Abstract
Developmental gene expression results from the orchestrated interplay between genetic and epigenetic mechanisms. Here we describe upSET, a transcriptional regulator encoding a SET domain-containing protein recruited to active and inducible genes in Drosophila. However, unlike other Drosophila SET proteins associated with gene transcription, UpSET is part of an Rpd3/Sin3-containing complex that restricts chromatin accessibility and histone acetylation to promoter regions. In the absence of UpSET, active chromatin marks and chromatin accessibility increase and spread to genic and flanking regions due to destabilization of the histone deacetylase complex. Consistent with this, transcriptional noise increases, as manifest by activation of repetitive elements and off-target genes. Interestingly, upSET mutant flies are female sterile due to up-regulation of key components of Notch signaling during oogenesis. Thus UpSET defines a class of metazoan transcriptional regulators required to fine tune transcription by preventing the spread of active chromatin.
Keywords: UpSET, Mixed Lineage Leukemia, histone acetylation, chromatin accessibility, Drosophila, Rpd3, Sin3
INTRODUCTION
Genome-wide analyses of chromatin accessibility, histone modifications, and factor/cofactor recruitment have led to the identification of regulatory elements and their associated protein components (modENCODE Consortium et al., 2010; Filion et al., 2010). For example, chromatin accessibility at promoters is usually higher than in transcribing regions and sharply localized to a few hundred base pairs from the transcriptional start site(TSS). In addition, while transcribed regions are enriched for histone marks such as H3K36m3 and H3K79m2, promoters are enriched for other histone modifications, including acetylation (H3K9Ac and H4K16Ac) and methylation (H3K4m2 and H3K4m3). Although the distribution of H3K4m2 is variable among species, H3K4m3 and histone acetylation are conserved landmarks for active promoters, and several reports suggest that they are functionally linked (Cai et al., 2010). However, it is still unclear whether these histone modifications are a cause or consequence of promoter accessibility and how chromatin accessibility and promoter remodeling are maintained in a localized fashion (Henikoff and Shilatifard, 2011).
In contrast, the correlation between the proteins establishing those modifications and transcriptional regulation is well accepted. Promoter-associated H3K4m3 is mainly established by SET (Su(var)3-9, Enhancer of Zeste, Trithorax) domain-containing proteins including the Trithorax family of proteins (TrxG). The SET domain is a 130-amino acid, evolutionarily conserved sequence motif present in chromosomal proteins that function in modulating gene activities from yeast to mammals (Dillon et al., 2005). SET domain containing proteins are part of large complexes and are usually involved in establishing or affecting post-translational histone modifications that correlate with chromatin remodeling, transcription initiation, and transcriptional elongation (Eissenberg and Shilatifard, 2010). For example, the yeast Set1 is part of a macromolecular H3K4 methyltransferase complex named COMPASS (Miller et al., 2001), and, in Drosophila, there are three COMPASS-like complexes that control H3K4me homeostasis (dSET1, Trx, and Trr) with dSET1 serving as the main H3K4 methyltransferase among these complexes (Ardehali et al., 2011; Mohan et al., 2011; Hallson et al., 2012; Eissenberg and Shilatifard, 2010). Proteomic analysis has revealed that histone acetyltransferases are critical components of COMPASS complexes, supporting the notion of a continuous crosstalk between histone acetylation and methylation (Schübeler et al., 2004; Bernstein et al., 2005). Therefore, it has been suggested that COMPASS complexes play a key role in establishing promoter architecture.
In mammals, the Mixed Lineage Leukemia (MLL) family of SET methyltransferase domain proteins includes at least six members (MLL1-4, Set1A, Set1B) and are involved in gene activation (Ruthenburg et al., 2007). Biochemical analysis has revealed that these proteins are also organized in COMPASS-like complexes that can methylate H3K4 (Eissenberg and Shilatifard, 2010). Recently, human MLL5 (KMT2E) was added to the MLL family due to its SET domain homology with MLL1 (Emerling et al., 2002). Interestingly, the mll5 gene is located in a region of chromosome 7 commonly deleted in a subset of leukemias associated with poor prognosis (Emerling et al., 2002). Unlike other MLL family members, MLL5 has a more centrally located SET domain, a single PHD domain, but lacks AT-hook domains. Putative MLL5 orthologs exist in other organisms, including CG9007 in Drosophila and Set3 and Set4 in yeast (Emerling et al., 2002; Eissenberg and Shilatifard, 2010). While the MLL5 SET domain is the primary region of conservation among MLL5 orthologs, it lacks key catalytic amino acids, bringing into question the catalytic activity of this SET domain (Sebastian et al., 2009; Madan et al., 2009). However, O-linked N-acetylglucosamine transferase (OGT)-mediated SET domain glycosylation of a small human MLL5 isoform may be required to mono- and di-methylate H3K4 (Fujiki et al., 2009). MLL5 knock out mice are viable, and surprisingly exhibit only mild hematological phenotypes and male sterility (Zhang et al., 2009; Madan et al., 2009; Heuser et al., 2009; Yap et al., 2011), which may reflect functional redundancy by a related gene (SETD5) with high protein homology with MLL5. The yeast MLL5 ortholog, Set3, lacks the proposed glycosylating residue and displays no histone methyltransferase activity (Pijnappel et al., 2001). Set3 is targeted to active genes and associated with a histone deacetylase (HDAC) complex, however, its precise function once recruited is unclear (Kim and Buratowski, 2009; Govind et al., 2010; Buratowski and Kim, 2011).
Here we describe the Drosophila gene whose protein’s SET domain most resembles MLL5. We have named this gene upSET (CG9007), due its genetic and biochemical features. DamID chromatin profiling reveals preferential recruitment of UpSET to transcriptionally active genes overlapping with RNA pol II, TrxG proteins and H3K4m3. UpSET interacts with a dRpd3-containing HDAC complex and promotes its recruitment to active regions. Knock down experiments reveal an increase in histone acetylation around the TSS, which can spread to neighboring regions and correlates with increased chromatin accessibility around UpSET bound regions. Knock down cells increase the expression of genes and repeat elements neighboring regions of UpSET binding. Consistent with this, oogenesis is impaired by the lack of UpSET, due to deregulated Notch signaling, causing alterations in egg chamber formation and anterior-posterior specification. This female sterility phenotype can be partially rescued by expressing the mouse MLL5 gene. In genetic interactions UpSET enhances phenotypes associated with PcG epigenetic repressors. Our results indicate that UpSET is part of the machinery recruited to promoter regions to regulate chromatin stability and transcription state by preventing the spread of active chromatin.
RESULTS
UpSET is a nuclear protein and localizes to gene rich euchromatic regions
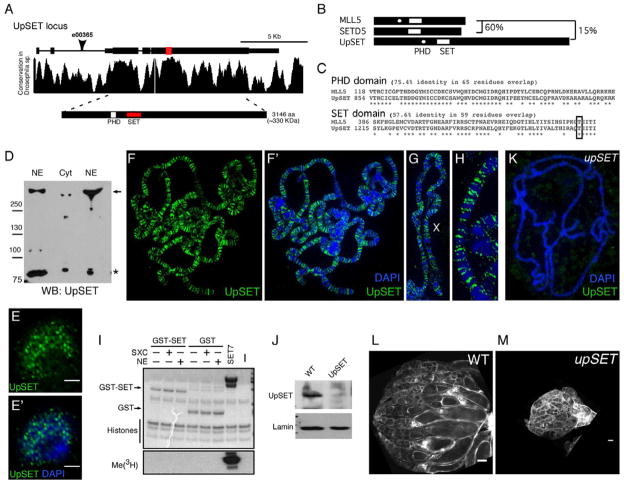
upSET encodes a ~330 KDa (3146aa) protein with a single PHD domain and centrally located H3K4 histone methyltransferase SET domain (Figure 1A). UpSET coding sequence is highly conserved among 12 Drosophila genomes, primarily over the 5’ end of the gene (Figure 1A). The UpSET SET domain shares high similarity among the Drosophilidae family (Figure S1A). UpSET shares restricted homology over the SET domain with two mammalian proteins, MLL5 and SETD5 (Figure 1B). UpSET and MLL5 share 58% identity in their SET domains and 75% identity in their adjacent PHD domains (Figure 1C). SETD5 SET domain similarly shares 60% identity with UpSET’s SET domain, but lacks a PHD domain. Other than these domain similarities, UpSET does not share significant homology with MLL5, SETD5 or other SET-containing proteins (Figure 1B–C, S1B). The presence of these chromatin regulatory domains suggests that UpSET function is nuclear. Thus we generated monoclonal antibodies to UpSET (Figures S1D) and performed Western (Figures 1D, S1C) and immunofluorescence (Figures 1E–E’) analyses in Drosophila Kc cells. As expected, UpSET is located mainly in the nucleus with a punctate distribution. To establish whether UpSET interacts with chromatin in vivo, we examined binding of endogenous UpSET to third instar larval salivary gland polytene chromosomes. Interestingly, we detect roughly 1000 strongly staining UpSET binding sites on polytene chromosomes (Figures 1F–H). These sites are mainly in gene rich euchromatic regions and generally excluded from DAPI-dense regions, supporting a global role of UpSET in gene regulation.
Figure 1. UpSET is a SET-containing nuclear protein required for Drosophila oogenesis.
(A) Schematic of the upSET locus on chromosome 3L. Location of the SET and PHD domains are highlighted. Arrowhead indicates P-element insertion in upSETe00365. Graph shows the locus conservation in the Drosophilidae family.
(B) UpSET shares only ~15% identity with mouse MLL5 in the SET domain. The mouse MLL5 SET domain shares 60% identity with that of mouse SETD5.
(C) Sequence alignment of the SET and PHD domains between Drosophila UpSET and mouse MLL5. % identity is indicated.
(D) UpSET is mainly a nuclear protein. Western blot analysis of nuclear (NE) and cytoplasmic (Cyt) extracts from Kc cells probed with anti-UpSET N-end monoclonal antibodies. Full-length UpSET protein is denoted by the arrow. *:Non-specific band.
(E–E’) UpSET forms foci in the nucleus. Confocal projection of 10 serial 0.2μm optical sections of a Kc cell stained with a mix anti-UpSET NH3-terminus monoclonal antibodies (green; F–F’) and DAPI to visualize DNA (F’).
(F–H) Third larval instar polytene chromosomes stained with a mix of antibodies to UpSET NH3- and COOH-terminus (green) and DNA counterstained with DAPI (blue). Higher magnification view of X chromosome (H). UpSET recruitment is enriched in euchromatic regions (I).
(I) UpSET does not exhibit histone methyltransferase activity (HMT). Radioactive HMT assays performed with bacterially purified GST or GST-UpSET SET domain and purified histones as substrate (J). Recombinant proteins were incubated with Super Sex Combs (SXC) or nuclear extracts (NE). SET7 was used as positive control. Loading controls for the histone methyltransferase assay are shown.
(K) Polytene chromosome from upSETe00365 mutant third instar larva stained with a mix of the 5 UpSET monoclonal antibodies (green) and DNA counterstained with DAPI (blue).
(J) Western blot analysis of larval nuclear extracts from wildtype and upSETe00365 mutants blotted with a mix of the 5 antibodies against UpSET NH3- and COOH-termini. Lamin Dm0 serves as loading control.
(L–M) Lack of UpSET disrupts oogenesis. Confocal projections of 5 serial 1μm optical sections of ovaries from wildtype (N) and upSETe00365 mutants (O) actin stained with phalloidin. Scale bars represent 100μm. See also Figure S1.
A hallmark of SET domain containing proteins is histone methyltransferase (HMT) activity. Interestingly, the UpSET SET domain lacks key amino acids required for this catalytic function (Emerling et al., 2002). Therefore, we performed an in vitro HMT assay to address whether UpSET’s SET domain is functional. We bacterially expressed and purified the UpSET SET domain, and found that by itself the SET domain does not have HMT activity (Figures 1I). The mouse MLL5 SET domain also lacks these key amino acids, however, whether it possesses catalytic activity is controversial (Madan et al., 2009; Eissenberg and Shilatifard, 2010): it has been suggested that a short human MLL5 isoform can exhibit HMT activity upon glycosylation of Threonine 440 (Fujiki et al., 2009). While this Threonine is conserved among Drosophila species (Figure 1C, S1A), pre-incubation of the UpSET SET domain with in vitro expressed Super sex combs (SXC; the main nuclear Drosophila OGT (Sinclair et al., 2009)) or nuclear extracts does not confer HMT activity (Figure 1I, S1E). Thus, while UpSET shares structural SET domain homology with HMTs, it does not appear to be catalytically active. It remains possible that the UpSET SET domain requires the context of the full-length protein to function, but due to its large size it has not yet been possible to express full-length UpSET in E. coli.
upSET mutants are female sterile
To address the biological functions of UpSET, we obtained an existing allele (upSETe00365), in which a P-element is inserted in the second intron of upSET upstream of the translation start site (PBac{RB}CG9007[e00365]; Figure 1A). upSETe00365 is a protein null allele: UpSET protein is absent in upSETe00365 mutants by western blot or on polytene chromosomes (Figures 1J–K). upSETe00365 homozygous mutants are viable, suggesting that UpSET is not required for survival. However, the resulting mutant females are sterile (98% of upSET females do not lay eggs; Table 1) and display severe oogenesis phenotypes: 80% of egg chambers do not progress beyond stage 8–9 and mature eggs are not produced (n=215; Figures 1L–M). Importantly, this female sterility can be rescued by a BAC transgene containing the upSET locus (Table 1). Interestingly, expression of the mouse MLL5 cDNA (similar to UpSET in that it does not show HMT activity), also partially rescues female sterility in ~20% of upSET females (n=50; Table 1, Figure S1F–G). The partial rescue is likely due to the limited functional conservation of UpSET with MLL5, as well as its similarity to the other SET domain protein SETD5.
Table 1. upSET.
mutants are female sterile, increase Polycomb phenotypes, and reduce Trithorax-associated homeotic transformations.
| Lack of UpSET causes female sterility
| |||||
|---|---|---|---|---|---|
| Genotype
|
|||||
| Female | Male | Sterile females % | No. sterile females N=50 | Rescue % | No. Egg |
| +/+ | +/+ | 0 | 0 | n/a | 1536 |
| +/ upSETe00365 | +/+ | 0 | 0 | n/a | 1330 |
| upSETe00365/ upSETe00365 | +/+ | 98 | 49 | n/a | 5 |
| BAC1X a; upSETe00365/ upSETe00365 | +/+ | 26 | 13 | 72 | 1035 |
| BAC2X b; upSETe00365/ upSETe00365 | +/+ | 5 | 3 | 93 | 1231 |
| mMLL5 c; upSETe00365/ upSETe00365 | +/+ | 78 | 39 | 20 | 408 |
| upSET enhances Pc phenotypes. | |||
|---|---|---|---|
| % with enhanced phenotype e, f (n)
|
|||
| Genotype*: | + | upSETe00365 | upSETMB8950 g |
| +/+; +/* d | 0 (350) | 0 (303) | 0 (239) |
| Pc3 /+; +/* e | 40 (403) | 89 (495) | 76 (184) |
| Pc1 Abd-B Mc /+; +/* e | 33 (320) | 75 (256) | 75 (122) |
| brm2 trxE2 /+ ; +/* f | 56 (465) | 9 (327) | 4 (409) |
BAC1X= w; P[acman](attA CH322.187J08 attB)/CyO.
BAC2X= w; P[acman](attA CH322.187J08 attB)/ P[acman](attA CH322.187J08 attB)
mMLL5= w;UAS_mMLL5/ActGAL4
wildtype is OregonR
For the Pc alleles Pc3 and Pc1, males of the genotypes indicated in the first column were scored for the presence of a sex comb on the 2nd and/or third leg (usually found only on the first leg).
brm2 trxE2 was scored for the presence of an apical bristle on the third leg (usually found on the second leg).
See also Figure S7A n/a = not applicable
UpSET binds to transcriptionally active regions
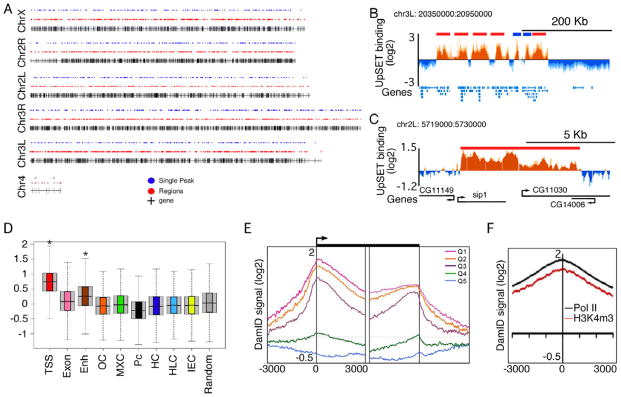
Based on the upSET female sterile phenotype and its presence in euchromatic regions, we addressed in more detail the genomic regions targeted by UpSET. DamID chromatin profiling in Kc cells (Greil et al., 2006; Orian et al., 2009) revealed that UpSET binds to ~3500 regions genome-wide and overlaps with gene rich regions (Figures 2A–C, S2A), corresponding to ~6000 genes. These results are consistent with the broad presence of UpSET observed on polytene chromosomes (Figure 1F–H).
Figure 2. UpSET is preferentially recruited to active genes.
(A) Sites of UpSET recruitment based on DamID. The black vertical lines represent the relative position on the chromosomes of the ~14000 Drosophila genes. Blue dots represent single peaks (3.5-6Kb) and the red dots (>6 Kb) UpSET binding sites.
(B–C) UpSET chromatin profile over a 600Kb region of chromosome 3R and a 11Kb region of chromosome 2L. Single peaks and regions are presented in blue and red, respectively. The mean signal was smoothed + whiskers. Coding regions are shown in blue (B) and black (C).
(D) UpSET is mainly enriched in promoter (TSS) regions. Box plots of the UpSET DamID-based chromatin profile over the 9 different chromatin states reported by the modENCODE Consortium. TSS: transcriptional start sites; Exon: elongating regions; Enh: Enhancers; OC: open chromatin; MXC: male X genes; Pc: Polycomb; HC: constitutive heterochromatin; HLC: heterochromatin-like in euchromatin; IEC: basal, intergenic euchromatin. Random: set of 6000 random sequences. *: p< 2.2X10−16
(E) Preferential association of UpSET over transcribed genes. End analysis of the UpSET chromatin profile to the 5’ and 3’ end of genes ranked by expression quintiles from high (Q1) to low (Q5) gene expression.
(F) End analysis of UpSET DamID signal to RNA Pol II (black) and H3K4m3 (red) enriched regions. See also Figure S2.
Recently, several different chromatin states have been characterized in the Drosophila genome by profiling the binding of 53 different proteins (Filion et al., 2010), as well as histone modifications, transcription factor binding, and/or gene expression (modENCODE Consortium et al., 2010). To determine whether UpSET associates with specific chromatin states, we examined the UpSET DamID-signal distribution for each regulatory region described by the modENCODE consortium. DamID signal distribution over each chromatin state showed a preferential recruitment of UpSET to TSS/promoter regions (p< 2.2x10−16), and to a lesser extent, enhancer and transcribed regions (Figure 2D). These enrichments are significant compared to other chromatin states, as well as a control cohort of 6000 random sequences (Figure 2D; see Supplemental Experimental Procedures). Analyses of UpSET enrichment in five chromatin types defined by Filion et al. revealed a similarly high overlap with active regions (Figure S2B). Thus, UpSET is preferentially targeted to active promoter regions.
To determine whether UpSET binding correlates with gene expression, we performed an end analysis to correlate UpSET binding relative to the 5’ and 3’ ends of genes arranged by expression-based quintiles (Deal et al., 2010). Our results show that a peak of UpSET binding mainly occurs 25–50 bp downstream of the TSS. This binding is dependent on gene expression, as quintiles with highly expressed genes have higher upSET signal (Figure 2E). As active TSS regions are marked by active chromatin marks (i.e., H3K4m3), as well as with high levels of RNA pol II, we performed an end analysis of RNA pol II and H3K4m3 enriched sites from S2 cells (Muse et al., 2007). Consistent with UpSET enrichment at active TSSs, we observed high UpSET signal in regions enriched for RNA pol II and H3K4m3 (Figure 2F). This overlap is not due to polymerase pausing, as promoters demonstrating high or low pausing indexes show similar enrichment for UpSET (Figure S2C) (Gilchrist et al., 2010). Together, our results suggest that UpSET targets promoter/TSS regions of transcriptionally active genes independently of the pausing features of the promoters.
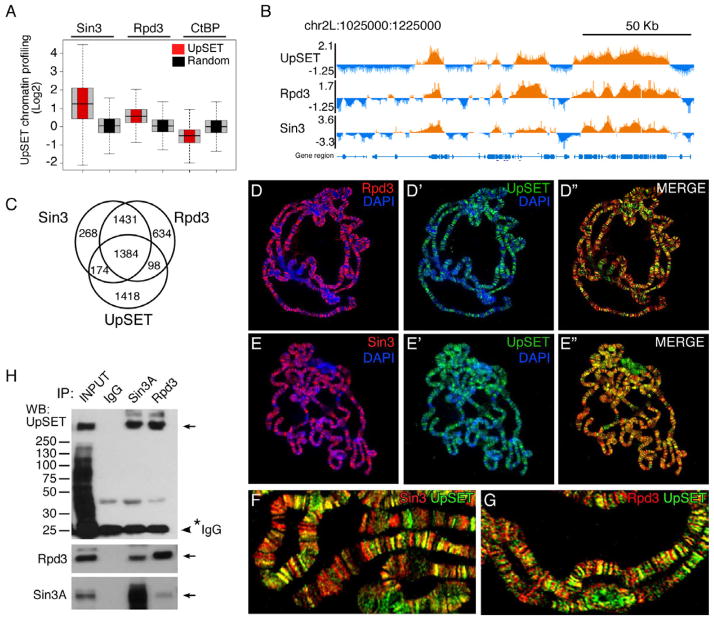
UpSET interacts with Sin3A/Rpd3 histone deacetylase complex
The histone deacetylase Rpd3 and its accessory subunit Sin3 can be recruited to transcriptionally active regions (Filion et al., 2010). Interestingly, the proposed yeast ortholog of UpSET, Set3, has also been shown to interact with an HDAC complex over transcribing regions, however its role once recruited is still unclear (Kim and Buratowski, 2009; Govind et al., 2010; Buratowski and Kim, 2011). Interestingly, the histone deacetylase Rpd3 and its accessory subunit Sin3 have recently been shown to be recruited to transcriptionally active regions (Filion et al., 2010). To determine if UpSET and the HDAC machinery occupy similar genomic regions, we compared the published Sin3 and Rpd3 chromatin profiles to the ~6000 UpSET bound promoter regions (Filion et al., 2010). This analysis revealed a high enrichment for Sin3 and Rpd3 in UpSET bound regions compared to 6000 random sequences (Figures 3A–B). This correlation is specific to these two co-repressors, as repressors present in other chromatin states (i.e., CtBP) are not enriched (Figure 3A). UpSET bound regions overlap with ~50% of Rpd3 and Sin3 bound sites (Figure 3C). Consistent with this, co-staining of polytene chromosomes with UpSET and Sin3 or Rpd3 antibodies shows a high degree of overlap (Figures 3D–G). This overlap is not generalized, as some regions are bound by UpSET, but not by the other two proteins and vice versa, suggesting that UpSET likely participates in additional protein complexes.
Figure 3. UpSET interacts with an HDAC complex.
(A) UpSET-bound regions are enriched for the Histone deacetylase Rpd3 and its associated cofactor Sin3. Box plot analysis of Rpd3 and Sin3 DamID signal overlapping with UpSET bound regions.
(B) Browser view of chromosome 2L highlighting the overlapping DamID signal for UpSET, Rpd3 and Sin3.
(C) Venn diagram showing the overlap among regions bound by UpSET, Rpd3, and Sin3. UpSET overlaps with only half of the loci co-bound by Rpd3 and Sin3.
(D–G) UpSET co-localizes with Sin3 and Rpd3 on chromosomes. Polytene chromosome sets stained with a mixture of antibodies to NH3- UpSET (D’-D”, E’-E”, F–G; green) and Rpd3 (D, D”, G; red) or Sin3 (E, E”, F; red). DNA was counterstained with DAPI (blue). (F–G) Higher magnification views showing co-localization of UpSET with Rpd3 (G) or Sin3 (F).
(H) UpSET interacts with Rpd3 and Sin3. Co-immunoprecipitation of UpSET with Rpd3 and Sin3 from Kc nuclear extracts. Arrows indicate the specified protein. 20% input is shown.
To determine if UpSET interacts with the Sin3 and Rpd3 deacetylase machinery directly in vivo, Kc cell nuclear extracts were incubated with antibodies against Sin3 and Rpd3, and then blotted with UpSET antibodies. As shown in Figure 3H, UpSET physically associates with both Sin3a and Rpd3.
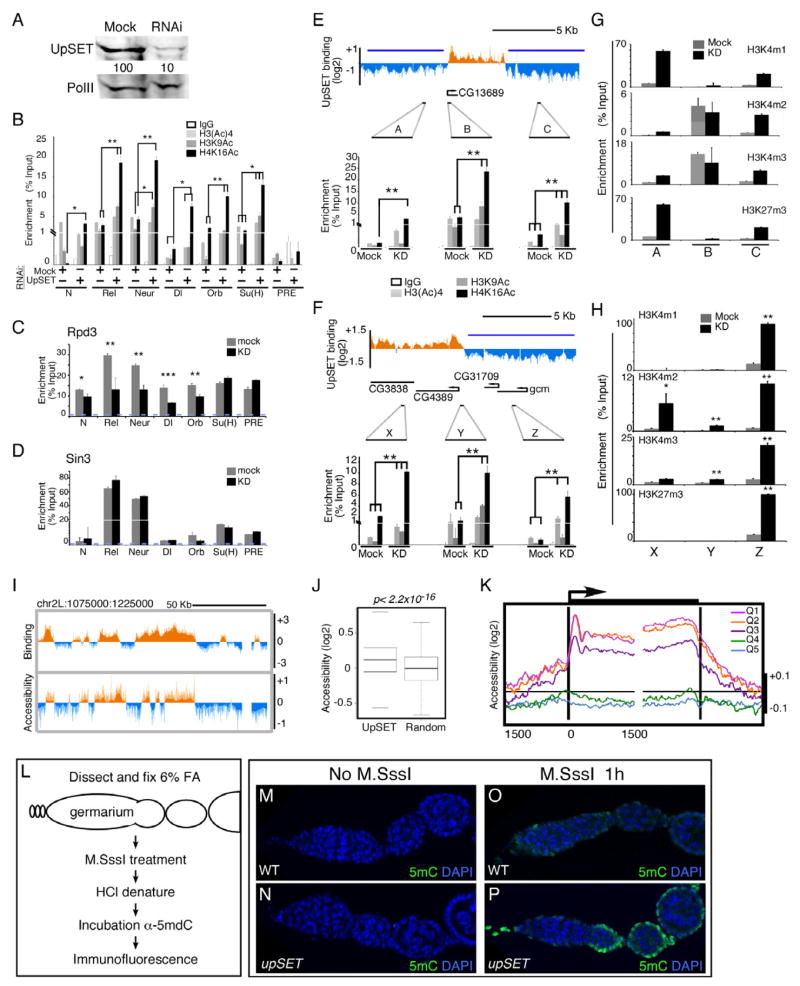
Lack of UpSET increases histone acetylation around promoter regions and causes a re- distribution of active marks
A recent genome-wide analysis revealed that the HDAC machinery binds at both active and inactive genes (Wang et al., 2009), consistent with results of DamID-based chromatin profiling of Sin3 and Rpd3 in insect cells (Filion et al., 2010). The recruitment of HDACs to active genes would be expected to control histone acetylation at promoters and transcribed regions. Therefore, we hypothesized that the lack of UpSET should increase acetylation levels at UpSET-regulated genes. We knocked down UpSET in Kc cells (Figure 4A) and performed chromatin immunoprecipitation (ChIP) with antibodies recognizing the H3-tetraAc, H3K9Ac and H4K16Ac histone marks (modENCODE Consortium et al., 2010). Primers targeting several promoter regions (+/−250 bp around TSS) of UpSET bound genes were used to evaluate acetylation levels. While UpSET-regulated genes exhibit acetylation marks due to their active transcription state, the absence of UpSET results in increased H3K9Ac and H4K16Ac levels (Figures 4B, S3A–B). This increase is not due to an increase in nucleosome density, as ChIP of histone H4 or H3 remains constant (Figures S3C–D). Our results reveal that upon UpSET depletion, acetylation levels increase at active promoter regions targeted by UpSET.
Figure 4. UpSET modulates active chromatin modifications of transcriptionally active genes.
(A) RNAi-based knock down of UpSET levels. Western blot for UpSET from nuclear extracts of Mock and UpSET-specific RNAi expressing cells same antibodies as in Figure 1J. RNA Pol II was used as loading control.
(B) UpSET knock down increases histone acetylation over UpSET bound promoters (TSS). Native ChIP of H3Ac, H3K9Ac and H4K16Ac from Mock and UpSET knock down cells as indicated. Data are represented as mean +/− standard error of the mean (SEM). N: Notch; Rel; Relish; Neur: neuralized; Dl: Delta; Orb: oo18 RNA-binding protein; Su(H): Suppressor of Hairless, and the PcG-silenced C15 gene, as negative control. (*: p<0.05; **: p<0.01; ***: p<0.001, non-significant p values are not marked).
(C–D) Cross-linked ChIP for Rpd3 and Sin3 from Mock and UpSET knock down cells. Bars represent mean +/− SEM.
(E–F) Histone acetylation increases around UpSET-depleted genes. Native ChIP of H3Ac, H3K9Ac and H4K16Ac from Mock and UpSET knock down cells as indicated. UpSET chromatin profiles show the location of the used primers. Repressive chromatin reported from (Bartkuhn et al., 2009); blue line). Bars represent mean +/− SEM.
(G–H) Native ChIP of H3K4m1, H3K4m2, H3K4m3 and H3K27m3 from Mock and UpSET knock down cells. Primers are same as in Figure 4E–F. Bars represent mean +/− SEM. (*: p<0.05; **: p<0.01; ***; p<0.001).
(I) UpSET depletion increases the accessibility of UpSET bound regions. Chromatin accessibility ratio in KD:mock treated cells from a 150Kb region of chromosome 2L.
(J) UpSET associated regions show increased accessibility. Chromatin accessibility signal distributions over UpSET bound regions or random sites. p<2.2x10−16
(K) UpSET modulates chromatin accessibility of transcriptionally active genes. End analysis of the chromatin accessibility signal in UpSET knock down cells over the 5’ and 3’ ends of genes clustered by expression quintiles from high (Q1) to low (Q5) gene expression as indicated.
(L–P) Chromatin accessibility in situ. Schematic of M.SssI DNA methyltransferase accessibility assay in ovaries. Low CpG methylation as detected by 5mC immunofluorescence in untreated Drosophila germarium from wildtype (M) and upSETe00365 mutants (N). Higher chromatin accessibility in M.SssI-treated upSETe00365 mutant ovaries (P) compared to wildtype (O). See also Figures S3-S5.
As UpSET is required for controlling histone acetylation at promoter regions, UpSET may be acting as a structural bridge to stabilize RPD3 containing complexes at promoter/transcribed regions. To test this hypothesis, we performed a ChIP analysis to investigate the presence of the RPD3-containing complex at UpSET targeted promoters in UpSET knock down cells. We find that Rpd3 and Sin3 are detected at most of the promoters tested, albeit enriched to different amounts (Figures 4C–D). Upon UpSET depletion, RPD3 levels are reduced at several UpSET regulated promoters (5 out of 6), but not in the C15 gene, which is targeted by Rpd3 and Sin3, but not UpSET (Figures 4C, S3A) (Schwartz et al., 2006). SIN3 is also detected in low levels over UpSET-regulated promoters (Figure 4D). Interestingly, and regardless of its interaction with UpSET, SIN3 binding was not dramatically affected in UpSET knock down cells, suggesting that recruitment of RPD3, but not SIN3, to active promoters is UpSET-dependent.
It has been suggested that HDAC recruitment to the promoter region functions to constrain active histone modifications to this region (Buratowski and Kim, 2011). Thus UpSET-dependent histone deacetylation may be responsible for restricting active marks to promoter regions. To test this hypothesis, we selected two genes (CG13689 and CG4389) that are flanked by repressive chromatin (H3K27m3) and reside in regions of low gene density (Bartkuhn et al., 2009). We chose genes with these characteristics to avoid the potentially confounding effect of active neighboring genes. We performed ChIP-qPCR using three sets of primers per gene that target ~2.5 Kb upstream, the promoter region, and ~2.5 Kb downstream of the gene. Upon UpSET depletion, the flanking repressed regions exhibit higher levels of histone acetylation, suggesting that histone acetylation is re-localized to neighboring regions in the absence of UpSET (Figures 4E–F). To investigate whether other histone marks associated with transcription also spread to flanking regions in the absence of UpSET, we performed ChIP of H3K4me1, H3K4me2, H3K4me3 and control H3K27m3 in UpSET knock down cells. In mock-treated cells, H3K4 methylation is reduced in repressive regions containing H3K27m3 (Figures 4G–H). Strikingly, UpSET knock down increases the levels of H3K4m2 and H3K4m3 in the flanking regions (Figures 4G–H). This increase in active marks in the absence of UpSET is not due to a global increase on histone acetylation and H3K4 methylation, as the levels of these modifications are not affected in knock down Kc cells or in upSET mutant ovaries, as determined by Western blot and polytene chromosome staining (Figures S3E–L). Interestingly, H3K27m3 levels are also increased, perhaps as a compensatory response to the presence of active marks (Figures 4G–H). Together, these results reveal that UpSET is required to restrict active chromatin marks to promoter/gene regions.
In mammals, the only currently known target of MLL5 is the Ccna2 gene, whose expression is regulated in C2C12 cells by binding of MLL5 to the cell cycle regulatory element located in the first exon, resembling the positioning of UpSET binding in Drosophila (Figure 2, S4A) (Sebastian et al., 2009). To determine whether the modulation of histone acetylation is a conserved feature of MLL5, we knocked down MLL5 in C2C12 cells. Interestingly, we find that histone acetylation, in particular H4K16Ac, increases at the MLL5 binding sites of Ccna2, as well as on neighboring regions located 1–2 Kb from the binding site (Figure S4B–C). Regions further away were not affected. Thus, our results suggest that the ability to modulate histone acetylation is conserved among these proteins.
UpSET modulates chromatin accessibility of active promoter regions
The UpSET-dependent targeting of HDAC machinery to active genes is required to maintain proper control of active histone marks. As histone acetylation usually correlates with a more open chromatin configuration, we hypothesized that the absence of UpSET must alter chromatin accessibility of active promoter regions. Using MNaseI digestion and nucleosome extractions at low salt concentrations (Figure S5A) (Henikoff et al., 2009), we assessed chromatin accessibility in UpSET knock down and mock RNAi-expressing control cells. As shown in Figures 4I–J, compared to 6000 random sequences, UpSET depletion increases chromatin accessibility of the ~6000 regions usually bound by this factor. To determine whether this increase in accessibility correlates with gene expression levels, we performed end analysis of genes clustered into quintiles by their expression levels. We find that the three main clusters (quintiles 1–3; genes with highest expression) of UpSET-bound genes are more accessible over 1.5 Kb upstream of the TSS (Figure 4K). Interestingly, this accessibility is higher over the first 1.5 Kb downstream of the TSS, as well as the 3’ end of the genes than over promoter regions (Figure 4K). Nucleosomes located just downstream of the TSS are more affected, which correlates with regions preferentially enriched for UpSET (Figure 2E). Consistent with this, genes that are hyper-acetylated in UpSET KD show different degrees of increased chromatin accessibility (Figure S5B). Our results indicate that UpSET is required to control chromatin accessibility around the promoter and transcribed regions.
upSET mutants are sterile suggesting a key role for this protein in oogenesis. To establish whether mutant ovary tissues possess a more open chromatin, we adapted a DNA methylation-based chromatin accessibility protocol for Drosophila tissues using the M.SssI methyltransferase (Figure 4L, S5C–G). The Drosophila genome has minimal cytosine methylation, thereby allowing efficient M.SssI modification of mCpG dinucleotides based on their chromatin accessibility. This methylation can subsequently be identified with a 5mC monoclonal antibody (Bell et al., 2010). 5mC methylation cannot be detected in genomic DNA from untreated wildtype or upSET mutant ovaries (Figures 4M–N, S5C), while extended incubation of genomic DNA with M.SssI reveals all CpG availability for methylation (Figures S5H–I). upSET mutant and wildtype ovaries were treated in situ with the M.SssI methyltransferase in non-saturating conditions. Detection of 5mC in mutant ovaries by immuno-fluorescence using the 5mC antibody shows higher chromatin accessibility in mutant ovaries (2-fold increase; p<0.01) (Figures 4O–P, S5J). Interestingly, this function does not seem to be conserved with its murine ortholog by assessing chromatin accessibility at the only described MLL5 binding site in the Ccna2 gene (Figure S4D–E). We do not know if this is a general property of MLL5 targets or if it is specific to this MLL5 target, as the region surrounding the Ccna2 gene is already highly accessible. Altogether, our results show that UpSET plays a key role in modulating chromatin accessibility, preferentially at actively transcribed genes.
UpSET modulates gene expression
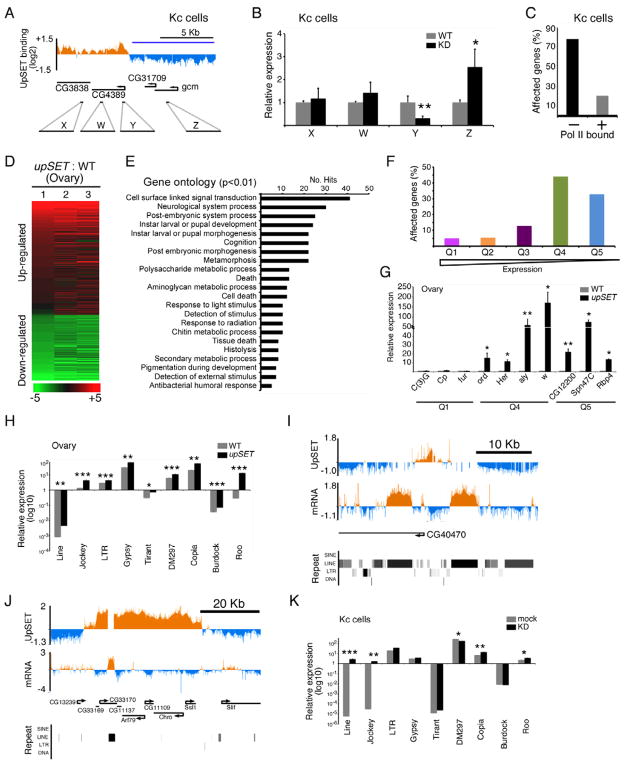
The spreading of histone acetylation and H3K4 methylation in the absence of UpSET suggests that UpSET-containing complexes may control intergenic transcription, as well as modulate gene expression levels of UpSET-regulated genes. To test this hypothesis, we analyzed the expression levels of coding and non-coding regions of the genes CG13689 and CG4389 by RT-qPCR in UpSET KD cells. While some UpSET-target genes display a subtle up-regulation in their levels, intergenic transcription remains the same (Figures 5A–B, S6A–B). Interestingly, silent genes neighboring UpSET target genes become up-regulated (Figure 5A–B). To establish whether this was a global feature due to UpSET, we performed a transcriptome analysis of KD cells and evaluated which genes are affected based on the presence or not of RNA polymerase II (Gilchrist et al., 2010). In Kc cells, UpSET KD affected approximately 438 genes, 80% of which are not transcribed in Kc cells (Figures 5C, S6C). We also analyzed upSET mutant and wildtype ovaries to determine whether upSET mutants have a similar transcriptional phenotype. These analyses revealed that 859 genes are differentially regulated (Figures 5D–E). By gene ontology analysis, the affected genes are preferentially involved in signal transduction pathways, as well as processes generally silent during oogenesis, but expressed during post-embryonic, larval and pupal development (Figure 5E). In addition, we categorized the modENCODE RNA-seq data for ovaries into expression quintiles (Q1,high to Q5,low) and evaluated the percentage of affected genes from each quintile. We found that low expressing or silent genes (Q4-5) are the most affected genes in the absence of UpSET (Figure 5F). These results were confirmed by RT-qPCR, using specific primers for genes from Q4-Q5 (Figure 5G). We also examined whether repeat elements are up-regulated in UpSET mutant ovaries and KD cells. In higher eukaryotes, repetitive elements including transposons and retroelements are the main component of heterochromatin, but they are also interspersed in euchromatic regions, including introns and intergenic regions (Kaminker et al., 2002). We found that transposable elements located near UpSET binding sites are consistently up-regulated (Figure 5H–K). Thus UpSET function is required to modulate gene expression of non-expressing genes and repeat elements.
Figure 5. UpSET modulates off-target gene.
(A–B) UpSET knock down up-regulates silent genes. Gene expression analysis of knock down Kc cells using primers (W–Z) targeting the genomic region from Figure 4F, as well as the coding sequence of the UpSET target gene CG4389. (*: p <0.05; **: p<0.01; Not significant p values are not marked).
(C) Non-expressed genes are more affected upon UpSET knock down. Genes differentially expressed in UpSET knock down cells were clustered by their association with RNA polymerase II.
(D–E) Transcriptome analysis of upSETe00365 mutant ovaries. Heat maps show the summary of three biological replicates of independent upSETe00365 mutant ovaries compared to wildtype (D). Up-regulated (red) and down-regulated (green) genes are shown. Gene ontology analysis of the differentially expressed genes (E). 22 categories were enriched in the dataset (p <0.001).
(F) RNA-seq from wildtype ovaries (modENCODE) was clustered in gene expression quintiles from high (Q1) to low (Q5) and upSETe00365-affected gene percentage for each quintile is shown.
(G) Low/silent genes are up-regulated in upSETe00365 mutants. RT-qPCR of low expressing genes (Q4-5) was compared to highly expressed genes (Q1). (*: p <0.05; **: p<0.01; Not significant p values are not marked).
(H–K) Loss of UpSET increases expression of transposons. RT-qPCR of transposons from 9 representative repetitive element families found near UpSET binding sites in upSETe00365 mutant ovaries (H) or UpSET knock down Kc cells (K). cDNA tiling analysis of transposon expression in upSET e00365 ovaries with DamID chromatin profile for a single peak (I) or region (J) of UpSET binding (see also Figure S2A). The middle track shows mRNA expression in ovaries. Repeat element locations are shown in lower panel. Bars represent mean +/− SEM. (*: p <0.05; **: p<0.01; Not significant p values are not marked)
See also Figure S6.
upSET mutants enhance Pc homeotic transformation phenotypes
Our results show that UpSET modulates gene expression by controlling histone acetylation and chromatin accessibility. As silent genes are up-regulated upon UpSET depletion, we hypothesized that lack of UpSET could enhance phenotypes associated with repressive complexes. Polycomb group (PcG) proteins are required for silencing homeotic and many other genes, and are key players of transcriptional memory (Ringrose and Paro, 2004). As the loss of PcG genes causes homeotic transformation due the ectopic expression of homeotic genes in tissues where their expression is repressed, we performed a genetic analysis to determine whether UpSET increases PcG-associated mutant phenotypes. Pc1 and Pc3 alleles show homeotic transformations of the second and third legs into the first leg in 33–40% of male Pc flies (Table 1). Strikingly, when these Pc mutants are also heterozygous for upSETe00365, the frequency of this transformation increased to 75–89% (Table 1). We confirmed this striking genetic interaction using a second upSET allele, upSETMB08950, a hypomorphic allele resulting from the insertion of a Minos transposon after amino acid T2599 (Figures S7A–C’). upSETMB08950 also enhances the homeotic transformation associated with the Pc alleles to ~75% (Table 1). These genetic interactions are not due to a direct protein-protein interaction as polytene chromosomes co-stained with Pc and UpSET antibodies show no overlap (Figure S7D), UpSET is not enriched at regions bound by PcG proteins (Figure 2D, S7E–F), and does not affect PcG-associated H3K27me3 mark (Figures S7G–H) (Schwartz et al., 2006; Nègre et al., 2010). These results genetically confirm that up-regulated transcription in upSET mutants enhances PcG associated phenotypes.
UpSET controls cell specification by modulating Notch signaling
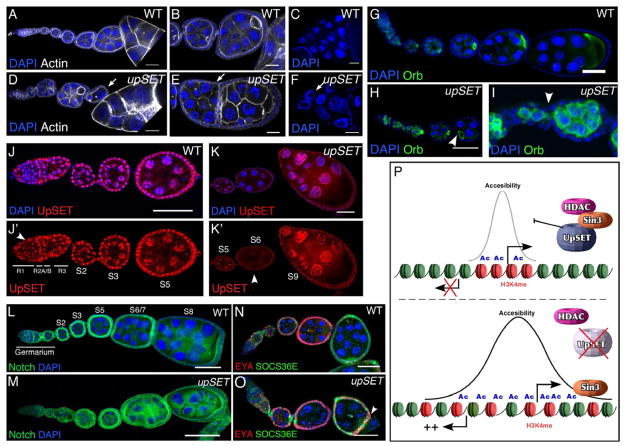
upSETe00365 mutant egg chambers exhibit several distinct phenotypes: anterior-posterior axis asymmetry (13%; n=164), reduced numbers of nurse cells (25%; n=164) and egg chamber fusions (16%; n=164) (Figures 6A–B, 6D–E). When egg chambers mature beyond stage 9 (S9) (15%; n=215), 80% of these display abnormally large nucleoli (Figures 6C, 6F). Staining with Orb, a marker for oocyte specification, confirmed that while UpSET mutants properly specify the oocyte (Figures 6G–I), egg chambers are not pinched off properly, resulting in a variable number of nurse cells (Figure 6I). UpSET is expressed in somatic and germ cell lineages and exhibits a dynamic temporal expression pattern in the germarium and during egg chamber development (Figure 6J–K’). Together, these phenotypes suggest that UpSET is required to establish proper signaling pathways within egg chambers.
Figure 6. UpSET modulates developmentally regulated gene silencing.
(A–F) upSET mutants show incomplete or fused egg chambers (arrows) in comparison to wildtype. Wildtype (A–C) and upSETe00365 mutant (D–F) ovarioles stained with DAPI (blue; to visualize DNA) and phalloidin (white). Confocal projections of 5 serial 1μm optical sections are shown.
(G–I) UpSET is required for the establishment of anterior-posterior axis asymmetry and proper egg chamber formation. Wildtype (G) or upSETe00365 mutant (H–I) ovarioles stained with the oocyte-specific marker Orb showing alterations on the oocyte position and improper pinching off of egg chambers (arrowhead) from the germarium. Confocal projections of 8 serial 1μm optical sections are shown.
(J–K’) UpSET protein levels are differentially regulated during oogenesis. Wildtype ovarioles were stained with a mix of antibodies against UpSET NH3- and COOH- termini and co-stained with DAPI (blue). Confocal projections of 5 serial 1μm optical sections are shown. Arrowhead indicates regions with differential UpSET levels.
(L–M) Wildtype (L) and upSETe00365 mutant (M) ovaries stained with antibodies against the Notch intracellular domain (green) and DAPI (blue).
(N–O) Co-staining of SOCS36 (green) and Eya (red) in wildtype (N) and upSETe00365 mutant (O) ovarioles. DAPI (blue) was used to visualize DNA. Overlapping expression is indicated with an arrowhead in upSET mutant ovary.
(P) UpSET restricts histone acetylation and chromatin accessibility around promoter/genic regions via interaction and stabilization of Rpd3/Sin3-containing HDAC machinery. Lack of UpSET increases chromatin accessibility that correlates with a higher histone acetylation level due to the loss of Rpd3 from transcriptionally active genes. Changes in the chromatin landscape increase the probability of activating transposon expression and off-target genes.
Posterior is oriented to the right in all images. Scale bars represent 50μm.
See also Figure S7.
Drosophila oogenesis is regulated by a fine interplay of signaling pathways that control asymmetric organization and cell specification. Our results show that UpSET functions are required to restrict activation of silent genes; thus we predicted that components of developmental pathways would be affected. The Notch pathway plays a key role in the specification of cells that control the asymmetry of the Drosophila ovary (Bastock and St Johnston, 2008, Assa-Kunik et al., 2007). Notch staining in upSET mutant ovarioles shows proper activation in the germarium, but Notch protein levels remain high in the maturing egg chambers compared to wildtype, indicating a role for UpSET in maintaining developmentally controlled Notch expression (Figure 6L–M). Consistent with this, we find that Notch target genes are likewise up-regulated (Figure 6N;S7I–J). upSET mutant flies exhibit higher levels of SOCS36 in polar cells compared to the rest of the follicle cells in early oogenesis (Figure 6O), and S5 egg chambers show high and heterogeneous accumulation of SOCS36 in follicle cells regardless of the presence of EYA. upSET S8 mutant egg chambers exhibit high levels of SOC36E and, unexpectedly, border cells retain higher levels of this protein compared to wildtype (Figure S7I–J). Up-regulation of these genes was confirmed by western blot (Figure S7K).
Notch signaling is also involved in a complicated interplay with other developmental signaling pathways, including the JAK/STAT pathway (Assa-Kunik et al., 2007). Consistent with up-regulation of the Notch pathway in upSET mutants, UpSET also antagonizes JAK/STAT signaling (Figure S7L–O). Together, our results suggest that uncontrolled Notch signaling, caused by the lack of UpSET, impairs polar cell establishment early in oogenesis and confirms that UpSET plays a key role in regulating gene expression in vivo.
DISCUSSION
upSET encodes an inactive SET domain most closely related to the mammalian protein MLL5. UpSET interacts with HDAC complexes to restrict active histone marks to promoter and genic regions. upSET mutant flies are female sterile due to uncontrolled up-regulation of developmentally important signaling pathways such as that mediated by Notch in oogenesis. Thus, UpSET defines a metazoan SET-containing epigenetic regulator: it functions to recruit the HDAC machinery to transcriptionally active regions to maintain proper chromatin opening and regulation of gene expression (Figure 6P).
UpSET modulates chromatin opening
In silenced loci, HDACs are recruited to chromatin via transcriptional repressors/co-repressors. However, recent genome-wide analyses of HDACs and associated proteins uncovered a complex binding pattern that also includes their recruitment to transcriptionally active loci in metazoans (Filion et al., 2010; Wang et al., 2009). In yeast, SET3 and SET4 recruit HDACs to the 5’ regions of active genes, resulting in the recruitment of Rpd3, which suppresses spurious transcription within the transcribing gene (Kim and Buratowski, 2009; Govind et al., 2010; Carrozza et al., 2005). Like SET3, UpSET is recruited to coding regions of transcriptionally active genes and interacts with HDAC machinery, and its depletion increases histone acetylation levels over transcribed regions. However, UpSET is also required to restrict active marks to promoters and transcribed regions via its direct interaction with an Rpd3/Sin3 containing complex, as well as to regulate chromatin accessibility. Our results also suggest that although UpSET binding peaks at the 5’ end of active genes, UpSET also targets the 3’ ends of active genes, albeit to a lower level. UpSET binding correlates with that of H2A.Z(H2Av)-containing nucleosomes, which are enriched mainly over coding sequences in metazoan and generally more accessible (Mavrich et al., 2008). In yeast, H2A.Z is enriched mainly at the 5’ regions of active genes, correlating with the preferential recruitment of SET3 to the same region (Raisner et al., 2005). Therefore, it is possible that UpSET-like machinery is required to modulate chromatin accessibility of H2A.Z enriched regions.
In the absence of UpSET, promoter-associated active marks, including H3K9Ac, H4K16 and H3K4m3, spread away from the gene into neighboring flanking regions, which may increase the probability of spurious transcription from cryptic TSSs or repetitive elements in the flanking regions (Carrozza et al., 2005). We have shown previously that mammalian MLL2, which contains an active SET domain, can spread along the chromatin fiber until reaching an open promoter where it tri-methylates H3K4 and activates gene expression (Demers et al., 2007). We speculate that further spreading of this active mark is restricted by the recruitment of UpSET and its associated HDAC.
An unusual SET domain containing protein
The UpSET SET domain is a well conserved domain (Emerling et al., 2002), and appears to be the Drosophila ortholog of yeast SET3 and SET4 and mammalian MLL5 (Eissenberg and Shilatifard, 2010; Buratowski and Kim, 2011). None of these SET domains encode H3K4 methyltransferase activity, other than a small MLL5 isoform whose histone methyltransferase activity may be activated by T440 glycosylation (Fujiki et al., 2009). While UpSET contains this residue, it does not exhibit methyltransferase activity in vitro, and H3K4 methylation is unaffected in upSET mutant flies and knock down cells. One possibility is that histones are not the main UpSET substrate, as the functions of different proteins including RNA pol II and PC2 are methylation-dependent (Sims et al., 2011; Yang et al., 2011). However, this seems unlikely as UpSET’s SET domain lacks several key amino acids required for methyltransferase activity. A second possibility is that the SET domain is used to identify H3K4 methylation as a docking site (Emerling et al., 2002; Madan et al., 2009). However, recent studies of the yeast ortholog SET3 suggest that the PHD domain performs this function (Kim and Buratowski, 2009). A third possibility is that the SET domain is required for protein-protein interactions or protein-RNA interactions. Consistent with this, PcG proteins require stable interaction with specific non-coding RNAs to promote silencing (Zhao et al., 2008).
Biologic Implications of UpSET function
Dysregulation of active chromatin marks could have broad implications for gene regulation and cell biology. We find that lack of UpSET has severe consequences in gametogenesis. Interestingly, deletion of SET3 and MLL5 also affect gametogenesis in yeast and mouse, respectively. mll5 knock out mice are viable, yet show aberrant terminal differentiation during spermatogenesis (Yap et al., 2011). Similarly, in yeast, deletion of SET3 results in normal growth, but deregulation of sporulation genes during meiosis results in reduced ascus formation (Pijnappel et al., 2001). Our results suggest that UpSET is required to silence developmentally regulated and inducible genes. The continuous presence of HDAC machinery near transcriptionally active genes would make gene silencing a more efficient process. In this regard, lack of UpSET impairs shut down of the Notch pathway during oogenesis, resulting in a cascade of non-controlled expression of Notch-target genes that ultimately affects cell fate specification in the ovary.
Repeat elements, including transposons, are often expressed at high levels during gametogenesis (Castañeda et al., 2011). Thus, a further increase in expression of repeat elements resulting from deregulation of chromatin architecture in the absence of UpSET may have severe consequences on genome stability, as well as on gene expression. In mammals, repeat elements are regulated by DNA methylation, as well as by co-repressors, including PcG proteins (Leeb et al., 2010). Based on our results, we hypothesize that deletion of UpSET ortholog proteins, including MLL5 or its sister gene SETD5, would increase histone acetylation and/or active methylation marks. Interestingly, loss of the human chromosome segment 7q22 encoding the MLL5 gene is among the most common recurring cytogenetic aberrations detected in myeloid malignancies with poor prognosis (Emerling et al., 2002; Zhang et al., 2009; Madan et al., 2009; Heuser et al., 2009). We suggest that the alterations in chromatin structure associated with MLL5 deletions would act synergistically with DNA demethylating agents often used to treat leukemias to generate a highly unstable chromatin structure. Consistent with this notion, hematopoietic cells from MLL5 knock out mice exhibit hypersensitivity to DNA demethylating agents (Heuser et al., 2009).
Taken together, our results suggest that UpSET and its orthologs represent a class of epigenetic regulators that restrict the location of active chromatin marks to control promoter architecture, and may provide a new therapeutic target in cancer. In addition, UpSET contributes to the stability of the genome by preventing the spread of active marks to flanking regions and the subsequent activation of cryptic promoters and repetitive elements.
EXPERIMENTAL PROCEDURES
Fly stocks
All fly stocks are maintained and crossed on yeast-cornmeal-molasses-malt extract medium at 25°C. The upSET alleles used in this study are: w; PBac{WH}CG9007e00365 (Exelixis Collection at Harvard), and w1118; Mi{ET1}CG9007MB08950 (Bloomington Stock Center). See Supplemental Experimental Procedures for additional alleles used in this study.
UpSET antibody production and characterization
Polyclonal mouse antiserum against the N-end and C-end of UpSET was generated by immunizing BALB/c BYJ Rb(8.12) 5BNR/J mice (Jackson Labs) with a GST- UpSET N-end (1-319aa) or UpSET C-end (2740-3155aa) fusion protein. See Supplementary Experimental Procedures for antibody characterization.
Chromatin accessibility assay
1x108 Kc cells were harvested and nuclei were prepared as described (Henikoff et al., 2009). Nuclei were treated with MNase I (1.6 U/ml, Sigma) for 10min at 37°C and the reaction stopped by adding EDTA to a final 20mM concentration. Nuclei were nutated for 2h in 80mM NaCl at 4°C in order to extract hypersensitive nucleosomes. Nuclei were spun at 1500xg for 10min and the supernatant was recovered and treated with 100 μg/ml proteinase K for 6h. Nucleosomal DNA was purified with a Qiagen PCR purification kit and labeled for hybridization according to Nimblegen’s protocol.
Supplementary Material
Acknowledgments
We thank Jorja Henikoff, Steve Henikoff and Gala Filipova for their interest, advice and comments on the manuscript. We thank Jorja Henikoff, Ryan Basom and Janet Young for bioinformatic help. We also thank Erika Bach, Al Courey, Barry Honda, Steven Hou, Lori Pile, Liz Wayner, the Harvard Exelixis Collection, the Bloomington/Kyoto Stock Centers, and the Developmental Studies Hybridoma Bank for antibodies, flies and other reagents used in this study. This work was supported by NIH grants GM073021 and GM097083 (to SMP) and DK44746 and HL65440 (to MG).
Footnotes
Accession codes. NCBI Gene Expression Omnibus: All datasets are deposited under accession number GSE34720.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J. 2011;30:2817–2828. doi: 10.1038/emboj.2011.194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assa-Kunik E, Torres IL, Schejter ED, St Johnston D, Shilo BZ. Drosophila follicle cells are patterned by multiple levels of Notch signaling and antagonism between the Notch and JAK/STAT pathways. Development. 2007;134:1161–1169. doi: 10.1242/dev.02800. [DOI] [PubMed] [Google Scholar]
- Bach EA, Ekas LA, Ayala-Camargo A, Flaherty MS, Lee H, Perrimon N, Baeg GH. GFP reporters detect the activation of the Drosophila JAK/STAT pathway in vivo. Gene Expr Patterns. 2007;7:323–331. doi: 10.1016/j.modgep.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Bartkuhn M, Straub T, Herold M, Herrmann M, Rathke C, Saumweber H, Gilfillan GD, Becker PB, Renkawitz R. Active promoters and insulators are marked by the centrosomal protein 190. EMBO J. 2009;28:877–888. doi: 10.1038/emboj.2009.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastock R, St Johnston D. Drosophila oogenesis. Curr Biol. 2008;18:R1082–7. doi: 10.1016/j.cub.2008.09.011. [DOI] [PubMed] [Google Scholar]
- Bell O, Schwaiger M, Oakeley EJ, Lienert F, Beisel C, Stadler MB, Schübeler D. Accessibility of the Drosophila genome discriminates PcG repression, H4K16 acetylation and replication timing. Nat Struct Mol Biol. 2010;17:894–900. doi: 10.1038/nsmb.1825. [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, McMahon S, Karlsson EK, Kulbokas EJ, Gingeras TR, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005;120:169–181. doi: 10.1016/j.cell.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Buratowski S, Kim T. The Role of Cotranscriptional Histone Methylations. Cold Spring Harbor symposia on quantitative biology. 2011 doi: 10.1101/sqb.2010.75.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Y, Jin J, Swanson SK, Cole MD, Choi SH, Florens L, Washburn MP, Conaway JW, Conaway RC. Subunit composition and substrate specificity of a MOF-containing histone acetyltransferase distinct from the male-specific lethal (MSL) complex. J Biol Chem. 2010;285:4268–4272. doi: 10.1074/jbc.C109.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- Castañeda J, Genzor P, Bortvin A. piRNAs, transposon silencing, and germline genome integrity. Mutat Res. 2011;714:95–104. doi: 10.1016/j.mrfmmm.2011.05.002. [DOI] [PubMed] [Google Scholar]
- Deal RB, Henikoff JG, Henikoff S. Genome-wide kinetics of nucleosome turnover determined by metabolic labeling of histones. Science. 2010;328:1161–1164. doi: 10.1126/science.1186777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demers C, Chaturvedi CP, Ranish JA, Juban G, Lai P, Morle F, Aebersold R, Dilworth FJ, Groudine M, Brand M. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol Cell. 2007;27:573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SC, Zhang X, Trievel RC, Cheng X. The SET-domain protein superfamily: protein lysine methyltransferases. Genome Biol. 2005;6:227. doi: 10.1186/gb-2005-6-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol. 2010;339:240–249. doi: 10.1016/j.ydbio.2009.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emerling BM, Bonifas J, Kratz CP, Donovan S, Taylor BR, Green ED, Le Beau MM, Shannon KM. MLL5, a homolog of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene. 2002;21:4849–4854. doi: 10.1038/sj.onc.1205615. [DOI] [PubMed] [Google Scholar]
- Filion GJ, van Bemmel JG, Braunschweig U, Talhout W, Kind J, Ward LD, Brugman W, de Castro IJ, Kerkhoven RM, Bussemaker HJ, et al. Systematic protein location mapping reveals five principal chromatin types in Drosophila cells. Cell. 2010;143:212–224. doi: 10.1016/j.cell.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, Kitagawa H, Kato S. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 2009;459:455–459. doi: 10.1038/nature07954. [DOI] [PubMed] [Google Scholar]
- Gilchrist DA, Santos, Dos G, Fargo DC, Xie B, Gao Y, Li L, Adelman K. Pausing of RNA polymerase II disrupts DNA-specified nucleosome organization to enable precise gene regulation. Cell. 2010;143:540–551. doi: 10.1016/j.cell.2010.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govind CK, Qiu H, Ginsburg DS, Ruan C, Hofmeyer K, Hu C, Swaminathan V, Workman JL, Li B, Hinnebusch AG. Phosphorylated Pol II CTD recruits multiple HDACs, including Rpd3C(S), for methylation-dependent deacetylation of ORF nucleosomes. Mol Cell. 2010;39:234–246. doi: 10.1016/j.molcel.2010.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil F, Moorman C, van Steensel B. DamID: mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase. Meth Enzymol. 2006;410:342–359. doi: 10.1016/S0076-6879(06)10016-6. [DOI] [PubMed] [Google Scholar]
- Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, Fitzpatrick KA, Sinclair DAR, Honda BM. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics. 2012;190:91–100. doi: 10.1534/genetics.111.135863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Shilatifard A. Histone modification: cause or cog? Trends Genet. 2011;27:389–396. doi: 10.1016/j.tig.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Henikoff JG, Sakai A, Loeb GB, Ahmad K. Genome-wide profiling of salt fractions maps physical properties of chromatin. Genome Res. 2009;19:460–469. doi: 10.1101/gr.087619.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuser M, Yap DB, Leung M, de Algara TR, Tafech A, McKinney S, Dixon J, Thresher R, Colledge B, Carlton M, et al. Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood. 2009;113:1432–1443. doi: 10.1182/blood-2008-06-162263. [DOI] [PubMed] [Google Scholar]
- Kaminker JS, Bergman CM, Kronmiller B, Carlson J, Svirskas R, Patel S, Frise E, Wheeler DA, Lewis SE, Rubin GM, et al. The transposable elements of the Drosophila melanogaster euchromatin: a genomics perspective. Genome Biol. 2002;3(12):RESEARCH0084. doi: 10.1186/gb-2002-3-12-research0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim T, Buratowski S. Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5' transcribed regions. Cell. 2009;137:259–272. doi: 10.1016/j.cell.2009.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leeb M, Pasini D, Novatchkova M, Jaritz M, Helin K, Wutz A. Polycomb complexes act redundantly to repress genomic repeats and genes. Genes Dev. 2010;24:265–276. doi: 10.1101/gad.544410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madan V, Madan B, Brykczynska U, Zilbermann F, Hogeveen K, Döhner K, Döhner H, Weber O, Blum C, Rodewald HR, et al. Impaired function of primitive hematopoietic cells in mice lacking the Mixed-Lineage-Leukemia homolog MLL5. Blood. 2009;113:1444–1454. doi: 10.1182/blood-2008-02-142638. [DOI] [PubMed] [Google Scholar]
- Mavrich TN, Jiang C, Ioshikhes IP, Li X, Venters BJ, Zanton SJ, Tomsho LP, Qi J, Glaser RL, Schuster SC, et al. Nucleosome organization in the Drosophila genome. Nature. 2008;453:358–362. doi: 10.1038/nature06929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A. COMPASS: a complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci USA. 2001;98:12902–12907. doi: 10.1073/pnas.231473398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- modENCODE Consortium. Roy S, Ernst J, Kharchenko PV, Kheradpour P, Nègre N, Eaton ML, Landolin JM, Bristow CA, Ma L, et al. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science. 2010;330:1787–1797. doi: 10.1126/science.1198374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A. The COMPASS Family of H3K4 Methylases in Drosophila. Mol Cell Biol. 2011;31:4310–4318. doi: 10.1128/MCB.06092-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nègre N, Brown CD, Shah PK, Kheradpour P, Morrison CA, Henikoff JG, Feng X, Ahmad K, Russell S, White RAH, et al. A comprehensive map of insulator elements for the Drosophila genome. PLoS Genet. 2010;6:e1000814. doi: 10.1371/journal.pgen.1000814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A, Abed M, Kenyagin-Karsenti D, Boico O. DamID: a methylation-based chromatin profiling approach. Methods Mol Biol. 2009;567:155–169. doi: 10.1007/978-1-60327-414-2_11. [DOI] [PubMed] [Google Scholar]
- Pijnappel WW, Schaft D, Roguev A, Shevchenko A, Tekotte H, Wilm M, Rigaut G, Séraphin B, Aasland R, Stewart AF. The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 2001;15:2991–3004. doi: 10.1101/gad.207401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raisner RM, Hartley PD, Meneghini MD, Bao MZ, Liu CL, Schreiber SL, Rando OJ, Madhani HD. Histone variant H2A.Z marks the 5' ends of both active and inactive genes in euchromatin. Cell. 2005;123:233–248. doi: 10.1016/j.cell.2005.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–443. doi: 10.1146/annurev.genet.38.072902.091907. [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell. 2007;25:15–30. doi: 10.1016/j.molcel.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Schübeler D, Macalpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Kahn TG, Nix DA, Li X-Y, Bourgon R, Biggin M, Pirrotta V. Genome-wide analysis of Polycomb targets in Drosophila melanogaster. Nat Genet. 2006;38:700–705. doi: 10.1038/ng1817. [DOI] [PubMed] [Google Scholar]
- Sebastian S, Sreenivas P, Sambasivan R, Cheedipudi S, Kandalla P, Pavlath GK, Dhawan J. MLL5, a trithorax homolog, indirectly regulates H3K4 methylation, represses cyclin A2 expression, and promotes myogenic differentiation. Proc Natl Acad Sci USA. 2009;106:4719–4724. doi: 10.1073/pnas.0807136106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Rojas LA, Beck D, Bonasio R, Schüller R, Drury WJ, Eick D, Reinberg D. The C-terminal domain of RNA polymerase II is modified by site-specific methylation. Science. 2011;332:99–103. doi: 10.1126/science.1202663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DAR, Syrzycka M, Macauley MS, Rastgardani T, Komljenovic I, Vocadlo DJ, Brock HW, Honda BM. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc) Proc Natl Acad Sci USA. 2009;106:13427–13432. doi: 10.1073/pnas.0904638106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L, Lin C, Liu W, Zhang J, Ohgi KA, Grinstein JD, Dorrestein PC, Rosenfeld MG. ncRNA- and Pc2 methylation-dependent gene relocation between nuclear structures mediates gene activation programs. Cell. 2011;147:773–788. doi: 10.1016/j.cell.2011.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yap DB, Walker DC, Prentice LM, McKinney S, Turashvili G, Mooslehner-Allen K, de Algara TR, Fee J, de Tassigny XD, Colledge WH, et al. Mll5 is required for normal spermatogenesis. PLoS ONE. 2011;6:e27127. doi: 10.1371/journal.pone.0027127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Wong J, Klinger M, Tran MT, Shannon KM, Killeen N. MLL5 contributes to hematopoietic stem cell fitness and homeostasis. Blood. 2009;113:1455–1463. doi: 10.1182/blood-2008-05-159905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Sun BK, Erwin JA, Song JJ, Lee JT. Polycomb proteins targeted by a short repeat RNA to the mouse X chromosome. Science. 2008;322:750–756. doi: 10.1126/science.1163045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.