Summary
The allosteric mechanism of Hsp70 molecular chaperones enables ATP binding to the N-terminal nucleotide-binding domain (NBD) to alter substrate affinity to the C-terminal substrate-binding domain (SBD), and substrate binding to enhance ATP hydrolysis. Cycling between ATP-bound and ADP-/substrate-bound states requires Hsp70s to visit a state with high ATPase activity and fast on/off kinetics of substrate binding. We have trapped this ‘allosterically active’ state for the E. coli Hsp70, DnaK, and identified how interactions between the NBD, the β-subdomain of the SBD, the SBD α-helical lid, and the conserved hydrophobic interdomain linker enable allosteric signal transmission between ligand-binding sites. Allostery in Hsp70s results from an energetic tug-of-war between domain conformations and formation of two orthogonal interfaces (between the NBD and SBD, and between the helical lid and the SBD). The resulting energetic tension underlies Hsp70 functional properties and enables them to be modulated by ligands and co-chaperones and ‘tuned’ through evolution.
INTRODUCTION
The 70-kDa heat-shock proteins (Hsp70s) are ubiquitous molecular chaperones that mediate a broad array of critical cellular functions integral to protein homeostasis (Calloni et al., 2012; Hartl et al., 2011; Mayer and Bukau, 2005; Mayer et al., 2001). Allosteric signals in Hsp70s initiate from ligand binding events—nucleotide and substrate—and are further modulated by co-chaperones (Figure 1A). Upon binding of ATP to the N-terminal 45-kDa actin-like nucleotide-binding domain (NBD), the affinity of the C-terminal 30-kDa substrate-binding domain (SBD) for substrates decreases by up to two orders of magnitude as compared to its affinity in the absence of nucleotide or when the NBD is bound to ADP (Figure S1A). In turn, substrate binding to the SBD stimulates the ATPase activity of the NBD (Figure S1B). These allosteric changes are reversible, and the resulting cycle of substrate binding/release, nucleotide hydrolysis, and nucleotide exchange repeats with a timing that is regulated by the availability and affinity of substrates, ATP/ADP balance, and co-chaperone interactions.
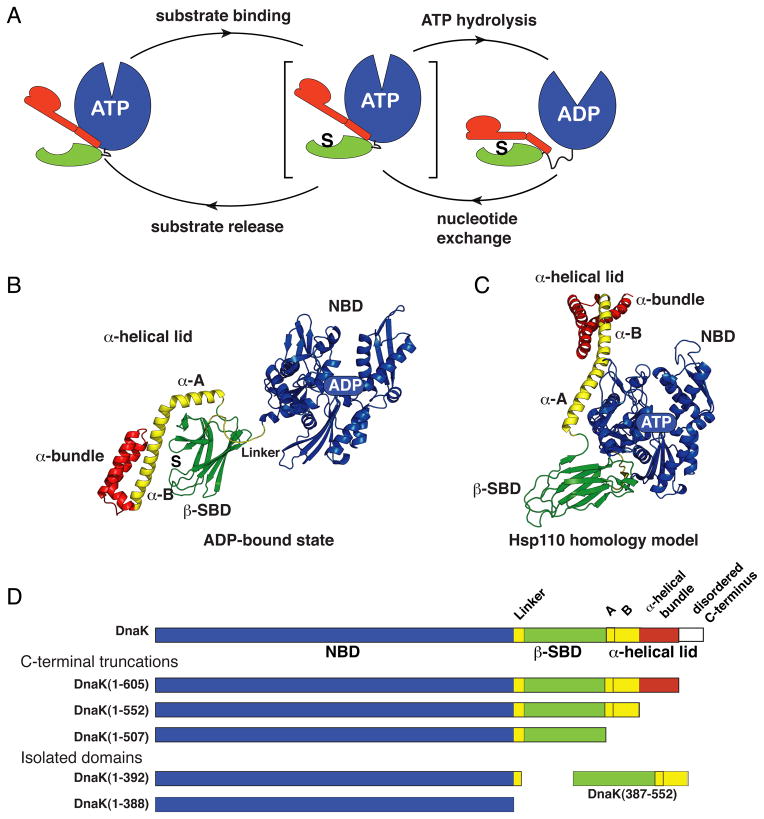
Figure 1. Conformational Insights into the Hsp70 Allosteric Cycle.
(A) The Hsp70 allosteric cycle.
(B) and (C) Ribbon representation of the structures of the two ‘end-point’ states of DnaK: the ADP-bound state (PDB ID code 2kho) and the Hsp110-based homology model of the ATP-bound state (Smock et al., 2010; coordinates available upon request); structural elements colored as in (D). (See also Figure S1.)
(D) DnaK constructs used for NMR studies.
The structural origin of allosteric signal transmission within and between Hsp70s domains remains poorly understood. In ADP-bound DnaK, the Escherichia coli Hsp70, which is the best studied Hsp70 from a biophysical perspective, the NBD and SBD are largely independent and behave as ‘beads on a string’ (Bertelsen et al., 2009; Swain et al., 2007), retaining the structures they adopt as isolated domains (Flaherty et al., 1990; Zhu et al., 1996) (Figure 1B). Upon ATP binding, the two domains rearrange to an intimately packed and dramatically reorganized domain-docked structure (Mapa et al., 2010; Marcinowski et al., 2011; Swain et al., 2007; Wilbanks et al., 1995). While the atomic structure of an ATP-bound domain-docked Hsp70 has not yet been reported, the recently described structure of an Hsp70 homolog, yeast Hsp110, Sse1, bound to ATP (Liu and Hendrickson, 2007) has provided a good model for the domain-docked ATP-bound conformation of an Hsp70 (Figure 1C). The domain rearrangements of ATP-bound DnaK deduced from a homology model based on the structure of ATP-bound Hsp110 are consistent with observed ATP-induced conformational changes (Buchberger et al., 1995; Mapa et al., 2010; Rist et al., 2006; Wilbanks et al., 1995), with the effects of several known mutations (Liu and Hendrickson, 2007), and with a recent analysis of residue co-evolution (Smock et al., 2010). The structural models for the ADP- and ATP-bound states implicate very large rigid body re-orientations and altered arrangements of the NBD, SBD and α-helical lid subdomains (Figure 1B and C), with relatively small changes in internal subdomain structures in the ATP-induced Hsp70 allosteric conformational change (Figure S1C).
Understanding how ligands mediate the Hsp70 allosteric structural rearrangement demands deeper insight into the mechanism of this conformational transition. Specifically, there must be at least one intermediate state visited between the two ‘end point’ states, undocked (ADP- and substrate-bound) and docked (ATP-bound with no substrate), in the allosteric cycle of an Hsp70 (Figure 1A). Substrate binding enhances the rate of ATP hydrolysis by the NBD; hence, both substrate and ATP must bind to the intermediate, allosterically active state. Moreover, this intermediate is a tipping point between two ‘end point’ states: On the one hand, ATP binding results in fast substrate unbinding and stabilizes the docked (substrate-unbound) state. On the other hand, substrate binding significantly enhances ATP hydrolysis, which then leads to the ADP–substrate bound (undocked) state. Provocatively, the presence of the SBD is not required for the NBD to adopt a conformation with stimulated ATPase activity. Indeed, binding of the highly conserved hydrophobic interdomain linker to the cleft beneath the crossing helices in an isolated NBD is necessary and sufficient for ATPase activation (Swain et al., 2007; Vogel et al., 2006).
How then does substrate binding lead to the population of an allosterically active conformation? And reciprocally, how does ATP-induced domain docking control substrate affinity? In this study, we address these questions through the use of a mutated DnaK that is impaired in ATP hydrolysis such that we can create a stable ATP-/substrate-bound DnaK sample. To elucidate the interdomain allosteric mechanism of DnaK we employed NMR spectroscopy as an extremely powerful tool to characterize the relationship between protein allostery and function (Manley and Loria, 2012; Tzeng and Kalodimos, 2011). The information we have obtained has allowed us to ‘dissect’ allostery in DnaK and define the roles of different allosteric units—viz., the NBD, the β-subdomain of the SBD or β-SBD, and the α-helical lid of the SBD (Figure 1). The tensions and balances among these allosteric units underlie the Hsp70 allosteric cycle and give rise to ‘tunability’ in the Hsp70 system. Our results, gleaned from study of DnaK, have implications for sequence and function relationships in other Hsp70s and shed light on how co-chaperones may modulate the Hsp70 allosteric cycle.
RESULTS
Methyl NMR Reveals Three Distinct Hsp70 Conformational Ensembles: Two End-Point States and an Intermediate
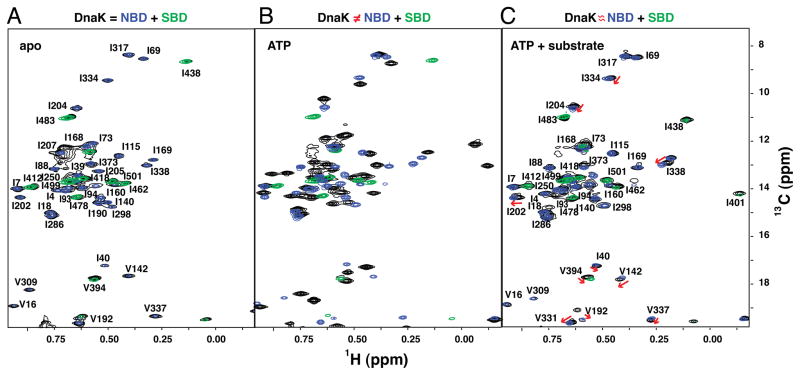
To explore ligand-induced allosteric transitions for the full-length 70-kDa DnaK, we employed methyl transverse relaxation optimized spectroscopy (methyl-TROSY) (Tugarinov et al., 2003), which is a powerful and sensitive method to probe protein conformations of large proteins with atomic resolution (Ruschak and Kay, 2010; Tugarinov and Kay, 2005). Consistent with previous results based on 1H-15N HSQC spectra (Swain et al., 2007), isoleucine methyl-TROSY spectra show few chemical shift changes between a two-domain DnaK construct, either apo (nucleotide-free) or ADP-bound, and isolated NBD and SBD (Figure 2A and S2C). These data support the conclusion that both nucleotide-free and ADP-bound DnaK lack significant interdomain interactions and behave as ‘beads on a string’ that are connected by a solvent-exposed, highly flexible interdomain linker.
Figure 2. NMR Fingerprints of DnaK Reveal Three Different Ligand-Bound States in Its Allosteric Cycle.
The isoleucine region of methyl-TROSY spectra of the two-domain DnaK constructs [black; DnaK(1-552) for (A, B) and full-length DnaK for (C), see Figure S2 for the full-length DnaK and ADP-bound state] are overlaid with the spectra of corresponding nucleotide and substrate-bound states of the individual domains, NBD, DnaK(1-388) (blue) and SBD, DnaK(387-552) (green): nucleotide-free (A), ATP-bound (B), and ATP-/substrate-bound (C). On (C), red arrows point to small but significant chemical shift differences between NBD resonances of the full-length ATP-/substrate-bound DnaK and those of its isolated NBD.
To ‘trap’ DnaK in the ATP-bound state, we incorporated the T199A mutation, which blocks ATP hydrolysis and thus stabilizes the ATP-bound state without disruption of ATP-induced conformational changes (McCarty and Walker, 1991). Large chemical shift changes are observed between ATP-bound DnaK-T199A (which we will refer to as DnaK) and its isolated domains in corresponding ligand-bound states (Figure 2B), consistent with widespread conformational rearrangements in both domains upon ATP binding as expected based on the Hsp110-based homology structure (Figure 1C). The perturbed chemical shifts may result from direct interactions between domains or/and indirect long-range conformational changes. Strikingly, substrate binding to ATP-bound DnaK results in additional conformational changes (Figure 2C), which we discuss in detail below and which we conclude are critical to the allosteric function of DnaK.
Our methyl-TROSY NMR data on the different DnaK ligand-bound states reveal that this protein samples at least three conformations during its allosteric cycle (Figure 1A); these arise from distinct domain arrangements: the ADP-bound domain-undocked conformation, (Figure 2A), the ATP-bound domain-docked conformation (Figure 2B), and the conformational ensemble populated in the present of ATP and substrate (Figure 2C). We performed detailed characterization of the ATP-bound states—both the domain-docked and the conformational ensemble created in the presence of both ATP and substrate—to gain deeper insight into how transitions between them affect Hsp70 functions (ATP hydrolysis and substrate binding) and how nucleotide and substrate binding control these conformational transitions.
The ATP-bound, Domain-Docked State of DnaK
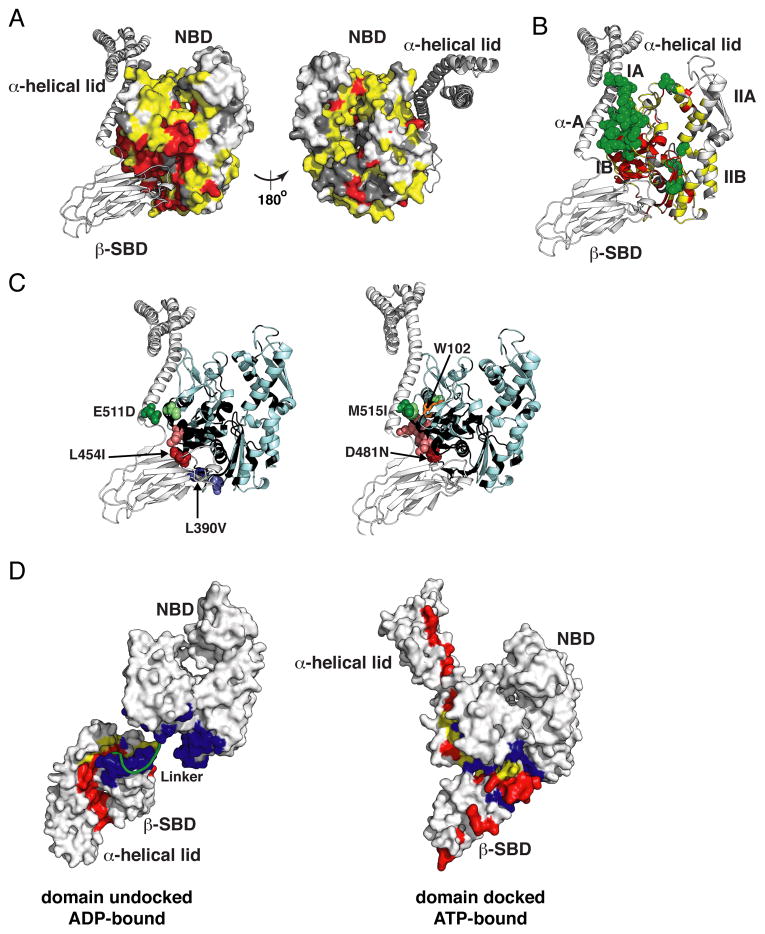
To validate the Hsp110-based homology model of ATP-bound DnaK we compared peak positions in HNCO spectra of the isolated NBD and two-domain DnaK constructs (Figures 3A and S3A). Figure 3A demonstrates that the NBD residues suffering significant changes in local environment between the ATP-bound NBD [DnaK(1-392)] and two-domain [DnaK(1-552)] constructs, as indicated either by chemical shift changes and/or altered μs-ms dynamics, are fully consistent with the NBD–SBD interfaces in the Hsp110-based model of ATP-bound DnaK.
Figure 3. Experimental Validation of the Hsp110-based Homology Model for the ATP-bound (Domain-Docked) State of DnaK.
(A, B) Mapping of the NBD residues significantly affected by interaction with the β-SBD onto the structure of the Hsp110-based homology model. Residues with significant chemical shift differences (see text) between DnaK(1-552) and either DnaK(1-392) or DnaK(1-507) are shown in yellow and green, respectively; residues showing enhanced μs-ms dynamics upon ATP binding, i.e., whose assignments were obtained for the isolated NBD but not for DnaK(1-552), are shown in red (see Supplemental Experimental Procedures); the unassigned residues in either constructs are shown in dark gray, the rest are shown in light gray (see also Figure S3A).
(C) Ribbon representation of the Hsp110-based homology model showing effects of ‘soft’ mutations on backbone (L454I, D481N, L390V, and E511D) and methyl (M515I) chemical shifts of ATP-bound (docked) DnaK(1-552). A site of mutation and the residues affected (see Supplemental Experimental Procedures) are shown as dark and light colored spheres, respectively (See also Figure S3B and C); Trp102, which becomes solvent-exposed in the ATP-bound conformation of DnaK (Buchberger et al., 1995), is shown in orange sticks on right; NBD residues without backbone assignments are shown in black.
(D) Interfaces between the NBD, β-SBD and α-helical lid are shown mapped onto the two endpoint Hsp70 states, domain-undocked/linker-unbound (PDB ID code 2kho) and domain-docked [Hsp110-based homology model (see Experimental Procedures)]: residues at the interface between the β-SBD and α-helical lid, which is stabilized by substrate binding are in red, and residues at the interface between the NBD and either the β-SBD or the α-helical lid, which is stabilized upon cooperative ATP and linker binding, are in blue; residues participating in both interfaces are in yellow.
A pairwise chemical shift comparison between ATP-bound DnaK(1-552) and DnaK(1-507) constructs, the latter of which is truncated so as to lack the α-helical lid sequence, revealed perturbations to the NBD interface upon interaction with the α-helical lid (Figures 3B, significant perturbations shown in green, and S3A) in full agreement with the Hsp110-based homology model of ATP-bound DnaK.
To obtain a more detailed description of the interfaces formed in the ATP-bound state without causing long-range changes we created ‘soft’ mutations of residues involved in the NBD–SBD and NBD–α-helical lid interfaces (Figure 3C). These mutations lead to local perturbations around the mutation site but do not perturb protein conformation (Figure S3B and C). Consequently, chemical shift perturbations in the NBD upon ‘soft’ mutations in the SBD or α-helical lid (Figure S3B and D) provide direct information about NBD–β-SBD or NBD–α-helical lid contacts in the ATP-bound states. The observed effects of several ‘soft’ mutations are completely consistent with the predicted packing of the interdomain interfaces based on the Hsp110-based model (Figure 3C).
The observation of random-coil-like chemical shifts (Figure S3E) and high peak intensities (Figure S3F) for residues 520-546 of helix B of the α-helical lid in ATP-bound DnaK(1-552) argues that this region is mobile and significantly destabilized, in contrast to the Hsp110 structure. These results argue that upon ATP-binding the α-helical bundle detaches from the β-SBD and forms only transient contacts with the NBD. Our conclusion is consistent with hydrogen exchange mass spectrometry results which showed that the proximal part of helical lid helix B (segment 512-532), but not the helical bundle, became unstructured upon ATP binding (Rist et al., 2006). To further check that lack of interaction of helix B with the NBD is real and not a truncation artifact arising from the absence of the α-helical bundle, we compared chemical shifts of the NBD and β-SBD in ATP-bound full-length DnaK and DnaK(1-605) (both with the full helical lid, the latter missing only the disordered C-terminal segment) and DnaK(1-552) (missing the helical bundle), and saw no significant differences (see Supplemental Experimental Procedures). The picture that emerges is that ATP binding is accompanied by a partial unfolding of the proximal part of helix B of the helical lid, mobility of the lid around this flexible site, loss of interaction with the β-SBD, and no new stable interactions with the NBD.
Intriguingly, chemical-shift differences between the ATP-bound two-domain protein, DnaK(1-552), and either the NBD [DnaK(1-392)] or SBD [DnaK(387-552)] are greater than 0.2, 1 and 0.5 ppm for 1HN, 15N, and 13CO atoms, respectively, and include residues distant from the interdomain interfaces, which is not consistent with solely rigid-body rearrangements of the NBD, β-SBD and α-helical bundle, as predicted from the Hsp110-based homology model. In particular, significant perturbations are observed at the nucleotide-binding site of the NBD (Figure S3A), and dramatic, widespread changes are observed throughout the β-SBD (Figures S3G and H). Such changes cannot be explained by local effects of interdomain docking or α-helical lid removal (Figure S3I) and point to extensive conformational changes everywhere in the β-SBD, including in the substrate-binding site.
Thus, our experimental data for ATP-bound DnaK argue that the Hsp110 homology model correctly describes relative arrangements of the NBD, β-SBD and α-helical bundle and define the NBD–β-SBD and NBD–α-helical lid interfaces in the docked conformation, but fails to capture long-range conformational changes inside Hsp70 domains. Intriguingly, the two ‘end-point’ Hsp70 conformations, domain-undocked (ADP-bound) and domain-docked (ATP-bound), have orthogonal patterns of interactions between the NBD, β-SBD and α-helical lid including: α-helical lid interaction with the β-SBD in the undocked state (red in Figure 3D), and NBD–β-SBD interaction in the docked state (blue in Figure 3D). These orthogonal interfaces can be expected to compete energetically when factors such as ligand binding differentially stabilize them (see below).
Binding of Substrate to ATP-bound DnaK Leads to Domain Dissociation with Interdomain Linker Binding: the Allosterically Active State
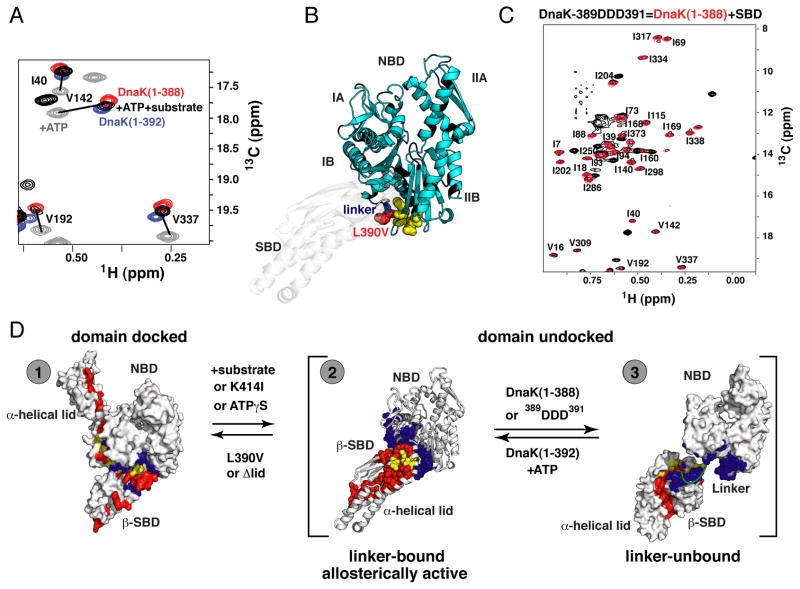
The key allosteric functions of an Hsp70 machine require that the binding of one ligand (ATP or substrate) influence the interaction of the chaperone with the other ligand (Figures 1A and S1A, B) and hence require the presence of both ligands. Our methyl TROSY data on the ATP/substrate-bound state of DnaK (Figure 2C) show that it is more similar to undocked (ADP-bound) DnaK than to the ATP-bound state, indicating that substrate binding induces domain undocking. Note in particular the lack of significant chemical shift perturbations in the SBD resonances relative to those of the substrate-bound isolated SBD (green in Figure 2C), including residues located on the interdomain interface. However, importantly, there are small but significant chemical shift differences between NBD resonances of the full-length ATP-/substrate-bound DnaK and those of the isolated NBD (in the ATP-bound state) (see arrows in Figure 2C). It is instructive to compare the NBD in ATP/substrate-bound DnaK to previously characterized ATP-bound conformations of isolated NBDs that retain the conserved hydrophobic interdomain linker (DnaK(1-392)) or not (DnaK(1-388)) (Zhuravleva and Gierasch, 2011) (Figure 4A). Resonances from the NBD of ATP/substrate-bound DnaK fall between those of DnaK(1-392) and DnaK(1-388). Based on our previous results, the chemical shift differences between these two constructs are a result of conformational changes in the NBD attributable to binding of the interdomain linker. To directly test the involvement of the linker in ATP/substrate-bound DnaK, we carried out chemical shift perturbation analysis of ATPγS-bound DnaK(1-552) [which populates a conformational ensemble similar to that populated by ATP-/substrate-bound full-length DnaK (see below)] using a ‘soft’ mutation of one of the conserved hydrophobic residues in the linker (L390V). The resulting pattern of chemical shift changes revealed that the linker in ATP/substrate-bound DnaK interacts with the β-strand of subdomain IIB (Figure 4B and Figure S4A)—the same linker-binding site that was previously found for the isolated NBD (Zhuravleva and Gierasch, 2011). Additionally, we mutated three consecutive conserved hydrophobic linker residues (389VLL391 to 389DDD391) to explore in greater depth the role of the linker in shifting the conformation of the undocked (ADP-bound) state of DnaK to its ATP/substrate-bound DnaK from. This mutation abolished the similarity of shifts for ATP/substrate-bound DnaK NBD resonances to those of ATP-bound DnaK(1-392), and the peaks now overlay on those of ATP-bound DnaK(1-388) (Figure 4C), indicating complete linker unbinding.
Figure 4. The Allosterically Active DnaK intermediate.
(A) Ligand-driven changes in the DnaK conformational ensemble: Blow-up of a representative region of methyl-TROSY spectra of ATP-bound DnaK in the absence of substrate (gray, the domain-docked state) and with substrate (black, the domain-undocked ensemble of linker-bound and –unbound conformations), overlaid on spectra of the isolated NBD, with linker [DnaK(1-392)] (blue, the linker-bound conformation) or without linker [DnaK(1-388)] (red, the linker-unbound, domain undocked conformation). Similar results were obtained for wild-type DnaK without the T199A mutation (see Figure S4).
(B) Linker binding site on the NBD: Residues with significant chemical-shift differences (more than 0.03 and 0.3 ppm for 1HN and 15N atoms, respectively; shown as yellow spheres) in the NBD of ATPγS-bound DnaK(1-552) [reflecting the ensemble of two domain-undocked conformations, linker-bound and linker unbound (see text)] between DnaK(1-552) and its ‘soft’ mutant L390V mapped onto the modeled structure of the allosterically active conformation [to schematically model this conformation the SBD of Hsp110-based homology model was replaced by the SBD from the ADP-bound DnaK (PBD ID code 2kho)] (see also Figure S4).
(C) The isoleucine region of the methyl-TROSY spectrum of the two-domain allosterically defective 389DDD391 DnaK(1-605) bound to ATP and substrate (black) showing near-perfect overlap with spectra of the individual, non-linker-bound NBD, DnaK(1-388) (red).
(D) Schematic illustration of two coupled conformational transitions in DnaK: (i) between the domain-docked conformation and the domain-undocked ensemble [corresponding to a transition between gray (ATP-bound) and black (ATP-/substrate-bound) peaks on left] and (ii) between linker-bound and –unbound conformations [corresponding to a transition between NBD+linker (blue) and NBD only (red) peaks (A); note in full-length DnaK that this transition is fast on the NMR time scale, and that black peaks on the left correspond to the dynamic domain-undocked ensemble of these two conformations]. Interdomain interfaces are colored as in Figure 3D.
Based on these results, we conclude that the interdomain linker binds to the NBD in ATP/substrate-bound DnaK, while the SBD has undocked from the NBD. However, several lines of evidence make it clear that both the linker-bound and linker-unbound conformations are significantly populated in ATP/substrate-bound DnaK. First, intensities of the unbound linker resonances are reduced in ATP/substrate-bound DnaK relative to apo-DnaK (domain-undocked, linker-unbound), but the resonances remain, arguing that the linker is unbound in a fraction of the population and bound in the rest (Figures S4B and C). Second, the NBD chemical shifts in ATP/substrate-bound DnaK, lie midway between the linker-unbound (DnaK(1-388)) and linker-bound (DnaK(1-392)) conformations (Figure 4A), which argues for a dynamic equilibrium between states with bound and unbound linker. Third, dynamic equilibration between linker-bound and linker-unbound states is also consistent with the observed overall decrease in peak intensities (more than 3-fold for most residues as compared with the apo, domain-undocked state) and line broadening in amide spectra of ATP/substrate-bound DnaK (Figure S2D).
The simultaneous action of ATP, which stabilizes linker-binding to the NBD and domain docking, and substrate, which favors domain undocking and stabilizes the β-SBD–α-helical lid interaction, subjects DnaK to two opposing driving forces. The result is an ensemble of fluctuating, interconverting domain-undocked conformations, with a fraction linker-bound and a fraction linker-unbound, in equilibrium with the domain-docked state (Figure 4D). The properties of this ensemble account for the allosteric functions of DnaK: enhanced NBD ATPase activity upon substrate binding, and rapid substrate association/dissociation upon ATP binding. ATPase activation upon substrate binding is explained by the action of linker binding on the NBD in the absence of domain docking, which is sufficient to activate NBD ATPase activity to an extent comparable to activation by substrate binding (Swain et al., 2007; Vogel et al., 2006).
A ‘Tug-of-War’ Between Two Orthogonal Interfaces Explains Modulation of Hsp70 Allosteric Activity by Ligands, Mutations, and Evolutionary Variation
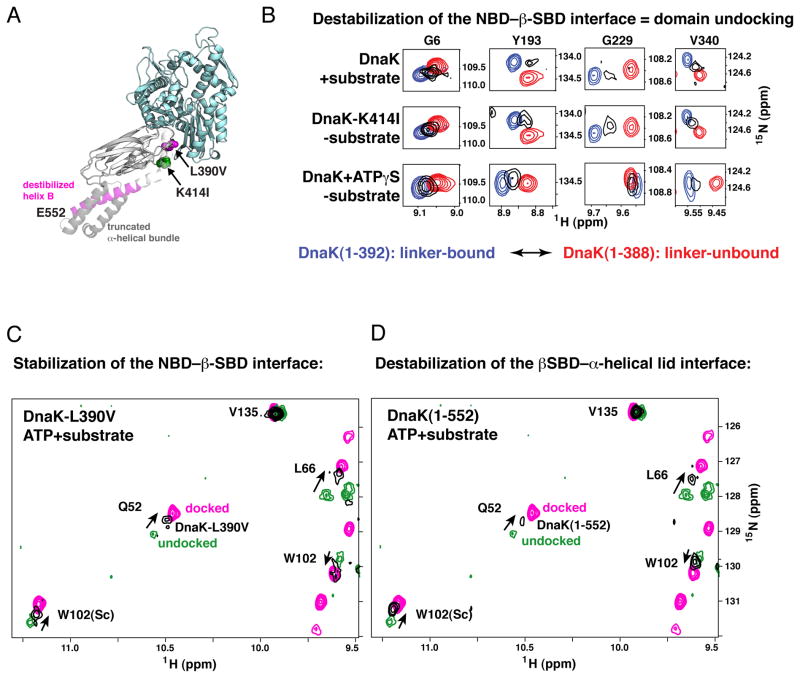
The two orthogonal interdomain sets of interactions in DnaK, the NBD–β-SBD interface, and the β-SBD–α-helical lid interface, emerge as ‘tunable’ elements that shape the allosteric landscape. Understanding the competitive energetic linkage of these interfaces provides a framework to explain several previous observations. For example, mutations that destabilize the NBD–β-SBD interface or stabilize the β-SBD–α-helical lid interface will decouple ATPase activation and substrate binding. Such interface variants will have significantly enhanced basal ATPase activity, and substrate binding will result in a minimal increase in the ATPase rate. As a specific example, the allosterically defective mutation K414I was identified in our lab (Montgomery et al., 1999); this residue substitution destabilizes the NBD–β-SBD interface as expected from the Hsp110 based model for the ATP-bound DnaK conformation, resulting in significant domain disengagement even without substrate, consistent with its red-shifted W102 fluorescence. Because of the weakening of the NBD–β-SBD interface, DnaK molecules harboring the K414I mutation (Figure 5A) populate the allosterically active, undocked, linker-bound DnaK conformation (Figure 5B, middle), which explains their enhanced basal ATPase activity. As would be predicted by this analysis of opposing energetic balances, perturbations of the NBD–β-SBD interface can also result in an opposite effect: The L390V mutation (Figure 5A) significantly stabilizes the docked state even in the presence of substrate (Figure 5C).
Figure 5. The Impact of Competition Between the NBD–β-SBD and β-SBD–α-Helical Lid Interfaces on the Hsp70 Allosteric Landscape.
(A) DnaK sequence modifications that result in perturbations in its conformational ensemble are mapped onto the modeled structure of the allosterically active conformation (modeled as for Figure 4): L390V and C-terminal truncations, which favor domain docking, are shown in magenta, K414I, which favors domain undocking, is shown in green.
(B) Destabilization of the NBD–β-SBD interface results in domain undocking even in the absence of substrate: Blow-up of the amide-TROSY spectra of DnaK(1-392) (blue, representing the NBD linker-bound state) and DnaK(1-388) (red, representing the NBD linker-unbound state) overlaid on the spectra of two-domain DnaK under conditions that stabilize domain undocking, either upon substrate binding to the ATP-bound DnaK(1-605) (top panels) or upon perturbation of the NBD–β-SBD interface, viz. ATP-bound DnaK(1-552)K414I (middle panels), or ATPγS-bound DnaK(1-552) (bottom panels). Resonances shown (Gly6, Tyr193, Gly229, and V340) report on long-range conformational changes in the nucleotide-binding site upon linker binding to the NBD (see Supplemental Experimental Procedures). The spectra of the isolated NBD constructs are with the corresponding nucleotide bound (ATP, top and middle panels) or ATPγS-bound (bottom panels).
(C) Stabilization of the NBD–β-SBD interface in DnaK(1-605)L390V or (D) destabilization of the β-SBD–α-helical lid interaction in DnaK(1-552) favors the domain-docked conformation even in the presence of substrate. A representative region of the amide-TROSY spectra of the ATP-bound state of DnaK(1-552) (magenta; the domain-docked conformation) and ATP-/substrate-bound DnaK(1-605) (green; the domain-undocked ensemble of linker-bound and linker-unbound conformations) overlaid with spectra of DnaK(1-605)L390V (C) and DnaK(1-552) (D), shown in black. Consistent with previous biochemical studies (Kumar et al., 2011; Swain et al., 2007) neither the L390V mutation on the NBD–β-SBD interface nor disruption of the β-SBD–α-helical lid interface in DnaK(1-552) affects the ATP-bound conformation in the absence of substrate.
The nucleotide analogue ATPγS does not mediate the same allosteric activities as the native ligand ATP (Theyssen et al., 1996). We now understand that ATPγS cannot shift the NBD fully to the conformation that forms a stable NBD–β-SBD interface (Figure 5B, bottom). Thus, ATPγS fails to stabilize the domain-docked conformation and results in significant domain disengagement. Remarkably, ATPγS binding does not perturb linker interaction with the NBD, but rather results in small changes to the interdomain interface (Zhuravleva and Gierasch, 2011) that perturb domain docking.
Modulating the strength of interactions between the β-SBD and the α-helical lid should also remodel the allosteric landscape in a way that is explained by the dueling interfaces. We compared amide-TROSY spectra of full-length DnaK and its C-terminally truncated construct DnaK(1-552), which lacks the helical bundle of the α-helical lid (Figure 5A) and as a result, has substantially less stable secondary structure in helix B (Swain et al., 2006), leading to a weaker β-SBD–α-helical lid interaction. In the absence of substrate, this C-terminal truncation does not affect the docked, ATP-bound state, reflected in the similar basal ATPase activities of the full-length DnaK and DnaK(1-552) (Swain et al., 2006). But these truncated variants display a reduction in the degree to which the substrate shifts the equilibrium toward the domain-undocked ensemble (Figure 5D). This interpretation fully explains why DnaK(1-552) displays a reduced substrate-activated ATPase rate (about two-times lower than that of full-length DnaK) (Swain et al., 2006).
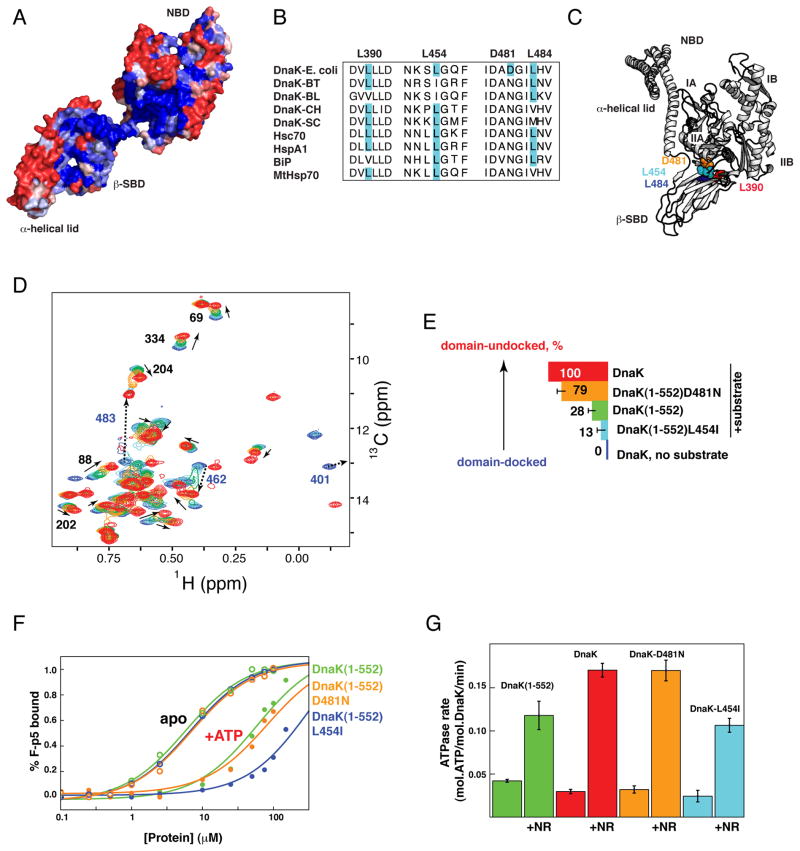
Given the functional significance of the NBD–β-SBD and β-SBD–α-helical lid interfaces, we anticipated high conservation of residues located on these interfaces and indeed found this to be true. The residues interacting across these interfaces are identical in 80 and 70% for the NBD–β-SBD and β-SBD–α-helical lid interfaces, respectively (Figure 6A) in a set of Hsp70s (see Experimental Procedure). In addition, statistical coupling analysis to reveal conservation of correlated variations found several linkages between these interfaces (Smock et al., 2010).
Figure 6. Evolutionary Variation at the NBD–β-SBD and β-SBD–α-Helical Lid Interfaces Modulates the Hsp70 Allosteric Landscape.
(A–C) Sequence conservation and diversity at the NBD–β-SBD interdomain interface in the Hsp70 family. (A) Sequence conservation between different Hsp70 family members colored from blue for fully conserved residues to red for residues with no conservations (see Experimental Procedures) shown on the structure of ADP-bound DnaK state (PDB ID code 2kho).
(B) Multiple sequence alignment (ClustalW) of DnaKs from E. coli, Bacteroides thetaiotaomicron VPI-5482 (BT), Bifidobacterium longum NCC2705 (BL), Cytophaga hutchinsonii (CH), and Streptomyces coelicolor A3(2) (SC), and four human Hsp70s (Hsc70, HspA1, endoplasmic reticulum-BiP, and mitochondrial-MtHsp70). (C) Observed amino acid substitutions identified in B are shown on the Hsp110-based homology model.
(D) The isoleucine region of methyl-TROSY spectra of ATP-bound full-length DnaK in the absence of substrate (blue, corresponds to the docked state) and with substrate (red, corresponds to the domain-undocked ensemble: the interconverting mixture of linker-bound and linker-unbound conformations) overlaid with ATP-/substrate-bound DnaK(1-552) (green) and its variants L454I (light blue; the DnaK(1-552)L484I shows very similar results, see Table S1) and D481N (orange). All experiments were performed at saturating substrate concentrations (2mM of NR peptide) (See also Figure S5).
(E) Histogram showing the degree of substrate-induced domain undocking for individual constructs, colored as in B and D. For each DnaK variant, the degree of domain undocking was estimated as described in the Experimental Procedures.
(F and G) Tuning of DnaK functionality. (F) Equilibrium binding of a fluorescently labeled peptide substrate to DnaK(1-552) (green) and its variants L454I (cyan) and D481N (orange) in the presence of ADP (open circles) and ATP (filled circles); KD values are 5.7±1.0 (60±20), 6.9±1.0 (80±20), and 6.8±1.0 (300±70) μM for the ADP-(ATP-)bound state of DnaK(1-552), DnaK(1-552)-D481N, and DnaK(1-552)-L454I, respectively (see Supplemental Experimental Procedures). (C) Stimulation of the ATPase activity of wild-type DnaK (red) and its variants DnaK-L454I (cyan), DnaK-D481N (orange), and DnaK(1-552) (green) by 200 μM NR peptide. (See Figure S5I and Supplemental Experimental Procedures).
Despite their high conservation, Hsp70s display some amino acid substitutions on the interdomain interfaces (Figure 6B). To explore whether evolutionarily-selected substitutions on the NBD-β-SBD interfaces affect Hsp70 conformational equilibrium and consequently functional properties, we tested several DnaK(1-552) variants based on known amino acid variations on the interdomain interfaces: L454I, D481N, and L484I (Figure 6B and C). While E. coli DnaK can tolerate these mutations in vivo (Figure S5A), these amino acid substitutions significantly affect the DnaK conformational ensemble by changing the degree to which substrate in the presence of ATP shifts the equilibrium between domain-docked and domain-undocked conformations (Figure 6D–E, Figure S5F-H, and Table S1). Upon substrate binding the tension and balances between the NBD, β-SBD, and α-helical lid couplings result in a highly tunable Hsp70 conformational ensemble, for which even minor changes on the interdomain interfaces [such as the L390V, L454I, D481N, and L484I substitutions (Figure 6C)] significantly affect the conformational equilibrium and, consequently, function, since stabilization of the docked conformation results in decrease of substrate affinity and ATPase activity (Figure 6F and G). In turn, substrate-binding affinity affords another tunable energetic contribution, as higher affinity substrates shift the equilibrium towards the allosterically active state (Figure S5C,F and G).
DISCUSSION
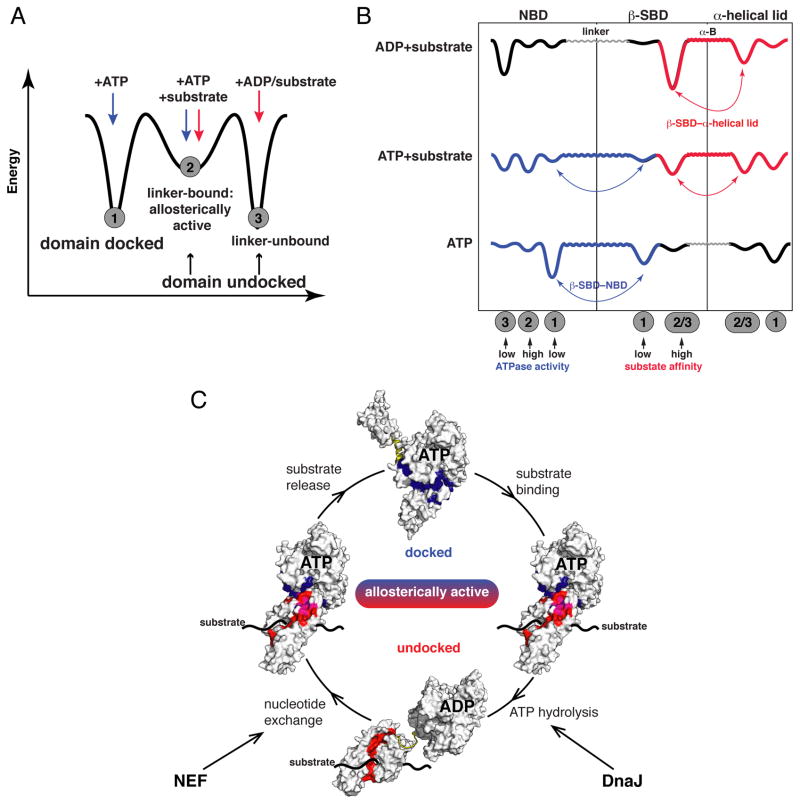
This study has revealed new insights into the fundamental mechanism of allostery of a paradigmatic Hsp70, DnaK (Figure 7 and Figure S6). We find that the Hsp70 allosteric landscape comprises three distinct protein conformations (Figure 7A): undocked (ADP-bound) and docked (ATP-bound) ‘end-point’ states, and a previously unidentified intermediate, the allosterically active, domain-dissociated linker-bound conformation that is partially populated in the presence of ATP and substrate. Each conformation is characterized by different arrangements of Hsp70 allosteric structural elements (NBD, β-SBD, α-helical lid), while two flexible regions–the interdomain linker and helix B of the helical lid–provide adjustable coupling connections between these units and create tunable interfaces between the structural elements (Figure 7B). This ‘Lego®’-like architecture creates a set of thermodynamic linkages that provide an explanation for the fundamental mystery of Hsp70 allostery: how events at each of the two domains can influence the other domain.
Figure 7. Mechanism of Hsp70 Allostery.
(A,B) Schematic illustration of Hsp70 allosteric landscapes showing how the allosterically active state serves as an intermediate between the two ‘end-point’ states (A) and (B) how thermodynamic coupling of Hsp70 domains determines the conformations the protein populates along its allosteric cycle (see also Figures 4D and S6): Binding of ADP and substrate favors interactions between the β-SBD and α-helical lid (red, in the domain-undocked conformation), ATP-induced linker binding to the NBD favors NBD–SBD docking (blue, in the domain-docked conformation). In the presence of both ATP and substrate, an interdomain energetic ‘tug-of-war’ results in a highly dynamic and tunable conformational ensemble. The interdomain linker and helix B provide flexible and ligand-adjustable connections between the NBD, the β-SBD, and the α-helical lid.
(C) Illustration of the roles of energetic interdomain coupling and ‘tunability’ in the allosteric cycle. The interfaces between the NBD and the β-SBD (blue) and between the β-SBD and the α-helical lid (red; magenta–residues participate in both interfaces) define the thermodynamics and kinetics of the allosteric cycle and can be modulated by either intrinsic (sequence changes) or extrinsic (binding to nucleotide, substrates, co-chaperones) factors.
Allostery in Hsp70s is achieved because binding of nucleotide and substrate ligands are thermodynamically linked so as to control the conformations of individual domains. In order for this allosteric machine to function, each separate domain must possess the capacity to sample (at least) two distinct conformations (Figure 7B): The result of linkages between domains is a novel state endowed with the properties required for active allostery (Csermely et al., 2010; del Sol et al., 2009; Smock and Gierasch, 2009): the ability to ‘breathe’ and sample multiple local conformations, including one with a catalytically active array of nucleotide ligands, and one with an un-lidded, disturbed substrate-binding site, which should have fast and reversible substrate binding/release. In the allosteric cycle of an Hsp70 depicted in Figure 7C, this state corresponds to the obligatory intermediate between the two end-point ADP- and ATP-bound states. For the isolated NBD, the binding of ATP perturbs the intradomain conformation so as to favor linker-binding and high ATPase activity (Bhattacharya et al., 2009; Revington et al., 2004; Zhuravleva and Gierasch, 2011). In full-length DnaK this ATP-induced linker binding transmits a signal to the SBD via stabilizing interactions between the NBD and SBD. Note that only minor changes in reorientations of NBD subdomains drastically affect ATPase activity of the protein (Zhuravleva and Gierasch, 2011), which explains how interactions between the linker and the NBD (in the allosterically active, domain-undocked conformation) and between the NBD and the SBD (in the docked conformation) significantly affect ATPase activity. Binding of substrate is coupled to these NBD conformational changes because of its direct stabilizing effect on the β-SBD-α-helical lid interface, and indirect destabilizing effect on the NBD-β-SBD interface. For the SBD, only one of its conformations has been described at atomic resolution. However, our results clearly demonstrate that domain docking stabilizes a very different un-lidded β-SBD conformation that we know has a markedly reduced capacity to bind substrates.
The delicate balance among conformational states created by thermodynamic coupling of opposing energetic contributions leads to exquisite ‘tunability’ of the Hsp70 system (Figure S7E). Each ‘end-point’ Hsp70 state is stabilized by only one major intrinsic interaction (Figure 7B): either the β-SBD–α-helical lid interaction in the undocked state (in the presence of ADP and substrate) or the NBD–β-SBD interaction in the docked (ATP-bound) state, while both interactions contribute to the allosterically active (ATP- and substrate-bound) conformation. Consequently, even minor perturbations of these interfaces result in redistributions in the Hsp70 conformational ensemble, and (through resulting ATPase activity and substrate affinity) define kinetics and thermodynamics of the Hsp70 allosteric cycle. Thus, the conformational distribution underlying the Hsp70 allosteric cycle can be readily shifted by either internal (sequence changes) or external factors (binding to co-chaperones, other chaperones, and different substrates).
The energetic tug-of-war in Hsp70s between intradomain interactions and interdomain interfaces provides an explanation for a number of previous observations, including the observation of a marked decrease in substrate affinity upon perturbation of the β-SBD–α-helical lid interaction (Fernandez-Saiz et al., 2006; Moro et al., 2004) and the fact that ATPase stimulation is proportional to substrate affinity (Mayer et al., 2000). Alterations in substrate binding affinity or kinetics clearly alter the allosteric reaction propensities. As a result, the behavior of the Hsp70 can be tuned to individual substrates, depending on their folding and aggregation properties, or on the physiological situation. DnaK is a ‘hub’ among chaperone networks, and forms complexes with at least 700 substrates (Calloni et al., 2012): Its tunability enables it to perform its allosteric cycle differently, depending on these extrinsic factors. Moreover, binding to a large substrate will significantly destabilize the interaction between the β-SBD and the α-helical lid (Schlecht et al., 2011), providing yet another way to affect the Hsp70 ensemble and result in substrate-dependent modulation of Hsp70 function.
Co-chaperones serve as extrinsic contributors to the allosteric balancing act in Hsp70s. We speculate that co-chaperone effects on Hsp70s will be clarified in terms of the balance of intra- and interdomain interactions. For example, shifting of the equilibrium between the linker-bound and linker-unbound conformations likely underlies the ability of the DnaJ-class of co-chaperone to enhance Hsp70 ATPase activities (Jiang et al., 2007). A recently discovered dynamic interface between DnaJ and DnaK (the segment 206-221 of the NBD) (Ahmad et al., 2011) overlaps with the NBD–SBD interdomain interfaces and provides another means to regulate Hsp70 ATPase activity.
The ‘tunability’ of the Hsp70 system offers an explanation for the striking functional diversity in the Hsp70 family (Kampinga and Craig, 2010; Sharma and Masison, 2011). Evolutionary tuning can occur via sequence changes at the key coupling interfaces. As illustrated above (Figure 6), even single conservative amino acid changes shift the equilibrium among docked, undocked (linker-unbound) and allosterically active (linker-bound) states and thus ‘tune’ conformational distributions to adjust kinetics and thermodynamic of the allosteric cycle to specific substrates, environment and function in different Hsp70 members. It will be of great interest to further explore the impact of sequence variations in these key interfaces among the Hsp70 family.
Taken together, our results provide new insights into the mechanism of Hsp70 allostery that explain many previous experimental observations, elucidate the basis of the striking functional diversity within the Hsp70 family, and reveal ‘tunable’ allosteric segments in Hsp70, which comprise potential binding sites for Hsp70 co-chaperones. The new insights into ‘tunability’ also provide a basis for design of small allosteric modulators of Hsp70 function, which are shown to have the great potential for therapeutic targeting of the Hsp70 system (Chang et al., 2011; Rousaki et al., 2011). Our data on Hsp70s also have implications more broadly, as allostery in other systems is likely to exploit analogous ligand-modulated changes in thermodynamic linkages between protein domains and allosteric interfaces. From an evolutionary standpoint, it is clear from the Hsp70 system that linking conformational equilibria within domains via interdomain interfaces is a blueprint to create allosteric signaling in multidomain protein systems. Indeed, recent work from the Ranganathan lab has illustrated successful creation of allosteric signaling by combining otherwise non-allosteric proteins (Halabi et al., 2009). We believe that the same mechanistic principles harnessed in the two-domain Hsp70s can also be extended as a general allosteric mechanism for another multidomain protein systems and for protein complexes with coupled allosteric functions.
EXPERIMENTAL PROCEDURES
Construct Design and NMR Experiments
We designed three C-terminally truncated constructs each including the T199A mutation (Figure 1D) including: DnaK(1-507), which comprises the NBD and β-SBD only; DnaK(1-552), containing the NBD and β-SBD plus helices A and B of the α-helical lid; and DnaK(1-605), including the NBD, β-SBD, and the whole α-helical lid. In the C-terminally truncated constructs, we incorporated L542Y and L543E mutations to disfavor self-binding (the helical lid back to the substrate-binding site) and ensure the same allosteric landscape as in full-length DnaK (Swain et al., 2006). Expression and purification of uniformly and ligated 2H-,13C-,15N-labeled and 2H-Methyl-13CH3-labeled samples were performed according to published methods (Tugarinov et al., 2006; Zhuravleva and Gierasch, 2011). NMR samples contained 300–500 μM (for backbone NMR analysis) or ~50–100 μM (for methyl NMR) of the protein, 10mM of potassium phosphate (pH 7.0), and, if needed, 5mM of appropriate nucleotide, 5mM MgCl2, 2mM NR (NRLLLTG) peptide as a substrate [saturating for all constructs (Figure S5B)]. All NMR spectra in this study were obtained at 26 °C on a 600-MHz Bruker Avance spectrometer using a TXI cryoprobe or 700-MHz Varian NMR system equipped with a cryogenically cooled triple-resonance probe. Spectra were processed using NMRpipe (Delaglio et al., 1995) and analyzed using Cara (Keller, 2004). Backbone assignments for the nucleotide-free and ATP-bound states of DnaK(1-388) and DnaK(1-392) and peptide-bound and -free SBD(387-552) were transferred from previous assignments (Swain et al., 2006; Zhuravleva and Gierasch, 2011) using transverse relaxation optimized spectroscopy (TROSY)-modified versions of HNCO and HNCA experiments (Weigelt, 1998). Methyl assignments of 1Hδ and 13Cδ of isoleucines were facilitated using the 3D HMCMCA and HMCMCB experiments (Tugarinov and Kay, 2003). To assign backbone spectra of ATP-bound DnaK, we applied a ‘divide-and-conquer’ strategy (Gelis et al., 2007; Ruschak and Kay, 2010), in which fragments of a protein are assigned, followed by the transfer of this assignment to the bigger constructs. The [1Hδ,13Cδ] methyl assignments for non-overlapping NBD peaks were transferred from the assignments of the isolated NBD. The partial assignments of SBD peaks were obtained using single-point mutagenesis. For more details see Supplemental Experimental Procedures.
Chemical Shift Analysis
To identify the residues that experience large structural or/and dynamic perturbations between different constructs, we performed a pairwise comparison of chemical shifts. For each residue we calculated a total chemical shift difference, Δδ=√(ΔδH)2 + (0.154ΔδN)2 + (0.341ΔδCO)2, where ΔδH, ΔδN, and ΔδCO are 1HN, 15N, and 13CO chemical shift differences between two constructs. Chemical shift differences were considered as significant if any of ΔδH, ΔδN, or ΔδCO were twofold larger than the corresponding chemical-shift errors, i.e., 0.08, 0.8, and 0.5 ppm for 1HN, 15N, and 13CO atoms, respectively.
To identify interdomain interfaces we constructed several ‘soft’ single-point β-SBD and α-helical lid mutations on the interdomain interfaces predicted from the Hsp110-based model: L390V, L454I, D481N, E511D, and M515I. Residues with backbone chemical-shift differences, ΔδHN > 0.03 ppm or/and ΔδN > 0.3 ppm (for the L390V, L454I, D481N, E511D DnaK constructs), or methyl chemical-shift, ΔδH > 0.01 ppm or/and ΔδC > 0.1 ppm (for the DnaK(1-552)-M515I) between DnaK(1-552) and a corresponding construct were considered to be affected by a given mutation.
To obtain estimates of the degree of domain undocking for different DnaK constructs in the ATP-/substrate-bound state, we used methyl chemical shifts of six NBD isoleucine residues (I40, I69, I88, I202, I204, and I334), which have well-resolved methyl peaks in NMR spectra of different constructs (Figure 6D). For these calculations, we assumed that for these residues, the domain-docking/undocking transition is fast on the NMR time scale; and the observed peak position is therefore a population-weighted average of the chemical shifts of the domain-docked conformation (ATP-bound DnaK) and domain-undocked ensemble (ATP-/substrate-bound DnaK). Further details are given in Table S1.
Functional DnaK assays
ATPase and substrate binding anisotropy assays were measured as previously described [(Chang et al., 2008) and (Montgomery et al., 1999), respectively]. For more details see Supplemental Experimental Procedures.
Homology Models and Definition of Interdomain Interfaces
We used the Hsp110-based homology model for DnaK developed previously (Smock et al., 2010) (coordinates are available upon request). For the undocked (PBD ID code 2kho) and docked (the Hsp110-based homology model) conformations, a residue was defined to reside on an interdomain interface if any of its atoms were located within 5 Å from any atoms belonging to the other allosteric units (the NBD, β-SBD, α-helical lid, or interdomain linker). To model the allosterically active (domain-undocked/linker-bound) conformation, the SBD was removed from the structure and the SBD from the ADP-bound DnaK (PBD ID code 2kho) was aligned on the β-SBD of the Hsp110-based homology model.
Evolutionary Conservation in the Hsp70 Family
Sequence conservation between different Hsp70 family members was estimated using the ConSurf Server (http://consurf.tau.ac.il, Glaser et al., 2003); a multiple sequence alignment was built using CLUSTALW; the homolog search algorithm used was CS-BLAST, with the minimal identity for homologs 60%. 150 homologs with the lowest E-values were used for analysis. Residue varieties for the highly conserved residues 390, 454, 481 and 484 were (L and V), (M,I, and L), (N and D), and (M, I, L, and V), respectively.
Supplementary Material
Highlights.
The allosterically active state explains roles of ligands in Hsp70 allostery.
Orthogonal Hsp70 domain-domain interfaces energetically compete.
The interdomain linker mediates Hsp70 allostery by binding the NBD.
Evolution can tune Hsp70 interdomain interfaces for a wide array of functions.
Acknowledgments
This work was supported by NIH grant GM027616. We thank Fabian Romano and Alejandro Heuck for assistance with the time-resolved fluorescence measurements.
Footnotes
Supplemental Information includes six figures, one table, and Supplemental Experimental Procedures and can be found with this article online.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad A, Bhattacharya A, McDonald RA, Cordes M, Ellington B, Bertelsen EB, Zuiderweg ERP. Heat shock protein 70 kDa chaperone/DnaJ cochaperone complex employs an unusual dynamic interface. Proc Natl Acad Sci USA. 2011;108:18966–18971. doi: 10.1073/pnas.1111220108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen EB, Chang L, Gestwicki JE, Zuiderweg ERP. Solution conformation of wild-type E. coli Hsp70 (DnaK) chaperone complexed with ADP and substrate. Proc Natl Acad Sci USA. 2009;106:8471–8476. doi: 10.1073/pnas.0903503106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertelsen EB, Zhou HJ, Lowry DF, Flynn GC, Dahlquist FW. Topology and dynamics of the 10 kDa C-terminal domain of DnaK in solution. Prot Sci. 1999;8:343–354. doi: 10.1110/ps.8.2.343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya A, Kurochkin AV, Yip GNB, Zhang Y, Bertelsen EB, Zuiderweg ERP. Allostery in Hsp70 chaperones is transduced by subdomain rotations. J Mol Biol. 2009;388:475–490. doi: 10.1016/j.jmb.2009.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchberger A, Theyssen H, Schroder H, McCarty JS, Virgallita G, Milkereit P, Reinstein J, Bukau B. Nucleotide-induced conformational-changes in the ATPase and substrate-binding domains of the DnaK chaperone provide evidence for interdomain communication. J Biol Chem. 1995;270:16903–16910. doi: 10.1074/jbc.270.28.16903. [DOI] [PubMed] [Google Scholar]
- Bukau B, Walker GC. Mutations altering heat shock specific subunit of RNA polymerase suppress major cellular defects of E. coli mutants lacking the DnaK chaperone. EMBO J. 1990;9:4027–36. doi: 10.1002/j.1460-2075.1990.tb07624.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calloni G, Chen T, Schermann SM, Chang H-c, Genevaux P, Agostini F, Tartaglia GG, Hayer-Hartl M, Hartl FU. DnaK functions as a central hub in the E. coli chaperone network. Cell Rep. 2012;1:251–264. doi: 10.1016/j.celrep.2011.12.007. [DOI] [PubMed] [Google Scholar]
- Chang L, Bertelsen EB, Wisén S, Larsen EM, Zuiderweg ER, Gestwicki JE. High-throughput screen for small molecules that modulate the ATPase activity of the molecular chaperone DnaK. Anal Biochem. 2008;372:167–76. doi: 10.1016/j.ab.2007.08.020. [DOI] [PubMed] [Google Scholar]
- Chang L, Miyata Y, Ung PMU, Bertelsen EB, McQuade TJ, Carlson HA, Zuiderweg ERP, Gestwicki JE. Chemical screens against a reconstituted multiprotein complex: myricetin blocks DnaJ regulation of DnaK through an allosteric mechanism. Chem Biol. 2011;18:210–221. doi: 10.1016/j.chembiol.2010.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clerico EM, Zhuravleva A, Smock RG, Gierasch LM. Segmental isotopic labeling of the Hsp70 molecular chaperone DnaK using expressed protein ligation. Biopolymers. 2010;94:742–752. doi: 10.1002/bip.21426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csermely P, Palotai R, Nussinov R. Induced fit, conformational selection and independent dynamic segments: an extended view of binding events. Trends Biochem Sci. 2010;35:539–546. doi: 10.1016/j.tibs.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Sol A, Tsai CJ, Ma B, Nussinov R. The origin of allosteric functional modulation: multiple pre-existing pathways. Structure. 2009;17:1042–1050. doi: 10.1016/j.str.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRpipe - a multidimensional spectral processing system based on Unix pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Fernandez-Saiz V, Moro F, Arizmendi JM, Acebron SP, Muga A. Ionic contacts at DnaK substrate binding domain involved in the allosteric regulation of lid dynamics. J Biol Chem. 2006;281:7479–7488. doi: 10.1074/jbc.M512744200. [DOI] [PubMed] [Google Scholar]
- Flaherty KM, DeLuca-Flaherty C, McKay DB. Three-dimensional structure of the ATPase fragment of a 70K heat-shock cognate protein. Nature. 1990;346:623–628. doi: 10.1038/346623a0. [DOI] [PubMed] [Google Scholar]
- Gelis I, Bonvin AMJJ, Keramisanou D, Koukaki M, Gouridis G, Karamanou S, Economou A, Kalodimos CG. Structural basis for signal-sequence recognition by the translocase motor SecA as determined by NMR. Cell. 2007;131:756–769. doi: 10.1016/j.cell.2007.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser F, Pupko T, Paz I, Bechor D, Martz E, Ben-Tal N. ConSurf: A server for the identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- Halabi N, Rivoire O, Leibler S, Ranganathan R. Protein sectors: evolutionary units of three-dimensional structure. Cell. 2009;138:774–786. doi: 10.1016/j.cell.2009.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU, Bracher A, Hayer-Hartl M. Molecular chaperones in protein folding and proteostasis. Nature. 2011;475:324–332. doi: 10.1038/nature10317. [DOI] [PubMed] [Google Scholar]
- Jiang J, Maes EG, Taylor AB, Wang L, Hinck AP, Lafer EM, Sousa R. Structural basis of J cochaperone binding and regulation of Hsp70. Mol Cell. 2007;28:422–433. doi: 10.1016/j.molcel.2007.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampinga HH, Craig EA. The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol. 2010;11:579–592. doi: 10.1038/nrm2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller R. PhD Dissertation. ETH; Zurich: 2004. Optimizing the process of nuclear magnetic resonance spectrum analysis and computer aided resonance assignment. [Google Scholar]
- Kumar DP, Vorvis C, Sarbeng EB, Ledesma VCC, Willis JE, Liu Q. The four hydrophobic residues on the Hsp70 inter-domain linker have two distinct roles. J Mol Biol. 2011;411:1099–1113. doi: 10.1016/j.jmb.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Hendrickson WA. Insights into Hsp70 chaperone activity from a crystal structure of the yeast Hsp110 Sse1. Cell. 2007;131:106–120. doi: 10.1016/j.cell.2007.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manley G, Loria JP. NMR insights into protein allostery. Arch Biochem Biophys. 2012;519:223–231. doi: 10.1016/j.abb.2011.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mapa K, Sikor M, Kudryavtsev V, Waegemann K, Kalinin S, Seidel CAM, Neupert W, Lamb DC, Mokranjac D. The conformational dynamics of the mitochondrial Hsp70 chaperone. Mol Cell. 2010;38:89–100. doi: 10.1016/j.molcel.2010.03.010. [DOI] [PubMed] [Google Scholar]
- Marcinowski M, Hoeller M, Feige MJ, Baerend D, Lamb DC, Buchner J. Substrate discrimination of the chaperone BiP by autonomous and cochaperone-regulated conformational transitions. Nat Struct Mol Biol. 2011;18:150–210. doi: 10.1038/nsmb.1970. [DOI] [PubMed] [Google Scholar]
- Mayer M, Bukau B. Hsp70 chaperones: Cellular functions and molecular mechanism. Cell Mol Life Sci. 2005;62:670–684. doi: 10.1007/s00018-004-4464-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer MP, Brehmer D, Gassler CS, Bukau B. Hsp70 chaperone machines. Adv Protein Chem. 2001;59:1–44. doi: 10.1016/s0065-3233(01)59001-4. [DOI] [PubMed] [Google Scholar]
- Mayer MP, Schroder H, Rudiger S, Paal K, Laufen T, Bukau B. Multistep mechanism of substrate binding determines chaperone activity of Hsp70. Nat Struct Biol. 2000;7:586–593. doi: 10.1038/76819. [DOI] [PubMed] [Google Scholar]
- McCarty JS, Walker GC. DnaK as a thermometer - threonine-199 is site of autophosphorylation and is critical for ATPase activity. Proc Natl Acad Sci USA. 1991;88:9513–9517. doi: 10.1073/pnas.88.21.9513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery DL, Morimoto RI, Gierasch LM. Mutations in the substrate binding domain of the Escherichia coli 70 kDa molecular chaperone, DnaK, which alter substrate affinity or interdomain coupling. J Mol Biol. 1999;286:915–932. doi: 10.1006/jmbi.1998.2514. [DOI] [PubMed] [Google Scholar]
- Moro F, Fernandez V, Muga A. Interdomain interaction through helices A and B of DnaK peptide binding domain. FEBS Lett. 2003;533:119–123. doi: 10.1016/s0014-5793(02)03752-3. [DOI] [PubMed] [Google Scholar]
- Moro F, Fernandez-Saiz V, Muga A. The lid subdomain of DnaK is required for the stabilization of the substrate-binding site. J Biol Chem. 2004;279:19600–19606. doi: 10.1074/jbc.M400921200. [DOI] [PubMed] [Google Scholar]
- Nashine VC, Hammes-Schiffer S, Benkovic SJ. Coupled motions in enzyme catalysis. Curr Opin Struct Biol. 2010;14:644–651. doi: 10.1016/j.cbpa.2010.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revington M, Holder TM, Zuiderweg ERP. NMR study of nucleotide-induced changes in the nucleotide binding domain of Thermus thermophilus Hsp70 chaperone DnaK - Implications for the allosteric mechanism. J Biol Chem. 2004;279:33958–33967. doi: 10.1074/jbc.M313967200. [DOI] [PubMed] [Google Scholar]
- Rist W, Graf C, Bukau B, Mayer MP. Amide hydrogen exchange reveals conformational changes in Hsp70 chaperones important for allosteric regulation. J Biol Chem. 2006;281:16493–16501. doi: 10.1074/jbc.M600847200. [DOI] [PubMed] [Google Scholar]
- Rousaki A, Miyata Y, Jinwal UK, Dickey CA, Gestwicki JE, Zuiderweg ERP. Allosteric drugs: The interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J Mol Biol. 2011;411:614–632. doi: 10.1016/j.jmb.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruschak AM, Kay LE. Methyl groups as probes of supra-molecular structure, dynamics and function. J Biomol NMR. 2010;46:75–87. doi: 10.1007/s10858-009-9376-1. [DOI] [PubMed] [Google Scholar]
- Schlecht R, Erbse AH, Bukau B, Mayer MP. Mechanics of Hsp70 chaperones enables differential interaction with client proteins. Nat Struct Mol Biol. 2011;18:345–351. doi: 10.1038/nsmb.2006. [DOI] [PubMed] [Google Scholar]
- Sharma D, Masison DC. Single methyl group determines prion propagation and protein degradation activities of yeast heat shock protein (Hsp)-70 chaperones Ssa1p and Ssa2p. Proc Natl Acad Sci USA. 2011;108:13665–13670. doi: 10.1073/pnas.1107421108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smock RG, Rivoire O, Russ WP, Swain JF, Leibler S, Ranganathan R, Gierasch LM. An interdomain sector mediating allostery in Hsp70 molecular chaperones. Mol Syst Biol. 2010;6:414. doi: 10.1038/msb.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain JF, Dinler G, Sivendran R, Montgomery DL, Stotz M, Gierasch LM. Hsp70 chaperone ligands control domain association via an allosteric mechanism mediated by the interdomain linker. Mol Cell. 2007;26:27–39. doi: 10.1016/j.molcel.2007.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swain JF, Schulz EG, Gierasch LM. Direct comparison of a stable isolated Hsp70 substrate-binding domain in the empty and substrate-bound states. J Biol Chem. 2006;281:1605–1611. doi: 10.1074/jbc.M509356200. [DOI] [PubMed] [Google Scholar]
- Theyssen H, Schuster HP, Packschies L, Bukau B, Reinstein J. The second step of ATP binding to DnaK induces peptide release. J Mol Biol. 1996;263:657–670. doi: 10.1006/jmbi.1996.0606. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Hwang PM, Ollerenshaw JE, Kay LE. Cross-correlated relaxation enhanced H-1-C-13 NMR spectroscopy of methyl groups in very high molecular weight proteins and protein complexes. J Am Chem Soc. 2003;125:10420–10428. doi: 10.1021/ja030153x. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Kanelis V, Kay LE. Isotope labeling strategies for the study of high-molecular-weight proteins by solution NMR spectroscopy. Nat Protoc. 2006;1:749–754. doi: 10.1038/nprot.2006.101. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Kay LE. Ile, Leu, and Val methyl assignments of the 723-residue malate synthase G using a new labeling strategy and novel NMR methods. J Am Chem Soc. 2003;125:13868–13878. doi: 10.1021/ja030345s. [DOI] [PubMed] [Google Scholar]
- Tugarinov V, Kay LE. Methyl groups as probes of structure and dynamics in NMR studies of high-molecular-weight proteins. ChemBioChem. 2005;6:1567–1577. doi: 10.1002/cbic.200500110. [DOI] [PubMed] [Google Scholar]
- Tzeng SR, Kalodimos CG. Protein dynamics and allostery: an NMR view. Curr Opin Struct Biol. 2011;21:62–67. doi: 10.1016/j.sbi.2010.10.007. [DOI] [PubMed] [Google Scholar]
- Vogel M, Mayer MP, Bukau B. Allosteric regulation of Hsp70 chaperones involves a conserved interdomain linker. J Biol Chem. 2006;281:38705–38711. doi: 10.1074/jbc.M609020200. [DOI] [PubMed] [Google Scholar]
- Weigelt J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. J Am Chem Soc. 1998;120:10778–10779. [Google Scholar]
- Wilbanks SM, Chen L, Tsuruta H, Hodgson KO, McKay DB. Solution small-angle X-ray scattering study of the molecular chaperone Hsc70 and its subfragments. Biochemistry. 1995;34:12095–12106. doi: 10.1021/bi00038a002. [DOI] [PubMed] [Google Scholar]
- Zhu XT, Zhao X, Burkholder WF, Gragerov A, Ogata CM, Gottesman ME, Hendrickson WA. Structural analysis of substrate binding by the molecular chaperone DnaK. Science. 1996;272:1606–1614. doi: 10.1126/science.272.5268.1606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuravleva A, Gierasch LM. Allosteric signal transmission in the nucleotide-binding domain of 70-kDa heat shock protein (Hsp70) molecular chaperones. Proc Natl Acad Sci USA. 2011;108:6987–6992. doi: 10.1073/pnas.1014448108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.