Abstract
The nematocyst is a complex intracellular structure unique to Cnidaria. When triggered to discharge, the nematocyst explosively releases a long spiny, tubule that delivers an often highly venomous mixture of components. The box jellyfish, Chironex fleckeri, produces exceptionally potent and rapid-acting venom and its stings to humans cause severe localized and systemic effects that are potentially life-threatening. In an effort to identify toxins that could be responsible for the serious health effects caused by C. fleckeri and related species, we used a proteomic approach to profile the protein components of C. fleckeri venom. Collectively, 61 proteins were identified, including toxins and proteins important for nematocyte development and nematocyst formation (nematogenesis). The most abundant toxins identified were isoforms of a taxonomically restricted family of potent cnidarian proteins. These toxins are associated with cytolytic, nociceptive, inflammatory, dermonecrotic and lethal properties and expansion of this important protein family goes some way to explaining the destructive and potentially fatal effects of C. fleckeri venom. Venom proteins and their post-translational modifications (PTMs) were further characterized using toxin-specific antibodies and phosphoprotein/glycoprotein-specific stains. Results indicated that glycosylation is a common PTM of the toxin family while a lack of cross-reactivity by toxin-specific antibodies infers there is significant divergence in structure and possibly function among family members. This study provides insight into the depth and diversity of protein toxins produced by harmful box jellyfish and represents the first description of a cubozoan jellyfish venom proteome.
Introduction
Cubozoan jellyfish, commonly known as box jellyfish, are members of the Phylum Cnidaria. Cnidarians represent some of the most ancient metazoans ( 500 million years old) and their defining feature is the nematocyst (cnidocyst); a nonliving organelle housed within a specialised cell, the nematocyte (cnidocyte). The nematocyst is formed within a large post-Golgi vesicle [1] and comprises a rigid proteinaceous capsule that contains a long spiny tubule and a complex mixture of proteins (often toxins) and other small molecular weight compounds. Upon stimulation of the nematocyte's sensory receptor (cnidocil), the nematocyst discharges explosively, expelling the tubule at high speed and releasing the capsular contents [2]. A number of distinct morphological forms of nematocysts are used for a variety of purposes, including prey capture, defence or locomotory functions [3]–[5].
500 million years old) and their defining feature is the nematocyst (cnidocyst); a nonliving organelle housed within a specialised cell, the nematocyte (cnidocyte). The nematocyst is formed within a large post-Golgi vesicle [1] and comprises a rigid proteinaceous capsule that contains a long spiny tubule and a complex mixture of proteins (often toxins) and other small molecular weight compounds. Upon stimulation of the nematocyte's sensory receptor (cnidocil), the nematocyst discharges explosively, expelling the tubule at high speed and releasing the capsular contents [2]. A number of distinct morphological forms of nematocysts are used for a variety of purposes, including prey capture, defence or locomotory functions [3]–[5].
C. fleckeri is the largest and most dangerous cubozoan jellyfish to humans and its occurrence in the tropical coastal waters of Australia is a problem, particularly in summer. Nematocysts containing potent venom are prolific along the tentacles of C. fleckeri and cause painful and potentially life-threatening stings to humans. Symptoms of major C. fleckeri stings include excruciating pain, rapid acute cutaneous inflammation, dermonecrosis, permanent scarring, hypertension, hypotension, shock, dyspnoea, impaired consciousness, cardiac dysfunction and pulmonary oedema (reviewed in [6]). The onset of symptoms is extremely rapid [7] and in severe cases, death from pulmonary and/or cardiac failure can occur within minutes [8]. At least 70 deaths due to C. fleckeri envenoming have occurred in Australia and numerous deaths from related species have been reported in the Philippines, Maldives islands, Japan, Papua New Guinea, South India, Java, Malaysia and Gulf of Thailand [9].
Several biological activities are associated with cubozoan venoms [6]. In particular, C. fleckeri whole tentacle and nematocyst extracts elicit lethal, dermonecrotic, nociceptive, cytotoxic, neurotoxic, myotoxic, cardiotoxic, haemodynamic and haemolytic effects [6]. Yet, despite the medical and pharmacological significance of box jellyfish venoms to humans, their compositions have not been extensively explored. To date only two C. fleckeri venom proteins, CfTX-1 and -2, have been formally identified; potent haemolysins that share sequence similarity to toxins from four related cubozoan species [10], [11]. However, the broad range of bioactivities in cubozoan venoms suggests a wealth of additional venom components remain to be found.
In this work we describe the proteomic characterisaton of C. fleckeri venom to identify proteins that may contribute to the deleterious effects of box jellyfish stings in humans. A major challenge in the study was the paucity of nucleotide sequence coverage for C. fleckeri or closely related species. Although the genomes of Hydra magnipapillata and the sea anemone, Nematostella vectensis, have been described, sequences specific to cubozoans in GenBank are limited (only 74 non-redundant, non-mitochondrial protein sequences). Accordingly, in the absence of genomic or transcriptomic data, we utilised a strategy combining traditional spectral matching using Mascot with de novo protein sequencing from tandem mass spectrometry (MS/MS) and homology searches. Using these approaches we identified 67 proteins from the nematocysts of C. fleckeri including toxins and proteins involved in nematocyst and nematocyte development. We report the expansion of an important family of toxins and examine their post-translational modifications and cross-reactivity with toxin-specific antibodies. Our study represents the first venom proteome of a cubozoan jellyfish and provides insight into the depth and diversity of proteins within the unique cnidarian attribute, the nematocyst.
Results and Discussion
Identification of C. fleckeri nematocyst proteins
C. fleckeri venom (CFV) was purified from nematocysts purified in a discontinuous Percoll gradient (Figure 1). Prior to MS/MS analysis, the CFV was separated by SDS-PAGE (Figure 2) and in-gel tryptic digests were performed on 40 gel fragments. In addition to SDS-PAGE, tryptic peptides from total CFV were subjected to OFFGEL electrophoresis (OGE). There are only 21 C. fleckeri protein sequences in GenBank and only 186 for Cubozoa (as of 13th August, 2012). Accordingly, a strategy incorporating both spectral searches using Mascot, with false discovery analysis conducted using X! Tandem and Scaffold, was combined with de novo analysis of generated spectra using PEAKS. Two different PEAKS searches were conducted, the first, analogous to a Mascot search, used only exact peptide matches derived from high quality de novo sequence tags. This was used to confirm and extend Mascot searches. The second search allowed for matching of high quality de novo sequence tags to homologous sequences in the target database, thus permitting the detection of proteins with similar sequence (see Figure 3 for a representative spectra). Both PEAKS search strategies utilised decoy databases to derive false positive rates. Following these searches, 46 proteins were identified using Mascot and PEAKS at a 0% FDR for Mascot identifications and a 1% FDR for PEAKS (Table 1, Information S1, S2 and S5). De novo homology searches conducted using PEAKS resulted in the identification of 46 proteins at a FDR of 1%, 16 of which had not been identified in the first search (Table 2, Information S3, S4, S5). Nine of these were single peptide identifications and annotated spectra for these are provided in Information S6. In total, sixty-one non-redundant protein identifications were made at a high level of significance using the two search strategies.
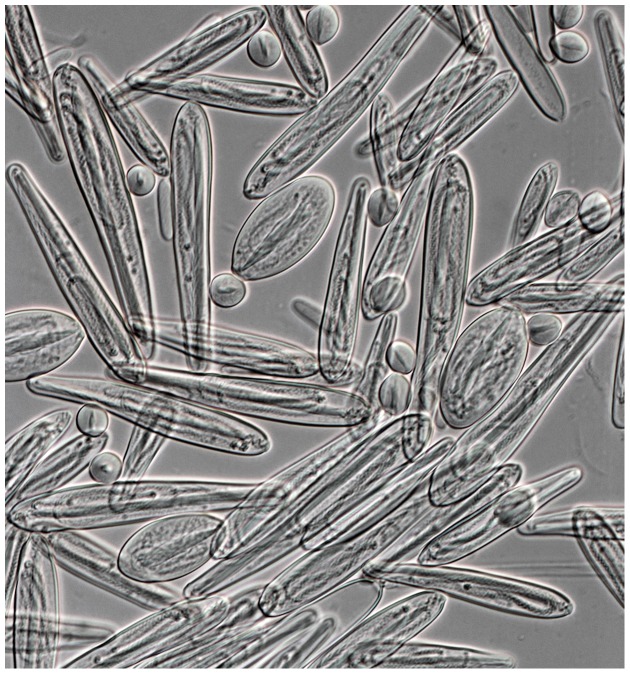
Figure 1. Chironex fleckeri nematocysts.
Light microscopy image of nematocysts isolated from C. fleckeri tentacles (magnification 400×).
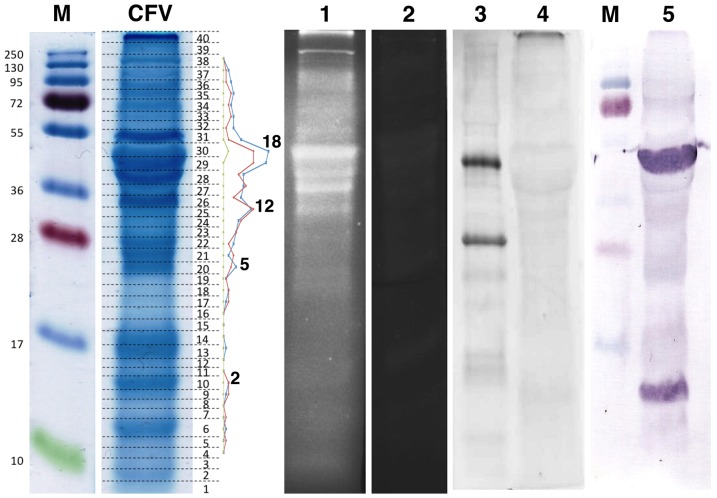
Figure 2. C. fleckeri venom proteins.
Total protein from CFV in a Coomassie-stained 15% SDS-PAGE gel. Lanes were divided into 40 gel slices (dotted lines) and subjected to in-gel tryptic digest before LC-MS/MS analysis. Markers (M) are indicated and the numbering system used for gel slices. CFV proteins separated using SDS-PAGE and stained in-gel with fluorescent dyes reactive to glycans or phosphate groups (Lanes 1–4). Glycan analysis showed fluorescence in bands corresponding to CfTX proteins (Lane 1) and no fluorescence in the negative control (Lane 2). No phosphorylation was observed except in band 40 (Lane 4; positive control in Lane 3). Western blot analysis using polyclonal antibodies for CfTX-1 and -2 showed hybridisation in two bands corresponding to the highest scoring Mascot identifications for these proteins and in the region corresponding to approximately 12 kDa (lane 5). No other bands were positive for these proteins despite their identification in MS/MS analysis. The spectral counts for proteins from this toxin family identified using Mascot are displayed adjacent to the band numbering (blue, red and green lines for CfTX-1, CfTX-2 and CqTX-A resp.). Actual spectral counts for a selection of CfTX-1 points are shown for reference.
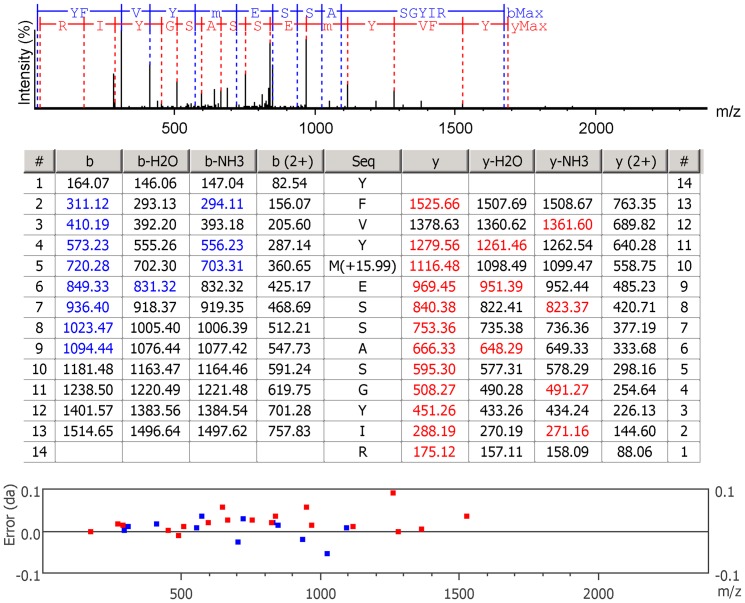
Figure 3. Representative example of a de novo sequence match to CaTX-A using a homology search.
Representative spectra from a de novo match to the toxin protein CaTX-A. The top panel shows the spectrum annotated with the y and b-ion series, the middle panel shows fragment ions detected and the bottom panel the errors, in daltons, for each fragment ion.
Table 1. Protein identifications using Mascot and PEAKS.
| Scaffold | Peaks | SP | H | ||||||
| ID | Description | USC | CO | −10lgP | SC | USC | CO | ||
| Toxins | |||||||||
| 160380600 | Toxin CfTX-1 | 26 | 41 | 359.39 | 42 | 21 | 54 | + | + |
| 160380601 | Toxin CfTX-2 | 16 | 43 | 327.90 | 31 | 23 | 63 | + | + |
| 18146993 | Toxin CqTX-A | 2 | 23 | 224.45 | 16 | 1 | 27 | + | + |
| Nematogalectins | |||||||||
| 219233028 | Nematogalectin-related | - | - | 148.12 | 3 | 1 | 14 | + | + |
| 156401683 | Nematogalectin-related | 5 | 17 | 124.05 | 4 | 4 | 12 | + | + |
| 308210726 | Nematogalectin | - | - | 93.89 | 2 | 1 | 14 | + | + |
| Structural | |||||||||
| 159254 | Actin | 12 | 38 | 190.23 | 7 | 1 | 28 | − | + |
| 26522784 | Actin | - | - | 194.30 | 9 | 1 | 23 | − | + |
| 282471169 | Actin | - | - | 137.66 | 3 | 1 | 16 | − | + |
| 162122963 | Actin | - | - | 108.89 | 3 | 1 | 22 | − | + |
| 294372631 | Non-muscle actin | - | - | 158.11 | 4 | 1 | 25 | − | + |
| 156223974 | Alpha tubulin | 16 | 44 | 77.48 | 4 | 1 | 12 | − | + |
| 221129327 | Beta tubulin | 16 | 39 | 206.15 | 15 | 3 | 45 | − | + |
| 221129323 | Beta tubulin | - | - | 129.01 | 5 | 1 | 11 | − | + |
| 156369515 | Tubulin beta-2C chain | - | - | 89.33 | 3 | 3 | 14 | − | + |
| 225547767 | Clathrin heavy chain | 2 | 7 | - | - | - | - | − | − |
| 221130531 | Clathrin heavy chain 1 | 2 | 2 | 124.69 | 3 | 3 | 2 | − | − |
| 221107857 | Histone H2B-1 | - | - | 69.50 | 2 | 1 | 23 | − | + |
| 156219637 | Histone H2B | - | - | 126.82 | 6 | 1 | 30 | − | + |
| 156219675 | Histone H3.2-like | - | - | 81.04 | 3 | 2 | 11 | − | + |
| 156198376 | Histone H2A | 2 | 12 | 137.98 | 7 | 3 | 36 | − | + |
| 163678147 | Histone 2 | - | - | 62.91 | 2 | 1 | 15 | − | + |
| Heat shock proteins | |||||||||
| 221132017 | Heat shock protein | 3 | 4 | 88.74 | 2 | 1 | 10 | + | − |
| 262478028 | 90 heat-shock protein | 2 | 12 | - | - | - | - | − | + |
| Oxido-reductive | |||||||||
| 100913623 | Cytochrome c oxidase subunit II | 2 | 11 | - | - | - | - | − | − |
| 156364605 | ATP synthase beta subunit | 2 | 10 | - | - | - | - | − | + |
| 156213313 | ATP synthase | 5 | 18 | - | - | - | - | − | + |
| 162106843 | ATP synthase subunit alpha | - | - | 103.04 | 3 | 3 | 13 | − | + |
| 82862750 | V-type proton ATPase | - | - | 52.02 | 2 | 1 | 20 | − | + |
| 221111162 | Thioredoxin peroxidase | 2 | 10 | - | - | - | - | − | + |
| Dickkopf | |||||||||
| 221115495 | Dickkopf protein 3 | 4 | 38 | 216.04 | 11 | 9 | 38 | + | + |
| 37498690 | Dickkopf-3 related protein | 5 | 27 | 237.58 | 12 | 11 | 53 | + | + |
| Proteases | |||||||||
| 221116853 | Neprilysin | 3 | 4 | 116.69 | 2 | 1 | 3 | + | + |
| 221128287 | Neprilysin | - | - | 102.93 | 2 | 1 | 3 | + | + |
| 221132105 | Endothelin-converting enzyme 2 | - | - | 85.94 | 2 | 2 | 3 | + | + |
| 156386196 | Endothelin-converting enzyme 2-like | - | - | 56.17 | 2 | 2 | 3 | + | + |
| Miscellaneous | |||||||||
| 221118656 | DNA-directed RNA polymerase II | 2 | 1 | 189.76 | 9 | 9 | 3 | + | + |
| 156223696 | Ras-related protein Rab-35 | 2 | 12 | - | - | - | - | − | − |
| 239736186 | Elongation factor-1 alpha | 8 | 22 | 64.17 | 2 | 2 | 5 | − | − |
| 221129496 | DNA-directed RNA polymerase, omega | - | - | 120.92 | 2 | 2 | 1 | − | + |
| 221119922 | Gamma-glutamyltranspeptidase 1 | - | - | 66.25 | 2 | 2 | 2 | + | + |
| 221120305 | Gamma-glutamyl hydrolase | - | - | 67.12 | 2 | 2 | 3 | − | + |
| Unknown | |||||||||
| 221123368 | Hypothetical protein [Hydra magnipapillata] | 2 | 2 | 135.12 | 4 | 4 | 3 | + | − |
| 221113955 | Hypothetical protein [Hydra magnipapillata] | 2 | 14 | 124.91 | 2 | 2 | 14 | + | − |
| 221116631 | Predicted protein [Hydra magnipapillata] | 2 | 1 | 107.93 | 2 | 2 | 1 | − | − |
Protein identifications using Mascot and PEAKS. Abbreviations used: ID — identification number corresponding to custom database supplied as S5; USC — number of unique and significant spectra contributing to the identification; SC — total number of significant spectra contributing to the identification; CO — percent cover; and −10lgP — the PEAKS probability based score. A ‘+’ in the SP column denotes the presence of a signal sequence and a ‘+’ in the ‘H’ column denotes the identification of a similar protein in the H. magnipapillata venom proteome.
Table 2. Additional proteins identified using a homology search of de novo sequence tags in PEAKS.
| ID | −10lgP | CO | SC | USC | Description | SP | H |
| Toxins | |||||||
| 20137964 | 45.69 | 3 | 1 | 1 | Toxin CaTX-A [Carybdea alata] | + | + |
| 20137965 | 56.47 | 4 | 1 | 1 | Toxin CrTX-A [Carybdea rastonii] | + | + |
| 260066272 | 46.78 | 2 | 1 | 1 | Cytotoxin A isoform 1 [Malo kingi] | − | + |
| Nematogalectins | |||||||
| 306408168 | 93.89 | 13 | 2 | 1 | Nematogalectin B [Hydra magnipapillata] | + | + |
| Structural | |||||||
| 221107145 | 90.25 | 10 | 2 | 2 | Collagen alpha-1(XII) chain [Hydra magnipapillata] | + | + |
| 156388143 | 61.02 | 3 | 1 | 1 | Collagen alpha-2(IX) chain [Nematostella vectensis] | − | + |
| 221121786 | 54.27 | 1 | 1 | 1 | Procollagen type XIV alpha 1 [Hydra magnipapillata] | − | + |
| Proteases | |||||||
| 156400736 | 54.28 | 8 | 2 | 2 | Carboxypeptidase [Nematostella vectensis] | − | + |
| 156226146 | 39.08 | 3 | 1 | 1 | Carboxypeptidase D [Nematostella vectensis] | − | + |
| Miscellaneous | |||||||
| 156401103 | 72.46 | 8 | 2 | 2 | ADP,ATP carrier protein-like [Nematostella vectensis] | − | − |
| 221112215 | 59.92 | 2 | 1 | 1 | Gamma-glutamyl transferase 1a [Hydra magnipapillata] | − | + |
| 221116631 | 119.78 | 1 | 3 | 3 | Viral A-type inclusion protein | − | − |
| 219231428 | 53.83 | 6 | 1 | 1 | Tyrosine kinase receptor [Anemonia viridis] | − | − |
| 223649562 | 34.95 | 5 | 1 | 1 | Nematoblast-specific protein nb012a [Hydra oligactis] | + | − |
| Unknown | |||||||
| 221116853 | 116.69 | 3 | 2 | 1 | Predicted protein [Hydra magnipapillata] | + | − |
| 50698431 | 60.51 | 17 | 2 | 2 | Hypothetical protein [Hydractinia echinata] | − | − |
In total 46 proteins were identified in homology searches, 17 of which were not identified in the Mascot and PEAKS searches. Abbreviations used: ID — identification number corresponding to custom database supplied as S5; −10lgP — PEAKS probability score; CO — percent cover; SC — total number of significant spectra contributing to the identification; USC — number of unique and significant spectra contributing to the identification. A ‘+’ in the SP column denotes the presence of a predicted signal sequence using SignalP and a ‘+’ in the ‘H’ column denotes the identification of a similar protein in the H. magnipapillata venom proteome. For complete list of proteins identified in de novo searches see Information S3.
Overall composition of CFV proteins
When categorised into functional groupings (Table 1 and 2) the most abundantly identified proteins reflect the known composition of the nematocyst, with toxins, collagens, dickkopf-3 proteins and nematogalectins all identified. The most commonly identified proteins were structural in nature, reflecting the composition of the nematocyst capsule, which is primarily composed of mini-collagens [5], and the tubule in which nematogalectin is a major component [12]. The detection of numerous structural proteins in C. fleckeri venom can be explained by the chemical method used for venom extraction. Dithiothreitol, a strong reducing agent, was used to partially disintegrate the nematocyst capsule and cause venom release, so it is likely that a proportion of capsular components and other structural proteins were solubilised during this process. In Hydra, a number of genes have been found to be specifically expressed in the nematocyte, including those encoding for toxins, proteins involved in the assembly of the nematocyst capsule, tubule and spines, as well as proteins associated with the cnidocil of the nematocyte [13], [14]. A variety of these proteins were also identified in CFV, including tubulin,  -glutamyltransferase and the nematogalectins. As found in proteomic studies of Hydra
[15], the proteins identified here are a mixture of secreted and non-secreted proteins with only 38% containing a classical secretory signal peptides targeting to the general secretory pathway. Many of these non-secreted proteins were likely present during nematocyst formation and do not have a specific function in the venom, although they may play an important role in nematocyst formation. Five proteins were identified that had no homology to proteins outside Cnidaria, three of which contained signal sequences, suggesting that some of the known bioactivities of CFV may be mediated by proteins unique to the phylum. A comparison of the protein families identified in this work with the recently published H. magnipapillata venom proteome showed that 75% of the proteins identified in the CFV were also present in the Hydra nematocyst.
-glutamyltransferase and the nematogalectins. As found in proteomic studies of Hydra
[15], the proteins identified here are a mixture of secreted and non-secreted proteins with only 38% containing a classical secretory signal peptides targeting to the general secretory pathway. Many of these non-secreted proteins were likely present during nematocyst formation and do not have a specific function in the venom, although they may play an important role in nematocyst formation. Five proteins were identified that had no homology to proteins outside Cnidaria, three of which contained signal sequences, suggesting that some of the known bioactivities of CFV may be mediated by proteins unique to the phylum. A comparison of the protein families identified in this work with the recently published H. magnipapillata venom proteome showed that 75% of the proteins identified in the CFV were also present in the Hydra nematocyst.
C. fleckeri toxin proteins and isoforms
A large number of toxic effects are attributed to C. fleckeri venom, but only two C. fleckeri toxins, CfTX-1 and -2, have been identified through peptide and cDNA sequencing [10], [11]. These proteins are related to box jellyfish toxins CrTX-A, CaTX-A and CqTX-A isolated from the venoms of Carybdea rastonii
[16], Carybdea alata
[17] and Chironex yamaguchii
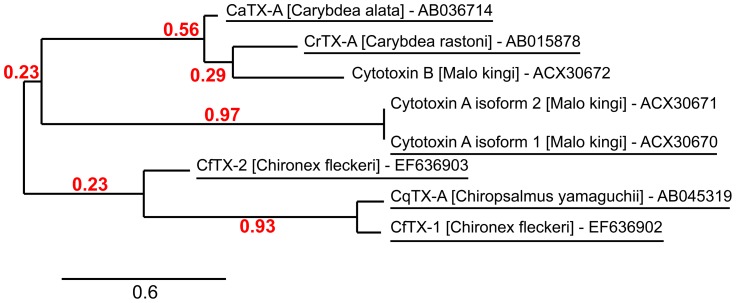
[18] (as Chiropsalmus quadrigatus; renamed 2009 [19]), respectively, as well as Cytotoxin A (isoforms 1 and 2) and Cytotoxin B (partial sequence), retrieved from a Malo kingi tentacle cDNA library [20] (see also [21] for disputed species identification). Using Mascot, three members of this family were identified, CfTX-1 and -2 and a homologue of CqTX-A (Table 1. A further three homologues of CaTX-A, CrTX-A and Cytotoxin A isoform 1, from M. kingi, were identified on the basis of homology (Table 2). Members of this toxin family are potently haemolytic and cause pain, inflammation, dermonecrosis and death in experimental animals [10], [11], [16]–[18], suggesting the toxins play an important functional role in box jellyfish envenoming. Computational analyses of the toxin sequences point to a pore-forming mechanism of action due to predictions of common transmembrane spanning regions and weak structural similarities to pore-forming insecticidal  -endotoxins [6]. Recently, proteins with sequence homology to the box jellyfish toxins have also been identified in the venoms of Cyanea capillata (Scyphozoa) [22] and H. magnipapillata (Hydrozoa) [15] inferring the toxin family is present throughout Cnidaria. Construction of a phylogenetic tree of currently available cubozoan protein sequences shows three groupings within the toxin family, suggesting structural and fn diversification has occurred between toxin groups during evolution. (Figure 4).
-endotoxins [6]. Recently, proteins with sequence homology to the box jellyfish toxins have also been identified in the venoms of Cyanea capillata (Scyphozoa) [22] and H. magnipapillata (Hydrozoa) [15] inferring the toxin family is present throughout Cnidaria. Construction of a phylogenetic tree of currently available cubozoan protein sequences shows three groupings within the toxin family, suggesting structural and fn diversification has occurred between toxin groups during evolution. (Figure 4).
Figure 4. Phylogenetic tree of Cubozoan toxin proteins.
Phylogenetic tree depicting the grouping of box jellyfish toxins found in GenBank into three broad classes. Proteins identified in this study are underlined and their GenBank accession number is indicated. The tree was produced using MUSCLE and PhyML for tree building and the aLRT statistical test [50] was used for branch support.
Both Mascot and de novo homology searches suggest that additional isoforms of the CfTX toxins are present in CFV. CfTX-1 and -2 were identified using Mascot in 29 of the 40 gel bands analysed yet the presence of well defined bands on SDS-PAGE suggests this was not the result of protein breakdown (Figure 2). This was supported by the identification of CqTX-A and the identification, on the basis of homology, of a further three examples of the toxins. Western blot analysis using polyclonal antibodies against CfTX-1 and -2 showed hybridisation to one major band (spanning gel bands 28 & 29) that provided the highest scoring Mascot identifications to CfTX-1 and -2 as well as a band, possibly a cleavage product, in the lower molecular weight region of the gel ( 12 kDa; gel band 10) (Figure 2). No other bands reacted positively towards the antibodies, suggesting that although a number of CfTX-like proteins are present in C. fleckeri venom, they do not contain common epitopes to which CfTX-1 and -2 specific antibodies can bind. Sequence divergence among toxin family members coupled with a lack of cross-reactivity by toxin-specific antibodies suggests that there are significant structural variations between the related toxins that could modulate their function and/or specificity. Although these toxins appear to be the major CFV toxin family, two isoforms of both neprilysin and endothelin-converting enzymes where also identified. Neprilysin, a metallo-endopeptidase, has been identified as a possible neurotoxic protein in snake venom [23] while the endothelin-converting enzymes may play a supporting role, as is the case in wasp venom [24], by processing pro-proteins into mature protein or peptide toxins.
12 kDa; gel band 10) (Figure 2). No other bands reacted positively towards the antibodies, suggesting that although a number of CfTX-like proteins are present in C. fleckeri venom, they do not contain common epitopes to which CfTX-1 and -2 specific antibodies can bind. Sequence divergence among toxin family members coupled with a lack of cross-reactivity by toxin-specific antibodies suggests that there are significant structural variations between the related toxins that could modulate their function and/or specificity. Although these toxins appear to be the major CFV toxin family, two isoforms of both neprilysin and endothelin-converting enzymes where also identified. Neprilysin, a metallo-endopeptidase, has been identified as a possible neurotoxic protein in snake venom [23] while the endothelin-converting enzymes may play a supporting role, as is the case in wasp venom [24], by processing pro-proteins into mature protein or peptide toxins.
Proteins involved in nematocyst structure and nematogenesis
Proteins identified in the Hydra nematocyst capsule are predominantly comprised of different species of mini-collagens that form a disulfide-linked polymer [25]. In this study no collagen was identified using Mascot, although three collagen isoforms were identified on the basis of homology. Given the limitations of available sequence for proteomic analysis, it is likely that sequence divergence between characterised cnidarians and C. fleckeri collagens resulted in fewer identifications in this study; the presence, in homology searches, of multiple collagen identifications at a level of significance below that permitted in the study supports this conjecture. Likewise, NOWA, another major protein constituent of the nematocyst capsule, was also identified at a lower significance level, suggesting that the protein is found in CFV but that its sequence has diverged from those NOWA sequences currently in the database. Multiple isoforms of nematogalectin, the main protein constituent of the tubule, were identified in CFV, suggesting that, like Hydra but unlike the anthozoans, C. fleckeri contains two copies of this gene [12], [26]. Spinalin, a spine protein, was not identified at any level of significance but is more resistant to dissolution in the presence of DTT than other structural nematocyst proteins [27].
The explosive potential of the nematocyst is due to an exceptionally high intracapsular pressure produced by a high concentration of poly- -glutamate (pG) that binds a 2M concentration of cations [28]. A key enzyme in the production of pG,
-glutamate (pG) that binds a 2M concentration of cations [28]. A key enzyme in the production of pG,  -glutamyltransferase, was identified in CFV confirming previous studies that showed enzymes necessary for pG synthesis are transferred into the capsule prior to the hardening of the capsule wall [29], [30]. Poly-
-glutamyltransferase, was identified in CFV confirming previous studies that showed enzymes necessary for pG synthesis are transferred into the capsule prior to the hardening of the capsule wall [29], [30]. Poly- -glutamate degradative activity has been reported in Hydra
[31] and a
-glutamate degradative activity has been reported in Hydra
[31] and a  -glutamyl hydrolase was identified in CFV. This molecule catalyses the hydrolysis of
-glutamyl hydrolase was identified in CFV. This molecule catalyses the hydrolysis of  -glutamyl bonds and may indicate that some regulation of pG biosynthesis occurs in the developing nematocyst. Similar to the Hydra nematocyst proteome, several isoforms of the nematogenesis-associated dickkopf protein 3 (Dkk3) [32], [33] were identified. Dkk3 belongs to the dickkopf family of developmental proteins that purportedly act via inhibition of the Wnt signalling pathway [33] and its presence in CFV suggests that nematogenesis in C. fleckeri proceeds along similar lines to other better characterised cnidarians.
-glutamyl bonds and may indicate that some regulation of pG biosynthesis occurs in the developing nematocyst. Similar to the Hydra nematocyst proteome, several isoforms of the nematogenesis-associated dickkopf protein 3 (Dkk3) [32], [33] were identified. Dkk3 belongs to the dickkopf family of developmental proteins that purportedly act via inhibition of the Wnt signalling pathway [33] and its presence in CFV suggests that nematogenesis in C. fleckeri proceeds along similar lines to other better characterised cnidarians.
Post-translation modifications
Both nematogalectins [12] and NOWA [34] are known to be glycosylated, and in snake venom the glycosylation of key venom proteins is thought to enhance protein stability and diffusion [35]. In silico analysis of the primary sequences of toxins CfTX-1 and -2 showed a number of conserved motifs, indicative of a range of post-translation modifications including phosphorylation and glycosylation. Therefore, to test for these modifications, total CFV was stained with fluorescent dyes after SDS-PAGE. For glycan staining, intense fluorescence was observed in bands confirmed to contain CfTX-1 and -2 by tandem MS (Figure 2; band 30); weaker staining was observed in areas corresponding to the identification of the nematogalectins (bands 34 and 26 and 27 respectively). Further staining was observed in areas thought to contain CfTX isoforms suggesting that glycosylation is a common post-translational modification of the C. fleckeri toxin proteins. Phosphorylation analysis indicated that no CFV proteins were phosphorylated, with the exception of a protein in band 40 which was identified as  -tubulin. In comparison, phosphorylation is common in snake venoms but restricted to glycoproteins containing phosphorylated carbohydrates [35]. Glycosylated carbohydrate moieties are common in nature and are often involved in lysosomal targeting by way of mannose 6-phosphate receptors [36] and their absence here suggests this pathway is not utilised during venom production in the nematocytes.
-tubulin. In comparison, phosphorylation is common in snake venoms but restricted to glycoproteins containing phosphorylated carbohydrates [35]. Glycosylated carbohydrate moieties are common in nature and are often involved in lysosomal targeting by way of mannose 6-phosphate receptors [36] and their absence here suggests this pathway is not utilised during venom production in the nematocytes.
Conclusions
The principle aim of this study was to identify proteins in the venom of C. fleckeri that could be responsible for its wide range of bioactivities and debilitating effects in humans. As our results show, the venom proteome of C. fleckeri contains a diverse array of proteins dominated by toxins and proteins involved in nematogenesis. The most abundant toxins in C. fleckeri venom are CfTX-1, CfTX-2 and a number of newly discovered CfTX-like isoforms. Toxicological data and bioinformatic information suggest that this expanding toxin family plays a significant functional role in envenoming and further research is necessary to elucidate the actions of these toxins at the molecular level. Due to the lack of genomic and transcriptomic sequences currently available for C. fleckeri, or closely related species, it is also likely that other toxins unique to Cubozoa are yet to be discovered.
The CfTX-like proteins belong to a family of potent toxins that is only found in Cnidaria. The evolutionary emergence of a particular protein family in venoms is a consistent theme in nature, with examples including the conotoxins, from cone snails [37], and the Ancylostoma secreted proteins from hookworms [38]. All of these organisms are currently being utilised as a source of therapeutic compounds [37], [39] and the expansion of the cnidarian toxin family promises to provide a rich source of novel bioactive compounds that could be utilised as prospective new drugs, novel research tools or other beneficial purposes. Finally, gaining a better understanding of the molecular, structural and functional diversity of this important toxin family will lead to the better understanding of and improvements in medical treatments of cnidarian stings.
Materials and Methods
Nematocyst Isolation
Nematocysts were isolated from excised tentacles [40] of a mature specimen (captured near Weipa, Queensland, Australia), except the nematocysts were not lyophilised. Isolated nematocysts were further purified in a discontinuous Percoll gradient [11]. The integrity of the undischarged nematocysts was verified using an Axioskop2 mot plus light microscope (Zeiss). No specific permits were required for the described field studies. No specific permissions were required as the animals collected are not protected and were collected from marine environments that are not protected or privately owned. C. fleckeri is not an endangered or protected species.
Electrophoresis and In-gel Digestion
Percoll-cleaned nematocysts were washed twice with 20 mM Tris-HCl (pH 7.5). Aliquots of nematocysts were resuspended 1∶6 (wet w/v) in reducing SDS-sample buffer [41] containing DTT and incubated at RT until  90% nematocyst discharge was observed microscopically. The samples were centrifuged (16 k
90% nematocyst discharge was observed microscopically. The samples were centrifuged (16 k g, 4°C, 10 min) to remove capsular debris and supernatants were transferred to clean tubes and heated (95°C, 5 min). Duplicate samples (10
g, 4°C, 10 min) to remove capsular debris and supernatants were transferred to clean tubes and heated (95°C, 5 min). Duplicate samples (10 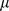 L) were applied to a 15% SDS-PAGE gel and electrophoresis performed according to Laemmli [41]. Proteins were stained with EZBlue G-250 colloidal Coomassie stain (Sigma) and each sample lane was divided into 40 gel slices. In-gel trypsin digestion was performed using established methods [42] and after digestion peptide mixtures were reduced to 12
L) were applied to a 15% SDS-PAGE gel and electrophoresis performed according to Laemmli [41]. Proteins were stained with EZBlue G-250 colloidal Coomassie stain (Sigma) and each sample lane was divided into 40 gel slices. In-gel trypsin digestion was performed using established methods [42] and after digestion peptide mixtures were reduced to 12 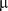 l in a vacuum centrifuge before mass spectral analysis.
l in a vacuum centrifuge before mass spectral analysis.
OFFGEL electrophoresis
Established methods [42] were used to reduce, alkylate and trypsinize two  2.5 mg samples of total protein from lyophilised C. fleckeri venom extracted from nematocysts using bead mill homogenisation. The resulting tryptic fragments were subjected to OFFGEL electrophoresis (OGE). The 3100 OFFGEL Fractionator and OFFGEL Kit pH 3–10 (Agilent Technologies) with a 24-well setup were prepared as per the manufacturers protocols. The tryptic digests were diluted in peptide-focusing buffer, without the addition of ampholytes, to a final volume of 3.6 ml and 150
2.5 mg samples of total protein from lyophilised C. fleckeri venom extracted from nematocysts using bead mill homogenisation. The resulting tryptic fragments were subjected to OFFGEL electrophoresis (OGE). The 3100 OFFGEL Fractionator and OFFGEL Kit pH 3–10 (Agilent Technologies) with a 24-well setup were prepared as per the manufacturers protocols. The tryptic digests were diluted in peptide-focusing buffer, without the addition of ampholytes, to a final volume of 3.6 ml and 150 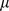 l was loaded into each well. The samples were focused with a maximum current of 50
l was loaded into each well. The samples were focused with a maximum current of 50 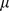 A until 50 kVh were achieved. Peptide fractions were harvested, lyophilised and resuspended in 5% formic acid before LC and mass spectral analysis.
A until 50 kVh were achieved. Peptide fractions were harvested, lyophilised and resuspended in 5% formic acid before LC and mass spectral analysis.
Western Blot Analysis
Polyclonal antibodies against CfTX-1 and -2 were commercially obtained from IMVS, Veterinary Services (Gilles Plains, Australia) under the aegis of the IMVS Animal Ethics Committee (license number 155) as previously described [11]. Venom proteins were separated by SDS-PAGE as described above and transferred to Immobilon-P PVDF membrane (Millipore). The membrane was blocked (5% (w/v) skim milk powder in TBST, 0.5 h) and incubated overnight with the rabbit antibodies diluted in blocking solution (1∶2000). The membrane was washed (3×10 min in TBST) then incubated (1 h) with goat anti-rabbit alkaline phosphatase-conjugated antibodies (Sigma) diluted in TBST (1∶5000). Following membrane washing, antibody-bound proteins were visualised using NBT/BCIP (Promega).
Glycoprotein and Phosphoprotein Staining
Following separation of nematocyst-derived proteins by SDS-PAGE, in-gel detection of glycoproteins and phosphoproteins was performed using a GlycoProfile III fluorescent glycoprotein detection kit (Sigma) or Pro-Q Diamond phosphoprotein gel stain (Invitrogen), respectively, according to the manufacturers' instructions. For glycoprotein analysis, a duplicate gel was processed omitting the oxidation step to detect any non-specific fluorescent staining. For phosphoprotein analysis, the ProteoProfile PTM marker (Sigma) containing phosphorylated ovalbumin (45 kDa) and 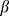 -casein (30 kDa) was included as a positive control. Fluorescently stained glycoproteins and phosphoproteins were visualised using a ChemiSmart 3000 image acquisition system (Viber Lourmat).
-casein (30 kDa) was included as a positive control. Fluorescently stained glycoproteins and phosphoproteins were visualised using a ChemiSmart 3000 image acquisition system (Viber Lourmat).
Protein Identification using MS/MS
OGE fractions and tryptic fragments from in-gel digests were chromatographically separated on a Dionex Ultimate 3000 HPLC using an Agilent Zorbax 300SB-C18 (3.5 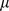 m, 150 mm×75
m, 150 mm×75 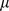 m) column and a linear gradient of 0–80% solvent B over 60 min. A flow rate of 300 nl/min was used for all experiments. The mobile phase consisted of solvent A (0.1% formic acid (aq)) and solvent B (80/20 acetonitrile/0.1% formic acid (aq)). Eluates from the RP-HPLC column were directly introduced into the NanoSpray II ionisation source of a QSTAR Elite Hybrid MS/MS System (Applied Biosystems) operated in positive ion electrospray mode. All analyses were performed using Information Dependant Acquisition. Analyst 2.0 (Applied Biosystems) was used for data analysis and peak list generation. Briefly, the acquisition protocol consisted of the use of an Enhanced Mass Spectrum scan as the survey scan. The three most abundant ions detected over the background threshold were subjected to examination using an Enhanced Resolution scan to confirm the charge state of the multiply charged ions. The ions with a charge state of
m) column and a linear gradient of 0–80% solvent B over 60 min. A flow rate of 300 nl/min was used for all experiments. The mobile phase consisted of solvent A (0.1% formic acid (aq)) and solvent B (80/20 acetonitrile/0.1% formic acid (aq)). Eluates from the RP-HPLC column were directly introduced into the NanoSpray II ionisation source of a QSTAR Elite Hybrid MS/MS System (Applied Biosystems) operated in positive ion electrospray mode. All analyses were performed using Information Dependant Acquisition. Analyst 2.0 (Applied Biosystems) was used for data analysis and peak list generation. Briefly, the acquisition protocol consisted of the use of an Enhanced Mass Spectrum scan as the survey scan. The three most abundant ions detected over the background threshold were subjected to examination using an Enhanced Resolution scan to confirm the charge state of the multiply charged ions. The ions with a charge state of 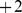 ,
, 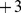 or with unknown charge were then subjected to collision-induced dissociation using a rolling collision energy dependent upon the m/z and the charge state of the ion. Enhanced Product Ion scans were acquired resulting in full product ion spectra for each of the selected precursors which were then used in subsequent database searches.
or with unknown charge were then subjected to collision-induced dissociation using a rolling collision energy dependent upon the m/z and the charge state of the ion. Enhanced Product Ion scans were acquired resulting in full product ion spectra for each of the selected precursors which were then used in subsequent database searches.
Mascot searches
Searches were performed using version 2.2.02 of Mascot with a 0.1 Da tolerance on the precursor, 0.1 Da tolerance on the product ions, allowing for methionine oxidation and carbamidomethylation as fixed and variable modifications respectively, two missed cleavages, charge states +2 and +3, trypsin as the enzyme and MudPIT scoring was used to derive protein scores. All experiments were searched against a custom-built protein database of 160,848 proteins comprised of protein sequences derived from all cnidarian nucleotide sequence in the NCBI non-redundant (nr) database. Nucleotide sequences were cleaned using SeqClean (http://compbio.dfci.harvard.edu/tgi/software/) in conjunction with the UniVec vector sequence database from the NCBI. Sequences were then clustered using CAP3 [43] and a set of predicted protein sequences generated using ESTScan [44]. Searches were also made against the SwissProt database to detect contamination. Mascot searches were further validated using Scaffold (version Scaffold_3_00_06, Proteome Software Inc). Using Scaffold X! Tandem searches were performed on a subset of the custom database using the same parameters as used for Mascot searches. Peptide identifications were accepted if they could be established at greater than 95.0% probability as specified by the PeptideProphet algorithm [45]. Protein identifications were accepted if they could be established at greater than 95.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the ProteinProphet algorithm [46]. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. A 0.0% false discovery rate (FDR) was calculated using Scaffold validated protein identifications.
PEAKS and de novo sequencing
De novo protein sequencing was achieved using PEAKS (version 4.5, Bioinformatics Solutions, Waterloo, Canada) [47]. De novo peptide sequences were derived from the combined MS/MS spectra from in-gel digests using 0.1 Da tolerance on the parent and fragment ions, digestion with trypsin, one missed cleavage and allowing for methionine oxidation and carbamidomethylation as fixed and variable modifications respectively. The de novo tags where then used in two searches, using PEAKS, of the custom database performed using the same parameters. The first utilised high quality de novo sequences to identify exactly matching peptides from the custom database. A decoy database was searched and spurious identifications were used to calculate a FDR for peptide identifications using the decoy-fusion method [47]. Using a desired false discovery rate of 1%, proteins were accepted only if they possessed at least two peptides scoring above the cutoff calculated for the desired FDR and if at least one significant peptide was unique to that protein. PEAKS was also used to conduct homology searches using the de novo sequence tags. In this search, the amino acid sequence of the de novo tags was used to search for homologous peptides, rather than exact matches, in the custom database. The same FDR procedure was used to estimate a peptide cutoff score and protein identifications were accepted only if they contained at least one unique and significant peptide. Identifications from homology searches containing only a single peptide were manually annotated and only high quality identifications, containing the majority of calculated fragment ions, were accepted.
Bioinformatic analysis
Protein descriptions were assigned to protein identifications using BLASTP on the non-redundant protein databases from NCBI (bit score  30). Classical secretory signal sequences were detected using a local version of SignalP [48]. The phylogenetic tree was produced using MUSCLE for multiple alignment, Gblocks for automatic alignment curation, PhyML for tree building and TreeDyn for tree drawing using the tree-generation pipeline at Phylogeny.fr website [49]. The aLRT statistical test [50] was used for branch support.
30). Classical secretory signal sequences were detected using a local version of SignalP [48]. The phylogenetic tree was produced using MUSCLE for multiple alignment, Gblocks for automatic alignment curation, PhyML for tree building and TreeDyn for tree drawing using the tree-generation pipeline at Phylogeny.fr website [49]. The aLRT statistical test [50] was used for branch support.
Supporting Information
Scaffold peptide and protein reports. Excel spreadsheet containing full protein and peptide information for C. fleckeri Mascot identifications.
(XLS)
Peaks peptide report. Excel spreadsheet containing full peptide information for C. fleckeri PEAKS identifications.
(XLSX)
Proteins identified during homology searches. Proteins identified using a homology search of de novo sequence tags in PEAKS. Abbreviations used: ID — identification number corresponding to custom database supplied as S5; −10lgP — PEAKS probability score; CO — percent cover; SC — total number of significant spectra contributing to the identification; USC — number of unique and significant spectra contributing to the identification. A ‘+’ in the SignalP column denotes the presence of a predicted signal sequence using SignalP and a ‘+’ in the Hydra column denotes the identification of a similar protein in the H. magnipapillata venom proteome.
(XLSX)
Homology peptide report. Excel spreadsheet containing full peptide information for C. fleckeri identifications made during homology searches of de novo peptide sequences.
(XLSX)
Sequences of identified proteins. Sequences of proteins identified during Mascot and PEAKS searches in fasta format.
(FASTA)
Annotated single peptide identifications. Annotated spectra, ion tables and error plots of all proteins identified by a single peptide during homology searches using PEAKS.
(PDF)
Funding Statement
This research was supported using infrastructure provided by the Australian Government through the Linkage Infrastructure, Equipment and Facilities scheme from the Australian Research Council. JM is supported by a Career Development Fellowship from the National Health and Medical Research Council, Australia (NHMRC). AL is supported by a senior research fellowship from NHMRC. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Slautterback D, Fawcett D (1959) The development of the cnidoblasts of Hydra . J Biophys Biochem Cy 5: 441–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nüchter T, Benoit M, Engel U, Özbek S, Holstein T (2006) Nanosecond-scale kinetics of nematocyst discharge. Curr Biol 16: 316–318. [DOI] [PubMed] [Google Scholar]
- 3. Kass-Simon G, Scappaticci A Jr (2002) The behavioral and developmental physiology of nematocysts. Can J Zoolog 80: 1772–1794. [Google Scholar]
- 4. Hidaka M (1993) Mechanism of nematocyst discharge and its cellular control. Advances in comparative and environmental physiology 15: 45–45. [Google Scholar]
- 5. Özbek S, Balasubramanian PG, Holstein TW (2009) Cnidocyst structure and the biomechanics of discharge. Toxicon 54: 1038–1045. [DOI] [PubMed] [Google Scholar]
- 6. Brinkman DL, Burnell JN (2009) Biochemical and molecular characterisation of cubozoan protein toxins. Toxicon 54: 1162–1173. [DOI] [PubMed] [Google Scholar]
- 7. Beadnell CE, Rider TA, Williamson JA, Fenner PJ (1992) Management of a major box jellyfish (Chironex eckeri) sting. lessons from the first minutes and hours. Med J Aust 156: 655–658. [DOI] [PubMed] [Google Scholar]
- 8. Lumley J, Williamson JA, Fenner PJ, Burnett JW, Colquhoun DM (1988) Fatal envenomation by Chironex fleckeri, the north Australian box jellyfish: the continuing search for lethal mechanisms. Med J Aust 148: 527–534. [DOI] [PubMed] [Google Scholar]
- 9. Fenner PJ, Williamson JA (1996) Worldwide deaths and severe envenomation from jellyfish stings. Med J Aust 165: 658–661. [DOI] [PubMed] [Google Scholar]
- 10. Brinkman D, Burnell J (2007) Identification, cloning and sequencing of two major venom proteins from the box jellyfish, Chironex fleckeri . Toxicon 50: 850–860. [DOI] [PubMed] [Google Scholar]
- 11. Brinkman D, Burnell J (2008) Partial purification of cytolytic venom proteins from the box jellyfish, Chironex fleckeri . Toxicon 51: 853–863. [DOI] [PubMed] [Google Scholar]
- 12. Hwang JS, Takaku Y, Momose T, Adamczyk P, Özbek S, et al. (2010) Nematogalectin, a nematocyst protein with glyxy and galectin domains, demonstrates nematocyte-specific alternative splicing in Hydra . Proc Natl Acad Sci USA 107: 18539–18544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hwang JS, Ohyanagi H, Hayakawa S, Osato N, Nishimiya-Fujisawa C, et al. (2007) The evolutionary emergence of cell type-specific genes inferred from the gene expression analysis of Hydra . Proc Natl Acad Sci USA 104: 14735–14740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Milde S, Hemmrich G, Anton-Erxleben F, Khalturin K, Wittlieb J, et al. (2009) Characterization of taxonomically restricted genes in a phylum-restricted cell type. Genome Biol 10: R8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Balasubramanian PG, Beckmann A, Warnken U, Schnölzer M, Schüler A, et al. (2012) Proteome of Hydra nematocyst. J Biol Chem 287: 9672–9681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nagai H, Takuwa K, Nakao M, Ito E, Miyake M, et al. (2000) Novel proteinaceous toxins from the box jellyfish (sea wasp) Carybdea rastoni . Biochem Biophys Res Commun 275: 582–588. [DOI] [PubMed] [Google Scholar]
- 17. Nagai H, Takuwa K, Nakao M, Sakamoto B, Crow GL, et al. (2000) Isolation and characterization of a novel protein toxin from the Hawaiian box jellyfish (sea wasp) Carybdea alata . Biochem Biophys Res Commun 275: 589–594. [DOI] [PubMed] [Google Scholar]
- 18. Nagai H, Takuwa-Kuroda K, Nakao M, Oshiro N, Iwanaga S, et al. (2002) A novel protein toxin from the deadly box jellyfish (sea wasp, Habu-kurage) Chiropsalmus quadrigatus . Biosci Biotech Bioch 66: 97–102. [DOI] [PubMed] [Google Scholar]
- 19. Lewis C, Bentlage B (2009) Clarifying the identity of the Japanese Habu-kurage, Chironex yam-aguchii, sp. nov.(Cnidaria: Cubozoa: Chirodropida). Zootaxa 2030: 59–65. [Google Scholar]
- 20.Ávila Soria G (2009) Molecular characterization of Carukia barnesi and Malo kingi, Cnidaria; Cubozoa; Carybdeidae. Ph.D. thesis, James Cook University.
- 21. Pereira P, Barry J, Corkeron M, Keir P, Little M, et al. (2010) Intracerebral hemorrhage and death after envenoming by the jellyfish Carukia barnesi . Clin Toxicol (Phila) 48: 390–392. [DOI] [PubMed] [Google Scholar]
- 22. Lassen S, Helmholz H, Ruhnau C, Prange A (2011) A novel proteinaceous cytotoxin from the northern Scyphozoa Cyanea capillata (L.) with structural homology to cubozoan haemolysins. Toxicon 57: 721–729. [DOI] [PubMed] [Google Scholar]
- 23. Casewell N, Harrison R, Wüster W, Wagstaff S (2009) Comparative venom gland transcriptome surveys of the saw-scaled vipers (Viperidae: Echis) reveal substantial intra-family gene diversity and novel venom transcripts. BMC genomics 10: 564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baek J, Woo T, Kim C, Park J, Kim H, et al. (2009) Differential gene expression profiles in the venom gland/sac of Orancistrocerus drewseni (Hymenoptera: Eumenidae). Arch Insect Biochem 71: 205–222. [DOI] [PubMed] [Google Scholar]
- 25. Kurz EM, Holstein TW, Petri BM, Engel J, David CN (1991) Mini-collagens in Hydra nematocytes. J Cell Biol 115: 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Steele R, David C, Technau U (2011) A genomic view of 500 million years of cnidarian evolution. Trends Genet 27: 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Koch AW, Holstein TW, Mala C, Kurz E, Engel J, et al. (1998) Spinalin, a new glycine- and histidine-rich protein in spines of Hydra nematocysts. J Cell Sci 111 (Pt 11) 1545–1554. [DOI] [PubMed] [Google Scholar]
- 28. Klug M, Weber J (1991) An extract from Hydra vulgaris (Cnidaria) nematocysts increases cytoplasmic Ca2+ levels in fibroblasts. Toxicon 29: 129–133. [DOI] [PubMed] [Google Scholar]
- 29. Szczepanek S, Cikala M, David CN (2002) Poly-γ-glutamate synthesis during formation of nematocyst capsules in Hydra . J Cell Sci 115: 745–751. [DOI] [PubMed] [Google Scholar]
- 30. Weber J (1995) The development of cnidarian stinging cells: maturation and migration of stenoteles of Hydra vulgaris. Dev Genes Evol 205: 171–181. [DOI] [PubMed] [Google Scholar]
- 31. Weber J (1994) The metabolism of poly(γ-glutamic acid)s of nematocysts in Hydra vulgaris: detection of two distinct hydrolytic enzymes in endoderm and in nematocysts. Comp Biochem Phys B 107: 21–32. [Google Scholar]
- 32. Seipel K, Yanze N, Schmid V (2004) The germ line and somatic stem cell gene Cniwi in the jellyfish Podocoryne carnea . Int J Dev Biol 48: 1–8. [DOI] [PubMed] [Google Scholar]
- 33. Fedders H, Augustin R, Bosch T (2004) A Dickkopf-3-related gene is expressed in differentiating nematocytes in the basal metazoan Hydra . Dev Genes Evol 214: 72–80. [DOI] [PubMed] [Google Scholar]
- 34. Engel U, Özbek S, Streitwolf-Engel R, Petri B, Lottspeich F, et al. (2002) Nowa, a novel protein with minicollagen cys-rich domains, is involved in nematocyst formation in Hydra . J Cell Sci 115: 3923–3934. [DOI] [PubMed] [Google Scholar]
- 35. Birrell GW, Earl ST, Wallis TP, Masci PP, de Jersey J, et al. (2007) The diversity of bioactive proteins in Australian snake venoms. Mol Cell Proteomics 6: 973–986. [DOI] [PubMed] [Google Scholar]
- 36. Sleat DE, Wang Y, Sohar I, Lackland H, Li Y, et al. (2006) Identification and validation of mannose 6-phosphate glycoproteins in human plasma reveal a wide range of lysosomal and non-lysosomal proteins. Mol Cell Proteomics 5: 1942–1956. [DOI] [PubMed] [Google Scholar]
- 37. Halai R, Craik DJ (2009) Conotoxins: natural product drug leads. Nat Prod Rep 26: 526–536. [DOI] [PubMed] [Google Scholar]
- 38. Datu BJ, Gasser RB, Nagaraj SH, Ong EK, O'Donoghue P, et al. (2008) Transcriptional changes in the hookworm, Ancylostoma caninum, during the transition from a free-living to a parasitic larva. PLoS Neglect Trop D 2: e130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ruyssers NE, De Winter BY, De Man JG, Loukas A, Pearson MS, et al. (2009) Therapeutic potential of helminth soluble proteins in TNBS-induced colitis in mice. Inamm Bowel Dis 15: 491–500. [DOI] [PubMed] [Google Scholar]
- 40. Bloom DA, Burnett JW, Alderslade P (1998) Partial purification of box jellyfish (Chironex fleckeri) nematocyst venom isolated at the beachside. Toxicon 36: 1075–1085. [DOI] [PubMed] [Google Scholar]
- 41. Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685. [DOI] [PubMed] [Google Scholar]
- 42. Mulvenna J, Hamilton B, Nagaraj SH, Smyth D, Loukas A, et al. (2009) Proteomics analysis of the excretory/secretory component of the blood-feeding stage of the hookworm, Ancylostoma caninum . Mol Cell Proteomics 8: 109–121. [DOI] [PubMed] [Google Scholar]
- 43. Huang X, Madan A (1999) Cap3: A dna sequence assembly program. Genome Res 9: 868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lottaz C, Iseli C, Jongeneel CV, Bucher P (2003) Modeling sequencing errors by combining Hidden Markov models. Bioinformatics 19 Suppl 2: ii103–12. [DOI] [PubMed] [Google Scholar]
- 45. Keller A, Nesvizhskii AI, Kolker E, Aebersold R (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392. [DOI] [PubMed] [Google Scholar]
- 46. Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658. [DOI] [PubMed] [Google Scholar]
- 47. Ma B, Zhang K, Hendrie C, Liang C, Li M, et al. (2003) PEAKS: powerful software for peptide de novo sequencing by tandem mass spectrometry. Rapid Commun Mass Spectrom 17: 2337–42. [DOI] [PubMed] [Google Scholar]
- 48. Emanuelsson O, Brunak S, von Heijne G, Nielsen H (2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc 2: 953–71. [DOI] [PubMed] [Google Scholar]
- 49. Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. (2008) Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res 36: W465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Anisimova M, Gascuel O (2006) Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol 55: 539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Scaffold peptide and protein reports. Excel spreadsheet containing full protein and peptide information for C. fleckeri Mascot identifications.
(XLS)
Peaks peptide report. Excel spreadsheet containing full peptide information for C. fleckeri PEAKS identifications.
(XLSX)
Proteins identified during homology searches. Proteins identified using a homology search of de novo sequence tags in PEAKS. Abbreviations used: ID — identification number corresponding to custom database supplied as S5; −10lgP — PEAKS probability score; CO — percent cover; SC — total number of significant spectra contributing to the identification; USC — number of unique and significant spectra contributing to the identification. A ‘+’ in the SignalP column denotes the presence of a predicted signal sequence using SignalP and a ‘+’ in the Hydra column denotes the identification of a similar protein in the H. magnipapillata venom proteome.
(XLSX)
Homology peptide report. Excel spreadsheet containing full peptide information for C. fleckeri identifications made during homology searches of de novo peptide sequences.
(XLSX)
Sequences of identified proteins. Sequences of proteins identified during Mascot and PEAKS searches in fasta format.
(FASTA)
Annotated single peptide identifications. Annotated spectra, ion tables and error plots of all proteins identified by a single peptide during homology searches using PEAKS.
(PDF)