Centrins are a family of small calcium-binding proteins found at basal bodies. The family can be separated into two groups based on sequence homology to the human centrins. The model organism Tetrahymena was used to show that the two centrin groups have similar but distinct functions at basal bodies.
Abstract
Centrins are a ubiquitous family of small Ca2+-binding proteins found at basal bodies that are placed into two groups based on sequence similarity to the human centrins 2 and 3. Analyses of basal body composition in different species suggest that they contain a centrin isoform from each group. We used the ciliate protist Tetrahymena thermophila to gain a better understanding of the functions of the two centrin groups and to determine their potential redundancy. We have previously shown that the Tetrahymena centrin 1 (Cen1), a human centrin 2 homologue, is required for proper basal body function. In this paper, we show that the Tetrahymena centrin 2 (Cen2), a human centrin 3 homologue, has functions similar to Cen1 in basal body orientation, maintenance, and separation. The two are, however, not redundant. A further examination of human centrin 3 homologues shows that they function in a manner distinct from human centrin 2 homologues. Our data suggest that basal bodies require a centrin from both groups in order to function correctly.
INTRODUCTION
The basal body is an important microtubule organizing center (MTOC) widely found in eukaryotes, except for yeast and higher plants, and is responsible for nucleating and organizing the ciliary axoneme and anchoring it at the surface of the cell. Cilia are microtubule‑based appendages that play key roles in cell locomotion, fluid flow, and mechanosensory, chemical sensory, or photosensory functions (Marshall and Nonaka, 2006). Aberrations in ciliary or basal body function have been associated with several human diseases, such as Meckel‑Gruber syndrome and polycystic kidney disease (Badano et al., 2006). The importance of basal bodies is further highlighted by their ability to be interconverted with centrioles, components of the centrosome, which is involved in mitotic spindle formation (Bornens and Azimzadeh, 2007).
The basal body has a key role in the regulation of ciliary function. For example, basal bodies in the multiciliated cells of the oviduct epithelium are specifically orientated to produce directional fluid flow that transports the egg from the ovaries to the uterus (Marshall and Kintner, 2008). Likewise, the maintenance of the basal body is important, and an incorrectly maintained basal body will likely disrupt ciliary functions. We have previously identified centrin as a protein involved in both basal body orientation and maintenance (Stemm-Wolf et al., 2005; Vonderfecht et al., 2011). Centrins are a ubiquitous family of small Ca2+-binding proteins found at MTOCs (Salisbury et al., 1984; Huang et al., 1988; Andersen et al., 2003; Keller et al., 2005; Kilburn et al., 2007; Liu et al., 2007; Hodges et al., 2010; Carvalho-Santos et al., 2011). Structurally, they consist of two domains connected by a short linker, with each domain containing a pair of EF hands, a Ca2+-binding motif (Veeraraghavan et al., 2002; Hu and Chazin, 2003; Matei et al., 2003; Yang et al., 2005; Li et al., 2006; Thompson et al., 2006). Alignments of centrins from a wide range of organisms show that centrins separate into two groups (Figure 1A). These two groups are annotated based on sequence similarity to the human centrins, centrin 2 and centrin 3 (Middendorp et al., 1997; Laoukili et al., 2000; Bornens and Azimzadeh, 2007). Despite this separation, the two groups share high sequence similarity (74.4% for the human centrins 2 and 3). Analyses of centriole or basal body composition in different species suggests that they contain a centrin isoform from each group (Figure 1A; Andersen et al., 2003; Keller et al., 2005; Kilburn et al., 2007; Liu et al., 2007). Most vertebrates have two centrins belonging to the human centrin 2 group, with one that appears to be ubiquitously expressed and the other expressed only in ciliated cells (Wolfrum and Salisbury, 1998; Laoukili et al., 2000). Budding yeast has only one centrin isoform (Cdc31p, a human centrin 3 homologue); however, the spindle pole body, the only MTOC in yeast, has no structural similarity to a basal body or centriole (Jaspersen and Winey, 2004), suggesting that having the two centrin isoforms at basal bodies or centrioles has been conserved throughout evolution (Bornens and Azimzadeh, 2007).
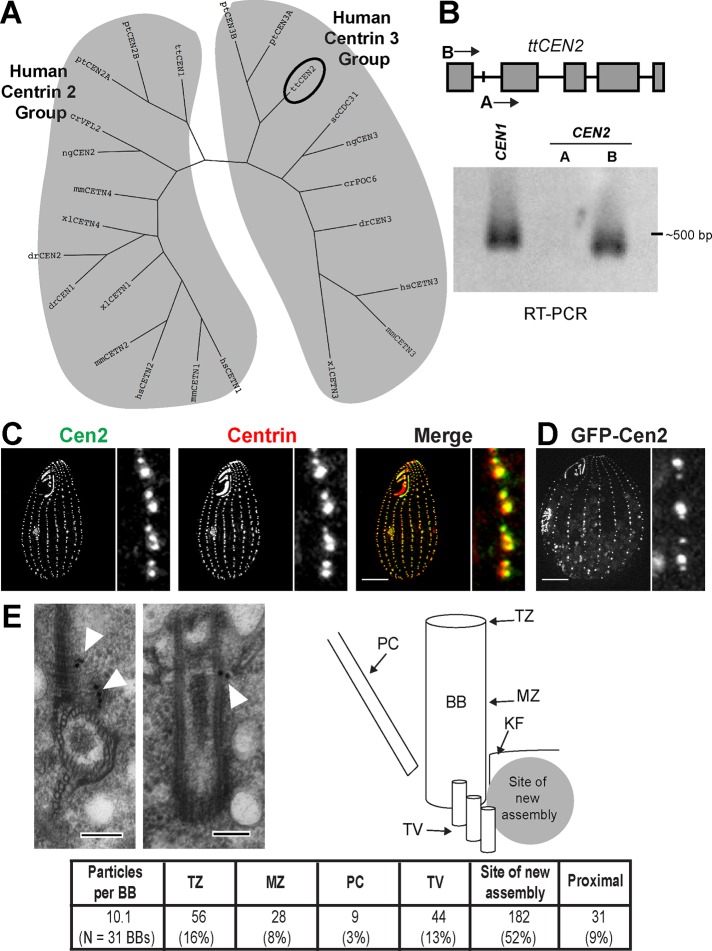
FIGURE 1:
Basal bodies have two centrin isoforms and the Tetrahymena Cen2 localizes to basal bodies. (A) A phylogenetic tree showing the two centrin groups as defined by homology to the human centrins 2 and 3. The circle indicates the location of the Tetrahymena CEN2. cr, Chlamydomonas reinhardtii; dr, Danio rerio; hs, Homo sapiens; mm, Mus musculus; ng, Naegleria gruberi; pt, Paramecium tetraurelia; sc; Saccharomyces cerevisiae; tt, Tetrahymena thermophila; xl, Xenopus laevis. (B) Top, the CEN2 locus. Gray boxes are the exons. “A” and “B” are the locations of the primers used for RT-PCR. Bottom, RT-PCR showing that CEN2 is expressed. CEN1 was used as a positive control. (C) Cen2 localizes to basal bodies. Green, anti-Cen2; red, centrin (20H5 antibody). Scale bar: 10 μm. (D) Localization of GFP-Cen2. Scale bar: 10 μm. (E) Immuno-EM showing localization of GFP‑Cen2. Left, cross-section; Right, longitudinal view. The table shows the particle distribution to various regions of the basal body. BB, basal body; KF, KD fiber; MZ, mid-zone; PC, postciliary microtubules; TV, transverse microtubules; TZ, transition zone. Scale bar: 100 nm.
To date, studies on centrin function at basal bodies and centrioles have largely focused on those belonging to the human centrin 2 group. Members of this group localize to the distal end of the basal body or centriole and sites of new assembly (Sanders and Salisbury, 1994; Paoletti et al., 1996; Geimer and Melkonian, 2005; Stemm-Wolf et al., 2005; Kilburn et al., 2007). Human centrin 3 is also found at these sites (Laoukili et al., 2000), suggesting that the two groups share similar localization. Analyses on the function of human centrin 2 and its homologues suggest that this group is involved in basal body or centriole maintenance, assembly, orientation of assembly, and separation of the daughter from its mother (Spang et al., 1993; Salisbury et al., 2002; Koblenz et al., 2003; Ruiz et al., 2005; Stemm-Wolf et al., 2005; Yang et al., 2010; Vonderfecht et al., 2011). Human centrin 3 may play a role in centrosome duplication (Middendorp et al., 2000), but the nature of its function remains unclear. Studies in Paramecium (Ruiz et al., 2005; Aubusson-Fleury et al., 2012) and Trypanosoma brucei (Selvapandiyan et al., 2012) have shown that knockdown of centrin isoforms yields differing effects on basal bodies, suggesting that the two centrin groups may have nonredundant functions; however, this hypothesis has not been fully tested. Cdc31p, the only yeast centrin, is the most-studied human centrin 3 homologue. It has been shown to be required for the duplication of the spindle pole body (Hartwell et al., 1973; Byers, 1981; Baum et al., 1986); however, since the yeast MTOC is structurally different from a basal body or centriole (Jaspersen and Winey, 2004), it remains to be seen whether the function of Cdc31p is conserved throughout the human centrin 3 group.
The role of centrins at vertebrate centrioles has been extensively studied, and the data suggest that centrins may play a role in regulating the rate of centriole assembly but are not required for completing centriole assembly (Kleylein-Sohn et al., 2007; Strnad et al., 2007; Yang et al., 2010; Dantas et al., 2011; Lukasiewicz et al., 2011). Studies examining the role of centrins at vertebrate basal bodies are sparse; however, it has been shown that the human centrins are up-regulated during ciliogenesis in epithelial tissue cultures (LeDizet et al., 1998). Furthermore, an RNA interference screen identified human centrin 2 as important for ciliogenesis (Graser et al., 2007). Morpholinos that target the human centrin 2 homologue in zebra fish lead to defects commonly associated with ciliopathies (Delaval et al., 2011). This accumulating body of evidence suggests that centrins function primarily at the basal body. The small number of studies on the human centrin 3 group at basal bodies and the fact that the two centrins share high sequence similarity raise two major questions: what is the function of this group and are the two centrin groups redundant, meaning do basal bodies need both centrin isoforms? To answer these two questions, we used the ciliate protist Tetrahymena thermophila as a model system for basal body biology. Basal bodies in Tetrahymena are found within cortical rows, which are rows of basal bodies that run along the posterior to anterior axis of the cell, and within the oral apparatus, a feeding structure at the anterior end of the cell (Frankel, 2000; Pearson and Winey, 2009). We have previously identified four centrins in Tetrahymena, with centrin 1 (Cen1) and centrin 2 (Cen2) localizing only to basal bodies (Stemm-Wolf et al., 2005). Like the human centrins, Cen1 and Cen2 share high sequence similarity (78.4%). We have shown that Cen1 is essential and is involved in basal body stability, assembly, orientation of assembly, and separation (Stemm-Wolf et al., 2005; Vonderfecht et al., 2011). We were unable to detect the expression of Cen2 (Stemm-Wolf et al., 2005); however, our Tetrahymena basal body proteome shows that Cen2 exists at basal bodies (Kilburn et al., 2007), and a Tetrahymena microarray study identified expression of the Cen2 gene, CEN2 (Miao et al., 2009).
We wished to gain a better understanding of the functions of the two centrin groups by determining Cen2’s role at basal bodies and whether Cen1 and Cen2 are redundant. We found that while Cen1 and Cen2 share similar functions, they are distinct from each other, suggesting that basal bodies require a centrin from each group to function correctly.
RESULTS
Cen2 is expressed and localizes to basal bodies
A Tetrahymena microarray study identified the expression of CEN2 (Miao et al., 2009), and our basal body proteome identified Cen2 as a component (Kilburn et al., 2007). However, we have been unable to detect expression of CEN2 by Northern blotting (Stemm-Wolf et al., 2005). At the start of this study, the Tetrahymena genome underwent an extensive reannotation, and the newly annotated CEN2 showed that the previously predicted start codon was an intron. We performed reverse transcriptase PCR (RT‑PCR) and were able to detect product only by using a primer that anneals to the newly annotated start codon (Figure 1B), confirming that CEN2 is expressed. The cDNA product was sequenced, and translation shows that it encodes a centrin protein. The only differences between the correct sequence and the misidentified sequence lie in the first 20 residues, which make up the N‑terminal tail of the protein (Supplemental Figure S1). The new protein sequence of Cen2 indicates that it is still a human centrin 3 homologue (Figures 1A and S1).
We generated a polyclonal peptide antibody that specifically recognizes the N-terminal extension of Cen2 (Figures S1, S2A, and S2B). With the αCen2 antibody, we were able to detect Cen2 at all basal bodies in the cortical rows and oral apparatus (Figure 1C), confirming that Cen2 is a basal body component. Immuno–electron microscopy (immuno-EM) was performed to determine Cen2’s localization within the basal body. The αCen2 antibody predominantly labeled regions at the site of new assembly at the proximal end of the basal body (Figure S2C). This was surprising because centrins generally localize to both the site of new assembly and the distal end of the basal body (Sanders and Salisbury, 1994; Laoukili et al., 2000; Geimer and Melkonian, 2005; Stemm-Wolf et al., 2005; Kilburn et al., 2007). We reasoned that the αCen2 antibody is not sensitive enough to label the distal end and generated a cell line expressing Cen2 tagged with green fluorescent protein (GFP), which localizes to basal bodies (Figure 1D). We observed labeling by a αGFP antibody at the distal end of the basal body, at or near the transition zone, with most of the labeling concentrated at the site of new assembly (Figure 1E). This localization slightly differs from Cen1, which also localizes to the midzone of the basal body (Stemm-Wolf et al., 2005; Kilburn et al., 2007). In all, our results show that Cen2 is expressed in Tetrahymena and localizes to the site of new assembly and the distal end of the basal body. Our results also indicate that Tetrahymena basal bodies have a centrin isoform from the two centrin groups, a characteristic that appears to be conserved at basal bodies throughout evolution.
Cen2 is required for basal body maintenance and orientation
Because the function of the human centrin 3 group at basal bodies is not well known, we wished determine the role of Cen2 at Tetrahymena basal bodies. To answer this question, we created a cen2 null allele (cen2Δ) strain (Figures S2A and S3A). Unlike the cen1 null allele, which is lethal, cen2Δ cells are viable and are able to divide for many generations, indicating that CEN2 is not essential. We found that the levels of Cen1 remained unchanged in the cen2Δ compared with wild-type cells (Figure S3B), suggesting that Cen1 is not compensating for the loss of Cen2 in the cen2Δ.
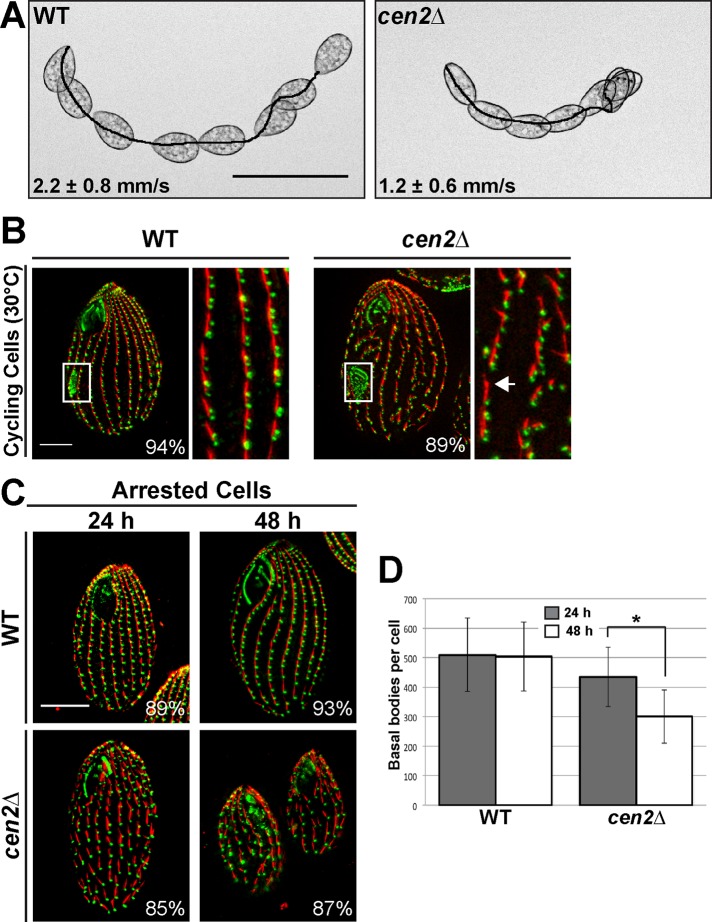
We examined cen2Δ cells by light microscopy and observed that they swim significantly slower than wild-type cells (Figure 2A), suggesting that the loss of Cen2 leads to a basal body or ciliary defect. This led us to perform immunofluorescence microscopy on cycling cells using αCen1 and anti-kinetodesmal (KD) fiber antibodies as basal body markers (Figure 2B). The KD fiber is a basal body accessory structure composed of striated rootlets that extend from the basal body, oriented toward the anterior of the cell (Allen, 1969). We observed that cen2Δ cycling cells have basal body defects (Figure 2B). The same basal body defects were observed using the Sas6a antibody, which labels basal bodies and has a background signal from the KD fibers (Figure S3C; Culver et al., 2009). The cen2Δ cells had gaps between basal bodies in their cortical rows (Figure 2B, arrow), suggesting that deletion of CEN2 leads to a loss of basal bodies. Quantification showed that there is almost a 27% reduction in the number of cortical row basal bodies per cell in the cen2Δ (461 ± 54) compared with wild-type cells (630 ± 86; p < 0.01, n = 25 cells). The loss of basal bodies is not likely due to the inability to assemble new basal bodies because of the existence of the oral primordium (Figure 2B, white boxes), which is indicative of new basal body assembly (Wolfe, 1970; Kaczanowski, 1978; Frankel, 2000).
FIGURE 2:
Deletion of CEN2 leads to swimming defects and basal body maintenance defects. (A) Images showing the swimming speed of wild-type and cen2Δ cells. Scale bar: 100 μm. (B) Immunofluorescence images showing gaps (arrow) in the cen2Δ. White box, oral primordium, which is indicative of new basal body assembly. (C) Cells arrested by starvation and fixed 24 and 48 h after cell arrest. (B and C) Green, Cen1; red, KD fibers. Scale bar: 10 μm. Width of insets: 10 μm. Percentages indicate the frequency of observed phenotype for 100 cells. (D) Plot showing the number of basal bodies per cell at the two different time points. *, p < 0.01; n = 25 cells.
Next we asked whether the loss of basal bodies is due to maintenance defects. Cells were arrested by starvation to inhibit new basal body assembly (Pearson et al., 2009), and we examined their ability to maintain their existing basal bodies by quantifying the number of cortical row basal bodies per cell after 24 and 48 h of cell arrest (Figure 2, C and D). The cen2Δ showed a 31% reduction in the number of basal bodies per cell between the two time points, whereas wild-type cells displayed no significant reduction in the number of basal bodies per cell (Figure 2D; p < 0.01, n = 25 cells), showing that deletion of CEN2 causes a loss of basal bodies due to maintenance defects.
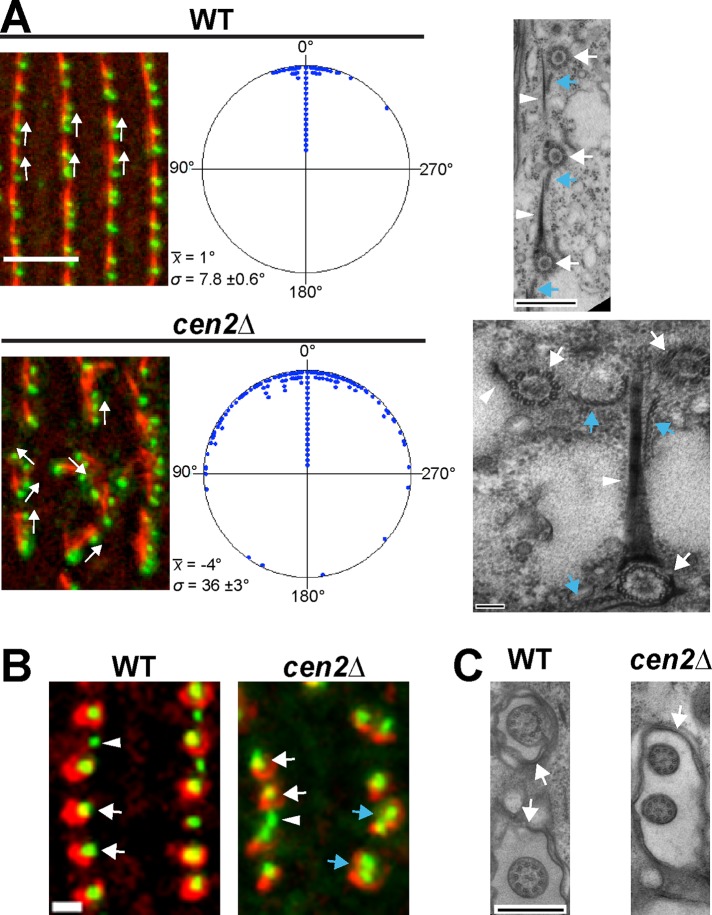
We also observed basal bodies branching off within the cortical rows in the cen2Δ, suggesting that there is a basal body orientation defect. Basal body angle was measured to confirm the orientation defect (Figure 3A). We found that the cen2Δ had a large variation in basal body angle, with an average angle SD of 36 ± 3 degrees (Figure 3A). This was significantly different from wild type, which had an average angle SD of 7.8 ± 0.6 degrees (Figure 3A), confirming that Cen2 has a role in basal body orientation. EM analysis of the cen2Δ found examples of basal bodies branching off from cortical rows (Figure 3A). The KD fibers and postciliary microtubules of these basal bodies were not aligned with those of correctly positioned basal bodies. This suggests that the orientation defect in the cen2Δ might be a result of aberrantly rotated basal bodies. We also observed what appeared to be basal body separation defects in the cen2Δ. Cells were stained with αCen1, which labels all basal bodies (Stemm-Wolf et al., 2005; Pearson et al., 2009), and K-like antigen, which labels only mature basal bodies (Williams et al., 1990; Shang et al., 2002). Some mature basal body pairs in the cen2Δ were able to separate (Figure 3B, white arrows); however, some did not show proper separation (Figure 3B, blue arrows). This defect was observed on average a little more than three times per cell in the cen2Δ but less than one time per cell in wild-type cells (n = 20 cells). EM analysis of the cen2Δ showed two cilia sharing a single ciliary pocket (Figure 3C), which may be a result of the separation defect. In all, the analysis of the cen2Δ shows that deletion of CEN2 leads to basal body maintenance, orientation, and separation defects.
FIGURE 3:
Deletion of CEN2 leads to basal body orientation and separation defects. (A) Left panels show the basal body angle distribution for wild type and the cen2Δ. Green, Cen1; red, KD fibers. Scale bar: 5 μm. x, average angle; σ, SD. In the circular plots, each point corresponds to five measurements in wild type and two measurements in the cen2Δ. n = 200 measurements. Right panels show electron micrographs of the basal body orientation defect. Basal bodies (white arrow) and their accessory structures (KD fiber, white arrowhead; postciliary microtubules, blue arrow) are rotated in cen2Δ cells. Scale bar: 500 or 100 nm (cen2Δ). (B) Close proximity of mature basal bodies suggests that there is a basal body separation defect in the cen2Δ (blue arrows). The average distance between a pair of old basal bodies is 1.5 ± 0.3 μm in wild type and 1.2 ± 0.4 μm in the cen2Δ, a significant difference (n = 150 basal body pairs). Green, Cen1; red, K-like antigen (mature basal body marker); white arrowhead, immature basal body; white arrow, mature basal body. Scale bar: 1 μm. (C) Electron micrographs showing that cilia in the cen2Δ share a ciliary pocket (arrow), whereas cilia in wild type do not. Scale bar: 500 nm.
To confirm that the observed basal body defects in the cen2Δ were due to deletion of CEN2, we introduced CEN2 under control of the metallothionein (MTT) promoter into the cen2Δ genome by integration at the RPL29 locus. The introduction of MTT-CEN2 rescues the basal body maintenance and separation defects in the cen2Δ (Figures S3, D and E, and S4), confirming that Cen2 is required for basal body maintenance and orientation.
Cen2 behaves differently from Cen1
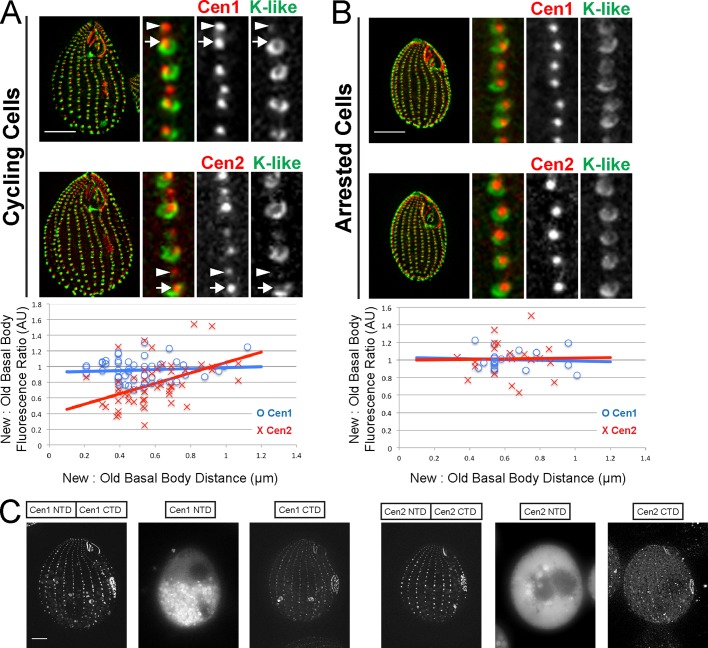
The analysis of the cen2Δ strain suggests that Cen2 has a similar function to Cen1, leading to the question of whether the two proteins behave similarly at basal bodies. It was previously shown that all basal bodies, new and old, have the same levels of Cen1 (Figure 4A; Pearson et al., 2009). With the αCen2 antibody, we observed that mature (old) basal bodies have brighter signals than immature (new) basal bodies (Figure 4A). The fluorescence of Cen1 or Cen2 labeling of new and old basal bodies was measured to calculate the new:old basal body fluorescence ratio (Figure 4A). The ratio was ∼1 (0.96 ± 0.14) for Cen1, which is expected, since all basal bodies have equal amounts of Cen1 (Pearson et al., 2009). As for Cen2, the ratio was 0.78 ± 0.28, and a Student's t test indicated that the two ratios are significantly different from each other (p < 0.01, n = 50 basal body pairs), showing that the old basal bodies have more Cen2 than the new basal bodies. To confirm this, we examined the labeling of Cen2 in arrested cells that were not undergoing new basal body assembly and whose basal bodies were all fully mature (Figure 4B; Pearson et al., 2009). We observed equal labeling by the αCen2 antibody, with a fluorescence ratio near 1 (1.01 ± 0.21). The data show that Cen2 continues to accumulate at the basal body as it fully matures, suggesting Cen2 may have a role in basal body maturation. This is in contrast to Cen1, whose protein levels remain constant throughout the basal body's life cycle.
FIGURE 4:
Cen2 behaves differently from Cen1. (A) Immunofluorescence images of cycling cells, showing that new (arrowhead) and old (arrow) basal bodies have equal levels of Cen1, while old basal bodies have higher levels of Cen2 than do new basal bodies. The plot shows the fluorescence ratio for the new and old basal body pairs vs. their distance. (B) Basal bodies in cells arrested by starvation have equal levels of Cen1 and Cen2, because all basal bodies are old (or mature). (A and B) Width of insets: 2.5 μm. Red, Cen1 (labels all basal bodies); green, K-like antigen (labels old basal bodies); AU, arbitrary units. (C) The CTD, but not the NTD, of Cen1 and Cen2 localize to basal bodies. Scale bars: 10 μm.
Next we wanted to determine whether the two domains of Cen2 behave in a manner similar to the domains of Cen1. We have previously shown that only the C-terminal domain (CTD) of Cen1 localizes to basal bodies (Vonderfecht et al., 2011). Cen2’s CTD and N-terminal domain (NTD) were tagged with GFP and expressed in a wild-type background containing endogenous wild-type Cen1 and Cen2. We found that, like Cen1, only the CTDs of Cen2 localize to basal bodies (Figure 4C). This suggests that the two centrin groups have similar modes of action, with the CTD involved in localization to basal bodies. In all, the data suggest that Cen1 and Cen2 have similar localization mechanisms but behave differently.
The two centrin groups are distinct from each other
Because Cen1 and Cen2 have similar functions, we wanted to determine whether the two proteins have redundant functions at the basal body. If the two proteins are redundant, then overexpression of Cen1 in the cen2Δ should rescue the basal body phenotypes by compensating for the loss of Cen2. CEN1 under control of the MTT promoter was introduced into cen2Δ, and it was observed that CEN1 is overexpressed upon induction with Cd2+ (Figure 5A). The cen2Δ MTT-CEN1 cell line was examined by immunofluorescence, and we found that overexpression of CEN1 is unable to compensate for the deletion of CEN2, as it was not capable of rescuing the loss of basal bodies and the orientation defects in the cen2Δ (Figures 5B and S4). The CEN1 overexpression data suggest the two Tetrahymena basal body centrins are distinct.
FIGURE 5:
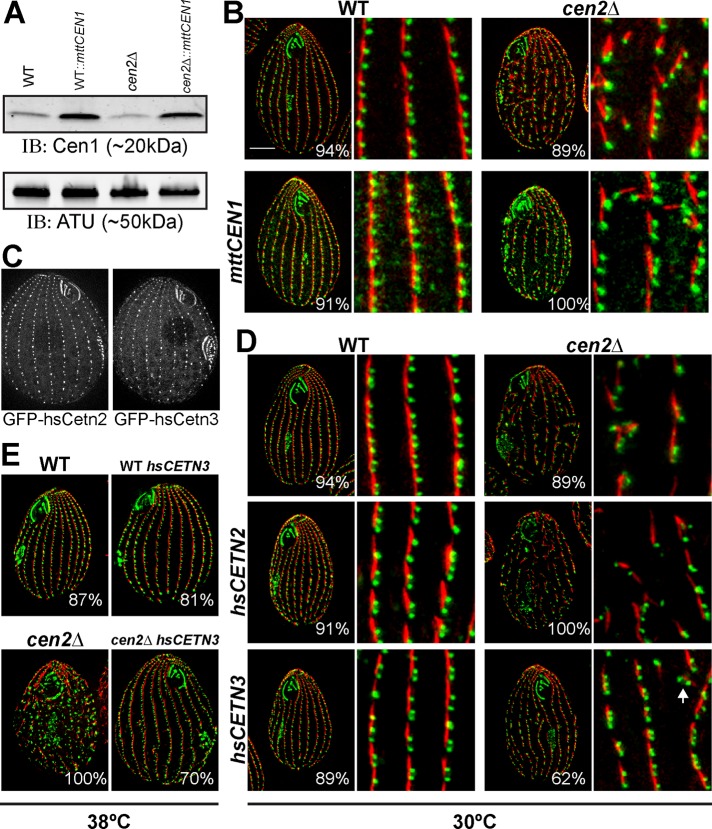
The two centrin groups are functionally distinct. (A) A Western blot showing Cen1 overexpression. α-Tubulin served as a loading control. (B) Immunofluorescence images showing that overexpression of CEN1 is unable to rescue the cen2Δ. (C) Both GFP-hsCen2 and GFP-hsCen3 localize to basal bodies in Tetrahymena. (D and E) Immunofluorescence images showing that only hsCEN3, not hsCEN2, can rescue the cen2Δ basal body phenotypes at 30°C (D) and 38°C (E). Green, Cen1; red, KD fibers. Scale bar: 10 μm; width of insets: 10 μm; Percentages indicate the frequency of observed phenotype for 100 cells.
Next we wanted to test whether that the two centrin groups are truly distinct by determining whether the human centrins can function similarly to their Tetrahymena centrin counterparts. To ensure that the human centrins 2 and 3 (hsCETN2 and hsCETN3, respectively) localize to basal bodies, we tagged them with GFP and observed their localization to basal bodies in a wild-type background (Figure 5C). Next hsCETN2 and hsCETN3 under control of the MTT promoter were introduced into the cen2Δ. We found that only hsCETN3 (the Tetrahymena CEN2 homologue) rescues the loss of basal bodies and the orientation defects caused by the deletion of CEN2, whereas hsCETN2 (the Tetrahymena CEN1 homologue) could not (Figures 5D and S4). The cen2Δ MTT‑hsCETN3 cell line does not display complete rescue for some minor disorganization of the basal body cortical rows remains; however, the basal body orientation defect is not as severe as seen in cen2Δ (Figure S4). An examination of cells grown at elevated temperatures (38°C) showed that the increase in temperature exacerbated the loss of basal bodies and orientation defects in the cen2Δ whereas the cen2Δ MTT-hsCETN3 cell line did not display any temperature-sensitive basal body phenotypes (Figures 5E and S4). In all, the data show that human centrin 3 functions similarly to Tetrahymena Cen2, confirming distinct functions for the two centrin groups.
The domains between the two centrin groups are also distinct from one another
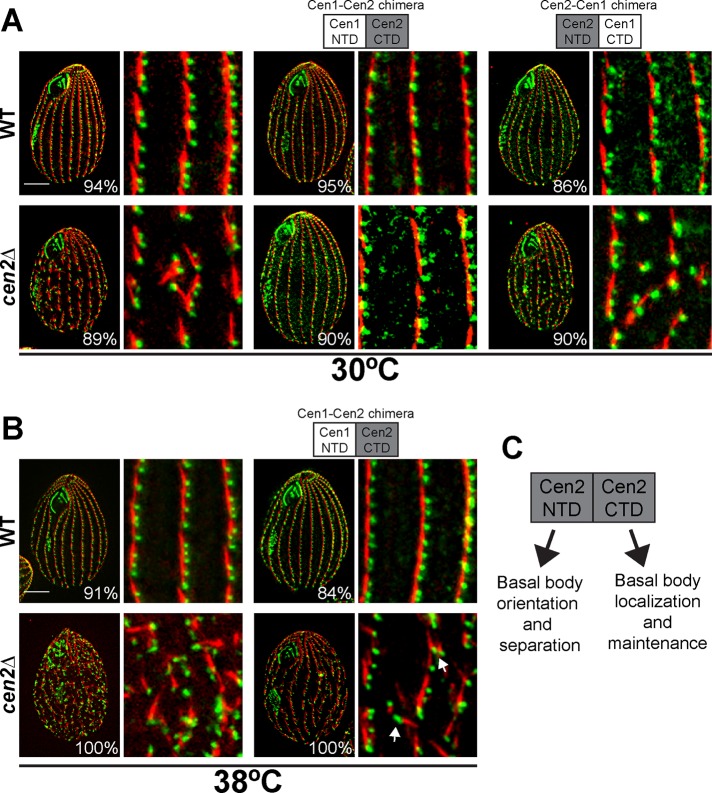
Because the two centrin groups have distinct functions at basal bodies despite a high degree of similarity, we wondered whether the NTD or the CTD was responsible for the separation of the two groups. An alignment the Cen1 and Cen2 domains showed that their CTDs were 91% similar, but their NTDs were 69% similar, suggesting that the NTD distinguishes between the two groups. To test this hypothesis, we introduced centrin chimeras composed of either Cen1’s NTD with Cen2’s CTD (the Cen1-Cen2 chimera) or Cen2’s NTD with Cen1’s CTD (the Cen2-Cen1 chimera) into the cen2Δ to determine which chimera would rescue the cen2Δ basal body phenotypes. Based on the alignments, it was expected that only the Cen2‑Cen1 chimera would rescue the cen2Δ, since it contains Cen2’s NTD. However, it was the Cen1-Cen2 chimera that unexpectedly rescued the loss of basal bodies and orientation defects in the cen2Δ at 30°C (Figures 6A and S4). For confirming that the Cen1-Cen2 chimera completely rescues the cen2Δ, cells were grown at 38°C to check for any temperature-sensitive basal body phenotypes. At this elevated temperature, we observed basal bodies branching from the cortical rows in the cen2Δ strain rescued by the Cen1-Cen2 chimera (Figure 6B, arrow). Quantification of the number of basal bodies per micrometer showed that there was no significant difference from wild type (Figure S4); however, there was more basal body angle variation in the cen2Δ strain rescued by the Cen1-Cen2 than in wild type (Figure S4). This indicates that the Cen1-Cen2 chimera in the cen2Δ is able to rescue the basal body loss defect but is unable to completely rescue the basal body orientation defect.
FIGURE 6:
The Cen1-Cen2 chimera can rescue the loss of basal bodies in the cen2Δ. (A) Immunofluorescence images showing that only the Cen1-Cen2 chimera, not the Cen2-Cen1 chimera, can rescue the cen2Δ basal body phenotypes at 30°C. (B) Immunofluorescence images showing that the Cen1-Cen2 chimera does not rescue the basal body orientation defects (arrow) in the cen2Δ at 38°C. (A and B) Green, Cen1; red, KD fibers. Scale bar: 10 μm; width of insets: 10 μm. Percentages indicate the frequency of observed phenotype for 100 cells. (C) Cen2’s domains have separate functions.
DISCUSSION
Centriole and basal body structures consisting of triplet microtubules have two distinct centrins, one each similar to human CENT2 and CENT3 (Andersen et al., 2003; Carvalho-Santos et al., 2011; Hodges et al., 2010; Keller et al., 2005; Kilburn et al., 2007; Liu et al., 2007). We have used the ciliate Tetrahymena to show that the two centrins have distinct functions at the basal body. Mutations in either CEN1 or CEN2 reveal similar functions in basal body orientation and maintenance (Stemm-Wolf et al., 2005; Vonderfecht et al., 2011); however, our rescue experiments and other analyses demonstrate that the two genes are not redundant. This separation is deeply rooted in the evolutionary history of these genes, since the distinct functions are maintained from Tetrahymena to humans, as shown by the specificity of rescue of the cen2Δ by human CENT3, but not CENT2. In all, our data indicate that the two Tetrahymena centrins have similar, but not redundant, functions at basal bodies and that Tetrahymena basal bodies require both centrins to perform properly. The human centrin 3 can function similarly to Cen2, suggesting that the same conclusions may hold for vertebrate centrins at basal bodies.
The fact that the two centrins are distinct raises the question of what is responsible for their differences. An alignment of the two Tetrahymena centrins shows that the degree of sequence conservation is much lower for the NTD than the CTD. This is also true for the human centrins. This suggests that the NTDs of centrins may be responsible for the separation and distinctness of the two groups. We tested this hypothesis, utilizing chimeric proteins, and discovered that it did not hold true. Our findings from this experiment suggest a mechanism for Cen2 function (Figure 6C). The data showing that the Cen1-Cen2 chimera was able to rescue the basal body maintenance defect in the cen2Δ and that only the CTD of Cen2 localizes to basal bodies suggest that the CTDs of all centrins are involved in basal body localization and maintenance. However, this chimera was unable to completely rescue the orientation defect in the cen2Δ, suggesting that basal bodies require Cen2’s distinctive NTD for proper basal body orientation. Thus it appears that both the NTD and CTD contribute to the distinct requirements for the two centrin groups.
Work with vertebrate centrins suggests that they are important for controlling the rate of centriole assembly (Yang et al., 2010; Lukasiewicz et al., 2011). We did not observe a change in basal body assembly rates but did see defects in basal body separation after assembly. It therefore may be possible that centrins do not function in the rate of assembly per se. Instead, defects in centriole separation may lead to defects in the rate of assembly. A centriole must first be able to properly separate from its paired centriole and undergo licensing before assembly occurs (Brownlee and Rogers, 2012). If a daughter centriole has difficulty separating from its mother in the absence of centrins, then the delay caused by this difficulty can lead to a delay in the licensing and the assembly of a new centriole. The cumulative effect would be a delay in the rate of centriole assembly. To test this notion, the rate of separation could be analyzed in vertebrate cells that contain centrin phospho-mutants (Yang et al., 2010; Lukasiewicz et al., 2011) or in cells depleted of centrins.
The next step in fully understanding centrin's role in basal body biogenesis will require analyzing the function of its binding partners. One of the centrin binding partners that exist at basal bodies or centrioles belongs to the Sfi1 family (Kilmartin, 2003; Li et al., 2006; Azimzadeh et al., 2009). The Sfi1 family consists of α-helical proteins that may contain 20 or more repeats that serve as centrin binding sites (Kilmartin, 2003; Li et al., 2006). The yeast Sfi1p has been shown to bind the human centrins 2 and 3 equally (Kilmartin, 2003; Martinez-Sanz et al., 2006, 2010), and the human Poc5, a small protein with only three centrin binding repeats, is able to bind both human centrins, but the affinity for each is not known (Azimzadeh et al., 2009). Thus the relationship of the two centrins at Sfi1 proteins is unclear. Careful studies on the interplay that occurs between the two domains of centrins and Sfi1 proteins will bring us much closer to truly understanding the functions and mechanisms of centrins and Sfi1 proteins at MTOCs.
MATERIALS AND METHODS
Strains and culture conditions
The wild-type Tetrahymena thermophila strain B2086 (Tetrahymena Stock Center, Cornell University, Ithaca, NY) was used in this study as the wild-type control strain. All cell lines were grown in 2% super-peptose (2% SPP) media (2% proteose peptone, 0.1% yeast extract, 0.2% glucose, and 0.003% Fe-EDTA) at 30°C or 38°C depending on the experiment. For cell-arrest experiments, cells were grown to mid–log phase in 2% SPP media, washed twice with 10 mM Tris-HCl (pH 7.4), and resuspended in 10 mM Tris-HCl (pH 7.4) at 30°C. For all experiments involving the inducible MTT promoter, induction was performed by adding CdCl2 to media to 0.25 μg/ml and incubating the cells at 30°C or 38°C for 8 h before fixing or live imaging of the cells.
DNA constructs and strain construction
Strains containing GFP-tagged full-length Cen1 or Cen2 were generated by cloning CEN1 or CEN2 cDNA into the pENTR-D Gateway Entry Vector (Invitrogen, Carlsbad, CA). The coding sequence was then cloned into pBSmttGFPgtw (Doug Chalker, Washington University, St. Louis, MO) using the Gateway cloning system (Invitrogen). The resulting construct adds the GFP tag to the N-terminus of Cen1 or Cen2, is under control of a MTT-inducible promoter, targets for integration into the rpl29 locus, and provides cycloheximide resistance. The first 276 bases of CEN1 were cloned into pENTR to make the Cen1 NTD, and the last 228 bases of CEN1 were cloned into pENTR to make the Cen1 CTD. The first 270 and last 228 bases of CEN2 were cloned into pENTR to make the Cen2 NTD and CTD, respectively. The constructs pECE-GFP-Cetn2 and pECE-GFP-Cetn3 (generous donations from H. Fisk, Ohio State University, Columbus, OH) were used to clone the human centrins into pENTR. The pENTR genes were then Gateway-cloned into pBSmttGFPgtw.
The expression of centrin genes without a tag was performed by cloning the desired gene into pBSmtt, which is under control of a MTT-inducible promoter, targets for integration into the rpl29 locus, and provides cycloheximide resistance. The Cen1-Cen2 chimera was created by PCR-stitching to create a nucleotide sequence that contained the first 276 bases of Cen1 and the last 228 bases of Cen2. Similarly, the Cen2-Cen1 chimera was created by PCR-stitching to create a nucleotide sequence with the first 270 bases of Cen2 and the last 228 bases of Cen1. The chimeras, CEN1, CEN2, and human centrin genes were all cloned into pBSmtt.
All DNA constructs were confirmed by sequencing (Macrogen USA, Rockville, MD) and were introduced into cells by biolistics and integrated into the Tetrahymena genome by homologous recombination (Cassidy-Hanley et al., 1997). Tetrahymena transformants were selected by growth in SPP containing cycloheximide (15 μg/ml).
Generation of cen2Δ
The cen2Δ was generated by creating a drug-selectable strain that has no functional CEN2 (Hai and Gorovsky, 1997). Briefly, p4T21-CEN2del, a construct containing the NEO2 cassette, which provides paromomycin resistance (Weide et al., 2007), flanked by 1.5 kb of the regions upstream and downstream of CEN2, was integrated at the CEN2 locus in the micronucleus of mating wild‑type Tetrahymena strains, B2068 or CU428 (Tetrahymena Stock Center) by biolistics. Strains of different mating types were made homozygous for the NEO2 cassette at the micronucleus by mating to strains with defective micronuclei (B*VI and B*VII; Tetrahymena Stock Center). The resulting strains homozygous for the NEO2 cassette at the CEN2 locus were mated to each other to generate a new macronucleus with the NEO2 cassette replacing CEN2 at the CEN2 locus. The cen2Δ was selected for resistance to paromomycin (100 μg/ml). Total genomic DNA was isolated by phenol:choloroform:isoamyl alcohol extraction and isopropyl alcohol precipitation (Gaertig et al., 1994). PCR confirmed proper integration of the NEO2 cassette at the CEN2 locus and deletion of CEN2 (Figure S3A).
Purification of recombinant Cen2
Because Tetrahymena stop codon usage differs from canonical stop codons, the CEN2 gene was resynthesized for expression in Escherichia coli by GenScript (Piscataway, NJ). The optimized CEN2 sequence was cloned into the E. coli expression vector pQE10 that adds an N-terminal 6xHis-tag to the protein. The resulting vector, pQE10-Cen2, was transformed into the E. coli strain M15.
The E. coli strain M15 containing pQE10-Cen2 was grown overnight at 37°C in Luria broth + 0.2% glucose with 50 μg/ml kanamycin and 100 μg/ml ampicillin. This culture was used to inoculate 500 ml of Luria broth, and cells were incubated at 37°C for 4 h, and then shifted to room temperature for 1 h. Expression of Cen2 was induced with 300 μM isopropyl β-d-1-thiogalactopyranoside. After 3 h, cells were pelleted, washed once with phosphate-buffered saline (PBS), and stored frozen.
Recombinant Cen2 was isolated on a nickel affinity column, as described by Stemm-Wolf et al. (2005). Briefly, pellets were resuspended in PBS with protease inhibitors (phenylmethylsulfonyl fluoride, leupeptin, aprotinin, and pepstatin) and lysozyme. Cells were sonicated and pelleted. The supernatant was loaded onto a Talon resin column (BD Biosciences Clontech, Palo Alto, CA), and elutions using PBS + 200 mM imidazole were collected. Fractions with Cen2 were pooled together and stored frozen in 10% glycerol. Recombinant Cen1 was purified in a similar manner, and the plasmid used to express Cen1 is described in Stemm-Wolf et al. (2005).
Generation of αCen2 peptide antibody
The rabbit αCen2 peptide antibody was developed by YenZym Antibodies (San Francisco, CA). The antibody epitope consisted of a peptide containing the first 17 amino acids of the N-terminal tail of Cen2 (MNYSPKANKMKRKLKQEC). The antibody was affinity-purified using an affinity column containing recombinant Cen2 chemically ligated to AminoLink gel (Pierce Biotechnologies, Rockford, IL). The column was washed by alternating between 0.2 M glycine (pH 2.8) and 0.1 M NaHCO3 (pH 8.5) + 0.5 M NaCl. The crude antibody was prepared by adding 10× TBS (Tris-buffered saline; 50 mM Tris, pH 7.5, 150 mM NaCl), 0.05% Tween, and 4 M NaCl and centrifuging the preparation. The crude preparation was incubated for 48 h in the affinity column at 4°C. Elutions using 0.2 M glycine (pH 2.8) + 0.02% NaN, were collected. Fractions containing the antibody were detected by absorbance at 280 nm and were pooled and dialyzed against PBS-azide at 4°C for 16 h.
Fluorescence microscopy
All cell imaging, both live cell and immunofluorescence, was conducted at room temperature using an Eclipse Ti inverted microscope (Nikon, Japan) fitted with a CFI Plan-Apo VC 60×/H 1.4 numerical aperture objective (Nikon, Japan) and a CoolSNAP HQ2 charge-coupled device camera (Photometrics, Tuscon, AZ). MetaMorph Imaging software (Molecular Devices, Sunnyvale, CA) was used to collect images. The imaging software was used to analyze images by subjecting them to the nearest neighbors’ deconvolution algorithm. Live-cell imaging was conducted to examine cells expressing GFP-tagged proteins. Cells were washed once with 10 mM Tris-HCl (pH 7.4), pelleted, and placed on microscope slides (VWR, Radnor, PA).
Cells examined by immunofluorescence were chemically fixed in paraformaldehyde for 10 min (Stuart and Cole, 2000). Cells were placed onto antibody slides coated with poly-l-lysine (Bellco Glass, Vineland, NJ). All primary antibodies were diluted in PBS + 1% bovine serum albumin (BSA). The affinity purified rabbit polyclonal Tetrahymena Centrin 2 antibody (this study) was diluted 1:100; the rabbit polyclonal Tetrahymena Centrin1 antibody (Stemm-Wolf et al., 2005) was diluted 1:1000; the mouse monoclonal 20H5 antibody raised against the Chlamydomonas centrin (provided by J. Salisbury, Mayo Clinic, Rochester, MN) was diluted 1:1000; the mouse monoclonal KD fiber antibody, F1-5D8, (provided by J. Frankel, University of Iowa, Iowa City, IA) was diluted 1:250; the mouse monoclonal K-like antigen antibody, 10D12 (provided by J. Frankel) was diluted 1:50; and the rabbit polyclonal Tetrahymena Sas6a antibody (Culver et al., 2009) was diluted 1:2500. Primary incubations were carried out overnight at 4°C, except for the K-like antigen antibody, which was incubated at 4°C over three nights. Following the primary incubation, cells were washed five times with PBS + 0.1% BSA. Cells were then incubated at room temperature for 1 h with secondary antibodies diluted 1:1000 in PBS + 1% BSA. The secondary antibodies used were anti-rabbit fluorescein isothiocyanate (FITC), anti-rabbit Texas Red, anti-mouse FITC, and anti-mouse Texas Red (Jackson ImmunoResearch Labs, West Grove, PA). After secondary antibody incubation, cells were washed five times with PBS + 0.1% BSA and mounted with Citifluor (Citfluor, London, United Kingdom).
Swimming analysis
Cells were grown to mid–log phase, washed in 10 mM Tris-HCl (pH 7.4), and incubated at 30°C for 30 min. Cells were diluted to 1 × 104 cells/ml, and 5 μl of cell solution was added to standard microscope slides (VWR). Images of the cells were taken for 0.16 s at 0.02-s intervals. The distance the cells swam was measured using ImageJ (National Institutes of Health, Bethesda, MD), and the speed was calculated by dividing the distance by the time (0.168 s). Twenty cells were measured for each condition.
Basal body new:old fluorescence ratio
Cells were grown to mid–log phase and washed in 10 mM Tris-HCl (pH 7.4). Cell arrest was carried out overnight at 30°C, and a portion of the cells was fixed. The remaining cells were released from arrest by washing them into fresh media, and after 4 h of release from arrest, cells were fixed. Fixed cells were stained with anti-Cen1 or anti-Cen2 antibodies.
The average fluorescence background signal (Fbkgrd) was measured by placing 5 × 5 pixel regions around the cell at areas without basal bodies and measuring the integrated fluorescence intensity for each region with MetaMorph. Then, a basal body pair was selected, with the anterior basal body considered to be “new” and the posterior basal body “old.” The integrated fluorescence intensity for each basal body (Fi) was measured by placing a 5 × 5 pixel region around the basal body, and the corrected integrated fluorescence intensity for each basal body was calculated (Fi − Fbkgrd). The distance between the two basal bodies was also measured. The basal body new:old fluorescence ratio was calculated by dividing the new basal body fluorescence by the old basal body fluorescence. For each condition, 50 basal body pairs were analyzed. For measuring all statistical differences, a Student's t test was performed using the Excel spreadsheet software (Microsoft, Redmond, WA).
Quantification of basal bodies per cell
Cells progressing through the cell cycle and cells arrested in starvation media (10 mM Tris-HCl, pH 7.4) were analyzed. Cells were stained with the anti-Cen1 antibody to label basal bodies. For cells progressing through the cell cycle, cells in oral primordium stages 1–2 were analyzed to ensure that the quantification was done at a consistent cell cycle stage (Bakowska et al., 1982). The Count Particles function of the program ImageJ was used to count the basal bodies across the entire cell surface area. A total of 25 cells were analyzed for each condition.
Quantification of basal bodies per micrometer
Cells progressing through the cell cycle at oral primordium stages 1–2 were stained with the anti-Cen1 antibody to label basal bodies. The number of basal bodies along a 10-μm length of a ciliary row was counted and subsequently divided by 10 μm to get the number of basal bodies per 1 μm. The mean cell length was measured to ensure that changes in basal body frequency were not a result of changes in cell length. A total of five measurements in 10 cells was analyzed to get 50 measurements for each condition.
Basal body angle
Cells were stained with the anti-Cen1 and KD fiber antibodies to label basal bodies and their orientation in the cell. Basal body angle was calculated by measuring the angle of two basal bodies from the anterior–posterior axis of the cell using the ImageJ program. A total of 100 measurements were made for each condition. Circular plots were constructed using the Oriana program (Kovach Computing Services, Pentraeth, Wales, UK).
EM
Tetrahymena cells were prepared for ultrastructural analysis and immunolocalization of Cen2 by high-pressure freezing followed by freeze substitution (Giddings et al., 2010; Meehl et al., 2010). Briefly, cells were centrifuged into a cryoprotectant solution consisting of 15% dextran (9–11 kDa; Sigma-Aldrich) and 5% BSA in 10 mM Tris-HCl (pH 7.4). The resulting loose pellet was high-pressure frozen in a Bal-Tec HPM-010 (Leica Microsystems, Wetzlar, Germany), then freeze-substituted in 0.25% glutaraldehyde and 0.1% uranyl acetate in acetone and embedded in Lowicryl HM20.
Nickel grids containing ribbons of 15–20 serial 60-nm–thick sections were prepared for immuno‑EM by incubating them in blocking solution (1% nonfat dry milk dissolved in PBS-Tween 20 [0.1%]) and then in blocking solution containing the rabbit polyclonal Cen2 antibody (diluted 1:5) or the rabbit polyclonal anti-GFP antibody (diluted 1:200; provided by C. Pearson, University of Colorado–Denver, Denver, CO). The 10- or 15-nm gold-conjugated anti-rabbit secondary antibody was applied to the grids (Ted Pella, Redding, CA). Grids were poststained with 2% uranyl acetate and lead citrate. Samples were imaged using a Philips CM 10 (FEI, Hillsboro, OR) equipped with a Gatan BioScan digital camera (Gatan, Pleasanton, CA) or a Philips CM 100 transmission electron microscope equipped with an AMT V600 digital camera (Advanced Microscopy Techniques, Danvers, MA). Six basal body domains were identified by morphological criteria (Kilburn et al., 2007). Gold particles on each serial cross-section through the basal bodies were counted and assigned to these domains.
PCR/RT-PCR
Genomic DNA was isolated from Tetrahymena cells by phenol:chloroform:isoamyl alcohol extraction, which was followed by precipitation with isopropanol, as described previously. PCR with Phusion polymerase (New England Biolabs, Ipswich, MA) was performed with 0.25 μg of genomic DNA. PCR was carried out according to the manufacturer's instructions. RT-PCR was performed using the SuperScript III One-Step RT-PCR System with Platinum Taq High Fidelity kit (Invitrogen).
Samples were run in a 1% Tris-acetate-EDTA agarose gel. The gel was stained with ethidium bromide and viewed through a FOTO/Analyst Investigator Eclipse workstation (FOTODYNE, Hartland, WI).
Western blot analysis
Whole-cell extracts were prepared by lysing ∼30,000 cells in sample buffer (20% glycerol, 2% SDS, 125 mM Tris, pH 6.8, 5% β-mercaptoethanol) and heating to 80°C for 4 min. Approximately 2250 cell equivalents were loaded in lanes in 4–20% Precise Protein precast gels (Pierce, Rockford, IL). For recombinant proteins, ∼0.5 mg/ml was added to each lane of the gel. Proteins were transferred to Immobilon membranes (Millipore, Billerica, MA) using a transfer apparatus (Bio-Rad Labs, Hercules, CA). Membranes were blotted in TBS containing 0.05% Tween and 2% BSA. Primary antibodies (rabbit polyclonal anti-Cen1 antibody, rabbit polyclonal Cen2 antibody, mouse polyclonal anti–6x histidine ascites [Babco International, Inc., Tucson, AZ], and mouse monoclonal B-5-1-2 anti–α-tubulin antibody [Sigma-Aldrich, St. Louis, MO]) were diluted 1:1000 into TBS containing 0.05% Tween and 2% BSA. Primary antibody was incubated overnight at 4°C, and membranes were washed three times with TBS containing 0.05% Tween. Secondary antibodies (anti–rabbit IR800 and anti–mouse IR680 [LI-COR Biosciences, Lincoln, NE]) were diluted 1:10,000 into TBS containing 0.05% Tween and 2% BSA. Secondary antibody incubations were performed at room temperature for 1 h and were followed by three washes with TBS containing 0.05% Tween. Blots were visualized on a LI-COR Odyssey scanner.
Supplementary Material
Acknowledgments
We thank Alex Stemm-Wolf, Chad Pearson, and Anne Aubusson-Fleury for advice and helpful discussions; Joe Frankel and Jeffrey Salisbury for antibodies; Harold Fisk for the human centrin plasmids; and Alex Stemm-Wolf and Melissa Hendershott for plasmids. This work was supported by National Institutes of Health grants GM074746 (to M.W.) and F31 GM095071 (to T.V.).
Abbreviations used:
- BSA
bovine serum albumin
- Cen1
Tetrahymena centrin 1
- Cen2
Tetrahymena centrin 2
- cen2Δ
cen2 null
- CTD
C-terminal domain
- FITC
fluorescein isothiocyanate
- GFP
green fluorescent protein
- immuno-EM
immuno–electron microscopy
- KD
kinetodesmal
- MTOC
microtubule organizing center
- MTT
metallothionein
- NTD
N-terminal domain
- PBS
phosphate-buffered saline
- RT-PCR
reverse transcriptase PCR
- SPP
super-peptose
- TBS
Tris‑buffered saline
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E12-06-0454) on October 19, 2012.
REFERENCES
- Allen RD. The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J Cell Biol. 1969;40:716–733. doi: 10.1083/jcb.40.3.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JR, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- Aubusson-Fleury A, Lemullois M, de Loubresse NG, Laligné C, Cohen J, Rosnet O, Jerka-Dziadosz M, Beisson J, Koll F. FOR20, a conserved centrosomal protein, is required for assembly of the transition zone and basal body docking at the cell surface. J Cell Sci. 2012;125:4395–4404. doi: 10.1242/jcs.108639. [DOI] [PubMed] [Google Scholar]
- Azimzadeh J, Hergert P, Delouvée A, Euteneuer U, Formstecher E, Khodjakov A, Bornens M. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J Cell Biol. 2009;185:101–114. doi: 10.1083/jcb.200808082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- Bakowska J, Frankel J, Nelsen EM. Regulation of the pattern of basal bodies within the oral apparatus of Tetrahymena thermophila. J Embryol Exp Morph. 1982;69:83–105. [PubMed] [Google Scholar]
- Baum P, Furlong C, Byers B. Yeast gene required for spindle pole body duplication: homology of its product with Ca2+-binding proteins. Proc Natl Acad Sci USA. 1986;83:5512–5516. doi: 10.1073/pnas.83.15.5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bornens M, Azimzadeh J. Origin and evolution of the centrosome. Adv Exp Med Biol. 2007;607:119–129. doi: 10.1007/978-0-387-74021-8_10. [DOI] [PubMed] [Google Scholar]
- Brownlee C, Rogers G. Show me your license, please: deregulation of centriole duplication mechanisms that promote amplification. Cell Mol Life Sci. doi: 10.1007/s00018-012-1102-6. DOI: 10.1007/s00018-012-1102-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B. Multiple roles of the spindle pole bodies in the life cycle of Saccharomyces cerevisiae. In: Wettstein DV, editor. Molecular Genetics in Yeast: Proceedings of the Alfred Benzon Symposium 16. Copenhagen, Denmark: Munksgaard; 1981. pp. 119–131. [Google Scholar]
- Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Tracing the origins of centrioles, cilia, and flagella. J Cell Biol. 2011;194:165–175. doi: 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassidy-Hanley D, Bowen J, Lee JH, Cole E, VerPlank LA, Gaertig J, Gorovsky MA, Bruns PJ. Germline and somatic transformation of mating Tetrahymena thermophila by particle bombardment. Genetics. 1997;146:135–147. doi: 10.1093/genetics/146.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Culver BP, Meehl JB, Giddings TH, Jr., Winey M. The two SAS-6 homologs in Tetrahymena thermophila have distinct functions in basal body assembly. Mol Biol Cell. 2009;20:1865–1877. doi: 10.1091/mbc.E08-08-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas TJ, Wang Y, Lalor P, Dockery P, Morrison CG. Defective nucleotide excision repair with normal centrosome structures and functions in the absence of all vertebrate centrins. J Cell Biol. 2011;193:307–318. doi: 10.1083/jcb.201012093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaval B, Covassin L, Lawson ND, Doxsey S. Centrin depletion causes cyst formation and other ciliopathy-related phenotypes in zebrafish. Cell Cycle. 2011;10:3964–3972. doi: 10.4161/cc.10.22.18150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel J. Cell biology of Tetrahymena thermophila. Methods Cell Biol. 2000;62:27–125. doi: 10.1016/s0091-679x(08)61528-9. [DOI] [PubMed] [Google Scholar]
- Gaertig J, Gu L, Hai B, Gorovsky MA. High frequency vector-mediated transformation and gene replacement in Tetrahymena. Nucleic Acids Res. 1994;22:5391–5398. doi: 10.1093/nar/22.24.5391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geimer S, Melkonian M. Centrin scaffold in Chlamydomonas reinhardtii revealed by immunoelectron microscopy. Eukaryotic Cell. 2005;4:1253–1263. doi: 10.1128/EC.4.7.1253-1263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings TH, Jr., Meehl JB, Pearson CG, Winey M. Electron tomography and immuno-labeling of Tetrahymena thermophila basal bodies. Methods Cell Biol. 2010;96:117–141. doi: 10.1016/S0091-679X(10)96006-8. [DOI] [PubMed] [Google Scholar]
- Graser S, Stierhof Y-D, Lavoie SB, Gassner OS, Lamla S, Le Clech M, Nigg EA. Cep164, a novel centriole appendage protein required for primary cilium formation. J Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hai B, Gorovsky MA. Germ-line knockout heterokaryons of an essential α-tubulin gene enable high-frequency gene replacement and a test of gene transfer from somatic to germ-line nuclei in Tetrahymena thermophila. Proc Natl Acad Sci USA. 1997;94:1310–1315. doi: 10.1073/pnas.94.4.1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell L, Mortimer R, Culotti J, Culotti M. Genetic control of the cell division cycle in yeast: V. genetic analysis of cdc mutants. Genetics. 1973;74:267–286. doi: 10.1093/genetics/74.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges ME, Scheumann N, Wickstead B, Langdale JA, Gull K. Reconstructing the evolutionary history of the centriole from protein components. J Cell Sci. 2010;123:1407–1413. doi: 10.1242/jcs.064873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu H, Chazin WJ. Unique features in the C-terminal domain provide caltractin with target specificity. J Mol Biol. 2003;330:473–484. doi: 10.1016/s0022-2836(03)00619-3. [DOI] [PubMed] [Google Scholar]
- Huang B, Mengersen A, Lee VD. Molecular cloning of cDNA for caltractin, a basal body-associated Ca2+-binding protein: homology in its protein sequence with calmodulin and the yeast CDC31 gene product. J Cell Biol. 1988;107:133–140. doi: 10.1083/jcb.107.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Kaczanowski A. Gradients of proliferation of ciliary basal bodies and the determination of the position of the oral primordium in Tetrahymena. J Exp Zool. 1978;204:417–430. doi: 10.1002/jez.1402040313. [DOI] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR, Marshall WF. Proteomic analysis of isolated Chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol. 2005;15:1090–1098. doi: 10.1016/j.cub.2005.05.024. [DOI] [PubMed] [Google Scholar]
- Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Jr., Culver BP, Yates JR, III, Winey M. New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol. 2007;178:905–912. doi: 10.1083/jcb.200703109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV. Sfi1p has conserved centrin-binding sites and an essential function in budding yeast spindle pole body duplication. J Cell Biol. 2003;162:1211–1221. doi: 10.1083/jcb.200307064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof Y-D, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Koblenz B, Schoppmeier J, Grunow A, Lechtreck KF. Centrin deficiency in Chlamydomonas causes defects in basal body replication, segregation and maturation. J Cell Sci. 2003;116:2635–2646. doi: 10.1242/jcs.00497. [DOI] [PubMed] [Google Scholar]
- Laoukili J, Perret E, Middendorp S, Houcine O, Guennou C, Marano F, Bornens M, Tournier F. Differential expression and cellular distribution of centrin isoforms during human ciliated cell differentiation in vitro. J Cell Sci. 2000;113:1355–1364. doi: 10.1242/jcs.113.8.1355. [DOI] [PubMed] [Google Scholar]
- LeDizet M, Beck JC, Finkbeiner WE. Differential regulation of centrin genes during ciliogenesis in human tracheal epithelial cells. Am J Physiol Lung Cell Mol Physiol. 1998;275:L1145–L1156. doi: 10.1152/ajplung.1998.275.6.L1145. [DOI] [PubMed] [Google Scholar]
- Li S, Sandercock AM, Conduit P, Robinson CV, Williams RL, Kilmartin JV. Structural role of Sfi1p-centrin filaments in budding yeast spindle pole body duplication. J. Cell Biol. 2006;173:867–877. doi: 10.1083/jcb.200603153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Tan G, Levenkova N, Li T, Pugh EN, Rux JJ, Speicher DW, Pierce EA. The proteome of the mouse photoreceptor sensory cilium complex. Mol Cell Proteomics. 2007;6:1299–1317. doi: 10.1074/mcp.M700054-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukasiewicz KB, Greenwood TM, Negron VC, Bruzek AK, Salisbury JL, Lingle WL. Control of centrin stability by Aurora A. PLoS One. 2011;6:e21291. doi: 10.1371/journal.pone.0021291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Kintner C. Cilia orientation and the fluid mechanics of development. Curr Opin Cell Biol. 2008;20:48–52. doi: 10.1016/j.ceb.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall WF, Nonaka S. Cilia: tuning in to the cell's antenna. Curr Biol. 2006;16:R604–R614. doi: 10.1016/j.cub.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanz J, Kateb F, Assairi L, Blouquit Y, Bodenhausen G, Abergel D, Mouawad L, Craescu CT. Structure, dynamics and thermodynamics of the human centrin 2/hSfi1 complex. J Mol Biol. 2010;395:191–204. doi: 10.1016/j.jmb.2009.10.041. [DOI] [PubMed] [Google Scholar]
- Martinez-Sanz J, Yang A, Blouquit Y, Duchambon P, Assairi L, Craescu CT. Binding of human centrin 2 to the centrosomal protein hSfi1. FEBS J. 2006;273:4504–4515. doi: 10.1111/j.1742-4658.2006.05456.x. [DOI] [PubMed] [Google Scholar]
- Matei E, Miron S, Blouquit Y, Duchambon P, Durussel I, Cox JA, Craescu CT. C-terminal half of human centrin 2 behaves like a regulatory EF-hand domain. Biochem. 2003;42:1439–1450. doi: 10.1021/bi0269714. [DOI] [PubMed] [Google Scholar]
- Meehl JB, Giddings TH, Winey M. High pressure freezing, electron microscopy, and immuno-electron microscopy of Tetrahymena thermophila basal bodies. In: Gavin RH, editor. In: Cytoskeleton Methods and Protocols. Vol. 586. New York: Humana Press; 20102010. pp. 227–241. [DOI] [PubMed] [Google Scholar]
- Miao W, Xiong J, Bowen J, Wang W, Liu Y, Braguinets O, Grigull J, Pearlman RE, Orias E, Gorovsky MA. Microarray analyses of gene expression during the Tetrahymena thermophila life cycle. PLoS One. 2009;4:e4429. doi: 10.1371/journal.pone.0004429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorp S, Kuntziger T, Abraham Y, Holmes S, Bordes N, Paintrand M, Paoletti A, Bornens M. A role for centrin 3 in centrosome reproduction. J Cell Biol. 2000;148:405–415. doi: 10.1083/jcb.148.3.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Middendorp S, Paoletti A, Schiebel E, Bornens M. Identification of a new mammalian centrin gene, more closely related to Saccharomyces cerevisiae CDC31 gene. Proc Natl Acad Sci USA. 1997;94:9141–9146. doi: 10.1073/pnas.94.17.9141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti A, Moudjou M, Paintrand M, Salisbury JL, Bornens M. Most of centrin in animal cells is not centrosome-associated and centrosomal centrin is confined to the distal lumen of centrioles. J Cell Sci. 1996;109:3089–3102. doi: 10.1242/jcs.109.13.3089. [DOI] [PubMed] [Google Scholar]
- Pearson CG, Winey M. Basal body assembly in ciliates: the power of numbers. Traffic. 2009;10:461–471. doi: 10.1111/j.1600-0854.2009.00885.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson CG, Giddings TH, Jr., Winey M. Basal body components exhibit differential protein dynamics during nascent basal body assembly. Mol Biol Cell. 2009;20:904–914. doi: 10.1091/mbc.E08-08-0835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz F, Garreau de Loubresse N, Klotz C, Beisson J, Koll F. Centrin deficiency in Paramecium affects the geometry of basal-body duplication. Curr Biol. 2005;15:2097–2106. doi: 10.1016/j.cub.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Salisbury JL, Baron A, Surek B, Melkonian M. Striated flagellar roots: isolation and partial characterization of a calcium-modulated contractile organelle. J Cell Biol. 1984;99:962–970. doi: 10.1083/jcb.99.3.962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury J, Suino K, Busby R, Springett M. Centrin-2 is required for centriole duplication in mammalian cells. Curr Biol. 2002;12:1287–1292. doi: 10.1016/s0960-9822(02)01019-9. [DOI] [PubMed] [Google Scholar]
- Sanders M, Salisbury J. Centrin plays an essential role in microtubule severing during flagellar excision in Chlamydomonas reinhardtii. J Cell Biol. 1994;124:795–805. doi: 10.1083/jcb.124.5.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvapandiyan A, Kumar P, Salisbury JL, Wang CC, Nakhasi HL. Role of centrins 2 and 3 in organelle segregation and cytokinesis in Trypanosoma brucei. PLoS One. 2012;7:e45288. doi: 10.1371/journal.pone.0045288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Li B, Gorovsky MA. Tetrahymena thermophila contains a conventional γ-tubulin that is differentially required for the maintenance of different microtubule-organizing centers. J Cell Biol. 2002;158:1195–1206. doi: 10.1083/jcb.200205101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Courtney I, Fackler U, Matzner M, Schiebel E. The calcium-binding protein division cycle 31 of Saccharomyces cerevisiae is a component of the half bridge of the spindle pole body. J Cell Biol. 1993;123:405–416. doi: 10.1083/jcb.123.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stemm-Wolf AJ, Morgan G, Giddings TH, Jr., White EA, Marchione R, McDonald HB, Winey M. Basal body duplication and maintenance require one member of the Tetrahymena thermophila centrin gene family. Mol Biol Cell. 2005;16:3606–3619. doi: 10.1091/mbc.E04-10-0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gönczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart KR, Cole ES. Nuclear and cytoskeletal fluorescence microscopy techniques. Methods Cell Biol. 2000;62:291–311. doi: 10.1016/s0091-679x(08)61538-1. [DOI] [PubMed] [Google Scholar]
- Thompson JR, Ryan ZC, Salisbury JL, Kumar R. The structure of the human centrin 2-xeroderma pigmentosum group C protein complex. J Biol Chem. 2006;281:18746–18752. doi: 10.1074/jbc.M513667200. [DOI] [PubMed] [Google Scholar]
- Veeraraghavan S, Fagan PA, Hu H, Lee V, Harper JF, Huang B, Chazin WJ. Structural independence of the two EF-hand domains of caltractin. J Biol Chem. 2002;277:28564–28571. doi: 10.1074/jbc.M112232200. [DOI] [PubMed] [Google Scholar]
- Vonderfecht T, Stemm-Wolf AJ, Hendershott M, Giddings TH, Meehl JB, Winey M. The two domains of centrin have distinct basal body functions in Tetrahymena. Mol Biol Cell. 2011;22:2221–2234. doi: 10.1091/mbc.E11-02-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weide T, Bockau U, Rave A, Herrmann L, Hartmann M. A recombinase system facilitates cloning of expression cassettes in the ciliate Tetrahymena thermophila. BMC Microbiol. 2007;7:12. doi: 10.1186/1471-2180-7-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams NE, Honts JE, Kaczanowska J. The formation of basal body domains in the membrane skeleton of Tetrahymena. Development. 1990;109:935–942. doi: 10.1242/dev.109.4.935. [DOI] [PubMed] [Google Scholar]
- Wolfe J. Structural analysis of basal bodies of the isolated oral apparatus of Tetrahymena pyriformis. J Cell Sci. 1970;6:679–700. doi: 10.1242/jcs.6.3.679. [DOI] [PubMed] [Google Scholar]
- Wolfrum U, Salisbury JL. Expression of centrin isoforms in the mammalian retina. Exp Cell Res. 1998;242:10–17. doi: 10.1006/excr.1998.4038. [DOI] [PubMed] [Google Scholar]
- Yang A, Miron S, Duchambon P, Assairi L, Blouquit Y, Craescu CT. The N-terminal domain of human centrin 2 has a closed structure, binds calcium with a very low affinity, and plays a role in the protein self-assembly. Biochem. 2005;45:880–889. doi: 10.1021/bi051397s. [DOI] [PubMed] [Google Scholar]
- Yang C-H, Kasbek C, Majumder S, Yusof AM, Fisk HA. Mps1 phosphorylation sites regulate the function of centrin 2 in centriole assembly. Mol Biol Cell. 2010;21:4361–4372. doi: 10.1091/mbc.E10-04-0298. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.