SUMMARY
Background
Cytoplasmic dynein motility along microtubules is critical for diverse cellular processes ranging from vesicular transport to nuclear envelope breakdown to mitotic spindle alignment. In yeast, we have proposed a regulated-offloading model to explain how dynein motility drives microtubule sliding along the cortex, powering transport of the nucleus into the mother-bud neck [1, 2]: the dynein regulator She1 limits dynein offloading by gating the recruitment of dynactin to the astral microtubule plus end, a prerequisite for offloading to the cortex. However, whether She1 subsequently affects cortically anchored dynein activity during microtubule sliding is unclear.
Results
Using single-molecule motility assays, we show that She1 strongly inhibits dynein movement along microtubules, acting directly on the motor domain in a manner independent of dynactin. She1 has no effect on the motility of either Kip2, a kinesin that utilizes the same microtubule track as dynein, or human kinesin-1, demonstrating the specificity of She1 for the dynein motor. At single-molecule resolution, She1 binds tightly to and exhibits diffusional behavior along microtubules. Diffusive She1 collides with and pauses motile dynein motors, prolonging their attachment to the microtubule. Furthermore, Aurora B/Ipl1 directly phosphorylates She1 and this modification appears to enhance the diffusive behavior of She1 along microtubules and its potency against dynein. In cells, She1 dampens productive microtubule-cortex interactions specifically in the mother compartment, polarizing spindle movements toward the bud cell.
Conclusions
Our data reveal how inhibitory microtubule-associated proteins selectively regulate motor activity to achieve unidirectional nuclear transport, and demonstrate a direct link between cell cycle machinery and dynein pathway activity.
INTRODUCTION
Dynein and kinesin function to position and organize a multitude of cargos in the cytoplasm as well as to build and breakdown diverse microtubule-based structures critical for cell growth and division. Both types of motors interact with the same microtubule cytoskeleton, often walk along the same microtubule track, and sometimes even occupy the same cargo simultaneously. Hence, selective activation and inhibition of these motors, spatially and/or temporally, are particularly important for the directional transport of specific cargos with respect to the microtubule cytoskeleton or to regions of the cell cortex.
Although much is known for how kinesin is activated or inhibited [3, 4], how dynein is regulated is less well understood. Interestingly, purified dynein seems to be fully active for processive movement [5, 6], suggesting that its activity in vivo might be controlled by inhibitory factors (as proposed in ref. [7]) or via an active targeting mechanism [1, 8]. One such inhibitory factor is the lissencephaly protein, LIS1, which interacts specifically with the dynein motor domain and reduces dynein motility in vitro [1, 9–11]. However, to date, the physiological relevance of LIS1-mediated inhibition of dynein remains controversial. While it appears to be required for high-load organelle transport [12], LIS1 has not been observed moving with cargo-bound dynein. In fact, in the fungi Ustilago maydis and Aspergillus nidulans, LIS1 does not colocalize with moving endosomes and appears to be released from early endosomes prior to their retrograde movement from the hyphal tip [13, 14]. Likewise, in Saccharomyces cerevisiae, LIS1/Pac1 is absent from the cell cortex [2, 15], where dynein generates pulling forces to move the nucleus through the bud neck. In the latter case, LIS1 is required for the targeting of dynein to the astral microtubule plus end, a prerequisite for dynein offloading to the cell cortex [1]. Consistent with this role, overexpression of LIS1 enhances dynein-mediated microtubule sliding along the yeast cell cortex due to an increase in cortical dynein [2]. Thus, these studies in fungi indicate that LIS1 plays a role in the steps preceding the initiation of minus end-directed transport, and is not required for dynein’s subsequent motility along microtubules.
Two recent studies showed that the tau family of MAPs is capable of differentially blocking the path of both dynein and kinesin motors, and thus has the potential to selectively inhibit dynein and kinesin along microtubules [16, 17]. Upon encountering tau, kinesin tends to detach from microtubules, whereas dynein tends to reverse direction [16]. Thus, due to their own distinct mechanical properties and stepping behaviors, dynein and kinesin respond differently to tau obstacles in their path. It remains an open question, however, whether there are MAPs that have an inherent selectivity for one motor over another.
Here we demonstrate that, in budding yeast, She1 is a MAP that exhibits the unique ability to specifically regulate dynein but not kinesin motility. Previous localization studies [1, 2, 18] have suggested that She1 inhibits dynein pathway function by restricting dynein offloading to the cortex: She1 limits the recruitment of dynactin to astral microtubule plus ends, a requirement for dynein offloading to the cortex [15, 19, 20]. Although offloading occurs preferentially at the daughter cell cortex [1], cortical dynein foci are observed predominantly along the mother cell cortex [21]; thus, it is unclear how dynein activity is regulated to achieve directional movement (i.e., bud-ward) and proper orientation of the mitotic spindle. We show that She1 strongly inhibits dynein movement along microtubules and acts directly on the motor domain in a manner that is independent of dynactin. Our results indicate that dynein activity is not only restricted by a targeting mechanism (i.e., offloading to the cortex), but also by a MAP along the astral microtubules that functions in polarizing dynein-mediated spindle movements.
RESULTS
Overexpression of She1 disrupts dynein pathway function and mislocalizes Dyn1 along astral microtubules
SHE1 (Sensitive to High Expression-1) causes growth arrest when overexpressed [22]. While the basis for this phenotype remains unclear, recent studies have implicated a role for She1 in regulating dynein pathway function [2, 18, 23]. We observed that overexpression of She1 (Fig. S1A) resulted in (1) a spindle orientation defect as severe as that exhibited by cells deficient for the dynein heavy chain gene (dyn1Δ; Fig. S1B), and (2) a mislocalization of Dyn1 along the length of astral microtubules (Fig. S1D and Movie S1). The same mislocalization phenotype was observed for Dyn1MOTOR-3YFP, a motor domain fragment of Dyn1 that localizes to the astral microtubule plus end (Fig. S1D) [24]. Furthermore, GAL1p-SHE1 kar9Δ, but not GAL1p-SHE1 dyn1 Δ cells exhibited growth defectsmore severe than GAL1p-SHE1 cells (Fig. S1C), suggesting that overexpression of She1 disrupts the dynein but not the Kar9 pathway. These data suggest that She1 directly affects the microtubule-binding behavior of the dynein motor, and that this effect is mediated via the motor domain in vivo.
She1 binds microtubules directly and exhibits diffusive movement along the microtubule lattice
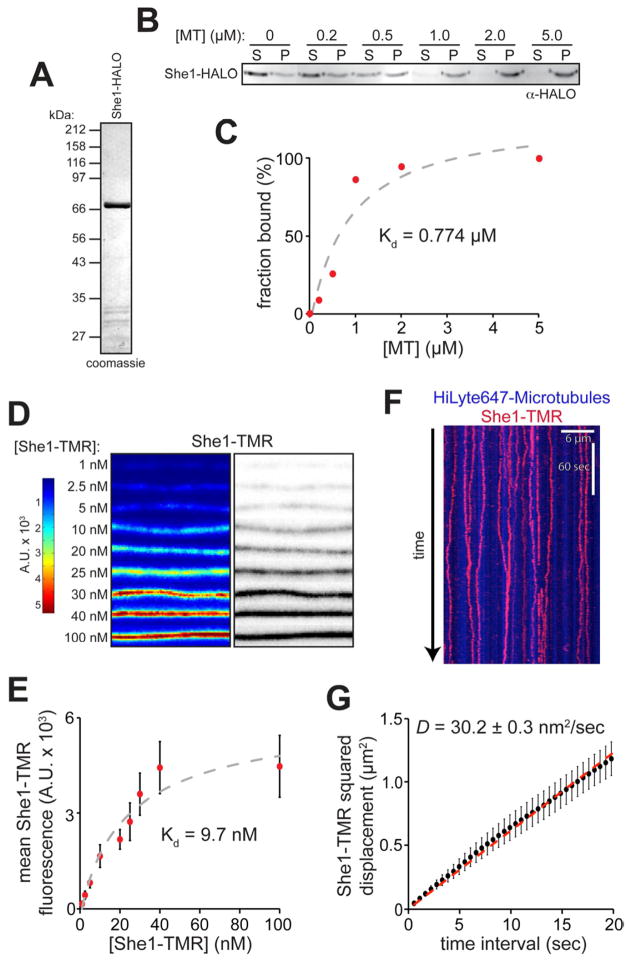
In cells, She1 associates with both spindle and astral microtubules [18, 25]. To characterize its microtubule-binding activity, we expressed and purified a functional (Fig. S1B, D and E) recombinant She1 from bacteria (She1-HALO; Fig. 1A). As shown in Figure 1B, She1-HALO cosediments with microtubules in vitro in a concentration dependent manner, with an apparent dissociation constant (Kd) of 0.77 μM (Fig. 1C). We next analyzed the binding of tetramethylrhodamine (TMR)-labeled She1-HALO (hereafter referred to as She1-TMR) to microtubules using quantitative total internal reflection fluorescence (TIRF) microscopy. We bound varying concentrations of She1-TMR to a fixed concentration of coverslip-immobilized HiLyte-647 microtubules (see Fig. S2A and B), and measured the fluorescence intensity of microtubule-bound She1 as a function of She1-TMR concentration (Fig. 1D and E). The resulting titration data showed saturable binding of She1 to microtubules, yielding an apparent microscopy-based Kd of 9.7 nM. The sensitivity of the microscopy-based assay allowed us to measure binding at microtubule concentrations below the detection limit of the copelleting assay, possibly accounting for the difference in the observed binding constants. Collectively, these data demonstrate that She1 can bind directly to microtubules with high affinity.
Figure 1. She1 binds microtubules in vitro and exhibits lattice diffusion.
(A) She1-HALO purified from E. coli. (B) Cosedimentation of 100 nM She1-HALO with varying concentrations of taxol-stabilized microtubules (S, supernatant; P, pellet). (C) Bound She1-HALO plotted against the concentration of polymerized tubulin. (D) Representative fluorescence images (shown as heat map, left, or inverted image, right) of She1-TMR bound to microtubules. (E) Average fluorescence intensity of microtubule-bound She1-TMR plotted against concentration (≥ 182 μm of microtubule length evaluated per condition). Error bars indicate standard deviation. (F) Kymograph showing single-molecule behavior of She1-TMR along microtubules (see Movie S2). (G) MSD is plotted against time interval for 30 pM She1-TMR on microtubules (n = 24 spots; r2 = 0.99487). Error bars indicate SEM. See Figure S2.
We found that the interaction of She1 with microtubules involves the acidic C-terminal tails of α- and β-tubulin (E-hooks), since She1-TMR did not bind microtubules treated with subtilisin, an enzyme that removes the E-hooks from microtubules (Fig. S2C and D). Consistent with the dependence on E-hooks, binding of She1 to microtubules was sensitive to high ionic strength (Fig. S2E and F).
At low concentrations of She1-TMR, we observed single molecules moving bidirectionally along the microtubule lattice (Movie S2 and Fig. 1F). Mean squared displacement (MSD) analysis indicated that this movement was consistent with a one-dimensional random walk with an average diffusion coefficient, D, of 30.2 nm2/s (Fig. 1G). No detectable biased diffusive component was observed for this motion (drift velocity ≈ 0; see Experimental Procedures). The dwell times for individual She1-TMR molecules on microtubules were remarkably long, often exceeding our image acquisition length (10 min), suggesting a low dissociation rate constant (i.e., slow off-rate). Additionally, photobleaching analysis revealed that She1-TMR particles underwent no more than one-step bleaching events (Fig. S2G; n = 17 spots), indicating that She1 diffuses along microtubules as a monomer.
She1 is a potent inhibitor of dynein motility in vitro
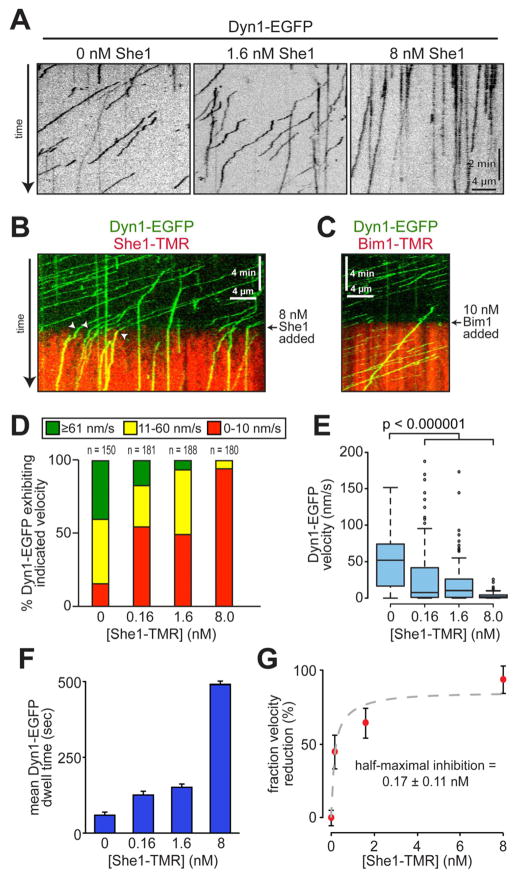
We next assessed the effects of She1 on dynein motility using TIRF microscopy and a GFP-tagged dynein heavy chain purified from yeast (Dyn1-EGFP). Compared to bare microtubules, single molecules of Dyn1-EGFP exhibited reduced velocities and longer dwell times on microtubules pre-bound with She1 (Fig. 2A and D – F; Fig. S3A – C), indicating that She1 not only slows the motor but also prolongs its encounter with the microtubule. Flow-in experiments revealed that moving Dyn1-EGFP motors abruptly reduced their velocity upon the introduction of She1 into the motility chamber (Fig. 2B, arrowheads; Movie S3), demonstrating that the effects of She1 on dynein motility are potent and rapid. In contrast, control experiments revealed that dynein motility was unaffected by Bim1 (Fig. 2C and Movie S4), the microtubule-binding EB1 homolog, confirming that She1 has a unique ability to inhibit dynein motility. The concentration-dependent inhibitory effects of She1 on dynein velocity yielded an apparent dose response curve (Fig. 2G) with half maximal inhibition occurring at 0.17 nM She1. The strength of inhibition suggests that there is a high affinity physical association between She1 and Dyn1 on the microtubule lattice. Quantitative western blotting revealed an approximate cellular She1 concentration of 1.96 nM (Fig. S1F and G). Thus, if She1 is equally distributed between the cytoplasm and nucleus, it is present at sufficient quantities to strongly inhibit dynein activity (see Discussion).
Figure 2. Recombinant She1 reduces the velocity and increases the dwell time of dynein isolated from cells.
(A) Representative kymographs showing dynein motility with varying concentrations of She1-TMR. (B – C) Kymograph of flow-in experiment in which motile Dyn1-EGFP motors were exposed to either She1-TMR (B) or Bim1-TMR (C) at the indicated time (arrow). White arrowheads in C indicate dynein motors that exhibit an abrupt reduction in velocity upon introduction of She1-TMR into the chamber. See Movies S3 and S4. (D) Fraction of dynein molecules moving at the indicated range of velocity as a function of She1-TMR concentration. (E) Box plot of Dyn1-EGFP velocity as a function of She1-TMR concentration. Whiskers define the range, boxes encompass 25th to 75th quartiles, lines depict the medians, and circles depict outlier values [defined as values > (upper quartile + 1.5*interquartile distance), or < (lower quartile – 1.5*interquartile distance)]. (F) Mean dwell time for Dyn1-EGFP molecules along microtubules as a function of She1-TMR concentration. Error bars indicate SEM. (G) Graph depicting the fraction velocity reduction of Dyn1-EGFP molecules as a function of She1-TMR concentration. Error bars indicate standard error of proportion. See Figure S3.
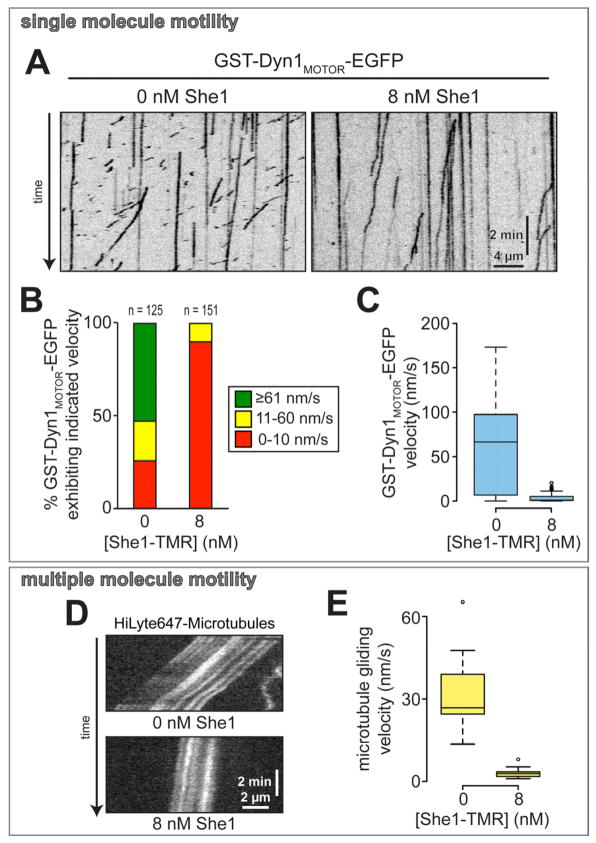
In cells, dynein activity depends on dynactin, a large (~1.2 MDa) multisubunit complex that enhances dynein processivity, supports high load dynein-mediated transport events, and is required for dynein offloading to the cell cortex [1, 26, 27]. We asked whether She1 mediates its inhibitory effects through the dynein tail domain, with which dynactin interacts indirectly, or the catalytic motor domain. To test this, we analyzed the effects of She1 on an artificially dimerized, tail-truncated Dyn1 construct, GST-Dyn1MOTOR-EGFP, which moves at a similar rate as native dynein but with a shorter run length, likely due to the absence of copurifying dynactin [5, 26]. We found that She1 inhibited GST-Dyn1MOTOR-EGFP motility to the same extent as the full-length motor (Fig. 3A – C; Fig. S3D and E). Furthermore, in ensemble motor assays, She1 inhibited GST-Dyn1MOTOR-mediated microtubule gliding (Fig. 3D – E). We conclude that She1 inhibits dynein motility through its C-terminal motor domain, in a manner that is independent of the dynactin complex.
Figure 3. Effect of She1 on dynein motility is independent of the dynein tail domain.
(A) Representative kymographs showing GST-Dyn1MOTOR-EGFP motility in the absence and presence of She1-TMR. (B) Graph depicting fraction of GST-Dyn1MOTOR-EGFP molecules moving at the indicated range of velocity in the absence and presence of She1-TMR. (C) Box plot of GST-Dyn1MOTOR-EGFP velocity. (D) Representative kymographs showing GST-Dyn1MOTOR-EGFP-mediated microtubule gliding in the absence (top) and presence (bottom) of She1. (E) Box plot of microtubule gliding velocity. See Figure S3.
She1 has no effect on kinesin motility
To investigate the specificity of She1 activity, we tested whether She1 affects the motility of the kinesin family of motors. We found that 20 nM She1, a saturating dose for Dyn1-EGFP, had no effect on the velocity, dwell time, or run length of a motor domain construct of human kinesin-1 (Kinesin-11-560-GFP; Fig. S4 and Movie S5).
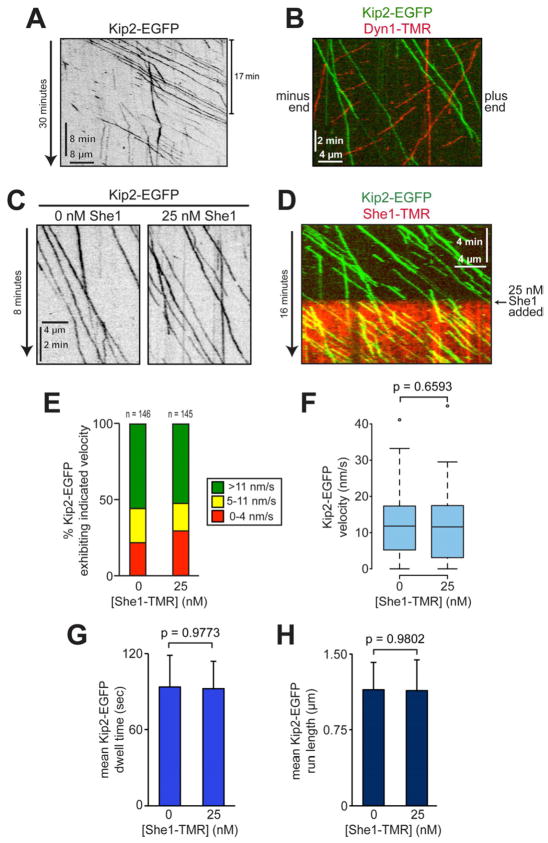
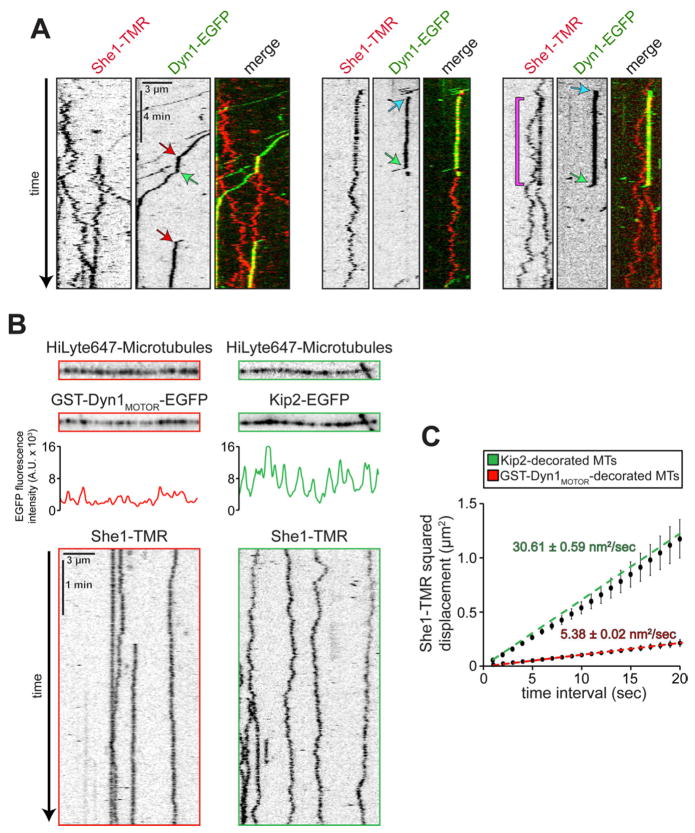
Next, we tested the effects of She1 on Kip2, a yeast kinesin-7 motor. Purified Kip2-EGFP molecules are dimers (Fig. S5A – C) that walk processively and unidirectionally toward the plus end of taxol-stabilized microtubules at 11.7 nm/s (Fig. 4A – B and Movie S6). We found that the velocity, dwell time, and run length of single Kip2-EGFP molecules on She1-decorated microtubules were indistinguishable from those on bare microtubules (Fig. 4C – H). Flow-in experiments revealed that moving Kip2-EGFP molecules proceeded to walk along their tracks upon the introduction of 25 nM She1-TMR into the motility chamber, without any apparent change in their velocity (Fig. 4D and Movie S7). Consistent with these in vitro data, we found that Kip2-3GFP speckles moved along astral microtubules toward the plus end at similar rates in wild-type (SHE1) and She1-overexpressing cells (GAL1p-SHE1; Fig. S5D – F). Taken together, these results indicate that She1 has no effect on the motility of either kinesin-1 or Kip2, demonstrating its specificity for dynein.
Figure 4. Recombinant She1 has no effect on Kip2 isolated from cells.
(A – B) Representative kymographs showing motility of Kip2-EGFP alone (A) or in the presence of Dyn1-TMR (B; see Movie S6). (C) Representative kymographs showing Kip2 motility in the absence and presence of She1-TMR. (D) Kymograph of flow-in experiment in which motile Kip2-EGFP motors were exposed to She1-TMR at the indicated time (see Movie S7). (E) Graph depicting fraction of Kip2 molecules moving at the indicated range of velocity in the absence and presence of She1-TMR. (F) Box plot of Kip2-EGFP velocity. (G – H) Mean dwell time (G) and run length (H) for Kip2-EGFP molecules in the absence and presence of She1-TMR. Error bars indicate SEM. See Figure S5.
Dynein, but not Kip2, interacts with She1 on the microtubule surface
Using TIRF microscopy and low concentrations of Dyn1-EGFP and She1-TMR, we observed collision events during which Dyn1-EGFP and She1-TMR apparently interacted on the microtubule lattice. The Dyn1-EGFP molecule stopped moving upon the collision (Fig. 5A, left panel, red arrows), whereas, coincidently, the She1-TMR molecule ceased its one-dimensional diffusion on the microtubule. We sometimes observed Dyn1-EGFP land on a She1-TMR molecule (Fig. 5A, middle and right panels, blue arrows), restricting the diffusive behavior of She1 on the microtubule (see purple bracket, right panel; compare two adjacent She1-TMR molecules). In most of the observed events, the two molecules eventually dissociate after a variable interval (1.2 – 7.1 min; mean = 3.1 ± 2.0 min; n = 13), with Dyn1-EGFP restarting its movement toward the minus end, and She1-TMR resuming its diffusive behavior. These observations contrast with the reported effects of tau on dynein motility [16], in which single dynein motors reversed direction upon encountering tau on the microtubule track. Furthermore, we found that the observed dynein-She1 interaction was specific, since single molecules of She1-TMR exhibited dramatically reduced diffusion along microtubules pre-decorated with GST-Dyn1MOTOR-EGFP but not Kip2-EGFP (Fig. 5B and C), despite lower levels of decoration by GST-Dyn1MOTOR -EGFP compared to Kip2-EGFP (see respective intensity traces in Fig. 5B).
Figure 5. Dynein, but not Kip2, interacts with She1 along microtubules.
(A) Representative kymographs showing Dyn1-EGFP encountering She1-TMR on the microtubule. Red arrows, Dyn1-She1 collision events; green arrows, Dyn1-She1 release events; blue arrows, events in which Dyn1-EGFP landed on microtubule-bound She1-TMR; purple bracket in the right panel delineates two adjacent She1-TMR molecules, only one of which is associated with a Dyn1-EGFP molecule. (B) Single molecule behavior of She1-TMR on GST-Dyn1MOTOR-EGFP or Kip2-EGFP-decorated microtubules, left and right, respectively. Inverse fluorescence images (top) depict HiLyte647-microtubules with bound GST-Dyn1MOTOR-EGFP or Kip2-EGFP. Plots (middle) indicate fluorescence intensity of GST-Dyn1MOTOR-EGFP or Kip2-EGFP along the respective microtubules, while kymographs (bottom) illustrate the diffusive behavior of She1-TMR molecules along the different motor-decorated microtubules. (C) MSD is plotted against time interval for She1-TMR on either GST-Dyn1MOTOR-EGFP or Kip2-EGFP-decorated microtubules (n ≥ 25 spots; r2 ≥ 0.98514). Error bars indicate SEM.
She1 phosphomutants differentially affect in vivo dynein pathway activity
In cells, She1 facilitates spindle disassembly during mitotic exit, and this function of She1 appears to be activated via phosphorylation by Ipl1/Aurora B [28]. We asked whether dynein-mediated spindle orientation is also regulated by Ipl1-mediated phosphorylation of She1. Using Phos-tag SDS-PAGE [29], we found that recombinant Ipl1 directly phosphorylates She1 (Fig. S6A). The phospho-She1 species were eliminated when ATP was omitted from the reaction (data not shown) or when all five Ipl1 consensus phosphorylation sites were mutated to alanine (She15A) [28]. In contrast to Bim1, which is phosphorylated by Ipl1 only in the presence of Sli15 (INCENP homolog) [30], phosphorylation of She1 by Ipl1 did not require Sli15.
To analyze the consequences of She1 phosphorylation in vivo, we mutated all five putative Ipl1 sites to either alanine (to prevent phosphorylation; she15A) or aspartate (to mimic constitutive phosphorylation; she15D) at the chromosomal SHE1 locus. In growth assays, overexpression of she15A or she15D resulted in synthetic growth defects with kar9Δ that were comparable to overexpression of SHE1 (Fig. S6B – D), indicating that, when overexpressed, both alleles are equally capable of impairing dynein pathway function.
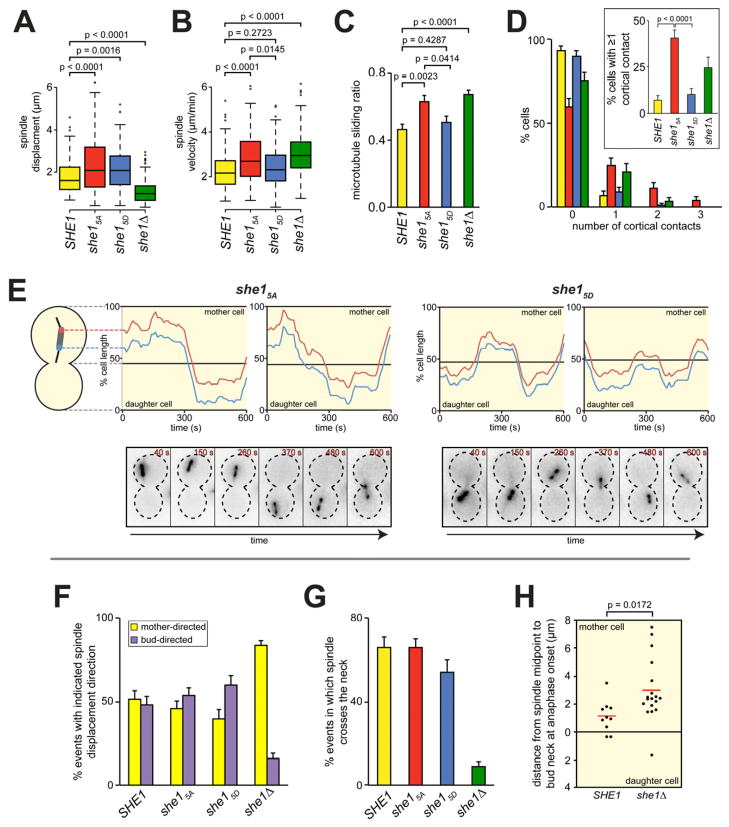
To further examine the effects of the phosphomutants on dynein pathway activity, we monitored the movement of preanaphase spindles in cells arrested with hydroxyurea, which eliminates spindle movements due to spindle elongation. Kar9 was deleted to dramatically enhance dynein-dependent spindle movements [27, 31]. As shown in Figure 6E, the short preanaphase spindles in these cells oscillated back and forth between the mother and daughter cell compartments. We tracked the position of the spindle poles over time and scored for the displacement and velocity of each unidirectional, persistent spindle movement (Fig. 6A and B). Compared to wild-type, she1Δ cells exhibited a significantly higher mean spindle velocity, consistent with an enhancement of cortical dynein activity. Whereas she15D cells exhibited spindle velocities similar to those of wild-type, the she15A mutants exhibited velocities indistinguishable from those of she1Δ (p=0.3127), suggesting that phosphorylation of She1 may function to prevent hyperactivity of cortical dynein in vivo.
Figure 6. she15A cells exhibit enhanced dynein activity in vivo.
(A and B) Box plots for the displacement and velocity of spindle movements observed for kar9Δ strains with the indicated she1 allele (n ≥ 48 cells; n ≥ 79 events). (C) Microtubule sliding ratios for indicated strains (n ≥ 18 cells; n ≥ 84 total events). Error bars indicate SEM. (D) Frequency of observing cells in which an SPB contacted the apex of either the mother or daughter cell cortex the indicated number of times for each indicated strain (n ≥ 57 cells; t = 40 minutes). Inset depicts percentage of cells exhibiting at least one contact. Error bars indicate standard error of proportion. (E) The relative position of each SPB was plotted over time for two representative kar9Δ she15A (left) and kar9Δ she15D (right) cells. Time-lapse images of GFP-Tub1 from the corresponding strains are shown below each plot. (F – G) Graphs depicting (F) the fraction of spindle movements in which the spindle moved either toward the mother (yellow) or daughter (purple) cell cortex, or (G) in which the spindle traversed the bud neck for each indicated strain (n ≥ 48 cells; n ≥ 79 events). Error bars indicate standard error of proportion. (H) Distance from the spindle midpoint to the bud neck at the moment of anaphase onset is plotted for wild-type and she1Δ cells (red bars indicate mean values). See Figures S6 and S7.
Dynein-dependent spindle movements rely on astral microtubule sliding along the cell cortex [32]. Therefore, as another measure of cortical dynein activity, we quantitated the frequency of observing astral microtubule-cortex interactions that resulted in a “productive” sliding event. The microtubule sliding ratio (productive contacts divided by total contacts) was significantly higher in both she1Δ and she15A cells than in either SHE1 or she15D cells (Fig. 6C). Furthermore, in she15A cells, we observed that the spindle translocation events often resulted in a spindle pole contacting the apex of the cell cortex (Fig. 6E). The frequency of finding these “cortical contacts” during a fixed imaging period (Fig. 6D) was higher in she1Δ and she15A cells than in SHE1 and she15D cells. Together, these results are consistent with an enhanced dynein activity in the she15A mutant and suggest that the spindle displacements observed in she15A cells (Fig. 6A) were likely underestimated since many of the spindle movements were prematurely terminated upon the pole contacting the cell tip.
She1 polarizes dynein activity toward the bud by inhibiting dynein in the mother
We observed that she1Δ cells exhibited significantly shorter spindle displacements than SHE1 cells (Fig. 6A). The majority of these abbreviated movements were biased toward the mother cell cortex (Fig. 6F). Furthermore, almost all of the “cortical contact” events observed for she1Δ were ones in which the spindle contacted the mother cell cortex (14 out of 15 events). We noted that, although astral microtubule-cortex interactions were significantly more “productive” in she1Δ cells, the resulting spindle movements rarely led to the spindle traversing the neck between the mother and daughter compartments (Fig. 6G and Fig. S7A). Thus, we posit that loss of She1 causes exaggerated dynein pulling forces at the mother cortex, thereby retarding movements of the spindle towards the bud in she1Δ cells.
Our posit predicts that spindle elongation would occur in the mother cell in a she1Δ mutant due to a delay in spindle movement to the bud neck. To test this, we assayed spindle dynamics in synchronized she1Δ and wild-type cells. We found that the spindle was significantly closer to the bud neck at anaphase onset in wild-type than in she1Δ cells (Fig. 6H and S7C). Additionally, the average spindle length at the time of initial neck penetration was longer in she1Δ than in wild-type cells (Fig. S7B), indicating that spindle penetration in the mutants occurred at a later point after anaphase onset. These data support the hypothesis that enhanced dynein activity in the mother cell prior to anaphase onset inhibits spindle movements toward the bud. Our results demonstrate a physiological role for She1 in polarizing dynein activity toward the daughter cell.
Phosphomimetic mutations of She1 weaken microtubule binding but enhance dynein inhibition
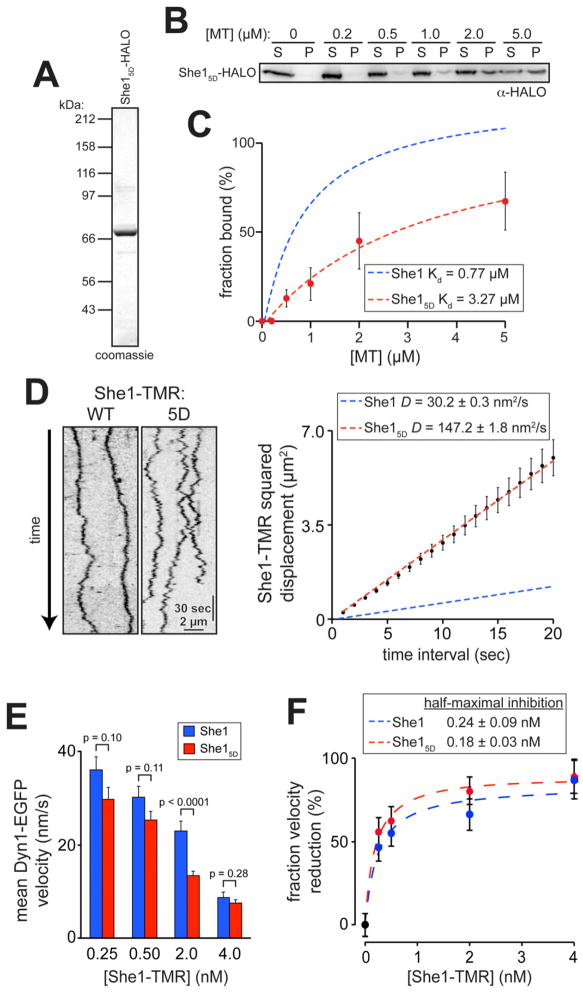
To investigate the consequences of phosphorylation on She1 activity in vitro, we expressed and purified recombinant phosphomimetic She15D-HALO (hereafter referred to as She15D) from bacteria (Fig. 7A). We found that She15D bound microtubules with a ~4-fold lower affinity than wild-type She1 (Fig. 7B and C) and exhibited a ~5-fold faster rate of diffusion along reconstituted microtubules in vitro (Fig. 7D).
Figure 7. She15D exhibits weaker microtubule-binding but enhanced dynein inhibition.
(A) She15D-HALO purified from E. coli. (B) Cosedimentation of 100 nM She15D-HALO with varying concentrations of taxol-stabilized microtubules (S, supernatant; P, pellet). (C) Bound She15D-HALO plotted against the concentration of polymerized tubulin. Error bars indicate standard deviation (n = 2 independent experiments). Binding curve for wild-type She1-HALO from Figure 1C is overlaid for comparison. (D) Kymographs showing single-molecule behavior of wild-type and She15D-TMR are depicted with MSD plots (n = 40 spots; r2 = 0.99751). Error bars indicate SEM. The fit for wild-type She1-HALO from Figure 1G is overlaid for comparison. (E) Mean velocity of Dyn1-EGFP motors in the presence of indicated concentration of either wild-type or She15D-TMR (n ≥ 111 for each data point). Error bars indicate SEM. (F) Graphs depicting the fraction velocity reduction of Dyn1-EGFP molecules as a function of She1 concentration. Error bars indicate standard error of proportion.
At high concentrations, She15D reduced dynein motility to a similar extent as wild-type She1 (i.e., “dephosphorylated” She1; see Experimental Procedures; data not shown), consistent with the in vivo overexpression data of she15D (Fig. S6B – D). However, at or near its physiological concentration (< 4 nM), She15D exhibited stronger inhibition of dynein motility than wild-type She1 (Fig. 7E), despite having an apparent weaker affinity for microtubules. The stronger inhibitory effects of She15D relative to the wild-type protein shifted the dose response curve to the left, reducing the half maximal inhibition by approximately 25% (Fig. 7F). Taken together with our in vivo data, these results suggest that phosphorylation of She1 by Ipl1/Aurora B negatively regulates dynein pathway activity within the mother cell, which may facilitate directional transport of the spindle toward the bud prior to anaphase.
DISCUSSION
In summary, we have identified She1 as a MAP that exhibits the unique property of specifically modulating dynein but not kinesin motility. Consistent with its role in dynein pathway activity [2, 18], She1 binds microtubules in vitro with high affinity, and also localizes along the length of astral and nuclear microtubules in vivo. Thus, She1 is appropriately situated to affect dynein activity in cells. The specificity of She1 activity ensures that, along She1-decorated astral microtubules, Kip2 is unhindered in its transport of Bik1/CLIP-170 towards the plus end, an event that is required for normal microtubule length and dynamics [33]. Thus, She1 activity provides cells with the ability to prevent unwanted (i.e., mother cell-directed) dynein-mediated spindle translocation events without compromising the important functions provided by Kip2. Although such specificity by a MAP has not yet been documented in animal cells, it would be advantageous for cells throughout evolution to be able to specifically modulate microtubule motor activity. Such ability could presumably alter the balance between minus and plus end-directed cargo transport, and could provide a switch to reorganize the internal architecture of a cell, for example, as in the response to external cues during pigment aggregation in Xenopus melanocytes [34].
How can cortically anchored dynein power spindle movements given the relatively high cellular concentration of She1 (~2 nM) and the apparent potency of She1 on dynein motility? Interestingly, Woodruff et al. [18] showed that, in cells, the localization of She1 to astral microtubules varies in a cell cycle-dependent manner: ~50% of G1 cells exhibited She1 along astral microtubules, while, as cells progressed through preanaphase to anaphase, the corresponding frequency dropped to ~10%. Thus, regulated targeting of She1 to astral microtubules may be sufficient to modulate cellular dynein function.
Our data suggest that She1’s ability to control dynein activity may also be regulated by the highly conserved Aurora B kinase, Ipl1, which is best known for its role in phosphorylating kinetochore components to ensure proper kinetochore-microtubule attachments [35, 36]. Phosphorylation reduces the affinity of kinetochore proteins [35] for spindle microtubules, thereby inducing the release of kinetochores that are improperly attached to both spindle poles. Similarly, the She1 mutant mimicking Ipl1 phosphorylation (She15D) exhibits lower affinity for microtubules (Fig. 7C) compared to wild-type She1 (“unphosphorylated”). However, counterintuitive to its apparent weaker microtubule affinity, She15D is a more potent inhibitor of dynein motility than unphosphorylated She1, suggesting that Ipl1 negatively regulates dynein pathway activity. Previous studies have demonstrated that Ipl1 kinase activity is lowest prior to spindle disassembly, but then peaks during spindle disassembly [37]. This suggests that when Ipl1 activity is low, i.e., prior to anaphase, unphosphorylated species of She1 predominate, and dynein activity is correspondingly high; in contrast, when Ipl1 activity is high, i.e., after anaphase, phosphorylated species of She1 accumulate and dynein activity is consequently low. This pattern of Ipl1 activity corresponds well with the time when dynein is needed to move the preanaphase spindle into the bud neck [38, 39], lending support for the proposed role of Ipl1 in the dynein pathway. Thus, conceivably, temporally-regulated phosphorylation of She1 may function to regulate dynein pathway activity as cells progress through mitosis.
Cells deficient for She1 appear morphologically normal and complete mitosis without any gross nuclear segregation errors. However, our analysis of spindle dynamics revealed that loss of She1 results in significantly fewer bud cell-directed spindle movements (Fig. 6F). Moreover, we observed that spindles in she1Δ cells initiate anaphase at a position that is more distal from the bud neck than wild-type cells (Fig. 6H). Thus, we propose that She1 plays a role in turning ‘off’ mother cell dynein activity, such that pulling forces are polarized towards the daughter cell. Since the She1 phosphomimetic mutant, She15D, is a more potent inhibitor of dynein motility, it is possible that localized Ipl1 activity leads to the phosphorylation of mother-localized, but not daughter-localized She1. This would result in the generation of asymmetric forces by dynein situated at the daughter cell cortex, and the consequent pulling of the spindle into the bud neck. Whether an asymmetry of Ipl1 localization or activity exists, however, is unknown.
How does She1 inhibit dynein motility? Our data indicate a mechanism of action for She1 that is unique from tau, the latter of which appears to function as a roadblock for dynein and kinesin motility [16]. In the presence of high levels of tau, dynein and kinesin binding to microtubules was significantly decreased [16]. In contrast, in the presence of high levels of She1, dynein binding to microtubules is enhanced (both in vivo and in vitro; Fig. S1D and 2A), likely as a result of its increased dwell time (Fig. 2F). We propose that She1 inhibits dynein motility as a result of their interaction along microtubules (Fig. 5). Thus, She1 behaves like molecular ‘glue’ that exhibits specificity for dynein. The fact that She1 has no apparent effect on the binding or motility of either Kip2 or kinesin-1 suggests that She1 does not compete with these kinesins for microtubule interaction. Additionally, since dynein and kinesin share a common microtubule-binding site – a groove at the interface between the α and β tubulin subunits [40] – She1 likely binds to a site on the microtubule lattice distinct from these two motors. Future ultrastructural and high-resolution mapping studies of the She1-binding site, and its relation to the dynein-binding site, will be required to further understand the molecular basis for the selective inhibition of dynein motility by She1.
EXPERIMENTAL PROCEDURES
Supplementary Material
HIGHLIGHTS.
She1 binds tightly to and diffuses along microtubules
She1 inhibits dynein but not kinesin motility
She1 polarizes cellular dynein activity to achieve unidirectional spindle movements
Aurora B phosphorylates She1, modulating its potency against dynein
Acknowledgments
We thank Dr. Sue Biggins for the GST-Ipl1 and GST-Sli15 expression plasmids, and Dr. Samara Reck-Peterson for the ZZ-TEV-GFP-3XHA-DYN1-GS-HALO yeast strain. We are grateful to Drs. Patricia Wadsworth, Tom Maresca, and Magdalena Bezanilla for sharing reagents and equipment. We thank Dr. Jennifer Ross for assistance with MSD analysis. This work was supported by an HHMI-sponsored Summer Research Internship, an American Heart Association Undergraduate Summer Fellowship, and a UMass Commonwealth College Undergraduate Research Assistantship to K. K., and an NIH grant (R01GM076094) to W.-L.L.
Footnotes
Supplemental Information found online with this article includes Supplemental Experimental Procedures, seven supplemental figures, and seven movies.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Markus SM, Lee WL. Regulated offloading of cytoplasmic dynein from microtubule plus ends to the cortex. Dev Cell. 2011;20:639–651. doi: 10.1016/j.devcel.2011.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Markus SM, Plevock KM, St Germain BJ, Punch JJ, Meaden CW, Lee WL. Quantitative analysis of Pac1/LIS1-mediated dynein targeting: Implications for regulation of dynein activity in budding yeast. Cytoskeleton (Hoboken) 2011;68:157–174. doi: 10.1002/cm.20502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hackney DD, Baek N, Snyder AC. Half-site inhibition of dimeric kinesin head domains by monomeric tail domains. Biochemistry. 2009;48:3448–3456. doi: 10.1021/bi8022575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blasius TL, Cai D, Jih GT, Toret CP, Verhey KJ. Two binding partners cooperate to activate the molecular motor Kinesin-1. J Cell Biol. 2007;176:11–17. doi: 10.1083/jcb.200605099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Reck-Peterson SL, Yildiz A, Carter AP, Gennerich A, Zhang N, Vale RD. Single-molecule analysis of dynein processivity and stepping behavior. Cell. 2006;126:335–348. doi: 10.1016/j.cell.2006.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ross JL, Wallace K, Shuman H, Goldman YE, Holzbaur EL. Processive bidirectional motion of dynein-dynactin complexes in vitro. Nat Cell Biol. 2006;8:562–570. doi: 10.1038/ncb1421. [DOI] [PubMed] [Google Scholar]
- 7.Kardon JR, Vale RD. Regulators of the cytoplasmic dynein motor. Nat Rev Mol Cell Biol. 2009;10:854–865. doi: 10.1038/nrm2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Whyte J, Bader JR, Tauhata SB, Raycroft M, Hornick J, Pfister KK, Lane WS, Chan GK, Hinchcliffe EH, Vaughan PS, et al. Phosphorylation regulates targeting of cytoplasmic dynein to kinetochores during mitosis. J Cell Biol. 2008;183:819–834. doi: 10.1083/jcb.200804114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McKenney RJ, Vershinin M, Kunwar A, Vallee RB, Gross SP. LIS1 and NudE induce a persistent dynein force-producing state. Cell. 2010;141:304–314. doi: 10.1016/j.cell.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yamada M, Toba S, Yoshida Y, Haratani K, Mori D, Yano Y, Mimori-Kiyosue Y, Nakamura T, Itoh K, Fushiki S, et al. LIS1 and NDEL1 coordinate the plus-end-directed transport of cytoplasmic dynein. EMBO J. 2008;27:2471–2483. doi: 10.1038/emboj.2008.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Roberts AJ, Leschziner AE, Reck-Peterson SL. Lis1 Acts as a “Clutch” between the ATPase and Microtubule-Binding Domains of the Dynein Motor. Cell. 2012;150:975–986. doi: 10.1016/j.cell.2012.07.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yi JY, Ori-McKenney KM, McKenney RJ, Vershinin M, Gross SP, Vallee RB. High-resolution imaging reveals indirect coordination of opposite motors and a role for LIS1 in high-load axonal transport. J Cell Biol. 2011;195:193–201. doi: 10.1083/jcb.201104076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lenz JH, Schuchardt I, Straube A, Steinberg G. A dynein loading zone for retrograde endosome motility at microtubule plus-ends. EMBO J. 2006;25:2275–2286. doi: 10.1038/sj.emboj.7601119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Egan MJ, Tan K, Reck-Peterson SL. Lis1 is an initiation factor for dynein-driven organelle transport. J Cell Biol. 2012 doi: 10.1083/jcb.201112101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee WL, Oberle JR, Cooper JA. The role of the lissencephaly protein Pac1 during nuclear migration in budding yeast. J Cell Biol. 2003;160:355–364. doi: 10.1083/jcb.200209022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dixit R, Ross JL, Goldman YE, Holzbaur EL. Differential regulation of dynein and kinesin motor proteins by tau. Science. 2008;319:1086–1089. doi: 10.1126/science.1152993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Samora CP, Mogessie B, Conway L, Ross JL, Straube A, McAinsh AD. MAP4 and CLASP1 operate as a safety mechanism to maintain a stable spindle position in mitosis. Nat Cell Biol. 2011;13:1040–1050. doi: 10.1038/ncb2297. [DOI] [PubMed] [Google Scholar]
- 18.Woodruff JB, Drubin DG, Barnes G. Dynein-driven mitotic spindle positioning restricted to anaphase by She1p inhibition of dynactin recruitment. Mol Biol Cell. 2009;20:3003–3011. doi: 10.1091/mbc.E09-03-0186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moore JK, Li J, Cooper JA. Dynactin function in mitotic spindle positioning. Traffic. 2008;9:510–527. doi: 10.1111/j.1600-0854.2008.00710.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sheeman B, Carvalho P, Sagot I, Geiser J, Kho D, Hoyt MA, Pellman D. Determinants of S. cerevisiae dynein localization and activation: implications for the mechanism of spindle positioning. Curr Biol. 2003;13:364–372. doi: 10.1016/s0960-9822(03)00013-7. [DOI] [PubMed] [Google Scholar]
- 21.Lee WL, Kaiser MA, Cooper JA. The offloading model for dynein function: differential function of motor subunits. J Cell Biol. 2005;168:201–207. doi: 10.1083/jcb.200407036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Espinet C, de la Torre MA, Aldea M, Herrero E. An efficient method to isolate yeast genes causing overexpression-mediated growth arrest. Yeast. 1995;11:25–32. doi: 10.1002/yea.320110104. [DOI] [PubMed] [Google Scholar]
- 23.Bergman ZJ, Xia X, Amaro IA, Huffaker TC. Constitutive dynein activity in she1 mutants reveals differences in microtubule attachment at the yeast spindle pole body. Mol Biol Cell. 2012;23:2319–2326. doi: 10.1091/mbc.E12-03-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markus SM, Punch JJ, Lee WL. Motor- and tail-dependent targeting of dynein to microtubule plus ends and the cell cortex. Curr Biol. 2009;19:196–205. doi: 10.1016/j.cub.2008.12.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wong J, Nakajima Y, Westermann S, Shang C, Kang JS, Goodner C, Houshmand P, Fields S, Chan CS, Drubin D, et al. A protein interaction map of the mitotic spindle. Mol Biol Cell. 2007;18:3800–3809. doi: 10.1091/mbc.E07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kardon JR, Reck-Peterson SL, Vale RD. Regulation of the processivity and intracellular localization of Saccharomyces cerevisiae dynein by dynactin. Proc Natl Acad Sci U S A. 2009;106:5669–5674. doi: 10.1073/pnas.0900976106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moore JK, Sept D, Cooper JA. Neurodegeneration mutations in dynactin impair dynein-dependent nuclear migration. Proc Natl Acad Sci U S A. 2009;106:5147–5152. doi: 10.1073/pnas.0810828106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woodruff JB, Drubin DG, Barnes G. Mitotic spindle disassembly occurs via distinct subprocesses driven by the anaphase-promoting complex, Aurora B kinase, and kinesin-8. J Cell Biol. 2010;191:795–808. doi: 10.1083/jcb.201006028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kinoshita E, Kinoshita-Kikuta E, Takiyama K, Koike T. Phosphate-binding tag, a new tool to visualize phosphorylated proteins. Mol Cell Proteomics. 2006;5:749–757. doi: 10.1074/mcp.T500024-MCP200. [DOI] [PubMed] [Google Scholar]
- 30.Zimniak T, Stengl K, Mechtler K, Westermann S. Phosphoregulation of the budding yeast EB1 homologue Bim1p by Aurora/Ipl1p. J Cell Biol. 2009;186:379–391. doi: 10.1083/jcb.200901036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yeh E, Yang C, Chin E, Maddox P, Salmon ED, Lew DJ, Bloom K. Dynamic positioning of mitotic spindles in yeast: role of microtubule motors and cortical determinants. Mol Biol Cell. 2000;11:3949–3961. doi: 10.1091/mbc.11.11.3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adames NR, Cooper JA. Microtubule interactions with the cell cortex causing nuclear movements in Saccharomyces cerevisiae. J Cell Biol. 2000;149:863–874. doi: 10.1083/jcb.149.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carvalho P, Gupta ML, Jr, Hoyt MA, Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 34.Ikeda K, Zhapparova O, Brodsky I, Semenova I, Tirnauer JS, Zaliapin I, Rodionov V. CK1 activates minus-end-directed transport of membrane organelles along microtubules. Mol Biol Cell. 2011;22:1321–1329. doi: 10.1091/mbc.E10-09-0741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, 3rd, Chan CS, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- 36.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, De Luca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Buvelot S, Tatsutani SY, Vermaak D, Biggins S. The budding yeast Ipl1/Aurora protein kinase regulates mitotic spindle disassembly. J Cell Biol. 2003;160:329–339. doi: 10.1083/jcb.200209018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moore JK, Cooper JA. Coordinating mitosis with cell polarity: Molecular motors at the cell cortex. Semin Cell Dev Biol. 2010;21:283–289. doi: 10.1016/j.semcdb.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moore JK, Stuchell-Brereton MD, Cooper JA. Function of dynein in budding yeast: mitotic spindle positioning in a polarized cell. Cell Motil Cytoskeleton. 2009;66:546–555. doi: 10.1002/cm.20364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Carter AP, Garbarino JE, Wilson-Kubalek EM, Shipley WE, Cho C, Milligan RA, Vale RD, Gibbons IR. Structure and functional role of dynein’s microtubule-binding domain. Science. 2008;322:1691–1695. doi: 10.1126/science.1164424. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.