This work investigates the residues of plastocyanin in close proximity to photosystem I and cytochrome b6f, in both the thylakoid vesicle–embedded and solubilized states. It demonstrates that electrostatic interactions are used to form a transient and loose contact of plastocyanin with photosystem I and cytochrome b6f, and then hydrophobic interactions are used to form electron transfer complexes of plastocyanin with photosystem I and cytochrome b6f.
Abstract
In the photosynthetic light reactions of plants and cyanobacteria, plastocyanin (Pc) plays a crucial role as an electron carrier and shuttle protein between two membrane protein complexes: cytochrome b6f (cyt b6f) and photosystem I (PSI). The rapid turnover of Pc between cyt b6f and PSI enables the efficient use of light energy. In the Pc-cyt b6f and Pc-PSI electron transfer complexes, the electron transfer reactions are accomplished within <10−4 s. However, the mechanisms enabling the rapid association and dissociation of Pc are still unclear because of the lack of an appropriate method to study huge complexes with short lifetimes. Here, using the transferred cross-saturation method, we investigated the residues of spinach (Spinacia oleracea) Pc in close proximity to spinach PSI and cyt b6f, in both the thylakoid vesicle–embedded and solubilized states. We demonstrated that the hydrophobic patch residues of Pc are in close proximity to PSI and cyt b6f, whereas the acidic patch residues of Pc do not form stable salt bridges with either PSI or cyt b6f, in the electron transfer complexes. The transient characteristics of the interactions on the acidic patch facilitate the rapid association and dissociation of Pc.
INTRODUCTION
Photosynthetic light reactions, in which light energy is converted into the chemical energy of ATP and NADPH, are essential for almost all life on the earth. ATP and NADPH are produced by light-driven electron transfer from water to NADP+, mediated by proteins such as cytochrome b6f (cyt b6f) and photosystem I (PSI), which are thylakoid membrane-embedded protein complexes, and plastocyanin (Pc). Pc is a soluble electron carrier protein that shuttles between cyt b6f and PSI. The heme molecules in the cytochrome f (cyt f) subunits of cyt b6f donate electrons to the copper ions in Pc, and the chlorophyll molecules, P700, in the PsaA and PsaB subunits of PSI accept electrons from Pc. Pc rapidly cycles through the following steps: binding to cyt b6f, accepting an electron from cyt b6f, dissociating from cyt b6f, binding to PSI, donating an electron to the photoexcited PSI, and then dissociating from PSI. These electron transport reactions are important in plant growth (Chida et al., 2007).
Pc forms separate electron transfer complexes with PSI and cyt b6f, and the distances between their reaction centers are sufficiently short to accomplish their electron transfer reactions in <10−4 s (Haehnel et al., 1980; Drepper et al., 1996; Hope, 2000). In addition, the Pc-PSI and Pc-cyt b6f electron transfer complexes rapidly associate and dissociate at rates of ∼108 M−1s−1 and ∼103 s−1, respectively (Drepper et al., 1996; Hope, 2000). The formation of such rapidly associating and dissociating electron transfer complexes facilitates efficient electron transport. Finazzi et al. elegantly demonstrated that the rate-limiting step in the cyt b6f-Pc-PSI electron transport in vivo is the release of Pc from PSI (Finazzi et al., 2005; Busch and Hippler, 2011). The importance of the Pc-cyt b6f interaction in vivo is less clear, although mutations in the Pc binding site of cyt f reportedly affect the growth rates of C. reinhardtii (Soriano et al., 1996).
The three-dimensional structures of the unbound forms of Pc (Xue et al., 1998; Musiani et al., 2005), cyt b6f (Kurisu et al., 2003; Stroebel et al., 2003; Yan et al., 2006; Yamashita et al., 2007; Baniulis et al., 2009), and PSI (Jordan et al., 2001; Ben-Shem et al., 2003; Jolley et al., 2005; Amunts et al., 2007, 2010) have been solved. Mutational analyses of Pc revealed that the residues in the hydrophobic patch that surrounds the copper ion, such as Leu-12 and Ala-90 (see Supplemental Figure 1 online), and those in the acidic patch located adjacent to the hydrophobic patch, such as Asp-42, Glu-43, Asp-44, Glu-45, Glu-59, Glu-60, and Asp-61 of spinach (Spinacia oleracea) Pc (see Supplemental Figure 1 online), are reportedly important for Pc-PSI (Haehnel et al., 1994; Lee et al., 1995; Hippler et al., 1996; Sigfridsson et al., 1996, 1997; Young et al., 1997) and Pc-cyt b6f (Modi et al., 1992b; Lee et al., 1995; Sigfridsson, 1998; Hope, 2000; Illerhaus et al., 2000) electron transport in eukaryotic plants. Mutational and cross-linking studies of the PSI from C. reinhardtii revealed that two Trp residues close to P700 stemming from the PsaA and PsaB subunits, PsaA-Trp-651 and PsaB-Trp-627 in Chlamydomonas reinhardtii (Sommer et al., 2002, 2004), and clusters of basic residues located adjacent to the Trp residues from the PsaF subunit, PsaF-Lys-16, PsaF-Lys-23, and PsaF-Lys-30 in C. reinhardtii (Hippler et al., 1989, 1996, 1997, 1998), participate in the electron transport (Busch and Hippler, 2011). The cyt b6f from eukaryotic plants also contains several hydrophobic residues surrounding the heme in cyt f and clusters of basic residues located adjacent to the hydrophobic residues, and these residues also participate in electron transport, based on mutational analyses of cyt b6f and isolated cyt f (Soriano et al., 1996, 1998; Gong et al., 2000a, 2000b). However, the roles of these residues in the rapid turnover of Pc remain unknown.
Structural analyses of the full-length Pc-PSI and Pc-cyt b6f complexes, which are required to clarify the mechanism of efficient electron transport, have been hampered by their large sizes and short lifetimes. Our group has developed the transferred cross-saturation (TCS) method, which enables the identification of the residues of protein ligands in close proximity to huge (>100 kD) and/or heterogeneous complexes under fast exchange conditions (Nakanishi et al., 2002; Shimada, 2005). The TCS method has been used to investigate the interactions of large proteins (Nakanishi et al., 2002; Impagliazzo and Ubbink, 2004; Yokogawa et al., 2011), membrane proteins (Takeuchi et al., 2003; Yokogawa et al., 2005; Malia and Wagner, 2007; Kofuku et al., 2009; Yoshiura et al., 2010), liposomes (Takeuchi et al., 2004), and insoluble biomolecules (Nishida et al., 2003; Ichikawa et al., 2007).
Here, we used the TCS method to identify the binding interfaces of Pc for PSI and cyt b6f, in both the thylakoid vesicle–embedded state and the solubilized and purified state. Based on these results, we discuss the roles of the hydrophobic and acidic patches of Pc in electron transport.
RESULTS
Outlines of the TCS Experiments
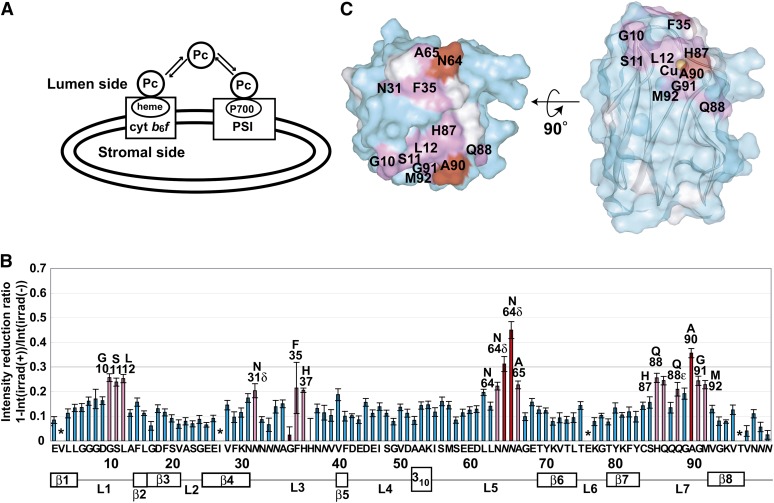
Schematic diagrams of the TCS experiments to assess the Pc-PSI and Pc- cyt b6f interactions are provided in Figure 1 (Shimada et al., 2009). The TCS experiments are performed with an excess amount of uniformly [2H,15N]-labeled Pc, relative to nonlabeled PSI or cyt b6f. Accordingly, Pc is a low proton density molecule with almost no aliphatic protons, whereas PSI and cyt b6f are high proton density molecules with aliphatic protons.
Figure 1.
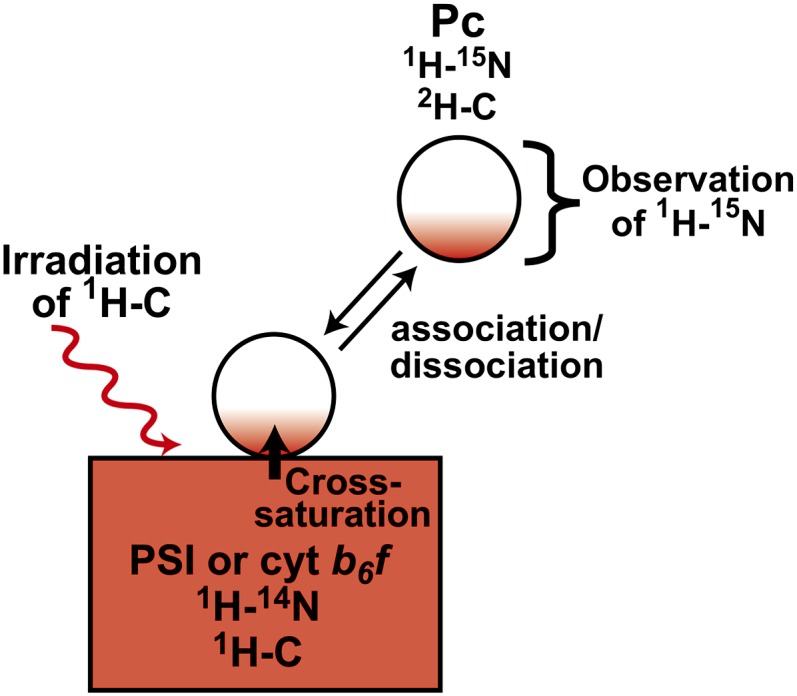
Schematic Diagram of the TCS Experiments with the Pc-PSI and Pc-cyt b6f Complexes.
The saturation of the interface residues of Pc is efficiently transferred to the free state. The saturation of each amide proton of unbound Pc can be observed as an intensity reduction in the 1H-15N shift correlation spectra. Therefore, the residues that are in close proximity to PSI can be identified by observations of their signal intensity reductions.
[See online article for color version of this figure.]
The complex is irradiated at a frequency corresponding to the aliphatic proton resonances, exclusively affecting either PSI or cyt b6f because almost no aliphatic protons exist in the deuterated Pc. The saturation in the aliphatic protons of PSI or cyt b6f is instantaneously transferred to all of the hydrogen atoms of PSI or cyt b6f, which has the high proton density. This phenomenon is known as the spin diffusion effect. Although deuterated Pc is not directly affected by the radiofrequency field, the saturation can be transferred from PSI or cyt b6f to Pc through the interface of the complex, in a phenomenon called the cross-saturation effect. The saturation transferred to Pc is limited to the interface, due to its low proton density. If the complexes have sufficiently large exchange rates between the free and bound states, then the saturation of the interface residues is efficiently transferred to the free state of Pc. The saturation of each amide proton of unbound Pc can be observed as an intensity reduction in the 1H-15N shift correlation spectra. Therefore, the residues that are in close proximity to PSI can be identified by observations of their signal intensity reductions. TCS enables the identification of the residues of protein ligands in close proximity to huge (>100 kD) and/or heterogeneous molecules because we do not need to directly observe the complex signals.
TCS Experiments Using Vesicles Containing Both PSI and cyt b6f
We prepared inside-out vesicles from spinach thylakoid membranes, in which the Pc binding sites of PSI and cyt b6f are exposed to the solvent, by sonication and subsequent aqueous two-phase partitioning (Andersson, 1986). The electron transport activities of the PSI embedded in the obtained vesicles were examined by the decrease of the absorption at 701 nm upon photoexcitation of P700 and its recovery upon electron acceptance from a substoichiometric amount of the reduced Pc (see Supplemental Figure 2A online). The electron transport activities of cyt b6f were examined by the decrease in the absorption at 600 nm, upon electron acceptance by oxidized Pc from a substoichiometric amount of cyt b6f (see Supplemental Figure 2B online). The sidedness of the inside-out vesicles was confirmed by light-induced proton extrusion by the photosynthetic light reaction of the vesicles, which was detected by the absorption change of bromocresol purple (Andersson and Akerlund, 1978).
NMR experiments were performed under conditions where both the copper ion in Pc and the heme in cyt f were reduced by ascorbic acid and PSI was in the resting state in the dark with reduced P700 to prevent the progression of the electron transfer reactions. The NMR signal assignments from the main chain atoms of free spinach Pc, in the reduced state, were established by conventional triple resonance experiments. The chemical shifts of the resonances from the amide protons of free Pc were almost identical to those of Pc isolated from spinach leaves (Driscoll et al., 1987; Musiani et al., 2005).
To investigate the binding modes of Pc with PSI and cyt b6f, we performed NMR analyses of Pc with substoichiometric amounts of PSI and cyt b6f embedded in the vesicles. The addition of RediGrad effectively suppressed the sedimentation of the vesicles during the NMR measurements (Pertoft, 2000). The chemical shifts of the resonances from uniformly [2H, 15N]-labeled Pc in the presence of a substoichiometric amount of vesicles (see Supplemental Figure 3 online) were almost identical to those for the free Pc molecule, whereas the intensities were significantly reduced upon the addition of the vesicles, suggesting that the Pc-PSI and Pc- cyt b6f interactions occur in the fast exchange mode on the NMR time scale. We confirmed these signal intensity reductions prior to all TCS experiments (see Supplemental Discussion 1 and Supplemental Table 1 online). We also verified that both the PSI and cyt b6f embedded in the vesicles still exhibited their full electron transport activities after the NMR measurements.
We performed the TCS experiments to identify the residues of Pc in close proximity to PSI and/or cyt b6f embedded in the thylakoid vesicles (Figure 2A). The TCS experimental conditions were optimized to observe the cross-saturation effect by relaxation matrix simulations (Matsumoto et al., 2010). The irradiation of the sample containing the vesicles with radio frequency pulses resulted in selective intensity losses of the NMR signals originating from the free Pc resonances, including the Asn-64 side chain resonances (see Supplemental Figure 3 online). Based on the spectra with and without irradiation, we calculated the reduction ratios of the peak intensities (Figure 2B). The residues significantly affected by irradiation, Gly-10, Ser-11, Leu-12, Asn-31, Phe-35, His-37, Asn-64, Ala-65, His-87, Gln-88, Ala-90, Gly-91, and Met-92, are mapped in Figure 2C. The affected residues formed a continuous surface on the hydrophobic patch of Pc, including Gly-10, Leu-12, and Ala-90.
Figure 2.
TCS Experiments with Excess Amounts of Pc Relative to the PSI and cyt b6f Embedded in Inside-Out Thylakoid Vesicles.
(A) Schematic diagram of the experiments. The spectra are shown in Supplemental Figure 3 online.
(B) Plots of the reduction ratios of the signal intensities originating from the amide groups, with and without presaturation. Red and pink plots represent the residues with signal intensity reduction ratios >0.3 and within the 0.2 to 0.3 range, and labeled. The residues with reduction ratios <0.2 are cyan. The error bars represent the root sum square of the reciprocal of the signal-to-noise ratio of the resonances with and without irradiation. Asterisks represent the residues with intensity reduction ratios that were not determined because of low signal intensity or spectral overlap. Side chains are denoted in italics.
(C) Mapping of the residues affected by the irradiation in the TCS experiments. The residues with signal intensity reduction ratios >0.3 and within the 0.2 to 0.3 range are colored red and pink, respectively. Pro residues and the residues with intensity reductions that were not determined because of low signal intensity or spectral overlap are white. In the middle view, the surface of Pc is transparent, and the copper atom and the ribbon diagram are simultaneously displayed. His-37, which is close to the copper atom, is hidden in these views. The molecular diagrams were generated with Web Lab Viewer Pro (Molecular Simulations).
TCS Experiments Using Solubilized PSI and cyt b6f
In order to observe the Pc–PSI and Pc–cyt b6f interactions separately, we also performed the TCS experiments using detergent-solubilized PSI and cyt b6f from spinach thylakoid membranes, according to the previously reported method (Zhang and Cramer, 2004; Amunts et al., 2005). We confirmed the electron transport activities of the solubilized PSI and cyt b6f in a similar manner (see Supplemental Figures 2A and 2B online). The slower reaction rate of the cyt b6f in vesicles compared with the solubilized cyt b6f (see Supplemental Figure 2B online) is probably due to the slower reduction of hemes by decylplastoquinol in vesicles. We also used the stopped-flow method to confirm the electron transport activities of the solubilized cyt b6f (see Supplemental Figure 2C online). In the flash photolysis and stopped flow experiments, the reaction rates were dependent on the Pc concentrations (see Supplemental Figures 2C to 2F online), and the calculated second-order rate constants of these reactions were 2.5 and 2.0 × 107 M−1 s−1, respectively, which are similar to the previously reported values (3.7 to 33 and 1.4 to 19 × 107 M−1 s−1, respectively) (Hope, 2000; Illerhaus et al., 2000; Sujak et al., 2004). The molar amounts of the polypeptide chains, as determined by the intensities of the SDS-PAGE bands, coincided with those of P700 and cyt f determined from their absorbance, suggesting that most of the solubilized PSI and cyt b6f retained their electron transport activities. We confirmed that both the solubilized PSI and cyt b6f still exhibited their full electron transport activities after the NMR measurements.
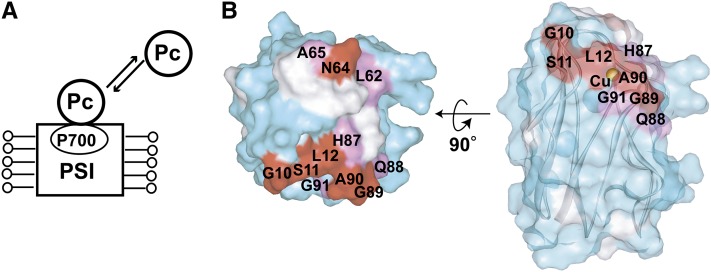
The residues of Pc in close proximity to the solubilized PSI were determined in TCS experiments (Figure 3A). Gly-10, Ser-11, Leu-12, His-37, Leu-62, Asn-64, Ala-65, His-87, Gln-88, Gly-89, Ala-90, and Gly-91 were significantly affected by the irradiation (Figure 3B; see Supplemental Figure 4A online).
Figure 3.
Determination of the Interface Residues of Pc for Solubilized PSI.
(A) Schematic diagram of the TCS experiments with excess amounts of Pc relative to the solubilized PSI.
(B) Mapping of the residues affected by the irradiation in the TCS experiments. The plots of the reduction ratios are shown in Supplemental Figure 4A online. The labeling and coloring schemes are the same as in Figure 2.
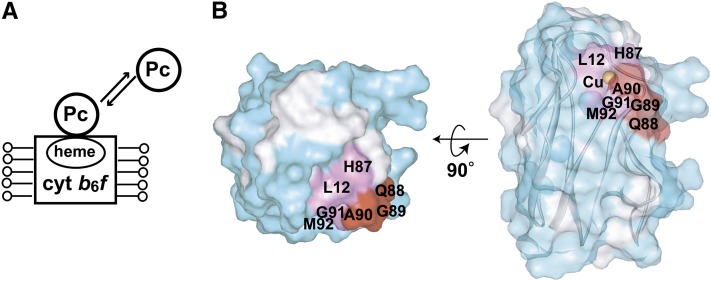
TCS experiments for the solubilized cyt b6f were also performed (Figure 4A). Leu-12, Asn-38, His-87, Asp-88, Gly-89, Ala-90, Gly-91, and Met-92 were affected by the irradiation (Figure 4B; see Supplemental Figure 4B online). The affected residues formed a continuous surface on the hydrophobic patch of Pc. It should be noted that in the TCS experiments, no effects of the irradiation were observed for the acidic patch residues.
Figure 4.
Determination of the Interface Residues of Pc for Solubilized cyt b6f.
(A) Schematic diagram of the experiments.
(B) Mapping of the residues affected by irradiation in the TCS experiments. The plots of the reduction ratios are shown in Supplemental Figure 4B online. The labeling and coloring schemes are the same as in Figure 2. Asn-38, which is close to the copper atom, is hidden in these views.
Investigation of the Effects of the Nonspecific Binding and the Residual Protons in Pc
The deuterated Pc contains residual aliphatic protons, which induce unfavorable intensity reductions for the nonbinding site residues in the TCS experiments. In addition, the irradiation causes the saturation of the nonlabeled additives in the sample, including detergents and RediGrad, and the saturation may transfer from these additives to Pc via water or nonspecific interactions, leading to other unfavorable intensity reductions. To estimate these unfavorable effects, we performed TCS experiments with Pc and the additives (see Supplemental Figure 5A online). The reduction ratios of the peak intensities in the TCS experiments are shown in Supplemental Figure 5B online. All of the residues exhibited similar intensity reductions, and none of them were affected by more than 25%, which is much less than that observed in the TCS experiments with PSI and/or cyt b6f (Figure 2B; see Supplemental Figure 4 online).
The thylakoid membrane vesicles contain various proteins and lipids; thus, nonspecific interactions between Pc and these molecules may cause intensity reductions in the TCS experiments using the inside-out vesicles. In addition, nonspecific interactions with the ferredoxin binding site of PSI may also be detected in the TCS experiments with solubilized PSI. To evaluate these effects, we used right-side-out vesicles, in which the lipid composition on the outer surface is similar to that of the inside-out vesicles (Gounaris et al., 1986), but the Pc binding sites of PSI and cyt b6f are inwardly directed and, thus, not accessible to the Pc in the solvent (see Supplemental Figure 5C online). The reduction ratios of the peak intensities of the TCS experiments are shown in Supplemental Figure 5D online. The intensity reduction was <20% for all residues, which is much smaller than that in the TCS experiments with the inside-out vesicles (Figure 2B).
Mutational Analyses
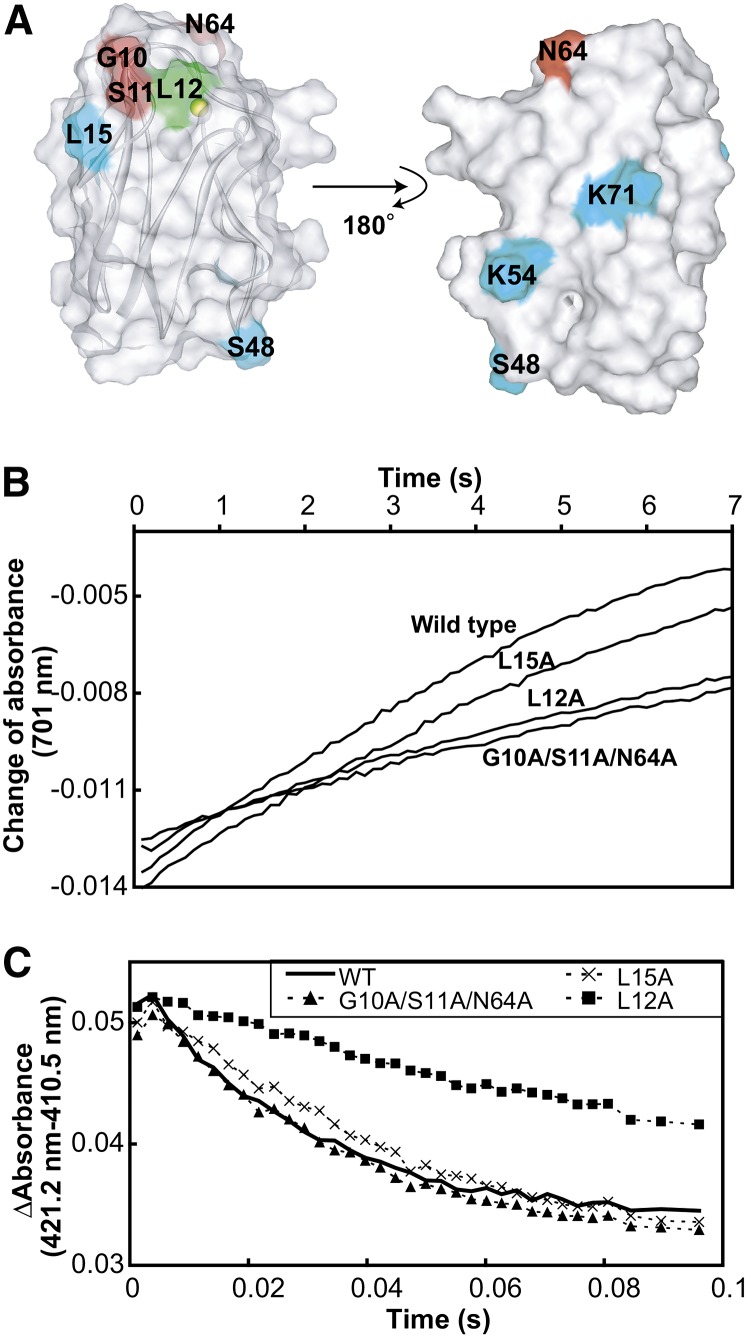
We performed mutational analyses to evaluate the residues in Pc identified as binding to PSI and cyt b6f (Figure 5, Table 1). The electron transport activities of several Pc mutants with alterations as specific residues, such as Gly-10, for PSI and cyt b6f were previously reported (Haehnel et al., 1994; Hope, 2000; Illerhaus et al., 2000). We introduced mutations in three residues that were shown to be close to PSI, but not to cyt b6f (G10A/S11A/N64A). As a control, we also prepared the L12A mutant, which exhibited reduced electron transport activity for both PSI and cyt b6f in previous reports (Sigfridsson et al., 1996; Illerhaus et al., 2000), and mutants for residues outside the binding site (L15A, S48V, K54V, and K71V) (Figure 5A). Their 1H-1H shift correlation spectra and UV/visible absorption spectra revealed that the global fold and the coordination of the copper ion of the mutants were not affected by the introduction of the mutations. The L12A mutant displayed a small shift to a longer wavelength in the UV/visible absorption spectra, as reported previously (Sigfridsson et al., 1996). The reaction rates of the Pc-PSI and Pc-cyt b6f electron transport in the wild type and the Pc mutants were determined by flash-photolysis and stopped-flow experiments, respectively. The reaction rate of the G10A/S11A/N64A mutant for PSI was significantly decreased (Figure 5B, Table 1). By contrast, its reaction rate for cyt b6f was almost identical to that of the wild type (Figure 5C, Table 1). On the other hand, the reaction rates of both the Pc-PSI and cyt b6f electron transport were significantly decreased for the L12A mutant (Figures 5B and 5C, Table 1), whereas for the other mutants, these rates were almost identical to those of the wild type (Figures 5B and 5C, Table 1).
Figure 5.
Electron Transport Activities of the Wild Type and Mutant Pcs for Solubilized PSI and cyt b6f.
(A) Mapping of the mutated residues on the structure of Pc. Gly-10, Ser-11, and Asn-64, which are included in the PSI binding site but not in the cyt b6f binding site, are colored red. Leu-12, the mutation of which reportedly led to diminished electron transport activity for both PSI and cyt b6f (Sigfridsson et al., 1996; Illerhaus et al., 2000), is colored green. Leu-15, Ser-48, Lys-54, and Lys-71, which are outside the binding site, are colored cyan.
(B) Time course of the absorbance change at 701 nm of the solution containing 200 nM solubilized PSI and 40 nM wild-type and mutant Pcs after the flash. The traces were averaged from eight flash-photolysis experiments.
(C) Time courses of the absorbance change at 421.2 minus 410.5 nm, after mixing 0.15 μM reduced and solubilized cyt b6f with 1 μM oxidized wild-type and mutant Pcs. The traces were averaged from eight stopped-flow experiments.
Table 1. Electron Transport Rates of the Wild Type and Mutant Pcs for PSI (kPSI) and cyt b6f (kb6f).
| Mutant Names | PSI Binding | cyt b6f Binding | kPSI (×10−1 s−1) | kb6f (×10 s−1) |
|---|---|---|---|---|
| Wild type | – | – | 2.72 ± 0.05 | 3.44 ± 0.13 |
| L15A | – | – | 2.43 ± 0.25 | 2.36 ± 0.20 |
| S48V | – | – | 2.26 ± 0.09 | 3.06 ± 0.08 |
| K54V | – | – | 2.59 ± 0.11 | 3.03 ± 0.10 |
| K71V | – | – | 2.28 ± 0.1 | 3.06 ± 0.13 |
| L12A | + | + | 0.84 ± 0.04 | 0.83 ± 0.05 |
| G10A/S11A/N64A | + | – | 0.68 ± 0.02 | 2.78 ± 0.08 |
Effect of Perturbation of the Interactions on the Hydrophobic Patch by Cadmium Substitution
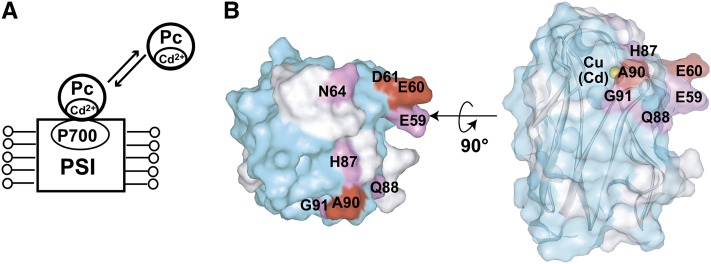
The substitution of the copper ion of Pc with a cadmium ion reportedly shifts the hydrogen bond network in the hydrophobic patch to a configuration resembling that in the oxidized Pc, without affecting the conformations of the other regions, including the acidic patch (Ubbink et al., 1996). To elucidate the PSI binding mode of Pc in more detail, the Pc–PSI interaction was further investigated under conditions where the interactions on the hydrophobic patch of Pc were perturbed by cadmium substitution.
A comparison of the 1H-15N shift correlation spectra of uniformly [15N]-labeled Pc and cadmium-substituted Pc (Cd-Pc) revealed that the signals that exhibited significantly different chemical shifts were all those from the hydrophobic patch residues, consistent with the previous report for Pcs from different species (Ubbink et al., 1996), suggesting that the Cd-Pc is correctly folded (see Supplemental Figures 6A and 6B online). Although the distance between the amide proton of Gly-8 and the Hβ of Phe-14, which are located at the stem of the loop1, is 9.5 Å in the structure of reduced Pc, a nuclear Overhauser effect, which is usually observed for hydrogen atom pairs with <5 Å distance, was observed between the corresponding hydrogen atoms in Cd-Pc, suggesting that the conformation of loop1 in the hydrophobic patch is perturbed by the cadmium substitution.
To examine whether Cd-Pc binds to PSI, a substoichiometric amount of solubilized PSI was added to uniformly [15N]-labeled Cd-Pc (see Supplemental Figure 6C online). As a result, the signal intensities of Cd-Pc remarkably decreased, and they were recovered by adding Monoraphidium braunii cytochrome c6, which reportedly binds to the Pc binding site of PSI, like Pc, and donates an electron to photoexcited PSI (Hervás et al., 1995; Hippler et al., 1997, 1998, 1999; Sommer et al., 2002, 2004, 2006), suggesting that Cd-Pc binds to PSI.
The residues of Cd-Pc in close proximity to the solubilized PSI were determined by the TCS experiments (Figure 6A). Supplemental Figure 7 online shows the calculated reduction ratios of the peak intensities from the TCS experiments using solubilized PSI, and the affected residues were mapped in Figure 6B. Glu-59, Glu-60, Asn-64, His-87, Gln-88, Ala-90, and Gly-91 were significantly affected by the irradiation. The affected residues formed continuous surfaces on the hydrophobic and acidic patches of Pc. As compared with the residues of Pc that are close to PSI (Figure 3), the residues of Cd-Pc that are close to PSI did not include several residues in loop1, such as Gly-10 and Ser-11, whereas they included the residues in the acidic patch, such as Glu-59 and Glu-60.
Figure 6.
Determination of the Interface Residues of Cd-Pc for Solubilized PSI.
(A) Schematic diagram of the TCS experiments with excess amounts of Cd-Pc, relative to the solubilized PSI.
(B) Mapping of the residues affected by the irradiation in the TCS experiments. The plots of the reduction ratios are shown in Supplemental Figure 7 online. The labeling and coloring schemes are the same as in Figure 2.
DISCUSSION
Validation of the Binding Site Determined by TCS
In this study, the PSI binding and cyt b6f binding sites of Pc were determined by TCS experiments, in both the thylakoid vesicle–embedded and solubilized states (Figures 2 to 4). The results from the experiments with the right-side-out vesicles revealed that the observed intensity reductions were not due to nonspecific interactions between Pc and the opposite stromal side of either PSI or cyt b6f (see Supplemental Figures 5C and 5D online). In addition, we confirmed that the intensity reductions were not derived from additives, such as detergents and so forth (see Supplemental Figures 5A and 5B online). Furthermore, the results of the mutational analyses were consistent with those of the TCS experiments (Figure 5, Table 1). Therefore, we concluded that the residues of Pc that are close to PSI and cyt b6f were accurately identified.
Under our experimental conditions, both the copper ion of Pc and the heme in cyt f were in the reduced state, and PSI was in the resting state with the reduced P700. Under the reactive conditions, Pc is oxidized in the Pc-cyt b6f complex, and PSI is photoexcited in the Pc-PSI complex. However, the differences in the states of Pc, cyt b6f, and PSI would hardly affect the binding modes of the Pc-cyt b6f and Pc-PSI complexes because the oxidized and reduced Pcs reportedly exhibit similar affinities for cyt f (Modi et al., 1992a; Kannt et al., 1996). In addition, previous flash photolysis analyses suggested that the electron transfer from Pc to PSI occurs in ∼10 μs upon photoexcitation of the ground state complex (Haehnel et al., 1989), whereas 10−5 to 100 s are usually required for a conformational change with >5 Å amplitude (McCammon and Harvey, 1987).
The previous laser flash photolysis and stopped-flow experiments suggested that the binding of Pc to PSI and cyt b6f leads to the formation of the electron transfer complexes, where electrons are efficiently transferred without rearrangement of the complex (Drepper et al., 1996; Hope, 2000). Therefore, the majority (>∼75%) of the Pc-PSI and Pc-cyt b6f complexes would exist as the electron transfer complexes. On the other hand, the minor binding modes with a slow electron transfer rate are estimated to be <∼25% of the total bound population and would not be unique (Hope, 2000). Therefore, our TCS experiments have identified the residues of Pc in close proximity to PSI and cyt b6f in their electron transfer complexes.
Comparison of the Binding Modes between the Solubilized Complexes and Those Embedded in Vesicles
There is a possibility that the Pc binding modes of solubilized PSI and cyt b6f are different from those embedded in the thylakoid membrane because the subunits of PSI or cyt b6f might be decomposed upon solubilization (Hiyama, 2004). Therefore, we compared the residues of Pc in close proximity to the thylakoid vesicles and solubilized PSI or cyt b6f, determined by our TCS experiments (Figures 2 to 4). The binding sites for the inside-out vesicles were mostly composed of those for the solubilized PSI and cyt b6f, indicating that the Pc binding modes of the solubilized PSI and cyt b6f and those embedded in the thylakoid vesicles are similar.
Roles of the Acidic and Hydrophobic Residues of Pc in the Pc–PSI and Pc–cyt b6f Interactions
The TCS experiments revealed that the acidic patch of Pc is not close to either PSI or cyt b6f in the electron transfer complexes (Figures 2 to 4). It should be noted that the salt concentration used here (20 mM potassium phosphate) is reportedly optimal for the electron transport of the Pc-PSI and Pc-cyt b6f complexes (Hope, 2000). In addition, in the TCS experiments using vesicles, no detergents were employed and, therefore, no decomposition of the subunits of PSI and/or cyt b6f caused by detergents occurred (Figure 2).
According to the classification proposed by Kumar and Nussinov, an ion pair is defined as a salt bridge if the side chain charged group centroids and at least one pair of side chain nitrogen and oxygen atoms are within 4 Å (Kumar and Nussinov, 2002a, 2002b). The definition of a stable salt bridge is based on the distance between the side chain atoms, whereas the distances between the main chain amide protons of Pc and protons of PSI or cyt b6f are reflected in the TCS experiments. However, our statistical analyses of the salt bridge structures revealed that most of the stable salt bridges could be identified by the TCS experiments; thus, the acidic patch residues of Pc do not form stable salt bridges with either PSI or cyt b6f in the electron transfer complexes (see Supplemental Discussion 1 and Supplemental Table 2 online).
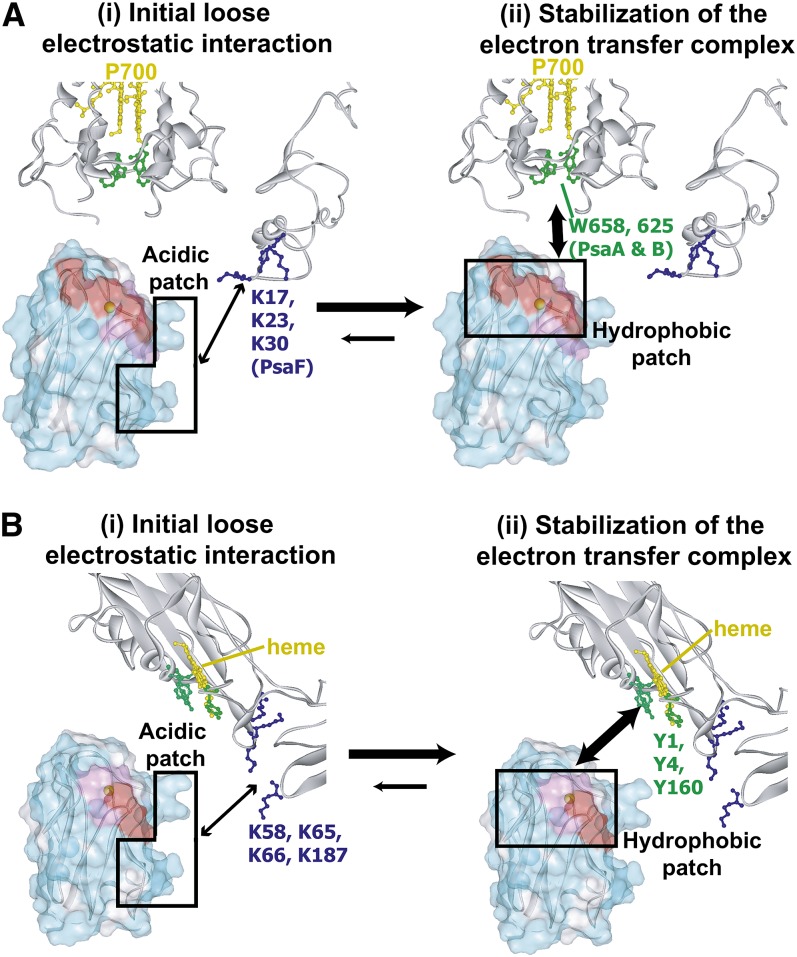
The previously reported mutational analyses, cross-linking studies, and ion strength dependencies of the Pc-PSI and Pc-cyt b6f electron transport rates have shown that the acidic patch residues of Pc and the basic residues of PSI and cyt b6f are responsible for the Pc-PSI (Hippler et al., 1989, 1996, 1997; Lee et al., 1995; Sigfridsson et al., 1996, 1997; Young et al., 1997) and Pc-cyt b6f electron transport reactions (Modi et al., 1992b; Lee et al., 1995; Hippler et al., 1998; Gong et al., 2000b; Illerhaus et al., 2000). Therefore, the acidic patch of Pc may increase its local concentration around PSI and cyt b6f, thus facilitating the rapid formation of the electron transfer complex by occasionally forming dynamic salt bridges (Bergkvist et al., 2001) or long-distance ion pairs (Figure 7, left). Most of these loose electrostatic interactions would not contribute to the stabilization of the complex (Kumar and Nussinov, 2002a) or be observable in our TCS experiments.
Figure 7.
Schematic Views of the Interaction Modes in the Pc-PSI and Pc-cyt b6f Complexes.
The structures of spinach Pc, pea (Pisum sativum) PSI, and turnip (Brassica rapa) cyt f were manually positioned. For simplicity, only the residues of the PsaA and PsaB subunits that are within 20 Å from Trp-658-PsaA and Trp-625-PsaB and the residues of PsaF are displayed in PSI. The residues of Pc in close proximity to PSI and cyt b6f, as determined by TCS, are labeled and colored in (A) and (B), respectively, according to the same scheme as in Figure 2. Hydrophobic and basic residues that were proposed to interact with Pc in previous mutational analyses are colored green and blue, respectively, in the structures of PSI (Hippler et al., 1996, 1997, 1998; Sommer et al., 2002, 2004; Busch and Hippler, 2011) and cyt f (Soriano et al., 1996, 1998; Gong et al., 2000a). The acidic and hydrophobic patch residues of Pc are enclosed in squares. First, Pc forms a transient and loose contact with PSI and cyt b6f by electrostatic interactions between the acidic residues in Pc and the basic residues in PSI and cyt b6f. Subsequently, Pc forms an electron transfer complex with PSI and cyt b6f by hydrophobic interactions. This model suggests that the basic residues of cyt f do not affect the conformation of the Pc-cyt b6f electron transfer complex, which is consistent with the previous finding that the mutations of the basic residues of cyt f do not affect the rate of cyt f oxidation in vivo (Soriano et al., 1996, 1998) The molecular diagrams were generated with Web Lab Viewer Pro (Molecular Simulations).
The TCS results also revealed that the residues in the hydrophobic patch are in close proximity to PSI and cyt b6f, in their electron transfer complexes (Figures 2 to 4). These results are consistent with those from previous mutational studies of Pc, PSI, and cyt b6f, which revealed that the hydrophobic patch residues of Pc (Gly-10 and Ala-90) (Haehnel et al., 1994) and the two Trp residues close to P700 stemming from the PsaA and PsaB subunits, PsaA-Trp-651 and PsaB-Trp-627 in C. reinhardtii (Sommer et al., 2002, 2004), are required for the efficient electron transport between Pc and PSI and that the hydrophobic patch residues of Pc (Illerhaus et al., 2000) and several hydrophobic residues surrounding the heme in cyt f (Gong et al., 2000a) are important for the electron transport between Pc and cyt b6f. These residues may be responsible for the formation of the electron transfer complex, which shortens the distances between their reaction centers to accomplish the electron transfer reaction in <10−4 s via the several hydrophobic residues surrounding P700 and the hemes in cyt f of PSI and cyt b6f, respectively (Figure 7, right).
The absence of stable salt bridges in the acidic patch is in agreement with the rapid dissociation of the complexes. In the case of the Pc–PSI interaction, Finazzi et al. elegantly demonstrated that the E613N mutation, which decreased the dissociation rate of the Pc-PSI complex in vitro, affected the total cyt b6f-Pc-PSI electron flow in vivo, whereas the W651 mutation, which decreased the affinity of the Pc-PSI complex in vitro, was no longer important in vivo (Finazzi et al., 2005; Busch and Hippler, 2011). These studies suggested that the rapid dissociation of Pc from PSI is important for efficient electron transport. Therefore, these studies support the importance of our finding that the lack of stable salt bridges in the Pc-PSI electron transfer complex enables rapid dissociation of the complexes.
The acidic patch residues of Pc can be bound to the isolated PsaF subunit or cross-linked with the basic residues of PsaF, and part of the cross-linked Pc-PSI complex can efficiently transfer electrons (Hippler et al., 1989, 1996, 1997; Farkas and Hansson, 2011). In order to further investigate the interaction between Pc and PSI, we analyzed Cd-Pc, in which the hydrophobic patches adopt a different conformation from that in Pc. Our TCS experiments for the complex between Cd-Pc and PSI revealed that in Cd-Pc, His-87, which coordinates the copper ion, and the residues in the acidic patch (Glu-59 and Glu-60) are close to PSI (Figure 6). Therefore, the Pc-PSI complex can assume a complex formation mode, in which the acidic patch residues of Pc are close to PSI but electrons can be efficiently transferred. The electron transfer rate of the cross-linked Pc-PSI complex is reportedly slightly lower than that of the in vivo Pc-PSI complex, suggesting that the relative positions of the hydrophobic patches, which contain the reaction centers, of the cross-linked Pc to PSI are not completely favorable for the electron transfer (Hippler et al., 1996). By contrast, the binding mode of the Pc-PSI electron transfer complex without the modification, which is clarified by our TCS experiments, is optimal for electron transfer. This is in agreement with the observations from the following reports: (1) The Pc-PSI complex exhibits low electron transport rates at salt concentrations <40 mM, suggesting that very strong electrostatic interactions are disadvantageous for the electron transport (Hope, 2000), and (2) the salt concentration under physiological conditions is 0.2 to 0.4 M, which is not optimal for salt bridge formation (Soriano et al., 1996; Hope, 2000).
In the case of the Pc–cyt b6f interaction, the fern (Dryopteris crassirhizoma) and spinach Pcs reportedly exhibited almost identical electron transfer activities for turnip (Brassica rapa) cyt f, although the locations of the acidic residues in fern Pc are totally different from those in spinach Pc (Sato et al., 2004) (see Supplemental Figures 1 and 8 online). The absence of a stable salt bridge is also in accordance with the pseudospecificity of the acidic patches in the fern and spinach Pcs because the relative orientations of Pc and cyt f are supposed to be flexible.
The structures of the Pc-cyt f complexes from vascular plants have been proposed based on NMR experiments of the chemical shift perturbations and intermolecular pseudocontact shifts on Pc, induced by the heme-containing cyt f (Ubbink et al., 1998; Lange et al., 2005). These structures are considered to represent the electron transfer complex because the timescales of the chemical and pseudocontact shifts (∼10 to 100 ms) are also much longer than the lifetime of the complex (Ubbink et al., 1998; Lange et al., 2005). For comparison with the results of our TCS experiments, these structures were also interpreted under the definition of the stable salt bridge proposed by Kumar and Nussinov (2002a, 2002b). In the proposed electron transfer complex structure of the spinach Pc-turnip cyt f complex (Ubbink et al., 1998), the side chain charged group centroid of Glu-43 is within 4 Å of that of Lys-187 in cyt f. Therefore, these residues form stable salt bridges. In addition, several other acidic patch residues on Pc are in close contact with cyt f; thus, according to these structures, Glu-59 and Ala-53, as well as Glu-43, are expected to be significantly affected by irradiation in the TCS experiments (see Supplemental Figure 9 online). Similar methods were used to construct another model of the electron transfer complex between Pc and cyt f from different species (Lange et al., 2005). These models also have the acidic patch residues in close contact with cyt f.
However, the residues in the acidic patch of Pc were not significantly affected by irradiation in our TCS experiments (Figures 2 and 4), revealing that the major states of the Pc-cyt b6f complexes lack stable salt bridges. The discrepancy may be due to the insufficient constraints in the construction of the Pc-cyt f model, especially in the acidic patch and/or the overestimation of the electrostatic interaction in the model building because the acidic patch residues are >20 Å away from the heme in cyt f, and few pseudocontact shifts were observed for the acidic patch residues (Ubbink et al., 1998; Lange et al., 2005).
Our TCS experiments revealed that the residues in loop 5, such as Asn-64, are close to PSI. Asn-64, which was not investigated in previous mutational studies, is conserved in vascular plants and green alga (M. braunii) but not in cyanobacteria (Anabaena variabilis and Synechocystis sp PCC 6803). Although the Pcs from cyanobacteria, green algae, and spinach share similar main chain structures, the Pcs from cyanobacteria are likely to have lower affinity for spinach PSI than the Pcs from spinach and green alga (Hervás et al., 1995). Hippler et al. (1999) demonstrated that the lack of basic residues in PsaF from cyanobacteria causes the lower affinity. The lack of conservation of Asn-64 in cyanobacteria would additionally cause a difference in the affinity.
In this study, we directly demonstrated that, in the Pc–PSI and Pc–cyt b6f interactions, electrostatic interactions are used to form a transient and loose contact of Pc with PSI and cyt b6f, and then hydrophobic interactions are used to form electron transfer complexes of Pc with PSI and cyt b6f (Figure 7). This binding mode explains the rapid dissociation of the complexes (Finazzi et al., 2005; Busch and Hippler, 2011) and the pseudospecificity of the acidic patches in fern and spinach Pcs (Sato et al., 2004). Rapid binding facilitated by transient electrostatic interactions has also been proposed in various systems (Schreiber and Fersht, 1996; Selzer et al., 2000; Miyashita et al., 2004; Alsallaq and Zhou, 2008; Schreiber et al., 2009), and evidence is accumulating for the formation of transient complexes, based on NMR studies using paramagnetic relaxation enhancement (Iwahara and Clore, 2006; Tang et al., 2006; Volkov et al., 2006; Clore et al., 2007; Bashir et al., 2011; Clore, 2011). Although the introduction of paramagnetic probes into specific sites of PSI and cyt b6f is a challenging task, paramagnetic relaxation enhancement analyses would provide further information about the encounter complex formation in the Pc-PSI and Pc-cyt b6f complexes.
The cyclic electron transport pathway, in which electrons in ferredoxin, an electron carrier protein, are transferred to plastoquinone, embedded in thylakoid membranes, functions in photosynthesis and photoprotection (Munekage et al., 2004; Shikanai, 2007). Although the mechanism of the cyclic electron transport remains largely unknown, subunits of cyt b6f and NAD(P)H dehydrogenase have been recently identified as candidates to be the primary electron acceptors from ferredoxin (Yamamoto et al., 2011). Application of the TCS method, along with genetic and biochemical approaches, to the ferredoxin–cyt b6f and ferredoxin–NAD(P)H dehydrogenase interactions would provide insight into the binding mode.
METHODS
Sample Preparation
Thylakoid membrane vesicles were prepared from the whole spinach (Spinacia oleracea) thylakoid membrane, according to the previously published procedure (Andersson, 1986; Zhang et al., 2001), with the substitution of the Yeda press step with a sonication step. Solubilized PSI and cyt b6f were prepared from market spinach as reported previously, with the substitution of the propyl-agarose chromatography with Macro-prep Methyl chromatography (Bio-Rad) (Zhang and Cramer, 2004; Amunts et al., 2005). The concentration of cyt b6f was determined from the absorbance difference between the reduced and oxidized proteins at 554 minus 543.3 nm using the difference extinction coefficient of 28 mM−1 cm−1 (Andersson, 1986; Metzger et al., 1997). PSI was quantified by flash photolysis (vide infra) using 2 μM dichlorophenolindophenol with εΔA700 = 64,000 (Hiyama and Ke, 1972; Metzger et al., 1997).
Spinach Pc and Monoraphidium braunii cyt c6 were expressed in Escherichia coli, and the purification was accomplished according to the published procedures (Hervás et al., 1992; Ejdebäck et al., 1997) with minor modifications. Double-stranded DNA encoding a signal peptide gene, the spinach Pc gene, and a transcription termination region (Ejdebäck et al., 1997) was prepared from ten oligonucleotides (see Supplemental Table 3 online) (Dillon and Rosen, 1990) and was amplified by PCR with following primers containing NdeI and HindIII recognition sites: 5′-CGAGTTTACGTAGCATATGGAATTCC-3′ and 5′-GCCGGGCTAAGCTTAAAA-3′. The double-stranded DNA was transferred into the pET43a vector (Novagen) using the NdeI and HindIII restriction sites. Mutagenesis of Pc was performed using a QuikChange site-directed mutagenesis kit (Stratagene). In all of the constructs, the Pc coding regions were completely sequenced using the T7 promoter primer for verification. The E. coli strain Tuner (DE3) (Novagen) was transformed with the expression constructs. For the unlabeled Pc, the transformed cells were grown in Luria-Bertani broth at 37°C in a shaker operating at 106 rpm. Uniformly 13C/15N-labeled Pc was prepared by growing cells in M9 medium containing 15NH4Cl (1 g/liter) and [U-13C] Glc (2 g/liter), supplemented with copper citrate (100 μM) and Celtone-CN powder (1 g/liter; Martek), at 37°C, in a fermenter with shaking at 350 rpm. The uniformly 2H/15N-labeled Pc was prepared using M9 medium in 99.9% deuterium oxide (2H2O) containing 15NH4Cl (1 g/liter) and [U-2H] Glc (98%; 2 g/liter) supplemented with copper citrate (100 μM) and Celtone-DN powder (1 g/liter), at 37°C, in a fermenter with shaking at 350 rpm. Protein production was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 30 μM when the culture reached an OD600 of 1.0. The cells were cultivated at 37°C for 5 h after induction. Purification of Pc was accomplished according to the published procedure (Ejdebäck et al., 1997), with the substitution of Sephacryl S-100 HR chromatography with Hiload 26/60 Superdex 75 prep grade chromatography (GE Healthcare Bio-Sciences). Double-stranded DNA containing a signal peptide gene and the M. braunii cyt c6 gene (constructed based on its amino acid sequence) was prepared from eight oligonucleotides (see Supplemental Table 3 online) (Dillon and Rosen, 1990) and was amplified by PCR with the following 5′ and 3′ primers: 5′-GACGACGACAATCATATG-3′ and 5′-GTGTCCGGGCTTCTCCTC-3′, respectively. The double-stranded DNA was transferred into the pET43a vector, with a modification of the nucleotides around the PshAI site from 5′-GACAAGAGTCCGGGAGC-3′ to 5′-GACAAGTGTCCGGGCTTCTCCTCAACGATATCTGAGC-3′, using T4 DNA polymerase and T4 DNA ligase. The gene encoding the NUS tag was removed by NdeI digestion and subsequent ligation. The cyt c6 coding regions were completely sequenced, using the T7 promoter primer, for verification. The E. coli strain Tuner (DE3) (Novagen) was transformed with the expression constructs and the pEC86 plasmid (Arslan et al., 1998). The transformed cells were grown in Luria-Bertani broth at 37°C, with shaking at 106 rpm. Protein production was induced by the addition of isopropyl β-d-thiogalactopyranoside to a final concentration of 30 μM, when the culture reached an OD600 of 1.0. The cells were cultivated at 37°C for 5 h after induction. The periplasmic fraction was obtained according to the same procedure used for Pc and was purified by ammonium sulfate precipitation and DEAE-cellulose column chromatography, as previously reported (Hervás et al., 1992). cyt c6 was further purified by Hiload 26/60 Superdex 75 prep grade chromatography and Resource Q chromatography (GE Healthcare Bio-Sciences).
The Cd-Pc was prepared according to the previously published procedure (Ubbink et al., 1996) and was purified by Hiload 26/60 Superdex 75 prep grade chromatography and Resource Q chromatography.
Electron Transfer Activity Assay
From 1 to 10 pmol of Pc and 100 pmol of PSI were dissolved in 500 μL of 50 mM Tricine-NaOH, pH 7.4, 40 mM KCl, 5 mM MgCl2, 0.05% n-dodecyl-β-d-thiomaltoside (DTM; Anatrace), 1.25 mM ascorbic acid, 0.5 mM methyl viologen, and 10 μM 3-(3′,4′-dichlorophenyl)-1,1-dimethylurea. PSI was activated by a camera flash at room temperature (∼298K), and the time course of the absorbance at 701 nm was recorded by a Shimadzu UV-2400 PC spectrometer. The time course was fitted to a two-exponential decay, using the Origin 5.0 software (OriginLab), and only the fast kinetic component was used for the determination of the reaction rate.
The electron transport activities of the obtained solubilized cyt b6f micelles and inside-out vesicles were examined according to the previously reported method (Pierre et al., 1995). Solubilized cyt b6f (0.5 pmol) or inside-out vesicles containing 1 pmol of cyt b6f were dissolved in 500 μL of 20 mM Tricine-NaOH, pH 8.0, 0.3 mM DTM, and 5 μM oxidized Pc. The reaction was initiated by adding 15 μM decylplastoquinol, and the reduction of Pc was monitored by the absorbance change at 600 nm using a Shimadzu UV-2400 PC spectrometer.
The electron transport activities of the wild type and mutant Pcs for the solubilized cyt b6f were compared by stopped-flow experiments (Illerhaus et al., 2000). Fifteen nanomoles of potassium ferricyanide and ascorbic acid were added to 1 to 50 nmol of Pc and 1.5 nmol of cyt b6f, respectively, and both samples were passed over short desalting columns (PD10; GE Healthcare Bio-Sciences). The Pc and cyt b6f were dissolved in 5 mL of 20 mM KPi, pH 6.5, 100 mM KCl, and 0.05% n-undecyl-β-d-maltopyranoside (UDM). Kinetic measurements were performed at 298K with a stopped-flow apparatus (SX. 18MV-R; Applied Photophysics) equipped with a diode array photometer. The absorbance difference of 421.2 minus 410.5 nm was extracted from the spectra as a function of time. The time course was fitted to a two-exponential decay using Origin 5.0 software (OriginLab), and only the fast kinetic component was used for the determination of the reaction rate.
NMR Spectroscopy
Sequential assignments of the backbone resonances of the spinach Pc and Cd-Pc were achieved by HNCACB and CBCA(CO)NH experiments, performed at 303K on Bruker Avance 500 or 600 spectrometers equipped with a cryogenic probe. For the analyses, 0.5 mM uniformly [13C,15N]-labeled Pc or 0.15 mM uniformly [13C,15N]-labeled Cd-Pc was dissolved in 20 mM phosphate buffer, pH 6.5, containing 2 mM ascorbic acid, 40 mM KCl, and 90% water/10% 2H2O. All spectra were processed by XWIN-NMR version 3.5 or Topspin version 2.1 and analyzed by SPARKY (T.D. Goddard and D.G. Kneller, University of California, San Francisco). All resonances from the backbone atoms and the side chain amide groups of Gln and Asn residues were assigned, except for Phe-35, with a reported amide proton chemical shift of amide proton of 5.5 ppm (Musiani et al., 2005).
Double quantum filtered correlation spectroscopy spectra for the characterization of Pc mutants were recorded on a Bruker Avance 400 spectrometer at 303K. A total of 500 μM of nonlabeled Pcs were dissolved in 20 mM potassium phosphate buffer, pH 6.5, containing 2 mM ascorbic acid, 1 mM 2,2-dimethyl-2-silapentane-5-sulfonate (DSS), and 90% water/10% 2H2O.
1H-15N heteronuclear single quantum coherence spectra for the characterization of Cd-Pc were recorded on a Bruker Avance 400 spectrometer at 303K. For the comparison of the chemical shifts of Pc and Cd-Pc, the Pcs were dissolved in 10 mM NaPi, pH 6.0, and 90% water/10% 2H2O. In the investigation of the PSI binding activity of Cd-Pc, 2.7 μM solubilized PSI and 19 μM cyt c6 were sequentially combined with 140 μM [μL-15N]Cd-Pc in 20 mM Bis-Tris, pH 6.5, 90% water/10% 2H2O, and 0.05% DTM.
The transverse relaxation-optimized spectroscopy for rotational correlation times (TRACT), pulsed field gradient (PFG), and TCS experiments were performed on a Bruker Avance 800 spectrometer equipped with a cryogenic probe. In the TRACT and PFG experiments, 10 μM solubilized PSI or 20 μM solubilized cyt b6f was combined with 100 μM 2H,15N-labeled Pc, in 20 mM potassium phosphate buffer, pH 6.5, containing 2 mM ascorbic acid, 1 mM DSS, and 20% water/80% 2H2O. The experiments with free Pc were also performed under the same conditions as those mentioned above, but in a buffer containing 0.05% DTM, 0.05% UDM, 30% RediGrad, and 50 μM EDTA. To remove the water in the RediGrad, the RediGrad was lyophilized and dissolved in 2H2O at 343K. The free Pc experiments were performed at both 283 and 303K, and the other experiments were performed at 303K. The one-dimensional TRACT experiments were performed using the reported pulse scheme (Lee et al., 2006). The relaxation periods (Δi) were set to 0.1, 22.2, 44.4, and 66.7 ms for the α-spin state and 0.1, 11.1, 16.7, and 33.3 ms for the β-spin state. The PFG experiments were performed using the reported pulse scheme (Nesmelova et al., 2004). The time delay for diffusion (Δ) was set to 75 ms, and the amplitude (g) and duration (δ) of the gradients for the diffusion measurements were set to 45 G/cm and 0.5 to 4.5 ms, respectively. In the TCS experiments, the inside-out vesicles with 5 μM PSI and 3.2 μM cyt b6f, or 9 μM solubilized PSI or 12 μM solubilized cyt b6f, or right-side-out vesicles with 5 μM PSI and 2.7 μM cyt b6f, were combined with 100 μM 2H,15N-labeled Pc in 20 mM potassium phosphate buffer, pH 6.5, containing 2 mM ascorbic acid, 1 mM DSS, and 20% water/80% 2H2O. For the experiments using solubilized PSI and cyt b6f, DTM and UDM were each added at a concentration of 0.05%, respectively. For the experiments using inside-out and right-side-out vesicles, RediGrad (GE Healthcare Bio-Sciences) was added at concentrations of 30 and 22.5%, respectively, and EDTA was added at a concentration of 50 μM. The control TCS experiments with additives were also performed under the same conditions as those mentioned above, but with buffer containing 0.05% DTM, 0.05% UDM, 30% RediGrad, and 50 μM EDTA. In the TCS experiments with Cd-Pc and solubilized PSI, 11 μM solubilized PSI was combined with 140 μM 2H,15N-labeled Cd-Pc under 20 mM potassium phosphate buffer, pH 6.5, containing 2 mM ascorbic acid, 1 mM DSS, 0.05% DTM, and 20% water/80% 2H2O. The experiments with solubilized cyt b6f and those with additives were performed at 283K, where the saturation effects were enhanced by the reduction of the Brownian motion. The other TCS experiments were performed at 303K. The TCS experiments were performed using the reported pulse scheme (Takahashi et al., 2000), with a minor modification from the 3-9-19 selective pulse to soft-90° hard-180° soft-90° pulse elements. The saturation for the aliphatic protons of PSI and cyt b6f was accomplished using the WURST-2 decoupling scheme. The saturation frequency was set at 1.5 ppm, and the maximum radiofrequency amplitude was 0.17 kHz for WURST-2 (adiabatic factor Q0 = 1). The saturation times were set to 0.5 s for the vesicles (Figures 2; see Supplemental Figure 5D online), 1.0 s for solubilized cyt b6f (Figure 4), and 1.5 s for solubilized PSI (Figure 3) and the control experiments with additives (see Supplemental Figure 5B online). The total relaxation delay (saturation time + relaxation delay) was set to 6.0 s.
Accession Numbers
Sequence and structure data from this article can be found in the GenBank/EMBL, UniProt, or Protein Data Bank (PDB) databases under the following accession numbers: spinach Pc nucleotide sequence (X04693.1), M. braunii cyt c6 amino acid sequence (Q09099.1), structure of spinach Pc (1YLB), structure of fern (Dryopteris crassirhizoma) Pc (1KDI), structure of pea (Pisum sativum) PSI (PDB: 2O01), structure of turnip (Brassica rapa) cyt f (PDB: 1CTM), and structure of spinach Pc-turnip cyt f complex: 2PCF.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Distribution of the Hydrophobic and Acidic Patches in Spinach Pc.
Supplemental Figure 2. Electron Transport Activities of the Obtained Thylakoid Vesicles and Solubilized PSI and cyt b6f.
Supplemental Figure 3. 1H-15N Shift Correlation Spectra Observed at 18.8 T for Excess Amounts of Uniformly [2H,15N]-Labeled Pc Relative to the PSI and cyt b6f Embedded in Thylakoid Vesicles, without and with Irradiation.
Supplemental Figure 4. Plots of the Reduction Ratios of the Signal Intensities Originating from the Amide Groups, with and without Presaturation, in the TCS Experiments with an Excess Amount of Pc Relative to the Solubilized PSI or cyt b6f.
Supplemental Figure 5. Investigation of the Effects of Residual Protons and Nonspecific Binding.
Supplemental Figure 6. Characterization and TCS Experiments of Cd-Pc.
Supplemental Figure 7. Plots of the Reduction Ratios of the Signal Intensities Originating from the Amide Groups, with and without Presaturation, in the TCS Experiments with an Excess Amount of Cd-Pc Relative to the Solubilized PSI.
Supplemental Figure 8. Distribution of the Acidic Residues in Spinach and Fern Pcs.
Supplemental Figure 9. Mapping of the Residues of Pc Close to cyt f in the Previously Proposed Structure of the Electron Transfer Complex between Pc and cyt f.
Supplemental Table 1. Amide 15N Transverse Cross-Correlated Relaxation Rate, ηxy, and Apparent Rotational Correlation Time, τcobs, of Uniformly [2H,15N]-Labeled Pc with Substoichiometric Amounts of PSI and cyt b6f Determined by TRACT Experiments, and the Diffusion Coefficient of DSS in the Sample, Ddss.
Supplemental Table 2. Sum(r) of the Salt Bridges in Various Protein-Protein Complex Structures.
Supplemental Table 3. Primers for the Construction of Spinach Pc and Monoraphidium braunii Cytochrome c6.
Supplemental Discussion 1. Analyses of the Intensity Reduction of the Resonances from Free Pc upon the Addition of PSI and/or cyt b6f and Statistical Analyses of Salt Bridge Structures.
Supplementary Material
Acknowledgments
We thank Hideo Takahashi, Masanori Osawa, Masayoshi Sakakura, and Noritaka Nishida for their helpful advice. This work was supported by grants from the Japan New Energy and Industrial Technology Development Organization and the Ministry of Economy, Trade, and Industry.
AUTHOR CONTRIBUTIONS
T.U. designed the research, performed the research, analyzed data, and wrote the article. N.N. performed the research, analyzed data, and wrote the article. M.K. performed the research and analyzed data. H.O. and Y.O. performed the research. M.M. contributed new computational tools. P.S. performed the research and analyzed data. K.S. contributed new tools. H.T. designed the research, analyzed data, and wrote the article. I.S. supervised the overall project, designed the research, analyzed data, and wrote the article.
Glossary
- PSI
photosystem I
- Pc
plastocyanin
- TCS
transferred cross-saturation
- Cd-Pc
cadmium-substituted Pc
- DTM
n-dodecyl-β-d-thiomaltoside
- UDM
n-undecyl-β-d-maltopyranoside
- D2O
to be defined
- DSS
2,2-dimethyl-2-silapentane-5-sulfonate
- TRACT
transverse relaxation-optimized spectroscopy for rotational correlation times
- PFG
pulsed field gradient
References
- Alsallaq R., Zhou H.X. (2008). Electrostatic rate enhancement and transient complex of protein-protein association. Proteins 71: 320–335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amunts A., Ben-Shem A., Nelson N. (2005). Solving the structure of plant photosystem I—Biochemistry is vital. Photochem. Photobiol. Sci. 4: 1011–1015 [DOI] [PubMed] [Google Scholar]
- Amunts A., Drory O., Nelson N. (2007). The structure of a plant photosystem I supercomplex at 3.4 A resolution. Nature 447: 58–63 [DOI] [PubMed] [Google Scholar]
- Amunts A., Toporik H., Borovikova A., Nelson N. (2010). Structure determination and improved model of plant photosystem I. J. Biol. Chem. 285: 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson B. (1986). Characterization of the thylakoid membrane by subfractionation analyses. Methods Enzymol. 118: 325–338 [Google Scholar]
- Andersson B., Akerlund H.E. (1978). Inside-out membrane vesicles isolated from spinach thylakoids. Biochim. Biophys. Acta 503: 462–472 [DOI] [PubMed] [Google Scholar]
- Arslan E., Schulz H., Zufferey R., Künzler P., Thöny-Meyer L. (1998). Overproduction of the Bradyrhizobium japonicum c-type cytochrome subunits of the cbb3 oxidase in Escherichia coli. Biochem. Biophys. Res. Commun. 251: 744–747 [DOI] [PubMed] [Google Scholar]
- Baniulis D., Yamashita E., Whitelegge J.P., Zatsman A.I., Hendrich M.P., Hasan S.S., Ryan C.M., Cramer W.A. (2009). Structure-function, stability, and chemical modification of the cyanobacterial cytochrome b6f complex from Nostoc sp. PCC 7120. J. Biol. Chem. 284: 9861–9869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir Q., Scanu S., Ubbink M. (2011). Dynamics in electron transfer protein complexes. FEBS J. 278: 1391–1400 [DOI] [PubMed] [Google Scholar]
- Ben-Shem A., Frolow F., Nelson N. (2003). Crystal structure of plant photosystem I. Nature 426: 630–635 [DOI] [PubMed] [Google Scholar]
- Bergkvist A., Ejdebäck M., Ubbink M., Karlsson B.G. (2001). Surface interactions in the complex between cytochrome f and the E43Q/D44N and E59K/E60Q plastocyanin double mutants as determined by (1)H-NMR chemical shift analysis. Protein Sci. 10: 2623–2626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A., Hippler M. (2011). The structure and function of eukaryotic photosystem I. Biochim. Biophys. Acta 1807: 864–877 [DOI] [PubMed] [Google Scholar]
- Chida H., et al. (2007). Expression of the algal cytochrome c6 gene in Arabidopsis enhances photosynthesis and growth. Plant Cell Physiol. 48: 948–957 [DOI] [PubMed] [Google Scholar]
- Clore G.M. (2011). Exploring sparsely populated states of macromolecules by diamagnetic and paramagnetic NMR relaxation. Protein Sci. 20: 229–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore G.M., Tang C., Iwahara J. (2007). Elucidating transient macromolecular interactions using paramagnetic relaxation enhancement. Curr. Opin. Struct. Biol. 17: 603–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon P.J., Rosen C.A. (1990). A rapid method for the construction of synthetic genes using the polymerase chain reaction. Biotechniques 9: 300. [PubMed] [Google Scholar]
- Drepper F., Hippler M., Nitschke W., Haehnel W. (1996). Binding dynamics and electron transfer between plastocyanin and photosystem I. Biochemistry 35: 1282–1295 [DOI] [PubMed] [Google Scholar]
- Driscoll P.C., Hill H.A., Redfield C. (1987). 1H-NMR sequential assignments and cation-binding studies of spinach plastocyanin. Eur. J. Biochem. 170: 279–292 [DOI] [PubMed] [Google Scholar]
- Ejdebäck M., Young S., Samuelsson A., Karlsson B.G. (1997). Effects of codon usage and vector-host combinations on the expression of spinach plastocyanin in Escherichia coli. Protein Expr. Purif. 11: 17–25 [DOI] [PubMed] [Google Scholar]
- Farkas D., Hansson O. (2011). An NMR study elucidating the binding of Mg(II) and Mn(II) to spinach plastocyanin. Regulation of the binding of plastocyanin to subunit PsaF of photosystem I. Biochim. Biophys. Acta 1807: 1539–1548 [DOI] [PubMed] [Google Scholar]
- Finazzi G., Sommer F., Hippler M. (2005). Release of oxidized plastocyanin from photosystem I limits electron transfer between photosystem I and cytochrome b6f complex in vivo. Proc. Natl. Acad. Sci. USA 102: 7031–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong X.S., Wen J.Q., Fisher N.E., Young S., Howe C.J., Bendall D.S., Gray J.C. (2000b). The role of individual lysine residues in the basic patch on turnip cytochrome f for electrostatic interactions with plastocyanin in vitro. Eur. J. Biochem. 267: 3461–3468 [DOI] [PubMed] [Google Scholar]
- Gong X.S., Wen J.Q., Gray J.C. (2000a). The role of amino-acid residues in the hydrophobic patch surrounding the haem group of cytochrome f in the interaction with plastocyanin. Eur. J. Biochem. 267: 1732–1742 [DOI] [PubMed] [Google Scholar]
- Gounaris K., Barber J., Harwood J.L. (1986). The thylakoid membranes of higher plant chloroplasts. Biochem. J. 237: 313–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haehnel W., Jansen T., Gause K., Klösgen R.B., Stahl B., Michl D., Huvermann B., Karas M., Herrmann R.G. (1994). Electron transfer from plastocyanin to photosystem I. EMBO J. 13: 1028–1038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haehnel W., Pröpper A., Krause H. (1980). Evidence for complexed plastocyanin as the immediate electron donor of P-700. Biochim. Biophys. Acta 593: 384–399 [DOI] [PubMed] [Google Scholar]
- Haehnel W., Ratajczak R., Robenek H. (1989). Lateral distribution and diffusion of plastocyanin in chloroplast thylakoids. J. Cell Biol. 108: 1397–1405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervás M., De la Rosa M.A., Tollin G. (1992). A comparative laser-flash absorption spectroscopy study of algal plastocyanin and cytochrome c552 photooxidation by photosystem I particles from spinach. Eur. J. Biochem. 203: 115–120 [DOI] [PubMed] [Google Scholar]
- Hervás M., Navarro J.A., Díaz A., Bottin H., De la Rosa M.A. (1995). Laser-flash kinetic analysis of the fast electron transfer from plastocyanin and cytochrome c6 to photosystem I. Experimental evidence on the evolution of the reaction mechanism. Biochemistry 34: 11321–11326 [DOI] [PubMed] [Google Scholar]
- Hippler M., Drepper F., Farah J., Rochaix J.D. (1997). Fast electron transfer from cytochrome c6 and plastocyanin to photosystem I of Chlamydomonas reinhardtii requires PsaF. Biochemistry 36: 6343–6349 [DOI] [PubMed] [Google Scholar]
- Hippler M., Drepper F., Haehnel W., Rochaix J.D. (1998). The N-terminal domain of PsaF: Precise recognition site for binding and fast electron transfer from cytochrome c6 and plastocyanin to photosystem I of Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 95: 7339–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippler M., Drepper F., Rochaix J.D., Mühlenhoff U. (1999). Insertion of the N-terminal part of PsaF from Chlamydomonas reinhardtii into photosystem I from Synechococcus elongatus enables efficient binding of algal plastocyanin and cytochrome c6. J. Biol. Chem. 274: 4180–4188 [DOI] [PubMed] [Google Scholar]
- Hippler M., Ratajczak R., Haehnel W. (1989). Identification of plastocyanin-binding subunit of photosystem-I. FEBS Lett. 250: 280–284 [Google Scholar]
- Hippler M., Reichert J., Sutter M., Zak E., Altschmied L., Schröer U., Herrmann R.G., Haehnel W. (1996). The plastocyanin binding domain of photosystem I. EMBO J. 15: 6374–6384 [PMC free article] [PubMed] [Google Scholar]
- Hiyama T. (2004). Isolation of photosystem I particles from spinach. Methods Mol. Biol. 274: 11–17 [DOI] [PubMed] [Google Scholar]
- Hiyama T., Ke B. (1972). Difference spectra and extinction coefficients of P 700. Biochim. Biophys. Acta 267: 160–171 [DOI] [PubMed] [Google Scholar]
- Hope A.B. (2000). Electron transfers amongst cytochrome f, plastocyanin and photosystem I: Kinetics and mechanisms. Biochim. Biophys. Acta 1456: 5–26 [DOI] [PubMed] [Google Scholar]
- Ichikawa O., Osawa M., Nishida N., Goshima N., Nomura N., Shimada I. (2007). Structural basis of the collagen-binding mode of discoidin domain receptor 2. EMBO J. 26: 4168–4176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illerhaus J., Altschmied L., Reichert J., Zak E., Herrmann R.G., Haehnel W. (2000). Dynamic interaction of plastocyanin with the cytochrome bf complex. J. Biol. Chem. 275: 17590–17595 [DOI] [PubMed] [Google Scholar]
- Impagliazzo A., Ubbink M. (2004). Mapping of the binding site on pseudoazurin in the transient 152 kDa complex with nitrite reductase. J. Am. Chem. Soc. 126: 5658–5659 [DOI] [PubMed] [Google Scholar]
- Iwahara J., Clore G.M. (2006). Detecting transient intermediates in macromolecular binding by paramagnetic NMR. Nature 440: 1227–1230 [DOI] [PubMed] [Google Scholar]
- Jolley C., Ben-Shem A., Nelson N., Fromme P. (2005). Structure of plant photosystem I revealed by theoretical modeling. J. Biol. Chem. 280: 33627–33636 [DOI] [PubMed] [Google Scholar]
- Jordan P., Fromme P., Witt H.T., Klukas O., Saenger W., Krauss N. (2001). Three-dimensional structure of cyanobacterial photosystem I at 2.5 A resolution. Nature 411: 909–917 [DOI] [PubMed] [Google Scholar]
- Kannt A., Young S., Bendall D.S. (1996). The role of acidic residues of plastocyanin in its interaction with cytochrome f. Biochim. Biophys. Acta 1277: 115–126 [PubMed] [Google Scholar]
- Kofuku Y., Yoshiura C., Ueda T., Terasawa H., Hirai T., Tominaga S., Hirose M., Maeda Y., Takahashi H., Terashima Y., Matsushima K., Shimada I. (2009). Structural basis of the interaction between chemokine stromal cell-derived factor-1/CXCL12 and its G-protein-coupled receptor CXCR4. J. Biol. Chem. 284: 35240–35250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nussinov R. (2002a). Relationship between ion pair geometries and electrostatic strengths in proteins. Biophys. J. 83: 1595–1612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S., Nussinov R. (2002b). Close-range electrostatic interactions in proteins. ChemBioChem 3: 604–617 [DOI] [PubMed] [Google Scholar]
- Kurisu G., Zhang H., Smith J.L., Cramer W.A. (2003). Structure of the cytochrome b6f complex of oxygenic photosynthesis: Tuning the cavity. Science 302: 1009–1014 [DOI] [PubMed] [Google Scholar]
- Lange C., Cornvik T., Díaz-Moreno I., Ubbink M. (2005). The transient complex of poplar plastocyanin with cytochrome f: Effects of ionic strength and pH. Biochim. Biophys. Acta 1707: 179–188 [DOI] [PubMed] [Google Scholar]
- Lee B.H., Hibino T., Takabe T., Weisbeek P.J., Takabe T. (1995). Site-directed mutagenetic study on the role of negative patches on silene plastocyanin in the interactions with cytochrome f and photosystem I. J. Biochem. 117: 1209–1217 [DOI] [PubMed] [Google Scholar]
- Lee D., Hilty C., Wider G., Wüthrich K. (2006). Effective rotational correlation times of proteins from NMR relaxation interference. J. Magn. Reson. 178: 72–76 [DOI] [PubMed] [Google Scholar]
- Malia T.J., Wagner G. (2007). NMR structural investigation of the mitochondrial outer membrane protein VDAC and its interaction with antiapoptotic Bcl-xL. Biochemistry 46: 514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto M., Ueda T., Shimada I. (2010). Theoretical analyses of the transferred cross-saturation method. J. Magn. Reson. 205: 114–124 [DOI] [PubMed] [Google Scholar]
- McCammon J.A., Harvey S.C. (1987). Dynamics of Proteins and Nucleic Acids. (Cambridge, UK: Cambridge University Press).
- Metzger S.U., Cramer W.A., Whitmarsh J. (1997). Critical analysis of the extinction coefficient of chloroplast cytochrome f. Biochim. Biophys. Acta 1319: 233–241 [DOI] [PubMed] [Google Scholar]
- Miyashita O., Onuchic J.N., Okamura M.Y. (2004). Transition state and encounter complex for fast association of cytochrome c2 with bacterial reaction center. Proc. Natl. Acad. Sci. USA 101: 16174–16179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modi S., He S.P., Gray J.C., Bendall D.S. (1992a). The role of surface-exposed Tyr-83 of plastocyanin in electron-transfer from cytochrome-c. Biochim. Biophys. Acta 1101: 64–68 [Google Scholar]
- Modi S., Nordling M., Lundberg L.G., Hansson O., Bendall D.S. (1992b). Reactivity of cytochromes c and f with mutant forms of spinach plastocyanin. Biochim. Biophys. Acta 1102: 85–90 [DOI] [PubMed] [Google Scholar]
- Munekage Y., Hashimoto M., Miyake C., Tomizawa K., Endo T., Tasaka M., Shikanai T. (2004). Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582 [DOI] [PubMed] [Google Scholar]
- Musiani F., Dikiy A., Semenov A.Y., Ciurli S. (2005). Structure of the intermolecular complex between plastocyanin and cytochrome f from spinach. J. Biol. Chem. 280: 18833–18841 [DOI] [PubMed] [Google Scholar]
- Nakanishi T., Miyazawa M., Sakakura M., Terasawa H., Takahashi H., Shimada I. (2002). Determination of the interface of a large protein complex by transferred cross-saturation measurements. J. Mol. Biol. 318: 245–249 [DOI] [PubMed] [Google Scholar]
- Nesmelova I.V., Idiyatullin D., Mayo K.H. (2004). Measuring protein self-diffusion in protein-protein mixtures using a pulsed gradient spin-echo technique with WATERGATE and isotope filtering. J. Magn. Reson. 166: 129–133 [DOI] [PubMed] [Google Scholar]
- Nishida N., Sumikawa H., Sakakura M., Shimba N., Takahashi H., Terasawa H., Suzuki E.I., Shimada I. (2003). Collagen-binding mode of vWF-A3 domain determined by a transferred cross-saturation experiment. Nat. Struct. Biol. 10: 53–58 [DOI] [PubMed] [Google Scholar]
- Pertoft H. (2000). Fractionation of cells and subcellular particles with Percoll. J. Biochem. Biophys. Methods 44: 1–30 [DOI] [PubMed] [Google Scholar]
- Pierre Y., Breyton C., Kramer D., Popot J.L. (1995). Purification and characterization of the cytochrome b6 f complex from Chlamydomonas reinhardtii. J. Biol. Chem. 270: 29342–29349 [DOI] [PubMed] [Google Scholar]
- Sato K., Kohzuma T., Dennison C. (2004). Pseudospecificity of the acidic patch of plastocyanin for the interaction with cytochrome f. J. Am. Chem. Soc. 126: 3028–3029 [DOI] [PubMed] [Google Scholar]
- Schreiber G., Fersht A.R. (1996). Rapid, electrostatically assisted association of proteins. Nat. Struct. Biol. 3: 427–431 [DOI] [PubMed] [Google Scholar]
- Schreiber G., Haran G., Zhou H.X. (2009). Fundamental aspects of protein-protein association kinetics. Chem. Rev. 109: 839–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selzer T., Albeck S., Schreiber G. (2000). Rational design of faster associating and tighter binding protein complexes. Nat. Struct. Biol. 7: 537–541 [DOI] [PubMed] [Google Scholar]
- Shikanai T. (2007). Cyclic electron transport around photosystem I: Genetic approaches. Annu. Rev. Plant Biol. 58: 199–217 [DOI] [PubMed] [Google Scholar]
- Shimada I. (2005). NMR techniques for identifying the interface of a larger protein-protein complex: Cross-saturation and transferred cross-saturation experiments. Methods Enzymol. 394: 483–506 [DOI] [PubMed] [Google Scholar]
- Shimada I., Ueda T., Matsumoto M., Sakakura M., Osawa M., Takeuchi K., Nishida N., Takahashi H. (2009). Cross-saturation and transferred cross-saturation experiments. Prog. Nucl. Magn. Reson. Spectrosc. 54: 123–140 [Google Scholar]
- Sigfridsson K. (1998). Plastocyanin, an electron-transfer protein. Photosynth. Res. 57: 1–28 [Google Scholar]
- Sigfridsson K., Young S., Hansson O. (1996). Structural dynamics in the plastocyanin-photosystem 1 electron-transfer complex as revealed by mutant studies. Biochemistry 35: 1249–1257 [DOI] [PubMed] [Google Scholar]
- Sigfridsson K., Young S., Hansson O. (1997). Electron transfer between spinach plastocyanin mutants and photosystem 1. Eur. J. Biochem. 245: 805–812 [DOI] [PubMed] [Google Scholar]
- Sommer F., Drepper F., Haehnel W., Hippler M. (2004). The hydrophobic recognition site formed by residues PsaA-Trp651 and PsaB-Trp627 of photosystem I in Chlamydomonas reinhardtii confers distinct selectivity for binding of plastocyanin and cytochrome c6. J. Biol. Chem. 279: 20009–20017 [DOI] [PubMed] [Google Scholar]
- Sommer F., Drepper F., Haehnel W., Hippler M. (2006). Identification of precise electrostatic recognition sites between cytochrome c6 and the photosystem I subunit PsaF using mass spectrometry. J. Biol. Chem. 281: 35097–35103 [DOI] [PubMed] [Google Scholar]
- Sommer F., Drepper F., Hippler M. (2002). The luminal helix l of PsaB is essential for recognition of plastocyanin or cytochrome c6 and fast electron transfer to photosystem I in Chlamydomonas reinhardtii. J. Biol. Chem. 277: 6573–6581 [DOI] [PubMed] [Google Scholar]
- Soriano G.M., Ponamarev M.V., Piskorowski R.A., Cramer W.A. (1998). Identification of the basic residues of cytochrome f responsible for electrostatic docking interactions with plastocyanin in vitro: Relevance to the electron transfer reaction in vivo. Biochemistry 37: 15120–15128 [DOI] [PubMed] [Google Scholar]
- Soriano G.M., Ponamarev M.V., Tae G.S., Cramer W.A. (1996). Effect of the interdomain basic region of cytochrome f on its redox reactions in vivo. Biochemistry 35: 14590–14598 [DOI] [PubMed] [Google Scholar]
- Stroebel D., Choquet Y., Popot J.L., Picot D. (2003). An atypical haem in the cytochrome b(6)f complex. Nature 426: 413–418 [DOI] [PubMed] [Google Scholar]
- Sujak A., Drepper F., Haehnel W. (2004). Spectroscopic studies on electron transfer between plastocyanin and cytochrome b6f complex. J. Photochem. Photobiol. B 74: 135–143 [DOI] [PubMed] [Google Scholar]
- Takahashi H., Nakanishi T., Kami K., Arata Y., Shimada I. (2000). A novel NMR method for determining the interfaces of large protein-protein complexes. Nat. Struct. Biol. 7: 220–223 [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Takahashi H., Sugai M., Iwai H., Kohno T., Sekimizu K., Natori S., Shimada I. (2004). Channel-forming membrane permeabilization by an antibacterial protein, sapecin: Determination of membrane-buried and oligomerization surfaces by NMR. J. Biol. Chem. 279: 4981–4987 [DOI] [PubMed] [Google Scholar]
- Takeuchi K., Yokogawa M., Matsuda T., Sugai M., Kawano S., Kohno T., Nakamura H., Takahashi H., Shimada I. (2003). Structural basis of the KcsA K(+) channel and agitoxin2 pore-blocking toxin interaction by using the transferred cross-saturation method. Structure 11: 1381–1392 [DOI] [PubMed] [Google Scholar]
- Tang C., Iwahara J., Clore G.M. (2006). Visualization of transient encounter complexes in protein-protein association. Nature 444: 383–386 [DOI] [PubMed] [Google Scholar]
- Ubbink M., Ejdebäck M., Karlsson B.G., Bendall D.S. (1998). The structure of the complex of plastocyanin and cytochrome f, determined by paramagnetic NMR and restrained rigid-body molecular dynamics. Structure 6: 323–335 [DOI] [PubMed] [Google Scholar]
- Ubbink M., Lian L.Y., Modi S., Evans P.A., Bendall D.S. (1996). Analysis of the 1H-NMR chemical shifts of Cu(I)-, Cu(II)- and Cd-substituted pea plastocyanin. Metal-dependent differences in the hydrogen-bond network around the copper site. Eur. J. Biochem. 242: 132–147 [DOI] [PubMed] [Google Scholar]
- Volkov A.N., Worrall J.A., Holtzmann E., Ubbink M. (2006). Solution structure and dynamics of the complex between cytochrome c and cytochrome c peroxidase determined by paramagnetic NMR. Proc. Natl. Acad. Sci. USA 103: 18945–18950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Okvist M., Hansson O., Young S. (1998). Crystal structure of spinach plastocyanin at 1.7 A resolution. Protein Sci. 7: 2099–2105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto H., Peng L., Fukao Y., Shikanai T. (2011). An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita E., Zhang H., Cramer W.A. (2007). Structure of the cytochrome b6f complex: Quinone analogue inhibitors as ligands of heme cn. J. Mol. Biol. 370: 39–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J., Kurisu G., Cramer W.A. (2006). Intraprotein transfer of the quinone analogue inhibitor 2,5-dibromo-3-methyl-6-isopropyl-p-benzoquinone in the cytochrome b6f complex. Proc. Natl. Acad. Sci. USA 103: 69–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa M., Osawa M., Takeuchi K., Mase Y., Shimada I. (2011). NMR analyses of the Gbetagamma binding and conformational rearrangements of the cytoplasmic pore of G protein-activated inwardly rectifying potassium channel 1 (GIRK1). J. Biol. Chem. 286: 2215–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokogawa M., Takeuchi K., Shimada I. (2005). Bead-linked proteoliposomes: A reconstitution method for NMR analyses of membrane protein-ligand interactions. J. Am. Chem. Soc. 127: 12021–12027 [DOI] [PubMed] [Google Scholar]
- Yoshiura C., Kofuku Y., Ueda T., Mase Y., Yokogawa M., Osawa M., Terashima Y., Matsushima K., Shimada I. (2010). NMR analyses of the interaction between CCR5 and its ligand using functional reconstitution of CCR5 in lipid bilayers. J. Am. Chem. Soc. 132: 6768–6777 [DOI] [PubMed] [Google Scholar]
- Young S., Sigfridsson K., Olesen K., Hansson O. (1997). The involvement of the two acidic patches of spinach plastocyanin in the reaction with photosystem I. Biochim. Biophys. Acta 1322: 106–114 [DOI] [PubMed] [Google Scholar]
- Zhang H., Cramer W.A. (2004). Purification and crystallization of the cytochrome b6f complex in oxygenic photosynthesis. Methods Mol. Biol. 274: 67–78 [DOI] [PubMed] [Google Scholar]
- Zhang H., Whitelegge J.P., Cramer W.A. (2001). Ferredoxin:NADP+ oxidoreductase is a subunit of the chloroplast cytochrome b6f complex. J. Biol. Chem. 276: 38159–38165 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.