SUMMARY
Regulation of intestinal dietary fat absorption is critical to maintaining energy balance. While intestinal microbiota clearly impact the host’s energy balance, their role in intestinal absorption and extra-intestinal metabolism of dietary fat is less clear. Using in vivo imaging of fluorescent fatty acid (FA) analogs delivered to gnotobiotic zebrafish hosts, we reveal that microbiota stimulate FA uptake and lipid droplet (LD) formation in the intestinal epithelium and liver. Microbiota increase epithelial LD number in a diet-dependent manner. The presence of food led to the intestinal enrichment of bacteria from the phylum Firmicutes. Diet-enriched Firmicutes and their products were sufficient to increase epithelial LD number, whereas LD size was increased by other bacterial types. Thus, different members of the intestinal microbiota promote FA absorption via distinct mechanisms. Diet-induced alterations in microbiota composition might influence fat absorption, providing mechanistic insight into how microbiota-diet interactions regulate host energy balance.
INTRODUCTION
Dietary fat contributes a significant caloric value to our diet. Dietary lipids supply 45-55% of the energy requirements in breastfed human infants (Boudry et al., 2010) and 40-55% of the calories in the Western diet (Meek et al., 2010). In vertebrates, dietary fats in the form of triglycerides are digested by lipases within the intestinal lumen and the released free fatty acids (FFAs) and monoglycerides are absorbed by enterocytes in the intestinal epithelium (Karasov and Hume, 1997). Fatty acid (FA) absorption at the brush border of enterocytes is enhanced by solubilization in bile salt micelles or liposomes (Kindel et al., 2010). Once absorbed by enterocytes, FAs are either oxidized to generate energy, reesterified into triglycerides and temporarily stored as cytoplasmic lipid droplets (LDs), incorporated into chylomicrons for secretion into the lymph, or released into circulation as free fatty acids (Iqbal and Hussain, 2009). These exogenously acquired FAs that enter circulation as chylomicrons or FFA are then available for oxidation or storage in extra-intestinal tissues such as liver. Many steps in the dynamic process of exogenous FA uptake into enterocytes and their subsequent assembly into LDs and chylomicrons remain unresolved. An improved understanding of factors controlling dietary FA absorption and LD formation could lead to new approaches for decreasing the efficiency of dietary energy harvest in the context of obesity and increasing efficiency in the context of malnutrition.
Environmental factors such as intestinal microorganisms and diet represent attractive targets for controlling dietary lipid absorption and energy balance. The digestive tract is colonized beginning at birth by complex assemblages of microorganisms (gut microbiota) that profoundly influence intestinal and extra-intestinal physiology (Sekirov et al., 2010). The ability of the gut microbiota to modify dietary nutrient metabolism has emerged as a key feature of host-microbe relationships in the gut (Musso et al., 2011). This capability has been most extensively described in the ruminant foregut and in the human and rodent hindgut (colon), where microbial fermentation of otherwise indigestible complex carbohydrates in the diet produces monosaccharides and short chain FAs that can then be absorbed by the host (Flint et al., 2012). The presence of a gut microbiota in mice increases fat storage in adipose tissue (Bäckhed et al., 2004; Bäckhed et al., 2007; Rabot et al., 2010) and causes significant alterations in secondary lipid metabolites in serum, liver, and adipose tissue (Martin et al., 2009; Velagapudi et al., 2010). However, these altered lipid profiles could be due to effects of microbiota on intestinal absorption or metabolism of exogenous dietary lipids or on hepatic production or metabolism of endogenous lipids. Therefore, the potential impact of the microbiota on intestinal absorption of dietary lipids remains unclear.
Although the microbiota can influence dietary nutrient harvest, the diet can also impact microbial community composition and function. Gut microbial community membership is correlated with diet composition in humans and other mammals (Ley et al., 2008; Wu et al., 2011; Muegge et al., 2011). Although not all studies have reported similar associations, an emerging pattern is that caloric intake can influence the relative abundance of the Firmicutes and Bacteroidetes phyla that dominate the intestines of vertebrates (Ley et al., 2008). For example, mice starved for 24 hours show a reduction of Firmicutes and increase of Bacteroidetes in their gut microbiota (Crawford et al., 2009). Conversely, obese humans (Ley et al., 2006), humans consuming a high-calorie diet (Jumpertz et al., 2011), hyperphagic obese ob/ob mice (Ley et al., 2005), mice fed high-calorie Western diets (Turnbaugh et al., 2008; Hildebrandt et al., 2009; Murphy et al., 2010), and postprandial pythons (Costello et al., 2010) assemble gut microbial communities with an enrichment of Firmicutes at the expense of Bacteroidetes and other major phyla. Diet-induced enrichment of Firmicutes in the gut microbiota has been associated with a positive energy balance in gnotobiotic mouse hosts (Turnbaugh et al., 2006; Turnbaugh et al., 2008). However, the mechanisms by which certain bacterial taxa such as Firmicutes become enriched in the gut as a function of caloric intake remain unresolved. Furthermore, the potential impact of diet-induced alterations in microbiota composition on dietary lipid absorption in the gut is unknown.
In this study, we use the zebrafish model to investigate how microbiota and diet interact to regulate lipid absorption in the gut epithelium. Digestive tract anatomy and physiology as well as lipid metabolism pathways in the zebrafish are similar to mammals and other vertebrates (Babin and Vernier, 1989; Carten and Farber, 2009). Zebrafish larvae begin exogenous feeding at ~5 days post-fertilization (dpf), and complete resorption of their endogenous yolk supply by ~6 dpf. Previous work comparing zebrafish raised germ free (GF) to those colonized with a normal zebrafish microbiota (conventionalized, CONVD) revealed roles for the microbiota on diverse aspects of host physiology (Kanther and Rawls, 2010; Kanther et al., 2011; Cheesman et al., 2011; Camp et al., 2012). The zebrafish gut microbiota, like that of humans and other mammals, are dominated by the bacterial phyla Proteobacteria, Firmicutes, and Bacteroidetes (Rawls et al., 2004; Rawls et al., 2006; Roeselers et al., 2011). However, the impact of diet on the zebrafish gut microbiota, and their relationship to the microbiota in the surrounding aqueous environment, is unknown. We recently developed a method to monitor FA absorption into the intestinal epithelium by incubating zebrafish in liposomes containing FA analogs fluorescently labeled with BODIPY (BODIPY-FL) (Carten et al., 2011; Walters et al., 2012). Here, we use this method to investigate the impact of microbial colonization and diet on intestinal FA absorption. Our work reveals a diet-dependent role for the microbiota in stimulating FA absorption into the intestine and extra-intestinal tissues, and provides an ecological explanation for the diet-dependent nature of this host-microbe interaction.
RESULTS
The microbiota promote intestinal fatty acid accumulation in a diet-dependent manner
To determine the impact of the microbiota on dietary lipid absorption in the intestine, we used BODIPY-labeled palmitic acid (BODIPY-C16), which represents the most common saturated long-chain FA found in triglycerides, and medium chain pentanoic acid (BODIPY-C5; the BODIPY fluorophore effectively adds 2-3 carbons in length to the C5:0 FA backbone), which is more rapidly absorbed in the intestine (Carten et al., 2011). We incubated 6 dpf GF and CONVD animals with BODIPY-C5 or BODIPY-C16 emulsified in egg yolk liposomes for 6 hrs (Figure 1A). To test whether the prior nutritional status of the animal can impact dietary lipid absorption, we performed this assay in the absence (starved) or presence of a sterile control diet from 3 dpf (C-fed; Table S1). Feeding was initiated at 3 dpf to provide a nutrient-rich environment for both zebrafish and microbiota; however, zebrafish do not begin consuming food until ~5 dpf. Therefore, zebrafish reared under starved conditions are only deprived of exogenous nutrition for ~1 day prior to the 6 dpf endpoint. Confocal imaging of the proximal intestine in live zebrafish revealed luminal and epithelial fluorescence under all tested conditions, indicating FA analog ingestion and absorption, respectively (Figure 1B,C). Quantification of intestinal epithelial fluorescence revealed higher levels of BODIPY-C5 and BODIPY-C16 fluorescence in CONVD compared to GF zebrafish (p<0.001 for C5, and p<0.0001 for C16), which was enhanced by prior exposure to diet (Figure 1D,F). In contrast, we observed a consistent trend of higher luminal fluorescence in GF than CONVD animals (p<0.05 for C5, n.s. for C16)(Figure 1E,G). These results suggest that the microbiota promote dietary FA accumulation in the intestinal epithelium in a diet-dependent manner.
Figure 1. Fatty acids accumulate in the intestinal epithelium in the presence of microbiota and diet.
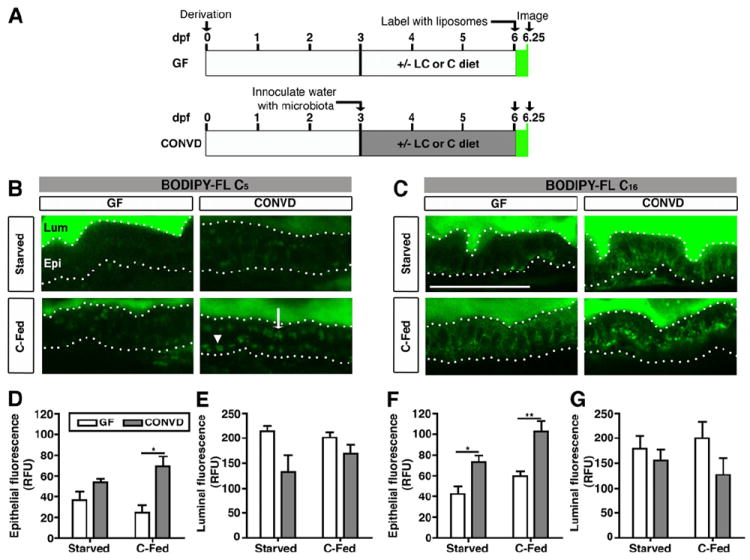
(A) Schematic of BODIPY-FL delivery assay in gnotobiotic zebrafish. Zebrafish derived germ-free (GF) at 0 days post-fertilization (dpf) were either reared GF (top) or inoculated at 3 dpf with normal microbiota (conventionalized, CONVD; bottom). From 3-6 dpf, fish were either starved, or fed a control (C) or low calorie (LC) diet (see Table S1). At 6 dpf, zebrafish were incubated with BODIPY-FL liposomes for 6 hrs and imaged or fixed for later imaging.
(B,C) Representative confocal images of the intestines of live 6 dpf GF and CONVD zebrafish incubated with BODIPY-FL C5 or C16. Scale bar, 50 μm.
(B) The intestinal lumen (Lum) and epithelium (Epi; bounded by dotted lines) of GF and CONVD zebrafish are indicated. The epithelium shows apical (white arrow) and basolateral accumulation of lipid droplets (white arrowhead) labeled with BODIPY-C5.
(C) Incubation with BODIPY-FL C16.
(D-G) Quantification of total epithelial (D,F) and luminal (E,G) fluorescence expressed in relative fluorescence units (RFU). Values represent the means ± SEM from 3 independent experiments: *, p<0.05; **, p<0.01.
The microbiota increase lipid droplet number and size in the intestinal epithelium
We next sought to determine if the observed BODIPY-labeled FA accumulation in the intestinal epithelium was due to alterations in LD formation in the enterocytes. Because microbial colonization and feeding appeared to have similar effects on epithelial accumulation of BODIPY-labeled medium- and long-chain FAs (Figure 1), we used the medium-chain FA analog BODIPY-C5 to develop a quantitative assay for LD formation in the intestinal epithelium (see Experimental Procedures and Figure S1A-F). A timecourse with BODIPY-FL C5 showed no difference in accumulated epithelial LD numbers between GF and CONVD fish after a 3 hr incubation (Figure S1G,H), and increased epithelial LD numbers after a longer 6 hr incubation only in animals that were both colonized with microbiota and fed (Figure 2A,B). This finding confirmed that interactions between microbiota and diet stimulate FA absorption (p<0.01)(Figure 2B). By analyzing the relative frequency of LD sizes, we observed a high percentage of small LDs after the 3 hr incubation in all conditions (Figure S1I). The percentage of large LDs was increased in all CONVD conditions after the 6 hr incubation (i.e., 5.6-11 μm2 area, p<0.01), whereas the percentage of small (i.e., <0.55 μm2 area, p<0.001) and medium LDs (i.e., 0.55-1.64 μm2 area, p<0.001) was strongly altered by diet (Figure 2C). These results suggest that the presence of microbiota promotes two distinct phenotypes of epithelial LD formation: increased LD size regardless of diet history and increased LD number only in fed animals. These phenotypes only appeared after 6 hrs of BODIPY-FL incubation, suggesting that the microbiota may increase enterocyte capacity for FA storage rather than accelerating initial LD formation. Notably, the microbiota also increased LD number and size in zebrafish fed a low calorie diet (LC-fed) that has 31.4% fewer calories than the control diet due to a 55.6% reduction in protein content (Table S1 and data not shown), suggesting that diet-dependent microbial stimulation of LD accumulation in the zebrafish gut is not determined by the caloric value or protein content of the diet.
Figure 2. The microbiota stimulate lipid absorption into intestinal epithelial lipid droplets and extra-intestinal tissues.
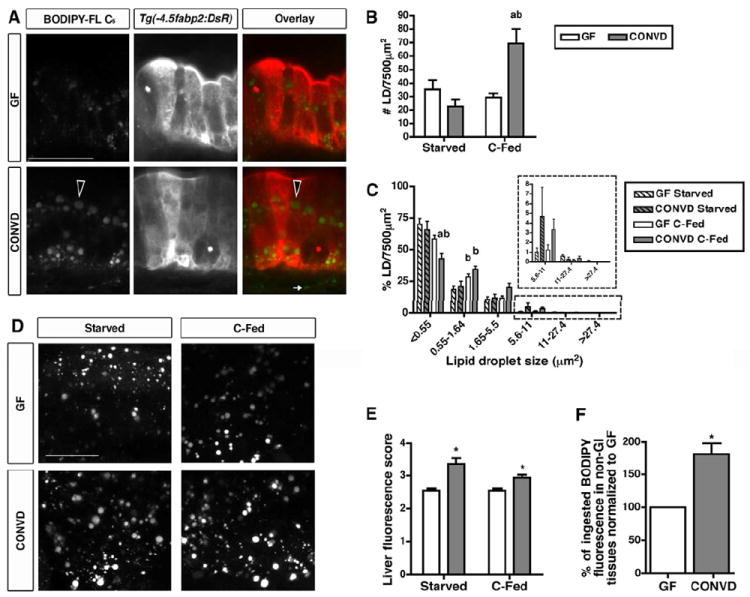
(A) Representative confocal images of fixed 6 dpf Tg(-4.5fabp2:DsRed) GF and CONVD zebrafish fed a control diet and incubated with BODIPY-FL C5 for 6 hrs. Scale bar, 20 μm. Intestinal epithelial cells show BODIPY-C5 accumulation as lipid droplets (LDs) in the epithelium and the lamina propria (white arrow). Large LDs are detected in the epithelium of CONVD zebrafish (black arrowheads).
(B,C) Lipid droplet quantification assay was developed using Volocity software (see Figure S1A-F) to determine LD number (B) and size frequency (C) in an epithelial region of interest (7500 μm2). The graphs depict the mean ± SEM of at least two independent experiments (3-15 fish/condition/experiment). Results of statistical significance analysis: a, significant vs. GF fed same diet; b, significant vs. starved in same microbial condition. See Figure S2 for data from a 3 hr timepoint.
(D) Representative confocal images of livers in 6 dpf GF and CONVD zebrafish incubated with BODIPY-FL C5 for 6 hrs. Scale bar, 20 μm.
(E) BODIPY-C5 fluorescence scores in livers of 6 dpf GF and CONVD zebrafish. The graph depicts the mean ± SD of two independent experiments (3-5 fish/condition/experiment) that were scored blindly (score scale 0-5).
(F) Non-GI BODIPY-C5 fluorescence in GF and CONVD C-fed zebrafish. The data represent mean ± SD of two experiments (20-30 carcasses and 9-10 whole larvae/condition/experiment). Significant differences are indicated: *, p<0.05 (E,F).
Microbial stimulation of dietary lipid absorption into extra-intestinal tissues
We postulated that the increased LD number and size in CONVD fed zebrafish could be due to either delayed export of chylomicrons or FFAs into the circulation or increased absorption of FAs from the lumen. Indeed, previous studies in mice have shown that accumulation of large LDs can be associated with reduced chylomicron exocytosis (Buhman et al., 2002). To test the possibility that chylomicron exocytosis is impaired in CONVD fed zebrafish, we performed a washout experiment to compare the ability of GF and CONVD zebrafish to clear LDs from their epithelium after BODIPY-FL C5 incubation. We incubated 6 dpf GF and CONVD C-fed zebrafish with BODIPY-FL C5 for 3 hrs and subsequently washed a subset of animals from each condition in sterile GZM for an additional 5 hrs (Figure 3A). Consistent with our prior observations (Figure 2), the 3 hr labeling resulted in similar LD numbers in the intestines of both GF and CONVD animals (Figure 3B,C). The washout reduced the number of LDs in the epithelium of both GF and CONVD zebrafish; however, this reduction was more prominent in CONVD fish (p<0.001)(Figure 3B,C). These data indicate that FA transport from the intestine of CONVD animals is more robust than GF zebrafish. Analysis of LD size frequency following the washout in GF and CONVD animals revealed similar reductions in small LDs (i.e., <0.55 μm2, p<0.01) and increases in medium (i.e., 1.65-5.5 μm2, p<0.01) and large LDs (i.e., 5.6-11 μm2, p<0.0001; 11-27.4 μm2, p<0.05) (Figure 3D). This result indicates that the significant reduction in LD number in post-wash CONVD animals (Figure 3C) is not due to enhanced LD fusion in the presence of microbiota. Furthermore, these data suggest that the observed increase in LD number and size in CONVD fish (Figure 2) is not due to impaired intestinal lipid export. The increase in epithelial LD accumulation in fed CONVD animals was also not associated with significant differences in ingestion rates, digestive organ size, or in vivo activity of digestive enzymes phospholipase and protease in the gut lumen (Figure S2).
Figure 3. Lipid droplet clearance is more efficient in the presence of microbiota.
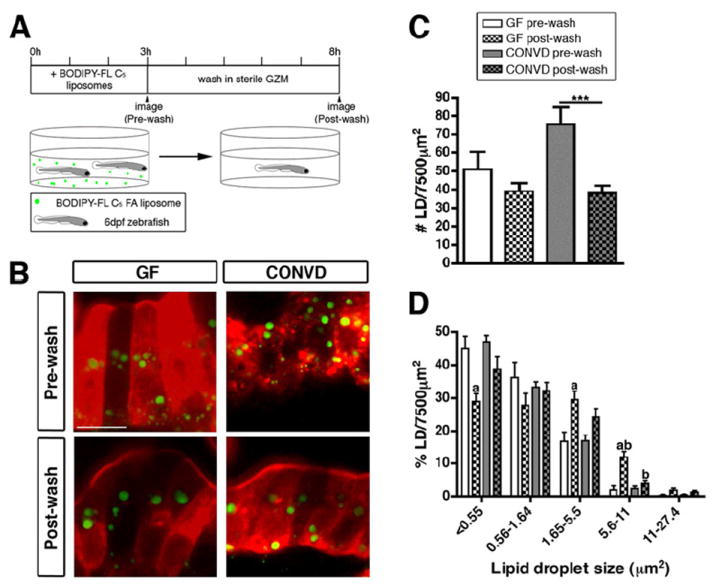
(A) Schematic representation of the BODIPY-FL C5 washout experiment.
(B) Representative confocal images of control-fed GF and CONVD zebrafish pre- and post-wash. Scale bar, 10 μm.
(C,D) Quantification of lipid droplet (LD) number (C) and relative size frequency (D), shown as the mean ± SEM from two independent experiments (4-14 fish/condition/experiment), and significant differences are identified: ***, p<0.001; a, significant vs. pre-wash in same microbial condition; b, significant vs. same wash in other microbial condition. See also Figure S2.
We next asked whether the increases in intestinal epithelial LD accumulation elicited by the microbiota are associated with increased FA export from the intestine to other tissues. After BODIPY-C5 molecules are absorbed and exported from the zebrafish gut, a primary site of accumulation is in LDs within the liver (Carten et al., 2011). We found that the presence of a microbiota resulted in increased BODIPY fluorescence in hepatic LDs after a 6 hr incubation in BODIPY-FL C5 (p<0.01) (Figure 2D,E). This increase was observed in both starved and C-fed CONVD zebrafish larvae (Figure 2E), suggesting that the microbiota promotes intestinal absorption and export of FA to the liver irrespective of diet history and diet-dependent effects of the microbiota on intestinal LD number. Additionally, CONVD animals displayed a significant increase in accumulation of BODIPY-C5 fluorescence in non-GI tissues compared to GF controls (82 ± 16% increase)(Figure 2F). Based on these results, we conclude that the ability of the microbiota to induce accumulation of LDs in the intestinal epithelium is associated with increased absorption of dietary FAs into extra-intestinal tissues.
Diet determines bacterial community composition in the zebrafish gut
Our results show that the microbiota promote intestinal epithelial LD number in fed but not in starved zebrafish. We hypothesized that this may be due to diet-induced alterations in gut microbiota composition. Diet-dependent differences in zebrafish gut microbiota composition could be due to direct effects of diet on the host gut habitat (e.g., alterations in host physiology or immunity). Alternatively, they could be due to indirect effects of diet on the surrounding aqueous environment (e.g., alterations in water chemistry and nutrient availability), which would modify the microbial community in the water that is available to colonize the zebrafish gut. We therefore analyzed the impact of diet on the bacterial communities that form in the zebrafish gut and in the surrounding water. GF zebrafish embryos were colonized at 3 dpf with a common conventional zebrafish microbiota (inoculum, Figure S3A) and subsequently either starved, or fed sterile C or LC diet until 6 dpf. Microbial genomic DNA was extracted from zebrafish guts and the housing water, and the respective bacterial communities were analyzed using 16S rRNA gene pyrosequence-based surveys (Figure S3A and Table S2). UniFrac principal coordinates analysis (PCoA) plots derived from both unweighted (an evaluation of community composition) and weighted (an evaluation of community structure) algorithms (Lozupone et al., 2007) provided several insights. First, biological replicates from each of the sample groups consistently clustered together (Figure 4A,B), suggesting that the processes determining the composition of these bacterial communities are reproducible. Second, we observed a striking separation of gut and water samples, revealing that the zebrafish gut selects or enriches a distinctive subset of bacteria from the surrounding water. Notably, the inoculum sample appeared central to all other samples, reflecting the fact that the majority of operational taxonomic units (OTUs) observed in the inoculum sample were also detected in other samples (108/111 or 97.3%; Figure S3B-E). Third, fed samples clustered together and were separated from the starved samples for both the gut and water environments, suggesting that feeding markedly alters the gut and water bacterial communities in distinctive ways. Furthermore, the clustering of LC-fed and C-fed samples indicates that the effect of diet on those communities is not strongly determined by the caloric and protein content of the diet.
Figure 4. 16S rRNA gene sequencing reveals distinct bacterial communities in the zebrafish gut and water that are strongly influenced by dietary status.
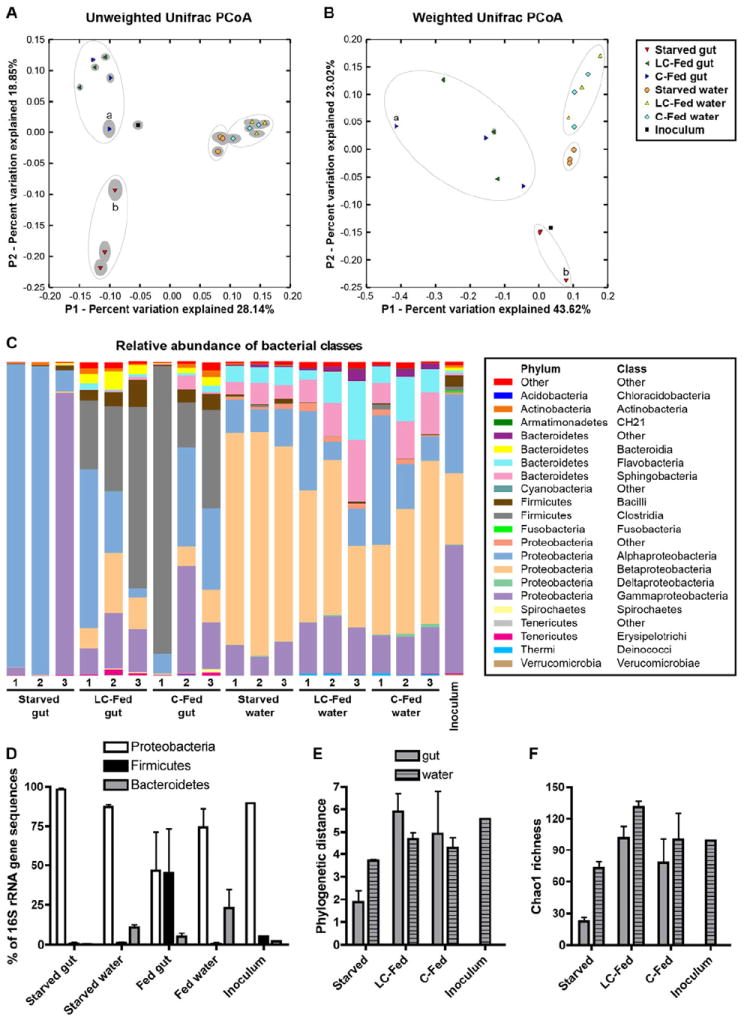
(A,B) UniFrac principal coordinates analysis (PCoA) plots using unweighted (A) and weighted (B) algorithms. Each replicate sample is represented by a single shape, with the solid grey ellipsoid around each shape indicating the confidence interval from 100 jackknife replicates of 500 sequences per sample. Apparent clusters of samples are indicated with open ovals. Samples C-Fed gut 1 (a) and Starved gut 3 (b) are labeled. See also Table S2.
(C) Stacked bar graph showing relative abundance (Y-axis) of 16S rRNA gene sequences from different bacterial classes (legend at right) observed in different samples (X-axis).
(D) Percentage of 16S rRNA gene sequences classified as Proteobacteria, Firmicutes, and Bacteroidetes, shown as the mean ± SD across different replicate sample groups and the inoculum sample. See also Table S3. Alpha diversity measures of (E) Phylogenetic distance and (F) Chao1 richness are shown as the mean ± SD across different replicate sample groups and the inoculum sample. See also Figure S3 and Table S4.
The distinct separation of gut and water samples observed in the PCoA analysis was accompanied by significant differences in the relative abundances of several bacterial taxa (Figure 4C,D and Table S3). Although all samples were dominated by Proteobacteria phylum sequences, sequences from the β-Proteobacteria class were enriched in the water (mean 49.7% vs. 6.4% of all water and gut samples, respectively; p<0.0001). Bacteroidetes phylum sequences were also enriched in water (mean 19% vs. 3.3% of all water and gut samples, respectively; p<0.01), through increases in classes Flavobacteria and Sphingobacteria. In contrast, Firmicutes phylum sequences were enriched in gut samples (mean 30.2% vs. 0.6% of all gut and water samples, respectively; p<0.05) through increases in classes Bacilli and Clostridia. Strikingly, gut-specific enrichment of Firmicutes occurred only in animals that had been fed, and not in starved animals (Figure 4C,D), indicating that feeding results in gut-specific enrichment of Firmicutes in zebrafish hosts.
In addition to affecting the relative abundance of specific bacterial taxa, the overall diversity of these bacterial communities also varied as a function of diet and environment. In the gut, bacterial diversity and richness were markedly lower in starved animals compared to those fed C or LC diet (e.g., mean phylogenetic distances of 1.87 vs. 5.41 in starved vs. C/LC-fed gut samples, respectively, p<0.005; Figure 4E,F and Table S4). In contrast, diversity and richness in water communities were not significantly different between fed and starved conditions. These diet-induced increases in gut microbiota diversity were not accompanied by significant alterations in the density of culturable microorganisms in the gut (Figure 5A). These results reveal that the presence of diet promotes diversity of bacterial communities in the zebrafish gut, without inducing similar alterations in the surrounding water.
Figure 5. Monoassociation with individual community members reveals diet-dependent colonization of a representative Firmicutes species.
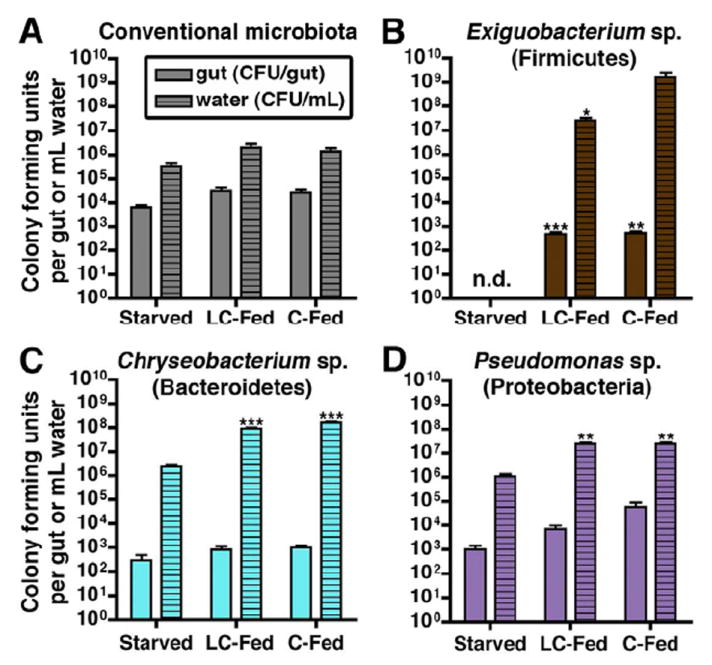
(A-D) Colony forming units (CFU) in the intestine (per dissected gut; n=4-5 per condition) or surrounding water (per mL; n=3 per condition) of 6 dpf zebrafish. The results represent the mean ± SEM of at least two independent experiments (n.d., not detected) with identified significant differences: *, p<0.05; **, p<0.01; ***, p<0.001.
(A) Density of the conventional microbiota in CONVD zebrafish.
(B-D) Bacterial densities in GF zebrafish monoassociated with (B) Exiguobacterium sp. ZWU0009 (Firmicutes), (C) Chryseobacterium sp. ZOR0023 (Bacteroidetes), or (D) Pseudomonas sp. ZWU0006 (γ-Proteobacteria). See also Table S5.
Although the trends among biological replicates were largely consistent, there were two instances in which one replicate deviated significantly from the others (C-Fed gut 1 and Starved gut 3; Figure 4A-C). The sequences in these samples grouped into OTUs that were also present in other samples (data not shown), suggesting that these deviations are not due to contamination from external sources but due to differences in relative OTU abundance. These two deviant samples may result from technical variation or may reflect stochasticity in gut microbial community assembly (Robinson et al., 2010).
A Firmicutes strain isolated from the zebrafish intestine is sensitive to exogenous nutrient levels in the absence of microbial competition
The ecological processes underlying diet-dependent Firmicutes enrichment in the intestines of vertebrate hosts remain unknown. We speculated that the Firmicutes enrichment is due in part to their improved ability to initiate or maintain colonization of the gut in the presence of dietary nutrients. To test this possibility, we inoculated GF zebrafish at 3 dpf with individual, culturable members of the zebrafish microbiota (a process called monoassociation; Table S5) and measured colonization efficiency in the zebrafish gut and surrounding water in the absence of competition from other microbes. Notably, colony forming unit (CFU) densities of a representative Firmicutes Exiguobacterium sp. ZWU0009 (class Bacilli) were below the level of detection in both gut and water under starved conditions but established robust colonization under fed conditions (Figure 5B). In contrast, the strains Chryseobacterium sp. ZOR0023 (phylum Bacteroidetes, class Flavobacteria; Figure 5C) and Pseudomonas sp. ZWU0006 (phylum Proteobacteria, class γ-Proteobacteria; Figure 5D) were able to colonize the gut and water under starved conditions, with only modest increases in CFU density under fed conditions. This finding indicates that even in a simple community without microbial competition, the Firmicutes strain we tested (Exiguobacterium sp.) requires nutrient-rich conditions to survive and successfully colonize the gut, whereas strains from other taxa (Chryseobacterium sp. and Pseudomonas sp.) can colonize even in the absence of dietary nutrients. These results suggest that diet-dependent Firmicutes enrichment in the CONVD (Figure 4D) gut might be due at least in part to an autonomous increase in Firmicutes colonization efficiency in the presence of diet.
Monoassociations reveal two distinct pathways for bacterial stimulation of intestinal fatty acid absorption
Our BODIPY-FL C5 labeling experiments revealed that the microbiota stimulated two phenotypes in enterocyte FA absorption: a diet-dependent increase in LD number and a diet-independent increase in LD size. This result raised the possibility that bacterial taxa that are enriched in a diet-dependent manner, such as Firmicutes, might promote LD number. To test this hypothesis, we performed monoassociation experiments on C-fed GF zebrafish using the same three bacterial strains as above and performed the BODIPY-FL C5 delivery assay (Figure 6A-D). Although there was an overall effect of colonization on enterocyte LD number (p<0.001), the only strain to induce a significant increase in LD number was the Firmicutes strain Exiguobacterium sp. (Figure 6B). In contrast, monoassociation with Exiguobacterium sp. had no effect on LD size frequency compared to GF controls, whereas the other two tested bacterial strains induced increases in the relative frequency of large LDs in the intestinal epithelium (Figure 6F). Analysis of BODIPY fluorescence in the livers of monoassociated zebrafish revealed a significant effect of colonization across all 3 tested species compared to GF controls (p<0.05; Figure 6G). However, the only individual strain sufficient to significantly increase BODIPY fluorescence in the liver was Exiguobacterium sp. (Figure 6G). These results suggest two mechanisms for bacterial stimulation of intestinal FA absorption: a Firmicutes-mediated induction of enterocyte LD number that is associated with robust FA export to the liver, and a non-Firmicutes bacterial induction of enterocyte LD size (Figure 7).
Figure 6. Monoassociations reveal distinct bacterial mechanisms for inducing fatty acid absorption in the intestinal epithelium.
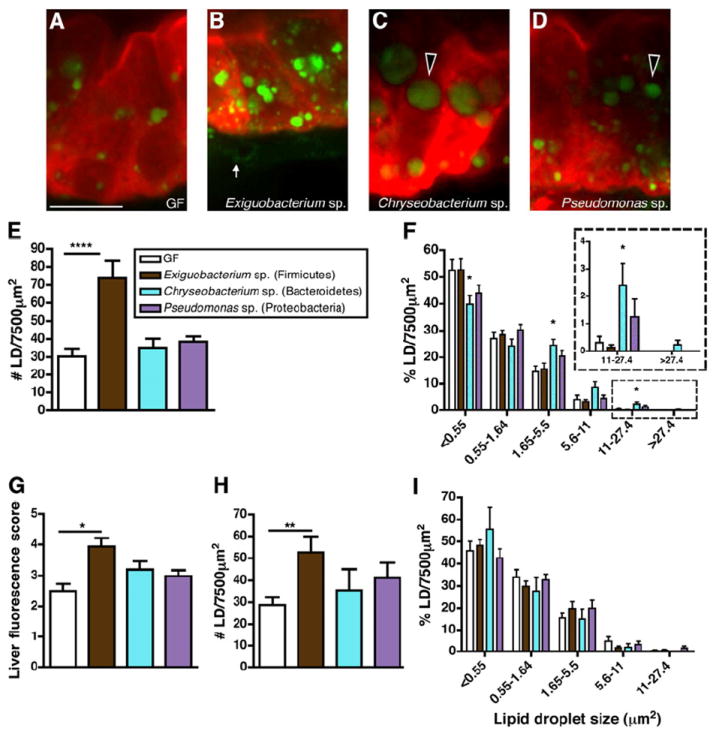
(A-D) Representative confocal images of the intestinal epithelium of 6 dpf C-fed zebrafish raised GF (A) or monoassociated with Exiguobacterium sp. ZWU0009 (B), Chryseobacterium sp. ZOR0023 (C), or Pseudomonas sp. ZWU0006 (D) incubated with BODIPY-FL C5 for 6 hrs. BODIPY-C5 accumulation in large epithelial lipid droplets (black arrowheads) and in the lamina propria (white arrow) is indicated.
(E) Lipid droplet quantification in the intestinal epithelium of monoassociated zebrafish compared to GF controls.
(F) Relative frequency of intestinal LD sizes in monoassociated zebrafish. The data represent the mean ± SEM of at least two independent experiments (5-16 fish/condition/experiment). Significant differences compared to GF controls are identified: *, p<0.05; ***, p<0.001.
(G) BODIPY-C5 fluorescence scores in the livers of monoassociated zebrafish compared to GF controls. The graph represents the mean ± SEM of at least two independent experiments (2-7 fish/condition/experiment) that were scored blindly (score scale 0-5). Significant differences to GF controls are identified: *, p<0.05.
(H,I) Intestinal LD number (H) and relative frequency of intestinal LD sizes (I) in GF zebrafish treated with filter-sterilized Exiguobacterium sp., Chryseobacterium sp., or Pseudomonas sp. conditioned media. Results are presented as mean ± SEM of at least two independent experiments (3-7 fish/condition/experiment) with identified significant differences compared to GF control: **, p<0.01.
Figure 7. Model for diet-dependent microbial regulation of intestinal fatty acid absorption.
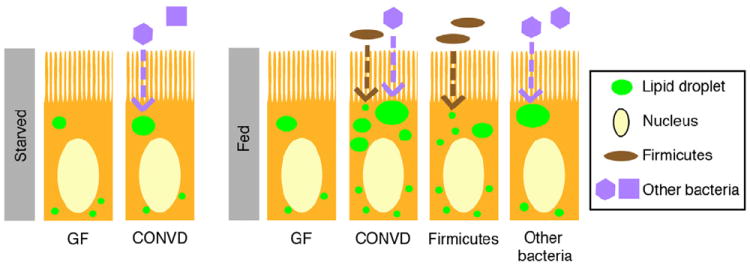
The microbiota promote LD size in intestinal enterocytes independent of diet (dashed violet arrow). The presence of diet promotes LD number in CONVD zebrafish and in those monoassociated with a Firmicutes strain (i.e., Exiguobacterium sp.; dashed brown arrow). Monoassociation with other bacterial strains Chryseobacterium sp. or Pseudomonas sp. promotes LD size. Although the extent to which these findings are generalizable to their respective phyla remains unclear, these data suggest two bacterial mechanisms that promote distinct LD accumulation phenotypes: a Firmicutes-induced increase in LD number and a non-Firmicutes bacterial induction in LD size.
We speculated that these distinct effects of individual bacterial strains on enterocyte FA absorption might be caused by distinct bacterial factors. To test this possibility, we introduced GF larvae at 3 dpf to filter-sterilized media that had been conditioned with one of the 3 tested bacterial strains, and then performed the BODIPY-FL C5 delivery assay at 6 dpf. We found that only conditioned media from Firmicutes Exiguobacterium sp. was sufficient to significantly increase enterocyte LD number (Figure 6H). This increase in LD number is similar to that observed in zebrafish monoassociated with live Exiguobacterium sp. (Figure 6E), indicating that this Firmicutes strain can increase enterocyte LD number via its bacterial products. In contrast, none of the conditioned medias were sufficient to increase enterocyte LD size (Figure 6I), suggesting that the increase in enterocyte LD size observed in animals monoassociated with Chryseobacterium sp. or Pseudomonas sp. (Figure 6F) might require the activities of live bacterial cells. Together, these results indicate that the distinct effects of bacterial strains on enterocyte LD accumulation are due to distinct molecular mechanisms, and that bacterial products from the Firmicutes strain Exiguobacterium sp. are sufficient to increase enterocyte LD number.
DISCUSSION
There is currently intense interest in understanding the gut microbiota’s contributions to vertebrate nutrient metabolism and energy balance (Musso et al., 2011). Although microbial contributions to degradation of complex dietary carbohydrates have been studied extensively (Flint et al., 2012), the impact of the microbiota on dietary lipid metabolism has received relatively little attention. Previous investigations of dietary lipid metabolism in gnotobiotic mammals evaluated serum lipid metabolites (Bäckhed et al., 2004; Bäckhed et al., 2007; Martin et al., 2009), which do not distinguish between exogenous and endogenous lipid sources, and serum chylomicrons (Velagapudi et al., 2010) or fecal crude fat (Yoshida et al., 1968; Rabot et al., 2010), which do not do not distinguish between dietary and microbe-produced lipids. Here, we used an in vivo imaging strategy in transparent zebrafish larvae to uncover a role for the microbiota in stimulating dietary FA absorption in the intestinal epithelium and extra-intestinal tissues. Our results identify the gut microbial community as a target for controlling dietary fat absorption and energy balance.
Fatty acid absorption, intracellular LD assembly in enterocytes, and subsequent secretion as chylomicrons and FFA have been extensively studied (Iqbal and Hussain, 2009). However, our mechanistic understanding of these physiologic processes in vivo remains incomplete, which poses challenges for understanding how they are regulated by environmental factors such as the microbiota. Our data reveal that colonization with a microbiota promotes epithelial absorption of FAs, resulting in accumulation of LDs in enterocytes and increased accumulation of dietary FAs in extra-intestinal tissues. We propose four nonexclusive mechanisms by which microbes might stimulate FA absorption and LD accumulation in enterocytes. First, microbes might increase bioavailability of FAs by modifying the production or composition of bile salts (Swann et al., 2011). Second, microbes could directly contribute to luminal lipolytic activity that promotes FA availability for potential absorption in the intestinal epithelium (Ringø et al., 1995). Third, microbes might enhance FA absorption indirectly by evoking physiologic responses in the intestinal epithelium that stimulate its inherent absorptive capabilities. Finally, the microbiota might reduce rates of fatty acid oxidation in intestinal epithelial cells permitting increased storage of FA in LDs. Identification of the underlying mechanisms could facilitate microbe-based strategies for controlling dietary fat absorption.
Our results show that the presence of a microbiota promotes two distinct phenotypes of LD formation within the enterocyte: increased LD number and increased LD size. The majority of enterocyte LDs evaluated in this study presumably represent temporary storage LDs that occur after high-fat feeding (Glatz et al., 2010). While enterocyte LD size and number reflect distinct and quantifiable aspects of FA absorption, they likely share common cell biological processes. Previous genetic analyses of LD formation have established that these dynamic organelles are under complex regulatory control and functionally linked to other cellular organelles and pathways (Guo et al., 2008; Beller et al., 2008). However, we anticipate that at least a subset of the ‘small LDs’ enumerated in this study (<0.55 μm2) represent chylomicrons, as this small size range is inclusive of the predicted chylomicron size in teleosts (Sire et al., 1981). Investigation of the mechanisms underlying the microbial and dietary regulation of these distinct lipid-rich organelles could provide insights into enterocyte lipid metabolism.
In addition to revealing a role for the microbiota in stimulating FA absorption, our data show that this host response to the microbiota is influenced by diet history. Although enterocyte LD size was increased by the microbiota regardless of diet history, consistent increases in enterocyte LD number were only observed in fed animals. These results are surprising since our data were collected at 6 dpf only ~1 day after they normally begin feeding. This suggests that the observed effects of starvation on zebrafish at 6 dpf might be determined primarily by diet-dependent effects on the microbiota rather than direct effects of diet on the host. Consistent with this notion, we discovered that the presence of diet results in alterations in the zebrafish gut microbiota including enrichment of Firmicutes bacteria. Furthermore, we find that monoassociation of GF zebrafish with a representative Firmicutes strain induced increased LD number, whereas two other non-Firmicutes strains induced increased LD size. Although it remains unknown if these findings are generalizable to other members of their respective phyla, these data are consistent with our observations that the intestines of fed CONVD animals enriched with Firmicutes display increased LD number while all CONVD animals display increased LD size regardless of diet history. Based on these results, we propose two distinct mechanisms to explain the observed diet-dependent interactions between gut microbial ecology and host FA absorption. First, Firmicutes are enriched in the intestines of fed animals, where they enhance the ability of host enterocytes to absorb FAs. Second, non-Firmicutes bacteria that colonize the gut irrespective of dietary status induce increased accumulation of large LDs within host enterocytes. In animals that are fed, these two bacterial signals combine to stimulate FA absorption through increases in both enterocyte LD size and number (Figure 7) and increased export to extra-intestinal tissues.
Our results provide insight into the impact of diet on the zebrafish gut microbiota, and the relationship between the microbiota of the zebrafish gut and the surrounding water. A frequently observed pattern in humans, mice, and pythons is that the relative abundance of Firmicutes in the gut is positively correlated with dietary caloric intake. Our evaluation of gut bacterial communities in fed and starved zebrafish revealed that this ecological principle also applies to bony fishes. Strikingly, diet-dependent enrichment of Firmicutes bacteria in the gut but not in the surrounding water provides strong evidence that the presence of diet exerts different selective pressures on bacteria in the zebrafish gut versus the surrounding water. Future longitudinal analyses of the zebrafish gut and surrounding water could help resolve temporal relationships between the respective diet-induced alterations in microbial community assembly and maintenance in these different habitats.
The mechanisms that promote Firmicutes abundance in nutrient-rich environments remain unresolved. Our monoassociation results suggest that the diet-dependent enrichment of Firmicutes may be due, at least in part, to an autonomous bacterial requirement for diet-derived nutrients to allow colonization. In contrast, the two non-Firmicutes bacterial strains tested here colonized in both starved and fed conditions, suggesting that nutritional niches provided by the host are sufficient for these microorganisms to colonize the zebrafish gut. It will be interesting to determine if diet-dependent enrichment of Firmicutes in the context of a more complex microbial community is also mediated in part by inter-microbe competitions. Furthermore, it will be important to determine whether the impact of diet on bacterial colonization of the gut is mediated by permitting initial gut colonization and/or maintenance of colonization over time. Firmicutes-enriched communities arising from genetic or diet-induced obesity have been shown to promote positive energy balance (Turnbaugh et al., 2006; Turnbaugh et al., 2008). Therefore, identification of the mechanisms underlying diet-dependent enrichment of Firmicutes could lead to new approaches for controlling energy balance in humans and other animals.
EXPERIMENTAL PROCEDURES
Gnotobiotic zebrafish husbandry
All experiments using zebrafish were conducted in conformity with the Public Health Service Policy on Humane Care and Use of Laboratory Animals using protocols approved by the Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill. All zebrafish were TL wild-type strain unless otherwise noted. Zebrafish embryos were derived GF and maintained in sterile conditions or colonized at 3 dpf with microbiota or individual bacterial strains (Table S5) according to published protocols (Pham et al., 2008). Zebrafish were maintained in gnotobiotic zebrafish media (GZM) at 28.5°C in 50 mL and 250 mL sterile tissue culture flasks (VWR International, LLC) on a 14 hr light cycle with daily 80% media changes. Control (C) and low-calorie (LC) diets (Table S1) were custom formulated and ground to a pellet size of 50-100 μm (Ziegler Brothers Inc.) and were then sterilized by irradiation (absorbed dose range 106.5-135.2 kGy; Neutron Products Inc.). Zebrafish were either fed once per day beginning 3 dpf with approximately 2.5 and 4 mg sterilized C or LC diet (for the 50 and 250 mL flasks, respectively). Conditioned medium was prepared fresh each day by filter sterilization of GZM incubated overnight with individual bacterial strains and Control diet. Sterility of GF zebrafish and zebrafish treated with conditioned media was assessed using culture-based methods as previously described (Pham et al., 2008). All zebrafish were treated with 0.015 mg/L 1-phenyl-2-thiourea (PTU)(Lancaster Synthesis, Inc.) to reduce melanin synthesis. At 6 dpf, homogenized gut and water samples were serially diluted and plated on Tryptic Soy (for the CONVD condition) or Brain Heart Infusion (for the monoassociated conditions) agar to determine colony forming units (CFU).
Liposome delivery assay
Fluorescent liposome solution preparation and incubation for live imaging was performed as described (Carten et al., 2011) using BODIPY-C5 (Invitrogen, D-3834) or BODIPY-C16 (Invitrogen, D-3821) fatty acids. For the lipid droplet measurements, zebrafish were euthanized in 0.67 g/L of tricaine (Argent Chemical Laboratories), fixed in freshly prepared 4% paraformaldehyde (PFA) (Acros) in phosphate buffered saline (PBS) for 1 hr at room temperature to preserve LD structure (DiDonato and Brasaemle, 2003), washed 3 times in PBS, and stored in a sterile 12-well plate at 4°C in the dark. For confocal imaging, fixed zebrafish were mounted on Superfrost Plus microscope slides (Fisher Scientific) in 1% low melting temperature agarose (Fisher Scientific) and sealed with high vacuum grease (Dow Corning).
Confocal imaging of BODIPY-FL labeled zebrafish
Live confocal imaging of BODIPY-FL labeled zebrafish was performed as previously described (Carten et al., 2011). Images were taken with a Leica TCS SP2 confocal microscope using HCX APO L W 40x (3.3 WD, 0.8 NA) and 63x (2.2 WD, 0.9 NA) oil-immersion objectives. Fluorescence quantification was performed in ImageJ by box analysis and was normalized to controls that were not incubated with liposomes. Imaging of fixed BODIPY-FL labeled zebrafish was performed on an Olympus FV1000 laser scanning confocal microscope. For these experiments, we used the Tg(-4.5fabp2:DsRed)pd1000 zebrafish line that expresses DsRed in the intestinal epithelium (Kanther et al., 2011). Imaging was performed with a 60x water-immersion objective (0.28 WD, 0.9 NA). We took Z-stacks through the same region of the proximal intestine in every fish using the multi Ar laser (488 nm excitation) and LD laser (559 nm excitation) at laser capacity of 3% and 6.9%, respectively. Z-stacks were collected using unidirectional laser scanning at 1.0–1.5 μm per slice. The image size was 640×640 pixels in the XY direction for quantified Z-stacks and 1600×1600 pixels for the images shown in Figures 2, 3, 6 and S1G.
Lipid droplet quantification
Three independent epithelial regions of interest in each fish were selected randomly from sagittal Z slices near the middle of the intestine that contained a single layer of DsRed-positive epithelial cells below a clear luminal space. Quantification of BODIPY-FL fluorescence in the intestinal epithelium was performed in three individual slices from the overlay Z-stacks using the Volocity Visualization+Quantification module (Improvision) (see Figure S1). For each slice, we identified a region of interest (ROI, 2500 μm2) and applied a measurement protocol (Figure S1F) to identify individual objects (lipid droplets; LDs) and their size (represented by area). The LD identification and measurement protocol consisted of the following steps, which are presented in order: find objects using % intensity (lower: 50%; upper: 100%); clip objects to ROIs; separate touching objects (object size guide: 5 μm2); and filter measurements (ID > 3). Collected measurement values were exported to Microsoft Excel to determine total LD number and the number of LDs for each size category. The relative frequency of LD size categories was determined as a percentage of the total LD number from each individual fish.
Statistical methods
The significance of diet and microbial status on lipid absorption, digestive enzyme activity, growth, and feeding behavior was determined by two-way ANOVA (p values reported in Results section). Statistical significance between individual conditions was based on individual Student’s t test adjusted for unequal variances (determined by the Bartlett’s test) and corrected by Bonferroni’s method for multiple comparisons (p values reported in Figures). We used ANOVA corrected for false discovery rate (Benjamini and Hochberg, 1995) to identify significant differences in relative abundance of bacterial taxa based on the Ribosomal Database Project (RDP) classifier version 2.2 (Wang et al., 2007) and in normalized alpha diversity distances (p values reported in Results). Any p values less than 0.05 with correction were considered significant.
Supplementary Material
Highlights.
Microbiota stimulate intestinal fat absorption in the zebrafish
Microbial stimulation of intestinal lipid droplet (LD) number is diet dependent
Diet promotes enrichment of Firmicutes bacteria in the zebrafish gut microbiota
Diet-enriched Firmicutes and their products stimulate intestinal LD number
Acknowledgments
The authors are grateful to Cathy Lozupone for assistance with pyrosequence data analysis, to James Bear and Stephen Jones for assistance with confocal imaging and fluorescence quantification, and to Michael Chua and James Minchin for assistance with Volocity software. We are also grateful to Gray Camp, Jordan Cocchiaro, Stephen Jones, and James Minchin for blindly scoring liver images. Chryseobacterium sp. ZOR0023 was a generous gift from Erika Mittge and Karen Guillemin at University of Oregon. We thank the UNC-Olympus Research Imaging Center for providing access to the Olympus FV1000 confocal microscope. We also thank the Core for Applied Genomics and Ecology (CAGE) at the University of Nebraska-Lincoln for 454 pyrosequencing services. This work was funded by grants from NIH (DK081426, GM095385 to J.F.R.; DK78669, HG4872, HG4866 to R.K.; DK093399, GM63904 to S.A.F.; DK091129 to J.D.C.). Additional funding was provided by the Pilot Project Award from the UNC Center for Environmental Health and Susceptibility (ES010126) and a Pew Scholars in the Biomedical Sciences award to J.F.R.; the Carnegie Institution for Science endowment and the G. Harold and Leila Y. Mathers Charitable Foundation to S.A.F.; and the Howard Hughes Medical Institute to R.K.. I.S. was funded in part through the Susan Fellner Physiology Graduate Student Fellowship (UNC-Chapel Hill).
Footnotes
Supplemental experimental procedures
Supplemental Information accompanying this manuscript includes Supplemental Results, Supplemental Experimental Procedures, Supplemental References, three Figures, and five Tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Babin PJ, Vernier JM. Plasma lipoproteins in fish. J Lipid Res. 1989;30(4):467–489. [PubMed] [Google Scholar]
- Bäckhed F, Ding H, Wang T, Hooper LV, Koh GY, Nagy A, Semenkovich CF, Gordon JI. The gut microbiota as an environmental factor that regulates fat storage. Proc Natl Acad Sci USA. 2004;101(44):15718–15723. doi: 10.1073/pnas.0407076101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104(3):979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Statist Soc B. 1995;57(1):289–300. [Google Scholar]
- Beller M, Sztalryd C, Southall N, Bell M, Jäckle H, Auld DS, Oliver B. COPI complex is a regulator of lipid homeostasis. PLoS Biol. 2008;6(11):e292. doi: 10.1371/journal.pbio.0060292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudry G, David ES, Douard V, Monteiro IM, Le Huërou-Luron I, Ferraris RP. Role of intestinal transporters in neonatal nutrition: carbohydrates, proteins, lipids, minerals, and vitamins. J Pediatr Gastroenterol Nutr. 2010;51(4):380–401. doi: 10.1097/MPG.0b013e3181eb5ad6. [DOI] [PubMed] [Google Scholar]
- Buhman KK, Smith SJ, Stone SJ, Repa JJ, Wong JS, Knapp FF, Burri BJ, Hamilton RL, Abumrad NA, Farese RV. DGAT1 is not essential for intestinal triacylglycerol absorption or chylomicron synthesis. J Biol Chem. 2002;277(28):25474–25479. doi: 10.1074/jbc.M202013200. [DOI] [PubMed] [Google Scholar]
- Camp JG, Jazwa AL, Trent CM, Rawls JF. Intronic cis-regulatory modules mediate tissue-specific and microbial control of angptl4/fiaf transcription. PLoS Genetics. 2012;8(3):e1002585. doi: 10.1371/journal.pgen.1002585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carten JD, Bradford MK, Farber SA. Visualizing digestive organ morphology and function using differential fatty acid metabolism in live zebrafish. Dev Biol. 2011;360(2):276–285. doi: 10.1016/j.ydbio.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carten JD, Farber SA. A new model system swims into focus: using the zebrafish to visualize intestinal metabolism in vivo. Clinical Lipidology. 2009;4(4):501–515. doi: 10.2217/clp.09.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheesman SE, Neal JT, Mittge E, Seredick BM, Guillemin K. Epithelial cell proliferation in the developing zebrafish intestine is regulated by the Wnt pathway and microbial signaling via Myd88. Proc Natl Acad Sci USA. 2011;108(Suppl 1):4570–4577. doi: 10.1073/pnas.1000072107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costello EK, Gordon JI, Secor SM, Knight R. Postprandial remodeling of the gut microbiota in Burmese pythons. ISME J. 2010;4(11):1375–1385. doi: 10.1038/ismej.2010.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford PA, Crowley JR, Sambandam N, Muegge BD, Costello EK, Hamady M, Knight R, Gordon JI. Regulation of myocardial ketone body metabolism by the gut microbiota during nutrient deprivation. Proc Natl Acad Sci USA. 2009;106(27):11276–11281. doi: 10.1073/pnas.0902366106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato D, Brasaemle DL. Fixation methods for the study of lipid droplets by immunofluorescence microscopy. J Histochem Cytochem. 2003;51(6):773–780. doi: 10.1177/002215540305100608. [DOI] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes. 2012 doi: 10.4161/gmic.19897. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatz JFC, Luiken JJFP, Bonen A. Membrane fatty acid transporters as regulators of lipid metabolism: implications for metabolic disease. Physiol Rev. 2010;90(1):367–417. doi: 10.1152/physrev.00003.2009. [DOI] [PubMed] [Google Scholar]
- Guo Y, Walther TC, Rao M, Stuurman N, Goshima G, Terayama K, Wong JS, Vale RD, Walter P, Farese RV. Functional genomic screen reveals genes involved in lipid-droplet formation and utilization. Nature. 2008;453(7195):657–661. doi: 10.1038/nature06928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt MA, Hoffmann C, Sherrill-Mix SA, Keilbaugh SA, Hamady M, Chen Y, Knight R, Ahima RS, Bushman F, Wu GD. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology. 2009;137(5):1716–1724.e1-2. doi: 10.1053/j.gastro.2009.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iqbal J, Hussain MM. Intestinal lipid absorption. Am J Physiol Endocrinol Metab. 2009;296:E1183–E1194. doi: 10.1152/ajpendo.90899.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jumpertz R, Le DS, Turnbaugh PJ, Trinidad C, Bogardus C, Gordon JI, Krakoff J. Energy-balance studies reveal associations between gut microbes, caloric load, and nutrient absorption in humans. Am J Clin Nutr. 2011;94(1):58–65. doi: 10.3945/ajcn.110.010132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Rawls JF. Host-microbe interactions in the developing zebrafish. Curr Opin Immunol. 2010;22:10–19. doi: 10.1016/j.coi.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanther M, Sun X, Mühlbauer M, Mackey LC, Flynn EJ, Bagnat M, Jobin C, Rawls JF. Microbial colonization induces dynamic temporal and spatial patterns of NF-κB activation in the zebrafish digestive tract. Gastroenterology. 2011;141(1):197–207. doi: 10.1053/j.gastro.2011.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasov WH, Hume ID. The vertebrate gastrointestinal system. In: Dantzler WH, editor. Handbook of Physiology, Section 13: Comparative Physiology. Vol. 1. New York: Oxford University Press; 1997. pp. 407–480. [Google Scholar]
- Kindel T, Lee DM, Tso P. The mechanism of the formation and secretion of chylomicrons. Atheroscler Suppl. 2010;11(1):11–16. doi: 10.1016/j.atherosclerosissup.2010.03.003. [DOI] [PubMed] [Google Scholar]
- Ley RE, Bäckhed F, Turnbaugh P, Lozupone CA, Knight RD, Gordon JI. Obesity alters gut microbial ecology. Proc Natl Acad Sci USA. 2005;102(31):11070–11075. doi: 10.1073/pnas.0504978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444(7122):1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- Ley RE, Hamady M, Lozupone C, Turnbaugh PJ, Ramey RR, Bircher JS, Schlegel ML, Tucker TA, Schrenzel MD, Knight R, et al. Evolution of mammals and their gut microbes. Science. 2008;320(5883):1647–1651. doi: 10.1126/science.1155725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Hamady M, Kelley ST, Knight R. Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl Environ Microbiol. 2007;73(5):1576–1585. doi: 10.1128/AEM.01996-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin FJ, Sprenger N, Yap IKS, Wang Y, Biblioni R, Rochat F, Rezzi S, Cherbut C, Kochhar S, Lindon JC, et al. Panorganismal gut microbiome-host metabolic crosstalk. J Proteome Res. 2009;8(4):2090–2105. doi: 10.1021/pr801068x. [DOI] [PubMed] [Google Scholar]
- Meek TH, Eisenmann JC, Garland T. Western diet increases wheel running in mice selectively bred for high voluntary wheel running. Int J Obes (Lond) 2010;34(6):960–969. doi: 10.1038/ijo.2010.25. [DOI] [PubMed] [Google Scholar]
- Muegge BD, Kuczynski J, Knights D, Clemente JC, González A, Fontana L, Henrissat B, Knight R, Gordon JI. Diet drives convergence in gut microbiome functions across mammalian phylogeny and within humans. Science. 2011;332(6032):970–974. doi: 10.1126/science.1198719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy EF, Cotter PD, Healy S, Marques TM, O’Sullivan O, Fouhy F, Clarke SF, O’Toole PW, Quigley EM, Stanton C, et al. Composition and energy harvesting capacity of the gut microbiota: relationship to diet, obesity and time in mouse models. Gut. 2010;59(12):1635–1642. doi: 10.1136/gut.2010.215665. [DOI] [PubMed] [Google Scholar]
- Musso G, Gambino R, Cassader M. Interactions between gut microbiota and host metabolism predisposing to obesity and diabetes. Annu Rev Med. 2011;62:361–380. doi: 10.1146/annurev-med-012510-175505. [DOI] [PubMed] [Google Scholar]
- Pham LN, Kanther M, Semova I, Rawls JF. Methods for generating and colonizing gnotobiotic zebrafish. Nat Protoc. 2008;3(12):1862–1875. doi: 10.1038/nprot.2008.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabot S, Membrez M, Bruneau A, Gérard P, Harach T, Moser M, Raymond F, Mansourian R, Chou CJ. Germ-free C57BL/6J mice are resistant to high-fat-diet-induced insulin resistance and have altered cholesterol metabolism. FASEB J. 2010;24(12):4948–4959. doi: 10.1096/fj.10-164921. [DOI] [PubMed] [Google Scholar]
- Rawls JF, Mahowald MA, Ley RE, Gordon JI. Reciprocal gut microbiota transplants from zebrafish and mice to germ-free recipients reveal host habitat selection. Cell. 2006;127(2):423–433. doi: 10.1016/j.cell.2006.08.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawls JF, Samuel BS, Gordon JI. Gnotobiotic zebrafish reveal evolutionarily conserved responses to the gut microbiota. Proc Natl Acad Sci USA. 2004;101(13):4596–4601. doi: 10.1073/pnas.0400706101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringø E, Strøm E, Tabachek JA. Intestinal microflora of salmonids: a review. Aquat Res. 1995;26:773–789. [Google Scholar]
- Robinson CJ, Bohannan BJM, Young VB. From structure to function: the ecology of host-associated microbial communities. Microbiol Mol Biol Rev. 2010;74(3):453–476. doi: 10.1128/MMBR.00014-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roeselers G, Mittge EK, Stephens WZ, Parichy DM, Cavanaugh CM, Guillemin K, Rawls JF. Evidence for a core gut microbiota in the zebrafish. ISME J. 2011;5(10):1595–1608. doi: 10.1038/ismej.2011.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Sire MF, Lutton C, Vernier JM. New views on intestinal absorption of lipids in teleostean fishes: an ultrastructural and biochemical study in the rainbow trout. J Lipid Res. 1981;22:81–94. [PubMed] [Google Scholar]
- Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, Nicholson JK, Holmes E. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci USA. 2011;108(1):4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Bäckhed F, Fulton L, Gordon JI. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe. 2008;3(4):213–223. doi: 10.1016/j.chom.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turnbaugh PJ, Ley RE, Mahowald MA, Magrini V, Mardis ER, Gordon JI. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature. 2006;444(7122):1027–1031. doi: 10.1038/nature05414. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi VR, Hezaveh R, Reigstad CS, Gopalacharyulu P, Yetukuri L, Islam S, Felin J, Perkins R, Borén J, Oresic M, et al. The gut microbiota modulates host energy and lipid metabolism in mice. J Lipid Res. 2010;51(5):1101–1112. doi: 10.1194/jlr.M002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters JW, Anderson JL, Bittman R, Pack M, Farber SA. Visualization of lipid metabolism in the zebrafish intestine reveals a relationship between NPC1L1-mediated cholesterol uptake and dietary fatty acid. Chemistry & Biology. 2012 doi: 10.1016/j.chembiol.2012.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Wang Q, Garrity GM, Tiedje JM, Cole JR. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–5267. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu GD, Chen J, Hoffmann C, Bittinger K, Chen Y, Keilbaugh SA, Bewtra M, Knights D, Walters WA, Knight R, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida T, Pleasants JR, Reddy BS, Wostmann BS. Efficiency of digestion in germ-free and conventional rabbits. Br J Nutr. 1968;22(4):723–727. doi: 10.1079/bjn19680082. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


