Abstract
The lack of markers to predict chemotherapy responses in patients poses a major handicap in cancer treatment. We searched for gene expression patterns that correlate with docetaxel or cisplatin response in a mouse model for breast cancer associated with BRCA1 deficiency. Array-based expression profiling did not identify a single marker gene predicting docetaxel response, despite an increase in Abcb1 (P-glycoprotein) expression that was sufficient to explain resistance in several poor responders. Intertumoral heterogeneity explained the inability to identify a predictive gene expression signature for docetaxel. To address this problem, we used a novel algorithm designed to detect differential gene expression in a subgroup of the poor responders which could identify tumors with increased Abcb1 transcript levels. In contrast, standard analytical tools, such as Significance Analysis of Microarrays (SAM), detected a marker only if it correlated with response in a substantial fraction of tumors. For example, low expression of the Xist gene correlated with cisplatin hypersensitivity in most tumors, and it also predicted long recurrence-free survival of HER2-negative, stage-III breast cancer patients treated with intensive platinum-based chemotherapy. Our findings may prove useful for selecting patients with high risk breast cancer who could benefit from platinum-based therapy.
Keywords: breast cancer, BRCA1, chemotherapy response prediction, gene expression profiling, XIST
INTRODUCTION
Most forms of cytotoxic cancer chemotherapy also hit normal tissues. This is acceptable when the tumor responds, but frustrating when the tumor is intrinsically resistant and the patient only suffers from the side effects of an unsuccessful treatment. A major goal of molecular oncology is therefore to identify biomarkers that predict the response of tumors before treatment is started. Such predictive markers have been found for some targeted therapies in which the target and its interaction with drugs are well defined (1-7). For classical cytotoxic chemotherapy with DNA damaging drugs or antimitotics, however, predictive biomarkers have been harder to find.
In an attempt to find new biomarkers many investigators have turned to the analysis of genome-wide gene expression profiles. These profiles have been successful for predicting prognosis, i.e. whether patients will require adjuvant chemotherapy after tumor removal (8). Prognostic and predictive biomarkers are fundamentally different, however (9). To detect predictive markers, considerable effort and money has been invested in the analysis of human breast cancer samples (10). In particular the neoadjuvant setting seemed attractive to correlate gene expression profiles with therapy outcome. No clear response profile was obtained, however (11;12). Other studies have gathered a number of unrelated signatures (9). These profiles either still await validation in an independent study; or the sensitivity and specificity was inadequate for clinical decision making; and some were based on flawed data (13-15). Moreover, cell line-based approaches to identify biomarkers suffer from the complication that the multidrug resistance transcriptome has been substantially altered during the long-term culture of these cell lines in vitro (16).
As progress in defining useful biomarkers using human tumor material has been limited, we have turned to a mouse model. In recent years chemotherapy responses have been investigated in a new generation of genetically engineered mouse models (GEMMs) (17). These models employ conditional, tissue-specific activation of oncogenes and/or deletion of tumor suppressor genes in a stochastic fashion. The resulting tumors closely mimic the epithelial cancers in humans. Using the K14cre;Brca1F/F;p53F/F model for hereditary breast cancer (18) we have shown that these tumors acquire resistance to classical and novel targeted anti-cancer drugs such as the topoisomerase I-targeting camptothecin topotecan, the topoisomerase II-inhibiting anthracycline doxorubicin, and the PARP inhibitor olaparib (19-22). We have observed that the initial response of these tumors is variable, as in human tumors, thus providing an attractive opportunity to correlate drug response with gene expression. The tumors are similar, as they start out with the ablation of the Brca1 and the p53 genes. Differences between tumors should make it comparatively easy to sort out which genes determine whether a tumor responds to drug or not. An advantage of this model is that tumors can be orthotopically transplanted into syngeneic, immunocompetent animals without losing their molecular fingerprint, morphologic phenotype or drug sensitivity (19). Using this orthotopically transplantable mouse model, we set out to find predictive markers of cisplatin or docetaxel response.
MATERIALS AND METHODS
Mice and drug treatments
KB1P mammary tumors were generated, genotyped, orthotopically transplanted and treated as described (18;19). Additional details including the generation of KB1PM mammary tumors can be found in Supplementary Materials. All experimental procedures on animals were approved by the Animal Ethics Committee of the Netherlands Cancer Institute.
Genome-wide expression profiling
RNA extraction, amplification, and microarray hybridization using dual channel MEEBO arrays (Illumina BV, Eindhoven) were performed as described (19;22;23). For the gene expression analysis using single channel 45K MouseWG-6 v2.0 BeadChips (Illumina, Eindhoven, The Netherlands) total RNA was processed according to the manufacture’s instructions (http://www.illumina.com/products/mousewg_6_expression_beadchip_kits_v2.ilmn). More details on the processing and analysis of the microarray data are presented in Supplementary Materials.
Quantitative RNA analyses using Reverse Transcription-Multiplex ligation-dependent probe amplification (RT-MLPA) or TaqMan low density arrays (TLDA)
These procedures were carried out as reported previously (20;21;24). Additional information is presented in Supplementary Materials.
ArrayCGH
ArrayCGH data was available from a recent study (25). Segmentation of the CGH profiles was performed with the CGHseg package (26). The CGHcall R package (v 2.12.0) was used to call aberrations in CGH profiles.
FISH
Three samples per individual tumor were investigated in a blinded fashion using tissue microarrays of the trial cohort. At least 100 nuclei per sample were assessed. More information on the protocol is presented in Supplementary Materials. If the number of cells with no XIST RNA clouds was >60%, the sample was classified 0 for “XIST RNA cloud”. In the presence of one X chromosome detected by the RNF12 DNA probe, XIST RNA was usually absent (loss of Xi). In the presence of two X chromosomes, loss of Xi and a XaXa configuration was defined as a more than 50% reduction in the number of expected XIST RNA clouds based on the RNF12 DNA FISH.
Patients
In a previous study stage-III HER2-negative breast cancer patients were randomly selected from a large randomized controlled trial (RCT) performed in the Netherlands between 1993 and 1999 (27) and analyzed for aCGH classification (25). Further details on these patients are presented in Supplementary Materials. All trials described in this manuscript were approved by the Institutional Review Board of the Netherlands Cancer Institute.
RESULTS
Brca1-/-;p53-/- (KB1P) mammary tumors show individual and reproducible differences in docetaxel or cisplatin sensitivity
We have previously shown that individual KB1P mammary tumors differ substantially in their response to docetaxel (19). The response to cisplatin varied as well: although all tumors were sensitive to cisplatin, the time until relapse differed between tumors (19). To exploit these inter-tumoral differences, we analyzed docetaxel or cisplatin responses of 43 individual tumors (Supplementary Fig. S1).
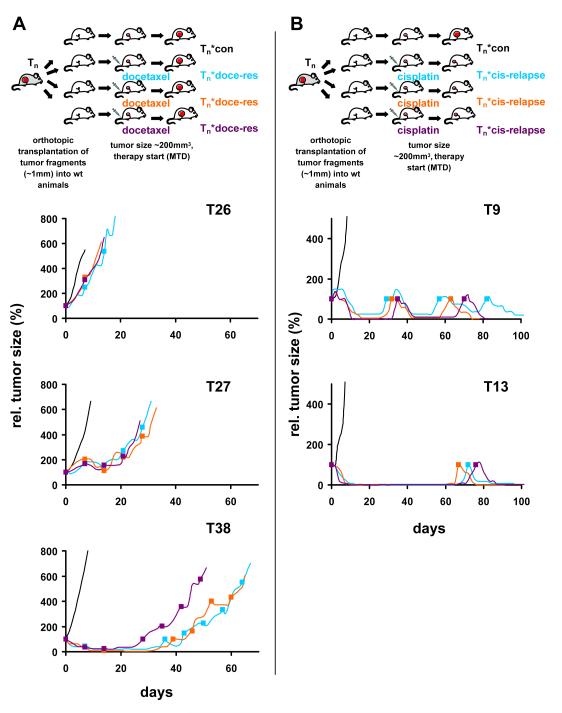
The correlation of drug sensitivities with characteristics of a particular tumor is only possible if the responses are reproducible. We therefore explored the heterogeneity within a single tumor by orthotopic transplantation of multiple tumor fragments (Fig. 1). For this purpose, 3 animals carrying orthotopically transplanted fragments of the same spontaneous tumor were treated with the maximum tolerable dose of docetaxel on days 0, 7 and 14. Fig. 1A shows that the docetaxel response was consistent for all 3 fragments derived from one tumor (T26 was consistently poor; T38 responded well; T27 fragments all had an intermediate response). The rate at which the tumors eventually become completely resistant to docetaxel differs somewhat between fragments from the same tumor (T38*docetaxel 3 vs. 1 or 2), as previously observed for doxorubicin (19). The initial drug response is reproducible, however. Also for cisplatin we confirmed that the time to relapse was reproducible (Fig. 1B; T9 tumor fragments all relapsed early, T13 fragments all relapsed late). Hence, this tumor model can be used to correlate initial docetaxel or cisplatin responses with other tumor characteristics, such as gene expression levels.
Figure 1.
Reproducibility of docetaxel or cisplatin sensitivity of individual KB1P tumors using orthotopic transplantation. Tumor fragments of spontaneous mammary tumors were transplanted orthotopically into syngeneic wild-type female mice as shown in (A) for docetaxel and (B) for cisplatin. When tumors reached a volume of 150–250mm3, animals were treated with docetaxel (A, filled squares) or cisplatin (B, filled squares). Treatment of tumors was resumed once the tumor relapsed to its original size (100%).
Supervised gene expression profiling does not yield a general signature that correlates with docetaxel response
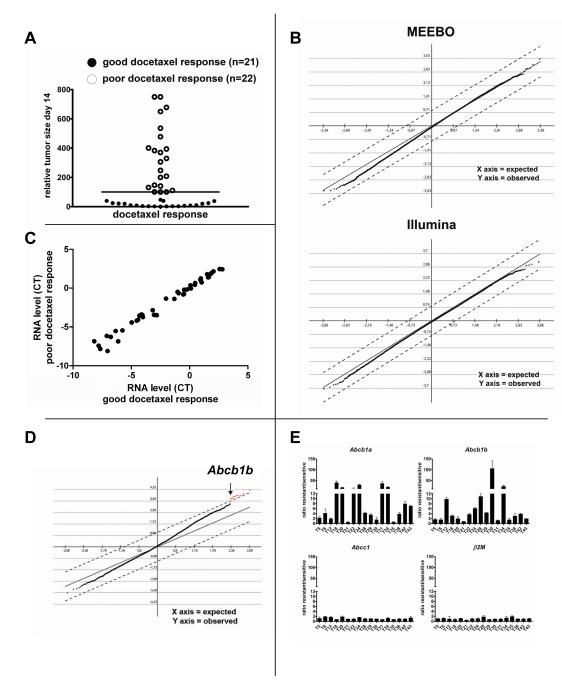
In our model we used the tumor volume as the basis for a response classifier. We found that after completion of the initial docetaxel treatment on day 14, 22 tumors did not shrink below their original size when treatment was started (100%), and eventually continued growing (‘poor response’). In contrast, 21 tumors regressed to a volume below 50% of the original size (‘good response’), and took on average 28 days (SD 11d) after the last docetaxel treatment to grow back to 100% (Fig. 2A and Supplementary Table S1). With such an obvious separation, we expected to identify genes that are differentially expressed between these 2 groups. To detect these, RNA of all 43 untreated tumors was analyzed using 39K Mouse Exonic Evidence Based Oligonucleotide (MEEBO) gene expression microarrays, and 45K Illumina Sentrix mouse V6 single-channel bead arrays. Unsupervised hierarchical cluster analysis did not separate good from poor responders (Supplementary Fig. S2). For the supervised analysis we used Significance Analysis of Microarrays (SAM) (28), which is frequently applied to detect differential gene expression. SAM uses non-parametric statistics to compute for each gene a delta that measures the strength of the relationship between gene expression and docetaxel sensitivity. Permutations of repeated measurements are employed to estimate the false discovery rate (FDR). Using this approach we expected to find several differentially expressed genes between good and poor docetaxel responders with a delta>0.7 (FDR<5%). Remarkably, this analysis did not detect a single gene that correlated with drug sensitivity with a meaningful delta (Fig. 2B).
Figure 2.
Analysis of intrinsic (A, B, C) or acquired (D, E) docetaxel resistance of KB1P tumors using gene expression profiling. A, Relative tumor size of 43 individual tumors after completion of initial treatment using 25mg docetaxel per kg i.v. on days 0, 7 and 14. Tumors with a relative volume below 100% (bar) were classified as good responders, the remaining as poor responders. B, SAM of untreated tumors of good vs. poor docetaxel responders (Δ=0.7) using the MEEBO or Illumina platforms. C, Average of median-normalized cycle threshold (CT) values determined by quantitative TLDA of 46 genes encoding ABC proteins. D, SAM of tumors that acquired docetaxel resistance (Δ=1.1, FDR=0). E, Ratios of Abcb1a or Abcb1b gene expression (RT-MLPA) of docetaxel-resistant tumors and samples from the matched drug-sensitive control tumors. Error bars indicate standard deviation of three independent reactions.
This negative result might be due to the lack of sensitivity of the gene expression platforms used for genes that are relevant for drug resistance. This is exemplified by the work of Orina et al (29) on drug transporters of the ATP-binding cassette (ABC) family. Using the NCI-60 panel of cell lines, they showed that TaqMan low density arrays (TLDA) are more precise and more sensitive in measuring the expression of these transporter genes than oligonucleotide arrays (29). Within this ABC family, a number of genes has been associated with docetaxel resistance, including ABCB1/P-glycoprotein (P-gp) (30), ABCC2 (31) and ABCC10 (32). We therefore examined whether the more quantitative TLDA analysis of the 49 genes that encode mouse ABC proteins would reveal differences between poor and good docetaxel responders. As shown in Fig. 2C and Supplementary Table S2, none was found at a significance level of P<0.01 (Mann-Whitney U test).
We note in passing that on both platforms used to analyze gene expression (Supplementary Fig. S2) two poor responders (T26*con and T41*con) form a separate branch which correlates with the sarcomatoid morphology (carcinosarcoma) of these tumors (Supplementary Table S1). Most likely, these 2 tumors have undergone an epithelial-mesenchymal transition (EMT), since in the K14cre;Brca1F/F;p53F/F model the Cre-mediated deletion of the Brca1 and p53 genes selectively occurs in epithelial cells driven by the K14 promoter. Whether such a morphologic change correlates with drug resistance is under investigation.
Increased gene expression of the Abcb1a and Abcb1b genes is frequently found in acquired docetaxel resistance
Since our analysis of gene expression did not turn up a single gene that correlated with intrinsic docetaxel resistance, we tested tumors with acquired docetaxel resistance. Genes responsible for the acquired resistance might also cause intrinsic docetaxel resistance. We therefore compared RNA from samples of the same tumor before treatment and after they had become resistant to docetaxel. Unsupervised hierarchical cluster analysis did not separate sensitive from resistant tumors. Instead, tumors derived from the same original tumor were found in close proximity (Supplementary Fig. S3A). Exceptions are tumors T20 and T38, but the docetaxel-resistant versions of these tumors (T20*doce-res or T38*doce-res) had a high content of stromal tissue (Supplementary Fig. S3B), explaining the unusual distance between resistant tumor and docetaxel-sensitive control.
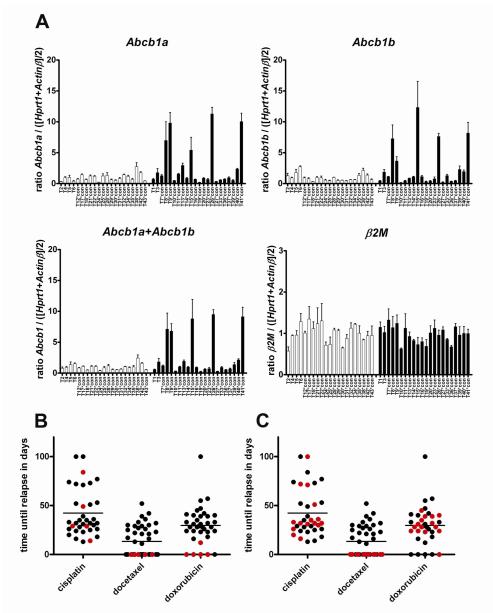
The SAM analysis of docetaxel-resistant tumors versus matched docetaxel-sensitive control tumors (Fig. 2D) yielded 9 genes that were significantly increased in docetaxel-resistant tumors (in red, see also Supplementary Table S3). Of these, only the Abcb1b gene -which encodes the mouse drug efflux transporter P-gp-can functionally explain docetaxel resistance. The other 8 genes (Gnγ10, Gp49a, lysozyme, Lzp-s, CD18, Trem2, Lilrb4, Slc11a1) appear to be linked to macrophages infiltrating drug-treated tumors to remove dead cells, as we have found previously for doxorubicin- or topotecan-resistant tumors (19;22). More precise quantification of the Abcb1a and Abcb1b transcripts that encode mouse P-gp by RT-Multiplex Ligation-dependent Probe Amplification (RT-MLPA) confirmed that one or both of the Abcb1 genes were upregulated at least 3-fold in 14 of the 17 tumors that acquired docetaxel resistance (Fig. 2E). We also investigated mouse Abcc1, which is a poor taxane transporter (33). Expression of this control gene was not altered in any of the docetaxel-resistant tumors.
Since the expression of Abcb1a was frequently found to be increased by RT-MLPA in the resistant tumors, it is surprising that it was not identified by the SAM analysis shown in Fig. 2D. This proved to be due to the poor sensitivity of the Abcb1a probe. When we investigated T18, T20, T22, T24, T31 and T34, the 6 tumors with a more than 10-fold increase in Abcb1a transcripts in the resistant tumors, as determined by RT-MLPA, Abcb1a was the top hit by SAM (Supplementary Fig. S4A). However, when we added 4 tumors with only ~4-fold increase in Abcb1a expression by RT-MLPA (T6, T28, T29 and T38), Abcb1a was lost as a significant gene (Supplementary Fig. S4B). This shows that the sensitivity of the Abcb1a probe is low in the MEEBO arrays.
Increased expression of the Abcb1a and Abcb1b genes can explain poor docetaxel response of 5/22 non-responders
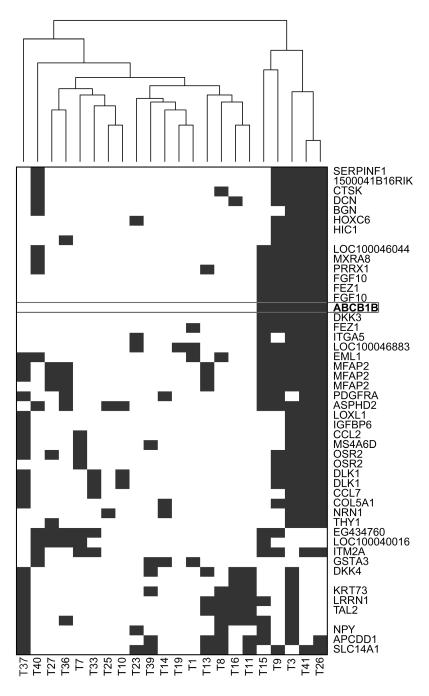
In addition to conventional SAM analyses we also tested an algorithm designed to specifically detect differential gene expression that only occurs in a subgroup of tumors within the non-responding group (34). This algorithm places a threshold on the gene expression corresponding to the highest expression level in the docetaxel responder group. For the docetaxel poor responders that exceed this threshold the sum of the differences of the expression is then calculated. Using this algorithm we found that Abcb1b was among the top outliers and formed a cluster with several other genes (Fig. 3 and Supplementary Table S4). This suggests that Abcb1b is not only involved in acquired docetaxel resistance, but may also contribute to upfront docetaxel resistance of some tumors. To further investigate whether an increased expression of the Abcb1a/b genes can explain the poor intrinsic docetaxel response of some of the 22 poor responder tumors (Fig. 2A), we quantified the RNA levels in the untreated tumors by RT-MLPA (Fig. 4A). In 5/22 tumors we found a 7- to 9-fold increase in Abcb1 RNA above the average level of the good responders. Abcb1a RNA was elevated as well in these 5 tumors (Fig. 4A). We have previously shown that a modest upregulation of Abcb1 by a factor 7-9 is sufficient to cause drug resistance in these tumors (21). Indeed, we found that the 5 tumors with increased Abcb1 gene expression also did not respond to the P-gp substrate doxorubicin (Fig. 4B and Supplementary Fig. S1), whereas the poor docetaxel responders without increased Abcb1 RNA usually shrank below 50% with doxorubicin (Fig. 4C and Supplementary Fig. S1). As expected, there was no correlation of Abcb1 transcript levels with cisplatin relapse-free survival (Fig. 4B,C), since cisplatin is not a substrate of P-gp.
Figure 3.
Identification of outliers present in subgroups of poor docetaxel responders using an algorithm developed by de Ronde et al. (34). Clustering analysis of top 50 ranked genes is shown. Each blue block represents a tumor with higher expression than the maximum of the expression of that particular gene in the responder group.
Figure 4.
Quantification of the expression of the mouse Abcb1 genes in untreated mouse mammary tumors. A, RT-MLPA analyses of Abcb1a or Abcb1b transcript levels of 43 individual KB1P tumors. Error bars indicate standard deviation of three independent reactions. B, time until relapse of KB1P tumors treated with the MTD of cisplatin, docetaxel or doxorubicin. The 5 tumors that showed increased Abcb1 gene expression are highlighted in red. In panel C the remaining 15 poor docetaxel responders that were also treated with cisplatin and doxorubicin are indicated in red.
P-gp-deficient mammary tumors are docetaxel hypersensitive
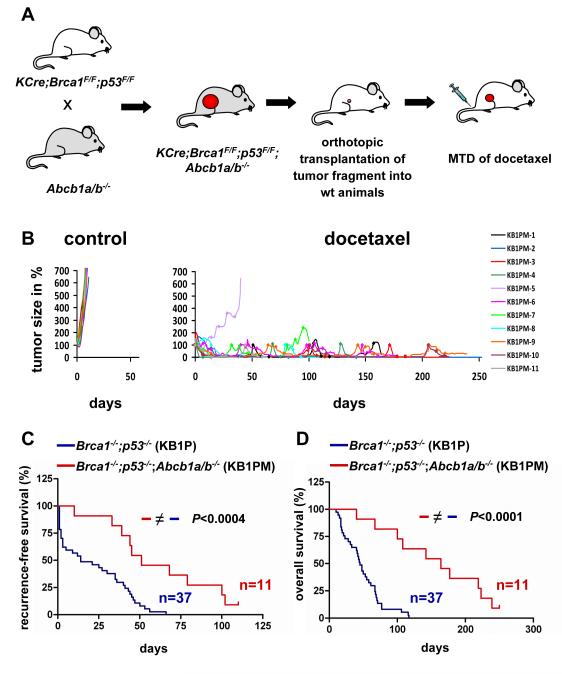
To improve our ability to detect P-gp-independent mechanisms of docetaxel resistance, we introduced the Abcb1a/b null alleles into the K14cre;Brca1F/F;p53F/F model. The lack of functional P-gp did not affect mammary tumor latency or morphology of the female K14cre;Brca1F/F;p53F/F;Abcb1a/b-/- mice (data not shown). P-gp-deficient mice carrying spontaneous mammary tumors cannot be treated with the docetaxel MTD, because P-gp contributes to the normal disposition of docetaxel in the mouse. We therefore grafted Brca1-/-;p53-/-;Abcb1a/b-/- tumors (KB1PM) orthotopically into syngeneic wild-type mice (Fig. 5A). In sharp contrast to Abcb1a/b wt tumors (KB1P), tumors unable to make P-gp were hypersensitive to the docetaxel MTD: only 1 out of 11 individual KB1PM tumors responded poorly to docetaxel and the mouse had to be sacrificed 40 days after the start of treatment (Fig. 5B). The median recurrence-free survival time increased significantly (P<0.0004) from 14 (T7-T43) to 51 days (KB1PM-1 and KB1PM-3 to -11), and for 1 tumor (KB1PM-2) no relapse occurred within 250 days, suggesting that this tumor was even eradicated (Fig. 5B,C). With the exception of KB1PM-5 none of the P-gp-deficient tumors acquired docetaxel resistance, and eventually the mice had to be killed due to cumulative docetaxel toxicity. The median survival of animals carrying orthotopically transplanted P-gp-deficient tumors increased significantly (P<0.0001) to 164 days (+/− 69 SD, n=11) compared with 45 days (+/− 28 SD, n=37) of animals with P-gp-proficient tumors (Fig. 5D). These data show that P-gp is a major contributor to docetaxel resistance of KB1P mammary tumors in vivo.
Figure 5.
Docetaxel responses of P-gp;BRCA1;p53-deficient (KB1PM) mammary tumors. A, Abcb1a/1b-/- alleles were crossed to homozygosity into the KCre;Brca1F/F;p53F/F model. KB1PM tumors were then orthotopically transplanted into female FVB/N animals and treated with docetaxel as indicated for KB1P tumors in Figure 1. B, 11 orthotopically transplanted KB1PM tumors were left untreated or received docetaxel (rhombi). Comparison of the time until tumors relapsed back to the original size of treatment start (C) or survival (D) of KB1PM-1 to -11 with the orthotopically transplanted P-gp-proficient KB1P tumors T7-T43 (logrank test).
Low expression of the Xist gene correlates with high cisplatin sensitivity of KB1P tumors and predicts benefit of platinum-based chemotherapy in patients with high risk primary breast cancer
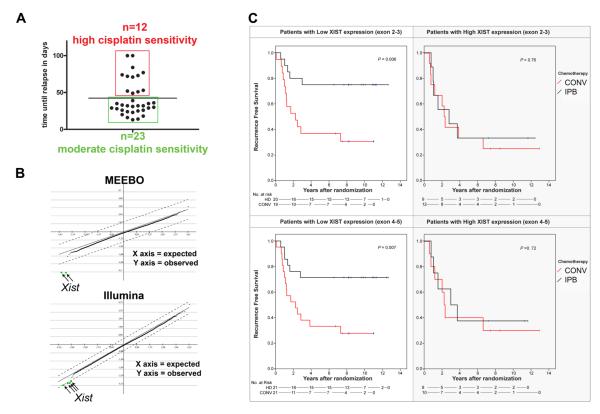
Since we found variation in the response to drug, not only for docetaxel, but also for cisplatin (Fig. 6A), we wondered whether standard gene expression analyses would also fail to identify predictive markers for this treatment. All KB1P tumors were cisplatin sensitive, but 23 tumors relapsed already within 39 days, whereas 12 tumors only grew back to 100% after 49 days. When we stratified the gene expression profiles of the untreated tumors based on their cisplatin sensitivity (above or below the mean time to relapse), we found a low expression of the Xist gene to correlate significantly with cisplatin hypersensitivity on 2 independent gene expression platforms (Fig. 6B).
Figure 6.
Correlation of gene expression with the response to platinum drugs. A, Time required for 35 individual KB1P tumors to grow back to 100% after cisplatin treatment. The bar indicates the mean. B, SAM of highly vs. moderately cisplatin-sensitive KB1P tumors using the MEEBO (Δ=1.5; FDR=0) or Illumina (Δ=0.7; FDR=0) platforms. C, KM survival curves according to XIST gene expression levels of patients who had been randomized between conventional (CONV, red) and intensive platinum-based chemotherapy (IPB, black). P values were calculated using the logrank test.
The physiological role of the non-coding RNA Xist is to coat one X chromosome of female cells in cis and subsequently trigger chromatin remodeling to form the heterochromatic Barr body (condensed inactivated X chromosome [Xi]). XIST is transcribed exclusively from the Xi to achieve equal X-linked gene dosage between the sexes. The analysis of genes correlating with a low Xist expression in our tumor model revealed a reduced expression of 3 other X-linked genes: Utx, Jarid1c, and Eif2s3x (Supplementary Fig. S5). As all of these genes are known to escape X inactivation (35), they are independent markers for the loss of the Xi.
Given the high frequency of reduced Xist expression in cisplatin hypersensitive mouse tumors, we tested whether XIST expression could serve as a biomarker to predict response to platinum-based chemotherapy in human breast cancer. For this purpose we took tumor samples of 60 stage-III, HER2-negative breast cancer patients who had been randomized between two treatment arms: intensive platinum-based chemotherapy, or a standard anthracycline-based regimen (conventional chemotherapy) (27). The patient information is summarized in Supplementary Table S5. To quantify XIST gene expression levels of FFPE material (>60% tumor cells), we used RT-MLPA including 2 independent probes hybridizing to the exon 2-3 or exon 4-5 boundary of XIST cDNA (Supplementary Table S6 and Supplementary Fig. S6). Analysis of the recurrence-free survival (RFS) showed that patients with a low XIST expression significantly benefited from the intensive platinum-based therapy compared to conventional chemotherapy: the 5-year RFS increased from 37% to 75% (Fig. 6C, adjusted hazard-ratio: 0.30, 95%CI: 0.11-0.82 for the probe of exon 4-5, Supplementary Table S5B). In patients with XIST gene expression above the cut-off no significant survival benefit was observed of platinum-based chemotherapy (5-year RFS 33% both treatment arms, Fig. 6C; adjusted hazard-ratio: 0.81, 95%CI: 0.23-2.89 for the exon 4-5 probe, Supplementary Table S5B). Analyses with the exon 2-3 probe confirmed those obtained with the exon 4-5 probe (Fig. 6C and Supplementary Table S5B).
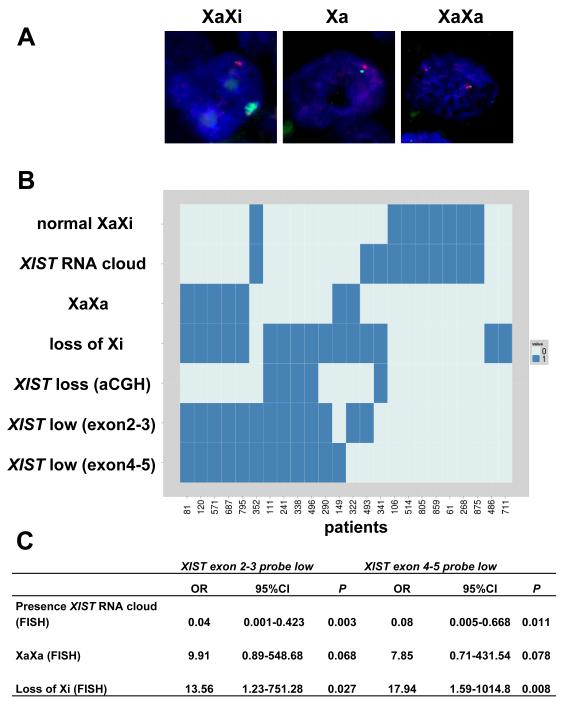
To determine whether loss of the XIST gene could explain the low XIST gene expression detected with both RT-MLPA probes, we analyzed the DNA of 37 patients with arrayCGH using probes flanking the XIST locus. Indeed, a XIST gene was lost in 16 out of 37 patients. Loss of XIST DNA correlated significantly with low RNA expression for all 60 patients (P<0.017 for the exon 2-3 probe, Fisher’s exact test). For 24 of the 60 samples we also managed to perform XIST RNA FISH analyses on the available FFPE material (Fig. 7). A DNA probe targeting RNF12 was taken along as X chromosome marker (Fig. 7A). RNA FISH confirmed that patients with low XIST gene expression had significantly fewer XIST clouds (Fig. 7B,C). Moreover, the combination of RNA and DNA FISH showed for all cases with aCGH-based XIST deletion that only the Xa was present (Fig. 7B). In several patients with low XIST gene expression, but no XIST gene deletion detectable by aCGH, we found two active X chromosomes and loss of Xi (Fig. 7B). Together, our data show that loss of Xi is the main cause of low XIST gene expression (P< 0.027, Fig. 7C).
Figure 7.
X chromosome aberrations investigated by FISH. A, examples of normal XaXi or abnormal cells with Xa or XaXa configuration (green: XIST RNA probe; red: RNF12 DNA). B, overview of 24 patients for which FISH results were obtained and their correlation to XIST RNA expression (0: XIST low, 1: XIST high) or aCGH (0: normal copy number, 1: XIST lost). Normal XiXa, XaXa and loss of Xi are also indicated categorically (0= no, 1= yes). XIST RNA cloud describes whether a normal XIST cloud is absent in >60% of cells (0) or not (1). (C) Associations between low XIST gene expression and aberrations identified by FISH (n=24, Fisher’s exact test). OR: Odds ratio, CI: Confidence interval
High prevalence of a predictive marker is required for its detection
Since Xist was readily identified as predictive marker for cisplatin sensitivity in our mouse model by SAM, it remains remarkable that our initial search to detect predictive markers for docetaxel sensitivity (Fig. 2) failed. When we analyzed only the tumors with an intrinsically high Abcb1 expression (T8*con, T9*con, T15*con, T26*con and T41*con) versus the 21 docetaxel-sensitive tumors as defined in Fig. 2A, Abcb1b was one of the most significantly increased genes on both the MEEBO and Illumina gene expression platforms (Supplementary Fig. S4C,D). Also the TLDA expression data showed a significant difference for Abcb1a and Abcb1b when only the 5 poor responders were compared with the docetaxel-sensitive tumors (Abcb1a: P < 0.0064; Abcb1b: P < 0.0043; Mann-Whitney U test). However, since increased expression of the Abcb1 genes is only found in a subgroup of the poor docetaxel responders, this significance is lost when samples with other docetaxel resistance mechanisms are added (Supplementary Fig. S7). In fact, addition of 5 samples without Abcb1 upregulation suffices to dilute the Abcb1 signal below significance.
In contrast to Abcb1b in the case of docetaxel treatment, the prevalence of low Xist expression was high in cisplatin hypersensitive tumors: 11 (MEEBO platform) or 10 (Illumina platform) out of the 12 showed Xist gene expression below the median (Supplementary Fig. S7).
DISCUSSION
We have investigated whether predictive markers for chemotherapy benefit can be identified in a GEMM using genome-wide expression profiling. GEMMs should be ideal for this purpose, as they lack the profound genetic heterogeneity of tumors from human patients. The tumors originate from the targeted deletion of Brca1 and p53, and all differences between tumors originate from random mutations in the period between the initiating deletions of Brca1 and p53 and the development of a mammary tumor. These additional mutations are responsible for the marked and stable differences in sensitivity to docetaxel and cisplatin that we find in individual tumors.
Even in this genetically homogeneous tumor system, we did not find a signature predicting docetaxel response, using genome-wide expression profiling. This negative result is instructive, however, because it has allowed us to delineate what is required to get useful predictive signatures. In our collection of 22 poor docetaxel responders, 5 tumors contained a substantial increase in Abcb1 RNA, known to be sufficient to cause drug resistance (21). Nevertheless, this increase in Abcb1 RNA was completely missed by 2 independent platforms measuring gene expression profiles. The Abcb1b transcript was readily detected in the 5 tumors with elevated transcript levels, as long as these tumors were analyzed as a group. However, when the results were pooled with those from only 5 tumors without elevated Abcb1b RNA, the positive result was completely lost. This shows why it is difficult to develop predictive markers, based on genome-wide expression arrays: only if the response to a drug is primarily determined by the expression level of a gene in most tumors, one can expect that gene to show up in the array-based gene expression analyses.
We found such a gene in analyzing the response of the mouse tumors to cisplatin. The low Xist expression associated with tumors hypersensitive to cisplatin was present in 10 out of 12 tumors and therefore detectable in our array analysis. The detection sensitivity can only be increased by the use of special algorithms that can identify subgroups within the samples. We show that such an algorithm is able to identify Abcb1b as outlier within the poor docetaxel responders. However, even with a more sophisticated analysis the problem remains that probes on the arrays are not sensitive enough to detect all relevant expression differences of genes causing therapy resistance. Gillet and co-workers found in a panel of cancer cell lines that the expression of the 380 “resistance–relevant” genes could only be reliably measured by quantitative PCR. For many genes the results obtained by microarrays were useless because of low sensitivity (29;36).
Given all these hurdles in finding predictive markers for chemotherapy, it is gratifying that we identified a gene that correlated with cisplatin response. It is encouraging that the low expression of XIST predicting high sensitivity to cisplatin in drug-naïve mouse tumors, also predicted an increased recurrence-free survival of high risk, primary breast cancer patients treated with intensive platinum-based chemotherapy. Although detected in a rather small group of 60 patients, the effect found is considerable. Intensive chemotherapy has largely been abandoned for the treatment of breast cancer, because for many patients the therapeutic benefit is limited (37). Nevertheless, several studies suggest that there are subgroups of patients that do benefit from this therapy, but the predictive tests to identify them are lacking (38;39). Hence, the analysis of XIST gene expression may be a useful tool to decide whether intensive platinum-based chemotherapy should be considered as alternative therapy for patients with HER2-negative, high risk breast cancer. Not all patients with a low XIST expression that we investigated benefited from the platinum-based therapy. An optimized cut-off for the level of XIST expression, validated in prospective clinical trials, may increase the positive predictive value, as may a combination with other classifiers, such as BRCA1-like CGH profiles (25).
Why tumors with a low expression of XIST are platinum hypersensitive is under investigation. Low XIST gene expression may be a flag for genomic instability as we found loss of Xi as the main cause underlying low transcript levels. The loss of Xi is most likely the consequence of chromosome segregation errors, which may be enhanced in BRCA1-defective cells due to a compromised spindle checkpoint (40). It has recently been found that missegregation stress induces a DNA damage response (41) and it was observed that aneuploid cells are more sensitive to anti-proliferative drugs (42). Mammary tumor cells with defects in DNA repair which are additionally stressed by improper execution of mitosis may therefore be hypersensitive to intensive platinum-based therapy.
The precise mechanism of XIST-mediated X inactivation is still under debate (43). It was suggested that BRCA1 supports the localization of XIST RNA to the Xi, as the BRCA1-deficient cells or tumors examined had lost localized XIST RNA (44-46). However, this hypothesis was challenged by others (47;48). The recent finding that BRCA1 maintains heterochromatin integrity (49) supports the idea that BRCA1 contributes to X inactivation after XIST-induced chromatin condensation. The contribution is not a simple one, however. Despite the large Brca1 deletion present in all mammary tumors of our mouse model, Xist gene expression varies considerably. Variability of XIST expression was also present in those human breast cancers in which a BRCA1 mutation was found, or which were classified as BRCA1-like by aCGH (25).
Our study shows that GEMMs that resemble breast cancer in humans are useful to investigate chemotherapy response prediction. Tools to identify predictive markers can be tested under controlled conditions, and targeted ablation of genes helps to dissect mechanisms of resistance. Ultimately, predictive markers identified in GEMMs may improve the clinical success rate for chemotherapy in humans.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank T. Fojo, H. te Riele, R. Bernards, A. Berns, S. Rodenhuis and G. Xu for critical reading of the manuscript. A. Velds and I. de Rink helped processing the gene expression profiling data.
Grant support: This work was supported by grants from the Dutch Cancer Society 2006-3566 (PB/SR/JJ), 2006-3706 (MV/SL), 2009-4303 (SR/JJ/PB), the Netherlands Organization for Scientific Research (NWO-VIDI-91711302 to SR), the European Union FP6 Integrated Project 037665-CHEMORES (PB/SR) and CTMM Breast Care (JJ/SR).
Footnotes
Disclosure of Potential Conflicts of Interest: M.J. and M.L. are employees of MRC-Holland BV which markets the RT-MLPA tests used in this article.
Reference List
- (1).McGuire WL. Steroid hormone receptors in breast cancer treatment strategy. Recent Prog Horm Res. 1980;36:135–56. doi: 10.1016/b978-0-12-571136-4.50010-3. [DOI] [PubMed] [Google Scholar]
- (2).Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344(11):783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- (3).Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, et al. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376(9737):235–44. doi: 10.1016/S0140-6736(10)60892-6. [DOI] [PubMed] [Google Scholar]
- (4).Gorre ME, Mohammed M, Ellwood K, Hsu N, Paquette R, Rao PN, et al. Clinical resistance to STI-571 cancer therapy caused by BCR-ABL gene mutation or amplification. Science. 2001;293(5531):876–80. doi: 10.1126/science.1062538. [DOI] [PubMed] [Google Scholar]
- (5).Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, et al. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17(1):77–88. doi: 10.1016/j.ccr.2009.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, Beelen K, et al. A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell. 2007;12(4):395–402. doi: 10.1016/j.ccr.2007.08.030. [DOI] [PubMed] [Google Scholar]
- (7).Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS Med. 2005;2(3):e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Van ’t Veer LJ, Dai H, Van der Vijver MJ, He YD, Hart AAM, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- (9).Borst P, Wessels L. Do predictive signatures really predict response to cancer chemotherapy? Cell Cycle. 2010;9(24):4836–40. doi: 10.4161/cc.9.24.14326. [DOI] [PubMed] [Google Scholar]
- (10).Nuyten DS, van de Vijver MJ. Using microarray analysis as a prognostic and predictive tool in oncology: focus on breast cancer and normal tissue toxicity. Semin Radiat Oncol. 2008;18(2):105–14. doi: 10.1016/j.semradonc.2007.10.007. [DOI] [PubMed] [Google Scholar]
- (11).Hannemann J, Oosterkamp H, Bosch C, Velds A, Wessels L, Loo C, et al. Changes in gene expression associated with response to neoadjuvant chemotherapy in breast cancer. J Clin Oncol. 2005;23(15):3331–42. doi: 10.1200/JCO.2005.09.077. [DOI] [PubMed] [Google Scholar]
- (12).Weigelt B, Pusztai L, Ashworth A, Reis-Filho JS. Challenges translating breast cancer gene signatures into the clinic. Nat Rev Clin Oncol. 2011 doi: 10.1038/nrclinonc.2011.125. [DOI] [PubMed] [Google Scholar]
- (13).Coombes KR, Wang J, Baggerly KA. Microarrays: retracing steps. Nat Med. 2007;13(11):1276–7. doi: 10.1038/nm1107-1276b. [DOI] [PubMed] [Google Scholar]
- (14).Baggerly KA, Coombes KR. Deriving chemosensitivity from cell lines: Forensic bioinformatics and reproducible research in high-throughput biology. Ann Appl Stat. 2009;3(4):1309–34. [Google Scholar]
- (15).Liedtke C, Wang J, Tordai A, Symmans WF, Hortobagyi GN, Kiesel L, et al. Clinical evaluation of chemotherapy response predictors developed from breast cancer cell lines. Breast Cancer Res Treat. 2010;121(2):301–9. doi: 10.1007/s10549-009-0445-7. [DOI] [PubMed] [Google Scholar]
- (16).Gillet JP, Calcagno AM, Varma S, Marino M, Green LJ, Vora MI, et al. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. Proc Natl Acad Sci U S A. 2011;108(46):18708–13. doi: 10.1073/pnas.1111840108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Michalak EM, Jonkers J. Studying therapy response and resistance in mouse models for BRCA1-deficient breast cancer. J Mammary Gland Biol Neoplasia. 2011;16(1):41–50. doi: 10.1007/s10911-011-9199-z. [DOI] [PubMed] [Google Scholar]
- (18).Liu X, Holstege H, van der Gulden H, Treur-Mulder M, Zevenhoven J, Velds A, et al. Somatic loss of BRCA1 and p53 in mice induces mammary tumors with pathologic and molecular features of human BRCA1-mutated basal-like breast cancer. Proc Natl Acad Sci U S A. 2007;104:12111–6. doi: 10.1073/pnas.0702969104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Rottenberg S, Nygren AOH, Pajic M, Van Leeuwen FWB, Van der Heijden I, Van de Wetering K, et al. Selective induction of chemotherapy resistance of mammary tumors in a conditional mouse model for hereditary breast cancer. Proc Natl Acad Sci U S A. 2007;104:12117–22. doi: 10.1073/pnas.0702955104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Rottenberg S, Jaspers JE, Kersbergen A, Van der Burg E, Nygren AO, Zander SA, et al. High sensitivity of BRCA1-deficient mammary tumors to the PARP inhibitor AZD2281 alone and in combination with platinum drugs. Proc Natl Acad Sci U S A. 2008;105(44):17079–84. doi: 10.1073/pnas.0806092105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Pajic M, Iyer JK, Kersbergen A, Van der Burg E, Nygren AO, Jonkers J, et al. Moderate increase in Mdr1a/1b expression causes in vivo resistance to doxorubicin in a mouse model for hereditary breast cancer. Cancer Res. 2009;69(16):6396–404. doi: 10.1158/0008-5472.CAN-09-0041. [DOI] [PubMed] [Google Scholar]
- (22).Zander SA, Kersbergen A, Van der Burg E, de Water N, Van Tellingen O, Gunnarsdottir S, et al. Sensitivity and acquired resistance of BRCA1;p53-deficient mouse mammary tumors to the topoisomerase I inhibitor topotecan. Cancer Res. 2010;70(4):1700–10. doi: 10.1158/0008-5472.CAN-09-3367. [DOI] [PubMed] [Google Scholar]
- (23).Pajic M, Kersbergen A, van Diepen F, Pfauth A, Jonkers J, Borst P, et al. Tumor-initiating cells are not enriched in cisplatin-surviving BRCA1;p53-deficient mammary tumor cells in vivo. Cell Cycle. 2010;9(18):3780–91. doi: 10.4161/cc.9.18.13002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Gillet JP, Wang J, Calcagno AM, Green LJ, Varma S, Bunkholt Elstrand M, et al. Clinical Relevance of Multidrug Resistance Gene Expression in Ovarian Serous Carcinoma Effusions. Mol Pharm. 2011 doi: 10.1021/mp200240a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Vollebergh MA, Lips EH, Nederlof PM, Wessels LF, Schmidt MK, van Beers EH, et al. An aCGH classifier derived from BRCA1-mutated breast cancer and benefit of high-dose platinum-based chemotherapy in HER2-negative breast cancer patients. Ann Oncol. 2011;22(7):1561–70. doi: 10.1093/annonc/mdq624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Picard F, Lebarbier E, Hoebeke M, Rigaill G, Thiam B, Robin S. Joint segmentation, calling, and normalization of multiple CGH profiles. Biostatistics. 2011;12(3):413–28. doi: 10.1093/biostatistics/kxq076. [DOI] [PubMed] [Google Scholar]
- (27).Rodenhuis S, Bontenbal M, Beex LV, Wagstaff J, Richel DJ, Nooij MA, et al. High-dose chemotherapy with hematopoietic stem-cell rescue for high-risk breast cancer. N Engl J Med. 2003;349(1):7–16. doi: 10.1056/NEJMoa022794. [DOI] [PubMed] [Google Scholar]
- (28).Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98(9):5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (29).Orina JN, Calcagno AM, Wu CP, Varma S, Shih J, Lin M, et al. Evaluation of current methods used to analyze the expression profiles of ATP-binding cassette transporters yields an improved drug-discovery database. Mol Cancer Ther. 2009;8(7):2057–66. doi: 10.1158/1535-7163.MCT-09-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (30).Coley HM. Mechanisms and strategies to overcome chemotherapy resistance in metastatic breast cancer. Cancer Treat Rev. 2008;34(4):378–90. doi: 10.1016/j.ctrv.2008.01.007. [DOI] [PubMed] [Google Scholar]
- (31).Huisman MT, Chhatta AA, Van Tellingen O, Beijnen JH, Schinkel AH. MRP2 (ABCC2) transports taxanes and confers paclitaxel resistance and both processes are stimulated by probenecid. Int J Cancer. 2005;116(5):824–9. doi: 10.1002/ijc.21013. [DOI] [PubMed] [Google Scholar]
- (32).Hopper-Borge E, Chen ZS, Shchaveleva I, Belinsky MG, Kruh GD. Analysis of the drug resistance profile of multidrug resistance protein 7 (ABCC10): resistance to docetaxel. Cancer Res. 2004;64(14):4927–30. doi: 10.1158/0008-5472.CAN-03-3111. [DOI] [PubMed] [Google Scholar]
- (33).Borst P, Evers R, Kool M, Wijnholds J. A family of drug transporters, the MRP’s. J Natl Cancer Inst. 2000;92:1295–302. doi: 10.1093/jnci/92.16.1295. [DOI] [PubMed] [Google Scholar]
- (34).de Ronde JJ, Lips E, Mulder L, Rigaill G, Vincent A, Wesseling J, et al. Gene expression analysis of a large cohort of neoadjuvantly treated breast cancer patients implicates SERPINA6, BEX1, AGTR1 and SLC26A3 in chemotherapy resistance. 2011. Submitted.
- (35).Lopes AM, Burgoyne PS, Ojarikre A, Bauer J, Sargent CA, Amorim A, et al. Transcriptional changes in response to X chromosome dosage in the mouse: implications for X inactivation and the molecular basis of Turner Syndrome. BMC Genomics. 2010;11:82. doi: 10.1186/1471-2164-11-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Gillet JP, Gottesman MM. Mechanisms of multidrug resistance in cancer. Methods Mol Biol. 2010;596:47–76. doi: 10.1007/978-1-60761-416-6_4. [DOI] [PubMed] [Google Scholar]
- (37).Crown J. Smart bombs versus blunderbusses: high-dose chemotherapy for breast cancer. Lancet. 2004;364(9442):1299–300. doi: 10.1016/S0140-6736(04)17207-3. [DOI] [PubMed] [Google Scholar]
- (38).Cheng YC, Ueno NT. Is high-dose chemotherapy with autologous hematopoietic stem cell transplantation in breast cancer patients a done deal? Womens Health (Lond Engl) 2010;6(4):481–5. doi: 10.2217/whe.10.31. [DOI] [PubMed] [Google Scholar]
- (39).Rodenhuis S. Is high-dose chemotherapy dead? Eur J Cancer. 2005;41(1):9–11. doi: 10.1016/j.ejca.2004.09.014. [DOI] [PubMed] [Google Scholar]
- (40).Wang RH, Yu H, Deng CX. A requirement for breast-cancer-associated gene 1 (BRCA1) in the spindle checkpoint. Proc Natl Acad Sci U S A. 2004;101(49):17108–13. doi: 10.1073/pnas.0407585101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (41).Janssen A, van der Burg M, Szuhai K, Kops GJ, Medema RH. Chromosome segregation errors as a cause of DNA damage and structural chromosome aberrations. Science. 2011;333(6051):1895–8. doi: 10.1126/science.1210214. [DOI] [PubMed] [Google Scholar]
- (42).Tang YC, Williams BR, Siegel JJ, Amon A. Identification of aneuploidy-selective antiproliferation compounds. Cell. 2011;144(4):499–512. doi: 10.1016/j.cell.2011.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Wutz A. Xist function: bridging chromatin and stem cells. Trends Genet. 2007;23(9):457–64. doi: 10.1016/j.tig.2007.07.004. [DOI] [PubMed] [Google Scholar]
- (44).Richardson AL, Wang ZC, De Nicolo A, Lu X, Brown M, Miron A, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9(2):121–32. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- (45).Silver DP, Dimitrov SD, Feunteun J, Gelman R, Drapkin R, Lu SD, et al. Further evidence for BRCA1 communication with the inactive X chromosome. Cell. 2007;128(5):991–1002. doi: 10.1016/j.cell.2007.02.025. [DOI] [PubMed] [Google Scholar]
- (46).Ganesan S, Silver DP, Greenberg RA, Avni D, Drapkin R, Miron A, et al. BRCA1 supports XIST RNA concentration on the inactive X chromosome. Cell. 2002;111(3):393–405. doi: 10.1016/s0092-8674(02)01052-8. [DOI] [PubMed] [Google Scholar]
- (47).Xiao C, Sharp JA, Kawahara M, Davalos AR, Difilippantonio MJ, Hu Y, et al. The XIST noncoding RNA functions independently of BRCA1 in X inactivation. Cell. 2007;128(5):977–89. doi: 10.1016/j.cell.2007.01.034. [DOI] [PubMed] [Google Scholar]
- (48).Pageau GJ, Hall LL, Lawrence JB. BRCA1 does not paint the inactive X to localize XIST RNA but may contribute to broad changes in cancer that impact XIST and Xi heterochromatin. J Cell Biochem. 2007;100(4):835–50. doi: 10.1002/jcb.21188. [DOI] [PubMed] [Google Scholar]
- (49).Zhu Q, Pao GM, Huynh AM, Suh H, Tonnu N, Nederlof PM, et al. BRCA1 tumour suppression occurs via heterochromatin-mediated silencing. Nature. 2011;477(7363):179–84. doi: 10.1038/nature10371. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.