Abstract
Ipl1/Aurora B is the catalytic subunit of a protein kinase complex required for chromosome segregation and nuclear division. Before anaphase, Ipl1 is required to establish proper kinetochore-microtubule associations and to regulate the spindle assembly checkpoint (SAC). The phosphatase Glc7/PP1 opposes Ipl1 for these activities. To investigate Ipl1 and Glc7 regulation in more detail, we isolated and characterized mutations in the yeast Saccharomyces cerevisiae that raise the restrictive temperature of the ipl-2 mutant. These suppressors include three intragenic, second-site revertants in IPL1; 17 mutations in Glc7 phosphatase components (GLC7, SDS22, YPI1); two mutations in SHP1, encoding a regulator of the AAA ATPase Cdc48; and a mutation in TCO89, encoding a subunit of the TOR Complex 1. Two revertants contain missense mutations in microtubule binding components of the kinetochore. rev76 contains the missense mutation duo1-S115F, which alters an essential component of the DAM1/DASH complex. The mutant is cold sensitive and arrests in G2/M due to activation of the SAC. rev8 contains the missense mutation ndc80-K204E. K204 of Ndc80 corresponds to K166 of human Ndc80 and the human Ndc80 K166E variant was previously shown to be defective for microtubule binding in vitro. In a wild-type IPL1 background, ndc80-K204E cells grow slowly and the SAC is activated. The slow growth and cell cycle delay of ndc80-K204E cells are partially alleviated by the ipl1-2 mutation. These data provide biological confirmation of a biochemically based model for the effect of phosphorylation on Ndc80 function.
Keywords: Aurora B, IPl1, kinetochore, NDC80, GLC7
The protein kinase Aurora B (Ipl1 in Saccharomyces cerevisiae, Ark1 in Schizosaccharomyces pombe, Air-2 in Caenorhabditis elegans) plays a pivotal role in ensuring bipolar attachment of chromosomes to the mitotic spindle. As the catalytic subunit of the tetrameric chromosome passenger complex, it associates with chromosomes early in mitosis and then accumulates at kinetochores, where it phosphorylates components of the kinetochore before anaphase (Ruchaud et al. 2007). After anaphase, Aurora B accumulates at the spindle midzone, where it has additional substrates involved in cytokinesis. The kinetochore is a large protein complex consisting of the inner kinetochore complex, which makes direct contact with centromeric chromatin, the outer kinetochore, which contains microtubule binding proteins that track the minus ends of growing and shrinking kinetochore microtubules, and a central domain that tethers the inner and outer kinetochore complexes [reviewed in (Santaguida and Musacchio 2009)]. Aurora B phosphorylates outer kinetochore proteins to regulate microtubule-binding dynamics, which is required to establish a bipolar arrangement of chromosomes on the mitotic spindle. Aurora B activity also is required for the spindle assembly checkpoint (SAC), which delays anaphase until all chromosomes are under bipolar attachment.
Tension at kinetochores, brought about by bipolar association of condensin-tethered chromosomes in the mitotic spindle, originally was proposed by Nicklas and Koch (Nicklas and Koch 1969) to regulate kinetochore microtubule dynamics. It is now thought that kinetochore-microtubule tension directly regulates the phosphorylation state of Aurora B kinetochore substrates. An attractive model for the coupling of tension to kinetochore substrate phosphorylation suggests that tension pulls the outer kinetochore away from the inner kinetochore (Andrews et al. 2004; Liu et al. 2009; Maresca and Salmon 2009; Uchida et al. 2009; Vanoosthuyse and Hardwick 2009). Indeed, a gradient of Aurora B activity is centered on inner centromeres of mammalian cells. Reduced Aurora B kinase activity at kinetochores under tension, in combination with a possible increase in protein phosphatase activity, leads to reduced phosphorylation and less dynamic kinetochore microtubule binding, and silencing of the SAC [reviewed by (Lampson and Cheeseman 2011)].
Multiple lines of evidence indicate that type 1 protein phosphatase (PP1 in mammals, Glc7 in S. cerevisiae, Dis2 and Sds21 in S. pombe, and Ceglc-7 in C. elegans) is largely responsible for the dephosphorylation of Aurora B substrates during mitosis. Loss of function mutations in PP1 genes induce mitotic arrest in S. pombe (Ishii et al. 1996), S. cerevisiae (Hisamoto et al. 1994; MacKelvie et al. 1995), Aspergillus nidulans (Doonan and Morris 1989), and Drosophila (Axton et al. 1990), and anti-PP1 antibodies induce mitotic arrest when injected into mammalian cells (Fernandez et al. 1992). PP1 mutations in S. cerevisiae suppress the temperature sensitivity of ipl1 mutants (Francisco et al. 1994; Hsu et al. 2000) and the phenotype of air-2(RNAi) can be suppressed by decreasing PP1 activity [Ceglc-7(RNAi)] in C. elegans (Hsu et al. 2000). In mammalian cells, PP1 localizes to kinetochores during mitosis (Trinkle-Mulcahy et al. 2003) and inhibition of PP1 activity suppresses defects associated with reduced Aurora B activity (Emanuele et al. 2008; Wang et al. 2008). The mitotic arrest of some glc7 mutants requires the SAC (Bloecher and Tatchell 1999; Sassoon et al. 1999), but PP1 is also required for SAC silencing (Pinsky et al. 2009; Vanoosthuyse and Hardwick 2009). Together, these results are consistent with the idea that PP1 acts on Aurora B substrates to regulate kinetochore microtubule dynamics and SAC silencing.
PP1 activity is regulated by a large number of regulatory/targeting subunits that direct PP1 catalytic activity toward specific substrate(s) [reviewed by (Virshup and Shenolikar 2009; Bollen et al. 2010)]. A degenerate motif, the so-called RVxF motif found on many targeting subunits, is an essential interaction motif required for PP1c binding and regulation (Egloff et al. 1997). The conserved outer kinetochore protein Spc105 (KNL1 in mammals, Spc7 in S. pombe) contains both an RVxF motif and a less frequent SILK motif found in a subset of PP1 binding proteins (Hendrickx et al. 2009). A KNL1 mutant whose product cannot bind PP1 (KNL1RVSF/AAAA) is lethal, and PP1 is not found at kinetochores in KNL1RVSF/AAAA cells (Liu et al. 2010). The mutant has enhanced Aurora B-dependent phosphorylation at the outer kinetochore and destabilized kinetochore microtubule attachments (Liu et al. 2010). Mutants in KNL1 orthologs in S. pombe (spc7) and S. cerevisiae (spc105) that cannot bind PP1 also are inviable due to activation of the SAC (Meadows et al. 2011; Rosenberg et al. 2011). Tethering PP1 directly to an Spc105 variant that cannot bind PP1 (Spc105RVSF-RASA) rescues cell lethality but, in contrast, tethering PP1 to wild-type Spc105 is lethal and cannot be rescued by disruption of the SAC (Rosenberg et al. 2011). These results suggest that the level of PP1 targeted to the outer kinetochore is under exquisite control. Serine residues in both PP1 binding motifs in KNL1 (RVSF and SILK) are phosphorylated by Aurora B in vitro and in vivo in human cells (Welburn et al. 2010). Phosphomimetic variants in KNL1 reduce microtubule and PP1 binding (Welburn et al. 2010). These findings suggest that KNL1 participates in a feed forward circuit regulating Aurora B substrate phosphorylation.
PP1 also binds to the kinesin-8 family members Klp5-Klp6 in S. pombe, where PP1 binding is thought to contribute to SAC silencing (Meadows et al. 2011). In S. cerevisiae, PP1 binds to the kinetochore/spindle protein Fin1, which assists in targeting PP1 to the kinetochore (Akiyoshi et al. 2009b). Misregulation of Fin1 results in premature silencing of the SAC in a PP1-dependent manner. However, Fin1 is not essential, suggesting that Fin1 activities overlap with those of other kinetochore proteins.
In addition to KNL1/Spc105/Spc7, Klp5-Klp6, and Fin1, several other PP1 regulators have been implicated with roles in opposing Ipl1/Aurora B activity at the kinetochore in S. cerevisiae. Mutations in GLC8 (Tung et al. 1995), SDS22 (Peggie et al. 2002), YPI1 (Bharucha et al. 2008), and SHP1 (Cheng and Chen 2010) suppress the temperature sensitivity of ipl1-2 or ipl1-321 mutations, implying that their products regulate PP1 activity opposing Ipl1. Although these proteins are evolutionarily conserved, they are not integral components of kinetochores and their precise roles are poorly understood. Sds22 and Ypi1 can form a ternary complex with Glc7 (Hazbun et al. 2003; Pedelini et al. 2007) and inhibit phosphatase activity in vitro (Garcia-Gimeno et al. 2003; Lesage et al. 2007; Pedelini et al. 2007) although both behave genetically as positive regulators of PP1 activity. Sds22 in fission yeast has been shown to change the specificity of PP1 activity, increasing PP1 activity toward histone H3 and reducing its activity toward phosphorylase a (Stone et al. 1993). In vertebrate cells, Sds22 has been proposed to have a more canonical targeting role. It associates with PP1 at kinetochores and appears to regulate the PP1 activity opposing Aurora B (Posch et al. 2010). Interestingly, an essential histone H2A variant from Tetrahymena thermophila that is necessary to dephosphorylate histone 3 serine 10 at the end of mitosis contains an Sds22-related domain that binds PP1 (Song et al. 2012). However, both Sds22 and Ypi1 have been reported to have additional biological activities. Sds22 in Drosophila has been implicated in regulating the actin cytoskeleton to control epithelial cell architecture (Grusche et al. 2009; Shao et al. 2010; Jiang et al. 2011) and mitotic exit (Kunda et al. 2012), and Ypi1 regulates ion homeostasis in S. cerevisiae independently of Glc7 (Marquina et al. 2012).
Because the assay for suppression of ipl1 temperature sensitivity has only been used to test mutants previously known to influence Glc7 activity, it is not known which additional gene products might act in Ipl1-dependent processes. We therefore conducted a classical genetic screen for suppressors of ipl1-2. In addition to new alleles of GLC7, SHP1, YPI1, and SHP1, the screen uncovered dominant alleles of IPL1, a mutant in the TOR complex 1 (TORC1) pathway, and mutants in DUO1 and NDC80, two components of the outer kinetochore.
Materials and Methods
Yeast strains and media
The yeast strains used in this study are listed in Supporting Information, Table S1 and are congenic to KT1112 [MATa leu2ura3his3 (Stuart et al. 1994)] and KT1113 (Frederick and Tatchell 1996). The ipl1-2 mutation (Chan and Botstein 1993) was introduced into the KT1112 background by seven serial backcrosses. Yeast strains were transformed by the method of Gietz et al. (Gietz et al. 1992). Growth of yeast was in 1% yeast extract, 2% Bacto peptone, and 2% glucose (Yeast Extract Peptone Dextrose, i.e., YPD) medium or in synthetic complete (SC) medium (Sherman et al. 1986), or modifications of these media as stated. Tetrad analysis was performed as described previously (Rose et al. 1990). Culture temperatures are indicated. 13myc-tagged alleles of PDS1 and IPL1 were generated using the method of Longtine et al. (Longtine et al. 1998) with F2 and R1 primers listed in Table S2. Polymerase chain reaction (PCR) products with the tagged alleles were introduced into the wild-type diploid strain KT1112/KT1113 by transformation; G418-resistant transformants were sporulated and protein extracts from G418-resistant meiotic progeny were tested for the presence of the tagged protein by immunoblot.
Bacterial growth and DNA manipulation
All plasmids were amplified in Escherichia coli strain DH5α. DH5α was grown in LB medium supplemented with ampicillin to select for plasmids. Transformation of chemically competent bacterial cells was as described (Fritsch et al. 1989). Restriction enzymes (Promega), DNA ligase (NEB), and high-fidelity DNA polymerase for PCR (Bio-Rad and Phenix) were used according to the manufacturer’s instructions. Yeast genomic DNA was isolated using the YeaStar kit (Zymo Research), and yeast plasmid DNA was isolated using the Zymoprep kit (Zymo Research). Plasmids were purified from E. coli using the Zyppy miniprep kit (Zymo Research) or the GenElute mimiprep kit (Sigma-Aldrich). DNA restriction fragments were purified from agarose gel slices using the ZymoClean kit (Zymo Research).
Mutant isolation
Strain KT1963 (ipl1-2) was grown overnight at 24°. Then, 250-μL aliquots of a 1/100 dilution were plated onto YPD plates and uncovered plates were irradiated with 5000 μJ/CM2 of ultraviolet (UV) light in a UVP-CL1000 UV crosslinker. After incubation in the dark at 24° for 24 hr, the plates were incubated for 3 d at 33°. Revertant colonies were tested for growth on YPD medium at temperatures ranging from 14° to 37° and on YPD medium containing 3% formamide, 0.1 M LiCl, 0.1 M CsCl, or 5 mM caffeine. Revertants with one or more growth defects and those that grew at 37° were backcrossed to an ipl1-2 strain, and tetrad analysis was performed to confirm that ipl1 suppression was due to a single Mendelian mutation and to test whether any growth defects were linked to suppression. Mutants were tested for complementation by strains carrying known ipl1 suppressor mutations or for genetic linkage to known ipl1 suppressor mutations. Tentative assignments were confirmed by DNA sequence analysis of the indicated suppressor locus from the mutant. In each case, the mutant locus was amplified by PCR and the DNA sequence of the ORF was obtained using the primers listed in Table S2. DNA sequences of PCR products and plasmids were determined by Macrogen USA (Rockville, MD). DNA sequences were compared with database sequences at the Saccharomyces Genome Database web site using the BLAST algorithm (Altschul et al. 1990).
Cloning of the suppressor loci
IPL1 meiotic segregants of KT2878 (rev8), KT2961 (rev76), and KT2967 (rev81) were transformed with a yeast genomic library in the CEN URA3 vector YCp50 (a generous gift from Doug S. Conklin), and transformants were selected by growth on SC-URA medium at 24°. Transformants were replica plated onto YPD medium containing 3% formamide and incubated for 2−5 d at 30°. Colonies that grew on formamide medium were transferred to SC-URA and retested for resistance to formamide. Positive transformants were grown in nonselective conditions (YPD), transferred to SC+5-FOA medium to select for plasmid loss, and retested for growth on formamide medium. Plasmid DNA was isolated from transformants whose formamide resistance required the URA3 plasmid, amplified in E. coli, and used to transform the original yeast strains. DNA sequences obtained from the genomic inserts of plasmids that conferred formamide resistance were compared to the Saccharomyces Genome Database using BLAST to identify the chromosome segment contained within each plasmid.
To ensure that the ndc80 and duo1 alleles are the suppressor mutations in rev8 and 76, respectively, we generated clones of these alleles and introduced these into diploid strains heterozygous for both ipl1-2 and deletion of one of the two genes. The diploid transformants were sporulated and tetrads dissected. The progeny of diploid transformants with vector alone showed two viable spore clones per tetrad, and roughly 1/2 of these were temperature-sensitive for growth. None of the viable spore clones was resistant to G418. In contrast, progeny of ndc80Δ/+ ipl1-2/+ transformants with pRS316:ndc80-8 include more than 2 viable spore clones per tetrad. We recovered 14 G418-resistant spore clones from 22 tetrads. All of these carry the plasmid marker, and those that have ipl1-2 grow at 30°. This confirms that the ndc80-8 allele is responsible for suppression of the ipl1-2 inability to grow at 30°. We observed similar results for ipl1-2/+ duo1Δ/+ diploid transformants with pRS303:duo1-76 (the duo1-76 fragment contains ARS717, so pRS303:duo1-76 is a replicating plasmid). In this case, we recovered 13 G418-resistant clones from 24 tetrads. All carry the plasmid marker and all grow at 30°. Figure S3 shows drop growth tests for four clones each of the ipl1-2 and ipl1-2 duo1-76 and ipl1-2 ndc80-8 genotypes.
To generate pRS316:ndc80-8 and pRS303:duo1-76, we used PCR with high-fidelity polymerase to amplify the genomic loci from strain KT3257 (ndc80-8) and strain KT3386 (duo1-76). Phenix HiFi polymerase was used to amplify ndc80-8, and HotStar (QIAGEN) was used to amplify duo1-76. Both polymerases were used according to manufacturer’s instructions. Genomic DNA was prepared using the YeaStar kit (Zymo Research). PCR primers were DUO1-Fa and DUO1-Ra, and NDC80-Fa and NDC80-Ra (Table S2). The NDC80 Fa and Ra primers contain Bam1H and XbaI sites, respectively. The ndc80-8 PCR product was purified using the Clean and Concentrate kit (Zymo Research), then digested with Bam1H and XbaI. The digestion product was again purified using the Clean and Concentrate kit, and was ligated to Bam1H and XbaI-digested pRS316. The duo1-76 PCR product was cloned via its A overhangs using the pGEM-T Easy kit (Promega). A SacII-Sal fragment containing duo1-76 was then cloned into pRS303. For both mutant alleles, the entire sequences of the cloned PCR products were determined to ensure the presence of the appropriate mutations and the absence of undesired changes.
Microscopy
We imaged Glc7-mCitrine and Sds22-mCitrine in live cells and Ypi1-13Myc by indirect immunofluorescence as described (Bharucha et al. 2008). The Myc-tagged YPI1 allele was chosen over an mCitrine-tagged allele due to a reduced activity of the mCitrine-tagged variant (Bharucha et al. 2008).
Immunoblot analysis
Total protein extracts were prepared from yeast cultures grown to log phase using glass bead lysis in TCA (Davis et al. 1993). Protein extracts were electrophoresed through 4–20% gradient gels (Criterion; Bio-Rad), gels were blotted to nitrocellulose filters (Protran BA83; Whatman), and filters were probed with indicated primary antibodies followed by horseradish peroxidase-conjugated secondary antibodies (Bio-Rad). The anti-GFP antibody is an affinity-purified rabbit antibody generously provided by J. Nathan Davis, LSUHSC-S. HA-tagged and myc-tagged proteins were detected using the 12CA5 and 9E10 mouse monoclonal antibodies, respectively. The antiphosphoglycerate kinase antibody used as a loading reference is a mouse monoclonal antibody to yeast Pgk1 (Molecular Probes). Antibody binding was detected using the Immobilon ECL reagents (Millipore) and the Chemidoc system (Bio-Rad). Quantity One software (Bio-Rad) was used to image and quantitate signal on immunoblots. Blots were stripped before reprobing with reference antibody using Western Reprobe (G Biosciences).
Results and Discussion
The ipl1-2 mutant was originally identified by Chan and Botstein (Chan and Botstein 1993) in a screen for mutants with increased ploidy. Ipl1 activity in this mutant, as measured by the phosphorylation levels of histone H3Ser10, is reduced at 24° but growth rate is not affected at 24° (Hsu et al. 2000). However, ipl1-2 cells lose chromosomes and viability rapidly at 30° (Tatchell et al. 2011). To survey in an unbiased way for mutations that suppress the temperature sensitivity of ipl1-2, we mutagenized an ipl1-2 strain (KT1963) with UV light and isolated revertants that grew at 33° (see Materials and Methods for details). In most cases, we focused on revertants in which ipl1-2 suppression segregated as a single Mendelian allele and cosegregated with a growth trait such as sensitivity to low temperature or to formamide, caffeine, or divalent cations in the growth medium. After placing suppressors into complementation groups, we showed that most are genetically linked to a predicted gene. In some cases, we cloned the loci by complementation of an associated growth defect. Finally, sequence analysis was performed on each mutant allele to identify the lesion responsible for suppression. Results for the 25 mutations that we identified as suppressors of ipl1-2 are summarized in Table 1.
Table 1. Summary of ipl1-2 suppressor mutations.
| Suppressor Locus Mutant Allele | Revertant No. | Mutation (S) | Residue in Human Ortholog |
|---|---|---|---|
| IPL1 | |||
| IPL1-R150K H352Y | 14,21 | G449A, C1054T | R139 H340 |
| IPL1-S167L H352Y | 16 | C500T, C1054T | M136 H340 |
| IPL1-G347E H352Y | 20 | G1040A, C1054T | K335 H340 |
| GLC7 (Type 1 protein phosphatase [PP1] catalytic subunit) | |||
| glc7-L15S | 106 | T44C | L16 |
| glc7-L37F | 85 | A111T, T175A | L38 |
| glc7-L71S | 82 | T737C | L72 |
| glc7-L74P | 39, 62, 89 | T746C | L75 |
| glc7-Y92N R141K | 55 | T799A, G947A | Y93 R142 |
| glc7-S99L | 84 | T820C, C821T | S100 |
| glc7-K112E | 107 | A859G | K113 |
| glc7-F118S | 102 | T878C | F119 |
| glc7-Y136N | 73 | T931A | Y137 |
| glc7-Q293D | 112 | A1403C | Q294 |
| YPI1 (Glc7/PP1 regulatory subunit) | |||
| ypi1-F74S | 52 | T221C | Y64 |
| ypi1-F74L | 95 | T222A | Y64 |
| SDS22 (Glc7/PP1 regulatory subunit) | |||
| sds22-F177S | 47 | T530C | I204 |
| sds22-W187R | 53 | T559A | F214 |
| sds22-D2N D119N | 74 | G4A, G355A | D148 |
| sds22-77 (C-terminal frameshift)a | 77 | ΔC1014 | |
| sds22-E163I L329P | 94 | 486AGA488GAT, T984C | E192 V350 |
| SHP1 (subunit of Cdc48/p97 AAA AATPase) | |||
| shp1-FS99b | 99 | A insertion after 368, T370C | |
| shp1-FS105c | 105 | A insertion after 873, G877A | |
| TCO89 (nonessential subunit of TOR Complex 1) | |||
| tco89-71 | 71 | four missense codons and a stop codond | not conserved |
| DUO1 (subunit of DAM/DASH complex) | |||
| duo1-S115F | 76, 81 | C344T | not conserved |
| NDC80 (subunit of Ndc80 complex) | |||
| ndc80-K204E | 8 | A610G | K166 |
Frameshift at final codon (G 338 underlined) resulting in stop after 22 missense codons: …YIRG* →…YIRGDLDQEMIFFLYTHTNIYIYIYI.*
Frameshift after codon 122 (underlined) resulting in stop after 15 missense codons: …GSGNN… →…GSGNKPQVYELFGYGKRSS*
Frameshift after codon 291 (underlined) resulting in stop after 2 missense codons: …NVYKKLD… →…NVYKKIK*
Intragenic IPL1 revertants
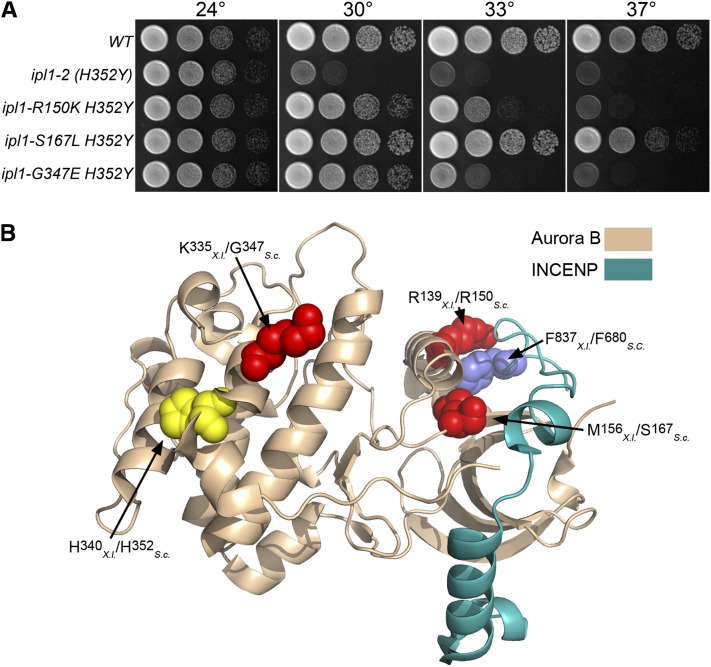
We recovered several strong and dominant suppressors, as shown by the ability of diploids between these revertants and ipl1-2 strains to grow at 33−37°. Spore clones with the ipl1-2 phenotype were never recovered from crosses between these revertants and a wild-type strain, indicating that the suppressor loci are tightly linked to IPL1. The sequence of the IPL1 locus from two such suppressor mutants is identical to that of the wild-type IPL1 locus, indicating that these two are true revertants. Sequence analysis of the other intragenic suppressor mutants revealed that these contain the original ipl1-2 missense mutation (H352Y) and an additional missense mutation: R150K, S167L or G347E. IPI1-S167L H352Y strains grow nearly as well as wild-type strains at 33°, IPI1-R150K H352Y mutants have an intermediate growth phenotype, whereas IPI1-G347E H352Y mutants grow poorly at 33° (Figure 1A).
Figure 1 .
Intragenic ipl1-2 suppressors. (A) Cultures of WT (KT1112), ipl1-2 (H352Y) (KT1829), ipl1-2 R150K H352Y (KT2865), ipl1-S167L H352Y (KT2867), and ipl1-G347E H352Y (KT2869) strains were serially diluted onto YPD medium and imaged after 40 hr at the designated temperatures. (B) The locations of intragenic ipl1-2 suppressor mutations are mapped on the X. laevis Aurora B- INCENP structure [2BFX.pdb (Sessa et al. 2005)]. The highlighted amino acid residues are represented as space-filling models. The location of the ipl1-2 mutation H352Y is shown in yellow. Residues altered by intragenic suppressor mutations are red, and the amino acid residue in INCENP that associates with R139 is blue. Note that both S167 and R150 are predicted to lie near the interface with INCENP/Sli15.
The C-terminal domain of Sli15, a positive regulator of Ipl1 (Kim et al. 1999), binds to and activates Ipl1 in vitro (Kang et al. 2001). Crystal structure of the Sli15-Ipl1 orthologous pair from Xenopus laevis (INCENP-Aurora B) reveals that R150 (R139 in Aurora B from X. laevis) lies at the Aurora B/INCENP interface and is part of a pocket on the surface of Aurora B that directly associates with the highly conserved residue F837 in INCENP [Figure 1B (Sessa et al. 2005)]. This pocket lies near the invariant E141 residue that is required for ATP binding and catalysis. It is likely that INCENP binding to Aurora B through this pocket allows for the proper orientation of E141. Interestingly, mutation of the adjacent residue R151K in Ipl1 results in reduced binding to Sli15 and reduced kinase activity (Kotwaliwale et al. 2007). Ipl1 S167 (M156 in X. laevis Aurora B) also lies close to the Aurora B/INCENP interface. One explanation for the suppressive effects of these mutations is that they increase the affinity of Ipl1 for Sli15, thereby effectively increasing the activity of the Ipl1-2 protein. G347 is in the protein kinase subdomain XI (Hanks and Hunter 1995), which is involved in stabilizing the large C-terminal lobe of the kinase. The substitution lies close to the ipl1-2 mutation (H352Y), but this glycine residue is not highly conserved in Aurora B proteins from metazoans.
Extragenic ipl1-2 suppressors in the GLC7 pathway
Most of the suppressor loci are unlinked to ipl1-2, as shown by the ability to recover strains with the original ipl1-2 phenotype from crosses between revertant and wild-type strains. We identified twenty-two different suppressor mutations in seven genes. The largest group of extragenic suppressors contains mutations altering Glc7 phosphatase components that were previously reported as ipl1 suppressor loci. We identified ten new alleles of GLC7, five SDS22 alleles, two YPI1 alleles, and two SHP1 alleles.
GLC7:
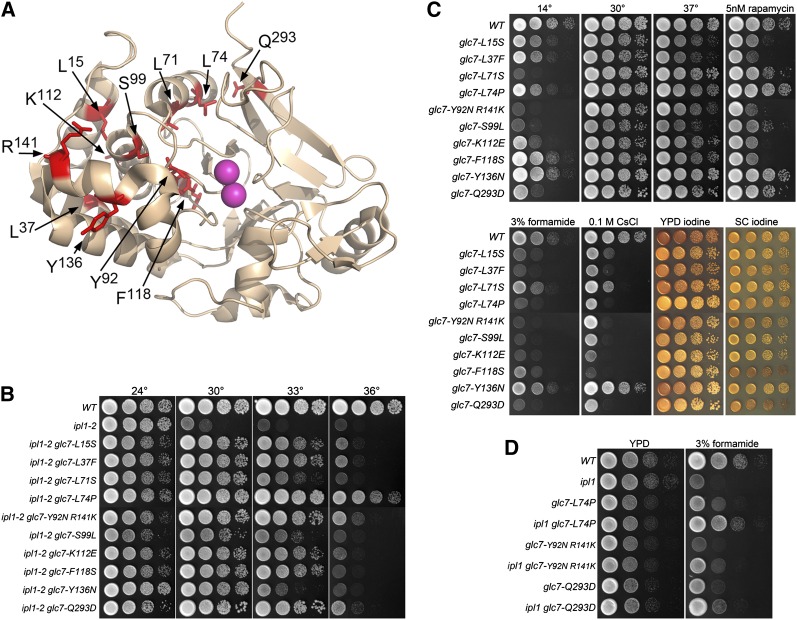
The 10 new GLC7 alleles are all missense mutations that alter residues identical to those in mammalian PP1 orthologs. One allele, Y92N R141K, is a double mutant. L74P was isolated three times, but each other allele was recovered once. We have highlighted the mutated residues on the crystal structure of rabbit PP1 shown in Figure 2A. Most mutated residues are in the N-terminal lobe of PP1, and a majority of these are buried in the interior of the protein. The surface residues altered in ipl1-2 suppressor strains are K112, Y137, R141, and Q293. It is worth noting that many glc7 mutations previously reported to suppress ipl1 mutants also are located in the N-terminal lobe of PP1 {glc7-10 [F135L] (Andrews and Stark 2000); glc7-127 [K110A, K112A], and glc7-129 [D137A and E138A] (Baker et al. 1997)}, although the significance is as yet unclear.
Figure 2 .
Extragenic ipl1-2 suppressors in GLC7. (A) Locations of GLC7 ipl1-2 suppressor changes mapped on the structure of rabbit PP1α [1FJM.pdb (Goldberg et al. 1995)]. The amino acid residues altered in the ipl1 suppressor mutants are stick representations in red. The two Mn2+ ions in the active site are in magenta. (B) Cultures of WT (KT1113), ipl1-2 (KT1829), ipl1-2 glc7-L15S (KT3062), ipl1-2 glc7-L37F (KT2940), ipl1-2 glc7-L71S (KT2938), ipl1-2 glc7-L74P (KT3361), ipl1-2 glc7-Y92N R141K (KT3365), ipl1-2 glc7-S99L (KT2939), ipl1-2 glc7-K112E (KT3064), ipl1-2 glc7-F118S (KT3059), ipl1-2 glc7-Y136N (KT3358), and ipl1-2 glc7-Q293D (KT3066) strains were serially diluted onto YPD medium and imaged after 40 hr at the designated temperatures. (C) Cultures of WT (KT1113), glc7-L15S (KT3304), glc7-L37F (KT2973), glc7-L71S (KT2969), glc7-L74P (KT3359), glc7-Y92N R141K (KT3363), glc7-S99L (KT2970), glc7-K112E (KT3308), glc7-F118S (KT3302), glc7-Y136N (KT3355), and glc7-Q293D (KT3310) strains were serially diluted onto the designated medium and imaged at the designated temperatures. All media, with the exception of SC, are either YPD or YPD supplemented with the indicated components. The YPD 14° plates were incubated for 14 d. All other plates were incubated for 40 hr. YPD iodine and SC iodine panels indicate cells grown on YPD or SC media for 40 hr and then stained with iodine vapor. (D) Cultures of WT (KT1113), ipl1-2 (KT1829), glc7-L74P (KT3359), ipl1-2 glc7-L74P (KT3361), glc7-Y92N R141K (KT3363), ipl1-2 glc7-Y92N R141K (KT3365), glc7-Q293D (KT3310), and ipl1-2 glc7-Q293D (KT3066) strains were serially diluted onto YPD medium and YPD medium containing 3% formamide and imaged after 24 and 72hr, respectively, at 24°.
Our new glc7 mutants are quite variable in their ability to raise the restrictive temperature of ipl1-2. As shown in Figure 2B, glc7-Y136N only weakly suppresses ipl1-2 at 33°, whereas glc7-L74P allows the ipl1-2 mutant to grow weakly at 36°. We also compared the phenotypes of our collection of glc7 mutants in a wild-type IPL1 background. Growth defects common to many of the mutants include cold sensitivity and sensitivity to formamide, CsCl, rapamycin, and caffeine (Figure 2C). However, each mutant strain has a unique phenotype, and no trait correlates perfectly with the ability to suppress ipl1-2. These phenotypes are due to the glc7 alleles rather than an accompanying mutation because each trait cosegregates with ipl1 suppression in backcrosses. We believe that the highly individual phenotypes reflect the large number of Glc7 binding proteins that regulate substrate specificity.
Although our preliminary screen indicated that the glc7 suppressor mutations are recessive for suppression, spot tests of diploid strains heterozygous for the glc7 mutant and homozygous for ipl1-2 revealed that all are partially dominant, as shown by some growth at 30° and 33° (Figure S1A). This finding is in contrast to suppressor mutants in SDS22, YPI1, and SHP1, which are completely recessive (Figure S1, B−D). One possibility is that the Glc7 variants sequester other cofactors required for phosphatase function, such as Sds22 or Ypi1. In this regard, it is worth noting that SDS22 was originally identified in S. pombe and S. cerevisiae as a dosage suppressor of dis2-11 (Ohkura and Yanagida 1991), glc7-12 (MacKelvie et al. 1995), and glc7-Y170 (Hisamoto et al. 1995).
If Glc7 directly opposes Ipl1 to dephosphorylate substrates involved in chromosome segregation, then we would expect that some traits associated with the GLC7 mutations would be more severe in an IPL1 background than in the ipl1-2 background. As shown in Figure 2D, several glc7 mutants containing the ipl-2 allele grow better on YPD medium containing 3% formamide than either the ipl1-2 or glc7 single mutants. The most striking results are with glc7-L74P. The ipl1-2 glc7-L74P strain grows nearly at the same rate as the wild type (Figure 2D, compare fourth row with first row). These results suggest that the formamide growth defect caused by the glc7-L74P allele is connected with Ipl1 activity.
SDS22:
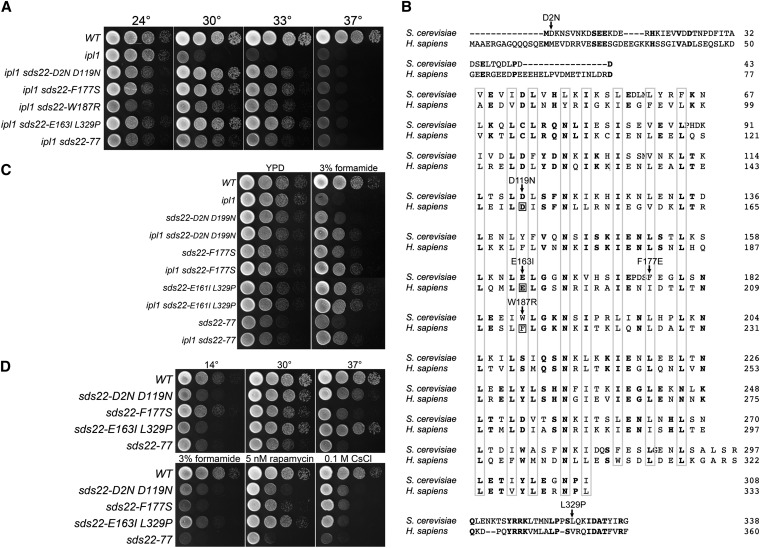
Five suppressors either fail to complement an sds22 mutant or are linked to SDS22-mCitrine::HIS3 (Figure 3A). SDS22 encodes a conserved protein whose PP1 binding domain consists of a tandem array of leucine-rich repeats rather than an RVxF motif. Sequence analysis of SDS22 from these mutants revealed that four contain missense mutations and one results in a frameshift mutation near the 3′ end of the gene. Sequence alignment of Sds22 from budding yeast and human shows that most of the missense mutations are in highly conserved residues (Figure 3B). Four of these, D119, E161, F177E, and W187, are within the leucine-rich repeats. Remarkably, three of these correspond to residues in human Sds22 (D148, E192, F214; boxed residues in Figure 2B) that are required for proper PP1 binding (Ceulemans et al. 2002). These observations suggest that Sds22 mutant proteins D119N, E163I L329P and W187R are defective for Glc7 binding.
Figure 3 .
Extragenic ipl1-2 suppressors in SDS22. (A) Cultures of WT (KT1113), ipl1-2 (KT1829), ipl1-2 sds22-D2N D119N (KT2934), ipl1-2 sds22-F177S (KT3353), ipl1-2 sds22-W187R (KT3381), ipl1-2 sds22-E163I L329P (KT3052), and ipl1-2 sds22-77 (KT2936) strains were serially diluted onto YPD medium and imaged after 40 hr at the designated temperatures. (B) ClustalW alignment of human and S. cerevisiae Sds22 proteins showing locations of ipl1 suppressor mutations. Boxed residues correspond to sites of mutations in human Sds22 that prevent proper binding of PP1 (Ceulemans et al. 2002). (C) Cultures of WT (KT1113), ipl1-2 (KT1829), sds22-D2N D119N (KT2963), ipl1-2 sds22-D2N D119N (KT2934), sds22-F177S (KT3351), ipl1-2 F177S (KT3353), sds22-E163I L329P (KT3292), ipl1-2 sds22-E163I L329P (KT3052), sds22-77 (KT2964), and ipl1-2 sds22-77 (KT2936) strains were serially diluted onto YPD medium and YPD medium containing 3% formamide and imaged after 24 hr and 72hr at 24°, respectively. (D) Cultures of WT (KT1113), sds22-D2N D119N (KT2963), sds22-F177S (KT3351), sds22-E163I L329P (KT3292), and sds22-77 (KT2964) strains were serially diluted onto the designated medium at the designated temperatures. Media, with the exception of SC, are either YPD or YPD supplemented as indicated. The YPD 14° plates were incubated for 14 d. All other plates were incubated for 40 hr.
We were unable to recover sds22-W187R in a wild-type IPL1 background. Spores corresponding to the sds22-W187R mutants germinated and grew into microscopic colonies but never formed colonies that could be characterized. Thus, a reduction of Ipl1 activity restores viability to the sds22-W187R mutant. This indicates that the inviability of sds22-W187R is due to a failure to balance the activity of Ipl1. The other sds22 mutant alleles are viable in an IPL1 background, but as observed for some glc7 mutant alleles, the sds22ipl1-2 mutants grow better on formamide medium than the corresponding sds22IPL1 mutants (Figure 3C).
As observed for the glc7 suppressor mutants, the sds22 mutants have diverse phenotypes. They exhibit wide variation in sensitivity to rapamycin and 0.1 M CsCl. sds22-D2N D199N and sds22-E163I-L329P and sds22-77 (C-terminal frame shift) confer cold sensitivity (Figure 3D), with accumulation of large budded cells at 14°. sds22-F117S and sds22-77 also cause temperature-sensitive growth. However, unlike the glc7 suppressor mutations, all sds22 mutant alleles are completely recessive for ipl1-2 suppression (Figure S1B).
YPI1:
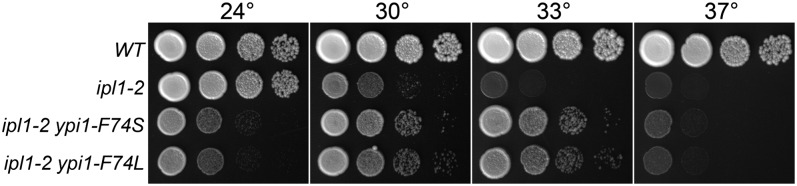
Two slow-growing ipl1 suppressors are tightly linked to an YPI1-13Myc::kan allele. Sequence analysis of the YPI1 gene in each of these mutants revealed a missense mutation at F74. Revertant 52 has an F74S missense mutation whereas revertant 95 has an F74L mutation. The residue corresponding to F74 in Ypi1/Inh3 orthologs in other eukaryotes is either a phenylalanine or tyrosine. Unlike the GLC7 mutants, ypi1-F74S and ypi1-F74L are completely recessive (Figure S1C). ypi1-F74S ipl1-2 and ypi1-F74L ipl1-2 mutants grow slowly at room temperature (Figure 4), and these alleles are lethal in a wild-type IPL1 background. We were unable to recover either ypi1 mutant in a wild-type IPL1 background from crosses between the two revertants and a wild-type strain. The spore clones corresponding to the ypi1IPL1 double mutants germinated and divided several times, but never generated viable colonies. All other genotypes from these crosses exhibited normal viability. The ability of ipl1-2 to rescue the inviability of ypi1-F74S and ypi1-F74L suggests that Ypi1, like Sds22, plays an essential function in regulating the Glc7 activity that opposes Ipl1.
Figure 4 .
Extragenic ipl1-2 suppressors in YPI1. Cultures of WT (KT1113), ipl1-2 (KT1829), ipl1-2 ypi1-F74S (KT3368), and ipl1-2 ypi1-F74L (KT3370) strains were serially diluted onto YPD medium and imaged after 40 hr at the designated temperatures.
SHP1:
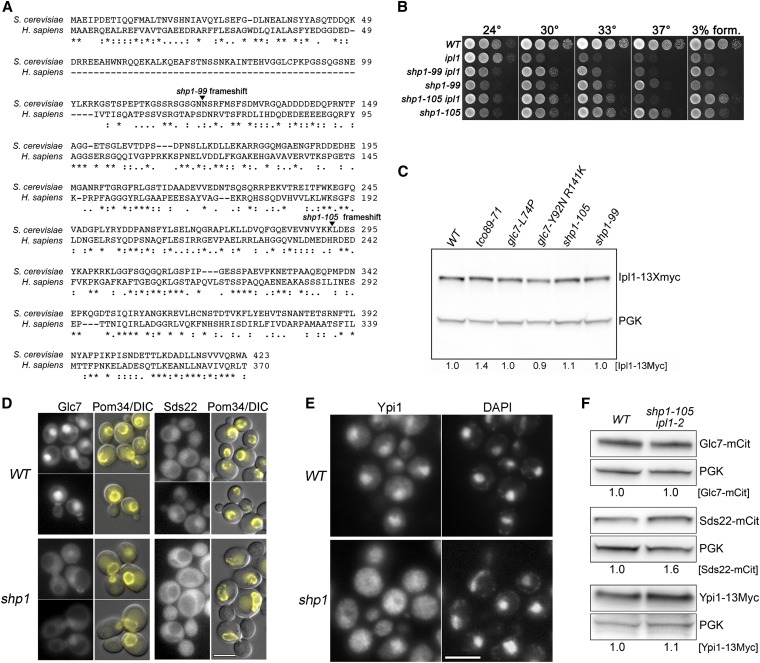
Revertants 99 and 105 define a complementation group separate from GLC7, SDS22, and YPI1. We assayed for genetic linkage between rev105 and SHP1 because SHP1 was initially identified as a mutant that suppresses the lethality caused by Glc7 overexpression (Zhang et al. 1995) and depletion of Shp1 has been shown to suppress the temperature sensitivity of ipl1-321 (Cheng and Chen 2010). Tetrad analysis between rev105 (KT3305) and GFP-CHS4::URA3 (KT2684) strains revealed tight linkage between rev105 and CHS4/SKT5 (23 PD, 0 TT, 0 NPD; ≤2CM). Because CHS4 is located 4 kb from SHP1, these linkage data strongly support the hypothesis that rev99 and rev105 are alleles of SHP1. Sequence analysis of the SHP1 locus from rev99 and rev105 revealed frameshift mutations after codons 122 and 291, respectively. The two mutants are completely recessive for ipl1-2 suppression (Figure S1D).
Shp1/p47 is a cofactor for Cdc48/p97 (also called VCP in humans), an AAA ATPase that acts as an ATP-dependent chaperone in widely divergent physiological processes. Cdc48 cofactors bind protein substrates that are usually ubiquitinated or have an ubiquitin-like structure and allow the hexameric Cdc48/p97 to structurally remodel the substrate [reviewed in (Jentsch and Rumpf 2007; Meyer et al. 2012)]. Shp1 contains an N-terminal 50 aa UBA-like domain that is thought to bind ubiquinated substrate and a C-terminal UBX domain that binds Cdc48. shp1-99 and shp1-105 both result in deletion of the UBX domain; thus, their products should not be able to associate with Cdc48. To determine whether shp1-99 and shp1-105 behave as null alleles, we constructed a heterozygous shp1Δ::kanMX mutant in a diploid strain and analyzed the meiotic progeny. Only G418-sensitive clones were recovered, indicating that SHP1 is an essential gene in our background, as reported for the W303 background (Cheng and Chen 2010). These results indicate that shp1-99 and shp1-105 products retain some activity, presumably Cdc48-independent.
As is the case for some alleles of GLC7, SDS22, and YPI1, the growth defects of shp1-99 and shp1-105 mutant cells are more severe in a wild-type than in an ipl1-2 background (Figure 5B). This is most striking on YPD medium at 30° or on media containing 3% formamide. Thus, as we have observed for some mutant alleles of GLC7, SDS22, and YPI1, a significant component of the shp1-99 and shp1-105 growth defects are associated with IPL1 function.
Figure 5 .
Extragenic ipl1-2 suppressors in SHP1. (A) Sequence alignment of human p47 and S. cerevisiae Shp1 proteins with the locations of ipl1 suppressor mutations indicated. (B) Cultures of WT (KT1113), ipl1-2 (KT1829), ipl1-2 shp1-99 (KT3413), shp1-99 (KT3412), ipl1-2 shp1-105 (KT3419), and shp1-105 (KT3416) strains were serially diluted onto YPD medium and imaged after 40 hr at the designated temperatures. (C) Immunoblot analysis of extracts from WT (KT3383), tco89-71 (KT3389), glc7-L74P (KT3391), glc7-Y92N R141K (KT3392), shp1-105 (KT105), and shp1-99 (KT3396) strains containing IPL1-13Myc. Ipl1-13Myc levels indicated under the lanes were calculated relative to the loading control (Pgk1) signal and normalized to the WT. (D) Fluorescence microscopy of Glc7-mCitrine and Sds22-mCitrine in WT (KT3242 and KT2856) and shp1-105 (KT3424 and KT3428) cells grown to log phase in YPD medium at 24°. The right panels of each set are overlays of Pom34-mCherry and DIC images to demarcate the nuclear periphery. Note that fluorescence images for the Glc7-mCitrine and Sds22-mCitrine are normalized independently. The actual fluorescence levels are much greater in the Glc7-mCitrine strains. Scale bar: 5 μm. (E) Indirect immunofluorescence of Ypi1-13Myc in WT (KT2881) and shp1-105 (KT3427) cells. Scale bar: 5 μm. (F) Immunoblot analysis of extracts from WT and shp1-105 ipl1-2 strains containing Glc7-mCitrine (KT3242 and KT3424), Sds22-mCitrine (KT2856 and KT3428), or Ypi1-13Myc (KT2881 and KT3427). Protein levels were calculated relative to the signal in the loading control (Pgk1) and normalized to the WT.
Cdc48/p97 has been directly implicated in acting on Aurora B in metazoans. It required for the extraction of Aurora B from mitotic chromatin in X. laevis egg extracts and in C. elegans embryos (Ramadan et al. 2007). In addition, RNAi treatment of two genes encoding Cdc48 (cdc-48.1 and cdc-48.2) alleviates the lethality of a temperature-sensitive Aurora B mutant [air-2(or207) (Ramadan et al. 2007)]. In contrast, Heallen et al. (Heallen et al. 2008) identified a Cdc48-related gene (cdc-48.3) in an RNAi feeding library screen for suppressors of air-2(or207). However, they found that RNAi of cdc-48.1 and cdc48.2, singly or in combination, did not suppress the lethality of air-2(or207). To determine whether our SHP1 mutants could be acting to increase Ipl1 levels, we assayed the steady-state levels of Ipl1 by immunoblot analysis in a collection of our ipl1-2 suppressor mutants. As shown in Figure 5C, Ipl1-13Myc levels are nearly identical between the wild-type strain and SHP1 mutants, arguing that the SHP1 suppressor mutants are not acting by increasing Ipl1 protein accumulation.
The levels of nuclear Glc7 are reduced in previously characterized SDS22, YPI1 and CDC48 mutants (Peggie et al. 2002; Pedelini et al. 2007; Bharucha et al. 2008; Cheng and Chen 2010) and in cells depleted of YPI and SHP1 (Pedelini et al. 2007; Bharucha et al. 2008; Cheng and Chen 2010). We also reported previously that nuclear Sds22 is decreased in cells depleted of Ypi1 (Bharucha et al. 2008). To determine whether loss of Shp1 has similar effects as loss of Ypi1, we imaged Glc7-mCitrine, Sds22-mCitrine, and Ypi1-13Myc in WT and shp1-105 mutants and found that nuclear levels of all three proteins are reduced in the shp1-105 mutant cells (Figure 5, D and E). Cytoplasmic levels of Sds22 are actually greater than nuclear levels in shp1 mutant cells (Figure 5D, lower right panels). These results cannot be explained by a decrease in total cellular levels of any of the three proteins. Immunoblot analysis of whole cell extracts (Figure 5F) revealed no obvious change in the total cellular levels of Glc7-mCitrine or Ypi1-13Myc in the shp1-105 mutant, and Sds22-mCitrine levels are reproducibly higher in shp1 mutant cells. Immunoblot analysis of two different shp1-105 strains revealed elevated levels of Sds22-mCitrine (1.6 ± 0.20- and 2.0 ± 0.25-fold, P < 0.003 according to the paired Student’s t-test).
SHP1, SDS22, and YPI1 mutants have remarkably similar effects on Glc7 localization. In all three cases, Glc7 is more uniformly distributed between the nucleus and cytoplasm. Nuclear Sds22 is also reduced in Ypi1 mutants (Bharucha et al. 2008), and we show here that shp1 mutations also affect Sds22 and Ypi1 localization. These effects on Glc7-Ypi1-Sds22 localization are likely through Cdc48 because cdc48-3 results in the delocalization of Glc7 (Cheng and Chen 2010), and our shp1 mutant products lack the Cdc48 binding domain. Cdc48 cofactors usually bind substrates that are ubiquitinated or have an ubiquitin-like structure [reviewed in (Jentsch and Rumpf 2007; Meyer et al. 2012)]. Glc7 is one possible target in our pathway because it is ubiquitinated in vivo (Peng et al. 2003; Starita et al. 2012). However, the immediate target of Cdc48-Shp1 need not be an ubiquitinated protein, since the ubiquitin-fold protein Atg8 is the substrate for Cdc48-Shp1 in autophagosome biogenesis (Krick et al. 2010). Together, these results suggest that the three proteins facilitate nuclear transport or retention of Glc7, a possibility supported by the observation that a screen for dosage suppressors of ipl1-321 uncovered cytoplasmic Glc7 binding proteins that reduce the nuclear accumulation of Glc7 when overexpressed (Pinsky et al. 2006).
Suppressors in TCO89, a component of TORC1
The ability of rev71 to suppress the temperature sensitivity of ipl1-2 at 33° does not segregate as a single Mendelian allele. Backcrosses between revertant 71 and an ipl1-2 strain revealed that, on average, only one of four spore clones grew at 33°, suggesting that two independent mutations are required for growth suppression at 33°. However, two spore clones in each tetrad were cold-sensitive and sensitive to 0.2 mM caffeine in the growth medium. These traits were invariably associated with growth of ipl1-2 mutants at 30°. We identified this cold and caffeine-sensitive suppressor as a null mutant in TCO89, which encodes a component of TORC1. A preliminary characterization of this mutant has been published (Tatchell et al. 2011).
We previously proposed that TORC1 acts as a positive regulator of the Glc7 activity that opposes Ipl1 (Tatchell et al. 2011). This was based on the observation that mutants with reduced TORC1 levels have lower levels of nuclear Glc7, which has correlated previously with suppression of ipl1 temperature sensitivity by SDS22, YPI1, and SHP1 mutants (Peggie et al. 2002; Pedelini et al. 2007; Bharucha et al. 2008; Cheng and Chen 2010). We also noted genetic interactions between mutants in the TORC1 pathway and GLC7 mutants that suppress ipl1-2. Among three alleles of GLC7 that were tested previously for suppression of ipl1-2 (Hsu et al. 2000), the two that acted as strong suppressors of ipl1-2 (glc7-127 and glc7-129) grow slowly in the presence of TORC1 inhibitors rapamycin or caffeine, whereas the glc7-109 mutation, which was previously shown to poorly suppress the temperature sensitivity of ipl1-2, does not confer strong sensitivity to rapamycin and caffeine (Tatchell et al. 2011). Our collection of 10 new GLC7 mutants that suppress the temperature sensitivity of ipl1-2 provided us with a more complete panel to test the relationship between Glc7 and TORC1. As shown in Figure 2C, there was significant variability in the sensitivity of the different GLC7 alleles to rapamycin. The sensitivity to rapamycin and caffeine conferred by GLC7 alleles correlates with negative genetic interactions with TORC1 mutants. For example, the rapamycin-resistant glc7-L74P mutant does not exhibit a strong genetic interaction with tco89Δ::kanMX, whereas the more rapamycin-sensitive glc7-L37F mutant is nearly lethal in combination with the tco89Δ::kanMX mutation (Figure S2). However, there is no strong correlation between the ability of a GLC7 allele to suppress ipl1-2 and the level of sensitivity to rapamycin conferred by that allele. For example, glc7-L74P is a strong suppressor of ipl1-2 (Figure 2B) but confers resistance to rapamycin (Figure 2C) and caffeine (data not shown). In contrast, many other GLC7 mutant alleles are less effective ipl1-2 suppressors but are more sensitive to rapamycin than glc7-L74P. The complexity of these genetic interactions suggests that TORC1 does not act simply as a positive regulator of total cellular Glc7 activity. Further characterization of these different GLC7 alleles may provide clues to the mechanism of TORC1 influence on nuclear Glc7 activity.
ipl1-2 suppressors in components of the kinetochore
DUO1, a component of the Dam1/DASH kinetochore complex:
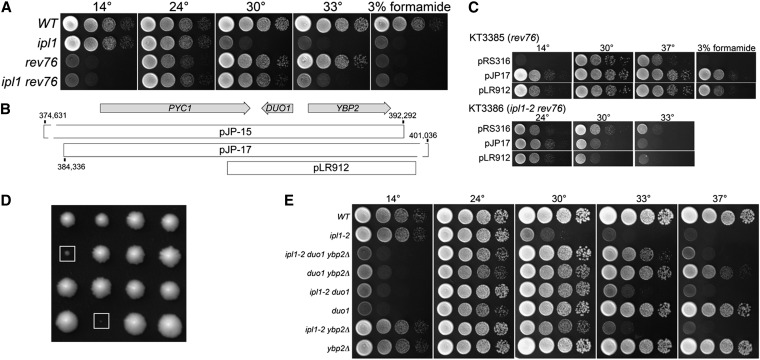
Revertants 76 and 81 are cold-sensitive, arresting as large budded cells at 14°, and are sensitive to 3% formamide at all temperatures (Figure 6A). The suppressor mutations in these two mutants are unlinked to GLC7, YPI1, SHP1, and SDS22. The cold and formamide sensitivities of revertants 76 and 81 failed to complement. To identify this locus, we transformed each mutant with a low-copy yeast genomic library and selected for growth in medium containing 3% formamide. The identities of the genomic inserts in plasmids conferring formamide resistance were determined by sequence analysis.
Figure 6 .
Extragenic ipl1-2 suppressors in DUO1. (A) Cultures of WT (KT1113), ipl-2 (KT1829), rev76 (KT3385), and ipl1-2 rev76 (KT3386) strains were serially diluted onto YPD medium and imaged after 40 hr at the indicated temperatures. (B) Genetic map of the PYC1-DUO1-YBP2 region of chromosome VII showing the locations of genomic DNA fragments that complement the rev76 suppressor phenotype. (C) Cultures of duo1-S115F (KT3385) and ipl1-2 duo1-S115F (KT3386) mutant strains transformed with the designated plasmids were serially diluted onto the following media: Top panels: YPD at the designated temperatures and YPD + 3% formamide at 30°. Bottom panels: Synthetic medium lacking uracil at the designated temperatures. (D) Images of tetrads from a cross between duo1-S115F and mad1::HIS3 strains (KT3386 X KT1688). The boxes identify the duo1-S115F mad1::HIS3 double mutants. These were the largest double mutant colonies observed; the majority of double mutants failed to grow into macroscopic colonies. Each column represents the four spore clones of a tetrad. (E) Cultures of WT (KT1113), ipl-2 (KT1829), ipl1-2 duo1-S115F ybp2Δ::kanMX6 (KT3409), duo1-S115F ybp2Δ::kanMX6 (KT3410), ipl1-2 duo1-S115F (KT3386), duo1-S115F (KT3385), ipl1-2 ybp2Δ::kanMX6 (KT3401), and ybp2Δ::kanMX6 (KT3403) strains were serially diluted onto YPD medium and imaged after 40 hr at the indicated temperatures with the exception of the plate at 14°, which was imaged after 7 d.
The genomic inserts in two clones that complement both cold and formamide sensitivities of revertants 76 and 81 contain overlapping regions of chromosome VII. The region of overlap contains PYC1, encoding pyruvate carboxylase, DUO1, encoding a component of the DAM/DASH kinetochore complex, and YBP2, a component of the kinetochore that is required for normal mitotic progression [Figure 6B (Ohkuni et al. 2008)]. To confirm that the rev76 and rev81 loci are genetically linked to this region of chromosome VII, we determined the map distance between rev76 and rev81 and two neighboring genes (pyc1Δ::kanMX and pkp2Δ::kanMX). In neither case was recombination observed between formamide sensitivity and the G418 resistance associated with pyc1Δ::kanMX and pkp2Δ::kanMX (≤ 2 map units for both crosses). Plasmid pLR912, containing DUO1 and YBP2 in yeast shuttle vector pRS316, fully complements the cold and formamide sensitivities as well as the ipl1-2 suppression phenotype of rev76. Sequence analysis of DUO1 from rev76 and rev81 revealed two base substitutions, C344T and C607T, that result in missense mutations S115F and P203S. C607T also was found in our wild-type strain. S115 is highly conserved in other sensu stricto yeasts and in Duo1 from S. pombe, but P203 is not conserved. In fact, other Duo1 proteins from sensu stricto yeasts contain an S or T at residue 203. The sequences of YBP2 obtained from rev76, rev81, and our wild-type strain are identical, but each differs from the S288C wild-type strain at six nucleotides, resulting in three missense mutations (H136Q, K140N, and I286T) and three silent mutations. These polymorphisms were previously reported to occur in a different genetic background (Ohkuni et al. 2008). H136, K140, and I286 are not conserved among sensu stricto yeasts, and Ohkuni et al. (Ohkuni et al. 2008) state that no functional significance of any was observed. Thus, the ipl1 suppressor mutation in rev76 and rev81 is most likely C344T in DUO1, causing the S115F change. Because rev76 and rev81 were isolated from the same round of mutagenesis and have identical phenotypes and identical DNA sequences at DUO1, the two revertants are likely from the same mutagenic event. Hereafter, we refer to the rev76 and rev81 ipl1 suppressors as duo1-S115F. To confirm that duo1-S115F is responsible for ipl1-2 suppression, we mapped the ipl1-2 suppressor in rev76 to the DUO1 region of chromosome VII as described in the Materials and Methods. We observed no recombination between the ipl1-2 suppression in rev76 and ybp2Δ::kan in 33 tetrads (<1.5 map units). We also replaced the wt DUO1 gene with the duo1-S115F allele from rev76 (Materials and Methods). All spore clones containing ipl1-2 and duo1-S115F grow at 30°, whereas ipl1-2 DUO1 clones cannot (Figure S3A).
duo1-S115F mutant cells arrest as large budded cells at 14°. A likely explanation for this arrest is that the duo1 mutation activates the SAC, delaying cells at the metaphase-anaphase transition. To test this possibility, we crossed a duo1-S115F mutant to the SAC mutant mad1::HIS3 and analyzed the progeny. Due to genetic linkage between MAD1 and DUO1, we identified only 27 tetrads predicted to contain the mad1::HIS3duo1-S115F double mutant from a total of 120 tetrads (map distance 11.2 map units). Only four of these predicted double mutants grew into small but visible colonies (Figure 6D). Sixteen formed microcolonies containing between 2 and ∼100 cells and seven failed to germinate or divide. These results are consistent with duo1-S115F activating the SAC.
Duo1 is 1 of 10 protein components of the microtubule binding Dam1 or DASH complex. The complex forms rings around microtubules in vitro (Miranda et al. 2005; Westermann et al. 2005) and can track the plus ends of microtubules (Asbury et al. 2006; Westermann et al. 2006; Gestaut et al. 2008), although the ring structure is not necessarily required for end tracking (Gestaut et al. 2008). Dam1/DASH also increases the processivity of the NDC80 complex (Lampert et al. 2010; Tien et al. 2010). The Duo1 and Dam1 subunits are thought to form the primary microtubule-binding domain of the complex (Miranda et al. 2007). Dam1 is phosphorylated by Ipl1/Aurora B, which decreases the affinity of the Dam1 complex for microtubules (Gestaut et al. 2008), but Duo1 is not known to be a substrate for Ipl1. Two previously characterized temperature-sensitive DUO1 mutants (duo1-1 and duo1-2) arrest with short spindles at the nonpermissive temperature due to activation of the SAC (Hofmann et al. 1998) and one of these (duo1-2) contains two missense mutations (A117T M124I) close to the mutation in duo1-S115F. Rather than suppress the temperature sensitivity of ipl1 mutations, however, the duo1-2 mutation was observed to be lethal in combination with ipl1-1 and showed no genetic interaction with ipl1-2 (Kang et al. 2001).
How is Duo1-S115F altering the function of the Dam1/DASH complex? Although Duo1 phosphorylation has not been reported, it is possible that S115 is phosphorylated, or that the mutation increases the phosphorylation state of other Dam1/DASH subunits. Alternatively, the altered Duo1 product may interact differently with the complex to affect the regulation of microtubule-binding.
ybp2Δ suppresses the temperature sensitivity of ipl1-2:
YBP2 lies adjacent to DUO1 and is transcribed from the opposite strand. YBP2 was originally identified because of its sequence similarity to YBP1, which is involved in the oxidative stress response (Gulshan et al. 2004). More recently, YBP2 was identified based on its negative genetic interaction with MAD2 (Ohkuni et al. 2008). Strains deleted for YBP2 show increased chromosome loss, and, in immune complexes, Ybp2 was reported to associate with Ndc80, MIND and COMA kinetochore components, as well as the centromere-specific histone Cse4 (Ohkuni et al. 2008). Paradoxically, the association of several kinetochore components with one another and with the centromere appears to be enhanced in a ybp2 null mutant (Ohkuni et al. 2008). In the course of our analysis of DUO1 and YBP2, we found that a ypb2Δ::kanMX mutation suppresses the temperature sensitivity of ipl1-2 at 30° (Figure 6E, row 7). Furthermore, the ybp2Δ::kanMX mutation enhances the ability of a duo1-S115F mutant to suppress ipl1-2 (Figure 6E, row 3) and ybp2Δ::kanMX duo1-S115F double mutants grow slowly at 37° (Figure 6E, row 4). We also observed genetic interactions between ybp2Δ::kanMX and mad1::HIS3. Analysis of a cross between ybp2Δ::kanMX and mad1::HIS3 strains (KT3400 × KT1687) revealed that all ybp2Δ::kanMX mad1::HIS3 double mutant spore clones were small or microcolonies (Figure S4). Spore clones of all other genotypes grew normally. Thus, as previously reported for another genetic background, loss of Ybp2 in our genetic background is not lethal, but results in SAC activation. Although Ybp2 apparently has a positive role in kinetochore function, the enhancement of interactions among NDC80, MIND, and COMA complex components in the absence of Ybp2 is puzzling. The suppression of ipl1-2 by loss of YBP2 is consistent with a positive role in kinetochore function, and suggests that whatever enhancement of complex interactions occurs in the deletion mutant, the net effect may be destabilization of kinetochore-microtubule interaction. This would be consistent with the chromosome loss phenotype of the deletion mutant.
Revertant 8 is a missense mutation in NDC80:
Revertant 8 (rev8) is hypersensitive to formamide and arrests with large buds at low temperature but is complemented by duo1-S155F. We isolated four clones from two genomic libraries that complement the formamide and cold sensitivity of rev8. The four clones contain overlapping regions of chromosome IX. The only full-length gene in common among the four clones is the evolutionarily conserved NDC80/TID3 gene. To confirm that the mutation in rev8 responsible for ipl1-2 suppression is genetically linked to the NDC80 locus, we determined the map distance between the ipl1-2 suppressor in rev8 and NDC80 tagged with 3XHA::KanMX and NDC80-3XFlag::KanMX. No recombination was observed between rev8 and the tagged NDC80 alleles in 56 tetrads (≤0.9 map units), indicating that rev8 is likely an NDC80 mutant. Ndc80/HEC1 is one of four subunits in the Ndc80 complex, the major microtubule binding and end tracking component of the kinetochore. The N-terminal portions of Ndc80/HEC1 and Nuf1 subunits bind microtubules, while their C-terminal coiled-coil domains associate with coiled-coil domains of the Spc24 and Spc25 subunits, which tether the complex to the inner kinetochore [reviewed by (DeLuca and Musacchio 2012)].
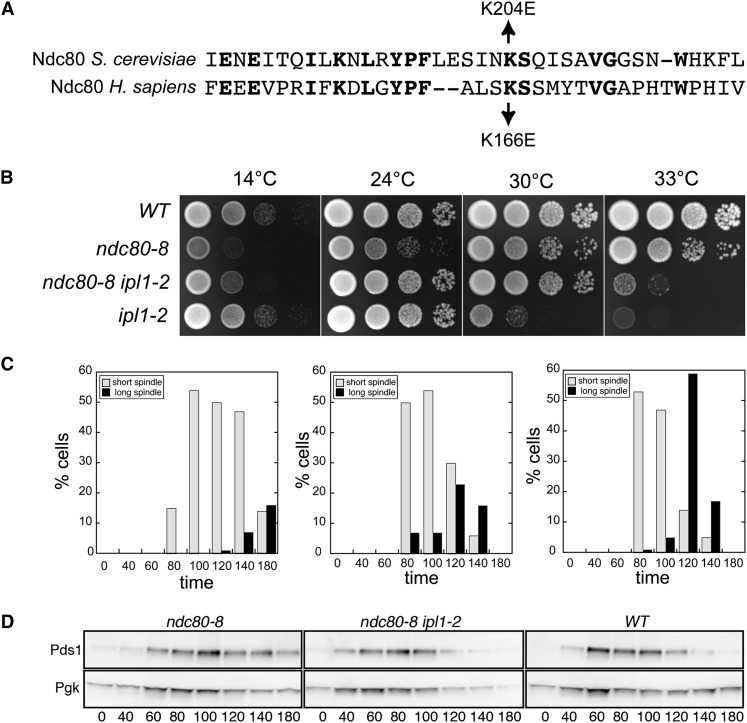
Sequence analysis of NDC80 from a rev8 strain revealed a single nucleotide substitution in NDC80, resulting in missense mutation K204E. K204 corresponds to K166 in human Ndc80/Hec1 (Figure 7A) and K89 in human EB1, which lies in the microtubule-binding calponin-homology (CH) domain. The K166E mutation in human Ndc80 and K89E in EB1 strongly impairs or eliminates in vitro microtubule binding, respectively (Hayashi and Ikura 2003; Ciferri et al. 2008), indicating that this conserved lysine residue in the N-terminal domain of Ndc80 is required for efficient microtubule binding. In vivo, Ndc80/Hec1 mutant cells containing K166D and K166E substitutions fail to accumulate in metaphase and have weak kinetochore-microtubule attachments (Sundin et al. 2011; Tooley et al. 2011). To confirm that the ipl1-2 suppressor in rev8 is ndc80-K204E, we replaced the wt NDC80 gene with a cloned version of NDC80 from rev8 see Materials and Methods). All ipl1-2 spore clones containing ndc80-K204E grew at 30° whereas those containing wt NDC80 did not (Figure S3B).
Figure 7 .
Extragenic ipl1-2 suppressors in NDC80. (A) Sequence alignment of yeast and human Ndc80 proteins. (B) Cultures of WT (KT1113), ndc80-K204E (ndc80-8) (KT3255), ndc80-K204E ipl1-2 (KT3257), and ipl1-2 (KT1963) strains were serially diluted, plated onto indicated media, and imaged after 44 hr. The first four panels show YPD medium incubated at the designated temperatures. (C) and (D) Cultures of ndc80-K204E(KT3317), ndc80-K204E ipl1-2 (KT3320), and WT (KT3319) cells were arrested with alpha factor, released into YPD medium at 24°, and monitored for spindle length and entrance into anaphase using Spc42-3XGFP and Pds1-13Myc fusions, respectively, at the designated time points. (C) Quantitation of the percentage of cells with short (≤2.0 μm) or long (>2.0 μm) spindles. At least 64 mitotic spindles were measured at each time point for each strain. (D) Immunoblot analysis of Pds1-13XMyc in whole cell extracts. Pgk serves as the loading control.
ndc80-K204E mutant cells arrest with large buds and short mitotic spindles at 14° (data not shown). Interestingly, the slow growth phenotype of ndc80-K204E at 14° and 24° is suppressed by ipl1-2 (Figure 7B). In a reciprocal manner, the temperature sensitivity of ipl1-2 is suppressed by ndc80-K204E at 30° and partially at 33° (Figure 7B). To characterize the slow growth phenotype of ndc80-K204E cells in more detail, we arrested ndc80-K204E, ipl1-2 ndc80-K204E, and WT strains in G1 with alpha mating pheromone, and monitored spindle formation and anaphase progression after cell cycle release in the absence of mating pheromone. Mating pheromone was added to the cultures after 60 min to prevent cells from re-entering the cell cycle. The strains contained SPC42-3XGFP and PDS1-13XMyc to facilitate monitoring spindle pole distances and entrance into anaphase, respectively. Wild-type cells duplicated their spindle pole bodies and formed spindles after 60 min. A majority of the cells were in anaphase by 120 min (Figure 7C, right panel), as shown by both spindle length and decreased Securin/Pds1 levels (Figure 7D, right panel). At 180 min, all wild-type cells had exited mitosis, as shown by unduplicated spindle pole bodies and the absence of Pds1. Although ndc80-K204E mutant cells started forming spindles after 60 min, less than 20% of the cells had entered anaphase at 180 min and substantial Pds1 remained in the cells by 180 min, consistent with an extended G2/M cell cycle delay (Figure 7, C and D, left panels). The delay is largely eliminated in ndc80-K204E ipl1-2 mutant cells (Figure 7, C and D, middle panels); all cells returned to G1 by the 180-minute time point.
Given the observation that the human Ndc80-K166E variant protein binds weakly to microtubules in vitro, it is likely that the cell cycle delay in ndc80-K204E cells is due to SAC activation. Consistent with this possibility, although we recovered slow growing ndc80-K204E mad1::HIS3ipl1-2 triple mutants, we failed to recover viable ndc80-K204E mad1::HIS3 progeny from a cross between ndc80-K204E ipl1-2 and mad1::HIS3 strains. These results strengthen the evidence that loss of Ipl1 activity can compensate for the loss of kinetochore-microtubule binding.
The identification of the ndc80-K201E mutant as an ipl1-2 suppressor represents a remarkable convergence of genetics, cell biology, biochemistry, and structural analysis. Previous studies have shown that basic residues in the unstructured N-terminus and in the CH domain of Ndc80 bind microtubules via electrostatic interactions between the Ndc80 complex and microtubules. Phosphorylation of the N-terminus of Ndc80 by AuroraB/Ipl1 is thought to reduce the negative charge of the microtubule binding domain and thus increase the microtubule/kinetochore dynamics necessary for the dissolution of improper kinetochore-microtubule interactions. Consistent with this model, the Ndc80-7A mutant, in which Aurora B phosphorylation sites in the N terminus of Ndc80 are mutated to alanine, is inviable under conditions of reduced Ipl1 activity [ipl1-321 background (Akiyoshi et al. 2009a)]. Our results provide direct biological support for the hypothesis that the phosphorylation of Ndc80 by Ipl1/Aurora B reduces the affinity of the Ndc80 complex for kinetochore microtubules, thereby increasing the kinetochore/microtubule dynamics required to effect bipolar chromosome attachment on the spindle.
In addition to three intragenic suppressors of ipl1-2, our classical genetic screen identified new alleles in previously identified components of the Glc7/PP1 pathway (GLC7, YPI1, SDS22, and SHP1), a component of the TORC1 complex (TCO89) and two mutants in components of the outer kinetochore (DUO1 and NDC80). Given the considerable attention IPL1 has received, and the plethora of genome-based screening strategies, we think that it is worth comparing our genetic screen with others, both directed and undirected. First, we did not recover mutants in GLC8, for which null alleles are known to suppress temperature sensitive IPL1 mutants (Tung et al. 1995). It is likely that the 33° temperature we set for the screen was too high to recover GLC8 mutants since glc8 null mutants in our background only suppress the ipl1-2 mutant allele weakly at 33° (Tatchell et al. 2011). It is possible that 33° was also too stringent to recover mutants in components of the Set1 protein methylation complex that methylates Dam1, which were previously shown to suppress ipl1-2 (Zhang et al. 2005; Latham et al. 2011). We note that tco89-71 also suppresses weakly at 33° but in this case, the original suppressor strain contained a second mutation that enhanced the suppression at 33°. In the future, it might be possible to sensitize the screen by using a genetic background that contains a weak suppressor.
With the exception of TCO89, all of the suppressor mutations identified in our screen are in evolutionarily conserved, essential genes, underscoring the continued value of traditional genetic screens. A high throughput screen of the deletion collection would have missed key components of the pathway. The screen is clearly not saturated, since only one allele each of DUO1 and NDC80 was identified, and we did not isolate any mutations in YBP2, which we found serendipitously to be an ipl1 suppressor locus. Given the potential importance of these novel alleles, it could be informative to repeat the screen with a starting strain in which GLC7 and SDS22 were duplicated, in order to reduce or eliminate the frequency of mutants in the Glc7 pathway.
Supplementary Material
Acknowledgments
We thank Nathan Davis for the affinity purified anti-GFP antibody and Eric First for other reagents. We also thank an anonymous reviewer for helpful comments. The work was supported in part by National Science Foundation grant MCB-0517204 (to L.C.R.) and grant-in-aid from Louisiana State University Health Sciences Center-Shreveport (to K.T.).
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Akiyoshi B., Nelson C. R., Ranish J. A., Biggins S., 2009a Analysis of Ipl1-mediated phosphorylation of the Ndc80 kinetochore protein in Saccharomyces cerevisiae. Genetics 183: 1591–1595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyoshi B., Nelson C. R., Ranish J. A., Biggins S., 2009b Quantitative proteomic analysis of purified yeast kinetochores identifies a PP1 regulatory subunit. Genes Dev. 23: 2887–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Andrews P. D., Stark M. J., 2000. Dynamic, Rho1p-dependent localization of Pkc1p to sites of polarized growth. J. Cell Sci. 113: 2685–2693 [DOI] [PubMed] [Google Scholar]
- Andrews P. D., Ovechkina Y., Morrice N., Wagenbach M., Duncan K., et al. , 2004. Aurora B regulates MCAK at the mitotic centromere. Dev. Cell 6: 253–268 [DOI] [PubMed] [Google Scholar]
- Asbury C. L., Gestaut D. R., Powers A. F., Franck A. D., Davis T. N., 2006. The Dam1 kinetochore complex harnesses microtubule dynamics to produce force and movement. Proc. Natl. Acad. Sci. USA 103: 9873–9878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axton J. M., Dombradi V., Cohen P. T. W., Glover D. M., 1990. One of the protein phosphatase 1 isoenzymes in Drosophila is essential for mitosis. Cell 63: 33–46 [DOI] [PubMed] [Google Scholar]
- Baker S. H., Frederick D. L., Bloecher A., Tatchell K., 1997. Alanine scanning mutagenesis of protein phosphatase type 1 in the yeast Saccharomyces cerevisiae. Genetics 145: 615–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharucha J. P., Larson J. R., Gao L., Daves L. K., Tatchell K., 2008. Ypi1, a positive regulator of nuclear protein phosphatase type 1 activity in Saccharomyces cerevisiae. Mol. Biol. Cell 19: 1032–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloecher A., Tatchell K., 1999. Defects in Saccharomyces cerevisiae protein phosphatase type I activate the spindle/kinetochore checkpoint. Genes Dev. 13: 517–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollen M., Peti W., Ragusa M. J., Beullens M., 2010. The extended PP1 toolkit: designed to create specificity. Trends Biochem. Sci. 35: 450–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceulemans H., Vulsteke V., De Maeyer M., Tatchell K., Stalmans W., et al. , 2002. Binding of the concave surface of the Sds22 superhelix to the alpha 4/alpha 5/alpha 6-triangle of protein phosphatase-1. J. Biol. Chem. 277: 47331–47337 [DOI] [PubMed] [Google Scholar]
- Chan C. S., Botstein D., 1993. Isolation and characterization of chromosome-gain and increase-in- ploidy mutants in yeast. Genetics 135: 677–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y. L., Chen R. H., 2010. The AAA-ATPase Cdc48 and cofactor Shp1 promote chromosome bi-orientation by balancing Aurora B activity. J. Cell Sci. 123: 2025–2034 [DOI] [PubMed] [Google Scholar]
- Ciferri C., Pasqualato S., Screpanti E., Varetti G., Santaguida S., et al. , 2008. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell 133: 427–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis N. G., Horecka J. L., Sprague G. F., Jr, 1993. Cis- and trans-acting functions required for endocytosis of the yeast pheromone receptors. J. Cell Biol. 122: 53–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLuca J. G., Musacchio A., 2012. Structural organization of the kinetochore-microtubule interface. Curr. Opin. Cell Biol. 24: 48–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doonan J. H., Morris N. R., 1989. The bimG gene of Aspergillus nidulans, required for completion of anaphase, encodes a homolog of mammalian phosphoprotein phosphatase 1. Cell 57: 987–996 [DOI] [PubMed] [Google Scholar]
- Egloff M. P., Johnson D. F., Moorhead G., Cohen P. T., Cohen P., et al. , 1997. Structural basis for the recognition of regulatory subunits by the catalytic subunit of protein phosphatase 1. EMBO J. 16: 1876–1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuele M. J., Lan W., Jwa M., Miller S. A., Chan C. S., et al. , 2008. Aurora B kinase and protein phosphatase 1 have opposing roles in modulating kinetochore assembly. J. Cell Biol. 181: 241–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez A., Brautigan D. L., Lamb N. J. C., 1992. Protein phosphatase type 1 in mammalian cell mitosis: chromosome localization and involvement in mitotic exit. J. Cell Biol. 116: 1421–1430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francisco L., Wang W., Chan C. S., 1994. Type 1 protein phosphatase acts in opposition to IpL1 protein kinase in regulating yeast chromosome segregation. Mol. Cell. Biol. 14: 4731–4740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick D. L., Tatchell K., 1996. The REG2 gene of Saccharomyces cerevisiae encodes a type1 protein phosphatase-binding protein that functions with Reg1p and the Snf1p protein kinase to regulate growth. Mol. Cell. Biol. 16: 2922–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritsch E. F., Sambrook J., Maniatis T., 1989. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York [Google Scholar]
- Garcia-Gimeno M. A., Munoz I., Arino J., Sanz P., 2003. Molecular characterization of Ypi1, a novel Saccharomyces cerevisiae type 1 protein phosphatase inhibitor. J. Biol. Chem. 278: 47744–47752 [DOI] [PubMed] [Google Scholar]
- Gestaut D. R., Graczyk B., Cooper J., Widlund P. O., Zelter A., et al. , 2008. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat. Cell Biol. 10: 407–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gietz D., St. Jean A., Woods R. A., Schiestl R. H., 1992. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20: 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg J., Huang H. B., Kwon Y. G., Greengard P., Nairn A. C., et al. , 1995. Three-dimensional structure of the catalytic subunit of protein serine/threonine phosphatase-1. Nature 376: 745–753 [DOI] [PubMed] [Google Scholar]
- Grusche F. A., Hidalgo C., Fletcher G., Sung H. H., Sahai E., et al. , 2009. Sds22, a PP1 phosphatase regulatory subunit, regulates epithelial cell polarity and shape [Sds22 in epithelial morphology]. BMC Dev. Biol. 9: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulshan K., Rovinsky S. A., Moye-Rowley W. S., 2004. YBP1 and its homologue YBP2/YBH1 influence oxidative-stress tolerance by nonidentical mechanisms in Saccharomyces cerevisiae. Eukaryot. Cell 3: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanks S. K., Hunter T., 1995. Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9: 576–596 [PubMed] [Google Scholar]
- Hayashi I., Ikura M., 2003. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1). J. Biol. Chem. 278: 36430–36434 [DOI] [PubMed] [Google Scholar]
- Hazbun T. R., Malmstrom L., Anderson S., Graczyk B. J., Fox B., et al. , 2003. Assigning function to yeast proteins by integration of technologies. Mol. Cell 12: 1353–1365 [DOI] [PubMed] [Google Scholar]
- Heallen T. R., Adams H. P., Furuta T., Verbrugghe K. J., Schumacher J. M., 2008. An Afg2/Spaf-related Cdc48-like AAA ATPase regulates the stability and activity of the C. elegans Aurora B kinase AIR-2. Dev. Cell 15: 603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickx A., Beullens M., Ceulemans H., Den Abt T., Van Eynde A., et al. , 2009. Docking motif-guided mapping of the interactome of protein phosphatase-1. Chem. Biol. 16: 365–371 [DOI] [PubMed] [Google Scholar]
- Hisamoto N., Sugimoto K., Matsumoto K., 1994. The Glc7 type 1 protein phosphatase of Saccharomyces cerevisiae is required for cell cycle progression in G2/M. Mol. Cell. Biol. 14: 3158–3165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hisamoto N., Frederick D. L., Sugimoto K., Tatchell K., Matsumoto K., 1995. The EGP1 gene may be a positive regulator of protein phosphatase type 1 in the growth control of Saccharomyces cerevisiae. Mol. Cell. Biol. 15: 3767–3776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann C., Cheeseman I. M., Goode B. L., McDonald K. L., Barnes G., et al. , 1998. Saccharomyces cerevisiae Duo1p and Dam1p, novel proteins involved in mitotic spindle function. J. Cell Biol. 143: 1029–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J.-Y., Sun Z.-W., Li X., Ruben M., Tatchell K., et al. , 2000. Mitotic phosphorylation of histone H3 is goverened by Ipl1/aurora kinase and Glc7p/PP1 phosphatase in budding yeast and nematodes. Cell 102: 279–291 [DOI] [PubMed] [Google Scholar]
- Ishii K., Kumada K., Toda T., Yanagida M., 1996. Requirement for PP1 phosphatase and 20S cyclosome/APC for the onset of anaphase is lessened by the dosage increase of a novel gene sds23+. EMBO J. 15: 6629–6640 [PMC free article] [PubMed] [Google Scholar]
- Jentsch S., Rumpf S., 2007. Cdc48 (p97): a “molecular gearbox” in the ubiquitin pathway? Trends Biochem. Sci. 32: 6–11 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Scott K. L., Kwak S. J., Chen R., Mardon G., 2011. Sds22/PP1 links epithelial integrity and tumor suppression via regulation of myosin II and JNK signaling. Oncogene 30: 3248–3260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Cheeseman I. M., Kallstrom G., Velmurugan S., Barnes G., et al. , 2001. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J. Cell Biol. 155: 763–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J. H., Kang J. S., Chan C. S., 1999. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J. Cell Biol. 145: 1381–1394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwaliwale C. V., Frei S. B., Stern B. M., Biggins S., 2007. A pathway containing the Ipl1/aurora protein kinase and the spindle midzone protein Ase1 regulates yeast spindle assembly. Dev. Cell 13: 433–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krick R., Bremer S., Welter E., Schlotterhose P., Muehe Y., et al. , 2010. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J. Cell Biol. 190: 965–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunda P., Rodrigues N. T., Moeendarbary E., Liu T., Ivetic A., et al. , 2012. PP1-mediated moesin dephosphorylation couples polar relaxation to mitotic exit. Curr. Biol. 22: 231–236 [DOI] [PubMed] [Google Scholar]
- Lampert F., Hornung P., Westermann S., 2010. The Dam1 complex confers microtubule plus end-tracking activity to the Ndc80 kinetochore complex. J. Cell Biol. 189: 641–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lampson M. A., Cheeseman I. M., 2011. Sensing centromere tension: Aurora B and the regulation of kinetochore function. Trends Cell Biol. 21: 133–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham J. A., Chosed R. J., Wang S., Dent S. Y., 2011. Chromatin signaling to kinetochores: transregulation of Dam1 methylation by histone H2B ubiquitination. Cell 146: 709–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage B., Beullens M., Pedelini L., Garcia-Gimeno M. A., Waelkens E., et al. , 2007. A complex of catalytically inactive protein phosphatase-1 sandwiched between Sds22 and inhibitor-3. Biochemistry 46: 8909–8919 [DOI] [PubMed] [Google Scholar]
- Liu D., Vader G., Vromans M. J., Lampson M. A., Lens S. M., 2009. Sensing chromosome bi-orientation by spatial separation of aurora B kinase from kinetochore substrates. Science 323: 1350–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Vleugel M., Backer C. B., Hori T., Fukagawa T., et al. , 2010. Regulated targeting of protein phosphatase 1 to the outer kinetochore by KNL1 opposes Aurora B kinase. J. Cell Biol. 188: 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine M. S., McKenzie A., 3rd, Demarini D. J., Shah N. G., Wach A., et al. , 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961 [DOI] [PubMed] [Google Scholar]
- MacKelvie S. H., Andrews P. D., Stark M. J. R., 1995. The Saccharomyces cerevisiae gene SDS22 encodes a potential regulator of the mitotic function of yeast type 1 protein phosphatase. Mol. Cell. Biol. 15: 3777–3785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maresca T. J., Salmon E. D., 2009. Intrakinetochore stretch is associated with changes in kinetochore phosphorylation and spindle assembly checkpoint activity. J. Cell Biol. 184: 373–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquina M., Gonzalez A., Barreto L., Gelis S., Munoz I., et al. , 2012. Modulation of yeast alkaline cation tolerance by ypi1 requires calcineurin. Genetics 190: 1355–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meadows J. C., Shepperd L. A., Vanoosthuyse V., Lancaster T. C., Sochaj A. M., et al. , 2011. Spindle checkpoint silencing requires association of PP1 to both Spc7 and kinesin-8 motors. Dev. Cell 20: 739–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer H., Bug M., Bremer S., 2012. Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14: 117–123 [DOI] [PubMed] [Google Scholar]
- Miranda J. J., De Wulf P., Sorger P. K., Harrison S. C., 2005. The yeast DASH complex forms closed rings on microtubules. Nat. Struct. Mol. Biol. 12: 138–143 [DOI] [PubMed] [Google Scholar]
- Miranda J. J., King D. S., Harrison S. C., 2007. Protein arms in the kinetochore-microtubule interface of the yeast DASH complex. Mol. Biol. Cell 18: 2503–2510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicklas R. B., Koch C. A., 1969. Chromosome micromanipulation. 3. Spindle fiber tension and the reorientation of mal-oriented chromosomes. J. Cell Biol. 43: 40–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuni K., Abdulle R., Tong A. H., Boone C., Kitagawa K., 2008. Ybp2 associates with the central kinetochore of Saccharomyces cerevisiae and mediates proper mitotic progression. PLoS One 3: e1617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkura H., Yanagida M., 1991. S. pombe gene sds22+ essential for a midmitotic transition encodes a leucine-rich repeat protein that positively modulates protein phosphatase-1. Cell 64: 149–157 [DOI] [PubMed] [Google Scholar]
- Pedelini L., Marquina M., Arino J., Casamayor A., Sanz L., et al. , 2007. YPI1 and SDS22 proteins regulate the nuclear localization and function of yeast type 1 phosphatase Glc7. J. Biol. Chem. 282: 3282–3292 [DOI] [PubMed] [Google Scholar]
- Peggie M. W., MacKelvie S. H., Bloecher A., Knatko E. V., Tatchell K., et al. , 2002. Essential functions of Sds22p in chromosome stability and nuclear localization of PP1. J. Cell Sci. 115: 195–206 [DOI] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J. E., Thoreen C. C., Cheng D., et al. , 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21: 921–926 [DOI] [PubMed] [Google Scholar]
- Pinsky B. A., Kotwaliwale C. V., Tatsutani S. Y., Breed C. A., Biggins S., 2006. Glc7/protein phosphatase 1 regulatory subunits can oppose the Ipl1/aurora protein kinase by redistributing Glc7. Mol. Cell. Biol. 26: 2648–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinsky B. A., Nelson C. R., Biggins S., 2009. Protein phosphatase 1 regulates exit from the spindle checkpoint in budding yeast. Curr. Biol. 19: 1182–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posch M., Khoudoli G. A., Swift S., King E. M., Deluca J. G., et al. , 2010. Sds22 regulates aurora B activity and microtubule-kinetochore interactions at mitosis. J. Cell Biol. 191: 61–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadan K., Bruderer R., Spiga F. M., Popp O., Baur T., et al. , 2007. Cdc48/p97 promotes reformation of the nucleus by extracting the kinase Aurora B from chromatin. Nature 450: 1258–1262 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Winston F., Hieter P., 1990. Methods in Yeast Genetics. A Laboratory Course Manual. Cold Spring Harbor Laboratory Press, Plainview, NY [Google Scholar]
- Rosenberg J. S., Cross F. R., Funabiki H., 2011. KNL1/Spc105 recruits PP1 to silence the spindle assembly checkpoint. Curr. Biol. 21: 942–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchaud S., Carmena M., Earnshaw W. C., 2007. Chromosomal passengers: conducting cell division. Nat. Rev. Mol. Cell Biol. 8: 798–812 [DOI] [PubMed] [Google Scholar]
- Santaguida S., Musacchio A., 2009. The life and miracles of kinetochores. EMBO J. 28: 2511–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sassoon I., Severin F. F., Andrews P. D., Taba M.-R., Kaplan K. B., et al. , 1999. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 13: 545–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa F., Mapelli M., Ciferri C., Tarricone C., Areces L. B., et al. , 2005. Mechanism of Aurora B activation by INCENP and inhibition by hesperadin. Mol. Cell 18: 379–391 [DOI] [PubMed] [Google Scholar]
- Shao W., Wu J., Chen J., Lee D. M., Tishkina A., et al. , 2010. A modifier screen for Bazooka/PAR-3 interacting genes in the Drosophila embryo epithelium. PLoS ONE 5: e9938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Fink G. R., Hicks J. B., 1986. Methods in Yeast Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Song X., Bowen J., Miao W., Liu Y., Gorovsky M. A., 2012. The nonhistone, N-terminal tail of an essential, chimeric H2A variant regulates mitotic H3–S10 dephosphorylation. Genes Dev. 26: 615–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starita L. M., Lo R. S., Eng J. K., von Haller P. D., Fields S., 2012. Sites of ubiquitin attachment in Saccharomyces cerevisiae. Proteomics 12: 236–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone E. M., Yamano H., Kinoshita N., Yanagida M., 1993. Mitotic regulation of protein phosphatases by the fission yeast sds22 protein. Curr. Biol. 3: 13–26 [DOI] [PubMed] [Google Scholar]
- Stuart J. S., Frederick D. L., Varner C. M., Tatchell K., 1994. The mutant type 1 protein phosphatase encoded by glc7–1 from Saccharomyces cerevisiae fails to interact productively with the GAC1-encoded regulatory subunit. Mol. Cell. Biol. 14: 896–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundin L. J., Guimaraes G. J., Deluca J. G., 2011. The NDC80 complex proteins Nuf2 and Hec1 make distinct contributions to kinetochore-microtubule attachment in mitosis. Mol. Biol. Cell 22: 759–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatchell K., Makrantoni V., Stark M. J., Robinson L. C., 2011. Temperature-sensitive ipl1–2/Aurora B mutation is suppressed by mutations in TOR complex 1 via the Glc7/PP1 phosphatase. Proc. Natl. Acad. Sci. USA 108: 3994–3999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien J. F., Umbreit N. T., Gestaut D. R., Franck A. D., Cooper J., et al. , 2010. Cooperation of the Dam1 and Ndc80 kinetochore complexes enhances microtubule coupling and is regulated by aurora B. J. Cell Biol. 189: 713–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooley J. G., Miller S. A., Stukenberg P. T., 2011. The Ndc80 complex uses a tripartite attachment point to couple microtubule depolymerization to chromosome movement. Mol. Biol. Cell 22: 1217–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkle-Mulcahy L., Andrews P. D., Wickramasinghe S., Sleeman J., Prescott A., et al. , 2003. Time-lapse imaging reveals dynamic relocalization of PP1gamma throughout the mammalian cell cycle. Mol. Biol. Cell 14: 107–117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tung H. Y., Wang W., Chan C. S., 1995. Regulation of chromosome segregation by Glc8p, a structural homolog of mammalian inhibitor 2 that functions as both an activator and an inhibitor of yeast protein phosphatase 1. Mol. Cell. Biol. 15: 6064–6074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida K. S., Takagaki K., Kumada K., Hirayama Y., Noda T., et al. , 2009. Kinetochore stretching inactivates the spindle assembly checkpoint. J. Cell Biol. 184: 383–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V., Hardwick K. G., 2009. A novel protein phosphatase 1-dependent spindle checkpoint silencing mechanism. Curr. Biol. 19: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virshup D. M., Shenolikar S., 2009. From promiscuity to precision: protein phosphatases get a makeover. Mol. Cell 33: 537–545 [DOI] [PubMed] [Google Scholar]
- Wang W., Stukenberg P. T., Brautigan D. L., 2008. Phosphatase inhibitor-2 balances protein phosphatase 1 and aurora B kinase for chromosome segregation and cytokinesis in human retinal epithelial cells. Mol. Biol. Cell 19: 4852–4862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welburn J. P., Vleugel M., Liu D., Yates J. R., 3rd, Lampson M. A., et al. , 2010. Aurora B phosphorylates spatially distinct targets to differentially regulate the kinetochore-microtubule interface. Mol. Cell 38: 383–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S., Avila-Sakar A., Wang H. W., Niederstrasser H., Wong J., et al. , 2005. Formation of a dynamic kinetochore- microtubule interface through assembly of the Dam1 ring complex. Mol. Cell 17: 277–290 [DOI] [PubMed] [Google Scholar]
- Westermann S., Wang H. W., Avila-Sakar A., Drubin D. G., Nogales E., et al. , 2006. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature 440: 565–569 [DOI] [PubMed] [Google Scholar]
- Zhang S., Guha S., Volkert F. C., 1995. The Saccharomyces SHP1 gene, which encodes a regulator of phosphoprotein phosphatase 1 with differential effects on glycogen metabolism, meiotic differentiation, and mitotic cell cycle progression. Mol. Cell. Biol. 15: 2037–2050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K., Lin W., Latham J. A., Riefler G. M., Schumacher J. M., et al. , 2005. The set 1 methyltransferase opposes ipl 1 aurora kinase functions in chromosome segregation. Cell 122: 723–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.