Misfolded protein studies of endoplasmic reticulum–associated degradation have relied on overexpressed model substrates, and the identities of endogenous substrates are largely unknown. This study used a proteomic approach to identify unassembled CD147 as an endogenous substrate of the OS-9/SEL1L/Hrd1 ubiquitin ligase.
Abstract
Degradation of folding- or assembly-defective proteins by the endoplasmic reticulum–associated degradation (ERAD) ubiquitin ligase, Hrd1, is facilitated by a process that involves recognition of demannosylated N-glycans by the lectin OS-9/XTP3-B via the adaptor protein SEL1L. Most of our knowledge of the machinery that commits proteins to this fate in metazoans comes from studies of overexpressed mutant proteins in heterologous cells. In this study, we used mass spectrometry to identify core-glycoslyated CD147 (CD147(CG)) as an endogenous substrate of the ERAD system that accumulates in a complex with OS-9 following SEL1L depletion. CD147 is an obligatory assembly factor for monocarboxylate transporters. The majority of newly synthesized endogenous CD147(CG) was degraded by the proteasome in a Hrd1-dependent manner. CD147(CG) turnover was blocked by kifunensine, and interaction of OS-9 and XTP3-B with CD147(CG) was inhibited by mutations to conserved residues in their lectin domains. These data establish unassembled CD147(CG) as an endogenous, constitutive ERAD substrate of the OS-9/SEL1L/Hrd1 pathway.
INTRODUCTION
The endoplasmic reticulum (ER) is the port of entry to the secretory pathway and is the site of production of ∼30% of all proteins synthesized in eukaryotic cells (Ghaemmaghami et al., 2003). Accordingly, this organelle is enriched in molecular chaperones, isomerases, and folding enzymes that are specialized for oxidative protein folding and assembly (Hurtley and Helenius, 1989). Because of the harmful systemic risks of deploying misfolded or incompletely assembled proteins, the ER contains elaborate quality control (QC) machinery that monitors the process of protein folding and assembly and ensures that only correctly folded proteins are released to distal compartments of the secretory pathway (Hammond and Helenius, 1995). Ultimately, proteins that fail to meet stringent criteria for deployment are degraded by cytoplasmic proteasomes in a process known as ER-associated protein degradation (ERAD; Smith et al., 2011).
Degradation of proteins by this system is a sequential process consisting of 1) recognition/commitment within the ER lumen or at the ER membrane, 2) dislocation across the ER membrane, and 3) degradation by the cytoplasmic ubiquitin–proteasome system (UPS). Many of the key components of the mammalian ERAD machinery have been identified and appear to be organized into a modular, distributed network that spans the ER bilayer and is centered around two membrane-embedded ubiquitin E3 ligases, Hrd1 and gp78 (Christianson et al., 2012). Tightly coupled reactions ensure that dislocated proteins emerging at the cytoplasmic surface of the ER are efficiently conjugated with ubiquitin, deglycosylated, and degraded by 26S proteasomes. Although considerable progress has been made in elucidating the identity and organization of the protein components that constitute the core ERAD “machinery,” a critical unanswered question is how proteins are recognized as substrates of this process and how they are delivered to the membrane-embedded complex for dislocation to the cytoplasm, and subsequent processing by the downstream UPS.
One prominent hypothesis posits that recognition of specific trimmed mannose structures on core N-glycans serves as a critical signal committing proteins bearing these marks to the degradative fate (Helenius and Aebi, 2004). The vast majority of proteins entering the secretory pathway are modified by a canonical triantennary N-linked glycan, and progressive removal of terminal mannose residues from this glycan by luminal mannosidases serves as a “timer” to monitor the processes of polypeptide folding and assembly. OS-9 and XTP3-B are ER luminal-resident lectins that bind selectively to N-glycans bearing a terminal C-branch α-1,6-linked mannose (Satoh et al., 2010; Yamaguchi et al., 2010) and to SEL1L, a subunit of the membrane-embedded Hrd1 E3 ubiquitin ligase complex (Christianson et al., 2008; Mueller et al., 2008). Accordingly, proteins that are slow to fold or fail to fold within the ER lumen become enzymatically demannosylated, causing them to be bound by OS-9/XTP3-B and subsequently delivered to the Hrd1 ligase complex via SEL1L. However, the functional relationship between these two lectins and their precise roles in the commitment of proteins to ERAD is unclear, in part because of the lack of an absolute requirement for N-glycans in ERAD (proteins lacking N-glycans are still degraded in a manner dependent on OS-9 or its yeast orthologue Yos9p [ Denic et al., 2006; Christianson et al., 2008]) and because the association of SEL1L with OS-9 and XTP3-B is blocked by mutations in their lectin domains that disrupt conserved contacts with the glycan (Christianson et al., 2008). Although there is compelling evidence to support a role for demannosylation in the commitment of proteins to ERAD, the relationship between OS-9 and XTP3-B and the role of N-glycans in the interaction of these two lectins with substrates and downstream machinery remains unclear.
Did ERAD evolve simply to eliminate proteins that, because of mutation or amino acid misincorporation, are unable to acquire native secondary or tertiary structures? Because most ERAD studies to date have focused on the degradation of mutant, folding- or assembly-defective proteins or orphan subunits of oligomeric complexes that are typically overexpressed in heterologous systems, little is known of “natural” or endogenous substrates of this QC system. In a previous study, we reported that the amount of an exogenous ERAD substrate, the folding-defective null Hong Kong (NHK) variant α-1 antitrypsin, copurifying with S peptide-tagged OS-9 was dramatically increased following short hairpin RNA (shRNA)-mediated depletion of SEL1L (Christianson et al., 2008). This finding, in addition to confirming a requirement for SEL1L in promoting the release of substrates from OS-9 to downstream ERAD components, such as Hrd1, provided an approach with which to identify endogenous substrates of the OS-9/SEL1L/Hrd1 ligase ERAD system. In the present study, we have exploited those observations and used affinity-capture liquid chromatography–tandem mass spectrometry (LC-MS/MS) to identify endogenous proteins that copurify with affinity-tagged OS-9.2 in the absence of SEL1L. Using this approach, we have identified and confirmed core-glycosylated CD147 to be a glycan-dependent substrate of the OS-9/SEL1L/Hrd1 ligase pathway.
RESULTS
Identification of endogenous glycan-dependent ERAD substrates
To identify endogenous substrates of the OS-9/SEL1L/Hrd1ligase ERAD pathway, we used LC-MS/MS analysis to identify proteins copurifying with S-protein affinity-purified (SAP) S peptide–tagged OS-9.2 (S-OS-9.2) from digitonin-solubilized HEK293 cells acutely infected with a lentivirus expressing shRNA targeting SEL1L or a control lentivirus (Table 1 and Supplemental Figure S1B). In lysates from control infected cells, S-OS-9.2 interacted robustly (judged by total spectral counts [TSC] and number of unique peptides) with GRP78/BiP, GRP94, and SEL1L—consistent with our previous findings (Christianson et al., 2008)—as well as with known (Mueller et al., 2008; Christianson et al., 2012) membrane and cytosolic interactors of the Hrd1 ligase complex, including Hrd1, Ubc6e, Derlin-2, and VCP/p97. SEL1L knockdown resulted in a complete loss of downstream interactions, confirming the essential role of SEL1L as a link between OS-9 and the membrane-embedded Hrd1 ligase and its associated membrane and cytoplasmic factors. We also observed a concomitant increase in the abundance of Hsp47 and the previously identified OS-9–interacting luminal chaperones, GRP78/BiP and GRP94, after SEL1L knockdown (Christianson et al., 2008).
TABLE 1:
LC-MS/MS analysis of S-OS-9.2 affinity-purified complexes from digitonin-solubilized cell lysates.
| Controlc | SEL1L shRNAd | ||||||
|---|---|---|---|---|---|---|---|
| Symbola | Uniprot ID | Protein name | Topologyb | TSCe | Peptides | TSCe | Peptides |
| OS9 | Q13438 | OS-9 | L | 165 | 58 | 455 | 108 |
| HSPA5 | P11021 | GRP78/BiP | L | 179 | 50 | 297 | 63 |
| HSP90B1 | P14625 | GRP94 | L | 149 | 54 | 200 | 62 |
| SEL1L | Q9UBV2 | SEL1L | TM | 21 | 10 | ||
| VCP | P55072 | p97/VCP | TMf | 16 | 10 | ||
| SYVN1 | Q86TM6 | Hrd1 | TM | 3 | 1 | ||
| DERL2 | Q9GZP9 | Derlin-2 | TM | 1 | 1 | ||
| UBE2J1 | Q9Y385 | UBC6e | TM | 1 | 1 | ||
| SERPINH1 | P50454 | Hsp47 | L | 7 | 4 | 33 | 14 |
| HYOU1 | Q9Y4L1 | GRP170 | L | 13 | 8 | ||
| UGCGL2 | Q9NYU1 | UGGT2 | L | 10 | 6 | ||
| FOXRED2 | Q8IWF2 | ERFAD | L | 3 | 2 | ||
| DNAJB11 | Q9UBS4 | Erdj3 | L | 2 | 1 | ||
| PPIB | P23284 | Cyclophilin B | L | 2 | 1 | ||
| PLOD2 | O00469 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | L | 5 | 3 | 24 | 15 |
| NUP210 | Q8TEM1 | POM210 | TM | 12 | 8 | ||
| EMMPRIN | P35613 | CD147 | TM | 7 | 3 | ||
| GLT25D1 | Q8NBJ5 | Glycosyltransferase 25 family member 1 | L | 2 | 2 | ||
| CERCAM | Q5T4B2 | Glycosyltransferase 25 family member 3 | L | 2 | 1 | ||
aGene name.
bL, lumenal protein; TM, transmembrane domain–containing protein.
cHEK293 cells infected with an empty lentivirus, not expressing an shRNA.
dHEK293 cells infected with a lentivirus targeting SEL1L.
eTSC, total spectral counts.
fp97/VCP is a cytoplasmic protein that is localized to the ER membrane.
This analysis also identified a number of proteins that were either increased in abundance or detected only in complex with OS-9 following SEL1L depletion. These proteins could be either SEL1L-dependent ERAD substrates or ERAD machinery components that act upstream of SEL1L. Of those proteins with an increased number of unique peptides and/or spectral counts in OS-9.2 complexes that were affinity-purified from SEL1L-depleted cells, we identified potential ERAD substrates using the following criteria: 1) the presence of a predicted ER-targeting signal sequence; 2) known or predicted N-linked glycosylation sites (since we were interested in identifying potential glycan-dependent ERAD substrates that rely on OS-9 for their degradation); and 3) persistent association with S-OS-9.2 following Triton X-100 solubilization (Table S1), since we have previously shown that Triton X-100 disrupts the interaction between SEL1L and the Hrd1 ligase complex (and downstream components), but not the interaction of S-OS-9.2 with SEL1L or with ectopically expressed ERAD substrates (Christianson et al., 2008).
Proteins identified in this analysis that have known associations with the protein folding machinery in the ER lumen include Hsp47, a collagen-specific chaperone (Ragg, 2007); GRP170/ORP150, an atypical chaperone of the Hsp70 family; uridine diphosphate (UDP)-glucose ceramide glucosyltransferase-like protein 2 (UGGT2); Erdj3, a GRP78/BiP cochaperone; and cyclophilin B, an ER peptidyl:prolyl isomerase. Several of these proteins—GRP78/BiP, GRP94, GRP170, Erdj3, and cyclophilin B—have been previously reported to reside in a large, ER-localized multiprotein complex together with another UDP-glucose ceramide glycosyltransferase, UGGT1, proposed to be involved in the folding and maturation of immunoglobulin (Ig) light chains (Meunier et al., 2002). Based on the above criteria, the group of potential ERAD substrates included procollagen-lysine, 2-oxoglutarate 5-dioxygenase (PLOD2), the nuclear pore protein NUP210, and the Ig superfamily member CD147, which is also known as basigin or EMMPRIN. We also detected two additional proteins, the glycosyltransferase 25 family members 1 (GLT25D1) and 3 (CERCAM). However, because of the low apparent abundance of the latter in the SAPs (estimated from TSC), and because the abundance of PLOD2 or NUP210 did not increase following treatment of cells with the proteasome inhibitor MG132 (unpublished data), we chose to focus our attention on CD147.
Inefficient biosynthetic processing and proteasome-mediated degradation of CD147
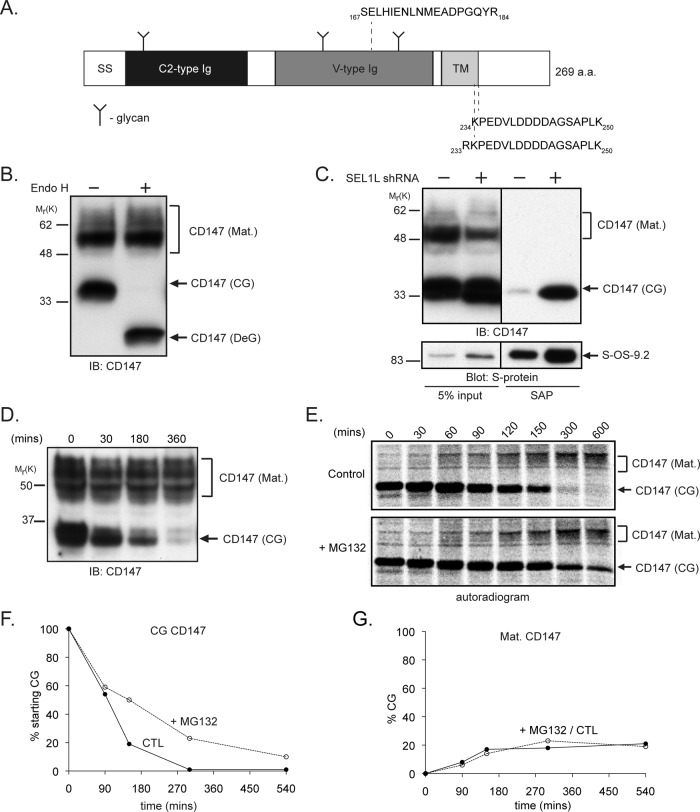
CD147 (Figure 1A) is a 269–amino acid, type I transmembrane (TM) glycoprotein, ubiquitously expressed in chordates, with reported roles in a multitude of cellular processes, including spermatogenesis, T-cell activation, cell adherence, metastasis, and neuronal development (reviewed in Iacono et al., 2007). Structurally, CD147 is composed of an ectodomain containing three confirmed N-glycans (Tang et al., 2004), an amino-terminal Ig C2–type domain, a membrane-proximal Ig V–type domain, a TM domain, and a short cytoplasmic tail of ∼60 residues. Immunoblot analysis (Figure 1B) revealed that endogenous CD147 is expressed at steady state in HEK293 cells, as in other cell types (Figure S2B; Tang et al., 2004; Yu et al., 2006), as a mixture of ∼35 kDa endoglycosidase H (Endo H)–sensitive and ∼45–60 kDa Endo H–insensitive glycoforms that correspond to core-glycosylated (CG) ER-localized and mature (Mat.) polylactosamine-modified (Tang et al., 2004), plasma membrane–localized (Gallagher et al., 2007) species, respectively. Only the CG glycoform was captured with S-OS-9.2; its abundance increased sharply after SEL1L depletion (Figure 1C), confirming our MS analysis and the localization of the CG form to the ER. The S-OS-9.1 splice variant also interacted with CD147(CG) upon SEL1L depletion (unpublished data), consistent with previous studies showing that both OS-9 splice variants interacted with the ERAD substrate NHK α-1 antitrypsin (Christianson et al., 2008).
FIGURE 1:
Identification of CD147 as a putative ERAD substrate. (A) Diagram depicting the human CD147 protein and important features (SS, signal peptide; Ig, immunoglobulin domain; TM, transmembrane domain). Peptides identified in LC-MS/MS analysis of S-OS-9.2 affinity purifications are shown. (B) Profile of CD147 expression in Triton X-100–solubilized whole-cell HEK293 lysates treated with or without Endo H (Mat., mature; CG, core-glycosylated; DeG, deglycosylated). (C) HEK293 cells expressing S-OS-9.2 and infected with lentivirus expressing an shRNA targeting SEL1L (+) or an empty virus as a control (−) were solubilized in 1% digitonin, and S-OS-9.2 complexes were affinity-purified from cell lysates and probed for the indicated proteins. (D) CD147 turnover was assessed by immunoblotting Triton X-100 cell extracts for CD147 after treating cells with cycloheximide for the indicated times. (E) Autoradiogram of a CD147 pulse–chase analysis in HEK293 cells treated with either dimethyl sulfoxide (DMSO; control) or 10 μM MG132 during the chase period. CD147 was isolated from cell lysates with anti-human CD147 8G6, and immunoprecipitates were separated by SDS–PAGE and analyzed with a phosphorimager. (F and G) Quantification of the pulse–chase data experiment from (E), in which degradation of CG and formation of Mat. glycoforms of CD147 are calculated as a percent of starting CG material. CTL, control; IB, immunoblot; SAP, S-peptide affinity purification; S-protein, S-protein-HRP.
We used translational shut-off assays to initially assess the stabilities of CD147 glycoforms. Immunoblot analysis of CD147 in HEK293 cells treated with cycloheximide (Figure 1D) indicated that while CD147(Mat.) was stable, CD147(CG) immunoreactivity disappeared over the 6-h treatment period. Metabolic pulse–chase analysis (Figure 1E) confirmed that CD147(CG) is unstable, with a half-life of ∼90 min (Figure 1F), while the mature glycoform remained stable over the 10-h chase period (Figure 1G). The proteasome inhibitor MG132 increased the stability of CD147(CG), leading to the accumulation of a slightly faster-migrating CD147 species, reflecting continued enzymatic demannosylation of this glycoform (see The role of ER lectins in CD147(CG) degradation.). MG132 treatment did not increase steady-state levels of CD147(CG) but did lead to the appearance of a deglycosylated (DeG) form of CD147 (Figure S2A), presumably representing a dislocated degradation intermediate. The incomplete stabilization of CD147(CG) by MG132 in the pulse–chase analysis could reflect a contribution of nonproteasomal degradation pathways; however, treatment of cells with inhibitors of lysosomal hydrolases failed to increase the stability of CD147(CG) or to lead to the appearance of CD147(DeG) (Figure S2, A and B). It is also possible that MG132 treatment leads to conversion of a fraction of stabilized CD147(CG) into high-molecular-weight, heterogeneous polyubiquitin conjugates that may not be well-resolved by SDS–PAGE, as has been observed for other integral membrane ERAD substrates (Gelman et al., 2002). However, enzymatic deubiquitination of lysates from MG132-treated cells (Figure S2C) did not result in increased abundance of either core-glycosylated or deglycosylated CD147, suggesting that a significant fraction of CD147 does not accumulate as high-molecular-weight polyubiquitinated species following exposure to proteasome inhibitor. Proteasome inhibition can also cause some dislocated ERAD substrates to become insoluble in nonionic detergents (Yu et al., 1997), which would lead to an underestimation of total CD147(CG) recovery. However, we observed no difference in the amount of either CD147 glycoform from MG132-treated cells solubilized with Triton X-100 or SDS (unpublished data). Interestingly, we did observe the appearance of an CD147 immunoreactive species that migrates on SDS–PAGE with faster mobility than the deglycosylated form (Figure S2C), raising the possibility that, in the presence of MG132, CD147 may be still be subject to limited proteolysis. We cannot rule out the possibility that the trypsin-like or postacidic activities of the 20S proteasome, which are not effectively inhibited by MG132 (or indeed by other proteasome inhibitors; Kisselev et al., 2012), could also contribute to the generation of these fragments in the presence of MG132. It is also possible that CD147 could be proteolytically processed by an intramembrane protease like the rhomboid protease RHBDL4, which has recently been described to be present in the ER, where it contributes to the degradation of ERAD substrates that, like CD147, contain a charged residue within the membrane-spanning segment (Fleig et al., 2012).
The gradual appearance of CD147(Mat.) coincident with CD147(CG) disappearance (Figure 1E) suggests a precursor–product relationship between the two major CD147 glycoforms typical of protein maturation into post-ER compartments. Quantification of the pulse–chase data revealed that endogenous CD147 (CG) is converted to the mature glycoform with only 25–30% efficiency (Figure 1G). Stabilization of immature CD147 by MG132 did not increase the fraction converted to the mature form, suggesting, perhaps, that CD147 maturation in the ER depends on the presence of one or more limiting factors. These data suggest that CD147(CG) is inefficiently processed in the ER and that the majority of newly synthesized CD147(CG) is degraded by the proteasome.
Degradation of CD147(CG) requires the Hrd1 ubiquitin ligase complex
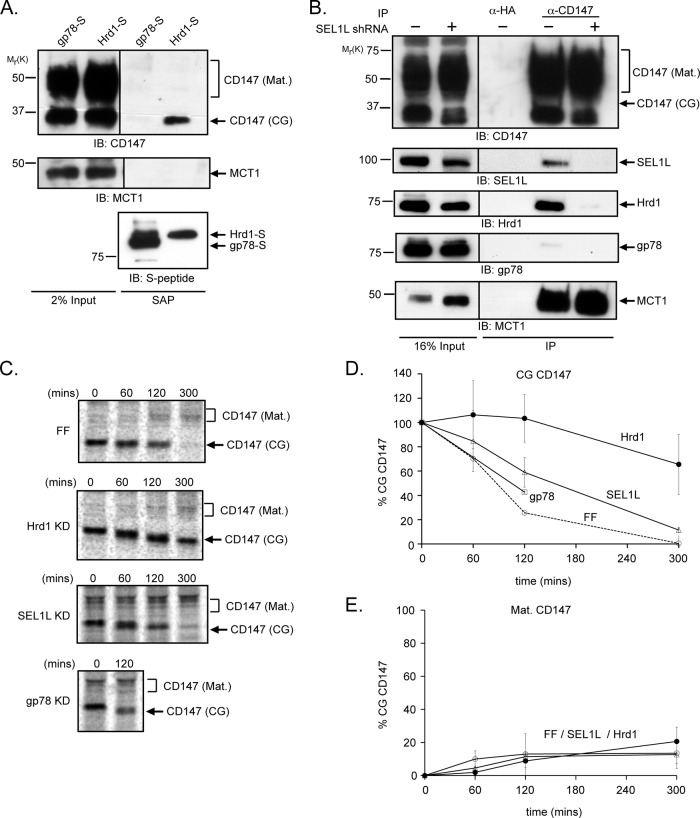
The mammalian ERAD network is organized around two key ubiquitin ligases, Hrd1 and gp78 (Ye et al., 2005; Chen et al., 2006; Christianson et al., 2012). These two ligases share a similar topological organization within the ER membrane, interact with an overlapping set of proteins, and exhibit distinct specificities for different ERAD substrates. Hrd1, but not gp78, forms a complex with SEL1L and OS-9 that facilitates the degradation of substrate proteins by the UPS (Mueller et al., 2008; Christianson et al., 2012). A role for Hrd1 in CD147 degradation was suggested by the detection of CD147 in LC-MS/MS analysis of Hrd1-S SAPs from digitonin-solubilized cell lysates (unpublished data; Christianson et al., 2012). Immunoblot analysis confirmed the presence of CD147(CG) in proteins captured with Hrd1-S, but not with gp78-S (Figure 2A), and endogenous Hrd1, but not gp78, was readily detected in CD147 immunoprecipitates from control HEK293 cells but not from those lacking SEL1L (Figure 2B). Thus endogenous CD147(CG) interacts specifically with endogenous Hrd1 in a SEL1L-dependent manner in HEK293 cells.
FIGURE 2:
Degradation of CD147(CG) is mediated by the Hrd1-SEL1L E3 ligase complex. (A) HEK293 cells expressing either gp78-S or Hrd1-S were lysed in 1% digitonin, and S-protein affinity-purified complexes were immunoblotted for the indicated proteins. (B) Cells infected with either control (−) or SEL1L (+) shRNA-expressing retrovirus were lysed in 1% digitonin, and anti-CD147 immunoprecipitates were probed for the indicated proteins. IB, immunoblot; SAP, S-peptide affinity purification; IP, immunoprecipitation. (C) Pulse–chase analysis of CD147 in HEK293 cells infected with retroviruses harboring shRNAs that target firefly luciferase (FF, control), Hrd1, SEL1L, or gp78. Anti-CD147 immunoprecipitates were separated by SDS–PAGE and analyzed on a phosphorimager. KD, knockdown. (D and E) Quantification of the data in (C) for CD147(CG) degradation (D) or formation of CD147 (Mat.). (E) Each time point represents the average of three independent experiments. The error bars represent the SEM.
Mature CD147 forms a stoichiometric complex with the monocarboxylic acid transporters MCT1, MCT3, and MCT4 (Kirk et al., 2000; Wilson et al., 2002; Fanelli et al., 2003), and previous studies have demonstrated that association of MCT1 and CD147(CG) in the ER is essential for the maturation of both proteins to post-ER compartments (Kirk et al., 2000; Gallagher et al., 2007). MCT1 was readily detected in immunoprecipitates with CD147 antibody (Figure 2B), but was undetectable following affinity capture with Hrd1-S (Figure 2A) or S-OS-9.2 (Figure S3). Thus the pool of CD147(CG) in cells partitions between degradation via OS-9/SEL1L/Hrd1 and maturation via association with MCT1.
Knockdown of endogenous Hrd1, but not gp78, strongly stabilized CD147(CG), but had no measurable effect on the stability of the mature glycoform or the efficiency of CD147(CG) maturation, as assessed by pulse–chase analysis (Figure 2, C–E). In the absence of Hrd1, CD147(CG) gradually became converted to a faster-migrating species, most likely reflecting a prolonged exposure to ER mannosidases (see The role of ER lectins in CD147(CG) degradation) and suggesting Hrd1 is essential for CD147(CG) dislocation to the cytoplasm. Knockdown of SEL1L also stabilized CD147(CG) and led to the accumulation of demannosylated glycoforms (Figure 2C), although the extent of CD147(CG) stabilization was less than that observed after Hrd1 knockdown.
The role of ER lectins in CD147(CG) degradation
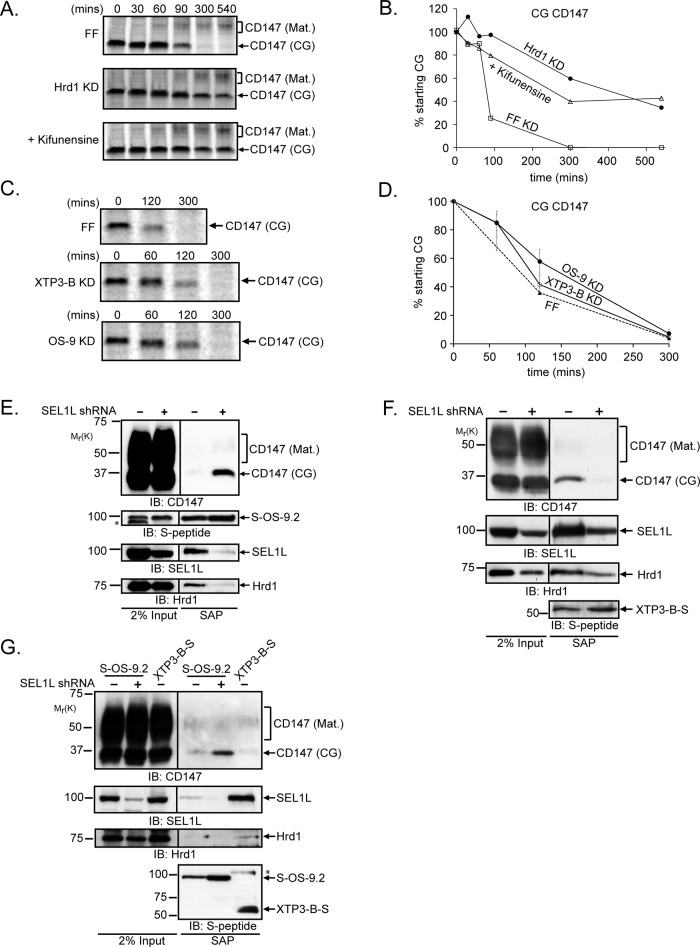
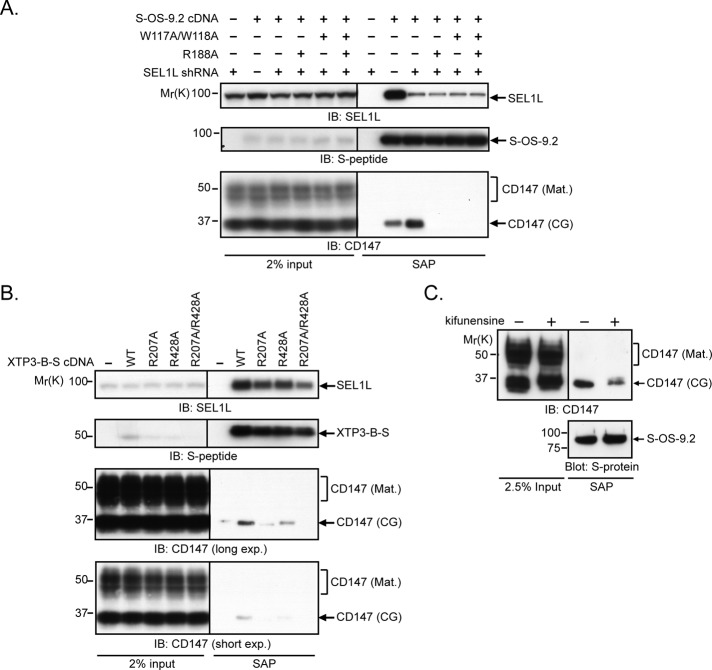
CD147(CG) was strongly stabilized by kifunensine, a selective inhibitor of type I α-mannosidases (Elbein et al., 1990), demonstrating that enzymatic demannosylation is essential for degradation of unassembled CD147(CG) subunits (Figure 3, A and B). The small increase in mobility of CD147(CG) observed during the pulse–chase analyses (Figures 2C and 3A) confirms that CD147 is itself a substrate of this mannosidase activity. We observed partial stabilization of CD147(CG) by depletion of the ER lectins OS-9 and XTP3-B (Figure 3, C and D), suggesting that these lectins may facilitate the delivery of immature CD147(CG) to the Hrd1 ligase complex. Although both lectins have mannose-6-phosphate receptor homology (MRH) domains that bind selectively to the same terminal α-1,6–linked mannose glycans (Satoh et al., 2010), SEL1L depletion had opposite effects on their association with CD147(CG), causing increased association with S-OS-9.2 (Figure 3E) but decreased interaction with XTP3-B-S (Figure 3F). In addition, the interaction of these lectins with CD147(CG) was differentially sensitive to detergents, with the S-OS-9.2–CD147(CG) interaction remaining stable in Triton X-100, and the XTP3-B-S–CD147(CG) interaction being Triton X-100 labile (Figure 3G). These data suggest that these two lectins interact with CD147(CG) in fundamentally different ways. Mutation of conserved residues identified in the crystal structure of the OS-9 MRH domain to make critical contacts with trimmed core glycans (Satoh et al., 2010) largely abolished the ability of both S-OS-9.2 (after SEL1L knockdown; Figure 4A) and XTP3-B-S (Figure 4B) to capture CD147(CG), confirming that the interaction between these ER lectins and CD147(CG) occurs between the lectin domain and a specific glycan structure containing a terminal C-branch α-1,6–linked mannose. This requirement for a mannosidase-trimmed glycan structure in the interaction of OS-9 with CD147(CG) was assessed by affinity purification of S-OS-9.2 from kifunensine-treated, SEL1L-depleted cells. As was seen in the pulse–chase analyses, the steady-state population of CD147(CG) from kifunensine-treated cells exhibited a slower mobility than control-treated cells, indicating an inhibition of mannosidase-mediated trimming (Figure 4C). The mannosidase inhibition resulted in a significant decrease in the amount of CD147(CG) copurifying with S-OS-9.2, demonstrating that OS-9’s interaction with CD147(CG) not only requires its MRH domain but also a mannosidase-trimmed glycan on CD147(CG).
FIGURE 3:
The role of N-glycans and the ER lectins OS-9 and XTP3-B in the degradation of CD147(CG). (A) Pulse–chase analysis of CD147 in cells stably transduced with retroviruses expressing shRNAs targeting firefly luciferase (FF, control), in the presence of vehicle control (DMSO) or 5 μg/ml kifunensine during the chase. Cells stably expressing shRNAs targeting Hrd1 were also analyzed as a positive control. Anti-CD147 immunoprecipitates were separated by SDS–PAGE and analyzed on a phosphorimager. (B) Quantification of the data in (A) for CD147(CG) turnover. (C) Pulse–chase analysis of CD147 in HEK293 cells infected with retroviruses expressing shRNAs that target firefly luciferase (FF, control) XTP3-B or OS-9. Anti-human CD147 immunoprecipitates were separated by SDS–PAGE and analyzed on a phosphorimager. (D) Quantification of the data in (C) for CD147(CG) turnover, with each time point representing the average of at least three independent experiments. Error bars represent SEM. (E) HEK293 cells were cotransfected with vectors expressing S-OS-9.2 and shRNAs targeting SEL1L (+) or firefly luciferase (−). S-OS-9.2 complexes were affinity-purified from 1% digitonin-solubilized cell lysates and analyzed by immunoblotting for the indicated proteins. (F) HEK293 cells stably expressing XTP3-B-S were transduced with lentiviruses harboring shRNAs targeting SEL1L (+) or firefly luciferase (−). S-protein-affinity purifications from 1% digitonin-solubilized lysates were processed as in (E). (G) S-OS-9.2 or XTP3-B-S affinity-purified complexes were isolated from cells expressing firefly luciferase (−) or SEL1L (+) shRNAs and lysed in 1% Triton X-100. * indicates nonspecific immunoreactive bands in (E) and (G). IB, immunoblot. SAP, S-peptide affinity purification; KD, knockdown.
FIGURE 4:
Glycan dependence of the interaction of OS-9 and XTP3-B with CD147(CG). (A) HEK293 cells were cotransfected with wild-type S-OS-9.2 or the indicated S-OS-9.2 MRH domain mutant together with shRNAs targeting firefly luciferase (−) or SEL1L (+). S-OS-9.2 complexes were affinity-purified from 1% digitonin-solubilized cell lysates and analyzed by immunoblotting for the indicated proteins. (B) HEK293 cells were transfected with wild-type XTP3-B-S or the indicated XTP3-B-S MRH domain mutants, and affinity-purified complexes from 1% digitonin-solubilized cell lysates were analyzed by immunoblotting for the indicated proteins. (C) S-OS-9.2 affinity-purified complexes from 1% Triton X-100–solubilized cell lysates were isolated from cells expressing shRNAs targeting SEL1L and treated with either DMSO (−) or 5 μg/ml kifunensine (+), and probed for the indicated proteins. IB, immunoblot. SAP, S-peptide affinity purification. S-protein, S-protein-HRP.
DISCUSSION
While the use of exogenous model ERAD substrates has led to important mechanistic insights into how misfolded and unassembled secretory proteins are degraded, our understanding of this pathway is substantially limited by the paucity of known bona fide endogenous substrates. In this study, we sought to identify novel endogenous ERAD substrates by using a proteomic strategy to identify proteins that accumulate in complex with the substrate recognition factor OS-9 following depletion of the downstream ERAD adaptor SEL1L. Among the proteins that increased in abundance in the absence of SEL1L were luminal chaperones, cochaperones, and proteins with known or inferred roles in protein folding, as well as potential substrates of OS-9– and SEL1L-dependent ERAD. This study identified CD147, a ubiquitously expressed TM protein that is a member of the Ig superfamily, as a constitutive endogenous ERAD substrate. CD147 is not the first described constitutive substrate of the OS-9/SEL1L/Hrd1 E3 ligase pathway. A carboxy-terminal fragment of the hedgehog ligand generated by an intein-like self-cleavage of the hedgehog ligand precursor (Chen et al., 2011) was recently shown to be a substrate of Hrd1-dependent ERAD and major histocompatibility complex (MHC) class I heavy chains synthesized in the absence of their oligomeric partner; β2 microglobulins are also degraded in a Hrd1-dependent manner (Burr et al., 2011). Chordates express two other closely related proteins, neuroplastin/SDR1 (Fossum et al., 1991) and the teratocarcinoma antigen GP70/embigin (Iacono et al., 2007), in addition to CD147, with a common domain organization and nearly identical TM domain sequences. The close similarity of these analogues to CD147 suggest they may also be degraded via ERAD and, along with CD147, may make up a new class of glycan-dependent endogenous substrates of the OS-9/SEL1L/Hrd1 E3 ligase degradation pathway.
CD147 is an obligatory assembly factor for several membrane transporters, of which the best characterized are the monocarboxylate cotransporters MCT1 and MCT4, which mediate proton-dependent efflux of lactate and pyruvate from cells and hence contribute to the regulation of intracellular pH in cells with high glycolytic activity (Kirk et al., 2000; Wilson et al., 2002; Gallagher et al., 2007). Previous studies have indicated that the mechanisms regulating the cellular levels of each of these proteins are quite distinct. MCT1 and MCT4 are transcriptionally up-regulated during cellular adaptation to increases in glycolytic flux and anaerobic conditions (Fanelli et al., 2003; Ullah et al., 2006; Le Floch et al., 2011), while CD147 transcription is relatively constitutive (Fanelli et al., 2003). This apparent “overcapacity” of constitutively expressed CD147 ensures a dynamic supply of CD147(CG) to drive the assembly of mature MCT complexes in response to changing metabolic demand, and is reminiscent of the proposed mechanism of assembly of the T-cell receptor, in which α-subunits that are synthesized in excess of their heterooligomeric partners are constitutively degraded (Chen et al., 1988).
Although the details of how MCT1/4 and CD147 coassemble in the ER membrane are not well understood, it has been suggested that a highly conserved glutamate residue in the middle of CD147’s single TM helix contributes to its assembly with MCT1 and MCT4, possibly via an intramembrane charge-pair interaction with a highly conserved arginine residue in the eighth TM domain in MCTs (Kirk et al., 2000; Manoharan et al., 2006). This paradigm is also reminiscent of the assembly of the T-cell receptor, in which intramembrane charge-pair interactions promote assembly of the heterooligomeric complex (Cosson et al., 1991). It has also been proposed that unpaired charges in TM domains serve as degrons to promote the ERAD of unassembled TCRα and CD3δ subunits (Bonifacino et al., 1990, 1991), but additional factors appear to influence the ability of such unpaired TM-charged residues to serve as degrons (Lankford et al., 1993; Shin et al., 1993). It is tempting to speculate that the putative dual functions in protein complex assembly and degradation attributed to intramembrane paired and unpaired charge interactions could contribute to both assembly of MCT1/4-CD147 complexes and direct excess unassembled CD147(CG) molecules to ERAD. However, this hypothesis has proven difficult to test because exogenously expressed, epitope-tagged versions of CD147 do not partition between assembly and ERAD in a manner reflecting the behavior of the endogenous protein (unpublished data).
Our data reveal that CD147(CG) is strongly stabilized by Hrd1 silencing and kifunensine treatment, indicating an essential role for this ubiquitin ligase and for enzymatic demannosylation in its degradation. It is therefore somewhat curious that knockdown of SEL1L and OS-9 did not stabilize CD147(CG) to a similar extent. This discrepancy could be due to incomplete knockdown or could indicate that some CD147(CG) can be delivered to Hrd1 independently of SEL1L and OS-9, as is the case for NHK α-1 antitrypsin (Christianson et al., 2008) and the integral membrane substrate GluR1 (Christianson et al., 2012) in HEK293 cells, and some ERAD substrates in yeast (Denic et al., 2006; Sato et al., 2009). It is also possible that XTP3-B and OS-9 have partially redundant functions in CD147(CG) degradation, as has been reported for the degradation of a truncated fragment of β-secretase (BACE476Δ; Bernasconi et al., 2010), but we did not observe enhanced stabilization of CD147(CG) in cells expressing a stable XTP3-B knockdown that were also transiently transfected with OS-9 shRNA or in cells transiently cotransfected with both OS-9 and XTP3-B shRNAs (unpublished data), suggesting that, in the case of CD147(CG), these two lectins do not operate in a redundant manner. This result is not altogether surprising in light of a previous study reporting that these two lectins act as interchangeable ERAD shuttles only for proteins such as BACE476Δ that lack TM domains, and not for proteins such as CD3δ that, similar to CD147(CG), contain destabilizing TM degrons (Bernasconi et al., 2010). Indeed, our observation that SEL1L depletion has opposing effects on the interaction between CD147(CG) and these two lectins, together with the observation that the XTP3-B–CD147(CG) complex is disrupted by Triton X-100, while the OS-9–CD147(CG) complex is not, argues against simple redundancy and suggests that the lectins interact with CD147(CG) in different ways. Although our data clearly demonstrate CD147(CG) to be a substrate for mannosidase trimming, to interact with OS-9 and XTP3-B, and to be stabilized by kifunensine, they do not provide evidence for (or against) a causal relationship between trimming of terminal mannose residues on CD147(CG) and its degradation. Many components of the ERAD machinery, including SEL1L, OS-9, and XTP3-B are themselves N-glycosylated, and recent data suggest that their glycans are also substrates for kifunensine-sensitive demannosylation (M.M.P.P. and R.R.K., unpublished data). Our observation that the amount of CD147(CG) copurifying with XTP3-B decreased following SEL1L depletion suggests that the interaction between CD147(CG) and XTP3-B may be mediated by a third protein, perhaps SEL1L, while the interaction between CD147(CG) and OS-9 is likely to occur through direct binding of OS-9’s MRH domain to a trimmed glycan on CD147(CG). Clearly, further study will be needed to fully understand how glycan trimming and luminal lectins collaborate with the SEL1L and Hrd1 complex for different ERAD substrates.
In SEL1L-depleted cells, OS-9 was found to be associated with a set of chaperones and folding enzymes, including GRP170/ORP150, UGGT2, ERFAD, Erdj3, and cyclophilin B that have previously been reported to form a protein complex associated with unassembled, incompletely folded Ig heavy chains (Meunier et al., 2002). We propose that SEL1L depletion creates a “logjam” of malfolded proteins in the ER lumen akin to the situation in cells expressing unassembled Ig heavy chains, leading to the recruitment of a complex rich in molecular chaperones and folding enzymes. Why this protein complex is associated with OS-9 under conditions of SEL1L depletion is less clear. In the absence of SEL1L, it is probable that the malfolded proteins associated with this complex are enriched in demannosylated glycans and hence bind OS-9. Furthermore, all of the members of this complex are glycoproteins and might themselves therefore be susceptible to demannosylation, enabling them to be bound by the OS-9 MRH domain. Finally the observed increase in total S-OS-9.2 abundance following SEL1L knockdown suggests that OS-9 may itself be degraded in a SEL1L/Hrd1-dependent manner, perhaps reflecting codegradation of OS-9, along with its bound substrates. Further study will be needed to elucidate the different roles that OS-9 and XTP3-B contribute to the recognition and commitment of unassembled or misfolded proteins in the early secretory pathway.
MATERIALS AND METHODS
Cell culture and transfection
HEK293 and 293T cells were cultured in DMEM containing 4.5 g/l glucose and l-glutamine (Mediatech, Manassas, VA), and supplemented with 10% FetalPlex animal serum complex (ASC; Gemini Bio-Products, West Sacramento, CA) at 37°C and 5% CO2. Transfection was carried out using a standard calcium-phosphate coprecipitation technique (Kingston et al., 2001). Stable cell lines were selected in 1 mg/ml G418 (Invitrogen, Carlsbad, CA), cloned by limiting dilution, and thereafter maintained in 500 ng/ml G418. Where indicated, cells were treated with MG132 (Enzo Life Sciences, Farmingdale, NY), cycloheximide (Sigma-Aldrich, St. Louis, MO), or kifunensine (Cayman Chemicals, Ann Arbor, MI). Endo H was from NEB (Ipswich, MA).
Plasmids and antibodies
Plasmids for expression of S-peptide epitope-tagged OS-9.1, OS-9.2, OS-9.2(R188A), Hrd1, XTP3-B, SEL1L, and gp78 have been described previously (Christianson et al., 2008, 2012). The W117A/W118A double point mutation was introduced into S-OS-9.2 cDNA by site-directed mutagenesis using the primers 5′-CTGCTTGCTGAAGACAAAGGACGCGGCGACATATGAATTCTGTTATG-3′ and 5′-CATAACAGAATTCATATGTCCACCAGTCCTTTGTCTTCAGCAAGCAG-3′, using the QuikChange Site-Directed Mutagenesis Kit (Agilent, Santa Clara, CA). pSUPERSTAR shRNA expression constructs targeting SEL1L have been described previously (Christianson et al., 2008, 2012).
Rabbit polyclonal antibodies used include: anti–CD147 “B10” (Sun and Hemler, 2001; a gift from Martin Hemler, Harvard University), anti–OS-9 and anti–XTP3-B (Christianson et al., 2008), anti-SEL1L (a gift from H. Ploegh, Whitehead Institute for Biomedical Research), anti-Hrd1 and anti-gp78 (gifts from R. Wojcikiewicz, SUNY Upstate Medical University), and anti-MCT1 (a gift from Nancy Philp, Thomas Jefferson University). Mouse monoclonal antibodies used included anti-CD147 hybridoma clone 8G6 (Berditchevski et al., 1997; a gift from Martin Hemler, Harvard University), anti-CD147 clone A-12 (Santa Cruz Biotechnology, Santa Cruz, CA), anti–S-peptide (EMD Millipore, Billerica, MA), anti-ubiquitin clone P4G7 (Covance, Princeton, NJ), and anti-GAPDH (EMD Millipore). Rabbit polyclonal anti–S-peptide was generated by immunizing rabbits with KLH-conjugated S-peptide, after which the sera were affinity-purified (Covance).
The S-protein-horseradish peroxidase (HRP) conjugate was purchased from Novagen. Anti-rabbit and anti-mouse secondary antibody-HRP conjugates were from GE Healthcare Life Sciences, Pittsburgh, PA.
Viral vectors and virus production
A list of the viruses and shRNA sequences used in these studies can be found in the table in Figure S1F. The lentivirus vector containing an shRNA targeting SEL1L was created by subcloning the ClaI/EcoRI fragment from the pSUPERSTAR-SEL1L construct into pLVTHM according to the Trono Lab (University of Geneva) protocol found at www.addgene.org (Wiznerowicz and Trono, 2003). Empty pLVTHM not containing an shRNA sequence was used as a negative control. The pRetroQ retrovirus vector targeting OS-9 and the control pRetroQ vector targeting firefly luciferase were generous gifts from Albert Koong (Stanford University). The pMSCV vectors targeting Hrd1, gp78, XTP3-B, or the control firefly sequence were generated by subcloning from pSM2 vectors (gifts from Wade Harper, Harvard University), as previously described (Schlabach et al., 2008). Both the lentivirus and retrovirus shuttle plasmids were cotransfected into 293T cells along with vectors pMD2.VSVG and pCMV-dR.74 or pCG-GagPol and pVSV-G, respectively, using the calcium-phosphate coprecipitation technique or Fugene6 (Roche, Indianapolis, IN). Infectious lentivirus or retrovirus particles were concentrated from tissue culture supernatants by either ultracentrifugation at 50,000 × g, 16°C, in an SW41 rotor (Beckman Coulter, Brea, CA) for 2 h or using 5X PEG-it (System Biosciences, Mountain View, CA) according to manufacturer's protocol, resuspended in 1X phosphate-buffered saline (PBS) and stored at −80°C. Lentiviral titers were determined by infecting HEK293 cells and measuring green fluorescent protein fluorescence by flow cytometry. Retroviral titers were determined by infecting HEK293 cells with various dilutions of virus and selecting with 1 μg/ml puromycin.
Cell lysis, SAP, immunoblotting, and MS analysis
cDNA-transfected or virus-transduced HEK293 cells were washed once in ice-cold 1X PBS; harvested; and lysed in buffer containing 50 mM Tris-HCl (pH 7.3), 150 mM NaCl, 5 mM EDTA, protease inhibitors (Roche), and either 1% (wt/vol) digitonin (Calbiochem, San Diego, CA) or 1% (vol/vol) Triton X-100; and incubated for 1 h at 4°C. The lysates were cleared by centrifugation at 20,000 × g for 15 min at 4°C. Protein concentration was measured by bicinchoninic acid protein assay (BCA; Pierce, Rockford, IL). Normalized lysates were incubated with S-protein agarose (Novagen) for 2–16 h at 4°C with mixing, and beads were washed three to five times in lysis buffer containing either 0.1% (wt/vol) digitonin or 1% (vol/vol) Triton X-100, depending on the lysis conditions. For immunoblot analysis, purified proteins were eluted in Laemmli buffer, separated by SDS–PAGE, and transferred to polyvinylidene fluoride (PVDF; GE Healthcare Life Sciences, Hercules, CA). The PVDF membranes were blocked in 5% nonfat milk and incubated with primary antibodies, which were followed by HRP-conjugated secondary antibodies, and immunoreactivity was detected using ECL+ chemiluminescence reagents (GE Healthcare, Waukesha, WI). For LC-MS/MS analysis, 1 mg of cell lysate was precleared with 50% (vol/vol) Sephadex G-100 beads (Sigma-Aldrich) for 60 min prior to incubation with S-protein agarose. The affinity-purified protein complexes were washed three times with lysis buffer and then twice in 50 mM ammonium bicarbonate (pH 8.0) eluted in 0.1% RapiGest SF (Waters, Milford, MA) for 20 h at 37°C, incubated with trypsin for 16 h at 37°C, and subjected to MS using a linear ion-trap mass spectrometer, as previously described (Christianson et al., 2008).
Immunoprecipitation of endogenous CD147
The mouse monoclonal antibodies to CD147 (8G6) or hemagglutinin (HA; 12C5; Sigma-Aldrich), as a control, were purified using the Pierce Melon Gel IgG Spin Purification Kit. Antibody (25 μg) was then cross-linked to a bead matrix using the Pierce Direct IP kit according to the manufacturer's protocol. After the cells were lysed in buffer containing 1% (wt/vol) digitonin, as described above, 250 μg soluble lysate was added to the antibody-conjugated matrix, and the lysate was incubated for 2 h at 4°C with gentle mixing. The immunoprecipitated complexes were washed five times in lysis buffer and then eluted with Pierce IgG Elution Buffer. After being neutralized with 1M Tris (pH 9.5) protein, the protein elutes were separated by SDS–PAGE in Pierce 5X Loading Buffer, transferred to PVDF, and analyzed by immunoblotting. Antibodies from previous blottings were stripped off by Pierce Restore Western Blot Stripping Buffer prior to subsequent immunoblot analysis.
Radiolabeling and pulse–chase analysis
Prior to radiolabeling, 1.8 × 106 HEK293 cells were plated on poly-l-lysine–coated plates in complete media. The next day, the cell monolayers were washed twice with “cold” labeling media containing DMEM, 4.5 g/l glucose and sodium pyruvate, l-methionine, l-cysteine, penicillin–streptomycin, and 10% dialyzed ASC. The cells were then starved in cold media lacking l-methionine and l-cysteine for 15 min; this was followed by radiolabeling in media containing 125 μCi/ml 35S-labeled cysteine/methionine (Easytag Express Protein Labeling Mix 35S; Perkin Elmer-Cetus, Waltham, MA) for 30 min. The cell monolayers were then quickly washed twice with 3 ml of 1X Hanks Buffered Saline Solution (Invitrogen) and chased in complete media supplemented with cold cysteine and methionine added to a final concentration of 2.5 mM each. At specified time points following this, the cells were harvested in ice-cold 1X PBS and collected by centrifugation, and the pellets were frozen and stored at −80°C. Cell pellets were lysed in buffer containing 50 mM Tris (pH 7.4), 150 mM NaCl, 5 mM EDTA, 0.5% (wt/vol) sodium deoxycholate, 1% (vol/vol) Triton X-100, and protease inhibitors (Roche) at 4°C for 1 h while mixing. The lysates were cleared by centrifugation at 20,000 × g for 15 min at 4°C, and normalized according to labeling specificity using trichloroacetic acid (TCA) precipitation of proteins, as previously described (Ward and Kopito, 1994). Lysates were precleared with protein G beads (GenScript, Piscataway, NJ) for 2 h at 4°C with mixing. CD147 was then immunoprecipitated by incubation with 5 μg of mouse 8G6 antibody and protein G beads overnight at 4°C with mixing. Immunoprecipitated proteins were washed three times in lysis buffer, eluted by boiling in Laemmli buffer, and separated by SDS–PAGE, and the gels were dried. The radioactive bands were detected and quantified on a phosphorimager (Molecular Dynamics, Sunnyvale, CA). The percent CG (%CG) for CD147(CG) and CD147(Mat.) was calculated by dividing the phophorimager CG or Mat. band intensity at a specific time by that of the CG at the t = 0 time point and then multiplying by 100%.
Deubiquitination of cellular extracts
HEK293 cells incubated in the presence or absence of MG132 for 12 h were washed in PBS and solubilized in a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl + 1% TX-100. The catalytic core of the deubiquitinating enzyme Usp2 (Usp2-cc; Catanzariti et al., 2004; Ryu et al., 2006) was purified and kindly provided by Stephen Kaiser (St. Jude Children's Research Hospital, Memphis, TN). Lysate (40 μg) was incubated in the presence or absence of 2 μg Usp2-cc for 1 h at 37°C in a buffer containing 50 mM Tris-HCl (pH 7.4), 150 mM NaCl + 1% TX100 + 5 mM EDTA + 1.4 mM DTT. Deubiquitinated proteins were separated by SDS–PAGE and analyzed by immunoblotting with anti-CD147 and anti-ubiquitin antibodies.
Supplementary Material
Acknowledgments
This work was supported by a grant from the National Institute of General Medical Science to R.R.K. R.E.T. was supported by a National Research Service Award (NRSA) postdoctoral award from the National Institute of Diabetes and Digestive and Kidney Diseases, M.M.P.P. by an NRSA postdoctoral award from the National Institute of Neurological Disorders and Stroke, J.A.O. by an NRSA postdoctoral award from NIGMS, and E.J.G. by a National Institutes of Health predoctoral training grant. We thank Nancy Philp and members of the Kopito laboratory for their insightful discussions, and J. Hwang for critical reading of the manuscript.
Abbreviations used:
- ASC
animal serum complex
- BACE476Δ
truncated fragment of β-secretase
- CG
core-glycosylated
- DeG
deglycosylated
- DMSO
dimethyl sulfoxide
- Endo H
endoglycosidase H
- ER
endoplasmic reticulum
- ERAD
ER-associated degradation
- HA
hemagglutinin
- HRP
horseradish peroxidase
- Ig
immunoglobulin
- LC-MS/MS
liquid chromatography–tandem mass spectrometry
- Mat.
mature
- MCT
monocarboxylic acid transporter
- MRH
mannose-6-phosphate receptor homology
- NHK
null Hong Kong variant
- PBS
phosphate-buffered saline
- PLOD2
procollagen-lysine, 2-oxoglutarate 5-dioxygenase
- PVDF
polyvinylidene fluoride
- QC
quality control
- SAP
S-protein affinity-purified
- shRNA
short hairpin RNA
- S-OS-9.2
S peptide–tagged OS-9.2
- TM
transmembrane
- TSC
total spectral counts
- UGGT2
UDP-glucose ceramide glucosyltransferase-like protein 2
- UPS
ubiquitin–proteasome system
- Usp2-cc
catalytic core of the deubiquitinating enzyme Usp2
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-06-0428) on October 24, 2012.
REFERENCES
- Berditchevski F, Chang S, Bodorova J, Hemler ME. Generation of monoclonal antibodies to integrin-associated proteins. J Biol Chem. 1997;272:29174–29180. doi: 10.1074/jbc.272.46.29174. [DOI] [PubMed] [Google Scholar]
- Bernasconi R, Galli C, Calanca V, Nakajima T, Molinari M. Stringent requirement for HRD1, SEL1L, and OS-9/XTP3-B for disposal of ERAD-LS substrates. J Cell Biol. 2010;188:223–235. doi: 10.1083/jcb.200910042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Klausner RD. Colocalized transmembrane determinants for ER degradation and subunit assembly explain the intracellular fate of TCR chains. Cell. 1990;63:503–513. doi: 10.1016/0092-8674(90)90447-m. [DOI] [PubMed] [Google Scholar]
- Bonifacino JS, Cosson P, Shah N, Klausner RD. Role of potentially charged transmembrane residues in targeting proteins for retention and degradation within the endoplasmic reticulum. EMBO J. 1991;10:2783–2793. doi: 10.1002/j.1460-2075.1991.tb07827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burr ML, Cano F, Svobodova S, Boyle LH, Boname JM, Lehner PJ. HRD1 and UBE2J1 target misfolded MHC class I heavy chains for endoplasmic reticulum-associated degradation. Proc Natl Acad Sci USA. 2011;108:2034–2039. doi: 10.1073/pnas.1016229108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti AM, Soboleva TA, Jans DA, Board PG, Baker RT. An efficient system for high-level expression and easy purification of authentic recombinant proteins. Protein Sci. 2004;13:1331–1339. doi: 10.1110/ps.04618904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen B, Mariano J, Tsai YC, Chan AH, Cohen M, Weissman AM. The activity of a human endoplasmic reticulum-associated degradation E3, gp78, requires its Cue domain, RING finger, and an E2-binding site. Proc Natl Acad Sci USA. 2006;103:341–346. doi: 10.1073/pnas.0506618103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Bonifacino JS, Yuan LC, Klausner RD. Selective degradation of T cell antigen receptor chains retained in a pre-Golgi compartment. J Cell Biol. 1988;107:2149–2161. doi: 10.1083/jcb.107.6.2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Tukachinsky H, Huang CH, Jao C, Chu YR, Tang HY, Mueller B, Schulman S, Rapoport TA, Salic A. Processing and turnover of the Hedgehog protein in the endoplasmic reticulum. J Cell Biol. 2011;192:825–838. doi: 10.1083/jcb.201008090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Olzmann JA, Shaler TA, Sowa ME, Bennett EJ, Richter CM, Tyler RE, Greenblatt EJ, Harper JW, Kopito RR. Defining human ERAD networks through an integrative mapping strategy. Nat Cell Biol. 2012;14:93–105. doi: 10.1038/ncb2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christianson JC, Shaler TA, Tyler RE, Kopito RR. OS-9 and GRP94 deliver mutant α1-antitrypsin to the Hrd1-SEL1L ubiquitin ligase complex for ERAD. Nat Cell Biol. 2008;10:272–282. doi: 10.1038/ncb1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosson P, Lankford SP, Bonifacino JS, Klausner RD. Membrane protein association by potential intramembrane charge pairs. Nature. 1991;351:414–416. doi: 10.1038/351414a0. [DOI] [PubMed] [Google Scholar]
- Denic V, Quan EM, Weissman JS. A luminal surveillance complex that selects misfolded glycoproteins for ER-associated degradation. Cell. 2006;126:349–359. doi: 10.1016/j.cell.2006.05.045. [DOI] [PubMed] [Google Scholar]
- Elbein AD, Tropea JE, Mitchell M, Kaushal GP. Kifunensine, a potent inhibitor of the glycoprotein processing mannosidase I. J Biol Chem. 1990;265:15599–15605. [PubMed] [Google Scholar]
- Fanelli A, Grollman EF, Wang D, Philp NJ. MCT1 and its accessory protein CD147 are differentially regulated by TSH in rat thyroid cells. Am J Phys Endocrinol Metab. 2003;285:E1223–E1229. doi: 10.1152/ajpendo.00172.2003. [DOI] [PubMed] [Google Scholar]
- Fleig L, Bergbold N, Sahasrabudhe P, Geiger B, Kaltak L, Lemberg MK. Ubiquitin-dependent intramembrane rhomboid protease promotes ERAD of membrane proteins. Mol Cell. 2012;47:558–569. doi: 10.1016/j.molcel.2012.06.008. [DOI] [PubMed] [Google Scholar]
- Fossum S, Mallett S, Barclay AN. The MRC OX-47 antigen is a member of the immunoglobulin superfamily with an unusual transmembrane sequence. Eur J Immunol. 1991;21:671–679. doi: 10.1002/eji.1830210320. [DOI] [PubMed] [Google Scholar]
- Gallagher SM, Castorino JJ, Wang D, Philp NJ. Monocarboxylate transporter 4 regulates maturation and trafficking of CD147 to the plasma membrane in the metastatic breast cancer cell line MDA-MB-231. Cancer Res. 2007;67:4182–4189. doi: 10.1158/0008-5472.CAN-06-3184. [DOI] [PubMed] [Google Scholar]
- Gelman MS, Kannegaard ES, Kopito RR. A principal role for the proteasome in endoplasmic reticulum-associated degradation of misfolded intracellular cystic fibrosis transmembrane conductance regulator. J Biol Chem. 2002;277:11709–11714. doi: 10.1074/jbc.M111958200. [DOI] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. Global analysis of protein expression in yeast. Nature. 2003;425:737–741. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- Hammond C, Helenius A. Quality control in the secretory pathway. Curr Opin Cell Biol. 1995;7:523–529. doi: 10.1016/0955-0674(95)80009-3. [DOI] [PubMed] [Google Scholar]
- Helenius A, Aebi M. Roles of N-linked glycans in the endoplasmic reticulum. Annu Rev Biochem. 2004;73:1019–1049. doi: 10.1146/annurev.biochem.73.011303.073752. [DOI] [PubMed] [Google Scholar]
- Hurtley SM, Helenius A. Protein oligomerization in the endoplasmic reticulum. Annu Rev Cell Biol. 1989;5:277–307. doi: 10.1146/annurev.cb.05.110189.001425. [DOI] [PubMed] [Google Scholar]
- Iacono KT, Brown AL, Greene MI, Saouaf SJ. CD147 immunoglobulin superfamily receptor function and role in pathology. Exp Mol Pathol. 2007;83:283–295. doi: 10.1016/j.yexmp.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingston RE, Chen CA, Rose JK. Calcium phosphate transfection. Curr Protoc Mol Biol. 2001;63:9.1.1–9.1.11. doi: 10.1002/0471142727.mb0901s63. [DOI] [PubMed] [Google Scholar]
- Kirk P, Wilson MC, Heddle C, Brown MH, Barclay AN, Halestrap AP. CD147 is tightly associated with lactate transporters MCT1 and MCT4 and facilitates their cell surface expression. EMBO J. 2000;19:3896–3904. doi: 10.1093/emboj/19.15.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kisselev AF, van der Linden WA, Overkleeft HS. Proteasome inhibitors: an expanding army attacking a unique target. Chem Biol. 2012;19:99–115. doi: 10.1016/j.chembiol.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankford SP, Cosson P, Bonifacino JS, Klausner RD. Transmembrane domain length affects charge-mediated retention and degradation of proteins within the endoplasmic reticulum. J Biol Chem. 1993;268:4814–4820. [PubMed] [Google Scholar]
- Le Floch R, Chiche J, Marchiq I, Naiken T, Ilc K, Murray CM, Critchlow SE, Roux D, Simon MP, Pouysségur J. CD147 subunit of lactate/H+ symporters MCT1 and hypoxia-inducible MCT4 is critical for energetics and growth of glycolytic tumors. Proc Natl Acad Sci USA. 2011;108:16663–16668. doi: 10.1073/pnas.1106123108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoharan C, Wilson MC, Sessions RB, Halestrap AP. The role of charged residues in the transmembrane helices of monocarboxylate transporter 1 and its ancillary protein basigin in determining plasma membrane expression and catalytic activity. Mol Membr Biol. 2006;23:486–498. doi: 10.1080/09687860600841967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller B, Klemm EJ, Spooner E, Claessen JH, Ploegh HL. SEL1L nucleates a protein complex required for dislocation of misfolded glycoproteins. Proc Natl Acad Sci USA. 2008;105:12325–12330. doi: 10.1073/pnas.0805371105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragg H. The role of serpins in the surveillance of the secretory pathway. Cell Mol Life Sci. 2007;64:2763–2770. doi: 10.1007/s00018-007-7157-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu KY, Baker RT, Kopito RR. Ubiquitin-specific protease 2 as a tool for quantification of total ubiquitin levels in biological specimens. Anal Biochem. 2006;353:153–155. doi: 10.1016/j.ab.2006.03.038. [DOI] [PubMed] [Google Scholar]
- Sato BK, Schulz D, Do PH, Hampton RY. Misfolded membrane proteins are specifically recognized by the transmembrane domain of the Hrd1p ubiquitin ligase. Mol Cell. 2009;34:212–222. doi: 10.1016/j.molcel.2009.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satoh T, Chen Y, Hu D, Hanashima S, Yamamoto K, Yamaguchi Y. Structural basis for oligosaccharide recognition of misfolded glycoproteins by OS-9 in ER-associated degradation. Mol Cell. 2010;40:905–916. doi: 10.1016/j.molcel.2010.11.017. [DOI] [PubMed] [Google Scholar]
- Schlabach MR, et al. Cancer proliferation gene discovery through functional genomics. Science. 2008;319:620–624. doi: 10.1126/science.1149200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin J, Lee S, Strominger JL. Translocation of TCR alpha chains into the lumen of the endoplasmic reticulum and their degradation. Science. 1993;259:1901–1904. doi: 10.1126/science.8456316. [DOI] [PubMed] [Google Scholar]
- Smith MH, Ploegh HL, Weissman JS. Road to ruin: targeting proteins for degradation in the endoplasmic reticulum. Science. 2011;334:1086–1090. doi: 10.1126/science.1209235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Hemler ME. Regulation of MMP-1 and MMP-2 production through CD147/extracellular matrix metalloproteinase inducer interactions. Cancer Res. 2001;61:2276–2281. [PubMed] [Google Scholar]
- Tang W, Chang SB, Hemler ME. Links between CD147 function, glycosylation, and caveolin-1. Mol Biol Cell. 2004;15:4043–4050. doi: 10.1091/mbc.E04-05-0402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullah MS, Davies AJ, Halestrap AP. The plasma membrane lactate transporter MCT4, but not MCT1, is up-regulated by hypoxia through a HIF-1alpha-dependent mechanism. J Biol Chem. 2006;281:9030–9037. doi: 10.1074/jbc.M511397200. [DOI] [PubMed] [Google Scholar]
- Ward CL, Kopito RR. Intracellular turnover of cystic fibrosis transmembrane conductance regulator. Inefficient processing and rapid degradation of wild-type and mutant proteins. J Biol Chem. 1994;269:25710–25718. [PubMed] [Google Scholar]
- Wilson MC, Meredith D, Halestrap AP. Fluorescence resonance energy transfer studies on the interaction between the lactate transporter MCT1 and CD147 provide information on the topology and stoichiometry of the complex in situ. J Biol Chem. 2002;277:3666–3672. doi: 10.1074/jbc.M109658200. [DOI] [PubMed] [Google Scholar]
- Wiznerowicz M, Trono D. Conditional suppression of cellular genes: lentivirus vector-mediated drug-inducible RNA interference. J Virol. 2003;77:8957–8961. doi: 10.1128/JVI.77.16.8957-8961.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi D, Hu D, Matsumoto N, Yamamoto K. Human XTP3-B binds to α1-antitrypsin variant nullHong Kong via the C-terminal MRH domain in a glycan-dependent manner. Glycobiology. 2010;20:348–355. doi: 10.1093/glycob/cwp182. [DOI] [PubMed] [Google Scholar]
- Ye Y, Shibata Y, Kikkert M, van Voorden S, Wiertz E, Rapoport TA. Recruitment of the p97 ATPase and ubiquitin ligases to the site of retrotranslocation at the endoplasmic reticulum membrane. Proc Natl Acad Sci USA. 2005;102:14132–14138. doi: 10.1073/pnas.0505006102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Kaung G, Kobayashi S, Kopito RR. Cytosolic degradation of T-cell receptor α chains by the proteasome. J Biol Chem. 1997;272:20800–20804. doi: 10.1074/jbc.272.33.20800. [DOI] [PubMed] [Google Scholar]
- Yu X-L, Jiang J-L, Li L, Feng Q, Xu J, Chen Z-N. The glycosylation characteristic of hepatoma-associated antigen HAb18G/CD147 in human hepatoma cells. Int J Biochem Cell Biol. 2006;38:1939–1945. doi: 10.1016/j.biocel.2006.05.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.