Abstract
The mechanisms by which some HIV-infected subjects resist disease progression, while others progress rapidly, are incompletely understood. Viral and host genetic factors, such as nef deletions and MHC alleles, explain a portion of the observed variability. However, it has been difficult to identify host immune functions that may be present before infection and that allow resistance to lentiviral disease progression. Here we show that SIV replication in the infected rhesus macaque is limited by the size of the pre-existing Th17 cell compartment: animals with a high representation of such cells in blood and intestinal tissue prior to infection experienced peak and set-point viral loads approximately one log unit lower than those with a lower representation of Th17 cells. Reciprocally, treatment of macaques with interleukin-2 (IL-2) and granulocyte colony stimulating factor (G-CSF) before infection led to depletion of Th17 cells, reduction of the ratio between Th17 cells and CD3+CD4+CD25+CD127low regulatory T cells (Tregs), and higher viral loads for six months after infection. These results demonstrate that the composition of the host immune system before infection has an influence on the course of disease after infection. Furthermore, to the extent that this influence shapes and interacts with T-cell-mediated responses to virus, our findings provide a new framework for understanding inter-individual variation in responses to therapies and vaccines against HIV.
Introduction
Th17 cells are CD4+ T cells that secrete the cytokine IL-17 upon stimulation (1). Th17 cells are thought to be important for maintenance of mucosal barriers because they home to intestinal tissue (2, 3), promote neutrophil recruitment by enhancing chemokine expression (e.g., that of CXCL1; 4, 5), and secrete IL-17, which contributes to strengthening mucosal epithelial tight junctions (6). In HIV and SIV infections, the intestinal mucosal barrier is compromised, allowing for translocation of bacterial products across the mucosal barrier and resulting in generalized T cell activation (7) that is associated with poor clinical outcomes (8). Recent studies have demonstrated that SIV and HIV infections lead to Th17 cell depletion, that this depletion is associated with increased permeability of the intestinal epithelium to bacteria and with induction of indoleamine 2,3-dioxygenase, and that the extent of Th17 cell depletion is predictive of disease progression (9–11).
Th17 and regulatory T cells (Tregs) develop from a common progenitor and have been shown to play opposing roles in murine models of inflammation and autoimmunity (1). For example, mice whose T cells produced very high levels of IL-17 after immunization with myelin oligodendrocyte glycoprotein peptide developed the most severe autoimmune encephalitis, whereas adoptive transfer of peptide-specific Tregs suppressed disease (12, 13). Similar associations between Th17 cells and inflammation have been noted in humans (13). By contrast, chronically HIV-infected individuals show signs of widespread T cell activation, but the frequency of Th17 cells is decreased and the proportion of Tregs is increased compared with uninfected controls (14). These observations suggest that HIV disease progression is facilitated by increased intestinal epithelial permeability (perhaps due to loss of IL-17; 15) and/or suppressed antiviral immune responses (caused at least in part by increased Treg activity; 16). We now show that the substantial inter-individual variability observed in the outcome of lentiviral infection (17) is related to inter-individual variation in the baseline Th17 cell compartment present at the time of infection with virus.
Results
Inter-individual variability in Th17 and Treg populations
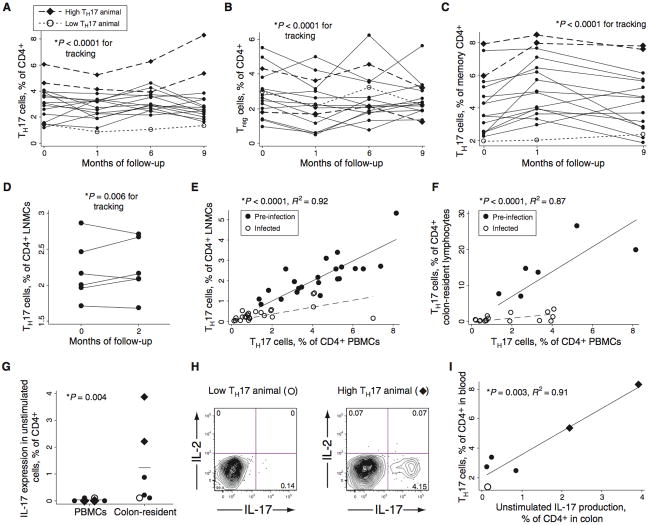
We examined the frequency of Th17 and Treg cells in the peripheral blood of 16 rhesus macaques over a period of nine months. This frequency was found to vary substantially (over 5-fold) between individuals but to remain stable within a given individual over time (Figure 1A). The “tracking” shown in the figure indicates significant longitudinal stability (p<0.0001; see ref. 18); in addition, the average inter-animal coefficient of variation (CV; 43%) was twice the average intra-animal CV (23%). Three of 16 individuals were statistical outliers (plotted in Figure 1A with diamonds or open circles; see legend for statistics): two demonstrated Th17 cell frequencies consistently higher than those of other animals whereas one demonstrated a lower frequency. Assessment of Treg frequencies also revealed substantial variation between individuals but stable frequencies within individuals over time (Figure 1B). There was no consistent trend to an inverse relationship between the frequency of Tregs and Th17 cells (Figure S1A). Variability in Th17 cells reflected different proportions within the memory cell compartment (CD3+CD4+CD45RA−), rather than differences in the overall frequency of memory cells. Thus, the fraction of Th17 cells among memory CD4+ T cells showed similar variability to the fraction of Th17 cells among all CD4+ T cells (Figures 1A and C; coefficients of variation are 42% and 44%, respectively). Th17 cell frequency among all CD4+ T cells was not correlated with the frequency of CD4+ memory cells (p = 0.22, R2=0.08; Figure S1B), but was strongly correlated with Th17 cell frequency among memory CD4+ T cells (p = 0.0003, R2=0.59; Figure S1C). Finally, we evaluated the stability of Th17 cell populations in lymph nodes, using two biopsies taken from each of six animals two months apart (Figure 1D). This experiment revealed stable frequencies within tissue samples from individuals over time (p=0.006).
Fig. 1.
Macaques display substantial inter-individual variability in Th17 cell populations. (A) Th17 cells in peripheral blood, as assessed by the fraction of cells containing intracellular IL-17 after stimulation with PMA and ionomycin in the presence of brefeldin A. Diamond symbols represent animals with unusually large Th17 populations, as defined below; open circles represent one animal with an unusually small Th17 population, also as defined below. These symbols are used for these same three animals throughout Figure 1. The p value shown for tracking (longitudinal stability of a certain measurement in an individual over time) was calculated using the procedure outlined by Twisk (18) and detailed in Methods. At a minimum of three out of four time points, the three animals marked with diamonds and open circles had Th17 cell levels that were outside the 99.5% confidence interval for the group mean. (B) Tregs in the peripheral blood, as assessed by flow cytometric detection of cells that were CD3+CD4+CD8−CD25+CD127low (see Figure S3C and refs. 16 and 41). (C) Th17 cells in peripheral blood as a fraction of CD4+ memory T cells (CD45RA−). (D) Th17 cells in CD4+ LNMC of six animals, sampled on two occasions that were two months apart. (E) Relationship between circulating and lymph node Th17 cell populations in uninfected and SIV-infected animals (filled and open circles, respectively). The regression lines, p value, and coefficient of determination shown are for one linear regression that accommodates the different slopes observed before and after infection; pre- and post-infection data were both included in the statistical analysis. Separate analyses on pre- and post-infection data also yielded significant correlations (p < 0.0001 in each case; R2=0.93 and 0.59). (F) Relationship between circulating and colon-resident Th17 cell populations in uninfected and SIV-infected animals (filled and open circles, respectively). Separate analyses on pre- and post-infection data also yielded significant correlations (p < 0.01 and p < 0.001, respectively; R2=0.88 and 0.53). (G) IL-17 expression in unstimulated CD4+ cells in PBMC or colon-resident lymphocyte populations, as assessed by intracellular cytokine staining and flow cytometry. The p value is from the Mann-Whitney U test. (H) Flow cytograms demonstrating IL-17 expression in unstimulated CD4+ T cells from colonic biopsy tissue. (I) Relationship between IL-17 production in unstimulated colon lymphocytes and in PBMC stimulated with PMA/ionomycin. The p value is for linear regression (p=0.003, R2=0.91).
In HIV and SIV infection, Th17 cells are depleted from blood, lymph node, and gut tissues (9–11). It is not known, however, if the circulating population is a transient subset that reflects the size of tissue-resident Th17 populations or if blood and tissue populations are distinct. To distinguish between these possibilities, lymph node and colon biopsies were taken from macaques both before and after SIV infection. Lymph node- and colon-resident Th17 cell populations were found to be strongly positively correlated with the circulating population, both before and after infection (Figures 1E and F; p < 0.0001 for both lymph node and colon tissue). In SIV infection, Th17 cells were preferentially depleted from both lymph node and colon tissues as compared to blood, reducing the slope of the observed relationship (Figures 1E and F, dotted lines). Nonetheless, these data show clearly that circulating Th17 cells are correlated with tissue-resident populations.
We next tested for IL-17 expression among unstimulated peripheral blood and colon-resident lymphocytes, incubating peripheral or colon-resident cells with brefeldin A immediately after isolation (Figures 1G and H). Surprisingly, we found that up to 4% (mean, 1.24%) of colon-resident CD4+ T cells expressed IL-17 without stimulation, whereas only ~0.05% of peripheral blood or lymph node CD4+ T cells expressed IL-17 under these conditions (Figure 1G). Nevertheless, the size of the stimulated peripheral Th17 cell population correlated with the fraction of unstimulated, colon-resident cells expressing IL-17 (Figures 1H and I; p=0.003). The gross physical and histological appearances of colon tissue from low-Th17 and high-Th17 animals were equivalent and unremarkable (Figure S2A).
Influence of pre-existing Th17 cells on the subsequent course of SIV infection
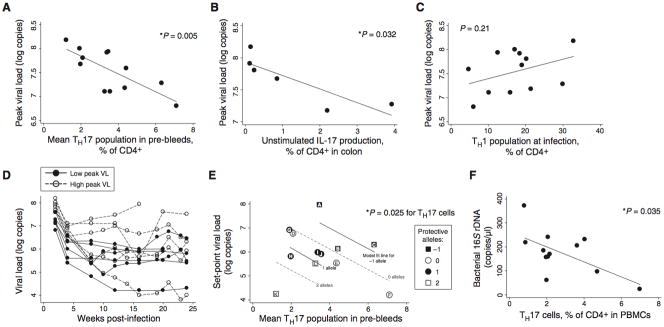
Twelve macaques with a broad range in Th17 cell populations (including the two animals with a larger pre-existing fraction of Th17 cells and the one animal with a smaller pre-existing fraction in Figure 1A) were infected with SIVmac251 (50–100 TCID50 IV), chosen because it is a very virulent uncloned isolate used extensively by us and other investigators. The size of the Th17 cell population observed before infection was found to be highly predictive of viral loads at early time points after infection (within the first month; Figure 2A; p=0.005; R2 = 0.56). Animals with higher fractions of Th17 cells demonstrated peak viral loads that were lower by one log unit for each 5.4% increment in Th17 cells. The Th17:Treg ratio and the absolute number of circulating Th17 cells were also correlated with peak viral load, although not more so than the circulating Th17 population as a fraction of CD4+ T cells (Figures S2B and S2C). Baseline IL-17 production in colon tissue, assessed by brefeldin A treatment of unstimulated colon lymphocytes in a subset of six animals, was also significantly associated with peak viral loads (Figure 2B; p = 0.032; R2 = 0.72). The relationship between pre-existing Th17 cell levels and viral loads lessened later in infection (exhibiting higher p values and lower coefficients of determination), presumably as other viral and host factors became more important (Figure S2D). Various other CD4+ T cell subsets demonstrated no consistent relationship with viral load, including Th1 cells (expressing IFN-γ but not IL-17 on stimulation; Figures 2C and S2E), Th2 cells (expressing IL-4), naïve cells (CD45RA+CD27+), central memory cells (CD45RA−CD27+), effector memory cells (CD45RA−CD27−), CCR5+ cells, and Ki-67+ cells (Figure S2F; p > 0.2 in all cases). Numbers of circulating neutrophils and monocytes were also not predictive of viral load (Figure S2G; p = 0.90 and 0.24, respectively).
Fig. 2.
Macaques with large pre-existing Th17 compartments develop lower viral loads. (A) The Th17 population measured in pre-bleeds is predictive of peak viral loads, reached two weeks after infection. The relationship remains significant when any single animal is eliminated from the analysis. (B) Production of IL-17 by unstimulated, colon-resident lymphocytes is also predictive of peak viral loads. Biopsies were taken before infection in six animals that were followed in Figure 1 (see Figure 1G). (C) The Th1 population (CD3+CD4+IFN-γ+IL-17−) present before infection was not predictive of viral loads. (D) Animals with a high peak viral load (peak viral load above the median; shown with open circles and dashed lines) did not always develop a high set-point; animals with a low peak viral load (below the median; closed circles and solid lines) did not always develop a low set point. (E) Th17 cells and MHC alleles were jointly predictive of set-point viral loads. Multiple regression of set-point viral loads against Th17 cells and protective MHC alleles (count of alleles; see Table S1) demonstrated significant predictive value overall (p=0.023 for multivariate F-test) and for each variable (p=0.025 and 0.009 for Th17 cells and MHC alleles, respectively). Letter labels provide a cross reference to MHC allele information detailed in Table S1. (F) The percentage of Th17 cells among CD4+ T cells is a significant predictor of bacterial 16S rDNA at 2 weeks after infection.
Set-point viral loads were influenced by MHC alleles, as has been previously reported (19, 20), and by Th17 cell populations. A plot of viral loads over time demonstrates that animals with lower peak viremia (Figures 2D and S2H, black dots and solid lines) often but not always experienced lower set-point viral loads, whereas those with higher peak viremia (open dots and dashed lines) sometimes experienced low set points. Neither the presence of “protective” MHC alleles (assessed with a simple scoring system; see Table S1) nor the percentage of Th17 cells present before infection were individually predictive of set-point viral loads. Two-variable regression analysis showed, however, that these two factors jointly predicted set-point viremia (Figure 2E; p = 0.023 overall, 0.025 for Th17 cells present before infection, and 0.009 for MHC allele score; adjusted R2 = 0.47). The apparent impact of Th17 cells on peak viral loads, independent of MHC (Figure 2A), is consistent with literature showing no effect of MHC alleles on acute phase viremia (21). The animals were genotyped for TRIM5 alleles but these genotypes were not predictive of peak or set-point viral loads, either alone or in combination with Th17 cells (Figure S2I).
To better understand the relationship between an abundant pre-existing Th17 cell population and lower viral loads after infection, we explored parameters reflective of bacterial translocation and T cell activation. Lower levels of bacterial 16S rDNA were circulating in animals with high fractions of Th17 cells or high Th17:Treg ratios (Figures 2F and S2J, respectively). In this 6-month experiment, however, there was no significant relationship between circulating bacterial 16S rDNA levels and generalized T cell activation, as assessed by Ki-67 expression in CD4+ and CD8+ T cells (Figure S2K). Furthermore, T cell activation within the time frame of this experiment did not generally predict viral load (Figure S2L).
Experimental modulation of Th17 and Treg populations with IL-2 and G-CSF
Although the above experiments demonstrated that a large pre-existing Th17 cell compartment is associated with lower peak and set-point viral loads, it remained unclear if Th17 cells were the cause of reduced virus replication. We accordingly designed an interventional study to alter the Th17 cell compartment in rhesus macaques before SIV infection. Interleukin-2 (IL-2) has been shown to inhibit the differentiation of Th17 cells in vitro and also to expand CD25-expressing cells, many of which are Tregs (22, 23). G-CSF has also been shown to expand or mobilize the Treg population in humans (24). We anticipated that long-term IL-2 and/or G-CSF treatment would simultaneously inhibit differentiation or maintenance of Th17 cells and expand Tregs, thereby decreasing both the fraction of Th17 cells and the Th17:Treg ratio. After cessation of drug treatment, treated animals with altered Th17 and Treg populations could then be infected with SIV. We hypothesized that a reduction in the fraction of Th17 cells and of the Th17:Treg ratio would lead to higher viral loads among treated animals than would have been otherwise expected.
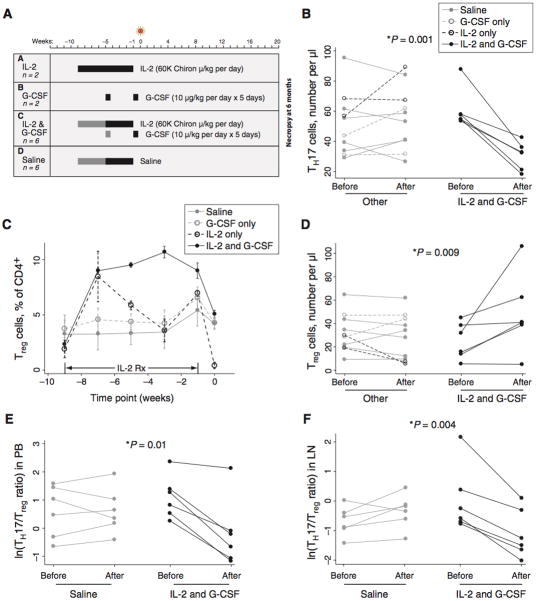
Juvenile rhesus macaques were assigned to four treatment groups (Figure 3A), including those treated with: (A) IL-2 alone, administered daily for eight weeks; (B) G-CSF alone, administered in two five-day courses separated by four weeks; (C) both agents together, administered in one or two consecutive treatment courses; or (D) saline administered daily for four to eight weeks. The fraction and number of T cells belonging to various subsets were then followed before, during, and after treatment.
Fig. 3.
Treatment with IL-2 and G-CSF causes expansion of Tregs, depletion of Th17 cells, and reduction in the Th17:Treg ratio. (A) Rhesus macaques were treated with IL-2 alone, G-CSF alone, both agents together, or saline only. In Groups C and D, two animals were given two cycles of treatment (black and gray bars) and four animals were given one cycle (black bars only). All drugs were withdrawn before infection with SIV. (B) IL-2 and G-CSF treatment causes depletion of Th17 cells. The patterns shown in the legend to this panel are also used in panels C–F. The label “Post” indicates the day of infection; i.e., time 0 in Figure 3A. Note that data for two of the six IL-2 and G-CSF-treated animals in this panel overlap closely. The p value shown (0.001) is for a Mann-Whitney U test of change in Th17 cell number after IL-2 and G-CSF treatment (difference of “Post” and “Pre” values) versus change after other treatment; a Kruskal-Wallis test of differences in groups A–D was also significant (p=0.01). Data in panels B–D are from PBMC. (C) Substantial expansion of phenotypic Tregs in macaques treated with IL-2 alone (black dashed line) or IL-2 and G-CSF (black solid line). Tregs plotted in this panel were identified as CD3+CD4+CD25+CD127low. Lines represent mean values observed in animals treated for nine weeks. Data are mean +/− SE. (D) Expansion of Tregs in animals treated with IL-2 and G-CSF. This panel shows the overall change in Treg number between the start and end of treatment. The p value shown (0.009) is for a Mann-Whitney U test of change in Treg cell number after IL-2 and G-CSF treatment versus change after other treatment; a Kruskal-Wallis test of differences in groups A–D was also significant (p=0.02). (E) Reduction of the Th17:Treg ratio in treated animals. The p value indicates that the reduction in the Th17:Treg ratio occurring with treatment is significantly greater in the IL-2- and G-CSF-treated group (Mann-Whitney U test of change occurring in treated animals of Group C compared to saline-treated animals in Group D). (F) Reduction of the Th17:Treg ratio in lymph node tissue after IL-2 and G-CSF treatment. Mann-Whitney (IL-2 and G-CSF vs. other treatment) and Kruskal-Wallis (groups A–D) tests both yielded significant p values (p=0.02 in both cases).
All animals receiving IL-2 and G-CSF together (Group C) demonstrated a pronounced decline in the circulating Th17 cell population (Figure 3B; p=0.001; see figure legend for statistics), accompanied by expansion of Tregs (Figures 3C and D; p=0.009). The expanded Treg population was maintained at a high level throughout the period of IL-2 administration, declining only when IL-2 was withdrawn in the final week before infection (Figure 3C). Nonetheless, five of six macaques in this group had a higher fraction and absolute number of Tregs at the end of treatment than before treatment (Figures 3D and S3A). By contrast, animals receiving IL-2 or G-CSF alone (Groups A and B, respectively) did not show durable expansion of Tregs or reduction of Th17 cells. Animals receiving IL-2 alone, for instance, demonstrated dramatic expansion of the Treg population two weeks after initiation of treatment (~5 fold over baseline; Figure 3C, dashed black lines); however, the cells declined precipitously from that point forward. By day 0 of the experiment (one week after drug administration was discontinued and at the time SIV was inoculated), these animals had the smallest fraction and amongst the lowest numbers of Tregs in the cohort. Administration of IL-2 alone was also ineffective in depleting Th17 cells (Figure 3B, dashed black lines).
Changes in the absolute number of Tregs after IL-2 and G-CSF treatment (Figure 3D) were mirrored by percentage changes (Figure S3A). These observations, combined with a dramatic increase in Ki-67 expression among CD4+ T cells (Figure S3B), suggest that this population was expanding rather than simply redistributing into peripheral blood. Furthermore, the expanded cells demonstrated both FoxP3 expression and the suppressive function that is characteristic of Tregs (Figures S3C and D, respectively). For example, CD25+ cells isolated before the start of treatment or at the point of peak expansion suppressed CD4+ T cell proliferation to an equivalent degree (Figure S3D).
We summarized these concerted changes in Th17 cells and Tregs using the Th17:Treg ratio, a measure that we used previously to follow changes in these cells after SIV and HIV infection (10, 11). Examination of this ratio seemed especially appropriate in the current study because IL-2 and G-CSF treatment caused simultaneous changes in the two populations, resulting in an inverse relationship between them (Figure S3E; p=0.047 among treated animals). Analysis of animals treated with either saline alone (n=6) or IL-2 and G-CSF together (n=6) showed that reduction in the Th17:Treg ratio in treated animals was statistically significant (Figure 3E; p=0.01). IL-2- and G-CSF-treated animals demonstrated parallel changes in lymph node tissue, including expansion of Tregs, decline in Th17 cells, and reduction in the Th17:Treg ratio (Figure 3F and S3F).
Finally, we evaluated neutrophil counts, cellular receptors used for viral entry, and CD8+ T cells to determine whether any effects of IL-2 and G-CSF treatment might be mediated by changes in these parameters. In all cases, blood was analyzed three days after completion of a five-day course of G-CSF. Consistent with the known dynamics of neutrophils after G-CSF administration (Figure S3G; see also ref. 25), neutrophil counts remained only modestly elevated three days after an initial course of G-CSF and were not significantly elevated three days after a second course (Figure S3G). By the time of infection with SIV, neutrophil counts were not significantly higher in IL-2- and G-CSF-treated animals with reduced Th17:Treg ratios than in saline-treated controls (Figure S3H). Neither the frequency of CD4- or CCR5-positive cells nor the expression level (mean fluorescence intensity on positive cells) for these markers was different in IL-2/G-CSF-treated animals as compared with untreated animals at the time of virus inoculation (Fig. S3I). The frequency and function of CD8+ T cells were also not different in treated vs. untreated animals (Fig. S3I).
Effect of changing the Th17:Treg ratio on SIV viral load
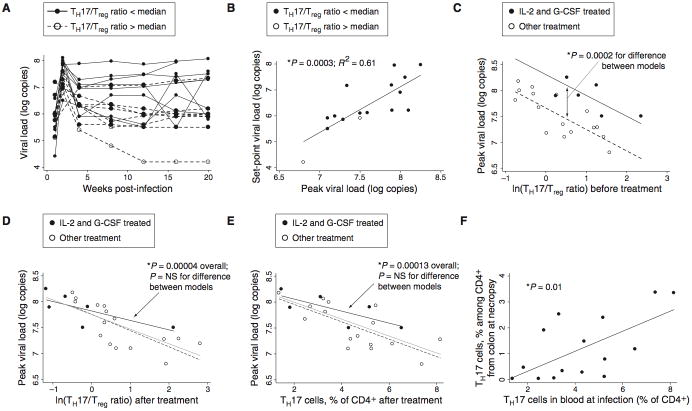
After cessation of all drug treatment (seven days after cessation of IL-2 and three days after cessation of G-CSF), animals in the interventional study were infected intravenously with SIVmac251 (Figure 4A; note that saline-treated animals from this study are included in Figure 2). Viral loads reached an average peak of 3.9 × 107 copies/ml two weeks after infection and declined to set points averaging 2.6 × 106 copies/ml. In this experiment, there was excellent correlation between peak and set-point viral loads, defined as the geometric mean viral load observed at or after 12 weeks (Figure 4B; p=0.0003, R2=0.61 for regression of log set-point copies vs. log peak copies). This finding suggests that much of the variability in disease course was determined by mechanisms operative within the first two weeks of infection.
Fig. 4.
Reduction of the Th17:Treg ratio leads to higher SIV viral loads. (A) Viral loads in infected animals from Groups A–D of Figure 3A (n=16). Macaques with high Th17:Treg ratios at the time of infection (the eight animals above the median value) are shown with dashed black lines; those with low Th17:Treg ratios (below the median) are shown with solid lines. (B) Peak viral load accurately predicts set-point viral load (p=0.0003 by linear regression, R2=0.61). (C) Viral loads observed in animals treated with IL-2 and G-CSF were higher than predicted based on Th17:Treg ratios measured before treatment. Viral loads in treated animals lie above the regression line for animals in other groups. The p value shown was calculated using a model including a dummy variable for animals receiving IL-2 and G-CSF; the coefficient for this variable was statistically significant (p=0.0002) and estimated at 0.6 log units, indicating that the intercept for a regression line appropriate to treated animals lies 0.6 log units above that for other animals. Values are plotted for six IL-2- and G-CSF-treated animals (filled markers) and 16 other animals (open markers) from Figure 3A (groups A, B, and D) and Figure 1G. The difference shown remains significant when any of these groups is removed from the analysis (p < 0.005 in all cases). (D) The Th17:Treg ratio established after treatment is correlated with the viral load that will develop after infection. Linear regression of peak viral load against the log Th17:Treg ratio, for all animals, indicates a significant trendline overall (p=0.00004). Use of a dummy variable for IL-2 and G-CSF treatment, as in Figure 4C, shows no significant difference between trendlines for treated and untreated animals (p=NS), suggesting that the Th17:Treg ratios established by treatment explain viral loads in treated animals. (E) The fraction of Th17 cells among CD4+ T cells correlates with viral load. Trendlines and statistical calculations are as in panel D. (F) Th17 cells in blood at the time of infection accurately predict the percentage of Th17 cells recovered from colon tissue six months later.
We tested whether viral loads in treated animals were best predicted by the Th17:Treg ratio measured before or after treatment with IL-2 and G-CSF. We reasoned that, if the association between higher Th17:Treg ratios and lower viral loads was causal in nature, then IL-2/G-CSF-mediated losses in Th17 cells should lead to higher viral loads; that is, the viral loads observed in treated animals should be higher than predicted by their pre-treatment Th17 cell populations. Indeed, a plot of pre-treatment Th17:Treg ratio against peak viral load shows that data from the group of animals treated with IL-2 and G-CSF lie significantly above the regression line established by other animals, indicating higher viral load (Figure 4C; p=0.0002; estimated increase of 0.6 log units). The Th17:Treg ratio established after IL-2 and G-CSF treatment, however, accurately predicted peak and set-point viral loads (Figures 4D and S4A), as did the fraction of Th17 cells (Figure 4E). The Th17-viral load relationship observed after IL-2 and G-CSF treatment was consistent with that found in animals not receiving IL-2 and G-CSF (Figures 4D and E). Absolute neutrophil counts were not significant predictors of peak viral load, whether tested alone or in models including Th17 cells or the Th17:Treg ratio (Figure S4B). Measures of immune activation in CD4+ T cells (HLA-DR+, CD38+, HLA-DR+CD38+, or Ki-67+) were not significantly greater in treated animals by the time of infection, one week after cessation of treatment, and also were not predictive of increased viral loads (Figure S4C). We conclude from these findings that IL-2 and G-CSF treatment most likely altered the outcome of infection by changing the Th17 and/or Treg compartments.
We also evaluated the gut-resident Th17 population at necropsy, six months after infection. Lymphocytes were isolated from tissue by collagenase digestion and then stimulated with PMA and ionomycin to induce IL-17 expression by Th17 cells. We found that the fraction of Th17 cells present in the peripheral blood at the time of infection was positively correlated with the fraction of Th17 cells found to be preserved in either colon (Figures 4F and S4D; p=0.01 and p=0.026, respectively) or ileum (Figure S4E; p=0.006) at necropsy.
The Th17:Treg ratio offered a convenient way for us to follow the joint effects of IL-2 and G-CSF on these two populations; however, as in the experiment shown in Figure 2, the fraction of Th17 cells among CD4+ T cells before infection was itself predictive of peak and set-point viral loads (Figures 4E and S4F). Furthermore, set point viral loads were again jointly influenced by Th17 cells and MHC allele inheritance. A linear regression model testing the relationship between set-point viral load and the fraction of Th17 cells at infection was improved by addition of the MHC score described above (Table S1), with the p value declining from 0.005 to 0.002 and adjusted R2 increasing from 0.40 to 0.55.
Effect of changing the Th17:Treg ratio on T cell activation and antiviral immune responses
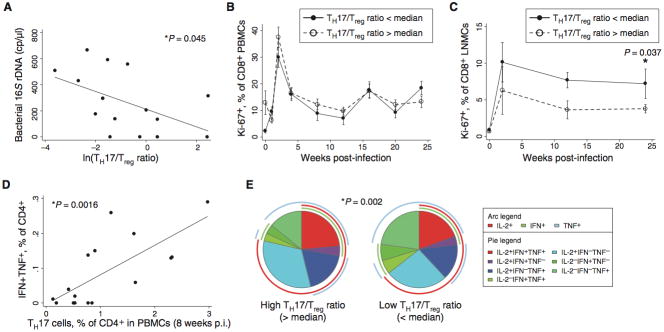
To better understand the mechanism by which Th17 cells affected viral load, we also followed 16S rDNA levels, generalized T cell activation as measured by Ki-67 levels, and virus-specific CD4+ T cell responses throughout the period of infection. Consistent with the trend seen in the observational study above, significantly lower 16S rDNA levels were observed in animals with high Th17:Treg ratios, both two weeks and eight weeks after infection (Figure S4G and Figure 5A). Among peripheral blood cells, this difference in microbial translocation did not result in lower generalized T cell activation, as assessed using Ki-67 (Figure 5B). However, T cell proliferation in lymph node cells was significantly lower in animals with high Th17:Treg ratios (Figure 5C). Furthermore, animals with high levels of Th17 cells had higher SIV-specific CD4+ and CD8+ T cell responses than those seen in animals with lower levels of Th17 cells (Figures 5D, S4H, and S4I). The robust CD4+ T cell responses observed in animals with high Th17:Treg ratios were also more highly functional, with a significantly greater proportion of trifunctional cells and cells expressing IL-2 (Figure 5E; p=0.002 for difference between pies, p=0.002 for t-test of difference in proportion of trifunctional cells, and p<0.001 for difference in IL-2-expressing cells).
Fig. 5.
Individuals with larger Th17 populations displayed reduced T cell activation and superior antiviral immune responses. (A) Bacterial 16S rDNA levels in plasma, assessed by real-time PCR, are correlated with the log Th17:Treg ratio at 8 weeks after infection. (B) Expression of Ki-67 in CD8+ T cells indicates a trend to lower T cell proliferation six months after infection, but no statistically significant difference between high Th17:Treg ratio and low Th17:Treg ratio groups. Data are mean +/− SE. (C) Animals with high Th17:Treg ratios before infection have reduced T cell activation in lymph node tissue. The p value shown indicates a significant difference between high- and low-Th17 groups at the 24-week timepoint, calculated using the Mann-Whitney U test. Data are mean +/− SE. (D) More virus-specific T cells develop in animals with more abundant Th17 cells after infection. Virus-specific T cells were measured by intracellular cytokine flow cytometry after stimulation with AT2-inactivated SIV virus. At 8 weeks after infection, animals demonstrating higher Th17 cells also demonstrated more robust antiviral T cell responses. (E) Virus-specific CD4+ T cells in high-Th17 animals are more highly functional (i.e., secrete a greater number of different cytokines) and secrete more IL-2 than CD4+ T cells in low-Th17 animals. The p value shown was calculated using a Student’s t test and a partial permutation test as described (40).
Discussion
The development and composition of the mucosal immune system has previously been invoked to explain local damage by opportunistic pathogens, repair of normal damage to the mucosal barrier, and susceptibility to systemic autoimmune conditions. For instance, microbial colonization of mice triggers production of RegIIIγ, a bactericidal lectin that mediates the early protective effects of IL-22 against Citrobacter rodentium, an attaching and effacing bacterial pathogen of mice (26, 27). More recently, Mazmanian et al. showed that presence of the commensal organism, Bacteroides fragilis, protects mice from experimental colitis induced by Helicobacter hepaticus (28), and we have shown that Trichuris trichiura colonization of the intestine can alleviate symptomatic ulcerative colitis (29). Because of these examples, as well as published evidence of the role of Th17 depletion in SIV and HIV disease pathogenesis (9–11), we wondered whether there might be substantial individual variability in intestinal immune cell populations at baseline that would, after acute lentiviral infection, affect the inflammatory stance of the host immune system and the subsequent course of disease.
Our data provide compelling evidence that the Th17 cell compartment varies widely between individual macaques and that, upon SIV infection, variation in the Th17 cell compartment may have an important influence on bacterial translocation and virus replication. Both observational (Figures 1 and 2) and prospective interventional (Figures 3–5) studies show that Th17 cells are correlated with protection against SIV virus replication, acutely and for months after infection. Additional cohort studies in humans will be needed to know if high levels of Th17 cell can reliably predict lower HIV viral loads. In addition to their effects on Th17 cells, IL-2 and G-CSF have effects on the proliferation and function of other immune cells. We attempted to control for these effects by discontinuing drug treatment before infection and by collecting data on cell populations of greatest interest (such as neutrophils and Tregs). However, it remains possible that lingering effects of these cytokines on the replication, activation, or migration of other cell populations contribute to higher viral loads. In the future, it will be of interest to identify drugs with more specific effects on Th17 cells.
The influence of Tregs on disease progression remains uncertain. In our observational study, Treg numbers were not associated with either viral load or the size of the Th17 cell population; however, after treatment with IL-2 and G-CSF, the Treg population was inversely correlated with the Th17 population and directly correlated with viral load (Figures S3E and S5A). Furthermore, in the interventional study, there was an association between higher Treg levels and lower generalized T cell activation at early time points, although this did not translate into a viral load benefit or into lower T cell activation levels at later time points (Figures S5A–C). Therefore, it is possible that the expanded or induced Tregs had a larger influence on course of disease than did Tregs present in untreated animals.
Th17 cells and MHC alleles were both influential in determining set-point viral loads. The importance of MHC alleles has been repeatedly demonstrated (21, 30, 31), but immunologic correlates of protection that must mediate MHC effects have not been conclusively identified. We propose that the status of Th17 cells in the infected host may be a key determinant of whether certain MHC alleles can exert their protective effects. Similarly, an adequate Th17 cell population may be important for deriving maximal benefit from prophylactic vaccine strategies. A sufficient complement of Th17 cells might allow antiviral T cell responses to control virus by conferring either a kinetic or functional advantage to host immune responses. A kinetic advantage would consist of establishing conditions within the intestine that slow viral replication sufficiently to allow control before extensive spread and generalized immune activation occur, as suggested by lower peak viral loads (Fig. 2A). A functional advantage would be conferred by establishment of conditions in which highly functional antiviral immune responses could be generated, perhaps because of reduced immune activation (Fig. 5E). It will be interesting to determine whether this hypothesis can explain the impressive but variable protection afforded by new experimental vaccines (32).
Considering the existing literature on Th17 cells, it is reasonable to ascribe their activity against SIV replication to effects on the mucosal epithelium of the gut (7, 10, 15). It has become increasingly clear that (i) mucosal surfaces are important not only in the transmission but also in the pathogenesis of lentiviral infections and (ii) development of the immune system at mucosal surfaces is heavily shaped by interaction with the outside world. Our study synthesized these findings and tested directly how the status of the mucosal immune system at the time of infection may impact upon the pathogenesis of disease. We found that circulating and intestinal Th17 cell populations were associated with each other and with lower viral loads, whether Th17 cells were assessed as a fraction of CD4+ T cells or as a ratio with CD25+ Tregs. Furthermore, circulating 16S rDNA was reduced in high Th17:Treg animals, although 16S rDNA itself was not a superior predictor of viral load.
It is interesting that SIV has such dramatic effects on gut-resident lymphocytes, particularly Th17 cells, regardless of whether the route of infection is intravenous (as in this study and ref. 10) or intrarectal (33). Microbial translocation has also been demonstrated following infection by either route. Therefore, despite clear kinetic differences between intravenous and intrarectal infections (34), it seems likely that lymphocytes of the intestinal mucosa provide a uniquely appropriate and perhaps required substrate for pathogenesis of lentiviral infection. It will be interesting to see whether the effects of altered Th17 cell compartments are different after intrarectal inoculation. One might imagine that the effects of small alterations in these cells, such as those reported here, would have greater impact in the face of a less overwhelming infection.
We also found that animals with high levels of Th17 cells generate more polyfunctional CD4+ T cells, including those expressing IL-2, than do animals with fewer Th17 cells. Because such polyfunctional cells have been shown to be more numerous in human controllers of HIV replication (35–37), Th17 cells may protect against SIV replication by promoting an environment in which polyfunctional cells can be produced. Further analysis of these parameters at early time points after infection should be useful in defining where Th17 cells exert their effects and via which mechanisms.
We have demonstrated here that macaques with large pre-existing Th17 cell compartments experience lower viral loads after SIV infection and that the Th17:Treg ratio can be manipulated to produce higher viral loads than would be otherwise expected. These observations raise a number of important questions: is there sufficient variability in human Th17 cell numbers to account for inter-individual differences in HIV replication and disease progression? Can Th17 compartment expansion be achieved in vivo and, if so, will it have a measurable effect on HIV replication in acute or chronic infection? To this end, it will be important to establish practical interventions to achieve such expansion.
Materials and Methods
Animals and infection
All rhesus macaques were housed in accordance with the regulations of the American Association for Accreditation of Laboratory Animal Care Standards (AAALAC) at the California National Primate Research Center (CNPRC). All animals were negative for HIV-2 (serology), SIV (serology), type D retrovirus (serology and PCR), and simian T-cell lymphotropic virus type 1 (serology). SIV-infected animals were housed in the infectious housing unit in pairs of two animals per cage; SIV-naïve monkeys were housed in outdoor corrals. Animals shown in Figure 1 were 5.9 to 7.9 years old at infection (mean, 6.8 years); those in Figure 3 were 2.4 to 5.1 years old (mean, 4.0 years). Animals were MHC typed by the MHC Typing Core of the UW AIDS Vaccine Research Lab (Madison, WI). SIV infection was performed by intravenous inoculation of 1 ml of 50–100 TCID50 of SIVmac251. The virus stock was grown in rhesus macaque PBMC and obtained from the Analytical Resource Core at the CNPRC (internal reference number: 06/04).
IL-2 and G-CSF treatment
G-CSF (Neupogen; Amgen) and IL-2 (Proleukin; Chiron, Inc.) were obtained from the San Francisco General Hospital pharmacy. G-CSF (10 μg/kg/d) was given in subcutaneous 0.5 mL bolus injections, once a day for 5 consecutive days. The 5 day treatment regimen with G-CSF was repeated in some animals, as shown in Figure 2. IL-2 (60,000 Chiron Units/kg/d) was given in subcutaneous 0.5-mL bolus injections daily for four weeks or eight weeks, as shown in Figure 3.
Virus load measurement
Plasma samples were analyzed for viral RNA (vRNA) by a quantitative branched DNA (bDNA) assay (38). Virus load in plasma samples is reported as log10 vRNA copy numbers per ml of plasma. The detection limit of this assay is 125 vRNA copies.
Phenotypic characterization of lymphocyte populations
Lymphocytes were isolated from whole blood by centrifugation onto a Ficoll step gradient. The cells were then stained with LIVE-DEAD marker (aqua amine-reactive dye, Invitrogen) and up to three panels of antibodies. Panel I included antibodies specific for FoxP3 (eBioscience, clone PCH101, Pacific Blue- or APC-labeled), Ki-67 (BD Biosciences, clone B56, FITC-labeled), CD4 (NIH Nonhuman Primate Reagent Resource or Invitrogen, clone 19Thy-5D7, QDot 605-labeled), CD127 (BD Biosciences, clone hIL-7R-M21, PE-labeled), CD45RA (Beckman Coulter, clone 2H4LDH11LDB9, ECD-labeled), CD27 (eBioscience, clone O323, APC-Cy7-labeled), CD25 (BD Biosciences, clone MA-251, PE-Cy7-labeled), and CD3 (BD Biosciences, clone SP34-2, Alexa700-labeled). Panel II included antibodies specific for CD3 (BD Biosciences, clone SP34-2, Pacific Blue-labeled), CD8 (Invitrogen, clone 3B5, PE-Cy5.5-labeled), CD38 (NIH Nonhuman Primate Reagent Resource, clone OKT10, PE-labeled), HLA-DR (Beckman Coulter, clone Immu-357, ECD-labeled), CD45RA (Beckman Coulter, clone 2H4LDH11LDB9, PE-Cy7-labeled), CCR5 (BD Biosciences, clone 3A9, APC-labeled), CD4 (NIH Nonhuman Primate Reagent Resource or Invitrogen, clone 19Thy-5D7, QDot655-labeled), and CD27 (eBioscience, clone O323, APC-Cy7-labeled). Panel III included antibodies specific for CCR6 (BD Biosciences, clone 11A9, PE-labeled), CD3 (BD Biosciences, clone SP34-2, Pacific Blue-labeled), CD4 (NIH Nonhuman Primate Reagent Resource or Invitrogen, clone 19Thy-5D7, QDot655-labeled), CD8 (Invitrogen, clone 3B5, PE-Cy5.5-labeled), CCR4 (eBioscience, clone 1G1, APC-labeled), CD25 (BD Biosciences, clone MA-251, PE-Cy7-labeled), CD45RA (Beckman Coulter, clone 2H4LDH11LDB9, PE-Cy7-labeled), and CD27 (eBioscience, clone O323, APC-Cy7-labeled). Staining with panel I was carried out according to instructions from eBioscience accompanying the company’s FoxP3 antibody. Staining with panels II and III was performed according to standard extracellular staining protocols. Briefly, the cells were collected by centrifugation, resuspended in diluted antibody mixture, washed in PBS containing 2% FBS (staining buffer), and finally resuspended in staining buffer containing 0.5% paraformaldehyde (PFA).
Cytokine flow cytometry assays
Up to 106 cells were placed in control or stimulation wells and resuspended in 120 μl of complete medium (RPMI containing 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, and 5 μM 2′,3′-dideoxy-3′-thiacytidine). Anti-CD28 (clone 28.2) and anti-CD49d (clone 9F10) were added to all wells at a concentration of 2 μg/ml each antibody. Various stimuli were then added to the experimental wells and plates were transferred to the incubator. A peptide pool comprising peptides from p27 of SIVmac239 was used at a final concentration of 5 μg/ml each peptide (NIH AIDS Research & Reference Reagent Program). AT2-inactivated SIVmac239 and control microvesicles (kindly provided by Dr. Jeff Lifson, NCI) were used at a final concentration of 70 ng protein per ml of medium. PMA and ionomycin were used at concentrations of 50 ng/ml and 1 μg/ml, respectively. Two hours after addition of stimuli, brefeldin A (GolgiPlug; BD Biosciences) was added to each well according to the instructions of the manufacturer. Twelve hours after addition of GolgiPlug, samples were harvested by centrifugation and stained using fixable live-dead stain (Aqua, Invitrogen) as well as antibodies reactive to CD3 (BD Biosciences, clone SP34-2, Pacific Blue-labeled), CD8 (Invitrogen, clone 3B5, PE-Cy5.5-labeled), CD4 (NIH Nonhuman Primate Reagent Resource or Invitrogen, clone 19Thy-5D7, QDot 605-labeled), IL-17 (eBioscience, clone eBio64DEC17, Alexa488-labeled), IL-10 (Miltenyi, clone B-T10, PE-labeled), IFN-γ (BD Biosciences, clone B27, PE-Cy7-labeled), IL-2 (BD Biosciences, clone MQ1-17H12, APC-labeled), and TNF-α (BD Biosciences, clone MAb11, Alexa700, BD-labeled). Live-dead staining and surface staining for CD4 and CD8 were carried out before permeabilization. The cells were fixed and permeabilized using Cytofix/Cytoperm (BD Biosciences), washed in Perm Buffer (BD Biosciences), incubated with appropriate antibodies in Perm Buffer, washed, and resuspended in PBS containing 2% FBS and 0.5% PFA. We normally stimulated one million cells and collected ~400,000 events on the cytometer, among which ~50,000 were CD4+ T cells. Figure S6 shows the gating tree and typical raw data for the IL-17 and IFN-γ parameters.
In vitro suppression assays
Anti-CD3 antibody (250 ng in 50 μl PBS; clone SP34-2, BD Biosciences) was added to wells of 96-well, U-bottom plates. The plates were incubated for at least four hours and then washed three times with PBS before use. Responder cells were prepared by depletion of purified PBMC using anti-CD25-labeled paramagnetic microbeads and LD columns, according to the instructions of the manufacturer (Miltenyi Biotec). The cells were resuspended in PBS containing 5% FBS at concentrations from 0.5–10 × 106 per ml. One-ninth volume of diluted carboxyfluorescein succinimidyl ester (CFSE) stock (50 μM) was then added and the cell suspension inverted several times to mix. The cells were incubated for five minutes at room temperature, 10 volumes of PBS containing 5% FBS were added, and the cells were washed at least three times in the same solution. Finally, labeled responder cells were resuspended in medium (RPMI 1640 supplemented with 2 nM L-glutamine, 5 mM HEPES, and 100 U/ml penicillin/mg/ml streptomycin). Candidate Tregs were prepared by positive selection on MS columns. These cells were not labeled with CFSE. Ninety thousand responder cells were then added to antibody-coated wells of a 96-well plate alone or together with a variable number of candidate regulatory cells. Plates were placed in the incubator for five days before harvesting, staining, and analysis. Percent suppression was calculated using the formula: (percent CFSElow in depleted sample - percent CFSElow in “addback” sample containing regulatory cells) / (percent CFSElow in depleted sample).
Bacterial 16S rDNA amplification
Amplification of bacterial 16S rDNA was performed exactly as described in ref. 39.
Statistical analysis
Results were analyzed using Stata/SE, version 11.1. All p values shown are for two-tailed tests. The p values shown for tracking in Figures 1A and B were calculated using the procedure outlined by Twisk (18), in which the value of the tracked variable at the initial time point is used as a predictor in a multivariable regression model fitted using generalized estimating equations. The p value shown for tracking in Figure 1D was calculated using simple linear regression of the final fraction of Th17 cells against the initial fraction of Th17 cells. Analysis and presentation of distributions was performed using SPICE version 5.1, downloaded from http://exon.niaid.nih.gov (40). Comparison of distributions was performed using a Student’s t test and a partial permutation test as described (40).
Supplementary Material
Figure S1. Relationships between Th17 cells, Tregs, and memory T cells in uninfected macaques.
Figure S2. Infection parameters and immune cells in the observational study.
Figure S3. Effects of IL-2 and G-CSF on various immune cells.
Figure S4. Infection parameters and immune cells in the interventional study.
Figure S5. Relationship of Tregs to viral load and T cell activation.
Figure S6. Gating tree and typical raw data for the IL-17 and IFN-γ parameters.
Table S1. MHC alleles of animals in the observational study.
Acknowledgments
We thank Jana Broadhurst, Din Lin, Shakun Aswani, Juliet Easlick, Joseph Moore, Yongzhi Geng, Kartika Jayashankar, and the staff of Colony Research Services, Primate Pathology, Primate Medicine, and Clinical Laboratories at the California National Primate Research Center (CNPRC) for their expert technical assistance with these studies. We thank Dr. Jeff Lifson for providing AT2-inactivated SIVmac239 and control microvesicles.
Funding: This work was supported in part by NIH award OD000329 to J. M. McCune, NIH award K23 AI081540 to D. J. Hartigan-O’Connor, a Bill and Melinda Gates Foundation “Grand Challenges Exploration” award (#52094) to D. J. Hartigan-O’Connor, a Pilot Award from the CNPRC to D. J. Hartigan-O’Connor as part of a program supported by NIH award RR-00169, NIH award R21 DE016541 to K. Abel, NIH award K08 DK83334 to B. Kanwar, grant RR-00169 from the National Center for Research Resources (NCRR; a component of the NIH) to the CNPRC, and by the Harvey V. Berneking Living Trust. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. J.M.M. is a recipient of the NIH Director’s Pioneer Award Program, part of the NIH Roadmap for Medical Research, through grant DPI OD00329.
Footnotes
Author contributions: D.J.H.-O’C. and J.M.M. conceived and designed the experiments. D.J.H.-O’C. performed most in vitro experiments. K.A. and K.K.A.V.R. performed or supervised most in vivo experiments. B.K. performed colonic biopsy procedures and performed some experiments with gut tissue specimens. D.J.H.-O’C. analyzed the data. D.J.H.-O’C. wrote the paper with substantial input from J.M.M., K.A., B.K., and K.K.A.V.R. J.M.M. supervised the project.
Competing interests: The authors declare that they have no competing interests.
References and Notes
- 1.Kanwar B, Favre D, McCune JM. Th17 and regulatory T cells: implications for AIDS pathogenesis. Curr Opin HIV AIDS. 2010;5:151–157. doi: 10.1097/COH.0b013e328335c0c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cook DN, et al. CCR6 mediates dendritic cell localization, lymphocyte homeostasis, and immune responses in mucosal tissue. Immunity. 2000;12:495–503. doi: 10.1016/s1074-7613(00)80201-0. [DOI] [PubMed] [Google Scholar]
- 3.Singh SP, Zhang HH, Foley JF, Hedrick MN, Farber JM. Human T cells that are able to produce IL-17 express the chemokine receptor CCR6. J Immunol. 2008;180:214–221. doi: 10.4049/jimmunol.180.1.214. [DOI] [PubMed] [Google Scholar]
- 4.Kolls JK, Linden A. Interleukin-17 family members and inflammation. Immunity. 2004;21:467–476. doi: 10.1016/j.immuni.2004.08.018. [DOI] [PubMed] [Google Scholar]
- 5.Onishi RM, Gaffen SL. Interleukin-17 and its target genes: mechanisms of interleukin-17 function in disease. Immunology. 2010;129:311–321. doi: 10.1111/j.1365-2567.2009.03240.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Blaschitz C, Raffatellu M. Th17 cytokines and the gut mucosal barrier. J Clin Immunol. 2010;30:196–203. doi: 10.1007/s10875-010-9368-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brenchley JM, et al. Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med. 2006;12:1365–1371. doi: 10.1038/nm1511. [DOI] [PubMed] [Google Scholar]
- 8.Giorgi JV, et al. Shorter survival in advanced human immunodeficiency virus type 1 infection is more closely associated with T lymphocyte activation than with plasma virus burden or virus chemokine coreceptor usage. J Infect Dis. 1999;179:859–870. doi: 10.1086/314660. [DOI] [PubMed] [Google Scholar]
- 9.Brenchley JM, et al. Differential Th17 CD4 T-cell depletion in pathogenic and nonpathogenic lentiviral infections. Blood. 2008;112:2826–2835. doi: 10.1182/blood-2008-05-159301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Favre D, et al. Critical loss of the balance between Th17 and T regulatory cell populations in pathogenic SIV infection. PLoS Pathog. 2009;5:e1000295. doi: 10.1371/journal.ppat.1000295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Favre D, et al. Tryptophan catabolism by indoleamine 2,3-dioxygenase 1 alters the balance of TH17 to regulatory T cells in HIV disease. Sci Transl Med. 2010;2:32ra36. doi: 10.1126/scitranslmed.3000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bettelli E, Carrier Y, Gao W, Korn T, Strom TB, Oukka M, Weiner HL, Kuchroo VK. Reciprocal developmental pathways for the generation of pathogenic effector TH17 and regulatory T cells. Nature. 2006;441:235–238. doi: 10.1038/nature04753. [DOI] [PubMed] [Google Scholar]
- 13.Iwakura Y, Nakae S, Saijo S, Ishigame H. The roles of IL-17A in inflammatory immune responses and host defense against pathogens. Immunol Rev. 2008;226:57–79. doi: 10.1111/j.1600-065X.2008.00699.x. [DOI] [PubMed] [Google Scholar]
- 14.Schulze Zur Wiesch J, et al. Comprehensive Analysis of Frequency and Phenotype of T Regulatory Cells in HIV Infection: CD39 expression of FoxP3+ T regulatory cells correlates with progressive disease. J Virol. 2010 doi: 10.1128/JVI.01758-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raffatellu M, et al. Simian immunodeficiency virus-induced mucosal interleukin-17 deficiency promotes Salmonella dissemination from the gut. Nat Med. 2008;14:421–428. doi: 10.1038/nm1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hartigan-O’Connor DJ, Abel K, McCune JM. Suppression of SIV-specific CD4+ T cells by infant but not adult macaque regulatory T cells: implications for SIV disease progression. J Exp Med. 2007;204:2679–2692. doi: 10.1084/jem.20071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lyles RH, Munoz A, Yamashita TE, Bazmi H, Detels R, Rinaldo CR, Margolick JB, Phair JP, Mellors JW. Natural history of human immunodeficiency virus type 1 viremia after seroconversion and proximal to AIDS in a large cohort of homosexual men. Multicenter AIDS Cohort Study. J Infect Dis. 2000;181:872–880. doi: 10.1086/315339. [DOI] [PubMed] [Google Scholar]
- 18.Twisk JW, Kemper HC, van Mechelen W, Post GB. Tracking of risk factors for coronary heart disease over a 14-year period: a comparison between lifestyle and biologic risk factors with data from the Amsterdam Growth and Health Study. Am J Epidemiol. 1997;145:888–898. doi: 10.1093/oxfordjournals.aje.a009048. [DOI] [PubMed] [Google Scholar]
- 19.Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Maness NJ, et al. Comprehensive immunological evaluation reveals surprisingly few differences between elite controller and progressor Mamu-B*17-positive Simian immunodeficiency virus-infected rhesus macaques. J Virol. 2008;82:5245–5254. doi: 10.1128/JVI.00292-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loffredo JT, et al. Mamu-B*08-positive macaques control simian immunodeficiency virus replication. J Virol. 2007;81:8827–8832. doi: 10.1128/JVI.00895-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurence A, et al. Interleukin-2 signaling via STAT5 constrains T helper 17 cell generation. Immunity. 2007;26:371–381. doi: 10.1016/j.immuni.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 23.Weiss L, Letimier FA, Carriere M, Maiella S, Donkova-Petrini V, Targat B, Benecke A, Rogge L, Levy Y. In vivo expansion of naive and activated CD4+CD25+FOXP3+ regulatory T cell populations in interleukin-2-treated HIV patients. Proc Natl Acad Sci U S A. 2010;107:10632–10637. doi: 10.1073/pnas.1000027107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zou Barnett, Safah Larussa, Evdemon-Hogan Mottram, Wei David, Curiel Zou. Bone Marrow Is a Reservoir for CD4+CD25+ Regulatory T Cells that Traffic through CXCL12/CXCR4 Signals. Cancer Res. 2004;64:8451–8455. doi: 10.1158/0008-5472.CAN-04-1987. [DOI] [PubMed] [Google Scholar]
- 25.van Der Auwera P, Platzer E, Xu ZX, Schulz R, Feugeas O, Capdeville R, Edwards DJ. Pharmacodynamics and pharmacokinetics of single doses of subcutaneous pegylated human G-CSF mutant (Ro 25–8315) in healthy volunteers: comparison with single and multiple daily doses of filgrastim. Am J Hematol. 2001;66:245–251. doi: 10.1002/ajh.1052. [DOI] [PubMed] [Google Scholar]
- 26.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zheng Y, Valdez PA, Danilenko DM, Hu Y, Sa SM, Gong Q, Abbas AR, Modrusan Z, Ghilardi N, de Sauvage FJ, Ouyang W. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat Med. 2008;14:282–289. doi: 10.1038/nm1720. [DOI] [PubMed] [Google Scholar]
- 28.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 29.Broadhurst MJ, Leung JM, Kashyap V, McCune JM, Mahadevan U, McKerrow JH, Loke P. IL-22+ CD4+ T cells are associated with therapeutic trichuris trichiura infection in an ulcerative colitis patient. Sci Transl Med. 2010;2:60ra88. doi: 10.1126/scitranslmed.3001500. [DOI] [PubMed] [Google Scholar]
- 30.Giraldo-Vela JP, et al. The major histocompatibility complex class II alleles Mamu-DRB1*1003 and -DRB1*0306 are enriched in a cohort of simian immunodeficiency virus-infected rhesus macaque elite controllers. J Virol. 2008;82:859–870. doi: 10.1128/JVI.01816-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mothe BR, Weinfurter J, Wang C, Rehrauer W, Wilson N, Allen TM, Allison DB, Watkins DI. Expression of the major histocompatibility complex class I molecule Mamu-A*01 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2003;77:2736–2740. doi: 10.1128/JVI.77.4.2736-2740.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen SG, et al. Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature. 2011;473:523–527. doi: 10.1038/nature10003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nigam P, Kwa S, Velu V, Amara RR. Loss of IL-17-producing CD8 T cells during late chronic stage of pathogenic simian immunodeficiency virus infection. J Immunol. 2011;186:745–753. doi: 10.4049/jimmunol.1002807. [DOI] [PubMed] [Google Scholar]
- 34.Pauza CD, Emau P, Salvato MS, Trivedi P, MacKenzie D, Malkovsky M, Uno H, Schultz KT. Pathogenesis of SIVmac251 after atraumatic inoculation of the rectal mucosa in rhesus monkeys. J Med Primatol. 1993;22:154–161. [PubMed] [Google Scholar]
- 35.Emu B, Sinclair E, Favre D, Moretto WJ, Hsue P, Hoh R, Martin JN, Nixon DF, McCune JM, Deeks SG. Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J Virol. 2005;79:14169–14178. doi: 10.1128/JVI.79.22.14169-14178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pereyra F, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J Infect Dis. 2008;197:563–571. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 37.Porichis F, Kaufmann DE. HIV-specific CD4 T cells and immune control of viral replication. Curr Opin HIV AIDS. 2011;6:174–180. doi: 10.1097/COH.0b013e3283454058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Urdea MS, Wilber JC, Yeghiazarian T, Todd JA, Kern DG, Fong SJ, Besemer D, Hoo B, Sheridan PJ, Kokka R, et al. Direct and quantitative detection of HIV-1 RNA in human plasma with a branched DNA signal amplification assay. Aids. 1993;7(Suppl 2):S11–14. doi: 10.1097/00002030-199311002-00004. [DOI] [PubMed] [Google Scholar]
- 39.Jiang W, et al. Plasma Levels of Bacterial DNA Correlate with Immune Activation and the Magnitude of Immune Restoration in Persons with Antiretroviral-Treated HIV Infection. J Infect Dis. 2009;199:1177–1185. doi: 10.1086/597476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roederer M, Nozzi JL, Nason MC. SPICE: exploration and analysis of post-cytometric complex multivariate datasets. Cytometry A. 2011;79:167–174. doi: 10.1002/cyto.a.21015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hartigan-O’Connor DJ, Poon C, Sinclair E, McCune JM. Human CD4+ regulatory T cells express lower levels of the IL-7 receptor alpha chain (CD127), allowing consistent identification and sorting of live cells. J Immunol Methods. 2007;319:41–52. doi: 10.1016/j.jim.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 42.Lim SY, Rogers T, Chan T, Whitney JB, Kim J, Sodroski J, Letvin NL. TRIM5alpha Modulates Immunodeficiency Virus Control in Rhesus Monkeys. PLoS Pathog. 2010;6:e1000738. doi: 10.1371/journal.ppat.1000738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Boyer JD, Kumar S, Robinson T, Parkinson R, Wu L, Lewis M, Weiner DB. Initiation of antiretroviral therapy during chronic SIV infection leads to rapid reduction in viral loads and the level of T-cell immune response. J Med Primatol. 2006;35:202–209. doi: 10.1111/j.1600-0684.2006.00179.x. [DOI] [PubMed] [Google Scholar]
- 44.Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, Lifson JD, O’Connor DH, Carrington M, Watkins DI. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Relationships between Th17 cells, Tregs, and memory T cells in uninfected macaques.
Figure S2. Infection parameters and immune cells in the observational study.
Figure S3. Effects of IL-2 and G-CSF on various immune cells.
Figure S4. Infection parameters and immune cells in the interventional study.
Figure S5. Relationship of Tregs to viral load and T cell activation.
Figure S6. Gating tree and typical raw data for the IL-17 and IFN-γ parameters.
Table S1. MHC alleles of animals in the observational study.