SUMMARY
The soil bacterium Bacillus subtilis is widely used in agriculture as a biocontrol agent able to protect plants from a variety of pathogens. Protection is thought to involve the formation of bacterial communities - biofilms - on the roots of the plants. Here we used confocal microscopy to visualize biofilms on the surface of the roots of tomato seedlings and demonstrated that biofilm formation requires genes governing the production of the extracellular matrix that holds cells together. We further show that biofilm formation was dependent on the sensor histidine kinase KinD and in particular on an extracellular CACHE domain implicated in small molecule sensing. Finally, we report that exudates of tomato roots strongly stimulated biofilm formation ex planta and that an abundant small molecule in the exudates, l-malic acid, was able to stimulate biofilm formation at high concentrations in a manner that depended on the KinD CACHE domain. We propose that small signaling molecules released by the roots of tomato plants are directly or indirectly recognized by KinD, triggering biofilm formation.
Keywords: Bacillus subtilis, biofilm, sensor kinase, CACHE domain, malic acid
INTRODUCTION
The soil bacterium Bacillus subtilis is a powerful model organism for laboratory studies of cell division, genetic competence, gene control, multicellularity, spore formation, swarming, swimming, and numerous other aspects of its biology and physiology (Aguilar et al., 2007, Kearns et al., 2004, Bramhill, 1997, Piggot & Hilbert, 2004, Stephenson & Hoch, 2002, Gonzalez-Pastor et al., 2003, Sonenshein, 2000, Dubnau, 1999). However, little is known about its behavior in the soil or indeed what niches it occupies in natural environments. One clue comes from its wide use as a biocontrol agent in agriculture. Various wild strains of B. subtilis and related species isolated from the soil are known to protect plants from diverse fungal, bacterial and helminthic pathogens (Ongena & Jacques, 2008, Lemessa & Zeller, 2007, Emmert & Handelsman, 1999). This protection is believed to be mediated in part by the formation of multicellular communities, biofilms, on the plant roots (Berg et al., 2005, Bais et al., 2004), although this has not been clearly demonstrated. Also, little is known about if and how plant roots trigger biofilm formation.
Biofilm formation was not studied in B. subtilis until recently because most laboratory strains are impaired in making these multicellular communities (Branda et al., 2001, McLoon et al., 2011a). However, wild (undomesticated) strains, such as NCIB3610, form robust, architecturally complex communities both at an air/liquid interface (such floating biofilms being known as pellicles) and on surfaces (Branda et al., 2001). A hallmark of these biofilms is a self-produced, extracellular matrix consisting of an exopolysaccharide and amyloid-like protein fibers (Chu et al., 2006, Branda et al., 2006, Kearns et al., 2005, Romero et al., 2010). The exopolysaccharide is synthesized by enzymes encoded by the epsA-O operon whereas the tapA-sipW-tasA operon is responsible for fiber production (Chu et al., 2006, Branda et al., 2006, Romero et al., 2010, Kearns et al., 2005). Expression of both operons is under the indirect control of the response regulator protein Spo0A (Chai et al., 2011, Hamon & Lazazzera, 2001), which is phosphorylated via a multicomponent phosphorelay (Jiang et al., 2000, Ireton et al., 1993). At least four histidine kinases, known as KinA, KinB, KinC, and KinD, feed phosphoryl groups into the relay to activate Spo0A and trigger biofilm formation (Jiang et al., 2000). The four kinases fall into two partially redundant pairs in regulating biofilm formation on standard biofilm-inducing medium (MSgg). These are the KinA and KinB pair and the KinC and KinD pair (McLoon et al., 2011b). Little is known about the environmental signals that these kinases recognize and which, if any, might be involved in triggering biofilm formation of B. subtilis on plant roots.
In this study, we established a system to study the interaction of B. subtilis with tomato plants. We demonstrate that B. subtilis forms robust biofilms on tomato plant roots in a manner that depends on matrix genes. We further show that biofilm formation can be elicited by an exudate from tomato plant roots and that this response is principally dependent on one of the four kinases, KinD. KinD is an integral membrane protein that exhibits an extracellular domain of similarity to the CACHE domain of small molecule sensing proteins (Wu et al., 2008, Zhang & Hendrickson, 2010). Biofilm formation on roots and the response to root exudates are blocked in a mutant of the KinD CACHE domain, which we infer is responsible for sensing one or more signals released by plant roots. Finally, we identify a group of small molecules that are abundantly present in tomato root exudates. Among them was l-malic acid, which was also identified previously as being abundantly secreted by plant roots (Kamilova et al., 2006a, Kamilova et al., 2006b, Rudrappa et al., 2008). We now show that l-malic acid, but not d-malic acid, stimulated biofilm formation in a KinD-dependent manner when provided at high concentrations. We propose that the CACHE domain is a mechanism for directly or indirectly sensing small molecule signals from the plant roots, which in turn elicit an intimate symbiotic relationship between the bacterium and its host.
MATERIALS AND METHODS
Bacterial strains, culture media, and culture conditions
B. subtilis strains were routinely cultured in Luria-Bertani (LB) medium or TY medium (LB medium with the addition of 10 mM MgSO4 and 100 µM MnSO4). For pellicle formation, LB medium, MSgg (Branda et al., 2001), or a minimal medium modified from MSgg [5 mM potassium phosphate, 100 mM morpholine propane sulfonic acid, pH 7 (MOPS), 2 mM MgCl2, 50 µM MnCl2, 50 µM FeCl3, 700 µM CaCl2, 1 µM ZnCl2, 2 µM thiamine, 0.5% glycerol, 0.5% NH4Cl, and 50 µg/ml (each) of threonine, tryptophan, and phenylalanine] was applied. The strains used in this study were listed in Table S3. Antibiotics were added, when necessary, at the following concentrations: 100 µg per ml of spectinomycin, 10 µg per ml of tetracycline, 5 µg per ml of chloramphenicol, 0.5 µg per ml of erythromycin, 2.5 µg per ml of lincomycin, and 10 µg per ml of kanamycin for B. subtilis strains. Chemicals were purchased from Sigma.
Strain construction
All deletion mutations were generated by long-flanking PCR mutagenesis (Wach, 1996). Transfer of antibiotic cassette-marked deletion mutations or reporter fusions among different strains were conducted using SSP1 phage-mediated transduction (Yasbin & Young, 1974). Other routine molecule manipulation techniques followed the published protocols (Sambrook & Russell, 2001). All primers used in the construction of strains and plasmids were listed in Tables S4 and S5.
To generate the mKate2 (Shcherbo et al., 2007) reporter fusion for visualization of cells in root-associated biofilms, the coding sequence of the mKate2 gene was amplified by PCR using the plasmid pMKate2 (a gift of T. Norman) as the template and primers mKate2-F1 and mKate2-R1. The PCR products were cloned into the plasmid pYC127 (Chai et al., 2008). The resulting plasmid contains the mKate2 gene under the control of a constitutively expressed promoter hyspank (Physpank-mKate2).
To complement the ∆kinD null mutant with the wild type copy of kinD, the promoter sequence and the open reading frame of the kinD gene were amplified by PCR using 3610 chromosomal DNA as the template and primers kinD-F1 and kinD-R1. The PCR products were cloned into the integration plasmid pDG1662 (Guérout-Fleury et al., 1996), resulting in the recombinant plasmid pYC240. To construct the CACHE domain mutant of KinD, overlapping PCR was applied. In brief, two DNA fragments were amplified by PCR using 3610 chromosomal DNA as the template and primers kinD-F1/ kinD-MR1 or kinD-MF1/ kinD-R1. The two PCR products were joined together by overlapping PCR. The overlapping PCR products containing the kinD allele with desired mutations in the CACHE domain were similarly cloned into the plasmid pDG1662, resulting in the recombinant plasmid pYC241. All the above recombinant plasmids were then introduced into PY79 by transformation (Gryczan et al., 1978). A double crossover recombination at the amyE locus on the chromosome of PY79 was confirmed. The amyE fragments were then introduced into different strain backgrounds by SSP1 phage-mediated transduction.
To construct KinD complementation strains (by wild type or the CACHE domain mutant allele of kinD) that harbor the constitutively expressed mKate2 reporter gene, the plasmids pYC240 and pYC241 were first introduced into a strain YC751 by transformation. YC751 bears a second copy of the amyE fragment in a mini-Tn10 transposon integrated at the 317° position on the chromosome in the laboratory strain PY79 and marked with a chloramphenicol resistance gene. Integration of amyE::kinD (wt or mut) via double crossover recombination at the amyE sequence at 317° was selected by SpcR CmS of the resulting transformants and carefully verified. The 317°Ω amyE::kinD (wt or mut) fragments were then introduced into the ∆kinD null mutant by SSP1 phage-mediated transduction. Subsequently, the mKate2 reporter fusion was introduced into the above strains at the native amyE locus on the chromosome, generating strains CY189 and CY190.
The reporter fusion sacA::PsdpA-lux was constructed by A McLoon by joining the promoter region of the sdpABC operon to the luciferase reporter genes in the plasmid pAH328, a modification of pAH321 (McLoon et al., 2011b). The reporter fusion was introduced into 3610 and various kinD strains by using SSP1-phage mediated transduction.
Preparation of tomato root exudates
The preparation procedure was modified from a published protocol (Graham et al., 1981). Tomato seeds (Lycopersicon esculentum Miller) were surface sterilized by a 30 second treatment in 70% ethanol followed by 15 min in sodium hypochloride (10% active chlorine) and by three subsequent washing steps with sterile water for at least 15 min each. Surface sterilized tomato seeds were planted into trays contained moist soil and incubated in a growth chamber (16:8 h light/dark photoperiod, 23°C) for germination. After two weeks, tomato plantlets were transplanted into pots filled with fertilizer soil and incubated in the growth chamber set up with the same parameters as above. After two more weeks of incubation, tomato roots were harvested and washed off attached soils with sterilized water. Ten plants were immediately placed in a beaker (500 ml) with the roots submerged in 200 ml of sterilized water containing 50 µg per ml of rifampicin and incubated for 2 hours to reduce bacterial contamination. After the antibiotic treatment, the roots were rinsed with sterilized water several times and placed back in a beaker (500 ml) containing 200 ml of sterilized water. The beaker was placed on a low speed shaker at the room temperature for 3–6 days. The plants were then removed from the beaker and the exudates were collected. The exudates were passed through a 0.22-µm filter and stored at 4 °C. Sterilized and filtered root exudates were dried down using a speed vacuum. Samples were finally suspended in 1-ml of distilled water for further analysis.
Assays of pellicle formation
For induction of pellicle biofilm by tomato root exudates or l-malic acid, cells were grown in 4 ml of LB broth or a minimal medium described above in 12-well plates at 22°C. Root exudate or l-malic acid was supplemented into the medium at indicated concentrations. The same volume of sterilized water was used as a negative control. Images of pellicle biofilms were taken after two or three days of incubation.
Assays of luciferase activities
Cells were grown in LB broth to mid-log-phase and diluted 1:100 with fresh LB broth. 1 ml of diluted culture was added into each well of a 24-well polystyrene Visiplate (white with a clear bottom; Wallac, USA). Root exudates (1%) or indicated concentrations of malic acid were added into the wells and samples were grown for 24 h at 30°C in a BioTek Synergy 2 plate reader (BioTek, USA) with slow continuous shaking and the optical density at 600 nm and luminescence (sensitivity setting, 200) were measured every 10 min. Data shown were representative results from at least three different experiments. Relative values of luminescence were normalized against cell optical density.
Biofilm formation on tomato plant roots
Various B. subtilis strains that all harbor a constitutively expressed mKate2 reporter gene were tested for biofilm formation on tomato root surfaces. To do so, tomato seeds (Lycopersicon esculentum Miller) were surface sterilized by a 30 sec treatment in 70% ethanol, followed by a 15 min treatment in sodium hypochloride (10% active chlorine) and by three subsequent washing steps with sterile water for at least 15 min each. Sterilized seeds were then transferred onto the 0.7% Murashige and Skoog (MS) agar plate (Murashige & Skoog, 1962) and incubated at 25°C for 4 days until the length of tomato roots reached about 3 cm. The seedlings were transplanted into 12-wells plates containing 4 ml of MS medium in each well, and incubated at 25°C for 48 hours in a shaker. The speed of the shaker was set at 60 rpm with photoperiod of 16 h of light and 8 h of dark. Two days later, the B. subtilis suspension, which was grown in TY medium to the mid-log-phase, was added into the tomato MS medium to reach an initial OD600 of 0.1. The co-cultured plates were statically incubated at 25°C for another 48 hours before examination of the bacterial biofilms on the root surfaces by CLSM. Seedlings without bacterial inoculation were used as a control. All treatments were repeatedly tested and representative results were shown.
For biofilm assays on heat-killed tomato plantlets, live tomato plantlets were prepared similarly to what was described above and were incubated in 80°C water bath for 10 min. Heat-killed tomato plantlets were then washed with sterilized water 5 times before testing. For cold-killing of tomato plantlets, live tomato plantlets were again prepared similarly and then stored at −80°C overnight. After treatment, these plantlets were then warmed up to room temperature and washed with sterilized water 5 times before testing. Incubation of B. subtilis cells with either heat-killed or cold-killed tomato plantlets and examination of root-surface associated biofilms followed the same procedures that were described above.
Confocal Laser Scanning Microscopy (CLSM)
To visualize cells in the root surface-associated biofilms by CLSM, cells were incubated with tomato roots for two days at room temperature. Roots were then harvested and gently washed three times with PBS buffer (10 mM NaH2PO4, 10 mM Na2HPO4, 130 mM NaCl, pH 7.2–7.4) to remove unattached cells from the roots. Roots were then fixed in a 4% paraformaldehyde solution in PBS at 4°C overnight. Single optical sections were captured with a Zeiss 20X AchroPlan objective lens using a Zeiss LSM 510 META. The HeNe1 laser (543 nm) and a 585-nm long-pass filter were used for excitation and emission, respectively. Experiments were repeated twice, each time with six replicas. About twenty field views were captured for each treatment and representative images were shown.
Competition assays
The competing strains (the ∆kinD null mutant and the two kinD complementation strains) and the wild type strain were grown to mid-log-phase to a same cell density (OD600=1.0) in TY medium. One of the competing strains and the wild type cells were mixed at a ratio of 1:1, and inoculated into the tomato-MS medium in 12-well plates to reach an initial OD600 of 0.1. The co-cultured plates were statically incubated at 25°C for 48 hours. Tomato roots were then generally rinsed three times with PBS buffer, cut into small pieces, and transferred to a 15-ml falcon tube containing PBS buffer. Samples were vortexed vigorously. Cell suspensions were serially diluted in PBS buffer and were plated on LB agar (for count of total cells) or on LB agar supplemented with of 0.5 µg per ml of erythromycin and 2.5 µg per ml of lincomycin (for count of cells of the kinD strain). Plates were incubated for 16 h at 37°C, and colony-forming unit (CFU) was calculated.
Identification of small chemical molecules in the root exudates
To identify the tomato root-released chemical(s) that induce biofilm formation, we first applied the root exudates to a Sep-Pak Plus C-18 column (Waters Corporation, MA, USA) and separated compounds by eluting them with aqueous methanol (from a gradient of 10% to 100%). We then tested each of the fractions for the ability to stimulate the PsdpA-lux reporter by using luciferase assays described above in 24-well plates. Fractions that exhibited activity were subjected to further separation by high performance liquid chromatography (HPLC). Finally, the subfractions from HPLC were tested similarly and those that still retained the activity were analyzed again by HPLC and mass spectrometry (MS). Commercially available l-malic acid (Sigma) was applied as a control and went through similar analyses.
Sequence alignment and structural analysis of proteins
Sequence alignment of the proteins was performed by using the program ClustalW (Thompson et al., 1994). Three-dimensional-structure-based searches for KinD and Med were conducted by using HHpred with default parameters (http://hhpred.tuebingen.mpg.de/hhpred). Three-dimensional ribbon structures of KinD and McpX were created by Swiss PDB Viewer (http://www.expasy.org/spdbv/) (Guex & Peitsch, 1997). All structural information and coordinates of proteins were obtained from the Protein Data Bank (http://www.rcsb.org/pdb/home/home.do).
RESULTS
B. subtilis forms biofilms on tomato roots
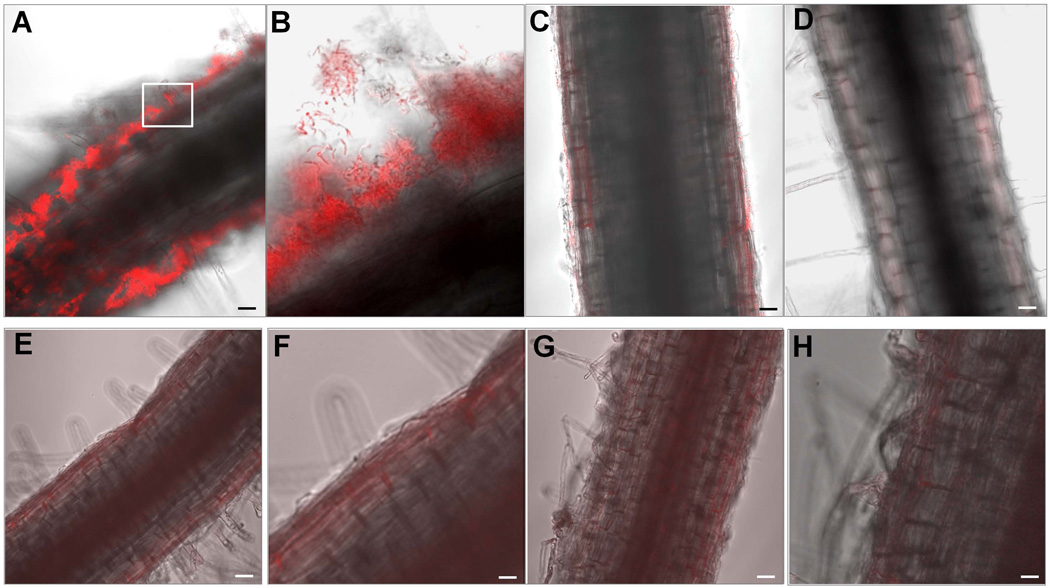
To study biofilm formation by B. subtilis on plant roots, we germinated tomato seedlings in Murashige and Skoog (MS) solid agar medium (Murashige & Skoog, 1962), allowing them to grow to young plantlets (see Materials and Methods). These young plantlets were transferred to liquid MS broth and grown for two days. We then applied B. subtilis cells to the medium and incubated the plantlets for two more days at room temperature to allow biofilm formation on the roots. (B. subtilis grew but did not form biofilms in Murashige and Skoog medium; data not shown.) The cells were a derivative of the wild strain 3610 that harbored a constitutively expressed red fluorescence reporter gene (mKate2). Confocal Laser Scanning Microscopy (CLSM) revealed large numbers of cells associated with the root surfaces (Fig. 1A), often more densely at the tips (data not shown). Many of these cells were in the form of chains of various lengths (Fig. 1B). Cells were also distributed three-dimensionally with the inner-most layer of cells appearing to be directly associated with the root surface. In contrast to the results obtained with the wild type, a similarly constructed reporter strain carrying a deletion of the epsA-O operon was largely unable to form multicellular structures on the root surfaces (Fig. 1C). Indeed, the roots of plantlets incubated with the epsA-O mutant cells were essentially indistinguishable from roots of plantlets that had been incubated without bacteria at all (Fig. 1D). This result indicates that biofilm formation was essential for efficient colonization of the root surfaces.
Figure 1. B. subtilis forms biofilms on the tomato root surfaces.
Panels A–D. B. subtilis cells that harbor a consistently expressed mkate2 reporter gene at amyE were co-cultured with tomato plantlets in a defined medium. After two days of incubation, root-associated cells were visualized by CLSM. Wild type cells (CY49) formed robust biofilms on the root surfaces (Panel A). A 50 × 50 µm framed area in Panel A was enlarged in Panel B to show details of the chained cells in the root-associated biofilms. A mutant of the epsA-O operon (CY209) exhibited severely decreased attachment of the cells to the root surfaces and virtually no biofilm-like structures (Panel C). Panel D shows a plant root grown in the absence of B. subtilis. Panels E-H show results of biofilm assays of the B. subtilis cells on root surfaces using either heat-killed (80°C for 10 min; Panels E and F) or cold-killed (−80°C overnight; Panels G and H) tomato plantlets. Wild type cells did not form biofilms on the surfaces of either heat-killed or cold-killed tomato plants. Images in panels F and H are enlarged ones from the panels E and G, respectively. Scale bars in Panels A, C, D, E, and G are 20 µm, and the scale bars in Panels F and H are 50 µm.
Finally, the experiment of Figs. 1E–H shows that biofilm formation required living plantlets. When heat-killed (80°C, 10 min; Fig. 1E–F) or cold-killed (−80°C, overnight; Fig. 1G–H) plantlets were incubated with biofilm-proficient cells (see Materials and Methods), no root surface-associated bacteria were observed. We conclude that B. subtilis is not simply forming biofilms on inert surfaces but rather that colonization of the roots requires active metabolic activity by the plant. It is known that B. subtilis does not ordinarily form biofilms on submerged surfaces even in biofilm-inducing medium, and thus its capacity to do so on plant roots is particularly striking.
Tomato root exudates induce pellicle formation and stimulate expression of matrix genes
We hypothesized that tomato plants release one or more chemical signals that induce biofilm formation on the root surface. To test this idea, we prepared exudates from tomato roots following the protocol described in the Materials and Methods. We then applied the root exudate at a final concentration of 1% (v/v) to a standing culture of LB medium in which B. subtilis normally does not form floating biofilms (pellicles). The root exudate significantly stimulated pellicle formation (Fig. 2A, WT). This was not due to a change of the growth rate upon addition of the root exudates because little or no alteration in the growth rate was observed in shaking cultures growing in either LB or defined minimal medium (Fig. S1).
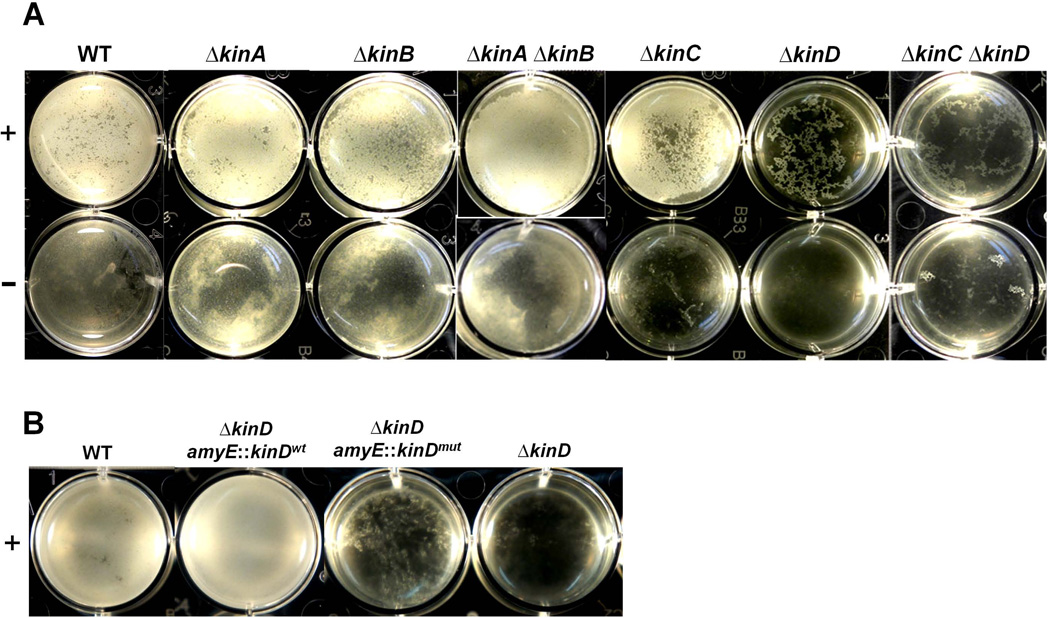
Figure 2. Tomato root exudates induce pellicle formation by B. subtilis in LB broth in a KinD-dependent manner.
In Panel A, wild type cells and various single or double kinase mutants [WT (3610); ∆kinA (RL4562); ∆kinB (RL4563); ∆kinA ∆kinB (RL4573); ∆kinC (RL4262); ∆kinD (RL4552); ∆kinC ∆kinD (RL5273) were inoculated into LB standing culture in 12-well plates with (top panels) or without (bottom panels) addition of 1% (v/v) tomato root exudates. Samples were incubated at room temperature for 2 days. Panel B compares the ability of wild type (3610), the kinD null mutant (RL1927), and two kinD complementation strains, ∆kinD amyE::kinDwt (CY78) and ∆kinD amyE::kinDmut (CY79) to form pellicles in LB medium with the addition of 1% root exudate.
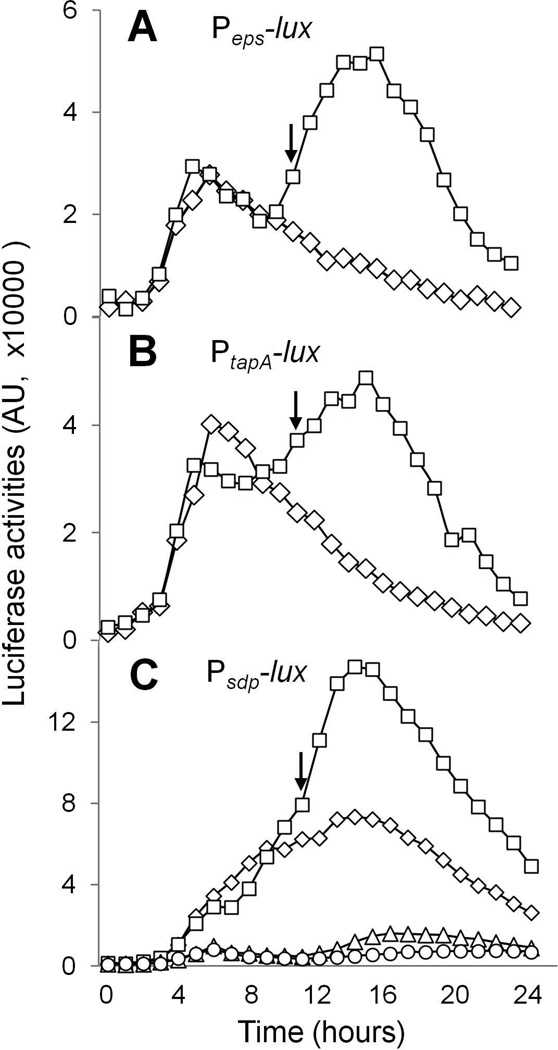
We also investigated whether the root exudate would stimulate the expression of matrix genes by using a derivative of 3610 that harbors a luciferase gene reporter fused to the regulatory region of the epsA-O operon (Peps-lux) or the tapA-sipW-tasA operon (PtapA-lux). In the absence of added root exudate, an early phase of gene expression was seen that peaked at about 5 hours post inoculation. In the presence of 1% root exudate, however, a second, higher phase of gene expression was seen that peaked at around 14 hours post inoculation (Figs. 3A and 3B). We interpret these results to indicate that in the absence of root exudate matrix gene expression is transiently turned on in LB medium whereas in the presence of root exudate expression is sustained for an extended period of time. Induction of the reporter fusions and stimulation of pellicle formation by the root exudate were also observed in a defined minimal medium which by itself does not support the formation of floating pellicles of B. subtilis (Figs. S2 and S3). This indicates that the observed response to addition of the root exudates and stimulation of pellicle formation were not medium-specific.
Figure 3. Tomato root exudates induce expression of the matrix genes and the sdpABC operon.
Panels A and B show luciferase activities for the two wild type reporter strains harboring either PepsA-lux (strain ALM89 in A) or PtapA-lux (strain ALM90 in B) that were grown in LB shaking culture with (squares) or without (diamonds) addition of 1% root exudate. Panel C shows luciferase activities of the wild type (CY136) and the ∆kinD mutant (CY137) that harbored the PsdpA-lux reporter fusion in the presence (squares for WT and triangles for ∆kinD) or absence (diamonds for WT and circles for ∆kinD) of 1% root exudate. The arrows indicate the start of stationary phase. The luciferase activities were presented in arbitrary units (AU).
The response to root exudate depends on KinD
The results so far are consistent with the idea that plant roots release a chemical signal(s) that triggers biofilm formation. If so, how does B. subtilis sense and respond to the signal(s)? Biofilm formation is controlled by the response regulator protein Spo0A, which is activated by phosphorylation via a phosphorelay (Sonenshein, 2000). At least four histidine kinases known as KinA, KinB, KinC, and KinD are capable of feeding phosphoryl groups into the relay in response to various environmental signals in triggering biofilm formation (Jiang et al., 2000). Mutants lacking individual kinases were mildly impaired in biofilm formation whereas double mutants lacking certain pairs of kinases (KinA and KinB or KinC and KinD) exhibited more pronounced defects in biofilm formation (McLoon et al., 2011b). (A fifth kinase KinE may also feed phosphoryl groups into the relay but no phenotype has been detected for mutants lacking KinE.)
When tested in LB medium, all of the single and double mutants responded robustly to the plant root exudate except for the ∆kinD single mutant and the ∆kinC ∆kinD double mutant (Fig. 2A). Also, the elevated expression of the matrix gene reporters in response to root exudate discussed above was found to depend strongly on KinD (Fig. S4). These results suggest that KinD directly or indirectly plays a critical role in sensing a plant-released signal(s) and activating Spo0A~P-mediated regulatory pathways for biofilm formation.
To investigate whether the root exudate was stimulating Spo0A~P activity, we used a luciferase gene fusion to the promoter for the sdpABC operon (PsdpA-lux), whose activity is known to be highly sensitive to Spo0A~P levels (Banse et al., 2011). Addition of 1% root exudate to the medium increased expression of the PsdpA-lux reporter fusion by about two-fold (Fig. 3C). Interestingly, expression was almost completely dependent on KinD, with or without added root exudate. We infer either that apo-KinD (KinD lacking the hypothetical signaling molecule) is partially active (but not enough to induce biofilms) or that a molecule present in LB medium is itself capable of partially activating KinD.
A KinD mutant is defective in forming biofilms on root surfaces
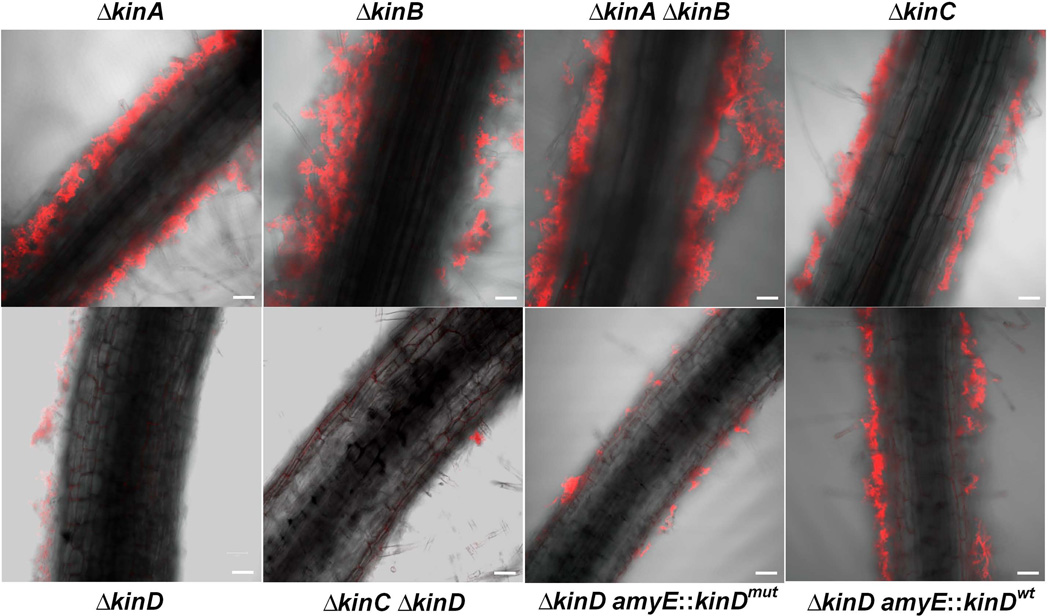
We next examined the ability of single and double kinase mutants to colonize and form biofilms on tomato root surfaces. The constitutively expressed red fluorescence reporter gene (mkate2) was introduced into the above mutants for visualization of the bacterial cells on the roots. All tested mutants except for the KinD single and the KinC KinD double mutants were able to colonize the root surfaces and form robust root-associated biofilms (Fig. 4). The KinC single mutant exhibited a mild decrease in colonization (Fig. 4). Thus, not only does KinD play an important role in inducing pellicles in LB medium in response to added root exudate but also in colonizing and forming biofilms on tomato root surfaces.
Figure 4. Biofilm formation by various kinase mutants on tomato roots.
Cells of the indicated mutants [∆kinA (CY204); ∆kinB (CY205); ∆kinA ∆kinB (CY206); ∆kinC (CY207); ∆kinD (CY127); ∆kinC ∆kinD (CY126); ∆kinD amyE::kinDmut (CY190); ∆kinD amyE::kinDwt (CY189)] that harbored a constitutively expressed mKate2 reporter gene were incubated with tomato plantlets for two days at room temperature and were visualized by CLSM. Scale bar, 50 µm.
The extracellular CACHE domain in KinD is required for signal-sensing
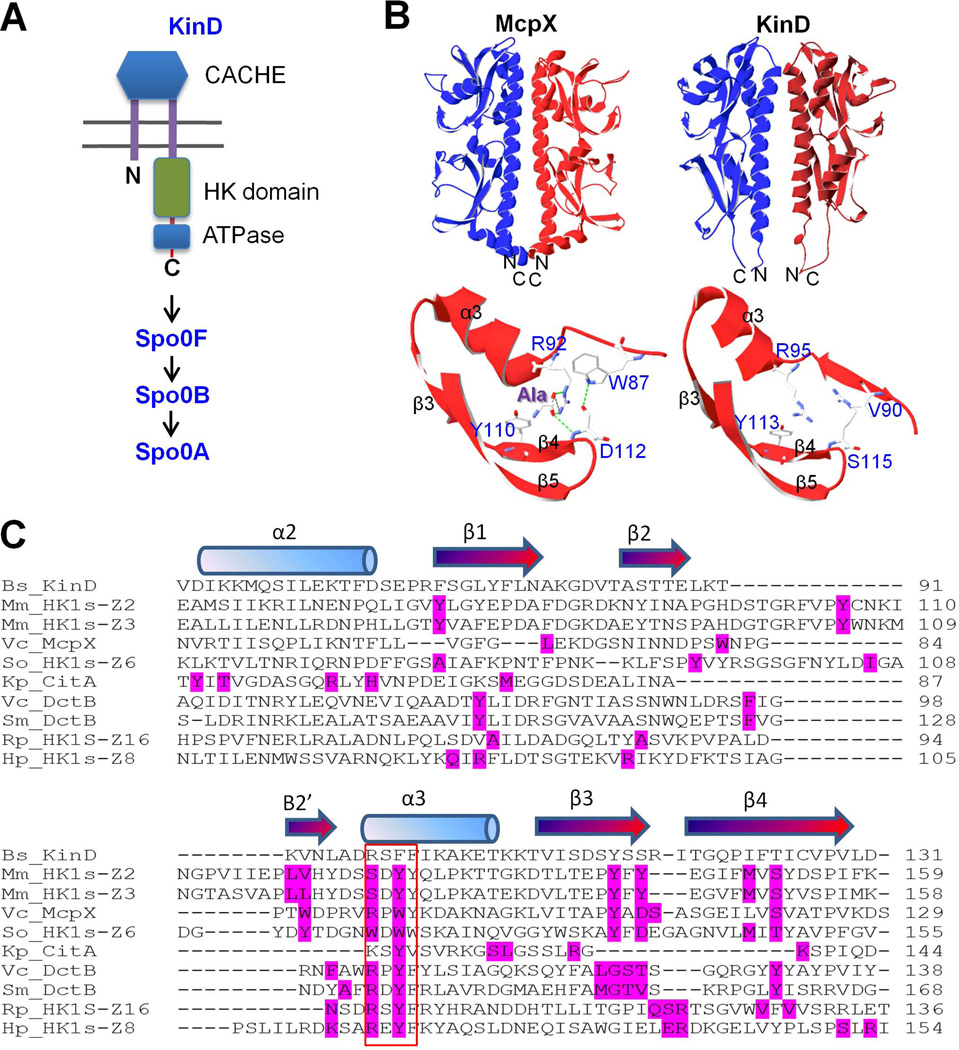
KinD has an extracellular domain, which resembles members of the CACHE family of sensing domains (Fig. 5A). (The name CACHE comes from the abbreviation of calcium channels and chemotaxis receptors.) Members of this family are widespread in both prokaryotic and eukaryotic organisms and are predicted to have a role in small molecule recognition (Anantharaman & Aravind, 2000). The small molecule-binding site is frequently found in the amino-terminal portion of the CACHE domain and consists of three beta-strands and an alpha helix (Fig. 5B) (Zhang & Hendrickson, 2010). This structural motif is also found in the X-ray crystal structure of the CACHE domain of KinD (Wu et al., 2008). A three-dimensional-structure-based homolog search (HHpred) revealed that the CACHE domain in KinD is closely related to signal-sensing domains from more than a dozen of bacterial chemotaxis proteins (Fig. 5C and Table S1), the closest match being the sensor domain of McpX, a methyl-accepting chemotaxis protein from Vibrio cholerae (Fig. 5B) (Patskovsky et al., 2008). In particular, the predicted small molecule-binding pocket in the CACHE domain in KinD is strikingly similar to that in McpX, which is known to bind alanine (Fig. 5B).
Figure 5. The CACHE domain of KinD.
Panel A is a cartoon representation of KinD. Panel B shows similarity between the three-dimensional structures of the CACHE domains of McpX (3C8C) from V. cholerae and of KinD (3FOS) from B. subtilis. In particular, the predicted small molecule-binding pocket in the CACHE domain in KinD is strikingly similar to that in the CACHE domain of McpX. Panel C shows the amino acid sequence alignment of the CACHE domains from KinD and nine related proteins. All residues in direct contact with a ligand are highlighted in red in the proteins that were known to bind ligands based on published protein structures. The red box indicates a four-amino-acid-residue sequence that is predicted as most critical residue in ligand binding. Amino acid substitutions of these residues were created in KinD. The protein IDs in the Protein Data Bank (PDB) for each of the sequences in Panel C are as follows: Bs_KinD (3FOS), Mm_HK1s-Z2 (3LI9), Mm_HK1s-Z3 (3LIB), Vc_McpX (3C8C), So_HK1s-Z6 (3LIC), Kp_CitA (1P0Z), Vc_DctB (3BY9), Sm_DctB (3E40), Rp_HK1s-Z16 (3LIF), and Hp_HK1s-Z8 (3LID).
To identify candidate residues critical for signal recognition, we analyzed nine proteins that are closely related to KinD based on our search and are known to bind to small molecules (Fig. 5C). All residues known to be in direct contact with the ligand based on crystal structures are highlighted in each of the nine proteins (residues in red, Fig. 5C). A four-amino-acid-residue sequence (indicated by the box in Fig. 5C) located in the third alpha helix in the CACHE domains was identified that mediates contact with ligands in all nine proteins (Fig. 5C). We therefore substituted these four amino acids (“RSFF”) in KinD with residues (“LLDS”) in the corresponding region of LuxQ from Vibrio harveyi. We did so because LuxQ is a membrane-spanning histidine kinase similar to KinD (Table S1) but has an unusual CACHE domain that is known not to directly bind a ligand (Neiditch et al., 2006, Slama & Hendrickson, 2008). Indeed, binding of the ligand, known as autoinducer 2 (AI-2, a bacterial interspecies quorum-sensing molecule), is mediated by a periplasmic partner protein called LuxP. LuxP, in turn, interacts with and activates LuxQ (Neiditch et al., 2006).
Next, we constructed two ∆kinD mutant strains that contained either a wild type copy of kinD at the amyE locus or the kinD mutant allele described above. We then tested the strains for their ability to respond to the root exudate and to form root-associated biofilms. Figure 2B shows that the ∆kinD strain complemented with the wild type copy of kinD exhibited a strong response to the added root exudate by forming pellicles. In contrast, the strain harboring the mutant allele of kinD responded weakly to the root exudate, with little pellicle formation (Fig. 2B).
To investigate the role of the CACHE domain in biofim formation on plant roots, we introduced a constitutively expressed red fluorescence reporter gene (mkate2) into the strains. Figure 4 shows that the strain harboring a wild-type copy of kinD at amyE robustly colonized plant roots whereas the strain carrying the mutant allele was markedly impaired in colonization.
We considered the possibility that rather than being specifically impaired in CACHE function, the CACHE mutant was proteolytically unstable. Three considerations suggest that this is unlikely. First, the amino acid substitutions that we created correspond to those found naturally in the CACHE domain of LuxQ and hence are unlikely to destabilize the protein. Second, we created fusions of the mutant and wild type genes at their 3’ termini to the gfp gene (see Supplemental Methods) and introduced the gene fusions (which were largely non-functional, data not shown) into B. subtilis. We then assessed the relative levels of accumulation of fusion proteins by immunoblot analysis using anti-GFP antibodies and found that the wild type and the mutant fusion proteins accumulated to approximately similar levels (data not shown). Thus alteration of the CACHE domain seemed not to have caused conspicuous proteolytic instability. However, we cannot exclude the possibility that fusion to GFP has protected the mutant protein from degradation. Third, in other work (see the Discussion) we have found that KinD has an independent function as an osmosensor. The CACHE mutant was not impaired in osmosensing (data not shown).
Finally, we carried out competition experiments on tomato roots (see the Materials and Methods) between the wild strain 3610 and either a ∆kinD mutant, a ∆kinD mutant harboring a wild type copy of kinD at amyE (∆kinD amyE::kinDwt), or a ∆kinD mutant harboring a CACHE domain mutant copy of kinD at amyE (∆kinD amyE::kinDmut). Cells of two competing strains were mixed at a 1:1 ratio and applied at a total concentration of ~108 cells/ml to the tomato plantlets grown in Murashige and Skoog (MS) medium. After two days, root-associated cells were collected and plated out. Cells of the ∆kinD strains were distinguished from 3610 cells by the presence of an antibiotic resistance gene. Colony-forming units for each tested strain were determined. When the competition was carried out with the strain harboring wild-type kinD at amyE about 65% of the recovered cells were the amyE::kinDwt cells (65.2±1.7%). However, when the competition was carried out against either the ∆kinD mutant strain or the ∆kinD strain harboring amyE::kinDmut, less than 10% of the recovered cells were the mutants (8.7±1.7% for the ∆kinD mutant strain and 9.8±3.5% for the ∆kinD strain harboring amyE::kinDmut). In a second version of the experiment, cells of the competing pairs of strains were mixed at 1:10 ratio representing one part 3610 cells to 10 parts of the other strains and applied to the plantlets at a concentration of ~107 cells/ml. (Thus, in this version, the initial concentration of 3610 cells was reduced ~50-fold compared to the first version.) Again, 3610 cells significantly out competed cells lacking kinD (~ 90% versus 10%) or harboring the CACHE mutant gene (~ 66% versus 34%). In toto, the results so far are consistent with the idea that the putative signal-recognition motif in the CACHE domain of KinD triggers biofilm formation by directly or indirectly responding to a plant-released chemical(s).
High concentrations of l-malic acid stimulate Spo0A activity and pellicle formation
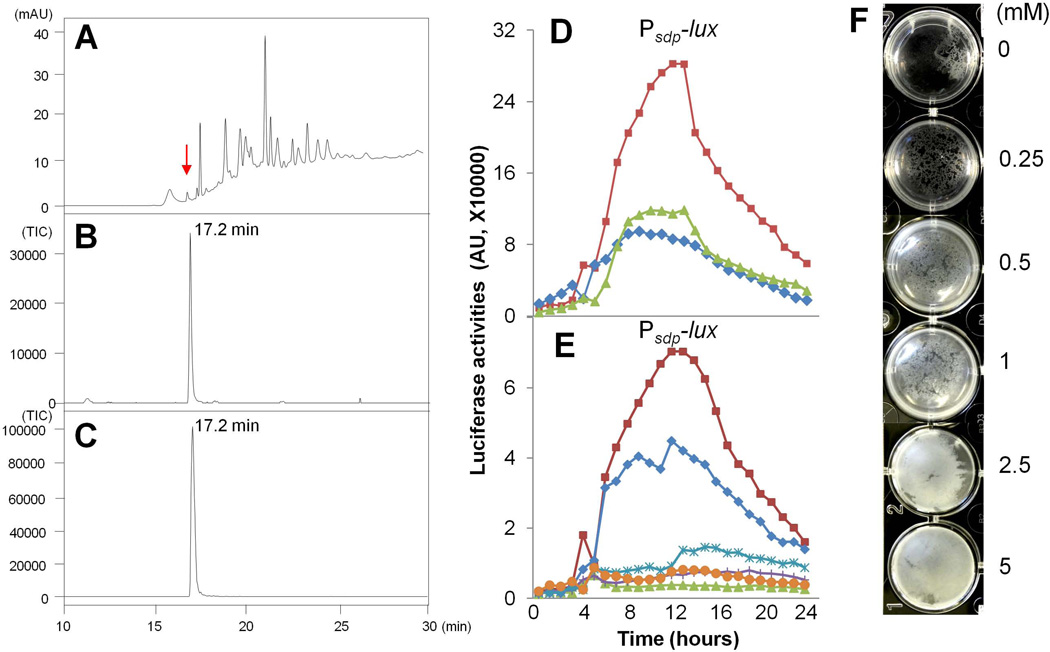
We sought to identify the tomato root-released chemical(s) that was inducing biofilm formation. To do so, we applied tomato root exudates to a Sep-Pak Plus C-18 column (Waters Corporation, MA, USA) and separated compounds by eluting them with various concentrations of methanol (see the Materials and Methods). We then tested the flow through and each of the methanol eluate fractions for the ability to stimulate the PsdpA-lux reporter. Fractions that exhibited activity were subjected to further separation by high performance liquid chromatography (HPLC) (data not shown), yielding eleven subfractions. Finally, the eleven subfractions were again tested for reporter stimulatory activity and two subfractions that had retained activity were analyzed again by HPLC. The elution profile of a subfraction from the flow through, which exhibited the majority of the stimulatory activity, is shown in Figure 6A. Based on mass spectrometry (MS) (Figs. 6B–6C) and NMR (data not shown) analyses, l-malic acid was identified as an abundant species in that subfraction, exhibiting a retention time of 17.2 min (indicated by arrow in Fig. 6A). (Note that l-malic acid had low UV absorption in HPLC analysis as seen in Fig. 6A). Our finding is also consistent with previous reports that l-malic acid is abundantly secreted by plant roots (Kamilova et al., 2006a, Kamilova et al., 2006b, Rudrappa et al., 2008). Through the above series of analyses, we also identified other small molecules that were abundant in the tomato root exudates (Table S2).
Figure 6. l-Malic acid stimulates biofilm formation.
Panels (A–C). Identification of l-malic acid in tomato root exudates by HPLC/MS. (A) Separation by HPLC of an active subfraction from tomato root exduate. mAU in Y-axis represents "milli-absorbance units".X-axis is the retention time. (B) An abundant molecule whose retention time is at 17.2 min (indicated by arrow in panel A) was identified as l-malic acid based on MS analysis (M-H Ion Extraction at 132). Values in Y-axis represent relative TIC (Total Ionization Current). (C) Commercially obtained l-malic acid (Sigma) was used as a standard in HPLC (not shown) and MS analysis (M-H Ion Extraction at 132). (D) Luciferase activities of the wild type strain (CY136) that harbored a PsdpA-lux reporter and was grown in LB shaking culture in the absence (diamonds), or presence of 5 mM l-malic acid (squares) or 5 mM d-malic acid (triangles). (E) Luciferase activities of the ∆kinD mutant (CY137) and the two kinD complementation strains, ∆kinD amyE::kinDwt (CY185) and ∆kinD amyE::kinDmut (CY186) that harbored the PsdpA-lux reporter and were grown in LB shaking culture in the presence or absence of 5 mM l-malic acid. Symbols are designated as follows: ∆kinD amyE::kinDwt, +malic acid (squares); ∆kinD amyE::kinDwt, −malic acid (diamonds); ∆kinD amyE::kinDmut, +malic acid (stars); ∆kinD amyE::kinDmut, −malic acid (crosses); ∆kinD, +malic acid (triangles); ∆kinD, −malic acid (circles). (F) 3610 cells were inoculated into a minimal medium (see Materials and Methods) that does not support pellicle formation. l-malic acid when added in a gradient from 250 µM to 5 mM, induced pellicle formation after 48 hours of incubation at the room temperature.
Among the molecules identified, only l-malic acid was able to stimulate expression of the PsdpA-lux reporter but it did so only at high concentrations. As shown in Fig. 6D, l-malic acid at 5 mM increased the activity of the reporter by about three-fold. Interestingly, when d-malic acid was provided at the same concentration, it had no activity in stimulating the expression of the PsdpA-lux reporter (Fig. 6D), indicating that the stereo isoform of malic acid is important. Induction of the PsdpA-lux reporter by l-malic acid was dependent on KinD and its CACHE domain since neither the KinD null mutant nor the KinD CACHE mutant responded to l-malic acid in the luciferase assays (Fig. 6E).
These results dovetail with those of Rudrappa et al. (Rudrappa et al., 2008) who found that l-malic acid was abundant in root exudates of Arabidopsis. (These workers also reported that l-malic acid (but not d-malic acid) stimulated the activity of a matrix gene promoter in B. subtilis but at a far lower concentration and far more robustly that we have been able to observe. We do not understand the basis for this discrepancy.)
We next asked whether l-malic acid would induce pellicle formation in LB medium inoculated with 3610 cells and found that it did so but only weakly (data not shown). However, when we applied l-malic acid to a minimal medium (see Materials and Methods for details) that like LB does not support biofilm formation, it strongly stimulated pellicle formation even at a concentration of 500 µM (Fig. 6F). Further, when we applied l-malic acid to MSgg, a defined medium that promotes biofilm formation (Branda et al., 2001), it caused pellicle formation to take place earlier than normal: robust pellicles were seen by 48 hours in the presence of malic acid as compared to 72 to 96 hours in its absence (Fig. S5). Thus, l-malic acid was able to stimulate pellicle formation albeit at high concentrations (500 µM to 5 mM). These effects were specific to l-malic acid as other organic acids and monosaccharides tested, such as succinic acid, fumaric acid, citric acid, glucose, and maltose (Table S2; data not shown), failed to stimulate pellicle formation under similar conditions. We infer that l-malic acid can contribute to stimulating biofilm formation in a specific manner but that it does so together with one or more additional and as yet unidentified factors present in plant root exudates.
DISCUSSION
We have shown that B. subtilis is capable of forming robust biofilms on the roots of tomato plants and that it does so in a manner that depends on production of extracellular matrix. We have also identified a particular sensor kinase, KinD, that is required for efficient biofilm formation. We hypothesize that KinD responds to one or more chemical signals released from plant roots that stimulates the activity of the kinase. KinD feeds phosphoryl groups into the phosphorelay that phosphorylates Spo0A, which in turn activates the regulatory circuitry leading to matrix production (Jiang et al., 2000). KinD differs from the other kinases contributing to biofilm formation in that it exhibits an extracellular domain that resembles CACHE domains of histidine kinases in other bacteria that are known to respond to small molecules such as succinic acid, alanine, and citric acid (Wu et al., 2008, Anantharaman & Aravind, 2000). Reinforcing the hypothesis that KinD senses one or more plant-released, chemical signals through its CACHE domain, we created amino acid substitutions in residues believed to be involved in ligand recognition and found that cells producing the mutant kinase were impaired in biofilm formation.
We do not know the identity of the signaling molecule(s) but an exudate from tomato plant roots was capable of strongly stimulating pellicle formation in a medium that otherwise does not support efficient biofilm formation. We surmise that the root exudates contain one or more signaling molecules that are recognized by KinD. We identified a variety of small molecules in the exudate, one of which, l-malic acid, was able to stimulate biofilm formation in a KinD-dependent manner but only at high concentrations. The stereo isoform of malic acid was important in that d-malic acid exhibited no activity in stimulating biofilm formation under similar conditions.
Because l-malic acid was active only at millimolar concentrations, we suspect that it principally functions as a carbon source and that growth on l-malic acid alters the metabolism of B. subtilis in a manner that favors biofilm formation. Indeed, in keeping with this idea, l-malic acid is one of the most abundant small molecules released from plant roots (Kamilova et al., 2006a, Kamilova et al., 2006b, Rudrappa et al., 2008). As a precedent, Photorhabdus and Xenorhabdus species enable their nematode hosts to parasitize insect larvae in response to high concentrations of l-proline in the insect hemolymph (Crawford et al., 2010). Growth on l-proline shifts the metabolism of the entomopathogenic bacteria in a manner that upregulates the production of various virulence factors important in antibiosis.
Conceivably, l-malic acid serves both as a carbon source and as a ligand that is recognized by the CACHE domain of KinD. Alternatively, l-malic acid and KinD might function independently with the sensor kinase recognizing and responding to an unrelated and yet-to-be-identified small molecule released from plant roots. It is also conceivable that the CACHE domain of KinD senses a small molecule produced by B. subtilis itself in response to a shift in metabolism induced by growth on l-malic acid and other nutrients in plant exudates. In an effort to investigate the possibility that l-malate serves as a signaling molecule independently of its role as a carbon source, we built single and double mutants of the putative malate transport genes maeN and mleN (Doan et al., 2003, Tanaka et al., 2003, Wei et al., 2000). In our hands neither the single nor the double mutants were substantially impaired in growth with l-malate as the sole carbon source (unpublished results). Evidently, an additional unknown transporter contributes to uptake of l-malic acid. Nonetheless, the double mutant responded to l-malate at 10-fold lower concentration (500 µM) than the wild type (5 mM).
Interestingly, in other work we have found that in addition to its inferred capacity to sense a small molecule, KinD functions as an osmosensor (S., Rubenstein, I. Kolodkin-Gal, L. Chai, A. McLoon, R.K., R.L. and D. Weitz, unpublished results). We have found that KinD activity is stimulated by increasing osmolarity from the production of exopolysaccharide during biofilm formation. Osmosensing occurs in a manner that is independent of the CACHE domain and instead requires on a transmembrane segment.
Finally, we note that KinD functions in conjunction with the membrane-anchored lipoprotein Med, and evidence suggests that KinD functions in a complex with Med (although this has not been shown directly) (Banse et al., 2011). Interestingly, Med is predicted to contain a ligand-binding domain that strongly resembles known sugar- or nucleoside-binding domains of other proteins (unpublished observation, Chen et al.), including LuxP from V. harveyi (a 99.9% probability based on HHpred search). In V. harveyi, LuxP binds directly to the quorum-sensing molecule autoinducer-2 (AI-2, a borate-containing analog of ribose) and interacts with the membrane histidine kinase LuxQ in quorum-sensing signal transduction (Ng & Bassler, 2009). LuxQ is a structural homolog of KinD as discussed above. Thus, Med and KinD may function in a somewhat analogous manner to that of LuxP and LuxQ, which act together in sensing the bacterial interspecies quorum-sensing signal AI-2. (A key difference is that LuxQ does not itself recognize a signal (Neiditch et al., 2006).) Although we do not know whether Med directly senses a plant-released signal, it is appealing to imagine that Med and the CACHE domain of KinD sense distinct signals, and together they activate KinD kinase activity in inducing biofilm formation. A key challenge for the future will be to identify signaling molecules released from tomato plant roots that activate KinD and to determine whether both Med and KinD are involved in signal recognition.
Supplementary Material
Acknowledgments
We wish to thank Dr. Elena Kramer for use of the plant growth chamber, T. Norman, A. McLoon A and A. Camp for strains and plasmids. We also thank members of the Losick and Kolter groups, especially Drs. Foulson L and Beauregard P for technical assistance and helpful suggestions. CLSM was performed at Harvard Center of Bioimaging. This work was in part supported by grants from National Natural Science Foundation of China (31171809, 30971956) and Specialized Research Fund for the Doctoral Program of Higher Education of China (20100097110010) to JG. Y. Chen was supported by China Scholarship Council (NO.2010685015) and Graduate Innovation Projects of Jiangsu Province (No. CX10B_3152). This work was also supported by NIH grants GM18568 to RL, GM58213 and GM82137 to RK, and GM086258 and AI057159 to JC.
REFERENCES
- Aguilar C, Vlamakis H, Losick R, Kolter R. Thinking about Bacillus subtilis as a multicellular organism. Curr. Opin. Microbiol. 2007;10:638–643. doi: 10.1016/j.mib.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharaman V, Aravind L. CACHE - a signaling domain common to animal Ca2+-channel subunits and a class of prokaryotic chemotaxis receptors. Trends Biochem. Sci. 2000;25:535–537. doi: 10.1016/s0968-0004(00)01672-8. [DOI] [PubMed] [Google Scholar]
- Bais HP, Fall R, Vivanco JM. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004;134:307–319. doi: 10.1104/pp.103.028712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banse AV, Hobbs EC, Losick R. Phosphorylation of Spo0A by the histidine kinase KinD requires the lipoprotein Med in Bacillus subtilis. J. Bacteriol. 2011;193:3949–3955. doi: 10.1128/JB.05199-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg G, Krechel A, Ditz M, Sikora RA, Ulrich A, Hallmann J. Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol. Ecol. 2005;51:215–229. doi: 10.1016/j.femsec.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Bramhill D. Bacterial cell division. Annu. Rev. Cell Dev. Biol. 1997;13:395–424. doi: 10.1146/annurev.cellbio.13.1.395. [DOI] [PubMed] [Google Scholar]
- Branda SS, Chu F, Kearns DB, Losick R, Kolter R. A major protein component of the Bacillus subtilis biofilm matrix. Mol. Microbiol. 2006;59:1229–1238. doi: 10.1111/j.1365-2958.2005.05020.x. [DOI] [PubMed] [Google Scholar]
- Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA. 2001;98:11621–11626. doi: 10.1073/pnas.191384198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Chu F, Kolter R, Losick R. Bistability and biofilm formation in Bacillus subtilis. Mol. Microbiol. 2008;67:254–263. doi: 10.1111/j.1365-2958.2007.06040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai Y, Norman T, Kolter R, Losick R. Evidence that metabolism and chromosome copy number control mutually exclusive cell fates in Bacillus subtilis. EMBO J. 2011;30:1402–1413. doi: 10.1038/emboj.2011.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu F, Kearns DB, Branda SS, Kolter R, Losick R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol. Microbiol. 2006;59:1216–1228. doi: 10.1111/j.1365-2958.2005.05019.x. [DOI] [PubMed] [Google Scholar]
- Crawford JM, Kontnik R, Clardy J. Regulating alternative lifestyles in entomopathogenic bacteria. Curr. Biol. 2010;20:69–74. doi: 10.1016/j.cub.2009.10.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan T, Servant P, Tojo S, Yamaguchi H, Lerondel G, Yoshida K-I, Fujita Y, Aymerich Sp. The Bacillus subtilis ywkA gene encodes a malic enzyme and its transcription is activated by the YufL/YufM two-component system in response to malate. Microbiology. 2003;149:2331–2343. doi: 10.1099/mic.0.26256-0. [DOI] [PubMed] [Google Scholar]
- Dubnau D. DNA uptake in bacteria. Annu. Rev. Microbiol. 1999;53:217–244. doi: 10.1146/annurev.micro.53.1.217. [DOI] [PubMed] [Google Scholar]
- Emmert EAB, Handelsman J. Biocontrol of plant disease: a (Gram-) positive perspective. FEMS Microbiol. Lett. 1999;171:1–9. doi: 10.1111/j.1574-6968.1999.tb13405.x. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- Graham JH, Leonard RT, Menge JA. Membrane-mediated decrease in root exudation responsible for phorphorus inhibition of vesicular-arbuscular mycorrhiza formation. Plant Physiol. 1981;68:548–552. doi: 10.1104/pp.68.3.548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan TJ, Contente S, Dubnau D. Characterization of Staphylococcus aureus plasmids introduced by transformation into Bacillus subtilis. J. Bacteriol. 1978;134:318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guérout-Fleury AM, Frandsen N, Stragier P. Plasmids for ectopic integration in Bacillus subtilis. Gene. 1996;180:57–61. doi: 10.1016/s0378-1119(96)00404-0. [DOI] [PubMed] [Google Scholar]
- Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hamon MA, Lazazzera BA. The sporulation transcription factor Spo0A is required for biofilm development in Bacillus subtilis. Mol. Microbiol. 2001;42:1199–1209. doi: 10.1046/j.1365-2958.2001.02709.x. [DOI] [PubMed] [Google Scholar]
- Ireton K, Rudner DA, Siranosian KJ, Grossman AD. Integration of multiple developmental signals in Bacillus subtilis through the Spo0A transcription factor. Genes Dev. 1993;7:283–294. doi: 10.1101/gad.7.2.283. [DOI] [PubMed] [Google Scholar]
- Jiang M, Shao W, Perego M, Hoch JA. Multiple histidine kinases regulate entry into stationary phase and sporulation in Bacillus subtilis. Mol. Microbiol. 2000;38:535–542. doi: 10.1046/j.1365-2958.2000.02148.x. [DOI] [PubMed] [Google Scholar]
- Kamilova F, Kravchenko LV, Shaposhnikov AI, Azarova T, Makarova N, Lugtenberg B. Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol. Plant-Microbe Int. 2006a;19:250–256. doi: 10.1094/MPMI-19-0250. [DOI] [PubMed] [Google Scholar]
- Kamilova F, Kravchenko LV, Shaposhnikov AI, Makarova N, Lugtenberg B. Effects of the tomato pathogen Fusarium oxysporum f. sp. radicis-lycopersici and of the biocontrol bacterium Pseudomonas fluorescens WCS365 on the composition of organic acids and sugars in tomato root exudate. Mol. Plant-Microbe Int. 2006b;19:1121–1126. doi: 10.1094/MPMI-19-1121. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Branda SS, Kolter R, Losick R. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 2005;55:739–749. doi: 10.1111/j.1365-2958.2004.04440.x. [DOI] [PubMed] [Google Scholar]
- Kearns DB, Chu F, Rudner R, Losick R. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol. Microbiol. 2004;52:357–369. doi: 10.1111/j.1365-2958.2004.03996.x. [DOI] [PubMed] [Google Scholar]
- Lemessa F, Zeller W. Screening rhizobacteria for biological control of Ralstonia solanacearum in Ethiopia. Biol. Control. 2007;42:336–344. [Google Scholar]
- McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. Tracing the domestication of a biofilm-forming bacterium. J. Bacteriol. 2011a;193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLoon AL, Kolodkin-Gal I, Rubinstein SM, Kolter R, Losick R. Spatial regulation of histidine kinases governing biofilm formation in Bacillus subtilis. J. Bacteriol. 2011b;193:679–685. doi: 10.1128/JB.01186-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiologia Plantarum. 1962;15:473–497. [Google Scholar]
- Neiditch MB, Federle MJ, Pompeani AJ, Kelly RC, Swem DL, Jeffrey PD, Bassler BL, Hughson FM. Ligand-Induced asymmetry in histidine sensor kinase complex regulates quorum sensing. Cell. 2006;126:1095–1108. doi: 10.1016/j.cell.2006.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng W-L, Bassler BL. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ongena M, Jacques P. Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 2008;16:115–125. doi: 10.1016/j.tim.2007.12.009. [DOI] [PubMed] [Google Scholar]
- Patskovsky Y, Ozyurt S, Freeman J, Hu S, Smith D, Bain K, Wasserman SR, Sauder JM, Burley SK, Almo SC. Crystal structure of Mcp_N and cache N-terminal domains of methyl-accepting chemotaxis protein from Vibrio cholerae. PDB. 2008 ID: 3c8c. [Google Scholar]
- Piggot PJ, Hilbert DW. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 2004;7:579–586. doi: 10.1016/j.mib.2004.10.001. [DOI] [PubMed] [Google Scholar]
- Romero D, Aguilar C, Losick R, Kolter R. Amyloid fibers provide structural integrity to Bacillus subtilis biofilms. Proc. Natl. Acad. Sci. USA. 2010;107:2230–2234. doi: 10.1073/pnas.0910560107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudrappa T, Czymmek KJ, Paré PW, Bais HP. Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 2008;148:1547–1556. doi: 10.1104/pp.108.127613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning. A Laboratory Manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Shcherbo D, Merzlyak EM, Chepurnykh TV, Fradkov AF, Ermakova GV, Solovieva EA, Lukyanov KA, Bogdanova EA, Zaraisky AG, Lukyanov S, Chudakov DM. Bright far-red fluorescent protein for whole-body imaging. Nat Meth. 2007;4:741–746. doi: 10.1038/nmeth1083. [DOI] [PubMed] [Google Scholar]
- Slama B, Hendrickson W. Crystal structure of the periplasmic domain of Vibrio Cholerae LuxQ. PDB. 2008 ID:3c38. [Google Scholar]
- Sonenshein AL. Control of sporulation initiation in Bacillus subtilis. Curr. Opin. Microbiol. 2000;3:561–566. doi: 10.1016/s1369-5274(00)00141-7. [DOI] [PubMed] [Google Scholar]
- Stephenson K, Hoch JA. Evolution of signalling in the sporulation phosphorelay. Mol. Microbiol. 2002;46:297–304. doi: 10.1046/j.1365-2958.2002.03186.x. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Kobayashi K, Ogasawara N. The Bacillus subtilis YufLM two-component system regulates the expression of the malate transporters MaeN (YufR) and YflS, and is essential for utilization of malate in minimal medium. Microbiology. 2003;149:2317–2329. doi: 10.1099/mic.0.26257-0. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in Saccharomyces cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wei Y, Guffanti AA, Ito M, Krulwich TA. Bacillus subtilis YqkI Is a novel malic/Na+-lactate antiporter that enhances growth on malate at low protonmotive force. J. Biol. Chem. 2000;275:30287–30292. doi: 10.1074/jbc.M001112200. [DOI] [PubMed] [Google Scholar]
- Wu R, Schiffer M, Gu M, Joachimiak A. The crystal structure of two-component sensor histidine kinase domain from Bacillus subtilis. PDB. 2008 doi: 10.1002/pro.2237. ID: 3FOS. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasbin RE, Young FE. Transduction in Bacillus subtilis by bacteriophage SPP1. J. Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Hendrickson WA. Structural characterization of the predominant family of histidine kinase sensor domains. J. Mol. Biol. 2010;400:335–353. doi: 10.1016/j.jmb.2010.04.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.