A third isoenzyme of Glu dehydrogenase (GDH) is expressed in mitochondria of Arabidopsis root companion cells. A GDH triple mutant differed greatly from the wild type in continuous darkness, suggesting that the main function of the enzyme is to provide 2-oxoglutarate for the tricarboxylic acid cycle, leading to an accumulation of Ala, γ-aminobutyrate, and Asp in both roots and leaves.
Abstract
The role of NADH-dependent glutamate dehydrogenase (GDH) was investigated by studying the physiological impact of a complete lack of enzyme activity in an Arabidopsis thaliana plant deficient in three genes encoding the enzyme. This study was conducted following the discovery that a third GDH gene is expressed in the mitochondria of the root companion cells, where all three active GDH enzyme proteins were shown to be present. A gdh1-2-3 triple mutant was constructed and exhibited major differences from the wild type in gene transcription and metabolite concentrations, and these differences appeared to originate in the roots. By placing the gdh triple mutant under continuous darkness for several days and comparing it to the wild type, the evidence strongly suggested that the main physiological function of NADH-GDH is to provide 2-oxoglutarate for the tricarboxylic acid cycle. The differences in key metabolites of the tricarboxylic acid cycle in the triple mutant versus the wild type indicated that, through metabolic processes operating mainly in roots, there was a strong impact on amino acid accumulation, in particular alanine, γ-aminobutyrate, and aspartate in both roots and leaves. These results are discussed in relation to the possible signaling and physiological functions of the enzyme at the interface of carbon and nitrogen metabolism.
INTRODUCTION
Although the function and regulation of Glu dehydrogenase (GDH) is well established in bacteria (Atkinson, 1969), yeast (Magasanik, 1992), animals (Frigerio et al., 2008), and ectomycorrhizal fungi (Morel et al., 2006), further research is still required to fully elucidate the physiological role of the NADH-dependent enzyme in plants (Dubois et al., 2003; Tercé-Laforgue et al., 2004a; Jaspard, 2006). Although it is widely accepted that over 95% of the ammonia available to higher plants is assimilated via the Gln synthetase/Glu synthase GS/GOGAT pathway (Lea and Miflin, 2011), some have argued that NADH-GDH could participate to a certain extent in ammonia assimilation. Such a reaction could occur when the ammonium ion concentration in the cell is considerably increased, either following external application (Yamaya and Oaks, 1987; Oaks, 1995; Melo-Oliveira et al., 1996; Skopelitis et al., 2007) or as the result of metabolic perturbations caused for example by salt stress (Skopelitis et al., 2006) or hypoxia (Limami et al., 2008). However, the majority of metabolic studies performed in vivo have clearly demonstrated that in higher plants, GDH operates in the direction of Glu deamination (Robinson et al., 1992; Fox et al., 1995; Stewart et al., 1995; Glevarec et al., 2004; Masclaux-Daubresse et al., 2006; Labboun et al., 2009).
Nevertheless, it is still unclear whether NADH-GDH provides C skeletons and reducing equivalents to the cell when there is a shortage in carbohydrates or an excess of N (Labboun et al., 2009). There is strong evidence that the NADH-GDH enzyme protein is mainly, if not exclusively, localized in the mitochondria of the phloem companion cells (Dubois et al., 2003; Fontaine et al., 2006). However, under certain physiological conditions when ammonium is present in high concentrations, a significant proportion of the protein has been detected in the cytosol of these cells (Tercé-Laforgue et al., 2004b). This finding led the authors to suggest that the presence of the enzyme in such a specialized tissue and cellular compartment has an important physiological meaning in terms of metabolic signaling in relation to the partitioning of C and N assimilates. It is therefore likely that the NADH-GDH enzyme, in conjunction with NADH-dependent Glu synthase, contributes to the control of the homeostasis of leaf Glu, an amino acid that plays a central signaling and metabolic role at the interface of the C and N assimilatory pathways (Forde and Lea, 2007; Labboun et al., 2009).
These findings are in line with the results published by Miyashita and Good (2008) and others (Melo-Oliveira et al., 1996), showing by means of GDH-deficient mutants of Arabidopsis thaliana that the enzyme serves as a major link between carbohydrate and amino acid metabolism. In particular, when these mutants were placed under continuous darkness, they exhibited an increase in sensitivity to C deficiency, accompanied by a modification of the amino acid profile, which probably lead to plant growth retardation, mimicking C starvation (Miyashita and Good, 2008).
The majority of recent studies performed on NADH-GDH in higher plants have focused on deciphering the role of the α- and β-subunits in the formation of seven isoenzymes (Thurman et al., 1965), which are encoded by two distinct nuclear genes, GDH2 and GDH1, respectively (Melo-Oliveira et al., 1996; Purnell et al., 2005; Miyashita and Good, 2008). More recently, it has been found that in Arabidopsis there is a third gene (GDH3) encoding a putative NADH-GDH that is actively transcribed and perhaps regulated by cytokinin (Yamada et al., 2003; Igarashi et al., 2009). However, there was no evidence that transcripts for this third gene were translated into active protein, although the gene was found to be expressed in the vasculature of root cells of control plants and in the rosette leaves in the presence of cytokinin. Similarly in rice (Oryza sativa), three genes encoding NADH-GDH are differentially expressed in various organs depending on N availability (Qiu et al., 2009), suggesting that the physiological functions of the GDH isoenzymes are more complex than originally described and may vary from one species to an other. The presence of a third active GDH isoenzyme preferentially expressed in the roots of dark-grown soybean (Glycine max) seedlings and composed of a single polypeptide is consistent with such a hypothesis (Turano et al., 1996).
A NADP(H)-dependent form of GDH (NADPH-GDH) activity has been well characterized in green algae (Jaspard, 2006). Low activities of NADPH-GDH have been demonstrated in a range of higher plants (Lea and Thurman, 1972; Turano et al., 1996; Grabowska et al., 2011), including Arabidopsis (Cammaerts and Jacobs, 1985; Miyashita and Good, 2008). However, since NADPH-GDH has been shown to be localized in the chloroplast, the role of the NADPH-dependent form is not clear (Leech and Kirk, 1968; Lea and Thurman, 1972). A fourth expressed gene, encoding a putative NADPH-GDH, has been identified in Arabidopsis (Yamada et al., 2003; Igarashi et al., 2009) and rice (Qiu et al., 2009). The predicted peptide of the NADPH-GDH enzyme encoded by the GDH4 gene is 50% longer than the NADH-GDH form (see Supplemental Figure 1 online). In rice, NADPH-GDH does not appear to have a specific NADP(H) binding site (Qiu et al., 2009), which provides an additional argument against the possible physiological function of a fourth GDH enzyme in vivo.
To further investigate the function of GDH in higher plants, we isolated and characterized mutants of Arabidopsis deficient in the expression of the GDH3 gene encoding NADH-GDH, and we demonstrated that this third gene encodes an active enzyme. In order to obtain plants totally impaired for NADH-GDH activity, we produced a triple mutant (gdh1-2-3) deficient in the expression of the three genes encoding the enzyme. We then conducted a detailed molecular, biochemical, and physiological characterization of the gdh triple mutant in which NADH-GDH activity was totally depleted, both in the roots and shoots.
RESULTS
Identification of a Third Active GDH Isoenzyme
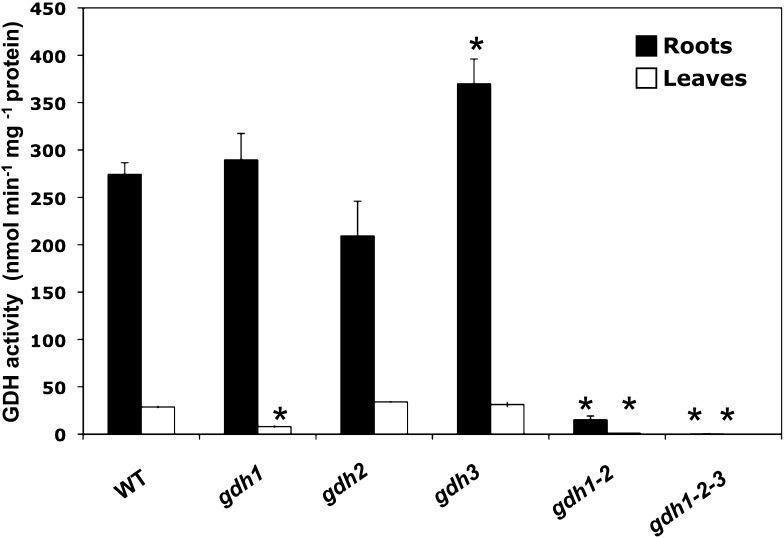
A sequence comparison of GDH1 and GDH2, the two genes encoding the subunits of NADH-GDH previously identified in the whole Arabidopsis genome (http://www.Arabidopsis.org/), uncovered the presence of a third gene (GDH3) exhibiting significant sequence similarity, a strong indicator of homology with the two genes (see Supplemental Figure 1 online). In order to determine if this third gene produced an active enzyme exhibiting NADH-GDH activity, a series of experiments were performed after the selection and production of backcrossed homozygous gdh1, gdh2, gdh3, gdh1-2, and gdh1-2-3 mutants. GDH aminating (NADH-dependent) activity was measured in the roots and leaves of the wild type and in the gdh1, 2, and 3 single mutants, gdh1-2 double mutant, and gdh1-2-3 triple mutant. In the wild type, the specific activity of the NADH-GDH enzyme was over 10 times higher in the roots compared with that measured in the leaves (Figure 1) and was similar to that measured in the roots of the gdh1 single mutant. NADH-GDH activity in the roots of the gdh2 mutant was ∼25% lower than the wild type, whereas in the roots of the gdh3 mutant, the enzyme activity was 30% higher. In the leaves, there was a 60% reduction in NADH-GDH activity in the gdh1 single mutant but not in the other two single mutants. By contrast, both in the roots and in the leaves of the gdh1-2 double mutant, a dramatic decrease in NADH-GDH activity was observed. Nevertheless, in the gdh1-2 double mutant, some remaining enzyme activity was still detected in the roots. No NADH-GDH enzyme activity was detected in either of the organs of the gdh1-2-3 triple mutant (Figure 1). In addition, no NADPH-GDH enzyme activity was detected in the wild type or in the gdh single, double, or triple mutants (data not shown).
Figure 1.
NADH-GDH Activity in Mutants Deficient for the Three Genes Encoding the Enzyme.
NADH-GDH aminating activity in the roots and leaves of the gdh1, gdh2, and gdh3 single mutants, a gdh1-2 double mutant, and a gdh1-2-3 triple mutant and in wild-type (WT) plants. Values are mean ± se of four individual plants. Asterisks indicate significant differences with a confidence interval at P < 0.05.
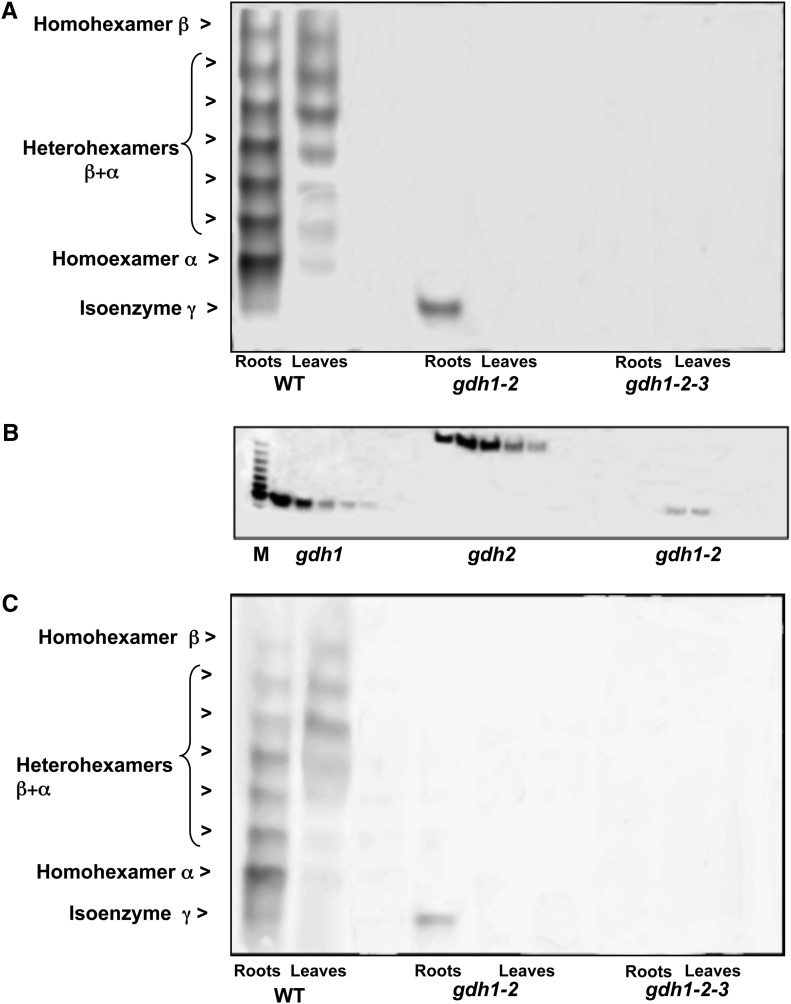
The GDH isoenzyme content was examined in roots and leaves of the wild type and in the gdh1-2 and gdh1-2-3 mutants by staining for NAD-dependent GDH activity following separation by nondenaturing PAGE. In the roots of the wild type, a faint band of enzyme activity was visible below the fastest moving most anodal form of the standard seven band isoenzyme pattern (Figure 2A) and was designated isoenzyme γ. Activity of the isoenzyme γ was not detected in leaves, while in the roots of the gdh1-2 mutant, only a band corresponding to the γ remained active. The isoenzyme γ was not detected in the gdh1-2-3 triple mutant (Figure 2A). An additional experiment consisted of the separation of the three different types of GDH isoenzymes by HPLC ion exchange chromatography, followed by nondenaturing PAGE and NAD-GDH activity staining. The experiment demonstrated unambiguously that in the roots of the gdh1-2 double mutant, isoenzyme γ activity was clearly visible in a number of fractions eluted from the column. Controls performed with root protein extracts showed that fractions from extracts of the gdh1 single mutant contained only homohexamer α, whereas those of the single mutant gdh2 contained only homohexamer β (Figure 2B). Protein gel blot analysis, confirmed that the different GDH isoenzymes detected using NAD-GDH enzyme activity staining, including the additional band of root GDH activity, correspond to a GDH protein recognized by two different antibodies raised against the NADH-GDH enzyme from grapevine (Vitis vinifera) or against a synthetic peptide common to the different GDH proteins (Figure 2C).
Figure 2.
NAD-GDH Isoenzyme Pattern in Mutants Deficient for the Three Genes Encoding the Enzyme.
(A) In-gel detection of NAD-GDH activity. Wild-type (WT), gdh1-2, and gdh1-2-3 mutants roots and leaves extracts were analyzed by nondenaturing PAGE followed by NAD-GDH in-gel activity staining. The positions of the homohexamers of isoenzymes α, β, and γ and of the heterohexamers (composed of mixture of α and β polypeptides) are indicated at the left.
(B) Separation of α, β, and γ NADH-GDH subunits. Subunits separation was performed by HPLC ion exchange chromatography using root extracts from the gdh1, gdh2, and gdh1-2 mutants. In each panel, eluted fractions exhibiting NADH-GDH enzyme activity were subjected to nondenaturing PAGE followed by NAD-GDH in-gel activity staining. M corresponds to a root extract from wild-type plants used as a marker to locate the eight active GDH isoenzymes on the gel.
(C) Immunodetection of α, β, and γ NADH-GDH subunits. The protein gel blot shows that all the polypeptides that positively stained for GDH activity in (A) were recognized by GDH antiserum raised against synthetic peptides. The position of the homohexamers of isoenzymes α, β, and γ and of the heterohexamers (composed of mixture of α and β polypeptides) is indicated at the left.
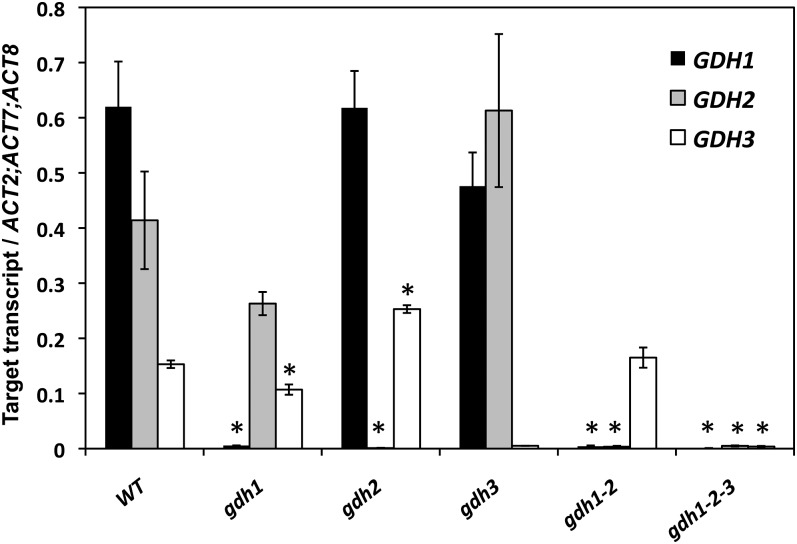
A quantitative RT-PCR (qRT-PCR) experiment showed that transcripts of the GDH3 gene were detected in the root tissues of the wild type and gdh1, gdh2, and gdh1-2 mutants. In the gdh1-2-3 mutant, no signal above the background level for any of the three GDH genes could be detected (Figure 3). In the gdh1 mutant, transcripts for GDH3 isoenzyme (GDH3) were downregulated, whereas in the gdh2 mutant, transcripts for GDH3 were upregulated in comparison to the wild type. In the gdh1-2 mutant, the steady state level of expression of GDH3 was similar to that of the wild type. Transcripts of GDH3 were not detected in the leaves of the wild type nor in any of the different gdh mutants, thus confirming that the expression of the third GDH isoenzyme was root specific (data not shown).
Figure 3.
Quantification of Transcript Abundance in Arabidopsis Mutants Deficient for the Genes Encoding GDH.
qRT-PCR reactions were performed on total RNA extracted from the roots of the wild type (WT) and the five mutants shown above. The level of expression of the GDH1, GDH2, and GDH3 genes was quantified relative to the expression of the β-actin genes ACT2, ACT7, and ACT8 for each of the three GDH genes. Results are presented as mean values for four plants with se. Asterisks indicate significant differences with a confidence interval at P < 0.05.
Cellular and Subcellular Localization of GDH3
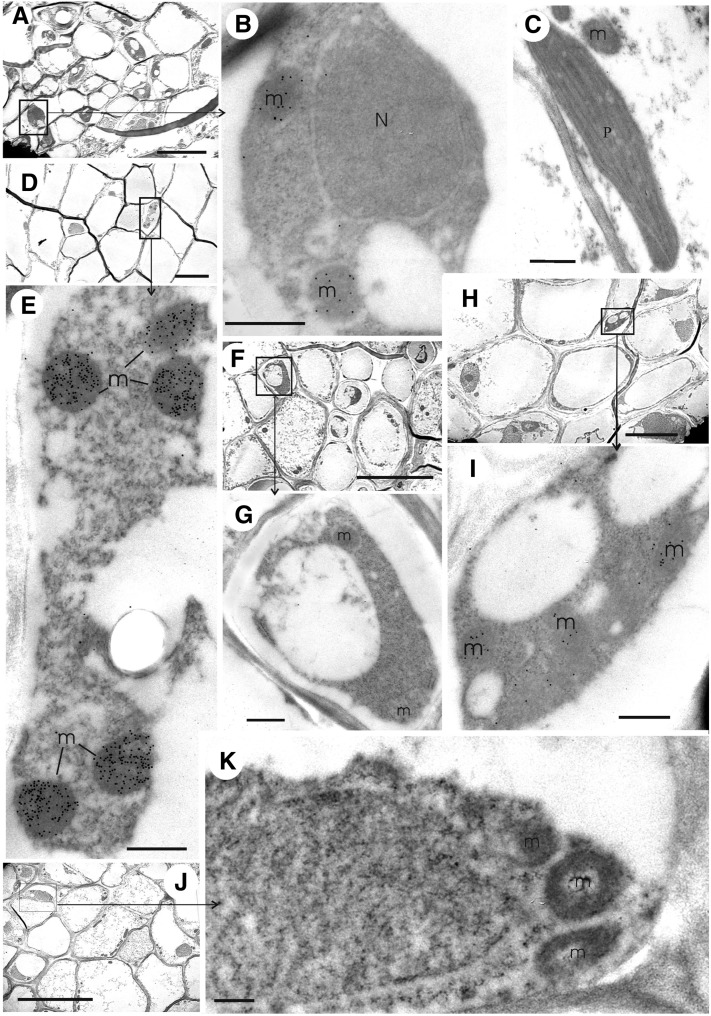
To determine the cellular and subcellular localization of the protein encoded by the GDH3 gene, immunogold localization experiments were conducted on root and leaf tissues of the wild type and of the gdh1-2 and gdh1-2-3 mutants using antibodies raised either against the NADH-GDH enzyme from grapevine or against a synthetic peptide, both of which recognized the homohexamers α, β, and γ and the heterohexamers containing α- and β-subunits with similar efficiency as shown in Figure 2C. In the wild-type plants, GDH protein was only detected in the mitochondria of the phloem companion cells of both leaves (Figures 4A and 4B) and roots, that of the latter being more strongly labeled (Figures 4D and 4E). No labeling was observed in the leaf parenchyma cells (Figure 4C) or in the rest of the root tissues (data not shown). In the leaf of the gdh1-2 double mutant, weak labeling similar to the background level obtained with preimmune serum (see Supplemental Table 1 online) was observed in the mitochondria of phloem companion cells (Figures 4F and 4G). By contrast, in the roots of the gdh1-2 mutant, some labeling remained in the mitochondria of the phloem companion cells (Figures 4H and 4I). In the roots of the gdh1-2-3 triple mutant, the number of gold particles detected in the mitochondria of the phloem companion cells (Figures 4J and 4K) was similar to that of the control performed with preimmune serum, confirming that NADH-GDH protein was totally lacking in the triple mutant (see Supplemental Table 1 online for quantification of the gold particles).
Figure 4.
Immunolocalization of NADH-GDH by Transmission Electron Microscopy in Roots and Leaves of the Wild Type and gdh1-2 and gdh1-2-3 Mutants.
Immunogold labeling experiments were performed on roots and leaves sections of the gdh1-2-3 mutant using the GDH antiserum raised against synthetic GDH polypeptides. m, mitochondrion; N, nucleus; P, plastid. Bars = 5 µm in (A), (D), (F), (H), and (J) and 0.5 µm in (B), (C), (E), (G), (I), and (K).
(A) Phloem tissues in wild-type leaves.
(B) Enlarged view of a companion cell in a leaf vascular bundle of the wild type (boxed area in [A]).
(C) Chloroplast and mitochondrion of wild-type parenchyma cell.
(D) Root phloem tissues in the wild type.
(E) Enlarged view of a companion cell in a root vascular bundle of the wild type (boxed area in [D]).
(F) Leaf phloem tissues in the gdh1-2 double mutant.
(G) Enlarged view of a companion cell in a leaf vascular bundle of the gdh1-2 mutant (boxed area in [F]).
(H) Root phloem tissues in the gdh1-2 double mutant.
(I) Enlarged view of a companion cell in a root vascular bundle of the gdh1-2 mutant (boxed area in [H]).
(J) Root phloem tissues in the gdh1-2-3 triple mutant.
(K) Enlarged view of a companion cell in a root vascular bundle of the gdh1-2-3 mutant (boxed area in [J]).
Metabolic Profile of the gdh1-2-3 Triple Mutant
Supplemental Figures 2A and 2B online show that the phenotype of the triple mutant was not visibly altered compared with the wild type, at either the rosette or the flowering stage. Thus, to evaluate the physiological impact of a total deficiency of NADH-GDH activity, plants were grown under short-day growth conditions and a comparative metabolomic analysis of the wild type and triple gdh1-2-3 mutant was performed at the rosette stage. Results of this study, in which samples of the roots and leaves of the wild type and triple mutant were harvested 2 h into the light period, are presented in Supplemental Data Sets 1 and 2 online. Among the large number of metabolites identified as exhibiting a different pattern of accumulation between the gdh triple mutant and the wild type, attention was focused on those involved in primary C and N metabolism. Among more than 150 identified compounds that showed statistically significant differences in the replicates (P ≤ 0.05), the concentration of a number of metabolites was increased in both the roots and leaves of the gdh triple mutant. These were notably Ala, Pro, γ-aminobutyrate (GABA), and Suc. A number of other amino acids and organic acids, such as Leu, His, pyruvate, and fumarate, were also present in higher amounts in the triple mutant, but their pattern of accumulation was different in the roots compared with the leaves (Table 1). Fewer metabolites were detected in lower concentrations in the triple mutant, in comparison to the wild type. For example, there were lower amounts of 2-oxoglutarate, Gln, Gly, and Glc-6-P in the mutant leaves, whereas in the roots, only aconitate and citrate were significantly decreased (Table 1). The nitrate and ammonium content of the roots was slightly lower in the triple mutant, whereas the concentrations in the leaves were not modified (Figure 5).
Table 1. Primary Metabolites Exhibiting an Increase or a Decrease in Roots and Leaves of the gdh1-2-3 Mutant Compared to the Wild Type and Following Prolonged Darkness.
| Organ T0 |
Organ D3 |
Organ D7 |
|||||||||
| Leaves |
Leaves |
Leaves |
|||||||||
| Increased | FC | Decreased | FC | Increased | FC | Decreased | FC | Increased | FC | Decreased | FC |
| Glcc,e | 3.2 | Gly | 0.5 | Glcc,e | 3.1 | 2-Oxoglutarated,e | 0.6 | Alac,e | 3.6 | 2-Oxoglutarated,e | 0.4 |
| Fruc,e | 2.1 | 2-Oxoglutarated,e | 0.6 | Alac | 1.9 | Cys | 0.7 | Aspc | 3.6 | Gln | 0.5 |
| His | 1.7 | Gln | 0.7 | Aspc,e | 1.8 | GABAc | 2.3 | Pyruvate | 0.5 | ||
| Pyruvate | 1.7 | Glc-6-P | 0.7 | Fruc,e | 1.6 | Pro | 2.0 | Fru-6-P | 0.6 | ||
| Suc | 1.6 | Aspd | 0.7 | Succinate | 1.4 | Glcc,e | 1.9 | Glc-6-P | 0.7 | ||
| GABAc | 1.4 | Fumaratec,e | 1.4 | Ser | 1.7 | ||||||
| Pro | 1.3 | Malate | 1.3 | Fruc,e | 1.7 | ||||||
| Fumaratec,e | 1.3 | GABAc,e | 1.2 | Tyr | 1.6 | ||||||
| Alac,e | 1.3 | Leu | 1.5 | ||||||||
| Lys | 1.4 | ||||||||||
| Fumaratec,e | 1.4 | ||||||||||
| Thr | 1.3 | ||||||||||
| Ile | 1.3 | ||||||||||
| Val | 1.3 | ||||||||||
| Roots | Roots | Roots | |||||||||
| Increased | FC | Decreased | FC | Increased | FC | Decreased | FC | Increased | FC | Decreased | FC |
| GABAc,e | 2.3 | Aconitate-trans | 0.6 | GABAd,e | 6.6 | 2-Oxoglutarated | 0.4 | Alac,e | 9.2 | 2-Oxoglutarated | 0.1 |
| Fumarate | 1.8 | Citrate | 0.7 | Alac,e | 5.8 | Malated | 0.5 | GABAd,e | 6.9 | Citrate | 0.1 |
| Pro | 1.7 | Aspc | 4.3 | Fru-6-Pd | 0.7 | Aspc | 4.4 | Fru-6-Pd | 0.1 | ||
| Suc | 1.5 | Leuc | 2.6 | Succinate | 0.7 | Ilec,e | 2.6 | Glc-6-Pd | 0.1 | ||
| Leu | 1.5 | Lysc | 2.2 | Glc-6-Pd | 0.7 | Valc,e | 2.2 | Malated | 0.1 | ||
| Trp | 1.5 | Ilec,e | 1.8 | Leuc | 1.9 | Gln | 0.2 | ||||
| Phe | 1.5 | Fumarate | 1.6 | Serc | 1.8 | Phe | 0.2 | ||||
| Ilec,e | 1.3 | Glyc | 1.5 | Pyruvatec | 1.8 | ||||||
| Alac,e | 1.3 | Pyruvatec | 1.5 | Asn | 1.7 | ||||||
| Valc,e | 1.3 | Valc,e | 1.4 | Glyc | 1.5 | ||||||
| Glu | 1.4 | Glc | 1.3 | ||||||||
| Fru | 1.4 | ||||||||||
| Suc | 1.3 | ||||||||||
| Serc | 1.3 | ||||||||||
Metabolomic analysis was performed on roots and leaves of plants grown under hydroponic conditions and harvested at the rosette stage as described in Methods. T0 = plants grown under short-day conditions and harvested 2 h after the beginning of the light period, D3 = after 3 d in the dark, and D7 = after 7 d in the dark. Fold change (FC) corresponds to the ratio of gdh1-2-3 mutant/wild type calculated by taking the average of the metabolite concentrations measured in the replicates of the metabolomic analyses (see Supplemental Data Sets 1 and 2 online). Those exhibiting a similar pattern of accumulation in both roots and leaves whatever the time point of the experiment are in bold characters. A schematic representation of the accumulation pattern of a number of selected metabolites exhibiting the most characteristic changes in roots after dark exposure is presented in Figure 9. The – indicates where the number of compounds is different from one column to the next.
Increased fold change (FC > 1.25).
Decreased fold change (FC < 0.75).
Metabolites present in higher concentration in the leaves of the mutant at T0, D3, and D7.
Metabolites present in higher concentration in the roots of the mutant at T0, D3, and D7.
Metabolites exhibiting a similar pattern of accumulation in roots and leaves at the three time points of the experiment.
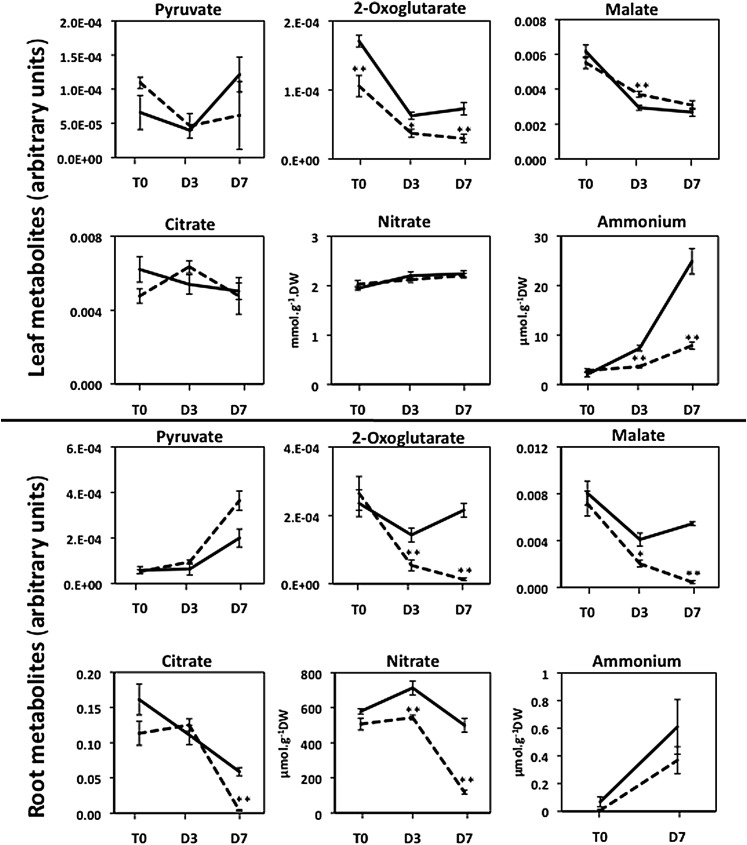
Figure 5.
Representative Arabidopsis Leaf and Root Organic Acids and Ions Exhibiting Differences in Concentration between the gdh1-2-3 Mutant and the Wild Type Following Prolonged Darkness.
T0 = plants grown under short-day conditions and harvested 2 h after the beginning of the light period, D3 = after three continuous days in the dark, and D7 = after seven continuous days in the dark. The dotted line corresponds to the gdh1-2-3 mutant and the solid line to the wild type. The data for leaf and root metabolomic analyses are presented in Supplemental Data Sets 1 and 2 online and were obtained as described in Methods. The metabolite content is expressed in arbitrary units, which correspond to normalized values of the peak area⋅standard ribitol−1⋅leaf dry weight−1. Bars indicate se. Magnification of the scale is shown when there were significant differences between the wild type and the mutant at T0 and T3. The name of the metabolite is in a white box when differences occurred both in the roots and leaves and in a pale-gray or dark-gray box when either in the leaves or roots, respectively. Results are presented as mean values for five plants with se. Asterisks indicate a bilateral t test P value <0.01 (**) and <0.05 (*) for statistically significant differences.
Transcriptome Analysis of gdh1-2-3 Triple Mutant
As shown above, complete loss of the NADH-dependent GDH enzyme activity in Arabidopsis plants leads to a number of significant differences in terms of metabolite accumulation. In order to determine if these changes might be linked to differences in gene expression, transcriptome profiling was performed on the roots and leaves of the Arabidopsis gdh1-2-3 mutant and wild-type plants using the Complete Arabidopsis Transcriptome Microarray (CATMA) system (Crowe et al., 2003; Hilson et al., 2004). Two biological replicates, each composed of a pool of 25 plants, were performed for each comparison, and for each biological replicate, two reverse-labeling technical replicates were performed. Supplemental Figure 3 online shows the number of differentially expressed genes in roots and leaves of the gdh1-2-3 mutant according to their false discovery rate (FDR). A low FDR indicates a good reproducibility of the results between the two biological replicates. Transcript abundances with a FDR lower than 0.0001% are presented in Table 2. Those with a FDR lower than 0.001% that showed lower but still statistically significant variations in their relative amounts are presented in Supplemental Data Sets 3 and 4 online. Transcript abundances with a FDR lower than 0.00001 or 0.0001% were only modified in the roots of the gdh1-2-3 triple mutant. Although more transcripts were upregulated (349) or downregulated (472) at a less stringent FDR of 0.0001%, differences in their relative amounts were still observed only in the roots (see Supplemental Figure 3 and Supplemental Data Sets 3 and 4 online for the detailed list of genes). Among these genes, those exhibiting putative key physiological or regulatory functions that may be related to the accumulation of metabolites in the mutant roots are presented in Tables 3 and 4. They belong to a limited number of functional categories including transport and metabolism of C and N compounds for those that were upregulated and to stress, defense, and proteolytic functions, for those that were downregulated. A qRT-PCR experiment performed on three selected genes exhibiting an increase, a decrease, or no change in the gdh1-2-3 mutant in comparison to the wild type confirmed that the level of root transcript accumulation was similar to that observed in the CATMA microarray experiment (see Supplemental Table 2 online).
Table 2. Transcript Abundance That Is Significantly Altered in the Arabidopsis gdh1-2-3 Mutants Compared with the Wild Type.
| Organ | Regulation | AG | Description | Average FC | FDR (%) |
|---|---|---|---|---|---|
| Root | Down | AT5G18170 | GDH1 | 0.07 | 1.5E-07 |
| AT2G14247 | Unknown protein | 0.21 | 4.4E-07 | ||
| AT4G10265 | Unknown protein,wound-responsive protein, putative | 0.27 | 9.8E-07 | ||
| AT1G72060 | Ser-type endopeptidase inhibitor heat shock protein binding | 0.30 | 2.6E-06 | ||
| AT3G48450 | Unknown protein,nitrate-responsive NOI protein, putative | 0.29 | 2.1E-06 | ||
| AT1G21520 | Unknown protein | 0.35 | 4.1E-06 | ||
| AT4G39675 | Unknown protein | 0.36 | 3.8E-06 | ||
| AT2G17850 | Unknown protein | 0.32 | 4.0E-06 | ||
| AT1G33055 | Unknown protein | 0.33 | 3.9E-06 | ||
| AT4G27110 | Cobra-like protein 11 precursor | 0.36 | 3.8E-06 | ||
| AT3G13520 | Arabinogalactan protein 12 | 0.38 | 4.8E-06 | ||
| AT2G15830 | Unknown protein | 0.35 | 4.9E-06 | ||
| AT1G12805 | Nucleotide binding | 0.34 | 4.6E-06 | ||
| AT3E49170 | EUGENE prediction | 0.35 | 5.7E-06 | ||
| AT2G38870 | Ser-type endopeptidase inhibitor | 0.39 | 5.9E-06 | ||
| AT2G01090 | Ubiquinol-cytochrome c reductase complex 7.8-kD protein, putative | 0.40 | 6.7E-06 | ||
| AT2G21640 | Unknown protein | 0.40 | 6.4E-06 | ||
| AT3G05880 | Rare cold-inducible 2A/low-temperature and salt-responsive protein | 0.41 | 6.4E-06 | ||
| AT3G11830 | ATP binding/protein binding, chaperonin, putative | 0.41 | 6.4E-06 | ||
| AT2G33585 | Unknown protein | 0.38 | 6.2E-06 | ||
| AT5G64790 | Glycosyl hydrolase family 17 protein | 0.38 | 6.1E-06 | ||
| AT3G02790 | Transcription factor, zinc finger (C2H2 type) family protein | 0.41 | 6.7E-06 | ||
| AT2G37750 | Unknown protein | 0.42 | 7.5E-06 | ||
| AT5G53650 | Unknown protein | 0.42 | 8.9E-06 | ||
| AT2G21185 | Unknown protein | 0.40 | 8.6E-06 | ||
| AT5G12880 | Unknown protein, Pro-rich family protein | 0.42 | 9.4E-06 | ||
| AT4G13520 | Small acidic protein 1 | 0.43 | 9.3E-06 | ||
| AT5G03545 | Unknown protein | 0.42 | 9.1E-06 | ||
| AT1G68450 | Unknown protein, VQ motif–containing protein | 0.41 | 8.9E-06 | ||
| AT5G10040 | Unknown protein | 0.36 | 8.6E-06 | ||
| AT3G15580 | Encodes APG8, a component of autophagy conjugation pathway | 0.44 | 9.6E-06 | ||
| AT2G05440 | Unknown protein,Gly-rich protein | 0.41 | 9.6E-06 | ||
| Root | Up | AT3G16400 | Nitrile specifier protein1 | 2.75 | 1.0E-06 |
| AT5G42020 | ATP binding, luminal binding protein 2 | 2.45 | 3.5E-06 | ||
| AT3G02470 | S-adenosylmethionine decarboxylase (polyamine biosynthesis) | 2.34 | 5.5E-06 | ||
| AT3G09440 | ATP binding, heat shock cognate 70-kD protein 3 | 2.36 | 4.2E-06 | ||
| AT1G20160 | Ser-type endopeptidase | 2.35 | 3.4E-06 | ||
| AT4G05230 | Ubiquitin family protein | 2.29 | 3.9E-06 | ||
| AT4G13770 | Cytochrome p450 83A1 | 2.26 | 3.4E-06 | ||
| AT3G16460 | Jacalin lectin family protein | 2.24 | 3.2E-06 | ||
| AT3G15950 | Unknown protein, similar to TSK-associating protein 1 (TSA1) | 2.20 | 4.6E-06 | ||
| mirspot706 | HYPMIR749706 | 2.15 | 6.4E-06 | ||
| AT3G30775 | Osmotic stress-responsive Pro dehydrogenase (POX; PRO1) | 2.20 | 6.2E-06 | ||
| AT4G24190 | Shepherd; ortholog of GRP94, an endoplasmic reticulum–resident HSP90-like protein | 2.06 | 9.3E-06 | ||
| mirspot705 | HYPMIR749706 | 2.07 | 9.8E-06 | ||
| AT5G02500 | ATP binding, heat shock cognate 70-kD protein 1 | 1.98 | 8.9E-06 | ||
| Leaf | Down | AT5G18170 | GDH1 | 0.09 | 1.5E-07 |
Arabidopsis gene identification.
Average fold change (FC) between two replicated experiments.
Transcripts exhibiting a FDR lower than 0.0001%.
Table 3. Selection of Transcripts Present in Higher Amounts in Roots of the Arabidopsis gdh1-2-3 Mutant Compared to the Wild Type Exhibiting Key Physiological Functions.
| Gene Description | Functional Category |
|---|---|
| PKP-ALPHA; pyruvate kinasea | Metabolism C |
| Transketolase, putative | Metabolism C |
| PYRUVATE DEHYDROGENASE E1 ALPHA | Metabolism C |
| GLYCERALDEHYDE-3-PHOSPHATE DEHYDROGENASE OF PLASTID1 | Metabolism C |
| PHOSPHOENOLPYRUVATE CARBOXYLASE2 | Metabolism C |
| HEXOKINASE1 | Metabolism C |
| Pyrophosphate-Fru-6-P 1-phosphotransferase-related | Metabolism C |
| NADP+ isocitrate dehydrogenase | Metabolism C |
| PHOSPHOGLYCERATE KINASE | Metabolism C |
| METHIONINE OVERACCUMULATION1; cystathionine γ-synthase | Metabolism N |
| EARLY RESPONSIVE TO DEHYDRATION5; Pro dehydrogenase | Metabolism N |
| GLUTAMATE DECARBOXYLASE2; calmodulin binding/Glu decarboxylase | Metabolism N |
| l-asparaginase, putative/l-Asn amidohydrolase, putative | Metabolism N |
| DELTA 1-PYRROLINE-5-CARBOXYLATE SYNTHASE2 | Metabolism N |
| ALANINE:GLYOXYLATE AMINOTRANSFERASE3 | Metabolism N |
| Arabidopsis Gly decarboxylase P-protein 1 | Metabolism N |
| O-phospho-l-Ser:2-oxoglutarate aminotransferase | Metabolism N |
| S-ADENOSYLMETHIONINE DECARBOXYLASE | Polyamine synthesis |
| ARGININE DECARBOXYLASE1; Arg decarboxylase | Polyamine synthesis |
| TONOPLAST MONOSACCHARIDE TRANSPORTER1 | Transport C |
| SUCROSE-PROTON SYMPORTER2 | Transport C |
| SUGAR TRANSPORTER4 | Transport C |
| Glc-6-P/phosphate translocator, putative | Transport C |
| Sugar transporter, putative | Transport C |
| AUXIN RESISTANT1; amino acid transmembrane transporter | Transport N |
| Amino acid permease, putative | Transport N |
| NRT1.1; nitrate transmembrane transporter/transporter | Transport N |
| Amino acid transporter family protein | Transport N |
| AAP3; amino acid transmembrane transporter | Transport N |
| Amino acid permease, putative | Transport N |
| Protein transport protein sec61, putative | Transport N |
| AMINO ACID PERMEASE2; amino acid transmembrane transporter | Transport N |
| DELTA-TIP; ammonia transporter | Transport N |
| Mitochondrial phosphate transporter | Transport P |
Transcripts for enzymes involved in the synthesis of metabolites exhibiting important differences in their accumulation are in bold. The gene selection was taken from Table 2 and Supplemental Data Set 4 online.
Table 4. Selection of Transcripts Present in Lower Amounts in Roots of the Arabidopsis gdh1-2-3 Mutant Compared to the Wild Type Exhibiting Key Physiological Functions.
| Gene Description | Functional Category |
|---|---|
| DEFENDER AGAINST APOPTOTIC DEATH1 | Cell death |
| DEFENDER AGAINST CELL DEATH2 | Cell death |
| Respiratory burst oxidase protein E/NADPH oxidase | Defense |
| Disease resistance response | Defense |
| Cold shock DNA binding family protein | Defense |
| Disease resistance–responsive protein-related/dirigent protein-related | Defense |
| ATP binding/ATP citrate synthase/citrate synthase | Metabolism C |
| Ser-type endopeptidase inhibitor | Proteolysis |
| AUTOPHAGY 8H; APG8 activating enzyme/APG8-specific protease | Proteolysis |
| CYSTATIN A; Cys-type endopeptidase inhibitor | Proteolysis |
| Identical protein binding/Ser-type endopeptidase | Proteolysis |
| AUTOPHAGY 8G; microtubule binding | Proteolysis |
| Protease inhibitor/seed storage/lipid transfer protein family protein | Proteolysis |
| UBIQUITIN-CONJUGATING ENZYME11; ubiquitin-protein ligase | Proteolysis |
| AUTOPHAGY 8G; microtubule binding | Proteolysis |
| Proteasome maturation factor UMP1 family protein | Proteolysis |
| Senescence-associated protein-related | Senescence |
| Senescence-associated protein, putative | Senescence |
| Senescence-associated protein-related | Senescence |
| Nitrate-responsive NOI protein, putative | N Sensing |
| Hydrophobic protein, putative/low-temperature and salt-responsive protein | Stress |
| Hypoxia-responsive family protein | Stress |
| Glutaredoxin family protein | Stress |
| 17.8-kD class I heat shock protein (HSP17.8-CI) | Stress |
| 23.5-kD mitochondrial small heat shock protein (HSP23.5-M) | Stress |
| ALCOHOL DEHYDROGENASE1; alcohol dehydrogenase | Stress |
| GLUTAMINE DUMPER1 | Transport N |
The gene selection was taken from Table 2 and Supplemental Data Set 3 online.
Metabolic Profile of the gdh1-2-3 Triple Mutant Placed under Continuous Darkness
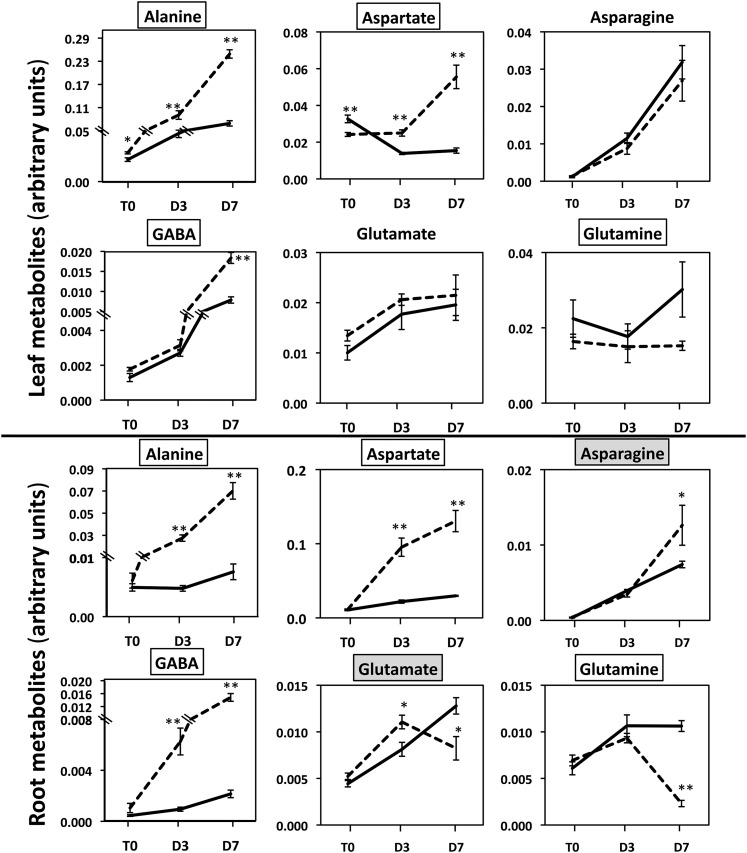
Further metabolomic analyses were performed on the gdh1-2-3 mutant and wild-type plants placed under continuous darkness for 3 and 7 d. An overall view of the significant differences in the content of the main metabolites involved in primary C and N assimilation is shown in Table 1. This view also provides a dynamic picture of the metabolite changes from short-day to dark growth conditions and as a function of the duration of the dark period. In addition, Figures 5 to 7 provide details of the most characteristic differences in terms of metabolite accumulation, starting from 2 h after the beginning of the light period of a dark/light cycle and followed by two time points of prolonged darkness. It was observed that a number of metabolites shared a common pattern of accumulation both in the roots and leaves, whereas others were detected at higher or lower concentrations in only one of the two organs. For example, there was a progressive increase in GABA, Ala, and Asp (Figure 6) as a function of the duration of the dark period in both the roots and leaves of the triple mutant.
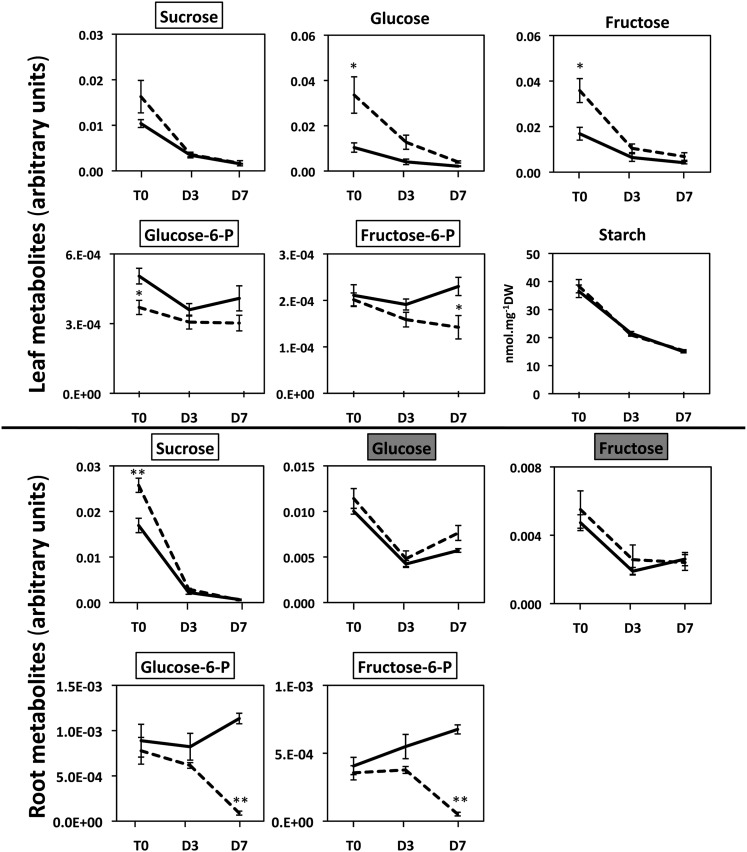
Figure 7.
Representative Arabidopsis Leaf and Root Carbohydrates Exhibiting Differences in Concentration between the gdh1-2-3 Mutant and the Wild Type Following Prolonged Darkness.
T0 = plants grown under short-day conditions and harvested 2 h after the beginning of the light period, D3 = after three continuous days in the dark, and D7 = after seven continuous days in the dark. The dotted line corresponds to the gdh1-2-3 mutant and the solid line to the wild type. The data for leaf and root metabolomic analyses are presented in Supplemental Data Sets 1 and 2 online and were obtained as described in Methods. The metabolite content is expressed in arbitrary units, which correspond to normalized values of the peak area⋅standard ribitol−1⋅leaf dry weight−1. Bars indicate se. Magnification of the scale is shown when there were significant differences between the wild type and the mutant at T0 and T3. The name of the metabolite is in a white box when differences occurred both in the roots and leaves and in a pale-gray or dark-gray box when either in the leaves or roots respectively. Results are presented as mean values for five plants with se. Asterisks indicate a bilateral t test P value <0.01 (**) and <0.05 (*) for statistically significant differences.
Figure 6.
Representative Arabidopsis Leaf and Root Amino Acids and N-Containing Molecules Exhibiting Differences in Concentration between the gdh1-2-3 Mutant and the Wild Type Following Prolonged Darkness.
T0 = plants grown under short-day conditions and harvested 2 h after the beginning of the light period, D3 = after three continuous days in the dark, and D7 = after seven continuous days in the dark. The dotted line corresponds to the gdh1-2-3 mutant and the solid line to the wild type. The data for leaf and root metabolomic analyses are presented in Supplemental Data Sets 1 and 2 online and were obtained as described in Methods. The metabolite content is expressed in arbitrary units, which correspond to normalized values of the peak area⋅standard ribitol−1⋅leaf dry weight−1. Bars indicate se. Magnification of the scale is shown when there were significant differences between the wild type and the mutant at T0 and T3. The name of the metabolite is in a white box when differences occurred both in the roots and leaves and in a pale-gray or dark-gray box when either in the leaves or roots, respectively. Results are presented as mean values for five plants with se. Asterisks indicate a bilateral t test P value <0.01 (**) and <0.05 (*) for statistically significant differences.
For a number of other amino acids, organic acids, and carbohydrates, the pattern of accumulation was different according either to the organ examined or to the period of dark exposure. This more complex metabolite accumulation pattern was found in leaves, in which, for example, Leu, Lys, Thr, and Tyr were more abundant in the triple mutant after 7 d in the dark. By contrast, the malate and succinate contents of the leaves were higher only after 3 d of darkness, whereas an accumulation of fumarate was observed after both 3 and 7 d of darkness. Higher amounts of Gly, Ser, and pyruvate were detected in the roots of the triple mutant after both 3 and 7 d of darkness. Interestingly, 2-oxoglutarate was always present at lower concentrations, in both the roots and leaves of the triple mutant, reaching a value close to its limit of detection after 7 d of dark treatment (Figure 5, Table 1). A progressive decrease in Glc-6-P and Fru-6-P was found in the roots of the triple mutant, whereas, in the leaves, reduced amounts of the two metabolites were only found after 7 d in the dark. In the leaves of the triple mutant, the amounts of Glc and Fru were always higher in comparison to the wild type, even though the concentration of these two metabolites decreased steadily when the plants were transferred to the dark. Additionally, reduced amounts of Gln were detected both in roots and in leaves of the triple mutant, but only after 7 d of darkness.
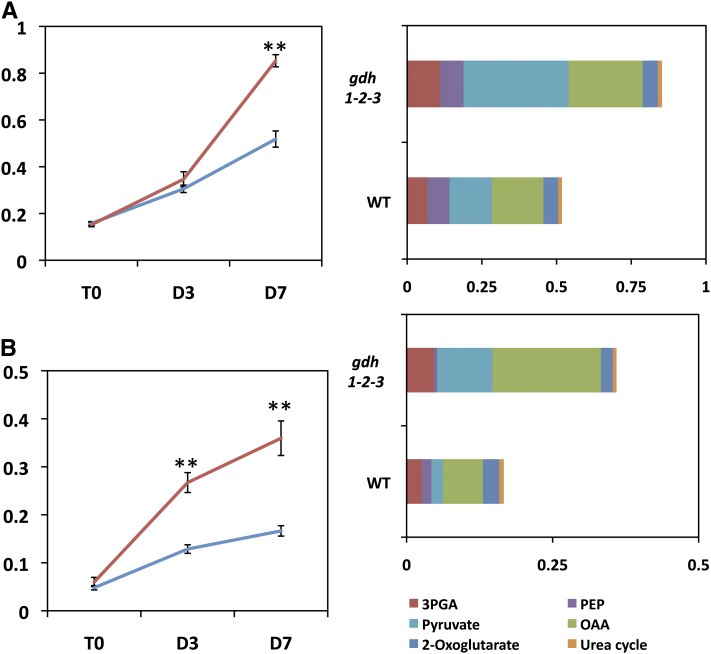
The starch concentration in the leaves decreased by about half after 7 d, in both the wild type and triple mutant, at almost exactly the same rate (Figure 7). In the roots of both the wild type and the gdh1-2-3 mutant, starch was at the limit of detection (data not shown). However, over the same time period, the concentration of ammonium increased in both the wild type and triple mutant in both the roots and leaves, although at a slower rate in the mutant (Figure 5). In the roots of the triple mutant, a decrease in nitrate content occurred at a higher rate, whereas in leaves, no marked differences were observed either due to the mutation or to dark exposure (Figure 5). A closer examination of the quantitative changes in amino acids indicated that both in the roots and leaves of the gdh triple mutant, the total amino acid concentration was almost twice as high as the wild type, with this difference reaching a maximum after 7 d in the dark. This was mainly due to an increase in the amino acids, such as Ala, Asp, GABA, Ile, Leu Pro, Ser, and Val in the gdh1-2-3 mutant (Figure 8). Although slightly decreased after 7 d in the dark, the protein contents of both the leaf and root of the triple mutant and the wild type were very similar (data not shown). In addition to the metabolomic study, dark respiration was measured in both the roots and leaves. As shown in Supplemental Figure 4 online, in the roots of the gdh1-2-3 mutant, O2 consumption remained constant even after 7 d in the dark. In the wild type, the O2 consumption of the roots decreased by 50% after 3 d of dark exposure and remained close to the same lower level after 7 d in the dark. In the leaves, the rate of O2 consumption was slightly higher in the gdh triple mutant and increased slowly in line with the wild type over the period of dark exposure.
Figure 8.
Differences in the Spectrum of Amino Acids in the Wild Type and gdh1-2-3 Mutant in the Dark.
In the left panels, differences in the total free amino acid content are shown. T0 = plants grown under short-day conditions and harvested 2 h after the beginning of the light period, D3 = plants harvested after three continuous days in the dark, D7 = plants harvested after seven continuous days in the dark. The red line corresponds to the gdh1-2-3 mutant and the blue line to the wild type (WT). Leaves (A) and roots (B). In the right panels, the main classes of amino acids accumulated after seven continuous days of darkness are shown. The amino acids derived from 3-phosphoglycerate (3PGA) are Cys, Gly, and Ser, from phosphoenolpyruvate (PEP) are Phe, Trp, and Tyr, from pyruvate are Ala, Leu, and Val, from oxaloacetate (OAA) are Asp, Asn, Ile, Lys, Met, and Thr, from 2-oxoglutarate are Arg, GABA, Gln, Glu, and Pro, from the urea cycle are Cit and Orn. Metabolite content is expressed in arbitrary units, which correspond to normalized values of the peak area⋅standard ribitol−1 leaf dry weight−1. Error bars correspond to se.
DISCUSSION
A Third GDH Isoenzyme Localized in Companion Cells Is Only Active in Roots
The availability of the Arabidopsis genome sequence database (http://www.Arabidopsis.org/) allowed the identification of a gene sequence (GDH3) exhibiting a strong homology with the two other previously identified genes (GDH1 and GDH2) encoding NADH-GDH (Turano et al., 1997). The occurrence of this gene was previously mentioned by Restivo (2004) and Purnell et al. (2005). Moreover, it has also been shown that this third gene encoding NADH-GDH was actively transcribed (Yamada et al., 2003; Igarashi et al., 2009). However, there was still no evidence that the GDH3 gene transcripts were able to produce an active enzyme protein and where this protein was located both at the organ or cellular level. As previously reported by several authors, NADH-GDH in Arabidopsis can be separated into seven isoenzymes composed of homohexamers of both α- or β-subunits and of heterohexamers containing different proportions of α and β subunits. Homohexamers of α or β are encoded by the two different genes named GDH2 and GDH1, respectively. In this investigation, it has been shown that the GDH3 transcripts are only expressed in the roots and that they are translated into a protein exhibiting NADH-GDH activity. This third NADH-GDH isoenzyme, termed GDH3, was identified by the detection of additional enzyme activity that could be separated by native PAGE and HPLC and is presumably assembled into a homohexameric γ form. These results clearly demonstrate that the GDH3 isoenzyme protein is active (at least when measured in vitro) and contributes to a low but significant proportion of total enzyme activity in the roots.
Interestingly, there would appear to be a compensatory mechanism for the enzyme activity in the gdh3 mutant. The GDH3 gene is not expressed, but the GDH2 gene is expressed at a higher level, with a resultant higher total NADH-GDH activity in the roots of the gdh3 mutant. Similar increases in GDH activity have been previously demonstrated in single mutants (Fontaine et al., 2006). The occurrence of such changes was also apparent in the roots of gdh1 and gdh2 single mutants, in which the level of GDH3 transcripts was lower and higher, respectively. However, the physiological significance of these compensatory mechanisms remains unresolved, and their occurrence could lead to an underestimation of the physiological impact of the different gdh mutations, if only studied individually or in a dual combination (Fontaine et al., 2006). The tissue and subcellular localization experiments clearly demonstrate that the GDH3 protein is localized in the mitochondria of companion cells in the roots, in a comparable manner to the isoenzyme counterparts GDH1 and GDH2, which are present in both the leaf and root vasculature (Fontaine et al., 2006). However, it is now clear that the three GDH isoenzymes, GDH1, 2, and 3 all contribute to the root enzyme activity, whereas in leaves, only GDH1 and GDH2 are involved. It will be also interesting to test if signal molecules, such as cytokinins (Igarashi et al., 2009), are able to modify the relative protein content and enzyme activity of the three GDH isoenzymes in order to differentiate their specific roles within the plant.
The gdh1-2-3 Mutation Alters Primary C and N Metabolism Preferentially in Roots
Recent studies have provided strong evidence that NADH-GDH is involved in regulating a range of pathways at the crossroads between C and N metabolism, particularly under abiotic stress conditions such as salinity (Skopelitis et al., 2006) or prolonged darkness mimicking C deficiency (Miyashita and Good, 2008). Despite this, it is still unclear whether the enzyme is only involved in the breakdown of Glu when there is a shortage of C or under certain physiological conditions in the process of ammonium detoxification (Dubois et al., 2003; Tercé-Laforgue et al., 2004b; Skopelitis et al., 2006). This uncertainty about the physiological role of NADH-GDH has arisen from a number of concerns that were not identified in most previously published works. One of these concerns is that the vast majority of the studies performed in an attempt to unravel the function of GDH were conducted on shoots, whereas the highest enzyme activity has been detected in the roots of most plant species, including Arabidopsis (Cammaerts and Jacobs, 1985; Turano et al., 1997; Miyashita and Good, 2008). Another concern was that in the studies on the single gdh1 and gdh2 mutants (Melo-Oliveira et al., 1996; Fontaine et al., 2006) or double gdh1-2 (Miyashita and Good, 2008), the presence of a third active root isoenzyme (GDH3) identified in this work was not taken into account. Thus, the presence of this third isoenzyme may well have misled the interpretation of the results obtained either with the single or double gdh1-2 mutants. It is possible that compensatory long-distance transport regulatory mechanisms could have operated between roots and shoots of either the single or the double gdh mutants. It is clear that the three NADH-GDH isoenzyme proteins are located in the mitochondria of the companion cells, thus reinforcing the hypothesis that the circulation of metabolites, such as Glu and 2-oxoglutarate, or a signal derived from them, occurs within the plant through the phloem stream.
In this study, a number of significant differences in the metabolite content of both the roots and leaves of the gdh1-2-3 mutant were identified when the plants were grown under short-day growth conditions. These results contrast with those previously published by Miyashita and Good (2008) using plants grown on Suc-free agar medium under long-day conditions and lower light intensity. These authors did not observe any major differences in the metabolite profile of the aboveground tissue of the gdh1-2 double mutant compared with the wild type except for the Orn and Arg contents, which were almost twice as high in the mutant. However, the authors did not analyze as a wide range of metabolites in the shoots as this study and presented no data for the roots, thus possibly missing signals involved in the translocation of metabolites from the root to shoot.
A higher Suc content was found in both the roots and leaves of the gdh1-2-3 mutant. An accumulation of Glc and Fru was also a characteristic feature of the triple mutant. In addition, there was a lower concentration of 2-oxoglutarate and a slightly higher Glu content in the gdh1-2-3 mutant. It is logical to think that the decrease in 2-oxoglutarate is due to the lack of GDH activity and that as a result of which there is a reduction in carboxylic acids to fuel the tricarboxylic acid (TCA) cycle. This decrease in 2-oxoglutarate would presumably also have some repercussions on the use of Glc and Fru that would be channeled to a lesser extent through the glycolytic pathway. This would indicate that the production of 2-oxoglutarate by the reaction catalyzed by NADH-GDH is of major importance for plant C metabolism. The pathways leading to the synthesis of 2-oxoglutarate have been the subject of considerable discussion (Foyer et al., 2011), and it has been demonstrated that the production of 2-oxoglutarate by the NAD-dependent isocitrate dehydrogenase is not limiting for N assimilation (Lemaitre et al., 2007).
Transcriptome Analysis Suggests That the Physiological Impact of the gdh1-2-3 Mutation Originates in the Roots
The major importance of the root system, with respect to the role of the high NADH-GDH activity, was revealed by means of transcriptome analysis. Using microarray technology, distinct differences in transcript accumulation of the gdh1-2-3 mutant in comparison to the wild type were identified. Interestingly, the most differentially expressed genes selected according to their lowest level of FDR (0.0001%) were found only in the roots. More surprisingly, the majority of the genes that were downregulated (21 out of 33 genes at FDR <0.00001%) do not have any known function, whereas those that were upregulated have a variety of metabolic, developmental, stress, and signaling functions. They corresponded to genes that encode proteins involved either in cell expansion (COBRA-like, arabinogalactan protein), nutrient recycling (Ser endopeptidase and APG8 autophagy component), or abiotic stress (cold-inducible protein 2A). The upregulated root genes in the gdh1-2-3 mutant were also involved in the control of cell expansion and plant development (SHEPERD ortholog and S-adenosylmethionine decarboxylase) and in a variety of stresses (myrosinate binding protein, ATP binding luminal protein, and osmotic stress–responsive Pro dehydrogenase). Two hypothetical miRNAs were also found to accumulate in the roots of the gdh1-2-3 mutant. These observations suggest that the cascade of regulatory events resulting from the lack of NADH-GDH activity originating from the roots could involve polyamines, microRNAs, and a number of stress-responsive elements, but also unknown signaling and metabolic regulatory processes (Table 2).
Even at a less stringent FDR (lower than 0.0001%), the differences observed in transcript accumulation were still confined to the roots. Due to the complexity of the transcriptomic data set and in order to integrate such data with that of the root and leaf metabolic profiling, attention was focused on the accumulation of transcripts of known physiological or regulatory function, which may be related directly or indirectly to the impairment of GDH activity in the mutant (Tables 3 and 4). Generally, it is not easy to find a strict correlation between metabolite accumulation and transcript abundance (Hirai et al., 2004; Fernie and Stitt, 2012). Nevertheless, the accumulation pattern of transcripts encoding proteins or enzymes involved in several metabolic processes was consistent with the differences observed in the metabolite concentrations. For example, in the roots of the gdh triple mutant, there was an accumulation of transcripts for NADP-isocitrate dehydrogenase, pyruvate kinase, and phosphoenolpyruvate carboxylase, three enzymes involved in providing C skeletons for the production of 2-oxoglutarate. Such upregulation was consistent with the finding that several metabolites involved in the reactions catalyzed by these enzymes or derived from them were present in higher amounts. These alterations in organic acid metabolism are likely to be linked to the decrease in the 2-oxoglutarate content. The finding that in the gdh1-2-3 triple mutant, a number of transcripts for carbohydrate transporters accumulate is in line with this hypothesis, if the role of GDH at the interface of C and N metabolism is taken into consideration (Aubert et al., 2001; Miyashita and Good, 2008).
Transcripts for genes encoding enzymes involved in N metabolism, notably those using Glu as a substrate, such as Glu decarboxylase and pyrroline-5-carboxylate synthase, were also more abundant in the roots of the gdh triple mutant. This observation suggests that the lack of NADH-GDH activity had important repercussions on N metabolism, leading to an accumulation of the N-containing molecules used for transient or long-term storage, such as GABA and Pro. This is probably because Glu metabolism was altered in the triple mutant as there was no deamination of the amino acid due to the lack of NADH-GDH activity. It would appear that other metabolic pathways are able to circumvent the deficiency in GDH in order to maintain the homeostasis of Glu, a regulatory process known to be of major importance during plant growth and development (Forde and Lea, 2007; Labboun et al., 2009). The perturbations in the Glu-derived metabolic pathways also fit in well with the finding that the transcripts for several amino acid permeases or amino acid transporters were more abundant in the gdh1-2-3 mutant. Such findings suggest that, as for C, the transport of a number of N-containing molecules was also altered. Similar perturbations in nitrate uptake may also occur, since an induction of the root high affinity nitrate transporter gene NRT1.1 was also detected. Interestingly, the accumulation of putrescine and spermidine, which have been shown to be involved in various stress and signaling processes, corresponded with the accumulation of transcripts for Arg decarboxylase, an enzyme involved in the synthesis of these two polyamines from Arg during which NADH-GDH may be involved (Skopelitis et al., 2006).
Intriguingly, a large proportion of the root transcripts present in lower amounts in the gdh1-2-3 mutant compared with the wild type corresponded to genes of unknown function and genes involved in plant ageing, nutrient recycling, and a variety of stress responses (Table 4). Although the exact function of NADH-GDH during protein degradation or during senescence is still a matter of debate (Tercé-Laforgue et al., 2004a; Masclaux-Daubresse et al., 2006; Miyashita and Good, 2008), the finding that a number of genes involved in the control of these two processes were downregulated in the mutant roots suggests that the enzyme activity itself could directly or indirectly trigger their induction.
Several classes of transcripts encoding proteins involved in signaling and in the regulation of transcriptional activity were also present in lower amounts in the triple mutant (see Supplemental Data Set 3 online), as has been frequently observed for a large number of mutations affecting either plant metabolism, such as photorespiration (Foyer et al., 2009), or development, such as floral identity (Causier et al., 2010). This suggests that there were probably modifications of a number of regulatory events that originate from the roots. These modifications may also involve the epigenetic control of root transcriptional activity since two putative miRNAs were upregulated in the roots of the gdh1-2-3 mutant (see Supplemental Data Set 4 online). Together, these results further strengthen the hypothesis that root transcriptional activity is directly or indirectly responsible for the impact of the gdh1-2-3 triple mutation on the physiology of the whole plant.
The Enzyme NADH-GDH Plays a Central Role at the Interface of C and N Metabolism, Primarily in Roots When C Becomes Limiting
In previous studies, it has been shown that placing plants under continuous darkness is a way of mimicking C deficiency (Miyashita and Good, 2008; Kunz et al., 2010). Although this procedure seems to be rather far from the standard day/night physiological conditions, it is known that plants can survive under prolonged darkness (Kunz et al., 2010). In agreement with this study, 28-d-old wild-type and gdh triple mutant plants at the rosette stage did not show any visible necrotic or senescent phenotype even after 7 d in the dark (see Supplemental Figure 2C online). In the study of Miyashita and Good (2008), necrotic lesions were already visible in the leaves of the double gdh1-2 mutant after 3 d in the dark. However, when compared with the plants described in this research, the Miyashita and Good (2008) mutant plants were grown under longer days and lower light intensity and were visible at a much earlier stage of rosette development. In this study, the starch content of the leaves of both the mutant and the wild type, although decreased by half, was still high, indicating that the C reserves were not totally exhausted. It is also unlikely that there was leaf N remobilization resulting from dark-induced leaf senescence (Kurt et al., 2006; Wingler et al., 2009), since the increase in the free amino content only concerned certain classes of amino acids derived from pyruvate and oxaloacetate. This suggests that N recycling from protein hydrolysis leading to the release of a larger spectrum of amino acids (Barneix, 2007) was limited.
When considering the differences in the metabolites involved in primary C and N metabolism, it is important to observe whether the changes that occurred in the dark took place in both the wild type and gdh1-2-3 mutant or only in the mutant. For example, regardless of the effect of the mutation, the accumulation of Asn and the decrease in soluble carbohydrates, such as Suc, Glc, and Fru (Figure 7), represent well-known physiological processes resulting from the absence of light, which appear to occur more in the leaves (Gibon et al., 2004; Lea et al., 2007; Stitt et al., 2010; Tcherkez et al., 2012). Considering the effect of the mutation alone, differences in the relative amounts of primary C and N metabolites occurred mainly in the roots. These differences concerned a number of amino acids (mostly Ala, Asp, and GABA and to a much lesser extent Asn, Glu, and Gln), organic acids (pyruvate, 2-oxoglutarate, and malate), soluble sugars (Glc-6-P, Fru-6-P, and to a lesser extent Suc), and ions such as ammonium in the leaves and nitrate in the roots. These findings further support the hypothesis that arose from the transcriptome study, in which it was shown that the main physiological impacts of the gdh1-2-3 mutations originate in the roots. The differences observed between the wild type and triple mutant in the leaves following dark exposure concerned fewer of the metabolites listed above, which could be either due to their selective translocation from the roots or to signals from the roots modifying their accumulation in the leaves. One explanation for this accumulation may be related to the perturbation of the TCA cycle in the roots, resulting either from the lack of 2-oxoglutarate production or from the accumulation of pyruvate.
Most interestingly, the dark period provides evidence that the metabolism of Glu is diverted through the GABA shunt (Fait et al., 2008) to regulate the homeostasis of Glu, which would be needed in the absence of NADH-GDH. It has been suggested that both NADH-GDH and Glu decarboxylase are involved in the homeostatic regulation of the cellular concentrations of Glu within the plant to avoid its accumulation (Forde and Lea, 2007; Labboun et al., 2009). Glu and GABA may also have an indirect signaling function on a number of plant metabolic and developmental processes (Fait et al., 2006; Forde and Lea, 2007). The operation of the GABA shunt allows for Glu to be decarboxylated to yield GABA, which can be subsequently converted to succinate, with the amino group being transferred to Ala (Fait et al., 2008; Shelp et al., 2012). In this way, a carboxylic acid can be introduced into the TCA cycle at succinate (Figure 9) rather than at 2-oxoglutarate, which would have occurred if NADH-GDH had been operating normally. Similar bypassing of enzymes in the TCA cycle via the GABA shunt have also been proposed when aconitase (Degu et al., 2011), 2-oxoglutarate dehydrogenase (Araújo et al., 2008), or succinyl-CoA ligase (Studart-Guimarães et al., 2007) have been inhibited. The reduction in the ammonium and nitrate content of the leaves and roots, respectively, of the triple mutant appears to be a consequence of the perturbation of the N primary assimilation pathway possibly involving feedback regulatory mechanisms in the uptake of NO3− and its subsequent reduction when there are changes in the N status of the plant (Girin et al., 2010).
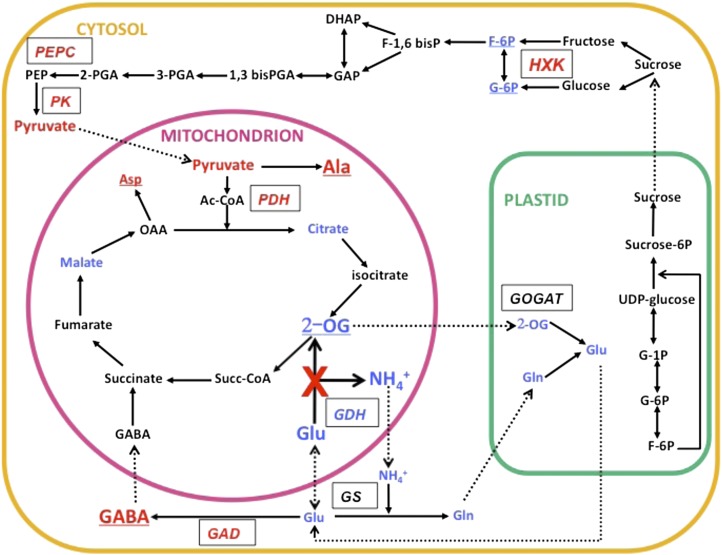
Figure 9.
Schematic Representation of the Physiological Changes that Occurred in the Roots of the Triple gdh1-2-3 Mutant.
A simplified representation of the main metabolic pathways involved in C and N metabolism is shown. Metabolites detected in lower or higher concentrations in the triple mutant are shown in blue and red, respectively. Dotted lines indicate transport of metabolites. Enzymes are shown in italics in boxes. Enzymes for which gene expression was upregulated in the triple gdh1-2-3 mutant are shown in red, and those that were downregulated are shown in blue. Metabolites underlined exhibit a similar pattern of accumulation in leaves. GAD, Glu decarboxylase; GOGAT, Glu synthase; GS, Glu synthetase; HXK, hexokinase; PDH, pyruvate dehydrogenase; PEPC, phosphoenolpyruvate carboxylase.
Surprisingly, the gdh1-2-3 mutant plants maintained a higher level of dark respiration in roots compared with the wild type even after 7 d of dark exposure. This observation further confirms that the physiological impact of the gdh mutation was mainly confined to the roots. In most investigations into transgenic plants or mutants with deficiencies in the TCA cycle, reductions in the rate of respiration have been observed (Araújo et al., 2012). However, an increase in the rate of respiration has been previously demonstrated in plants with reduced activity of the leaf enzymes citrate synthase (Sienkiewicz-Porzucek et al., 2008) and malate dehydrogenase (Tomaz et al., 2010). This increase in respiration, although not fully understood, could be attributed to the occurrence of compensatory respiratory mechanisms resulting from the lower flux of C going through the TCA cycle. The hypothesis that the enzyme malate dehydrogenase is a significant regulator of respiration in plants was also put forward (Tomaz et al., 2010); thus, the same situation may occur in roots when GDH activity is impaired. The data confirm that 2-oxoglutarate and the reactions involved in its production and use play a critical role in controlling the rate of respiration (Araújo et al., 2008, 2012). However, further work is necessary to assess the role of GDH in root energy production and to determine if alternate pathways, such as the GABA shunt, are able to bypass 2-oxoglutarate production via the synthesis of succinate.
The main differences between the metabolite content of the roots of the gdh1-2-3 mutant compared with the wild type following 7 d in the dark are presented in a metabolic scheme in Figure 9 and may be interpreted as follows: (1) As there is no NADH-GDH activity, the deamination of Glu does not take place and the amount of 2-oxoglutarate in the mutant decreases to a level close to the limit of detection. (2) The low concentrations of 2-oxoglutarate and Gln reduce the flux through the GS/GOGAT cycle. (3) Glu is metabolized through the GABA shunt, probably to avoid its accumulation (Forde and Lea, 2007; Fait et al., 2008). The amino group of GABA is transaminated to pyruvate to yield Ala (Clark et al., 2009), which may be then transaminated to Asp if oxaloacetate is available. (4) There is a reduction in the flux through TCA cycle due to the reduced availability of 2-oxoglutarate, although this may be compensated to a certain extent by the synthesis of succinate via the GABA shunt. Lower concentrations of malate and citrate and an accumulation of pyruvate would again suggest that the TCA cycle does not function normally. Pyruvate may alternatively be used for the synthesis of the various amino acids, in particular Ala, which also accumulates. When pyruvate is metabolized in the TCA cycle, it gives rise to CO2 and NADH but is not able to replenish metabolites. This can be performed by the anaplerotic reaction of phosphoenolpyruvate carboxylase, which synthesizes oxaloacetate (Nunes-Nesi et al., 2010) and is also the transamination substrate for the formation of Asp. A gene encoding phosphoenolpyruvate carboxylase is upregulated in the roots of the triple mutant, as shown in Table 3.
METHODS
Plant Material and Production of gdh1-2-3 Triple Mutants
Seeds of Arabidopsis thaliana background Columbia-0 (N60000), the wild type, and the three NADH-GDH T-DNA insertion mutants, gdh1 (SALK_042736), gdh2 (SALK_102711), and gdh3 (SALK_137670), were obtained from the Nottingham Arabidopsis Stock Centre. Before performing crosses to obtain the gdh1-2 double mutant and the gdh1-2-3 triple mutant, six backcrosses were performed for each mutant line. Homozygous mutant plants for the T-DNA insertion were isolated from the seed stocks using specific pairs of primers for each insertion of the T-DNA (see Supplemental Table 3 online for primer sequences). The gdh1-2 double mutant was obtained by crossing the gdh1 and gdh2 single mutants. Since the genes encoding GDH1 and GDH2 are both located on chromosome 5, the gdh1 gdh2 double mutants were first selected by producing a F2 population of double mutants homozygous for one of the two genes and heterozygous for the other. Homozygous plants for the two mutations were selected following the production of a F3 double mutant population. A cross between the double mutant gdh1-2 and the single mutant gdh3 allowed the production of a F2 population homozygous for gdh1-2 and heterozygous for gdh3, which was followed by the production of a F3 population, from which one line corresponding to a homozygous triple mutant gdh1-2-3 was selected. The occurrence of knockout mutations for the three genes encoding GDH in the gdh1-2-3 mutant was checked by qRT-PCR for transcript levels by staining for NAD-GDH activity following PAGE and by measuring extractable enzyme activity as described below. Characterization of gdh1 and gdh2 single mutants was performed previously (Fontaine et al., 2006).
Plant Growth Conditions
Arabidopsis wild-type or mutant seeds were surface sterilized for 20 min in a 0.25% NaOCl solution containing 0.1% Tween 20, washed six times in sterile water, and placed on Petri dishes containing a complete Murashige and Skoog (Murashige and Skoog, 1962) medium (Duchefa) solidified with 0.8% agar (Sigma-Aldrich), and supplemented with 2% Suc. After 48 to 72 h of incubation at 4°C in the dark, the plates were transferred to a controlled environment growth chamber (8 h light, 350 to 400 μmol photons m−2 s−1, 16 h dark at 25°C). After 2 to 3 weeks, seedlings were removed from the plates and transferred to hydroponic units consisting of plastic containers containing 10 liters of complete nutrient solution containing 6 mM NO3− as sole N source (Orsel et al., 2004). Each hydroponic unit contained 59 plants. Plants were grown for 35 d on a nutrient solution containing 3 mM KNO3, 1.5 mM Ca(NO3)2, 1 mM MgSO4, 1 mM KH2PO4, 0.5 mM K2SO4, 0.07 mM CaCl2, 10 mM MnSO4, 24 mM H3BO3, 3 mM CuSO4, 0.9 mM ZnSO4, 0.04 mM (NH4)6Mo7O24, and Fe-EDTA 10 mg L−1 (Sequestrene; Syngenta). The nutrient solution was replaced every day. For the metabolome and transcriptome analyses, roots and leaves were harvested 2 h after the beginning of the light period (T0). To induce flowering, plants were grown on the same hydroponic system and transferred to 16 h light until they reached the 10 open flowers stage. For the dark-induced experiments, 25 wild-type plants and 25 triple mutants were grown for 28 d under hydroponic conditions as described above in a controlled environment growth chamber (8 h light, 350 to 400 μmol photons-m−2 s−1, 25°C; 16 h dark, 18°C). They were then placed in complete darkness for 7 d at 18°C, and the roots and shoots were harvested at 3 d (D3) and 7 d (D7).
Enzyme Assays and Acrylamide Gel Analysis
For soluble protein extraction, leaves and roots of 28-d-old Arabidopsis plants were harvested. Soluble proteins were extracted from frozen root and leaf material stored at −80°C. All extractions were performed at 4°C. NADH-GDH activity was measured as described by Turano et al. (1996). Results for the NADH-GDH activities are presented as mean values for four plants with standard errors (se = sd/√n-1, where sd is the standard deviation and n the number of replicates). For ion exchange chromatography, all operations were performed at 4°C. Soluble proteins were extracted from 2 g of frozen root material in 10 mL of a buffer containing 25 mM Tricine, 10 mM CaCl2, 1mM EDTA, 0.05% (v/v) Triton X-100, 10 mM β-mercaptoethanol, and 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride hydrochloride, pH 8.0, at 4°C. Extracts were then centrifuged at 15,000g for 15 min. The supernatant was filtered (0.2-µm Gelman Sciences filter) and injected onto a MonoQ anion exchange column (5/50 GL; GE Healthcare) connected to a fast protein liquid chromatography system (ÄKTApurifier; GE Healthcare). The Mono Q column had been pre-equilibrated with 30 mL of buffer containing 25 mM Tricine, 10 mM CaCl2, 1 mM EDTA, and 0.05% (v/v) Triton X-100, pH 8. Fractions were eluted from the column using a linear gradient between 0.04 and 0.4 M NaCl (gdh1 and gdh2 mutants) or 0 and 1 M NaCl (gdh1-2 mutants) at a flow rate of 1.0 mL min−1. Fractions (500 µL) were collected and assayed for soluble protein concentration, NADH-GDH activity, and subjected to nondenaturing PAGE followed by NAD-GDH in-gel activity staining as described by Restivo (2004). Staining for NAD-GDH activity revealed an isoenzyme profile in agreement with the work of Loulakakis and Roubelakis-Angelakis (1996). Soluble protein was determined using a commercially available kit (Coomassie Protein assay reagent; Bio-Rad) using BSA as a standard. Leaf and root soluble proteins were transferred to nitrocellulose membranes for immunoblot analysis. Antibodies against GDH protein were obtained by immunization of rabbits (Eurogentec X2 protocol) with two synthetic peptides (NH2-VQHDNARGPMKGGIRC-CONH2 and NH2-PIDLGGSLGRDAATGR-CONH2) corresponding to a highly conserved motif in Arabidopsis GDH1 and GDH2 proteins (Fontaine et al., 2006). Antibodies raised against GDH from grapevine (Vitis vinifera) were also used as a control (Loulakakis and Roubelakis-Angelakis, 1990).
Quantitative Real-Time Reverse Transcription Experiments
Total RNA was extracted from leaves and roots using TRIzol Reagent (Invitrogen) according to the manufacturer’s protocol. Genomic DNA was removed from the total RNA by treatment with amplification-grade DNase I (Sigma-Aldrich) according to the manufacturer’s protocol. RNA was subsequently purified by two phenol washes, precipitated overnight in ethanol, and resuspended in water. Two micrograms of total RNA were reverse-transcribed for 1 h at 37°C, using 200 units of M-MLV reverse transcriptase (Promega) and 2 µg of pd(N)6 Random Hexamer (Amersham Biosciences) in the presence of 40 units of Recombinant Rnasin Ribonuclease Inhibitor (Promega) in a 50-μL final volume. The cDNAs were used for PCR experiments using gene-specific primers designed for GDH1, GDH2, and GDH3 transcripts (listed in Supplemental Table 4 online). qRT-PCR was performed on a Roche LightCycler using the ABsolute QPCR SYBR Green Capillary Mix (Applied Biosystems). Each reaction was performed using 5 μL of a cDNA solution diluted twice (v/v) in a final volume of 20 µL. Amplification was conducted according to the manufacturer’s protocol. All reactions were duplicated using RNA extracted from two independent plant samples. Correction of the quantification efficiency was performed using RealQuant (LightCycler software 3.5; Roche) based on relative standard curves established for each target and reference transcripts. The relative standard curves were determined using cDNA dilutions and were used for each analysis. The expression of three β-actin genes of Arabidopsis, ACT2 (At3g18780), ACT7 (At5g09810), and ACT8 (At1g49240), were used as combined internal standards to normalize template amounts (Schenk et al., 2003). Primers are listed in Supplemental Table 4 online. For mRNA quantification, results are presented as mean values for four plants with standard errors (se = sd/√n-1, where sd is the standard deviation and n the number of replicates). For the qRT-PCR experiment used to validate the CATMA microarray experiment, the three genes PP2AA3 encoding the 65-kD regulatory subunit of protein phosphatase 2A subunit (At1g13320); Q-TIP41, a TIP41-like family protein (At4g34270); and UBI-10, one of five polyubiquitin genes (At4g05320) were used to normalize the amount of templates. The three selected upregulated root genes were those that encoded the enzyme S-adenosylmethionine decarboxylase (SAMDC; At3g02470), the tonoplast monosaccharide transporter 1 (TMT1; At1g20840), and the enzyme Glu decarboxylase 2 (GAD2; At1g65960). The three selected downregulated root genes corresponded to a nitrate responsive NOI protein (NOI; At3g48450), a Ser-type endopeptidase inhibitor (ENDOP; At1g72060), and a Gln dumper 1 (GDU1; At4g31730). The three genes YSL8 (At5g08290); HAK5, a K transporter (At4g13420); and a G6Ta Gluc6PT (GPT2; At1g61800), which did not show any significant variation in the accumulation of their transcripts, were also used as controls in the qRT-PCR experiment. The specific primers designed for the genes are listed in Supplemental Table 5 online. Three technical replicates were performed on the two biological replicates used in the microarray experiment.
Cytoimmunochemical Studies
Cytoimmunochemical studies were conducted on roots and leaves of the gdh1-2-3 mutant using the GDH antiserum raised against synthetic GDH polypeptides diluted 300-fold in a 0.05 M Tris-HCl buffer containing 2.5% (w/v) NaCl, 0.1% (w/v) BSA, and 0.05% (v/v) Tween 20, pH 7.4, essentially as described by Fontaine et al. (2006). Antibodies raised against GDH purified from grapevine were also used as a control (Loulakakis and Roubelakis-Angelakis, 1990).
Microarray Analysis
Microarray analysis was performed at the Unité de Recherche en Génomique Végétale under the CATMA project. The CATMA project is being used to build a compendium of Arabidopsis gene expression profiles. The main objective of the European Union Framework project entitled Compendium of Arabidopsis Gene Expression (http://www.psb.ugent.be/CAGE) is to build a gene expression reference database containing 24,576 gene-specific tags corresponding to 22,089 genes from Arabidopsis (Crowe et al., 2003; Hilson et al., 2004). In addition, CATdb is a free resource available at http://urgv.evry.inra.fr/CATdb that provides public access to a large collection of transcriptome data for Arabidopsis produced by the CATMA Micro Array platform. Data in CATdb are entirely processed with the same standardized protocol, from microarray printing to data analyses. CATdb also gives easy access to a complete description of the experiments and experimental design (Gagnot et al., 2008). Two independent biological replicates were performed for each comparison and for each biological replicate, and two reverse-labeling technical replicates were performed. For each biological repetition and each time point, RNA samples were obtained by pooling RNA from 25 plants. Total RNA was extracted from leaves and roots using TRIzol reagent (Invitrogen) according to the supplier’s instructions. For each comparison, one technical replication with fluorochrome reversal was performed for each biological replicate (i.e., four hybridizations per comparison). The labeling of cRNAs with Cy3-dUTP or Cy5-dUTP (Perkin-Elmer-NEN Life Science Products), the hybridization to the slides, and the scanning were performed as described by Lurin et al. (2004).
Statistical Analysis of Microarray Data
Experiments were designed in collaboration with the Statistics Group of the Unité de Recherche en Génomique Végétale. Normalization and statistical analyses were based on two dye swaps (i.e., four arrays, each containing 24,576 gene-specific sequence tags and 384 controls) as described by Gagnot et al. (2008). To determine differentially expressed genes, a paired t test on the log ratios was performed, assuming that the variance of the log ratios was the same for all genes. Spots displaying extreme variance (too small or too large) were excluded. The spots that were excluded are those with a “specific variance/common variance” ratio smaller than the “alpha-quantile of a chi-squared distribution of one degree of liberty” or greater than the “1-alpha-quantile of a chi-squared distribution of one degree of liberty” with alpha equal to 0.0001 (Gagnot et al., 2008). The raw P values were adjusted by the Bonferroni method, which controls the family-wise error rate (with a type I error equal to 5%), in order to keep a strong control of the false positives in a multiple-comparison context (Ge et al., 2003). Genes with a Bonferroni P value ≤ 0.05 were considered as being differentially expressed, as described by Gagnot et al. (2008). In addition to the paired t test approach, the rank product method was used to detect differentially expressed genes according to different levels of FDR (Storey, 2003; Breitling et al., 2004a). We have chosen to present transcripts satisfying both the above family-wise error rate level and a FDR <0.0001% for an optimal interpretation of the transcriptome. Sorting genes by descending rank product values provided a hierarchical list based on both strength and reproducibility, which was used as an input to identify groups of genes with the same or related annotated function (iterative group analysis; Breitling et al., 2004b).
Metabolomic Studies Using Gas Chromatography–Mass Spectrometry Analysis
All steps were adapted from the original protocol described by Fiehn (2006). All extraction steps were performed in Safelock Eppendorf tubes. Five individual plants were used for the study, and for each plant, the ground frozen leaf and root samples were resuspended in 1 mL of frozen (−20°C) water:chloroform:methanol (1:1:2.5) and extracted for 10 min at 4°C with shaking at 1400 rpm in an Eppendorf Thermomixer. Insoluble material was removed by centrifugation, and 900 μL of the supernatant was mixed with 20 μL of 200 µg/mL ribitol in methanol. Water (360 µL) was then added and after mixing and centrifugation, 50 μL of the upper polar phase was collected and dried for 3 h in a Speed-Vac and stored at −80°C. Four blank tubes were subjected to the same steps as the samples. For derivatization, samples were removed from −80°C storage, warmed for 15 min before opening, and dried in a Speed-Vac for 1 h before the addition of 10 μL of 20 mg/mL methoxyamine in pyridine. The reactions with the individual samples, blanks, and amino acid standards were performed for 90 min at 28°C under continuous shaking in an Eppendorf thermomixer. Ninety microliters of N-methyl-N-trimethylsilyl-trifluoroacetamide were then added and the reaction continued for 30 min at 37°C. After cooling, 50 μL of the reaction mixture were transferred to an Agilent vial for injection. For the analysis, 3 h and 20 min after derivatization, 1 μL of the derivatized samples was injected in the splitless mode onto an Agilent 7890A gas chromatograph coupled to an Agilent 5975C mass spectrometer. The column used was an Rxi-5SilMS from Restek (30 m with 10 m Integra-Guard column). The liner (Restek #20994) was changed before each series of analyses, and 10 cm of the column was removed. The oven temperature ramp was 70°C for 7 min, then 10°C⋅min−1 up to 325°C, which was maintained for 4 min. Overall, the total run time was 36.5 min. A constant flow of helium was maintained at 1.5 mL min−1. Temperatures in the gas chromatograph were the following: injector, 250°C; transfer line, 290°C; source, 250°C; and quadripole, 150°C. Samples and blanks were randomized. Amino acid standards were injected at the beginning and end of the analyses for monitoring of the derivatization stability. An alkane mix (C10, C12, C15, C19, C22, C28, C32, and C36) was injected in the middle of the analyses for external retention index calibration. Five scans per second were acquired. For data processing, raw Agilent data files were converted into the NetCDF format and analyzed with AMDIS (http://chemdata.nist.gov/mass-spc/amdis/). A home retention indices/mass spectra library built from the National Institute of Standards and Technology, Golm, and Fiehn databases (Hummel et al., 2007) and standard compounds was used for metabolite identification. Peak areas were then determined using the QuanLynx software (Waters) after conversion of the NetCDF file into the MassLynx format. Statistical analyses were performed with TMEV (http://www.tm4.org/mev.html). Univariate analyses by permutation (one-way and two-way analysis of variance) were first used to select the metabolites exhibiting significant changes in their concentration. Starch and ammonium contents were determined separately as described by Tercé-Laforgue et al. (2004a). Nitrate was determined by the method of Cataldo et al. (1975).
Measurement of Respiration
The rate of respiration was determined over a 30- to 45-min period in the roots and leaves of the wild type and of the gdh1-2-3 triple mutant by measuring oxygen uptake. Respiration rate was linear after 3 min. Five individual plants were used for the study. Between 50 and 75 mg of fresh roots and leaves was harvested from the five individual plants grown as described above in the dark-induced experiment. The plant material was placed in a phosphate buffer, pH 6.8, at 25°C, with constant shaking in the dark. Oxygen consumption was measured with a YSI model 5300 biological oxygen monitor equipped with a Clark electrode and expressed in nmol O2 min−1 g−1 fresh weight.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database or the Arabidopsis Genome Initiative database under the following accession numbers: GDH1 (At5g18170), GDH2 (At5g07440), GDH3 (At3g03910), ACT2 (At3g18780), ACT7 (At5g09810), ACT8 (At1g49240), PP2AA3 (At1g13320), Q-TIP41 (At4g34270), and UBI-10 (At4g05320). Genes used for the qRT-PCR experiment to validate the CATMA microarray experiment were three selected upregulated root genes, SAMDC (At3g02470), TMT1 (At1g20840), and GAD2 (At1g65960); three selected downregulated root genes, NOI (At3g48450), ENDOP (At1g72060), and GDU1 (At4g31730), and three control genes that did not show any significant variation in the accumulation of their transcripts, YSL8 (At5g08290), HAK5 (At4g13420), and GPT2 (At1g61800). Germplasm used was as follows: gdh1 (SALK_042736), gdh2 (SALK_102711), and gdh3 (SALK_137670). Microarray data from this article have been deposited at Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE14614 and at CATdb (http://urgv.evry.inra.fr/CATdb/; Project AU2007-06_GDH).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Alignment of Deduced Protein Sequences of Arabidopsis GDH Genes Encoding the Three NADH-Dependent Enzymes and the NAD(P)H-Dependent Enzyme.
Supplemental Figure 2. Phenotype of the Wild Type and of the gdh1-2-3 Mutant.
Supplemental Figure 3. Overall Changes in Transcript Accumulation in Roots and Leaves of gdh1-2-3 Mutants.
Supplemental Figure 4. Respiration in Leaves and Roots of the Wild Type and gdh1-2-3 Mutant Plants Following Prolonged Darkness.
Supplemental Table 1. Quantification of GDH Protein in Different Tissue Sections of NADH-GDH Mutant Plants.
Supplemental Table 2. Comparison of the Results Obtained with the CATMA Microarray and a qRT-PCR Experiment for Transcript Abundance in the Arabidopsis gdh1-2-3 Mutant Compared with the Wild Type.
Supplemental Table 3. Oligonucleotides Used for Screening Arabidopsis GDH Mutant Plants.
Supplemental Table 4. Oligonucleotides Used for Amplification of RNA to Obtain DNA Fragments Specific for the Three Genes Encoding NADH-GDH and for the β-Actin Genes Used as Controls.
Supplemental Table 5. Oligonucleotides Used for Amplification of RNA to Obtain DNA Fragments for the qRT-PCR Used to Validate the Microarray Experiment.
Supplemental Data Set 1. Metabolites Exhibiting an Increase or Decrease in Roots of the gdh1-2-3 Mutants Compared with the Wild Type and Following Prolonged Darkness.
Supplemental Data Set 2. Metabolites Exhibiting an Increase or Decrease in Leaves of the gdh1-2-3 Mutants Compared with the Wild Type and Following Prolonged Darkness.
Supplemental Data Set 3. Transcripts Present in Lower Quantity in Roots of the Arabidopsis gdh1-2-3 Mutants Compared with the Wild Type.
Supplemental Data Set 4. Transcripts Present in Higher amounts in Roots of the Arabidopsis gdh1-2-3 Mutants Compared with the Wild Type.
Supplementary Material
Acknowledgments
This work was supported by the Conseil Régional de Picardie and the Fonds Social Européen. We thank Sophie Bouton and Karine Pageau for their helpful assistance and Jacques Legouis for fruitful discussions.
AUTHOR CONTRIBUTIONS
J.-X.F. produced the gdh triple mutant and performed its molecular and biochemical characterization. T.T.-L. performed the physiological characterization of the mutant under different growth conditions. P.A. performed the statistical analysis of the microarray data and contributed to the analysis in the metabolome data. G.C. performed the metabolome analysis and its statistical treatment. J.-P.R. and S.P. performed the CATMA microarray experiments and statistical analysis. M.C. performed metabolites measurements and respiration experiments. M.A. performed the HPLC analysis and the in-gel detection of GDH activity. Y.G. contributed to the physiological analysis of the mutant. F.D. performed the immunolocalization experiments. P.J.L., B.H., and F.D. designed the experiment, discussed the study, and wrote the article.
Glossary
- GDH
Glu dehydrogenase
- qRT-PCR
quantitative RT-PCR
- GABA
γ-aminobutyrate
- CATMA
Complete Arabidopsis Transcriptome Microarray
- FDR
false discovery rate
- TCA
tricarboxylic acid
References
- Araújo W.L., Nunes-Nesi A., Trenkamp S., Bunik V.I., Fernie A.R. (2008). Inhibition of 2-oxoglutarate dehydrogenase in potato tuber suggests the enzyme is limiting for respiration and confirms its importance in nitrogen assimilation. Plant Physiol. 148: 1782–1796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araújo W.L., Tohge T., Osorio S., Lohse M., Balbo I., Krahnert I., Sienkiewicz-Porzucek A., Usadel B., Nunes-Nesi A., Fernie A.R. (2012). Antisense inhibition of the 2-oxoglutarate dehydrogenase complex in tomato demonstrates its importance for plant respiration and during leaf senescence and fruit maturation. Plant Cell 24: 2328–2351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aubert S., Bligny R., Douce R., Gout E., Ratcliffe R.G., Roberts J.K. (2001). Contribution of glutamate dehydrogenase to mitochondrial glutamate metabolism studied by (13)C and (31)P nuclear magnetic resonance. J. Exp. Bot. 52: 37–45 [PubMed] [Google Scholar]
- Atkinson D.E. (1969). Regulation of enzyme function. Annu. Rev. Microbiol. 23: 47–68 [DOI] [PubMed] [Google Scholar]
- Barneix A.J. (2007). Physiology and biochemistry of source-regulated protein accumulation in the wheat grain. J. Plant Physiol. 164: 581–590 [DOI] [PubMed] [Google Scholar]
- Breitling R., Amtmann A., Herzyk P. (2004b). Iterative Group Analysis (iGA): A simple tool to enhance sensitivity and facilitate interpretation of microarray experiments. BMC Bioinformatics 5: 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitling R., Armengaud P., Amtmann A., Herzyk P. (2004a). Rank products: A simple, yet powerful, new method to detect differentially regulated genes in replicated microarray experiments. FEBS Lett. 573: 83–92 [DOI] [PubMed] [Google Scholar]
- Cammaerts D., Jacobs M. (1985). A study of the role of glutamate dehydrogenase in the nitrogen metabolism of Arabidopsis thaliana. Planta 163: 517–526 [DOI] [PubMed] [Google Scholar]
- Cataldo D.A., Haroon M., Schrader T.E., Youngs V.L. (1975). Rapid colorimetric determination of nitrate in plant tissue by nitration of salicylic acid. Commun. Soil Sci. Plant Anal. 6: 71–80 [Google Scholar]
- Causier B., Schwarz-Sommer Z., Davies B. (2010). Floral organ identity: 20 years of ABCs. Semin. Cell Dev. Biol. 21: 73–79 [DOI] [PubMed] [Google Scholar]
- Clark S.M., Di Leo R., Dhanoa P.K., Van Cauwenberghe O.R., Mullen R.T., Shelp B.J. (2009). Biochemical characterization, mitochondrial localization, expression, and potential functions for an Arabidopsis γ-aminobutyrate transaminase that utilizes both pyruvate and glyoxylate. J. Exp. Bot. 60: 1743–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowe M.L., et al. (2003). CATMA: A complete Arabidopsis GST database. Nucleic Acids Res. 31: 156–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Degu A., Hatew B., Nunes-Nesi A., Shlizerman L., Zur N., Katz E., Fernie A.R., Blumwald E., Sadka A. (2011). Inhibition of aconitase in citrus fruit callus results in a metabolic shift towards amino acid biosynthesis. Planta 234: 501–513 [DOI] [PubMed] [Google Scholar]
- Dubois F., Tercé-Laforgue T., Gonzalez-Moro M.B., Estavillo M.B., Sangwan R., Gallais A., Hirel B. (2003). Glutamate dehydrogenase in plants; is there a new story for an old enzyme? Plant Physiol. Biochem. 41: 565–576 [Google Scholar]
- Fait A., Fromm H., Walter D., Galili G., Fernie A.R. (2008). Highway or byway: The metabolic role of the GABA shunt in plants. Trends Plant Sci. 13: 14–19 [DOI] [PubMed] [Google Scholar]
- Fait A., Yellin A., Fromm H. (2006). GABA and GHB neurotransmitters in plants and animals. In Communication in Plants: Neuronal Aspects of Plant Life, F. Baluska, S. Mancuso, and D. Volkmann, eds (Dusseldorf, Germany: Springer), pp 171–185 [Google Scholar]
- Fernie A.R., Stitt M. (2012). On the discordance of metabolomics with proteomics and transcriptomics: Coping with increasing complexity in logic, chemistry, and network interactions scientific correspondence. Plant Physiol. 158: 1139–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O. (2006). Metabolite profiling in Arabidopsis. In Arabidopsis Protocols: Methods in Molecular Biology, 2nd ed., J. Salinas and J.J. Sanchez-Serrano, eds (Totowa, NJ: Humana Press), pp 439–447 [DOI] [PubMed] [Google Scholar]
- Fontaine J.X., Saladino F., Agrimonti C., Bedu M., Tercé-Laforgue T., Tétu T., Hirel B., Restivo F.M., Dubois F. (2006). Control of the synthesis and subcellular targeting of the two GDH genes products in leaves and stems of Nicotiana plumbaginifolia and Arabidopsis thaliana. Plant Cell Physiol. 47: 410–418 [DOI] [PubMed] [Google Scholar]
- Forde B.G., Lea P.J. (2007). Glutamate in plants: metabolism, regulation, and signalling. J. Exp. Bot. 58: 2339–2358 [DOI] [PubMed] [Google Scholar]
- Fox G.G., Ratcliffe R.G., Robinson S.A., Stewart G.R. (1995). Evidence for deamination by glutamate dehydrogenase in higher plants. Commentary. Rev. Can. Bot. 73: 1112–1115 [Google Scholar]
- Foyer C.H., Bloom A.J., Queval G., Noctor G. (2009). Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 60: 455–484 [DOI] [PubMed] [Google Scholar]
- Foyer C.H., Noctor G., Hodges M. (2011). Respiration and nitrogen assimilation: targeting mitochondria-associated metabolism as a means to enhance nitrogen use efficiency. J. Exp. Bot. 62: 1467–1482 [DOI] [PubMed] [Google Scholar]
- Frigerio F., Casimir M., Carobbio S., Maechler P. (2008). Tissue specificity of mitochondrial glutamate pathways and the control of metabolic homeostasis. Biochim. Biophys. Acta 1777: 965–972 [DOI] [PubMed] [Google Scholar]
- Gagnot S., Tamby J.P., Martin-Magniette M.L., Bitton F., Taconnat L., Balzergue S., Aubourg S., Renou J.P., Lecharny A., Brunaud V. (2008). CATdb: A public access to Arabidopsis transcriptome data from the URGV-CATMA platform. Nucleic Acids Res. 36(Database issue): D986–D990 (Database issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y., Dudoit S., Speed T.P. (2003). Resampling-based multiple testing for microarray data analysis. Math. Stat. 12: 1–77 [Google Scholar]
- Gibon Y., Bläsing O.E., Palacios-Rojas N., Pankovic D., Hendriks J.H.M., Fisahn J., Höhne M., Günther M., Stitt M. (2004). Adjustment of diurnal starch turnover to short days: Depletion of sugar during the night leads to a temporary inhibition of carbohydrate utilization, accumulation of sugars and post-translational activation of ADP-glucose pyrophosphorylase in the following light period. Plant J. 39: 847–862 [DOI] [PubMed] [Google Scholar]
- Girin T., El-Kafafi S., Widiez T., Erban A., Hubberten H.M., Kopka J., Hoefgen R., Gojon A., Lepetit M. (2010). Identification of Arabidopsis mutants impaired in the systemic regulation of root nitrate uptake by the nitrogen status of the plant. Plant Physiol. 153: 1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glevarec G., Bouton S., Jaspard E., Riou M.T., Cliquet J.B., Suzuki A., Limami A.M. (2004). Respective roles of the glutamine synthetase/glutamate synthase cycle and glutamate dehydrogenase in ammonium and amino acid metabolism during germination and post-germinative growth in the model legume Medicago truncatula. Planta 219: 286–297 [DOI] [PubMed] [Google Scholar]
- Grabowska A., Nowicki M., Kwinta J. (2011). Glutamate dehydrogenase of the germinating triticale seeds: Gene expression, activity distribution and kinetic characteristics. Acta Physiol. Plant. 33: 1981–1990 [Google Scholar]
- Hilson P., et al. (2004). Versatile gene-specific sequence tags for Arabidopsis functional genomics: Transcript profiling and reverse genetics applications. Genome Res. 14: 2176–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirai M.Y., Yano M., Goodenowe D.B., Kanaya S., Kimura T., Awazuhara M., Arita M., Fujiwara T., Saito K. (2004). Integration of transcriptomics and metabolomics for understanding of global responses to nutritional stresses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 101: 10205–10210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel J., Selbig S., Walther D., Kopla J. (2007). The Golm Metabolome Database: A database for GC-MS based metabolite profiling. Topics in Current Genetics. Metabolomics 18: 75–95 [Google Scholar]
- Igarashi D., Izumi Y., Dokiya Y., Totsuka K., Fukusaki E., Ohsumi C. (2009). Reproductive organs regulate leaf nitrogen metabolism mediated by cytokinin signal. Planta 229: 633–644 [DOI] [PubMed] [Google Scholar]
- Jaspard E. (2006). A computational analysis of the three isoforms of glutamate dehydrogenase reveals structural features of the isoform EC 1.4.1.4 supporting a key role in ammonium assimilation by plants. Biol. Direct 1: 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz H.-H., Scharnewski M., von Berlepsch S., Shahi S., Fulda M., Flügge U.-I., Gierth M. (2010). Nocturnal energy demand in plants: Insights from studying mutants impaired in β-oxidation. Plant Signal. Behav. 5: 842–844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurt M., Peeters U., van Laere A.J. (2006). Ammonium and amino acid metabolism in excised leaves of wheat (Triticum aestivum) senescing in the dark. Physiol. Plant. 84: 243–249 [Google Scholar]
- Labboun S., Tercé-Laforgue T., Roscher A., Bedu M., Restivo F.M., Velanis C.N., Skopelitis D.S., Moschou P.N., Roubelakis-Angelakis K.A., Suzuki A., Hirel B. (2009). Resolving the role of plant glutamate dehydrogenase. I. In vivo real time nuclear magnetic resonance spectroscopy experiments. Plant Cell Physiol. 50: 1761–1773 Erratum. Plant Cell Physiol. 50: 1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lea P.J., Miflin B.J. (2011). Nitrogen assimilation and its relevance to crop improvement. In Nitrogen Metabolism in Plants in the Post-Genomic Era, C.H. Foyer and H. Zhang, eds (Chichester, UK: Wiley-Blackwell), pp 1–40 [Google Scholar]
- Lea P.J., Sodek L., Parry M.A.J., Shewry P.R., Halford N.G. (2007). Asparagine in plants. Ann. Appl. Biol. 150: 1–26 [Google Scholar]
- Lea P.J., Thurman D.A. (1972). Intracellular location and properties of plant L-glutamate dehydrogenases. J. Exp. Bot. 23: 440–449 [Google Scholar]
- Leech R.M., Kirk P.R. (1968). An NADP-dependent L-glutamate dehydrogenase from chloroplasts of Vicia faba L. Biochem. Biophys. Res. Commun. 32: 685–690 [DOI] [PubMed] [Google Scholar]
- Lemaitre T., Urbanczyk-Wochniak E., Flesch V., Bismuth E., Fernie A.R., Hodges M. (2007). NAD-dependent isocitrate dehydrogenase mutants of Arabidopsis suggest the enzyme is not limiting for nitrogen assimilation. Plant Physiol. 144: 1546–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limami A.M., Glévarec G., Ricoult C., Cliquet J.B., Planchet E. (2008). Concerted modulation of alanine and glutamate metabolism in young Medicago truncatula seedlings under hypoxic stress. J. Exp. Bot. 59: 2325–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulakakis C.A., Roubelakis-Angelakis K.A. (1990). Immunocharacterization of NADH-glutamate dehydrogenase from Vitis vinifera L. Plant Physiol. 94: 109–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loulakakis K.A., Roubelakis-Angelakis K.A. (1996). The seven NAD(H)-glutamate dehydrogenase isoenzymes exhibit similar anabolic activities. Physiol. Plant. 96: 29–35 [Google Scholar]
- Lurin C., et al. (2004). Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magasanik B. (1992). Regulation of nitrogen assimilation. In The Molecular and Cell Biology of the Yeast Saccharomyces cerevisiae: Gene Expression, Vol. II, E.W. Jones, J.R. Pringle, and J.R. Broach, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp 283–317 [Google Scholar]
- Melo-Oliveira R., Oliveira I.C., Coruzzi G.M. (1996). Arabidopsis mutant analysis and gene regulation define a nonredundant role for glutamate dehydrogenase in nitrogen assimilation. Proc. Natl. Acad. Sci. USA 96: 4718–4723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masclaux-Daubresse C., Reisdorf-Cren M., Pageau K., Lelandais M., Grandjean O., Kronenberger J., Valadier M.H., Feraud M., Jouglet T., Suzuki A. (2006). Glutamine synthetase-glutamate synthase pathway and glutamate dehydrogenase play distinct roles in the sink-source nitrogen cycle in tobacco. Plant Physiol. 140: 444–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyashita Y., Good A.G. (2008). NAD(H)-dependent glutamate dehydrogenase is essential for the survival of Arabidopsis thaliana during dark-induced carbon starvation. J. Exp. Bot. 59: 667–680 [DOI] [PubMed] [Google Scholar]
- Morel M., Buée M., Chalot M., Brun A. (2006). NADP-dependent glutamate dehydrogenase: a dispensable function in ectomycorrhizal fungi. New Phytol. 169: 179–189 [DOI] [PubMed] [Google Scholar]
- Murashige J., Skoog F. (1962). A revised medium for rapid growth and bioassay with tobacco tissue culture. Physiol. Plant. 115: 473–497 [Google Scholar]
- Nunes-Nesi A., Fernie A.R., Stitt M. (2010). Metabolic and signaling aspects underpinning the regulation of plant carbon nitrogen interactions. Mol. Plant 3: 973–996 [DOI] [PubMed] [Google Scholar]
- Oaks A. (1995). Evidence for deamination by glutamate dehydrogenase in higher plants. Reply Can. J. Bot. 73: 1116–1117 [Google Scholar]
- Orsel M., Eulenburg K., Krapp A., Daniel-Vedele F. (2004). Disruption of the nitrate transporter genes AtNRT2.1 and AtNRT2.2 restricts growth at low external nitrate concentration. Planta 219: 714–721 [DOI] [PubMed] [Google Scholar]
- Purnell M.P., Skopelitis D.S., Roubelakis-Angelakis K.A., Botella J.R. (2005). Modulation of higher-plant NAD(H)-dependent glutamate dehydrogenase activity in transgenic tobacco via alteration of beta subunit levels. Planta 222: 167–180 [DOI] [PubMed] [Google Scholar]
- Qiu X., Xie W., Lian X., Zhang Q. (2009). Molecular analyses of the rice glutamate dehydrogenase gene family and their response to nitrogen and phosphorous deprivation. Plant Cell Rep. 28: 1115–1126 [DOI] [PubMed] [Google Scholar]
- Restivo F.M. (2004). Molecular cloning of glutamate dehydrogenase genes of Nicotiana plumbaginifolia: Structure and regulation of their expression by physiological and stress conditions. Plant Sci. 166: 971–982 [Google Scholar]
- Robinson S.A., Stewart G.R., Phillips R. (1992). Regulation of glutamate dehydrogenase activity in relation to carbon limitation and protein catabolism in carrot cell suspension cultures. Plant Physiol. 98: 1190–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk P.M., Kazan K., Manners J.M., Anderson J.P., Simpson R.S., Wilson I.W., Somerville S.C., Maclean D.J. (2003). Systemic gene expression in Arabidopsis during an incompatible interaction with Alternaria brassicicola. Plant Physiol. 132: 999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelp B.J., Mullen R.T., Waller J.C. (2012). Compartmentation of GABA metabolism raises intriguing questions. Trends Plant Sci. 17: 57–59 [DOI] [PubMed] [Google Scholar]
- Sienkiewicz-Porzucek A., Nunes-Nesi A., Sulpice R., Lisec J., Centeno D.C., Carillo P., Leisse A., Urbanczyk-Wochniak E., Fernie A.R. (2008). Mild reductions in mitochondrial citrate synthase activity result in a compromised nitrate assimilation and reduced leaf pigmentation but have no effect on photosynthetic performance or growth. Plant Physiol. 147: 115–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitis D.S., Paranychiankis N.V., Kouvarakis A., Spyros A., Stephanou E.G., Roubelakis-Angelakis K.A. (2007). The isoenzyme 7 of tobacco NADH-dependent glutamate dehydrogenase exhibits high deaminating and low aminating activity. Plant Physiol. 145: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skopelitis D.S., Paranychianakis N.V., Paschalidis K.A., Pliakonis E.D., Delis I.D., Yakoumakis D.I., Kouvarakis A., Papadakis A.K., Stephanou E.G., Roubelakis-Angelakis K.A. (2006). Abiotic stress generates ROS that signal expression of anionic glutamate dehydrogenases to form glutamate for proline synthesis in tobacco and grapevine. Plant Cell 18: 2767–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart G.R., Shatilov V.R., Turnbull M.H., Robinson S.A., Goodall R. (1995). Evidence that glutamate dehydrogenase plays a role in the oxidative deamination of glutamate in seedlings of Zea mays. Funct. Plant Biol. 22: 805–809 [Google Scholar]
- Stitt M., Lunn J., Usadel B. (2010). Arabidopsis and primary photosynthetic metabolism - More than the icing on the cake. Plant J. 61: 1067–1091 [DOI] [PubMed] [Google Scholar]
- Storey J.D. (2003). The positive false discovery rate: A Bayesian interpretation and the q-value. Ann. Stat. 31: 2013–2035 [Google Scholar]
- Studart-Guimarães C., Fait A., Nunes-Nesi A., Carrari F., Usadel B., Fernie A.R. (2007). Reduced expression of succinyl-coenzyme A ligase can be compensated for by up-regulation of the γ-aminobutyrate shunt in illuminated tomato leaves. Plant Physiol. 145: 626–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tcherkez G., Boex-Fontvieille E., Mahé A., Hodges M. (2012). Respiratory carbon fluxes in leaves. Curr. Opin. Plant Biol. 15: 308–314 [DOI] [PubMed] [Google Scholar]
- Tercé-Laforgue T., Dubois F., Ferrario-Mery S., Pou de Crecenzo M.A., Sangwan R., Hirel B. (2004b). Glutamate dehydrogenase of tobacco is mainly induced in the cytosol of phloem companion cells when ammonia is provided either externally or released during photorespiration. Plant Physiol. 136: 4308–4317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tercé-Laforgue T., Mäck G., Hirel B. (2004a). New insights towards the function of glutamate dehydrogenase revealed during source-sink transition of tobacco (Nicotiana tabacum) plants grown under different nitrogen regimes. Physiol. Plant. 120: 220–228 [DOI] [PubMed] [Google Scholar]
- Thurman D.A., Palin C., Laycock M.V. (1965). Isoenzymatic nature of L-glutamic dehydrogenase of higher plants. Nature 207: 193–194 [DOI] [PubMed] [Google Scholar]
- Tomaz T., Bagard M., Pracharoenwattana I., Lindén P., Lee C.P., Carroll A.J., Ströher E., Smith S.M., Gardeström P., Millar A.H. (2010). Mitochondrial malate dehydrogenase lowers leaf respiration and alters photorespiration and plant growth in Arabidopsis. Plant Physiol. 154: 1143–1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turano F.J., Dashner R., Upadhyaya A., Caldwell C.R. (1996). Purification of mitochondrial glutamate dehydrogenase from dark-grown soybean seedlings. Plant Physiol. 112: 1357–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turano F.J., Thakkar S.S., Fang T., Weisemann J.M. (1997). Characterization and expression of NAD(H)-dependent glutamate dehydrogenase genes in Arabidopsis. Plant Physiol. 113: 1329–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingler A., Masclaux-Daubresse C., Fischer A.M. (2009). Sugars, senescence, and ageing in plants and heterotrophic organisms. J. Exp. Bot. 60: 1063–1066 [DOI] [PubMed] [Google Scholar]
- Yamada K., et al. (2003). Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yamaya T., Oaks A. (1987). Synthesis of glutamate by mitochondria - An anaplerotic function for glutamate dehydrogenase. Physiol. Plant. 70: 749–756 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.