10.1186/bcr3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ascierto ML, Kmieciak M, Idowu MO, Manjili R, Zhao Y, Grimes M, Dumur C, Wang E, Ramakrishnan V, Wang XY, Bear HD, et al. A signature of immune function genes associated with recurrence-free survival in breast cancer patients. Breast Cancer Res Treat. 2012;131:871–880. doi: 10.1007/s10549-011-1470-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cronin M, Sangli C, Liu ML, Pho M, Dutta D, Nguyen A, Jeong J, Wu J, Langone KC, Watson D. Analytical validation of the oncotype DX genomic diagnostic test for recurrence prognosis and therapeutic response prediction in node-negative, estrogen receptor-positive breast cancer. Clin Chem. 2007;53:1084–1091. doi: 10.1373/clinchem.2006.076497. [DOI] [PubMed] [Google Scholar]
- 50.Knauer M, Mook S, Rutgers EJ, Bender RA, Hauptmann M, van de Vijver MJ, Koornstra RH, Bueno-de-Mesquita JM, Linn SC, van't Veer LJ. The predictive value of the 70-gene signature for adjuvant chemotherapy in early breast cancer. Breast Cancer Res Treat. 2010;120:655–661. doi: 10.1007/s10549-010-0814-2. [DOI] [PubMed] [Google Scholar]
- 51.Gianni L, Baselga J, Eiermann W, Guillem Porta V, Semiglazov V, Lluch A, Zambetti M, Sabadell D, Raab G, Llombart Cussac A, Bozhok A, et al. Feasibility and tolerability of sequential doxorubicin/paclitaxel followed by cyclophosphamide, methotrexate, and fluorouracil and its effects on tumor response as preoperative therapy. Clin Cancer Res. 2005;11:8715–8721. doi: 10.1158/1078-0432.CCR-05-0539. [DOI] [PubMed] [Google Scholar]
- 52.Tsai MH, Cook JA, Chandramouli GV, DeGraff W, Yan H, Zhao S, Coleman CN, Mitchell JB, Chuang EY. Gene expression profiling of breast, prostate, and glioma cells following single versus fractionated doses of radiation. Cancer Res. 2007;67:3845–3852. doi: 10.1158/0008-5472.CAN-06-4250. [DOI] [PubMed] [Google Scholar]
- 53.Ma Y, Aymeric L, Locher C, Mattarollo SR, Delahaye NF, Pereira P, Boucontet L, Apetoh L, Ghiringhelli F, Casares N, Lasarte JJ, et al. Contribution of IL-17-producing gamma delta t cells to the efficacy of anticancer chemotherapy. J Exp Med. 2011;208:491–503. doi: 10.1084/jem.20100269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.West NR, Milne K, Truong PT, Macpherson N, Nelson BH, Watson PH. Tumor-infiltrating lymphocytes predict response to anthracycline-based chemotherapy in estrogen receptor-negative breast cancer. Breast Cancer Res. 2011;13:R126. doi: 10.1186/bcr3072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Denkert C, Loibl S, Noske A, Roller M, Muller BM, Komor M, Budczies J, Darb-Esfahani S, Kronenwett R, Hanusch C, von Torne C, et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2010;28:105–113. doi: 10.1200/JCO.2009.23.7370. [DOI] [PubMed] [Google Scholar]
- 56.Apetoh L, Ghiringhelli F, Tesniere A, Obeid M, Ortiz C, Criollo A, Mignot G, Maiuri MC, Ullrich E, Saulnier P, Yang H, et al. Toll-like receptor 4-dependent contribution of the immune system to anticancer chemotherapy and radiotherapy. Nat Med. 2007;13:1050–1059. doi: 10.1038/nm1622. [DOI] [PubMed] [Google Scholar]
- 57.Ghiringhelli F, Apetoh L, Tesniere A, Aymeric L, Ma Y, Ortiz C, Vermaelen K, Panaretakis T, Mignot G, Ullrich E, Perfettini JL, et al. Activation of the NLRP3 inflammasome in dendritic cells induces IL-1beta-dependent adaptive immunity against tumors. Nat Med. 2009;15:1170–1178. doi: 10.1038/nm.2028. [DOI] [PubMed] [Google Scholar]
- 58.Ignatiadis M, Singhal SK, Desmedt C, Haibe-Kains B, Criscitiello C, Andre F, Loi S, Piccart M, Michiels S, Sotiriou C. Gene modules and response to neoadjuvant chemotherapy in breast cancer subtypes: a pooled analysis. J Clin Oncol. 2012;30:1996–2004. doi: 10.1200/JCO.2011.39.5624. [DOI] [PubMed] [Google Scholar]
- 59.Van der Burg SH, Kalos M, Gouttefangeas C, Janetzki S, Ottensmeier C, Welters MJ, Romero P, Britten CM, Hoos A. Harmonization of immune biomarker assays for clinical studies. Sci Transl Med. 2011;3:108ps144. doi: 10.1126/scitranslmed.3002785. [DOI] [PubMed] [Google Scholar]
- 60.O'Shea JJ, Paul WE. Mechanisms underlying lineage commitment and plasticity of helper CD4+ T cells. Science. 2010;327:1098–1102. doi: 10.1126/science.1178334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Emamian ES, Leon JM, Lessard CJ, Grandits M, Baechler EC, Gaffney PM, Segal B, Rhodus NL, Moser KL. Peripheral blood gene expression profiling in Sjogren's syndrome. Genes Immun. 2009;10:285–296. doi: 10.1038/gene.2009.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Julia A, Erra A, Palacio C, Tomas C, Sans X, Barcelo P, Marsal S. An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. PLoS One. 2009;4:e7556. doi: 10.1371/journal.pone.0007556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.McKinney EF, Lyons PA, Carr EJ, Hollis JL, Jayne DR, Willcocks LC, Koukoulaki M, Brazma A, Jovanovic V, Kemeny DM, Pollard AJ, et al. A CD8+ T cell transcription signature predicts prognosis in autoimmune disease. Nat Med. 2010;16:586–591. doi: 10.1038/nm.2130. p. 581 following p. 591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baine MJ, Chakraborty S, Smith LM, Mallya K, Sasson AR, Brand RE, Batra SK. Transcriptional profiling of peripheral blood mononuclear cells in pancreatic cancer patients identifies novel genes with potential diagnostic utility. PLoS One. 2011;6:e17014. doi: 10.1371/journal.pone.0017014. [DOI] [PMC free article] [PubMed] [Google Scholar]