Abstract
We investigated circuits for object recognition in macaque anterior (TE) and posterior inferotemporal cortex (TEO), using a two-step method with in vivo anatomical imaging. In step 1, red fluorescent tracer was injected into TE to reveal and Pre-target patches of feedforward neurons in TEO. In step 2, these were visualized on the cortical surface in vivo, and injected with green fluorescent tracer. Histological processing revealed that patches >500 μm from the injection site in TEO consisted of intermingled green TEO intrinsically projecting neurons and red TEO-to-TE neurons, with only few double-labeled neurons. In contrast, patches near the injection site in TEO contained many double-labeled neurons. Two parallel, spatially intermingled circuits are suggested: (1) TEO neurons having very local intrinsic collaterals and projection to TE (2) TEO neurons projecting more widely in the intrinsic network, but not to TE. These parallel systems might be specialized for, respectively, fast vs. highly processed signals.
Pre-selection of identified domains targeted for anatomical or physiological manipulations is a significant aid, and even a prerequisite, in investigating functionally related, but widely distributed network components; for example, tracer injections guided by wide-field electrophysiological mapping or electrical stimulation of specific sites pre-determined by fMRI (e.g., colour and face processing)1,2. For anatomical experiments in particular, the difficulty in visualizing and targeting functionally identified domains in vivo has been a serious hindrance. Small injections of retrograde tracers within approximately one functional column domain of 500 μm diameter typically result in patches of retrogradely labeled neurons both within and outside the boundary of injected area (respectively, intrinsic and extrinsic connections)3,4,5,6. But, how these distributed patches relate to functional networks is incompletely understood.
To address this issue at the level of neural circuits, we developed a two-step fluorescent in vivo surface connection imaging (in vivo SCI) method for visualizing patches of cortical neurons projecting to a designated target, such as a single cortical column (approximately 500 μm diameter). The visualized patches serve to guide a second injection, and thus allow a rigorous examination of the spatial relationship between two patchy systems of projections. In this study, we used in vivo SCI to further investigate inferotemporal cortical connections involved in object and face recognition7. In particular, we asked, what is the relationship between patches of feedforward neurons projecting from area TEO to TE and patches of intrinsically projecting neurons in TEO?
Our results demonstrated two patterns within TEO: (1) feedforward projecting TEO neurons having very local intrinsic collaterals that appear not to extend beyond the home column of the parent neurons and (2) TEO neurons projecting more widely within the network of intrinsic patches, but not to the injected column in TE. These two subpopulations are intermingled within the same patch in TEO, but as reported below, appear to constitute two parallel systems. The two subpopulations as thus defined may also correspond to different types of pyramidal neurons.
There are two hypotheses compatible with these results, in the context of information transfer in the ventral visual pathway. One possibility is selective feedforward convergence, where feedforward integration of object aspects encoded by discrete clusters of neurons in the TEO would facilitate rapid object recognition in the injected column in TE8. An alternative is that recurrent processing within the intrinsic TEO network may iteratively refine feedforward stimulus signals that are initially ambiguous9,10,11. Since, however, neurons in what we are proposing as two TEO systems are intermingled, some degree of interaction is likely.
Results
Feedforward-projecting neurons in TEO are visualized
Visualization of transported tracer in vivo was initiated by injections of red fluorescence tracer CTB-Alexa555 into cortical area TE in three macaque monkeys (M1, M2, and M3: Fig. 1(c),(f),(i)). The maximum diameters of the three injections were 400 μm (M1), 350 μm (M2), and 400 μm (M3) as determined by a previously described method12.
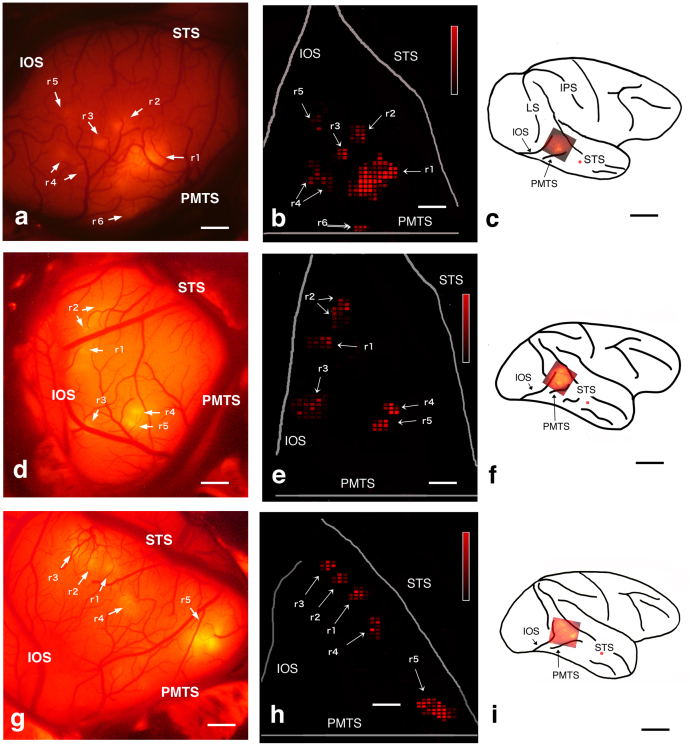
Figure 1. In vivo SCI and comparison with a color-coded flat map of CTB-Alexa555 staining.
(a), (d), (g): In vivo SC images of the TEO of monkeys M1, M2, and M3, respectively. Arrows indicate areas of fluorescence. (b), (e), (h): two-dimensional “unfolded” labeled neuron density map of the cortical surface, which was constructed from coronal sections of TEO obtained from M1, M2, and M3, respectively. A program (gifted by Dr. Eiji Hoshi: current position is written in acknowledgements) was used to make these map. This program allowed us to load and display digitized data (i.e., plotted neurons and drown contours), to put landmarks on the displayed section for future alignment, and to draw the middle line of layer 4 onto which labeled neurons in all layers were projected. Using this program, all TEO lines of layer 4 with projected neurons were unfolded and aligned vertically so that the dorsal bank of PMTS could be horizontal. Distance between lines were set to 250 μm, since we examined each section (50 μm thickness) in every 5 section (total 250 μm). To display the density of labeled neurons, we divided the unfolded map into square pixels (250 μm×250 μm), and the number of neurons contained in one left line of a pixel was assigned to neuronal density of the pixel. Note that fluorescence spots and flat map patches can be found at similar locations, shape and size ((a) and (b): (d) and (e):(g) and (h)), (c), (f), (i): Brain surface schematics showing the positions of the injections (red spots) and the locations shown in the (a), (d), and (g), respectively. Abbreviations: IOS, inferior occipital sulcus; IPS, intraparietal sulcus; LS, lunate sulcus; PMTS, posterior middle temporal sulcus; r+Arabic number, red spot or patch + Arabic number; STS, superior temporal sulcus. Scale bar represent 1 mm (a),(b),(d),(e),(g) and 10 mm (c),(f),(i). Heat map scale of neuron density of a pixel represents 0–80/250 × 250 μm2 (minimum-maximum) in (b),(d),(e). Brighter color indicates higher neuron density. When density of a pixel is lower than 19/250 × 250 μm2, color is black.
The cortical distribution of transported red signal was examined 10–14 days later by a fluorescence stereomicroscope with appropriate filter (in vivo surface connection imaging, SCI: see Methods), within a triangular area corresponding to area TEO dorsal to PMTS (Fig. 1(a),(d),(g)). This is bounded by the superior temporal sulcus (STS), inferior occipital sulcus (IOS), and posterior middle temporal sulcus (PMTS). Six patches were evident in this field in M1, and five patches in M2 and M3.
Intrinsic connections within TEO are labeled
To investigate the relationship between feedforward and intrinsic projections in TEO, in three monkeys schematically illustrated in Fig. 1, green fluorescent tracer CTB-Alexa488 was injected into one of the red patches, as visualized in a second surgical procedure (Fig. 2(a),(b),(c)). In addition, CTB-gold was injected 2 mm apart from the injection of CTB-Alexa488, and positioned so as not to fall in the red patches (Fig. 2(a),(b), 4, Supplementary Fig. S2,S4). The CTB-gold injection was intended to provide further data on the organization of patches of intrinsic connections. Fourteen days after these injections, the three monkeys were sacrificed. Patches of green (CTB-Alexa488 labeled) and silver (CTB-gold labeled as revealed by a silver enhancement kit) neurons within TEO were observed in coronal sections (Fig. 2(d),(e), Supplementary Fig. S1). In addition, higher magnification microscopic examination revealed green terminal-like structures and labeled cell bodies within green patches (Fig. 3(d)).
Figure 2. Triple-color injection based on in vivo SCI: Explanation of method and photographs of results.
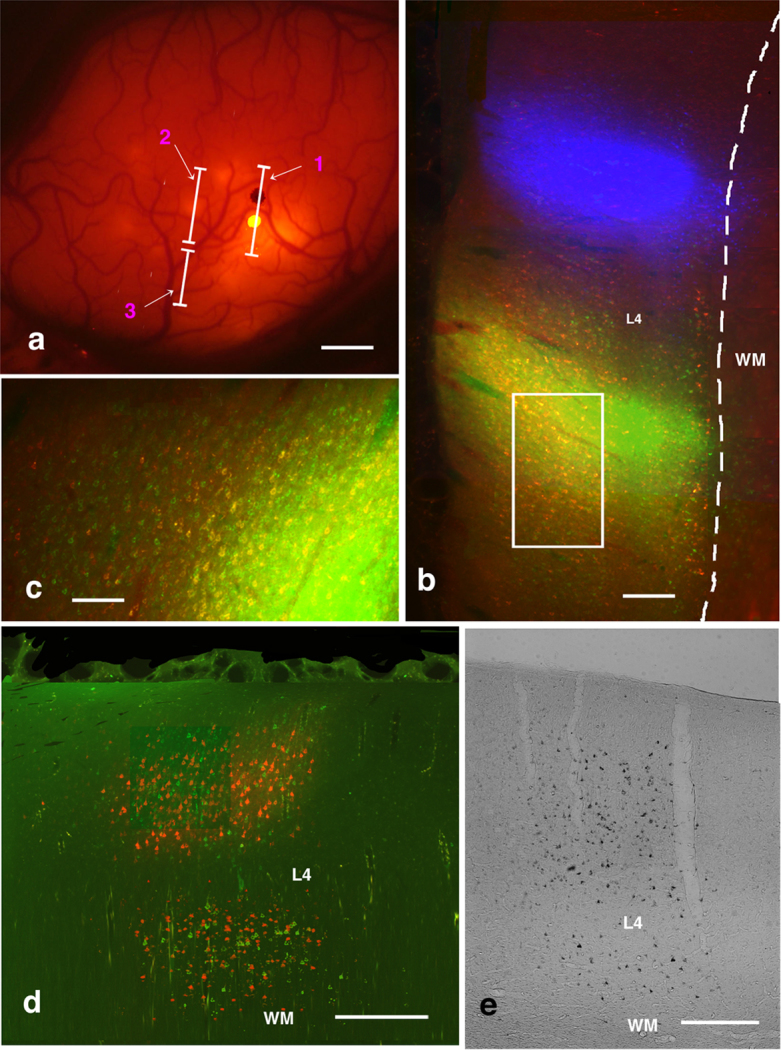
(a) In vivo SC imaging of M1 after injection of CTB-Alexa488 (a bright yellow spot on line 1) and CTB-gold (a black spot on line 1). Yellow spots demarcate red-stained patches, and black dots areas without red-stained patches. Lines 1, 2, and 3 indicate positions of the coronal sections in b, d and e, respectively. (c) Coronal section along line b in (a). This image confirms CTB-Alexa488 (green) injection was within patches of red-stained neurons, whereas the CTB-gold (pseudo-color blue) injection site was outside patches of red-stained neurons. (c) Enlarged image of rectangular area in (b). There were many yellow fluorescent (double-labeled) neurons close to CTB-Alexa488 injection sites. (d) Coronal section along line 2 in (a). Patches of red- and green-stained neurons were intermingled, but it was difficult to find double-labeled neurons. (e) Coronal section along line 3 in (a). Example of a patch of silver-stained neurons. Even using a fluorescence microscope, it was difficult to find double- or triple-labeled neurons. Abbreviations: L4, layer 4; WM, white matter. Scale bar represent 1 mm (a), 500 μm (b), 100 μm (c), and 250 μm (d and e).
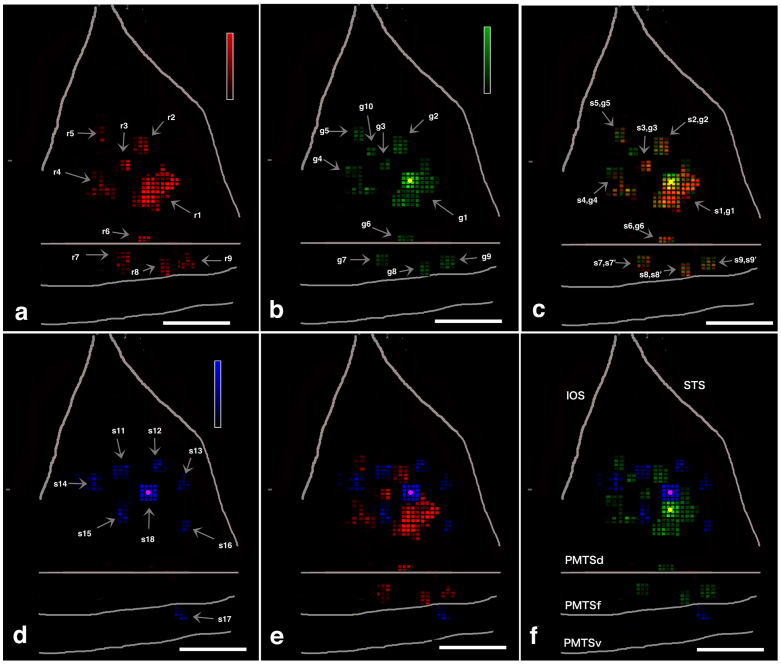
Figure 4. Spatial relationship among one feedforward projection (red) and two intrinsic networks (green and silver) in M1.
Color-coded flat maps of red-, green-, blue-, silver-, and double-stained neurons. Color-coded flat maps of red-stained pixels (a), green-stained pixels (b) red- and green-merged pixels (c), silver-stained pixels (d), red- and silver-merged pixels (e), and green- and silver-merged pixels (f). CTB-Alexa488 injection sites are yellow filled circle in (b), (c), and (f), and CTB-Gold are purple dots in (d), (e), and (f). Abbreviations: g+Arabic number, green spot + Arabic number; IOS, inferior occipital sulcus; OTSl, lateral bank of occipitotemporal sulcus; OTSf, fundus of occipitotemporal sulcus; PMTSd, dorsal bank of posterior middle temporal sulcus, PMTSf, fundus of posterior middle temporal sulcus, PMTSv, ventral bank of posterior middle temporal sulcus, r+Arabic number, red spot + Arabic number, s+Arabic number, silver spot+Arabic number, when two color spots are spatially well-matched, we called as spot + number, spot' + number as if one spot. STS, superior temporal sulcus. Scale bar represents 1 mm (a–f). Heat map scale of density of a pixel represents 0–80/250 × 250 μm2 (minimum-maximum) in (b, e, d). Brighter color indicates higher neuronal density. When density of a pixel is lower than 19/250 × 250 μm2, color is black.
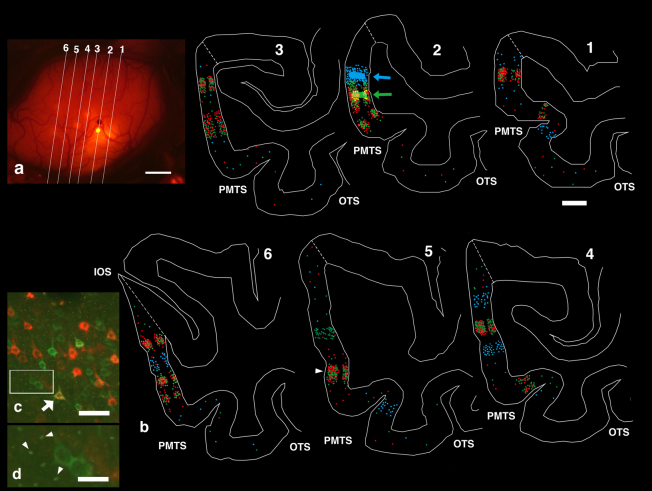
Figure 3. Plotted coronal sections of M1.
(a) Lines demarcate the coronal sections shown in (b). (b) Location of labeled neurons (color-coded) within the coronal sections in (a). Red, green, and blue circles represent red-, green-, and silver neurons. Plotting was limited to TEO. Dotted lines close to STS and IOS was dorsal borders of TEO. Ventral borders were omitted because they did not contain labeled neurons. Arrows (Section 2) point to injection sites of CTB-Alexa488 and CTB-gold tracers. Arrowhead (section 5) points to the overlapping patch shown in detail in (c) and (d). (c) Enlarged images of layer 3 within the overlapping patch demarcated by the arrowhead in (b). White arrow marks a double-labeled cell intermingled with other red and green neurons. (d) Enlarged image from rectangular area in (c). Arrowheads point to structures resembling synaptic terminals. Abbreviations: Arabic number, section number in a and b; IOS, inferior occipital sulcus; OTS, Occipital temporal sulcus; PMTS, posterior middle temporal sulcus. Scale bar represent 1 mm (a), 2 mm (b), 50 μm (c), 15 μm (d).
Confirmation of in vivo SCI by a two-dimensional “unfolded” labeled neuron density map based on histological sections
Two weeks following injection of the tracers in the TEO field, animals were sacrificed, serial coronal sections were histologically processed (Fig. 2,3(b)(c)(d), Supplementary Fig. S1,S3) and the sections were used to construct a two-dimensional “unfolded” labeled neuron density map of the cortical surface2,13,14,15 (Fig. 1(b),(e),(g),4, Supplementary Fig. S2,S4). This map was used since TEO has a convoluted and distorted shape due to the deep posterior middle temporal sulcus (PMTS)16. The map consisted of pixels (250 μm×250 μm), where pixels consisted of flattened cuboidals with top and bottom corresponding to the given position. Pixel height represented the collapsed distance from the pia surface to the white matter. The number of labeled neurons in each pixel was colour-coded (detailed methods, see legend of Fig. 1). In our convention, pixels corresponding to a defined “patch” should have a high neural density (>20) of neurons, and interpatch pixels should have 0–19 neurons. Thus, the unfolded map can show at a glance the distribution of labeled neuron density and pattern of patches. Detailed layer information can be found in a subsequent section (Cell and layer level analysis of overlapping patches).
In the unfolded map, note that it was mainly the dorsal part of TEO, dorsal to PMTS, that could be visualized by in vivo SCI (Fig. 1(a),(d),(g)), since the fluorescence light source could not reach within PMTS or to the tissue ventral to PMTS. Because of this, the number of in vivo SCI visualized patches was slightly less than the full, histologically visualized set. The histological patches as plotted on the unfolded map dorsal to PMTS were of the same position, size, and brightness as the images taken by in vivo SCI (Fig. 1(a), (b),(d),(e),(f),(g)). The correspondence of the patch pattern identified by in vivo SCI and by reconstruction of the histological sections is discussed in more detail in the Supplementary Information A.
Analysis of patches of intrinsic and projection neurons in TEO by unfolded maps
An “unfolded” labeled neuron density map of TEO13,14,15 was created for red, green and silver neurons (see legend of Fig. 1 for methods: Fig. 4, Supplementary Fig. S2,S4). This revealed nine red patches in M1, six patches in M2, and eight patches in M3 (Fig. 4a, Supplementary Fig. S2,S4). The dimensions of red patches are detailed in Supplementary Table S1 and S2 and summarized below. In brief, the long axis × short axis ranged from 0.50–2.75 mm (median, 1.25 mm) × 0.50–2.50 mm (median, 0.75 mm) and the patch area ranged from 0.25–6.75 mm2 (median, 1.10 mm2). The distances between individual red patches are detailed in Supplementary Table S3. In brief, the mean center-to-center distance between a patch and all surrounding patches was 3.05 ± 1.02 mm (mean ± SD), and the mean distance to the nearest neighbouring patch was 1.67 ± 0.51 mm.
The same convention for red neurons is applied to patches of green and silver TEO-TEO intrinsically projecting neurons. Ten green patches were observed in M1, eight in M2, and nine in M3 (Fig. 4(b), Supplementary Fig. S2,S4). Eight silver patches were observed in M1 and six in M2 and M3 (Fig. 4(d), Supplementary Fig. S2,S4). The dimensions of green and silver patches are given in again Supplementary Table S1 and S2. In brief, the long axis × short axis of green patches ranged from 0.50–2.50 mm (median, 1.00 mm) × 0.50–2.00 mm (median, 0.75 mm), and the long axis × short axis of silver patches ranged from 0.50–2.00 mm (median, 1.00 mm) × 0.50–1.00 μm (median, 0.75 mm). The area of the green patches ranged from 0.25–6.75 mm2 (median, 1.00 mm2), and the area of patches ranged from 0.25–1.31 mm2 (median, 0.60 mm2). The distances between green and silver patches are presented in detail in Supplementary Table S3. In brief, the mean center-to-center distance between a green patch and all of its surrounding neighbours was 3.15 ± 1.12 mm (mean ± SD) and that for silver patch was 3.01 ± 0.95 mm. The mean distance to the nearest neighboring patch was 1.72 ± 0.58 mm for green patches and 1.82 ± 0.52 mm for silver patches. These dimensions and inter-patch distances seemed to depend on whether anterograde or retrograde tracers are used when comparing with this study and previous studies4,17 as detailed in Supplementary Information B.
Spatial relationships between patches of red, green, and silver neurons
Three patterns were identified. 1) In the immediate vicinity of green injection sites, most of the red neurons (TEO-to-TE) were doubled-labeled with green (see below in detail, Fig. 2(b),(c)). This showed that all extrinsically projecting cortical neurons concurrently have home column, intrinsic projections18,19. 2) In an 1 × 1 mm area (16 pixels) surrounding injection needle tracks (but beyond the inner 4 pixels), the number of green neurons gradually decreased; and, accordingly, the double-labeled neurons (DL(r+g))/r also decreased (about 6%: see below in detail). 3) Close to a CTB-Alexa488 injection site, one or two green patches were identified as distinct from red patches (Fig. 4(b),(c), Supplementary Fig. S2,S4). These are interpreted as consisting of neurons with very local intrinsic connections, but without feedforward projections to the injected column in TE.
The spatial relationships of red, green and silver neurons were determined by using Spearman's rho correlation coefficient (CC) test between 3 pairs of two color neurons throughout the entire TEO in pixel level, except for the zone (16 pixels) immediately around injection sites. The CC between red and green patches over the entire TEO revealed a strongly positive spatial correlation (mean r = 0.96 for M1–M3; p < 0.001, Spearman's rho CC test; Supplementary Table S4). In contrast, the CCs between red and silver patches and between green and silver patches showed significant (p < 0.001, Spearman's rho CC test) negative spatial correlations (mean of all animals: red and silver: r = −0.61; green and silver: r = −0.59; Supplementary Table S4). That is, green patches largely overlapped with the pre-targeted red patches but interdigitated with the silver patches.
Cell- and layer-level analysis of overlapping patches
Finer examination of histological sections confirmed that red and green neurons intermingled, although the neuronal distributions exhibited some degree of segregation (Fig. 2 (d), 4, Supplementary Fig. S2,S4). We first analyzed the distribution of red and green neurons around CTB-Alexa488 (green) injection sites (Fig. 1(b),(c)) in both the supragranular layers (SGL or L2, 3) and infagranular layers (IGL or L5, 6). Within a 0.5 × 0.5 mm2 area (4 pixels) around CTB-Alexa488 injection sites, there were many double-labeled (DL(r+g)) neurons (DL(r+g)/r: 79–97%, median, 88%; Supplementary Table S5). Within a 1 × 1 mm area surrounding CTB-Alexa488 injection sites (12 pixels of 4 × 4 pixel array excluding the centralmost four pixels), the number of DL neurons was substantially lower, and DL neurons (DL(r+g)/r) comprised approximately 5–6% (Supplementary Table S5). No obvious difference was observed in the distribution of green and red neurons between SGL and IGL within this area. The probability of double labeling decreased with distance, again suggesting that feedforward red neurons have few long intrinsic collaterals within TEO.
We then analyzed the neuronal distribution pattern of red and green overlapping patches outside the 1 × 1 mm area surrounding CTB-Alexa488 injection sites (Supplementary Table S5). All of the pixels which have more than 20 red and green neurons were overlapped with exception of a few green neuron pixels (Fig. 4(c), Supplementary Fig. S2,S4, see above). In other words, red and green patches were almost completely overlapped. As these overlapping patches contained very few silver neurons, we did not include these in the analysis. Notably, although red and green overlapping patches contained many intermingled red or green neurons (n = 72,601 in 456 pixels examined), but there were surprisingly few DL neurons (n = 100 or 0.14% in the red and green neurons). This low frequency of DL neurons was observed in all cortical layers examined (illustrated in Fig. 3(c); details in Supplementary Table S6, S7, S8).
The injection halo is estimated as a 50–100 μm doughnut surrounding the injection, where the core of obscurely dense uptake detectably changes and features of neuropil can be distinguished, even against a high background. This halo zone contains a high proportion of DL neurons, but it cannot be distinguished with confidence whether label is from diffusion of the tracer or from active transport along short axon collaterals (Supplementary Table S5). At 250–500 μm from the injection center, the number of intermingled neurons is large, but the proportion of DL(r+g)/total is only 2–3% (Supplementary Table S5). If tracer diffusion were still effective at this distance, we think there would have been a distinctly higher proportion of DL neurons
Discussion
The patchy distribution of both intrinsic and extrinsic cortical connections has been repeatedly demonstrated, but much less information is available concerning the spatial organization of the two systems. In vivo SCI allowed us to visualize and selectively target patches of TEO-to-TE projecting neurons, and to investigate how these related to the subsequently visualized pattern of intrinsically projecting TEO-TEO neurons. By this means, we determined a close overlap of the two systems. Surprisingly, however, we further found that, although extrinsic and intrinsic pyramidal neurons intermingled in the same set of patches, only a very few neurons were actually double labeled. A notable exception was the high degree of double labeling for intrinsic neurons in the immediate vicinity of the TEO-injected patch; that is, these neurons also projected to TE.
This result implies two populations of intrinsic neurons within TEO. One has local terminations mainly within the home column of the parent neuron and also projects extrinsically. A second has more widespread intrinsic collaterals, but does not seem to project extrinsically. This is consistent with previous studies that have identified pyramidal neurons without extrinsic projections18,19. The small pyramids in non-primary cortical areas in the macaque are another, abundant pyramidal population that does not have extrinsic projections.
Laminar and tangential heterogeneity in the pattern of local pyramidal collaterals has been abundantly demonstrated in in vitro experiments, and correlated with different extrinsically projecting populations20,21. While more data about specific patterns are needed, one interesting example is the population of Meynert cells in area V1. These have both extrinsic projections and widespread local collaterals, but there is a relative lack of local collaterals in the vicinity of the parent neuron (“home column”; see fig. 6 in Rockland and Knutson)22.
Important to keep in mind, however, is the possibility that a proportion of the green (putative intrinsic-only) neurons do have extrinsic projections, but that these are directed to targets other than the injected tissue in area TE. These targets could be to other, non-injected parts of TE, other cortical areas (V4, FST), or, for the infragranular neurons, to subcortical targets23. Ultimately, the question of how intrinsic and extrinsic collaterals are arranged needs to be addressed by mapping the entire axonal tree for individual pyramidal neurons; for example, by intra- or juxtacellular fills in in vivo or other comparable methods24,25.
In vivo SCI is a relatively simple and reliable method for pre-targeting connectionally identified populations (and see Supplemental Infromation C for other applications). In this report, we investigated two relatively close together areas (TEO and TE), but the method can easily be applied to more distant interconnected regions; for example, prefrontal cortex and inferotemporal or parietal cortex. Potential limitations include: the need for relatively large craniotomy (but, this can be repaired post-surgically by artificial dura or other means), the need for a strongly fluorescent tracer in the first experiment, and the obvious need for a lissencephalic brain or at least a relatively large gyral expanse to visualize transported tracer and target the second injection.
Theoretical models suggest two alternative mechanisms for synthesizing feature information into global shape representations by the ventral visual pathway. One is selective feedforward convergence, where purely feedforward summation of object aspects by TE neurons would facilitate rapid object recognition7. An alternative model is that recurrent processing iteratively refines feedforward stimulus signals that are initially weak or ambiguous9,10,11. In TEO, Brincat and Connor found that neural responses occurring immediately after presentation of visual stimuli consisting of multiple and dissociable features carried information about individual features formed in V410. Subsequently, information about specific multipart configurations emerged, building gradually over the course of 60 ms to produce a more explicit representation of object shape. This gradual transformation can result from a recurrent network process that effectively compares aspect signals across neurons to generate inferences about multi-aspect shape configurations.
Our anatomical findings provide evidence for a recurrent network of at least two parallel systems. One system is part of a feedforward chain that can send information on salient aspects of an object rapidly to TE. These neurons interact with other TEO neurons only within the range of small clusters (<250 μm)26. A separate system in TEO does not send information directly to TE but may help compute multipart shape configurations via an intrinsic network within TEO. These two systems may engage in multiple potential interactions in the overlapping patches of intermixed feedforward and intrinsic TEO neurons11. The intrinsic system itself consists of what can be viewed as interdigitating patches (i.e., the neurons labeled by the green and silver injections) or as a patch and matrix pattern, broadly analogous to the pattern of cytochrome oxidase patches and their surrounding matrix in the primary visual cortex27.
Visual responses in TEO are more sensitive to changes in position, size, and point-of-view than those in TE, while the TE response is object specific but less invariant8. Maintaining this robust invariance in TE may require that signals encoding ambiguous features of an object not be transmitted rapidly to TE by TEO feedforward projections, but rather be redundantly processed via recurrent networks of intrinsic neurons in TEO. In the future, we hope to test the specific functions of these parallel circuits using an in vivo connection approach combined with two-photon calcium imaging28.
Methods
Animals
Five male rhesus monkeys (Macaca mulatta) weighing 4.3, 4.0, 4.2, 5.5, and 4.5 kg were used in this study (Supplementary Table S9). Cortical connections in three monkeys (M1, M2, and M3) were examined by in vivo SCI (Supplementary Table S9) using retrograde tracers. The fourth monkey (M4) was used for in vivo SCI plus postmortem DiI labeling to confirm that bright spots visualized from the cortical surface were aggregates of neurons labeled by the tracers. The fifth monkey (M5) was used to compare the labeling efficiency of CTB-conjugated tracers (Supplementary Table S9). Surgery was performed under sterile conditions and deep barbiturate anesthesia (35 mg/kg Nembutal i.v., 11 mg/kg ketamine i.m.)2,12. All animal procedures were performed in accordance with official Japanese regulations for research on animals, the US National Institutes of Health Guide for the Care and Use of Laboratory Animals (NIH Publication No. 80–23), and institutionally approved protocols (RIKEN, Brain Science Institute).
Retrograde tracing
CTB-Alexa488 (Invitrogen-Molecular Probes, Eugene, OR), CTB-Alexa555 (Invitrogen-Molecular Probes), and 7-nm colloidal gold (CTB-gold; List Biological Laboratories, Inc., Campbell, CA) were used as retrograde tracers. Tracers were diluted to 1% in 0.1 M PBS. For cortical injection, a 0.12 μL volume of tracer solution was pressure injected through a 50-μm glass micropipette attached to a 10-μL Hamilton syringe12.
Injection sites were visualized immediately after injection, using a fluorescence stereomicroscope (VB-G05, Keyence Corporation, Osaka, Japan) with a filter for red fluorescent protein (RFP, emission: 540/25, absorption: 572; Fig. 1 of ref. 15) in order to confirm delivery of the tracer.
Tracer injection
In all monkeys, the cortical areas of interest were localized by direct visualization subsequent to craniotomy and durotomy, using STS and AMTS as landmarks. M1–M4 were injected with CTB-Alexa555 and M5 was injected with a mixture of CTB-Alexa555, CTB-Alexa448, and CTB-gold. Injections were administered into TE between STS and AMTS (Fig. 1(c),(f),(g)). After the injection, the dura was sutured and the wound was closed.
Tracer injection after in vivo SCI
Ten (M1), 11 (M2), or 14 days (M3) after the first injection, the animals were deeply anesthetized with barbiturate (35 mg/kg Nembutal i.v. after a tranquilizing dose of 11 mg/kg ketamine i.m.). Craniotomy and durotomy were performed caudal to the first injection to expose dorsal TEO (bounded by STS, PMTS, and IOS; Fig. 1(a),(d),(g)). The targeted injection field was visualized using a fluorescence stereomicroscope with filters for RFP and an exposure time of 10–60 s. Three to seven bright red patches were usually distinguishable, with patch sizes varying from 500–1000 µm in diameter. CTB-Alexa448 was then injected at the edge of a selected bright patch and as close as possible to the middle of the field of patches. CTB-gold was injected into an area that was approximately 2 mm away from the centre of the CTB-Alexa448 injected patch, in a zone that was devoid of CTB-Alexa555 fluorescence (Fig. 2(a), (b)).
Fixation and tissue preparation
After an additional survival period of 14 days (M1–M3) (a survival period of 13 days was used after the single injection in M4 and M5), monkeys were tranquilized with 11 mg/kg ketamine i.m., sacrificed with an overdose of Nembutal (75 mg/kg, i.p.), and perfused transcardially, in sequence, with saline containing 0.5% sodium nitrite (for 2 min), 4 L of 4% paraformaldehyde in PBS (30 min), and ice-cold 0.1 M PBS with 10%, 20%, and 30% sucrose. For processing of the M4 brain, see Supplemental information A.
Brains were removed and coronal sections were prepared at 50 μm in a series of five sections. The CTB-Alexa488 signal was enhanced by immunofluorescence, and sections reacted for CTB-gold were silver enhanced. Other sections were used for Nissl staining and/or immunoperoxidase staining to visualize CTB-Alexa488-loaded neurons.
Enhancement of CTB-Alexa488 signal by immunofluoresence
Sections were blocked in 0.1 M PBS (pH 7.4) containing 0.5% Triton X-100 and 5% normal goat serum (PBS-TG) for 1 h at room temperature, and subsequently incubated with rabbit anti-Alexa488 antibody (1:1000; Invitrogen-Molecular Probes) in PBS-TG for 2 days at 4°C. After washing with 0.1 M PBS, the sections were incubated for 1.5 h at room temperature with Alexa488-conjugated anti-rabbit polyclonal goat antibody (1:200; Invitrogen-Molecular Probes). To combine this procedure with the visualization of CTB-gold (see next section), silver enhancement of CTB-gold was performed first.
Visualization of CTB-gold by silver enhancement
Sections were washed with 0.1 M PBS, followed by 0.01 M PBS. The IntenSE M silver enhancement kit (Amersham International, Little Chalfont, Bucks, UK) was used to visualize CTB-gold signals13 using a 1:1 cocktail of IntenSE M kit solution and 33% gum arabic solution. Reactions were monitored under a microscope and terminated by rinsing the sections in 0.01 M PBS followed by several rinses in 0.1 M PBS. The combined incubation times were approximately 2 h.
Immunoperoxidase reaction for CTB-Alexa488
To mitigate the problem of fluorescence fading after prolonged light exposure, immunoperoxidase staining of CTB-Alexa488 was performed on the third section in each series. The sections were blocked in PBS-TG for 1 h at room temperature, and subsequently incubated with an Alexa488-conjugated anti-rabbit antibody (1:1000; Invitrogen-Molecular Probes) in PBS-TG for 2 days at 4°C. After washing with 0.1 M PBS, the sections were incubated with biotinylated anti-rabbit polyclonal goat antibody (1:200; Vector Laboratories Inc., Burlingame, CA) for 1.5 h at room temperature. Immunoreactivity was visualized using the ABC Elite kit (Vector Laboratories Inc.), followed by diaminobenzidine histochemistry with 0.03% nickel ammonium sulfate.
All sections were mounted on gelatin-coated glass slides, air dried, dehydrated in graded EtOH solutions, immersed in xylene, and coverslipped in DPX (Sigma-Aldrich Co., Buchs, Switzerland).
Analysis
Fluorescent signal was analyzed by using standard sets of filters: fluorescein (for CTB-Alexa488) or rhodamine (for CTB-Alexa555). CTB-Alexa488 and CTB-Alexa555 labeled neurons were identified by a green and red granular fluorescence in the cytoplasm, respectively. In the one section of each 5, reacted for CTB-gold, labeled cells were identified in dark field by the granular precipitate in the cytoplasm. In the coronal sections, the distribution of retrogradely labeled cells was analyzed and plotted in every 250 μm. Material was analyzed with a Nikon Eclipse E-800 microscope (Nikon Co,, Tokyo, Japan), at 100x-200x-400x, and plotting of neurons and drawing outer suface, white matter and the middle of layer 4 were carried out by using Neurolucida (MBF Bioscience, Williston, VT), through a MicroFIRE digital camera (MicroFire Technology Company, Ltd. Shenzhen, China) incorporated in the microscope.
Injection sites were defined according to previous studies13. The core of the injection site for CTB-Alexa 488 and CTB-Alexa 555, considered as the effective tracer uptake area, included the intensely fluorescent area around the needle track. The core of the injection site for CTB-gold was identified as the densely stained region surrounding the needle track.
Identification of areas is not straightforward in the occipitotemporal region. There is considerable individual variability, and the borders between areas are ambiguous or gradual16,29,30,31,32,33. To identify the location of projection foci and injection sites, we carefully compared sulcal landmarks in our study with those of published physiological and anatomical maps16,29,30,31,32,33.
For quantitative analysis, in M1–M3, we manually scored the number of green-, red-, silver-, double- and triple-labeled neurons and plotted the counts at 250 μm intervals using Neurolucida software. In Supplementary Table S2, S3, the total number of labeled neurons is given for each area or cortical region. The percentage of double-labeled (DL(g+r)) neurons was calculated with respect to the total number of labeled cells after adding the two tracers together (DL(g+r)/total) and then recalculated for each tracer separately (DL/g and DL/r). The calculations were performed and tables were constructed using Microsoft Office Excel (Microsoft Corporation, Redmond, WA).
In the unfolded map, distances between patches were calculated using NeuroExplorer (MBF Bioscience, Williston, VT) after images were uploaded in the computer.
Photographic presentation
Images were captured using an AxioCam HRc (Carl Zeiss AG, Oberkochen, Germany) and an AxioScope 2 plus microscope (Carl Zeiss AG), since images were clearer than Neurolucida set up. Individual images were processed in Adobe Photoshop (Adobe Systems Incorporated, San Jose, CA) and assembled into digital montages. Image adjustments were limited to brightness and contrast when required.
Author Contributions
NI and KR designed the experiments; NI performed the experiments; NI and EB performed data analysis; NI and KR wrote the manuscripts with input from EB.
Supplementary Material
Supplementary Information
Acknowledgments
This work was supported by a Funding Program for World-Leading Innovative R&D on Science and Technology (FIRST Program), by Strategic Research Program for Brain Science, by a Grant-in-Aid for Scientific Research on Innovative Areas “Face perception and recognition” and “Shitsukan” by the Ministry of Education, Sciences, Sports and Culture, Japan, and by an Intramural Research Grant (grant number 23–7) for Neurological and Psychiatric Disorders from the National Center of Neurology and Psychiatry. We thank Drs. Manabu Tanifuji, Tohru Kurotani and Mr. Takayuki Sato for helping to develop in vivo SCI in early stage, Ms. Hiromi Mashiko and Yoshiko Abe for their excellent technical support, and Dr. Eiji Hoshi (Tokyo Metropolitan Institute of Medical Science) for generously providing a program to create two-dimensional “unfolded” neuron density maps from Neurolucida data.
References
- Moeller S. Patches with links: a unified system for processing faces in the macaque temporal lobe. Science 320, 1355–1359 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banno T., Ichinohe N., Rockland K. S. & Komatsu H. Reciprocal connectivity of identified color-processing modules in the monkey inferior temporal cortex. Cereb. Cortex 21, 1295–1310 (2011). [DOI] [PubMed] [Google Scholar]
- Rockland K. S., Lund J. S. & Humphrey A. L. Anatomical banding of intrinsic connections in striate cortex of tree shrews (Tupaia glis). J. Comp. Neurol. 209, 41–58 (1982). [DOI] [PubMed] [Google Scholar]
- Fujita I. & Fujita T. Intrinsic Connections in the macaque inferior temporal cortex. J. Comp. Neurol. 368, 467–486 (1996). [DOI] [PubMed] [Google Scholar]
- Pucak M. L., Levitt J. B., Lund J. S. & Lewis D. A. Patterns of intrinsic and associational circuitry in monkey prefrontal cortex. J. Comp. Neurol. 376, 614–630 (1996). [DOI] [PubMed] [Google Scholar]
- Lund J. S., Yoshioka T. & Levitt J. B. Comparison of intrinsic connectivity in different areas of macaque monkey cerebral cortex. Cereb Cortex. 3, 148–162 (1993). [DOI] [PubMed] [Google Scholar]
- Tanaka K. Inferotemporal cortex and object vision. Annu. Rev. Neurosci. 19, 109–139 (1996). [DOI] [PubMed] [Google Scholar]
- Panzeri S., Rolls E. T., Battaglia F. & Lavis R. Speed of feedforward and recurrent processing in multilayer networks of integrate-and-fire neurons. Network 12, 423–440 (2001). [PubMed] [Google Scholar]
- Schendan H. E. & Kutas M. Neurophysiological evidence for the time course of activation of global shape, part, and local contour representations during visual object categorization and memory. J Cogn. Neurosci. 19, 734–749 (2007). [DOI] [PubMed] [Google Scholar]
- Brincat S. L. & Connor C. E. Dynamic shape synthesis in posterior inferotemporal cortex. Neuron 49, 17–24 (2006). [DOI] [PubMed] [Google Scholar]
- van Rossum M. C., van der Meer M. A., Xiao D. & Oram M. W. Adaptive integration in the visual cortex by depressing recurrent cortical circuits. Neural. Comput. 20, 1847–1872 (2008). [DOI] [PubMed] [Google Scholar]
- Borra E., Ichinohe N., Sato T., Tanifuji M. & Rockland K. S. Cortical connections to area TE in monkey: hybrid modular and distributed organization. Cereb. Cortex 20, 257–270 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dum R. P. & Strick P. L. An unfolded map of the cerebellar dentate nucleus and its projections to the cerebral cortex. J. Neurophysiol. 89, 634–639 (2003). [DOI] [PubMed] [Google Scholar]
- Saga Y., Hirata Y., Takahara D., Inoue K., Miyachi S., Nambu A., Tanji J., Takada M. & Hoshi E. Origins of multisynaptic projections from the basal ganglia to rostrocaudally distinct sectors of the dorsal premotor area in macaques. Eur. J. Neurosci. 33, 285–297 (2011). [DOI] [PubMed] [Google Scholar]
- Suzuki W. A. & Amaral D. G. Topographic organization of the reciprocal connections between the monkey entorhinal cortex and the perirhinal and parahippocampal cortices. J. Neurosci. 14, 856–1877 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boussaoud D., Desimone R. & Ungerleider L. G. Visual topography of area TEO in the macaque. J. Comp. Neurol. 306, 554–575 (1991). [DOI] [PubMed] [Google Scholar]
- Felleman D. J., Xiao Y. & McClendon E. Modular organization of occipito-temporal pathways: cortical connections between visual area 4 and visual area 2 and posterior inferotemporal ventral area in macaque monkeys. J. Neurosci. 17, 3185–3200 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs F. & Callaway E. M. Layer-specific input to distinct cell types in layer 6 of monkey primary visual cortex. J. Neurosci. 21, 3600–3608 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs F. & Callaway E. M. Laminar patterns of local excitatory input to layer 5 neurons in macaque primary visual cortex. Cereb Cortex. 205, 479–488 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz L. C. Local circuitry of identified projection neurons in cat visual cortex brain slices. J. Neurosci. 7, 1223–1249 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas R. J. & Martin K. A. Neuronal circuits of the neocortex. Annu. Rev. Neurosci. 27, 419–451(2004). [DOI] [PubMed] [Google Scholar]
- Rockland K. S. & Knutson T. Axon collaterals of Meynert cells diverge over large portions of area V1 in the macaque monkey. J. Comp. Neurol. 441, 134–147 (2001). [DOI] [PubMed] [Google Scholar]
- Webster M. J., Bachevalier J. & Ungerleider L. G. Subcortical connections of inferior temporal areas TE and TEO in macaque monkeys. J Comp Neurol. 335, 73–91 (1993). [DOI] [PubMed] [Google Scholar]
- Johnson D. M., Illig K. R., Behan M. & Haberly L. B. New features of connectivity in piriform cortex visualized by intracellular injection of pyramidal cells suggest that “primary” olfactory cortex functions like “association” cortex in other sensory systems. J. Neurosci. 20, 6974–6982 (2000) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiyama F., Sohn J., Nakano T., Furuta T., Nakamura K. C., Matsuda W. & Kaneko T. Exclusive and common targets of neostriatofugal projections of rat striosome neurons: a single neuron-tracing study using a viral vector. Eur J Neurosci. 33, 668–677 (2011). [DOI] [PubMed] [Google Scholar]
- Perin R., Berger T. K. & Markram H. A synaptic organizing principle for cortical neuronal groups. Proc. Natl. Acad. Sci. U S A. 108, 5419–5424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sincich L. C. & Horton J. C. The circuitry of V1 and V2: integration of color, form, and motion. Annu. Rev. Neurosci. 28, 303–326 (2005) [DOI] [PubMed] [Google Scholar]
- Jarosiewicz B., Schummers J., Malik W. Q., Brown E. N. & Sur M. Functional biases in visual cortex neurons with identified projections to higher cortical targets. Curr. Biol. 22, 269–277 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobatake E. & Tanaka K. Neuronal selectivities to complex object features in the ventral visual pathway of the macaque cerebral cortex. J. Neurophysiol. 71, 856–867 (1994). [DOI] [PubMed] [Google Scholar]
- Zeki S. Are areas TEO and PIT of monkey visual cortex wholly distinct from the fourth visual complex (V4 complex)? Proc. Biol. Sci. 263, 1539–1544 (1996). [DOI] [PubMed] [Google Scholar]
- Seltzer B. & Pandya D. N. Afferent cortical connections and architectonics of the superior temporal sulcus and surrounding cortex in the rhesus monkey. Brain Res. 149, 1–24 (1978). [DOI] [PubMed] [Google Scholar]
- Yukie M., Takeuchi H., Hasegawa Y. & Iwai E. Differential connectivity of inferotemporal area TE with the amygdala and the hippocampus in the monkey. .In: Iwai E., Mishkin M. editors. Vision, memory, and the temporal lobe. New York: Elsevier; pp. 129–135 (1990).
- Saleem K. S. & Tanaka K. Divergent projections from the anterior inferotemporal area TE to the perirhinal and entorhinal cortices in the macaque monkey. J Neurosci. 16, 4757–4775 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information