Abstract
Mutations in phosphoribosyl pyrophosphate synthetase 1 (PRPS1) are associated with a spectrum of non-syndromic to syndromic hearing loss. PRPS1 transcript levels have been shown to be regulated by the microRNA-376 genes. The long primary RNA transcript of the miR-376 RNA cluster members undergo extensive and simultaneous A → I editing at one or both of two specific sites (+4 and +44) in particular human and mouse tissues. The PRPS1 gene, which contains target sites for the edited version of miR-376a-5p within its 3′UTR, has been shown to be repressed in a tissue-specific manner. To investigate whether the transcription of Prps1 is regulated by miR-376 cluster members in the mouse inner ear, we first quantified the expression of the mature miR-376 RNAs by quantitative real-time-PCR. The spatio-temporal patterns of miR-376 expression were assessed by in situ hybridization. Finally, we examined whether A →I editing of pri-miR-376 RNAs occurs in mouse inner ear by direct sequencing. Our data showed that the miR-376a-3p, b-3p, c-3p are present in mouse embryonic inner ears and intensive expression of miR-376a-3p/b-3p was detected in the sensory epithelia and ganglia of both auditory and vestibular portions of the inner ear. In adult inner ear, the expression of miR-376a-3p/b-3p is restricted within ganglion neurons of auditory and vestibular systems as well as the cells in the stria vascularis. Only unedited pri-miR-376 RNAs were detected in the cochlea suggesting that the activity of PRPS1 in the inner ear may not be regulated through the editing of miR-376 cluster.
Keywords: A → I editing, expression analysis, hearing loss, inner ear, microRNA-376, PRPS1
MicroRNAs (miRNAs) are an abundant class of small non-coding RNAs that regulate diverse developmental and physiological processes (Kapranov et al. 2007). This group of RNAs can negatively regulate protein expression at the post-transcriptional level by translational inhibition and/or mRNA degradation, mostly through base pairing with the 3′-UTR of their target mRNAs (Ambros 2003). Bioinformatic data indicate that a single miRNA could potentially bind to hundreds of mRNA targets, thus controlling various cellular functions (Bartel 2004). MicroRNAs genes (known as Mir genes) appear as single genes or in cluster of several Mir genes that are cotranscribed as a single transcription unit.
MiRNAs are transcribed as parts of longer molecules called pri-miRNAs, which are processed in the nucleus into hairpin RNAs of 70–100 nucleotides (nt), called pre-miRNAs, by the double-stranded RNA (dsRNA)-specific ribonuclease Drosha (Lee et al. 2004). These hairpins are then transported to the cytoplasm, where they act as substrates for a second double-strand specific ribonuclease called Dicer and miRNA duplex of 20–22nt is obtained (Bernstein et al. 2003; Lee et al. 2004, Kim 2005). Studies have shown that pri-miRNAs of certain miRNA genes are subject to RNA editing that converts adenosine to inosine (A→I) (Luciano et al. 2004; Blow et al. 2006; Yang et al. 2006). This A-to-I RNA editing is catalysed by members of the ADAR (adenosine deaminases acting on RNA) gene family (Bass 2002; Nishikura 2006). Editing of pri-miRNAs could alter their processing including suppression of the Dicer or inhibition of the Drosha cleavage step. For instance, editing of pri-miR-151 by ADAR1 interferes with Dicer processing (Kawahara et al. 2007a), whereas that of pri-miR-142 by ADAR1 and ADAR2 suppresses Drosha processing and consequently affects the expression of the mature miRNA levels (Yang et al. 2006). Furthermore, editing of pri-miRNAs could also lead to the expression of edited mature miRNAs that are likely to repress genes different from those targeted by unedited miRNAs (Kawahara et al. 2007b).
The miR-376 cluster of miRNA genes is located on human chromosome 14 and syntenic regions of the distal end of mouse chromosome 12 (Seitz et al. 2004). Expression of miR-376 RNAs has been reported in the developing embryos, and adult tissues, including the cochlea (Poy et al. 2004; Seitz et al. 2004; Friedman et al. 2009a; Wang et al. 2010). The four human miR-376 RNAs (miR-376a1, miR-376a2, miR-376b, miR-376c previously known as miR-368) and three mouse miR-376a-c RNAs share a high degree of sequence identity (Kawahara et al. 2007b; miRBase 18; genome.ucsc.edu/). It has also been shown that the miR-376 cluster members are transcribed into one long primary RNA transcript that can be subject to extensive and simultaneous editing by adenosine deaminase at one or two specific sites (+4 and +44) in select human and mouse tissues and subregions of the brain tissue (Kawahara et al. 2007b) during the processing of pri-miRNAs to mature miRNA (Luciano et al. 2004; Blow et al. 2006; Yang et al. 2006). The +4 site is edited by ADAR2, whereas the +44 site is selectively edited by ADAR1. Finding of the edited forms of mature miR-376 RNAs in certain tissues, such as brain, implicated that, unlike the case of pri-miR-142 and pri-miR-151, editing of pri-miR-376 RNAs at the two sites does not interfere with the Drosha and Dicer cleavage steps. A→I editing of miR-376 cluster transcripts is tissue specific, leading to the silencing of a different set of genes and tissue-specific regulation of certain gene products. Some miR-376 cluster members (pri-miR-376a2, pri-miR-376b and pri-miR-376c) are nearly 100% edited at the +44 site in the human cortex and medulla, whereas no editing was detected at the +4 site of human pri-miR-376a1 in liver and at the +44 site of mouse pri-miR-376a in all tissues (Kawahara et al. 2007b).
Missense mutations in phosphoribosylpyrophosphate synthetase 1 (PRPS1) are associated with a spectrum of non-syndromic to syndromic hearing loss (de Brouwer et al. 2010; Liu et al. 2010). Both of the two editing sites in pri-miR-376 RNAs (+4 and + 44) are located within the functionally critical 5′ proximal ‘seed’ sequences of miRNA-376-5p and −3p strands, suggesting that the edited mature miR-376 RNAs may target genes different from those targeted by the unedited miR-376 isoform RNAs. The transcript of PRPS1 contains multiple binding sites for the edited version of miR-376a-5p within its 3′UTR that controls its transcription levels. This editing of miR-376a-5p is tissue specific. In the brain cortex, heart and kidney, it is edited, whereas in the liver almost no editing occurs. Because only the edited miR-376a-5p silences the expression of PRPS1, the activity of this target gene is repressed in a tissue-specific manner (Kawahara et al. 2007b). However, it is still unknown whether the pri-miRNAs editing of miR-376 cluster members occurs in the inner ear. In this study, we examined the pri-miRNAs editing of miR-376 cluster members in the mouse inner ear. We used quantitative real-time PCR (q-RT-PCR) and in situ hybridization to examine the expression pattern of miR-376a, b in the developing and adult mouse inner ear.
Materials and methods
Animals
Adult C57BL/6 mice were bred in-house in a low-noise environment at the Animal Research Facility of the Medical University of South Carolina with original breeding pairs purchased from The Jackson Laboratory (Bar Harbor, ME, USA). All mice received food and water ad libitum and were maintained on a 12-h light/dark cycle. Throughout the paper, the term ‘adult mice’ applies to all of the 8–12-week-old mice. Mice with any symptoms of ear infection were excluded from the study.
Ethical Approval
All aspects of the animal research were conducted in accordance with the guidelines of the Institutional Animal Care and Use Committee of the Medical University of South Carolina.
RNA preparation
Cochleae were obtained from embryonic day (E) 15, E18 and postnatal day 3 (P3) C57BL/6 mice. Total RNA from cochlear tissues was purified using the miRVana™ miRNA isolation kit according to the manufacturer's instructions (Ambion) and was DNase-treated using a Turbo DNA-free kit (Ambion, Austin, TX, USA) according to manufacturer's protocol.
Quantitative real-time PCR quantification of microRNAs
For detection of mature miRNAs, predesigned TaqMan MicroRNA Assays including the primer set and TaqMan probe were obtained from Applied Biosystems. Fifty nanograms of RNA was reverse transcribed in 15 μl at 42 °C for 30 min using a miRNA- or mouse U6B-specific oligonucleotide. All PCRs were performed in 20-μl aliquots containing 1.33 μl of reverse transcribed reactions and run in triplicate on the 7500 real-time PCR system (Applied Biosystems). Thermal cycling was initiated with a first denaturation step at 95 °C for 10 min, followed by 40 cycles of 95 °C for 15 s and 60 °C for 1 min. Relative expression was calculated using the cycle threshold (Ct) values for each miR, and a small RNA, mouse U6B, was used as the endogenous control for data normalization. Data were analysed for significant differences by t-test.
Determination of pri-miRNA editing
First-strand cDNA was synthesized using 1 μg of total DNase-treated RNA from cochlear tissues and each previously reported mouse miRNA 376a (mir-376a-5p, miR-376a-3p), 376b (miR-376b-5p, miR-376b-3p), c (miR-376c-3p)-specific RT primer (Kawahara et al. 2007b). The resultant cDNA was then amplified by PCR using each miRNA-specific PCR primer pairs. The no template and no enzymes control samples were included in the assay to ensure that any amplification that occurs in the sample is derived from the synthesized cDNA and not genomic DNA or other amplicon contamination. RT-PCR products were subsequently sequenced from both directions.
LNA probes, tissue sections and in situ hybridization (ISH)
Double digoxigenin (DIG)-labelled miRCURY LNA™ (locked nucleic acid) 5′-DIG and 3′-DIG labelled microRNA probes (Exiqon, Vedbæk, Denmark) for mmu-miR-376a-3p and mmu-miR-376b-3p were employed in this study. Inner ears from E9.5, E17, P0 and adult C57BL/6 mice were carefully and quickly resected and placed in fresh 4% paraformaldehyde in RNase-free PBS for 1–2 h. The tissue was then cryprotected in 30% sucrose in PBS (phosphate-buffered saline), embedded in Tissue-Tek OCT compound (Electron Microscopy Science, FT, Washington, PA, USA), cryostat sectioned at 10 μm and mounted on Superfrost Plus slides (Fisher; Thermo Fisher Scientific, Portsmouth, NH, USA). Sections were air-dried for 30 min and postfixed in 4% paraformaldehyde for 10 min. Hybridization was performed as previously described (Jørgensen et al. 2010).
Results
Quantitative real-time PCR
To investigate the expression of the members of the miRNA 376 family in the mouse inner ear, we performed RT-PCR for detection of mature miRNAs, using predesigned TaqMan MicroRNA Assays for mmu-miR-376a-3p, mmu-miR-376b-3p, mmu-miR-376c-3p. Cochlear tissues from developmental stage E15, 18 and P3 were analysed. Our data showed an approximately threefold increase in the expression level in mature miR-376a and miR-376b and twofold in miR-376c, from E15 to E18 (Figure 1). However, the expression levels of mature miR-376a, b, c genes significantly decrease at P3 (data not shown).
Figure 1.
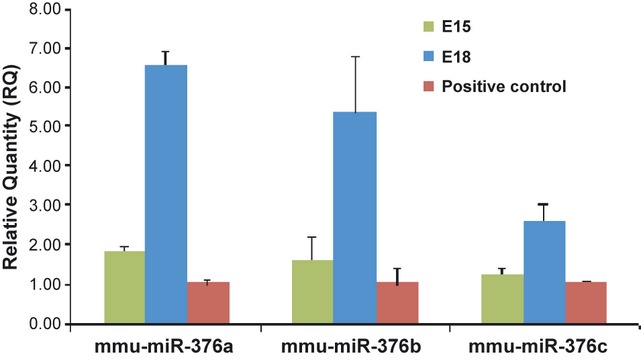
Quantitative real-time PCR detection of mature miR-376a-3p, miR-376b-3p, miR-376c-3p expression in the developing mouse inner ear. PCRs were performed in triplicate using cDNA products derived from total RNA purified from embryonic day (E) 15 and E18 mouse cochlear tissues. Data are normalized using mouse U6 small nuclear RNA as an endogenous control for RNA input. Mouse total RNA pool from Ambion was used as a positive control for validation of the TaqMan® miRNA assays. The relative expression levels of endogenous are shown.
In situ hybridization of inner ear sections for the miRNA 376 family
We next applied an in situ hybridization procedure to assess the cellular and subcellular distribution of miR-376 RNA cluster members in inner ear tissues using LNA miR-376a-3p and miR-376b-3p probes. LNA-ISH data for miR-376a-3p and miR-376b-3p in the sensory epithelium and spiral ganglion (SG) of the developing and adult inner ear are shown in Figure 2. MiR-376a and miR-376b are detected in the otic placode at E9.5 (transverse section, Figure 2a,b). MiR-376b is present in the sensory epithelium and SG at E17 (Figure 2c). MiR-376a (E) and miR-376b (D) are expressed in the organ of Corti (OCT), SG and stria vascularis (StV) at P0 (Figure 2d,e,f). MiR-376b (F) is also present in the SG neurons of an adult ear.
Figure 2.
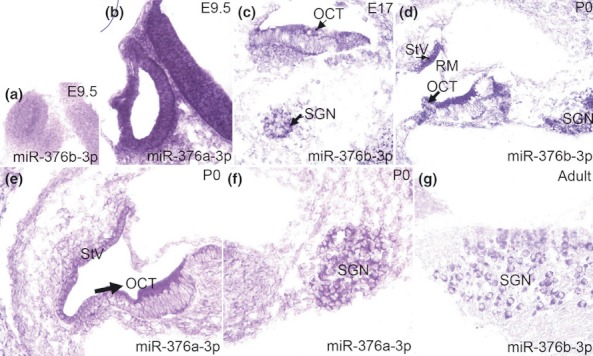
Expression of miR-376a-3p and miR-376b-3p in the sensory epithelium and spiral ganglion (SG) of the inner ear. MiR-376a (b) and miR-376b (a) are expressed in the otic placode at E9.5 (transverse section). MiR-376b is present in the sensory epithelium and SG at E17 (c). MiR-376a (e, f) and miR-376b (d) are expressed in the organ of Corti (OCT), SG and stria vascularis (StV) at P0. MiR-376b (g) is present in the SG neurons of an adult ear.
Location of miR-376b-3p in the vestibular portion of the developing inner ear is shown in Figure 3. At E17, miR-376b is expressed in the ampullea of vestibular organs (Figure 3a) and at P0, and it is restricted within in the sensory epithelia of the maculae and ampullae (Figure 3b) as well as in the vestibular ganglion (VG) neurons (Figure 3c). In the lateral wall of the inner ear, miR-376a-3p is expressed in the stria vascular (StV) at P0 (Figure 4a) and is present in the marginal cells of the StV of adult ear (Figure 4b). At P0, the expression of miR-376b-3p appears in all the cells in the StV (Figure 4c) and is also present in the marginal (arrows; right panel), intermediate and basal (arrowheads; right panel) cells of the StV in an adult ear (Figure 4d).
Figure 3.
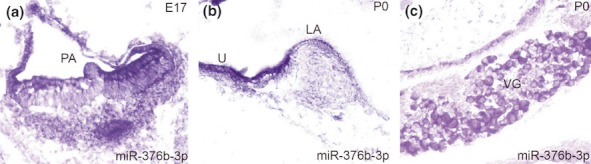
Expression of miR-376b-3p in the vestibular portion of the inner ear. MiR-376b is expressed in the posterior ampulla (PA) at E17 (a). MiR-376b is present in the sensory epithelium of the utricle (U) and lateral ampulla (LA) at P0 (b). MiR-376b is expressed the vestibular ganglion (VG) neurons at P0 (c).
Figure 4.
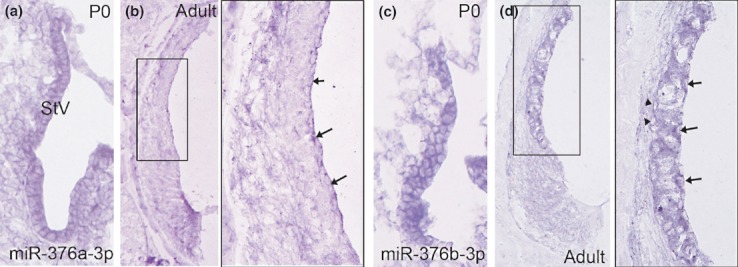
Expression of miR-376a-3p and miR-376b-3p in the stria vasculari of the inner ear. MiR-376a is expressed in the StV at P0 (a). MiR-376a is present in the marginal cells (arrows; right panel) of the StV of an adult ear (b). The expression of miR-376b appears in the StV at P0 (c). D: miR-376a is present in the marginal (arrows; right panel), intermediate and basal (arrowheads; right panel) cells of the StV in an adult ear. The right panel is an enlarged image of corresponding boxed area in the left panel.
Analysis of editing of pri-miR-376 cluster RNAs in mouse cochlea
A→I editing of a miRNA may result in an alteration of the subsequent miRNA target. This has been demonstrated in the case of miR-376a-5p, which undergoes A→I editing at a site within the 5′ seed region (Kawahara et al. 2007a; b). PRPS1, which contains multiple target sites for the edited version of miR-376-5p within its 3′UTR, is repressed in a tissue-specific manner (Kawahara et al. 2007b). Thus, editing of miR-376 appears to be one of the mechanisms that ensure tight regulation of PRPS1 in select tissues such as the brain cortex. However, the miR-376 editing and its possible involvement in PRPS1 transcripts regulation in the cochlea remain to be established.
The inosine residue converted from adenosine in RNA is detected as an A → G change in the cDNA sequence. To determine whether RNA editing is present at sites known to be edited within the pri-mir-376 hairpins in the cochlea, we analyse cDNA corresponding to pri-mir-376a (−5p and 3p), pri-mir-376b (−5p and 3p), pri-mir-376c (−5p and 3p) hairpins amplified using primers located around 100 nucleotides before and after each hairpin (Figure 5a,b,c). Our results indicate that only unedited pri-miR-376 RNAs were present in the cochlea suggesting that PRPS1 levels and activity in the inner ear may not be regulated through the editing of miR-376.
Figure 5.
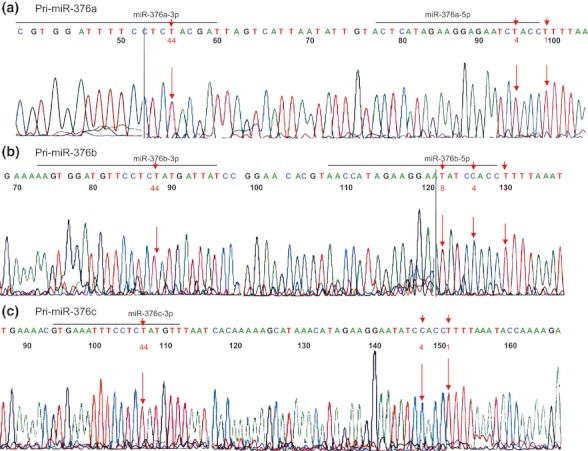
Analysis of pri-miR-376a, b, c in mouse inner ear. The reverse transcription polymerase chain reaction (RT-PCR) products obtained from E15 embryos are sequenced. The nucleotides reported to undergo editing are shown.
Discussion
MiRNA genes represent approximately 1% of the predicted genes in the genome of different species. Currently, over 1500 different human miRNAs have been deposited in the publicly available miRNA database Release 18.0 (Kozomara & Griffiths-Jones 2011). These miRNAs are often expressed in a cell-specific manner or during different stages of development (Landgraf et al. 2007). MiRNAs have the potential to regulate hundreds of genes at the post-transcriptional level by either repressing translation or decreasing mRNA stability (Valencia-Sanchez et al. 2006; Yoda et al. 2010). It is now estimated that approximately 30 to 60% of human protein-coding genes are post-transcriptionally regulated by miRNAs (Lewis et al. 2005; Rajewsky 2006; Friedman et al. 2009).
Inner ear development requires regulation of expression of genes in a coordinated manner (Fritzsch et al. 2006; Kelley 2006). Several studies suggest that the genetic regulatory properties of miRNAs are essential to the proper establishment of sensory neurons and sensory epithelial supporting and hair cells. For example, Dicer knockout (KO) in zebrafish has a demonstrated effect on ear organogenesis and otoconia formation (Giraldez et al. 2005). Dicer KO mice exhibit early embryonic lethality preceding otocyst development (Bernstein et al. 2003), whereas the conditional Dicer KO mice, under the control of the Pax2 promoter, showed gross inner ear malformations, with loss or reduction of the lateral and anterior semicircular canals and cristae. The cochlea is also malformed, lacking coils and with aberrant hair bundle formation and stereocilia defects, and impaired hearing function (Soukup et al. 2009). A hair-cell-specific Dicer-Pou4f3-Cre-induced conditional knockout mouse has also been created. At E18 and at birth, the inner ear sensory epithelia have normal morphology, but by P38, the hair cells showed signs of degeneration (Friedman et al. 2009a,b). Moreover, a Foxg1-cre-mediated conditional deletion of Dicer1 and microRNA (miRNA) depletion on mouse embryos have been shown to abolish inner ear development which in turn led to a truncated development of the otic capsule (Kersigo et al. 2011). Furthermore, it has been reported that mutations within the seed region of mature miR-96 are associated with autosomal dominant, progressive hearing loss in humans and mice. The homozygous mutant mice were profoundly deaf, exhibiting no cochlear responses (Lewis et al. 2009; Mencia et al. 2009). MiR-96 belongs to a triad, together with MiR-182 and MiR-183, which has been shown to be expressed in the inner ear sensory epithelia and ganglia neurons (Pierce et al. 2008) and is required for hair cell maintenance and survival (Weston et al. 2011).
To investigate the possible regulation of the transcription of Prps1 by miR-376 cluster members in the mouse inner ear, we first quantified the expression of the mature miR-376a-c RNAs by quantitative real-time PCR using TaqMan® MicroRNA Assays probes for mmu-miR-376a-3p, mmu-miR-376b-3p, mmu-miR-376c-3p. Our data showed that the miR-376a, b and c are present at E15 and E18 and an approximately threefold increase in the expression level in mature miR-376a and miR-376b and twofold in miR-376c are observed from E15 to E18. Expression seemed to be decreased during the postnatal stages. By in situ hybridization, we detected robust expression of miR-376a-3p and miR-376b-3p in the sensory epithelia and ganglia of both auditory and vestibular portions in the embryonic inner ears. A significant decrease in miR-376a-3p and miR-376b-3p expression with age was seen in sensory epithelia. In adult inner ear, the expression of miR-376a-3p and miR-376b-3p is restricted within ganglion neurons of auditory and vestibular systems as well as in the cells in the stria vascularis. Together, these findings suggest that miR-376 cluster members may function in regulating the development of the sensory epithelia and ganglia in the embryonic inner ear as well as in maintaining normal function of spiral ganglion neurons and cells in the cochlear lateral wall of adult mouse. It was interesting to note that there was a difference in expression patterns of miR-376a-3p and miR-376b-3p in the stria vascularis of adult inner ear (Figure 4b,d), although these molecules appear to have similar expression profile at P0 (Figure 4a,c). These results suggest that these two miRNAs may play a different role in maintaining the homoeostasis of inner ear microenvironment, that is, miR-376a but not miR-376b may be involved in directing transepithelial movement of fluid and ions transport activity by marginal cells and intermediate cells (Schulte & Adams 1989). Additionally, it is worthy of notice that all the miR-376-3p are more abundant than those with 5p and that the mature mmu-miR-376a-3p is the most highly expressed cloned form. We have not included mmu-miR-376c-3p in our in Situ analysis, but its expression level seems very low (miRBase 18).
The tissue-specific adenosine (A)-to-inosine (I) editing of miR-376 cluster transcripts leads to predominant expression of edited miR-376 isoform RNAs. This editing resulted in selection of a set of target genes different from those targeted by the unedited miRNAs (Kawahara et al. 2007b). The two active mammalian ADAR enzymes (ADAR 1 and 2) responsible for the A to I catalysis are ubiquitously expressed. Nevertheless, in mammals, most of the ADAR-edited transcripts are expressed in the central nervous system (CNS). The relevance of ADARs is clearly shown by the phenotype of knock-out mice. ADAR2−/− mice are viable, exhibit profound epileptic seizures and die prematurely between day 0 and day 20 (Higuchi et al. 2000), whereas the deletion of ADAR1 in mice is embryonically lethal, and mice die between E11.5 and 12.5 because of a severe liver defect that involves cells of both the haematopoietic and hepatic lineages as well as apoptosis in heart, liver and vertebra. The examination of the neuroepithelium failed to detect abnormalities (Hartner et al. 2004). Thus, although ADAR1 clearly edits neuronal RNA templates (Burns et al. 1997), it appears to have a critical function in non-nervous tissues. Another intriguing aspect on the regulation of A to I editing is that ADAR expression and localization in conjunction with the developmental and cell-type-specific modulation of RNA editing studies have revealed that the presence of ADAR mRNA and proteins rarely correlates with the observed intracellular RNA editing activity (Bass 2002; Mightdanovych & Beal 2006; Jacobs et al. 2009; Tan et al. 2009; Nishikura 2010). While In vitro RNA editing studies have not revealed involvement of any proteins other than ADARs for site-selective RNA editing, the discordance between ADAR expression levels and editing patterns suggests that additional regulatory factors may be required.
Analysis of RNA editing of the pri-miR-376 family members in the brain cortices of ADAR2−/− mice and ADAR1−/− mouse E11.5 embryos revealed that editing of the −1 site of pri-miR-376a, pri-miR-376b and pri-miR-376c, as well as the +4 site of pri-miR-376a, is almost absent in the cortex of ADAR2−/− mice. However, an increase in editing frequency at the +44 site of pri-miR-376b and pri-miR-376c is found in ADAR2−/− mice, whereas editing of the +44 site was eliminated in ADAR1−/− embryos. These findings suggest that the −1 and +4 sites are mainly edited by ADAR2 and that the +44 site is selectively edited by ADAR1 (Kawahara et al. 2007a; b). In selected tissues such as the human medulla and the brain cortex, only the edited version of mature miR-376a2-5p was detected. Both of the major editing sites in pri-miR-376 RNAs (+4 and +44) are located within the functionally critical 5′-proximal ‘seed’ sequences of miR-376-5p and miR-376-3p. The observed editing events in miR-376 appears to be one of the mechanisms that ensure regulation of transcript levels PRPS1, which contains target sites for the edited version of miR-376 within its 3′UTR.
By in situ hybridization, we previously showed expression of Prps1 in murine vestibular and cochlea hair cells, with continuous expression in hair cells and postnatal expression in the spiral ganglion (Liu et al. 2010). This expression pattern of prps1 is very similar to that for the miR-376. In this study, we analysed mouse inner ear–amplified pri-miRNA-376a (mir-376a-5p, miR-376a-3p), 376b (miR-376b-5p, miR-376b-3p), c (miR-376c-3p) cDNAs for A to I RNA editing. However, our data show that only unedited pri-miR-376 RNAs were detected in the cochlea, suggesting that the activity of PRPS1 in the inner ear may not be regulated through the editing of miR-376.
Acknowledgments
This work was supported by National Institutes of Health (NIH DC012546); Grant number: DC05575 (L.X.); Grant number: DC00422 (H.L.); Grant number: DC07506 (H.L.); Office of Research & Development, Medical Research Services. This work also benefitted from the South Carolina Clinical and Translational Research Institute (SCTR) Clinical and Translational Science Award (NIH/NCRR UL1RR029882). The funders had no role in study design, data collection and analysis.
Conflict of interest
The authors declare no conflict of interest.
References
- Ambros V. MicroRNA pathways in flies and worms: growth, death, fat, stress, and timing. Cell. 2003;113:673–676. doi: 10.1016/s0092-8674(03)00428-8. [DOI] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu. Rev. Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, et al. Dicer is essential for mouse development. Nat. Genet. 2003;35:215–217. doi: 10.1038/ng1253. [DOI] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, et al. RNA editing of human microRNAs. Genome Biol. 2006;7:R27. doi: 10.1186/gb-2006-7-4-r27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Brouwer APM, van Bokhover H, Nabuurs SB, et al. PRPS1 mutations: four distinct syndromes and potential treatment. Am. J. Human. Gen. 2010;86:506–518. doi: 10.1016/j.ajhg.2010.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns CM, Chu H, Rueter SM, et al. Regulation of serotonin-2C receptor G-protein coupling by RNA editing. Nature. 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- Friedman LM, Dror AA, Mor E, et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc. Natl. Acad. Sci. USA. 2009a;106:7915–7920. doi: 10.1073/pnas.0812446106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of miRNAs. Genome Res. 2009b;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritzsch B, Pauley S, Beisel KW. Cells, molecules and morphogenesis: the making of the vertebrate ear. Brain Res. 2006;1091:151–171. doi: 10.1016/j.brainres.2006.02.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308:833–838. doi: 10.1126/science.1109020. [DOI] [PubMed] [Google Scholar]
- Hartner JC, Schmittwolf C, Kispert A, Muller AM, Higuchi M, Seeburg PH. Liver disintegration in the mouse embryo caused by deficiency in the RNA-editing enzyme ADAR1. J. Biol. Chem. 2004;279:4894–4902. doi: 10.1074/jbc.M311347200. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Maas S, Single FN, et al. Point mutation in an AMPA receptor gene rescues lethality in mice deficient in the RNAediting enzyme ADAR2. Nature. 2000;406:78–81. doi: 10.1038/35017558. [DOI] [PubMed] [Google Scholar]
- Jacobs MM, Fogg RL, Emeson RB, Stanwood GD. ADAR1 and ADAR2 expression and editing activity during forebrain development. Dev. Neurosci. 2009;31:223–237. doi: 10.1159/000210185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen S, Baker A, Møller S, Nielsen BS. Robust 1-day in situ hybridization protocol for detection of microRNAs in paraffin samples using LNA probes. Methods. 2010;52:375–381. doi: 10.1016/j.ymeth.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Chendrimada TP, Shiekhattar R, Nishikura K. RNA editing of microRNA-151 blocks cleavage by the Dicer-TRBP complex. EMBO Rep. 2007a;8:763–769. doi: 10.1038/sj.embor.7401011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, Zinshteyn B, Sethupathy P, Iizasa H, Hatzigeorgiou AG, Nishikura K. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science. 2007b;315:1137–1140. doi: 10.1126/science.1138050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley MW. Hair cell development: commitment through differentiation. Brain Res. 2006;1091:172–185. doi: 10.1016/j.brainres.2006.02.062. [DOI] [PubMed] [Google Scholar]
- Kersigo J, D'Angelo A, Gray BD, Soukup GA, Fritzsch B. The role of sensory organs and the forebrain for the development of the craniofacial shape as revealed by Foxg1-cre-mediated microRNA loss. Genesis. 2011;49:326–341. doi: 10.1002/dvg.20714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat. Rev. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucl. Acids Res. 2011;39:D152–D157. doi: 10.1093/nar/gkq1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim M, Han J, et al. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B, Burge C, Bartel D, Burge C, Bartel D. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Lewis MA, Quint E, Glazier AM, et al. An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat. Genet. 2009;41:614–618. doi: 10.1038/ng.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Han D, Li J, et al. Loss-of function mutations in the PRPS1 gene cause a type of nonsyndromic X-linked sensorineural deafness, DFN2. Am. J. Hum. Genet. 2010;86:65–71. doi: 10.1016/j.ajhg.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luciano DJ, Mirsky H, Vendetti NJ, Maas S. RNA editing of a miRNA precursor. RNA. 2004;10:1174–1177. doi: 10.1261/rna.7350304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mencia A, Modamio-Hoybjor S, Redshaw N, et al. Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 2009;41:609–613. doi: 10.1038/ng.355. [DOI] [PubMed] [Google Scholar]
- Mightdanovych O, Beal PA. Breaking the central dogma by RNA editing. Chem. Rev. 2006;106:3397–3411. doi: 10.1021/cr050314a. [DOI] [PubMed] [Google Scholar]
- Nishikura K. Editor meets silencer: crosstalk between RNA editing and RNA interference. Nat. Rev. 2006;7:919–931. doi: 10.1038/nrm2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishikura K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 2010;79:1–29. doi: 10.1146/annurev-biochem-060208-105251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce ML, Weston MD, Fritzsch B, Gabel HW, Ruvkun G, Soukup GA. MicroRNA-183 family conservation and ciliated neurosensory organ expression. Evol. Dev. 2008;10:106–113. doi: 10.1111/j.1525-142X.2007.00217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- Rajewsky N. MicroRNA target predictions in animals. Nat. Genet. 2006;38:S8–S13. doi: 10.1038/ng1798. [DOI] [PubMed] [Google Scholar]
- Schulte BA, Adams JC. Distribution of immunoreactive Na+, K+-ATPase in gerbil cochlea J. Histochem. Cytochem. 1989;37:127–134. doi: 10.1177/37.2.2536055. [DOI] [PubMed] [Google Scholar]
- Seitz H, Royo H, Bortolin ML, Lin SP, Ferguson-Smith AC, Cavaillé J. A Large Imprinted microRNA Gene Cluster at the Mouse Dlk1-Gtl2 Domain. Genome Res. 2004;14:1741–1748. doi: 10.1101/gr.2743304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup GA, Fritzsch B, Pierce ML, et al. Residual microRNA expression dictates the extent of inner ear development in conditional Dicer knockout mice. Dev. Biol. 2009;328:328–341. doi: 10.1016/j.ydbio.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan BZ, Huang H, Lam R, Soong TW. Dynamic regulation of RNA editing of ion channels and receptors in the mammalian nervous system. Mol. Brain. 2009;29:2–13. doi: 10.1186/1756-6606-2-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valencia-Sanchez MA, Liu J, Hannon GJ, Parker R. Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 2006;20:515–524. doi: 10.1101/gad.1399806. [DOI] [PubMed] [Google Scholar]
- Wang XR, Zhang XM, Zhen J, Zhang PX, Xu G, Jiang H. MicroRNA expression in the embryonic mouse inner ear. NeuroReport. 2010;21:611–617. doi: 10.1097/WNR.0b013e328338864b. [DOI] [PubMed] [Google Scholar]
- Weston MD, Pierce ML, Jensen-Smith HC, et al. MicroRNA-183 family expression in hair cell development and requirement of microRNAs for hair cell maintenance and survival. Dev. Dyn. 2011;240:808–819. doi: 10.1002/dvdy.22591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W, Chendrimada TP, Wang Q, et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 2006;13:13–21. doi: 10.1038/nsmb1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda M, Kawamata T, Paroo Z, Ye X, Iwasaki S. ATP-dependent human RISC assembly pathways Nat. Struct. Mol. Biol. 2010;17:17–23. doi: 10.1038/nsmb.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]


