This study probes the maintenance of enhancer modifications. Shilatifard and colleagues show that Trr functions as a major H3K4 monomethyltransferase on enhancers. Using in vitro data along with genome-wide analyses, the authors propose that Trr and the H3K27 demethylase UTX cooperate to maintain the H3K4me1 and H3K27ac chromatin signature and regulate the transition from poised to active enhancers in Drosophila.
Keywords: Trithorax-like, H3K4 monomethylation, COMPASS-like complexes, enhancers
Abstract
Monomethylation of histone H3 on Lys 4 (H3K4me1) and acetylation of histone H3 on Lys 27 (H3K27ac) are histone modifications that are highly enriched over the body of actively transcribed genes and on enhancers. Although in yeast all H3K4 methylation patterns, including H3K4me1, are implemented by Set1/COMPASS (complex of proteins associated with Set1), there are three classes of COMPASS-like complexes in Drosophila that could carry out H3K4me1 on enhancers: dSet1, Trithorax, and Trithorax-related (Trr). Here, we report that Trr, the Drosophila homolog of the mammalian Mll3/4 COMPASS-like complexes, can function as a major H3K4 monomethyltransferase on enhancers in vivo. Loss of Trr results in a global decrease of H3K4me1 and H3K27ac levels in various tissues. Assays with the cut wing margin enhancer implied a functional role for Trr in enhancer-mediated processes. A genome-wide analysis demonstrated that Trr is required to maintain the H3K4me1 and H3K27ac chromatin signature that resembles the histone modification patterns described for enhancers. Furthermore, studies in the mammalian system suggested a role for the Trr homolog Mll3 in similar processes. Since Trr and mammalian Mll3/4 complexes are distinguished by bearing a unique subunit, the H3K27 demethylase UTX, we propose a model in which the H3K4 monomethyltransferases Trr/Mll3/Mll4 and the H3K27 demethylase UTX cooperate to regulate the transition from inactive/poised to active enhancers.
Gene expression by RNA polymerase II (Pol II) is regulated at multiple levels in order to allow for the faithful transmission of genomic information throughout development and upon external stimuli (Cheung and Cramer 2012; Zhou et al. 2012). In Drosophila, the Trithorax (Trx) and Polycomb (Pc) groups of proteins have long been known as crucial regulators of development and are required for the maintenance of active transcription or transcriptional repression, respectively (Poux et al. 2002; Klymenko and Muller 2004). The subsequent identification of Trx (a Trx group member) as a histone H3K4 methyltransferase and Enhancer of zeste [E(z); a Pc group representative] as a H3K27 methyltransferase exemplifies the importance of developmental gene regulation within a chromatin context (Simon and Tamkun 2002; Simon and Kingston 2009; Beisel and Paro 2011; Schuettengruber et al. 2011; Shilatifard 2012).
Our molecular understanding of the implementation of H3K4 methylation activity was aided by the biochemical purification of Set1, the yeast homolog of Trx (Miller et al. 2001). Set1 was found within the macromolecular complex COMPASS (complex of proteins associated with Set1) and was identified as the first H3K4 methylase capable of catalyzing mono-, di- and trimethylation of histone H3K4 (Briggs et al. 2001; Miller et al. 2001; Roguev et al. 2001; Krogan et al. 2002; Schneider et al. 2005; Shilatifard 2012). In contrast to yeast, where Set1 within COMPASS mediates all of the H3K4 methylation patterns, Drosophila contains three H3K4 methyltransferases—dSet1, Trx, and Trithorax-related (Trr)—that are each related to yeast Set1 and can be found in COMPASS-like complexes (Fig. 1A; Mohan et al. 2011). Mammals possess two representatives for each of the three H3K4 methyltransferases found in Drosophila, resulting in a total of six COMPASS-like complexes: Set1a and Set1b (related to dSet1), Mll1 and Mll2 (related to Trx), and Mll3 and Mll4 (related to Trr) (Shilatifard 2012). All COMPASS-like complexes contain a set of core components: Wdr5, Ash2, Rbbp5, and Dpy30 (Fig. 1A, highlighted in green). The three classes of the COMPASS family—dSet1/Set1a/Set1b, Trx/Mll1/Mll2, and Trr/Mll3/Mll4—each contain additional class-specific subunits providing functional diversity for the complexes (Fig. 1A, highlighted in blue; Hughes et al. 2004; Yokoyama et al. 2004; JH Lee et al. 2007; Lee and Skalnik 2008; Wu et al. 2008; Eissenberg and Shilatifard 2010; Mohan et al. 2011, 2012; Schuettengruber et al. 2011; Shilatifard 2012).
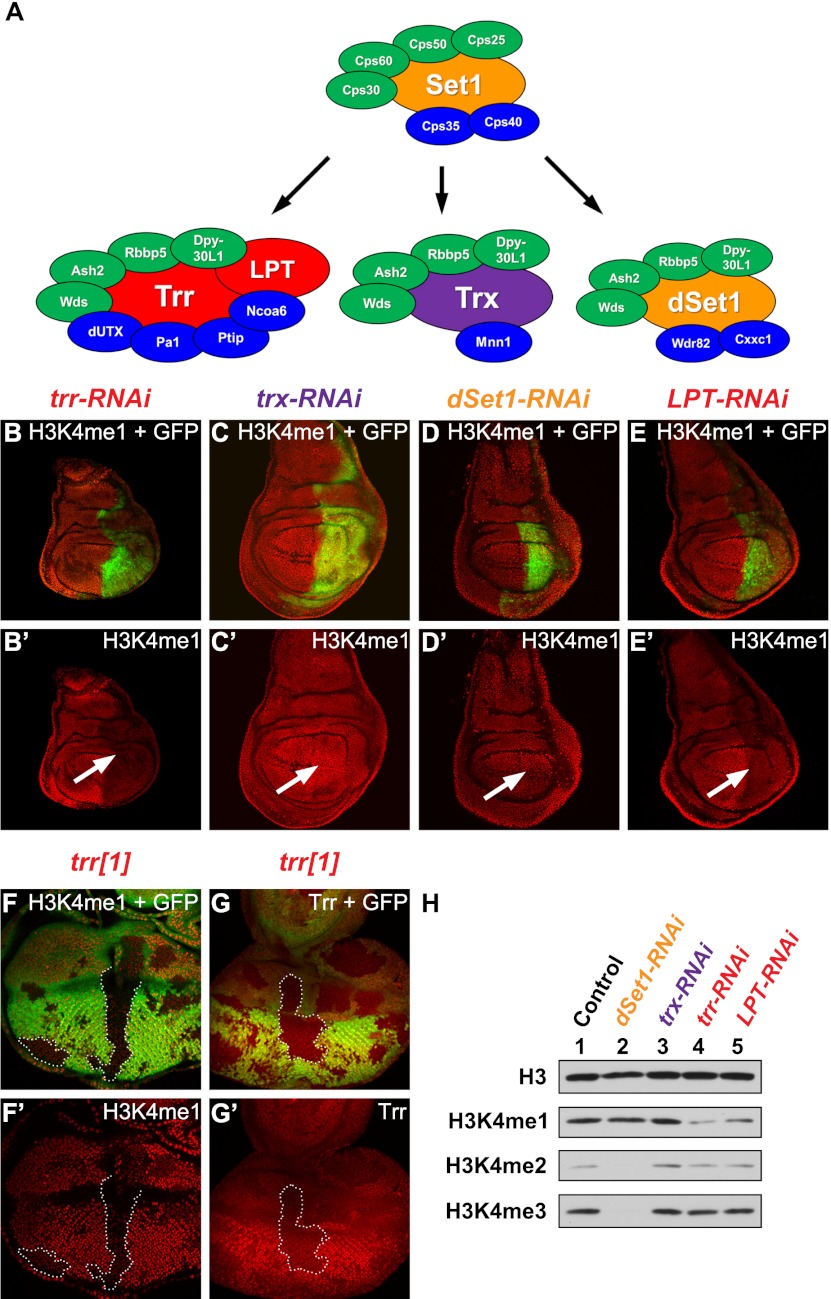
Figure 1.
The Drosophila Mll3/Mll4 homolog Trr is a major H3K4 monomethyltransferase in vivo. (A) The COMPASS family of H3K4 methyltransferase complexes in yeast and Drosophila (Mohan et al. 2011, 2012). Trr and LPT are highlighted in red, Trx is in purple, dSet 1 is in orange, core complex subunits are in green, and complex-specific subunits are in blue. (B–E) RNAi-mediated knockdown of Drosophila H3K4 methyltransferases in the posterior compartment of the wing imaginal disc. GFP expression in green marks the posterior part (highlighted by a white arrow in the antibody-only channel) where the knockdown occurs. Knockdown of Trr (B′) or LPT (E′) results in a global reduction of H3K4me1. No bulk changes in H3K4me1 were observed when Trx (C′) or dSet1 (D′) levels were reduced by RNAi. (F,G) Flipase-catalyzed induction of trr mutant clones (no GFP expression) with the eye-specific eyeless (ey) promoter. Wild-type tissue is marked in green (GFP expression). Representative trr mutant clones are outlined by white dashed lines. Mutant clones of trr1, a trr-null allele, display decreased levels of H3K4me1 (F′) and loss of Trr (G′). (H) Western blot of lysates from RNAi-treated Drosophila S2 cells. (Lane 1) Control lacZ-RNAi. (Lane 2) dSet1-RNAi. (Lane 3) trx-RNAi. (Lane 4) trr-RNAi. (Lane 5) LPT-RNAi. (Panel 1) α-H3. (Panel 2) α-H3K4me1. (Panel 3) α-H3K4me2. (Panel 4) α-H3K4me3. Genotypes used were as follows: UAS-Dcr-2/+; en-GAL4 UAS-EGFP/+; UAS-trr-RNAi/+ (B), UAS-Dcr-2/+; en-GAL4 UAS-EGFP/UAS-trx-RNAi (C), UAS-Dcr-2/+; en-GAL4 UAS-EGFP/UAS-dSet1-RNAi (D), UAS-Dcr-2/+; en-GAL4 UAS-EGFP/+; UAS-LPT-RNAi/+ (E), and trr1 FRT19A/ubi-GFP FRT19A; ey-FLP/+ (F,G).
In agreement with what had already been reported for mammalian Set1a and Set1b (Wu et al. 2008), recent studies have ascribed Drosophila Set1 within COMPASS a predominant role in bulk H3K4 dimethylation (H3K4me2) and trimethylation (H3K4me3) (Ardehali et al. 2011; Mohan et al. 2011; Hallson et al. 2012). Nonetheless, Trx/Mll1/Mll2 is required gene-specifically to implement H3K4me3 at the promoters of the Hox genes (Wang et al. 2009). Similarly, the Trr/Mll3/Mll4 COMPASS-like complexes play an important role in hormone receptor-induced signaling. Upon hormonal induction, they are recruited to target promoters by nuclear hormone receptors, resulting in increased H3K4me3 and gene activation (Goo et al. 2003; Sedkov et al. 2003; Lee et al. 2006; Mo et al. 2006; Johnston et al. 2011; Vicent et al. 2011; Chauhan et al. 2012). However, RNAi-mediated knockdown of Trr, the Drosophila homolog of mammalian Mll3/Mll4, showed only a modest reduction in H3K4me2 and H3K4me3 levels in wing imaginal discs (Mohan et al. 2011). These subtle changes in H3K4me2 and H3K4me3 are consistent with Trr regulating the activation of a subset of genes, such as the role of Trr in the transcriptional induction of ecdysone receptor target genes at promoter regions (Sedkov et al. 2003; Johnston et al. 2011). Depending on the developmental context and the requirement for Trr-mediated processes, these effects on H3K4me2 and H3K4me3 might be more pronounced at certain stages of development (Sedkov et al. 2003; Chauhan et al. 2012).
Genome-wide mapping of various histone modifications has provided a means to identify signatures of different chromatin states. For example, while H3K4me3 is found to be enriched at transcription start sites (TSSs), monomethylation of H3K4 (H3K4me1) was shown to be enriched at enhancers (Heintzman et al. 2007; Wang et al. 2008). Enhancers constitute promoter-distally located genomic elements that, in many instances, are necessary for the induction and maintenance of gene expression. They are often bound by developmental transcription factors, which, through looping, bring these distal regulatory elements in close proximity to the promoter-proximal regions regulating their transcriptional activities. Thus, enhancers provide an important regulatory cog for optimal transcriptional coherence to allow for tissue- and context-specific transcription of important developmental genes in a time-sensitive and optimized manner (Levine 2010; Bulger and Groudine 2011; Ong and Corces 2011). The ability of enhancers to work at long distances is mediated by cohesins, which were first identified to function in this process through a genetic screen for factors required for communication between the cut locus and its ∼80-kb distally located wing margin enhancer (Rollins et al. 1999, 2004; Dorsett et al. 2005).
Recently, it has been shown that chromatin signatures can be used to further classify enhancers as being in either active or inactive/“poised” states. Active enhancers are dually marked with H3K4me1 and H3K27 acetylation (H3K27ac), which allows them to be distinguished from inactive/poised enhancers that are characterized by the presence of H3K4me1 and H3K27me3 (Heintzman et al. 2009; Creyghton et al. 2010; Rada-Iglesias et al. 2011; Zentner et al. 2011; Bonn et al. 2012). H3K27ac is catalyzed by CBP and p300 in mammals and by a single CBP-related enzyme in Drosophila (Tie et al. 2009; Pasini et al. 2010; Jin et al. 2011). As lysine residues cannot be simultaneously modified by both methylation and acetylation, it has been proposed that the histone demethylase UTX can facilitate CBP-mediated H3K27ac through its ability to remove methyl groups from H3K27 (Tie et al. 2012).
During a recent study of dUTX's function in Notch signaling, we observed that dUTX mutant tissue exhibits a global decrease in H3K4me1 (Herz et al. 2010) but does not show significant changes in H3K4me2 or H3K4me3 (Herz et al. 2010). Thus, dUTX appears to link the removal of H3K27me3 with the implementation of both H3K4me1 and H3K27ac. dUTX is a complex-specific subunit of the Drosophila Trr complex (Fig. 1A; Mohan et al. 2011), while mammalian UTX is found in the homologous mammalian Mll3/Mll4 H3K4 methyltransferase complexes (Cho et al. 2007; Issaeva et al. 2007; MG Lee et al. 2007; Patel et al. 2007). Other complex-specific subunits of the Trr/Mll3/Mll4 complexes include Pa1, Ptip, and Ncoa6 (Fig. 1A). One feature of the Drosophila Trr complex that distinguishes it from mammalian Mll3/Mll4 complexes is that in Drosophila, two proteins—Trr and LPT (Lost PHDs of Trr)—constitute the functional homolog of either Mll3 or Mll4 in mammals. While Trr corresponds to the C-terminal portion of Mll3/Mll4, which includes the catalytic SET domain, LPT is homologous to the Mll3/Mll4 N terminus and contains several PHD domains and an HMG domain (Mohan et al. 2011; Chauhan et al. 2012).
Here, we report a novel function for the Trr/COMPASS-like complex as a major H3K4 monomethyltransferase at enhancers. Our results suggest that in various tissues, Trr or LPT loss coincides with a strong decrease in bulk H3K4me1 and a reduction in H3K27ac levels. Furthermore, Trr can function in enhancer-mediated processes, as it modulates enhancer–promoter interactions at the cut locus in wing imaginal discs. Genome-wide, Trr is required for the maintenance of the H3K4me1 and H3K27ac chromatin signature previously described for enhancers. Analysis of the role of Mll3 (one of the mammalian homologs of Trr) also suggests a role for Mll3/Mll4–COMPASS-like complexes in enhancer monomethylation. We propose a model in which UTX, as a complex-specific subunit of the Trr and Mll3/4 complexes, demethylates H3K27 at inactive/poised enhancers to prime them for H3K27ac by CBP.
Results
Trr and its partner, LPT, are required for H3K4me1 in vivo
In order to investigate the function of H3K4me1 in Drosophila, we used RNAi by the Gal4/UAS system in wing imaginal discs directed toward the Drosophila H3K4 methyltransferases that might regulate this modification. We found that expression of dsRNA against Trr under the control of the engrailed promoter results in a strong decrease in global H3K4me1 in the posterior half of the wing imaginal disc (Fig. 1B; Supplemental Fig. S1A). No bulk changes in H3K4me1 were observed with trx-RNAi (Fig. 1C; Supplemental Fig. S1B) or dSet1-RNAi (Fig. 1D; Supplemental Fig. S1C). We and others have previously shown that Trr is only homologous to the C-terminal portion of mammalian Mll3/Mll4 and that the functional equivalent of the Mll3/Mll4 N terminus is represented by another protein—LPT—in Drosophila (Mohan et al. 2011; Chauhan et al. 2012). Likewise, in the absence of LPT function (Supplemental Fig. S1D), H3K4me1 is strongly reduced in wing imaginal discs (Fig. 1E), suggesting that Trr and LPT work together to mediate global H3K4me1. A role for Trr in H3K4me1 was further confirmed with a trr-null allele (trr1) in eye-antenna imaginal discs, as trr mutant clones show reduced H3K4me1 staining compared with surrounding wild-type tissue (Fig. 1F,G).
Western blot analysis in S2 cells after RNAi-mediated knockdown of the three H3K4 methyltransferases or LPT confirmed our findings from imaginal discs (Fig. 1H; Supplemental Fig. S1E–H). Whereas dSet1-RNAi results in almost complete bulk losses of H3K4me2 (Fig. 1H, panel 3) and H3K4me3 (Fig. 1H, panel 4), a global reduction in H3K4me1 can only be observed with trr-RNAi and LPT-RNAi (Fig. 1H, panel 2). Since Set1 and related enzyme activities are stimulated by core COMPASS subunits (Dou et al. 2006; Steward et al. 2006; Shilatifard 2012), we used a baculovirus system to reconstitute the SET domains of Trr and dSet1 with the core subunits of COMPASS and observed that in the context of the core subunits, both SET domains of Trr and dSet1 were able to mono-, di- and trimethylate H3K4 (Supplemental Fig. S2). However, Trr showed a preference for H4K4me1, while dSet1 showed a tendency toward H3K4me3. Taken together, these results support the in vivo data showing that Trr/LPT is a major H3K4 monomethyltransferase.
Trr/COMPASS-like core and complex-specific subunits are required for H3K4me1 in vivo
Recently, all three H3K4 methyltransferases in Drosophila—dSet1, Trx, and Trr—have been shown to exist in independent COMPASS-like complexes (Mohan et al. 2011). Each COMPASS-like complex can be further subdivided into core members that are common to each complex and complex-specific subunits that are unique to only one of the complexes. Wds, Ash2, Rbbp5, and Dpy-30L1 constitute the core components of all COMPASS family members, while dUTX, Ptip, Pa1, and Ncoa6 are found only in the Trr/LPT complex (Fig. 1A). Hcf is another component that is often copurified with the COMPASS family (Mohan et al. 2011).
In order to identify subunit-specific requirements for bulk H3K4me1, we systematically depleted core members of the COMPASS family (Fig. 2A–C), Hcf (Fig. 2D), or Trr complex-specific subunits (Fig. 2E–G) in the posterior part of wing imaginal discs by RNAi. ash2-RNAi and Rbbp5-RNAi resulted in drastic losses of H3K4me1 (Fig. 2A,B; Supplemental Fig. S3A,D,E), H3K4me2 (data not shown), and H3K4me3 (data not shown). The loss of H3K4me1 is not the result of Trr destabilization, as Trr levels are unaltered when Ash2 or Rbbp5 are knocked down (data not shown), thus suggesting a fundamental requirement for these core components in Trr-mediated H3K4me1. Other core components, such as Dpy-30L1 (Fig. 2C; Supplemental Fig. S3F) or Hcf (Fig. 2D; Supplemental Fig. S3B,G), do not seem to be required for implementing bulk H3K4me1. Dpy-30L1-RNAi also did not affect H3K4me2 or H3K4me3 (data not shown). Hcf-RNAi did not affect H3K4me2 levels (data not shown) but did show reduced H3K4me3 (data not shown), in accordance with a recent study (Hallson et al. 2012).
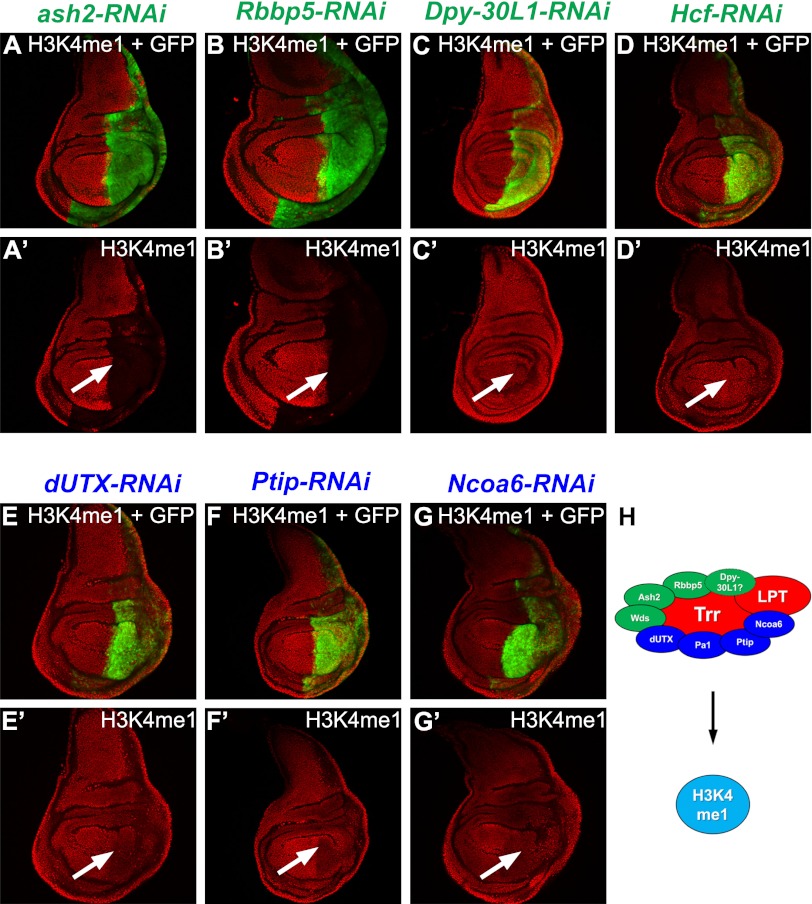
Figure 2.
Trr/COMPASS-like core and complex-specific subunits are required for H3K4me1. (A–G) RNAi-mediated knockdown of core members of COMPASS-like complexes (A–D) and Trr complex-specific subunits (E–G) in the posterior compartment of the wing imaginal disc. GFP expression in green marks the posterior part (highlighted by a white arrow in the antibody-only channel) where the knockdown occurs. ash2-RNAi (A′) and Rbbp5-RNAi (B′) result in strongly decreased H3K4me1, whereas other core members such as Dpy-30L1 (C′) and Hcf (D′) are not required for bulk H3K4me1. RNAi-mediated knockdowns of the Trr complex-specific subunits dUTX (E′), Ptip (F′), and Ncoa6 (G′) only weakly affect bulk levels of H3K4me1. (H) Cartoon representing the implementation of bulk H3K4me1 by the Trr/LPT complex. The SET domain-containing catalytic subunit Trr and LPT are highlighted in red, core complex subunits are in green, and complex-specific subunits are in blue. Genotypes used were as follows: UAS-Dcr-2/+; en-GAL4 UAS-EGFP/UAS-ash2-RNAi (A), UAS-Dcr-2/+; en-GAL4 UAS-EGFP/UAS-Rbbp5-RNAi (B), UAS-Dcr-2/+; en-GAL4 UAS-EGFP/+; UAS-Dpy-30L1-RNAi/+ (C), UAS-Dcr-2/+; en-GAL4 UAS-EGFP/+; UAS-Hcf-RNAi/+ (D), UAS-Dcr-2/+; en-GAL4 UAS-EGFP/+; UAS-dUTX-RNAi/+(E), UAS-Dcr-2/+; en-GAL4 UAS-EGFP/+; UAS-Ptip-RNAi/+ (F), and UAS-Dcr-2/+; en-GAL4 UAS-EGFP/+; UAS-Ncoa6-RNAi/+ (G).
Depletion of Trr complex-specific subunits such as dUTX (Fig. 2E; Supplemental Fig. S3C,H), Ptip (Fig. 2F; Supplemental Fig. S3I), or Ncoa6 (Fig. 2G; Supplemental Fig. S3J) resulted in decreased global H3K4me1 levels. In summary, our data support a model in which the Trr/LPT COMPASS-like complex is required for the maintenance of the majority of H3K4me1 levels.
Trr mediates stabilization of Trr/COMPASS-like complex-specific subunits and H3K27ac
The knockdown efficiencies of Trx, Trr, and LPT were confirmed for all components of the COMPASS family; namely, dSet1 (Fig. 3A, panel 1), Trx (Fig. 3A, panel 2), Trr (Fig. 3A, panel 3), and LPT (Fig. 3A, panel 4). LPT levels were strongly reduced not only with LPT-RNAi (Fig. 3A, lane 5, panel 4), but also with trr-RNAi (Fig. 3A, lane 4, panel 4). dUTX levels were also diminished upon trr-RNAi (Fig. 3A, lane 4, panel 5), reminiscent of what has been observed for Caenorhabditis elegans UTX-1 when SET-16 (Trr homolog) is knocked down (Vandamme et al. 2012). However, contrary to the findings in C. elegans, where reduction in SET-16 levels altered transcription of utx-1, we did not observe transcriptional changes for dUTX or LPT with trr-RNAi in Drosophila (data not shown). We further tested LPT and dUTX levels in Trr-depleted wing imaginal discs either by RNAi or when trr mutant clones were induced in eye-antenna imaginal discs (Fig. 3B–E). Both LPT (Fig. 3B) and dUTX (Fig. 3C) were reduced in trr-RNAi wing imaginal discs. A similar effect was seen in trr mutant clones for LPT (Fig. 3D) and dUTX (Fig. 3E) levels in eye-antenna imaginal discs. Conversely, LPT-RNAi or dUTX-RNAi in wing imaginal discs does not show significant effects on Trr stability (data not shown). Thus, in Drosophila, Trr is required for stabilization of at least some of the Trr/COMPASS-like complex-specific subunits.
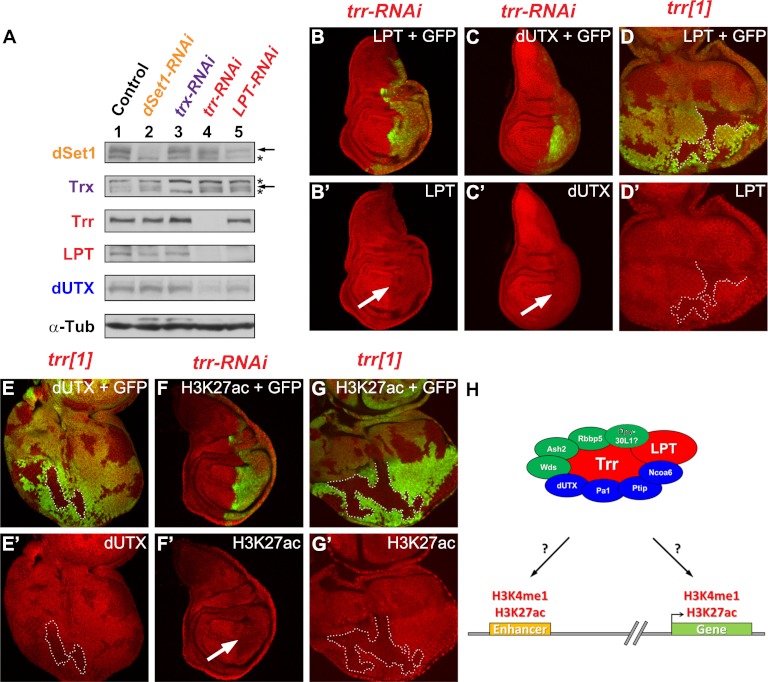
Figure 3.
Trr stabilizes Trr complex-specific subunits and promotes H3K27 acetylation. Trr is required for LPT and dUTX stability (A–E) and proper H3K27ac (F,G). (A) Western blots of lysates from RNAi-treated Drosophila S2 cells. (Lane 1) Control lacZ-RNAi. (Lane 2) dSet1-RNAi. (Lane 3) trx-RNAi. (Lane 4) trr-RNAi. (Lane 5) LPT-RNAi. (Panel 1) α-dSet1; the arrow marks the dSet1 band, and the asterisk indicates an unspecific band. (Panel 2) α-Trx; the arrow marks the Trx band, and the asterisks indicate unspecific bands. (Panel 3) α-Trr. (Panel 4) α-LPT. (Panel 5) α-dUTX. (Panel 6) αα-Tub. (B,C,F) RNAi-mediated knockdown of Trr in the posterior compartment of the wing imaginal disc. GFP expression in green marks the posterior part (highlighted by a white arrow in the antibody-only channel) where the knockdown occurs. LPT (B′) and dUTX (C′) are destabilized in the absence of Trr. (F′) H3K27ac levels are decreased when Trr function is removed. (D,E,G) Flipase-catalyzed induction of trr mutant clones (no GFP expression) with the eye-specific eyeless (ey) promoter. Wild-type tissue is marked in green (GFP expression). Representative clones are outlined by white dashed lines. Mutant clones of trr1, a trr-null allele, display decreased levels of LPT (D′), dUTX (E′), and H3K27ac (G′). (H) Possible functions for the Trr/LPT complex in implementing H3K4me1. The SET domain-containing catalytic subunit Trr and LPT are highlighted in red, core complex subunits are in green, and complex-specific subunits are in blue. Genotypes used were as follows: UAS-Dcr-2/+; en-GAL4 UAS-EGFP/+; UAS-trr-RNAi/+ (B,C,F) and trr1 FRT19A/ubi-GFP FRT19A; ey-FLP/+ (D,E,G).
The destabilization of dUTX in the absence of Trr is interesting in light of the recent finding that dUTX, by binding to CBP, modulates H3K27ac (Tie et al. 2012). This finding links the H3K27 demethylase dUTX (Smith et al. 2008; Herz et al. 2010) to H3K27ac and provides a model for how a transition could occur from a silenced H3K27-methylated chromatin state to an activated state that is marked by H3K27ac. Comparable with the results obtained with dUTX-RNAi (Tie et al. 2012), trr-RNAi in wing imaginal discs results in a global reduction of H3K27ac (Fig. 3F), and similar effects can be observed in trr mutant clones in eye-antenna imaginal discs (Fig. 3G). In conclusion, our data suggest that Trr is required to stabilize Trr/COMPASS-like complex-specific subunits to regulate H3K27ac. Based on genome-wide studies, it has been proposed that the distribution pattern for H3K4me1 is very broad and particularly enriched on enhancers and over the body of actively transcribed genes (Fig. 3H; Barski et al. 2007; Heintzman et al. 2007). Likewise, H3K27ac has been reported to mark active enhancers but can also be found on promoters and gene bodies (Fig. 3H; Heintzman et al. 2009; Creyghton et al. 2010; Rada-Iglesias et al. 2011; Zentner et al. 2011; Bonn et al. 2012).
Trr is required for enhancer function at the cut (ct) locus in wing imaginal discs
Based on our findings that Trr is globally required for proper H3K4me1 and H3K27ac, we turned to the well-characterized ct locus (Jack et al. 1991; Rollins et al. 2004; Dorsett et al. 2005) in order to functionally verify a possible role of Trr in enhancer-mediated processes. The ct wing margin enhancer is required for proper expression of ct at the dorso–ventral boundary in the wing imaginal disc (Jack et al. 1991; Rollins et al. 2004; Dorsett et al. 2005). In the ctk allele, a gypsy transposon is inserted between the ct wing margin enhancer and the ct promoter, interfering with enhancer–promoter communication and resulting in a characteristic wing-notching phenotype (Fig. 4A; Dorsett et al. 2005). The ctk allele has been deployed for the characterization of cohesin subunits such as Nipped-B, which are important regulators of enhancer–promoter interaction at the ct gene (Rollins et al. 2004). The wing-notching phenotype of the ctk allele can be quantified by counting the number of wing nicks per fly in homozygous ctk mutants. ctk control flies average 10.6 wing nicks per fly (Fig. 4B, left box plot). Removing 50% of the trr gene dosage by crossing in a null allele of trr, trr1 (Sedkov et al. 1999), increases the number of wing nicks to an average of 12.3 (Fig. 4B, right box plot). This represents a statistically significant enhancement (P < 0.0005, Welch two-sample t-test) and supports a requirement for Trr in enhancer-mediated processes.
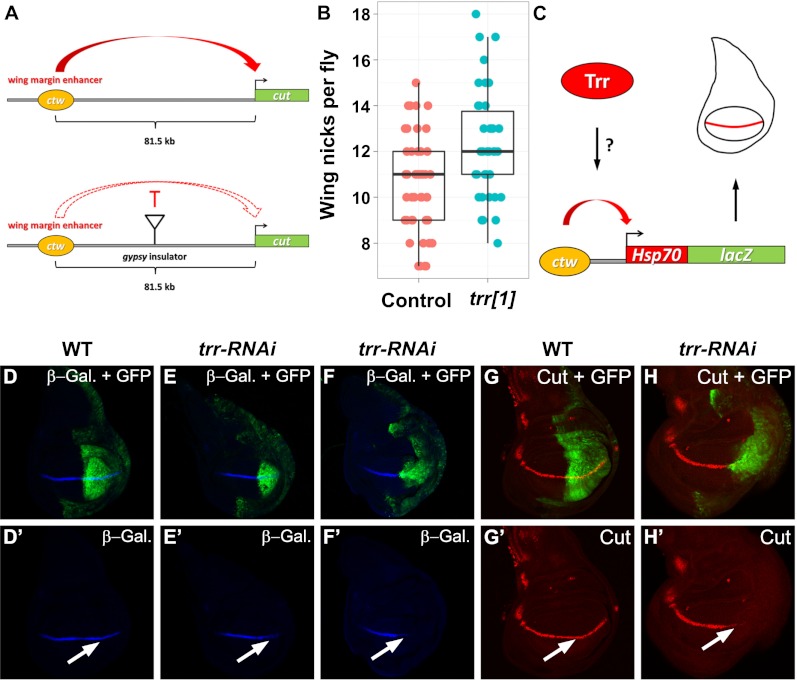
Figure 4.
Trr interferes with the enhancer–promoter interaction at the cut locus in wing imaginal discs. (A, top panel) The cut (ct) wing margin enhancer (ctw) is located 81.5 kb from the cut TSS. Insertion of a gypsy transposon between ctw and the ct promoter in the ctK allele interferes with the enhancer–promoter interaction (A, bottom panel) and results in a wing-nicking phenotype (B, left box plot) (n = 53). (B, right box plot) This wing-nicking phenotype is significantly enhanced (P < 0.0005, Welch two-sample t-test) when the trr gene dosage is reduced by 50% with a null allele of trr (trr1) (n = 38). The boxes indicate the two middle quartiles, the horizontal line within the box indicates the median, and the vertical extending lines indicate 1.5 times the interquartile range. (C) A 2.7-kb ctw sequence was inserted into a plasmid containing a heterologous Hsp70 promoter fused to the lacZ gene. In transgenic flies, this construct mimics the expression pattern of ct at the wing margin (horizontal red line) in wing imaginal discs. (D–H) Trr is required for proper ct expression at the dorso–ventral boundary of the wing imaginal disc. (E,F,H) RNAi-mediated knockdown of Trr in the posterior compartment of the wing imaginal disc. GFP expression in green marks the posterior part (highlighted by a white arrow in the antibody-only channel) where the knockdown occurs. (D,G) Wild-type (WT) control. β-Galactosidase (β-gal.), which serves as a readout for ctw activity is reduced in trr-RNAi (E′,F′) compared with wild type (D′). Similarly, Ct protein levels are also strongly reduced in the absence of Trr (H′) in comparison with the wild-type pattern (G′). Genotypes used were as follows: UAS-Dcr-2/+; en-GAL4 UAS-EGFP/ctw-Hsp70-lacZ (D), UAS-Dcr-2/+; en-GAL4 UAS-EGFP/ctw-Hsp70-lacZ; UAS-trr-RNAi/+ (E,F), UAS-Dcr-2/+; en-GAL4 UAS-EGFP/+ (G), and UAS-Dcr-2/+; en-GAL4 UAS-EGFP/+; UAS-trr-RNAi/+ (H).
To further test whether Trr directly regulates the wing margin enhancer, we used a transgenic fly line in which the ct wing margin enhancer (ctw) is inserted upstream of the Hsp70 promoter fused to the lacZ gene (Fig. 4C). This transgene recapitulates the endogenous ct expression pattern at the dorso–ventral boundary in wing imaginal discs (Fig. 4C,D). Trr knockdown in the posterior half of the wing imaginal disc results in reduced lacZ expression (Fig. 4E,F), suggesting a direct role for Trr in enhancer–promoter communication. Furthermore, upon RNAi-mediated knockdown of Trr, the dorso–ventral stripe of Ct, which can be detected across the whole antero–posterior axis in wild-type wing imaginal discs (Fig. 4G), is now confined to the anterior portion of the disc (Fig. 4H), providing further evidence for an enhancer-mediated function of Trr at the endogenous ct locus. Protein levels of other regulators of ct, such as Wingless (Wg) (Supplemental Fig. S4A,B) or Notch (N) (Supplemental Fig. S4C,D), were not changed when Trr function was removed.
The Trr/COMPASS-like complex localizes to promoters and enhancers
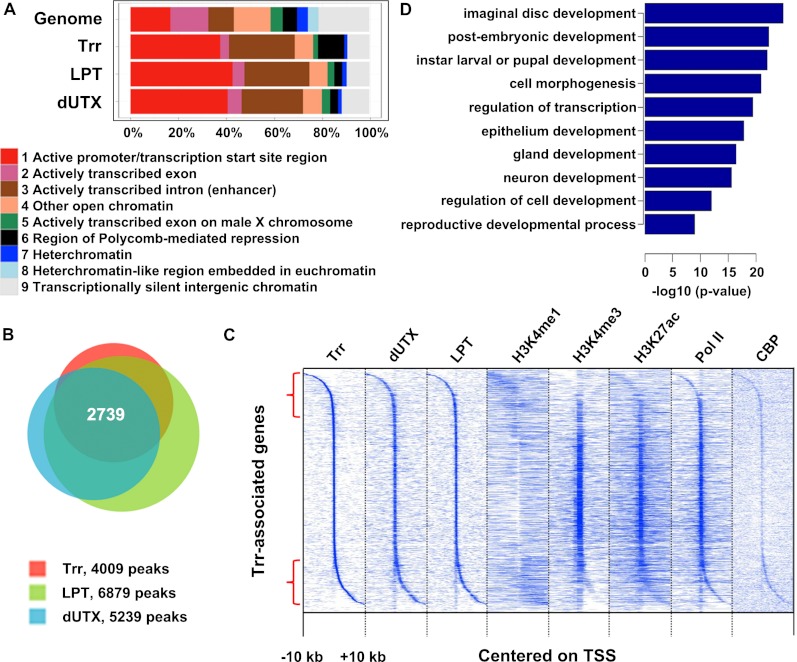
In order to determine whether the Trr/LPT complex generally acts as a H3K4 monomethyltransferase on enhancers, we performed chromatin immunoprecipitation (ChIP) combined with deep sequencing (ChIP-seq) studies in S2 cells to identify the genome-wide localization patterns of Trr, LPT, dUTX, H3K4me1, H3K4me3, H3K27ac, and RNA Pol II (Fig. 5). Preprocessed CBP data were obtained from Tie et al. (2012). The specificity of antibodies used for ChIP-seq against H3K4me1 and H3K4me3 was validated with yeast extracts (Supplemental Fig. S5). Genomic regions enriched for Trr were sorted into the nine previously proposed chromatin states that have been determined by combining the patterns of 18 different histone modifications (Kharchenko et al. 2011). Compared with the genomic distribution, Trr, LPT, and dUTX are particularly enriched at or near TSSs (Fig. 5A, chromatin state 1, red) but also significantly accumulate on actively transcribed introns (Fig. 5A, chromatin state 3, brown). Interestingly, state 3 has been described to be highly enriched for H3K4me1 and H3K27ac, which is characteristic of active enhancers (Kharchenko et al. 2011).
Figure 5.
The Trr complex localizes to promoters and enhancers. (A) The stacked bar plot shows the genome-wide percentage of each chromatin state as defined by Kharchenko et al. (2011). The top row shows the whole-genome distribution of the nine chromatin states in S2 cells, the second row shows the distribution for Trr-enriched regions, the third row shows the distribution for LPT-enriched regions, and the fourth row shows the distribution for dUTX-enriched regions. (Red) State 1, promoter-proximal regions; (purple) state 2, transcriptional elongation; (brown) state 3, intronic regions highly enriched for H3K4me1 and H3K27ac; (coral) state 4, related to state 3; (green) state 5, X chromosome, highly enriched for H4K16ac and H3K36me3; (dark gray) state 6, Polycomb-repressed regions; (dark blue) state 7, heterochromatin, high levels of H3K9me2/me3; (light blue) state 8, heterochromatin-like, moderate levels of H3K9me2/me3; (light gray) state 9, silent domains. (B) Venn diagram displaying co-occupancy between Trr (red), LPT (green), and dUTX (blue) peaks. Trr-enriched (4009 peaks), LPT-enriched (6879 peaks), and dUTX-enriched (5239 peaks) regions overlap significantly within 100 base pairs (bp) (2739 peaks). (C) Cluster analysis of Trr-occupied regions centered on the TSS of the nearest gene. Each column extends over 20 kb (±10 kb from the TSS). (Column 1) Trr. (Column 2) dUTX. (Column 3) LPT. (Column 4) H3K4me1. (Column 5) H3K4me3. (Column 6) H3K27ac. (Column 7) Pol II. (Column 8) CBP. CBP data were obtained from Tie et al. (2012). (D) Gene ontology (GO) term analysis of overlapping Trr, LPT, and dUTX peaks (from B). GO terms were ascribed based on the 2134 closest genes to overlapping peaks. P-values shown are Benjamini-corrected.
Trr, LPT, and dUTX co-occupy a large number of target loci (2739 peaks) (Fig. 5B). A cluster analysis confirmed that Trr-enriched regions that are centered to the nearest TSS (Fig. 5C, column 1) overlap very well with dUTX (Fig. 5C, column 2) and LPT (Fig. 5C, column 3). Many Trr-, LPT- and dUTX-bound regions localize at or close to TSS regions and are highly enriched for H3K4me3 (Fig. 5C, column 5), H3K27ac (Fig. 5C, column 6), and Pol II (Fig. 5C, column 7), confirming a promoter-proximal role for the Trr/LPT complex in transcriptional regulation. A significant number of Trr, LPT, and dUTX peaks are also found at promoter-distal elements (Fig. 5C, highlighted by red brackets), accompanied by high enrichment for H3K4me1 (Fig. 5C, column 4), CBP (Fig. 5C, column 8), and, to a lesser extent, H3K27ac (Fig. 5C, column 6) and Pol II (Fig. 5C, column 7). Overlapping Trr, LPT, and dUTX peaks linked to the closest gene display a prominent enrichment for developmental genes and transcription factors (Fig. 5D). In summary, the Trr/LPT COMPASS-like complex localizes not only to TSSs, but can also be found at promoter-distal locations that co-occur with enrichment for H3K4me1.
Trr regulates H3K4me1, H3K27ac, and H3K27me3 predominantly on chromatin regions containing enhancer-like histone modification signatures
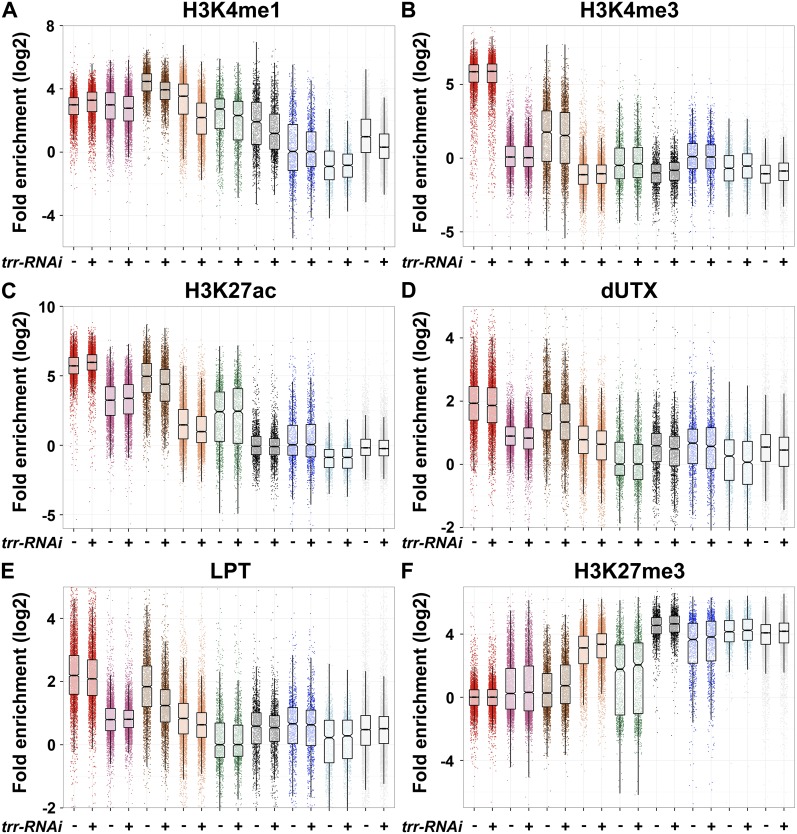
In order to assess which chromatin regions are most strongly depleted for H3K4me1 in the absence of Trr, we compared changes in H3K4me1, H3K4me3, and H3K27ac levels of the nine chromatin states between control and trr-RNAi S2 cells (Fig. 6A–C). Chromatin regions represented by state 7 (heterochromatin, highly enriched for H3K9me2/3) are strongly depleted for H3K4me1, H3K4me3, and H3K27ac (Kharchenko et al. 2011). Therefore, state 7 was used to normalize the samples against background read levels. When Trr was depleted, the strongest losses in H3K4me1 were observed in chromatin states 3 (Fig. 6A, brown box plots) and 4 (Fig. 6A, coral box plots), which are characterized by the highest levels of H3K4me1 enrichment and simultaneously most closely mimic enhancer-like histone modification patterns. Additionally, a significant reduction in H3K4me1 was also evident in Pc-repressed regions (Fig. 6A, dark-gray box plots) and silent chromatin (Fig. 6A, light-gray box plots) and, to a lesser degree, on actively transcribed exons (Fig. 6 A, purple and green box plots). Similar changes in H3K4me1 were observed in LPT-RNAi S2 cells (Supplemental Fig. S6A). Unexpectedly, H3K4me1 increases with trr-RNAi and LPT-RNAi on TSSs (Fig. 6A; Supplemental Fig. S6A, red box plots), which might be the result of an indirect effect of freeing up more substrate for other H3K4 methyltransferases. No comparable changes in H3K4me3 were observed within highly enriched H3K4me3 states such as TSS regions (Fig. 6B, red box plots), with the exception of a weak reduction of H3K4me3 in state 3 (Fig. 6B, coral box plots).
Figure 6.
Trr/COMPASS regulates H3K4me1, H3K27ac, and H3K27me3 on chromatin regions that resemble enhancer-like histone modification signatures. (A–F) Box plot diagrams displaying fold enrichment (log2) of various histone modifications (H3K4me1, H3K4me3, H3K27ac, and H3K27me3) and dUTX and LPT in wild-type (left box plots) and trr-RNAi (right box plots) S2 cells over the nine chromatin states as defined by Kharchenko et al. (2011). Characteristics of the different chromatin states are summarized in Figure 5A. Enrichment for each chromatin state is represented by the left box plot for wild-type S2 cells and by the right box plot in trr-RNAi S2 cells. (A) H3K4me1 enrichment. (B) H3K4me3 enrichment. (C) H3K27ac enrichment. (D) dUTX enrichment. (E) LPT enrichment. (F) H3K27me3 enrichment. Fold enrichments were calculated for each region of the genome by taking the read density per base pair of each region divided by the median read density per base pair of the appropriate background regions: all state 7 regions for A–C, all state 5 regions for D,E, and all state 1 regions for F. The boxes indicate the two middle quartiles, the horizontal line within the box indicates the median, and the vertical extending lines indicate 1.5 times the interquartile range. The notches shown around the median indicate a 95% confidence interval of the median.
Decreases in H3K27ac levels after trr-RNAi paralleled the losses of H3K4me1 in states 3 (Fig. 6C, brown box plots) and 4 (Fig. 6C, coral box plots). Not surprisingly, changes in H3K27ac could not be detected in Pc-repressed regions (Fig. 6C, dark-gray box plots) or in silent chromatin (Fig. 6C, light-gray box plots), as they do not contain significant levels of H3K27ac in wild-type S2 cells. Thus, the observed global decreases of H3K27ac in trr-RNAi wing imaginal discs (Fig. 3F) and trr mutant clones (Fig. 3G) likely correspond to H3K27ac losses on active enhancer-like chromatin regions. The decrease of H3K27ac at enhancers when removing Trr function is consistent with previous reports that H3K4me1 precedes H3K27ac (Bonn et al. 2012). As observed for H3K4me1, H3K27ac is also increased at TSS regions in trr-RNAi S2 cells (Fig. 6C, red box plots), which further suggests that some indirect effects on these histone modifications are occurring by reducing Trr levels.
dUTX was recently shown to modulate the balance between H3K27ac and H3K27me3 (Tie et al. 2012). We found that dUTX and LPT levels are most strongly reduced on enhancer-like chromatin states 3 and 4 upon trr-RNAi (Fig. 6D,E). These are also the regions that show an increase in H3K27me3 after trr-RNAi (Fig. 6F). Similarly, knockdown of LPT in S2 cells shows significant increases in H3K27me3 on states 3 and 4 (Supplemental Fig. S6B). In summary, our data suggest an interdependency of H3K4me1, H3K27ac, and H3K27me3 levels on enhancer-like chromatin.
The mammalian homolog of Trr, Mll3, is also a major monomethyltransferase
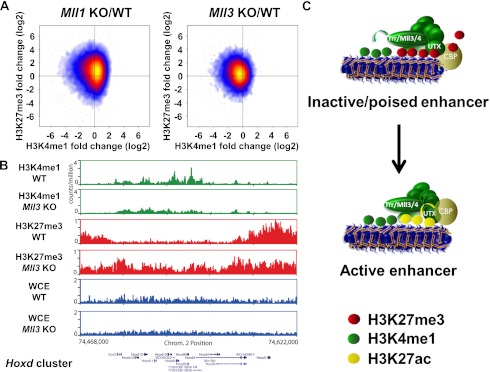
To verify that the mammalian homologs of Trr have a similar function in implementing H3K4me1 on enhancers, we performed ChIP-seq studies in mouse embryonic fibroblasts (MEFs) (Fig. 7A,B). Many genomic regions that are located outside of genes and are enriched for H3K4me1 show decreased H3K4me1 levels with a concomitant increase in H3K27me3 in Mll3 knockout MEFs (Fig. 7A, right scatter plot). In contrast, Mll1 knockout MEFs display an increase in H3K4me1 and H3K27me3 (Fig. 7A, left scatter plot). The modest shift into the upper left quadrant, representing decreased H3K4me1 and increased H3K27me3, might be even greater in Mll3/Mll4 double-knockout cells, as Mll3 and Mll4 are found in similar complexes and may share some functional redundancy toward H3K4me1 in the mammalian system. Nonetheless, many promoter-distal regions with a considerable decrease in H3K4me1 can be identified in Mll3 knockout MEFs. For example, the Hoxd cluster is strongly enriched for H3K4me1 in Mll3 wild-type MEFs and is known to contain many enhancers (Fig. 7B; Montavon et al. 2011). Removal of Mll3 function results in a decrease of H3K4me1 at many sites within the Hoxd cluster and a broad increase in H3K27me3 throughout this cluster (Fig. 7B). Thus, at least the role of Mll3 as a H3K4 monomethyltransferase appears to be functionally conserved in the mammalian system.
Figure 7.
The mammalian homolog of Trr, Mll3, is one of the major monomethyltransferases in mammalian cells. (A) Many promoter-distal elements (those H3K4me1 peaks not occurring within a gene) in Mll3 knockout (KO) MEFs exhibit a reduction in the H3K4me1 levels and an increase in H3K27me3 levels (right scatter plot), whereas Mll1 knockout MEFs display an increase in H3K4me1 and H3K27me3 levels (left scatter plot). The quotient of fold enrichment (immunprecipitation/whole-cell extract) in mutant MEFs versus wild-type (WT) MEFs was plotted for both the X-axis and Y-axis for H3K4me1 and H3K27me3, respectively. The scatter plot point density is indicated by color. (Blue) Low; (red) medium; (yellow) high. Mll1 and Mll3 wild-type and knockout MEFs were previously described in Hanson et al. (1999), Hughes et al. (2004), and Wang et al. (2009). (B) Decrease of H3K4me1 and increase of H3K27me3 over the Hoxd cluster in Mll3 knockout MEFs. Whole-cell extracts (WCE) represent the input chromatin. (C) Model describing the role of Mll3/4/Trr COMPASS-like complexes in the transitioning of enhancer activation. Mll3/4 in mammals and Trr in Drosophila participate in complexes joining the H3K27 demethylase UTX with the H3K4 monomethyltransferase activity of Mll3/4 and Trr to allow for H3K4me1 by the Trr/COMPASS and H3K27ac by CBP on enhancers. The transition to the active state of enhancers is promoted by activation of the Trr/Mll3/Mll4 COMPASS complex, which is able to achieve demethylation of H3K27me3 through UTX and H3K4me1 through Trr/Mll3/Mll4. CBP is activated via its interaction with UTX.
Based on these findings, we propose a model in which the Trr/Mll3/Mll4 COMPASS-like complexes form a hub combining the H3K27 demethylase function of UTX with the H3K4 monomethyltransferase activity of Trr/Mll3/Mll4 to allow for H3K27ac by CBP/p300 on enhancers (Fig. 7C). The combination of all of the histone-modifying activities provides an explanation for how enhancers can transition from an inactive/poised state to an activated state (Fig. 7C). The transition to the active state is promoted by activation of Trr/Mll3/Mll4 COMPASS-like complexes, which are able to achieve demethylation of H3K27me3 through UTX and implementation of H3K4me1 through Trr/Mll3/Mll4. It is also possible that the recruitment of UTX through Trr/Mll3/Mll4 COMPASS-like complexes to inactive enhancers provides context dependency for the activation of CBP, as many of the inactive enhancer regions are not modified by H3K27me3 and H3H27ac but do have CBP present (Fig. 7C).
Discussion
Our previous studies of the H3K27 demethylase dUTX had shown reduced levels of H3K4me1 in dUTX mutant tissue (Herz et al. 2010). As dUTX is a subunit of the Trr/COMPASS-like complex (Mohan et al. 2011), we investigated Trr's role in H3K4me1. In this study, we describe a function for Trr as a major H3K4 monomethyltransferase functioning at enhancer regions. Mutant trr clones in eye-antenna imaginal discs or knockdown of Trr or its partner, LPT, in wing imaginal discs and in S2 cells show a global decrease in both H3K4me1 and the enhancer-associated mark of H3K27ac (Figs. 1–3). Assays at the cut locus confirm a role for Trr in enhancer-mediated processes (Fig. 4). Using ChIP-seq in cells depleted of Trr by RNAi, we found that chromosomal regions most affected for H3K4me1 and H3K27ac share chromatin properties with enhancers (Fig. 6). Furthermore, the deletion of one of the mammalian Trr homologs, the Mll3 gene, results in a reduction in H3K4me1 levels with a concomitant increase in H3K27me3 (Fig. 7A). Since loss of Trr or LPT leads to an increase of H3K27me3 at enhancer regions, we propose a role for Trr/COMPASS in regulating the transition from inactive/poised enhancers to active enhancers. As Trr/LPT and dUTX are conserved in mammals, our findings have implications for understanding the transitions among various enhancer states during development and their misregulation in disease.
Trr/LPT and Mll3/Mll4 COMPASS-like complexes were previously shown to be major activators of hormone-responsive genes and are required for the establishment of H3K4me3 at promoter regions of these genes (Sedkov et al. 2003; Lee et al. 2006; Chauhan et al. 2012). A promoter-proximal function for Trr is in agreement with our observation that Trr, LPT, and dUTX localize to many TSSs (Fig. 5A,C). However, it is somewhat surprising that the knockdown of Trr or LPT does not significantly affect H3K4me3 at active promoters in S2 cells, as many Trr peaks localize near TSS regions (Fig. 5C). This is not specific to S2 cells, as the loss of Trr by RNAi does not result in global reduction in H3K4me3 levels in fly tissue (Mohan et al. 2011). Trr has been reported to be required for bulk H3K4me3 in embryos and prepupae (Sedkov et al. 2003; Chauhan et al. 2012); however, Trr localization at TSSs does not necessarily imply a functional role for Trr in H3K4me3 at promoters in all developmental contexts. Trr could provide a “priming” mechanism without functional consequences for H3K4me3. Additionally, dSet1 and/or Trx might be able to compensate for a loss of Trr function at some TSSs. We did observe an increase in H3K4me1 at TSSs after Trr or LPT RNAi (Fig. 6A; Supplemental Fig. S6A), which may be an indication of a change in competition among the different Set1-related enzymes or other, as yet unknown, factors for binding to TSS regions.
Our finding that the Trr/COMPASS-like complex is responsible for H3K4me1 at enhancers suggests a context-dependent difference in the activity of this complex. In vitro data from our group (Supplemental Fig. S2) and others (Ardehali et al. 2011) suggest that Trr/COMPASS may have a tendency that is skewed toward H3K4me1 compared with dSet1/COMPASS, which appears to be a better H3K4 trimethyltransferase (Supplemental Fig. S2). However, at the promoters of hormone-inducible genes, the binding of the ecdysone receptor and its partner, ultraspiracle, could target and stabilize Trr long enough to establish a trimethylated state. Alternatively, the cobinding of hormone receptors with the Trr/COMPASS-like complex might induce structural changes to open the catalytic pocket of Trr to transform Trr from a H3K4 monomethylase to a di- and trimethylase at target promoters, similar to what has been proposed for yeast Set1/COMPASS and its interactions with the Cps40 subunit of the complex (Takahashi et al. 2009). At enhancers, H3K4me1 is found over very broad regions, which is in contrast to the sharp peaks of H3K4me3 seen at TSSs. Therefore, in the context of enhancers, the Trr/LPT complex could be less constrained, interacting more transiently with nucleosomes within a certain domain. Investigating how the Trr/COMPASS complex is recruited and regulated at enhancers is an important area for future investigation.
Recently, studies in differentiated and embryonic stem (ES) cells have led to the hypothesis that enhancers can exist in inactive/poised and active states (Heintzman et al. 2009; Creyghton et al. 2010; Rada-Iglesias et al. 2011; Zentner et al. 2011; Bonn et al. 2012). The inactive/poised enhancer state is marked by H3K4me1, and it is proposed that transition to dually marked H3K4me1 and H3K27ac at active enhancers can facilitate gene activation. We found that the knockdown of Trr led to decreases in H3K4me1 in several S2 cell chromatin states described by Kharchenko et al. (2011), with the most pronounced effects for states 3, 4, and 6, each of which is likely to contain enhancers. State 6 (Pc-repressed) displays features of inactive/poised enhancers with lower enrichment of H3K4me1 (Fig. 6A) and no H3K27ac (Fig. 6C). State 3 has characteristics of active enhancers with the highest levels of H3K4me1 and high levels of H3K27ac. State 4 also has high levels of H3K4me1 but only moderate levels of H3K27ac. This state could include enhancers that are in an intermediate state between the active and inactive/poised states. The H3K27 demethylase UTX (MG Lee et al. 2007; Smith et al. 2008) could play an important role in the transition of these enhancer states, since removal of the H3K27me3 mark at enhancers would allow for H3K27ac by CBP/p300 (Heintzman et al. 2009; Tie et al. 2009, 2012; Visel et al. 2009; Kim et al. 2010). Together with the fact that UTX is a subunit-specific component of the Drosophila Trr/LPT and mammalian Mll3/Mll4 complexes (Cho et al. 2007; Issaeva et al. 2007; MG Lee et al. 2007; Patel et al. 2007; Mohan et al. 2011), this provides an attractive model in which the transition from inactive/poised enhancers to active enhancers is controlled by a functional module that combines H3K4me1 by Trr/Mll3/Mll4 with the removal of H3K27me3 by UTX, allowing for H3K27ac by CBP/p300 (Fig. 7C). Alternatively, the recruitment of UTX via Trr/Mll3/Mll4 complexes to enhancer regions could activate CBP function by context dependency and the presence of the interaction of UTX and CBP on these enhancer regions.
It is possible that the Trr/COMPASS-like complex could function in H3K4me1 in other contexts besides enhancers. One function previously proposed for dUTX and its mammalian homologs, UTX and JMJD3, was facilitating transcription elongation by maintaining H3K27 in an unmethylated state, thereby preventing Pc repression on gene bodies (Smith et al. 2008; Seenundun et al. 2010; Chen et al. 2012). H3K4me1 and H3K27ac, in addition to being found at enhancers, are significantly enriched on gene bodies of actively transcribed genes. The largest reductions in these modifications that we observed after Trr knockdown were in states 3 and 4, which Kharchenko et al. (2011) noted are chromatin states more likely to be found in long genes in Drosophila than short ones. Shorter genes are predominantly comprised of state 1 (TSSs, marked by H3K4me3) and state 2 (exons, marked by H3K36me3) (Kharchenko et al. 2011). Shorter genes might be less dependent on elongation factors. However, due to the compact nature of the Drosophila genome, in which genes are closely packed, many enhancer elements are embedded within introns. Thus, Kharchenko et al. (2011) summarized state 3 as an “actively transcribed intron (enhancer)” largely due to the enrichment of H3K27ac and H3K4me1. Furthermore, this state was enriched for DNase-hypersensitive sites, a feature consistent with active enhancers. Therefore, since in Drosophila enhancers are frequently located within introns and since the transcribed exon states 2 and 5 show smaller changes in H3K4me1 or H3K27ac after trr-RNAi, at least some of the changes that we observed in H3K4me1 and H3K27ac are likely due to enhancer functions of Trr. Another argument in favor of Trr functioning at enhancers is that chromatin state 6 (Pc-repressed), which has high levels of H3K27me3 and is transcriptionally repressed, also shows significant losses in H3K4me1 levels after Trr and LPT knockdown (Fig. 6A; Supplemental Fig. S6A). Pc-repressed domains include developmentally regulated genes as well as their enhancers, known as Polycomb response elements (PREs).
PREs enriched for H3K27me3 and H3K4me1 could be the Drosophila equivalent of some of the inactive/poised enhancers described in mammalian cells. An interesting question is how an enhancer transitions from an inactive/poised state to an active state. In mammalian ES cells, CBP/p300 is present at enhancers in the inactive/poised state, yet H3K27 is methylated, preventing acetylation. A signaling event activating UTX within the Mll3/Mll4 complex and/or inactivating the H3K27 methyltransferases E(z) homolog 1/2 would allow H3K27 to be demethylated and then acetylated. However, at this point, it is not clear whether the H3K27 demethylase function of UTX is generally required for this transition from an inactive/poised state to an active enhancer state. In Drosophila, dUTX mutants, while generally lethal, do survive into adulthood (Herz et al. 2010). Similarly, mouse ES cells require UTX for differentiation into mesoderm but not its demethylase activity (Wang et al. 2012), indicating that dilution of H3K27me3 through replication, chromatin remodeling, and histone replacement could also lead to the transitioning from an inactive/poised state to an active enhancer state. Studies in Drosophila, with less redundancy of some of these factors, and established genetics for assaying enhancer function will provide a useful system to answer some of these questions.
Recently, UTX and MLL3/4 have emerged as major regulators of tumorigenesis, being frequently mutated in different cancers (van Haaften et al. 2009; Ashktorab et al. 2010; Morin et al. 2011; Parsons et al. 2011; Mar et al. 2012). Some of the effects on tumorigenesis might be exerted through the direct role of UTX and MLL3/4 in gene activation by establishing H3K4me3 at promoters. However, a recent report of changes in histone modification patterns at enhancers highlights the importance of properly maintaining enhancer activity to prevent carcinogenesis (Akhtar-Zaidi et al. 2012). Our findings reported here raise the possibility that mutations in UTX or MLL3/4 might alter the enhancer landscape by misregulating H3K4me1 and H3K27ac levels, and this could contribute to tumorigenesis through changes in enhancer activity of genes, including tumor suppressor genes.
Materials and methods
Antibodies
Histone antibodies
Rabbit α-H3 (1791; Abcam) was used at 1:50,000 (S2 cells and yeast extract) for Westerns. Rabbit α-H3K4me1 (8895, Abcam) was used at 1:1000 for imaginal disc stainings, 1:20,000 (S2 cells) and 1:50,000 (yeast extract) for Westerns, and 30 μL (10 μg) was used for ChIP-seq. Rabbit α-H3K4me1 (39297, Active Motif) was used at 1:25,000 (yeast extract) for Westerns, and 25 μL was used for ChIP-seq. Rabbit α-H3K4me2 (32456, Abcam) was used at 1:5000 (S2 cells) and 1:10,000 (yeast extract) for Westerns, and 25 μL was used for ChIP-seq. Rabbit α-H3K4me3 (8850, Abcam) was used at 1:1000 for imaginal disc stainings and 1:100,000 (yeast extract) for Westerns. Rabbit α-H3K4me3 (#528, Shilatifard laboratory) was used at 1:2000 (S2 cells) and 1:25,000 (yeast extract) for Westerns, and 25 μL was used for ChIP-seq. Rabbit α-H3K27ac (4729, Abcam) was used at 1:4500 for imaginal disc stainings, and 10 μL (10 μg) was used for ChIP-seq. Rabbit α-H3K27me3 (39155, Active Motif): Twenty micrograms was used for ChIP-seq.
Other antibodies
Mouse α-Cut (2B10, supernatant, Developmental Studies Hybridoma Bank [DSHB]) was used at 1:50 for imaginal disc stainings. Rabbit α-Hcf (a kind gift from Jerry Workman) was used at 1:400 for imaginal disc stainings. Rabbit α-LPT (#470, Shilatifard laboratory) was used at 1:400 for imaginal disc stainings, and 50 μL was used for ChIP-seq. Rabbit α-LPT (#471, Shilatifard laboratory) was used at 1:4000 (S2 cells) for Westerns. Rabbit α-N (C458.2H, supernatant, DSHB) was used at 1:20 for imaginal disc stainings. Rabbit α-RBBP5 (300A-109A, Bethyl Laboratories) was used at 1:200 for imaginal disc stainings (cross-reacts to some degree with Drosophila Rbbp5). Rabbit α-Rpb1 (#828+829, Shilatifard laboratory): Twenty micrograms was used for ChIP-seq. Rabbit α-dSet1 (#868, Shilatifard laboratory) was used at 1:500 for imaginal disc stainings and 1:1000 (S2 cells) for Westerns. Mouse α-α-Tub (#E7 supernatant, DSHB) was used at 1:2000 (S2 cells) for Westerns. Rabbit α-Trr (#570, Shilatifard laboratory) was used at 1:500 for imaginal disc stainings and 1:4000 (S2 cells) for Westerns, and 50 μL was used for ChIP-seq. Rabbit α-Trx (#567, Shilatifard laboratory) was used at 1:500 for imaginal disc stainings. Rabbit α-Trx (#568, Shilatifard laboratory) was used at 1:1000 (S2 cells) for Westerns. Rabbit α-dUTX (#662, Shilatifard laboratory) was used at 1:500 for imaginal disc stainings and 1:4000 (S2 cells) for Westerns. Rabbit α-dUTX (#663, Shilatifard laboratory): Fifty microliters was used for ChIP-seq. Rabbit α-Wg (4D4, supernatant; DSHB) was used at 1:50 for imaginal disc stainings.
ChIP-seq
Drosophila
S2 cells (one T75 flask per histone antibody and two T75 flasks for other antibodies) were cross-linked in 1% formaldehyde (by adding 37% formaldehyde to medium) on a nutator for 15 min at room temperature. Samples were quenched by adding 2.5 M glycine to a final glycine concentration of 225 mM and incubated on a nutator for 5 min at room temperature. Following centrifugation at 2000g for 5 min at 4°C, the supernatant was aspirated. The cell pellet was resuspended in 5 mL of Orlando/Paro buffer (10 mM Tris HCl at pH 7.5, 10 mM EDTA, 0.5 mM EGTA, 0.25% Triton X-100, 0.5 mM DTT, protease inhibitors [complete, EDTA-free, catalog no. 05056489, Roche]) and centrifuged at 2000g for 5 min at 4°C. The supernatant was aspirated, and the wash step with Orlando/Paro buffer was repeated for another two times. The cell pellet (∼100 μL for one T75 flask) was resuspended in 1.5 mL of RIPA buffer (25 mM Tris at pH 7.5, 140 mM NaCl, 1% Triton X-100, 1 mM EDTA, 0.1% SDS, 0.1% Na-deoxycholate, 0.5% N-lauroylsarcosine, 0.5 mM DTT, protease inhibitors [complete, EDTA-free; catalog no. 05056489, Roche]) per 100 μL of cell pellet, and 1.5-mL samples were sonicated for 15 min (50% on/off cycle, high) (Bioruptor, Diagenode) in 15-mL hard plastic tubes (430055, Corning). Sonicated chromatin was centrifuged at 14,000 rpm for 20 min at 4°C, and the supernatant was kept. Ten microliters of the sonicated chromatin was kept for gel analysis (to check sizing pattern), and 50 μL was used as an input control. Sizing samples were reverse-cross-linked overnight at 65°C by adding 90 μL of RIPA and 3 μL of proteinase K (30 mg/μL), and input samples were reverse-cross-linked by adding 50 μL of RIPA and 5 μL of proteinase K and processed/purified in the same way as the ChIP samples (see below). The remaining chromatin was diluted twofold with RIPA buffer (without N-lauroylsarcosine) and incubated overnight at 4°C with the respective antibody on a nutator. Sixty microliters of protein A agarose (15918-014, Invitrogen) was washed in 5 mL of RIPA buffer and centrifuged at 1000 rpm for 2 min at 4°C. The supernatant was aspirated, and the chromatin sample was added and incubated for 2 h at 4°C on a nutator. After centrifugation at 1000 rpm for 2 min at 4°C, the protein A agarose was transferred into a 1.5-mL tube, washed with 1 mL of RIPA buffer, incubated for 5 min at room temperature on a nutator, and centrifuged at 2500 rpm for 2 min at 4°C. The supernatant was aspirated, and the previous washing steps were repeated another five times. Elution was performed on a nutator for 20 min at room temperature with 300 μL of elution buffer (0.1 M NaHCO3, 1% SDS) containing proteinase K (1 mL of elution buffer, 5 μL proteinase K [30 mg/μL]), and the sample was centrifuged at 2500 rpm for 2 min at room temperature. The supernatant was kept, and the elution step was repeated. Elution fractions were pooled and reverse-cross-linked overnight at 65°C. One microliter of RNase A (R4642, Sigma) was added to reverse-cross-linked samples followed by incubation for 1 h at 37°C. DNA was isolated with the Qiagen PCR purification kit and eluted in 50 μL of H2O, and DNA concentration was determined by Pico Green assay (Invitrogen).
MEFs
ChIP was performed according to a previously described protocol (Wang et al. 2009). Briefly, ∼107 MEFs for each assay were cross-linked with 1% formaldehyde and sonicated. Ten micrograms of antibody and 50 μL of protein A agarose were used in the ChIP assays.
ChIP DNA was amplified using an adapted version of the RNA TrueSeq sample prep kit (single end primers; Illumina) to allow for different barcoding of individual ChIP libraries. DNA libraries were validated on a 2100 Bioanalyzer (Agilent Technologies) before submission to sequencing. Up to six libraries were combined per lane. Reads were generated on the HiSeq 2000 using default Illumina standards for base calling and read filtering. Reads were aligned to the fly genome, University of California at Santa Cruz (UCSC) dm3, using Bowtie version 0.12.7, allowing unique reads only and up to three mismatches of the 50-base-pair (bp) read length. Enrichment for all ChIP-seq samples was determined by MACS version 1.4.1 with a threshold of P < 1 × 10−5 and a fold change greater than five, or false discovery rate (FDR) <5%. Gene and transcript annotations for flies are from Ensembl 67 and were used to determine the distance to the nearest TSSs for Trr-enriched regions (Fig. 5C). Gene ontology (GO) term analysis (Fig. 5D) was performed using DAVID (accessed July 2012) for the unique gene identifiers associated with overlapping Trr, LPT, and dUTX peaks that are nearest to any isoform TSS. The ChIP-seq profile diagram (Fig. 5C) displays only the canonical start site for each gene and is sorted based on the minimum distance and orientation (upstream/downstream) of a Trr peak to the canonical start site. The analyses for Figures 5 and 6 and Supplemental Figure S6 were carried out with one replicate each. Additional biological replicates of H3K4me1 and H3K27ac after trr-RNAi are available at Gene Expression Omnibus (GEO). Mouse sequencing data (Fig. 7) were aligned using the same parameters above and the UCSC mm9 genome. Scatter plots show fold changes of enrichment as calculated by first determining wild-type peaks of H3K4me1 at FDR <5% using MACS and then by calculating the total read sums for the input and immunoprecipitation samples, respectively, within these areas of enrichment. The ratio of immunoprecipitation over input for the knockout sample was divided by the ratio of immunoprecipitation over input for the wild-type sample, resulting in a fold change for each wild-type peak region. The same calculation of fold changes was performed for H3K27me3 levels within the H3K4me1 peak regions. The points shown are wild-type H3K4me1 peak regions occurring upstream of or downstream from a gene with a RefSeq mRNA identifier from Ensembl 66 (Fig. 7A). All sequencing data are available at the GEO accession number GSE41440
Processed ChIP–chip CBP data for the enrichment profile shown (Fig. 5C) were obtained from Tie et al. (2012). Enriched regions were determined at a fold change >1.1.
Chromatin state analysis
Regions for the nine colors of chromatin resulting from the analysis by Kharchenko et al. (2011) were downloaded from http://intermine.modencode.org/release-30/report.do?id=69000052. In order to determine fold enrichment distributions for each chromatin state, a background normalization algorithm was applied. First, the average level of background read density per base pair for each region of heterochromatin (state 7) was computed, where the expected level of real signal for H3K4me1, H3K4me3, and H3K27ac is to be the lowest (Kharchenko et al. 2011). The average read density per base pair for each state 7 region was computed by taking the total number of unprocessed reads within each state 7 region divided by the width of the region. The median value of the resulting distribution of average read densities for state 7 was selected as a background measurement value. Using this background value, the average read density per base pair for all state regions was computed by dividing each region's average read density per base pair by the background value. Therefore, each region of the genome is represented as a single fold enrichment value over background, and each state is represented as the distribution of individual fold enrichment values from the corresponding regions of the given state. The distribution of fold enrichment values was plotted in order to determine gains and losses for various histone modifications (H3K4me1, H3K4me2, and H3K27ac) between wild-type and RNAi-treated samples with respect to their chromatin states. Wild-type and trr-RNAi region analysis for each histone modification tested used its own level of background, respectively. For H3K27me3 analysis, state 1, having the lowest levels of this modification, was chosen for normalization. For analysis of dUTX and LPT levels, state 5, having the lowest levels of these factors, was chosen for normalization.
Fly lines
RNAi lines were as follows: UAS-ash2-RNAi (100718, Vienna Drosophila RNAi center [VDRC]), UAS-Dpy-30L1-RNAi (27626, VDRC), UAS-Hcf-RNAi (36799, Bloomington Drosophila Stock Center), UAS-LPT-RNAi (25994, Bloomington Drosophila Stock Center), UAS-Ncoa6-RNAi (34964, Bloomington Drosophila Stock Center), UAS-Ptip-RNAi (32133R-1, National Institute of Genetics Japan), UAS-Rbbp5-RNAi (106139, VDRC), UAS-dSet1-RNAi (40682, VDRC), UAS-trr-RNAi (29563, Bloomington Drosophila Stock Center), UAS-trx-RNAi (108122, VDRC), and UAS-dUTX-RNAi (34076, Bloomington Drosophila Stock Center).
All RNAi crosses were performed at 27°C to increase knockdown efficiency.
ctk was a kind gift from Dale Dorsett. trr1 FRT19A/FM7, act-GFP was a kind gift from Alexander Mazo. ubi-GFP FRT19A; ey-FLP was a kind gift from Ishwar Hariharan.
Histone methyltransferase assay
The SET domains of Trr (amino acids 193–421 from AAL39418) and dSet1 (amino acids 1403–1641 from NP_001015221) and full-length human WDR5, ASH2L, RBBP5, and DPY30 (WARD) were each cloned into N-terminally Flag-tagged pBacPAK8. Baculoviruses were grown according to the manufacturer's protocol (BacPAKBaculovirus expression system, Clontech). Sf9 cells were cotransfected with a virus mixture of Trr and human WARD, dSet1 and human WARD, or human WARD followed by α-Flag purification, respectively. Purified COMPASS-like complexes were incubated with 0.5 μg of free histone H3 and 200 μM S-adenosylmethionine in methyltransferase buffer (50 mM Tris-HCl at pH 8.8, 20 mM KCl, 5 mM MgCl2, 0.5 mM dithiothreitol) for 2 h at 30°C. The methylation status of histone H3 was examined by Western blot with the specified antibodies.
Immunofluorescence labeling of imaginal discs
Antibody labeling of wing imaginal discs was performed as described in Herz et al. (2012). Generally, an average of 30 discs was dissected per experiment, and fluorescence intensities were compared between the posterior and anterior compartments or between wild-type and mutant clones.
Primers
Primers for RNAi in S2 cells were as follows: lacZ forward, TAATACGACTCACTATAGGGAGGAATGCTTAATCAGTGAGGCACC; lacZ reverse, TAATACGACTCACTATAGGGAGGAAAGCCATACCAAACGACGAGC; LPT 1 forward, TAATACGACTCACTATAGGGCGACGAGGAGCACTAACTCC; LPT 1 reverse, TAATACGACTCACTATAGGGCTTCTTGAGACCTCGGTTGC; LPT 2 forward, TAATACGACTCACTATAGGGTTGTTGTGAGCATGGAGGAG; LPT 2 reverse, TAATACGACTCACTATAGGGCCGTAGCGTCCACAAAAGTT; dSet1 1 forward, TAATACGACTCACTATAGGGAGCGAAGAAAAGACGACGAA; dSet1 1 reverse, TAATACGACTCACTATAGGGATTTCGTCTGCAGCTATGGG; dSet1 2 forward, TTAATACGACTCACTATAGGGAGAAGAGATTCAGATTCACGTCCTCG; dSet1 2 reverse, TTAATACGACTCACTATAGGGAGAGCTTCATTTGGCTGATGGAGAAC; trr 1 forward, TAATACGACTCACTATAGGGCGGAGACTCGCCTGGCAGCTTCTGC; trr 1 reverse, TAATACGACTCACTATAGGGCCTGGTTGGTGACAAGCGCTACACG; trr 2 forward, TTAATACGACTCACTATAGGGAGAAAGACGGAGCTGCTTCTCGGA; trr 2 reverse, TTAATACGACTCACTATAGGGAGACATCAGCTGGGTTTTCATCTTGG; trx 1 forward, TAATACGACTCACTATAGGGGCCAGTGTGTCCAAGTGCTATGCCC; trx 1 reverse, TAATACGACTCACTATAGGGGCGCTGGCATCCACTTCCATCGTCG; trx 2 forward, TAATACGACTCACTATAGGGGCAATGCAGCAGATCAAAAA; and trx 2 reverse, TAATACGACTCACTATAGGGTCGATTCATCACCAACAGGA.
Quantitative RT–PCR primers were as follows: ash2 forward, TTTATGCCGGCAGCTATTTC; ash2 reverse, GAGCACCTCGGGATACTTGA; Dpy-30L1 forward, GCCGTGGACAACAACTCCTA; Dpy-30L1 reverse, TTCACTGTTCAATTATACAACTAAGGA; Hcf forward, ATCTGCCTCTGCCCTGTTTA; Hcf reverse, ACGTCAGTTGTGCTGCTCAC; LPT forward, CGACAAGAAGCTCAGGAACC; LPT reverse, TGGGTGTGTTCTGCATCATT; Ncoa6 forward, TCCGTTCTGGATACCCTCAC; Ncoa6 reverse, GCGTCTTGCAGGAGAGATTC; Ptip forward, CACCTCATGTCATTGGATGC; Ptip reverse, ACCTGTTGGCGAAGCATTAC; Rbbp5 forward, CGGACAAAACTACCCAGAGG; Rbbp5 reverse, CCTCGTCAGAAAATCCCAGA; dSet1 forward, TCCGGGTTACAATGAGGAAG; rp49 forward, CCAGTCGGATCGATATGCTAA; rp49 reverse, GTTCGATCCGTAACCGATGT; dSet1 reverse, TCCTCGGAGTCGCTGTAAAT; trr forward, TAAGGTGCACAAGTGGTTGC; trr reverse, CTTTCCAGGATTCGCACAAT; trx forward, AAAGGATCAAAACGGTGACG; trx reverse, ACTTGCTCAAGGCTTTTCCA; dUTX forward, AATGTTGGACCCTTGACTGC; and dUTX reverse, TCCTTGCAAGATTCCAGCTT.
RNAi in S2 cells
We used 10 μg of dsRNA per 1.5 × 106 cells per 60-mm well (six-well plate) or 80 μg of dsRNA per 12 × 106 cells per T75 flask with 2.5 mL of SFX medium (1% penicillin/streptomycin) per 60-mm well and 20 mL of SFX medium (1% penicillin/streptomycin) per T75 flask. dsRNA was mixed with the corresponding volume of S2 cells (0.6 × 106 S2 cells per milliliter) and distributed in either 60-mm wells or T75 flasks. RNAi-mediated knockdown was performed for 5 d.
Acknowledgments
We thank Drs. Dale Dorsett, Ishwar Hariharan, Alexander Mazo, and Jerry Workman, the Bloomington Drosophila Stock Center, and the Developmental Studies Hybridoma Bank (DSHB) for providing antibodies and fly stocks. We thank Dr. Edwin Smith for helpful discussions, comments, suggestions, and critical reading of the manuscript. We also thank Jeff Johnston and Sam Meier of the Zeitlinger laboratory for computational analysis advice, and the Molecular Biology core facility at Stowers Institute—namely, Anoja Perera, Rhonda Egidy, and Allison Peak—for creating and sequencing libraries for next-generation sequencing. The work was performed to fulfill, in part, requirements for the Ph.D. thesis research of K.L. as a student registered with the Open University. This investigation has been aided by a grant from The Jane Coffin Childs Memorial Fund for Medical Research to H.-M.H., and funds provided from the National Institute of Health R01CA150265 to A.S.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.201327.112.
References
- Akhtar-Zaidi B, Cowper-Sal-lari R, Corradin O, Saiakhova A, Bartels CF, Balasubramanian D, Myeroff L, Lutterbaugh J, Jarrar A, Kalady MF, et al. 2012. Epigenomic enhancer profiling defines a signature of colon cancer. Science 336: 736–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ardehali MB, Mei A, Zobeck KL, Caron M, Lis JT, Kusch T 2011. Drosophila Set1 is the major histone H3 lysine 4 trimethyltransferase with role in transcription. EMBO J 30: 2817–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashktorab H, Schaffer AA, Daremipouran M, Smoot DT, Lee E, Brim H 2010. Distinct genetic alterations in colorectal cancer. PLoS ONE 5: e8879 doi: 10.1371/journal.pone.0008879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K 2007. High-resolution profiling of histone methylations in the human genome. Cell 129: 823–837 [DOI] [PubMed] [Google Scholar]
- Beisel C, Paro R 2011. Silencing chromatin: Comparing modes and mechanisms. Nat Rev Genet 12: 123–135 [DOI] [PubMed] [Google Scholar]
- Bonn S, Zinzen RP, Girardot C, Gustafson EH, Perez-Gonzalez A, Delhomme N, Ghavi-Helm Y, Wilczynski B, Riddell A, Furlong EE 2012. Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44: 148–156 [DOI] [PubMed] [Google Scholar]
- Briggs SD, Bryk M, Strahl BD, Cheung WL, Davie JK, Dent SY, Winston F, Allis CD 2001. Histone H3 lysine 4 methylation is mediated by Set1 and required for cell growth and rDNA silencing in Saccharomyces cerevisiae. Genes Dev 15: 3286–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulger M, Groudine M 2011. Functional and mechanistic diversity of distal transcription enhancers. Cell 144: 327–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauhan C, Zraly CB, Parilla M, Diaz MO, Dingwall AK 2012. Histone recognition and nuclear receptor co-activator functions of Drosophila Cara Mitad, a homolog of the N-terminal portion of mammalian MLL2 and MLL3. Development 139: 1997–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Ma J, Wu F, Xiong LJ, Ma H, Xu W, Lv R, Li X, Villen J, Gygi SP, et al. 2012. The histone H3 Lys 27 demethylase JMJD3 regulates gene expression by impacting transcriptional elongation. Genes Dev 26: 1364–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung AC, Cramer P 2012. A movie of RNA polymerase II transcription. Cell 149: 1431–1437 [DOI] [PubMed] [Google Scholar]
- Cho YW, Hong T, Hong S, Guo H, Yu H, Kim D, Guszczynski T, Dressler GR, Copeland TD, Kalkum M, et al. 2007. PTIP associates with MLL3- and MLL4-containing histone H3 lysine 4 methyltransferase complex. J Biol Chem 282: 20395–20406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsett D, Eissenberg JC, Misulovin Z, Martens A, Redding B, McKim K 2005. Effects of sister chromatid cohesion proteins on cut gene expression during wing development in Drosophila. Development 132: 4743–4753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dou Y, Milne TA, Ruthenburg AJ, Lee S, Lee JW, Verdine GL, Allis CD, Roeder RG 2006. Regulation of MLL1 H3K4 methyltransferase activity by its core components. Nat Struct Mol Biol 13: 713–719 [DOI] [PubMed] [Google Scholar]
- Eissenberg JC, Shilatifard A 2010. Histone H3 lysine 4 (H3K4) methylation in development and differentiation. Dev Biol 339: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goo YH, Sohn YC, Kim DH, Kim SW, Kang MJ, Jung DJ, Kwak E, Barlev NA, Berger SL, Chow VT, et al. 2003. Activating signal cointegrator 2 belongs to a novel steady-state complex that contains a subset of trithorax group proteins. Mol Cell Biol 23: 140–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallson G, Hollebakken RE, Li T, Syrzycka M, Kim I, Cotsworth S, Fitzpatrick KA, Sinclair DA, Honda BM 2012. dSet1 is the main H3K4 di- and tri-methyltransferase throughout Drosophila development. Genetics 190: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RD, Hess JL, Yu BD, Ernst P, van Lohuizen M, Berns A, van der Lugt NM, Shashikant CS, Ruddle FH, Seto M, et al. 1999. Mammalian Trithorax and polycomb-group homologues are antagonistic regulators of homeotic development. Proc Natl Acad Sci 96: 14372–14377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintzman ND, Stuart RK, Hon G, Fu Y, Ching CW, Hawkins RD, Barrera LO, Van Calcar S, Qu C, Ching KA, et al. 2007. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39: 311–318 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Madden LD, Chen Z, Bolduc C, Buff E, Gupta R, Davuluri R, Shilatifard A, Hariharan IK, Bergmann A 2010. The H3K27me3 demethylase dUTX is a suppressor of Notch- and Rb-dependent tumors in Drosophila. Mol Cell Biol 30: 2485–2497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herz HM, Mohan M, Garrett AS, Miller C, Casto D, Zhang Y, Seidel C, Haug JS, Florens L, Washburn MP, et al. 2012. Polycomb repressive complex 2-dependent and -independent functions of Jarid2 in transcriptional regulation in Drosophila. Mol Cell Biol 32: 1683–1693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes CM, Rozenblatt-Rosen O, Milne TA, Copeland TD, Levine SS, Lee JC, Hayes DN, Shanmugam KS, Bhattacharjee A, Biondi CA, et al. 2004. Menin associates with a trithorax family histone methyltransferase complex and with the hoxc8 locus. Mol Cell 13: 587–597 [DOI] [PubMed] [Google Scholar]
- Issaeva I, Zonis Y, Rozovskaia T, Orlovsky K, Croce CM, Nakamura T, Mazo A, Eisenbach L, Canaani E 2007. Knockdown of ALR (MLL2) reveals ALR target genes and leads to alterations in cell adhesion and growth. Mol Cell Biol 27: 1889–1903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack J, Dorsett D, Delotto Y, Liu S 1991. Expression of the cut locus in the Drosophila wing margin is required for cell type specification and is regulated by a distant enhancer. Development 113: 735–747 [DOI] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K 2011. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J 30: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston DM, Sedkov Y, Petruk S, Riley KM, Fujioka M, Jaynes JB, Mazo A 2011. Ecdysone- and NO-mediated gene regulation by competing EcR/Usp and E75A nuclear receptors during Drosophila development. Mol Cell 44: 51–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharchenko PV, Alekseyenko AA, Schwartz YB, Minoda A, Riddle NC, Ernst J, Sabo PJ, Larschan E, Gorchakov AA, Gu T, et al. 2011. Comprehensive analysis of the chromatin landscape in Drosophila melanogaster. Nature 471: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. 2010. Widespread transcription at neuronal activity-regulated enhancers. Nature 465: 182–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klymenko T, Muller J 2004. The histone methyltransferases Trithorax and Ash1 prevent transcriptional silencing by Polycomb group proteins. EMBO Rep 5: 373–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Khorrami S, Greenblatt JF, Schneider J, Johnston M, Shilatifard A 2002. COMPASS, a histone H3 (lysine 4) methyltransferase required for telomeric silencing of gene expression. J Biol Chem 277: 10753–10755 [DOI] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG 2008. Wdr82 is a C-terminal domain-binding protein that recruits the Setd1A histone H3-Lys4 methyltransferase complex to transcription start sites of transcribed human genes. Mol Cell Biol 28: 609–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Lee DK, Dou Y, Lee J, Lee B, Kwak E, Kong YY, Lee SK, Roeder RG, Lee JW 2006. Coactivator as a target gene specificity determinant for histone H3 lysine 4 methyltransferases. Proc Natl Acad Sci 103: 15392–15397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Tate CM, You JS, Skalnik DG 2007. Identification and characterization of the human Set1B histone H3-Lys4 methyltransferase complex. J Biol Chem 282: 13419–13428 [DOI] [PubMed] [Google Scholar]
- Lee MG, Villa R, Trojer P, Norman J, Yan KP, Reinberg D, Di Croce L, Shiekhattar R 2007. Demethylation of H3K27 regulates polycomb recruitment and H2A ubiquitination. Science 318: 447–450 [DOI] [PubMed] [Google Scholar]
- Levine M 2010. Transcriptional enhancers in animal development and evolution. Curr Biol 20: R754–R763 doi: 10.1016/j.cub.2010.06.070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mar BG, Bullinger L, Basu E, Schlis K, Silverman LB, Dohner K, Armstrong SA 2012. Sequencing histone-modifying enzymes identifies UTX mutations in acute lymphoblastic leukemia. Leukemia 26: 1881–1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller T, Krogan NJ, Dover J, Erdjument-Bromage H, Tempst P, Johnston M, Greenblatt JF, Shilatifard A 2001. COMPASS: A complex of proteins associated with a trithorax-related SET domain protein. Proc Natl Acad Sci 98: 12902–12907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo R, Rao SM, Zhu YJ 2006. Identification of the MLL2 complex as a coactivator for estrogen receptor α. J Biol Chem 281: 15714–15720 [DOI] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Smith ER, Zhang Y, Jackson J, Washburn MP, Florens L, Eissenberg JC, Shilatifard A 2011. The COMPASS family of H3K4 methylases in Drosophila. Mol Cell Biol 31: 4310–4318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Herz HM, Shilatifard A 2012. SnapShot: Histone lysine methylase complexes. Cell 149: 498–498e1 doi: 10.1016/j.cell.2012.03.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montavon T, Soshnikova N, Mascrez B, Joye E, Thevenet L, Splinter E, de Laat W, Spitz F, Duboule D 2011. A regulatory archipelago controls Hox genes transcription in digits. Cell 147: 1132–1145 [DOI] [PubMed] [Google Scholar]
- Morin RD, Mendez-Lago M, Mungall AJ, Goya R, Mungall KL, Corbett RD, Johnson NA, Severson TM, Chiu R, Field M, et al. 2011. Frequent mutation of histone-modifying genes in non-Hodgkin lymphoma. Nature 476: 298–303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong CT, Corces VG 2011. Enhancer function: New insights into the regulation of tissue-specific gene expression. Nat Rev Genet 12: 283–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons DW, Li M, Zhang X, Jones S, Leary RJ, Lin JC, Boca SM, Carter H, Samayoa J, Bettegowda C, et al. 2011. The genetic landscape of the childhood cancer medulloblastoma. Science 331: 435–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasini D, Malatesta M, Jung HR, Walfridsson J, Willer A, Olsson L, Skotte J, Wutz A, Porse B, Jensen ON, et al. 2010. Characterization of an antagonistic switch between histone H3 lysine 27 methylation and acetylation in the transcriptional regulation of Polycomb group target genes. Nucleic Acids Res 38: 4958–4969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel SR, Kim D, Levitan I, Dressler GR 2007. The BRCT-domain containing protein PTIP links PAX2 to a histone H3, lysine 4 methyltransferase complex. Dev Cell 13: 580–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poux S, Horard B, Sigrist CJ, Pirrotta V 2002. The Drosophila trithorax protein is a coactivator required to prevent re-establishment of polycomb silencing. Development 129: 2483–2493 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Schaft D, Shevchenko A, Pijnappel WW, Wilm M, Aasland R, Stewart AF 2001. The Saccharomyces cerevisiae Set1 complex includes an Ash2 homologue and methylates histone 3 lysine 4. EMBO J 20: 7137–7148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Morcillo P, Dorsett D 1999. Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152: 577–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollins RA, Korom M, Aulner N, Martens A, Dorsett D 2004. Drosophila nipped-B protein supports sister chromatid cohesion and opposes the stromalin/Scc3 cohesion factor to facilitate long-range activation of the cut gene. Mol Cell Biol 24: 3100–3111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider J, Wood A, Lee JS, Schuster R, Dueker J, Maguire C, Swanson SK, Florens L, Washburn MP, Shilatifard A 2005. Molecular regulation of histone H3 trimethylation by COMPASS and the regulation of gene expression. Mol Cell 19: 849–856 [DOI] [PubMed] [Google Scholar]
- Schuettengruber B, Martinez AM, Iovino N, Cavalli G 2011. Trithorax group proteins: Switching genes on and keeping them active. Nat Rev Mol Cell Biol 12: 799–814 [DOI] [PubMed] [Google Scholar]
- Sedkov Y, Benes JJ, Berger JR, Riker KM, Tillib S, Jones RS, Mazo A 1999. Molecular genetic analysis of the Drosophila trithorax-related gene which encodes a novel SET domain protein. Mech Dev 82: 171–179 [DOI] [PubMed] [Google Scholar]
- Sedkov Y, Cho E, Petruk S, Cherbas L, Smith ST, Jones RS, Cherbas P, Canaani E, Jaynes JB, Mazo A 2003. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature 426: 78–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seenundun S, Rampalli S, Liu QC, Aziz A, Palii C, Hong S, Blais A, Brand M, Ge K, Dilworth FJ 2010. UTX mediates demethylation of H3K27me3 at muscle-specific genes during myogenesis. EMBO J 29: 1401–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A 2012. The COMPASS family of histone H3K4 methylases: Mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon JA, Kingston RE 2009. Mechanisms of polycomb gene silencing: Knowns and unknowns. Nat Rev Mol Cell Biol 10: 697–708 [DOI] [PubMed] [Google Scholar]
- Simon JA, Tamkun JW 2002. Programming off and on states in chromatin: Mechanisms of Polycomb and trithorax group complexes. Curr Opin Genet Dev 12: 210–218 [DOI] [PubMed] [Google Scholar]
- Smith ER, Lee MG, Winter B, Droz NM, Eissenberg JC, Shiekhattar R, Shilatifard A 2008. Drosophila UTX is a histone H3 Lys27 demethylase that colocalizes with the elongating form of RNA polymerase II. Mol Cell Biol 28: 1041–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steward MM, Lee JS, O'Donovan A, Wyatt M, Bernstein BE, Shilatifard A 2006. Molecular regulation of H3K4 trimethylation by ASH2L, a shared subunit of MLL complexes. Nat Struct Mol Biol 13: 852–854 [DOI] [PubMed] [Google Scholar]
- Takahashi YH, Lee JS, Swanson SK, Saraf A, Florens L, Washburn MP, Trievel RC, Shilatifard A 2009. Regulation of H3K4 trimethylation via Cps40 (Spp1) of COMPASS is monoubiquitination independent: Implication for a Phe/Tyr switch by the catalytic domain of Set1. Mol Cell Biol 29: 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ 2009. CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Conrad PA, Scacheri PC, Harte PJ 2012. The histone demethylase UTX and chromatin remodeler BRM bind directly to CBP and modulate acetylation of histone H3 lysine 27. Mol Cell Biol 32: 2323–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandamme J, Lettier G, Sidoli S, Di Schiavi E, Norregaard Jensen O, Salcini AE 2012. The C. elegans H3K27 demethylase UTX-1 is essential for normal development, independent of its enzymatic activity. PLoS Genet 8: e1002647 doi: 10.1371/journal.pgen.1002647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Haaften G, Dalgliesh GL, Davies H, Chen L, Bignell G, Greenman C, Edkins S, Hardy C, O'Meara S, Teague J, et al. 2009. Somatic mutations of the histone H3K27 demethylase gene UTX in human cancer. Nat Genet 41: 521–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicent GP, Nacht AS, Font-Mateu J, Castellano G, Gaveglia L, Ballare C, Beato M 2011. Four enzymes cooperate to displace histone H1 during the first minute of hormonal gene activation. Genes Dev 25: 845–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, et al. 2009. ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Lin C, Smith ER, Guo H, Sanderson BW, Wu M, Gogol M, Alexander T, Seidel C, Wiedemann LM, et al. 2009. Global analysis of H3K4 methylation defines MLL family member targets and points to a role for MLL1-mediated H3K4 methylation in the regulation of transcriptional initiation by RNA polymerase II. Mol Cell Biol 29: 6074–6085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Lee JE, Cho YW, Xiao Y, Jin Q, Liu C, Ge K 2012. UTX regulates mesoderm differentiation of embryonic stem cells independent of H3K27 demethylase activity. Proc Natl Acad Sci 109: 15324–15329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol 28: 7337–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokoyama A, Wang Z, Wysocka J, Sanyal M, Aufiero DJ, Kitabayashi I, Herr W, Cleary ML 2004. Leukemia proto-oncoprotein MLL forms a SET1-like histone methyltransferase complex with menin to regulate Hox gene expression. Mol Cell Biol 24: 5639–5649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentner GE, Tesar PJ, Scacheri PC 2011. Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21: 1273–1283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q, Li T, Price DH 2012. RNA polymerase II elongation control. Annu Rev Biochem 81: 119–143 [DOI] [PMC free article] [PubMed] [Google Scholar]