Abstract
Endohedral metallofullerenol Gd@C82(OH)22 has recently been shown to effectively inhibit tumor growth; however, its potential adverse bioeffects remain to be understood before its wider applications. Here, we present our study on the interaction between Gd@C82(OH)22 and WW domain, a representative protein domain involved in signaling and regulatory pathway, using all-atom explicit solvent molecular dynamics simulations. We find that Gd@C82(OH)22 has an intrinsic binding preference to the binding groove, particularly the key signature residues Y28 and W39. In its binding competition with the native ligand PRM, Gd@C82(OH)22 is shown to easily win the competition over PRM in occupying the active site, implying that Gd@C82(OH)22 can impose a potential inhibitory effect on the WW domain. Further analyses with binding free energy landscapes reveal that Gd@C82(OH)22 can not only directly block the binding site of the WW domain, but also effectively distract the PRM from its native binding pocket.
Carbone-based nanomaterials such as fullerenes and their derivatives have been widely used in de novo designs of nanoelectronics and nanomechanics due to their unique optoelectronic and physicochemical properties1,2,3. Advances in the biomedical science are particularly impressive in recent years, including applications in diagnostics as well as therapeutics for fatal diseases such as pancreatic cancer4,5. Although such success based on the nanotechnology has been referred to as a new medical paradigm, there is a growing concern on their potential adverse biological effects, which deserves an equal attention6,7. For example, CNTs can penetrate the cell membranes and accumulate in the cytoplasm, causing the cell death8. Meanwhile, functionalized CNTs can interact with the protein enzyme α-chymotrypsin and inhibit its enzymatic activity9. More recently, we find that the blood serum protein coated CNTs (protein-corona) are still toxic to various cell lines, though the cytotoxicity is reduced somewhat10. To make the situation more complicated, different experiments often show very different and controversial bioeffects, depending on their action context (i.e., in vitro, in vivo, target organs, interacting molecules, etc.). A fullerenol C60(OH)24 has been shown to be less toxic than its pristine form C60 with in vitro keratinocyte and hepatocyte cell lines11, as it is generally perceived that a fullerene decreases its toxicity with the increase of its surface functionalization. While a following study observed no toxicity with both fullerene and its fullerenol-derivative in rats exposed via intratracheal instillation12. Furthermore, another study showed that the fullerenol (i.e., C60(OH)18) was more toxic than C60 although both elicite membrane damage in isolated rat liver microsomes13. These studies demonstrate that the pharmacokinetics of nanoparticles are more complicated than previously thought, and are often difficult to extrapolate from one evidence, thus necessitating molecular level understanding of the detailed mechanism14.
Recently, endohedral metallofullerenol Gd@C82(OH)22 has been shown to have an antineoplastic effect as it effectively confines the tumor cells within their extracellular matrix (ECM)15,16. The in vitro biochemical assays show that Gd@C82(OH)22 downregulates various pro-angiogenic factors such as matrix metalloproteinases MMP-2/-917, which are used to breakdown the ECM and thus promote the metastasis of cancers18. In the subsequent study with pancreatic cancer, we have explored its metastasis inhibition mechanism using molecular dynamics simulations, where Gd@C82(OH)22 was shown to preferentially bind to MMP-9 near its S1′ ligand specificity loop, thus potentially interfering with the enzymatic activity via allosteric modulation19. Originally, Gd@C82(OH)22 was developed as a contrasting agent for the magnetic resonance imaging (MRI)20 due to its high proton relaxivity21. The encapsulation of toxic lanthanide metals, such as Gd ion, in the functionalized fullerene cages has endowed versatile potentials in the biomeidical field; however, the underling bioeffects of the nanoparticle, particularly its cytotoxicity, remain to be understood before its wider applications.
Here, we present our study on the interaction between Gd@C82(OH)22 and a small protein domain, using all-atom explicit solvent molecular dynamics (MD) simulations. We selected the WW domain as our target protein due to its seminal role and ubiquitous involvement in signalling and regulatory pathway. The WW domain recognizes the proline-rich motifs (PRMs) in signal transduction and is involved in the control of epithelial sodium channels. It also has implications in several human diseases including Alzheimer's disease22,23. We aim to address this nanoparticle interaction of Gd@C82(OH)22 from two different perspectives: one on its intrinsic recognition and binding property to the WW domain; and the other, more importantly, on its binding competition with the native ligand proline-rich motif (PRM), to the WW domain, and thus its potential inhibition effect on the WW domain.
Results
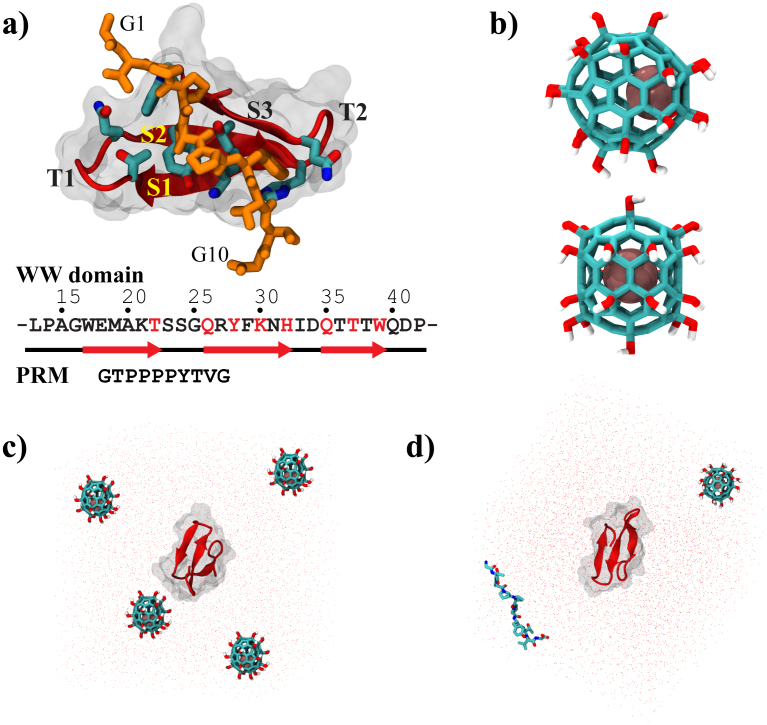
Figure 1 shows the human Yes-associated protein WW domain24 (hYAP65, L30K mutant) along with its co-crystallized PRM (Fig. 1a), and the metallofullerenol Gd@C82(OH)22 (Fig. 1b). In our simulation, the residues of the functional unit between L13 and P42 were chosen for better understanding of the nanoparticle effect on the functional unit of WW domain, following previous studies25,26,27. The intrinsic binding property of Gd@C82(OH)22 was evaluated with the binary system of WW domain and Gd@C82(OH)22, where four Gd@C82(OH)22 molecules are arranged at tetrahedral corners of the simulation box (Fig. 1c). The inhibitory dynamics of Gd@C82(OH)22 was approached by simultaneously simulating both the native ligand (i.e. GTPPPPYTVG) and Gd@C82(OH)22 in their binding competition with the WW domain (i.e., a ternary binding system, Fig. 1d). For each system, we have performed five simulations, each with at least 200-ns long, using NAMD2 program28 with the explicit TIP3P waters29 and CHARMM22 force field30 (see more details in Methods section).
Figure 1.
(a) Complex structures of the WW domain and proline rich motif (PRM); (b) Molecular structure of Gd@C82(OH)22; (c) Initial simulation system for the intrinsic binding dynamics of Gd@C82(OH)22 on the WW domain; and (d) Initial system for the inhibitory dynamics of Gd@C82(OH)22 in competition with PRM.
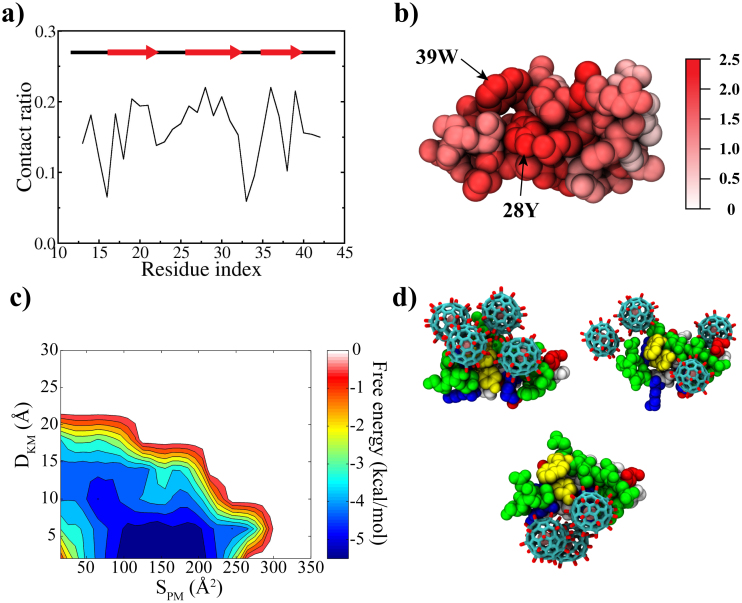
For the binary system, the backbone root-mean-square deviation (RMSD) fluctuates between 1 to 3 Å, indicating that the WW domain is stable overall, despite the frequent adsorption and binding with Gd@C82(OH)22 molecules (Fig. S1.a). The site-specific contact map shows that the three β-sheets contribute highly to its interaction with Gd@C82(OH)22 (Fig. 2a and b). These key contacts that Gd@C82(OH)22 “attacks” are all important contacts for the native ligand PRM, which include: i) the two “signature” residues Y28 and W39 at the hydrophobic groove, into which the first two Pro's of the PPxY motif of PRM are packed (Fig. S2); ii) T37, with which the backbone carbonyl group of P5, the second Pro of the PPxY makes a hydrogen bond; and iii) a shallow hydrophobic groove by K30, H32 and Q35, where the Y7 of the PPxY motif is accommodated (Fig S2)25,31. These interactions are critical for the WW domain to recognize the PRM with high affinity24. Even with this large number of contacts, the beta-strands seem relatively tolerant (or insensitive) to the binding of Gd@C82(OH)22 except for the C-terminal end of the first sheet (Fig. S1.b). This is in contrast to the result of a single-wall carbon nanotube (SWCNT) to the same WW domain, where the SWCNT was plugged into the hydrophobic ligand binding groove, deforming the WW domain and hence disrupting its biological function26. This was attributed to the strong hydrophobic and π−π stacking interactions between the SWCNT and key residues Y28 and W39 of the WW domain.
Figure 2.
(a) Site-specific contact ratio of WW domain, where the contact ratio of a residue was obtained by counting the number of frames of the residue in contact with Gd@C82(OH)22 over all time frames; (b) Residues involved in contact with Gd@C82(OH)22; (c) The binding free energy surface, where DKM is the minimum distance between Gd@C82(OH)22 and the signature residues (Y28 and W39) of the WW domain and SPM is the contacting surface area; and (d) Representative binding modes found in the global minimum. Yellow: key residues, white: hydrophobic, green: non-charged polar, red: negatively charged and blue: positively charged residues.
On the contrary, Gd@C82(OH)22 might not be as hydrophobic as SWCNT due to its surface hydroxylation and surface charges induced by the encapsulated Gd ion, which causes less impact on the structural integrity of the WW domain. However, our above analysis indicates that Gd@C82(OH)22 can have specific interactions with the WW domain, especially the important residues for the PRM binding. For a more quantitative understanding, we constructed the binding free energy landscape along two reaction coordinates (Fig. 2c): the minimum distance (DKM) between Gd@C82(OH)22 and the two “signature” residues (Y28 and W39) of the WW domain, and the contacting surface area (SPM). The potential of mean force (PMF) was calculated by the histogramming analysis26,32, using the equation W(SPM, DKM) = −RT ln p(SPM, DKM), where the probability p(SPM, DKM) was obtained by counting events in a bin (SPM, DKM). The global minimum (−5.44 kcal/mol) is located in close vicinity to the key residues (Y28 and W39) with a contact area of ~150 Å2, indicating that Gd@C82(OH)22 has an energetically favorable interaction with the key binding site residues of the WW domain. Figure 2d displays representative binding modes found near the global minimum. They all show that Gd@C82(OH)22 interacts directly with the two key “signature” residues Y28 and W39 which are supposedly “designed” for the PRM binding.
We rationalize the binding dynamics of Gd@C82(OH)22 with the WW domain as following. The nanoparticle Gd@C82(OH)22 has an amphiphilic nature. It utilizes the long-range electrostatic interaction between the negative surface charges of the mellalofullerenol (i.e. Gd3+@[C82(OH)22]3− in our simulation) and the basic residues (such as K21 or R27) on the WW domain surface33. Although the WW domain (hYAP65 L30K mutant) is neutral overall, it forms a macro-dipole from the second turn (T2) (partially negative) to the first turn (T1) (partially positive) (Fig. S2). Thus, the long-range electrostatic interactions guide the mellalofullerenol to favour the partially positive T1-turn region. Once it approaches to the protein surface, it can have both specific and non-specific interactions with the protein surface, by making hydrogen bonds with the exposed polar/charged residues and/or backbones, and also interacting with hydrophobic residues through the fullerene carbon cage. It should be noted that Gd@C82(OH)22 is still largely hydrophobic despite its surface charge and hydroxylation, as shown in its clustering in aqueous solution in experiments15. This was also confirmed in our simulations where Gd@C82(OH)22 molecules often form an aggregated cluster (which also frequently interact with the hydrophobic residues in the middle of the macro-dipole of the WW domain; see representative binding modes in Fig. 2d). In addition, the aromaticity of the fullerenol cage, and the imperfect shielding of the fullereneol cage for the encapsulated Gd3+ ion also facilitate the contacts with surface exposed aromatic residues (i.e. Y28 and W39) via the π–π34 and/or π-cation35 interactions, which explains why the global minimum binding modes all involve these two signature residues.
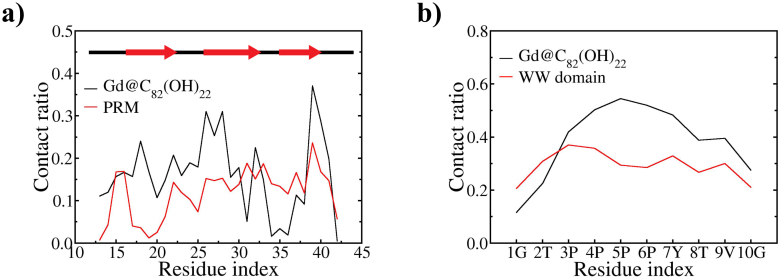
As shown above, the relatively strong and specific binding of Gd@C82(OH)22 on the WW domain surface might interfere with the ligand PRMs' binding. Thus, we turn our attention to the inhibitory dynamics with the ternary system of Gd@C82(OH)22, PRM, and the WW domain. Here, the WW domain structure is again stable, in fact it is even slightly more stable than the previous binary system (Fig. S1.c and d). This seems consistent with the previous experimental results, where the WW domain folding was enhanced by adding the PRM ligand27. Figure 3a and b shows the site-specific contacts on the WW domain by Gd@C82(OH)22 and PRM, as well as on PRM by Gd@C82(OH)22 and the WW domain, respectively. For the WW domain (Fig. 3a), it is clear that Gd@C82(OH)22 generally wins the competition with PRM except for the N-terminal region of the third strand, which has residues (i.e. Q35 & T36) in specific contacts with Y7 of the PPxY motif. Overall, as mentioned above, many of the key residues of the WW domain interact strongly with Gd@C82(OH)22 instead, including the two signature residues Y28 and W39, and other residues such as H32. These key residues are all known to be critical in PRM's binding affinity and specificity to the WW domain27.
Figure 3.
(a) Site-specific contact ratio of WW domain by Gd@C82(OH)22 and PRM and (b) site-specific contact of PRM by Gd@C82(OH)22 and WW domain.
Interestingly, the PRM itself also displays a higher contact preference to Gd@C82(OH)22 rather than to the WW domain. In Fig. 3b, PRM's contacts with the WW domain are rather evenly distributed among all the 10 residues of the PRM, whereas the contacts with Gd@C82(OH)22 are more focused on the middle of PRM from P3 to Y7 (i.e. PPPPY) with significantly higher probability than the WW domain. We observe that the PPxY motif has a specific interaction with Gd@C82(OH)22, which may be partly explained by entropy reduction associated with the likelihood of the rigid intramolecular poly-proline helix formation of the four consecutive prolines and intermolecular hydrophobic interaction between Gd@C82(OH)22 and the (P)PPPY motif of PRM. Similar interaction mechanism but to a lesser degree is also utilized in PPPY motif's binding to the WW domain36.
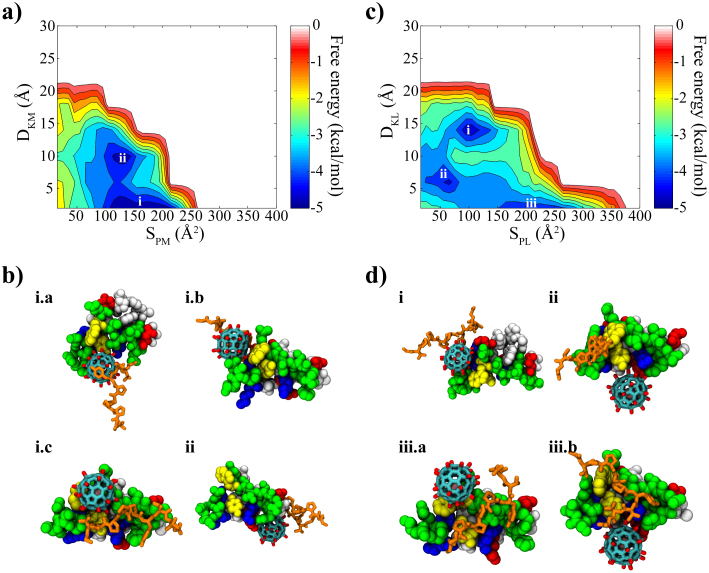
These findings imply that Gd@C82(OH)22 can potentially inhibit the binding between the WW domain and its native PRM ligand. More evidences are found from the binding free energy analyses among the WW domain, Gd@C82(OH)22 and PRM. Although the free energy landscape between the WW domain and Gd@C82(OH)22 in the presence of the PRM has changed somewhat from that of the binary system (Fig. 2c vs. Fig. 4a), the direct binding to the key residues of the WW domain remains predominant. Figure 4a shows that the global minimum (designated by symbol “i”) is −4.78 kcal/mol, and the local minimum (indicated by symbol “ii”) is −4.57 kcal/mol (formed away from the binding site with a slightly smaller contacting surface area and a larger distance from the signature residues, named as “off-site” binding mode). The selected structures (i.a, i.b & i.c of Fig. 4b) in the global minimum reveal that Gd@C82(OH)22 is directly interacting with the key residues of the binding groove and effectively blocking the incoming PRM. Even in the “off-site” binding mode (Fig. 4b.ii), we can see that the PRM is distracted by Gd@C82(OH)22. This again shows that the accurate and specific ligand binding could be hampered by the presence of Gd@C82(OH)22.
Figure 4.
(a) Binding free energy landscape between WW domain and Gd@C82(OH)22 and (b) representative structures found in local energy minima. (c) Binding free energy landscape between WW domain and PRM and (d) representative structures found in local energy minima. D and S of two axes in the free energy diagrams indicate distance and contact area, respectively. The subscripts P, K, L and M represent protein, key residues, PRM ligand and Gd@C82(OH)22, respectively. The local energy minima are rank-ordered starting from the global minimum in the a) and (c).
On the other hand, the off-site bindings become dominant for PRM and the WW domain under the presence of Gd@C82(OH)22 with three different binding modes, indicated as “i”, “ii” and “iii” in Fig. 4c (ranked by their binding affinity, with their representative structures shown in Fig. 4d). The potential of mean force for the global minimum (indicated by “i” in Fig 4c) is −4.67 kcal/mol. The representative structures indicate that this off-site “binding mode i” is formed mainly by the distraction of Gd@C82(OH)22 (Fig. 4d.i). Even though the accurate binding of PRM was found near the region indicated by “iii” in Fig. 4c (“binding mode iii”), its binding free energy of −4.06 kcal/mol is not as strong as the direct binding mode of Gd@C82(OH)22 found at the same region (−4.78 kcal/mol). This indicates that Gd@C82(OH)22 wins the competition over the native ligand PRM in its binding with WW domain by approximately ΔΔG ≈ −0.72 kcal/mol. Even for this accurate “binding mode iii”, the PRM binding is still somewhat interrupted by the competition from Gd@C82(OH)22 (iii.a and iii.b of Fig. 4d), emphasizing the potential inhibitory effect of Gd@C82(OH)22 on the function of the WW domain. We also noticed another binding mode indicated by “ii” with −4.35 kcal/mol in Fig. 4c, an intermediate state between the off-site binding mode “i” and the interrupted on-site binding mode “iii”. In this binding mode, the PRM interacts with the WW domain, while searching for the native binding position, with no direct contact with Gd@C82(OH)22 (ii of Fig. 4d).
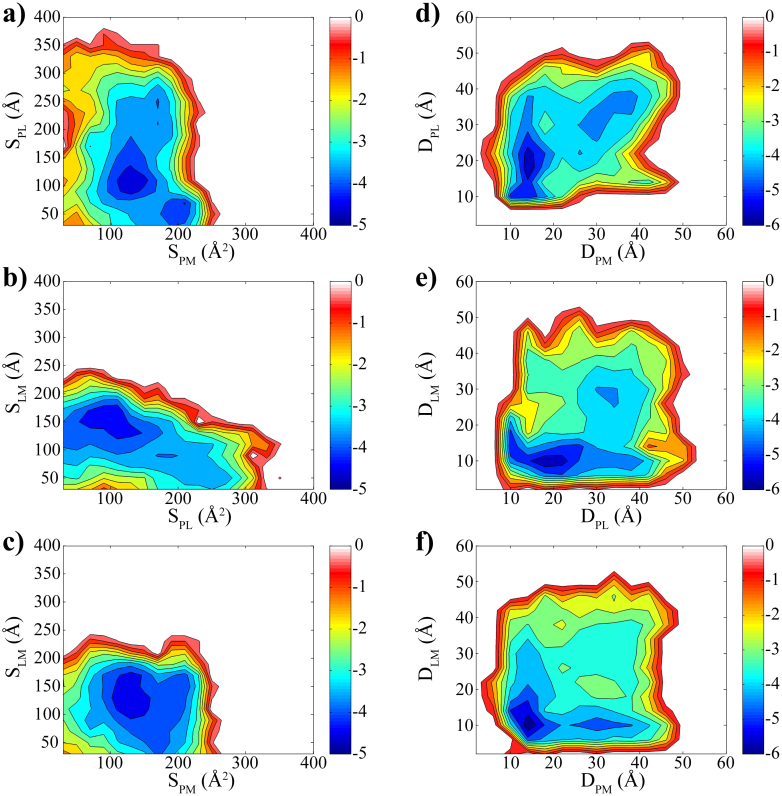
Furthermore, as another perspective about the inhibitory dynamics, we try to compare the relative binding preference among the WW domain, PRM and Gd@C82(OH)22 using another two sets of reaction coordinates (i.e. center-of-mass distances and contact areas). For example, Fig. 5a and d explain the binding preference of the WW domain between Gd@C82(OH)22 and PRM. In the free energy surface made of contact areas (Fig. 5a), Gd@C82(OH)22 is making more contacts with the WW domain (i.e. large contact area of 200 Å2), while PRM is making relatively much less contacts (i.e. about 50 Å2). Complementarily, the free energy landscape of center-of-mass distances shows a binding mode that Gd@C82(OH)22 approaches closer to the WW domain than PRM (Fig. 5d). Therefore, the WW domain can be thought to have a higher preference to Gd@C82(OH)22 than to PRM. In a similar manner, Fig. 5b and e show that PRM favours Gd@C82(OH)22 more than the WW domain, indicating that Gd@C82(OH)22 can not only directly block the binding site of the WW domain but also effectively distract the incoming ligand PRM. Therefore, both the WW domain and PRM prefer to interact with Gd@C82(OH)22 directly rather than to interact between themselves, which is also reflected in Fig. 5c and f, where Gd@C82(OH)22 seems to be well preferred by both PRM and the WW domain. That is, Gd@C82(OH)22 can inhibit the WW domain function not only by directly blocking the ligand binding site, but also by indirectly distracting the binding pathway of the PRM.
Figure 5. Relative binding preference.
(a) and (d) Binding preference for WW domain toward PRM and Gd@C82(OH)22. (b) and (e) Binding preference for PRM toward WW domain and Gd@C82(OH)22. (c) and (f) Binding preference of Gd@C82(OH)22 toward WW domain and PRM. The contact areas were used for panel (a), (b) and (c) and the centre-of-mass distances for panel (d), (e) & (f).
Finally, we also simulated the ternary system with the full sequence of WW domain (all 46 residues) to see how the appended terminal residues alter the binding competition (see Fig. 3S for more details). Despite the more flexible terminal residues, our results show that Gd@C82(OH)22 still wins the competition over the native ligand PRM by approximately ΔΔG ≈ −0.53 kcal/mol, consistent with the results from the functional unit (L13-P42) only, indicating Gd@C82(OH)22 can potentially inhibit the WW domain function.
Discussion
We have investigated the interaction between Gd@C82(OH)22, a recently developed potential nanomedicine for cancers, and the WW domain, a representative seminal protein involved in the signal transduction. With the binary system, our explicit solvent molecular dynamics simulations reveal that Gd@C82(OH)22 binds favorably to the PRM binding groove of the WW domain, interacting particularly with the two signature residues Y28 and W39. Whereas the structural integrity of the WW domain is little affected by the binding, the preferential binding of metallofullerenol possibly causes an inhibitory effect on the WW domain in mediating the protein-protein interaction. More directly with the ternary system involving the native ligand PRM, we found that Gd@C82(OH)22 competitively occupies the putative binding site for the PRM compared to the PRM. The binding free energy landscapes between the interacting pairs suggest that the metallofullerenol Gd@C82(OH)22 can directly block the binding site of the WW domain while it is effectively interrupting the approach of the PRM to the WW domain.
Our current study with the WW domain as an example consistently indicates that metallofullerenol Gd@C82(OH)22 might cause undesired side effect on protein functions. The blockage of signalling proteins might not immediately cause cell or animal deaths37, but it may induce latent malfunctioning of related cellular processes including RNA transcription38 and apoptotic regulation39 of damaged cells in the long run. The accumulative damage might be implicated in various diseases such as neurodegenerative diseases as well as cancers40,41. For a more general conclusion, it requires further studies on other signalling transduction domains involving proline-rich-motifs, such as the SH3 domain, and it needs experimental validations.
Methods
The human Yes-associated protein WW domain (hYAP65, L30K) along with the co-crystallized 10-residue-long proline rich motif (PRM) was chosen as our target system (PDB code: 1JMQ)24. Following previous studies26,42,43, only residues from L13 to P42 of the WW domain (the functional unit) were employed in the majority of our simulations, which have been shown to be critical in the function and stability of the WW domain from the site-directed mutagenesis studies25,27 (the full length of the protein including the flexible N- and C-terminals was also simulated for comparison, see Fig. 3S in Supporting Information). As described in the main text, we prepared two different configurations for i) the intrinsic binding mode and ii) the inhibitory dynamics of Gd@C82(OH)22. For the intrinsic binding mode of Gd@C82(OH)22, the WW domain was prepared with multiple Gd@C82(OH)22 to facilitate the binding site search on the protein. In our simulations, four Gd@C82(OH)22 molecules were placed at the tetrahedral corners of a cubic simulation box at least 30 Å away from the central protein to avoid contact between each Gd@C82(OH)22 and the WW domain as well as other Gd@C82(OH)22 in the periodic boundary condition. In the inhibitory dynamics study, the co-crystallized PRM was included as a competitor of Gd@C82(OH)22 for binding to the WW domain. The PRM and Gd@C82(OH)22 were arranged at the dihedral corner of the simulation box so as not to see each other in the initial configuration. Each molecular system was then immersed in a cubic 60 Å × 60 Å × 60 Å water box with 100 mM NaCl. The CHARMM22 (c32b1 parameter set) force field30 was used for the WW domain and the PRM. The atomic charges for Gd@C82(OH)22 was obtained by the DFT level quantum mechanics calculation with the frozen core approximation on the heavy atoms as described in our recent study with MMP-919. The dispersion terms were employed from the similar atom types in CHARMM22 force field. The long range electrostatic interactions were enumerated with the particle-mesh Ewald method44 and the van der Waals interactions were considered within a cutoff distance of 12 Å. All molecular dynamics simulations were done with NAMD245 software compiled in a massive parallelization environment of the IBM Bluegene machine28. The whole system of ~20,000 atoms with about 6,300 TIP3P29 waters was energy minimized for 20,000 steps followed by a 250 ps equilibration. Then, each production simulation was performed with a 2-fs time step in NPT ensemble of 1 atm and 310 K, generating at least 200 ns long trajectory. For each system, five independent trajectories were obtained by starting from different orientation of the WW domain and different random velocity.
Author Contributions
R.Z. designed and supervised the research. S.G.K. and R.Z. prepared the manuscript. S.G.K. and T.H. carried out the molecular dynamics simulations. S.G.K., R.Z. and T.H. analyzed the molecular dynamics data.
Supplementary Material
Supporting figures
Acknowledgments
We would like to thank Bruce Berne, Yuliang Zhao, Guanghong Zuo, Mike Pitman, and Haiping Fang for helpful discussions. This work was supported by the IBM Blue Gene Program.
References
- Kroto H. W., Heath J. R., Obrien S. C., Curl R. F. & Smalley R. E. C60: Buckminsterfullerene. Nature 318, 162–163 (1985). [Google Scholar]
- Guldi D. M., Rahman A., Sgobba V. & Ehli C. Multifunctional molecular carbon materials - from fullerenes to carbon nanotubes. Chem. Soc. Rev. 35, 471–487 (2006). [DOI] [PubMed] [Google Scholar]
- Mateo-Alonso A., Guldi D. M., Paolucci F. & Prato M. Fullerenes: Multitask components in molecular machinery. Angew. Chem., Int. Ed. 46, 8120–8126 (2007). [DOI] [PubMed] [Google Scholar]
- Anilkumar P. et al. Fullerenes for Applications in Biology and Medicine. Curr. Med. Chem. 18, 2045–2059 (2011). [DOI] [PubMed] [Google Scholar]
- Davis M. E., Chen Z. G. & Shin D. M. Nanoparticle therapeutics: an emerging treatment modality for cancer. Nat. Rev. Drug Discovery 7, 771–782 (2008). [DOI] [PubMed] [Google Scholar]
- Xia T., Li N. & Nel A. E. Potential health impact of nanoparticles. Annu. Rev. Public Health 30, 137–150 (2009). [DOI] [PubMed] [Google Scholar]
- Gilbert N. Nanoparticle safety in doubt. Nature 460, 937 (2009). [DOI] [PubMed] [Google Scholar]
- Porter A. E. et al. Direct imaging of single-walled carbon nanotubes in cells. Nat. Nanotechnol. 2, 713–717 (2007). [DOI] [PubMed] [Google Scholar]
- Zhang B. et al. Functionalized carbon nanotubes specifically bind to alpha-chymotrypsin's catalytic site and regulate its enzymatic function. Nano Lett. 9, 2280–2284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge C. et al. Binding of blood proteins to carbon nanotubes reduces cytotoxicity. Proc. Natl. Acad. Sci. U. S. A. 108, 16968–16973 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayes C. M. et al. The differential cytotoxicity of water-soluble fullerenes. Nano Lett. 4, 1881–1887 (2004). [Google Scholar]
- Sayes C. M., Marchione A. A., Reed K. L. & Warheit D. B. Comparative pulmonary toxicity assessments of C-60 water suspensions in rats: Few differences in fullerene toxicity in vivo in contrast to in vitro profiles. Nano Lett. 7, 2399–2406 (2007). [DOI] [PubMed] [Google Scholar]
- Kamat J. P., Devasagayam T. P., Priyadarsini K. I. & Mohan H. Reactive oxygen species mediated membrane damage induced by fullerene derivatives and its possible biological implications. Toxicology 155, 55–61 (2000). [DOI] [PubMed] [Google Scholar]
- Johnston H. J., Hutchison G. R., Christensen F. M., Aschberger K. & Stone V. The Biological Mechanisms and Physicochemical Characteristics Responsible for Driving Fullerene Toxicity. Toxicol. Sci. 114, 162–182 (2010). [DOI] [PubMed] [Google Scholar]
- Chen C. et al. Multihydroxylated [Gd@C82(OH)22]n nanoparticles: antineoplastic activity of high efficiency and low toxicity. Nano Lett. 5, 2050–2057 (2005). [DOI] [PubMed] [Google Scholar]
- Meng H. et al. Gadolinium metallofullerenol nanoparticles inhibit cancer metastasis through matrix metalloproteinase inhibition: imprisoning instead of poisoning cancer cells. Nanomedicine 8, 136–146 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H. et al. Potent angiogenesis inhibition by the particulate form of fullerene derivatives. ACS Nano 4, 2773–2783 (2010). [DOI] [PubMed] [Google Scholar]
- Carmeliet P. & Jain R. K. Angiogenesis in cancer and other diseases. Nature 407, 249–257 (2000). [DOI] [PubMed] [Google Scholar]
- Kang S.-G. et al. Molecular mechanism of pancreatic tumor metastases inhibition by Gd@C82(OH)22: Implication for de novo design of nanomedicine. Proc. Natl. Acad. Sci. U. S. A. 109, 15431–15436 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikawa M. et al. Paramagnetic water-soluble metallofullerenes having the highest relaxivity for MRI contrast agents. Bioconjugate Chem. 12, 510–514 (2001). [DOI] [PubMed] [Google Scholar]
- Kato H. et al. Lanthanoid endohedral metallofullerenols for MRI contrast agents. J. Am. Chem. Soc. 125, 4391–4397 (2003). [DOI] [PubMed] [Google Scholar]
- Sudol M., Sliwa K. & Russo T. Functions of WW domains in the nucleus. FEBS Lett. 490, 190–195 (2001). [DOI] [PubMed] [Google Scholar]
- Ingham R. J. et al. WW domains provide a platform for the assembly of multiprotein networks. Mol. Cell. Biol. 25, 7092–7106 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pires J. R. et al. Solution structures of the YAP65 WW domain and the variant L30 K in complex with the peptides GTPPPPYTVG, N-(n-octyl)-GPPPY and PLPPY and the application of peptide libraries reveal a minimal binding epitope. J. Mol. Biol. 314, 1147–1156 (2001). [DOI] [PubMed] [Google Scholar]
- Macias M. J. et al. Structure of the WW domain of a kinase-associated protein complexed with a proline-rich peptide. Nature 382, 646–649 (1996). [DOI] [PubMed] [Google Scholar]
- Zuo G., Huang Q., Wei G., Zhou R. & Fang H. Plugging into proteins: poisoning protein function by a hydrophobic nanoparticle. ACS Nano 4, 7508–7514 (2010). [DOI] [PubMed] [Google Scholar]
- Toepert F., Pires J. R., Landgraf C., Oschkinat H. & Schneider-Mergener J. Synthesis of an Array Comprising 837 Variants of the hYAP WW Protein Domain. Angew. Chem., Int. Ed. 40, 897–900 (2001). [DOI] [PubMed] [Google Scholar]
- Kumar S. et al. Scalable molecular dynamics with NAMD on the IBM Blue Gene/L system. IBM J. Res. Dev. 52, 177–188 (2008). [Google Scholar]
- Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W. & Klein M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983). [Google Scholar]
- MacKerell A. D. et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J. Phys. Chem. B 102, 3586–3616 (1998). [DOI] [PubMed] [Google Scholar]
- Macias M. J., Wiesner S. & Sudol M. WW and SH3 domains, two different scaffolds to recognize proline-rich ligands. FEBS Lett. 513, 30–37 (2002). [DOI] [PubMed] [Google Scholar]
- Zhou R., Berne B. J. & Germain R. The free energy landscape for beta hairpin folding in explicit water. Proc. Natl. Acad. Sci. U. S. A. 98, 14931–14936 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade R. C., Gabdoulline R. R., Ludemann S. K. & Lounnas V. Electrostatic steering and ionic tethering in enzyme-ligand binding: insights from simulations. Proc. Natl. Acad. Sci. U. S. A. 95, 5942–5949 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wang Z., Tian X., Xiu P. & Zhou R. Amino acid analogues bind to carbon nanotube via pi-pi interactions: comparison of molecular mechanical and quantum mechanical calculations. J. Chem. Phys. 136, 025103 (2012). [DOI] [PubMed] [Google Scholar]
- Woolf T. B., Grossfield A. & Pearson J. G. Indoles at interfaces: Calculations of electrostatic effects with density functional and molecular dynamics methods. Int. J. Quantum Chem. 75, 197–206 (1999). [Google Scholar]
- Ball L. J., Kuhne R., Schneider-Mergener J. & Oschkinat H. Recognition of proline-rich motifs by protein-protein-interaction domains. Angew. Chem., Int. Ed. 44, 2852–2869 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang W. et al. Biosafety assessment of Gd@C82(OH)22 nanoparticles on Caenorhabditis elegans. Nanoscale 3, 2636–2641 (2011). [DOI] [PubMed] [Google Scholar]
- Yagi R., Chen L. F., Shigesada K., Murakami Y. & Ito Y. A WW domain-containing yes-associated protein (YAP) is a novel transcriptional co-activator. EMBO J. 18, 2551–2562 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy D., Adamovich Y., Reuven N. & Shaul Y. Yap1 phosphorylation by c-Abl is a critical step in selective activation of proapoptotic genes in response to DNA damage. Mol. Cell 29, 350–361 (2008). [DOI] [PubMed] [Google Scholar]
- Sudol M. Structure and function of the WW domain. Prog. Biophys. Mol. Biol. 65, 113–132 (1996). [DOI] [PubMed] [Google Scholar]
- Salah Z., Alian A. & Aqeilan R. I. WW domain-containing proteins: retrospectives and the future. Front. Biosci. 17, 331–348 (2012). [DOI] [PubMed] [Google Scholar]
- Ibragimova G. T. & Wade R. C. Stability of the beta-sheet of the WW domain: A molecular dynamics simulation study. Biophys. J. 77, 2191–2198 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkan S. B., Wu G. A., Chodera J. D. & Dill K. A. Protein folding by zipping and assembly. Proc. Natl. Acad. Sci. U. S. A. 104, 11987–11992 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darden T., York D. & Pedersen L. Particle mesh Ewald: An Nlog(N) method for Ewald sums in large systems. J. Chem. Phys. 98, 10089–10092 (1993). [Google Scholar]
- Phillips J. C. et al. Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting figures