Abstract
RNA aptamers that bind human immunodeficiency virus 1 (HIV-1) reverse transcriptase (RT) also inhibit viral replication, making them attractive as therapeutic candidates and potential tools for dissecting viral pathogenesis. However, it is not well understood how aptamer-expression context and cellular RNA pathways govern aptamer accumulation and net antiviral bioactivity. Using a previously-described expression cassette in which aptamers were flanked by two “minimal core” hammerhead ribozymes, we observed only weak suppression of pseudotyped HIV. To evaluate the importance of the minimal ribozymes, we replaced them with extended, tertiary-stabilized hammerhead ribozymes with enhanced self-cleavage activity, in addition to noncleaving ribozymes with active site mutations. Both the active and inactive versions of the extended hammerhead ribozymes increased inhibition of pseudotyped virus, indicating that processing is not necessary for bioactivity. Clonal stable cell lines expressing aptamers from these modified constructs strongly suppressed infectious virus, and were more effective than minimal ribozymes at high viral multiplicity of infection (MOI). Tertiary stabilization greatly increased aptamer accumulation in viral and subcellular compartments, again regardless of self-cleavage capability. We therefore propose that the increased accumulation is responsible for increased suppression, that the bioactive form of the aptamer is one of the uncleaved or partially cleaved transcripts, and that tertiary stabilization increases transcript stability by reducing exonuclease degradation.
Introduction
Aptamers are structured nucleic acids generated through systematic evolution of ligands by exponential enrichment, or SELEX, to bind molecular targets with high affinity and specificity.1,2,3,4,5 Several aptamers selected to recognize viral proteins have been demonstrated to impair the replication of the corresponding viruses, such as human immunodeficiency virus (HIV),6,7,8,9,10,11 hepatitis B virus,12,13 human cytomegalovirus (CMV),14 hepatitis C virus,13,15,16,17 and influenza virus.18 Aptamers are therefore promising therapeutic candidates for use as adjuvants to conventional drug-based therapies such as highly active antiretroviral therapy.3,4,19,20 However, little is known about how the design of the expression system influences aptamer-mediated antiviral effects, or how it determines the aptamers' access to, and influence on, cellular RNA biology. Aptamer-mediated inhibition is expected to require achieving a suitable accumulation level of aptamer RNA and colocalization of the aptamer with its target, in addition to other biological factors that can vary from one aptamer to the next. Furthermore, aptamer libraries generated by SELEX can be populated with tremendous molecular diversity and exhibit extensive variation in molecular structure, target binding specificity and affinity.1,2,4,5,21 Thus, it would be valuable to screen the individual members of such libraries for their respective net inhibitory potential.
With respect to HIV, aptamers selected to bind the viral reverse transcriptase (RT) have attracted extensive interest. Many of these aptamers strongly inhibit RT function in biochemical assays at low nanomolar concentrations by competing with nucleic acid substrates for access to the protein.22,23,24,25,26 Importantly, a few of these aptamers have also been reported to inhibit viral replication in cell culture when infectious HIV was passaged in aptamer-expressing stable cell lines.6,8,10 Inhibition has been suggested to occur via an aptamer-mediated blockage of viral cDNA synthesis by being copackaged with the RT in nascent virions,10 although the mechanism of such packaging is not known. The generation of clonal, stable cell lines for these inhibition assays and the subsequent serial passage of infectious HIV are time-intensive activities that constitute bottlenecks in screening aptamers and their expression contexts. In contrast, single-cycle assays using pseudotyped virus can provide a rapid, transfection-based approach that enables high throughput screening of many inhibitors. Additionally, pseudotyped virus cannot propagate, increasing safety associated with the assay. To accelerate preclinical development of anti-RT aptamers as potential therapeutics and as reagents for studying viral pathogenesis, it is important to develop streamlined assays for evaluation of candidate aptamers and to explore how components of the aptamer-expression platform influence antiviral bioactivity.
The present study analyzes modifications of an expression cassette that has been utilized in several previous studies of potentially therapeutic RNA,6,10,11,27,28 and in an investigation of an in vitro method to produce hepatitis C virus.29 Two of the aforementioned studies investigated aptamer-mediated inhibition of HIV-1 replication by using the expression cassette driven by a CMV immediate early promoter. In this cassette, the catalytic cores of two identical minimal, self-cleaving hammerhead ribozyme (mcHHRz) are situated on the 5′ and 3′ flanking regions of the aptamer.10,11,28 While the affinity between an aptamer and its target is clearly an important determinant of bioactivity, cis-acting components of the expression cassette can also affect the observed inhibitor potency through their effects on transcript stability, processing and localization. The potential contribution of HHRz cleavage to net viral suppression is not immediately apparent. On the one hand, efficient cleavage could facilitate aptamer folding by liberating it from the transcript and reducing potential interference from flanking sequences. Cleavage would also reduce the size of the RNA-protein particles that need to get packaged into the virus. On the other hand, HHRz cleavage of mRNA induces rapid degradation of the cleaved products.30,31 Ribozyme cleavage could therefore accelerate aptamer degradation and even prevent escape from the nucleus by removing 5′ cap and polyA tail, thereby reducing cytoplasmic availability and limiting their availability for inhibiting the virus. Apart from these considerations, fully cleaved (released) fragments have been observed in cotranscriptional cleavage assays in vitro, but larger fragments corresponding to uncleaved and partially cleaved transcripts were also observed.30,31 Any of these fully cleaved, partially cleaved, or uncleaved aptamer-containing fragments could potentially have been responsible for suppressing viral replication. Furthermore, studies during the past decade have established that tertiary stabilizing elements in extended hammerhead ribozymes (eHHRz) are important determinants of cleavage efficiency in the low concentrations of Mg2+ that are characteristic of intracellular environments (0.1–2.0 mmol/l).31,32,33 The absence of such interactions in the mcHHRz within this vector may result in relatively poor HHRz cleavage in cells, again raising the question of which fragment(s) constitute the bioactive form(s) of the aptamer.
To ascertain the importance of HHRz-mediated cleavage and the effects of tertiary stabilization, we modified the original construct to contain eHHRz in which cleavage ability was very efficient (denoted “active”) or was inactivated through mutation (“inactive”). We found that the eHHRz modifications improved inhibition of viral replication and increased the levels of aptamer RNA accumulation relative to the mcHHRz construct. Importantly, these effects did not require ribozyme cleavage, thereby establishing that partially cleaved and noncleaved transcripts are among the bioactive forms of the aptamer. Furthermore, these modifications yielded an efficient screening system that accurately predicted outcomes in the far more labor-intensive assays involving replication-competent infectious virus in clonal, aptamer-expressing cell lines. These findings will accelerate molecular understanding of aptamer-mediated viral suppression and will help to identify the best aptamers to be developed for downstream therapeutic applications.
Results
The mcHHRz expression cassette inhibits well in multiple-cycle, but not single-cycle inhibition assays
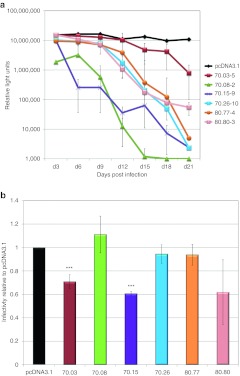
For analysis of aptamer-mediated inhibition in multiple-cycle assays, six full-length aptamers [118–134 nucleotide (nt)] were cloned between two mcHHRz (Supplementary Table S1 and Supplementary Figure S1), and the resulting plasmids were transfected into TZM-bl reporter cells for G418 selection. These cells contain β-galactosidase and luciferase reporter genes under the control of the HIV-1 long-terminal repeats, both of which are induced by Tat expression upon infection by HIV-1. For each plasmid construct, multiple aptamer-expressing cell lines were generated by seeding single G418-resistant cells on 96-well plates and then expanding to confluency. Clonal cell lines were then plated in quadruplicate with the same number of cells in each well and infected with HIV-1NL4-3 at an initial multiplicity of infection (MOI) of 0.6, retaining one well as a noninfected control to measure background luciferase activity. Culture supernatants containing replicating virus were passaged onto fresh cells in series every 3 days for 12–30 days, and luciferase activity was measured at each passage to monitor viral propagation. Background luciferase activity in the absence of virus was subtracted for each sample. Infectivity remained steady throughout the analysis in cell lines expressing an arbitrary control RNA (a 70-nt fragment of the luciferase gene flanked by aptamer constant regions) and in cell lines carrying either the parental vector (pcDNA3.1) or the empty mcHHRz cassette (Supplementary Figure S2). In contrast, infection by HIV-1NL4-3 was inhibited up to 104-fold in the aptamer-expressing cells (Figure 1a). Stable cell lines within a given set of clones expressing the same aptamer demonstrated differential inhibitory ability. As shown in Supplementary Figure S3a, only three of five 70.26-expressing cell lines inhibited HIVNL4-3. AptamerRNA was readily detectable by real-time RT-PCR for each of the threeclonal cell lines that inhibited HIV-1NL4-3, but was within twofold of background for the two cell lines that failed to inhibit (Supplementary Figure S3b). Inhibition was therefore dependent upon the level of accumulated aptamer RNA, which varied across stable cell lines propagated from individual cell colonies within a given set. These effects may be due to differences in integration. Aptamer RNA expression was stable over the time required for these assays, as demonstrated for aptamer 70.05 monitored at day 3, 9, and 18 in five independent expression contexts (Supplementary Figure S4a).34 To rule out the possibility that the observed inhibition was due to loss of cellular receptor or reporter gene, stable aptamer 70.26-expressing cell lines for which infectivity had dropped essentially to background levels (passaged to day 21) were re-infected with fresh HIV-1NL4-3 at an MOI of 0.6. The degree of infection of these day 21 (high-passage) cells was very similar to the initial infection at day 3 (Supplementary Figure S4b). These same cells retained the ability to suppress replication, as evidenced by the observation that viral inhibition during further passages occurred with similar kinetics to those observed for the initial (day 3) infection (Supplementary Figure S4c). Thus, the mcHHRz plasmids expressing anti-RT aptamers clearly support robust and reproducible suppression of HIV-1 replication in the context of clonal, stable cell lines and in multiple-passage assays, and the observed inhibition was dependent upon the expressed aptamer.
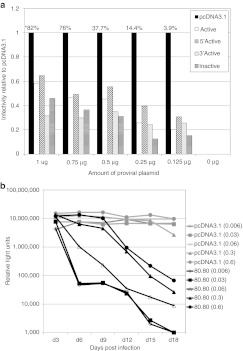
Figure 1.
Aptamer-mediated inhibition of human immunodeficiency virus (HIV) replication in multiple-cycle and single-cycle assays when aptamers are expressed from the mcHHRz cassette. (a) Multiple-cycle assays. HIV-1NL4-3 serial passage using an initial multiplicity of infection (MOI) of 0.6 in clonal, stable TZM-bl cell lines made by transfection and subsequent G418 selection with the mcHHRz aptamer-expression constructs expressing aptamers 70.03, 70.08, 70.15, 70.26, 80.77, and 80.80. The same number of cells for each line were plated in quadruplicate for each passage, using three wells for infection and one well as a noninfected control for background luciferase activity. One-half of the virus-containing supernatant was transferred in series to fresh cells every 3 days, and infectivity was determined using BetaGlo reagent to quantify luciferase activity. Background luciferase activity in the absence of virus was subtracted and values are shown ±SD for three replicates. (b) Single-cycle assays. mcHHRz aptamer-expression constructs were transfected into 293FT cells in triplicate, followed four hours later by media change and cotransfection of pNL4-3-CMV-EGFP and pMD-G. Virus-containing supernatants were harvested 48 hours after the final media change and 100 µl was used to infect fresh 293FT cells at a density of 50%. Infectivity was determined using flow cytometry and is shown as % EGFP-positive cells normalized to the negative control, pcDNA3.1. One-way ANOVA analysis and Student's t-test were used to determine significance compared to pcDNA3.1 controls (P < 0.001). Values are shown as the mean ±SD for three experiments. CMV, cytomegalovirus; EGFP, enhanced green fluorescent protein.
To evaluate the predictive value of the more rapid, high throughput, single-cycle infectivity assays, 293FT cells were transiently transfected with the same mcHHRz aptamer-expressing plasmids as above, followed by cotransfection with plasmids pNL4-3-CMV- enhanced green fluorescent protein (EGFP) (reporter provirus) and pMD-G (VSV-G glycoprotein), so that pseudotyped virus production took place in the presence of aptamer RNA. In the proviral plasmid, the env, vif, vpr, vpu, and nef genes were deleted from the NL4-3 genome, and the nef gene was replaced with a gene for EGFP transcribed from the CMV promoter. Similar levels of transfection and virus production were observed for controls and aptamer-expressing cells, as measured by flow cytometry and p24 ELISA, respectively (data not shown). However, only modest inhibition was observed for two of the six aptamers tested in single-cycle assays, and little or no inhibition was observed for the other four aptamers (Figure 1b), even though all six of these aptamers strongly suppressed replication when expressed from identical platforms in the time-intensive, multiple-cycle assay (Figure 1a). Thus, expressing the aptamers between the mcHHRz did not provide adequate sensitivity to demonstrate aptamer-mediated inhibition over one round of replication, and the single-cycle infectivity assays using the mcHHRz cassettes were unable to predict the performance of these aptamers against infectious virus.
Tertiary-stabilized expression cassettes improve single-cycle antiviral bioactivity
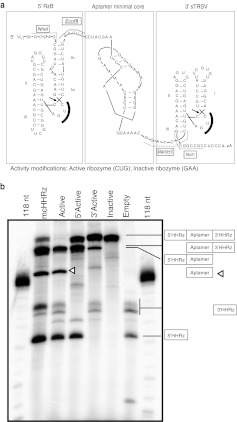
To test the effect of HHRz tertiary stabilization and cleavage on aptamer-mediated inhibition and RNA accumulation, the two identical mcHHRz were replaced with two different eHHRz. Ribozyme RzB,35 an in vitro-selected variant of the HHRz from peach latent mosaic virus, was inserted on the 5′ side of the aptamer, and the HHRz from satellite tobacco ring spot virus (sTRSV) was inserted on the 3′ side. Separately, inactivated forms of each eHHRz were also inserted by mutating the first 3 nt of the highly conserved CUGAUGA element within the catalytic core to GAA (Figure 2a and Supplementary Figure S1). In cotranscriptional cleavage assays, only transcripts that included mcHHRz or active eHHRz on both the 5′ and 3′ ends of the aptamer yielded fully excised aptamer (Figure 2b). Ribozyme RzB cleaved at especially high efficiency, while the mcHHRz and sTRSV ribozymes cleaved less efficiently, even in the high Mg2+ concentrations associated with in vitro transcription. The reactions for all five constructs yielded fragments that included aptamer within uncleaved or partially cleaved transcripts.
Figure 2.
Modified aptamer-expression cassettes with 5′-flanking and 3′-flanking extended hammerhead ribozymes. (a) Oligonucleotides corresponding to the intended sequences were annealed, PCR amplified, digested with NheI and NotI restriction enzymes, and ligated in place of the mcHHRz aptamer-expression cassette. Individual aptamers were cloned into modified cassettes using the EcoRI and BamHI restriction sites. Modified extended hammerhead ribozymes (eHHRz) cassettes include inactive or highly active RzB (5′) and inactive or highly active satellite tobacco ring spot virus (sTRSV) (3′). Active eHHRz contain a CUG within the catalytic core, indicated by a solid black arc. The CUG was mutated to GAA in the inactive variants of the eHHRz. (b) In vitro cotranscriptional cleavage activity of the mcHHRz and eHHRz aptamer-expression cassettes expressing aptamer 70.05. Expected cleavage products and locations are illustrated by the boxes on the right, with lines directed toward the expected products. Product sizes are given in Supplementary Table S3. The aptamer-containing fragment is denoted by an open triangle.
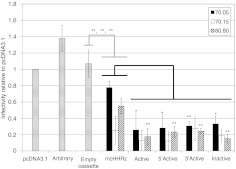
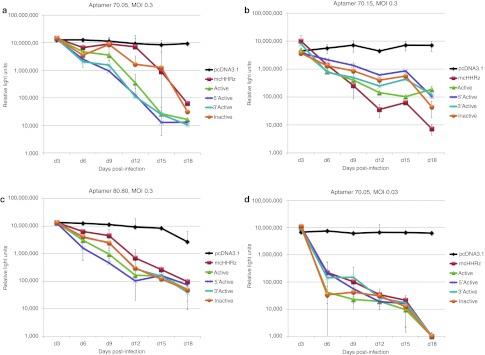
Aptamers 70.05, 70.15, and 80.80 were cloned into each of the four eHHRz cassettes, and these plasmids were used in single-cycle infectivity assays to assess their antiviral bioactivity. Infectivity of virus generated in the presence of empty HHRz cassettes was indistinguishable from the pcDNA3.1 control, indicating that these HHRz have no direct effect on the virus. Plasmids expressing the arbitrary RNA control were slightly stimulatory (Figure 3), reminiscent of the modest stimulation of viral replication observed for some aptamers to HIV Gag.11 In contrast, anti-RT aptamers expressed from the eHHRz cassette were all inhibitory, with five- to tenfold suppression observed for the 12 eHHRz constructs. Most of the eHHRz constructs, in particular the 70.05 and 80.80 constructs, demonstrated increased inhibition relative to the mcHHRz (Figure 3). Aptamer 70.15 was already inhibitory in the mcHHRz construct, as noted above, and inhibition was not improved by modifying the ribozymes to eHHRz. However, the most striking feature of these data is that there was no observable differences among the modified constructs based on whether they carried active or inactivated eHHRz. Next, clonal, stable cell lines were generated as above to express each of the three aptamers in all four combinations of eHHRz variants (denoted Active, 5′Active, 3′Active, and Inactive) (Figure 4). These cell lines were then challenged with infectious HIV-1NL4-3 at an initial MOI of 0.3, and viral propagation was monitored by measuring Tat-induced luciferase activity at each passage (every 3 days) as above. Again, all 12 of these constructs suppressed viral propagation by two to three orders of magnitude, essentially independent of whether the construct carried active or inactivated eHHRz. The eHHRz constructs expressing aptamers 70.05 and 80.80 were more inhibitory than the corresponding mcHHRz constructs in early passages (Figure 4a,b), while inhibition by the eHHRz construct expressing aptamer 70.15 was similar to that observed using the mcHHRz construct (Figure 4c). Thus, viral suppression in clonal, stable cell lines does not require processing by HHRz cleavage. Importantly, the HIV inhibition observed in single-cycle assays using the eHHRz cassettes, but not the mcHHRz cassette, accurately predicted performance against infectious virus in multiple-cycle assays.
Figure 3.
Aptamer-mediated human immunodeficiency virus-1 (HIV-1) inhibition in a single-cycle infectivity assay: extended hammerhead ribozymes (eHHRz) aptamer-expression constructs display increased inhibition as compared to the mcHHRz aptamer-expression constructs. 293FT cells were transfected with aptamer-expression constructs or controls (pcDNA3.1, arbitrary RNA, or empty cassette) followed 4 hours later by cotransfection of pNL4-3-CMV-EGFP and pMD-G. Virus was collected and infectivity was determined as in Figure 1b. The controls, “arbitrary RNA” and “empty cassette”, shown as bars with horizontal lines, represent averages of data obtained for the two indicated control fragments inserted into each of the five expression contexts (mcHHRz, Active, 5′Active, 3′Active, and Inactive). One-way ANOVA and Student's t-test were used to determine statistical significance between samples (*P <0.05; **P < 0.01; ***P < 0.001). Statistical comparisons to arbitrary RNA (gray lines) and to mcHHRz (black lines) were considered separately. Values are shown as the mean ± SD for three experiments. CMV, cytomegalovirus; EGFP, enhanced green fluorescent protein.
Figure 4.
Aptamer-mediated inhibition in a multiple-cycle infectivity assay using clonal, stable cell lines expressing aptamers from the mcHHRz or extended hammerhead ribozymes (eHHRz) cassettes. Aptamer-expressing stable cell lines were made by transfecting TZM-bl cells with the indicated expression cassettes and subsequent G418 selection to form clonal, stable cell lines from single cells. Cells were plated in quadruplicate for each passage, using three wells for infection and one well as a noninfected control for background luciferase activity. Cells were infected with HIV-1NL4-3 using an initial multiplicity of infection (MOI) of 0.3 or 0.03. One-half of the virus-containing supernatant was transferred to fresh cells in series every 3 days, and infectivity was measured using BetaGlo reagent to quantify luciferase activity. (a) Aptamer 70.05, MOI = 0.3, (b) Aptamer 80.80, MOI = 0.3, (c) Aptamer 70.15, MOI = 0.3, and (d) Aptamer 70.05, MOI = 0.03. Background luciferase activity in the absence of virus was subtracted and values are shown as the mean ± SD for three replicates. HIV-1, human immunodeficiency virus-1.
Differences in viral inhibition at early passage are a function of input viral MOI
The effect of viral load was investigated to further explore the mechanism of aptamer-mediated suppression of HIV replication. Aptamers 70.05 and 80.80 were used because they demonstrated significantly improved inhibition compared to the mcHHRz in the single-cycle experiment reported in Figure 3. When tenfold less virus was used in the initial infection (MOI = 0.03), inhibition of HIV propagation in the infectious virus assays was strikingly more potent and more rapid for all of the constructs expressing aptamers 70.05 (Figure 4d) and 80.80 (Supplementary Figure S5). These observations raised the possibility that the ratio of aptamer to virus may contribute to the magnitude of the observed inhibition. Two approaches were used to test this hypothesis systematically. First, in a single-cycle assay, a proviral titration was performed in which a constant amount of aptamer-expressing plasmid (1 µg) was transfected into 293FT cells along with a constant amount of pMD-G and variable amounts of proviral plasmid. As expected, virus production (as measured by p24 ELISA and endpoint PCR amplification of HIV Gag, data not shown) and total observed infection (EGFP-positive cells) were reduced by decreasing the amount of input proviral plasmid (Figure 5a, raw percentages over graph). Yet even after normalizing to the infectivity observed in the presence of the pcDNA3.1 control, viral suppression by the aptamer-expressing plasmids clearly increased as the viral load was reduced (Figure 5a). The most informative range for the assay used 250 ng of v550. In the second approach, a stable 80.80-expressing cell line was challenged with HIV-1NL4-3 at initial MOIs ranging from 0.006 to 0.6 and monitored through multiple passages as above. Although all five conditions eventually led to at least 100-fold viral suppression, replication was most potently suppressed (104-fold) at lower MOI (Figure 5b and data not shown). Similarly, the effect was also observed for each of the expression contexts as shown in Supplementary Figure S5, with the most potent suppression occurring at lower MOI for each context. The magnitude of observed viral suppression is therefore determined in part by viral load.
Figure 5.
The ratio of aptamer to virus dictates observed magnitude of suppression. (a) Aptamer-expressing plasmids were transiently transfected into 293FT cells, followed by cotransfection 4 hours later with decreasing amounts of pNL4-3-CMV-GFP (1,000, 750, 500, 250, and 125 ng) and a constant 125 ng pMD-G. Virus-containing supernatants were harvested after 48 hours and 100 µl was used to infect fresh 293FT cells. Infectivity was determined by flow cytometry and is displayed as percent enhanced green fluorescent protein (EGFP) positive cells relative to pcDNA3.1. Raw data values for pcDNA3.1 infections are listed above the corresponding bars in the graph (*). Experiments were performed three times, and a representative experiment is shown. (b) In multiple-cycle assays, clonal, stable TZM-bl cells expressing pcDNA3.1 (gray lines) or aptamer 80.80 between mcHHRz (black lines) were infected with HIV-1NL4-3 in triplicate. Numbers in parenthesis indicate the initial multiplicity of infection (MOI). One-half of the virus-containing supernatant was transferred to fresh cells in series after 3 days and infectivity was measured using BetaGlo reagent to quantify luciferase activity. Background luciferase activity in the absence of virus was subtracted and values are shown as the mean ± SD for three replicates. CMV, cytomegalovirus; HIV-1, human immunodeficiency virus-1.
Intracellular aptamer localization
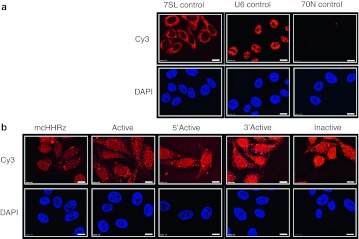
The patterns of antiviral bioactivity observed above could potentially result from differences in RNA accumulation or trafficking as a function of the context of aptamer expression. To explore this hypothesis, fluorescent in situ hybridization (FISH) was used to visualize aptamer RNA directly within transfected 293FT cells (data not shown) or in stable TZM-bl cell lines-expressing pcDNA3.1 (Figure 6a) or aptamer 70.15 (Figure 6b). Controls (Figure 6a) were probed with Cy3-labeled oligonucleotides complementary to the 5′ constant region of the 70N aptamers, Cy3-7SL (cytoplasmic), or Cy3-U6 (nuclear) and stained as expected for each, with appropriately low levels of staining for 70N, indicating low background. When aptamer-expressing stable cell lines were probed with the aptamer-specific oligonucleotides (Figure 6b), localization was observed throughout the cytoplasmic and nuclear compartments, and a clear distinction between cytoplasmic and nuclear compartments was not evident. Similarly, the aptamer distribution in transfected 293FT cells was also uniform, with little distinction between cytoplasm and nucleus (data not shown). However, the relative signal of aptamer RNA within the stable cell lines (Figure 6b) was not as intense as compared to the transfected 293FT cells (data not shown). Consistent with this observation, stable cell lines are well known to express lower levels of RNA relative to transfected cells. Thus, aptamer RNA can be readily detected in both transiently and stably transfected aptamer-expressing cells.
Figure 6.
Expression and localization of aptamer 70.15 in clonal, stable TZM-bl cells. RNA fluorescent in situ hybridization (FISH) was performed on stable aptamer-expressing cell lines and transfected 293FT cells (data not shown) using Cy3-tagged oligonucleotides to detect the cytoplasmic marker, 7SL, the nuclear marker U6, or the 5′ constant region of aptamer 70.15 (orange). Oligonucleotide sequences can be found in Supplementary Table S2. DAPI staining was performed to detect nuclei (blue). (a) pcDNA3.1-transfected cells were used to detect 7SL staining (cytoplasmic control), U6 staining (nuclear control), and 70N aptamer constant region staining (to establish nonspecific background of the probe used in b). (b) Aptamer expression and intracellular localization were detected for aptamer 70.15 in five stable cell lines expressing the aptamer in each of the five different expression contexts.
Bioactivity correlates to increased RNA accumulation and viral packaging
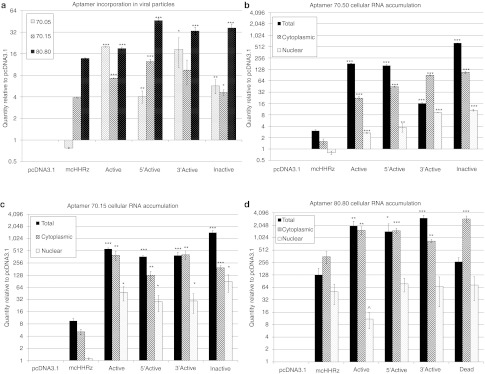
The observation of diffuse cytoplasmic and nuclear aptamer localization by FISH (Figure 6), together with the demonstration that RNA accumulation in stable cell lines above a threshold concentration is required for the observed viral inhibition (Supplementary Figure S3), suggest that quantitative assessment of aptamer RNA localization could help define cellular mechanisms contributing to aptamer-mediated inhibition of HIV replication. To gain a quantitative understanding of aptamer distribution, RNA accumulation levels in viral (Figure 7a), total (Figure 7b–d), cytoplasmic (Figure 7b–d), and nuclear (Figure 7b–d) RNA preparations from single-cycle assays were compared by real time PCR. “No RT” controls were included for each sample and cycle threshold (CT) values obtained for these controls were at least 10 cycles greater than the +RT experimental sample CT values. CT values for each sample were normalized to a corresponding reference RNA (HIV genomic gag for viral RNA; unspliced GAPDH for total RNA; spliced GAPDH for cytoplasmic RNA; and U6 snRNA for nuclear RNA), to control for amounts of input RNA, followed by normalization to the pcDNA3.1 negative control. Thus, experimental sample RNA accumulation is shown relative to pcDNA3.1 (set at 1). RNA levels for all three aptamers were strikingly higher in the four eHHRz constructs than in the corresponding mcHHRz constructs, potentially suggesting that the enhanced accumulation of aptamer RNA accounted for the increased degree of inhibition by these constructs in the single-cycle assay. Notably, these results are consistent with the observed correlation between aptamer accumulation and inhibition in multiple-cycle assays shown in Supplementary Figure S3. HIV is known to package random cellular RNAs along with its genome,36 although the mechanism of random RNA incorporation has not been elucidated. While it is not yet known whether aptamer packaging is due to a direct interaction with the RT component of the HIV Gag-Pol polyprotein or due to random cellular RNA incorporation, expression constructs that increase the overall amount of aptamer RNA in the cell may result in a greater incorporation of inhibitory aptamers during viral assembly, so that they are available to inhibit reverse transcription upon infection.
Figure 7.
Aptamer RNA accumulation in viral, total, cytoplasmic, and nuclear RNA preparations. (a) Viral RNA was harvested from virus produced by transfection in the presence of aptamer. cDNA was synthesized from 200 ng of RNA and subjected to quantitative real-time-PCR using primers specific for 70N or 80N aptamers or reference RNAs (unspliced GAPDH for total RNA; spliced GAPDH for cytoplasmic RNA; U6snRNA for nuclear RNA; or human immunodeficiency virus-1 (HIV-1) gag for viral RNA). “No RT” controls were included for each sample, and each sample was assayed in triplicate. Each quantity was determined using the relative quantity (2–ΔΔCT) method where samples were first normalized to the corresponding reference RNAs, then to the pcDNA3.1 control (set to 1), and averaged. Values are shown as the mean ± SD. One-way ANOVA and Student's t-test were performed to determine statistical significance as compared to the original expression context (*P < 0.05; ** P < 0.01; ***P < 0.001). Experiments were performed three times, and a representative experiment is shown. (b–d) Total RNA and cytoplasmic and nuclear RNA from fractionated cells were collected from 293FT cells transfected with aptamers (b) 70.05, (c) 70.15, or (d) 80.80. cDNA, qPCR, and data analysis were performed as in (a) using the corresponding reference RNAs.
Discussion
Aptamers are potent inhibitors of HIV RT enzymatic activities and of viral replication. While there is increasing understanding of the molecular interactions underlying biochemical inhibition in vitro, there is little understanding of the biological mechanisms by which they encounter their molecular targets in a cellular context. This study addresses the roles of ribozyme-mediated cleavage and tertiary stabilization in determining aptamer-mediated suppression of HIV-1 replication. We observed that modifying an existing aptamer-expressing cassette to include two tertiary-stabilized HHRz led to significantly greater inhibition of HIV-1 infection in a single-cycle assay format without any requirement for self-cleavage by the embedded ribozymes, and that the inhibition correlated with increased accumulation of aptamer RNA in viral, total, cytoplasmic, and nuclear RNA preparations. Potent suppression of infectious virus was also observed, further supporting the utility and predictive value of this design. The single-cycle assay system therefore allows for much higher throughput testing of potential new inhibitory RNAs—singly and in combination—to take advantage of differences in specificities, structures, binding characteristics, cytotoxicity and net inhibitory potential, and to identify the most promising inhibitors for therapeutic development. This marks a significant advance over previous infectivity assays that involve generating clonal, stable cell lines, and serial viral passage,6,8,10 both of which are not conducive to high throughput screening assays. In principle, the same expression platform may provide a convenient means of delivering and screening aptamer inhibitors of other HIV targets, such as integrase, protease, and Gag. Notably, these results also resolve a problematic feature of adapting the HHRz-cassette design for gene therapy using viral vectors. Specifically, self-cleavage of the primary transcript of such a viral vector by a functional HHRz could seriously limit assembly of the gene-transducing particles and thereby impede effective transgene delivery. However, in our study, HHRz self-cleavage is not a requirement for aptamer-mediated inhibition; hence, these findings expand the potential application of this cassette in viral vectors for aptamer delivery.
Flanking sequences can be very important for proper expression and folding of aptamers expressed in a cellular context.37 These sequences can interfere with aptamer folding into active conformations, as well as overall stability and localization of the transcript, resulting in the inability of the aptamer to bind its target. The mcHHRz ribozymes were incorporated into the design of the expression cassette to prevent flanking sequences from interfering with folding of the delivered aptamer RNA into its conformationally active secondary structure. The first generation of these cassettes delivered a trans-cleaving hammerhead ribozyme (trans-mcHHRz) targeted to the retinoblastoma (Rb) gene. The trans-mcHHRz was expressed between two identical cis-cleaving mcHHRz and resulted in strong suppression of the target RbmRNA.28 A similar cis-trans-cis ribozyme design resulted in specific cleavage of an mRNA encoding a 2-oxo-glutarate-dependant dioxygenase in the wild potato Solanum.30 Initial adaptation of that design for aptamer delivery to mammalian cells yielded some of the first clear demonstrations that intracellularly expressed aptamers targeted to RT could suppress HIV replication in stable cell lines.6,10 However, those publications did not evaluate the importance of the flanking ribozymes, which are clearly not required for the purposes of their self-cleavage activity. We speculate that the increased structural stability per se of the RNA modules flanking the aptamer increased aptamer potency primarily by providing better protection of uncleaved and partially cleaved aptamer products from cellular exonucleases than the protection afforded by non-stabilized mcHHRz. Under this model, the structurally stabilized transcripts are less susceptible to degradation, accumulate to higher levels, package more effectively into assembling virions, and are more available to inhibit RT during subsequent infection. Consistent with this model, improved accumulation of a trans-cleaving HHRz was previously demonstrated upon pairing the 5′ and 3′ ends of the transcript used in that study.38 A second, minor route for increasing bioactivity may have been that the compact, stable structures at each end of the transcript may have enhanced aptamer folding into the active, RT-binding conformation by isolating the aptamer more effectively from the flanking sequences. This may also have reduced any misfolding caused by potential interactions between identical mcHHRz on either side of the aptamer.
A complete molecular understanding of the processing accompanying anti-HIV aptamer expression must include identification of the bioactive forms. Aptamers expressed from the cassette in which both eHHRz ribozymes were inactivated strongly inhibited the virus in both single-cycle and multiple-cycle formats. This strongly suggests that the unprocessed (uncleaved) form of the aptamer-containing transcript is clearly bioactive. Indeed, to remove self-cleavage as a variable, we recently chose this eHHRz-inactivated cassette for further screens of several newly identified aptamers (M.A. Ditzler, M.J. Lange, D. Bose, C. Bottoms, K. Virkler, A.W. Sawyer et al., manuscript submitted, and A.S. Whatley, M.A. Ditzler, M.J. Lange, J. Chang, A.W. Sawyer, E. Biondi et al., manuscript submitted). The uncleaved, aptamer-containing transcript is effectively a small mRNA, for which the presence of a 5′ cap and 3′ polyA tail may aid in translocation out of the nucleus.39,40 Furthermore, because HHRz cleavage products are often very quickly degraded,30,31,34,38 the uncleaved or partially cleaved form of the transcript may be the major form that accumulates. It may be that the polyA tail plays a more important role than the 5′ cap in stabilizing the aptamer transcript, since the RzB ribozyme on the 5′ side of the aptamer cleaved much more efficiently in vitro than the sTRSV ribozyme on the 3′ side (Figure 2). Although previous studies that utilized cis-cleaving HHRz for delivering bioactive RNAs have shown that the fully cleaved products were generated during cotranscriptional cleavage in vitro, uncleaved and partially cleaved transcripts were also present in the products of those reactions.10,38 Therefore, engineering transcripts for optimal delivery of aptamers into HIV may be more closely tied to nuclease protection, localization, and packaging than to excision and release from the transcript.
Materials and Methods
Unless otherwise noted, all chemicals were purchased from Sigma-Aldrich (St Louis, MO). Restriction enzymes and T4 DNA ligase used for cloning purposes were purchased from New England Biolabs (Ipswitch, MA).
Plasmids. The CMV-driven plasmid containing the functional pseudoknot core of aptamer 70.15 (40 nt) between two identical mcHHRz10 was generously provided by Vinayaka R. Prasad. This plasmid was digested by EcoRI and ApaI to remove the minimal 70.15 aptamer. Full length aptamers 70.03, 70.05, 70.08, 70.15, 70.26, 80.77, and 80.80 were then inserted into these sites. Additionally, a 70-nt fragment of the luciferase gene was amplified with primers to append the aptamer constant regions and was inserted into the plasmid as an arbitrary RNA control. Aptamer and arbitrary RNA sequences are given in Supplementary Table S1. Aptamer-expression cassettes carrying eHHRz were prepared by replacing the mHHRz with either an inactive or highly active form of hammerhead ribozyme RzB35 (5′) or an inactive or highly active form of a hammerhead ribozyme sTRSV (3′). Ribozyme sequences and structures are given in Supplementary Table S1 and Figure 2, respectively. The eHHRz cassettes were made by annealing synthetic oligonucleotides with the desired sequences (Supplementary Table S3) in separate reactions for each eHHRz combination, PCR amplifying with primers carrying sites for restriction enzyme sites NheI and NotI, and cloning the digested inserts in place of the full original mcHHRz cassette using NheI and NotI restriction sites (Supplementary Figure S1). The resulting cassettes, also expressing the 70.15 functional pseudoknot core, carried one of the four possible combinations of HHRz: two highly active ribozymes (denoted “Active”), a 5′ active and 3′ inactive ribozyme (“5′ Active”), a 5′ inactive and 3′ active ribozyme (“3′ Active”) or two inactive ribozymes (“Inactive”). Full-length aptamers 70.05, 70.15, and 80.80, and the 70-nt arbitrary luciferase fragment were inserted into the eHHRz cassettes via EcoRI and BamHI restriction sites. Empty ribozyme cassettes for each of the ribozyme combinations were also made by ligation that joined the EcoRI and BamHI restriction sites. All plasmids were confirmed by DNA sequencing using primers upstream of the ribozyme cassette.
The HIV-1NL4-3-derived CMV-EGFP plasmid (pNL4-3-CMV-EGFP) used in single-cycle infectivity assays was kindly provided by Vineet KewalRamani (National Cancer Institute [NCI]-Fredrick). This proviral vector lacks the genes encoding vif, vpr, vpu, nef, and env, and has a CMV immediate early promoter-driven EGFP in place of nef. The vesticular stomatitis virus (VSV) glycoprotein-expressing plasmid, pMD-G, used for viral pseudotyping, was obtained from Invitrogen (Carlsbad, CA).
Cell lines and viruses. The human cell line, 293FT (Invitrogen), was maintained in standard culture media containing Dulbecco's minimum essential medium (Sigma-Aldrich), 10% fetal bovine serum (Sigma-Aldrich), 2 mmol/l L-glutamine (Gibco, Life Technologies, Grand Island, NY), 1 mmol/l non-essential amino acids (Gibco, Life Technologies), 1 mmol/l sodium pyruvate (Gibco, Life Technologies), and 0.5 mg/ml G418 (Sigma-Aldrich). The following reagents were obtained through the NIH AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: TZM-bl cells from Dr John C. Kappes, Dr Xiaoyun Wu and Tranzyme Inc., MT2 cells from Dr Douglas Richmond, and the pNL4-3 molecular clone from Dr Malcolm Martin. TZM-bl cells, which are a HeLa-derived luciferase/β-galactosidase reporter cell line-expressing CD4, CCR5, and CXCR4, and MT2 cells, which are human T cell leukemia cells isolated from cord blood, were also cultured in standard culture media, but without G418. All cell lines were maintained at 37 °C in 5% carbon dioxide with splitting twice per week.
Clonal aptamer-expressing stable cell lines were made by transfecting TZM-bl cells with aptamer-expressing plasmids followed by G418 drug selection. G418-resistant cells were seeded as single cells in 96-well plates. Six to twelve individual lines were cultured from the single seeded cells for each aptamer-expressing construct. Stable cells were maintained in standard media containing G418 except during infection, when G418 was omitted.
Infectious HIV-1NL4-3 was produced by transfecting 293FT cells using polyethylenimine41 with 1 µg of the pNL4-3 molecular clone. Supernatant was harvested 48 hours post-transfection and transferred to MT2 cells for viral propagation. Virus was harvested after 5 days, when syncytia formation was apparent. Virus-containing supernatant was syringe filtered through a 0.45-µm filter, aliquotted, and frozen at –80 °C. Viral titer was determined by β-galactosidase assay using the TZM-bl cells. Briefly, cells were fixed with 0.1% glutaraldehyde 72 hours postinfection and stained with X-Gal (5-bromo-4-chloro-3-indolyl-b-D-galactopyranoside). Viral titer was determined by counting blue-forming units and determining blue-forming units/ml.
Transfections. All transfections were performed on cells plated the previous day (~50% confluence) using polyethylenimine at 3 µl/µg of DNA, as previously described.41 Medium was changed on each transfection after 4 hours. After 48 hours, virus-containing supernatant was harvested by syringe filtration using a 0.45-µm filter. Cells were trypsinized, fixed in 4% paraformaldehyde, washed with 1× phosphate-buffered saline (PBS), and analyzed by flow cytometry to determine transfection efficiency by EGFP expression.
In vitro cotranscriptional cleavage assays. Ribozyme cassettes for all five expression contexts were PCR amplified with primers that appended a T7 promoter and confirmed by size using agarose gel electrophoresis. In vitro transcription was performed by incubating the PCR product in buffer containing 120 mmol/l HEPES–KOH, pH 7.5, 30 mmol/l MgCl2, 2 mmol/l spermidine, 40 mmol/l DTT, 0.01% Triton X-100 (vol/vol) with 4 mmol/l NTPs including [α-32P]CTP (Perkin Elmer, Waltham, MA) to internally radiolabel the transcripts, and T7 RNA polymerase for 3 hours at 37 °C. Products were resolved on a 10% denaturing polyacrylamide gel and applied to a phosphor screen to determine cotranscriptional cleavage activity for each of the cassettes using a Typhoon 9000 FLA (GE Healthcare Life Sciences, Piscataway, NJ).
Single-cycle infectivity assays. Single-cycle infectivity assays using aptamer-expressing plasmids, pNL4-3-CMV-GFP and pMD-G, were performed by transfecting 293FT cells with polyethylenimine in 6-well cell culture dishes. Aptamer-expressing plasmids were transfected first (1 µg), followed 4 hours later by a mixture of pNL4-3-CMV-GFP (250 ng) and pMD-G (125 ng). The medium was changed between transfections and again four hours after the second transfection. Supernatant was harvested 48 hours post-transfection, syringe filtered through 0.45-µm filters, and 100 µl was added to fresh 293FT cells to determine infectivity. Viral concentration was determined by p24 ELISA. Infected cells were collected after 48 hours, fixed with 4% paraformaldehyde, washed with 1× PBS, and analyzed on an Accuri Flow Cytometer to determine the percentage of infected (EGFP-positive) cells (BD Biosciences, San Jose, CA).
Proviral titration assays were performed as described above in the presence of 1 µg aptamer-expression plasmid, except that the transfections were carried out with 1000, 750, 500, 250, or 125 ng of pNL4-3-CMV-EGFP combined with 125 ng of pMD-G. Filler plasmid (p202, in which a large portion of the CMV promoter was deleted from pcDNA3.1) was included to maintain a total of 1.125 µg plasmid DNA in each transfection.
Multiple-cycle infectivity assays. Multiple-cycle infectivity assays were performed using clonal, stable TZM-bl cell lines expressing the vector (pcDNA3.1), an arbitrary RNA (70-nt fragment of luciferase), the empty ribozyme cassettes, and all 15 aptamer-plus-ribozyme combinations (three aptamers in each of the five different expression contexts). Cell lines were plated in black-walled 96-well plates (BD Falcon, Bedford, MA) with 10,000 cells/well in quadruplicate, including one well as a noninfected control to account for background luciferase activity. Cells were infected with HIV-1NL4-3 produced by 293FT transfection and expansion in MT2 cells at an MOI of 0.006, 0.03, 0.06, 0.3, or 0.6 in a total volume of 200 µl/well with 8 µg/ml polybrene. Every 3 days, 100 µl of the supernatant was transferred to fresh aptamer-expressing cells in 100 µl fresh medium containing polybrene. The BetaGlo Assay System (Promega, Madison, WI) was used to determine infectivity by luminescence, with quantities expressed in relative light units. Multiple-cycle experiments were carried out for at least 12 days for each aptamer-expressing cell line and infectivity was assayed every 3 days.
Cellular fractionation and RNA isolation. Transfected 293FT cells were collected by scraping into 1× PBS, washed with 1× PBS, and divided into aliquots for isolation of total RNA and cytoplasmic and nuclear fractionation. For total RNA isolation, TRIzol reagent (Invitrogen) was used as per the manufacturer's instructions. Cytoplasmic and nuclear fractions were separated essentially as described by chemical disruption and centrifugation.42 Briefly, cells were resuspended in 500 µl TKM buffer (10 mmol/l KCl, 10 mmol/l Tris–HCl pH 7.5, 1 mmol/l MgCl2,) and incubated on ice for 5 minutes. Then 15 µl of 10% Triton X-100 was added to lyse the cells while keeping the nuclei intact; cells were incubated for 10 minutes on ice during lysis. Samples were then centrifuged at 500g for 5 minutes at 4 °C. The supernatant containing the cytoplasmic material was transferred to a fresh tube being careful not to disrupt the nuclear pellet. The lysis process was repeated once for the nuclear pellet to ensure that there was no cytoplasmic contamination. The nuclear pellet was then washed in TKM buffer once before RNA extraction. RNA was extracted from the fractionated samples using TRIzol, as per the manufacturer's instructions. Isolated RNA was subjected to DNase treatment using Turbo DNase (Ambion, Life Technologies, Grand Island, NY) and quantified on a NanoDrop Spectrophotometer (Thermo-Fisher Scientific, Waltham, MA). For isolation of viral RNA, 1 ml of virus-containing supernatant was centrifuged at 45,000g for 1 hour at 4 °C using a Beckman-Coulter Ultra Centrifuge. Supernatant was discarded and viral RNA was isolated from the viral pellet using the TRIzol-LS reagent (Invitrogen) as per the manufacturer's instructions and was subjected to Turbo DNase treatment and quantification.
Real-time quantitative PCR. Total, cytoplasmic, nuclear, and viral RNA (200 ng) were used to synthesize cDNA using random hexamer primers with the ImProm II Reverse Transcription System (Promega) as per the manufacturer's instructions. Real time quantitative PCR was performed on eachset of cDNA using PowerSybr Green (Applied Biosystems, Foster City, CA) and primers specific to 70N aptamers, 80N aptamers, GAPDH (non-spliced; total endogenous control), GAPDHsp (spliced; cytoplasmic endogenous control), U6 (nuclear endogenous control), or HIV-1 Gag (viral genome endogenous control). Primers are listed in Supplementary Table S3. Primer pair amplification efficiencies were determined using 1:10 cDNA dilutions and test and housekeeping gene primer pairs with similar efficiencies were used for the qPCR. Quantities were determined using the relative quantity (2–ΔΔCT) method and were verified using the Pfaffl method.43 In brief, CT values for the housekeeping controls were subtracted from the sample CT values (e.g., ΔCT = CTaptamer – CTGAPDH) to determine the ΔCT. Then, the ΔCT values for the negative control (pcDNA3.1) were subtracted from the ΔCT values for the samples (ΔΔCT = ΔCTsample – ΔCTcontrol). The ΔΔCT values were then used to calculate relative quantity, using the equation relative quantity =2–ΔΔCT. Thus, graphs are represented as quantity relative to the specified endogenous control, with normalization to pcDNA3.1 (set to 1).
RNA FISH. RNA FISH was performed essentially as described.44 293FT cells were transfected with 1 µg of aptamer-expressing plasmid in 6-well plates. After 24 hours, cells were replated on sterilized coverslips, allowed to adhere for an additional 24 hours, and fixed in 4% paraformaldehyde. Stable aptamer-expressing cell lines were also plated on coverslips and fixed. After fixation, cells were permeabilized by submersion in 70% ethanol and stored at 4 °C until use. Aptamer expression and localization was determined using oligonucleotides tagged with Cy3 (Integrated DNA Technologies, Coralville, IA) that recognize the 5′ constant region of the 70N aptamer sequence (Supplementary Table S2). Probes for endogenous RNA controls, Cy3-U6 (nuclear) and Cy3-7SL (cytoplasmic), were obtained from the literature.45 Hybridization was performed overnight at 37 °C using 2× SSC (300 mmol/l sodium chloride, 30 mmol/l sodium citrate), 50% formamide, 0.02% bovine serum albumin, 40 µg yeast tRNA, and 30 ng probe. After hybridization, cells were washed twice at 37 °C for 30 minutes using 2× SSC, 50% formamide, and once in PBS. Coverslips were mounted on slides using VectaShield Hardset mounting medium containing 4′,6′-diamidino-2-phenlindole (DAPI; Vector Laboratories, Burlingame, CA). Images were obtained using an Olympus IX81 microscope using CellSens software with deconvolution (Olympus, Center Valley, PA).
SUPPLEMENTARY MATERIAL Figure S1. Design schematic for extended hammerhead ribozyme modifications. Figure S2. Control RNAs—arbitrary RNA, empty HHRz cassette, or pcDNA3.1—are not inhibitory in clonal TZM-bl stable cell lines. Figure S3. Aptamer-mediated inhibition is dependent upon the level of intracellular aptamer RNA accumulation. Figure S4. Clonal, stable cell lines retain aptamer expression, susceptibility to HIV infection, reporter gene activity, and inhibitory capability throughout passage. Figure S5. Increased magnitude of aptamer-mediated inhibition in clonal, stable cell lines expressing aptamer 80.80 from the mcHHRz or eHHRz cassettes at lower MOI. Table S1. Full-length aptamer and extended hammerhead ribozyme sequences cloned into aptamer-expression cassettes. Table S2. Oligonucleotide and primer sequences used for engineering and cloning of the modified aptamer-expression cassettes, RNA FISH, and qRT-PCR. Table S3. Expected cleavage products for mcHHRz and eHHRz cassettes.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Disease, R01AI074389 (D.H.B.) and F32AI085627 (M.J.L.). We would like to acknowledge Vinayaka R. Prasad for the generous contribution of the mcHHRz aptamer-expression plasmid. Additionally, we would like to acknowledge the following individuals for their intellectual contributions, assistance with methodology and interpretation, scientific discussion, and feedback: Stefan G. Sarafianos (University of Missouri), Atsuko Hachiya (National Center for Global Health and Medicine, Tokyo), Terri D. Lyddon (University of Missouri), Rebecca Chitima-Matsiga (University of Missouri), and members of the Burke Laboratory at the University of Missouri (Elisa Biondi, Mark Ditzler, Raghav Poudyal, Adam Maxwell, Andrew Sawyer, Mackenzie Callaway). This work was completed at the University of Missouri, Columbia, Missouri. All authors declared no conflict of interest.
Supplementary Material
Design schematic for extended hammerhead ribozyme modifications.
Control RNAs—arbitrary RNA, empty HHRz cassette, or pcDNA3.1—are not inhibitory in clonal TZM-bl stable cell lines.
Aptamer-mediated inhibition is dependent upon the level of intracellular aptamer RNA accumulation.
Clonal, stable cell lines retain aptamer expression, susceptibility to HIV infection, reporter gene activity, and inhibitory capability throughout passage.
Increased magnitude of aptamer-mediated inhibition in clonal, stable cell lines expressing aptamer 80.80 from the mcHHRz or eHHRz cassettes at lower MOI.
Full-length aptamer and extended hammerhead ribozyme sequences cloned into aptamer-expression cassettes.
Oligonucleotide and primer sequences used for engineering and cloning of the modified aptamer-expression cassettes, RNA FISH, and qRT-PCR.
Expected cleavage products for mcHHRz and eHHRz cassettes.
REFERENCES
- Burke DH, Scates L, Andrews K., and, Gold L. Bent pseudoknots and novel RNA inhibitors of type 1 human immunodeficiency virus (HIV-1) reverse transcriptase. J Mol Biol. 1996;264:650–666. doi: 10.1006/jmbi.1996.0667. [DOI] [PubMed] [Google Scholar]
- Tuerk C, MacDougal S., and, Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc Natl Acad Sci USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopinath SC. Antiviral aptamers. Arch Virol. 2007;152:2137–2157. doi: 10.1007/s00705-007-1014-1. [DOI] [PubMed] [Google Scholar]
- Held DM, Kissel JD, Patterson JT, Nickens DG., and, Burke DH. HIV-1 inactivation by nucleic acid aptamers. Front Biosci. 2006;11:89–112. doi: 10.2741/1782. [DOI] [PubMed] [Google Scholar]
- Burnett JC., and, Rossi JJ. RNA-based therapeutics: current progress and future prospects. Chemistry & Biology. 2012;19:60–71. doi: 10.1016/j.chembiol.2011.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi PJ, North TW., and, Prasad VR. Aptamers directed to HIV-1 reverse transcriptase display greater efficacy over small hairpin RNAs targeted to viral RNA in blocking HIV-1 replication. Mol Ther. 2005;11:677–686. doi: 10.1016/j.ymthe.2005.01.013. [DOI] [PubMed] [Google Scholar]
- Neff CP, Zhou J, Remling L, Kuruvilla J, Zhang J, Li H.et al. (2011An aptamer-siRNA chimera suppresses HIV-1 viral loads and protects from helper CD4(+) T cell decline in humanized mice Sci Transl Med 366ra6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaloin L, Lehmann MJ, Sczakiel G., and, Restle T. Endogenous expression of a high-affinity pseudoknot RNA aptamer suppresses replication of HIV-1. Nucleic Acids Res. 2002;30:4001–4008. doi: 10.1093/nar/gkf522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb G, Reigadas S, Castanotto D, Faure A, Ventura M, Rossi JJ.et al. (2006Endogenous expression of an anti-TAR aptamer reduces HIV-1 replication RNA Biol 3150–156. [DOI] [PubMed] [Google Scholar]
- Joshi P., and, Prasad VR. Potent inhibition of human immunodeficiency virus type 1 replication by template analog reverse transcriptase inhibitors derived by SELEX (systematic evolution of ligands by exponential enrichment) J Virol. 2002;76:6545–6557. doi: 10.1128/JVI.76.13.6545-6557.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam D, Duclair S, Datta SA, Ellington A, Rein A., and, Prasad VR. RNA aptamers directed to human immunodeficiency virus type 1 Gag polyprotein bind to the matrix and nucleocapsid domains and inhibit virus production. J Virol. 2011;85:305–314. doi: 10.1128/JVI.02626-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Ke W, Wu SS, Gan L, Zhou R, Sun CY.et al. An adenovirus-delivered peptide aptamer C1-1 targeting the core protein of hepatitis B virus inhibits viral DNA replication and production in vitro and in vivo Peptides 301816–1821.2009 [DOI] [PubMed] [Google Scholar]
- Feng H, Beck J, Nassal M., and, Hu KH. A SELEX-screened aptamer of human hepatitis B virus RNA encapsidation signal suppresses viral replication. PLoS One 6. 2011;e27862 doi: 10.1371/journal.pone.0027862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser N, Lischka P, Wagenknecht N., and, Stamminger T. Inhibition of human cytomegalovirus replication via peptide aptamers directed against the nonconventional nuclear localization signal of the essential viral replication factor pUL84. J Virol. 2009;83:11902–11913. doi: 10.1128/JVI.01378-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umehara T, Fukuda K, Nishikawa F, Sekiya S, Kohara M, Hasegawa T.et al. (2004Designing and analysis of a potent bi-functional aptamers that inhibit protease and helicase activities of HCV NS3 Nucleic Acids Symp Ser (Oxf) 195–196. [DOI] [PubMed]
- Romero-López C, Díaz-González R, Barroso-delJesus A., and, Berzal-Herranz A. Inhibition of hepatitis C virus replication and internal ribosome entry site-dependent translation by an RNA molecule. J Gen Virol. 2009;90 Pt 7:1659–1669. doi: 10.1099/vir.0.008821-0. [DOI] [PubMed] [Google Scholar]
- Tomai E, Butz K, Lohrey C, von Weizsäcker F, Zentgraf H., and, Hoppe-Seyler F. Peptide aptamer-mediated inhibition of target proteins by sequestration into aggresomes. J Biol Chem. 2006;281:21345–21352. doi: 10.1074/jbc.M604258200. [DOI] [PubMed] [Google Scholar]
- Cheng C, Dong J, Yao L, Chen A, Jia R, Huan L.et al. (2008Potent inhibition of human influenza H5N1 virus by oligonucleotides derived by SELEX Biochem Biophys Res Commun 366670–674. [DOI] [PubMed] [Google Scholar]
- Syed MA., and, Pervaiz S. Advances in aptamers. Oligonucleotides. 2010;20:215–224. doi: 10.1089/oli.2010.0234. [DOI] [PubMed] [Google Scholar]
- Joshi PJ, Fisher TS., and, Prasad VR. Anti-HIV inhibitors based on nucleic acids: emergence of aptamers as potent antivirals. Curr Drug Targets Infect Disord. 2003;3:383–400. doi: 10.2174/1568005033481060. [DOI] [PubMed] [Google Scholar]
- Fichou Y., and, Férec C. The potential of oligonucleotides for therapeutic applications. Trends Biotechnol. 2006;24:563–570. doi: 10.1016/j.tibtech.2006.10.003. [DOI] [PubMed] [Google Scholar]
- Li N, Wang Y, Pothukuchy A, Syrett A, Husain N, Gopalakrisha S.et al. (2008Aptamers that recognize drug-resistant HIV-1 reverse transcriptase Nucleic Acids Res 366739–6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditzler MA, Bose D, Shkriabai N, Marchand B, Sarafianos SG, Kvaratskhelia M.et al. (2011Broad-spectrum aptamer inhibitors of HIV reverse transcriptase closely mimic natural substrates Nucleic Acids Res 398237–8247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held DM, Kissel JD, Thacker SJ, Michalowski D, Saran D, Ji J.et al. (2007Cross-clade inhibition of recombinant human immunodeficiency virus type 1 (HIV-1), HIV-2, and simian immunodeficiency virus SIVcpz reverse transcriptases by RNA pseudoknot aptamers J Virol 815375–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held DM, Kissel JD, Saran D, Michalowski D., and, Burke DH. Differential susceptibility of HIV-1 reverse transcriptase to inhibition by RNA aptamers in enzymatic reactions monitoring specific steps during genome replication. J Biol Chem. 2006;281:25712–25722. doi: 10.1074/jbc.M604460200. [DOI] [PubMed] [Google Scholar]
- Kissel JD, Held DM, Hardy RW., and, Burke DH. Single-stranded DNA aptamer RT1t49 inhibits RT polymerase and RNase H functions of HIV type 1, HIV type 2, and SIVCPZ RTs. AIDS Res Hum Retroviruses. 2007;23:699–708. doi: 10.1089/aid.2006.0262. [DOI] [PubMed] [Google Scholar]
- Braun SE, Shi X, Qiu G, Wong FE, Joshi PJ, Prasad VR.et al. (2007Instability of retroviral vectors with HIV-1-specific RT aptamers due to cryptic splice sites in the U6 promoter AIDS Res Ther 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benedict CM, Pan W, Loy SE., and, Clawson GA. Triple ribozyme-mediated down-regulation of the retinoblastoma gene. Carcinogenesis. 1998;19:1223–1230. doi: 10.1093/carcin/19.7.1223. [DOI] [PubMed] [Google Scholar]
- Heller T, Saito S, Auerbach J, Williams T, Moreen TR, Jazwinski A.et al. (2005An in vitro model of hepatitis C virion production Proc Natl Acad Sci USA 1022579–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussière F, Ledû S, Girard M, Héroux M, Perreault JP., and, Matton DP. Development of an efficient cis-trans-cis ribozyme cassette to inactivate plant genes. Plant Biotechnol J. 2003;1:423–435. doi: 10.1046/j.1467-7652.2003.00039.x. [DOI] [PubMed] [Google Scholar]
- Khvorova A, Lescoute A, Westhof E., and, Jayasena SD. Sequence elements outside the hammerhead ribozyme catalytic core enable intracellular activity. Nat Struct Biol. 2003;10:708–712. doi: 10.1038/nsb959. [DOI] [PubMed] [Google Scholar]
- Burke DH., and, Greathouse ST. Low-magnesium, trans-cleavage activity by type III, tertiary stabilized hammerhead ribozymes with stem 1 discontinuities. BMC Biochem. 2005;6:14. doi: 10.1186/1471-2091-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De la Pena M, Gago S., and, Flores R. Peripheral regions of natural hammerhead ribozymes greatly increase their self-cleavage activity. EMBO J. 2003;22:5561–5570. doi: 10.1093/emboj/cdg530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andäng M, Maijgren-Steffensson C, Hinkula J., and, Ahrlund-Richter L. Cis-cleavage affects hammerhead and hairpin ribozyme steady-state levels differently and has strong impact on trans-targeting efficiency. Oligonucleotides. 2004;14:11–21. doi: 10.1089/154545704322988012. [DOI] [PubMed] [Google Scholar]
- Saksmerprome V, Roychowdhury-Saha M, Jayasena S, Khvorova A., and, Burke DH. Artificial tertiary motifs stabilize trans-cleaving hammerhead ribozymes under conditions of submillimolar divalent ions and high temperatures. RNA. 2004;10:1916–1924. doi: 10.1261/rna.7159504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muriaux D., and, Rein A. Encapsidation and transduction of cellular genes by retroviruses. Front Biosci. 2003;8:d135–d142. doi: 10.2741/950. [DOI] [PubMed] [Google Scholar]
- Martell RE, Nevins JR., and, Sullenger BA. Optimizing aptamer activity for gene therapy applications using expression cassette SELEX. Mol Ther. 2002;6:30–34. doi: 10.1006/mthe.2002.0624. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Ayers DF, Malmstrom TA, McKenzie TL, Ganousis L, Chowrira BM.et al. (1995Improved accumulation and activity of ribozymes expressed from a tRNA-based RNA polymerase III promoter Nucleic Acids Res 232259–2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart M. Nuclear export of mRNA. Trends Biochem Sci. 2010;35:609–617. doi: 10.1016/j.tibs.2010.07.001. [DOI] [PubMed] [Google Scholar]
- Cullen BR. Nuclear RNA export pathways. Mol Cell Biol. 2000;20:4181–4187. doi: 10.1128/mcb.20.12.4181-4187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SE, Staley EM, Mayginnes JP, Pintel DJ., and, Tullis GE. Transfection of mammalian cells using linear polyethylenimine is a simple and effective means of producing recombinant adeno-associated virus vectors. J Virol Methods. 2006;138:85–98. doi: 10.1016/j.jviromet.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Donald, C, Rio GJH, Manuel Ares Jr., and, Nilsen TW. John Inglis, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York; 2011. RNA: A Laboratory Manual. [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenklusen D., and, Singer RH. Analyzing mRNA expression using single mRNA resolution fluorescent in situ hybridization. Meth Enzymol. 2010;470:641–659. doi: 10.1016/S0076-6879(10)70026-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul CP, Good PD, Li SX, Kleihauer A, Rossi JJ., and, Engelke DR. Localized expression of small RNA inhibitors in human cells. Mol Ther. 2003;7:237–247. doi: 10.1016/s1525-0016(02)00038-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Design schematic for extended hammerhead ribozyme modifications.
Control RNAs—arbitrary RNA, empty HHRz cassette, or pcDNA3.1—are not inhibitory in clonal TZM-bl stable cell lines.
Aptamer-mediated inhibition is dependent upon the level of intracellular aptamer RNA accumulation.
Clonal, stable cell lines retain aptamer expression, susceptibility to HIV infection, reporter gene activity, and inhibitory capability throughout passage.
Increased magnitude of aptamer-mediated inhibition in clonal, stable cell lines expressing aptamer 80.80 from the mcHHRz or eHHRz cassettes at lower MOI.
Full-length aptamer and extended hammerhead ribozyme sequences cloned into aptamer-expression cassettes.
Oligonucleotide and primer sequences used for engineering and cloning of the modified aptamer-expression cassettes, RNA FISH, and qRT-PCR.
Expected cleavage products for mcHHRz and eHHRz cassettes.