This work shows that the evolutionarily conserved DUF59 family gene AE7 encodes a central member of the cytosolic iron-sulfur cluster assembly pathway and demonstrates that this pathway is required for maintaining nuclear genome integrity and, thus, normal mitotic cell cycle in Arabidopsis thaliana. The results link plant mitochondria to nuclear genome integrity through assembly of Fe-S proteins.
Abstract
Eukaryotic organisms have evolved a set of strategies to safeguard genome integrity, but the underlying mechanisms remain poorly understood. Here, we report that ASYMMETRIC LEAVES1/2 ENHANCER7 (AE7), an Arabidopsis thaliana gene encoding a protein in the evolutionarily conserved Domain of Unknown Function 59 family, participates in the cytosolic iron-sulfur (Fe-S) cluster assembly (CIA) pathway to maintain genome integrity. The severe ae7-2 allele is embryo lethal, whereas plants with the weak ae7 (ae7-1) allele are viable but exhibit highly accumulated DNA damage that activates the DNA damage response to arrest the cell cycle. AE7 is part of a protein complex with CIA1, NAR1, and MET18, which are highly conserved in eukaryotes and are involved in the biogenesis of cytosolic and nuclear Fe-S proteins. ae7-1 plants have lower activities of the cytosolic [4Fe-4S] enzyme aconitase and the nuclear [4Fe-4S] enzyme DNA glycosylase ROS1. Additionally, mutations in the gene encoding the mitochondrial ATP binding cassette transporter ATM3/ABCB25, which is required for the activity of cytosolic Fe-S enzymes in Arabidopsis, also result in defective genome integrity similar to that of ae7-1. These results indicate that AE7 is a central member of the CIA pathway, linking plant mitochondria to nuclear genome integrity through assembly of Fe-S proteins.
INTRODUCTION
The genome of eukaryotic organisms is under continuous assault from various genotoxic stresses, originating from exogenous environmental factors and endogenous causes, and these stresses often cause DNA damage. In response to DNA damage in eukaryotes, highly conserved ATM (for ataxia telangiectasia-mutated) and ATR (for ATM and Rad3-related) kinase signaling cascades are induced to switch on DNA repair machineries and to arrest or delay the progression of the mitotic cell cycle (Zhou and Elledge, 2000; Cools and De Veylder, 2009; Branzei and Foiani, 2010). In plants, the ATM/ATR-mediated DNA damage response activates the WEE1 kinase to arrest the cell cycle, allowing cells to repair damaged DNA before proceeding into mitosis (Garcia et al., 2003; Culligan et al., 2004; De Schutter et al., 2007).
Several proteins required for DNA repair and/or replication are found to contain iron-sulfur (Fe-S) clusters as cofactors, including Rad3/XPD (UVH6 in Arabidopsis thaliana); FancJ and RTEL1, which are DNA helicases for nucleotide excision repair (Rudolf et al., 2006; Uringa et al., 2011); Ntg2 in yeast and ROS1 in Arabidopsis, which are DNA glycosylases for base excision repair (Alseth et al., 1999; Kim et al., 2001; Gong et al., 2002; Mok et al., 2010); Pri2, a DNA primase subunit for DNA synthesis and double-strand break (DSB) repair (Klinge et al., 2007; Vaithiyalingam et al., 2010); and Polα, δ, ℇ, and ζ, the eukaryotic DNA polymerases for DNA replication and repair (Netz et al., 2012). Biogenesis of Fe-S proteins requires the function of dedicated Fe-S cluster assembly pathways, which are located in the mitochondria, the cytosol, and the plastids in eukaryotes. In yeast, maturation of nuclear and cytosolic Fe-S proteins is mediated by a pathway termed cytosolic iron-sulfur cluster assembly (CIA) that exclusively depends on the mitochondrial Fe-S cluster assembly machinery and the ATP binding cassette (ABC) transporter of the mitochondria, Atm1p (Kispal et al., 1997, 1999; Lill, 2009; Sharma et al., 2010). However, the substrate of Atm1p, or its plant (ATM3/ABCB25) and human (ABCB7) orthologs, is as yet unknown.
The first CIA proteins to be identified in yeast were two related P loop NTPases, Cfd1 (Roy et al., 2003) and Nbp35 (Hausmann et al., 2005), which form a protein complex serving as a scaffold in Fe-S cluster assembly (Netz et al., 2007). The Fe-S clusters are thought to be transferred from Cfd1-Nbp35 to apoproteins with the help of the CIA targeting complex consisting of Nar1, Cia1, Cia2/YHR122W, and Met18 in yeast (IOP1, CIAO1, MIP18, and MMS19 in humans) (Gari et al., 2012; Stehling et al., 2012). Nar1 has sequence similarity to Fe-Fe hydrogenases and binds Fe-S clusters (Balk et al., 2004); Cia1 is a WD40-repeat protein that presumably functions as a platform for protein interactions (Balk et al., 2005; Srinivasan et al., 2007); Cia2/YHR122W is a small protein belonging to the conserved domain of unknown function 59 (DUF59) family (Weerapana et al., 2010), and Met18/Mms19 is a highly conserved HEAT repeat protein previously implicated in DNA repair (Prakash and Prakash, 1979; Lauder et al., 1996; Hatfield et al., 2006) that was recently found to have a function in Fe-S cluster assembly (Gari et al., 2012; Stehling et al., 2012). The Arabidopsis genome contains homologs with high sequence similarity to all known components of the yeast CIA pathway except for Cfd1 (Balk and Pilon, 2011), but only ATM3 and NBP35 have been functionally characterized (Bych et al., 2008; Bernard et al., 2009; Kohbushi et al., 2009).
We previously reported that ASYMMETRIC LEAVES1/2 ENHANCER7 (AE7) is required for leaf polarity formation by promoting cell proliferation and cell cycle progression in Arabidopsis (Yuan et al., 2010). AE7 encodes a DUF59 protein with 45% identity and 61% similarity to yeast Cia2/YHR122W and 58% identity and 73% similarity to human MIP18 (MMS19-interacting protein 18 kD). The DUF59 domain is also found at the N terminus of HCF101, which belongs to the same family of P loop NTPases as NBP35 but is localized in the plastids and required for the assembly of a subset of Fe-S proteins (Lezhneva et al., 2004; Schwenkert et al., 2010).
In this study, we show that plants with a partial loss of AE7 function due to a conserved Glu-51 to Lys substitution exhibited highly accumulated DNA damage, activation of the DNA damage response, and decreased activities of cytosolic and nuclear Fe-S enzymes. Together with similar defective genome integrity in the atm3-4 mutant, our results indicate that a pathway including the mitochondria and a protein complex with AE7 is essential for Fe-S protein biogenesis and genome integrity in plants.
RESULTS
The ae7 Mutation Causes DNA Damage
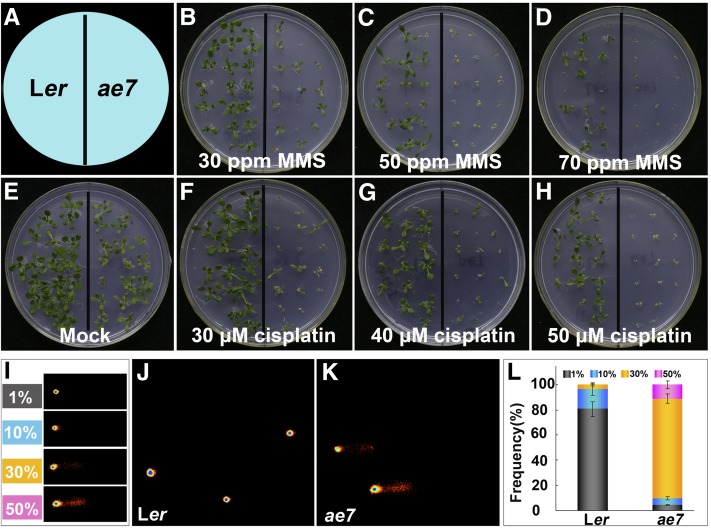
We previously showed that AE7 expression was associated with proliferating cells in a pattern similar to that of the cell cycle marker gene HISTONE4 and that the DNA repair gene RAD51 was upregulated in the ae7 mutant (Yuan et al., 2010). To test whether AE7 is involved in DNA repair, we first examined the sensitivity of ae7 seedlings to DNA damage agents methyl methanesulfonate (MMS) and cisplatin. MMS is a DNA-alkylating agent and can cause DSBs (Yin et al., 2009), whereas cisplatin is a cross-linking reagent that induces DNA lesions (Abe et al., 2005). Compared with the wild type, the growth of ae7 seedlings was strongly compromised in the presence of MMS and cisplatin (Figures 1A to 1H). This sensitivity to DNA damage agents suggests that AE7 is involved in DNA replication and/or repair.
Figure 1.
The ae7 Mutation Resulted in Increased Sensitivity of Seedlings to DNA-Damaging Agents and Accumulation of DNA Damage.
(A) A diagram indicates the positions of wild-type Ler and ae7 seedlings grown on media.
(B) to (D) Treatment with different concentrations of MMS.
(E) Mock treatment.
(F) to (H) Treatment with different concentrations of cisplatin.
(I) to (L) Quantification of DNA damage in ae7 using the comet assay.
(I) Representative images of four arbitrarily determined classes of DNA damage: 1, 10, 30, and 50% DNA present in the tail compared with the nucleus, as quantified.
(J) and (K) Typical comet images of nuclei from 8-d-old leaves of Ler (J) and ae7 (K).
(L) Frequency distribution of different classes of DNA damage in wild-type Ler and ae7 nuclei. Bars indicate means ± sd based on at least 200 comets from five independent gels.
To gain direct evidence that ae7 carries increased DNA damage, we performed a comet assay to measure DNA breakage, using the first-pair leaves of 13-d-old ae7 seedlings. Because the extent of DNA damage differed from cell to cell even in the same plant, we classified the comet signals into four grades based on a ratio of signal intensities from the comet tail over the entire nucleus (Figure 1I). At the developmental stage examined, only weak DNA damage signals were detected in the wild-type leaves (Figures 1J and 1L), whereas not only was DNA damage more severe but also the number of cells containing damaged DNA markedly increased in the leaves of ae7 seedlings (Figures 1K and 1L). The results further indicate the role of AE7 in DNA replication and/or repair.
A Partial Loss of AE7 Activates the DNA Damage Response to Arrest the Cell Cycle
In eukaryotic cells, DNA damage usually leads to activation of ATM/ATR-mediated signaling response cascades that turn on DNA repair machineries and arrest or delay cell cycle progression (Zhou and Elledge, 2000; De Schutter et al., 2007; Cools and De Veylder, 2009; Branzei and Foiani, 2010). Since DNA damage highly accumulated in the ae7 mutant, we next examined expression of several genes that are often induced in the DNA damage response. The tested genes included PARP genes that encode poly (ADP-ribose) polymerases required for the maintenance of DNA integrity during replication (Doucet-Chabeaud et al., 2001), BREAST CANCER SUSCEPTIBILITY1 (BRCA1) and GAMMA RESPONSE1 (GR1) that are involved in DSB repair by the homologous recombination (HR) pathway (Lafarge and Montané, 2003; Bleuyard et al., 2005), KU70, KU80, and LIGASE4 (LIG4) that are required for DSB repair by the nonhomologous end joining pathway (Tamura et al., 2002; van Attikum et al., 2003), TSO2 that is activated in response to DSBs, and RIBONUCLEOTIDE REDUCTASE 2A (RNR2A) and RNR2B that are specifically induced by hydroxyurea (Wang and Liu, 2006; Roa et al., 2009). Our real-time quantitative RT-PCR (qRT-PCR) showed that among the genes tested, transcript levels of PARP1, PARP2, BRCA1, GR1, and TSO2 were elevated in ae7 (Figure 2A).
Figure 2.
The ae7 Mutation Activated the DNA Damage Response to Arrest the Cell Cycle.
(A) qRT-PCR analyses of DNA damage-inducible genes in Ler and ae7 seedlings. Total RNA prepared from 13-d-old seedlings was amplified by RT-PCR. All values were normalized against the expression level of the ACTIN genes. Triplicate repeats and three biological replicates were performed, and the data are shown as the averages with sd. Asterisk indicates significant difference from Ler by t test (P < 0.05).
(B) to (E) Phenotypes of 21-d-old seedlings of Ler (B), ae7 (C), Col-0 (D), and ae7-1 (E). Bars = 1 cm.
(F) to (I) GUS staining of 13-d-old seedlings of pPARP2:GUS/Col-0 (F), pPARP2:GUS/ae7-1 (G), pCYCB1;1:Dbox-GUS/Col-0 (H), and pCYCB1;1:Dbox-GUS/ae7-1 (I). Bars = 1 mm.
To understand better the functions of AE7, we backcrossed the ae7 mutant (Landsberg erecta [Ler]) to wild-type Columbia-0 (Col-0) five times and then introduced several marker lines that are in the Col-0 background into ae7-1 (for convenience, the backcrossed ae7 mutant was renamed ae7-1). Compared with wild-type plants (Figures 2B and 2D), the ae7 (Figure 2C) and ae7-1 (Figure 2E) plants displayed overall similar phenotypic abnormalities, with a smaller size and narrower, serrated leaves. Activation of the DNA damage response in the ae7-1 background was also confirmed by β-glucuronidase (GUS) staining of the pPARP2:GUS transgenic plants (Takahashi et al., 2008): GUS staining appeared strongly in the shoot apex and young leaves of the ae7-1 seedlings (Figure 2G) but was barely detectable in the wild-type Col-0 (Figure 2F). To investigate whether the activated DNA damage response in ae7-1 leads to cell cycle arrest, we introduced a pCYCB1;1:Dbox-GUS construct (CYCB1;1-GUS) into ae7-1 plants by crossing. CYCB1;1 is activated in the G2 phase, and the CYCB1;1-GUS protein is degraded at metaphase; thus, GUS staining marks cells in the G2-to-M transition (Colón-Carmona et al., 1999; Criqui et al., 2001). Compared with the wild type (Figure 2H), GUS signals strongly accumulated in the shoot apex of ae7-1 (Figure 2I), indicating that ae7-1 mutation resulted in delayed or arrested transition of G2 to M phase. All of these results strongly suggest that the accumulated DNA damage causes activation of DNA damage responses in the ae7 mutants, leading to arrest of the cell cycle.
Mutation of AE7 Results in Increased Frequency of Intrachromosomal HR
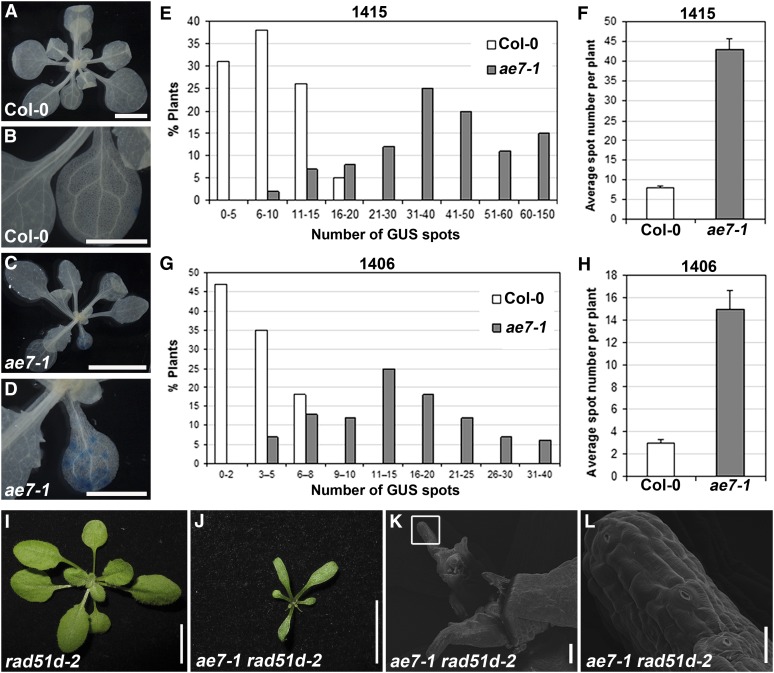
In eukaryotes, DSBs are repaired in part via the HR pathway, which is important for maintaining genome integrity (Bleuyard et al., 2006). Given that ae7 seedlings are hypersensitive to DNA damaging agents, exhibited increased levels of DNA damage, and upregulated genes involved in HR-mediated DSB repair, we employed an HR repair assay, which allows recombination events to be visualized and scored by GUS histochemical staining (Gherbi et al., 2001).The ae7-1 mutant was crossed to two HR reporter lines 1415 and 1406 (Col-0), which carry a T-DNA construct containing parts of the GUS gene with an overlapping sequence in a direct or inverted orientation, respectively. If an intrachromosomal recombination event occurs between the overlapping sequences, a functional GUS gene will be reconstituted and expressed; therefore, GUS staining in these lines reflects the frequency of intrachromosomal HR.
Compared with line 1415 in the wild-type background (Figures 3A and 3B), a much greater number of GUS spots/patches were observed in the 1415/ae7-1 plants (Figures 3C and 3D). About 80% of the 1415/ae7-1 plants carried more than 20 GUS staining spots (Figure 3E) with an average of ∼43 per plant (Figure 3F). For 1406/ae7-1, ∼80% plants showed more than nine GUS spots (Figure 3G) with an average of ∼15 per plant (Figure 3H). By contrast, 1415 and 1406 in the wild-type background yielded far fewer GUS spots, with approximately eight and three per plant on average, respectively (Figures 3F and 3H). These results indicate that the ae7-1 mutation severely increased DSB events, leading to a higher frequency of intrachromosomal HR.
Figure 3.
Partial Loss of AE7 Function Results in Increased Frequency of Intrachromosomal HR.
(A) and (C) GUS staining of line 1415 in the wild-type (A) and ae7-1 (C) seedlings.
(B) and (D) Close-ups of one leaf in (A) and (C), respectively.
(E) to (H) Quantitative analysis of GUS-staining spots in wild-type and ae7-1 seedlings carrying the HR reporter line 1415 ([E] and [F]) or 1406 ([G] and [H]), showing the distribution of GUS spots in the population ([E] and [G]) and the average number of GUS spots per plant ([F] and [H]). About 100 plants were analyzed for each assay. The average number of GUS spots per seedling for 1415 (F) or 1406 (H) was calculated from two independent experiments. Bars show sd.
(I) and (J) The rad51d-2 single mutant showed no obvious phenotypic changes (I), whereas the ae7-1 rad51d-2 double mutant markedly enhanced the ae7-1 phenotypes (J).
(K) and (L) Scanning electron microscopy analysis showed that ae7-1 rad51d-2 plants contained arrested leaf primordia.
Bars = 0.5 cm in (A) and (C), 2.5 mm in (B) and (D), 1 cm in (I) and (J), 100 μm in (K), and 20 μm in (L).
To provide additional evidence that accumulation of DNA damage stimulated intrachromosomal HR repair in ae7-1, we introduced the rad51d-2 mutation into the ae7-1 mutant by crossing. The RAD51D gene encodes a RecA/Rad51 family protein functioning in DNA recombination and repair and is required for somatic HR (Durrant et al., 2007). Although the rad51d-2 single mutant was indistinguishable from the wild-type plants (Figure 3I), the ae7-1 rad51d-2 double mutant displayed much stronger developmental defects than ae7-1, as shown by the reduced plant size (cf. Figures 3J and 2E). Compared with the wild-type plants, ∼20% of the ae7-1 rad51d-2 plants (33 out of 162) carried arrested leaf primordia (Figures 3K and 3L). Because RAD51D functions in the intrachromosomal HR repair pathway, it is likely that the enhanced ae7-1 rad51d-2 phenotypes are due to a strongly decreased efficiency of intrachromosomal HR repair.
AE7 Forms a Complex with CIA1, NAR1, and MET18
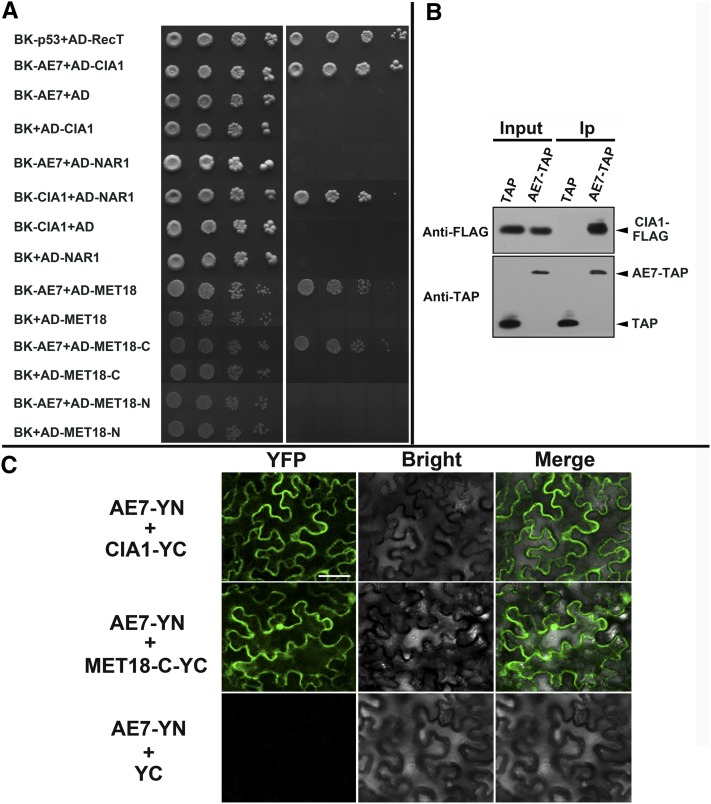
To test whether AE7 interacts with CIA1, NAR1, and MET18, as reported for the yeast and human homologs, we performed yeast two-hybrid assays with the Arabidopsis proteins. Yeast cells that coexpressed the AE7 bait and the CIA1 prey, the CIA1 bait and the NAR1 prey, or the AE7 bait and MET18 prey promoted the expression of the HIS3 and ADE2 reporter genes, allowing cells to grow on media lacking His and adenine (Figure 4A). However, an interaction between AE7 and NAR1was not detected using this assay (Figure 4A).
Figure 4.
Interactions between the Arabidopsis CIA Members.
(A) Yeast two-hybrid assays showed interactions between AE7 and CIA1, CIA1 and NAR1, AE7 and MET18, or its C-terminal domain (MET18-C). Yeast cells carrying different fusion protein combinations are listed on the left. Serial dilutions (10×) of yeast cells expressing the indicated proteins from the pGBK-T7 (BK) and pGAD-T7 (AD) vectors were plated onto standard medium (SD/+Ade/+His) (left) or medium lacking adenine and His (SD/-Ade/-His) (right). Interaction between BK-p53 and AD-RecT served as positive control.
(B) Coimmunoprecipitation assays. AE7-TAP but not TAP alone coprecipitated with FLAG-CIA1. Arrowheads indicate the bands corresponding to FLAG-CIA1, AE7-TAP, and TAP alone, respectively. Ip, immunoprecipitation.
(C) BiFC assays showing that AE7 interacts with CIA1 and the C-terminal domain of MET18 (MET18-C) in planta. Bar = 50 μm.
[See online article for color version of this figure.]
To confirm the protein–protein interaction between AE7 and CIA1, we performed a coimmunoprecipitation (Co-IP) assay. Tandem affinity protein (TAP)-tagged AE7 and FLAG-tagged CIA1 proteins were coexpressed transiently in the leaves of Nicotiana benthamiana, and the protein extracts were immunoprecipitated with IgG beads followed by immunoblotting analysis. The AE7-TAP protein but not TAP alone coprecipitated the FLAG-CIA1 protein (Figure 4B), suggesting that the AE7 protein was associated with CIA1 in planta. The AE7-CIA1 and AE7-MET18 interactions were also confirmed by bimolecular fluorescence complementation (BiFC) assays. Yellow fluorescent protein (YFP) fluorescence was observed in cells coinfiltrated with AE7-YN and CIA1-YC, or AE7-YN and the C-terminal domain of MET18 (MET18-C-YC), but not in cells with AE7-YN and YC, CIA1-YC and YN, or MET18-C-YC and YN (Figure 4C; data not shown for latter two combinations). Additionally, subcellular localization experiments revealed that the AE7-YFP, CIA1-mCherry, and MET18-mCherry fusion proteins were present in both nucleus and cytoplasm (see Supplemental Figure 1 online), further supporting the possibility that AE7, CIA1, and MET18 function in the same protein complex.
AE7, CIA1, and NAR1 Are All Indispensable for Plant Viability
The direct interactions between the Arabidopsis CIA members prompted us to investigate further whether the corresponding mutants display similar developmental defects. One Ds transposon insertional mutant heterozygous for AE7 (ae7-2), two T-DNA insertional mutants heterozygous for CIA1 (cia-1 and cia1-2), and another two T-DNA insertional mutants heterozygous for NAR1 (nar1-1 and nar1-2) were obtained (see Supplemental Figure 2 online). Because no homozygous plants were identified from the segregating population of any of these mutants, we examined seeds in the siliques of heterozygous plants. Aborted seeds were found among normal ones in a 1:3 ratio in each of these mutant alleles (104:308 for ae7-2+/−, 104:282 for cia1-1+/−, 111:331 for cia1-2+/−, 77:234 for nar1-1+/−, and 117:359 for nar1-2+/−) (Figures 5A to 5G).
Figure 5.
Early Developmental Defects in Mutants of the Arabidopsis CIA Components.
(A) to (G) Siliques of the wild-type Ler (A), ae7-2+/− (B), wild-type Col-0 (C), cia1-1+/− (D), cia1-2+/− (E), nar-1+/− (F), and nar1-2+/− (G) plants.
(H) to (L) Differential interference contrast microscopy observation of seeds of wild-type Ler (H), ae7-1 (I), wild-type Col-0 (J), cia1-1 (K), and nar1-1 (L).
(M) to (P) Phenotypes of 21-d-old met18-1 (M), met18-2 (N), met18-1 ae7-1+/− (O), and met18-2 ae7-1+/− (P).
(Q) to (T) Siliques of the ae7-1 (Q), met18-1 (R), ae7-1 met18-1+/− (S), and ae7-1 met18-2+/− (T) plants. Note that plant phenotypes of both met18-1ae7-1+/− and met18-2 ae7-1+/− were enhanced compared with ae7-1 (Figure 2E), while met18 ae7-1 double homozygous mutations were lethal.
Bars = 1 mm in (A) to (G) and (Q) to (T), 50 μm in (H) to (L), and 1 cm in (M) to (P).
To investigate the possible roles of AE7, CIA1, and NAR1 in embryonic development, we analyzed embryo phenotypes of the defective seeds from siliques of heterozygous mutant plants using Nomarski differential interference contrast microscopy. Different from those in the wild type (Figure 5H), the ae7-2 embryos displayed an irregular cell division pattern and arrested at the globular stage (Figure 5I). Compared with the wild-type embryos (Figure 5J), embryos of cia1-1 were arrested even earlier, resulting in a smaller apical part with abnormal shapes (Figure 5K). No embryo structure was observed in the aborted seeds of nar1-1 (Figure 5L), possibly due to defective cell division at the very early embryogenesis stage or gametophytic sterility.
In contrast with AE7, CIA1, and NAR1, which are indispensable for Arabidopsis, phenotypes of two knockout mutant alleles of the single MET18 gene were indistinguishable from those of the wild type (Figures 5M and 5N compared with Figure 2D; see Supplemental Figure 2 online). To investigate a genetic interaction between AE7 and MET18, we crossed ae7-1 to met18-1 or met18-2. In the F2 progeny, plants with enhanced ae7-1 phenotypes were found, but genotyping analysis demonstrated that all the ae7-1 enhancer mutants were met18-1 ae7-1+/− or met18-2 ae7-1+/− plants (Figures 5O and 5P). In addition, the plants with ae7-1 phenotypes included ae7-1, met18-1+/− ae7-1, or met18-2+/− ae7-1, whereas met18 ae7-1 double mutant plants were not found in the F2 populations. We further analyzed the seeds in the siliques of met18-1+/− ae7-1 and met18-2+/− ae7-1 plants and found that about a quarter of seeds were aborted (Figures 5Q to 5T), indicating that met18 ae7-1 double homozygotes were not viable.
At-CIA1 Rescued the Yeast cia1 Mutant
To start our investigation into whether AE7 and its interaction partners in Arabidopsis play a role in Fe-S protein biogenesis, we performed yeast complementation assays. First, we tested whether AE7 is able to rescue a yeast cia2/yhr122w mutant, but introduction of AE7 into the yeast cells failed to rescue their growth on selective medium (see Supplemental Figure 3 online). At-CIA1 shares 37% identity and 53% similarity over the entire amino acid sequence with yeast Cia1, with seven conserved WD-40 repeats (see Supplemental Figure 4A online). Next, we tested At-CIA1 for complementation of the inducible Gal-CIA1 mutant in which the endogenous CIA1 promoter is replaced by the Gal-inducible GAL1-10 promoter. Because CIA1 is essential for yeast viability, the Gal-CIA1 cells were able to grow only on the Gal-containing medium (see Supplemental Figure 4B online, left panel). However, by shifting the cells from Gal to Glc as a sole carbon source, the endogenous Cia1 protein was depleted, and the cells carrying the empty vector p416 stopped growing, unless another copy of the yeast CIA1 gene, driven by a different promoter (p416-ScCIA1), was expressed. Growth of the Cia1-depleted cells could also be rescued by expression of the Arabidopsis homolog (p416-AtCIA1), although the growth was slower than with yeast CIA1 (see Supplemental Figure 4B online, right panel). Further biochemical analysis showed that the activities of two cytosolic [4Fe-4S] enzymes, isopropylmalate isomerase (Leu1) and sulfite reductase (SiR), were partially restored by expression of At-CIA1 (see Supplemental Figures 4C and 4D online). These results indicate that At-CIA1 is a functional ortholog of yeast Cia1.
AE7 Is Required for Activities of Selected Cytosolic and Nuclear Fe-S Enzymes
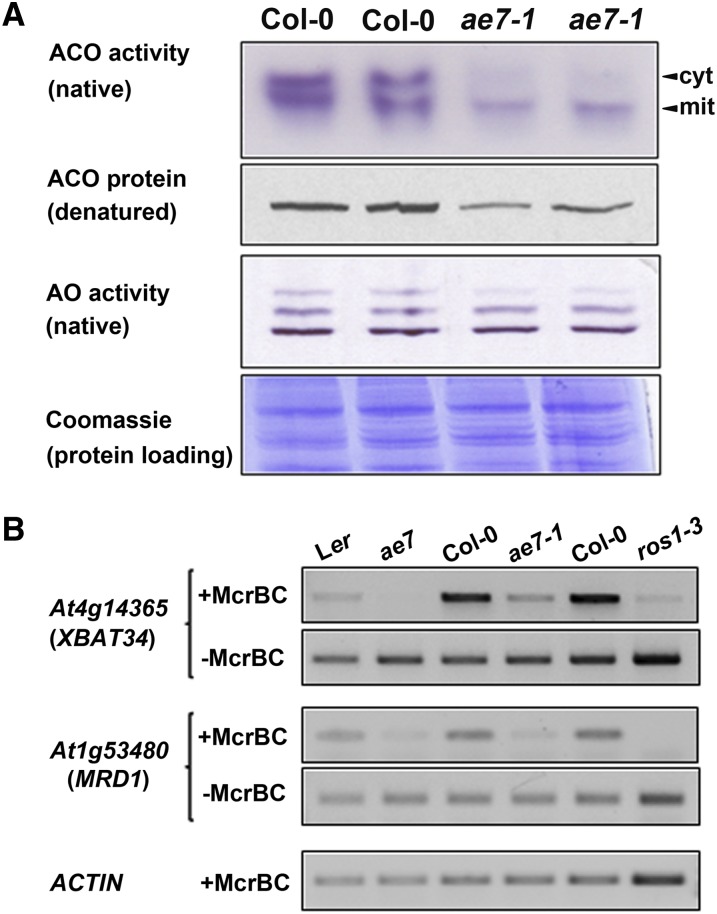
To investigate whether AE7 plays a role in the assembly of Fe-S proteins like its yeast homolog Cia2/YHR122W (Weerapana et al., 2010), we analyzed activities of aconitases (ACOs) and aldehyde oxidases (AOs), which carry [4Fe-4S] and 2[2Fe-2S] clusters, respectively, in ae7-1 seedlings. Two prominent ACO activities were visualized by in-gel staining in the wild type, corresponding to the cytosolic and one of the mitochondrial isozymes (Bernard et al., 2009). Both activities were decreased in ae7-1 seedling extracts (Figure 6A, top panel), the cytosolic activity by 75% and the mitochondrial activity by 55% (see Supplemental Figure 5A online). The decreased levels of ACO activity were confirmed using cell fractionation and a photospectrometric assay (see Supplemental Figure 5B online). Levels of ACO protein were also diminished in ae7-1 (Figure 6A, second panel), as shown by protein blot analysis with specific antibodies against ACO (Bernard et al., 2009), but the transcript levels of ACO genes were not affected (see Supplemental Figure 6 online). These results indicate a posttranscriptional defect in ACO biogenesis, most likely a lack of Fe-S cluster assembly, which is known to destabilize the protein (Bernard et al., 2009). By contrast, the activities of AO isozymes were unchanged in the ae7-1 mutant, although a slight decrease of the upper activity was noticeable (Figure 6A, third panel).
Figure 6.
AE7 Is Required for Activities of Selected Cytosolic and Nuclear Fe-S Enzymes.
(A) Activities of ACO isozymes and AO isozymes in leaves from wild-type and ae7-1 seedlings, visualized by in-gel activity staining. Levels of ACO protein were analyzed by immunoblotting of protein extract separated by SDS-PAGE and labeled with antibodies that cross-react with all three ACO isozymes, which cannot be distinguished by size under these conditions (Bernard et al., 2009). Total protein amount in the leaf samples was revealed by the Coomassie blue staining. cyt, cytosolic; mit, mitochondrial.
(B) McrBC-PCR assay on genomic DNA from wild-type, ae7 mutants, and the ros1-3 mutant as control. Two selected loci (XBAT34 and MRD1) generated little or no PCR product in the ae7 and ros1-3 mutants due to digestion by the methylation-sensitive McrBC enzyme. This indicates that the loci are hypermethylated because the [4Fe-4S]-dependent ROS1 glycosylase is inactive.
[See online article for color version of this figure.]
Next, we performed a PCR assay to assess the demethylation activity of ROS1, a DNA glycosylase closely related to DEMETER for which 5-methylcytosine excision activity has been shown to depend on the [4Fe-4S] cluster ligands (Mok et al., 2010). Genomic DNA from wild-type and ae7-1 leaves was digested with McrBC, which only cuts when methylated cytosines are present, followed by PCR with primer sets spanning two different loci known to be methylated (Figure 6B). The ROS1-dependent locus 3′ of At4g14365 (Penterman et al., 2007) was hypermethylated in ae7-1 compared with the wild type, based on the absence or presence, respectively, of a PCR product (Figure 6B, top panel). Similarly, the MRD1 coding sequence (At1g53480) was hypermethylated in ae7 mutants and in ros1-3, but not in the wild type (Figure 6B, third panel; Yu et al., 2010). Control PCR reactions for undigested DNA or ACTIN confirmed that differences in the levels of PCR product were due to methylation status rather than DNA template concentration (Figure 6B, second, fourth, and fifth panels). Also, the levels of ROS1 expression were not affected at the mRNA level (see Supplemental Figure 6 online). The requirement of AE7 for the activity of at least two Fe-S enzymes indicates that the function of the CIA pathway is conserved in plants.
The Mitochondrial Transporter ATM3 Is Also Required for Maintaining Genome Integrity
To extend our knowledge of the Arabidopsis CIA pathway, we further analyzed phenotypes of atm3, as the Arabidopsis ortholog of yeast Atm1p is known to be critical for the activity of cytosolic Fe-S enzymes (Kushnir et al., 2001; Bernard et al., 2009). We chose to focus our study on the weak atm3-4 allele, which has a similar reduction in growth as the weak ae7 alleles. Compared with that of the wild type, the number of palisade cells was reduced markedly, and CYCB1;1-GUS accumulated strongly in the atm3-4 leaves (see Supplemental Figures 7A to 7E online), similar to those of ae7-1 in which cell proliferation was defective. In addition, the ae7-1 atm3-4 double mutant displayed enhanced phenotypic abnormalities compared with either ae7-1 or atm3-4 single mutant, including diminutive plant size and serrated leaves (see Supplemental Figures 7F to 7I online). The phenotypic analysis strongly indicates a genetic interaction between AE7 and ATM3.
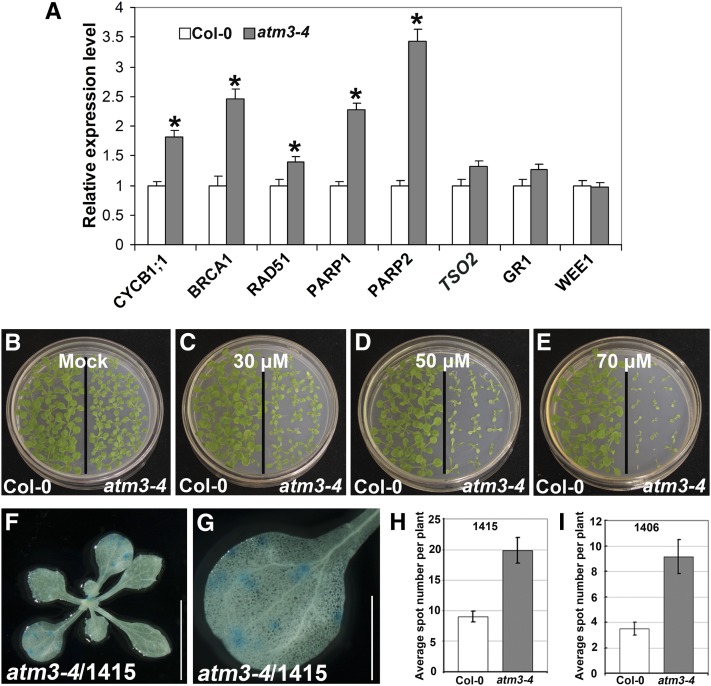
Our expression analysis revealed that transcript levels of the DNA damage-inducible genes, including BRCA1, RAD51, PARP1, PARP2, and CYCB1;1, were elevated significantly in atm3-4 (Figure 7A), consistent with upregulation of RAD51 and PARP1 in sta1, another atm3 allele (Kushnir et al., 2001). Additionally, atm3-4 seedlings were also hypersensitive to cisplatin (Figures 7B to 7E), similar to ae7 seedlings (Figures 1E to 1H). We also introduced atm3-4 into the HR marker lines 1415 and 1406. Compared with wild-type plants, the number of GUS-stained spots was increased for both the 1415/atm3-4 (Figures 7F to 7H) and 1406/atm3-4 (Figure 7I) plants. These results indicate that ATM3 function is also required for maintaining genome integrity.
Figure 7.
The atm3 Mutation Resulted in Genome Instability.
(A) qRT-PCR analysis of expression of DNA damage-inducible genes in 13-d-old seedlings of Col-0 and atm3-4. Asterisk indicates significant difference by t test (P < 0.05).
(B) to (E) Compared with Col-0, the atm3-4 mutant plants were hypersensitive to cisplatin treatment.
(F) and (G) HR assays in atm3-4 mutants. Representative images of GUS staining of a 1415/atm3-4 plant grown under normal conditions. Bars = 1 cm in (F) and 2.5 mm in (G).
(H) and (I) The average number of GUS spots per seedling was analyzed in the 1415/atm3-4 (H) and 1406/atm3-4 (I) plants. About 100 plants were analyzed for each assay. The average number of GUS spots per seedling for 1415 (F) or 1406 (H) was calculated from two independent experiments. Error bars show sd.
DISCUSSION
To ensure genome integrity during the mitotic cell cycle, eukaryotic organisms have evolved elaborate systems to safeguard the faithful duplication of nuclear genomes and to respond to various kinds of stresses. In this study, we characterized four Arabidopsis genes, AE7, CIA1, NAR1, and MET18, belonging to the highly conserved CIA pathway and show that the CIA pathway is required for maintaining nuclear genome integrity. A weak allele of the essential AE7 gene was particularly informative in our study. The ae7 mutant had induced expression of DNA damage response–related genes, and GUS staining was enhanced in CYCB1;1-GUS/ae7-1 transgenic plants. In addition, the accumulation of damaged DNA was accompanied by the induction of DNA repair and DNA damage checkpoint genes in ae7-1, which in turn arrest the cell cycle (Yuan et al., 2010). Furthermore, the ae7-1 seedlings were hypersensitive to DNA damaging agents, and the ae7-1 mutant displayed increased frequency of intrachromosomal HR. All of these results indicate that the DUF59 family member AE7, which acts with other partners CIA1, NAR1, and MET18 of the CIA protein complex, is essential for the maintenance of nuclear genome integrity.
Although the yeast DUF59 protein Cia2/YHR122W shares high sequence similarity in most parts of the protein with other DUF59 members, the N-terminal domain differs considerably from those of many plant and animal species (Yuan et al., 2010). This could be one reason for AE7 being unable to rescue the yeast cia2/yhr122w strain. By contrast, the moss (Physcomitrella patens) and rice (Oryza sativa) AE7 orthologs could fully rescue the developmental defects of ae7 (see Supplemental Figure 8 online), indicating conservation of the DUF59 protein function in distantly related plant species. In addition to AE7, the Arabidopsis genome carries two other AE7-like genes, At3g50845 and At3g09380, which are referred to here as AEL1 and AEL2, respectively. Transformation with either AEL1 or AEL2 failed to rescue the ae7 mutant (see Supplemental Figure 9 online), suggesting that these two genes and AE7 have different biological functions.
Whereas very little is known about their functions, the DUF59 proteins have been implicated several times in Fe-S protein biogenesis (Lezhneva et al., 2004; Schwenkert et al., 2010; Weerapana et al., 2010). We found that AE7 is required for activity of the cytosolic Fe-S enzyme ACO and the nuclear DNA glycosylase ROS1, similar to Cia2/YHR122W, which is required for activity of the Fe-S enzyme Leu1 in yeast (Weerapana et al., 2010). These data suggest that AE7, like its yeast homolog Cia2/YHR122W, acts in the CIA pathway for Fe-S protein assembly in Arabidopsis. Additionally, the DUF59 domain is also found at the N terminus of HCF101, which is related to NBP35 but is localized in the plastids and required for the assembly of a subset of Fe-S proteins (Lezhneva et al., 2004; Schwenkert et al., 2010). Thus, it appears that the DUF59 domain has an evolutionarily conserved function relating to the biogenesis of Fe-S proteins in eukaryotic species.
The yeast proteins Nar1, Cia1, Cia2/YHR122W, and Met18 and the human orthologs IOP1, CIAO1, MIP18, and MMS19 are present in their respective complexes proposed to transfer Fe-S clusters to target proteins (Lill, 2009; Weerapana et al., 2010; Gari et al., 2012; Stehling et al., 2012). In this study, we show that the Arabidopsis homologs also form a complex, with positive protein interactions for the AE7-CIA1, CIA1-NAR1, and AE7-MET18 pairs. It is therefore likely that all four proteins participate in the same biological process, which appears to be conserved across eukaryotes. In support of this, we found that CIA1 could rescue the yeast cia1 mutant. Also, the Nar1 homolog of Chlamydomonas reinhardtii is known to be required for the activities of at least two cytosolic Fe-S enzymes, AO and xanthine dehydrogenase (Godman et al., 2010). In addition, insertion mutants of AE7, CIA1, and NAR1 all died at early reproductive or embryo developmental stages.
By contrast, the function of Met18/MMS19 may be different from Cia2/YHR122W, CIA1, and NAR1. First of all, knockout mutants of MET18 are viable in yeast (Kou et al., 2008) and without obvious phenotypic changes in Arabidopsis (Figures 5M and 5N), although in mouse MMS19 knockout is embryonic lethal (Gari et al., 2012). It was reported that Met18 and MMS19 have the ability to bind directly to target apoproteins for Fe-S cluster assembly (Gari et al., 2012; Stehling et al., 2012), and depletion of MMS19 caused a decrease in protein level of MIP18, suggesting that MMS19 is critical for the stability of MIP18 (Gari et al., 2012). We found that in Arabidopsis plants lacking MET18 but containing one copy of the ae7-1 allele (E51K) caused developmental defects, and the double homozygous met18 ae7-1 mutations were lethal. These observations suggest that AE7 has overlapping functions with MET18 and thus may partially compensate for MET18 deficiency in a dosage-dependent manner.
In yeast, Met18 is required for multiple target Fe-S proteins during the Fe-S assembly process. By contrast, human MMS19 was found to be required for only a subset of Fe-S proteins (Gari et al., 2012; Stehling et al., 2012). Interestingly, we found that ae7 mutation only affected some, but not all, cytosolic/nuclear Fe-S enzymes in Arabidopsis. Also, we found that mitochondrial ACO activity was decreased. The mitochondria contain their own Fe-S cluster assembly machinery, which functions upstream of the CIA machinery in yeast and plants (Frazzon et al., 2007; Lill, 2009). However, regulatory feedback mechanisms may exist: For example, if the cytosolic Fe-S cluster assembly pathway is impaired, the production of precursors by the mitochondrial iron-sulfur cluster machinery may be downregulated to prevent accumulation of the substrate of ATM3 or other intermediates. This will be the focus of future studies.
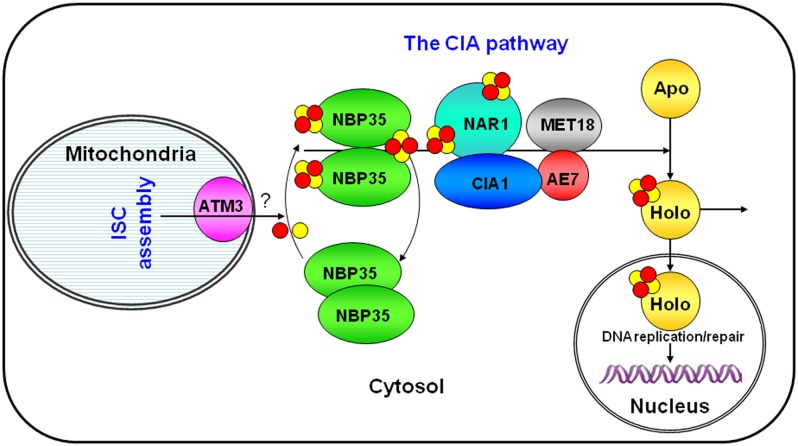
It has recently been discovered that Fe-S clusters are essential for the activity of several important DNA replication/repair-related proteins, which are conserved in all eukaryotes, including plants (Balk and Pilon, 2011). These include DNA helicases Rad3/XPD, FancJ, and RTEL1, DNA glycosylases Ntg2 and ROS1, DNA primase Pri2, and DNA polymerases Polα, δ, ℇ, and ζ (see the Introduction). We found in this study that mutations in AE7 and ATM3 resulted in highly accumulated DNA damage and elevated HR frequency. Therefore, it is likely that the defective genome integrity in ae7 and atm3 is due to inefficient Fe-S cluster assembly of important DNA replication/repair-related proteins. A connection between intact mitochondria, Nar1 of the CIA targeting complex, and genome stability was previously observed in yeast (Veatch et al., 2009). Based on functional studies on Fe-S cluster assembly in yeast (Lill, 2009) and the results that we obtained, we propose an action model for the Arabidopsis CIA pathway in maintaining nuclear genome integrity through Fe-S proteins involved in DNA metabolism (Figure 8). ATM3 exports an as yet unknown substance from the mitochondria to the cytosol for forming the initial Fe-S clusters (Bernard et al., 2009). The NBP35 dimer may act as a scaffold for assembly of Fe-S clusters in the cytosol (Bych et al., 2008). MET18 may select specific target apoproteins while the AE7-CIA1-NAR1 complex facilitates the transfer of Fe-S clusters to the target apoproteins. These targets include not only the cytosolic Fe-S enzymes, such as ACO, but also the nuclear DNA metabolism-related Fe-S proteins, such as ROS1. Not included in this model are TAH18 and DRE2, which form a short electron transfer chain in the CIA pathway (Zhang et al., 2008; Netz et al., 2010). Both proteins are also conserved in Arabidopsis. TAH18 (ATR3) is essential for embryo development, and its expression pattern suggests a role in cell cycle progression (Varadarajan et al., 2010). The Arabidopsis TAH18 and DRE2 genes combined can complement the yeast dre2 mutant (Delphine G. Bernard and Janneke Balk, unpublished data). Therefore, we conclude that because of the importance of the Fe-S proteins in DNA replication and repair, the CIA pathway is essential for the maintenance of genome integrity.
Figure 8.
A Model for the Arabidopsis CIA Pathway, with a Focus on Proteins Discussed in This Article.
In this model, the mitochondrial membrane-localized ABC transporter ATM3/ABCB25 exports an as yet unknown sulfur compound from the mitochondria to the cytosol, which is required for the assembly of Fe-S clusters. The NBP35 dimer in the cytosol may act as a scaffold for Fe-S cluster maturation, while the AE7-CIA1-NAR1-MET18 complex facilitates the transfer of Fe-S clusters to target apoproteins localized in the cytosol or nucleus. Given the importance of the Fe-S proteins in DNA replication and repair, the CIA pathway is essential for the maintenance of genome integrity in Arabidopsis.
[See online article for color version of this figure.]
METHODS
Plant Materials and Growth Conditions
The ae7 mutant in Ler accession has been described previously (Yuan et al., 2010). This allele was backcrossed to wild-type Col-0 five times to generate ae7-1. Seeds of ae7-2 (GT6615), cia1-1 (SALK_060584), cia1-2 (CS16106), nar1-1 (GK_674D01), nar1-2 (GK_462G04), met18-1 (SALK_121963), met18-2 (SALK_147068), and rad51d-2 (SAIL_690_A08) were obtained from the European Arabidopsis Stock Centre or the ABRC. The primers used in PCR to identify insertional mutations are listed in Supplemental Table 1 online. Seeds of pCYCB1;1:Dbox-GUS/Col-0 (Colón-Carmona et al., 1999), pPARP2:GUS/Col-1 (Takahashi et al., 2008), and HR marker lines 1415 and 1406 (Gherbi et al., 2001) were kindly provided by Peter Doerner (University of Edinburgh, UK), Lieven De Veylder (Ghent University, Belgium), and Barbara Hohn (Friedrich Miescher Institute for Biomedical Research, Switzerland), respectively.
Plants were grown in soil according to our previous conditions (Chen et al., 2000). For seedlings grown on plates, surface-sterilized seeds were germinated in half-strength Murashige and Skoog medium supplemented with 1% Suc, kept at 4°C for 2 d, and then moved into a growth chamber with a 16-h-light and 8-h-dark regime at 22°C.
Microscopy and Histology
Fresh seedlings and siliques from wild-type and mutant plants were examined using a SZH10 dissecting microscope (Nikon) and photographed using a Nikon E995 digital camera. Scanning electron microscopy, differential interference contrast microscopy, and GUS staining for pCYCB1;1:Dbox-GUS/Col-0 and pPARP2:GUS /Col-0 transgenic plants were performed according to our previous methods (Li et al., 2005; Yuan et al., 2010; Wang et al., 2011; Gu et al., 2012). GUS staining to detect intrachromosomal HR was performed as described previously (Swoboda et al., 1994).
Yeast Two-Hybrid, Co-IP, and BiFC Assays
For yeast two-hybrid assays, the coding sequences of AE7, CIA1, NAR1, MET18, and the N-terminal (MET18-N; amino acids 1 to 600) or C-terminal (MET18-C; amino acids 601 to 1134) sequence of MET18 were amplified by PCR. After verification of the sequences, the genes were subcloned into pGBK-T7 or pGAD-T7 (Clontech) to generate DNA binding or activation domain fusion constructs, respectively. The EcoRI and BamHI sites were used for cloning AE7, CIA1, and NAR1 into pGBK-T7 or pGAD-T7, but the NdeI and BamHI sites for MET18 and the NdeI and XhoI sites for MET18-N and MET18-C into pGAD-T7. For protein interaction analysis, two combinatory constructs were transformed simultaneously into the yeast strain AH109 (Clontech) and tested for Ade-, His-, Trp-, and Leu- auxotrophy according to the manufacturer’s protocols.
For Co-IP, the coding regions of AE7 and CIA1 were fused to a DNA sequence encoding the TAP tag or the FLAG tag, respectively, and placed under the control of the 35S promoter. The resultant constructs p35S:AE7-TAP and p35S:FLAG-CIA1 were introduced into Agrobacterium tumefaciens strain GV3101 and then coinfiltrated into the leaves of 4-week-old Nicotiana benthamiana plants. IgG beads (Sigma-Aldrich) were used for capturing the protein complex from the total proteins prepared from the infiltrated leaves. The protein samples were analyzed on blots probed with the monoclonal anti-FLAG M2-peroxidase (horseradish peroxidase) antibody (Sigma-Aldrich; A8592) and peroxidase antiperoxidase soluble complex antibody (Sigma-Aldrich; P1291). Immunoreactive signals were revealed by chemiluminescence using Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific).
For the BiFC assays, the coding sequence of AE7 was fused in frame with the N-terminal fragment of YFP to generate p35S:AE7-YN. CIA1 or the C-terminal sequence (amino acids 601 to 1134) of MET18 was fused with the C-terminal fragment of YFP to generate p35S:CIA1-YC and p35S:MET18-C-YC, respectively. The resultant constructs were introduced into the leaves of N. benthamiana. After infiltration for 2 d, the epidermis of infiltrated leaves was examined for YFP signals under a Zeiss 510META confocal laser scanning microscope at a wavelength of 488 nm.
PCR
For qRT-PCR, total RNA was extracted from 13-d-old seedlings using TRIzol reagent (Invitrogen), and reverse transcription was performed with 3 μg total RNA using a kit (Fermentas). qRT-PCR was performed according to our previous methods (Wang et al., 2011).
For the McrBC-PCR assay, genomic DNA was extracted from leaf samples using the Qiagen Pure Gene kit. One microgram of DNA was mixed into a volume of 100 μL with restriction buffer, then split into 2 × 50 μL with either 2 μL McrBC enzyme (New England Biolabs) or 2 μL 2% (v/v) glycerol as control. After overnight digestion, a standard PCR was performed on 2 μL sample. The sequences of primers used for PCR are listed in Supplemental Table 1 online.
Comet Assay and Treatment with DNA Damaging Agents
The comet assay was performed as described previously (Wang and Liu, 2006). The percentage of DNA in each comet tail was evaluated by comet-scoring software (http://www.autocomet.com). For chemical treatment, surface-sterilized seeds were germinated on agar containing different concentrations of MMS or cisplatin, kept at 4°C for 2 d, and then grown at 22°C for additional 16 d.
Enzyme Assays and Protein Blot Analysis
The in-gel activity assays for AOs and ACO were performed as described previously (Koshiba et al., 1996; Bernard et al., 2009). Protein samples were separated by standard SDS-PAGE (12%), transferred to nitrocellulose, and labeled with antisera against ACO (Bernard et al., 2009) or the mitochondrial marker protein Cox2 (Agrisera).
Yeast Strains, Cell Growth, and Plasmids
The GAL-CIA1 mutant strain and the plasmids p416 and p416-ScCIA1 (Balk et al., 2005) were kindly provided by R. Lill (University of Marburg, Germany). The coding sequence of At-CIA1 was subcloned into the EcoRI and SalI sites of the plasmid p416 to generate p416-AtCIA1. Cells were grown in rich (YP) or minimal (SC) medium containing Glc or Gal as carbon sources at a concentration of 2%. The yeast strain harboring a conditional (doxycycline-dependent) disruption in the CIA2/YHR122W gene (Weerapana et al., 2010) was provided by Z. Li (Zhejiang University, China). The coding sequence of CIA2/YHR122W or AE7 was subcloned into the EcoRI and SpeI sites of the pESC-HIS plasmid (Stratagene) to generate FLAG fusion constructs and transformed into yeast. The expression of the endogenous CIA2/YHR122W gene was switched off by the addition of 10 μM/mL doxycycline.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: AE7 (At1g68310), AEL1 (At3g50845), AEL2 (At3g09380), CIA1 (At2g26060), NAR1 (At4g16440), MET18 (At5g48120), ATM3 (At5g58270), ROS1 (At2g36490), BRCA1 (At4g21070), GR1 (At3g52115), KU70 (At1g16970), KU80 (At1g48050), LIG4 (At5Gg57160), PARP1 (At4g02390), PARP2 (At2g31320), CYCB1;1 (At4g37490), WEE1 (At1g02970), RAD51D (At1g07745), MRD1 (At1g53480), RAD51 (At5t20850), ACO1 (At4g35830), ACO2 (At4g26970), and ACO3 (At2g05710).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Subcellular Localization of AE7, CIA1, and MET18.
Supplemental Figure 2. Diagram of Gene Structures of AE7, CIA1, NAR1, and MET18.
Supplemental Figure 3. Arabidopsis AE7 Failed to Rescue the Growth of a Yeast Strain with a Doxycycline-Repressible CIA2/YHR122W Gene.
Supplemental Figure 4. Arabidopsis CIA1 Rescued the Yeast Gal-CIA1 Mutant.
Supplemental Figure 5. Aconitase Activity in Wild-Type and ae7-1 Seedlings.
Supplemental Figure 6. qRT-PCR Analysis of Expressions of the ACO and ROS1 Genes.
Supplemental Figure 7. The atm3-4 Mutant Showed Cell Proliferation Defects and a Genetic Interaction with ae7-1.
Supplemental Figure 8. The ae7 Phenotype Was Rescued by the Moss (Physcomitrella patens) and Rice (Oryza sativa) Orthologs of AE7.
Supplemental Figure 9. The ae7 Phenotype Could Not Be Rescued by Arabidopsis AE7-Like Genes, AEL1 (At3g50845) and AEL2 (At3g09380), Respectively.
Supplemental Table 1. Sequences of Oligonucleotide Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank Peter Doerner, Lieven De Veylder, Barbara Hohn, the ABRC, and the European Arabidopsis Stock Centre for seeds of Arabidopsis mutants and marker lines, R. Lill for the yeast GAL-CIA1 mutant strain and the p416 and p416-ScCIA1 plasmids, Z. Li for the doxycycline-repressible cia2/yhr122w yeast strain and pESC-HIS plasmid, X. Gao for scanning electron microscopy, X. Gao for confocal microscopy, and L. Taylor for advise and primers for the McrBC-PCR assay. This work was supported by grants from the Chinese National Scientific Foundation (31070163), the Chinese Academy of Sciences (KSCX2-EW-Q-1-04), and the National Basic Research Program of China (973 Program, 2012CB910503). D.G.B. was supported by a Marie Curie Intra-European Fellowship (EU FP7), and J.B. was supported by a University Research Fellowship from the Royal Society.
AUTHOR CONTRIBUTIONS
D.L., J.B., H.H., and X.C. designed the research. D.L., D.G.B., J.B., and X.C. performed the experiments. D.L., J.B., H.H., and X.C. analyzed the data and wrote the article.
Glossary
- Fe-S
iron-sulfur
- DSB
double-strand break
- CIA
cytosolic iron-sulfur cluster assembly
- MMS
methyl methanesulfonate
- HR
homologous recombination
- qRT-PCR
quantitative RT-PCR
- Ler
Landsberg erecta
- Col-0
Columbia-0
- GUS
β-glucuronidase
- Co-IP
coimmunoprecipitation
- BiFC
bimolecular fluorescence complementation
- ACO
aconitase
- AO
aldehyde oxidase
- TAP
tandem affinity protein
- YFP
yellow fluorescent protein
References
- Abe K., Osakabe K., Nakayama S., Endo M., Tagiri A., Todoriki S., Ichikawa H., Toki S. (2005). Arabidopsis RAD51C gene is important for homologous recombination in meiosis and mitosis. Plant Physiol. 139: 896–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseth I., Eide L., Pirovano M., Rognes T., Seeberg E., Bjørås M. (1999). The Saccharomyces cerevisiae homologues of endonuclease III from Escherichia coli, Ntg1 and Ntg2, are both required for efficient repair of spontaneous and induced oxidative DNA damage in yeast. Mol. Cell. Biol. 19: 3779–3787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J., Aguilar Netz D.J., Tepper K., Pierik A.J., Lill R. (2005). The essential WD40 protein Cia1 is involved in a late step of cytosolic and nuclear iron-sulfur protein assembly. Mol. Cell. Biol. 25: 10833–10841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J., Pierik A.J., Netz D.J., Mühlenhoff U., Lill R. (2004). The hydrogenase-like Nar1p is essential for maturation of cytosolic and nuclear iron-sulphur proteins. EMBO J. 23: 2105–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J., Pilon M. (2011). Ancient and essential: The assembly of iron-sulfur clusters in plants. Trends Plant Sci. 16: 218–226 [DOI] [PubMed] [Google Scholar]
- Bernard D.G., Cheng Y., Zhao Y., Balk J. (2009). An allelic mutant series of ATM3 reveals its key role in the biogenesis of cytosolic iron-sulfur proteins in Arabidopsis. Plant Physiol. 151: 590–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleuyard J.-Y., Gallego M.E., Savigny F., White C.I. (2005). Differing requirements for the Arabidopsis Rad51 paralogs in meiosis and DNA repair. Plant J. 41: 533–545 [DOI] [PubMed] [Google Scholar]
- Bleuyard J.Y., Gallego M.E., White C.I. (2006). Recent advances in understanding of the DNA double-strand break repair machinery of plants. DNA Repair (Amst.) 5: 1–12 [DOI] [PubMed] [Google Scholar]
- Branzei D., Foiani M. (2010). Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell Biol. 11: 208–219 [DOI] [PubMed] [Google Scholar]
- Bych K., Netz D.J., Vigani G., Bill E., Lill R., Pierik A.J., Balk J. (2008). The essential cytosolic iron-sulfur protein Nbp35 acts without Cfd1 partner in the green lineage. J. Biol. Chem. 283: 35797–35804 [DOI] [PubMed] [Google Scholar]
- Chen C., Wang S., Huang H. (2000). LEUNIG has multiple functions in gynoecium development in Arabidopsis. Genesis 26: 42–54 [DOI] [PubMed] [Google Scholar]
- Colón-Carmona A., You R., Haimovitch-Gal T., Doerner P. (1999). Technical advance: Spatio-temporal analysis of mitotic activity with a labile cyclin-GUS fusion protein. Plant J. 20: 503–508 [DOI] [PubMed] [Google Scholar]
- Cools T., De Veylder L. (2009). DNA stress checkpoint control and plant development. Curr. Opin. Plant Biol. 12: 23–28 [DOI] [PubMed] [Google Scholar]
- Criqui M.C., Weingartner M., Capron A., Parmentier Y., Shen W.H., Heberle-Bors E., Bögre L., Genschik P. (2001). Sub-cellular localisation of GFP-tagged tobacco mitotic cyclins during the cell cycle and after spindle checkpoint activation. Plant J. 28: 569–581 [DOI] [PubMed] [Google Scholar]
- Culligan K., Tissier A., Britt A. (2004). ATR regulates a G2-phase cell-cycle checkpoint in Arabidopsis thaliana. Plant Cell 16: 1091–1104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter K., Joubès J., Cools T., Verkest A., Corellou F., Babiychuk E., Van Der Schueren E., Beeckman T., Kushnir S., Inzé D., De Veylder L. (2007). Arabidopsis WEE1 kinase controls cell cycle arrest in response to activation of the DNA integrity checkpoint. Plant Cell 19: 211–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doucet-Chabeaud G., Godon C., Brutesco C., de Murcia G., Kazmaier M. (2001). Ionising radiation induces the expression of PARP-1 and PARP-2 genes in Arabidopsis. Mol. Genet. Genomics 265: 954–963 [DOI] [PubMed] [Google Scholar]
- Durrant W.E., Wang S., Dong X. (2007). Arabidopsis SNI1 and RAD51D regulate both gene transcription and DNA recombination during the defense response. Proc. Natl. Acad. Sci. USA 104: 4223–4227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frazzon A.P., Ramirez M.V., Warek U., Balk J., Frazzon J., Dean D.R., Winkel B.S. (2007). Functional analysis of Arabidopsis genes involved in mitochondrial iron-sulfur cluster assembly. Plant Mol. Biol. 64: 225–240 [DOI] [PubMed] [Google Scholar]
- Garcia V., Bruchet H., Camescasse D., Granier F., Bouchez D., Tissier A. (2003). AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell 15: 119–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gari K., León Ortiz A.M., Borel V., Flynn H., Skehel J.M., Boulton S.J. (2012). MMS19 links cytoplasmic iron-sulfur cluster assembly to DNA metabolism. Science 337: 243–245 [DOI] [PubMed] [Google Scholar]
- Gherbi H., Gallego M.E., Jalut N., Lucht J.M., Hohn B., White C.I. (2001). Homologous recombination in planta is stimulated in the absence of Rad50. EMBO Rep. 2: 287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godman J.E., Molnár A., Baulcombe D.C., Balk J. (2010). RNA silencing of hydrogenase(-like) genes and investigation of their physiological roles in the green alga Chlamydomonas reinhardtii. Biochem. J. 431: 345–351 [DOI] [PubMed] [Google Scholar]
- Gong Z., Morales-Ruiz T., Ariza R.R., Roldán-Arjona T., David L., Zhu J.K. (2002). ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylase/lyase. Cell 111: 803–814 [DOI] [PubMed] [Google Scholar]
- Gu X.L., Wang H., Huang H., Cui X.F. (2012). SPT6L encoding a putative WG/GW-repeat protein regulates apical-basal polarity of embryo in Arabidopsis. Mol. Plant 5: 249–259 [DOI] [PubMed] [Google Scholar]
- Hatfield M.D., Reis A.M., Obeso D., Cook J.R., Thompson D.M., Rao M., Friedberg E.C., Queimado L. (2006). Identification of MMS19 domains with distinct functions in NER and transcription. DNA Repair (Amst.) 5: 914–924 [DOI] [PubMed] [Google Scholar]
- Hausmann A., Aguilar Netz D.J., Balk J., Pierik A.J., Mühlenhoff U., Lill R. (2005). The eukaryotic P loop NTPase Nbp35: An essential component of the cytosolic and nuclear iron-sulfur protein assembly machinery. Proc. Natl. Acad. Sci. USA 102: 3266–3271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.E., You H.J., Choi J.Y., Doetsch P.W., Kim J.S., Chung M.H. (2001). Ntg2 of Saccharomyces cerevisiae repairs the oxidation products of 8-hydroxyguanine. Biochem. Biophys. Res. Commun. 285: 1186–1191 [DOI] [PubMed] [Google Scholar]
- Kispal G., Csere P., Guiard B., Lill R. (1997). The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 418: 346–350 [DOI] [PubMed] [Google Scholar]
- Kispal G., Csere P., Prohl C., Lill R. (1999). The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18: 3981–3989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klinge S., Hirst J., Maman J.D., Krude T., Pellegrini L. (2007). An iron-sulfur domain of the eukaryotic primase is essential for RNA primer synthesis. Nat. Struct. Mol. Biol. 14: 875–877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohbushi H., Nakai Y., Kikuchi S., Yabe T., Hori H., Nakai M. (2009). Arabidopsis cytosolic Nbp35 homodimer can assemble both [2Fe-2S] and [4Fe-4S] clusters in two distinct domains. Biochem. Biophys. Res. Commun. 378: 810–815 [DOI] [PubMed] [Google Scholar]
- Koshiba T., Saito E., Ono N., Yamamoto N., Sato M. (1996). Purification and properties of flavin- and molybdenum-containing aldehyde oxidase from coleoptiles of maize. Plant Physiol. 110: 781–789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou H., Zhou Y., Gorospe R.M., Wang Z. (2008). Mms19 protein functions in nucleotide excision repair by sustaining an adequate cellular concentration of the TFIIH component Rad3. Proc. Natl. Acad. Sci. USA 105: 15714–15719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir S., Babiychuk E., Storozhenko S., Davey M.W., Papenbrock J., De Rycke R., Engler G., Stephan U.W., Lange H., Kispal G., Lill R., Van Montagu M. (2001). A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13: 89–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafarge S., Montané M.H. (2003). Characterization of Arabidopsis thaliana ortholog of the human breast cancer susceptibility gene 1: AtBRCA1, strongly induced by gamma rays. Nucleic Acids Res. 31: 1148–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder S., Bankmann M., Guzder S.N., Sung P., Prakash L., Prakash S. (1996). Dual requirement for the yeast MMS19 gene in DNA repair and RNA polymerase II transcription. Mol. Cell. Biol. 16: 6783–6793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezhneva L., Amann K., Meurer J. (2004). The universally conserved HCF101 protein is involved in assembly of [4Fe-4S]-cluster-containing complexes in Arabidopsis thaliana chloroplasts. Plant J. 37: 174–185 [DOI] [PubMed] [Google Scholar]
- Li H., Xu L., Wang H., Yuan Z., Cao X., Yang Z., Zhang D., Xu Y., Huang H. (2005). The Putative RNA-dependent RNA polymerase RDR6 acts synergistically with ASYMMETRIC LEAVES1 and 2 to repress BREVIPEDICELLUS and MicroRNA165/166 in Arabidopsis leaf development. Plant Cell 17: 2157–2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill R. (2009). Function and biogenesis of iron-sulphur proteins. Nature 460: 831–838 [DOI] [PubMed] [Google Scholar]
- Mok Y.G., Uzawa R., Lee J., Weiner G.M., Eichman B.F., Fischer R.L., Huh J.H. (2010). Domain structure of the DEMETER 5-methylcytosine DNA glycosylase. Proc. Natl. Acad. Sci. USA 107: 19225–19230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz D.J., Pierik A.J., Stümpfig M., Mühlenhoff U., Lill R. (2007). The Cfd1-Nbp35 complex acts as a scaffold for iron-sulfur protein assembly in the yeast cytosol. Nat. Chem. Biol. 3: 278–286 [DOI] [PubMed] [Google Scholar]
- Netz D.J., Stith C.M., Stümpfig M., Köpf G., Vogel D., Genau H.M., Stodola J.L., Lill R., Burgers P.M., Pierik A.J. (2012). Eukaryotic DNA polymerases require an iron-sulfur cluster for the formation of active complexes. Nat. Chem. Biol. 8: 125–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Netz D.J., Stümpfig M., Doré C., Mühlenhoff U., Pierik A.J., Lill R. (2010). Tah18 transfers electrons to Dre2 in cytosolic iron-sulfur protein biogenesis. Nat. Chem. Biol. 6: 758–765 [DOI] [PubMed] [Google Scholar]
- Penterman J., Zilberman D., Huh J.H., Ballinger T., Henikoff S., Fischer R.L. (2007). DNA demethylation in the Arabidopsis genome. Proc. Natl. Acad. Sci. USA 104: 6752–6757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L., Prakash S. (1979). Three additional genes involved in pyrimidine dimer removal in Saccharomyces cerevisiae: RAD7, RAD14 and MMS19. Mol. Gen. Genet. 176: 351–359 [DOI] [PubMed] [Google Scholar]
- Roa H., Lang J., Culligan K.M., Keller M., Holec S., Cognat V., Montané M.H., Houlné G., Chabouté M.E. (2009). Ribonucleotide reductase regulation in response to genotoxic stress in Arabidopsis. Plant Physiol. 151: 461–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Solodovnikova N., Nicholson T., Antholine W., Walden W.E. (2003). A novel eukaryotic factor for cytosolic Fe-S cluster assembly. EMBO J. 22: 4826–4835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolf J., Makrantoni V., Ingledew W.J., Stark M.J., White M.F. (2006). The DNA repair helicases XPD and FancJ have essential iron-sulfur domains. Mol. Cell 23: 801–808 [DOI] [PubMed] [Google Scholar]
- Schwenkert S., Netz D.J., Frazzon J., Pierik A.J., Bill E., Gross J., Lill R., Meurer J. (2010). Chloroplast HCF101 is a scaffold protein for [4Fe-4S] cluster assembly. Biochem. J. 425: 207–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A.K., Pallesen L.J., Spang R.J., Walden W.E. (2010). Cytosolic iron-sulfur cluster assembly (CIA) system: Factors, mechanism, and relevance to cellular iron regulation. J. Biol. Chem. 285: 26745–26751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan V., Netz D.J., Webert H., Mascarenhas J., Pierik A.J., Michel H., Lill R. (2007). Structure of the yeast WD40 domain protein Cia1, a component acting late in iron-sulfur protein biogenesis. Structure 15: 1246–1257 [DOI] [PubMed] [Google Scholar]
- Stehling O., Vashisht A.A., Mascarenhas J., Jonsson Z.O., Sharma T., Netz D.J., Pierik A.J., Wohlschlegel J.A., Lill R. (2012). MMS19 assembles iron-sulfur proteins required for DNA metabolism and genomic integrity. Science 337: 195–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swoboda P., Gal S., Hohn B., Puchta H. (1994). Intrachromosomal homologous recombination in whole plants. EMBO J. 13: 484–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi N., Lammens T., Boudolf V., Maes S., Yoshizumi T., De Jaeger G., Witters E., Inzé D., De Veylder L. (2008). The DNA replication checkpoint aids survival of plants deficient in the novel replisome factor ETG1. EMBO J. 27: 1840–1851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Adachi Y., Chiba K., Oguchi K., Takahashi H. (2002). Identification of Ku70 and Ku80 homologues in Arabidopsis thaliana: Evidence for a role in the repair of DNA double-strand breaks. Plant J. 29: 771–781 [DOI] [PubMed] [Google Scholar]
- Uringa E.J., Youds J.L., Lisaingo K., Lansdorp P.M., Boulton S.J. (2011). RTEL1: An essential helicase for telomere maintenance and the regulation of homologous recombination. Nucleic Acids Res. 39: 1647–1655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaithiyalingam S., Warren E.M., Eichman B.F., Chazin W.J. (2010). Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc. Natl. Acad. Sci. USA 107: 13684–13689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Attikum H., Bundock P., Overmeer R.M., Lee L.Y., Gelvin S.B., Hooykaas P.J. (2003). The Arabidopsis AtLIG4 gene is required for the repair of DNA damage, but not for the integration of Agrobacterium T-DNA. Nucleic Acids Res. 31: 4247–4255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadarajan J., Guilleminot J., Saint-Jore-Dupas C., Piégu B., Chabouté M.E., Gomord V., Coolbaugh R.C., Devic M., Delorme V. (2010). ATR3 encodes a diflavin reductase essential for Arabidopsis embryo development. New Phytol. 187: 67–82 [DOI] [PubMed] [Google Scholar]
- Veatch J.R., McMurray M.A., Nelson Z.W., Gottschling D.E. (2009). Mitochondrial dysfunction leads to nuclear genome instability via an iron-sulfur cluster defect. Cell 137: 1247–1258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Liu Z. (2006). Arabidopsis ribonucleotide reductases are critical for cell cycle progression, DNA damage repair, and plant development. Plant Cell 18: 350–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Gu X., Xu D., Wang W., Wang H., Zeng M., Chang Z., Huang H., Cui X. (2011). miR396-targeted AtGRF transcription factors are required for coordination of cell division and differentiation during leaf development in Arabidopsis. J. Exp. Bot. 62: 761–773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weerapana E., Wang C., Simon G.M., Richter F., Khare S., Dillon M.B., Bachovchin D.A., Mowen K., Baker D., Cravatt B.F. (2010). Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 468: 790–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H., Zhang X., Liu J., Wang Y., He J., Yang T., Hong X., Yang Q., Gong Z. (2009). Epigenetic regulation, somatic homologous recombination, and abscisic acid signaling are influenced by DNA polymerase ε mutation in Arabidopsis. Plant Cell 21: 386–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Haberer G., Matthes M., Rattei T., Mayer K.F., Gierl A., Torres-Ruiz R.A. (2010). Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 17809–17814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Z., Luo D., Li G., Yao X., Wang H., Zeng M., Huang H., Cui X. (2010). Characterization of the AE7 gene in Arabidopsis suggests that normal cell proliferation is essential for leaf polarity establishment. Plant J. 64: 331–342 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Lyver E.R., Nakamaru-Ogiso E., Yoon H., Amutha B., Lee D.W., Bi E., Ohnishi T., Daldal F., Pain D., Dancis A. (2008). Dre2, a conserved eukaryotic Fe/S cluster protein, functions in cytosolic Fe/S protein biogenesis. Mol. Cell. Biol. 28: 5569–5582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou B.B., Elledge S.J. (2000). The DNA damage response: Putting checkpoints in perspective. Nature 408: 433–439 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.