Summary
Telomeres play crucial roles in the maintenance of genome integrity and control of cellular senescence. Most eukaryotic telomeres can be transcribed to generate a telomeric repeat-containing RNA (TERRA) that persists as a heterogeneous nuclear RNA and can be developmentally regulated. However, the precise function and regulation of TERRA in normal and cancer cell development remains poorly understood. Here, we show that TERRA accumulates in highly proliferating normal and cancer cells, and forms large nuclear foci, which are distinct from previously characterized markers of DNA damage or replication stress. Using a mouse model for medulloblastoma driven by chronic Sonic hedgehog (SHH) signaling, TERRA RNA was detected in tumor, but not adjacent normal cells using both RNA fluorescence in situ hybridization (FISH) and northern blotting. RNA FISH revealed the formation of TERRA foci (TERFs) in the nuclear regions of rapidly proliferating tumor cells. In the normal developing cerebellum, TERRA aggregates could also be detected in highly proliferating zones of progenitor neurons. SHH could enhance TERRA expression in purified granule progenitor cells in vitro, suggesting that proliferation signals contribute to TERRA expression in responsive tissue. TERRA foci did not colocalize with γH2AX foci, promyelocytic leukemia (PML) or Cajal bodies in mouse tumor tissue. We also provide evidence that TERRA is elevated in a variety of human cancers. These findings suggest that elevated TERRA levels reflect a novel early form of telomere regulation during replication stress and cancer cell evolution, and the TERRA RNA aggregates may form a novel nuclear body in highly proliferating mammalian cells.
Key words: Telomere repeat-containing non-coding RNA, TERRA, Neuronal progenitors, Cancer, Cerebellum, Medulloblastoma, Sonic hedgehog
Introduction
Telomeres are repetitive DNA structures at the ends of linear chromosomes required for chromosome maintenance and genome stability (Blackburn et al., 2006; Cech, 2004). Mammalian telomere DNA consists of variable length double stranded TTAGGG repeats ending in a single stranded 3′ overhang that can form complex higher-order nucleoprotein structures to cap chromosome ends (de Lange, 2002; de Lange, 2004). The telomere-associated proteins, termed shelterin or telosome, are important for telomere end-protection and length regulation (de Lange, 2005a; Liu et al., 2004; Palm and de Lange, 2008). In proliferating cells, telomere repeat length is maintained by an elaborate mechanism involving DNA replication machinery, telomerase and its associated factors, and nucleolytic end-processing enzymes (Jain and Cooper, 2010; Ye et al., 2010). In the absence of telomerase activity, telomeres shorten by attrition, and critically short telomeres elicit a DNA damage signal that can lead to cell cycle arrest and replicative senescence (Sahin and Depinho, 2010). Loss of telomere capping function leads to cell cycle arrest in normal senescing cells, but this protective mechanism is bypassed in normal proliferating cells and potentially further deregulated in human cancers (Blasco, 2005; Blasco, 2007; De Lange, 2005b).
TERRA is a heterogeneous length non-coding RNA transcribed through the terminal telomere repeats of eukaryotic chromosomes (Azzalin et al., 2007; Schoeftner and Blasco, 2008). In fission yeast, transcripts can be detected for both sense and antisense strands of the telomere repeat tract and adjacent telomere sequence (Bah et al., 2012; Greenwood and Cooper, 2012). TERRA RNA can interact with shelterin proteins to regulate telomere heterochromatin formation (Deng et al., 2009) and capping by the telomere single strand DNA binding protein Pot 1 (Flynn et al., 2011; López de Silanes et al., 2010). TERRA can also interact with the catalytic subunit of telomerase (TERT) and inhibit telomerase enzyme activity in vitro (Redon et al., 2010; Schoeftner and Blasco, 2008). TERRA RNA expression can be regulated by several mechanisms, including developmental status, cellular stress, and telomere epigenetic state (Azzalin et al., 2007; Caslini et al., 2009; Deng et al., 2009; Schoeftner and Blasco, 2008). In addition, TERRA levels are elevated in both human and mouse iPS cells, suggesting that TERRA expression correlates with proliferative capacity and contributes to nuclear reprogramming (Marion et al., 2009; Yehezkel et al., 2011). However, the expression and function of TERRA in the context of cancer remains poorly understood.
To investigate the regulation and function of TERRA in normal and cancer cell development, we examined TERRA expression in human cancer biopsies and in a mouse model for medulloblastoma. Medulloblastoma is a malignancy of the cerebellum with the highest incidence of all human pediatric malignant brain tumors (Ellison, 2010). A subset of these tumors is thought to arise from deregulation of granule neuron progenitors (GNPs) development in the cerebellum (Ellison, 2002; Ellison, 2010; Gilbertson, 2004; Gilbertson and Ellison, 2008). GNPs are subject to very high rates of proliferation during the early postnatal period. Sonic hedgehog (SHH) is required for the rapid proliferation of these neuronal progenitors during normal development (Dahmane and Ruiz i Altaba, 1999; Wallace, 1999; Wechsler-Reya and Scott, 1999). Patched 1 (Ptch1) is a receptor for the SHH ligand and an important negative regulator of SHH signaling. Mutations in Ptch1 can lead to medulloblastoma in human (Hahn et al., 1996; Johnson et al., 1996) and mouse models (Goodrich et al., 1997) (reviewed by Corcoran and Scott, 2001; Ruiz i Altaba et al., 2002). In this work, we show that normal and cancer proliferating granule neuron progenitors express high level of TERRA and exhibit formation of TERRA foci. These foci (TERFs) are distinct from γH2AX DNA damage foci, but occur in cells where the telomere repeat DNA has shortened. TERRA foci can also be found in highly proliferating progenitor cells during normal mouse development. Finally, we show that TERRA is elevated in various types of human cancers originating in diverse organs.
Results
TERRA form foci in a mouse model for medulloblastoma
To analyze the expression of TERRA in a mouse model of human cancer, we employed Ptch1+/− mice, a widely used genetic model for human SHH-positive subtype medulloblastoma (Ellison, 2010; Goodrich et al., 1997). These tumors are composed of proliferating GNPs marked by Math1 (also known as Atoh1) (Oliver et al., 2005), and express Gli1, a target and mediator of the SHH signaling pathway (Goodrich et al., 1997) (Fig. 1A; supplementary material Fig. S1). Tumor can be readily distinguished from normal/non-tumor cerebellar tissue based on histology and in situ expression analysis of various markers (Fig. 1A; supplementary material Fig. S1). To examine TERRA expression in mouse normal and cancer tissue, we first employed RNA fluorescence in situ hybridization (FISH) using methods that have been optimized for detection of rare and unstable RNA (Deng et al., 2009; Flynn et al., 2011). A TAMRA-conjugated PNA probe was used under non-denaturing conditions to selectively distinguish telomere RNA from telomere DNA. RNA-FISH revealed that TERRA forms discrete foci (TERFs) in the tumor cells, but not in the adjacent non-tumor cells of the same cerebellum (Fig. 1B). TERFs were not detected in sections pre-treated with RNase A, indicating that the signal detected with the TERRA probe indeed corresponds to RNA expression (Fig. 1B, lower panels; supplementary material Fig. S1B). As an additional specificity control, a FAM-conjugated PNA probe for antisense TERRA failed to detect any distinct foci (supplementary material Fig. S2B). Quantification of multiple RNA FISH experiments using computer imaging software indicated that ∼80% of tumor cells have a ∼7.5-fold greater mean fluorescence intensity relative to normal cells in adjacent non-tumor tissue (Fig. 1C–E). These findings were further confirmed by RNA FISH using a DNA oligonucleotide probe (TAACCC)7, which unlike the PNA probe, has very low capacity for binding duplex DNA. The (TAACCC)7 DNA oligonucleotide probe also revealed elevated TERFs in tumor cells with no detectable signal in the normal part of the same cerebellum (Fig. 1F). No signal for TERRA expression was observed with a mutated (TAACAC)7 version of this DNA oligo probe (Fig. 1F), further indicating that these foci are TERRA-specific and that TERRA levels are selectively elevated in tumor cells.
Fig. 1.
TERRA foci formation in mouse medulloblastoma. (A) (Top panel) Hematoxylin and Eosin staining of a region of a Ptch1+/− mouse cerebellum with non-tumor (left panel; indicated by an arrow) and tumor tissue (right panel; no. 6850). The normal morphology of cerebellar folia with the IGL (containing the mature granule neurons) and the Purkinje layer containing the Purkinje neurons is seen in left panel. Scale bar: 200 µm. (Lower panel) Sections of cerebellum containing both normal and tumor tissue showing the results of in situ hybridizations with specific digoxigenin-labeled RNA probes for Gli1 and Math1. Note that Gli1 and Math1 are highly expressed in the tumor part of the cerebellum. Asterisk in left panel denotes the normal expression of Gli1 in the normal Bergmann glial cells that form a layer in this, still organized, non-tumor part of the cerebellum. Scale bars: 100 µm. (B) Confocal images of FISH analyses of TERRA expression in sections of the cerebellum of the same mouse as above containing both normal and tumor tissue. Note the presence of TERRA expression (strong red labeling) in the tumor as compared to the low or undetectable expression in the non-tumor cerebellar tissue. Asterisk denote non-specific TERRA signal in the Purkinje layer. RNase A treatment leads to an elimination of the tumor-specific TERRA signal (middle panels). Images were taken with 40× lens (lower panels) to reveal details of TERRA localization in the nuclei. Scale bar: 20 µm. Other images were taken with 20× lens. (C) Histogram comparing relative TERRA fluorescence signal intensity (FU) in non-tumor (black) vs tumor (red) tissue, analyzed by RNA FISH and ImageProPlus software. (D) TERRA expression measured as total mean fluorescence intensity (FU). Values are means ± standard error from three independent experiments. (E) Quantification of cells with ≧1 TERRA signals of mean intensity >40 FU. More than 800 nuclei from at least three independent experiments were counted for quantification. The P-value was calculated using a two-tailed Student's t-test in all cases. (F) Confocal images of FISH analyses of TERRA expression in tumor (top panel) or non-tumor (middle panel) using an Alexa-Fluor-488-conjugated DNA oligonucleotide probe (TAACCC)7 (top two panels) or a control mutant probe (TAACAC)7 as a negative control (lower panel). Sections of the cerebellum of the same mouse (no. 7040) as in supplementary material Fig. S1 containing both non-tumor and tumor tissue were used. Scale bar: 20 µm.
Elevated TERRA expression in mouse medulloblastoma tissue
To confirm and validate RNA FISH results with other independent techniques, we next examined TERRA expression by northern blot and quantitative (q) RT-PCR using RNA isolated from dissected normal (non visible tumor) and tumor regions of Ptch1+/− cerebella (Fig. 2; supplementary material Fig. S3). In agreement with the in situ data (Fig. 1; supplementary material Fig. S1), northern blot analysis indicated that the tumor contained significantly higher levels (∼4-fold; P = 0.0301) of TERRA RNA than the non-tumor counterpart (Fig. 2A,B; supplementary material Fig. S3). RNase A treatment eliminated TERRA detection, indicating that TERRA signal consists of RNA, and not fragmented telomere DNA (Fig. 2A, right lanes). Comparable results were observed with three other matched non-tumor and tumor cerebella from different mice (supplementary material Fig. S3A). Quantitative RT-PCR analysis of TERRA expression from individual subtelomeres revealed an increase of TERRA expression from various chromosomes (Fig. 2C; supplementary material Fig. S3B,C). For example, one of the tumors presented a large increase at 11q (∼6-fold), but a more moderate increase at 2q (1.8-fold; Fig. 2C, upper panel). Marker mRNAs for medulloblastoma tumor identity (Math1 and Gli1) and post-mitotic neuronal marker (Map2) were used to confirm the accuracy of dissection (Fig. 2C, bottom panel; supplementary material Fig. S3B,C). We also observed that mRNA for the telomerase subunit TERT was not increased in tumor samples (Fig. 2C, lower panel; supplementary material Fig. S3). These results validate findings by RNA FISH, indicating that TERRA is elevated in mouse cancer tissue, and that some, but not all telomeres express high TERRA levels (Fig. 2B,C).
Fig. 2.
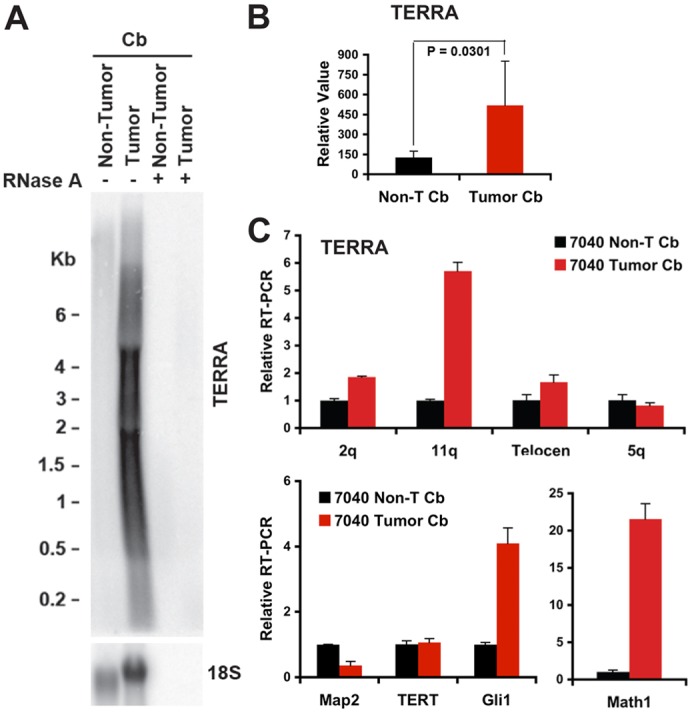
TERRA is highly expressed in mouse primary medulloblastoma. (A) Total RNA from dissected mouse medulloblastoma tumor and non-tumor tissue (no. 7040) were assayed by northern blot and the blot was first probed with 32P-labeled (TAACCC)4 for TERRA RNA expression. 18S RNA expression is shown as a quantification control. Numbers on the left show the position of RNA markers (in kb). (B) Quantification of TERRA levels from at least three independent northern blot analyses using RNA isolated from matched non-tumor and tumor cerebella in different mice, one of which is shown in A. Bar graph represents TERRA signal intensity relative to 18S signal, and relative intensity for non-tumor cerebella was set at 100. The P-value was calculated using a two-tailed Student's t-test. (C) Top panel: quantitative RT-PCR analysis of TERRA using primers specific for subtelomeres of chromosome 2q, 11q, 5q and telocentric chromosomes (Telocen). Lower panel: expression of Map2, Tert, Gli1 and Math1 in non-tumor and tumor cerebellum (no. 7040) to validate the accuracy of dissection. ΔΔCT method relative to non-T cerebellum and Gapdh were used to calculate relative RT-PCR between non-tumor and tumor cerebellum. Bar graph represents the average value from at least three independent PCR reactions (means ± s.d.).
TERRA is elevated in highly proliferating progenitor cells in the developing mouse brain
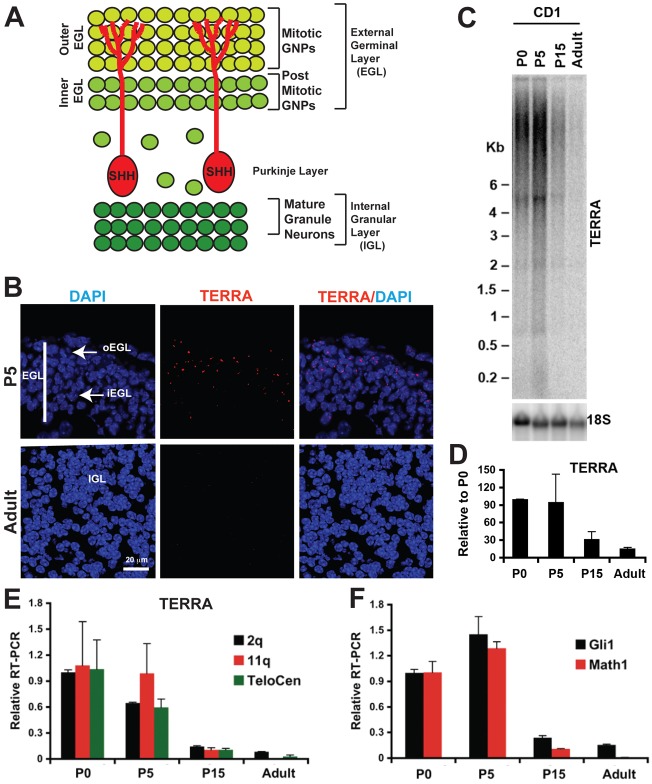
Transcriptional profiling analysis have shown that mouse postnatal (P1–P10) GNPs resemble a subtype of both mouse and human SHH-dependent medulloblastoma (Kho et al., 2004). The expression of TERRA in primary SHH-subtype medulloblastoma suggested that it might also be expressed in the rapidly dividing postnatal GNPs. To investigate whether TERRA expression correlates with normal proliferating GNPs, we first used RNA FISH on mouse 5-day postnatal (P5) cerebellar sections; this stage corresponds to a peak of proliferation during the amplification of the GNPs pool, and a time when GNPs respond to mitogenic SHH (Fig. 3A) (Dahmane and Ruiz i Altaba, 1999). We detected elevated TERRA levels in the outer external germinal layer (oEGL) containing the proliferating GNPs, while TERRA expression decreases as GNPs become SHH unresponsive and postmitotic in the inner EGL (iEGL; Fig. 3B, top panel). In the adult cerebellum only very few mature granule neurons in the internal granule layer (IGL) show low levels of TERRA expression (Fig. 3B, lower panel). TERRA foci were not observed in RNase A-treated cells, indicating that these are indeed RNA foci (data not shown). Northern blot and qRT-PCR analyses confirmed the RNA FISH results and showed that TERRA RNA levels are highest during the proliferative expansion phases of GNPs development (P0 and P5; Fig. 3C–E) and then decrease as GNPs differentiate (P15 and adult; Fig. 3C–E), in a similar manner to Gli1 and Math1 (Fig. 3F).
Fig. 3.
TERRA is elevated in highly proliferating progenitor cells. (A) Schematic representation of the mouse cerebellar cortex during the first postnatal week. Granule neurons, their progenitors (GNPs) and Purkinje neurons (red) are shown. Proliferation of GNPs occurs in the outer external germinal layer (oEGL). Postmitotic GNPs accumulate in the inner external germinal layer (iEGL) and migrate to the internal granular layer (IGL) through the Purkinje layer. Cycling GNPs in the oEGL respond to SHH secreted form Purkinje neurons. (B) RNA FISH analysis of TERRA expression on wild-type mouse P5 and adult cerebella sagittal sections. The iEGL and outer oEGL are indicated. Note the expression of TERRA in the P5 EGL, with a stronger expression in the oEGL where most cells are cycling. All nuclei are counterstained with DAPI (blue). TERRA expression is low in mature granule neurons in the adult IGL. Scale bar: 20 µm. (C) Northern blot analysis of TERRA levels in normal mouse cerebellum at various stages of development. 18S RNA expression is shown as quantification control. Numbers on the left show the position of markers in kb. Highest TERRA levels are observed at the highest peak of GNPs proliferation at P5. Levels of TERRA are downregulated as GNPs start their differentiation program. (D) Quantification of northern blot analyses for TERRA levels for each cerebellar stages from three independent experiments, one of which is shown in C. (E) Quantitative RT-PCR of TERRA RNA from different cerebellar stages using TERRA-specific primers for subtelomeres of chromosomes 2q, 11q, and telocentric chromosomes (TeloCen). ΔΔCT methods relative to P0 cerebellum and Gapdh were used to calculate relative RT-PCR between different cerebellar stages. Bar graph represents the average value from two independent experiments and at least three independent PCR reactions (means ± s.d.). (F) Quantitative RT-PCR analysis of Gli1 and Math1 expression in the developing and adult cerebellum as described in E.
SHH growth factor stimulation increases TERRA levels
To determine whether the growth factor SHH contributes to elevated TERRA expression, we assayed the effect of recombinant SHH on purified primary P5 mouse GNPs in vitro (Fig. 4). Using northern blot analysis, we found that high level of SHH activation (indicated by a large increase in Gli1 expression) following GNPs treatment with recombinant SHH, results in a moderate (∼1.3-fold) increase in bulk TERRA levels in GNP cells, as measured by northern blotting (Fig. 4A,B; supplementary material Fig. S4). By qRT-PCR analysis, we found that TERRA from several different chromosomes increase ∼2- to 3-fold (Fig. 4D). To control for SHH activity, we show that SHH treatment of GNPs leads to ∼30-fold increase in Gli1 and ∼2.5-fold increase in Math1 RNA levels, two bona fide targets of SHH activation in these cells (Fig. 4C). These findings indicate that TERRA expression can be elevated by high level of SHH signaling in normal GNPs derived from the developing cerebellum.
Fig. 4.
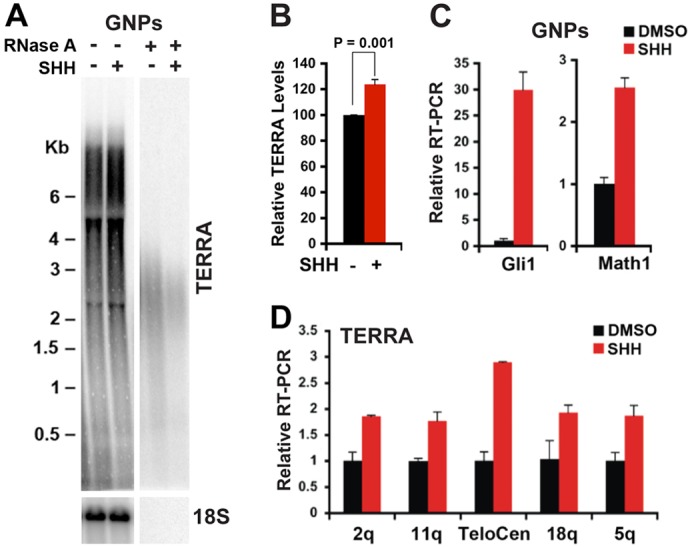
TERRA expression can be induced in purified progenitor cells stimulated with high SHH signaling activation in vitro. (A) GNPs were purified from wild-type mouse P5 cerebella and cultured for 12 hours with or without SHH (600 ng/ml). TERRA RNA was analyzed by northern blotting using a 32P-labeled (TAACCC)4 oligonucleotide probe. 18S RNA expression is shown as an internal control for RNA loading. RNase A treatment is shown in the right panel. (B) Quantification of TERRA levels from at least three independent northern blot analyses using RNA isolated from GNPs treated with SHH for 12 hours or left untreated, one of which is shown in A. Bar graph represents TERRA signal intensity relative to 18S signal, and relative intensity for SHH (−) was set at 100. The P-value was calculated using a two-tailed Student's t-test. (C) qRT-PCR analysis for expression of Gli1 and Math1 is shown as a control for SHH activity. ΔΔCT methods relative to untreated GNPs and Gapdh were used to calculate relative RT-PCR by SHH treatment. Bar graph represents mean ± standard deviations from three independent experiments. (D) qRT-PCR analysis of individual TERRA expression using subtelomere-specific primers for chromosomes 2q, 11q, TeloCen, 18q or 5q as described in C.
TERFs form in cells with shortened telomeres
Telomere shortening has been reported to occur in highly proliferative normal and cancer cells (Raynaud et al., 2010; Wentzensen et al., 2011; Xu and Blackburn, 2007). To compare the average telomere DNA length in tumor relative to normal cerebellar tissue, we compared the mean average fluorescence intensity of telomere DNA FISH signals. This method can be used to assess the average length of telomere repeat DNA (Baerlocher et al., 2006). Computer analyzed images of FISH signals showed that in medulloblastoma, telomere repeat signal intensity in tumor tissue was significantly reduced relative to normal tissue (supplementary material Fig. S5). These results indicate that TERRA foci form in tumor cells where telomere length has shortened.
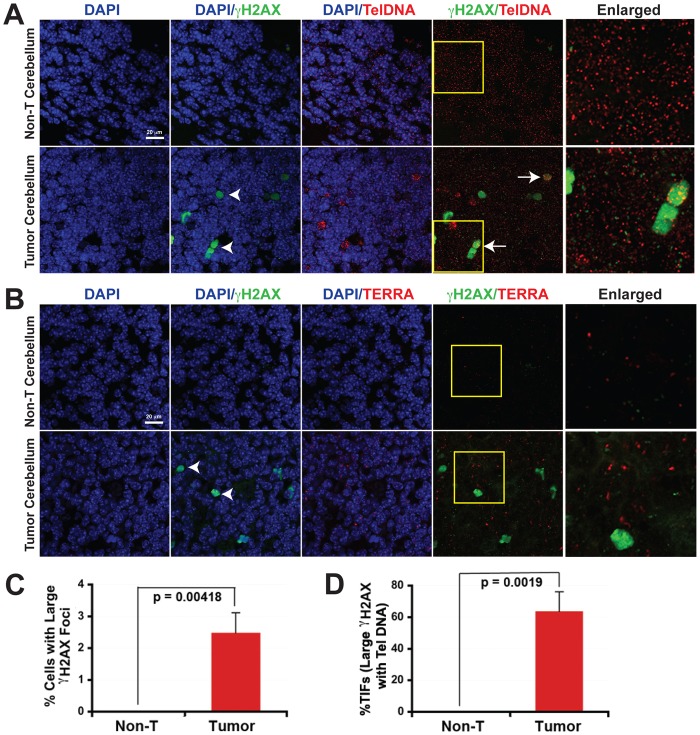
TERFs are distinct from γH2AX DNA damage foci
DNA damage foci are commonly observed in rapidly dividing and precancerous tissue (Sedelnikova and Bonner, 2006). Telomere dysfunction-induced foci (TIFs) are thought to arise from the critical short length and/or uncapping of telomere ends, and can be examined by colocalization of γH2AX foci with telomeric DNA (TelDNA) (d'Adda di Fagagna et al., 2003; Takai et al., 2003). To determine if TERRA expression correlates with the appearance of TIFs, we examined the immunofluorescence staining pattern of γH2AX and in situ hybridization signals for TelDNA (Fig. 5A; supplementary material Fig. S6A, Fig. S7A) or TERRA RNA (Fig. 5B; supplementary material Fig. S6B,C, Fig. S7B,C) in mouse medulloblastoma. Medulloblastoma tumors accumulate heterogeneous staining for γH2AX (with ∼2.5% of cells forming pan-nuclear foci, referred as large γH2AX foci), while non-tumor sections of the adult cerebellum show essentially no γH2AX signals. Pan-nuclear γH2AX foci have been observed in certain types of DNA damage conditions, including viral infection (Fragkos et al., 2009) and inhibition of ATR signaling (Fragkos et al., 2009; Toledo et al., 2011). TERFs were also detected in most tumor cells but not in normal adjacent tissue (Fig. 5B and Fig. 1). Remarkably, TERFs did not appear in cells with pan-nuclear γH2AX staining (Fig. 5B; supplementary material Figs S6,S7). Quantification of at least three independent experiments indicated that ∼2.5% of tumor cells contain pan-nuclear γH2AX staining (Fig. 5C). This is in contrast to the ∼80% of tumor cells that contain TERFs (Fig. 1E). Among the 2.5% of γH2AX positive cells, we found 80% of these colocalized with strong telomere DNA FISH signals (Fig. 5D). While small γH2AX foci could also be detected in tumor sections, these foci did not overlap with TERRA RNA or Tel DNA to a significant extent (supplementary material Figs S6, S7, S12). Similarly, TERFs did not overlap with smaller γH2AX foci observed in rapidly dividing normal neurons of developing cerebellum in P6 mice (supplementary material Fig. S8). Taken together, these findings indicate that TERFs are distinct from TIFs and other γH2AX signals, yet form among the same population of highly proliferative normal and tumor tissue.
Fig. 5.
Tumor-associated TERRA foci do not colocalize with γH2AX. (A) Confocal microscopy images of immunofluorescence of γH2AX foci (green) combined with DNA FISH for telomere repeat DNA (TelDNA; red) in non-tumor (non-T) or tumor sections. DAPI stain is in blue. Arrowheads indicate large γH2AX foci. Arrows indicate nuclei containing intense telomere DNA signals colocalizing with large γH2AX foci. Enlarged (5×) images of the regions in the yellow boxes are shown in the right panels. (B) Confocal microscopy images of immunofluorescence of γH2AX foci (green) combined with RNA FISH for TERRA (red) in non-tumor or tumor sections of the cerebellum. Arrowheads indicate large γH2AX foci. Enlarged (5×) images of the regions in the yellow boxes are shown in the right panels. (C) Quantification of cells containing large γH2AX foci. More than 1000 nuclei from at least three independent experiments were counted for quantification. The P-value was calculated using a two-tailed Student's t-test. (D) Quantification of large γH2AX foci that colocalize with intense telomere DNA FISH signals to form TIFs. More than 1000 nuclei from at least three independent experiments were counted for quantification. The P-value was calculated using a two-tailed Student's t-test.
TERFs are nuclear foci distinct from PML and Cajal bodies
To further investigate the subcellular localization of TERFs we examined at higher magnification the confocal images of mouse medulloblastoma tumors (Fig. 6A; supplementary material Fig. S9A) or normal P5 cerebellar (Fig. 6B; supplementary material Fig. S9B) sections after in situ hybridization with a TERRA PNA probe. We observed that TERRA foci typically appear as one to three bright foci in the nuclear region of tumor tissue (Fig. 6A). Fewer and less bright foci were observed in the nucleus of normal P5 cerebellar tissues known to be rapidly dividing (Fig. 6B). To determine whether TERFs colocalized with other well-characterized nuclear structures, we performed immuno-RNA-FISH with antibodies to either promyeolocytic leukemia (PML) protein (Fig. 6C), coilin (Fig. 6D), or phospho-histone H3 (H3pS10; Fig. 6E). While TERRA can colocalize with PML in some ALT cell lines (data not shown), we found only few TERFs that colocalize with PML in mouse medulloblastoma tumor tissue (Fig. 6C). Similarly, TERFs rarely colocalized with coilin, which is a marker of Cajal bodies where the telomerase holoenzyme is assembled (Fig. 6D). Additionally, TERFs were not observed in H3pS10 positive cells, which is a marker for mitotic cells. Thus, TERFs do not colocalize with several well-characterized nuclear structures, and may represent a unique nuclear structure that forms during the interphase of rapidly dividing cells.
Fig. 6.
TERFs are nuclear, interphase-associated foci distinct from PML and Cajal bodies. (A) Confocal microscopy images of TERRA RNA FISH on tissue sections derived from mouse medulloblastoma (no. 6850). (B) Same as in A, except in normal cerebellar tissue derived from a stage P5 mouse. (C) TERRA RNA FISH (red) combined with immunofluorescence with antibody to PML (green) on tissue sections derived from mouse medulloblastoma (no. 7171). All nuclei were counterstained with DAPI (blue). Enlarged (zoomed) merged images are shown in the right panels. (D) Same as in C, except TERRA RNA FISH (red) combined with coilin immunofluorescence (green). (E) Same as in C, except TERRA RNA FISH (red) combined with immunofluorescence with antibody to phosphorylated histone H3S10 (green).
TERRA expression is elevated in many different human cancer types
To explore whether TERRA levels are elevated in human cancers, we first examined RNA derived from human ovarian tissue diagnosed as either normal, advanced primary tumor, or metastatic tumor (Fig. 7A–D). Northern blot analysis indicated that most of the ovarian primary and metastatic tumors expressed higher levels of TERRA than otherwise normal ovarian tissue (Fig. 7A,B). Chromosome-specific qRT-PCR analysis also confirmed that primary and metastatic tumors had higher levels of TERRA at most, but not all subtelomeres tested relative to the normal tissue (Fig. 7C,D). In one sample derived from normal ovarian tissue, we observed very high levels of TERRA by northern blotting (supplementary material Fig. S10A). However, upon further molecular characterization of these tissues, this sample (HR406) was found to express levels of the proliferation marker Ki67 that were >50-fold relative to two other normal ovarian tissue, and 3- to 5-fold greater than advanced primary or metastatic tumors (supplementary material Fig. S10B), indicating that this ovarian tissue was highly proliferative. This further supports the correlation between cell proliferation state and elevated TERRA levels. To further investigate whether TERRA was elevated in other human cancer tissues, we compared TERRA expression in primary human tumors and matched normal tissue controls derived from various cancer biopsies, including those derived from stomach, lung and colon (Fig. 7E–H). Northern blot analyses showed that TERRA is expressed at higher levels in tumor-derived tissues relative to matched control tissues (Fig. 7E; supplementary material Fig. S10A). Antisense TERRA was not detected when the same northern blot was stripped and reprobed with G-rich probe, indicating that the telomere RNA species is strand specific (Fig. 7E, right panel; supplementary material Fig. S11). TERRA expression was also analyzed by chromosome-specific qRT-PCR for several different telomeres (Fig. 7F–H). The cell proliferation marker Ki67 was used as a control for tumor dissection, and was elevated ∼6- to 100-fold relative to normal matched tissue RNA. Consistent with northern blotting data, we found that TERRA RNA was elevated in tumor tissue relative to matched control tissue for most subtelomeres tested. However, only a few subtelomeres showed significantly (>4-fold) higher TERRA levels in tumor tissue relative to normal, and no particular subtelomere showed elevated TERRA consistently for all the tumor samples. For example, only telomere 13q expressed high (>4-fold) TERRA levels in one stomach cancer biopsy, while 17q and 2q were elevated in lung, and 10q and 17q elevated in a colon carcinoma biopsy. No one subtelomere was consistently elevated for TERRA expression, even when tumors were derived from the same organ (data not shown). These results indicate that TERRA expression is elevated in various human cancer tissues, and can be expressed from a few chromosomes, which may vary among cell and cancer types.
Fig. 7.
TERRA is overexpressed in human tumor tissues. (A) Northern blot analysis of TERRA RNA isolated from normal ovarian tissue, advanced primary ovarian cancer and metastatic ovarian cancer tissue. Numbers at the bottom show the average value of TERRA signals relative to 18S RNA signals in tumor versus normal tissues from two independent northern blots. Asterisks indicate that the sample is not included in the analysis because of the degradation of the 18S signal. (B) Quantification of average TERRA levels from many northern blot analyses using RNA isolated from tissues derived from two normal ovaries (540071 and 709152), eight primary and nine metastatic ovarian cancers, a representative of which is shown in A. Bar graph represents TERRA signal intensity relative to the 18S signal, and average relative intensity for two normal ovary tissues was set at 100. (C) qRT-PCR analysis of Ki-67 expression and TERRA levels in the indicated ovarian cancers and normal ovary tissue (709152). TERRA levels were assayed using primers specific for TERRA RNA transcribed from subtelomeres of human chromosome 2q, 10q, 13q, as indicated. ΔΔCT methods relative to normal ovary and Gapdh were used to calculate relative RT-PCR between normal and tumor samples. Bar graph represents the average value from three independent PCR reactions (means ± s.d.). (D) The same as in C, except using primers specific for TERRA RNA transcribed from subtelomeres of human chromosome XqYq, 15q, 16p, as indicated. (E) TERRA expression in dissected primary human tumor tissue from carcinoma of the stomach, lung and colon (normal and tumor) analyzed by northern blotting. 18S RNA expression is shown as quantification control. Equal intensity of 18S* signals indicate that RNA from each sample were subject to similar levels of degradation during the preparation process. The same northern blot was stripped, and reprobed with 32P-labeled (TTAGGG)4 probe for anti-sense TERRA (right panel). Numbers on the left show the position of markers (in kb). Numbers at the bottom show the value of TERRA signals relative to the 18S RNA signals in tumor versus normal tissues. (F) qRT-PCR analysis of Ki-67 expression (left panel) and TERRA levels (right panel) in the tumor and patient matched control tissues from stomach. TERRA levels were assayed using primers specific for TERRA RNA transcribed from subtelomeres of human chromosome 2q, 17q, 10q, 13q, 15q, 16q and XqYq, as indicated. Bar graph represents the average value from at least three independent PCR reactions (means ± s.d.). (G) The same as in F, except in the tumor and patient matched control tissues from lung. (H) The same as in F, except in the tumor and patient matched control tissues from the colon.
Discussion
We show here that in mammalian cells in vivo, TERRA expression is linked to the proliferative and tumorigenic state of the cell. We found that TERRA levels were elevated in highly proliferating normal and cancer cells in both mouse tissue sections and human cancer biopsies. Using a well-established mouse model of brain cancer (i.e. medulloblastoma), we found that TERRA RNA was elevated in tumor, but not in normal cerebellar tissue (Figs 1, 2; supplementary material Figs S1–S3). RNA FISH revealed pronounced TERRA foci, termed TERFs, in cells overexpressing TERRA RNA. TERFs were sensitive to RNase A and not detected with control probes containing a mutation in the telomere repeat sequence, indicating that these were indeed TERRA-containing foci. Furthermore, TERFs appeared only in tissues where high level TERRA expression was validated by northern blotting and qRT-PCR (Fig. 2; supplementary material Fig. S3). TERFs generally formed as one or few major foci in the nuclear region of proliferating cells. TERRA and TERFs were also elevated in rapidly dividing progenitor cells of the developing cerebellum, indicating that normal proliferating cells can also form TERFs (Fig. 3). TERRA levels were also enhanced by addition of recombinant SHH growth factor to purified progenitor cells in vitro, suggesting that strong and chronic growth factor stimulation promotes TERRA accumulation (Fig. 4; supplementary material Fig. S4). TERFs formed in the same cell populations where γH2AX DNA damage foci form (Fig. 5; supplementary material Figs S6, S7, S12) and telomere DNA shortening has occurred (supplementary material Fig. S5), but TERFs did not colocalize with γH2AX foci (Fig. 5), nor with PML or Cajal nuclear bodies (Fig. 6). Finally, we provide evidence that TERRA is expressed at higher levels in many human cancer tissues relative to matched controls (Fig. 7; supplementary material Fig. S10). These findings support the model that TERRA expression is coupled to cellular proliferation and that TERFs are a novel indicator of proliferative stress.
Our results are different from several previous reports that described either a decrease in TERRA expression between normal and colon cancer (Schoeftner and Blasco, 2008), diminished TERRA in advanced astrocytoma (Sampl et al., 2012) or a lack of expression in cancer cell lines (Ng et al., 2009; Zhang et al., 2009). The differences may reside in the use of tumor tissue compared to cancer cell lines (Ng et al., 2009; Zhang et al., 2009) as well as in the techniques used to detect TERRA expression [a comprehensive use of RNA FISH, northern blotting and qRT-PCR in our study as opposed to dot blotting in Schoeftner and Blasco or quantitative RT-PCR from few chromosomes (supplemental material Figs. S13-15) (Schoeftner and Blasco, 2008; Sampl et al., 2012)]. It is also likely that TERRA expression in human cancers may be regulated in a more complex manner corresponding to the patient history, cancer type, and tumor tissue integrity. In our experiments, human tumor RNA was purified from freshly isolated unfixed biopsies to avoid RNA degradation. Our use of a mouse cancer model has also enabled us to examine adjacent non-tumor and tumor tissue on the same section. In mouse model of medulloblastoma, we were able to utilize methods developed for RNA FISH that reduce the time of fixation and denaturation that commonly degrade or modify RNA. Together, these methods have allowed us to observe TERRA expression and localization in tissue sections and isolates that better preserve the in vivo physiology of cellular proliferation and cancer.
The function and regulation of TERRA has been explored in several previous studies (Azzalin et al., 2007; Deng et al., 2009; Redon et al., 2010; Schoeftner and Blasco, 2008). We have previously shown that TERRA can protect telomere ends from DNA damage signaling by promoting telomeric heterochromatin formation (Deng et al., 2009). TERRA has also been shown to inhibit telomerase activity in vitro (Redon et al., 2010; Schoeftner and Blasco, 2008). TERRA levels can be elevated in cell lines lacking DNA methyltransferase 3b (DNMT3b) (Yehezkel et al., 2008), as well as in response to developmental and stress signals (Schoeftner and Blasco, 2008). Our data suggests that TERRA expression can be enhanced by growth factor signaling (e.g. SHH) and cell proliferation in vivo. Moreover, chronic proliferation signals associated with carcinogenic mutations (like Ptch1+/−) lead to elevated TERRA and TERF formation. We also observed that telomere DNA signal decreases (supplementary material Fig. S5) and that TERT mRNA levels are not elevated (Fig. 2; supplementary material Fig. S3) in tumor tissue. We propose that TERRA is required for telomere end processing and protection during normal cell proliferation, and that elevated TERRA and TERF protects telomeres from eliciting a DNA damage response during conditions of proliferative stress (Fig. 8).
Fig. 8.
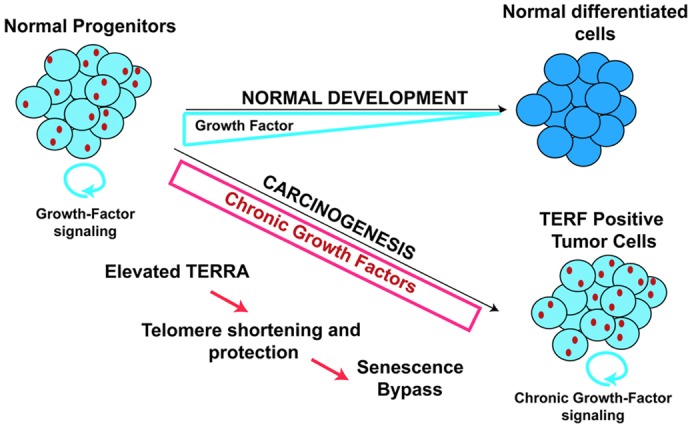
TERRA foci (TERF) formation during growth-factor-induced proliferation in progenitor and cancer cells. Normal progenitor cells subjected to high level of proliferation (such as granule neuron progenitors in the mammalian postnatal cerebellum) through the activity of growth factors (such as SHH) express high level of TERRA RNA (red dots). Elevated chronic growth factor signaling, as occurs in the Ptch1+/− model of medulloblastoma, results in high TERRA expression and stabilization of shortened telomeres in tumor cells. TERFs form at high level in early stage cancer cells in the absence of γH2AX DNA damage foci. Stabilization of short telomeres is predicted to promote cancer cell evolution.
γH2AX-positive DNA damage foci have been shown to be elevated in highly proliferating cells and precancerous lesions (Bartkova et al., 2005; Gorgoulis et al., 2005; Jackson and Bartek, 2009). We found that small γH2AX foci accumulate in mouse medulloblastoma, with ∼2.5% of the tumor cells containing pan-nuclear γH2AX foci (Fig. 5). In contrast, ∼80% of these cells were TERF positive. Importantly, most γH2AX foci did not overlap with TERFs, but did colocalize with telomere DNA foci (TIFs). Others have shown that telomere DNA forms aggregates in highly proliferating cancer cells (Mai and Garini, 2006). Unfortunately, we were unable to colocalize TERFs with telomere DNA signals, so can not exclude the possibility that TERFs are expressed at telomere aggregates in highly proliferating cells. We did find that telomeres were generally shorter in mouse cancer cells that express TERRA and form TERFs (supplementary material Fig. S5), suggesting that short telomeres may promote TERRA transcription activation. Elevated TERRA expression in highly proliferating cancer and normal progenitor cells may stabilize these abnormally short telomeres, perhaps through inhibition of the DNA damage response. This is consistent with the observed lack of DNA damage associated γH2AX foci colocalizing with TERFs (Fig. 5; supplementary material Figs S6 and S7). TERRA may prevent DNA damage response at short telomeres by stabilizing the shelterin complex, heterochromatin formation, or protection of single stranded telomeric DNA (Deng et al., 2009; Flynn et al., 2011).
TERRA foci and telomeric transcript accumulations (Tacs) have been described previously (Azzalin et al., 2007; Marion et al., 2009; Schoeftner and Blasco, 2008; Zhang et al., 2009). In human cell lines, TERRA can colocalizes to metaphase telomeres and form variable numbered foci that can be upregulated by depleting components of the nonsense-mediated decay (NMD) pathway (Azzalin et al., 2007). In undifferentiated mouse ES cells, Tacs associate with both sex chromosomes, but upon ES cell differentiation, Tacs relocalize to the inactive X-chromosome in females or the Y-chromosome in males (Schoeftner and Blasco, 2008). In telomerase-deficient TRF2-overexpressing (K5TRF2/TERC−/−) mouse models of telomere dysfunction, Tacs delocalize from the inactive X-chromosome, reflecting a novel form of telomere dysfunction (Schoeftner et al., 2009). While we did not directly examine the localization of TERFs with respect to the inactive X chromosome, the TERFs that we observe in rapidly proliferating cells are likely to be very similar to Tacs. However, since Tacs have not been reported to be elevated in rapidly proliferating neuronal progenitor or medulloblastoma, we refrain from concluding that these TERFs and Tacs are identical structures.
RNA mediated nuclear body formation has been observed for a variety of other RNA species (Shevtsov and Dundr, 2011). Transcription of the tandem arrays of satellite III (sat III) repetitive DNA is known to form nuclear stress bodies located at pericentromeric DNA (Biamonti and Vourc'h, 2010). Recent genome-wide sequencing studies have found that satellite repeat transcripts were aberrantly overexpressed in various human and mouse epithelial cancers (Ting et al., 2011). While, Sat III repeat DNA and RNA have some intriguing similarities with TERRA DNA and RNA, we did not observe that TERFs had similar patterns of granulation commonly seen with SatIII nuclear stress bodies in tissue culture cell lines. However, it will be important to determine if TERF positive tumor tissue is also enriched in Sat III nuclear bodies, and whether these structures share common features with TERFs. TERFs may also share features with other RNA-nucleated bodies, including the miRNA processing P-bodies (Parker and Sheth, 2007) and cytoprotective aggresomes (Wileman, 2007). In some cancer cells, telomere DNA has been shown to aggregate in conjunction with nuclear lamina (Mai and Garini, 2006; Raz et al., 2008). In contrast to TERFs, telomere aggregates colocalize with γH2AX DNA damage foci, suggesting that these are distinct nuclear structures (Raz et al., 2008). Nevertheless, it is possible that TERFs reflect a cluster of several highly transcribed telomeres that correspond to the telomere aggregates observed in some cancer types. TERFs did not show significant colocalization with other well-characterized nuclear structures, including coilin-containing Cajal bodies or PML-containing ND10 bodies (Fig. 6). Thus, TERFs appear to represent a unique nuclear body formed by TERRA accumulation from one or more telomeres in highly proliferating cells.
An important unanswered question is how TERRA expression, processing, and localization may differ in normal differentiated, normal proliferating, and abnormal cancer cells. TERRA levels and TERFs can be detected in both proliferating cell types, although we did observe greater TERRA processing to several smaller RNA species (Fig. 7; supplementary material Fig. S10) and an average greater density of TERFs in cancer tissue relative to progenitor cells (Figs 2,6). It may not be surprising that progenitor cells have similar TERRA expression patterns as cancer cells, since they are known to share common gene expression profile as in the case of medulloblastoma (Kho et al., 2004). We suggest that the major difference between cancer and progenitor cells is the abnormal prolonged exposure to proliferation signals and the potential stabilization of shortened and damaged telomeres through the action of TERRA. We also observed TERF expression is highly elevated in human cancer cell lines with ALT elongated telomeres, as observed in U2OS cells (supplementary material Fig. S15). It is therefore possible that TERRA expression in mouse medulloblastoma correlates with the ALT phenotype, but additional experiments are needed to confirm this possibility. While the precise function, processing, and localization of TERFs remains unknown, these details will be essential for understanding the regulation of telomeres during normal and cancer cell proliferation. In summary, our data demonstrates that TERRA accumulates and forms TERFs in highly proliferating progenitors and cancer tissue, and suggests that TERFs may provide a novel and sensitive biomarker for telomere dysfunction in cancer.
Materials and Methods
Mouse breeding
Medulloblastoma used in our study formed in the Ptch1+/− colony either alone or in combination with p53+/−. The tumor 6850 (Fig. 1A) originated in a Ptch1+/−;RP58fl/fl mouse obtained during the breeding between Ptch1+/− and RP58fl/fl;nestinCreER. The RP58fl/fl does not present any abnormality (Xiang et al., 2012).
All procedures were approved by the Wistar Institute Institutional Animal Care and Use Committee.
Telomeric RNA FISH on tissue section
Tissue sections (7–12 µm) were prepared as described previously (Fernandez et al., 2010). Fresh frozen sections were fixed in 4% paraformaldehyde (PFA) for 10 minutes on ice, washed twice with cold PBS, and twice with RIPA (150 mM NaCl, 50 mM Tris-HCl, pH 8.0, 1% NP-40, 0.5% Sodium Deoxycholate, 0.1% SDS, 1 mM EDTA) buffer for 10 mins each. The sections were fixed again in 4% PFA for 10 mins at 25°C, washed three times with cold PBS, and acetylated in acetylation buffer for 10 mins at 25°C. After acetylation, the sections were washed three times with cold PBS containing 0.05% Tween-20 (PBST), and prehybridized in hybridization buffer (50% formamide, 5 × SSC, 5 × Denhardts, 25 mg/ml yeast RNA, 0.5 mg/ml salmon sperm DNA) for 1 hr at 37°C. RNase A treatment was performed in PBST with 100 μg/ml RNase A for 30-60 mins at 37°C before prehybridization. Hybridization was performed overnight at 37°C, and the following probes were used in the hybridization: a Tamra-(CCCTAA)3 PNA probe (Panagene Inc.) or an Alexa-Fluor-488-(TAACCC)7 oligonucleotide probe (IDT) for TERRA RNA, a Fam-(TTAGGG)3 PNA probe for TERRA antisense, and an Alexa-Fluor-488-(TAACAC)7 probe as a control for specificity. After hybridization, slides were washed as follows: 2 × SSC, 50% formamide, three times at 39°C for 5 mins; 2 × SSC, three times at 39°C for 5 mins each; 1 × SSC, 10 mins at room temperature; 4 × SSC once at room temperature. Slides were counterstained with 0.1 g/ml DAPI in 4 × SSC, 0.1% Tween-20, washed in 4 × SSC, and mounted with mounting media. Images for H/E staining were taken with a Nikon E600 Upright microscope (Nikon Instruments) with ImageProPlus software (media Cybemetrics) and Adobe PhotoShop CS5 for image processing. Confocal images were taken with Zeiss LSM510META NLO laser scanning confocal system on a Zeiss Axiovert 200M inverted microscope with Zeiss AIM Ver.4.0 software and Adobe PhotoShop CS5 for image processing. For quantification purpose, the same technical settings were applied to the capture of all images (Ferlicot et al., 2003; Ourliac-Garnier and Londono-Vallejo, 2011). The mean fluorescence intensity of TERRA spots (FI/spot) from unmodified black and white images was automatically quantified by ImageProPlus software and expressed in fluorescence units (FU). The data was exported into an Excel worksheet for frequency and statistical analysis. P-values were calculated using two-tailed Student's t-tests.
RNA preparation and analysis
Total RNA for normal ovarian tissue samples was purchased from Biochain (R1234183, lot no. 709152), Agilent Technologies (540071), and Zyagen (HR406). Total RNA (540045) for normal human breast was purchased from Agilent Technologies. All other human tissues were collected according to the guidelines and policies of the Hospital of the University of Pennsylvania Institutional Review Board. Stage III–IV human ovarian carcinoma specimens were procured through Research Pathology Services at Dartmouth–Hitchcock Medical Center under institutional approval (CPHS17702). For ovarian cancer samples, primary means from the initial mass in the ovary and metastatic means from anywhere else in the peritoneal cavity. Total RNA was purified with Trizol reagent (Invitrogen) as manufacturer's instructions. The RNA samples were treated with DNase I for 45 min at 37°C, followed by DNase I inactivation in the presence of EDTA at 65°C for 5 min prior to further analysis. For northern blotting, about 3–7.5 µg of total RNA were used. Hybridizations were performed in Church buffer (0.5N Na-phosphate, pH 7.2, 7% SDS, 1mM EDTA, 1% BSA) for 16-18 hrs at 50°C. The membrane was washed twice in 0.2N Na-phosphate, 2% SDS, 1 mM EDTA at room temperature, once in 0.1N Na-phosphate, 2% SDS, 1 mM EDTA at 50°C, and analyzed by phosphor-imager (Amersham Biosciences). The blots were first hybridized with a 32P-labeled (TAACCC)4 probe, then stripped, and probed with either a 32P-labeled (TTAGGG)4 or 18S probe. When indicated, RNA samples were treated with RNase A (Roche) at a final concentration of 100 µg/ml for 30–60 mins at 37°C. Images were processed with a Typhoon 9410 Imager (GE Healthcare) and quantified with ImageQuant 5.2 software (Molecular Dynamics). TERRA RNA levels were calculated as percentages relative to signals from control samples and 18S internal control. Reverse transcriptions were performed with SuperScript III (Invitrogen) using 1 µg of total RNA as per the manufacturer's instructions. A specific primer (CCCTAA)6 was used to obtain TERRA cDNA at 55°C. Real-time PCR experiments were performed as described (Deng et al., 2009). Relative RT-PCR was determined using ΔΔCT methods relative to control samples and internal control Actin and Gapdh for human samples or Gapdh for mouse samples. Primer sequences used for real-time PCR are listed in supplementary material Table S1.
Telomere DNA FISH
Tissue sections were prepared essentially as described in RNA FISH method section. Telomeric DNA FISH was performed as followings. After acetylation, sections were subject to RNase A treatment by the incubation in PBST with 100 µg/ml RNase A for 30–60 mins at 37°C. Sections were washed three times in PBS, and dehydrated in cold ethanol series 5 mins for each (70%, 95%, 100%). Slides with sections were denatured on a 80–85°C hot plate for 5 mins in the presence of about 120 µl of hybridization mix (70% formamide, 10 mM Tris-HCl, pH 7.2, 0.5% blocking reagent) containing telomeric PNA-Tamra-(CCCTAA)3 probe, and hybridization was performed in the dark for overnight at room temperature. The slides were washed two times for 15 mins each in 70% formamide, 10 mM Tris-HCl, pH 7.2, 0.1% BSA followed by three washes of 5 mins each in 0.1 M Tris-HCl, pH 7.2, 0.15 M NaCl, 0.08% Tween-20, stained with 0.1 µg/ml DAPI, and mounted in mounting medium. Confocal images were taken with Zeiss LSM510META NLO laser scanning confocal system on a Zeiss Axiovert 200M inverted microscope with Zeiss AIM Ver.4.0 software and Adobe PhotoShop CS5 for image processing. Image capture and quantification process were performed using the same methods as those used for RNA FISH.
For combined γH2AX staining and FISH analysis, we performed RNA or DNA FISH as described above. After washes, the sections were fixed again in 4% paraformaldehyde for 15 mins, blocked in blocking solution (1 mg/ml BSA, 3% goat serum, 0.1% Triton X-100, 1 mM EDTA in PBS) for at least 30 mins, and incubated with γH2AX monoclonal antibody (Upstate) diluted in blocking solution (1:100) for 1 hr at room temperature. The sections were washed three times with PBST for 5 mins each, and incubated with Alexa-Fluor-488-conjugated secondary antibody (2 mg/ml stock solution diluted 1:600 in PBST) in blocking solution for 30 mins. The sections were further washed with PBST, counter stained with DAPI, and mounted in mounting medium before confocal microscope. Rabbit polyclonal antibodies to Coilin (H-300) and phospho-histone H3 (Ser10) were purchased from Santa Cruz and Millipore, respectively. Monoclonal antibody to mouse PML was a gift from Olga Vladimirova at the Wistar Institute.
Granule neuron progenitor purification
Granule neuron progenitors (GNPs) were purified from P5 mouse cerebella as described previously (Fernandez et al., 2010). SHH (600 ng/ml, R&D) was added to the media for 12 hours.
Supplementary Material
Acknowledgments
We would like to thank Federico Valdivieso, Michael Feldman, the Tumor tissue bank and the Abramson Cancer Center for the tumor samples. We also thank the Department of Pathology at Dartmouth University for ovarian cancer tissue RNA, and Fred Keeney and James Hayden in the Wistar Institute Microscopy Core.
Footnotes
Funding
This work was supported by the Wistar Institute Cancer Center Core Grant [grant number P30 CA10815]; the American Cancer Society [grant number RSG-08-045-01-DDC to N.D.]; the National Brain Tumor Society to N.D., National Institutes of Health [grant number RO1CA140652 to P.M.L.); and a Scientist Development grant from the American Heart Association [grant number 11SDG5330017 to Z.D.]. Deposited in PMC for release after 12 months.
Supplementary material available online at http://jcs.biologists.org/lookup/suppl/doi:10.1242/jcs.108118/-/DC1
References
- Armentano M., Filosa A., Andolfi G., Studer M. (2006). Coup-TFI is required for the formation of commisural projections in the forebrain by regulating axonal growth. Development 133, 4151–4162 10.1242/dev.02600 [DOI] [PubMed] [Google Scholar]
- Azzalin C. M., Reichenbach P., Khoriauli L., Giulotto E., Lingner J. (2007). Telomeric repeat containing RNA and RNA surveillance factors at mammalian chromosome ends. Science 318, 798–801 10.1126/science.1147182 [DOI] [PubMed] [Google Scholar]
- Baerlocher G. M., Vulto I., de Jong G., Lansdorp P. M. (2006). Flow cytometry and FISH to measure the average length of telomeres (flow FISH). Nat. Protoc. 1, 2365–2376 10.1038/nprot.2006.263 [DOI] [PubMed] [Google Scholar]
- Bah A., Wischnewski H., Shchepachev V., Azzalin C. M. (2012). The telomeric transcriptome of Schizosaccharomyces pombe. Nucleic Acids Res. 40, 2995–3005. 10.1093/nar/gkr1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartkova J., Horejsí Z., Koed K., Krämer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J. M., Lukas C., et al. (2005). DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 10.1038/nature03482 [DOI] [PubMed] [Google Scholar]
- Biamonti G., Vourc'h C. (2010). Nuclear stress bodies. Cold Spring Harb. Perspect. Biol. 2, a000695 10.1101/cshperspect.a000695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn E. H., Grieder C W., Szostak J. W. (2006). Telomeres and Telomerase: the path from maize, Tetrahymena, and yeast to human cancer and aging. Nat. Med. 12, 1133–1338 10.1038/nm1006-1133 [DOI] [PubMed] [Google Scholar]
- Blasco M. A. (2005). Telomeres and human disease: ageing, cancer and beyond. Nat. Rev. Genet. 6, 611–622 10.1038/nrg1656 [DOI] [PubMed] [Google Scholar]
- Blasco M. A. (2007). Telomere length, stem cells and aging. Nat. Chem. Biol. 3, 640–649 10.1038/nchembio.2007.38 [DOI] [PubMed] [Google Scholar]
- Caslini C., Connelly J. A., Serna A., Broccoli D., Hess J. L. (2009). MLL associates with telomeres and regulates telomeric repeat-containing RNA transcription. Mol. Cell. Biol. 29, 4519–4526 10.1128/MCB.00195-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R. (2004). Beginning to understand the end of the chromosome. Cell. 116, 273–279 10.1016/S0092-8674(04)00038-8 [DOI] [PubMed] [Google Scholar]
- Corcoran R. B., Scott M. P. (2001). A mouse model for medulloblastoma and basal cell nevus syndrome. J. Neurooncol. 53, 307–318 10.1023/A%3A1012260318979 [DOI] [PubMed] [Google Scholar]
- d'Adda di Fagagna F., Reaper P. M., Clay–Farrace L., Fiegler H., Carr P., Von Zglinicki T., Saretzki G., Carter N. P., Jackson S. P. (2003). A DNA damage checkpoint response in telomere-initiated senescence. Nature 426, 194–198 10.1038/nature02118 [DOI] [PubMed] [Google Scholar]
- Dahmane N., Ruiz i Altaba A. (1999). Sonic hedgehog regulates the growth and patterning of the cerebellum. Development 126, 3089–3100 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2002). Protection of mammalian telomeres. Oncogene 21, 532–540 10.1038/sj.onc.1205080 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2004). T-loops and the origin of telomeres. Nat. Rev. Mol. Cell Biol. 5, 323–329 10.1038/nrm1359 [DOI] [PubMed] [Google Scholar]
- de Lange T. (2005a). Shelterin: the protein complex that shapes and safeguards human telomeres. Genes Dev. 19, 2100–2110 10.1101/gad.1346005 [DOI] [PubMed] [Google Scholar]
- De Lange T.(2005b). Telomere-related genome instability in cancer. Cold Spring Harb. Symp. Quant. Biol. 70, 197–204 10.1101/sqb.2005.70.032 [DOI] [PubMed] [Google Scholar]
- Deng Z., Norseen J., Wiedmer A., Riethman H., Lieberman P. M. (2009). TERRA RNA binding to TRF2 facilitates heterochromatin formation and ORC recruitment at telomeres. Mol. Cell 35, 403–413 10.1016/j.molcel.2009.06.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellison D. (2002). Classifying the medulloblastoma: insights from morphology and molecular genetics. Neuropathol. Appl. Neurobiol. 28, 257–282 10.1046/j.1365-2990.2002.00419.x [DOI] [PubMed] [Google Scholar]
- Ellison D. W. (2010). Childhood medulloblastoma: novel approaches to the classification of a heterogeneous disease. Acta Neuropathol. 120, 305–316 10.1007/s00401-010-0726-6 [DOI] [PubMed] [Google Scholar]
- El–Zaatari M., Tobias A., Grabowska A. M., Kumari R., Scotting P. J., Kaye P., Atherton J., Clarke P. A., Powe D G., Watson S A. (2007). <<pub-tl>De–regulation of the sonic hedgehog pathway in the InsGas mouse model of gastric carcinogenesis. </pub-tl> 96, 1855–1861 10.1038/sj.bjc.6603782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlicot S., Durrbach A., Ba N., Desvaux D., Bedossa P., Paradis V. (2003). The role of replicative senescence in chronic allograft nephropathy. Hum. Pathol. 34, 924–928 10.1016/S0046-8177(03)00340-X [DOI] [PubMed] [Google Scholar]
- Fernandez C., Tatard V. M., Bertrand N., Dahmane N. (2010). Differential modulation of Sonic-hedgehog-induced cerebellar granule cell precursor proliferation by the IGF signaling network. Dev. Neurosci. 32, 59–70 10.1159/000274458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynn R. L., Centore R. C., O'Sullivan R. J., Rai R., Tse A., Songyang Z., Chang S., Karlseder J., Zou L. (2011). TERRA and hnRNPA1 orchestrate an RPA-to-POT1 switch on telomeric single-stranded DNA. Nature 471, 532–536 10.1038/nature09772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkos M., Jurvansuu J., Beard P. (2009). H2AX is required for cell cycle arrest via the p53/p21 pathway. Mol. Cell. Biol. 29, 2828–2840 10.1128/MCB.01830-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbertson R. J. (2004). Medulloblastoma: signalling a change in treatment. Lancet Oncol. 5, 209–218 10.1016/S1470-2045(04)01424-X [DOI] [PubMed] [Google Scholar]
- Gilbertson R. J., Ellison D. W. (2008). The origins of medulloblastoma subtypes. Annu. Rev. Pathol. 3, 341–365 10.1146/annurev.pathmechdis.3.121806.151518 [DOI] [PubMed] [Google Scholar]
- Goodrich L. V., Milenković L., Higgins K. M., Scott M. P. (1997). Altered neural cell fates and medulloblastoma in mouse patched mutants. Science 277, 1109–1113 10.1126/science.277.5329.1109 [DOI] [PubMed] [Google Scholar]
- Gorgoulis V. G., Vassiliou L. V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R. A., Jr, Kastrinakis N. G., Levy B., et al. (2005). Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 10.1038/nature03485 [DOI] [PubMed] [Google Scholar]
- Greenwood J., Cooper J. P. (2012). Non-coding telomeric and subtelomeric transcripts are differentially regulated by telomeric and heterochromatin assembly factors in fission yeast. Nucleic Acids Res.40, 2956–2963 10.1093/nar/gkr1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn H., Wicking C., Zaphiropoulous P. G., Gailani M. R., Shanley S., Chidambaram A., Vorechovsky I., Holmberg E., Unden A. B., Gillies S., et al. (1996). Mutations of the human homolog of Drosophila patched in the nevoid basal cell carcinoma syndrome. Cell 85, 841–851 10.1016/S0092-8674(00)81268-4 [DOI] [PubMed] [Google Scholar]
- Jackson S. P., Bartek J. (2009). The DNA-damage response in human biology and disease. Nature 461, 1071–1078 10.1038/nature08467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain D., Cooper J. P. (2010). Telomeric strategies: means to an end. Annu. Rev. Genet. 44, 243–269 10.1146/annurev-genet-102108-134841 [DOI] [PubMed] [Google Scholar]
- Johnson R. L., Rothman A. L., Xie J., Goodrich L. V., Bare J. W., Bonifas J. M., Quinn A. G., Myers R. M., Cox D. R., Epstein E. H., Jr, et al. (1996). Human homolog of patched, a candidate gene for the basal cell nevus syndrome. Science 272, 1668–1671 10.1126/science.272.5268.1668 [DOI] [PubMed] [Google Scholar]
- Kho A. T., Zhao Q., Cai Z., Butte A. J., Kim J. Y., Pomeroy S. L., Rowitch D. H., Kohane I. S. (2004). Conserved mechanisms across development and tumorigenesis revealed by a mouse development perspective of human cancers. Genes Dev. 18, 629–640 10.1101/gad.1182504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., O'Connor M. S., Qin J., Songyang Z. (2004). Telosome, a mammalian telomere-associated complex formed by multiple telomeric proteins. J. Biol. Chem. 279, 51338–51342 10.1074/jbc.M409293200 [DOI] [PubMed] [Google Scholar]
- López de Silanes I., Stagno d'Alcontres M., Blasco M. A. (2010). TERRA transcripts are bound by a complex array of RNA-binding proteins. Nat. Commun. 1, 33. [DOI] [PubMed] [Google Scholar]
- Mai S., Garini Y. (2006). The significance of telomeric aggregates in the interphase nuclei of tumor cells. J. Cell. Biochem. 97, 904–915 10.1002/jcb.20760 [DOI] [PubMed] [Google Scholar]
- Marion R. M., Strati K., Li H., Tejera A., Schoeftner S., Ortega S., Serrano M., Blasco M. A. (2009). Telomeres acquire embryonic stem cell characteristics in induced pluripotent stem cells. Cell Stem Cell 4, 141–154 10.1016/j.stem.2008.12.010 [DOI] [PubMed] [Google Scholar]
- Ng L. J., Cropley J. E., Pickett H. A., Reddel R. R., Suter C. M. (2009). Telomerase activity is associated with an increase in DNA methylation at the proximal subtelomere and a reduction in telomeric transcription. Nucleic Acids Res. 37, 1152–1159 10.1093/nar/gkn1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver T. G., Read T. A., Kessler J. D., Mehmeti A., Wells J. F., Huynh T. T., Lin S. M., Wechsler–Reya R. J. (2005). Loss of patched and disruption of granule cell development in a pre-neoplastic stage of medulloblastoma. Development 132, 2425–2439 10.1242/dev.01793 [DOI] [PubMed] [Google Scholar]
- Palm W., de Lange T. (2008). How shelterin protects mammalian telomeres. Annu. Rev. Genet. 42, 301–334 10.1146/annurev.genet.41.110306.130350 [DOI] [PubMed] [Google Scholar]
- Parker R., Sheth U. (2007). P bodies and the control of mRNA translation and degradation. Mol. Cell 25, 635–646 10.1016/j.molcel.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Raynaud C. M., Hernandez J., Llorca F. P., Nuciforo P., Mathieu M. C., Commo F., Delaloge S., Sabatier L., André F., Soria J. C. (2010). DNA damage repair and telomere length in normal breast, preneoplastic lesions, and invasive cancer. Am. J. Clin. Oncol. 33, 341–345 10.1097/COC.0b013e3181b0c4c2 [DOI] [PubMed] [Google Scholar]
- Raz V., Vermolen B. J., Garini Y., Onderwater J. J., Mommaas–Kienhuis M. A., Koster A. J., Young I. T., Tanke H., Dirks R. W. (2008). The nuclear lamina promotes telomere aggregation and centromere peripheral localization during senescence of human mesenchymal stem cells. J. Cell Sci. 121, 4018–4028 10.1242/jcs.034876 [DOI] [PubMed] [Google Scholar]
- Redon S., Reichenbach P., Lingner J. (2010). The non-coding RNA TERRA is a natural ligand and direct inhibitor of human telomerase. Nucleic Acids Res. 38, 5797–5806 10.1093/nar/gkq296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz i Altaba A., Sánchez P., Dahmane N. (2002). Gli and hedgehog in cancer: tumours, embryos and stem cells. Nat. Rev. Cancer 2, 361–372 10.1038/nrc796 [DOI] [PubMed] [Google Scholar]
- Sahin E., Depinho R. A. (2010). Linking functional decline of telomeres, mitochondria and stem cells during ageing. Nature 464, 520–528 10.1038/nature08982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampl S., Pramhas S., Stern C., Preusser M., Marosi C., Holzmann K. (2012). Expression of telomeres in astrocytoma WHO Grade 2 to 4: TERRA level correlates with telomere lenght, telomerase activity, and advanced clinical grade. Transl. Oncol. 5, 56–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoeftner S., Blasco M. A. (2008). Developmentally regulated transcription of mammalian telomeres by DNA-dependent RNA polymerase II. Nat. Cell Biol. 10, 228–236 10.1038/ncb1685 [DOI] [PubMed] [Google Scholar]
- Schoeftner S., Blanco R., Lopez de Silanes I., Muñoz P., Gómez–López G., Flores J. M., Blasco M. A. (2009). Telomere shortening relaxes X chromosome inactivation and forces global transcriptome alterations. Proc. Natl. Acad. Sci. USA 106, 19393–19398 10.1073/pnas.0909265106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedelnikova O. A., Bonner W. M. (2006). GammaH2AX in cancer cells: a potential biomarker for cancer diagnostics, prediction and recurrence. Cell Cycle 5, 2909–2913 10.4161/cc.5.24.3569 [DOI] [PubMed] [Google Scholar]
- Shevtsov S. P., Dundr M. (2011). Nucleation of nuclear bodies by RNA. Nat. Cell Biol. 13, 167–173 10.1038/ncb2157 [DOI] [PubMed] [Google Scholar]
- Takai H., Smogorzewska A., de Lange T. (2003). DNA damage foci at dysfunctional telomeres. Curr. Biol. 13, 1549–1556 10.1016/S0960-9822(03)00542-6 [DOI] [PubMed] [Google Scholar]
- Ting D. T., Lipson D., Paul S., Brannigan B. W., Akhavanfard S., Coffman E. J., Contino G., Deshpande V., Iafrate A. J., Letovsky S., et al. (2011). Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science 331, 593–596 10.1126/science.1200801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toledo L. I., Murga M., Zur R., Soria R., Rodriguez A., Martinez S., Oyarzabal J., Pastor J., Bischoff J. R., Fernandez–Capetillo O. (2011). A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat. Struct. Mol. Biol. 18, 721–727 10.1038/nsmb.2076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace V. A. (1999). Purkinje-cell-derived Sonic hedgehog regulates granule neuron precursor cell proliferation in the developing mouse cerebellum. Curr. Biol. 9, 445–448 10.1016/S0960-9822(99)80195-X [DOI] [PubMed] [Google Scholar]
- Wechsler–Reya R. J., Scott M. P. (1999). Control of neuronal precursor proliferation in the cerebellum by Sonic Hedgehog. Neuron 22, 103–114 10.1016/S0896-6273(00)80682-0 [DOI] [PubMed] [Google Scholar]
- Wentzensen I. M., Mirabello L., Pfeiffer R. M., Savage S. A. (2011). The association of telomere length and cancer: a meta-analysis. Cancer Epidemiol. Biomarkers Prev. 20, 1238–1250 10.1158/1055-9965.EPI-11-0005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wileman T. (2007). Aggresomes and pericentriolar sites of virus assembly: cellular defense or viral design? Annu. Rev. Microbiol. 61, 149–167 10.1146/annurev.micro.57.030502.090836 [DOI] [PubMed] [Google Scholar]
- Xiang C., Baubet V., Pal S., Holderbaum L., Tatard V., Jiang P., Davuluri R. V., Dahmane N. (2012). RP58/ZNF238 directly modulates proneurogenic gene levels and is required for neuronal differentiation and brain expansion. Cell Death Differ. 19, 692–702 10.1038/cdd.2011.144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L., Blackburn E. H. (2007). Human cancer cells harbor T-stumps, a distinct class of extremely short telomeres. Mol. Cell 28, 315–327 10.1016/j.molcel.2007.10.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J., Wu Y., Gilson E. (2010). Dynamics of telomeric chromatin at the crossroads of aging and cancer. Essays Biochem. 48, 147–164 10.1042/bse0480147 [DOI] [PubMed] [Google Scholar]
- Yehezkel S., Segev Y., Viegas–Péquignot E., Skorecki K., Selig S. (2008). Hypomethylation of subtelomeric regions in ICF syndrome is associated with abnormally short telomeres and enhanced transcription from telomeric regions. Hum. Mol. Genet. 17, 2776–2789 10.1093/hmg/ddn177 [DOI] [PubMed] [Google Scholar]
- Yehezkel S., Rebibo–Sabbah A., Segev Y., Tzukerman M., Shaked R., Huber I., Gepstein L., Skorecki K., Selig S. (2011). Reprogramming of telomeric regions during the generation of human induced pluripotent stem cells and subsequent differentiation into fibroblast-like derivatives. Epigenetics 6, 63–75 10.4161/epi.6.1.13390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L. F., Ogawa Y., Ahn J. Y., Namekawa S. H., Silva S. S., Lee J. T. (2009). Telomeric RNAs mark sex chromosomes in stem cells. Genetics 182, 685–698 10.1534/genetics.109.103093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J. L., Gao W. Q. (2000). Overexpresion of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat. Neurosci. 3, 580–586 10.1038/75753 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.