Background: The Werner syndrome protein (WRN) stimulates specialized Y-family DNA polymerase activity through an unknown mechanism.
Results: A bipartite interaction between WRN and hpol η increases the efficiency and fidelity of polymerization.
Conclusion: Direct interactions between hpol η and WRN increase kpol 4-fold and decrease the misinsertion frequency 2.5- to 4.5-fold.
Significance: WRN-mediated enhancement of polymerase accuracy and efficiency may suppress mutations and genomic instability.
Keywords: DNA Damage Response, DNA Helicase, DNA Polymerase, DNA Replication, Enzyme Kinetics
Abstract
We have investigated the interaction between human DNA polymerase η (hpol η) and the Werner syndrome protein (WRN). Functional assays revealed that the WRN exonuclease and RecQ C-terminal (RQC) domains are necessary for full stimulation of hpol η-catalyzed formation of correct base pairs. We find that WRN does not stimulate hpol η-catalyzed formation of mispairs. Moreover, the exonuclease activity of WRN prevents stable mispair formation by hpol η. These results are consistent with a proofreading activity for WRN during single-nucleotide additions. ATP hydrolysis by WRN appears to attenuate stimulation of hpol η. Pre-steady-state kinetic results show that kpol is increased 4-fold by WRN. Finally, pulldown assays reveal a bipartite physical interaction between hpol η and WRN that is mediated by the exonuclease and RQC domains. Taken together, these results are consistent with alteration of the rate-limiting step in polymerase catalysis by direct protein-protein interactions between WRN and hpol η. In summary, WRN improves the efficiency and fidelity of hpol η to promote more effective replication of DNA.
Introduction
Genomic maintenance, including timely and efficient DNA replication, is a fundamental biological process and an important barrier to tumorigenesis in higher eukaryotes (1, 2). Successful replication of the genome requires the spatial and temporal coordination of many different proteins and enzymes (3). The physical interactions and coupling of enzymatic properties that leads to efficient copying of damaged and/or structured DNA is an area that is still under investigation (4). Specialized DNA polymerases, such as the Y-family members, are among the downstream targets of ATR and other damage response kinases (5). Y-family DNA polymerases appear to be an important means of bypassing the DNA adducts and structured DNA (i.e. G4 quadruplex DNA) in an efficient manner (6–11). The participation of Y-family DNA polymerases in replication fork progression and in the cellular replication stress response has revealed their importance in maintaining genomic stability under basal conditions and following exposure to DNA damaging agents (12). However, much remains unclear regarding interactions that either facilitate polymerase switching or modulate the efficiency of specialized replication events.
There are four mammalian Y-family DNA polymerases: Rev1, pol η, pol ι, and pol κ (13). The structural and functional properties of the Y-family polymerases have been studied extensively (14). The Y-family enzymes are distinct in their ability to replicate bulky DNA lesions and distorted template structures. Although the overall domain architecture of the polymerase core is retained in the Y-family (Fig. 1A), each member possesses unique structural attributes that produce a fascinating set of different mechanisms for nucleotide selection, which results in diverse functional properties (7). The Y-family member under investigation in the current work is human DNA polymerase η (hpol η).2 The skin cancer-prone disease xeroderma pigmentosum variant results from mutations in the RAD30 gene encoding hpol η (15). In vitro assays and structural work have shown that hpol η is the most efficient and accurate means of bypassing cyclobutane pyrimidine dimers formed by exposure to UV-B irradiation (16–19). It is clear from in vitro analyses that Y-family members, including hpol η, are stimulated by protein-protein interactions, such as those that occur with the sliding clamp PCNA and that PCNA ubiquitination plays an important role in governing the localization of these specialized polymerases to sites of replication stress (12, 20–23). It is of great importance that Y-family DNA polymerase activity be studied in complex with proteins and enzymes that modulate the replication stress response inside of cells.
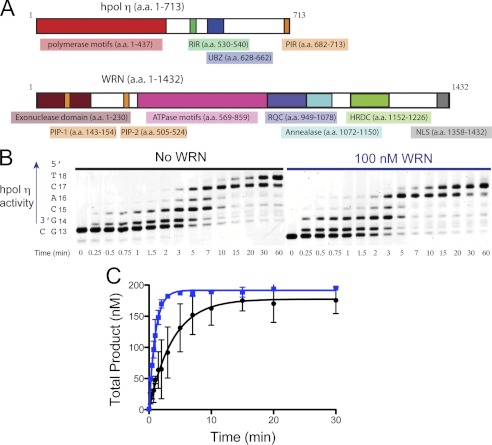
FIGURE 1.
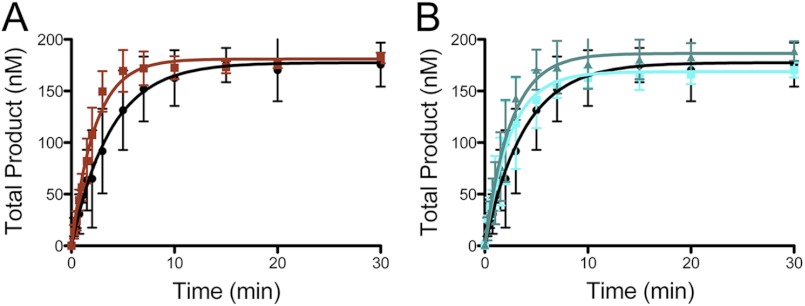
Full-length WRN1–1432 stimulates hpol η1–437 core polymerase domain. A, an overview of the hpol η and WRN proteins showing domains with either structural or catalytic properties for each enzyme. PIR, PCNA interacting region; RIR, Rev1-interacting region; UBZ, ubiquitin binding zinc-finger domain; HRDC, Helicase and RNaseD C-terminal; PIP, PCNA interacting peptide; NLS, nuclear localization signal. B, hpol η1–437 (2 nm) polymerization was allowed to proceed on a DNA substrate (200 nm) possessing a 5-nucleotide ssDNA overhang (13/18-mer) in either the presence (●) or absence (○) of full-length WRN (100 nm). C, DNA synthesis by hpol η1–437 (2 nm) was monitored over time using a 13/18-mer primer-template DNA substrate (200 nm) in the absence of WRN and in the presence of full-length WRN1–1432 (100 nm). Total product formation (i.e. all of the product bands from panel B) is shown. The experiments were performed in triplicate and the mean ± S.D. is shown. The single-turnover results for each experiment were fit to a single-exponential equation to yield the following kinetic parameters: No WRN (●): A = 178 ± 6 nm, kobs = 0.26 ± 0.03 min−1; WRN1–1432 (■): A = 192 ± 2 nm, kobs = 0.98 ± 0.04 min−1.
In vitro assays have shown that the Y-family DNA polymerases η, ι, and κ interact functionally with the Werner syndrome protein (WRN) (24). However, a mechanistic understanding of WRN-mediated alterations in replication efficiency and/or fidelity is lacking. The ability of WRN to modulate the activity of DNA polymerases and the presence of WRN at actively replicating forks inside cells provides a strong impetus for understanding WRN-mediated alteration of replication efficiency and fidelity at the molecular level. Although a general increase in activity for three of the four human Y-family members was reported when these enzymes were incubated with WRN (24), the mechanism of action still remains obscure. In the current study, we have investigated the functional interaction between hpol η and WRN in an effort to reveal mechanistic features associated with modulation of polymerase activity. Our experiments not only show that WRN can increase the efficiency and fidelity of DNA synthesis but we also detail the enzymatic properties and physical interactions that are involved in stimulation of hpol η activity. These results provide new insights into specialized DNA replication events that participate in the resolution of replication stress.
EXPERIMENTAL PROCEDURES
A detailed description of materials and methods used to express and purify the recombinant proteins can be found in the supplemental “Materials and Methods”.
Full-length Extension Polymerase Assays
Fluorescein-labeled primer was annealed to template oligonucleotide by heating a 1:2 molar ratio of primer:template DNA to 95 °C for 5 min and then slow cooling to room temperature. The primer/template DNA was then incubated with hpol η prior to extension in the presence or absence of WRN. Each reaction was initiated by adding dNTP·MgCl2 (0.5 mm of each dNTP and 5 mm MgCl2) solution to a preincubated hpol η·DNA complex (2 nm hpol η and 200 nm DNA). For experiments measuring the effect of WRN-catalyzed ATP hydrolysis on polymerase activity, ATP was added to a final concentration of 100 μm. Unless otherwise stated, all enzymatic reactions were carried out at 37 °C in 50 mm HEPES buffer (pH 7.5) containing 60 mm KCl, 5 mm dithiothreitol (DTT), 100 μg ml−1 of bovine serum albumin (BSA), and 10% (v/v) glycerol. At the indicated time points, 4-μl aliquots were quenched with 16 μl of a 95% formamide (v/v), 20 mm EDTA, 0.1% bromphenol blue (w/v) solution and were separated by electrophoresis on a 16% polyacryamide (w/v), 7 m urea gel. The products were then visualized using a Typhoon imager (GE Healthcare) and quantified using ImageQuantTM software (GE Healthcare). For the polymerase extension assays we quantified the amount of each product band (i.e. 13-mer substrate, 14-mer product, 15-mer product, etc.) at each time point. For “multiple hit” polymerase reactions, the summation of all the products formed at a given point in time follows a Poisson distribution (25). A plot of individual product bands formed over time reveals that there is a “lag” in product formation for the 15-mer, 16-mer, 17-mer, and 18-mer products. This allowed us to calculate the fraction of each product band at a given time point. To obtain an estimate of “total” product formation over time (i.e. all of the product bands) we fit the results to a single exponential equation (Equation 1, where n = 1), which provides an estimate of the amplitude of product formed (A) and the single-turnover rate constant (kobs) governing the reaction. To obtain estimates for the rate of single-turnover product formation of individual product bands (i.e. 15-mer, 16-mer, 17-mer, and 18-mer) we used an expression derived from a simple n-step sequential reaction (Scheme 1). Where product formation at each step is defined by the rate constant kobs, which describes the single-turnover rate of product formation for a given species. In the simplest reaction scheme kobs is equal for each step. However, it is clear that polymerases incorporate different dNTPs with slightly different rates. In an effort to analyze the rate of product formation at each insertion step, we plotted the amount of individual product bands as a function of time and fit the resulting curve to Equation 1, which has been derived for sequential n-step reaction schemes, such as the one shown above, and previously (26, 27).
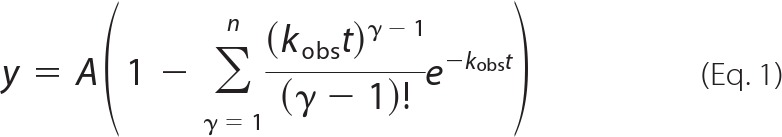 |
Where A = amplitude of product formation, n = number of steps to form product, kobs = observed rate of product formation.
SCHEME 1.
Steady-state Kinetic Analysis of Polymerase Activity
Single-nucleotide incorporation by hpol η was measured over a range of dNTP concentrations. The products were analyzed as described for full-length extension assays. The initial portion of the velocity curve was fit to a linear equation in the program GraphPad Prism (GraphPad, San Diego, CA). The resulting velocity was plotted as a function of dNTP concentration and then fit to a hyperbolic equation, correcting for enzyme concentration, to obtain estimates for the turnover number (kcat) and Michaelis constant (Km,dNTP).
Transient-state Kinetic Analysis of hpol η Polymerase Activity
All pre-steady-state experiments were performed using a KinTek RQF-3 model chemical quench-flow apparatus (KinTek Corp., Austin, TX). hpol η1–437 (25 nm) was preincubated with fluorescein-labeled primer-template DNA (substrate 2, supplemental Table S1, 50 nm). The reaction was initiated by rapid mixing of the enzyme·DNA solution with a solution containing MgCl2 (5 mm) and varying concentrations of dCTP (1–250 μm). Polymerase catalysis was stopped by the addition of 500 mm EDTA (pH 9.0). Product formation was fit to a single-exponential equation with a second linear phase (Equation 2).
Where A = product formed in first binding event, kobs = rate constant defining polymerization under the conditions used for the experiment being analyzed, k2 = the observed second-order rate of product formation, and t = time.
Measurement of WRN 3′-5′ Exonuclease Activity
Exonucleolytic degradation of polymerase products was measured by incubating full-length WRN with hpol η1–437 (50:1, WRN:hpol η1–437) and 13/18-mer DNA (substrate 2). The reaction was initiated by the addition of either dCTP (100 μm) or dTTP (1 mm) and MgCl2 (5 mm). Reactions were quenched, products were separated and quantified in a manner identical to that described above for the polymerase assays. WRN-catalyzed degradation of fluorescein-labeled primer was quantified and the rate of degradation was estimated by fitting the results to a linear equation.
GST-hpol η Pulldown Experiments
GST-tagged hpol η1–437 (30 μg) was incubated with different WRN constructs (30 μg for the HRDC deletion and 100 μg for WRN1–333 and WRN949–1078) in 50 mm HEPES buffer (pH 7.5) containing 40 mm KCl and 10% glycerol (v/v) for 16 h at 4 °C. The binding solutions were then mixed with glutathione-Sepharose beads with agitation for an additional 16 h at 4 °C. The beads were centrifuged at 500 relative centrifugal force for 5 min and the solution was pipetted off the beads. The beads were then washed two more times with 75 μl of binding buffer. The bound proteins were eluted by boiling the beads in SDS-PAGE loading buffer for 10 min followed by centrifugation at 500 RCF for an additional 10 min. The samples were separated by 4–20% gradient SDS-PAGE. Proteins were transferred to polyvinylidene difluoride membranes (200 mA; 3 h) and probed with antibodies against WRN (1:10,000 dilution; overnight incubation). Membranes were washed and incubated with HRP-conjugated secondary antibodies (1:2,000; 1 h) prior to visualization with ECL-Prime (GE Healthcare). The membranes were then stained with Ponceau S (Sigma) and scanned to create electronic images.
RESULTS
Full-length WRN Stimulates hpol η Core Polymerase Activity
Both hpol η and WRN possess multiple structured domains that perform distinct functions related to the chemistry and biology of each enzyme (Fig. 1A). WRN is a multifaceted protein with at least four independently structured domains and several catalytic functions, including 3′ to 5′ helicase and exonuclease activities (Fig. 1A) (28). The domains of relevance to the current study include: 1) the helicase and RNase D C-terminal (HRDC) domain, 2) the RecQ C-terminal (RQC) domain, 3) the N-terminal exonuclease (Exo) domain, and 4) the ATPase domain (Fig. 1A). In addition to the core polymerase domains, hpol η has a C-terminal region that is primarily involved in recruitment to sites of replication stress through protein-protein interactions and post-translational modifications (29). We first tested the ability of full-length WRN1–1432 to modulate the activity of both full-length hpol η (amino acids 1–713) and the core polymerase domains (amino acids 1–437) by monitoring product formation over time. In all instances, recombinant WRN was tested for contaminating polymerase activity to be certain that any enhancement observed was due to WRN (data not shown). DNA-dependent ATPase and DNA helicase activities, as well as strand-annealing properties for recombinant full-length WRN were also verified (supplemental Fig. S1). Similar to a previous report (24), full-length hpol η DNA synthesis activity is stimulated ∼2-fold by full-length WRN (supplemental Fig. S2) on a DNA substrate possessing a 12-nucleotide ssDNA template (substrate 1, supplemental Table S1). Likewise, the core polymerase hpol η1–437 is stimulated by WRN1–1432 to a similar extent as the full-length polymerase (supplemental Fig. S3). At longer time points we observe WRN-dependent exonuclease activity (supplemental Figs. S2 and S3). These results indicate that the C-terminal region of hpol η is not required to observe the stimulatory effect of WRN. We, therefore, used the core polymerase domain (amino acids 1–437) for all subsequent experiments.
DNA synthesis by hpol η1–437 was then allowed to proceed on a 13/18-mer DNA substrate (substrate 2, supplemental Table S1) either in the presence or absence of full-length WRN1–1432 (Fig. 1B). The experiments were performed with different batches of purified full-length WRN and repeated in triplicate to ensure reproducibility. We quantified each individual product band to determine the fraction of the total product for each time point (Fig. 1C). Analyzing the rate of formation for each product band (i.e. 14-, 15-, 16-, 17-, and 18-mer) allowed us to observe whether WRN-mediated stimulation occurs at each step in the polymerase extension assays (Fig. 2). The observed single-turnover rate constant defined by Equation 1 is assumed to be equal for each step in the polymerase reaction. This is an oversimplification, as different dNTPs are clearly incorporated at slightly different rates. In an effort to clarify this issue, we analyzed each product band separately to determine the rate of product formation as the polymerase progresses through the extension reaction (Fig. 2).
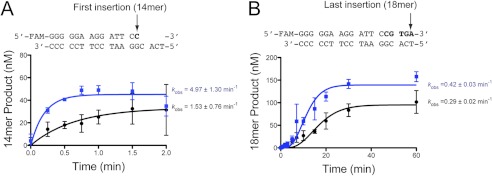
FIGURE 2.
Stepwise analysis of WRN-mediated stimulation of hpol η activity. hpol η1–437 (2 nm) was incubated with 13/18-mer DNA substrate (200 nm) in either the absence (●) or presence (■) of full-length WRN1–1432 (100 nm). The reaction was initiated by the addition of dNTP (500 μm each dNTP) and MgCl2 (5 mm). Each product band was quantified. The rate of product formation was determined by fitting the data to a stepping equation (Equation 1), where n = 1 or 5 for A the first, and B, the last dNTP insertion event, respectively. The experiments were performed in triplicate and the mean ± S.D. is shown. The single-turnover rate constant for each experiment is shown to the right of the plot.
Plotting the total product formed as a function of time reveals that the single-turnover rate constant defining the sum of all the product bands is increased 3.8-fold by the addition of full-length WRN1–1432 (Fig. 1C). Plotting the formation of individual product bands shows that WRN stimulation of hpol η1–437 occurs most efficiently at the initial insertion point, with the effect upon the observed rate constant being less prominent at the end of the template (Fig. 2). It may be that the initial stimulatory effect is carried over to subsequent insertions, rather than through a processive coupling of hpol η and WRN. The effective stimulation of hpol η1–437 allowed us to then map the functional interaction between the Y-family core polymerase domains and WRN in greater detail.
The RQC Domain of WRN Is Necessary for Full Stimulation of hpol η Activity
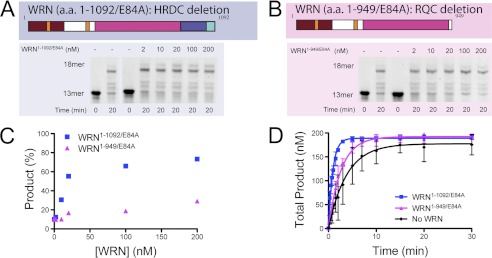
Next, we wanted to assess the domain requirements for WRN-mediated stimulation of polymerase activity. We prepared four WRN mutants to quantify the contributions of different domains to the functional interaction with hpol η1–437 (supplemental Fig. S4). The first WRN mutant (WRN1–1092/E84A, HRDC-deletion) truncates the protein just beyond the RQC domain. The second deletion mutant (WRN1–949/E84A, RQC-deletion) removes both the RQC and HRDC domains from the polypeptide. Finally, we prepared the exonuclease (WRN1–333) and RQC (WRN949–1078) domains of WRN to test for stimulation of hpol η1–437 activity by isolated domains. It should be noted here that the E84A mutation in WRN abolishes exonuclease activity by removing the acidic side chain of Glu-84, which coordinates catalytic divalent cations in the WRN exonuclease domain (30). Where indicated, E84A mutant forms of WRN were used to test for modulation of hpol η1–437 catalysis in the absence of any contribution from WRN exonuclease activity.
We initially measured the hpol η1–437-catalyzed formation of the full-length product in the presence of increasing amounts of the two truncated WRN constructs, the HRDC- and RQC-deletion mutants (Fig. 3). Varying amounts of WRN HRDC- and RQC-deletion mutants were incubated with hpol η1–437 and the amount of product formed at 20 min was quantified (Fig. 3, A and B). The WRN HRDC-deletion mutant increased full-length product formation from about 10 to nearly 75% at 20 min (Fig. 3C). The ratio at which product formation was half-maximal was ∼5:1, WRN1–1092/E84A:hpol η1–437 (Fig. 3C). The WRN RQC-deletion mutant was less effective at stimulating product formation under these conditions (Fig. 3C). Subsequent experiments were performed with higher ratios of WRN to hpol η to maximize the effect upon DNA polymerase activity. These results suggest that the WRN HRDC domain is not required for stimulation of hpol η1–437 activity and that loss of the RQC domain impairs functional interaction between WRN and hpol η1–437. However, we wanted to examine the kinetics of product formation more closely for these two WRN mutants. Therefore, we quantified the rate of hpol η1–437-catalyzed formation of full-length DNA product over time in the presence of our different WRN deletion constructs. Of all the WRN mutant constructs tested, the WRN1–1092/E84A HRDC-deletion mutant most effectively stimulated hpol η1–437 activity in a time course reaction (Fig. 3D). The observed single-turnover rate of product formation was increased 4.3-fold from 0.26 ± 0.03 min−1 without WRN to 1.13 ± 0.05 min−1 upon addition of WRN1–1092/E84A to the reaction mixture. The observed single-turnover rate of product formed in the presence of the WRN1–949/E84A RQC-deletion mutant was 0.38 ± 0.05 min−1, which is 1.5-fold faster than product formation by hpol η1–437 alone (Fig. 3D, magenta trace). These results support the idea that the RQC domain is largely responsible for the effective stimulation of hpol η activity but that some residual stimulatory interaction resides in the first 949 amino acids of WRN.
FIGURE 3.
The RQC domain of WRN is necessary for full stimulation of hpol η1–437 activity. A, hpol η1–437 (2 nm) DNA synthesis was allowed to proceed for 20 min in the presence of increasing amounts of WRN1–1092/E84A (blue panel) and the products were separated by 16% PAGE, 7 m urea. B, hpol η1–437 (2 nm) DNA synthesis was allowed to proceed for 20 min in the presence of increasing amounts of WRN1–949/E84A (magenta panel) and the products were separated by 16% PAGE, 7 m urea. C, full-length product formation shown in panels A and B was quantified. D, DNA synthesis by hpol η1–437 (2 nm) was monitored over time in the presence of either the HRDC-deletion mutant WRN1–1092/E84A (100 nm) or the RQC-deletion mutant WRN1–949/E84A (100 nm). The experiments were performed in triplicate and the mean ± S.D. is shown. The single-turnover results for each experiment were fit to a single-exponential equation to yield the following kinetic parameters: WRN1–1092/E84A (■): A = 189 ± 2 nm and kobs = 1.13 ± 0.05 min−1; WRN1–949/E84A (▴): A = 192 ± 3 nm and kobs = 0.38 ± 0.02 min−1. The control experiment with hpol η alone from Fig. 1C was re-plotted for comparison (●).
The Addition of ATP Attenuates WRN-mediated Stimulation of hpol η
WRN is an ATP-dependent helicase that unwinds double-stranded nucleic acids to form single-stranded intermediates (31, 32). We wanted to test whether addition of ATP to our polymerase reaction mixture would further modulate the interaction between WRN and hpol η1–437. When the polymerase is alone, the addition of ATP (100 μm) to the reaction mixture modestly perturbs DNA synthesis by hpol η1–437, decreasing the rate of product formation presumably because ATP competes with dATP for binding to the polymerase active site (supplemental Fig. S5). The observed single-turnover rate of 18-mer product formation was decreased from 0.26 ± 0.03 to 0.16 ± 0.01 min−1 for hpol η1–437 alone. Adding ATP to the reaction mixture with WRN1–1092/E84A decreases the observed single-turnover rate of product from 1.13 ± 0.05 to 0.26 ± 0.02 min−1 (supplemental Fig. S5). Thus, upon addition of ATP the rate of DNA synthesis is decreased ∼40% for hpol η1–437 alone and ∼75% for experiments with WRN1–1092/E84A. These results suggest that ATP hydrolysis by WRN may induce a change in the RecQ enzyme that inhibits functional interaction with hpol η1–437.
The Isolated Exonuclease and RQC Domains Provide Intermediate Levels of Stimulation to hpol η
We then assessed the role of individual WRN domains in the enhancement of polymerase activity by adding the recombinant exonuclease (WRN1–333) and strand separating RQC winged helix (WRN949–1078) domains to the reaction mixture. The exonuclease domain showed a modest stimulation of polymerase activity when the molar ratio of WRN exo:hpol η1–437 was 50:1 (Fig. 4A). The observed single-turnover rate of product was increased 1.7-fold from 0.26 ± 0.02 to 0.44 ± 0.03 min−1 upon addition of 100-fold excess WRN1–333 (Fig. 4A). The isolated WRN RQC domain was able to effect a noticeable stimulation of polymerase activity (1.6-fold) when the molar ratio was 50:1, WRN RQC:hpol η1–437 (Fig. 4B). Increasing the amount of RQC in the reaction did not stimulate the polymerase activity beyond ∼1.6-fold (Fig. 4B, dark cyan triangles). These results suggest that stimulation of hpol η1–437 activity can occur through interactions with either the exonuclease or the RQC domains. However, the full stimulatory effect is only observed when both domains are housed in the same polypeptide, such as observed for the WRN HRDC-deletion mutant (Fig. 3, results with WRN1–1092/E84A).
FIGURE 4.
The isolated exonuclease and RQC domains of WRN stimulate hpol η1–437 activity. A, DNA synthesis by hpol η1–437 (2 nm) was monitored over time in the presence of the 100 nm (■) WRN1–333 exonuclease domain. The no WRN control experiment (●) from Fig. 1 was re-plotted for comparison. Formation of total product is shown. The single turnover results for each experiment were fit to a single exponential equation to yield the following kinetic parameters: WRN1–333 (■): A = 181 ± 3 nm, kobs = 0.44 ± 0.03 min−1. B, DNA synthesis by hpol η1–437 (2 nm) was monitored over time in the presence of 100 nm (■) and 1 μm (▴) WRN949–1078 RQC domain. The no WRN control experiment (●) from Fig. 1 was re-plotted for comparison. Formation of the total product is shown. The single turnover results for each experiment were fit to a single exponential equation to yield the following kinetic parameters: WRN949–1078 (■): A = 169 ± 5 nm, kobs = 0.41 ± 0.04 min−1; WRN949–1078 (▴): A = 186 ± 5 nm and kobs = 0.40 ± 0.04 min−1.
Stimulation of Accurate Base Pair Formation by hpol η1–437 Is Dependent Upon the RQC Domain of WRN
Next, we wanted to assess the ability of WRN to modulate the kinetic parameters that define hpol η catalytic efficiency and fidelity. As a first measure of these properties we performed steady-state kinetic analysis of hpol η-catalyzed formation of “accurate” Watson-Crick base pairs (i.e. insertion of dCTP opposite dG). Both full-length WRN and the HRDC-deletion mutant stimulated hpol η1–437 catalysis 3–4-fold, as measured by comparing the specificity constants (supplemental Table S2). The increased catalytic efficiency is primarily due to an increase in the turnover number (kcat), as the Michaelis constant changes very little. Abolishing the exonuclease activity of WRN did not improve or diminish the kinetic parameters (compare WRN1–1092 to WRN1–1092/E84A). Importantly, the WRN RQC-deletion mutant (WRN1–949/E84A) does not stimulate hpol η1–437 activity to the same extent as the HRDC-deletion mutant (supplemental Table S2). These results indicate that the RQC domain is required to enhance the activity of hpol η1–437 during insertion of dCTP opposite dG.
Transient-state Kinetic Analysis Reveals That WRN Increases the Rate Constant kpol for hpol η-catalyzed dNTP Insertion Almost 4-fold during Formation of dCTP:dG Base Pairs
Determination of the specificity constant (kcat/Km,dNTP) by steady-state kinetic analysis can provide an estimate of the apparent second-order rate constant for dNTP binding, which is the summation of multiple kinetic steps following binding of the substrate (34). Alterations to specific kinetic steps in the polymerase catalytic cycle are usually not isolated by Michaelis-Menten kinetics, which can limit the conclusions derived from the kinetic constants. Using pre-steady-state kinetic analysis to measure the concentration dependence of hpol η1–437-catalyzed formation of accurate base pairs (i.e. dCTP:dG) provides a measurement of the ground state binding affinity for the incoming nucleotide triphosphate (Kd,dNTP) and the maximum rate of polymerization (kpol), which for pol η is limited by a noncovalent step occurring prior to phosphoryl transfer (35, 36). Previous work has shown that the partially rate-limiting step in hpol η catalysis occurs prior to phosphoryl transfer (36, 37). The structural explanation for what actually comprises the rate-limiting step is less clear but may involve coordination of the divalent metal ions in the hpol η active site or some other noncovalent conformational rearrangement (35, 36).
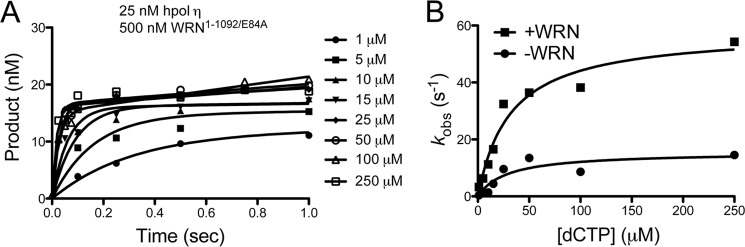
Our results show that inclusion of the WRN HRDC-deletion mutant (WRN1–1092/E84A) increases the rate constant kpol nearly 4-fold from 15.5 ± 3.0 s−1 in the absence of WRN to 58.1 ± 5.2 s−1 with WRN present in the reaction mixture (Fig. 5A). The ground state binding affinity for the incoming dCTP (Kd,dCTP) is not changed by the addition of WRN but remains near 30 μm (Fig. 5B). Construction of a conventional Gibbs free-energy diagram (34) reveals that WRN reduces the free-energy change for insertion of dCTP opposite dG by ∼0.7 kcal/mol from −13.2 kcal/mol without WRN to −13.9 kcal/mol in the presence of WRN. These results are consistent with the notion that WRN may directly alter the structure of the hpol η1–437 active site in a manner that increases the rate-limiting step in the polymerase catalytic cycle.
FIGURE 5.
Pre-steady-state kinetic analysis of hpol η1–437 catalysis reveals that kpol is four times greater in the presence of the WRN1–1092/E84A HRDC-deletion mutant. A, pre-steady-state analysis of hpol η1–437 (25 nm) catalyzed insertion of dCTP (1–250 μm; shown to the right of the plot) opposite dG in the presence of WRN1–1092/E84A (500 nm). Product formation was plotted as a function of time and fit to Equation 2. B, the observed rate of product formation (kobs) derived from panel A was plotted as a function of dCTP concentration for pre-steady-state experiments with hpol η1–437 alone and in the presence of WRN1–1092/E84A. Fitting the data to a hyperbolic equation yields an estimate of kpol and Kd,dCTP: hpol η1–437 alone (●): kpol = 15.5 ± 3.0 s−1 and Kd,dCTP = 28.5 ± 16.2 μm; hpol η1–437 + WRN1–1092/E84A (■): kpol = 58.1 ± 5.2 s−1 and Kd,dCTP = 31.8 ± 8.2 μm.
WRN Improves the Efficiency and Fidelity of hpol η DNA Synthesis in an Exonuclease-independent Manner
We then went on to measure the steady-state parameters for mispair formation by hpol η1–437 in the presence and absence of WRN. In our assays, WRN does not stimulate hpol η1–437-catalyzed formation of mispairs (Table 1). The calculated misinsertion frequency for hpol η1–437 catalysis is improved roughly 2.5–4.5-fold by the inclusion of the WRN HRDC-deletion mutant (compare the misinsertion frequencies, ƒ, for each mispair with and without WRN). It should be noted here that the WRN mutant protein used in the mispair assays does not possess exonuclease activity (E84A mutation) but does possess the RQC domain. Thus, WRN improves the fidelity of DNA synthesis by hpol η1–437 at the level of single-nucleotide insertion events.
TABLE 1.
Steady-state kinetic parameters for hpol η-catalyzed formation of mispairs in the presence of WRN
| kcat | Km,dNTP | kcat/Km,dNTP | ƒ (misinsertion frequency)a | |
|---|---|---|---|---|
| min−1 | μm | min−1 μm−1 | ||
| dTTP:dG | 23 ± 1 | 157 ± 25 | 0.15 | 36 |
| WRN1–1092/E84A | 27 ± 4 | 168 ± 54 | 0.16 | 12 |
| dATP:dG | 7 ± 3 | 260 ± 100 | 0.027 | 6.4 |
| WRN1–1092/E84A | 11 ± 2 | 570 ± 200 | 0.019 | 1.4 |
| dGTP:dG | 6 ± 1 | 50 ± 9 | 0.12 | 28 |
| WRN1–1092/E84A | 9 ± 1 | 60 ± 7 | 0.15 | 11 |
a The frequencies of misinsertion were calculated relative to insertion of dCTP opposite dG using the ratio (kcat/Km, mispaired dNTP)/(kcat/Km,dCTP) × 103. The smaller the value of ƒ, the more accurate the polymerase.
The Exonuclease Activity of WRN Prevents the Stable Formation of Mispairs by hpol η but Does Not Perturb Polymerase-catalyzed Insertion of dCTP Opposite dG
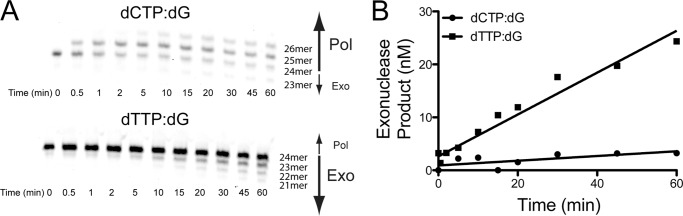
We further tested the ability of WRN to improve hpol η fidelity by measuring exonucleolytic degradation of polymerase reaction products catalyzed by the WRN exonuclease domain. hpol η1–437 was allowed to insert either dCTP or dTTP opposite dG in the presence of full-length WRN1–1432, which has exonuclease activity (Fig. 6). The concentration of dNTP in the reaction mixture was saturating (100 μm for dCTP and 1 mm for dTTP). At these concentrations, the rate of dNTP insertion in the absence of exonuclease activity should be comparable for both correct and incorrect base pairs, based on our steady-state kinetic parameters (Table 1 and supplemental Table S2). However, only dCTP is incorporated over the time course of the experiment, with dTTP apparently stimulating exonucleolytic degradation by WRN (Fig. 6A). Quantification of the rate of primer degradation shows that the mispair reaction is degraded ∼10-fold faster than the accurate base pair reaction (Fig. 6B). These results are consistent with our analysis of accurate single-nucleotide insertion events, where versions of WRN possessing exonuclease activity were still able to increase the rate of dCTP insertion opposite template dG (supplemental Table S2). Previous structural work has noted the close similarity between the WRN exonuclease domain and the exonuclease domain of Escherichia coli Pol I Klenow fragment (30). Notably, the exonuclease domain of the Klenow fragment is positioned in an orientation relative to the core polymerase domains that are similar to that of the little finger domain of the Y-family polymerases. The little finger (or palm-associated) domain of the Y-family is tethered to the polymerase core domains by a flexible linker and has been shown to undergo large conformational rearrangements (38, 39). The fact that WRN impairs hpol η1–437-catalyzed insertion of dTTP opposite dG to a larger extent than dCTP insertion is further suggestive of a role for WRN as polymerase “proofreader.”
FIGURE 6.
WRN exonuclease activity prevents mispair formation by hpol η1–437. A, hpol η1–437 single nucleotide insertion in the presence of 50-fold excess wild-type WRN1–1432 was measured for dCTP (100 μm) and dTTP (1 mm) and the products were resolved by 16% PAGE, 7 m urea. Pol, polymerase activity; Exo, WRN 3′-5′ exonuclease activity. B, the rate of WRN1–1432-catalyzed exonuclease activity was estimated from the slopes of the linear dependence of product formation as a function of time: dCTP (●): v0 = 44.5 ± 18.8 nm min−1 × 103; dTTP (■): v0 = 395 ± 30 nm min−1 × 103.
WRN Physically Interacts with the hpol η Core Polymerase
Finally, we examined the possibility that WRN and hpol η1–437 are physically interacting with one another. Such an interaction has not been reported previously. To test this possibility, we performed affinity enrichment (“pulldown”) experiments with a version of hpol η1–437 that retained the GST tag (Fig. 7). WRN proteins were incubated with either GST-hpol η1–437 bound or empty glutathione-Sepharose beads. After washing the resin, the proteins that remained bound to the beads were boiled off into SDS-PAGE loading buffer. The fraction of each WRN construct retained at the elution step was visualized following SDS-PAGE separation of the proteins (Fig. 7A). The HRDC-deletion mutant is retained in the pulldown experiment (Fig. 7A). Immunoblotting clearly shows that WRN is specifically retained on the GST-hpol η1–437-bound resin (Fig. 7A). WRN was not enriched on the empty resin, which indicates that the retention is specific. Likewise, both the exonuclease and RQC domains were retained in our pulldown experiments, which is consistent with the notion that each domain can physically interact with hpol η1–437 in an independent manner (Fig. 7, B and C).
FIGURE 7.
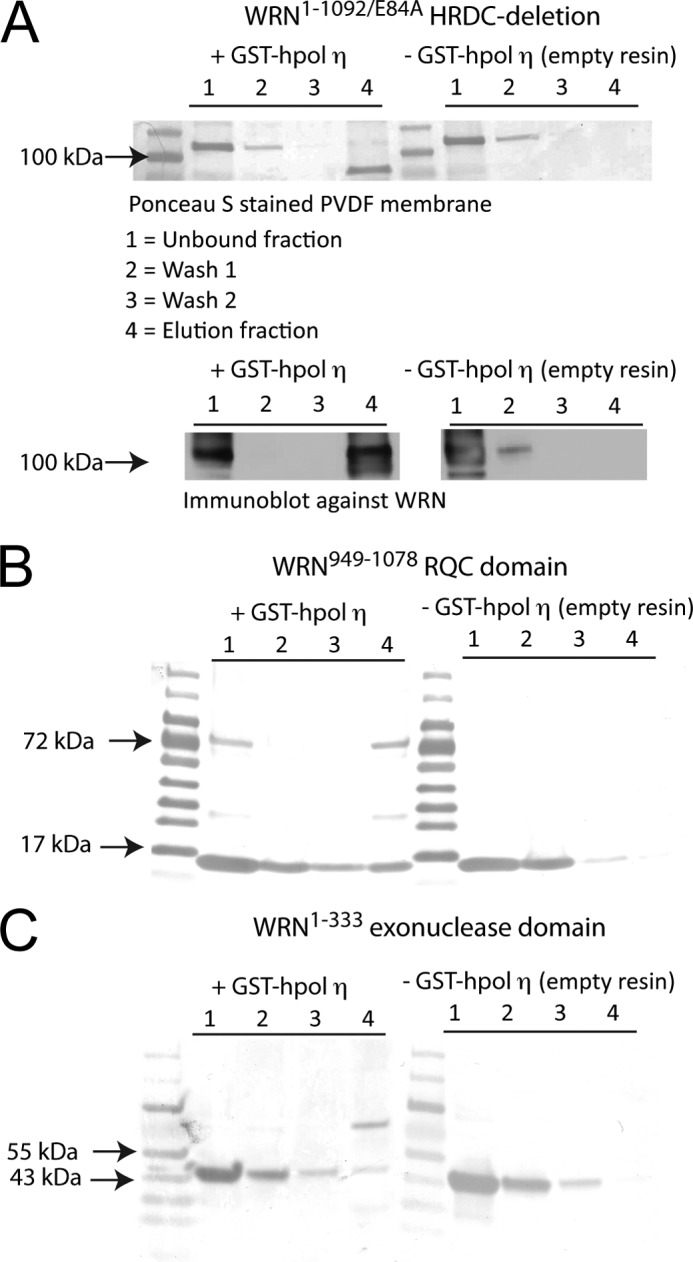
WRN interacts with hpol η1–437 physically through the exonuclease and RQC domains. A, WRN1–1092/E84A was incubated with either GST-hpol η1–437 coupled or empty glutathione-Sepharose resin and subjected to a subsequent wash and elution steps. Aliquots from each fraction were separated by 4–20% SDS-PAGE, transferred to PVDF membranes, and either stained with Ponceau S or subjected to immunoblot analysis. Note that GST-hpol η1–437 migrates near the 72-kDa marker. The WRN1–1092/E84A HRDC-deletion mutant remains bound to the resin only when GST-hpol η1–437 is present. B, the WRN949–1078 RQC domain was incubated with either GST-hpol η1–437 coupled or empty glutathione-Sepharose resin and subjected to subsequent wash and elution steps. Aliquots from each fraction were separated by 4–20% SDS-PAGE, transferred to PVDF membranes, and stained with Ponceau S. C, the WRN1–333 exonuclease domain was incubated with either GST-hpol η1–437 coupled or empty glutathione-Sepharose resin and subjected to subsequent wash and elution steps. Aliquots from each fraction were separated by 4–20% SDS-PAGE, transferred to PVDF membranes, and stained with Ponceau S.
DISCUSSION
Cellular responses to perturbed replication involves the coordination of many different proteins and enzymes s(40). The need for enzymes that can process covalently modified or structured nucleic acids is apparent from the participation of Y-family DNA polymerases in DNA damage tolerance pathways (41). Accumulation of these specialized enzymes at sites of stress is dependent upon post-translational modifications and protein-protein interactions (5, 20, 21, 29, 42–45). Despite the often generalized association of Y-family polymerases with “error-prone response” mechanisms there is evidence that DNA polymerase η can assist normal replication fork progression in the absence of damage signaling (12, 46). Deciphering the protein-protein interactions that might alter the catalytic properties of pol η is an important part of understanding how this enzyme contributes to mechanisms that suppress genomic instability.
Like hpol η, the RecQ helicase WRN also participates in the alleviation of replication stress and continued fork progression through interactions with several different proteins, including Y-family DNA polymerases (24, 47–52). The role of WRN in the modulation of Y-family DNA polymerase activity and the potentially important ramifications upon genomic stability led us to investigate the mechanistic features of this interaction. The experiments reported herein contribute several important and previously unrecognized insights into how WRN modifies the activity of a specialized human DNA polymerase.
First, full-length extension by hpol η1–437 is stimulated ∼4-fold by the addition of full-length WRN (Fig. 1C). Careful examination of full-length extension experiments reveals that WRN-mediated stimulation of hpol η1–437 activity is greatest at the first insertion event (∼3–4-fold), diminishing slightly as the polymerase extends the primer (Fig. 2). Additional experiments were performed to elucidate the mechanism promoting WRN-mediated stimulation of hpol η catalysis.
We set out to map the domains of WRN that functionally interact with hpol η in the hopes that this information would provide some indication regarding the molecular mechanism involved in stimulating polymerase activity. We began by deleting two domains in the C-terminal half of WRN, namely the HRDC and RQC domains (Fig. 3). Similar to full-length WRN1–1432, the WRN1–1092/E84A HRDC-deletion mutant enhances the rate of full-length product formation ∼4-fold (Fig. 3D). However, extension experiments indicated that deleting the RQC domain of WRN largely (but not completely) impairs the stimulatory effect upon hpol η1–437 activity, as WRN1–949/E84A only stimulates polymerase activity ∼1.5-fold (Fig. 3). Using the HRDC-deletion mutant we then observed that the addition of ATP attenuates WRN-mediated stimulation of hpol η1–437 (supplemental Fig. S5). The mechanistic basis for this attenuation by ATP is unclear but previous studies with the WRN homologue, human RecQ1, have illustrated very nicely how binding of ATP can alter the oligomeric state of the enzyme (53, 54). The previous study with RecQ1 showed that it forms a tetramer in the absence of ATP and that the addition of ATP/Mg2+ shifted the equilibrium to a lower order species, which is most likely dimeric in nature (53, 54). WRN has been shown to exist as a tetramer when bound to replication intermediates (55). It may be that WRN undergoes a similar alteration in the oligomeric state upon binding of ATP and that this change has a negative effect upon WRN-mediated stimulation of hpol η1–437. Experiments are ongoing to assess changes in the oligomeric state of WRN and the potential ramifications upon its functions. From these results, we concluded that the RQC domain of WRN is playing a major role in the functional interaction with hpol η and that ATP hydrolysis by WRN negatively impacts the functional interaction with the polymerase.
We next tested the ability of isolated WRN domains to enhance hpol η DNA synthesis. The N-terminal exonuclease domain of WRN only increases polymerase activity ∼1.7-fold (Fig. 4A). Similarly, the WRN949–1078 RQC domain by itself fails to stimulate hpol η1–437 to the same extent as the WRN1–1092/E84A HRDC-deletion mutant (compare Figs. 2B and 4B). Rather, the effect of both the exonuclease and RQC domains appears to be intermediate to that observed for the full-length and HRDC-deletion proteins. Consistent with a role for the exonuclease domain in the stimulation of hpol η1–437 activity, an intermediate effect for the WRN1–949/E84A RQC-deletion is observed for full-length product formation (Fig. 3D). Thus, the polymerase extension experiments are supportive of an interaction between hpol η1–437 and WRN that is greatest when both the RQC and the exonuclease domains are present and is, thus, bipartite in nature.
Steady-state kinetic analysis of hpol η1–437-catalyzed insertion of dCTP opposite dG shows that WRN increases the specificity of the polymerase reaction by increasing the turnover number (kcat) with essentially no effect upon the Km,dNTP (supplemental Table S2). Similar increases in polymerase activity were observed for full-length WRN and in both exonuclease proficient (WRN1–1092) and deficient (WRN1–1092/E84A) versions of the WRN HRDC-deletion mutant (supplemental Table S2). Importantly, mispair insertion by hpol η1–437 in the steady-state is not modulated by WRN at the level of single nucleotide insertions (Table 1). We observed a clear enhancement of hpol η1–437 fidelity upon addition of WRN1–1092/E84A at the level of single nucleotide additions. We also show that mispair formation by hpol η1–437 is largely inhibited by full-length WRN possessing exonuclease activity (Fig. 6). These results indicate that the accuracy of specialized DNA synthesis events may be improved by WRN in a manner that is both exonuclease-dependent (i.e. active degradation of mispairs) and exonuclease-independent (i.e. modulation of only accurate dNTP insertion). The initial report showing a functional interaction between WRN and Y-family polymerases observed a WRN-dependent increase in misinsertion rates using qualitative single nucleotide insertion experiments and a plasmid-based assay (24). The reason for this difference is not clear. However, previous experiments where hpol η was allowed to extend across a template with only three of the four dNTPs present, the most pronounced defect in WRN-mediated stimulation was observed when the dNTP that correctly pairs with the first template residue is absent (i.e. WRN does not stimulate pol η when the correct dNTP for the first insertion is absent from the reaction mixture). We would contend that this is consistent with WRN failing to stimulate mispair formation. The plasmid-based experiments used previously required pol η to copy a 407-nucleotide gap and used an exonuclease-deficient version of WRN. Our results suggest that interactions between these two proteins will be less pronounced over such a span of dNTP insertion events (Fig. 2).
In addition to steady-state kinetic analysis, we measured the effect of WRN upon polymerase catalysis in the first binding event between enzyme and substrate. Pre-steady-state kinetic analysis of hpol η1–437 activity reveals that the maximum forward rate of polymerization (kpol) is increased ∼4-fold by WRN (Fig. 5). Previous kinetic studies indicate that the rate-limiting step (or steps) in the hpol η catalytic cycle occurs prior to phosphodiester bond formation (37). Structural studies have failed to reveal obvious conformational changes as candidates for the rate-limiting step in either yeast or human pol η catalysis (16, 18). A comparison of binary and ternary hpol η crystal structures is consistent with the notion that the enzyme active site is pre-aligned for catalysis and that binding of the metal ions is the rate-limiting step in catalysis (35). Recently, an elegant series of time-resolved structures of hpol η provides convincing evidence that deprotonation of the primer 3′-OH, with a concomitant change in the sugar pucker from South-type to North-type is the rate-limiting step in the polymerase η catalytic cycle (56). Interestingly, a third metal ion is observed following phosphodiester bond formation and was proposed to stabilize the intermediate state during phosphoryl transfer. There are other studies with Y-family DNA polymerases that indicate that subtle conformational dynamics may indeed play a role in nucleotide selectivity (57, 58). The alteration of hpol η1–437 pre-steady-state kinetic parameters in our experiments indicates that WRN is modulating the rate-limiting step in the polymerase catalytic cycle and the recent crystal structures of hpol η suggest specific active site dynamics, including coordination of a third metal ion and primer-template conformational transitions, may be important in this regard.
We then calculated the ΔGbind,obsTS from the measured pre-steady-state parameters kpol and Kd,DNA as described previously (59) and obtained estimates of −13.2 and −13.9 kcal/mol for dCTP binding/insertion to the hpol η1–437 active site in the absence and presence of WRN, respectively. The diminishment in the free energy associated with dCTP binding/insertion is ∼0.7 kcal/mol, which is a modest change but perhaps a relevant one given the fact that pol η exhibits a free energy barrier between accurate and inaccurate insertion events that is 2.6 and 3.0 kcal/mol for yeast and human pol η, respectively. These values are ∼2–2.4 kcal/mol less than that of Klenow fragment (36). The A-family member, Klenow fragment, is ∼50 times more accurate than hpol η when discriminating between correct and incorrect base pairs (36). A 0.7 kcal/mol change in the free energy barrier to misinsertion by hpol η1–437 makes up about 30% of the 2.4 kcal/mol difference between Klenow and hpol η in thermodynamic terms. A strict translation of this value to polymerase fidelity would mean that hpol η should be about 15-fold more accurate. We observe a 2.5–4.5-fold increase in hpol η1–437 accuracy in the steady-state (Table 1), which is not as large as the value predicted from thermodynamic estimates. A caveat to this interpretation of our results in that we are comparing thermodynamic values derived from pre-steady-state analysis to steady-state kinetic parameters.
Pulldown experiments show that GST-hpol η1–437 physically interacts with the WRN HRDC-deletion mutant, as well as isolated exonuclease and RQC domains (Fig. 7). Although co-localization to damaged replication foci in human cells has been reported (24), a direct physical interaction between WRN and the Y-family polymerases has not been shown. We show that this interaction is minimally bipartite in nature. The intermediate stimulation of hpol η by WRN exonuclease and RQC domains is consistent with such a notion. Several studies have reported that WRN interacts with its binding partners through more than one domain (60). Indeed, a similar bipartite interaction between WRN and the Bloom syndrome protein (BLM) was shown previously and mapped to residues 50–120 and 949–1092 (61), strikingly similar to our results. The bipartite interaction with hpol η is, therefore, not without precedent. Whether the physical or functional interaction occurs within cells remains to be determined. Experiments are ongoing to elucidate the specific amino acid residues and structural features that facilitate the physical and functional interactions between hpol η and WRN.
It can be deduced from our results that the interaction between WRN and hpol η is not a simple one. Our results suggest that the interaction between WRN and hpol η is minimally bipartite in nature, consisting of both physical and functional molecular determinants existing in the exonuclease and RQC domains of WRN. The biological relevance of these observations remains unclear. The amount of polymerase stimulation imparted by WRN is not as great as that observed for proteins such as PCNA. Still, when one considers the number of protein-protein interactions and post-translational modifications associated with DNA replication, it seems clear that fork progress and accuracy depends upon the sum of multiple interactions at sites of active replication. In this regard, even relatively small alterations in polymerase efficiency/accuracy may be important. The conclusions reported herein provide new insight into the management of specialized DNA polymerase activity.
This work was supported, in whole or in part, by National Institutes of Health Grants R00 GM084460 (to R. L. E.) and Award Number UL1RR029884 from the National Center for Research Resources (to C. Lowery).

This article contains supplemental ”Materials and Methods,“ Tables S1 and S2, and Figs. S1–S5.
- hpol η
- human DNA polymerase η
- pol
- (DNA) polymerase
- PCNA
- proliferating cell nuclear antigen
- WRN
- Werner syndrome protein
- HRDC
- helicase and RNase D C-terminal
- RQC
- RecQ C-terminal.
REFERENCES
- 1. Bartkova J., Horejsi Z., Koed K., Krämer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J. M., Lukas C., Ørntoft T., Lukas J., Bartek J. (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 [DOI] [PubMed] [Google Scholar]
- 2. Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L. V., Kolettas E., Niforou K., Zoumpourlis V. C., Takaoka M., Nakagawa H., Tort F., Fugger K., Johansson F., Sehested M., Andersen C. L., Dyrskjot L., Ørntoft T., Lukas J., Kittas C., Helleday T., Halazonetis T. D., Bartek J., Gorgoulis V. G. (2006) Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633–637 [DOI] [PubMed] [Google Scholar]
- 3. Langston L. D., Indiani C., O'Donnell M. (2009) Whither the replisome: emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle 8, 2686–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sirbu B. M., Couch F. B., Feigerle J. T., Bhaskara S., Hiebert S. W., Cortez D. (2011) Analysis of protein dynamics at active, stalled, and collapsed replication forks. Genes Dev. 25, 1320–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chen Y. W., Cleaver J. E., Hatahet Z., Honkanen R. E., Chang J. Y., Yen Y., Chou K. M. (2008) Human DNA polymerase η activity and translocation is regulated by phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 16578–16583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bétous R., Rey L., Wang G., Pillaire M. J., Puget N., Selves J., Biard D. S., Shin-ya K., Vasquez K. M., Cazaux C., Hoffmann J. S. (2009) Role of TLS DNA polymerases η and κ in processing naturally occurring structured DNA in human cells. Mol. Carcinog. 48, 369–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eoff R. L., Choi J.-Y., Guengerich F. P. (2010) Mechanistic studies with DNA polymerases reveal complex outcomes following bypass of DNA damage. J. Nucleic Acids, Sept. 26;2010. pii: 830473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Friedberg E. C., Wagner R., Radman M. (2002) Specialized DNA polymerases, cellular survival, and the genesis of mutations. Science 296, 1627–1630 [DOI] [PubMed] [Google Scholar]
- 9. Goodman M. F. (2002) Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu. Rev. Biochem. 71, 17–50 [DOI] [PubMed] [Google Scholar]
- 10. Sarkies P., Murat P., Phillips L. G., Patel K. J., Balasubramanian S., Sale J. E. (2012) FANCJ coordinates two pathways that maintain epigenetic stability at G-quadruplex DNA. Nucleic Acids Res., 40(4), 1485–1498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sarkies P., Reams C., Simpson L. J., Sale J. E. (2010) Epigenetic instability due to defective replication of structured DNA. Mol. Cell 40, 703–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Edmunds C. E., Simpson L. J., Sale J. E. (2008) PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell 30, 519–529 [DOI] [PubMed] [Google Scholar]
- 13. Guo C., Kosarek-Stancel J. N., Tang T. S., Friedberg E. C. (2009) Y-family DNA polymerases in mammalian cells. Cell. Mol. Life Sci. 66, 2363–2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yang W., Woodgate R. (2007) What a difference a decade makes. Insights into translesion DNA synthesis. Proc. Natl. Acad. Sci. U.S.A. 104, 15591–15598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Masutani C., Kusumoto R., Yamada A., Dohmae N., Yokoi M., Yuasa M., Araki M., Iwai S., Takio K., Hanaoka F. (1999) The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature 399, 700–704 [DOI] [PubMed] [Google Scholar]
- 16. Biertümpfel C., Zhao Y., Kondo Y., Ramón-Maiques S., Gregory M., Lee J. Y., Masutani C., Lehmann A. R., Hanaoka F., Yang W. (2010) Structure and mechanism of human DNA polymerase η. Nature 465, 1044–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Johnson R. E., Prakash S., Prakash L. (1999) Efficient bypass of a thymine-thymine dimer by yeast DNA polymerase, Polη. Science 283, 1001–1004 [DOI] [PubMed] [Google Scholar]
- 18. Silverstein T. D., Johnson R. E., Jain R., Prakash L., Prakash S., Aggarwal A. K. (2010) Structural basis for the suppression of skin cancers by DNA polymerase η. Nature 465, 1039–1043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Washington M. T., Johnson R. E., Prakash S., Prakash L. (2000) Accuracy of thymine-thymine dimer bypass by Saccharomyces cerevisiae DNA polymerase η. Proc. Natl. Acad. Sci. U.S.A. 97, 3094–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bienko M., Green C. M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A. R., Hofmann K., Dikic I. (2005) Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science 310, 1821–1824 [DOI] [PubMed] [Google Scholar]
- 21. Bienko M., Green C. M., Sabbioneda S., Crosetto N., Matic I., Hibbert R. G., Begovic T., Niimi A., Mann M., Lehmann A. R., Dikic I. (2010) Regulation of translesion synthesis DNA polymerase η by monoubiquitination. Mol. Cell 37, 396–407 [DOI] [PubMed] [Google Scholar]
- 22. Haracska L., Kondratick C. M., Unk I., Prakash S., Prakash L. (2001) Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell 8, 407–415 [DOI] [PubMed] [Google Scholar]
- 23. Haracska L., Unk I., Johnson R. E., Phillips B. B., Hurwitz J., Prakash L., Prakash S. (2002) Stimulation of DNA synthesis activity of human DNA polymerase κ by PCNA. Mol. Cell Biol. 22, 784–791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamath-Loeb A. S., Lan L., Nakajima S., Yasui A., Loeb L. A. (2007) Werner syndrome protein interacts functionally with translesion DNA polymerases. Proc. Natl. Acad. Sci. U.S.A. 104, 10394–10399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fygenson D. K., Goodman M. F. (1997) Appendix. Gel kinetic analysis of polymerase fidelity in the presence of multiple enzyme DNA encounters. J. Biol. Chem. 272, 27931–27935 [DOI] [PubMed] [Google Scholar]
- 26. Ali J. A., Lohman T. M. (1997) Kinetic measurement of the step size of DNA unwinding by Escherichia coli UvrD helicase. Science 275, 377–380 [DOI] [PubMed] [Google Scholar]
- 27. Lucius A. L., Maluf N. K., Fischer C. J., Lohman T. M. (2003) General methods for analysis of sequential “n-step” kinetic mechanisms. Application to single turnover kinetics of helicase-catalyzed DNA unwinding. Biophys. J. 85, 2224–2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bohr V. A., Cooper M., Orren D., Machwe A., Piotrowski J., Sommers J., Karmakar P., Brosh R. (2000) Werner syndrome protein. Biochemical properties and functional interactions. Exp. Gerontol. 35, 695–702 [DOI] [PubMed] [Google Scholar]
- 29. van der Kemp P. A., de Padula M., Burguiere-Slezak G., Ulrich H. D., Boiteux S. (2009) PCNA monoubiquitylation and DNA polymerase eta ubiquitin-binding domain are required to prevent 8-oxoguanine-induced mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 37, 2549–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Perry J. J., Yannone S. M., Holden L. G., Hitomi C., Asaithamby A., Han S., Cooper P. K., Chen D. J., Tainer J. A. (2006) WRN exonuclease structure and molecular mechanism imply an editing role in DNA end processing. Nat. Struct. Mol. Biol. 13, 414–422 [DOI] [PubMed] [Google Scholar]
- 31. Choudhary S., Sommers J. A., Brosh R. M., Jr. (2004) Biochemical and kinetic characterization of the DNA helicase and exonuclease activities of werner syndrome protein. J. Biol. Chem. 279, 34603–34613 [DOI] [PubMed] [Google Scholar]
- 32. Orren D. K., Brosh R. M., Jr., Nehlin J. O., Machwe A., Gray M. D., Bohr V. A. (1999) Enzymatic and DNA binding properties of purified WRN protein. High affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res. 27, 3557–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Deleted in proof
- 34. Tsai Y. C., Johnson K. A. (2006) A new paradigm for DNA polymerase specificity. Biochemistry 45, 9675–9687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ummat A., Silverstein T. D., Jain R., Buku A., Johnson R. E., Prakash L., Prakash S., Aggarwal A. K. (2012) Human DNA polymerase η is pre-aligned for dNTP binding and catalysis. J. Mol. Biol. 415, 627–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Washington M. T., Johnson R. E., Prakash L., Prakash S. (2003) The mechanism of nucleotide incorporation by human DNA polymerase η differs from that of the yeast enzyme. Mol. Cell Biol. 23, 8316–8322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Choi J. Y., Guengerich F. P. (2005) Adduct size limits efficient and error-free bypass across bulky N2-guanine DNA lesions by human DNA polymerase η. J. Mol. Biol. 352, 72–90 [DOI] [PubMed] [Google Scholar]
- 38. Lone S., Townson S. A., Uljon S. N., Johnson R. E., Brahma A., Nair D. T., Prakash S., Prakash L., Aggarwal A. K. (2007) Human DNA polymerase κ encircles DNA. Implications for mismatch extension and lesion bypass. Mol. Cell 25, 601–614 [DOI] [PubMed] [Google Scholar]
- 39. Xing G., Kirouac K., Shin Y. J., Bell S. D., Ling H. (2009) Structural insight into recruitment of translesion DNA polymerase Dpo4 to sliding clamp PCNA. Mol. Microbiol. 71, 678–691 [DOI] [PubMed] [Google Scholar]
- 40. Osborn A. J., Elledge S. J., Zou L. (2002) Checking on the fork. The DNA-replication stress-response pathway. Trends Cell Biol. 12, 509–516 [DOI] [PubMed] [Google Scholar]
- 41. Chang D. J., Cimprich K. A. (2009) DNA damage tolerance. When it's OK to make mistakes. Nat. Chem. Biol. 5, 82–90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kim S. H., Michael W. M. (2008) Regulated proteolysis of DNA polymerase η during the DNA-damage response in C. elegans. Mol. Cell 32, 757–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Garg P., Burgers P. M. (2005) Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases η and REV1. Proc. Natl. Acad. Sci. U.S.A. 102, 18361–18366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kannouche P. L., Wing J., Lehmann A. R. (2004) Interaction of human DNA polymerase η with monoubiquitinated PCNA. A possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 45. Yuasa M. S., Masutani C., Hirano A., Cohn M. A., Yamaizumi M., Nakatani Y., Hanaoka F. (2006) A human DNA polymerase eta complex containing Rad18, Rad6 and Rev1. Proteomic analysis and targeting of the complex to the chromatin-bound fraction of cells undergoing replication fork arrest. Genes Cells 11, 731–744 [DOI] [PubMed] [Google Scholar]
- 46. Rey L., Sidorova J. M., Puget N., Boudsocq F., Biard D. S., Monnat R. J., Jr., Cazaux C., Hoffmann J. S. (2009) Human DNA polymerase η is required for common fragile site stability during unperturbed DNA replication. Mol. Cell Biol. 29, 3344–3354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sidorova J. M., Li N., Folch A., Monnat R. J., Jr. (2008) The RecQ helicase WRN is required for normal replication fork progression after DNA damage or replication fork arrest. Cell Cycle 7, 796–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ammazzalorso F., Pirzio L. M., Bignami M., Franchitto A., Pichierri P. (2010) ATR and ATM differently regulate WRN to prevent DSBs at stalled replication forks and promote replication fork recovery. EMBO J. 29, 3156–3169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Machwe A., Karale R., Xu X., Liu Y., Orren D. K. (2011) The Werner and Bloom syndrome proteins help resolve replication blockage by converting (regressed) holliday junctions to functional replication forks. Biochemistry 50, 6774–6788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Phillips L. G., Sale J. E. (2010) The Werner's Syndrome protein collaborates with REV1 to promote replication fork progression on damaged DNA. DNA Repair 9, 1064–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kamath-Loeb A. S., Shen J. C., Schmitt M. W., Loeb L. A. (2012) The Werner syndrome exonuclease facilitates DNA degradation and high fidelity DNA polymerase by human DNA polymerase δ. J. Biol. Chem. 287, 12480–12490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Murfuni I., Nicolai S., Baldari S., Crescenzi M., Bignami M., Franchitto A., Pichierri P. (2012) The WRN and MUS81 proteins limit cell death and genome instability following oncogene activation. Oncogene, DOI: 10.1038/onc.2012.80 [DOI] [PubMed] [Google Scholar]
- 53. Cui S., Arosio D., Doherty K. M., Brosh R. M., Jr., Falaschi A., Vindigni A. (2004) Analysis of the unwinding activity of the dimeric RECQ1 helicase in the presence of human replication protein A. Nucleic Acids Res. 32, 2158–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Muzzolini L., Beuron F., Patwardhan A., Popuri V., Cui S., Niccolini B., Rappas M., Freemont P. S., Vindigni A. (2007) Different quaternary structures of human RECQ1 are associated with its dual enzymatic activity. PLoS Biol. 5, e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Compton S. A., Tolun G., Kamath-Loeb A. S., Loeb L. A., Griffith J. D. (2008) The Werner syndrome protein binds replication fork and holliday junction DNAs as an oligomer. J. Biol. Chem. 283, 24478–24483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Nakamura T., Zhao Y., Yamagata Y., Hua Y. J., Yang W. (2012) Watching DNA polymerase η make a phosphodiester bond. Nature 487, 196–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Beckman J. W., Wang Q., Guengerich F. P. (2008) Kinetic analysis of correct nucleotide insertion by a Y-family DNA polymerase reveals conformational changes both prior to and following phosphodiester bond formation as detected by tryptophan fluorescence. J. Biol. Chem. 283, 36711–36723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Eoff R. L., Sanchez-Ponce R., Guengerich F. P. (2009) Conformational changes during nucleotide selection by Sulfolobus solfataricus DNA polymerase Dpo4. J. Biol. Chem. 284, 21090–21099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Rucker R., Oelschlaeger P., Warshel A. (2010) A binding free energy decomposition approach for accurate calculations of the fidelity of DNA polymerases. Proteins 78, 671–680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vindigni A., Hickson I. D. (2009) RecQ helicases. Multiple structures for multiple functions? HFSP J. 3, 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. von Kobbe C., Karmakar P., Dawut L., Opresko P., Zeng X., Brosh R. M., Jr., Hickson I. D., Bohr V. A. (2002) Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem. 277, 22035–22044 [DOI] [PubMed] [Google Scholar]