This work examines the chloroplast ribonucleoproteins CP31A and CP29A by plastome-wide analysis of chloroplast RNA metabolism, finding that these proteins associate with a large set of chloroplast transcripts and are required for cold stress resistance, targeting multiple chloroplast mRNA processing steps.
Abstract
Chloroplast RNA metabolism is mediated by a multitude of nuclear encoded factors, many of which are highly specific for individual RNA processing events. In addition, a family of chloroplast ribonucleoproteins (cpRNPs) has been suspected to regulate larger sets of chloroplast transcripts. This together with their propensity for posttranslational modifications in response to external cues suggested a potential role of cpRNPs in the signal-dependent coregulation of chloroplast genes. We show here on a transcriptome-wide scale that the Arabidopsis thaliana cpRNPs CP31A and CP29A (for 31 kD and 29 kD chloroplast protein, respectively), associate with large, overlapping sets of chloroplast transcripts. We demonstrate that both proteins are essential for resistance of chloroplast development to cold stress. They are required to guarantee transcript stability of numerous mRNAs at low temperatures and under these conditions also support specific processing steps. Fine mapping of cpRNP–RNA interactions in vivo suggests multiple points of contact between these proteins and their RNA ligands. For CP31A, we demonstrate an essential function in stabilizing sense and antisense transcripts that span the border of the small single copy region and the inverted repeat of the chloroplast genome. CP31A associates with the common 3′-terminus of these RNAs and protects them against 3′-exonucleolytic activity.
INTRODUCTION
The chloroplast genome of land plants contains roughly 80 genes, most of which encode proteins that play roles in photosynthesis or gene expression. Chloroplast mRNAs require the actions of numerous nuclear-encoded factors before they can serve as templates for the translational apparatus. Dozens of factors required for the cleavage, splicing, or editing of specific mRNAs have been described in recent years (Stern et al., 2010; Tillich et al., 2010; Barkan, 2011; Jacobs and Kück, 2011). Not surprisingly, many of these factors are RNA binding proteins (RBPs). For example, RBPs from the pentatricopeptide repeat (PPR) protein family have been shown to support specific RNA processing events in chloroplasts (Jacobs and Kück, 2011). By contrast, the chloroplast ribonucleoproteins (cpRNPs) are suspected to regulate larger sets of chloroplast RNAs.
The cpRNP protein family consists of 10 members in Arabidopsis thaliana (Ohta et al., 1995; Lorković and Barta, 2002; Ruwe et al., 2011). All cpRNPs share a similar domain structure in which an acidic residue–rich domain precedes two RNA recognition motifs (RRMs) separated by a short spacer. Furthermore, all cpRNPs carry an N-terminal signal peptide that is predicted to direct their import into the chloroplast after their translation in the cytosol. Chloroplast localization has been confirmed in vivo for tobacco (Nicotiana tabacum; Nakamura et al., 1999) and Arabidopsis (Raab et al., 2006) cpRNPs. The cpRNPs are abundant (Nakamura et al., 2001) and highly regulated proteins that react to various external and internal signals, particularly light (Ruwe et al., 2011), which controls their expression levels and protein modification states (Li and Sugiura, 1990; Schuster and Gruissem, 1991; Churin et al., 1999; Wang et al., 2006; Kleffmann et al., 2007).
The functional analyses of cpRNPs performed to date have mostly depended on in vitro techniques. The results have implicated cpRNPs in the RNA editing of specific sites (Hirose and Sugiura, 2001), in helping to generate the 3′-ends of several chloroplast mRNAs (Schuster and Gruissem, 1991; Hayes et al., 1996; Nakamura et al., 2001), and in protecting the chloroplast psbA mRNA against degradation (Nakamura et al., 2001). Certain chloroplast mRNAs have been found to coimmunoprecipitate with tobacco cpRNPs (Nakamura et al., 1999). Recently, mutants of Arabidopsis CP31A and its paralog, CP31B, were isolated and analyzed for RNA processing defects on a genome-wide scale (Tillich et al., 2009). The results in cp31a mutants demonstrated that multiple editing sites were only partially edited and the levels of multiple mRNAs were slightly reduced (Tillich et al., 2009). The most severely affected was the ndhF mRNA, which was reduced to the brink of the detection limit in RNA gel blot experiments. NdhF encodes a subunit of the plastid NAD dehydrogenase (NDH), and the NDH complex was shown to be defective in CP31A mutants (Tillich et al., 2009). However, we do not yet fully understand how CP31A guarantees the RNA stability of ndhF.
Here, we examine overlapping sets of RNAs associated with Arabidopsis CP31A and CP29A proteins. Plants devoid of CP31A or CP29A show bleaching of newly emerging tissues at low temperatures. Analysis of RNA ligands under these conditions reveals defects in RNA stabilization, processing, and splicing, as well as loss of chloroplast proteins. Finally, we focus on the severe effects on the ndhF message and show that CP31A is required to protect the ndhF mRNA against 3′-exonucleolytic degradation.
RESULTS
CP29A and CP31A Associate with Largely Overlapping Sets of Chloroplast mRNAs
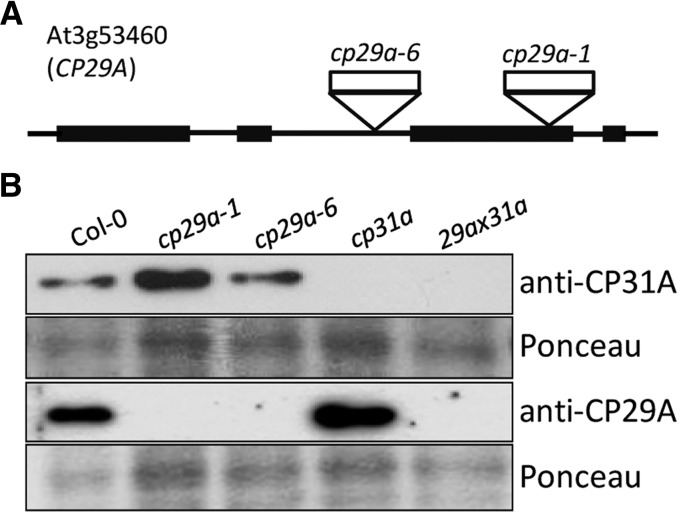
We previously isolated and analyzed null mutants of the Arabidopsis CP31A and CP31B genes (Tillich et al., 2009). Here, we further isolated T-DNA insertion lines for CP29A (Figure 1A), which had no macroscopic defects under standard growth conditions. We raised antibodies against CP29A (At3g53460) and CP31A (At4g24770) and used them to probe total protein extracts from wild-type and T-DNA insertion lines (Figure 1B; see Supplemental Figure 1 online). The signals observed in wild-type extracts were in the range of 30 kD, consistent with the predicted protein size (Ohta et al., 1995). These signals were absent from the mutant lines analyzed. These results show that (1) the antibodies were specific for their cognate proteins, and (2) the T-DNA insertion lines do represent null alleles of the respective genes.
Figure 1.
Isolation of cp29a Null Mutants.
(A) Gene map of CP29A (At3g53460) with four exons (black bars) and three introns (black lines). Positions of the T-DNA insertions cp29a-1 (SALK_003066) and cp29a-6 (001G06) are marked by triangles.
(B) Immunoblot analyses using anti-CP29A and anti-CP31A antibodies demonstrate the absence of CP29A and CP31A in cp29a and cp31a single mutants, respectively, and the lack of both proteins in cp29a × cp31a double mutants. A quantitative analysis with dilutions of wild-type protein extracts demonstrates that loss of CP29A does not affect the accumulation of CP31A and vice versa (see Supplemental Figure 1 online).
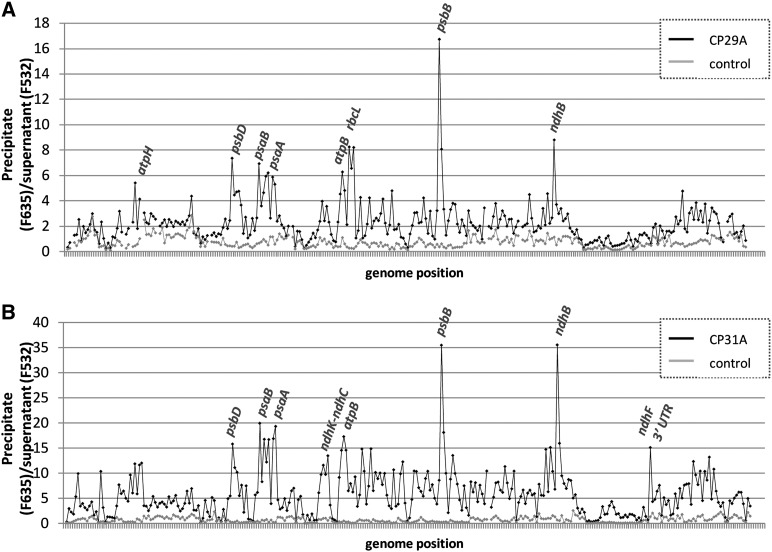
As an initial survey to determine the RNA ligands of CP29A and CP31A on a transcriptome-wide scale, we used the RIP-chip technique, in which RNAs immunoprecipitated with antibodies to an RBP are chemically labeled and analyzed by microarray (Schmitz-Linneweber et al., 2005). We extracted stroma from purified chloroplasts as a source for immunoprecipitating the two cpRNPs (see Supplemental Figure 2 online). RNA was extracted from the pellet and supernatant fractions of each immunoprecipitation experiment and labeled with either red or green fluorescent dyes. The dye-labeled RNAs from corresponding pellet and supernatant RNAs were hybridized onto a tiled microarray representing the Arabidopsis chloroplast chromosome (see Supplemental Data Set 1 online). Control assays were performed with preimmune serum or with stromal extracts from cpRNP-deficient plants. The results were unexpectedly complex. We observed strong enrichment of selected RNAs for both cpRNPs, including peaks for psbB, psbD, psaA/B, atpB, and ndhB (Figure 2). Lower-level enrichment was observed for several tRNAs and rRNAs, and intermediate enrichment was found for almost all of the remaining chloroplast mRNAs. The enrichment curves of CP31A and CP29A were generally similar, indicating that their binding specificities may overlap.
Figure 2.
Identification of RNAs Associated with CP29A and CP31A in Chloroplast Stroma.
The enrichment ratios (pellet F635/supernatant F532) were normalized between three assays involving wild-type stroma, one control assay with stroma from the corresponding null mutants (from cp29a [A] or cp31a [B] seedlings, respectively), and one control assay involving wild-type stroma, but assayed with preimmune serum of the corresponding antibody. The median normalized values for replicate spots from the mutant data and from the control assays were plotted according to chromosomal position. The data used to generate these graphs are provided in Supplemental Data Set 1 online.
It is rare for our group and others to find high enrichment values for numerous RNAs in chloroplast RIP-chip assays for nonenzymatic RBPs (Schmitz-Linneweber et al., 2005; Beick et al., 2008; Kroeger et al., 2009; Pfalz et al., 2009; Zoschke et al., 2010). Visual inspection of the microarray scans also revealed strong signals in the pellet (red channel) for many probes, which is something we have not encountered previously in RIP-chip analyses (see Supplemental Figure 3 online). For example, when we previously assayed PPR proteins, very few probes showed strong pellet signals (Schmitz-Linneweber et al., 2005). This indicates that the cpRNPs have a remarkably large RNA target range.
To complement and confirm the RIP-chip data, we analyzed cpRNP-associated RNAs with dot blot assays (see Supplemental Figure 4 online). We examined the top hits obtained in our RIP-Chip analysis (atpH, atpB, psaA, psbB, psbD, and rbcL), RNAs with intermediate enrichment (ycf3 and rpl33), and selected RNAs with low enrichment ratios (trnI and rrn16). The dot blots corroborated the qualitative findings from our RIP-chip analysis. Also, except for rbcL, far more RNA was coprecipitated with CP31A compared with CP29A, when pellet-to-supernatant ratios were considered (see Supplemental Figure 4 online). This difference was not due to a more efficient precipitation of CP31A than CP29A (see Supplemental Figure 2 online); CP29A was cleared completely from stromal preparations, whereas <50% of CP31A was pelleted in our immunoprecipitations (see Supplemental Figure 2 online). Nonspecific precipitation of RNA in dot blot assays was examined in control experiments using an antibody against the Haemophilus influenzae hemagglutinin (HA) epitope. As expected, only minimal amounts of signal were obtained in our HA dot blot assays. In sum, these results demonstrate that CP29A and CP31A associate with large chloroplast mRNA pools.
RNA Metabolism Is Largely Unaffected in cp29a and cp31a Mutants at Standard Growth Conditions
Since there are multiple lines of evidence that cpRNPs act in different RNA maturation events (Ruwe et al., 2011), we decided to analyze the cp29a null mutants for mRNA accumulation and RNA editing, as was done before for cp31a mutants (Tillich et al., 2009). In contrast with the defects observed for cp31a, none of the 32 chloroplast editing sites in cp29a showed deviations from the wild type (see Supplemental Figure 5 online). Also, mRNA levels in cp29a mutants largely mirror the wild-type situation as judged by genome-wide quantitative RT-PCR (qRT-PCR) analyses (see Supplemental Figure 6 and Supplemental Table 2 online). Furthermore, we decided to investigate mRNA splicing efficiencies of plastid mRNAs in cp29a and cp31a mutants, as the contribution of cpRNPs to this particular step in RNA processing has not been addressed to date. For that, we used a qRT-PCR platform that measures the abundance of spliced versus unspliced transcripts (de Longevialle et al., 2008). Splicing was not affected in cp29a mutants, but our analysis indicated that there was reduced accumulation of spliced ndhB and ycf3 mRNAs in the cp31a mutants (see Supplemental Figure 7 online). The corresponding unspliced transcripts accumulated normally, whereas spliced forms are reduced; this suggests that there may be selective destabilization of spliced transcripts in cp31a rather than a true splicing defect because the latter is usually characterized by overaccumulation of the unspliced precursor RNA.
CP29A and CP31A Are Required for Normal Chloroplast Development under Cold Stress Conditions
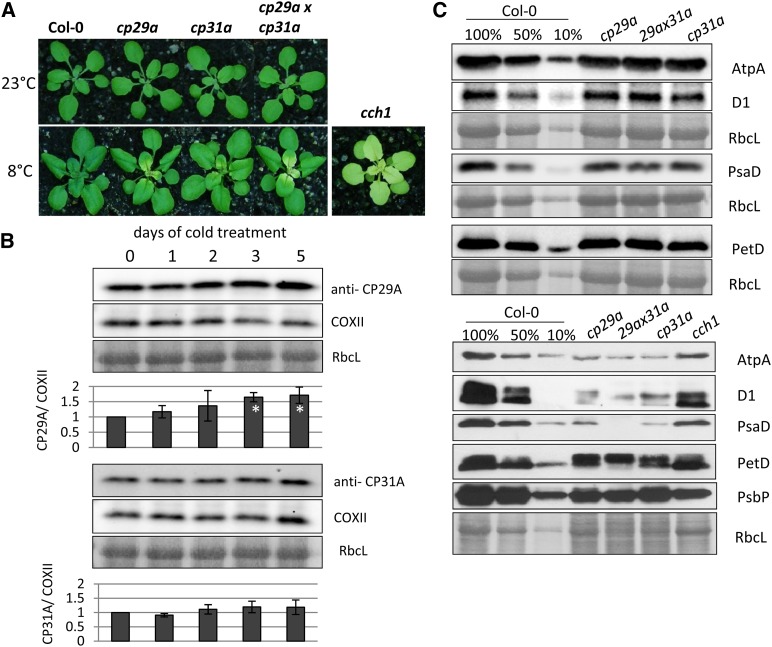
We did not observe phenotypic deviations among the wild type, cp29a and cp31a mutants, or the double mutant under standard growth conditions (Tillich et al., 2009; Figure 3A), even at the very early stages of plant development, when chloroplast gene expression is crucial for the proper setup of the photosynthetic machinery (Rogalski et al., 2006; Fleischmann et al., 2011). We therefore examined the phenotypes of cp29a and cp31a mutants under different stress conditions. Because cpRNPs are known to undergo light-dependent regulation, we shifted 12-d-old plants grown under standard light intensity (180 µmol⋅m−2⋅s−1) to high light (500 to 600 µmol⋅m−2⋅s−1) or low light (20 to 30 µmol⋅m−2⋅s−1) conditions for 3 weeks. However, we did not observe any phenotypic alterations relative to the wild type (see Supplemental Figure 8 online). Next, we examined the effects of cold stress because several cpRNPs (including CP29A) reportedly showed increased mRNA levels under cold conditions (Ruwe et al., 2011). The 3-week-old mutant plantlets transferred to 8°C for 18 d showed bleaching of newly emerging tissues at the bases of the youngest leaves (Figure 3A), but the fully expanded leaves and older leaf tip tissues remained green. Seeds from mutant plants germinated at 8°C displayed pale primary leaves (see Supplemental Figure 9A online), and germination was drastically delayed under cold conditions. Shifting the cold-stressed tissue back to 23°C did trigger regreening of the bleached tissue, demonstrating that the phenotype is reversible (see Supplemental Figure 9B online). The phenotypes of plants at both ages were aggravated in cp31a × cp29a double mutants, indicating that the functions of the two proteins are partially redundant. We also tested the accumulation of CP29A and CP31A during cold treatment of 2-week old seedlings and found that CP31A did not show any change, but CP29A increased after 1 d of cold treatment and gradually increased later when grown at 8°C (Figure 3B). Together, these data demonstrate that CP29A and CP31A appear to function in chloroplast development under cold stress conditions.
Figure 3.
Phenotype and Protein Accumulation in cp29a and cp31a Single and Double Mutants.
(A) Macroscopic effects of cold stress on cpRNP mutants. Top tier: Plants grown for 3 weeks at 23°C. Bottom tier: Plants grown for 18 d at 23°C, then shifted to 8°C for additional 3 weeks. Phenotype of the cch1 mutant grown after cold treatment is included as control for photosynthetic deficiency.
(B) Expression of CP29A (top part) and CP31A (bottom part) during cold treatment. Two-week-old Arabidopsis plants were exposed to cold stress (8°C), and tissue samples were taken after 1, 2, 3, and 5 d of cold treatment. For immunoblot analysis, the four youngest leaves of every plant were harvested. Equal amounts of total protein were separated by SDS-PAGE, blotted to nitrocellulose membranes, and hybridized with anti-CP29A and anti-CP31A antibodies. Equal loading is demonstrated by Ponceau staining of RbcL and by reprobing the blots after stripping with an antibody against the mitochondrial COXII protein. Signals were measured using a chemiluminoimager. The maximum signal intensity measured at 0 d was set to 1. Signals were normalized to COX2 signals. Standard deviations are based on three biological replicates. Significance of changes relative to day 0 was calculated with a two-tailed, paired Student’s t test. White asterisks denote P values below 0.05 (i.e., a significant induction in the cold).
(C) Immunological analysis of photosynthetic enzyme accumulation in cp29a and cp31a single and double mutants. The accumulation of representative subunits of the ATP synthase (AtpA), photosystem II (D1), photosystem I (PsaD), and the cytochrome b6f complex (PetD) were analyzed by probing immunoblots with specific antisera. Blots were prepared with total leaf proteins from the wild type with dilutions indicated and from cp29a, cp31a, and cp29axcp31a mutant seedlings grown at normal temperatures for 3 weeks (top part) or from the bleached tissue from plants grown for 18 d at 23°C and then cold-stressed for an additional 3 weeks at 8°C (bottom panels). The blots were stained with Ponceau S to visualize the large subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (RbcL; corresponding Ponceau stains and antibody probing are shown as blocks of panels; i.e., several blots were probed with multiple antibodies).
Since the pale phenotype of the emerging tissues suggested that there was a failure in setting up the photosynthetic apparatus, we used immunoblot analysis to examine subunits of the photosynthetic complexes in cpRNP mutants. To test the effects of cold stress, we used only the emerging tissues from wild-type and cpRNP-deficient plants (i.e., the leaf areas that were pale under cold stress conditions in the mutants). Our results revealed that the subunits of photosystem II, the cytochrome b6f complex, and the plastid ATP synthase accumulated to wild-type levels in cpRNP single and double mutants under normal growth conditions (Figure 3C, top panels). At 8°C, however, the levels of all tested proteins were reduced to 50% or less compared with the wild-type levels (Figure 3C, bottom panels). Since chlorophyll deficiency could lead secondarily to a reduction in photosynthetic proteins, we used the cch1 (conditional chlorina1) mutant as a control. CCH1 encodes the H subunit of the Mg-chelatase and is required for chlorophyll synthesis, and loss of CCH1 leads to pale plants (Figure 3A; Mochizuki et al., 2001). We found that the chloroplast-encoded proteins accumulated to higher levels in cch1 mutants compared with cold-stressed cpRNP mutants. This suggests that the defects seen in the pale tissues of cpRNP mutants are not entirely due to the loss of photosynthetic ability or chlorophyll production but instead represent a more direct consequence of the mutations.
CP29A and CP31A Are Required for the Accumulation of Target RNAs under Cold Stress Conditions
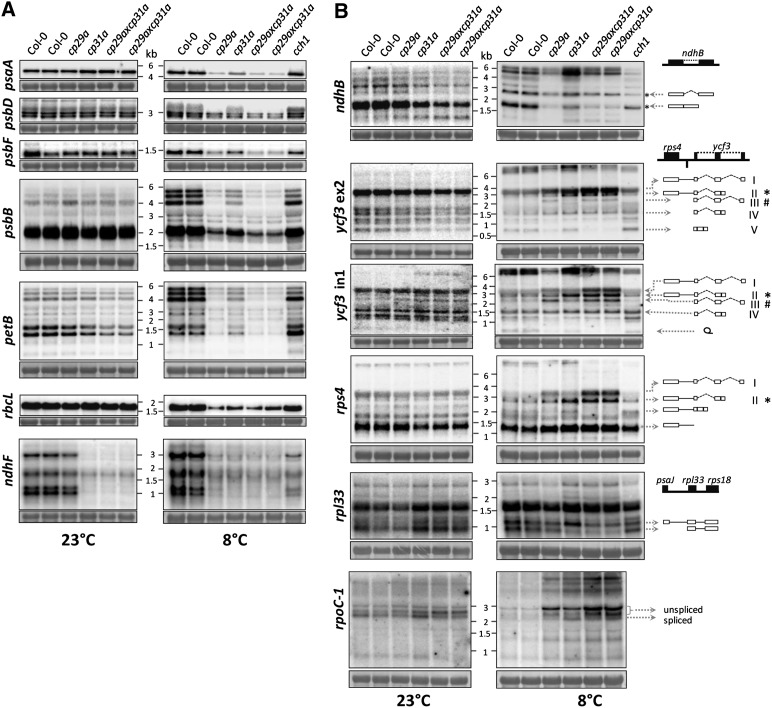
Our findings that cpRNP mutants show a global reduction in chloroplast protein accumulation under cold stress and associate with multiple mRNAs suggest that they may promote the expression of many chloroplast mRNA species. Given that cpRNPs have been suggested to stabilize chloroplast messages (Nakamura et al., 2001), we used RNA gel blot hybridization to test whether the loss of cpRNPs altered mRNA accumulation under cold stress conditions. We found two classes of changes: Class I genes displayed a general reduction in RNA levels (Figure 4A), whereas class II genes showed alterations of RNA accumulation for specific transcripts (Figure 4B).
Figure 4.
Analysis of Transcript Accumulation in cp29a and cp31a Mutants at 23 and 8°C.
Four micrograms of total leaf RNA from the indicated genotypes was fractionated on 1.2% agarose gels and analyzed by hybridization to probes for the plastid RNAs indicated (see Supplemental Table 1 online for primer sequences). Equal loading was controlled by staining the cytosolic 25S rRNA (25S) with methylene blue. Autoradiographs shown on the left are prepared with plants grown at normal temperatures; those on the right from plants cold-treated for 3 weeks.
(A) Transcript patterns with reductions (i.e., with quantitative but not qualitative changes in the transcript pattern) after cold treatment relative to wild-type controls.
(B) Genes with altered transcript patterns under cold stress conditions. Cartoons of the assessed operons are shown at the right. Selected transcripts are represented as well. Open boxes, cistrons; exons; lines, intergenic regions; dotted lines, introns.
For class I genes, overall steady state transcript levels were reduced after cold treatment, but the transcript pattern (i.e., transcript processing) is identical or very similar to those found under normal conditions and in the wild type (Figure 4A). This group includes psaA, psbD, psbF, psbB, petB, rbcL, and ndhF. The decreases in mRNA for these genes (with the exception of ndhF) were much stronger than those in the cold-treated, pale cch1 mutants that were used as a phenocopy control, indicating that the reductions were not merely caused by a loss of photosynthetic capacity. No mRNA reduction was observed under standard conditions.
Notably, there are quantitative differences in class I when comparing cp31a and cp29a mutants. At 23°C, the ndhF mRNA was reduced below the detection limit in cp31a mutants under normal conditions (Tillich et al., 2009; Figure 4A), but it was only slightly reduced compared with the wild type in the cp29a mutants (Figure 4A). At 8°C, the ndhF mRNA was strongly decreased to similar residual levels in all of the tested cpRNP mutants, but also in cch1 mutants, suggesting that for ndhF, this effect could be largely attributed to a general photosynthetic deficiency. Other gene-specific effects were also noted in psaA, psbF, and psbB. In these cases, the loss of CP29A decreased the mRNA levels more than the loss of CP31A (Figure 4A). In most cases, the mRNA levels were further reduced in the double mutants than in single mutants, suggesting that CP29A and CP31A have additive functions in terms of mRNA accumulation (i.e., are partially redundant). This is consistent with the observation that double mutants display a more severe chlorotic effect than single mutants (Figure 3A). In all, class I defects demonstrate that cpRNPs are required for normal chloroplast mRNA accumulation at lower temperatures.
Decreased RNA levels can be caused by diminished transcription or increased RNA degradation. Assays to determine chloroplast transcription rates depend on isolated chloroplasts. We were unable to isolate chloroplasts from the bleached tissues of cold-stressed material; under standard growth conditions, however, cpRNPs do not display transcription defects (Tillich et al., 2009; see Supplemental Figure 10 and Supplemental Table 3 online). Since cpRNPs associate with RNA (Figure 2; see Supplemental Figure 4 online) and stabilize transcripts in vivo and in vitro under normal conditions (Nakamura et al., 2001; Tillich et al., 2009), a direct effect of cpRNPs on RNA stability seems more likely than an effect on transcription.
CP29A and CP31A Foster Transcript-Specific Processing under Cold Stress Conditions
After cold treatment, RNA preparations from pale tissue are characterized by aberrant transcript patterns, including overaccumulation of partially or nonprocessed precursor transcripts. This includes transcripts from the genes ycf3, rps4, rpl33, accD, ndhB, rpoC1, and clpP (Figure 4B; see Supplemental Figure 11 online). For ndhB, we found that unspliced precursors are only moderately reduced, whereas spliced transcripts are almost lost (Figure 4B). For ycf3, we found a strong genotype-dependent overaccumulation of several unspliced precursor RNAs (Figures 4B and 4C; cp31a-dependent transcript II and cp29a-dependent transcript III). Spliced transcripts were strongly reduced only in cp29a mutants (transcript V) and the doubly unspliced rps4-ycf3 RNA overaccumulated strongly in double mutants, suggesting that both cpRNPs contribute additively in this case (transcript I). Overaccumulation of precursors was also observed for accD, rpoC1, and clpP transcripts in both mutants; a dicistronic transcript containing rpl33 and rps18 is cp31a dependently reduced (Figure 4B; see Supplemental Figure 11 online). These findings suggest both redundant and individual functions of CP29A and CP31A in generating correctly processed chloroplast transcripts.
Since CP31A was previously found to affect the RNA editing efficiencies of multiple sites in chloroplast transcripts (Tillich et al., 2009), we herein also investigated the editing of selected RNAs in null mutants of cpRNPs under cold stress conditions. However, the reductions in RNA editing efficiencies observed in mutants were similar at 23 and 8°C (see Supplemental Figure 12 online) and therefore cannot account for the pale phenotype observed in cold-treated plants. Only the editing sites within the psbZ and ndhB mRNAs showed a minor decrease of editing efficiency in cp31a and cp29a mutants under cold stress conditions (see Supplemental Figure 12 online). Compared with the drastic effects on RNA steady state levels in the cold, editing defects appear minor. In sum, these data show that CP31A and CP29A differentially influence the processing pattern of many mRNAs in the cold.
Reduced Processing of rRNA in the cp31a and cp29a Mutants
Given that a global loss of plastid proteins under cold stress conditions might indicate a general problem in plastid translation (e.g., in ribosome biogenesis), we tested the accumulation of chloroplast rRNAs. rRNAs and a number of ribosomal proteins are encoded in the chloroplast genome, and partially defective ribosomes that lack nonessential ribosomal proteins have been shown to be sensitive to cold stress (Rogalski et al., 2008; Fleischmann et al., 2011). Although rRNAs were not strongly enriched in cpRNP immunoprecipitation (IPs) (Figure 2; Nakamura et al., 1999), we note that the RIP-chip assays would be unable to detect the binding of cpRNPs to rare rRNA precursor molecules if there was no binding to the much more abundant mature rRNAs (i.e., because there would be only minor changes in the pellet-to-supernatant ratios). We therefore analyzed rRNAs in the cold-stressed tissues of cpRNP mutants in the absence of RIP-chip support for rRNA binding.
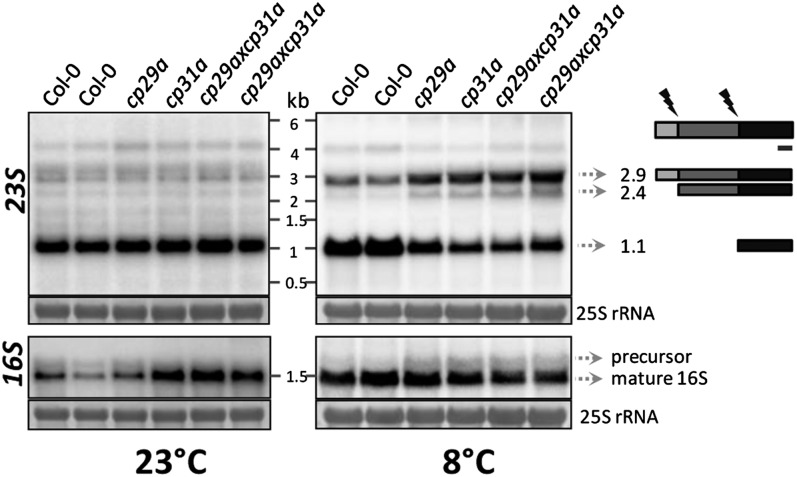
In contrast with bacteria, the 23S rRNA of chloroplasts is cleaved internally at two sites, yielding fragments of 0.5, 1.1, and 1.3 kb (Nishimura et al., 2010). In cold-stressed cpRNP mutants, the 2.9-kb full-length 23S RNA and the 2.4-kb processed transcript strongly overaccumulate, and the mature processing products (except for the 0.5-kb form) are reduced (Figure 5). This suggests that cleavage at the 5′-end, which separates the 23S rRNA into the 2.4- and 0.5-kb fragments, is not affected in the mutants, whereas disjunction of the 1.1-kb 3′-fragment is strongly reduced. The 16S rRNA precursor shows only a mild overaccumulation after cold treatment. Overall, our analysis indicates that rRNA processing is impaired in both of the cpRNP mutants under cold stress.
Figure 5.
Analysis of rRNA Accumulation in cp29a and cp31a Mutants at 23 and 8°C.
Four micrograms of total leaf RNA from wild-type and mutant plants before and after 3 weeks of cold stress were subjected to RNA gel blot analyses using probes for the 16S and 23S rRNAs (see Supplemental Table 1 online for primer sequences). Equal loading was controlled by staining the cytosolic 25S rRNA (25S) with methylene blue. Autoradiographs shown at left were prepared with plants grown at 23°C; those on the right from plants cold-treated (8°C) for 3 weeks. Cartoon: The 23S rRNA is matured into three fragments on chloroplast ribosomes by cleavage of the precursor (flashes). The probe used detects the 3′-end of the 23S rRNA and is indicated by a black bar.
Multiple Points of Association between CP29A, CP31A, and Their Target mRNAs
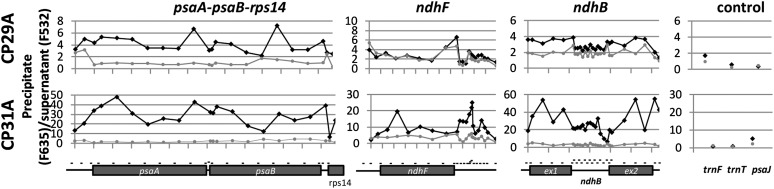
To get insights into how cpRNPs mediate RNA processing steps, we decided to investigate the binding sites of CP29A and CP31A on selected mRNAs in more detail. We developed a microarray of oligonucleotides distributed along the length of the psaA/psaB operon, the ndhF mRNA, and the ndhB mRNA. The use of multiple probes has the potential to narrow down binding sites because RNA is fragmented during stromal extraction procedures (Zoschke et al., 2010). If the breakdown products are considerably smaller than the full-length messages on average, the limits of binding sites can be determined. We chose the tricistronic psaA/B-rps14 mRNA for this analysis because it was a prime peak in our RIP-chip analysis. We chose ndhB because it contains an intron, and it was another top target identified in our initial RIP-chip analysis. Finally, we chose ndhF because this mRNA is drastically reduced under normal growth conditions in cp31a mutants (Tillich et al., 2009). As controls, we used oligonucleotides against RNAs that were not found to be enriched in the initial RIP-chip survey. Three experiments with antisera against CP29A and CP31A and three control experiments using the corresponding preimmune sera, null mutants, or anti-HA antibodies were performed (see Supplemental Data Set 2 online). For the tricistronic psaA/B-rps14 mRNA, enrichment was higher for all probes analyzed in CP29A or CP31A immunoprecipitations compared with mock precipitations (Figure 6). The peaks and valleys for this transcript were similar between the CP29A and CP31A immunoprecipitations, albeit with stronger enrichment in the CP31A IP experiments. Overall, the entire mRNA seemed to be precipitated by both proteins, and only the 5′ and 3′ ends of the tricistronic RNA showed declines in enrichment (Figure 6). This suggests that the RNA contains multiple RNA binding sites for both proteins. In the case of the ndhB mRNA, we also observed stronger overall RNA enrichment in CP31A IPs compared with those of CP29A (Figure 6). The patterns of enrichment were similar for the two proteins, with higher enrichment ratios in exons compared with introns. This could potentially indicate that cpRNPs have a higher affinity for linear exonic RNA segments compared with the more convoluted, structure-rich intronic sequences. This is in line with previous findings that cpRNPs prefer single-stranded over double-stranded RNAs (Li and Sugiura, 1991).
Figure 6.
Mapping of cpRNP Binding Sites on Selected Chloroplast mRNAs.
RIP-chip assays using stroma extracts and antisera against CP29A (top) or CP31A (bottom) were performed using an array with oligonucleotide probes antisense to three transcription units, psaA-psaB-rps14, ndhF, and intron containing ndhB. Oligos against nontargets of the two cpRNPs (trnF, trnT, and psaJ) were included as well. The oligos were distributed to represent all areas of the target transcripts of the two cpRNPs as indicated by short thin lines in the schematic representation drawn to scale below the charts. The enrichment ratios (pellet F635/supernatant F532) were normalized between three assays from wild-type extracts with CP29A- and CP31A-specific antibodies (black lines) and controlled with two assays from mutant extracts and one assay using preimmune serum of the antibody in case of CP29A (gray lines). For CP31A, two experiments were controlled using the preimmune serum of the antibody and one is controlled using an HA antibody. We did not use mutant serum in the latter case because it lacks the ndhF mRNA (Tillich et al., 2009), which is one target to be analyzed in this approach. The median values for three experiments and three controls are provided in Supplemental Data Set 2 online.
We obtained similar coprecipitation results for the psaA/B and ndhB RNAs, but we noted striking differences with the ndhF message. There was no enrichment of ndhF in CP29A immunoprecipitations, but there was clear enrichment when CP31A was precipitated (Figures 2 and 6). Moreover, we found two distinct peaks of enrichment in the CP31A IPs: the 5′-part of the coding region, which harbors the ndhF editing site, and a region downstream of the stop codon, which most likely represents the 3′-untranslated region (UTR). This demonstrates that CP31 is a specific ligand of the ndhF mRNA with at least two sites of attachment. As expected, the enrichment ratios for the control probes were low and similar in experiments with antisera and preimmune sera (Figure 6).
CP31A Is Required for the Accumulation of a Set of Transcripts Originating at the Borders of the Chloroplast Small-Single-Copy Region and Overlapping with the Inverted Repeat
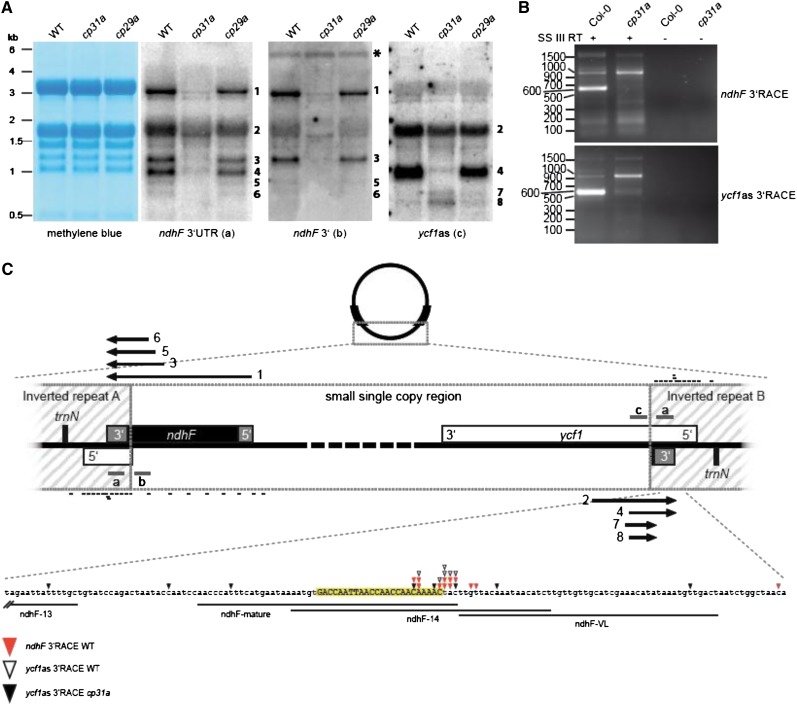
CP31A is essential for accumulation of the ndhF message (Figure 4A; Tillich et al., 2009) and, thus, for the expression and activity of the chloroplast NDH complex (Tillich et al., 2009). The results of our oligo-chip analysis suggested that the main binding site of CP31A lies within the 3′-UTR of ndhF (Figure 6). Accordingly, we analyzed the maturation of transcripts ending in the 3′-UTR of ndhF by RNA gel blot hybridization using a single-stranded RNA probe located in the region suggested to be a target of CP31A in our RIP-chip analysis (Figure 2). We observed at least six major transcripts corresponding to the 3′-UTR of ndhF; they ranged from 3.0 to 0.7 kb and were assigned consecutive numbers according to their size (Figure 7A). The largest band, which was the only one that could contain the complete ndhF reading frame, was previously detected with probes against the coding region of ndhF (Tillich et al., 2009). All of the detected bands were strongly reduced in the cp31a mutants, but not in the cp29a mutants (Figure 7A); this is consistent with our RIP-chip data, suggesting that ndhF is not a target of CP29A (Figures 2 and 6). However, the identities of the additional, smaller transcripts were unclear, in part because the probe we used for the 3′-UTR of ndhF lies within the inverted repeat (IR) and thus can detect transcripts originating from both edges of the small-single-copy (SSC) region of the chloroplast genome (Figure 7C). To differentiate between these two possible transcript types, we used strand-specific probes located within the SSC region of either the ndhF or ycf1 coding regions (Figure 7A; probes ndhF 3′ and ycf1as, respectively). This hybridization demonstrated that bands 1, 3, 5, and 6 were derived from the ndhF site of the SSC region, whereas bands 2 and 4 originated from within the ycf1 region. Notably, transcripts 1, 3, 5, and 6 were sense to ndhF, whereas 2 and 4 were antisense to ycf1. All of these transcripts terminated within the IR and thus shared common 3′-sequences. Two additional bands were detected with the ycf1as probe, which did not include IR sequences. These bands (no. 7 and 8) were the only ones that overaccumulated in cp31a mutants relative to the wild type and the cp29a mutants. In sum, our RNA gel blot hybridizations demonstrate that CP31A is required for the accumulation of a swarm of transcripts originating close to the SSC/IR borders and terminating within the IR.
Figure 7.
CP31A Stabilizes ndhF and Antisense Transcripts of ycf1 under Normal Growth Conditions.
(A) RNA gel blots using strand-specific RNA probes showing that ndhF and ycf1as transcripts are strongly reduced in cp31a mutants. ndhF 3′-UTR probe (a) lies within the IR region and detects ndhF and ycf1as transcripts. ndhF3′ (b) and ycf1as (c) probes are located in the SSC region and selectively detect ndhF and ycf1as transcripts, respectively. Methylene blue staining of membranes is shown as a loading control. A signal corresponding to the tricistronic transcript psaA-psaB-rps14 is marked by an asterisk. WT, the wild type.
(B) 3′-RACE detects transcript termini, which are strongly reduced in cp31a mutants. Primers inside the SSC region were used to amplify 3′ ends of ndhF and ycf1as transcripts. The band at 600 bp was gel eluted and cloned. The corresponding 3′ ends are shown in (C).
(C) Map of the two borders between the SSC and IR regions. ndhF and ycf1 genes are transcribed from right to left. ndhF and ycf1as transcripts are shown as arrows with numbers corresponding to the bands detected in the RNA gel blots in (A). The probes used in RNA gel blots are shown as gray bars. Positions of the oligonucleotides used to determine the binding sites of CP31A (Figure 6) are marked with black bars. The 3′-ends identified in (B) are shown as color-coded triangles. The 3′-ends from ndhF wild-type RNA are shown with closed red triangles, those for ycf1as wild-type RNA with open triangles. The 3′-ends from ycf1as transcripts obtained from cp31a RNA are shown by closed triangles. A short noncoding RNA found at the position of mature 3′-ends is shown in uppercase letters and highlighted in yellow.
CP31A Defines a 3′-Exonuclease Block for a Host of Transcripts at the Border of the Chloroplast IR
The intriguing role of CP31A for several transcripts that terminate within the same sequence area prompted us to determine their 3′-end by 3′-rapid amplification of cDNA ends (RACE) experiments. To obtain transcripts from both IR borders, we used two oligonucleotides: one sense to the ndhF coding region and one antisense to the ycf1 coding region. The amplification products differed markedly between RACE experiments from wild-type and mutant RNA extracts (Figure 7B). Wild-type amplifications contained a dominant band of ∼600 nucleotides, but very little signal of the same size was seen in the mutant cDNAs. After gel purification, sequencing of cloned amplification products corresponding to the band in wild-type experiments identified a single narrow hot spot of transcript termini within the IR. We previously evaluated deep-sequencing data sets and reported that a small noncoding RNA accumulates in this area (Ruwe and Schmitz-Linneweber, 2012). The ends determined in our present wild-type RACE experiments clustered around the 3′-end of this small RNA (sRNA), which was located in the middle of the oligonucleotide that showed peak enrichment in our CP31A RIP-chip experiments (Figure 6). By contrast, the 3′-ends were randomly distributed within the cp31a mutant (Figures 7B and 7C). Together with our results from the RNA gel blot hybridizations, this shows that CP31A is required for the correct generation of the 3′-ends of transcripts originating in the SSC and terminating in the IR.
If CP31A acts to protect the 3′-end of the ndhF mRNA, then we would expect to see accumulation of degradation products that are shortened in their 3′-regions. Only two bands (numbers 7 and 8 in Figure 7A) were detected by a probe corresponding to the ycf1 gene (probe c) but not by probes corresponding to IR sequences. Both bands were <700 nucleotides and therefore could not reach the 3′-ends of the other ycf1 antisense RNAs detected in the wild type. However, transcripts 7 and 8 were also found in the wild type (albeit at much lower levels), suggesting that this may be a standard degradation pathway for ycf1 antisense transcripts and, therefore, likely also for the ndhF message (Figure 7A).
CP31A Is Required to Stabilize a Small Noncoding RNA That Represents the 3′-End of the ndhF mRNA
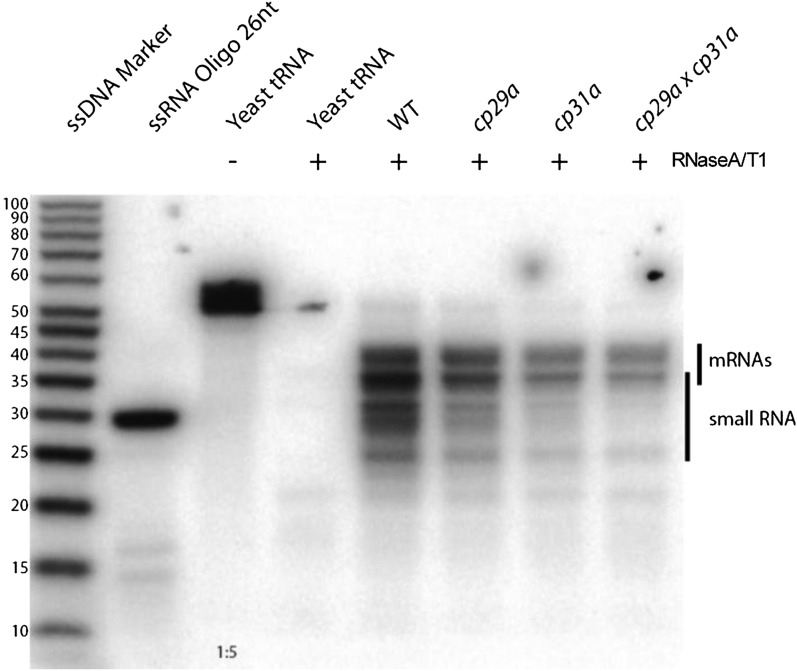
The ndhF sRNA belongs to a recently identified group of small RNAs that accumulate in chloroplasts and have been shown to be footprints of RBPs, typically PPR proteins (Ruwe and Schmitz-Linneweber, 2012; Zhelyazkova et al., 2012). These proteins protect mRNAs against exonucleolytic degradation, eventually leading to the generation of small protected fragments that accumulate as sRNAs. If CP31A participates in protecting the ndhF RNA and leaves the sRNA as a footprint, then we should see a loss of this sRNA in cp31a mutants. This was assayed by RNase protection experiments (Figure 8), in which RNA extracts from mutants and wild-type samples were hybridized against a probe that is several nucleotides longer and antisense to the sRNA. Degradation of single-stranded RNA yields RNAs that correspond in size to the processed mRNA/antisense RNA and the sRNA. Both RNAs were readily discernible in wild-type extracts; the signal for the sRNA was greatly diminished in cp31a mutants but not cp29a mutants (Figure 8). This demonstrates that CP31A supports the accumulation of the ndhF sRNA and further implies that CP31A is involved in protecting the ndhF mRNA against 3′-exonucleolytic degradation.
Figure 8.
A Small RNA Corresponding to the 3′-End of ndhF Is Strongly Reduced in cp31a Mutants.
Mature ndhF and ycf1as transcripts and the small RNA located at the mature 3′-end are detected by an RNase protection assay. A radioactive probe complementary to the small RNA is hybridized to total RNA from the wild type and mutants as indicated. Double-stranded RNA is protected from degradation by single-strand specific RNases A and T1. Protected fragments were separated in a denaturing polyacrylamide gel. In negative control experiments without chloroplast RNA, yeast tRNAs were added to arrive at similar final RNA concentrations. This serves as control for degradation of the probe during the procedure (-RNase) or self-protection of the probe (+RNase). End-labeled RNA oligo and single-strand DNA (ssDNA) ladder are shown as size markers. WT, the wild type.
DISCUSSION
CpRNPs Protect Chloroplast RNA Metabolism against Cold Stress
How could cpRNPs support cold resistance of chloroplast biogenesis? One process long suspected to be sensitive to cold stress is chloroplast translation. For example, chilling stress impedes protein biosynthesis in plastids by eliciting frequent ribosome pausing (Grennan and Ort, 2007). Ribosomal biogenesis and RNA processing is chilling sensitive in different mutant backgrounds (Millerd et al., 1969; Hopkins and Elfman, 1984; Barkan, 1993). Vice versa, the loss of ribosomal subunits reduces the ability of plants to recover from cold-induced injuries after prolonged chilling periods (5 weeks at 4°C; Rogalski et al., 2008; Fleischmann et al., 2011), suggesting that translation can become limiting in the cold. Also, loss of the translation elongation factor SVR3, the rRNA methylase PCF1, and the rRNA maturation factor NUS1 leads to increased cold sensitivity (Tokuhisa et al., 1998; Liu et al., 2010; Kusumi et al., 2011).
cpRNP mutants also displayed rRNA processing defects in the cold, but we did not observe a strong association between cpRNPs and the rRNAs in our RIP-chip or dot blot assays. Therefore, it seems unlikely that loss of cpRNPs causes cold stress via a direct effect on rRNA maturation. More likely, effects on a ribosome-related mRNA and, thus, a ribosomal protein could lead to the observed rRNA processing defects in the cpRNP mutants, ultimately reducing translation levels and leading to cold sensitivity. The rpl33 mRNA, which has been reported to code for an essential component of intact ribosomes under cold stress conditions (Rogalski et al., 2008), showed a 10-fold enrichment with CP31A and two- to threefold enrichment with CP29A in our RIP-chip experiments. The mature rpl33 mRNA was strongly reduced in the cp29a × cp31a double mutants in the cold (Figure 4B). Further effects on other mRNAs for ribosomal proteins can be expected. Thus, defective ribosome biogenesis likely contributes to the observed chilling sensitivity of cpRNP mutants.
In addition, our findings suggest that other functions of cpRNPs, particularly the reduced ycf3 intron splicing in cp29a mutants, may contribute to the cold shock syndrome. Loss of ycf3 splicing in tobacco was associated with a chlorotic phenotype of tissues that emerged in cold conditions due to reduced photosystem I assembly followed by enhanced reactive oxygen species accumulation (Petersen et al., 2011). An impact on the expression of proteins essential for photosynthesis (e.g., psaA and psbB) might also contribute to cold sensitivity, as photosynthetic electron transport is known to be adapted to cold stress (Yamori et al., 2010). In sum, our results indicate that the cold syndrome displayed by cpRNP mutants is likely to be due to a variety of detrimental effects on chloroplast gene expression.
CpRNPs Are Global Modulators of Chloroplast RNA Metabolism
We herein demonstrated that the cpRNPs, CP29A and CP31A, associated with multiple chloroplast mRNAs and prefer mRNAs over tRNAs and rRNAs. This extends a previous study in tobacco demonstrating that cpRNPs preferred certain mRNAs over rRNAs (Nakamura et al., 2001). Also, our analysis of the ndhB mRNA demonstrated that their association is stronger with exonic sequences than with introns. This suggests that cpRNPs discriminate against structured RNAs on a transcriptome-wide scale. This observation is consistent with the known preference of cpRNPs for single-stranded over double-stranded RNAs (Li and Sugiura, 1991) and the failure of cpRNPs to coprecipitate rRNAs, corroborating that they are not associated with ribosomes or polysomes (Nakamura et al., 2001).
RBPs with multiple RNA targets have so far rarely been described in chloroplasts but are common in the nucleocytosolic compartment, where they associate with large pools of messages and modulate the processing and localization of their RNA ligands (Krecic and Swanson, 1999; Dreyfuss et al., 2002). Such pools are called posttranscriptional operons because the bound RNAs are coregulated in a manner analogous to the genes in a bacterial operon (Keene, 2007). A classical example would be the nucleocytosolic heterogeneous nuclear ribonucleoproteins (hnRNPs), which bind and coregulate hundreds to thousands of RNAs in humans and other systems (Dreyfuss et al., 2002). The hnRNPs are the closest relatives of the cpRNPs (Maruyama et al., 1999). Could cpRNPs be coregulators of sets of chloroplast genes? Cluster analyses of microarray data across hundreds of conditions showed that most of the photosynthesis-related chloroplast genes are joined in a coregulated cluster together with more than 900 nuclear genes (Mentzen and Wurtele, 2008; Cho et al., 2009). Interestingly, the coregulated genes include seven genes for cpRNPs (among them, CP29A and CP31A) that could act as regulatory integrators for chloroplast mRNAs. Given the many modifications experienced by cpRNP proteins (reviewed in Ruwe et al., 2011), their ability to bind and process multiple RNAs, their coexpression with photosynthesis-related chloroplast genes, and their phylogenetic clustering with hnRNPs, we hypothesize that cpRNPs are required to coregulate mRNAs to form a posttranscriptional operon in the chloroplast. The observed overlap in the target ranges of CP31A and CP29A, together with the lack of phenotype in their mutants under normal conditions, suggests that we can expect redundancy between cpRNPs. Therefore, future efforts to construct n-tuple mutants of the 10 known cpRNPs, possibly using RNA interference approaches, are needed to functionally test the coregulation of multiple chloroplast genes by these proteins.
Direct and Indirect Effects of cpRNPs on Chloroplast RNA Processing
We found that the loss of the tested cpRNPs led to the reduction of several chloroplast mRNAs, but only under cold stress conditions. Below, we discuss how loss of cpRNPs could lead to this reduction.
The cpRNPs may stabilize target RNAs directly, as evidenced by their binding characteristics. For example, we found that cpRNPs associated with the entire transcripts of psaA/psaB/rps14 and ndhB. By contrast, analogous mapping studies of PPR proteins and the chloroplast splicing factor MatK used the same technical approach to narrow down the binding sites to much shorter sequence elements (Schmitz-Linneweber et al., 2006; Beick et al., 2008; Pfalz et al., 2009; Zoschke et al., 2010). We interpret these data as indicating that chloroplast mRNAs have multiple binding sites for cpRNPs, suggesting that they have low target specificities. RRMs vary in the number of nucleobases that they bind, but a typical number would be two to three bases, or, if two RRMs are combined, four to 10 bases (Maris et al., 2005; Cléry et al., 2008). Their affinities and specificities vary dramatically (Maris et al., 2005; Cléry et al., 2008) and can depend on unconserved sequences flanking the RRM domain (Kenan et al., 1991). Different cpRNPs are expected to have different RNA binding specificities. Consistent with this, we found that the enrichment patterns of CP31A and CP29A for psaA/B and ndhB were similar but not identical. If other members of the cpRNP family behave similarly, we can expect that they exert their stabilizing effects in part through the sheer number of entry sites on chloroplast messages. They would simply cover and package messages, passively protecting them against nucleolytic degradation. Future by-base mapping of the cpRNP binding sites and proteomic analyses of the complex compositions of cpRNPs will be required to test this. Such work will also help explain how cpRNPs can distinguish between precursors and processed transcripts to perform selective stabilization, as seen in the cases of ycf3 and psbB.
The loss of CP29A and CP31A may lead to a defective transcription of PEP (plastid-encoded RNA polymerase)-dependent transcripts in cold-stressed tissue either via reduced translation of PEP or via reduced splicing of rpoC1, coding for a subunit of PEP. Reduced translation could be caused by the observed defect in rRNA maturation (see above) and/or by failed processing of rpl33, which encodes a ribosomal subunit essential under cold conditions (Fleischmann et al., 2011). Any defect in overall plastid translation will reduce the expression of PEP. Loss of PEP activity leads to a reduction in PEP-dependent mRNAs, whereas the genes dependent on the nuclear-encoded plastid RNA polymerase become overexpressed (Hajdukiewicz et al., 1997). This phenotype is mirrored by cold-shocked cp29a and (to a lesser extent) cp31a mutants, where most (but not all; see 16S rRNA) of the PEP-dependent genes show reduced transcript levels, while many NEP (nuclear encoded plastid RNA polymerase)-dependent genes appear overexpressed (e.g., rpoC1, ycf3, rps4, and clpP; Figure 4B; see Supplemental Figure 11 online).
CP31A Stabilizes the ndhF mRNA by Protecting its 3′-End
We observed a strong reduction of ndhF mRNA in cp31a mutants under normal conditions and a seemingly nonspecific loss of ndhF mRNA under cold stress conditions in both mutants. The latter effect is likely to be secondary, as we did not find any association of CP29A with the ndhF mRNA, and the effect was also seen in the cch1 mutant. By contrast, the effect of CP31A appears to be specific, as the protein binds to the ndhF mRNA, in particular to the 3′-terminus. Obviously, functional redundancy is not an issue for this message, and the loss of CP31A (but not CP29A) has dramatic consequences for the stability of ndhF.
The key to understanding the effect of CP31A was the strong reduction not only of ndhF but also of several RNA species antisense to ycf1. We identified informative degradation products for these antisense RNAs, which were unfortunately lacking for ndhF. The degradation fragments were shortened at their 3′-termini, as evidenced by RNA gel blot hybridizations. These findings suggest that CP31A protects the as-ycf1 RNAs (and probably also the ndhF mRNA) from 3′-exonucleolytic degradation. But how does it achieve this protection?
We recently identified short noncoding RNAs located downstream of ndhF (Ruwe and Schmitz-Linneweber, 2012). Mapping of the ndhF 3′-end demonstrated that it colocalized with the 3′-end of this sRNA. Located in the IR of the chloroplast genome, the entire 3′-UTR is also found antisense to ycf1. Here, the sRNAs colocalized with mapped ends of the as-ycf1 RNA species. Notably, sRNAs have been shown to be footprints of RBPs (e.g., PPR proteins) that bind to mRNAs and temporarily protect them against exonucleolytic degradation (Ruwe and Schmitz-Linneweber, 2012; Zhelyazkova et al., 2012). Eventually, however, the mRNA is degraded until only the fragment directly bound by the RBP remains. Here, we found that the sRNAs were reduced (but still detectable) in cp31a null mutants. Thus, CP31A is not solely responsible for generating the sRNA, and we suspect that at least one additional RBP binds to this particular RNA segment. CP31A could help to recruit this RBP or stabilize the entire complex, making it critical for protecting the RNA. A similar scenario could explain the editing defects we observed in the CP31A mutant. CP31A might be needed for the recruitment of a specificity factor, likely a PPR protein. Similar considerations may apply to other processing steps influenced by cpRNPs. Both direct effects, for example, by modulating local RNA structures, as well as indirect effects as recruiters of other RBPs are possible and will need further testing.
Finally, we can address the question of which RNase degrades these CP31A-dependent transcripts. Recently, RNAs antisense to ycf1 were detected in a deep-sequencing study of the Arabidopsis chloroplast transcriptome and in mutants of the chloroplast PNPase, which is a 3′-to-5′ exonuclease (Hotto et al., 2011). RNA gel blot analyses demonstrated that this PNPase is needed for the accumulation of processed as-ycf1 transcripts (Hotto et al., 2011). In the future, studies on the interplay among CP31A, PNPase, and other factors will allow us to understand how chloroplast 3′-degradation of ndhF (and potentially other mRNAs) is regulated.
METHODS
Plant Growth Conditions
Arabidopsis thaliana Columbia-0 and cpRNP T-DNA insertion lines (obtained from ABRC and GABI-Kat) were grown on soil with a 16-h-light/8-h-dark cycle at 23°C; for cold treatments, plants were transferred to 8°C for 3 weeks. cch1 mutants were ordered from the ABRC as well (seed stock: CS6499).
Antibody Production
Anti-CP31A
ClustalW alignments with all Arabidopsis cpRNP protein sequences were performed to find peptides, which are unique for CP31A and CP29A. A polyclonal antibody was generated by immunization of rabbits with a cocktail of two CP31A peptides: NH2-CEGRAIRVNVAEERPPRRGY-CONH2 (P1) and NH2-CVNKAAPRGSRPERAPRVYE-CONH2 (P2). The antibody was affinity purified using P2 immobilized onto a Sepharose 6B resin.
CP29A has a 32-nucleotide-long Ser- and Gly-rich sequence stretch, consisting of four repeats of SERGGGYG, which is unique not only within the cpRNP protein family, but among the whole Arabidopsis genome. Therefore, part of this sequence was used for antibody production. A polyclonal antibody eluate (in the following named anti-CP29A) was generated by immunization of rabbits with the CP29A peptide NH2-CGGYGSERGGGYGSER-CONH2.
Chloroplast Stroma Preparation
Intact chloroplasts were prepared from 2-week-old Arabidopsis plants according to Kunst (1998), with the following modifications: After homogenization, chloroplasts were pelleted at 500g for 6 min, resuspended in 2 mL resuspension buffer, loaded onto a 40%/80% Percoll gradient, and centrifuged for 30 min with breaks off. The lower of the two resulting bands, containing the intact chloroplasts, was collected, washed with 30 mL resuspension buffer, and repelleted at 500g for 6 min.
Chloroplasts then are disrupted in 200 μL extraction buffer (2 mM DTT, 200 mM KOAC, 30 mM HEPES, pH 8.0, 10 mM MgOAc, and proteinase inhibitor cocktail) per 20 g leaf material and pulled through a syringe (0.4 mm × 20 mm) 40 times.
Coimmunoprecipitation
Stroma protein (200 to 400 µg) in extraction buffer was diluted with one volume of coimmunoprecipitation (CoIP) buffer (150 mM NaCl, 20 mM Tris-HCl, 1 mM EDTA, 5 mM MgCl2, and 0.5% Nonidet P-40), incubated with 8 μL of affinity-purified anti-CP29A or affinity-purified anti-CP31A antibody for 1 h at 12 rpm, followed by 1 h rotation with 50 μL Dynabeads ProteinG (Invitrogen). Each CoIP was controlled by either using stroma of cp29a or cp31a null mutants, using preimmune serum of the appropriate antibodies, or using a monoclonal HA antibody.
Beads containing the immunoprecipitated protein and its bound RNAs were collected with a magnet, and supernatants were recovered and pellets were washed three times with CoIP buffer.
RIP-Chip Analyses
Tiling Microarray Construction
Overlapping PCR products covering the whole Arabidopsis chloroplast genome (see Supplemental Table 1 online for primers) were generated using a self-made Taq-polymerase and were purified via the QIAquick 96 PCR purification kit (Qiagen). A total of 500 ng of each PCR product was transferred into a 348-well plate, dried in a vacuum concentrator, and resuspended in 5 μL of 1 M betaine in 3× SSC (1× SSC is 0.15 M NaCl and 0.015 M sodium citrate). Each DNA was spotted on silanated glass slides (Vantage Silanated Amine Slides; CEL Associates) using an OMNIGRID ACCENT microarrayer (GeneMachines).
Oligonucleotide Microarray
The 50-nucleotide-long oligonucleotides representing selected RNA targets of CP29A and CP31A (see Supplemental Data Set 2 online) were used to generate a microarray as described previously (Zoschke et al., 2010).
RNA Extraction and Labeling
RNA from both pellet and supernatant fractions of immunoprecipitations was phenol/chloroform extracted after incubation in the presence of 1% SDS and 5 mM EDTA to allow dissociation of RNA-protein complexes. RNA was ethanol precipitated, washed with 75% ethanol, air-dried, and resuspended in 20 μL RNase-free water.
Labeling reactions were performed by adding 2 μL of 10× labeling buffer and 0.5 μL Cy5 or 1 μL Cy3 fluorescent dye (aRNA labeling kit; Kreatech Diagnostics) to 17.5 μL of pellet RNA or 17 μL of supernatant RNA, respectively. The mixture was incubated at 85°C for 20 min and stopped on ice. Unincorporated dye was removed according to the manufacturer’s instructions by the use of KREApure purification columns provided in the kit.
The labeled RNAs were concentrated in a vacuum centrifuge until the volume is reduced to 5 to 10 μL and mixed with 1 volume of hybridization buffer (25 parts formamide, 25 parts 20× SSC, and one part 10% SDS).
Microarray Hybridization
Microarrays were cross-linked at 250 mJ/cm2 in a UV cross-linker (GS Gene Linker; Bio-Rad) and blocked with a BSA buffer (1% BSA, 0.1% SDS, and 5× SSC) for 1 h to avoid unspecific binding of RNA to the slide surface. The labeled RNAs of pellet and supernatant (∼15 μL each) were mixed, pipetted onto the array, and sealed with a cover slip. Hybridization was performed overnight in a 42°C water bath in Corning microarray hybridization chambers. The slides were then soaked in 2× SSC and 0.2% SDS to remove the cover slip, washed in 1× SSC, 0.2× SSC, and 0.05× SSC for 8 min each on a horizontal shaker at 180 rpm. Finally, slides were dried for 2 min in a plate centrifuge at 1000 rpm and scanned using the ScanArrayGx Plus microarray scanner (Perkin-Elmer).
RNA Gel Blot Analyses
Total RNA was extracted with TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. For each RNA sample, a pool of at least three individual plants was used. For analyzing transcript accumulation under standard conditions, whole plants grown at 23°C for 2 weeks were harvested; for investigation of cold stress effects, plants were grown for 2 weeks at 23°C, and after an additional 18 d at 8°C, the leaf bases of the youngest five leaves were harvested for RNA extraction. Four micrograms of RNA per lane were separated on an agarose gel containing 1.2% formaldehyde and transferred to uncharged nylon membranes (Hybond N; GE Healthcare). After transfer, the blots were UV cross-linked (150 mJ/cm2) and then stained with methylene blue to check RNA integrity and equal loading.
Riboprobes were generated from PCR products via T7 polymerase (Fermentas) by in vitro transcription in the presence of 32P-UTP according to the manufacturer’s instructions. Primers used are listed in Supplemental Table 1 online. After preincubating the membranes in Church buffer (0.5 M sodium phosphate buffer [342 mM Na2HPO4 and 158 mM NaH2PO4], pH 7.2, and 7% SDS) for 1 h at 68°C, hybridizations were performed overnight at 68°C in the same buffer, followed by three 15-min washes in 0.2× SSC/0.1% SDS, 0.1× SSC/0.1% SDS, and 0.05× SSC/0.1% SDS, respectively. Signals were detected by autoradiography with the Personal Molecular Imager system (Bio-Rad).
Immunoblot Analyses
Proteins were separated by SDS-PAGE and transferred to Hybond-C Extra Nitrocellulose membranes (GE Healthcare). Integrity and loading of the proteins was tested by Ponceau S staining. Antibody hybridization was performed for 1 h in 2% skim milk powder in TBST (10 mM Tris-HCl, pH 7.5, 150 mM NaCl, and 0.1% Tween 20) for primary antibodies and in TBST for secondary horseradish peroxidase antibody (horseradish peroxidase coupled to anti-rabbit antibody).
Transcript End Mapping
The 3′-RACE was performed as described (Ruwe and Schmitz-Linneweber, 2012). Primers used are listed in Supplemental Table 1 online.
RNase Protection Assay
RNase protection was performed as described (Ruwe and Schmitz-Linneweber, 2012). The oligonucleotides for antisense RNA production are listed in Supplemental Table 1 online.
qRT-PCR
qRT-PCR to determine chloroplast transcript levels was performed as described previously (de Longevialle et al., 2008).
Editing Analyses
Total RNA was prepared from 3-week-old wild-type and cp29a mutants. cDNA was generated by reverse transcription using the SuperScript III first-strand synthesis system (Invitrogen). cDNAs harboring one or several editing sites were amplified by PCR and sequenced (Services in Molecular Biology). Excerpts of sequencing electropherograms for all 34 known RNA editing sites within the Arabidopsis chloroplast are shown with the edited C/T in the middle of the triplets.
Dot Blot Hybridization
RNAs from both CoIP pellet and supernatant fraction were used for dot blot analyses. Twenty microliters of RNA was mixed with 300 μL denaturing buffer (66% deionized formamide, 7.7% formaldehyde, and 1.3× MOPS buffer, pH 7), incubated for 15 min at 75°C, and applied to a nylon membrane (Hybond N) using the Bio-Dot SF microfiltration apparatus (Bio-Rad). Membranes were washed with 10× SSC, UV cross-linked, and stained with methylene blue. Hybridization with riboprobes was performed as described for RNA gel blot analyses.
Run-on Transcription
Run-on transcription was performed as previously described (Zubo and Kusnetsov, 2008).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: cch/Gun5, At5g13630; CP29A, At3g53460; CP31A, At4g24770; CP31B, At5g50250; 18S rRNA, At2g01010; and Arabidopsis chloroplast genome, NC_000932.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. CP29A and CP31A Protein Levels Are Not Altered by the Presence or Absence of CP31A and CP29A, Respectively.
Supplemental Figure 2. Immunoprecipitation of CP29A and CP31A.
Supplemental Figure 3. Excerpt of a Representative CP31A RIP-Chip.
Supplemental Figure 4. Dot Blot Analyses for Validation of RIP-Chip Data.
Supplemental Figure 5. Editing in cp29A.
Supplemental Figure 6. Plastid Transcript Level in Two cp29a Mutants.
Supplemental Figure 7. Splicing Analyses in cpRNP Mutants.
Supplemental Figure 8. Light Stress Phenotypes
Supplemental Figure 9. Phenotypes of cp29a and cp31a Single and Double Mutants Germinated under Cold Stress Conditions.
Supplemental Figure 10. Run-on Transcription Assay of Selected Chloroplast Genes in a cp29a Knockout Mutant.
Supplemental Figure 11. Transcript Accumulation of Selected NEP-Dependent mRNAs under Standard (23°C) and Cold Stress (8°C) Conditions.
Supplemental Figure 12. Analysis of Chloroplast RNA Editing in cpRNP Mutants.
Supplemental Table 1. Oligonucleotides.
Supplemental Table 2. qRT-PCR Data for cp29a Mutants.
Supplemental Table 3. Run-on Data.
Supplemental Table 4. Splicing Analysis in cpRNP Mutants by qRT-PCR.
Supplemental Data Set 1. RIP-Chip Data for Figure 2.
Supplemental Data Set 2. RIP-Chip Data for Figure 6.
Supplementary Material
Acknowledgments
We thank Etienne Dellanoy for support with qRT-PCR analyses and Masha V. Yamburenko for providing technical help and Col reference experiments for the run-on experiments. This work was funded by Deutsche Forschungsgemeinschaft grants to M.T. (Ti 605/2-1) and C.S.-L. (SCHM 1698/4-1).
AUTHOR CONTRIBUTIONS
C.K. performed research. H.R. performed research. S.G. performed RNA editing analyses. M.T. designed research. I.S. contributed access and expertise for qRT-PCR platforms. C.S.-L. designed research and wrote the article.
Glossary
- RBP
RNA binding protein
- PPR
pentatricopeptide repeat
- cpRNP
chloroplast ribonucleoprotein
- RRM
RNA recognition motif
- NDH
NAD dehydrogenase
- HA
to be defined
- qRT-PCR
quantitative RT-PCR
- UTR
untranslated region
- IR
inverted repeat
- SSC
small-single-copy
- RACE
rapid amplification of cDNA ends
- sRNA
to be defined
- hnRNP
to be defined
- PEP
to be defined
- CoIP
coimmunoprecipitation
References
- Barkan A. (1993). Nuclear mutants of maize with defects in chloroplast polysome assembly have altered chloroplast RNA metabolism. Plant Cell 5: 389–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A. (2011). Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 155: 1520–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beick S., Schmitz-Linneweber C., Williams-Carrier R., Jensen B., Barkan A. (2008). The pentatricopeptide repeat protein PPR5 stabilizes a specific tRNA precursor in maize chloroplasts. Mol. Cell. Biol. 28: 5337–5347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho W.K., Geimer S., Meurer J. (2009). Cluster analysis and comparison of various chloroplast transcriptomes and genes in Arabidopsis thaliana. DNA Res. 16: 31–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churin Y., Hess W.R., Börner T. (1999). Cloning and characterization of three cDNAs encoding chloroplast RNA-binding proteins from barley (Hordeum vulgare L.): Differential regulation of expression by light and plastid development. Curr. Genet. 36: 173–181 [DOI] [PubMed] [Google Scholar]
- Cléry A., Blatter M., Allain F.H. (2008). RNA recognition motifs: Boring? Not quite. Curr. Opin. Struct. Biol. 18: 290–298 [DOI] [PubMed] [Google Scholar]
- de Longevialle A.F., Hendrickson L., Taylor N.L., Delannoy E., Lurin C., Badger M., Millar A.H., Small I. (2008). The pentatricopeptide repeat gene OTP51 with two LAGLIDADG motifs is required for the cis-splicing of plastid ycf3 intron 2 in Arabidopsis thaliana. Plant J. 56: 157–168 [DOI] [PubMed] [Google Scholar]
- Dreyfuss G., Kim V.N., Kataoka N. (2002). Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell Biol. 3: 195–205 [DOI] [PubMed] [Google Scholar]
- Fleischmann T.T., Scharff L.B., Alkatib S., Hasdorf S., Schöttler M.A., Bock R. (2011). Nonessential plastid-encoded ribosomal proteins in tobacco: A developmental role for plastid translation and implications for reductive genome evolution. Plant Cell 23: 3137–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grennan A.K., Ort D.R. (2007). Cool temperatures interfere with D1 synthesis in tomato by causing ribosomal pausing. Photosynth. Res. 94: 375–385 [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz P.T., Allison L.A., Maliga P. (1997). The two RNA polymerases encoded by the nuclear and the plastid compartments transcribe distinct groups of genes in tobacco plastids. EMBO J. 16: 4041–4048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes R., Kudla J., Schuster G., Gabay L., Maliga P., Gruissem W. (1996). Chloroplast mRNA 3′-end processing by a high molecular weight protein complex is regulated by nuclear encoded RNA binding proteins. EMBO J. 15: 1132–1141 [PMC free article] [PubMed] [Google Scholar]
- Hirose T., Sugiura M. (2001). Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: Development of a chloroplast in vitro RNA editing system. EMBO J. 20: 1144–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins W.G., Elfman B. (1984). Temperature-induced chloroplast ribosome deficiency in virescent maize. J. Hered. 75: 207–211 [Google Scholar]
- Hotto A.M., Schmitz R.J., Fei Z., Ecker J.R., Stern D.B. (2011). Unexpected diversity of chloroplast noncoding RNAs as revealed by deep sequencing of the Arabidopsis transcriptome. G3 1: 559–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J., Kück U. (2011). Function of chloroplast RNA-binding proteins. Cell. Mol. Life Sci. 68: 735–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keene J.D. (2007). RNA regulons: Coordination of post-transcriptional events. Nat. Rev. Genet. 8: 533–543 [DOI] [PubMed] [Google Scholar]
- Kenan D.J., Query C.C., Keene J.D. (1991). RNA recognition: Towards identifying determinants of specificity. Trends Biochem. Sci. 16: 214–220 [DOI] [PubMed] [Google Scholar]
- Kleffmann T., von Zychlinski A., Russenberger D., Hirsch-Hoffmann M., Gehrig P., Gruissem W., Baginsky S. (2007). Proteome dynamics during plastid differentiation in rice. Plant Physiol. 143: 912–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krecic A.M., Swanson M.S. (1999). hnRNP complexes: Composition, structure, and function. Curr. Opin. Cell Biol. 11: 363–371 [DOI] [PubMed] [Google Scholar]
- Kroeger T.S., Watkins K.P., Friso G., van Wijk K.J., Barkan A. (2009). A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. USA 106: 4537–4542 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunst L. (1998). Preparation of physiologically active chloroplasts from Arabidopsis. Methods Mol Biol. 82: 43–48 [DOI] [PubMed] [Google Scholar]
- Kusumi K., Sakata C., Nakamura T., Kawasaki S., Yoshimura A., Iba K. (2011). A plastid protein NUS1 is essential for build-up of the genetic system for early chloroplast development under cold stress conditions. Plant J. 68: 1039–1050 [DOI] [PubMed] [Google Scholar]
- Li Y.Q., Sugiura M. (1990). Three distinct ribonucleoproteins from tobacco chloroplasts: Each contains a unique amino terminal acidic domain and two ribonucleoprotein consensus motifs. EMBO J. 9: 3059–3066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y.Q., Sugiura M. (1991). Nucleic acid-binding specificities of tobacco chloroplast ribonucleoproteins. Nucleic Acids Res. 19: 2893–2896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Rodermel S.R., Yu F. (2010). A var2 leaf variegation suppressor locus, SUPPRESSOR OF VARIEGATION3, encodes a putative chloroplast translation elongation factor that is important for chloroplast development in the cold. BMC Plant Biol. 10: 287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorković Z.J., Barta A. (2002). Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res. 30: 623–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maris C., Dominguez C., Allain F.H. (2005). The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. FEBS J. 272: 2118–2131 [DOI] [PubMed] [Google Scholar]
- Maruyama K., Sato N., Ohta N. (1999). Conservation of structure and cold-regulation of RNA-binding proteins in cyanobacteria: probable convergent evolution with eukaryotic glycine-rich RNA-binding proteins. Nucleic Acids Res. 27: 2029–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mentzen W.I., Wurtele E.S. (2008). Regulon organization of Arabidopsis. BMC Plant Biol. 8: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millerd A., Goodchild D.J., Spencer D. (1969). Studies on a maize mutant sensitive to low temperature II. Chloroplast structure, development, and physiology. Plant Physiol. 44: 567–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki N., Brusslan J.A., Larkin R., Nagatani A., Chory J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. USA 98: 2053–2058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T., Ohta M., Sugiura M., Sugita M. (1999). Chloroplast ribonucleoproteins are associated with both mRNAs and intron-containing precursor tRNAs. FEBS Lett. 460: 437–441 [DOI] [PubMed] [Google Scholar]
- Nakamura T., Ohta M., Sugiura M., Sugita M. (2001). Chloroplast ribonucleoproteins function as a stabilizing factor of ribosome-free mRNAs in the stroma. J. Biol. Chem. 276: 147–152 [DOI] [PubMed] [Google Scholar]
- Nishimura K., Ashida H., Ogawa T., Yokota A. (2010). A DEAD box protein is required for formation of a hidden break in Arabidopsis chloroplast 23S rRNA. Plant J. 63: 766–777 [DOI] [PubMed] [Google Scholar]
- Ohta M., Sugita M., Sugiura M. (1995). Three types of nuclear genes encoding chloroplast RNA-binding proteins (cp29, cp31 and cp33) are present in Arabidopsis thaliana: Presence of cp31 in chloroplasts and its homologue in nuclei/cytoplasms. Plant Mol. Biol. 27: 529–539 [DOI] [PubMed] [Google Scholar]
- Petersen K., Schöttler M.A., Karcher D., Thiele W., Bock R. (2011). Elimination of a group II intron from a plastid gene causes a mutant phenotype. Nucleic Acids Res. 39: 5181–5192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz J., Bayraktar O.A., Prikryl J., Barkan A. (2009). Site-specific binding of a PPR protein defines and stabilizes 5′ and 3′ mRNA termini in chloroplasts. EMBO J. 28: 2042–2052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab S., Toth Z., de Groot C., Stamminger T., Hoth S. (2006). ABA-responsive RNA-binding proteins are involved in chloroplast and stromule function in Arabidopsis seedlings. Planta 224: 900–914 [DOI] [PubMed] [Google Scholar]
- Rogalski M., Ruf S., Bock R. (2006). Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 34: 4537–4545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogalski M., Schöttler M.A., Thiele W., Schulze W.X., Bock R. (2008). Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20: 2221–2237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruwe H., Kupsch C., Teubner M., Schmitz-Linneweber C. (2011). The RNA-recognition motif in chloroplasts. J. Plant Physiol. 168: 1361–1371 [DOI] [PubMed] [Google Scholar]
- Ruwe H., Schmitz-Linneweber C. (2012). Short non-coding RNA fragments accumulating in chloroplasts: footprints of RNA binding proteins? Nucleic Acids Res. 40: 3106–3116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R., Barkan A. (2005). RNA immunoprecipitation and microarray analysis show a chloroplast pentatricopeptide repeat protein to be associated with the 5′ region of mRNAs whose translation it activates. Plant Cell 17: 2791–2804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz-Linneweber C., Williams-Carrier R.E., Williams-Voelker P.M., Kroeger T.S., Vichas A., Barkan A. (2006). A pentatricopeptide repeat protein facilitates the trans-splicing of the maize chloroplast rps12 pre-mRNA. Plant Cell 18: 2650–2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuster G., Gruissem W. (1991). Chloroplast mRNA 3′ end processing requires a nuclear-encoded RNA-binding protein. EMBO J. 10: 1493–1502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern D.B., Goldschmidt-Clermont M., Hanson M.R. (2010). Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 61: 125–155 [DOI] [PubMed] [Google Scholar]
- Tillich M., Beick S., Schmitz-Linneweber C. (2010). Chloroplast RNA-binding proteins: repair and regulation of chloroplast transcripts. RNA Biol. 7: 172–178 [DOI] [PubMed] [Google Scholar]
- Tillich M., Hardel S.L., Kupsch C., Armbruster U., Delannoy E., Gualberto J.M., Lehwark P., Leister D., Small I.D., Schmitz-Linneweber C. (2009). Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. USA 106: 6002–6007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuhisa J.G., Vijayan P., Feldmann K.A., Browse J.A. (1998). Chloroplast development at low temperatures requires a homolog of DIM1, a yeast gene encoding the 18S rRNA dimethylase. Plant Cell 10: 699–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B.C., Wang H.X., Feng J.X., Meng D.Z., Qu L.J., Zhu Y.X. (2006). Post-translational modifications, but not transcriptional regulation, of major chloroplast RNA-binding proteins are related to Arabidopsis seedling development. Proteomics 6: 2555–2563 [DOI] [PubMed] [Google Scholar]
- Yamori W., Noguchi K., Hikosaka K., Terashima I. (2010). Phenotypic plasticity in photosynthetic temperature acclimation among crop species with different cold tolerances. Plant Physiol. 152: 388–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhelyazkova P., Hammani K., Rojas M., Voelker R., Vargas-Suárez M., Börner T., Barkan A. (2012). Protein-mediated protection as the predominant mechanism for defining processed mRNA termini in land plant chloroplasts. Nucleic Acids Res. 40: 3092–3105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoschke R., Nakamura M., Liere K., Sugiura M., Börner T., Schmitz-Linneweber C. (2010). An organellar maturase associates with multiple group II introns. Proc. Natl. Acad. Sci. USA 107: 3245–3250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubo Y. O., et al. (2008). Cytokinin stimulates chloroplast transcription in detached barley leaves. Plant Physiol 148: 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.