Abstract
Heteroallelic and homo- or hemiallelic Complementary sex determiner (Csd) proteins determine sexual fate in the honeybee (Apis mellifera) by controlling the alternative splicing of the downstream gene fem (feminizer). Thus far, we have little understanding of how heteroallelic Csd proteins mediate the splicing of female fem messenger RNAs (mRNAs) or how Fem proteins direct the splicing of honeybee dsx (Am-dsx) pre-mRNAs. Here, we report that Am-tra2, which is an ortholog of Drosophila melanogaster tra2, is an essential component of female splicing of the fem and Am-dsx transcripts in the honeybee. The Am-tra2 transcripts are alternatively (but non-sex-specifically) spliced, and they are translated into six protein isoforms that all share the basic RNA-binding domain/RS (arginine/serine) domain structure. Knockdown studies showed that the Am-tra2 gene is required to splice fem mRNAs into the productive female and nonproductive male forms. We suggest that the Am-Tra2 proteins are essential regulators of fem pre-mRNA splicing that, together with heteroallelic Csd proteins and/or Fem proteins, implement the female pathway. In males, the Am-Tra2 proteins may enhance the switch of fem transcripts into the nonproductive male form when heteroallelic Csd proteins are absent. This dual function of Am-Tra2 proteins possibly enhances and stabilizes the binary decision process of male/female splicing. Our knockdown studies also imply that the Am-Tra2 protein is an essential regulator for Am-dsx female splice regulation, suggesting an ancestral role in holometabolous insects. We also provide evidence that the Am-tra2 gene has an essential function in honeybee embryogenesis that is unrelated to sex determination.
Keywords: sex determination, insects, evolution of genetic pathways, Apis mellifera
IN contrast to the well-studied sex chromosome system in Drosophila melanogaster (Cline and Meyer 1996; Erickson and Quintero 2007), sex in the honeybee (Apis mellifera) is determined by heterozygosity at the complementary sex determiner (csd) gene (Beye et al. 2003). Bees that are heterozygous at the csd locus develop into females, whereas bees that are homozygous or hemizygous at csd develop into males. Queens in honeybee colonies lay unfertilized eggs to produce fertile males (drones) and fertilized eggs to produce females that differentiate into either workers or queens; queen fate is determined by specific feeding of the queen larvae with royal jelly (Kucharski et al. 2008; Kamakura 2011). Diploid males, homozygous for the csd gene, do not survive in a colony because they are eaten by worker bees shortly after they hatch from the egg (Woyke 1963). The csd gene translates into an SR-type protein that has at least 15 major allelic variants (Beye 2004; Hasselmann et al. 2008) that differ at an average of ∼3% of their amino acid residues in the putative specifying domain (Hasselmann and Beye 2004; Hasselmann et al. 2008). Females express heteroallelic Csd proteins that direct female splicing of feminizer (fem) pre-messenger RNAs (pre-mRNAs) (Hasselmann et al. 2008; Gempe et al. 2009). These female fem transcripts are also translated into SR-type proteins that are required for female differentiation. The Fem proteins promote the female splicing of the A. mellifera dsx (Am-dsx) transcripts, which express a transcription factor of the DM type, a protein with a female-specific carboxy-terminal end (Dearden et al. 2006; Cho et al. 2007). In addition, Fem proteins direct splicing of their own pre-mRNAs into the productive female form, which generates an autoregulatory feedback loop that maintains the female state throughout development (Gempe et al. 2009). In the absence of Csd protein activity in males (homo- or hemiallelic Csd proteins), fem transcripts are spliced into the male form, which contains a translational stop codon in exon 3 that causes premature translation termination (Gempe et al. 2009). As a consequence, Am-dsx pre-mRNAs are spliced into the male variant expressing a Dsx protein (Gempe et al. 2009), which has a male-specific carboxy-terminal end as part of the oligomerization domain 2 (Cho et al. 2007).
The csd gene is thus the primary signal of sex determination in the honeybee. The csd gene evolved recently in the honeybee lineage by gene duplication of an ancestral copy of the fem gene (Hasselmann et al. 2008). Although substantially diverged in sequence, the fem gene is the ortholog of the sex-determining gene transformer (tra) of D. melanogaster. Thus far, we have little understanding of how the heteroallelic Csd proteins mediate fem splicing or how Fem proteins direct Am-dsx pre-mRNA splicing (Gempe and Beye 2011). Both proteins lack an RNA-binding domain (RBD), which is suggestive of a cofactor that can directly interact with the respective RNA sequence.
In this study, we explored the role of the tra2 gene of the honeybee [Am-tra2 (Dearden et al. 2006)] in regulating the sex-specific splicing of the fem and Am-dsx transcripts. In D. melanogaster, the RNA-binding protein Tra2 acts together with the Tra protein to promote the female splicing of dsx pre-mRNAs (Burtis and Baker 1989; Amrein et al. 1990; Hedley and Maniatis 1991; Inoue et al. 1992; Lynch and Maniatis, 1996, 1995; Sciabica and Hertel 2006). D. melanogaster females express in somatic tissues two major protein isoforms of the Tra2 proteins (Tra2264 and Tra2226) that, together with the Tra proteins, activate a weak 3′ splice acceptor site in dsx pre-mRNAs by binding to the six repeats of a 13-nucleotide exonic splicing enhancer (ESE) sequence and a single purine-rich element. This activation leads to the inclusion of the female exon 4 in female dsx mRNAs (Burtis and Baker 1989; Hedley and Maniatis 1991; Inoue et al. 1992; Lynch and Maniatis 1996, 1995; Sciabica and Hertel 2006). Splicing of honeybee Am-dsx has putatively evolved in that respect compared to D. melanogaster, as Am-dsx pre-mRNAs lack the canonical binding sites of Tra/Tra2 proteins.
The tra2 gene has also been characterized in other dipteran species, including Musca domestica, Anastrepha obliqua, Ceratitis capitata, Lucilia cuprina, and Sciara ocellaris (Burghardt et al. 2005; Concha and Scott 2009; Salvemini et al. 2009; Sarno et al. 2010; Martín et al. 2011), as well as in the lepidopteran insect Bombyx mori (Niu et al. 2005). All Tra2 proteins share the same domain structure of a single RBD that is flanked by two arginine-/serine-rich (RS-rich) regions. The RBD consists of 80–90 amino acids that form a βαββαβ barrel-like topology. One side of the β-sheet surface (β1 and β3) of the RBD exposes two sequence elements, RNP-1 and RNP-2, which are directly involved in RNA recognition (Dreyfuss et al. 1988; Merrill et al. 1988; Nagai et al. 1990; Amrein et al. 1994).
In the dipteran insects M. domestica, C. capitata, Anastrepha suspensa, and A. obliqua, RNA interference (RNAi) knockdown studies of the tra2 gene showed that Tra2 proteins are also involved in female splicing of tra mRNAs (Burghardt et al. 2005; Concha and Scott 2009; Salvemini et al. 2009; Sarno et al. 2010; Martín et al. 2011). Due to the presence of the canonical 13-nucleotide Tra/Tra2-binding motif in transformer of M. domestica, A. suspensa, and C. capitata, the authors suggest that Tra2 proteins act as cofactors in the autoregulatory splicing loop in which Tra/Tra2 proteins direct the female splicing of tra transcripts and thus the expression of Tra protein (Salvemini et al. 2009; Hediger et al. 2010; Schetelig et al. 2012). In the lepidopteran insect B. mori, the function of Tra2 proteins in sexual regulation of the Bm-dsx transcripts is not known. In this species, male splicing of Bm-dsx transcripts requires the splicing inhibitor (Bm-PSI) and the male-specific IMP (Bm-IMP) proteins. The activation of the female exon splicing is repressed in males by the binding of the Bm-PSI and the male-specific Bm-IMP proteins to the 20-nucleotide CE1 motif of the female exon (Suzuki et al. 2001, 2008, 2010).
In the male germline of D. melanogaster, the Tra2226 protein isoform has an additional function in spermatogenesis in controlling the splicing of the exuperantia (exu) and alternative-testes-transcript (att) transcripts (Hazelrigg and Tu 1994; Madigan et al. 1996; Mattox et al. 1996). In the testes, Tra2226 proteins negatively affect their own expression by promoting the splicing of tra2179 mRNAs, which produce no functional protein (Mattox and Baker 1991; Mattox et al. 1996; McGuffin et al. 1998). This negative feedback loop controls the level of Tra2 expression, which is critical for proper spermatogenesis.
In this study, we report the cloning and functional analysis of the A. mellifera Am-tra2 gene. Our study showed that the Am-tra2 gene serves as a regulator in female-specific splicing of fem and Am-dsx transcripts. Furthermore, we show that Am-tra2 has a vital function in embryogenesis that differs from its reported functions in other species.
Materials and Methods
Bee sources
Diploid female embryos were derived from the progeny of queens inseminated by semen from a single drone having a different sex allele than that of the queen. Haploid male embryos were collected from colonies that were headed by a virgin queen. These non-mated queens laid unfertilized male eggs that we induced by repeated CO2 treatments of virgin queens.
RNA extraction, cDNA synthesis, and PCR
Total RNA was extracted using the TRIzol protocol (GIBCO BRL Life Technologies, Darmstadt, Germany). The first-strand complementary DNA (cDNA) from mRNA was generated by reverse transcription using an oligo(dT) primer following the protocol of the supplier (Fermentas, St. Leon-Rot, Germany). We quantified the amount of cDNA in our samples in a NanoDrop ND-1000 spectral photometer and adjusted the amount of cDNA prior to PCR amplification. PCR was performed using GoTaq Flexi DNA Polymerase (RNAi experiments) according to the protocol of the supplier (Promega, Mannheim, Germany) and Taq polymerase (RACE and transcriptional analysis of Am-tra2 throughout development). All RT-PCR fragments were resolved by agarose gel electrophoresis and stained with ethidium bromide. The identity of the fem, Am-dsx, csd, and Am-tra2 amplicons was verified by sequencing. We amplified cDNA fragments of the ef-1α gene using oligonucleotides #EM033 and #EM034 for the semiquantitative studies across samples (Supporting Information, Table S1).
Characterization of Am-tra2 gene
To determine the entire sequences of the Am-tra2 transcripts, we performed 5′ and 3′ RACE experiments following the manufacturer’s instructions (FirstChoice RLM-RACE kit; Ambion). The cDNAs were generated from male and female RNA samples of honeybee embryos. Gene-specific primers for RACE reactions were designed from the sequence of the Am-tra2 gene model at the NCBI web site (NCBI Reference Sequence: XM_001121070.2) (Table S1). All RACE products were cloned into the pGEM-T vector (Promega), and both strands were sequenced. We translated the mRNA sequences into the amino acid sequence, and we predicted the protein domains by the similarity to domains in the PROSITE database (http://www.expasy.org/prosite/). The GenBank accession numbers are JQ518311 (Am-tra2285), JQ518314 (Am-tra2284), JQ518312 (Am-tra2253), JQ518314 (Am-tra2252), JQ518313 (Am-tra2234), and JQ518316 (Am-tra2233).
Transcriptional studies of the Am-tra2 gene throughout development
Total RNA was extracted from male and female eggs (at 0–6, 9–24, 33–48, and 72 hr), larvae (L1 and L4 instar), pupae (3 days before hatching from comb), adult heads, and germline tissue (testes of L4 larvae and ovaries of virgin queens). We amplified cDNA fragments using oligonucleotides #359 and #421 (Table S1) that span the complete open reading frame (ORF) of all six Am-tra2 splice variants. The identity of the amplicons of the male and female L1 larvae and pupae were verified by sequencing.
Functional studies of the Am-tra2 gene
RNAi knockdown was induced in early embryogenesis at the syncytial stage (0–4 hr after egg deposition) in females and males (Beye et al. 2003, 2002). Am-tra2 double-stranded RNA-1 (dsRNA-1), encompassing the region from 322 to 767 bp (446 bp long), was generated using oligonucleotides #22M and #23M (Table S1) from cloned cDNAs of the Am-tra2285 transcript following the protocol previously described (Beye et al. 2003, 2002; Hasselmann et al. 2008). Am-tra2 dsRNA-2, encompassing the region from 108 to 499 bp (392 bp long), was generated using oligonucleotides #591 and #592 (Table S1 and Figure S1) from cloned cDNAs of transcript Am-tra2285. The dsRNAs were dissolved in ddH2O and injected at a concentration of 4–200 pg/embryo (Tables 1 and 2). In the control samples, we injected only ddH2O (Roth, Karlsruhe, Germany).
Table 1 . Development of Am-tra2-dsRNA-treated female embryos.
| Embryos injected | No. of embryos showing normal development after ∼70 hr |
Hatched L1 larvae |
|||
|---|---|---|---|---|---|
| Treatment/amount of injected dsRNA per embryo | N | N | % | N | % |
| Nontreated | 683 | 594 | 86.9 | 365 | 53.4 |
| ddH20 | 406 | 182 | 44.8 | 97 | 23.9 |
| Am-tra2 dsRNA-1 | |||||
| 200 pg | 92 | 0 | 0 | 0 | 0 |
| 100 pg | 25 | 0 | 0 | 0 | 0 |
| 75 pg | 60 | 0 | 0 | 0 | 0 |
| 40 pg | 20 | 0 | 0 | 0 | 0 |
| 20 pg | 53 | 0 | 0 | 0 | 0 |
| 4 pg | 40 | 5 | 12.5 | 2 | 5.0 |
| Am-tra2 dsRNA-2 | |||||
| 224 pg | 27 | 0 | 0 | 0 | 0 |
| 96 pg | 100 | 15 | 15 | 0 | 0 |
| 67 pg | 104 | 38 | 36.5 | 0 | 0 |
| 56 pg | 105 | 14 | 13.3 | 4 | 3.8 |
| 33 pg | 146 | 31 | 21.2 | 6 | 4.1 |
| 4 pg | 155 | 29 | 18.7 | 9 | 5.8 |
The number of individuals (N) and the relative proportions (%) with respect to the total number of initially injected embryos are shown.
Table 2 . Development of Am-tra2-dsRNA-treated male embryos.
| Embryos injected | No. of embryos showing normal development after ∼70 hr |
Hatched L1 larvae |
|||
|---|---|---|---|---|---|
| Treatment/amount of injected dsRNA per embryo | N | N | % | N | % |
| Nontreated | 55 | 36 | 65.5 | 36 | 65.5 |
| ddH20 | 78 | 30 | 38.5 | 26 | 33.3 |
| Am-tra2 dsRNA-2 | |||||
| 96 pg | 73 | 8 | 11.0 | 0 | 0 |
The number of individuals (N) and the relative proportions (%) with respect to the total number of initially injected embryos are shown.
We counted the number of embryos showing normal development ∼70 hr after egg deposition and the number of hatched L1 larvae 77–80 hr after egg deposition. All embryos that were malformed, showed necrotic tissue, or lacked the segmentation pattern 70 hr after egg deposition were classified as aberrant.
To study the effect of Am-tra2 knockdown on the splice patterns of fem and Am-dsx transcripts, we collected embryos and L1 larvae from dsRNA-treated and nontreated samples 48 or 77–80 hr after egg deposition. Treated embryos and controls were reared in the incubator at 35° until an age of 48 or 77–80 hr or until reaching the L1 larval stage. The samples were directly frozen in liquid nitrogen.
Fragments corresponding to the female fem mRNAs were amplified using oligonucleotides #412 and #523 (Table S1), which are composed of a part of exon 3 and exon 6 (size: 177 bp). We amplified fragments (size: 458 bp; exons 3–4 and part of exon 5) corresponding to the male fem mRNAs using oligonucleotides #410 and #566 (Table S1). Because the predetermined state in the early embryos is male splicing of fem transcripts (Gempe et al. 2009), the presence of female-specific fragments indicates that female splicing is induced. We used oligonucleotides #417 and #418 (Table S1) to amplify exons 4–5–6 (size: 1.2 kb) and exons 4–6 (size: 392 bp), which correspond to the female and male Am-dsx transcripts, respectively. The female Am-dsx transcripts were also specifically amplified by oligonucleotides #417 and #419 (Table S1) (size: 188 bp), which encompass exon 4–5. Fragments corresponding to the csd mRNAs were amplified using oligonucleotides #CS-1 and #CS-2 (Table S1), which are composed of a part of exon 6, exons 7 and 8, and a part of exon 9. As exons 7 and 8 include the hypervariable region of the csd gene, the length of the amplified fragments can vary substantially between the alleles.
Phylogenetic and molecular evolutionary sequence analyses
We utilized nine tra-2 sequences from different insect species to compare the phylogenetic and molecular relationships: Tribolium castaneum (GenBank:XP_968550.2), Acromyrmex echinatior (GenBank: EGI70155.1), D. melanogaster (NCBI Reference Sequence: NP_476764.1), B. mori (GenBank: NP_001119705.1), Nasonia vitripennis (GenBank: XP_001601106.1), S. ocellaris (GenBank: CBX45935.1), C. capitata (GenBank: ACC68674.1), M. domestica (GenBank: AAW34233.1) and Anastrepha obliqua (GenBank: CBJ17280.1).
Results
Genomic organization of the tra2 gene in the honeybee
The existence of the tra2 gene in the honeybee genome was predicted by the similarity of its RBD to those of other insects (Dearden et al. 2006). We isolated Am-tra2 transcripts that included the 5′ and 3′ untranslated regions (UTRs) and three different polyadenylation sites (Figure 1) using RACE experiments with cDNA preparations from both male and female embryos. Using cDNA preparations from embryos and pupae of males and females, we performed RT-PCRs to amplify the entire ORF of Am-tra2 with oligonucleotide primers that bound the 5′ and the 3′ ends of the ORF. As a result, we detected up to six splice variants (Figure 1) that were not sex-specific in embryos or pupae. The six splice variants in embryos were Am-tra2285, Am-tra2284, Am-tra2253, Am-tra2252, Am-tra2234, and Am-tra2233, and the four transcript variants in pupae were Am-tra2285, Am-tra2284, Am-tra2253, and Am-tra2252 (Figure 1). Three splice variants showed major sequence differences in exon 2. The other three are minor variants of the other three major variants that differ in three nucleotides in exon 4. All transcripts express essentially the same protein that harbors a RBD flanked by two RS-rich domains but differ in the length in the first RS domain (RS1) (Figure 1, A–C). The RBD amino acid sequence has the strongest similarity (61–85% sequence identity) to the Tra2 proteins of a variety of insects (N. vitripennis, T. castaneum, A. echinatior, B. mori, S. ocellaris, D. melanogaster, M. domestica, C. capitata and A. obliqua) (Amrein et al. 1990; Mattox et al. 1996; Burghardt et al. 2005; Niu et al. 2005; Tribolium Genome Sequencing Consortium 2008; Salvemini et al. 2009; Sarno et al. 2010; Nygaard et al. 2011), supporting our notion that we have identified the Tra2 ortholog of the honeybee (Figure 2).
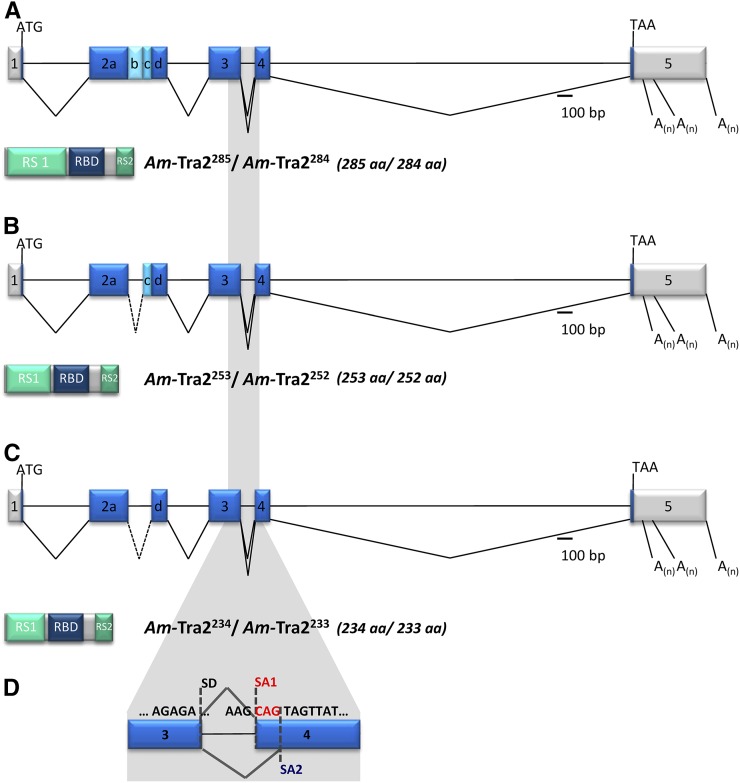
Figure 1 .
Genomic organization of the Am-tra2 gene and the alternatively spliced mRNAs. Schematic representation of the intron and exon organization (presented as lines and boxes, respectively). The alternatively spliced transcripts are indicated by the connecting lines between exons. The three alternative polyadenylation sites are labeled “A(n).” The scale denotes the relative size of the introns and exons. The 5′- and 3′-UTRs are presented in gray and the ORF in blue boxes. Below the genomic organization, the domain structure and relative size of the predicted Am-Tra2 proteins are shown (RS, arginine/serine-rich domain; RBD, RNA-binding domain). Superscript of Am-Tra2 proteins denotes the number of amino acids in that particular protein isoform. (A) Am-Tra2285 and Am-Tra2284. (B) Am-Tra2253 and Am-Tra2252. (C) Am-Tra2234 and Am-Tra2233. (D) Alternatively spliced variants in exon 4 producing trinucleotide and single-amino-acid differences. This splicing affected all three transcripts shown in A–C that are denoted as the Am-tra2284, Am-tra2252, and Am-tra2233 transcripts. SA1 and SA2 label the alternatively splice acceptors in exon 4.
Figure 2 .
The RBD domain of the honeybee Am-Tra2 protein and its relation to the Tra2 RBD domain of other holometabolous insects. (A) The phylogenetic relationship of the members of the different insect orders used in this comparison (Savard et al. 2006). (B) Amino acid sequence alignment of Tra2 RBDs of Apis mellifera, N. vitripennis, A. echinatior, T. castaneum, B. mori, S. ocellaris, D. melanogaster, M. domestica, C. capitata, and A. obliqua. Dots indicate amino acids identical to the predicted Am-Tra2 RBD of the honeybee. Shaded areas denote the RNP sequence elements.
The largest transcript, Am-tra2285, consists of 1401 nt and five exons and harbors an ORF coding for 285 amino acids (Figure 1A). The other two major transcript variants (Am-tra2253 and Am-tra2234) consist of six exons in which different parts of exon 2 are spliced out. These two variants are produced by sharing the same splice donor but using two alternative splice acceptors. They are 1248 and 1305 nt long and are putatively translated into 253- and 234-amino-acid proteins, respectively (Figure 1, B and C). All three major splice variants have minor splice variants that utilize an alternative splice acceptor site at exon 4, producing a single-serine amino acid deletion in the RS2 domain of the putative protein (Figure 1D).
Am-Tra2 has a typical Tra2 RBD that has evolved in the RNP-1 element
We compared our deduced amino acid sequence with those of dipteran (D. melanogaster, M. domestica, S. ocellaris, and A. obliqua), coleopteran (T. castaneum), lepidopteran (B. mori), and other hymenopteran [N. vitripennis (wasp) and A. echinatior (ant)] insects to identify shared structural features of Tra2 proteins in holometabolous insects. We were able to unambiguously align the amino acid sequences only for the RBD and neighboring regions, but not for other parts of the protein (Figure S2), suggesting that the RBD is evolutionarily constrained. All Tra2 proteins in the different organisms share the two RS domains, but the arginine- and serine-rich sequence is highly diverged (Figure S2), suggesting that these domains are faster-evolving and evolutionarily less constrained than the RBD.
In D. melanogaster, the RBD domain of Tra2 protein binds to ESEs, which are composed of six nearly identical 13-nucleotide-long sequences, the dsx repeat elements (RE). Similar motifs have also been detected in the dsx gene sequences of other dipteran insects, but are lacking in the Am-dsx pre-mRNAs of the honeybee (Crampton et al. 1998; Hediger et al. 2004; Lagos et al. 2005; Ruiz et al. 2005, 2007; Cho et al. 2007; Saccone et al. 2008; Concha et al. 2010; Permpoon et al. 2011). We next studied the sequence similarities within the RBDs of Tra2 proteins of different holometabolous insects (Figure 2). The RBD amino acid sequence diverges in relation to phylogenetic distance. The RBD domains of the hymenopteran species honeybees, N. vitripennis, and A. echinatior show a pairwise sequence identity of 82–85%, whereas RBDs of the honeybee and dipteran species have pairwise sequence identity of 61–68%. Within the RBD of the Am-Tra2 protein, there are two putative ribonucleoprotein consensus peptide (RNP) elements (Figure 2). These elements can directly interact with the dsx pre-mRNAs in D. melanogaster (Dreyfuss et al. 1988; Merrill et al. 1988; Nagai et al. 1990; Amrein et al. 1994). In the eight-amino-acid-long honeybee RNP-1 sequence element, we detected three amino acid differences compared with that of D. melanogaster. A mutation in the first amino acid of RNP-1 has been shown in D. melanogaster to be essential for female dsx splicing (Amrein et al. 1994). Interestingly, this functionally important first amino acid has especially evolved from amino acid R to K within the honeybee and the Nasonia lineage (Figure 2). We propose that the target RNA sequences of the RNP-1 element in the Am-Tra2 protein may have correspondingly evolved and thus differ from those found in D. melanogaster.
Am-tra2 transcript variants are not sex-specifically regulated and are transcribed throughout development
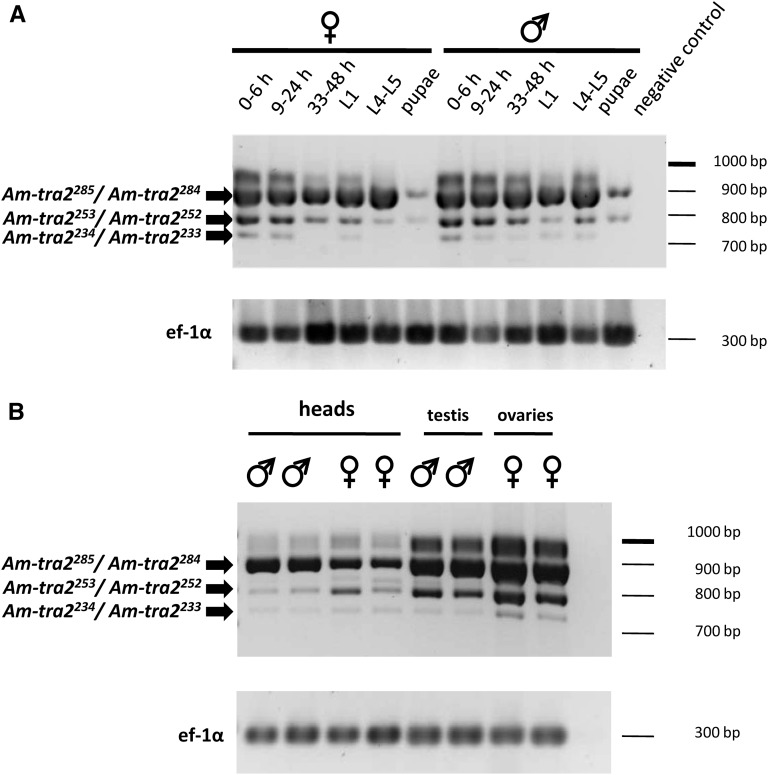
In D. melanogaster, the tra2 gene produces three transcript variants that translate into three Tra2 protein isoforms (Tra-2264, Tra-2226, and Tra-2179), which differ in the length of the first RS domain (Amrein et al. 1990; Mattox et al. 1996). Tra-2264 and Tra-2226 are expressed in somatic tissues in both sexes and are involved in dsx regulation. To identify sex-specific transcripts and the relative abundance of transcripts at different stages of development and sex determination in the honeybee, we simultaneously amplified all Am-tra2 transcript variants in 0- to 6-, 9- to 24-, and 33-to 48-hr-old embryos; L1 and L4–L5 larvae; and pupae by semiquantitative RT-PCR (Figure 3A). At the stage of 0- to 6-h-old embryos, the primary signal csd does not yet determines the sexual fate. The csd gene is not transcribed at this syncytial embryonic stage (Beye et al. 2003; Gempe et al. 2009). The transcripts are most abundant in the embryonic stages before and after the primary signal is active, but also at the larval stages and in both sexes. However, the level of expression substantially decreases at the pupal stage. The Am-tra2285 and Am-tra2284 transcripts, which are translated into the largest RS domain-containing proteins of all the transcripts, have the highest level of expression (Figure 3A). The Am-tra2253/Am-tra2252 transcripts are present in sizable amounts in early embryos, whereas the Am-tra2234/Am-tra2233 transcripts are apparently minor splice products.
Figure 3 .
Semiquantitative transcriptional profile of Am-tra2 transcripts throughout male and female development (A) and in somatic and gonadal tissue (B). Fragments of Am-tra2 transcripts spanning the entire ORF were amplified by RT-PCR, resolved by agarose gel electrophoresis, and stained with ethidium bromide. We used amplification of the cDNAs of the gene ef-1α as a relative control to semiquantify Am-tra2 transcripts across samples (Beye et al. 2003). The fragments in 0- to 6-hr-old embryos and pupae were cloned and sequenced. Ovaries were dissected from virgin queens as were testes from L4 larvae; heads are from adult bees. L1, larval stage 1; L4–L5, larval stages 4 and 5. For the embryonic stages, the hours (h) after egg deposition are indicated.
In D. melanogaster, there is a male-specific tra2 transcript, tra-2179, whose splicing is mediated by Tra-2226 proteins derived from the general tra-2226 transcript. This negative feedback loop at the level of splicing regulates proper Tra2 protein expression in the germline, which is essential for correct sperm formation (McGuffin et al. 1998). To identify a similar role of the Am-tra2 gene in sperm formation of the honeybee, we specifically searched for sex-specific Am-tra2 transcripts in the female (ovary tissue) and male (testis tissue) germlines (Figure 3B). We detected no sex-specific transcripts, suggesting that Am-tra2 mRNAs are not sexually spliced in male germ cells.
Taken together, these results indicate that Am-tra2 transcripts are not sex-specifically spliced and present before the primary decision of sexual fate is made by the csd gene. The Am-tra2285 and Am-tra2284 are the dominant transcripts, and the relative amount of Am-tra2 decreases at the pupal stage.
Knockdown of the Am-tra2 gene affects embryonic viability and female splicing of the fem and Am-dsx transcripts
In the honeybee, the primary signal csd mediates, in the heteroallelic condition, the female splicing of fem transcripts. The downstream target of csd, the fem gene, is required to direct splicing of Am-dsx pre-mRNAs and its own fem transcripts (Gempe et al. 2009), the latter of which establishes a positive feedback loop of self-regulated fem female splicing (Pane et al. 2002; Gempe et al. 2009). Although substantially diverged in sequence, the fem gene and the tra gene of D. melanogaster are orthologs, whereas the csd gene was derived by gene duplication of an ancestral copy of the fem gene (Hasselmann et al. 2008). As Csd and Fem proteins harbor no RBD but have similar sex-determining and splice regulation functions as the Tra protein in D. melanogaster (Gempe et al. 2009), we proposed that Am-Tra2 is the RNA-binding cofactor that is essential for fem and Am-dsx splicing. To study the role of the Am-tra2 gene in female splicing, we induced Am-tra2 knockdown by RNAi in 0- to 3-hr-old female embryos. We injected two dsRNAs in which the first (dsRNA-1) targets the region expressing the RS1 and the RBD domains and the second (dsRNA-2) targets the entire RS1 domain (Figure S1). Both dsRNAs overlap in a small segment with no stretches of sequence identity over the small interfering RNA length of 20–22 bp to other genes in the honeybee genome.
No female embryo reached larval stage L1 when we injected ∼200 pg of dsRNA-1 or dsRNA-2 per embryo (Table 1). This is an amount that is substantially below the ∼900 pg of dsRNA per embryo that we repeatedly used in previous studies, in which we observed no lethal effect (Beye et al. 2003; Hasselmann et al. 2008). Phenotype data from null mutants suggest that the tra2 gene in D. melanogaster is not essential for viability (Watanabe 1975; Fujihara et al. 1978). We further reduced the amount of Am-tra2-dsRNAs until we observed fully developed L1 female larvae (Table 1). At concentrations of 56 pg of dsRNA-2 and 4 pg of dsRNA-1 per embryo, we obtained the first viable L1 female larvae, but at a very low frequency (5% compared to 24% in our ddH2O-treated controls). When we further reduced the dsRNA-2 concentration from 56 to 4 pg per embryo, the hatching rate still did not substantially improve. We studied whether the lethality is sex-specific and injected 96 pg of dsRNA-2 into the male embryos for comparison (Table 2). Also, no male embryos reached larval stage L1, suggesting that knockdown of Am-tra2 caused some non-sex-specific lethality during embryogenesis. Because none of the hatched L1 female larvae reached the L4 stage (data not shown), we were not able to further study the role of Am-tra2 in morphological sexual differentiation. Taken together, our knockdown results suggest that the Am-tra2 gene is essential for embryogenesis in the honeybee.
We proposed that, in addition to a vital role for Am-tra2 in embryogenesis, Am-tra2 possibly has another function in sex determination, specifically in promoting female-specific splicing of the fem and Am-dsx transcripts. The knockdown of the fem and the csd genes, which regulate female splicing and sex determination, had no general effects on lethality, suggesting that the putative role of Am-tra2 in activation of the female pathway did not cause the embryonic lethality (Beye et al. 2003; Gempe et al. 2009). To study the sex-determining role of the Am-tra2 gene, we injected 4 or 33 pg of dsRNA-2 in embryos and after 77–80 hr studied the splice patterns, irrespective of whether the larvae hatched. If Am-tra2 promotes female splicing, we expected that knockdown of this gene would induce male-like splice patterns in these females.
The injection of 4 pg of dsRNA-2 (Table 3) induced male splicing of Am-dsx mRNAs in females (Figure 4A, lanes 1–10), which is entirely absent in the control embryos (Figure 4A, lanes 21–30), which produce only the Am-dsx female splice product. This result suggests that the Am-tra2 gene is essential to promote female splicing of Am-dsx transcripts. The female splicing of fem mRNAs is obviously not sizably affected as we observed comparable amounts of the corresponding female fem fragments in treated and nontreated female embryos in our semiquantitative PCR analysis (Figure 4B, lanes 1–10 and lanes 21–30).
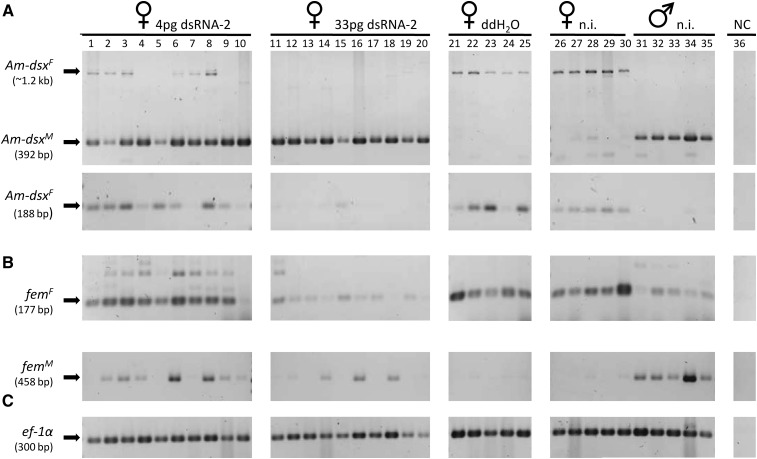
Table 3 . Production of female and male fem and Am-dsx transcripts in 72- to 80-hr-old individuals in response to embryonic Am-tra2 dsRNA-2 treatment.
| No. of individuals with transcripts |
|||||||
|---|---|---|---|---|---|---|---|
|
fem |
Am-dsx |
||||||
| Treatment | No. of embryos | Solely male | Solely female | Male and female | Solely male | Solely female | Male and female |
| Nontreated controls | |||||||
| Males | 10 | 9 | 0 | 0 | 10 | 0 | 0 |
| Females | 14 | 1 | 8 | 5 | 1 | 12 | 1 |
| Treated females | |||||||
| ddH20 | 15 | 0 | 8 | 15 | 1 | 13 | 1 |
| dsRNA-2 (33 pg) | 12 | 6 | 0 | 1 | 12 | 0 | 0 |
| dsRNA-2 (4 pg) | 11 | 0 | 4 | 7 | 1 | 1 | 9 |
Figure 4 .
Sex-specific splicing of the fem and Am-dsx transcripts in Am-tra2 dsRNA-2-treated embryos. The male and female Am-dsx (A) and fem (B) mRNAs of individuals 77–80 hr after egg laying were studied using semiquantitative RT-PCR. Early embryos were injected with 4 pg of Am-tra2 dsRNA-2 (lanes 1–10), 33 pg of Am-tra2 dsRNA-2 (lanes 11–20), or ddH20 (lanes 21–25). The untreated (labeled as “n.i.”) female and male controls are shown in lanes 26–30 and 31–35, respectively. “NC” denotes our control PCR in which no cDNA was added (lane 36). Fragments corresponding to the fem female (size of 177 bp) and male (size of 458 bp) mRNAs and the Am-dsx female (size of 1.2 kb and 188 bp) and male (size of 392 bp) mRNAs were resolved by agarose gel electrophoresis and stained with ethidium bromide. We used amplification of the cDNAs of the gene ef-1α (C) as a relative control to semiquantify Am-tra2 transcripts across embryonic samples.
The treatment of female embryos with the higher concentration of 33 pg of dsRNA-2 per embryo compromised female splicing of the fem mRNAs (Figure 4B, lanes 11–20) compared to our control embryos (Figure 4B, lanes 26–35; Table 3), indicating that Am-Tra2 protein is also required for fem female transcript splicing. To provide further evidence that the effect on splicing is specific for dsx and fem transcripts, we also assayed the splice products of our control gene ef-1α (Figure 4C) and that of the csd gene (Figure S3) in the dsRNA-treated and nontreated embryos. We detected in embryos that were treated with 4 or 33 pg Am-tra2 dsRNA-2 the csd and the ef-1α exon spanning fragments of cDNA, suggesting that our treatment or the knockdown of Am-tra2 has not generally affected the splice process or degradation of embryonic mRNAs. This latter result supports our notion that the Am-Tra2 protein is specifically involved in the sex-specific splice regulation of Am-dsx and fem transcripts.
In contrary to our expectation, the knockdown of the Am-tra2 gene in females did not produce the alternative, male splice form of the fem transcripts. We consistently observed in the 33-pg-treated females the absence of the male fem transcript (Figure 4B, lanes 11–20). We thus studied in male (haploid) embryos that only produce the male splice variant the influence of the Am-tra2 gene on male fem splicing. The injection of 33 and 67 pg dsRNA-2 repeatedly produced males that lacked the male fem transcript in 48-hr-old embryos (Figure S4, lanes 1–19) whereas the transcript of our control gene ef-1α was present. This result suggests a role of Am-Tra2 protein in splicing the fem pre-mRNAs into the male form.
Taken together, these results suggest that Am-Tra2 promotes female splicing of the productive female fem mRNAs and also of the nonproductive male fem mRNAs.
Discussion
Heteroallelic Csd proteins determine honeybee femaleness and set the downstream regulator of the sex determination cascade, fem, into the female mode by alternative splicing (Beye et al. 2003; Hasselmann et al. 2008). The Fem proteins in females maintain the female-determined state by promoting female splicing of the fem mRNAs (positive autoregulation) and direct female splicing of the Am-dsx transcripts (Hasselmann et al. 2008; Gempe et al. 2009). In this study, we showed that the Am-tra2 gene, an ortholog of the tra2 gene of D. melanogaster, is a component of the honeybee sex determination hierarchy. The Am-tra2 proteins are required to regulate female and male splicing of fem mRNAs and female splicing of the Am-dsx mRNAs. In addition, we showed that the Am-tra2 gene has an essential role in embryogenesis that is not related to sex determination.
We characterized the Am-tra2 gene in the honeybee and showed that the deduced Am-Tra2 proteins share the same domain structure as other Tra2 orthologs described thus far (Amrein et al. 1988; Bandziulis et al. 1989; Goralski et al. 1989; Mattox et al. 1996; Burghardt et al. 2005; Salvemini et al. 2009; Sarno et al. 2010). Am-Tra2 protein contains a RBD that is supposed to directly interact with the pre-mRNA and two flanking RS-rich domains that provide a potential surface for an interaction with other proteins, such as Tra proteins (Amrein et al. 1988; Hoshijima et al. 1991; Graveley 2000; Sciabica and Hertel 2006). We identified six splice variants of Am-tra2 mRNAs that translate into proteins that differ in the length of the first RS domain and in the absence/presence of one amino acid (serine) in the second RS domain. The six splice variants are not sex-specifically regulated throughout development, suggesting that Am-Tra2 proteins are constitutively expressed. The tra2 transcripts are present in early embryos before the primary signal csd is transcribed and thus before the primary decision of sexual fate is made, which is after blastoderm formation (Beye et al. 2003; Gempe et al. 2009). However, we observed that the level of Am-tra2 transcription substantially decreases at the pupal stage, possibly at a stage when sexual signals of the primary sex determination cascade are less important. We also showed that the Am-tra2 gene is not sex-specifically spliced in the gonadal tissues (Figure 3B). This finding is in contrast to the germline-specific control of tra2 transcripts in males of D. melanogaster. Here, the Tra-2226 protein directs splicing of the tra-2179 transcript in the fruit fly germline, thereby regulating the level of Tra-2226 protein expression that is critical for proper sperm formation (McGuffin et al. 1998; Mattox et al. 1990).
When we repressed the Am-tra2 gene by injecting 4-pg dsRNAs into early embryos, we observed that female splicing of Am-dsx switched into male splicing. This Am-tra2 knockdown had no sizable effect on female splicing of fem transcripts, suggesting that the Am-dsx switch of splicing was not caused by affecting splice regulation of the upstream regulator fem. We also showed that the knockdown did not affect the splicing of control genes, the csd, and the ef-1α gene, suggesting that embryonic lethality has not compromised our testing. Taken together, these results suggest that the Am-tra2 gene plays a role in the regulation of female Am-dsx mRNA splicing (Figure 5). Our result suggests a conserved role of the Am-Tra2 protein in Am-dsx regulation, although the canonical Tra/Tra2-binding motifs that have been reported in different dipteran insects are absent in the Am-dsx gene. This finding suggests that the Tra2 protein-binding sites have evolved. In D. melanogaster, Tra2, together with the Tra proteins, binds to six repeats of a 13-nucleotide motif [TC(T/A)(A/T)C(A/G)ATCAACA] on the dsx pre-mRNA and promotes the activation of the weak female splice acceptor that directs the production of the female dsx transcripts. In other dipteran species (M. domestica, C. capitata, Bactrocera oleae, Bactrocera dorsalis, Bactrocera correcta, Bactrocera tyroni, and different Anastrepha species), the canonical Tra/Tra2-binding motifs are consistently present in the dsx genes (Crampton et al. 1998; Hediger et al. 2004; Lagos et al. 2005; Ruiz et al. 2005, 2007; Saccone et al. 2008; Concha et al. 2010; Permpoon et al. 2011) and are proposed to be utilized in promoting female splicing (Burghardt et al. 2005; Salvemini et al. 2009; Sarno et al. 2010). We propose that the Am-Tra2 protein, like its ortholog in D. melanogaster, is an essential, constitutively expressed cofactor that, together with the female-specific Fem protein, directs the female processing of the Am-dsx transcript. The honeybees are a member of the hymenopteran insects and are at the base of the phylogeny of holometabolous insects (Savard et al. 2006). The shared function in dsx regulation across the different insect orders thus suggests that the role of Tra2 proteins in regulating female dsx splicing is the ancestral state in holometabolous insects.
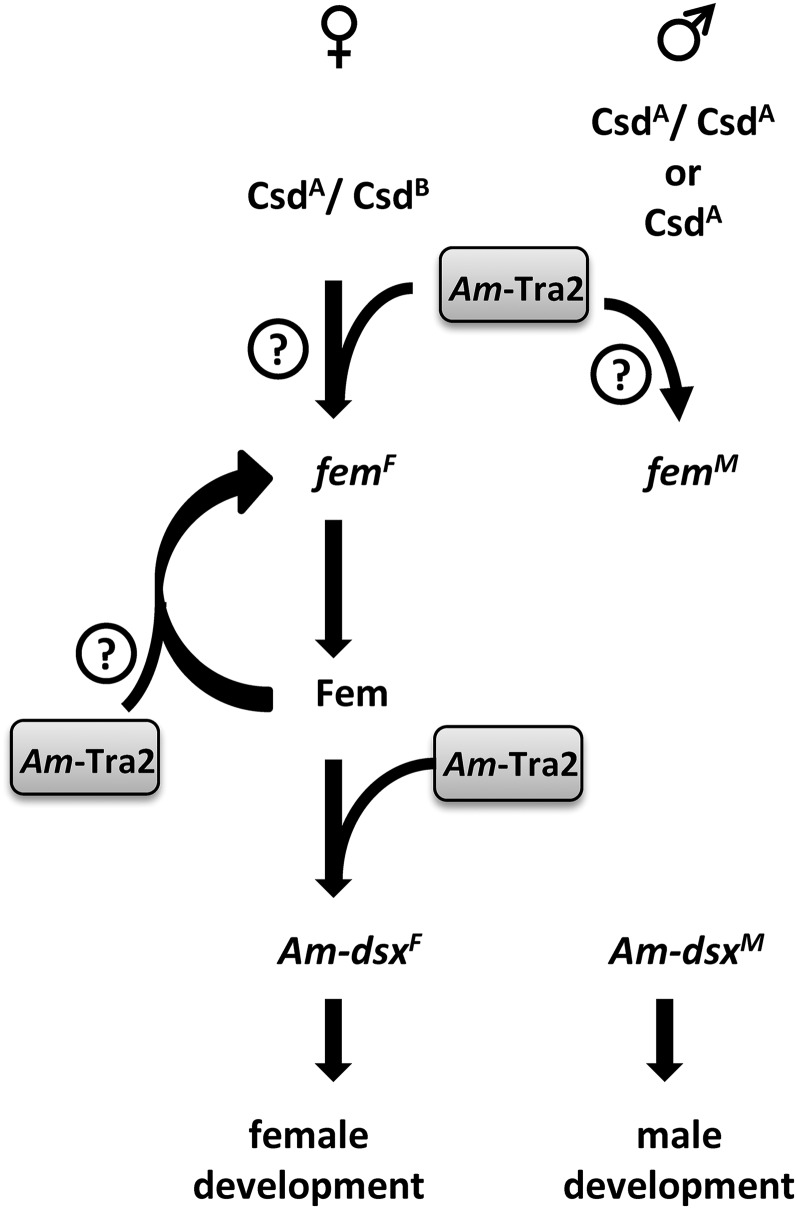
Figure 5 .
Model of the role of the Am-Tra2 protein in honeybee sex determination. Am-Tra2 protein is necessary for splicing of the productive female and fem and Am-dsx mRNAs. It is also required to splice the nonproductive male fem splice form. In females, the Am-Tra2 proteins either act together with heterozygous Csd proteins (CsdA/CsdB) and/or together with Fem proteins (femF-positive autoregulatory loop) to promote female processing of the fem pre-mRNAs (femF). Am-Tra2, together with with Fem proteins, directs female-specific splicing of the Am-dsx pre-mRNA (Am-dsxF). In males, Am-Tra2 protein directs splicing of male fem mRNAs in the presence of inactive Csd proteins (that are homoallelic CsdA/CsdA or CsdA proteins) and Fem proteins.
Consistent with an evolved binding site of the Fem/Am-Tra2 proteins, we identified several amino acid replacements in the RBD that affect the designated binding nucleotide sequence of the RNA. We found three amino acid sites in the RNP-1 sequence element that diverged with respect to the D. melanogaster sequence. The RNP-1 and RNP-2 sequence elements are part of the RBD, which has a βαββαβ barrel-like structure. The RNP sequence elements have exposed positions at the surface of the β-sheets β1 and β3 and are used to bind directly to the ribonucleotide sequence (Dreyfuss et al. 1988; Merrill et al. 1988; Nagai et al. 1990; Amrein et al. 1994). Mutation of the first amino acid arginine of the RNP-1 sequence element in D. melanogaster abolishes the female processing of dsx pre-mRNAs (Amrein et al. 1994). This critical arginine amino acid residue in the RNP-1 sequence element is replaced in the honeybee by a lysine. These findings support our conclusion that the corresponding Fem/Am-Tra2 protein-binding sites diverge from that of the fruit fly.
When we repressed the Am-tra2 gene by injecting higher amounts of dsRNAs (33 pg) into female embryos, we also observed a reduction of the productive female and the nonproductive male fem splice variants. This amount of dsRNA did not affect splicing of the paralogous gene csd and the ef-1α gene, suggesting that embryonic lethality has not compromised our testing of splicing. We also confirmed that the Am-tra2 gene is essential for the male splicing of fem mRNAs by knockdown of the Am-tra2 gene in males. These results together indicate a function of the Am-tra2 gene also on the level of splicing of the fem gene. We propose that the Am-Tra2 protein is a required cofactor of heteroallelic Csd proteins in females that mediates the binding and splicing of female fem pre-mRNAs (Figure 5). Am-Tra2 proteins may together with Fem proteins direct female splicing of the fem transcripts, which are maintaining the female determined state through development by a positive feedback loop (Figure 5). In males, where the active, heteroallelic Csd proteins and Fem proteins are absent, the Am-Tra2 proteins may enhance the switch of the fem transcripts into the nonproductive male form. In the absence of the Am-tra2-dependent splicing, the fem RNAs may undergo RNA decay machinery and are removed. We speculate that the dual function of the Am-tra2 gene as a regulator in male and female splicing may enhance the binary decision when the female-specific proteins (Fem, heteroallelic Csd) are present or absent. The proper male and female regulation of fem splicing is important to implement and maintain the sexual fate as the analysis of gynandromorphs indicates that they were induced by knockdowns of the fem gene (Gempe et al. 2009).
The fem gene is an ortholog of the tra gene of dipteran insects. In D. melanogaster, Tra2 proteins are not deployed in regulating female splicing of tra transcripts. Female tra mRNA processing in this species is regulated by the Sxl proteins (Bell et al. 1988; Sosnowski et al. 1989; Bell et al. 1991; Valcárcel et al. 1993). In contrast, Tra2 proteins in the dipteran insects M. domestica and C. capitata are, as in the honeybee, required to splice tra transcripts into the female form (Burghardt et al. 2005; Salvemini et al. 2009; Hediger et al. 2010). In these dipteran species, Tra2 proteins are presumably regulators of an autoregulatory loop in females in which the maternally provided Tra proteins mediate female tra mRNAs. The presence of a male-determining factor, M, apparently impairs this tra-positive regulatory loop, resulting in male tra pre-mRNA splicing and male differentiation (Pane et al. 2002; Hediger et al. 2010). The role of Am-tra2 gene in controlling the male splicing of the fem gene has not been reported in other insects so far.
Our knockdown studies in early embryos also suggest that the Am-tra2 gene is essential and has a vital role in embryogenesis. We suggest that this role is independent of the sex determination process as no sex-specific lethality has been observed and the other components (fem and csd) regulating sex determination produce no lethal phenotype (Beye et al. 2003; Hasselmann et al. 2008; Gempe et al. 2009). We suggest that this lethal effect during embryogenesis is not caused by unspecific effects due to our dsRNA method as (i) different regions of the transcript with our dsRNAs were targeted, (ii) lethal effects with dsRNA concentrations were observed that were substantially below that of previous experiments (4–40 times) that showed no lethal embryonic effects (Beye et al. 2003; Hasselmann et al. 2008), and (iii) the viability of embryos did not further increase above ∼5% when we further decreased the dsRNA concentration by 1/10th (Tables 1 and 2). This additional role in embryogenesis is absent in other dipteran insects (Watanabe 1975; Fujihara et al. 1978; Burghardt et al. 2005; Salvemini et al. 2009), suggesting that this role evolved in insects.
Taken together, our results suggest that the Am-tra2 gene is a non-sex-specifically expressed regulator that is essential for generating the productive female and nonproductive male fem transcripts. We propose that the Am-Tra2 protein acts together with heteroallelic Csd proteins and/or Fem proteins to mediate female fem splicing by binding to fem pre-mRNAs in accordance with the function of its ortholog in D. melanogaster. This predicted role, however, needs to be further tested in splice assays and protein-binding studies. The Am-tra2 gene thus may have a central role in initiating the primary signal csd in females and in maintaining the female determined state by the positive regulatory loop at the level of the fem gene (Figure 5). In males with hemi- or homoallelic Csd proteins, Am-Tra2 may enhance the switch of fem transcripts into the nonproductive male form. The use of the Am-Tra2 proteins in male and female splicing may enhance and stabilize the male and female splicing state at the level of the fem gene. We also provided evidence that the Am-Tra2 protein is an essential regulator of female Am-dsx splice regulation, a feature that is shared with other dipteran insects, suggesting an ancestral role in dsx splice regulation in holometabolous insects. In addition, the Am-tra2 gene has an essential function in honeybee embryogenesis that is unrelated to sex determination and has thus far not been reported in other insects.
Supplementary Material
Acknowledgments
We thank two anonymous reviewers for helpful comments on the manuscript; Eva-Maria Theilenberg, Marion Mueller-Borg, Isabel Röös, and Jessica Langer for technical support; Dalibor Titera for providing bee crosses; and Michael Griese for beekeeping support. This work was supported by grants from the Deutsche Forschungsgemeinschaft.
Footnotes
Communicating editor: T. Schupbach
Literature Cited
- Amrein H., Gorman M., Nöthiger R., 1988. The sex-determining gene tra-2 of Drosophila encodes a putative RNA binding protein. Cell 55: 1025–1035. [DOI] [PubMed] [Google Scholar]
- Amrein H., Maniatis T., Nöthiger R., 1990. Alternatively spliced transcripts of the sex-determining gene tra-2 of Drosophila encode functional proteins of different size. EMBO J. 9: 3619–3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amrein H., Hedley M. L., Maniatis T., 1994. The role of specific protein-RNA and protein-protein interactions in positive and negative control of pre-mRNA splicing by transformer-2. Cell 76: 735–746. [DOI] [PubMed] [Google Scholar]
- Bandziulis R. J., Swanson M. S., Dreyfuss G., 1989. RNA-binding proteins as developmental regulators. Genes Dev. 3: 431–437. [DOI] [PubMed] [Google Scholar]
- Bell L. R., Maine E. M., Schedl P., Cline T. W., 1988. Sex-lethal, a Drosophila sex determination switch gene, exhibits sex-specific RNA splicing and sequence similarity to RNA binding proteins. Cell 55: 1037–1046. [DOI] [PubMed] [Google Scholar]
- Bell L. R., Horabin J. I., Schedl P., Cline T. W., 1991. Positive autoregulation of sex-lethal by alternative splicing maintains the female determined state in Drosophila. Cell 65: 229–239. [DOI] [PubMed] [Google Scholar]
- Beye M., 2004. The dice of fate: the csd gene and how its allelic composition regulates sexual development in the honey bee, Apis mellifera. Bioessays 26: 1131–1139. [DOI] [PubMed]
- Beye M., Härtel S., Hagen A., Hasselmann M., Omholt S. W., 2002. Specific developmental gene silencing in the honey bee using a homeobox motif. Insect Mol. Biol. 11: 527–532. [DOI] [PubMed] [Google Scholar]
- Beye M., Hasselmann M., Fondrk M. K., Page R. E., Omholt S. W., 2003. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell 114: 419–429. [DOI] [PubMed] [Google Scholar]
- Burghardt G., Hediger M., Siegenthaler C., Moser M., Dübendorfer A., et al. , 2005. The transformer2 gene in Musca domestica is required for selecting and maintaining the female pathway of development. Dev. Genes Evol. 215: 165–176. [DOI] [PubMed] [Google Scholar]
- Burtis K. C., Baker B. S., 1989. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell 56: 997–1010. [DOI] [PubMed] [Google Scholar]
- Cho S., Huang Z. Y., Zhang J., 2007. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics 177: 1733–1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline T. W., Meyer B. J., 1996. Vive la différence: males vs. females in flies vs. worms. Annu. Rev. Genet. 30: 637–702. [DOI] [PubMed] [Google Scholar]
- Concha C., Scott M. J., 2009. Sexual development in Lucilia cuprina (Diptera, Calliphoridae) is controlled by the transformer gene. Genetics 182: 785–798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Concha C., Li F., Scott M. J., 2010. Conservation and sex-specific splicing of the doublesex gene in the economically important pest species Lucilia cuprina. J. Genet. 89: 279–285. [DOI] [PubMed] [Google Scholar]
- Crampton J. M., James A. A., Shearman D. C. A., Frommer M., 1998. The Bactrocera tyroni homologue of the Drosophila melanogaster sex-determination gene doublesex. Insect Mol. Biol. 7: 355–366. [DOI] [PubMed] [Google Scholar]
- Dearden P. K., Wilson M. J., Sablan L., Osborne P. W., Havler M., et al. , 2006. Patterns of conservation and change in honey bee developmental genes. Genome Res. 16: 1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfuss G., Swanson M. S., Pinol-Roma S., 1988. Heterogeneous nuclear ribonucleoprotein particles and the pathway of mRNA formation. Trends Biochem. Sci. 13: 86–91. [DOI] [PubMed] [Google Scholar]
- Erickson J. W., Quintero J. J., 2007. Indirect effects of ploidy suggest X chromosome dose, not the X:A ratio, signals sex in Drosophila. PLoS Biol. 5: e332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara T., Kawabe M., Oishi K., 1978. A sex-transformation gene in Drosophila. J. Hered. 69: 229–236. [DOI] [PubMed] [Google Scholar]
- Gempe T., Beye M., 2011. Function and evolution of sex determination mechanisms, genes and pathways in insects. Bioessays 33: 52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gempe T., Hasselmann M., Schiøtt M., Hause G., Otte M., et al. , 2009. Sex determination in honeybees: two separate mechanisms induce and maintain the female pathway. PLoS Biol. 7: e1000222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goralski T. J., Edstrom J.-E., Baker B. S., 1989. The sex determination locus transformer-2 of Drosophila encodes a polypeptide with similarity to RNA binding proteins. Cell 56: 1011–1018. [DOI] [PubMed] [Google Scholar]
- Graveley B. R., 2000. Sorting out the complexity of SR protein functions. RNA 6: 1197–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmann M., Beye M., 2004. Signatures of selection among sex-determining alleles of the honey bee. Proc. Natl. Acad. Sci. USA 101: 4888–4893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasselmann M., Gempe T., Schiøtt M., Nunes-Silva C. G., Otte M., et al. , 2008. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature 454: 519–522. [DOI] [PubMed] [Google Scholar]
- Hazelrigg T., Tu C., 1994. Sex-specific processing of the Drosophila exuperantia transcript is regulated in male germ cells by the tra-2 gene. Proc. Natl. Acad. Sci. USA 91: 10752–10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hediger M., Burghardt G., Siegenthaler C., Buser N., Hilfiker-Kleiner D., et al. , 2004. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev. Genes Evol. 214: 29–42. [DOI] [PubMed] [Google Scholar]
- Hediger M., Henggeler C., Meier N., Perez R., Saccone G., et al. , 2010. Molecular characterization of the key switch F provides a basis for understanding the rapid divergence of the sex-determining pathway in the housefly. Genetics 184: 155–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedley M. L., Maniatis T., 1991. Sex-specific splicing and polyadenylation of dsx pre-mRNA requires a sequence that binds specifically to tra-2 protein in vitro. Cell 65: 579–586. [DOI] [PubMed] [Google Scholar]
- Hoshijima K., Inoue K., Higuchi I., Sakamoto H., Shimura Y., 1991. Control of Doublesex alternative splicing by transformer and transformer-2 in Drosophila. Science 252: 833–836. [DOI] [PubMed] [Google Scholar]
- Inoue K., Hoshijima K., Higuchi I., Sakamoto H., Shimura Y., 1992. Binding of the Drosophila transformer and transformer-2 proteins to the regulatory elements of doublesex primary transcript for sex-specific RNA processing. Proc. Natl. Acad. Sci. USA 89: 8092–8096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamakura M., 2011. Royalactin induces queen differentiation in honeybees. Nature 473: 478–483. [DOI] [PubMed] [Google Scholar]
- Kucharski R., Maleszka J., Foret S., Maleszka R., 2008. Nutritional control of reproductive status in honeybees via DNA methylation. Science 319: 1827–1830. [DOI] [PubMed] [Google Scholar]
- Lagos D., Ruiz M. F., Sánchez L., Komitopoulou K., 2005. Isolation and characterization of the Bactrocera oleae genes orthologous to the sex determining Sex-lethal and doublesex genes of Drosophila melanogaster. Gene 348: 111–121. [DOI] [PubMed] [Google Scholar]
- Lynch K. W., Maniatis T., 1995. Synergistic interactions between two distinct elements of a regulated splicing enhancer. Genes Dev. 9: 284–293. [DOI] [PubMed] [Google Scholar]
- Lynch K. W., Maniatis T., 1996. Assembly of specific SR protein complexes on distinct regulatory elements of the Drosophila doublesex splicing enhancer. Genes Dev. 10: 2089–2101. [DOI] [PubMed] [Google Scholar]
- Madigan S. J., Edeen P., Esnayra J., McKeown M., 1996. att, a target for regulation by tra2 in the testes of Drosophila melanogaster, encodes alternative RNAs and alternative proteins. Mol. Cell. Biol. 16: 4222–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín I., Ruiz M. F., Sánchez L., 2011. The gene transformer-2 of Sciara (Diptera, Nematocera) and its effect on Drosophila sexual development. BMC Dev. Biol. 11: 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattox W., Baker B. S., 1991. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev. 5: 786–796. [DOI] [PubMed] [Google Scholar]
- Mattox W., Palmer M. J., Baker B. S., 1990. Alternative splicing of the sex determination gene transformer-2 is sex-specific in the germ line but not in the soma. Genes Dev. 4: 789–805. [DOI] [PubMed] [Google Scholar]
- Mattox W., McGuffin M. E., Baker B. S., 1996. A negative feedback mechanism revealed by functional analysis of the alternative isoforms of the Drosophila splicing regulator transformer-2. Genetics 143: 303–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGuffin M. E., Chandler D., Somaiya D., Dauwalder B., Mattox W., 1998. Autoregulation of transformer-2 alternative splicing is necessary for normal male fertility in Drosophila. Genetics 149: 1477–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrill B. M., Stone K. L., Cobianchi F., Wilson S. H., Williams K. R., 1988. Phenylalanines that are conserved among several RNA-binding proteins form part of a nucleic acid-binding pocket in the A1 heterogeneous nuclear ribonucleoprotein. J. Biol. Chem. 263: 3307–3313. [PubMed] [Google Scholar]
- Nagai K., Oubridge C., Jessen T. H., Li J., Evans P. R., 1990. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature 348: 515–520. [DOI] [PubMed] [Google Scholar]
- Niu B.-L., Meng Z.-Q., Tao Y.-Z., Lu S.-L., Weng H.-B., et al. , 2005. Cloning and alternative splicing analysis of Bombyx mori Transformer-2 gene using silkworm EST database. Acta Biochim. Biophys. Sin. (Shanghai) 37: 728–736. [DOI] [PubMed] [Google Scholar]
- Nygaard S., Zhang G., Schiøtt M., Li C., Wurm Y., et al. , 2011. The genome of the leaf-cutting ant Acromyrmex echinatior suggests key adaptations to advanced social life and fungus farming. Genome Res. 21: 1339–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pane A., Salvemini M., Delli Bovi P., Polito C., Saccone G., 2002. The transformer gene in Ceratitis capitata provides a genetic basis for selecting and remembering the sexual fate. Development 129: 3715–3725. [DOI] [PubMed] [Google Scholar]
- Permpoon R., Aketarawong N., Thanaphum S., 2011. Isolation and characterization of Doublesex homologues in the Bactrocera species: B. dorsalis (Hendel) and B. correcta (Bezzi) and their putative promoter regulatory regions. Genetica 139: 113–127. [DOI] [PubMed] [Google Scholar]
- Ruiz M. F., Stefani R. N., Mascarenhas R. O., Perondini A. L. P., Selivon D., et al. , 2005. The gene doublesex of the fruit fly Anastrepha obliqua (Diptera, Tephritidae). Genetics 171: 849–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruiz M. F., Eirín-López J. M., Stefani R. N., Perondini A. L. P., Selivon D., et al. , 2007. The gene doublesex of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insects. Dev. Genes Evol. 217: 725–731. [DOI] [PubMed] [Google Scholar]
- Saccone G., Salvemini M., Pane A., Polito L. C., 2008. Masculinization of XX Drosophila transgenic flies expressing the Ceratitis capitata DoublesexM isoform. Int. J. Dev. Biol. 52: 1051–1057. [DOI] [PubMed] [Google Scholar]
- Salvemini M., Robertson M., Aronson B., Atkinson P., Polito L. C., et al. , 2009. Ceratitis capitata transformer-2 gene is required to establish and maintain the autoregulation of Cctra, the master gene for female sex determination. Int. J. Dev. Biol. 53: 109–120. [DOI] [PubMed] [Google Scholar]
- Sarno F., Ruiz M. F., Eirín-López J. M., Perondini A. L. P., Selivon D., et al. , 2010. The gene transformer-2 of Anastrepha fruit flies (Diptera, Tephritidae) and its evolution in insects. BMC Evol. Biol. 10: 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard J., Tautz D., Richards S., Weinstock G. M., Gibbs R. A., et al. , 2006. Phylogenomic analysis reveals bees and wasps (Hymenoptera) at the base of the radiation of Holometabolous insects. Genome Res. 16: 1334–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schetelig M. F., Milano A., Saccone G., Handler A. M., 2012. Male only progeny in Anastrepha suspensa by RNAi-induced sex reversion of chromosomal females. Insect Biochem. Mol. Biol. 42: 51–57. [DOI] [PubMed] [Google Scholar]
- Sciabica K. S., Hertel K. J., 2006. The splicing regulators Tra and Tra2 are unusually potent activators of pre-mRNA splicing. Nucleic Acids Res. 34: 6612–6620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnowski B. A., Belote J. M., McKeown M., 1989. Sex-specific alternative splicing of RNA from the transformer gene results from sequence-dependent splice site blockage. Cell 58: 449–459. [DOI] [PubMed] [Google Scholar]
- Suzuki M. G., Ohbayashi F., Mita K., Shimada T., 2001. The mechanism of sex-specific splicing at the doublesex gene is different between Drosophila melanogaster and Bombyx mori. Insect Biochem. Mol. Biol. 31: 1201–1211. [DOI] [PubMed] [Google Scholar]
- Suzuki M. G., Imanishi S., Dohmae N., Nishimura T., Shimada T., et al. , 2008. Establishment of a novel in vivo sex-specific splicing assay system to identify a trans-acting factor that negatively regulates splicing of Bombyx mori dsx female exons. Mol. Cell. Biol. 28: 333–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M. G., Imanishi S., Dohmae N., Asanuma M., Matsumoto S., 2010. Identification of a male-specific RNA binding protein that regulates sex-specific splicing of Bmdsx by increasing RNA binding activity of BmPSI. Mol. Cell. Biol. 30: 5776–5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium, 2008. The genome of the model beetle and pest Tribolium castaneum. Nature 452: 949–955. [DOI] [PubMed] [Google Scholar]
- Valcárcel J., Singh R., Zamore P. D., Green M. R., 1993. The protein Sex-lethal antagonizes the splicing factor U2AF to regulate alternative splicing of transformer pre-mRNA. Nature 362: 171–175. [DOI] [PubMed] [Google Scholar]
- Watanabe T. K., 1975. A new sex-transforming gene on the second chromosome of Drosophila melanogaster. Jpn. J. Genet. 50(3): 269–271. [Google Scholar]
- Woyke J., 1963. Drone larvae from fertilized eggs of the honeybee. J. Apic. Res. 2: 19–24. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.