Abstract
Engineered polyethylene glycol-maleimide matrices for regenerative medicine exhibit improved reaction efficiency and wider range of Young’s moduli by utilizing maleimide cross-linking chemistry. This hydrogel chemistry is advantageous for cell delivery due to the mild reaction that occurs rapidly enough for in situ delivery, while easily lending itself to “plug-and-play” design variations such as incorporation of enzyme-cleavable cross-links and cell-adhesion peptides.
Keywords: hydrogel, cell encapsulation, regenerative medicine, biomaterial, PEG, peptide, maleimide, Michael addition
Hydrogels, highly hydrated cross-linked polymer networks, have emerged as powerful synthetic analogs of extracellular matrices for basic cell studies as well as promising biomaterials for regenerative medicine applications.[1] A critical advantage of these artificial matrices over natural networks is that bioactive functionalities, such as cell adhesive sequences and growth factors, can be incorporated in precise densities while the substrate mechanical properties are independently controlled.[2–7] Polyethylene glycol (PEG) hydrogels represent the ‘gold standard’ in this field due to their intrinsic low-protein adsorption properties, minimal inflammatory profile and history of safe in vivo use, ease in incorporating various functionalities, and commercial availability of reagents. In the present study, we explored the use of an alternative reactive cross-linking moiety for PEG hydrogels, the maleimide functional group. We demonstrate several advantages over other cross-linking chemistries, namely stoichiometric hydrogels with improved cross-linking efficiency, bioligand incorporation and reaction time scales appropriate for clinical use for in situ gelation. The maleimide reactive group is extensively used in peptide bioconjugate chemistry because of its fast reaction kinetics and high specificity for thiols at physiological pH.[8] For these experiments, we used a 4-arm PEG-maleimide (PEG-4MAL) macromer and compared it to 4-arm PEG-acrylate (PEG-4A), 4-arm PEG-vinylsulfone (PEG-4VS), and UV photo-cross-linked PEG-DA.
Various cross-linking chemistries have been described to create bioactive hydrogel networks of PEG macromers, with Michael-type addition reactions and acrylate polymerization being the most widely utilized.[4] Cross-linking chemistry, gelation time, polymer network structure, and buffer conditions are important considerations when selecting a hydrogel cross-linking format for basic cell biology studies or regenerative medicine applications. In PEG-diacrylate (PEG-DA) hydrogels, PEG-DA macromers are cross-linked via free-radical initiated polymerization of acrylate end groups. Free radicals are created either by chemical activation or UV cleavage of a photoinitiator with the added ability to spatially control the presentation of incorporated ligands or mechanical properties through additive[9, 10] or subtractive[11] photo-patterning. A major drawback of free-radical cross-linking is that it can significantly reduce encapsulated cell viability and is unwieldy for in vivo delivery of hydrogels cross-linked in situ. In contrast, for hydrogels cross-linked by Michael-type addition, functionalized end groups on branched PEG macromers are reacted with bi-functional or branched cross-linking molecules. Michael-addition PEG hydrogels based on 4- or 8-arm PEG macromers with acrylate[12–14], vinyl-sulfone[15–19], and thiol[20–24] end-groups have been extensively investigated. Michael-type addition cross-linking avoids the use of cytotoxic free-radicals and UV light, but instead require a nucleophilic buffering reagent[25], such as triethanolamine (TEA) or HEPES[26], to facilitate the addition reaction. However, hydrogels formed in the presence of high concentrations of TEA have cytotoxic effects on sensitive cell types such as endothelial cells, and cells in ovarian follicles[27] and pancreatic islets.
PEG-DA, PEG-4MAL, and PEG-4A macromers (20 kDa, >95% end-group substitution) were obtained from Laysan Bio. PEG-4VS (20 kDa) was synthesized as previously reported.[28] Michael-type addition hydrogels incorporating cell adhesive ligands were formed in two steps. First, end-functionalized 20 kDa four-arm PEG macromers (PEG-4A, PEG-4MAL, PEG-4VS) were reacted with a thiol-containing adhesive peptide GRGDSPC in PBS with 4 mM or 400 mM TEA at pH 7.4 for 1 hour (Figure 1). Physical conjugation of the peptide to PEG-macromer was confirmed by increased molecular weight of fluorescein-labeled RGD visualized by SDS-PAGE (Figure S1). RGD-functionalized PEG macromers were subsequently cross-linked into a hydrogel by addition of the dithiol protease-cleavable peptide cross-linker GCRDVPMSMRGGDRCG[29, 30] at a 1:1 molar ratio of remaining PEG reactive end groups to peptide thiols. The protease-cleavable peptide cross-linker is necessary for cell encapsulation studies (see below).[28, 31] It has been reported that RGD concentrations ranging between 25 μM and 3.5 mM support 3D cell adhesion and spreading in PEG hydrogels.[29, 32–37] We used an RGD concentration of 2.0 mM to maximize adhesion sites while retaining cross-linking ability in tetra-functionalized PEG macromers. To determine the incorporation efficiency of RGD peptide and the conjugation efficiency of the different Michael-addition reagents, we measured unreacted/free thiols in the reaction buffer over time with the Measure-iT thiol quantification kit (Invitrogen). We observed rapid reaction of RGD with PEG-4MAL and nearly 100% incorporation at MAL:RGD molar ratios 1:1 and higher in both 4 mM and 400 mM TEA as early as 10 min (Figure 2A). Both PEG-4VS and PEG-4A exhibited poor RGD incorporation in 4 mM TEA. At 400 mM TEA, PEG-4VS showed complete RGD incorporation only at VS:peptide molar ratios 4:1 and higher and after 60 min incubation, whereas PEG-4A required a A:peptide molar ratio of 8:1 for complete RGD incorporation at 60 min. For all 4-arm PEG hydrogels incorporating 2.0 mM RGD, the reactive group to RGD molar ratio was 2.73:1 or greater. PEG-DA functionalization with RGD required a separate 1 hour reaction with acrylate-PEG-NHS with excess RGD peptide followed by purification and concentration of the product.
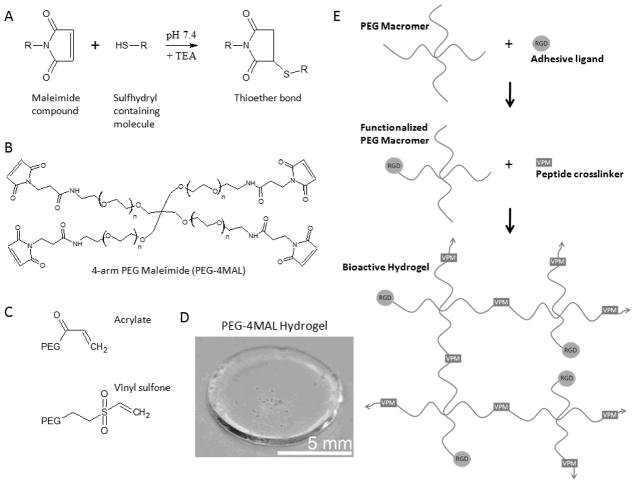
Figure 1.
PEG-maleimide hydrogel chemistry. A) Maleimide Michael-type addition reaction. B) 4-arm PEG Maleimide macromer. C) Acrylate and vinyl sulfone reactive groups. D) Sample cross-linked PEG-4MAL hydrogel. E) Michael-type addition hydrogel reaction scheme: PEG-macromers are first functionalized with RGD adhesive ligand followed by cross-linking with a thiol-flanked enzyme-degradable peptide.
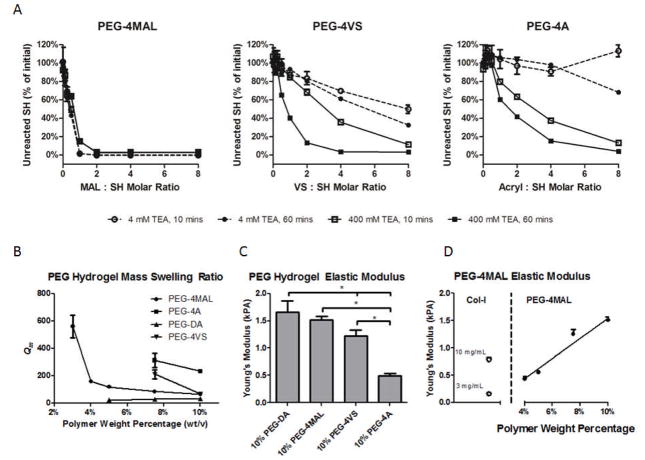
Figure 2.
Thiol reactivity and material properties comparisons between different end groups. A) Quantification of thiols remaining after 4-arm PEG macromer functionalization with RGDS peptide at 10 and 60 mins, 4 and 400 mM TEA in PBS with varying end-group to thiol molar ratio. B) Influence of polymer weight percentage on equilibrium swelling ratio (Qm) for networks made from PEG-4MAL, PEG-4A, PEG-4VS, or linear PEG-DA. C) Young’s modulus measured by AFM for 10% (wt/v) PEG gels. D) Young’s modulus measured by AFM for PEG-4MAL gels with varying polymer weight percentage compared with collagen-I gels.
The lower reaction efficiency of PEG-4VS and PEG-4A macromers is also reflected in the inability to form low weight percentage gels. Whereas PEG-4MAL forms gels as low as 3%, we observed lower polymer weight percentage limits of 7.5% for PEG-4A, 4% for PEG-4VS, and 7.5% for PEG-DA. Additionally, the time to gelation was significantly shorter in PEG-4MAL (~1–5 min) in 4 mM TEA compared to PEG-4VS (~ 30–60 min) or PEG-4A (~ 60 min) in 400 mM TEA and 10 minutes for PEG-DA under 10 mW/cm2 365 nm UV light. Fast gelation times are critically important for uniform distribution of encapsulated cells in 3D cultures and allow for in situ gelation of conformal gels in regenerative medicine applications. For the remaining studies, PEG-4MAL gels were formed in 4 mM TEA while PEG-4VS and PEG-4A were formed in 400 mM TEA.
Hydrogel swelling ratio is related to the average distance between cross-links by the modified Flory-Rehner equations as described by Peppas[38, 39] and is an indication of overall hydrogel network structure.[6, 16, 17, 40, 41] A higher mass swelling ratio indicates a more loosely cross-linked network. We measured the equilibrium mass swelling ratio for PEG-4MAL, PEG-4VS, PEG-4A, and PEG-DA gels containing 2.0 mM RGD at multiple polymer weight percentages (wt/v) (Figure 2B). For PEG-4MAL hydrogels, the equilibrium mass swelling ratio (Qm) was greater than 500 for 3.0% gels indicating a very loose network. Qm was dramatically lower in 4.0% PEG-4MAL gels (~150) with swelling decreasing only moderately in higher percent gels. For both PEG-4A and PEG-4VS gels, the swelling ratio was higher compared to PEG-4MAL and decreased as polymer weight percentage increased from 7.5% to 10.0%. PEG-4A swelling remained 2–3 fold higher than PEG-4MAL swelling between 7.5% and 10%. PEG-4VS swelling was 2-fold higher than PEG-4MAL swelling at 7.5% but the swelling ratio was identical at 10.0%. The PEG-4MAL swelling curve features an inflection that suggests a transition from a non-ideal, high-swelling network at 3.0% to a more robust, moderate-swelling network (Qm <200) at polymer weight percentage greater than or equal to 4.0%. In contrast, PEG-4VS network swelling ratio remains above this threshold until the polymer weight percentage exceeds 7.5%. PEG-4A gels remain in a high-swelling regime even at 10%. PEG-DA had a low mass swelling ratio for all polymer weight percentages. This low-swelling ratio results from the acrylate polymerization cross-linking reaction which generates in a highly entangled and cross-linked network structure. These mass swelling ratio measurements suggest that PEG-4MAL gels offer superior network-forming characteristics compared to PEG-4VS and PEG-4A. The very low mass swelling ratio of the PEG-DA gel suggests that degradation by host-tissue or encapsulated cells would be more difficult due to denser bulk properties.
We next measured hydrogel Young’s modulus using atomic force microscopy indentation testing. PEG hydrogel samples with 2 mM RGD were prepared and allowed to swell overnight in H2O prior to mechanical testing. Measurements for hydrogels below 10% (wt/v) for PEG-DA, PEG-4A, and PEG-4VS or below 4% (wt/v) PEG-4MAL were not obtained as the material was too viscous for the AFM probe to accurately measure. Comparison among the Michael-type addition hydrogels of the same polymer weight percentage indicated the highest modulus for PEG-4MAL (Figure 2C), consistent with a more fully cross-linked network. Young’s modulus measurement for a variety of polymer weight percentages of PEG-4MAL (Figure 2D) varied linearly with polymer weight percentage (R2 = 0.96). Importantly, because PEG-4MAL was able to form robust networks at low polymer weight percentages, mechanical properties in the range of natural collagen gels used for 3D cell culture and in vivo delivery was possible with 4% and 5% PEG-4MAL gels while other higher percentage gels were much stiffer.
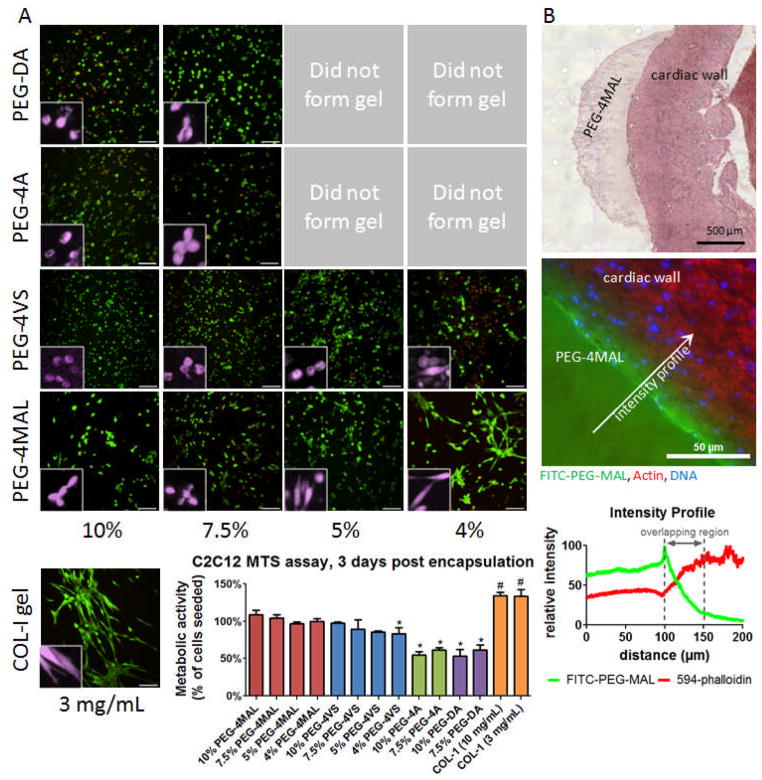
To examine the ability of these hydrogels to support cellular activities, murine C2C12 myoblast cells were encapsulated in 3 mg/mL collagen-I and 10%, 7.5%, 5%, 4%, and 3% (wt/v) PEG hydrogels (50 μL) incorporating 2.0 mM RGD peptide at 3 × 106 cells/mL and cultured for 3 days followed by Live/Dead staining (Invitrogen) (Figure 3A). Due to differential swelling ratios and gelation times (cells settling out of gel to bottom of well), post-encapsulation cell density varied among groups. PEG-4A and PEG-DA with cells did not form gels at polymer weight percentages below 7.5%. Three percent PEG-4MAL gels containing cells dissipated before 3 days due to cell-mediated proteolysis and were not imaged. A large fraction of cells in all conditions stained positive for viability. More dead cells were visible in 4% PEG-4VS gels and in 10% PEG-DA gels than other conditions. Cell spreading was the highest in 4% PEG-4MAL and was the most comparable to the natural collagen matrix. Encapsulated cells were also assayed for metabolic activity at 3 days by MTS assay. C2C12 cells encapsulated in PEG-4MAL and PEG-4VS had metabolic activities similar to controls consisting of samples with same number of cells seeded at day 0. Collagen gels had metabolic activity significantly higher than the initial seeding density indicating signs of cell proliferation. 4% PEG-4VS, PEG-DA, PEG-4A, gels had significantly lower metabolic activities compared to controls, indicating cell loss attributed to cyto-toxicity. Notably metabolic activity was only 60% of the control in PEG-DA and PEG-4A gels. Exposure to high TEA concentrations, free radicals, and UV light could be responsible for the lower metabolic activity/viability. We observed that high TEA concentrations (400 mM) alone had negative effects on sensitive cells types such as endothelial cells (HUVEC) assayed by MTS assay (Figure S2).
Figure 3.
Cellular encapsulation and in situ polymerization of PEG-4MAL hydrogel. A) Live/Dead staining of C2C12 murine myoblasts at 3 days after encapsulation in PEG hydrogels of varying polymer weight percentage compared to collagen-I gel (3 mg/mL). Cross-linked hydrogels could not be generated for low percentage PEG-4A and PEG-DA gels. Scale bar = 100 μm. Inset false color higher magnification showing individual cell spreading. MTS metabolic activity assay of encapsulated C2C12 cells indicates viability similar to number of cells seeded for PEG-4MAL and PEG-4VS, with PEG-4A and PEG-DA approximately 60%. B) H&E stain of PEG-4MAL matrix cross-linked directly on mouse myocardial wall. PEG-4MAL matrix incorporating 1% polymer substitution FITC-PEG-MAL cross-linked directly on mouse myocardial wall, counterstained with Alexa-Fluor 594 phalloidin and DAPI. Fluorescence intensity profiles for FITC-PEG-MAL and 594-phalloidin illustrate a physical incorporation depth of hydrogel into tissue of approximately 50 μm.
Lastly, we examined the potential for in situ application of PEG-4MAL as a myocardial surface patch. A 5% PEG-4MAL precursor solution with addition of labeled FITC-PEG-MAL for visualization was mixed and pipetted directly onto the pericardium of an excised rat heart. The hydrogel formed rapidly and was observed to bond with the contacting tissue. Hematoxylin and eosin staining showed a continuous interfacial surface between the hydrogel and the myocardial wall (Figure 3B). Fluorescence microscopy of the hydrogel-tissue interface revealed excellent hydrogel incorporation into the tissue with a penetration depth of approximately 50 μm. The FITC signal penetration was not due to diffuse or unbonded FITC-PEG-MAL which would have washed out during the multiple wash steps of the tissue fixation, processing, and staining procedures.
The PEG-4MAL, PEG-4VS, and PEG-4A 4-arm macromers are identical in structure aside from the reactive end groups and should form similar networks if all the available reactive groups are able to combine with the dithiol cross-linker at 100% efficiency. However, the considerable differences in RGD incorporation, swelling behavior, gelation time, Young’s modulus, and cell viability observed among the different Michael-type addition reactive groups indicates that hydrogel cross-linking efficiency and gelation are markedly different among the reactive macromers. Taken together, our results indicate that PEG-4MAL exhibits faster reaction kinetics and tighter network structure than PEG-4A or PEG-4VS. Additionally, we found that the PEG-4MAL cross-linking reaction requires two orders of magnitude less TEA than either PEG-4A or PEG-4VS. Furthermore, we were able to create hydrogels of lower polymer weight-percentage and with a wider range of Young’s moduli than gels based on PEG-DA, PEG-4A, or PEG-4VS. Importantly, low polymer weight percentage PEG-4MAL gels could be formed with mechanical properties that ranged in the low modulus environment of naturally-derived extracellular matrices such as type I collagen. These lower polymer weight percentage PEG-4MAL networks promoted increased spreading of encapsulated cells which could not be recapitulated in the other macromers. Many published articles report gelation times on the order of 15–60 minutes for Michael-addition cross-linking [13, 26, 42, 43] which is unwieldy for in situ clinical application where the gel must set up quickly and not flow from the administration site or be diluted with fluid. We found that PEG-4MAL hydrogels had significantly faster cross-linking times of 1–5 minutes depending on the weight percentage and hold strong potential for clinical use with in situ gelation. These results establish PEG-4MAL hydrogels with improved cross-linking efficiency, bioligand incorporation, encapsulated cell viability and reaction time scales appropriate for in situ gelation as versatile synthetic analogs of extracellular matrices for basic cell studies and regenerative medicine applications.
Experimental
Measure-iT Thiol Assay
Serial dilutions of end-group functionalized PEG macromers were added to a standard concentration of 10 mM GRGDSPC in 4 or 400 mM TEA in PBS. At specified time points, the reaction was quenched by 1:100 dilution in water. PEG-RGD (100 μL) plus thiol-quantitation reagent (10 μL) was added per well of a black 96-well plate and read using a microplate reader. Dilutions of RGD in TEA (4 or 400 mM) in PBS were used as standards. All samples were measured in triplicate.
PEG macromer synthesis
For PEG-4VS, 4-arm PEG (Sunbright PTE-20000, MW=19858; NEKTAR Therapeutics, San Carlos, CA) was functionalized at the OH-termini with divinyl sulfone (Sigma-Aldrich) as described previously.[28, 42] In brief, PEG-4VS was synthesized by reacting a dichloromethane solution of the PEG-OH (previously dried over molecular sieves) with NaH under argon gas and then, after hydrogen evolution, with diVS (molar ratios: OH 1/NaH 5/diVS 50), at room temperature for 3 days, under argon with stirring. The resulting solution was neutralized with acetic acid and filtered through filter paper until clear. VS-functionalized PEG was then precipitated in ice-cold diethyl ether, washed, and re-dissolved in dichloromethane; this cycle was repeated twice to remove all excess diVS, and the PEG-4VS was finally dried under vacuum. The success of VS conversion on the OH-termini was confirmed by NMR spectroscopy to be 90–95% as previously described.[28]
For PEG-DA hydrogels, the protease degradable peptide cross-linker GCRDVPMSMRGGDRCG (VPM) was incorporated into the backbone of the PEG-DA macromer as previously reported.[31] PEG-diacrylate (3400 MW, Laysan Bio) was reacted with the VPM peptide at a 2:1 PEG to peptide molar ratio in PBS + TEA (400 mM) for 6 hours to create the macromer acrylate-PEG-VPM-PEG-acrylate. The reaction product was dialyzed three times against di-H2O and lyophilized for storage. The cell adhesion peptide GRGDSP was similarly conjugated to PEG-acrylate with the amine-reactive molecule acrylate-PEG-NHS (Laysan Bio) in sodium biocarbonate (50 mM) at 1:2 molar ratio for 6 hours followed by dialysis against di-H2O and lyophilization. PEG-peptide conjugates were confirmed by molecular weight increases of products vs. reactants on SDS-PAGE.
PEG Hydrogel Formation
PEG-DA gels were formed by adding acrylate-PEG-RGD (2 mM) with various polymer weight percentage solutions of acrylate-PEG-VPM-PEG-acrylate in PBS + 0.05% Irgacure 2959 (Ciba) photoinitiator and exposure to UV light (10 mW cm−2) for 10 minutes. Michael-type addition PEG hydrogels (PEG-4A, PEG-4VS, PEG-4MAL) were formed by reacting 4-arm functionalized PEG-macromer with the cell-adhesion peptide GRGDSPC followed by cross-linking with the protease degradable peptide VPM at stoichiometrically balanced 1:1 cysteine to remaining reactive group molar ratio. For most experiments, PEG-4A and PEG-4VS were reacted in PBS + TEA (400 mM, pH 7.4), whereas PEG-4MAL was reacted in PBS + TEA (4 mM, pH 7.4).
Mass Swelling Ratio
Six hydrogels per condition were formed in 9 mm diameter × 1 mm deep silicone isolator wells sandwiched between two coverslips coated in Sigmacote® (Sigma- Aldrich). After cross-linking, hydrogels were allowed to freely swell in di-H2O for 24 hours and the swollen hydrogel mass measured. Hydrogels were then snap-frozen in liquid N2 and lyophilized followed by dry mass measurement. The mass swelling ratio is reported as the ratio of swollen mass to dry mass.
AFM Modulus Testing
Using an MFP-3D-BIO atomic force microscope (Asylum Research; Santa Barbara, CA), samples were probed under fluid conditions (ultrapure-H2O) using a pyramidal tipped-silicon nitride cantilever (Bruker, Camarillo, CA). Cantilever spring constants were measured prior to sample analysis using the thermal fluctuation method,[44] with nominal values of 20–30 mN/m. The force-indentation curve was obtained for each measurement and then analyzed with a Hertzian model for a pyramidal tip (Wavemetrics, IgorPro software routines) from which the Young’s modulus values were calculated. The sample Poisson’s ratio was assumed as 0.33, and a power law of 2.0 for the sample indentation distance was used to model tip geometry. All AFM measurements were made using a cantilever deflection set point of 100 nm, and the rate of indentation was 22.86 μm/s. 100 nm was chosen as the cantilever deflection set point for mechanical testing, as this corresponds to loading forces of approximately 5–10 nN. This scale of probe conditions is within the range of the traction forces applied through single adhesions to the ECM, and is thus a relevant range for mechanical testing.[45, 46] Furthermore, similar testing forces have been recently published for PEG-containing hydrogels.[47] A minimum of 9 independent measurements was obtained and analyzed for each sample condition.
Cell Encapsulation
Murine C2C12 myoblast cells (ATCC) were encapsulated in collagen-I (3 mg/mL) or PEG matrices (10%, 7.5%, 5%, and 4% (wt/v)) with RGD (2 mM) at 3 × 106 cells/mL and cultured in DMEM + 20% fetal bovine serum + 1% penicillin/streptomycin. PEG-4A and PEG-4VS were cross-linked with VPM peptide for 1 hour in TEA (400 mM). PEG-4MAL was cross-linked with VPM peptide for 10 minutes in 4 mM TEA. PEG-DA was cross-linked by addition of 0.05% Irgacure 2959 and 10 min exposure to a UV lamp (10 mW cm−2). At 3 days post-encapsulation, culture media was replaced with PBS containing calcein AM (2 μM) and ethidium homodimer-1 (4 μM) for Live/Dead staining. Hydrogels were incubated in Live/Dead stain for 30 minutes and visualized on a Nikon-C1 laser scanning confocal microscope with a 20X air objective. Z-stack projections through a 100 μm thick section of the swollen hydrogel were rendered. For MTS measurements, gels containing cells were degraded with 1 mg/mL collagenase 1 in PBS to eliminate differences due to diffusivity in the gels followed by incubation for 4 hours with MTS reagent (Promega). For direct effects of TEA on cell viability, human umbilical vein endothelial cells (HUVEC, Lonza) were suspended in PBS containing TEA (0, 4, 40, or 400 mM, pH 7.4) for one hour. The cells were subsequently examined for differences in metabolism by MTS assay (Promega).
Myocardial Patch
Fifty microliters of 5% PEG-4MAL with RGD (2 mM) hydrogel and 1% dry weight substation of labeled FITC-PEG-MAL for visualization in TEA (4 mM) was mixed and pipetted directly onto the pericardium of a freshly excised rat heart. The tissue was fixed and processed for preservation in Immuno-Bed (Polysciences) plastic resin, sectioned at 2 μm, and counterstained with Alexa-Fluor 594-phalloidin (Invitrogen) and DAPI.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health Grants R01-EB004496 and R01-EB011566; Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services, under Contract No. HHSN268201000043C; National Science Foundation under the Science and Technology Center Emergent Behaviors of Integrated Cellular Systems (EBICS) Grant No. CBET-0939511; Georgia Tech/Emory Center for the Engineering of Living Tissues and the Atlanta Clinical and Translational Science Institute; the Center for Pediatric Healthcare Technology Innovation at Georgia Tech and Children’s Hospital of Atlanta; an American Heart Association Predoctoral Fellowship (E.A.P.); a NASA-Jenkins Predoctoral Fellowship (N.O.E.), and National Science Foundation Graduate Research Fellowships (V.F.F., J.C.S.)
Footnotes
Supporting Information is available online from Wiley InterScience or from the author
Contributor Information
Dr. Edward A. Phelps, Woodruff School of Mechanical Engineering, Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, GA 30332-0363, USA
Mr. Nduka O. Enemchukwu, Woodruff School of Mechanical Engineering, Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, GA 30332-0363, USA
Mr. Vincent F. Fiore, Coulter Department of Biomedical Engineering, Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, GA 30332-0363, USA
Dr. Jay C. Sy, Coulter Department of Biomedical Engineering, Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, GA 30332-0363, USA
Prof. Niren Murthy, Coulter Department of Biomedical Engineering, Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, GA 30332-0363, USA
Prof. Todd A. Sulchek, Woodruff School of Mechanical Engineering, Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, GA 30332-0363, USA
Prof. Thomas H. Barker, Coulter Department of Biomedical Engineering, Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, GA 30332-0363, USA
Prof. Andrés J. García, Email: andres.garcia@me.gatech.edu, Woodruff School of Mechanical Engineering, Petit Institute for Bioengineering and Bioscience, Georgia Institute of Technology, Atlanta, GA 30332-0363, USA.
References
- 1.Lutolf MP, Hubbell JA. Nature Biotechnology. 2005;23:47. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 2.Lutolf MP, Lauer-Fields JL, Schmoekel HG, Metters AT, Weber FE, Fields GB, Hubbell JA. Proc Natl Acad Sci U S A. 2003;100:5413. doi: 10.1073/pnas.0737381100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kloxin AM, Kloxin CJ, Bowman CN, Anseth KS. Advanced Materials. 2010;22:3484. doi: 10.1002/adma.200904179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhu J. Biomaterials. 2010;31:4639. doi: 10.1016/j.biomaterials.2010.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hou Y, Schoener CA, Regan KR, Munoz-Pinto D, Hahn MS, Grunlan MA. Biomacromolecules. 2010;11:648. doi: 10.1021/bm9012293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Munoz-Pinto DJ, Bulick AS, Hahn MS. Journal of biomedical materials research Part A. 2009;90:303. doi: 10.1002/jbm.a.32492. [DOI] [PubMed] [Google Scholar]
- 7.Zieris A, Prokoph S, Levental KR, Welzel PB, Grimmer M, Freudenberg U, Werner C. Biomaterials. 2010;31:7985. doi: 10.1016/j.biomaterials.2010.07.021. [DOI] [PubMed] [Google Scholar]
- 8.Hermanson GT. Bioconjugate techniques. Academic Press; San Diego: 1996. [Google Scholar]
- 9.Hahn MS, Miller JS, West JL. Advanced Materials. 2006;18:2679. [Google Scholar]
- 10.Nemir S, Hayenga HN, West JL. Biotechnol Bioeng. 2010;105:636. doi: 10.1002/bit.22574. [DOI] [PubMed] [Google Scholar]
- 11.Kloxin AM, Tibbitt MW, Anseth KS. Nat Protoc. 2010;5:1867. doi: 10.1038/nprot.2010.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rizzi SC, Hubbell JA. Biomacromolecules. 2005;6:1226. doi: 10.1021/bm049614c. [DOI] [PubMed] [Google Scholar]
- 13.Rizzi SC, Ehrbar M, Halstenberg S, Raeber GP, Schmoekel HG, Hagenmuller H, Muller R, Weber FE, Hubbell JA. Biomacromolecules. 2006;7:3019. doi: 10.1021/bm060504a. [DOI] [PubMed] [Google Scholar]
- 14.Elbert DL, Hubbell JA. Biomacromolecules. 2001;2:430. doi: 10.1021/bm0056299. [DOI] [PubMed] [Google Scholar]
- 15.Fu Y, Kao WJ. Journal of biomedical materials research Part A. 2011 doi: 10.1002/jbm.a.33106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Metters A, Hubbell J. Biomacromolecules. 2005;6:290. doi: 10.1021/bm049607o. [DOI] [PubMed] [Google Scholar]
- 17.Lutolf MP, Hubbell JA. Biomacromolecules. 2003;4:713. doi: 10.1021/bm025744e. [DOI] [PubMed] [Google Scholar]
- 18.Seliktar D, Zisch AH, Lutolf MP, Wrana JL, Hubbell JA. J Biomed Mater Res A. 2004;68:704. doi: 10.1002/jbm.a.20091. [DOI] [PubMed] [Google Scholar]
- 19.Kim J, Wacker BK, Elbert DL. Biomacromolecules. 2007;8:3682. doi: 10.1021/bm700756z. [DOI] [PubMed] [Google Scholar]
- 20.Hiemstra C, van der Aa LJ, Zhong Z, Dijkstra PJ, Feijen J. Macromolecules. 2007;40:1165. [Google Scholar]
- 21.Yu H, Feng Z-g, Zhang A-y, Sun L-g, Qian L. Soft Matter. 2006;2:343. doi: 10.1039/b517206c. [DOI] [PubMed] [Google Scholar]
- 22.Hiemstra C, van der Aa LJ, Zhong Z, Dijkstra PJ, Feijen J. Biomacromolecules. 2007;8:1548. doi: 10.1021/bm061191m. [DOI] [PubMed] [Google Scholar]
- 23.Hu BH, Su J, Messersmith PB. Biomacromolecules. 2009;10:2194. doi: 10.1021/bm900366e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Su J, Hu B-H, Lowe WL, Jr, Kaufman DB, Messersmith PB. Biomaterials. 2010;31:308. doi: 10.1016/j.biomaterials.2009.09.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mather BD, Viswanathan K, Miller KM, Long TE. Progress in Polymer Science. 2006;31:487. [Google Scholar]
- 26.Pratt AB, Weber FE, Schmoekel HG, Muller R, Hubbell JA. Biotechnology and bioengineering. 2004;86:27. doi: 10.1002/bit.10897. [DOI] [PubMed] [Google Scholar]
- 27.Shikanov A, Smith RM, Xu M, Woodruff TK, Shea LD. Biomaterials. 2011;32:2524. doi: 10.1016/j.biomaterials.2010.12.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chung IM, Enemchukwu NO, Khaja SD, Murthy N, Mantalaris A, Garcia AJ. Biomaterials. 2008;29:2637. doi: 10.1016/j.biomaterials.2008.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bott K, Upton Z, Schrobback K, Ehrbar M, Hubbell JA, Lutolf MP, Rizzi SC. Biomaterials. 2010;31:8454. doi: 10.1016/j.biomaterials.2010.07.046. [DOI] [PubMed] [Google Scholar]
- 30.Patterson J, Hubbell JA. Biomaterials. 2010;31:7836. doi: 10.1016/j.biomaterials.2010.06.061. [DOI] [PubMed] [Google Scholar]
- 31.Phelps EA, Landazuri N, Thule PM, Taylor WR, Garcia AJ. Proc Natl Acad Sci U S A. 2010;107:3323. doi: 10.1073/pnas.0905447107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moon JJ, Saik JE, Poche RA, Leslie-Barbick JE, Lee SH, Smith AA, Dickinson ME, West JL. Biomaterials. 2010;31:3840. doi: 10.1016/j.biomaterials.2010.01.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zisch AH, Lutolf MP, Ehrbar M, Raeber GP, Rizzi SC, Davies N, Schmokel H, Bezuidenhout D, Djonov V, Zilla P, Hubbell JA. The FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2003;17:2260. doi: 10.1096/fj.02-1041fje. [DOI] [PubMed] [Google Scholar]
- 34.Raeber GP, Lutolf MP, Hubbell JA. Biophys J. 2005;89:1374. doi: 10.1529/biophysj.104.050682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Adelow C, Segura T, Hubbell JA, Frey P. Biomaterials. 2008;29:314. doi: 10.1016/j.biomaterials.2007.09.036. [DOI] [PubMed] [Google Scholar]
- 36.Lee SH, Miller JS, Moon JJ, West JL. Biotechnology Progress. 2005;21:1736. doi: 10.1021/bp0502429. [DOI] [PubMed] [Google Scholar]
- 37.Wacker BK, Alford SK, Scott EA, Das Thakur M, Longmore GD, Elbert DL. Biophysical journal. 2008;94:273. doi: 10.1529/biophysj.107.109074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Peppas NA, Huang Y, Torres-Lugo M, Ward JH, Zhang J. Annual Review of Biomedical Engineering. 2000;2:9. doi: 10.1146/annurev.bioeng.2.1.9. [DOI] [PubMed] [Google Scholar]
- 39.Canal T, Peppas NA. Journal of Biomedical Materials Research. 1989;23:1183. doi: 10.1002/jbm.820231007. [DOI] [PubMed] [Google Scholar]
- 40.Lu SX, Anseth KS. Macromolecules. 2000;33:2509. [Google Scholar]
- 41.Cruise GM, Scharp DS, Hubbell JA. Biomaterials. 1998;19:1287. doi: 10.1016/s0142-9612(98)00025-8. [DOI] [PubMed] [Google Scholar]
- 42.Lutolf MP, Raeber GP, Zisch AH, Tirelli N, Hubbell JA. Advanced Materials. 2003;15:888. [Google Scholar]
- 43.Patterson J, Hubbell JA. Biomaterials. 2011;32:1301. doi: 10.1016/j.biomaterials.2010.10.016. [DOI] [PubMed] [Google Scholar]
- 44.Lévy R, Maaloum M. Nanotechnology. 2002;13:33. [Google Scholar]
- 45.Sabass B, Gardel ML, Waterman CM, Schwarz US. Biophysical Journal. 2008;94:207. doi: 10.1529/biophysj.107.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tan JL. Proceedings of the National Academy of Sciences. 2003;100:1484. [Google Scholar]
- 47.Ouasti S, Donno R, Cellesi F, Sherratt MJ, Terenghi G, Tirelli N. Biomaterials. 2011;32:6456. doi: 10.1016/j.biomaterials.2011.05.044. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.