This work examines the role of a nonhomologous end-joining pathway gene, X-RAY CROSS COMPLEMENTATION GROUP4 (XRCC4), in T-DNA integration, finding that XRCC4 limits T-DNA integration and interacts directly with Agrobacterium VirE2 protein. These results suggest that Agrobacterium may modulate host DNA repair functions to increase T-DNA integration.
Abstract
Agrobacterium tumefaciens is a soilborne pathogen that causes crown gall disease in many dicotyledonous plants by transfer of a portion of its tumor-inducing plasmid (T-DNA) into the plant genome. Several plant factors that play a role in Agrobacterium attachment to plant cells and transport of T-DNA to the nucleus have been identified, but the T-DNA integration step during transformation is poorly understood and has been proposed to occur via nonhomologous end-joining (NHEJ)–mediated double-strand DNA break (DSB) repair. Here, we report a negative role of X-RAY CROSS COMPLEMENTATION GROUP4 (XRCC4), one of the key proteins required for NHEJ, in Agrobacterium T-DNA integration. Downregulation of XRCC4 in Arabidopsis and Nicotiana benthamiana increased stable transformation due to increased T-DNA integration. Overexpression of XRCC4 in Arabidopsis decreased stable transformation due to decreased T-DNA integration. Interestingly, XRCC4 directly interacted with Agrobacterium protein VirE2 in a yeast two-hybrid system and in planta. VirE2-expressing Arabidopsis plants were more susceptible to the DNA damaging chemical bleomycin and showed increased stable transformation. We hypothesize that VirE2 titrates or excludes active XRCC4 protein available for DSB repair, thus delaying the closure of DSBs in the chromosome, providing greater opportunity for T-DNA to integrate.
INTRODUCTION
Agrobacterium tumefaciens is well known as a natural transformation agent because of its ability to transfer a fragment of its Ti (tumor-inducing) plasmid known as T-DNA into its hosts. T-DNA–encoded oncogene expression leads to opine and plant hormone production, followed by tumor induction, causing crown gall disease in many dicotyledonous plants. Certain plant phenolic compounds, such as acetosyringone, induce Ti plasmid–encoded virulence (vir) genes, leading to production of T-strands that associate with various Vir proteins to form a proposed T-complex, which is transferred to the host via a type IV secretion system (Gelvin, 2010; Pitzschke and Hirt, 2010). Vir proteins associated with the proposed T-complex interact with various plant factors to help direct the import of the T-complex from the cytoplasm into the plant nucleus. These proteins may eventually be stripped from the T-DNA by the SCFvirF complex (Zaltsman et al., 2010; Anand et al., 2012). The T-DNA then presumably is converted into a double-stranded moiety and integrates into the host genome (Tzfira et al., 2004). T-DNA integrates at random sites (Kim et al., 2007) and preferentially into double-strand breaks (DSBs) (Salomon and Puchta, 1998; Chilton and Que, 2003; Tzfira et al., 2003). T-DNA integration into DSBs was inferred from increased foreign gene insertions into plants irradiated by x-rays (Köhler et al., 1989) and from plants whose genomes were cut by rare-cutting endonucleases (Salomon and Puchta, 1998; Chilton and Que, 2003; Tzfira et al., 2003). The process of T-DNA integration is not well understood. Studies in yeast have identified yeast genes required for T-DNA integration both via nonhomologous recombination and homologous recombination (van Attikum et al., 2001, 2003). Several Arabidopsis thaliana ecotypes and mutants deficient in T-DNA integration have also been identified and characterized (Nam et al., 1997, 1998; Mysore et al., 2000a, 2000b; Anand et al., 2007a).
The DNA repair machinery of higher eukaryotes comprises five pathways of which homologous recombination and nonhomologous end joining (NHEJ) are the major ones. Unlike in yeast, plants predominantly direct DSB repair via NHEJ (Ray and Langer, 2002; Britt and May, 2003). One of the models proposed for T-DNA integration in plants suggests the involvement of NHEJ-mediated DSB repair, which is inherent to eukaryotes (Tzfira et al., 2004). The principal components of the classical NHEJ pathway are the KU70-KU80 (Ku 70/80-kD autoantigen) heterodimer, DNA-activated protein kinase catalytic subunit, Ataxia telangiectasia mutated (ATM) kinase, ATM and Rad3-related kinase, and the X-RAY CROSS COMPLEMENTATION PROTEIN4 (XRCC4)-DNA ligase IV complex (Mladenov and Iliakis, 2011). Except for KU80 and DNA ligase IV, the role of the NHEJ proteins in T-DNA integration has not been studied. There are conflicting reports about the role of KU80 in T-DNA integration. Gallego et al. (2003) found no deficiency in T-DNA integration in Arabidopsis ku80 mutants, whereas Friesner and Britt (2003) showed a reduction in T-DNA integration in Arabidopsis ku80 mutants. Subsequently, Li et al. (2005) showed that Arabidopsis ku80 mutants showed decreased stable transformation, whereas Ku80 overexpression showed increased T-DNA integration and increased resistance to methyl methanesulfonate (MMS).
An important step during NHEJ of DSBs is mediated by the XRCC4-DNA ligase IV complex. West et al. (2000) described the isolation and characterization of the Arabidopsis homologs of DNA ligase IV and XRCC4. They show a tight interaction in a yeast two-hybrid system between DNA ligase IV and XRCC4 via their BRCA1 C terminus domains and go on to show that DNA ligase IV is induced by gamma irradiation.
Since DNA ligase IV was shown to be not required for T-DNA integration (van Attikum et al., 2003), we focused on the role of XRCC4 in T-DNA integration in this study. Using overexpression and gene silencing studies in two models systems, Nicotiana benthamiana and Arabidopsis, we demonstrate that XRCC4 activity limits T-DNA integration. We also demonstrate that XRCC4 directly interacts with Agrobacterium VirE2 protein, both in a yeast-two-hybrid assay and in planta. Furthermore, we show that VirE2-expressing Arabidopsis plants are more sensitive to a DNA-damaging agent and exhibit increased stable transformation. Our study suggests that Agrobacterium may delay the plant DNA repair machinery for its benefit.
RESULTS
A Virus-Induced Gene Silencing–Based Reverse Genetics System Identifies a Role for XRCC4 in Agrobacterium-Mediated Plant Transformation
To investigate the role of XRCC4 in Agrobacterium-mediated plant transformation, we used a well-established virus-induced gene silencing (VIGS)-based reverse genetics system (Anand et al., 2007a, 2007b, 2008). A partial cDNA of N. benthamiana XRCC4 (see Supplemental Figure 1A online) cloned into the vector pTRV2 was used to silence the XRCC4 gene in N. benthamiana plants. Leaf disks from XRCC4-silenced (see Supplemental Figure 2A online) and control (TRV:GFP [for green fluorescent protein] inoculated) plants were inoculated with tumorigenic strain Agrobacterium A348 (Nam et al., 1999). Surprisingly, XRCC4-silenced leaf disks were more transformable than were leaf disks from control plants, as indicated by the fresh weight of leaf disks (Figure 1A) and the average number of leaf disks that showed tumors (Figure 1B; see Supplemental Figure 2B online).
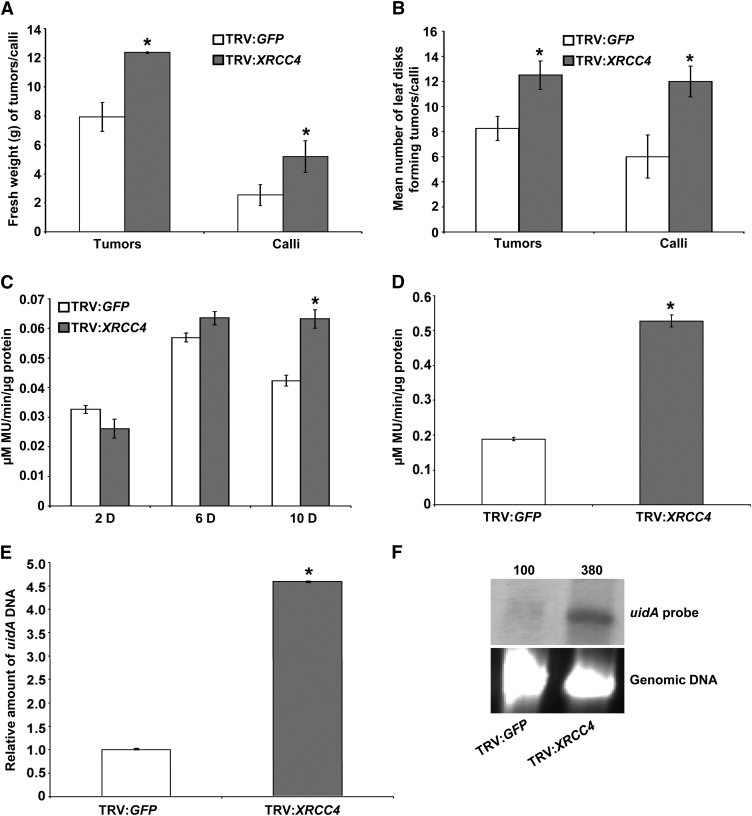
Figure 1.
VIGS of XRCC4 Suggests Its Negative Role in Stable Transformation and T-DNA Integration.
(A) Fresh weight quantification of TRV:GFP (control) and TRV:Nb-XRCC4–silenced leaf disks challenged with low concentration (1 × 107 cfu/mL) of Agrobacterium A348 for tumor assay or Agrobacterium GV2260 harboring pCAS1 for stable transformation assay to generate PPT-resistant calli.
(B) Quantification of the average number of leaf disks forming tumors or calli from the samples in (A) per 15 leaf disks. In (A) and (B), data are presented as mean ± sd (n = 10) from 10 replicates with 15 leaf disks per treatment.
(C) Quantification of GUS activity by recording the fluorescence of 4-methylumbelliferone (MU) from TRV:GFP and TRV:Nb-XRCC4–silenced leaf disks infected with Agrobacterium GV2260 harboring pBISN1 at 2, 6, and 10 DAI.
(D) Quantification of GUS activity 3 weeks after infection of TRV:GFP and TRV:Nb-XRCC4–silenced leaf disks infected with Agrobacterium GV2260 harboring pKM1. In (C) and (D), data represent the average of three biological replicates with sd (n = 5) values shown as error bars.
(E) qPCR-mediated quantification of relative T-DNA integration in callus suspension cultures raised from TRV:GFP and TRV:Nb-XRCC4–silenced leaf disks challenged with Agrobacterium GV2260 harboring pBISN1.
(F) T-DNA integration assay in TRV:Nb-XRCC4–silenced and TRV:GFP-infected plants. Undigested genomic DNA isolated from 2-month-old culture was blotted on to a nylon membrane and subjected to hybridization with a DIG-labeled uidA gene probe (top panel). The intensity of uidA-specific signals in the top panel was quantified using a densitometer, and values were assigned relative to that of TRV:GFP control (100%). Ethidium bromide staining shows equal loading of genomic DNA (bottom panel).
Asterisks in (A) to (E) indicate a significant difference between TRV:GFP and TRV:Nb-XRCC4–silenced samples according to Student’s t test (P < 0.05; see Supplemental Figures 1 and 2 online).
To rule out the possibility that VIGS of XRCC4 leads to upregulation of phytohormones, thus resulting in increased numbers of tumors on leaf disks, another independent stable transformation assay was performed using a nontumorigenic Agrobacterium strain. Leaf disks from XRCC4-silenced and control plants were cocultivated with the disarmed strain Agrobacterium GV2260 harboring pCAS1, which contains the phosphinothricin (PPT) resistance gene within the T-DNA (Nam et al., 1999). Herbicide-resistant calli were selected on callus induction medium (CIM) containing PPT. In this assay, the calli generated from XRCC4-silenced leaf disks showed a 56% increase in fresh weight compared with calli generated from control plants (Figure 1A). In addition, the average number of leaf disks producing PPT-resistant calli was significantly higher in XRCC4-silenced leaf disks when compared with control plants (Figures 1B; see Supplemental Figure 2B online). To rule out the possibility that the rate of cell division in XRCC4-silenced plants exceeds that of wild-type plants, uninfected leaf disks from control and XRCC4-silenced plants were cultured on CIM without selection, and no significant differences in fresh weights of calli were observed between the control and XRCC4-silenced leaf disks. XRCC4-silenced plants did not have any obvious developmental defects and looked similar to control plants.
XRCC4-Silenced Plants Integrate More T-DNA
Since T-DNA can integrate into DSBs and XRCC4 is one of the key proteins involved in NHEJ repair of DSBs, we investigated the role of XRCC4 in T-DNA integration. Control and XRCC4-silenced (see Supplemental Figure 2A online) N. benthamiana leaf disks were inoculated with the disarmed strain Agrobacterium GV2260 harboring the binary plasmid pBISN1 (contains a uidA-intron gene within the T-DNA) (Narasimhulu et al., 1996). Leaf disks were assayed for β-glucuronidase (GUS) activity at 2, 6, and 10 d after inoculation, with the first two time points representing transient expression from the T-DNA, while the 10 d time point likely represents stable transformation. Interestingly, the XRCC4-silenced plants did not show any significant difference in transient GUS activity at 2 and 6 d compared with control plants but showed a statistically significant increase in GUS activity (∼50% increase compared with control) at 10 d (Figure 1C).
To demonstrate that XRCC4 downregulation leads to increased stable transformation due to increased T-DNA integration, we inoculated control and XRCC4-silenced leaf disks with Agrobacterium GV2260 harboring the binary vector pKM1 (Mysore et al., 1998) that has a promoterless uidA-intron gene within the T-DNA. T-DNA integration downstream of a plant promoter is therefore needed for uidA expression. Calli generated on nonselective CIM from XRCC4-silenced leaf disks showed significantly higher GUS activity (∼60% more) than did calli from control plants (Figure 1D). This experiment suggests that downregulation of XRCC4 favors T-DNA integration.
The negative role of XRCC4 in T-DNA integration was further confirmed by quantitative PCR (qPCR) and a biochemical assay (Mysore et al., 2000a; Anand et al., 2007a). Control and XRCC4-silenced N. benthamiana leaf disks were infected with Agrobacterium GV2260 harboring pBISN1. Two days after cocultivation, the leaf disks were incubated on nonselective CIM to form calli. Four weeks later, the calli were transferred to nonselective liquid culture medium in the presence of antibiotics to remove Agrobacterium and subcultured for 3 months. Intact high molecular weight genomic DNA extracted from the suspension cultures derived from XRCC4-silenced and control plants was subjected to qPCR as described (Anand et al., 2007a). The relative amount of uidA DNA, corresponding to integrated T-DNA, was 4.5-fold greater in the XRCC4-silenced lines than in control (Figure 1E). We confirmed that there was no contaminating Agrobacterium DNA in plant genomic DNA by qPCR with primers specific to the Agrobacterium chromosomal gene Atu0972. Furthermore, we conducted DNA gel blot analysis on one of these lines. DNA from the XRCC4-silenced line showed a stronger hybridization signal (380% more signal) for a uidA probe compared with DNA from control plant (Figure 1F). Taken together, our results indicate that silencing of XRCC4 leads to increased T-DNA integration and, thus, stable transformation.
XRCC4 Downregulation in Arabidopsis Leads to Increased Stable Transformation
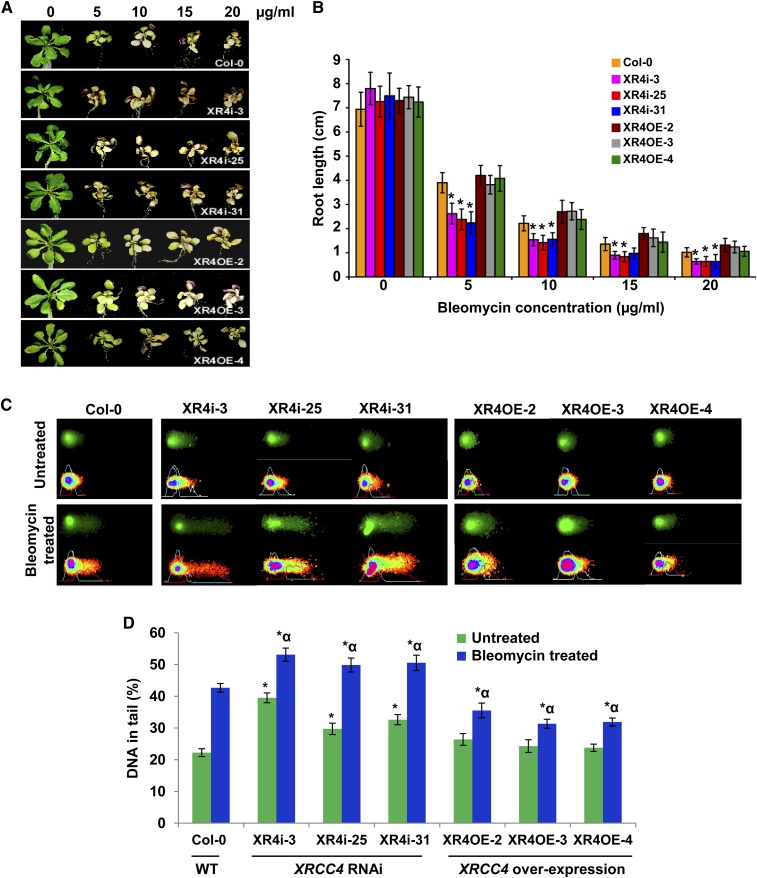
To test whether XRCC4 plays a role in T-DNA integration in more than one plant species, we developed Arabidopsis RNA interference (RNAi) lines that downregulate XRCC4. Arabidopsis contains one full-length copy of the XRCC4 gene (At-XRCC4) on chromosome 3 (At3G23100; see Supplemental Figure 1B online) in the coding strand and another partial sequence (annotated as DNA DSB repair and VJ recombination XRCC4, At1G61410) on chromosome 1 that shares 93% nucleotide sequence homology to At-XRCC4 (At3G23100; see Supplemental Figure 1C online). We generated RNAi lines using the 3′ untranslated region sequence of At-XRCC4 (see Supplemental Figure 1D online). Most of the XRCC4 RNAi lines that we developed appeared developmentally normal. However, a few lines were stunted and produced fewer seeds. We speculate that downregulation of XRCC4 beyond a certain baseline level is deleterious to plant health and survival. Real-time quantitative RT-PCR analysis of several transgenic lines showed more than 80% lower XRCC4 transcripts when compared with controls (see Supplemental Figure 3A and Supplemental Table 1 online). We selected three healthy-looking RNAi lines that had ∼80% downregulation of XRCC4 and confirmed that they were defective in DNA repair by exposing them to various concentrations of bleomycin, a powerful and well-known radiomimetic drug that induces DSBs in eukaryotes (Povirk, 1996). As shown in Figures 2A and Supplemental Figure 3C online, XRCC4 RNAi lines were hypersensitive, relative to the wild type, to bleomycin at both low (2 and 3 µg/mL) and high (5 and 10 µg/mL) concentrations. We also observed significant inhibition of root growth in the XRCC4 RNAi lines compared with the wild type (Figure 2B). In addition to these data, a neutral comet assay (Singh et al., 1988; Olive et al., 1991) confirmed that Arabidopsis XRCC4 RNAi lines contain more DSBs in their nuclei. The fraction of DNA in comet tails of XRCC4 RNAi lines was ∼1.5-fold to twofold higher than in the wild type after bleomycin (5 µg/mL) treatment (Figures 2C and D). Interestingly, we observed more DNA damage in XRCC4 RNAi lines even without bleomycin treatment (Figures 2C and 2D).
Figure 2.
Bleomycin Sensitivity and Comet Assays Show Hypersensitivity of XRCC4 RNAi Lines and Tolerance of XRCC4 Overexpression Lines to a DNA Damaging Chemical.
(A) Response of representative samples of Col-0, XRCC4 RNAi (XR4i), and overexpression (XR4OE) lines after exposure to bleomycin at noted concentrations. Photographs were taken at 10 d after exposure.
(B) Root length of above lines after exposure to bleomycin at various concentrations. Data are presented as mean ± sd (n = 5). Asterisks denote significant difference in the root length between XRCC4 RNAi lines versus Col-0 as determined by Student’s t test (P < 0.05).
(C) Microphotographs of nuclei in XRCC4 RNAi and overexpression lines by neutral comet assay with or without bleomycin (5 μg/mL) treatment. Nuclei with loss of round shape and smearing DNA fragments in the tail of the comet are considered damaged. The fraction of DNA in comet tails was visualized by SYBR Green staining (top panel), and quantitative assessment of DNA damage was performed using CometScore software (bottom panel)
(D) Quantification of the DNA fragments in tail (%) of 150 randomly selected cells per slide. Data are presented as mean ± sd. Asterisks indicate a significant difference between Col-0 and XRCC4 RNAi or overexpression lines without any bleomycin treatment, and asterisks with an alpha indicate a significant difference between Col-0 and XRCC4 RNAi or overexpression lines with 5 μg/mL bleomycin treatment according to Student’s t test (P < 0.05). WT, the wild type.
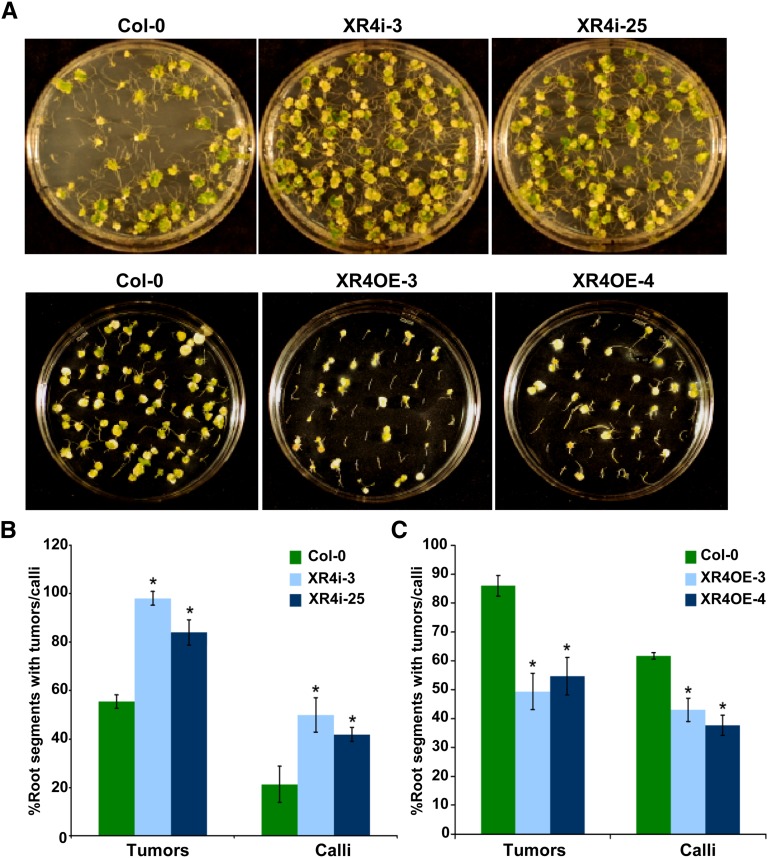
We performed root tumor assays (Nam et al., 1999; Mysore et al., 2000a) using a low concentration (5 × 106 colony-forming units [cfu]/mL) of the oncogenic strain Agrobacterium A208 on two XRCC4 RNAi lines. Roots from RNAi lines produced significantly more tumors (50% ± 6%) than did roots of wild-type plants (Figures 3A, top panel, and 3B). Stable herbicide resistance transformation assays using disarmed Agrobacterium GV3101 harboring pCAS1 also yielded similar results (Figures 3B; see Supplemental Figure 4A online). We confirmed that the transgenic lines used in this study did not have enhanced cell division by incubating the noninoculated root segments on nonselective CIM. Taken together, the results from XRCC4-silenced Arabidopsis and N. benthamiana suggest both that XRCC4 is not required for Agrobacterium-mediated plant transformation and that its activity limits transformation.
Figure 3.
Downregulation or Overexpression of Arabidopsis XRCC4 Results in Increased or Decreased Agrobacterium-Mediated Transformation Efficiencies, Respectively.
(A) Root segments from wild-type Col-0, representative XRCC4 RNAi lines (top panel), and overexpression lines (bottom panel) were infected with tumorigenic strain Agrobacterium A208 and photographs were taken 4 weeks after infection.
(B) Frequency of tumor or transgenic calli formation in XRCC4 RNAi lines after challenging the root segments with Agrobacterium A208 for the tumorigenesis assay or disarmed Agrobacterium GV3101 harboring pCAS1 for the stable transformation assay that produces PPT-resistant calli.
(C) Efficiency of tumor or transgenic calli formation in XRCC4 overexpression lines. In (B) and (C), data are presented as mean ± sd (n = 3) from three replicates. Asterisks indicate a significant difference between Col-0 and XRCC4 RNAi or overexpression lines according to Student’s t test (P < 0.05; see Supplemental Figures 3 and 4 online).
XRCC4 Overexpression in Arabidopsis Leads to Attenuation of Agrobacterium-Mediated Stable Transformation
To explore further the negative effect of XRCC4 in transformation, we overexpressed At-XRCC4 in Arabidopsis. XRCC4-overexpressing plants had an increase in XRCC4 transcript levels ranging from three- to sixfold relative to those in wild-type plants (see Supplemental Figure 3B online). The overexpression lines were slightly more tolerant to bleomycin, indicated by greener and more rosette leaves; however, they did not show any quantitative difference in root length when compared with wild-type plants (Figures 2A and 2B). Using the comet assay as described above, we showed that XRCC4 overexpression lines had significantly less DNA damage than wild-type plants following bleomycin treatment (Figures 2C and 2D). Root segments from two independent transgenic lines were infected with the tumorigenic strain Agrobacterium A208 (2.5 × 107 cfu/mL). Figure 3A (bottom panel) shows that root segments of overexpression lines showed ∼60% decrease in tumor formation frequency compared with roots from wild-type plants (Figure 3C). Root segments from these transgenic plants were also assayed for transformation susceptibility using Agrobacterium GV3101 (pCAS1). Root segments from XRCC4 overexpression lines produced 33% fewer PPT-resistant calli than did root segments from wild-type plants (Figures 3C; see Supplemental Figure 4B online). These results further indicate that XRCC4 plays a negative role in plant transformation.
XRCC4 Downregulated and Overexpressing Arabidopsis Lines Are Not Affected in Transient Transformation but Have Altered T-DNA Integration
To evaluate the effect of XRCC4 downregulation and overexpression on transient transformation, root segments of RNAi lines, overexpression lines, and the wild type, Columbia-0 (Col-0), were infected with a disarmed strain Agrobacterium GV3101 (pBISN1) and assayed for GUS activity at various times after inoculation. There was no significant difference in the number of blue spots on root segments among any of these lines 2 d after infection (DAI; Figure 4A). However, at 12 DAI, the RNAi lines showed a significant increase in the number of blue spots, and overexpression lines exhibited a significant reduction in blue spots compared with Col-0 (Figure 4A). These observations suggest that alteration in XRCC4 expression does not influence transient transformation but plays a negative role in stable transformation.
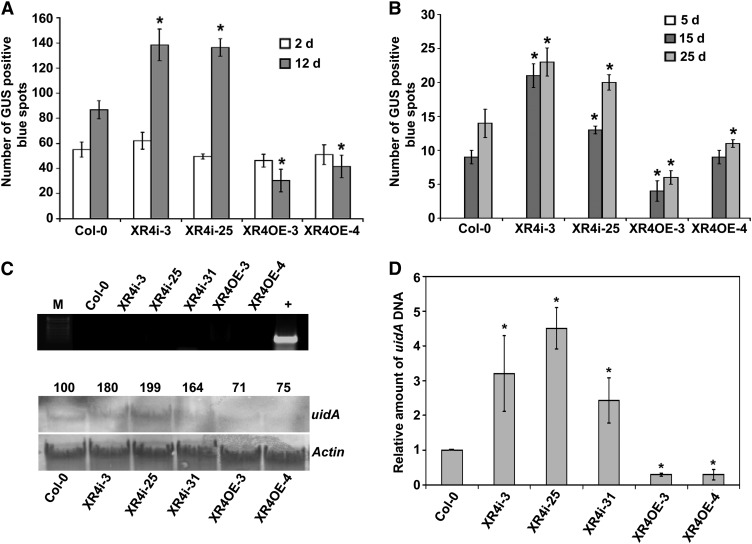
Figure 4.
Arabidopsis XRCC4 Expression Does Not Affect Transient Transformation but Affects T-DNA Integration.
(A) Root segments of Col-0, XRCC4 RNAi (XR4i), and XRCC4 overexpression lines (XR4OE) were inoculated with Agrobacterium GV3101 harboring pBISN1, and blue spots on roots were counted at 2 and 12 d after staining with X-gluc.
(B) Root segments of Col-0, XRCC4 RNAi, and overexpression lines were inoculated with Agrobacterium GV3101 harboring pKM1 for promoter trap assay. Root segments were stained with X-gluc, and blue spots were counted at 5, 15, and 25 DAI. In (A) and (B), data are presented as mean ± sd (n = 3) from three replicates with 50 root segments per treatment. Asterisks indicate a significant difference between Col-0 and XRCC4 RNAi or overexpression lines according to Student’s t test (P < 0.05).
(C) PCR to check contamination of Agrobacterium DNA in plant DNA samples used for DNA gel blot analysis (top panel). Quantification of T-DNA integration using uidA probe on DNA gel blot containing intact high molecular weight genomic DNA isolated from suspension cell cultures of Col-0, XRCC4 RNAi, and overexpression lines. Root segments from these samples were challenged with Agrobacterium GV3101 harboring pBISN1 and calli raised on nonselective media were subcultured to raise suspension cultures. Actin was used as a probe to show equal loading. Band intensities were quantified using a densitometer, and values were assigned relative to that of Col-0 (100%; bottom panel). M, DNA marker; +, positive control showing amplification of Atu0972 gene from Agrobacterium genomic DNA.
(D) DNA isolated from samples in (C) was subjected to a qPCR assay using uidA primers, and relative amounts of integrated T-DNA are shown compared with Col-0 control. The data represent the average of three biological replicates with sd values shown as error bars. Asterisks indicate a significant difference between Col-0 and XRCC4 RNAi or overexpression lines according to Student\x{2019}s t test (P < 0.05).
To provide evidence that the T-DNA integration was reduced in roots of the overexpression lines and enhanced in the RNAi lines, we inoculated the root segments from Col-0, XRCC4 RNAi lines, and overexpression lines with a disarmed strain Agrobacterium GV3101 (pKM1). At 5 DAI, a time point indicating transient expression, no GUS staining was observed in all the lines tested (Figure 4B). However, we detected a significantly higher number of GUS-positive blue spots, due to integrated T-DNA, in root segments of the RNAi lines XR4i-3 and XR4i-25 relative to Col-0 at 15 and 25 DAI (Figure 4B). By contrast, we observed a significant reduction in the number of blue spots in the overexpression lines for both the time points analyzed (Figure 4B).
We further provide direct evidence to show that the difference in transformation efficiency is linked to T-DNA integration using a biochemical approach as described earlier for N. benthamiana. DNA from all the XRCC4 RNAi lines tested showed 64 to 99% more signal, whereas DNA from At-XRCC4 overexpression lines showed 25 to 29% less signal when compared with DNA from Col-0 (Figure 4C, bottom panel). An Arabidopsis Actin gene was used as a normalization control to demonstrate equal loading of DNA in all lanes (Figure 4C, bottom panel). No Agrobacterium contamination was detected in the plant DNA samples that were used for DNA gel blot analysis (Figure 4C, top panel), indicating that the observed hybridization signals were derived from T-DNA integrated into plant DNA. These DNA gel blot hybridization results were confirmed by quantifying the relative amount of T-DNA integrated into the genome by qPCR as described (Anand et al., 2007a, 2007b). The amount of PCR products specific to the uidA gene, determined by qPCR, was two- to fourfold more in XRCC4 RNAi lines and threefold less in overexpressor lines compared with Col-0 plants (Figure 4D). Taken together, our results in Arabidopsis and N. benthamiana provide evidence for a negative effect of XRCC4 in T-DNA integration.
XRCC4 Localizes to Both the Nucleus and Cytoplasm
We fused the full-length open reading frame (ORF) of GFP to the N terminus of At-XRCC4 and delivered the construct under the control of 2X Cauliflower mosaic virus 35S promoter into N. benthamiana leaves via agroinfiltration. After 48 h, confocal microscopy revealed the presence of the GFP-XRCC4 fusion in the cytoplasm and the nucleus (see Supplemental Figure 5 online). This localization pattern is similar to that of human XRCC4 (Yurchenko et al., 2006).
At-XRCC4 Interacts with Agrobacterium VirE2 Protein in the Plant Nucleus
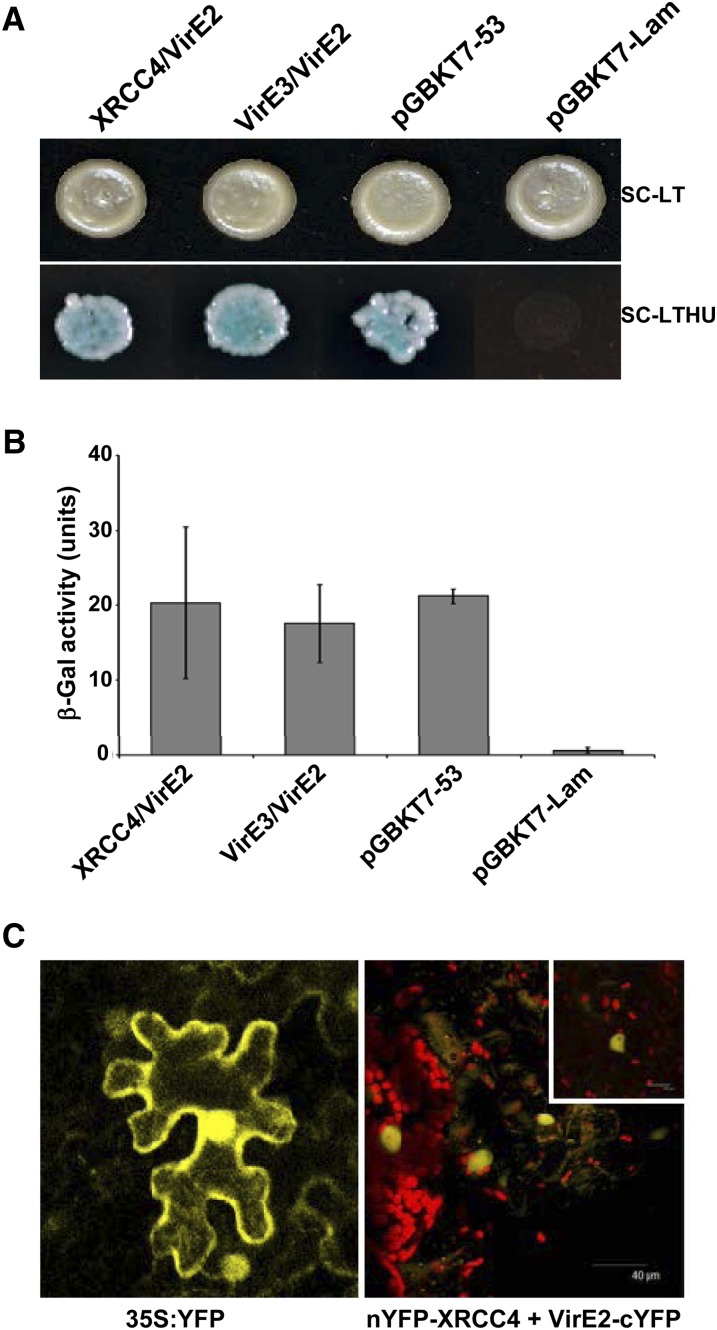
To determine if Agrobacterium takes advantage of the plant DNA repair machinery for transformation, we tested whether XRCC4 interacted with any of the Agrobacterium virulence proteins that are shuttled into the plant cell via the type IV secretion apparatus. A yeast two-hybrid (Y2H) analysis using At-XRCC4 as a bait and Agrobacterium Vir proteins VirD2, VirE2, or the plant VirE2 INTERACTOR PROTEIN2 (VIP2) (Anand et al., 2007a) as prey, revealed a very tight interaction between VirE2 and At-XRCC4 (Figure 5A, bottom panel). We also observed a strong intermolecular interaction for XRCC4 (see Supplemental Figure 6 online, left panel). The quantitative β-galactosidase assay also confirmed these results (Figures 5B; see Supplemental Figure 6 online, right panel). To confirm the VirE2-XRCC4 interaction in planta, we used a bimolecular fluorescence complementation assay (BiFC) (Walter et al., 2004). The interaction between N-YFP-XRCC4 and VirE2-C-YFP was observed in the nuclear compartment of agroinfiltrated N. benthamiana leaf epidermal cells (Figure 5C, right panel). A clear reconstitution of the YFP signal (not seen in the leaves infiltrated with either construct alone) was seen in the nucleus but excluded from the nucleolus (inset). Full-length YFP expressed from the Cauliflower mosaic virus 35S promoter localizes to both the nucleus and cytoplasm (Figure 5C, left panel), while coinfiltration of N-YFP-At-XRCC4 and Vip2-C-YFP or VirD2-C-YFP showed no interaction.
Figure 5.
Arabidopsis XRCC4 Exhibits a Strong Interaction with VirE2 in Yeast and Plant Cells.
(A) Y2H analysis using At-XRCC4 as bait and VirE2 as prey shows a strong interaction (bottom panel, selective medium without Leu, Trp, His, Ura): XRCC4 (bait) + VirE2 (prey), VirE3 (bait) + VirE2 (prey), positive control (pGADT7-T encoding the Gal4 AD fused with SV40 large T-antigen and pGBKT7-53 encoding the Gal4 DNA binding domain fused with murine p53), and negative control (pGADT7-T and pGBKT7-Lam encoding the Gal4 binding domain fused with lamin).
(B) β-Galactosidase assay shows quantification of the Y2H interactions shown in (A). Data represent the average of three biological replicates and three technical replicates with sd shown as error bars.
(C) YFP reconstitution observed during the in planta interaction of nYFP-At-XRCC4 and VirE2-cYFP in the nucleus of agroinfiltrated N. benthamiana leaves using a BiFC assay 48 h after infiltration of both cultures (right panel). Interaction within the nucleus is clearly visible as YFP expression in a magnified view of nucleus (inset). A full-length 35S-YFP agroinfiltrated into N. benthamiana leaves acts as a positive control (left panel; see Supplemental Figures 5 and 6 online).
Arabidopsis Plants Expressing VirE2 Are Hypersensitive to Bleomycin and Show Increased Stable Transformation
To gain better understanding on the functional significance of the VirE2 interaction with XRCC4 in relation to Agrobacterium-mediated transformation, we took advantage of transgenic Arabidopsis plants expressing VirE2-YFP (Bhattacharjee et al., 2008). We hypothesized that VirE2 can interact with XRCC4 in plants expressing VirE2-YFP, thereby delaying the NHEJ-mediated DSB repair and providing greater opportunity for T-DNA integration. If our hypothesis that Agrobacterium disrupts host DNA repair processes to favor T-DNA integration is true, the transgenic plants expressing VirE2-YFP should be more sensitive to bleomycin (DSB-inducing agent) and show increased stable transformation.
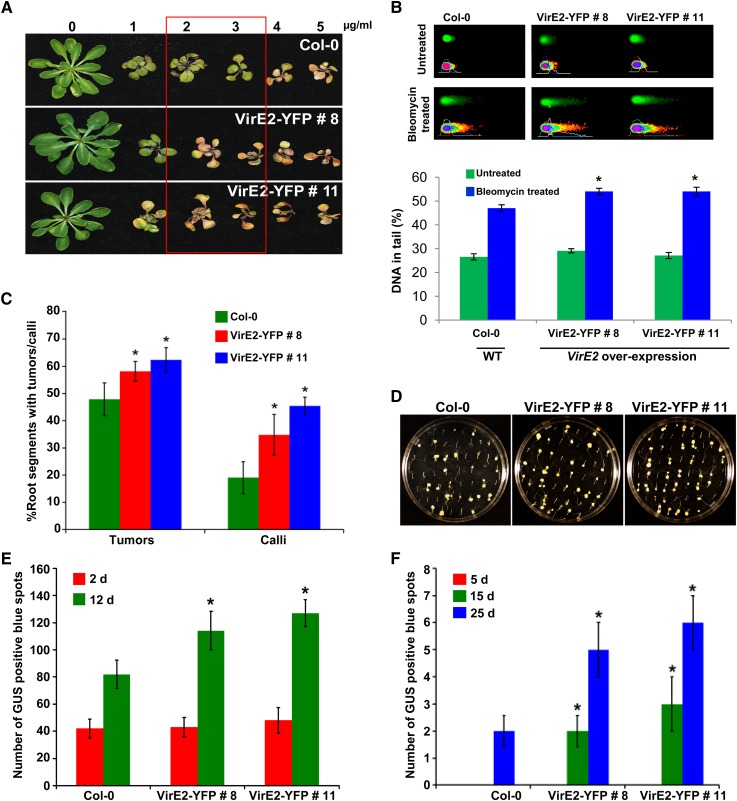
Indeed, both VirE2 expressing lines (8 and 11) were hypersensitive to bleomycin compared with a wild type, Col-0 (Figure 6A). Using a comet assay, we further showed that the VirE2-expressing lines had more DNA damage than the wild type after bleomycin treatment (Figure 6B). To rule out the possibility that VirE2-expressing lines are more prone to general stress that might have contributed to increased sensitivity to bleomycin, the plants were exposed to MMS. MMS is an alkylating agent that causes heat-labile DNA damage but not DSBs (Lundin et al., 2005). As shown in Supplemental Figure 7 online, there was no significant difference in the response of Col-0 and VirE2-expressing lines to MMS at the two different concentrations tested. Root transformation assays (tumor and PPT-resistant calli) with low concentration of Agrobacterium as described above showed a slight but significant increase in stable transformation for both the VirE2-expressing lines when compared with Col-0 (Figures 6C and 6D). Interestingly, line 11 showed slightly higher transformation efficiency than did line 8, and this correlated well with the higher VirE2 gene expression in line 11 (43 and 22% more than line 8 in leaf and root tissues, respectively). The enhanced transformation efficiency displayed by VirE2-expressing lines was consistent and reproducible in an independent experiment.
Figure 6.
Arabidopsis Plants Expressing VirE2 Are Hypersensitive to Bleomycin and Show Enhanced Stable Transformation Efficiency.
(A) Response of transgenic VirE2-YFP–expressing lines to bleomycin. One-week-old healthy seedlings were transferred to MS medium containing varying concentrations of bleomycin, and the response was photographed after 21 d. The box indicates the hypersensitivity of VirE2-YFP–expressing lines 8 and 11 over Col-0 control.
(B) Quantification of DNA damage by comet assay. Micrographs of nuclei in Col-0 and VirE2-YFP–expressing lines by neutral comet assay with or without bleomycin (5 μg/mL) treatment (top panel). The fraction of DNA in comet tails was visualized by SYBR Green staining and quantified with CometScore software (bottom panel). Data are presented as mean ± sd of 150 randomly selected cells per slide. Asterisks indicate a significant difference between Col-0 and VirE2-YFP–expressing lines according to Student’s t test (P < 0.05). WT, the wild type.
(C) Frequency of tumor and transgenic calli formation from root segments infected with low concentration (1 × 107 cfu/mL) of tumorigenic strain Agrobacterium A208 and disarmed strain Agrobacterium GV3101 (pCAS1), respectively. Data are presented as mean ± sd (n = 3) from three replicates with 50 root segments per treatment. Asterisks indicate a significant difference between Col-0 and VirE2-YFP–expressing lines according to Student’s t test (P < 0.05).
(D) Representative plates of root tumor transformation assay from Col-0 versus VirE2-YFP–expressing lines. Root segments of wild-type Col-0 and VirE2-YFP–expressing lines were infected with a low concentration (1 × 107 cfu/mL) of tumorigenic strain Agrobacterium A208. Tumors on root segments were photographed 4 weeks after infection.
(E) Root segments of Col-0 and VirE2-YFP–expressing lines were inoculated with strain Agrobacterium GV3101 harboring pBISN1, and blue spots on roots were counted at 2 and 12 d after staining with X-gluc.
(F) Root segments of Col-0 and VirE2-YFP–expressing lines were inoculated with strain Agrobacterium GV3101 harboring pKM1. Root segments were stained with X-gluc, and blue spots were counted at 5, 15, and 25 DAI. In (E) and (F), data are presented as mean ± sd (n = 3) from three replicates with 50 root segments per treatment. Asterisks indicate a significant difference between Col-0 and VirE2-YFP–expressing lines according to Student’s t test (P < 0.05; see Supplemental Figure 7 online).
To determine the effect of VirE2 expression on transient and stable transformation, root segments of Col-0 and VirE2-expressing lines were infected with a disarmed strain, Agrobacterium GV3101, containing the binary vector pBISN1. X-gluc staining to see transient GUS expression at 2 DAI revealed no significant difference in the number of blue spots on root segments between Col-0 and VirE2-expressing lines (Figure 6E). However, at 12 DAI, which likely represents stable transformation, both the VirE2-expressing lines showed a significant increase in the number of blue spots compared with Col-0 (Figure 6E). We further showed that T-DNA integration was significantly enhanced in the VirE2-expressing lines by inoculating the root segments with a disarmed strain Agrobacterium GV3101 containing the binary vector pKM1 that has promoterless uidA-intron gene in the T-DNA. Upon X-gluc staining, a significantly higher number of blue spots due to integrated T-DNA were detected in root segments of VirE2-expressing lines relative to Col-0 at 15 and 25 DAI (Figure 6F). These results suggest that VirE2 expression in plants does not influence transient transformation but enhances T-DNA integration and, thus, stable transformation.
DISCUSSION
Based on the assumption that DSBs in the host genome are insertion sites for T-DNA molecules, a host DSB repair mechanism, the NHEJ pathway, may play a role in T-DNA integration (Tzfira et al., 2004). XRCC4 is an important protein in the NHEJ pathway. Its primary function is to bring the processed DSB ends in close proximity and stimulate ligase IV activity (Grawunder et al., 1997). XRCC4 gene knockouts have been found to be embryo lethal in mammalian systems (Gao et al., 1998; Soulas-Sprauel et al., 2007). In this report, we show that XRCC4 expression in plants reduces Agrobacterium T-DNA integration.
Our initial observation showing that XRCC4-silenced N. benthamiana plants were more susceptible to stable transformation but not to transient transformation suggested that XRCC4 may limit T-DNA integration. We hypothesized that downregulation of XRCC4 delays DNA repair and this leaves DSBs open long enough for T-DNA to get integrated and that the overexpression of XRCC4 should have the opposite effect (i.e., DSB repair would become more efficient than normal and, therefore, T-DNA integration would be drastically reduced). The results obtained in Arabidopsis XRCC4 RNAi and overexpression lines (Figure 3) corroborated the results of N. benthamiana and strengthened our hypothesis. In addition, data from transient and stable GUS activity assays, the promoterless uidA expression assay, and the T-DNA integration assay in both N. benthamiana and Arabidopsis lines downregulated for XRCC4 added credence to our hypothesis and demonstrated the negative role of XRCC4 in T-DNA integration (Figures 1 and 4).
Because XRCC4 is a DSB repair protein, it was predicted to localize to the nucleus (AtSubP) (Kaundal et al., 2010). However, our results surprisingly showed localization in both cytoplasmic and nuclear compartments (see Supplemental Figure 5 online). This result corroborates an earlier report, which showed that human XRCC4 partitions from the cytoplasm to the nucleus after a SUMO modification at Lys-210 (Yurchenko et al., 2006). At-XRCC4 contains this conserved Lys residue (Lys-206) in that domain, indicating the possibility of a similar spatial function at the subcellular level. We observed an interaction of At-XRCC4 with VirE2 in an Y2H assay, and in planta interaction in the nucleus was confirmed by a BiFC assay (Figures 4A and 4C). At-XRCC4 could interact with itself in the Y2H assay, and human XRCC4 has been shown to dimerize for binding to ligase IV during DSB repair (Sibanda et al., 2001). The interaction of At-XRCC4 with VirE2 suggests that Agrobacterium may interfere with NHEJ machinery to favor T-DNA integration.
Based on our results, we propose a model for a negative effect of XRCC4 in T-DNA integration. An efficient NHEJ pathway does not allow DSBs to exist for long time. Because T-DNA can integrate into DSBs (Chilton and Que, 2003; Tzfira et al., 2003), an efficient NHEJ mechanism may limit T-DNA integration. We demonstrated this by increasing T-DNA integration efficiency by downregulating XRCC4 in both N. benthamiana and Arabidopsis. Many molecules of Agrobacterium VirE2 protein likely enter the plant cell independent of T-DNA (Sundberg et al., 1996; Gelvin, 1998) and may localize into the nucleus (Citovsky et al., 1992). VirE2 localization in plant cells, either to the nucleus or perinuclear vicinity, is controversial (Citovsky et al., 1992; Bhattacharjee et al., 2008). We propose that free VirE2 molecules can interact with XRCC4 and titrate/exclude active XRCC4 protein available for DSB repair in the cell. Binding of VirE2 to XRCC4 would therefore disrupt/delay normal kinetics of DSB repair, thus allowing more opportunity for T-DNA to integrate. This model was supported by our XRCC4 overexpression studies in which incoming VirE2 proteins from Agrobacterium may have failed to titrate XRCC4 and resulted in decreased T-DNA integration efficiency. Additional evidence in support of this model comes from our data showing VirE2-expressing Arabidopsis plants were more sensitive to a DSB-inducing chemical and showed increased stable transformation efficiency due to possible titration of XRCC4. Our results are supported by a previous study where a 10% increase in tumor formation was recorded in transgenic tobacco (Nicotiana tabacum) expressing VirE2 compared with the wild type (Gelvin, 1998). However, Citovsky et al. (1992) showed that the frequency of tumor formation in transgenic tobacco expressing VirE2 plants upon inoculation with a tumorigenic Agrobacterium strain was same as that of wild-type plants. Contrary to this, Citovsky et al. (1994) in a similar experiment showed that there was a slight reduction in the frequency of tumor formation in transgenic lines expressing VirE2 when compared with wild-type plants. It should be noted that the experiments in the above-mentioned publications were not designed to quantify the transformation efficiency, and higher bacterial concentrations were used for inoculation. Therefore, the previous results pertaining to transformation efficiency are inconclusive.
An alternative explanation for increased transformation in XRCC4-silenced plants would be the stimulation of an alternative NHEJ pathway (Mladenov and Iliakis, 2011). The alternative NHEJ pathway is only stimulated upon sensing a depressed state of classical NHEJ machinery (Mladenov and Iliakis, 2011). Supporting data for stimulation of other DNA repair pathways also comes from a study that showed that when XRCC4 was downregulated by RNAi in human somatic cells, random integration was reduced but targeted insertions were up by 33% using an homologous recombination–like mechanism (Bertolini et al., 2009). To determine the roles and functional hierarchy of specific DNA repair pathways, a recent study used various single and multiple Arabidopsis mutants of the classical NHEJ (ku80), alternate/backup NHEJ (xrcc1), microhomology-mediated end joining (xpf) and homologous recombination (xrcc2) pathways to study DSB repair induced by gamma irradiation (Charbonnel et al., 2011). They concluded that the first three DNA repair processes are all active in DSB repair with varying kinetics with classical NHEJ acting first and then alternate NHEJ followed by microhomology-mediated end joining. These data support our hypothesis that reduction of efficiency of the classical NHEJ, which exhibits the fastest repair kinetics, leads to the takeover of the DSB repair process by other pathways with reduced repair kinetics, thereby enabling increased T-DNA capture into DSBs. The data presented here provide another example of Agrobacterium manipulating the host machinery for the purpose of T-DNA integration, thereby providing it with a competitive advantage for survival.
METHODS
Plasmid Construction
Heterologous primers based on tomato (Solanum lycopersicum) sequence were used to amplify partial sequence of XRCC4 from Nicotiana benthamiana (see Supplemental Table 1 online). VIGS vectors pTRV1 and pTRV2 (Liu et al., 2002) were used to clone the Nb-XRCC4 fragment for silencing. Constructs were introduced into Agrobacterium tumefaciens GV2260 for assays in N. benthamiana or into Agrobacterium GV3101 for assays with Arabidopsis thaliana by electroporation. The pMDC vector (Curtis and Grossniklaus, 2003) was used for XRCC4 overexpression construct, and pK7WIWG2(II) vector (Karimi et al., 2007) was used for making RNAi constructs. Primer sets used to amplify partial and full-length At-XRCC4 sequence for RNAi and overexpression, respectively, are shown in Supplemental Table 1 online. BiFC constructs were made using Gateway ready pSPYNE and pSPYCE vectors (Walter et al., 2004; Anand et al., 2007a).
VIGS and Transformation Assays
VIGS of Nb-XRCC4 and leaf disk transformation assays were performed as described (Anand et al., 2007a) with107 cfu/mL of appropriate Agrobacterium strains. For leaf disk tumorigenesis assays, leaf disks were infected with an oncogenic strain, Agrobacterium A348. Tumors produced per leaf disk were counted 3 weeks after infection. The total biomass of the leaf (including tumors) was measured by weighing the fresh and dry weights (incubated at 37°C for 5 d) of a minimum of 150 disks (15 disks per treatment were pooled). For transient transformation assays, the leaf disks were infected with the nontumorigenic strain Agrobacterium GV2260 containing the binary vector pBISN1 (Narasimhulu et al., 1996) or with the binary plasmid pKM1 (Mysore et al., 1998) for promoterless uidA gene expression analyses. Leaf disks were either immediately stained with X-gluc staining solution or analyzed for GUS activity using fluorometric assays (Jefferson et al., 1987). For stable transformation assays, leaf disks were inoculated with the nontumorigenic strain Agrobacterium GV2260 containing pCAS1 (Nam et al., 1999). After 2 d, the leaf disks were transferred onto CIM containing PPT at 5 μg/mL concentration and were incubated at 25°C for 1 month. The number of leaf disks with PPT-resistant calli was scored 4 weeks after Agrobacterium infection and calli biomass was quantified by measuring fresh and dry weight.
Arabidopsis root transformation assays were performed as described earlier (Nam et al., 1999; Zhu et al., 2003). A low concentration of 5 × 106 cfu/mL and 2 × 107 cfu/mL was used for root assays with At-XRCC4 RNAi and overexpression lines, respectively. In vitro tumorigenesis assays were performed on the axenic root segments by infecting with an oncogenic strain Agrobacterium A208 containing a nopaline-type Ti plasmid (pTiT37), cocultivated for 48 h in dark at room temperature, transferred to a hormone-free Murashige and Skoog (MS) media supplemented with cefotaxime (250 μg/mL) and tricarcillin (100 μg/mL), and the tumor numbers and phenotypes were recorded 4 to 5 weeks after infection. For transient/stable GUS expression assays, promoter trap assay, and another stable transformation assay (PPT-resistant calli production), the root segments were infected with a disarmed strain Agrobacterium GV3101 containing either pBISN1 (Narasimhulu et al., 1996), pKM1 (Mysore et al., 1998), or pCAS1 (Nam et al., 1999). For transient and promoter trap assays, after 2 d of cocultivation, the root segments were transferred to CIM with cefotaxime and tricarcillin. To measure GUS expression, root segments were sampled at various time points and stained with X-gluc. For stable transformation, the root segments were incubated on CIM plates containing cefotaxime (250 μg/mL), tricarcillin (100 μg/mL), and PPT (10 μg/mL). The number of root segments forming PPT-resistant calli was counted 4 to 5 weeks after infection.
T-DNA Integration and qPCR Assays
The T-DNA integration assay was performed as described previously (Mysore et al., 2000a; Anand et al., 2007b; Li et al., 2005) on the genomic DNA extracted from suspension cell lines generated from the calli produced on nonselective CIM by leaf disks of N. benthamiana control and Nb-XRCC4 silenced lines or Arabidopsis root segments of Col-0, XRCC4 RNAi lines (XR4i-3, XR4i-25, and XR4i-31), and overexpressing lines (XR4OE-3 and XR4OE-4) infected with either disarmed strain Agrobacterium GV2260 (for N. benthamiana) or GV3101 (for Arabidopsis) carrying pBISN1.
Prior to DNA gel blot analysis, PCR was performed on genomic DNA with Agrobacterium chromosomal gene (Atu0972)-specific primers (forward, 5′-GCGTTCGCTGGTGTCACGCC-3′, and reverse, 5′-GATCAGCGGAGACCAGCTTC-3′) to check for the bacterial DNA contamination in the plant DNA samples. For DNA gel blot analysis, 10 μg of undigested genomic DNA was electrophoresed through a 0.8% agarose gel and transferred to a Hybond N+ nylon membrane (GE Healthcare). Digoxygenin (DIG)-labeled probes (437 bp at the 3′ of uidA gene sequence and 410 bp Arabidopsis Actin) were prepared by PCR according to the manufacturer’s labeling kit system (Roche Diagnostics) employing DIG-conjugated deoxyuridine triphosphates, primers specific for uidA (forward, 5′-ACTCCTACCGTACCTCGCATTACC-3′, and reverse, 5′-GTAATAACGGTTCAGGCACAGCAC-3′) or Actin (forward, 5′-CACAACAGCAGAGCGGGAAATTGT-3′, and reverse, 5′-TCTTCATGCTGCTTGGTGCAAGTG-3′) and pBISN1 plasmid DNA or Arabidopsis genomic DNA as templates. The DNA gel blot was hybridized with DIG-labeled uidA probe for detection of the integrated T-DNA. The intensity of the signals was quantified using the LabWorks Image Acquisition and Analysis Software (UVP). Quantification of integrated T-DNA was also performed by qPCR as outlined (Anand et al., 2007b) using uidA (5′-ATGAAGATGCGGACTTACGTGGCA-3′ and 5′-ATCTGCCCAGTCGAGCATCTCTTC-3′) and EF-1α (5′-TTCACCCTTGGTGTCAAGCA-3′ and 5′-TTTCATCGTACCTGGCCTTGCA-3′) primers. Normalization of templates was performed using EF-1α amplification levels. The amount of integrated DNA in XRCC4 downregulated or overexpression lines was calculated relative to the amount of integrated DNA in the calli derived from control plants.
Y2H, Subcellular Localization, and BiFC Assays
Y2H assays were performed as described (Ciftci-Yilmaz et al., 2007). Full-length cDNA clones were first cloned into pDONR207 and, subsequently, the full-length gene was introduced into the Y2H vector pXDGATCY86 as bait or pGADT-Rec7 as prey (Ciftci-Yilmaz et al., 2007). Positive interactions were quantified using the β-galactosidase assay kit (Pierce Biotechnology). To examine interactions between fusion proteins, both bait and prey plasmids were cotransformed into MaV203 yeast strain carrying three GAL4-inducible reporter genes (lacZ, HIS3, and URA3). Bait–prey interactions were selected on the synthetic dropout medium lacking Leu and Trp (SC-Leu-Trp). The yeast colony grown in SC-Leu-Trp was streaked on the medium lacking Leu, Trp, His, and Ura supplemented with 10 mM 3-amino-1,2,4-triazole with X-Gal (20 µg/mL). Plasmids pGBKT7-53, pGBKT7-Lam, and pGADT7-T from Clontech (Takara Bio) were included as positive and negative controls for the interaction. To determine autoactivation, yeast clones containing only prey were cotransformed with empty bait vector (pXDGATCY86), and the growth of the yeast cells on SC-Leu-His with 10 mM 3-amino-1,2,4-triazole was examined.
For localization predictions, XRCC4 sequence was processed with At-SubP (Kaundal et al., 2010), a species-specific predictor for protein localization. For localization assay, At-XRCC4 ORF was fused to GFP at the N terminus, and the construct was subsequently agroinfiltrated into N. benthamiana leaves, followed by observation at 48 HAI under a fluorescence microscope. Cells expressing the GFP-XRCC4 fusion protein were photographed using a ×60 immersion lens in a spinning disc confocal microscope.
For BiFC assay, the ORFs of XRCC4 and VirE2 were cloned via Gateway cloning into BiFC vectors (Walter et al., 2004) pSPYNE-35S and pSPYCE-35S, and constructs were agroinfiltrated into N. benthamiana leaves. Leaf discs were examined using a Leica TCS SP2 AOBS confocal laser scanning microscope with the samples excited at 514 nm for YFP reconstitution. 35S-YFP in pCAMBIA1390 was used as a positive control. Individual constructs were also infiltrated, and constructs known to have no interactions were used as negative controls (e.g., VIP2).
Bleomycin Tolerance and Comet Assay
Seeds of Arabidopsis ecotype Col-0 were germinated on Petri plates with MS media solidified with 0.2% phytagel (Sigma-Aldrich). Seeds of XRCC4 RNAi lines and overexpression lines were germinated on MS medium supplemented with kanamycin and hygromycin at a concentration of 50 and 20 µg/mL, respectively, for a week during a 16/8-h day/light regime at 24°C. Seeds of VirE2-YFP–expressing Arabidopsis lines were germinated on hygromycin containing MS medium. Healthy seedlings were transferred to Petri plates containing MS media supplemented with varying concentrations of bleomycin (0, 1, 1.5, 2, 3, 4, 5, 10, 15, and 20 µg/mL). Sensitivity to the bleomycin treatment was determined after 10 d for higher concentrations (5, 10, 15, and 20 µg/mL) and after 3 weeks for lower concentrations (1, 1.5, 2, 3, and 4 µg/mL).
DSBs were detected by a neutral comet assay using CometAssay reagent kit (Trevigen) with minor modifications. Briefly, 1-week-old Arabidopsis seedlings grown in half-strength MS medium were transferred to solid half-strength MS medium with or without bleomycin (5 µg/mL). Arabidopsis seedlings were harvested after 7 d and stored at −80°C. Approximately 100 mg of frozen leaf tissue was cut into small pieces with a razor blade in 300 μL PBS buffer (20 mM EDTA) on ice. Nuclei suspension was collected into Eppendorf tubes on ice after removing tissue debris with 50-µm nylon mesh. Thirty microliters of nuclei suspension was combined with 300 μL of molten low melting agarose at 37°C, and 50 μL was immediately pipetted onto coated microscope slides provided in the kit. After chilling the slides at 4°C in the dark for 10 min, slides were immersed in prechilled lysis solution on ice for 2 h. After lysis, comet slides were placed in prechilled 1× tris-borate-EDTA electrophoresis buffer for 10 min and then subjected to electrophoresis at 1 V per cm for 10 min. After electrophoresis, slides were immersed in distilled water for 5 min and 70% ethanol for 5 min and dried at 40°C for 15 min. SYBR Green I (100 μL; Trevigen) was added onto each sample and incubated at room temperature in the dark for 10 min. Comets were imaged by epifluorescence microscopy using the green channel (excitation at 494 nm and emission at 521 nm). The comet data analysis was performed by comet scoring software (CometScore; TriTek). DNA damage was measured as the fraction of DNA in comet tails (percentage of DNA in tail).
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number JN687989 for the Nb-XRCC4 sequence. The Arabidopsis XRCC4 gene characterized in this article has accession number At3G23100.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Nicotiana benthamiana XRCC4 Sequence Used for Virus-Induced Gene Silencing.
Supplemental Figure 2. Effect of Nb-XRCC4 Downregulation on Stable Transformation in Nicotiana benthamiana.
Supplemental Figure 3. Estimation of At-XRCC4 Transcript Level by Quantitative RT-PCR in RNAi and Overexpression Lines and Sensitivity of These Lines to Bleomycin.
Supplemental Figure 4. Root Callus Assay to Determine the Effect of At-XRCC4 Downregulation and Overexpression on Stable Transformation.
Supplemental Figure 5. Localization of At-XRCC4.
Supplemental Figure 6. Interaction of At-XRCC4 with Agrobacterium VirE2 Protein.
Supplemental Figure 7. Methyl Methanesulfonate Sensitivity Assay for VirE2-Expressing Lines.
Supplemental Table 1. List of Primers Used in VIGS and Arabidopsis thaliana Studies.
Supplementary Material
Acknowledgments
This work was supported by The Samuel Roberts Noble Foundation and in part by the National Science Foundation (Award IOB-0445799 to K.S.M.). The Leica AOBS confocal system used in this study was purchased with a National Science Foundation equipment grant (DBI 0722635). We thank Elison Blancaflor for his assistance with BiFC and localization experiments, Stanton Gelvin for providing the seeds of Arabidopsis VirE2-YFP–expressing lines, Yasuhiro Ishiga for help with DNA gel blot analysis, Stanton Gelvin for critical reading of the article, and Ajith Anand for stimulating discussions.
AUTHOR CONTRIBUTIONS
Z.E.V. and K.S.M. designed the experiments. Z.E.V. and B.V. executed the majority of the experiments. M.R.M. and S.L. executed the Y2H experiments and comet assay. K.S.M. coordinated the research. Z.E.V., B.V., and K.S.M. wrote the article.
Glossary
- DSB
double-strand break
- NHEJ
nonhomologous end joining
- MMS
methyl methanesulfonate
- VIGS
virus-induced gene silencing
- PPT
phosphinothricin
- CIM
callus induction medium
- GUS
β-glucuronidase
- qPCR
quantitative PCR
- RNAi
RNA interference
- cfu
colony-forming units
- Col-0
Columbia-0
- DAI
days after infection
- ORF
open reading frame
- Y2H
yeast two-hybrid
- BiFC
bimolecular fluorescence complementation assay
- MS
Murashige and Skoog
- DIG
digoxygenin
References
- Anand A., Krichevsky A., Schornack S., Lahaye T., Tzfira T., Tang Y.H., Citovsky V., Mysore K.S. (2007a). Arabidopsis VIRE2 INTERACTING PROTEIN2 is required for Agrobacterium T-DNA integration in plants. Plant Cell 19: 1695–1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A., Rojas C.M., Tang Y., Mysore K.S. (2012). Several components of SKP1/Cullin/F-box E3 ubiquitin ligase complex and associated factors play a role in Agrobacterium-mediated plant transformation. New Phytol. 195: 203–216 [DOI] [PubMed] [Google Scholar]
- Anand A., Uppalapati S.R., Ryu C.-M., Allen S.N., Kang L., Tang Y., Mysore K.S. (2008). Salicylic acid and systemic acquired resistance play a role in attenuating crown gall disease caused by Agrobacterium tumefaciens. Plant Physiol. 146: 703–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand A., Vaghchhipawala Z., Ryu C.-M., Kang L., Wang K., del-Pozo O., Martin G.B., Mysore K.S. (2007b). Identification and characterization of plant genes involved in Agrobacterium-mediated plant transformation by virus-induced gene silencing. Mol. Plant Microbe Interact. 20: 41–52 [DOI] [PubMed] [Google Scholar]
- Bertolini L.R., Bertolini M., Maga E.A., Madden K.R., Murray J.D. (2009). Increased gene targeting in Ku70 and Xrcc4 transiently deficient human somatic cells. Mol. Biotechnol. 41: 106–114 [DOI] [PubMed] [Google Scholar]
- Bhattacharjee S., Lee L.-Y., Oltmanns H., Cao H., Veena, Cuperus J., Gelvin S.B. (2008). IMPa-4, an Arabidopsis importin alpha isoform, is preferentially involved in agrobacterium-mediated plant transformation. Plant Cell 20: 2661–2680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britt A.B., May G.D. (2003). Re-engineering plant gene targeting. Trends Plant Sci. 8: 90–95 [DOI] [PubMed] [Google Scholar]
- Charbonnel C., Allain E., Gallego M.E., White C.I. (2011). Kinetic analysis of DNA double-strand break repair pathways in Arabidopsis. DNA Repair (Amst.) 10: 611–619 [DOI] [PubMed] [Google Scholar]
- Chilton M.D., Que Q. (2003). Targeted integration of T-DNA into the tobacco genome at double-stranded breaks: new insights on the mechanism of T-DNA integration. Plant Physiol. 133: 956–965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciftci-Yilmaz S., Morsy M.R., Song L., Coutu A., Krizek B.A., Lewis M.W., Warren D., Cushman J., Connolly E.L., Mittler R. (2007). The EAR-motif of the Cys2/His2-type zinc finger protein Zat7 plays a key role in the defense response of Arabidopsis to salinity stress. J. Biol. Chem. 282: 9260–9268 [DOI] [PubMed] [Google Scholar]
- Citovsky V., Warnick D., Zambryski P. (1994). Nuclear import of Agrobacterium VirD2 and VirE2 proteins in maize and tobacco. Proc. Natl. Acad. Sci. USA 91: 3210–3214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citovsky V., Zupan J., Warnick D., Zambryski P. (1992). Nuclear localization of Agrobacterium VirE2 protein in plant cells. Science 256: 1802–1805 [DOI] [PubMed] [Google Scholar]
- Curtis M.D., Grossniklaus U. (2003). A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner J., Britt A.B. (2003). Ku80- and DNA ligase IV-deficient plants are sensitive to ionizing radiation and defective in T-DNA integration. Plant J. 34: 427–440 [DOI] [PubMed] [Google Scholar]
- Gallego M.E., Bleuyard J.-Y., Daoudal-Cotterell S., Jallut N., White C.I. (2003). Ku80 plays a role in non-homologous recombination but is not required for T-DNA integration in Arabidopsis. Plant J. 35: 557–565 [DOI] [PubMed] [Google Scholar]
- Gao Y., et al. (1998). A critical role for DNA end-joining proteins in both lymphogenesis and neurogenesis. Cell 95: 891–902 [DOI] [PubMed] [Google Scholar]
- Gelvin S.B. (1998). Agrobacterium VirE2 proteins can form a complex with T strands in the plant cytoplasm. J. Bacteriol. 180: 4300–4302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelvin S.B. (2010). Finding a way to the nucleus. Curr. Opin. Microbiol. 13: 53–58 [DOI] [PubMed] [Google Scholar]
- Grawunder U., Wilm M., Wu X., Kulesza P., Wilson T.E., Mann M., Lieber M.R. (1997). Activity of DNA ligase IV stimulated by complex formation with XRCC4 protein in mammalian cells. Nature 388: 492–495 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Depicker A., Hilson P. (2007). Recombinational cloning with plant Gateway vectors. Plant Physiol. 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaundal R., Saini R., Zhao P.X. (2010). Combining machine learning and homology-based approaches to accurately predict subcellular localization in Arabidopsis. Plant Physiol. 154: 36–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.-I., Veena, Gelvin S.B. (2007). Genome-wide analysis of Agrobacterium T-DNA integration sites in the Arabidopsis genome generated under non-selective conditions. Plant J. 51: 779–791 [DOI] [PubMed] [Google Scholar]
- Köhler F., Cardon G., Pöhlman M., Gill R., Schieder O. (1989). Enhancement of transformation rates in higher plants by low-dose irradiation: Are DNA repair systems involved in the incorporation of exogenous DNA into the plant genome? Plant Mol. Biol. 12: 189–199 [DOI] [PubMed] [Google Scholar]
- Li J.X., Vaidya M., White C., Vainstein A., Citovsky V., Tzfira T. (2005). Involvement of KU80 in T-DNA integration in plant cells. Proc. Natl. Acad. Sci. USA 102: 19231–19236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh-Kumar S.P. (2002). Virus-induced gene silencing in tomato. Plant J. 31: 777–786 [DOI] [PubMed] [Google Scholar]
- Lundin C., North M., Erixon K., Walters K., Jenssen D., Goldman A.S.H., Helleday T. (2005). Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 33: 3799–3811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mladenov E., Iliakis G. (2011). Induction and repair of DNA double strand breaks: The increasing spectrum of non-homologous end joining pathways. Mutat. Res. 711: 61–72 [DOI] [PubMed] [Google Scholar]
- Mysore K.S., Bassuner B., Deng X.B., Darbinian N.S., Motchoulski A., Ream W., Gelvin S.B. (1998). Role of the Agrobacterium tumefaciens VirD2 protein in T-DNA transfer and integration. Mol. Plant Microbe Interact. 11: 668–683 [DOI] [PubMed] [Google Scholar]
- Mysore K.S., Kumar C.T.R., Gelvin S.B. (2000b). Arabidopsis ecotypes and mutants that are recalcitrant to Agrobacterium root transformation are susceptible to germ-line transformation. Plant J. 21: 9–16 [DOI] [PubMed] [Google Scholar]
- Mysore K.S., Nam J., Gelvin S.B. (2000a). An Arabidopsis histone H2A mutant is deficient in Agrobacterium T-DNA integration. Proc. Natl. Acad. Sci. USA 97: 948–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J., Matthysse A.G., Gelvin S.B. (1997). Differences in susceptibility of Arabidopsis ecotypes to crown gall disease may result from a deficiency in T-DNA integration. Plant Cell 9: 317–333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nam J., Mysore K.S., Gelvin S.B. (1998). Agrobacterium tumefaciens transformation of the radiation hypersensitive Arabidopsis thaliana mutants uvh1 and rad5. Mol. Plant Microbe Interact. 11: 1136–1141 [DOI] [PubMed] [Google Scholar]
- Nam J., Mysore K.S., Zheng C., Knue M.K., Matthysse A.G., Gelvin S.B. (1999). Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by Agrobacterium. Mol. Gen. Genet. 261: 429–438 [DOI] [PubMed] [Google Scholar]
- Narasimhulu S.B., Deng X.B., Sarria R., Gelvin S.B. (1996). Early transcription of Agrobacterium T-DNA genes in tobacco and maize. Plant Cell 8: 873–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olive P.L., Wlodek D., Banath J.P. (1991). DNA double-strand breaks measured in individual cells subjected to gel electrophoresis. Cancer Res. 51: 4671–4676 [PubMed] [Google Scholar]
- Pitzschke A., Hirt H. (2010). New insights into an old story: Agrobacterium-induced tumor formation in plants by plant transformation. EMBO J. 29: 1021–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Povirk L.F. (1996). DNA damage and mutagenesis by radiomimetic DNA-cleaving agents: bleomycin, neocarzinostatin and other enediynes. Mutat. Res. 355: 71–89 [DOI] [PubMed] [Google Scholar]
- Ray A., Langer M. (2002). Homologous recombination: ends as the means. Trends Plant Sci. 7: 435–440 [DOI] [PubMed] [Google Scholar]
- Salomon S., Puchta H. (1998). Capture of genomic and T-DNA sequences during double-strand break repair in somatic plant cells. EMBO J. 17: 6086–6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibanda B.L., Critchlow S.E., Begun J., Pei X.Y., Jackson S.P., Blundell T.L., Pellegrini L. (2001). Crystal structure of an Xrcc4-DNA ligase IV complex. Nat. Struct. Biol. 8: 1015–1019 [DOI] [PubMed] [Google Scholar]
- Singh N.P., McCoy M.T., Tice R.R., Schneider E.L. (1988). A simple technique for quantitation of low levels of DNA damage in individual cells. Exp. Cell Res. 175: 184–191 [DOI] [PubMed] [Google Scholar]
- Soulas-Sprauel P., Le Guyader G., Rivera-Munoz P., Abramowski V., Olivier-Martin C., Goujet-Zalc C., Charneau P., de Villartay J.P. (2007). Role for DNA repair factor XRCC4 in immunoglobulin class switch recombination. J. Exp. Med. 204: 1717–1727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg C., Meek L., Carroll K., Das A., Ream W. (1996). VirE1 protein mediates export of the single-stranded DNA-binding protein VirE2 from Agrobacterium tumefaciens into plant cells. J. Bacteriol. 178: 1207–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T., Frankman L.R., Vaidya M., Citovsky V. (2003). Site-specific integration of Agrobacterium tumefaciens T-DNA via double-stranded intermediates. Plant Physiol. 133: 1011–1023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T., Li J.X., Lacroix B., Citovsky V. (2004). Agrobacterium T-DNA integration: Molecules and models. Trends Genet. 20: 375–383 [DOI] [PubMed] [Google Scholar]
- van Attikum H., Bundock P., Hooykaas P.J.J. (2001). Non-homologous end-joining proteins are required for Agrobacterium T-DNA integration. EMBO J. 20: 6550–6558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Bundock P., Overmeer R.M., Lee L.Y., Gelvin S.B., Hooykaas P.J.J. (2003). The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 31: 4247–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter M., Chaban C., Schütze K., Batistic O., Weckermann K., Näke C., Blazevic D., Grefen C., Schumacher K., Oecking C., Harter K., Kudla J. (2004). Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J. 40: 428–438 [DOI] [PubMed] [Google Scholar]
- West C.E., Waterworth W.M., Jiang Q., Bray C.M. (2000). Arabidopsis DNA ligase IV is induced by γ-irradiation and interacts with an Arabidopsis homologue of the double strand break repair protein XRCC4. Plant J. 24: 67–78 [DOI] [PubMed] [Google Scholar]
- Yurchenko V., Xue Z., Sadofsky M.J. (2006). SUMO modification of human XRCC4 regulates its localization and function in DNA double-strand break repair. Mol. Cell. Biol. 26: 1786–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaltsman A., Krichevsky A., Loyter A., Citovsky V. (2010). Agrobacterium induces expression of a host F-box protein required for tumorigenicity. Cell Host Microbe 7: 197–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y.M., et al. (2003). Identification of Arabidopsis rat mutants. Plant Physiol. 132: 494–505 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.