Abstract
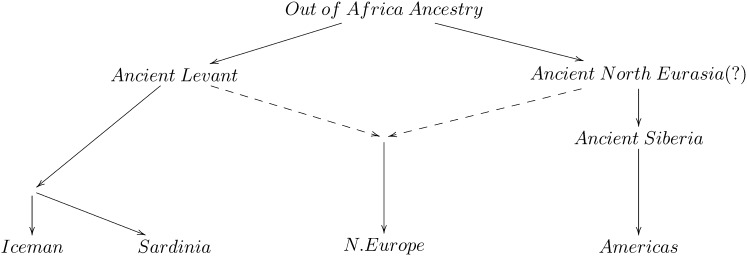
Population mixture is an important process in biology. We present a suite of methods for learning about population mixtures, implemented in a software package called ADMIXTOOLS, that support formal tests for whether mixture occurred and make it possible to infer proportions and dates of mixture. We also describe the development of a new single nucleotide polymorphism (SNP) array consisting of 629,433 sites with clearly documented ascertainment that was specifically designed for population genetic analyses and that we genotyped in 934 individuals from 53 diverse populations. To illustrate the methods, we give a number of examples that provide new insights about the history of human admixture. The most striking finding is a clear signal of admixture into northern Europe, with one ancestral population related to present-day Basques and Sardinians and the other related to present-day populations of northeast Asia and the Americas. This likely reflects a history of admixture between Neolithic migrants and the indigenous Mesolithic population of Europe, consistent with recent analyses of ancient bones from Sweden and the sequencing of the genome of the Tyrolean “Iceman.”
Keywords: population genetics, admixture, single nucleotide polymorphism (SNP) array
ADMIXTURE between populations is a fundamental process that shapes genetic variation and disease risk. For example, African Americans and Latinos derive their genomes from mixtures of individuals who trace their ancestry to divergent populations. Study of the ancestral origin of the admixed individuals provides an opportunity to infer the history of the ancestral groups, some of whom may no longer be extant. The two main classes of methods in this field are local ancestry-based methods and global ancestry-based methods. Local ancestry-based methods such as LAMP (Sankararaman et al. 2008), HAPMIX (Price et al. 2009), and PCADMIX (Brisbin 2010) deconvolve ancestry at each locus in the genome and provide individual-level information about ancestry. While these methods provide valuable insights into the recent history of populations, they have reduced power to detect older events. The most commonly used methods for studying global ancestry are principal component analysis (PCA) (Patterson et al. 2006) and model-based clustering methods such as STRUCTURE (Pritchard et al. 2000) and ADMIXTURE (Alexander et al. 2009). While these are powerful tools for detecting population substructure, they do not provide any formal tests for admixture (the patterns in data detected using these methods can be generated by multiple population histories). For instance, Novembre et al. (2008) showed that isolation-by-distance can generate PCA gradients that are similar to those that arise from long-distance historical migrations, making PCA results difficult to interpret from a historical perspective. STRUCTURE/ADMIXTURE results are also difficult to interpret historically, because these methods work either without explicitly fitting a historical model or by fitting a model that assumes that all the populations have radiated from a single ancestral group, which is unrealistic.
An alternative approach is to make explicit inferences about history by fitting phylogenetic tree-based models to genetic data. A limitation of this approach, however, is that many of these methods do not allow for the possibility of migrations between groups, whereas most human populations derive ancestry from multiple ancestral groups. Indeed there is only a handful of examples of human groups in which there is no evidence of genetic admixture today. In this article, we describe a suite of methods that formally test for a history of population mixture and allow researchers to build models of population relationships (including admixture) that fit genetic data. These methods are inspired by the ideas by Cavalli-Sforza and Edwards (1967), who fit phylogenetic trees of population relationships to the Fst values measuring allele frequency differentiation between pairs of populations. Later studies by Thompson (1975); Lathrop (1982); Waddell and Penny (1996); and Beerli and Felsenstein (2001) are more similar in spirit to our methods, in that they describe frameworks for fitting population mixture events (not just simple phylogenetic trees) to the allele frequencies observed in multiple populations, although the technical details are quite different from our work. In what follows we describe five methods: the three-population test, D-statistics, F4-ratio estimation, admixture graph fitting, and rolloff. These have been introduced in some form in earlier articles (Reich et al. 2009; Green et al. 2010; Durand et al. 2011; Moorjani et al. 2011) but not coherently together and with the key material placed in supplementary sections, making it difficult for readers to understand the methods and their scope. We also release a software package, ADMIXTOOLS, that implements these five methods for users interested in applying them to studies of population history.
The first four techniques are based on studying patterns of allele frequency correlations across populations. The three-population test is a formal test of admixture and can provide clear evidence of admixture, even if the gene flow events occurred hundreds of generations ago. The four-population test implemented here as D-statistics is also a formal test for admixture, which not only can provide evidence for admixture but also can provide some information about the directionality of the gene flow. F4-ratio estimation allows inference of the mixing proportions of an admixture event, even without access to accurate surrogates for the ancestral populations. However, this method demands more assumptions about the historical phylogeny. Admixture graph fitting allows one to build a model of population relationships for an arbitrarily large number of populations simultaneously and to assess whether it fits the allele frequency correlation patterns among populations. Admixture graph fitting has some similarities to the TreeMix method of Pickrell and Pritchard (2012) but differs in that TreeMix allows users to automatically explore the space of possible models and to find the one that best fits the data (our method does not), while our method provides a rigorous test for whether a proposed model fits the data (TreeMix does not).
It is important to point out that all four of the methods described in the previous paragraph measure allele frequency correlations among populations using the f-statistics and D-statistics that we define precisely in what follows. The expected values of these statistics are functions not just of the demographic history relating the populations, but also of the way that the analyzed polymorphisms were discovered (the so-called ascertainment process). In principle, explicit inferences about the demographic history of populations can be made using the magnitudes of allele frequency correlation statistics, an idea that is exploited to great advantage by Durand et al. (2011); however, for this approach to work, it is essential to analyze sites with rigorously documented ascertainment, as are available, for example, from whole-genome sequencing data. Here our approach is fundamentally different in that we are focusing on tests for a history of admixture that assess whether particular statistics are consistent with 0. The expectation of zero in the absence of admixture is robust to all but the most extreme ascertainment processes, and thus these methods provide valid tests for admixture even using data from SNP arrays with complex ascertainment. We show this robustness both by simulation and with application to real data. In some simple scenarios, we also demonstrate this robustness theoretically. Furthermore, we show that ratios of f-statistics can provide precise estimates of admixture proportions that are robust to both details of the ascertainment and to population size changes over the course of history, even if the f-statistics in the numerator and denominator themselves have magnitudes that are affected by ascertainment.
The fifth method that we introduce in this study, rolloff, is an approach for estimating the date of admixture which models the decay of admixture linkage disequilibrium in the target population. Rolloff uses different statistics from those used by haplotype-based methods such as STRUCTURE (Pritchard et al. 2000) and HAPMIX (Price et al. 2009). The most relevant comparison is to the method of Pool and Nielsen (2009), who like us are specifically interested in learning about history, and who estimate population mixture dates by studying the distribution of ancestry tracts inherited from the two ancestral populations. A limitation of the Pool and Nielsen (2009) approach, however, is that it assumes that local ancestry inference is perfect, whereas in fact most local ancestry methods are unable to accurately infer the short ancestry tracts that are typical for older dates of mixture. Precisely for these reasons, the HAPMIX article cautions against using HAPMIX for date estimation (Price et al. 2009). In contrast, rolloff does not require accurate reconstruction of the breakpoints across the chromosomes or data from good surrogates for the ancestors, making it possible to interrogate older dates. Simulations that we report in what follows show that rolloff can produce unbiased and quite accurate estimates for dates up to 500 generations in the past.
Materials and Methods
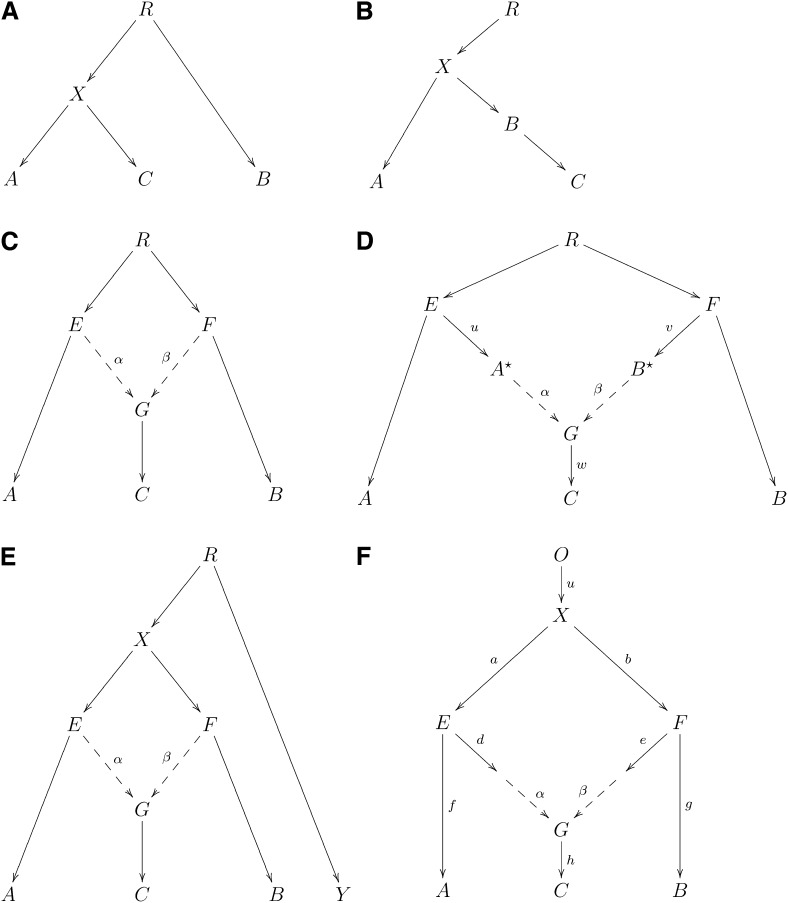
Throughout this article, unless otherwise stated, we consider biallelic markers only, and we ignore the possibility of recurrent or back mutations. Our notation in this article is that we write f2 (and later f3, f4) for statistics: empirical quantities that we can compute from data, and F2 (and later F3, F4) for corresponding theoretical quantities that depend on an assumed phylogeny (and the ascertainment). We define “drift” as the frequency change of an allele along a graph edge (hence drift between two populations A and B is a function of the difference in the allele frequency of polymorphisms in A and B).
The three-population test and introduction of f-statistics
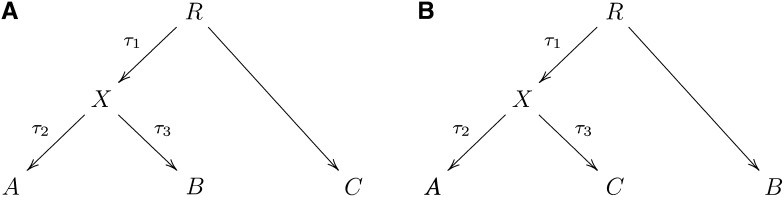
We begin with a description of the three-population test. First we give some theory. Consider the tree of Figure 1A. We see that the path from C to A and the path from C to B just share the edge from C to X. Let a′, b′, c′ be allele frequencies in the populations A, B, C, respectively, at a single polymorphism. Define
We, similarly, in an obvious notation define
Choice of the allele does not affect any of F2, F3, F4 as choosing the alternate allele simply flips the sign of both terms in the product. We refer to F2(A, B) as the branch length between populations A and B. We use these branch lengths in admixture graph fitting for graph edges.
Figure 1 .
f-statistics: (A) A simple phylogenetic tree, (B) the additivity of branch lengths; the genetic drift between (A, B) computed using our f-statistic-based methods is the same as the sum of the genetic drifts between (A, B) and (B, C), regardless of the population in which SNPs are ascertained, (C) phylogenetic tree with simple admixture, (D) a more general form of Figure 1C, (E) example of an outgroup case, and (F) example of admixture with an outgroup.
Our F-values should be viewed as population parameters, but we note that they depend both on the demography and choice of SNPs. In Appendix A we give formulae that use sample frequencies and that yield unbiased estimates of the corresponding F parameters. The unbiased estimates of F computed using these formulae at each marker are then averaged over many markers to form our f-statistics.
The results that follow hold rigorously if we identify the polymorphisms we are studying in an outgroup (that is, we select SNPs based on patterns of genetic variation in populations that all have the same genetic relationship to populations A, B, C). Since only markers with variation in A, B, C are relevant to the analysis, then by ascertaining in an outgroup we ensure that our markers are polymorphic in the root population of A, B, C. Later on, we discuss how other strategies for ascertaining polymorphisms would be expected to affect our results. In general, our tests for admixture and estimates of admixture proportion are strikingly robust to the ascertainment processes that are typical for human SNP array data, as we verify both by simulations and by empirical analysis.
Suppose the allele frequency of a SNP is r at the root. In the tree of Figure 1A, let a′, b′, c′, x′, r′ be allele frequencies in A, B, C, X, R. Condition on r′. Then
since E[a′|x′] = x′, and E[x′ − b′] = E[r′ − b′ − (r′ − x′)] = 0. If the phylogeny has C as an outgroup (switching B, C in Figure 1A), then a similar argument shows that
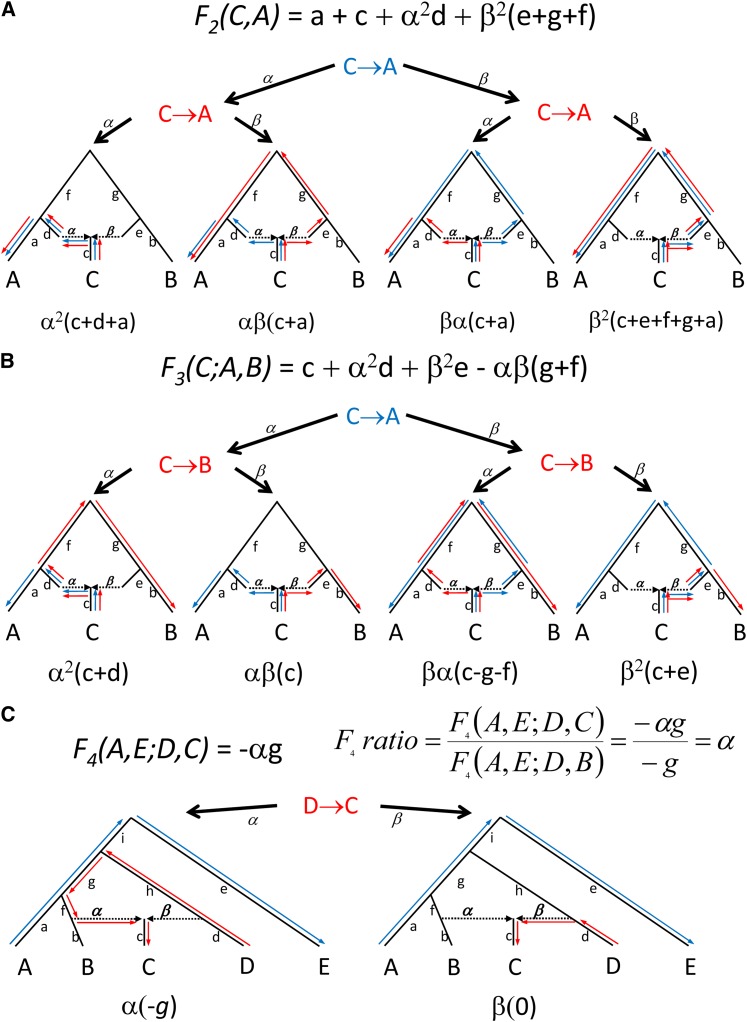
There is an intuitive way to think about the expected values of f-statistics, which relies on tracing the overlap of genetic drift paths between the first and second terms in the quadratic expression, as illustrated in Figure 2 and discussed further in Appendix B. For example, E[(c′ − a′)(c′ − b′)] can be negative only if population C has ancestry from populations related to both A and B. Only in this case are there paths between C and A and C and B that also take opposite drift directions through the tree (Figure 1C and Figure 2), which contributes to a negative expectation for the statistics. The observation of a significantly negative value of f3(C; A, B) is thus evidence of complex phylogeny in C. We prove this formally in Appendix C (Theorem 1). In Appendix D, we also relax our assumptions about the ascertainment process, showing that F3 is guaranteed to be positive if C is unadmixed under quite general conditions, for example, polymorphic in the root R and in addition ascertained as polymorphic in any of A, B, C. It is important to recognize, however, that a history of admixture does not always result in a negative f3(C; A, B)-statistic. If population C has experienced a high degree of population-specific drift (perhaps due to founder events after admixture), it can mask the signal so that f3(C; A, B) might not be negative.
Figure 2 .
Visual computation of expected values of F2, F3, and F4 statistics. See Appendix 2 for a discussion of this figure.
An important feature of this test is that it definitively shows that the history of mixture occurred in population C; a complex history for A or B cannot produce negative F3(C; A, B). To explain why this is so, we recapitulate material from Reich et al. (2009). If population A is admixed then if we pick an allele of A, it must have originated in one of the admixing populations. Pick alleles α, β from populations A and B and γ1, γ2 independently from C, coding 1 for a reference allele, 0 for a variant, etc. Thus, F3(C; A, B) = E[(γ1 − α)](γ2 − β)]. Suppose population A is admixed; B and C are not admixed. The allele α sampled from population A can take more than one path through the ancestral populations. F3(C; A, B) can then be computed as a weighted average over the possible phylogenies, in all of which the quantity has a positive expectation because A and B are now unadmixed (Appendix B and Figure 2). In conclusion, the diagram makes it visually evident that if F3(C; A, B) < 0 then population C itself must have a complex history.
Additivity of F2 along a tree branch
In this article we consider generalizations of phylogenetic trees where graph edges indicate that one population is a descendant of another. Consider the phylogenetic tree in Figure 1B, and a marker polymorphic at the root. Drift on a given edge is a random variable with mean 0. For if A → B is a graph edge, with corresponding allele frequencies a′, b′,
This is the martingale property of allele frequency diffusion. Drifts on two distinct edges of a tree are orthogonal, where orthogonality of random variables X, Y simply means that E[XY] = 0. In our context this means that the drifts on distinct edges have mean 0 and are uncorrelated.
A valuable feature of our F-statistics definition is that branch lengths on the tree (as defined by F2) are additive. We illustrate this with an example from human history (Figure 1B). (We note that all examples in this article refer to human history, although the methods should apply equally well to other species.) In this example, A, and C are present-day populations that split from an ancestral population X. B is an ancestral population to C. For instance, A might be modern Yoruba, C a European population, and B an ancient population, perhaps a sample from archeological material of a population that existed thousands of years ago. We assume here that we ascertain in an outgroup (implying polymorphism at the root) and again assume neutrality and that we can ignore recurrent or backmutations. Then we mean by additivity that
for
but the last term is 0 since the change in allele frequencies (drifts) X → A, X → B, B → C are all uncorrelated.
We remark that our F2-distance resembles the familiar Fst, but is not the same. In particular, parts of a graph that are far from the root (in genetic drift distance) have F2 reduced. Some insight into this effect is given by considering the simple graph
where τ1, τ2 are drift times on the standard diffusion timescale (two random alleles of B have probability that they have not coalesced in the ancestral population A).
If r′, a′, b′ are allele frequencies in R, A, B, respectively, then F2(A, B) = E[(a′ − b′)2]. Write Er′, Ea′ for expectations conditional on population allele frequencies r′, a′. Then (Nei 1987, Chap. 13). Moreover . Hence
Informally the drift from R → A shrinks F2(A, B) by a factor . Thus expected drift is additive,
but the drift does depend on ascertainment. For a given edge, the more distant the root, the smaller the drift. A loose analogy is projecting a curved surface, such as part of the globe, into a plane. Locally all is well, but any projection will cause distortion in the large. Additivity in F2 distances is all we require in what follows. We note that there is no assumption here that population sizes are constant along a branch edge, and so we are not assuming linearity of branch lengths in time.
Expected values of our f-statistics
We can calculate expected values for our f-statistics, at least for simple demographic histories that involve population splits and admixture events. We assume that genetic drift events on distinct edges are uncorrelated, which as mentioned before will be true if we ascertain in an outgroup, and our alleles are neutral.
We give an illustration for f3-statistics. Consider the demography shown in Figure 1C. Populations E, F split from a root population R. G then was formed by admixture in proportions α: β (β = 1 − α). Modern populations A, B, C are then formed by drift from E, F, G. We want to calculate the expected value of f3(C; A, B). Assume that our ascertainment is such that drifts on distinct edges are orthogonal, which will hold true if we ascertained the markers in an outgroup.
We recapitulate some material from (Reich et al. 2009, Supplementary S2, Sect. 2.2). As before let a′, b′, c′ be population allele frequencies in A, B, C, and let g′ be the allele frequency in G and so on:
We see by orthogonality of drifts that
which we write as
| (1) |
Now, label alleles at a marker 0, 1. Then picking chromosomes from our populations independently we can write
where a1, b1 are alleles chosen randomly in populations A, B and g1, g2 are alleles chosen randomly and independently in population G. Similarly, we define e1, e2, f1, and f2. However, g1 originated from E with probability α and so on. Thus
where a1, a2 are independently picked from E and b1, b2 from F. The first three terms vanish. Further
This shows that under our assumptions of orthogonal drift on distinct edges,
| (2) |
It might appear that Figure 1C is too restricted, as it assumes that the admixing populations E, F are ancestral to A, B and that we should consider the more general graph shown in Figure 1D. But it turns out that using our f-statistics alone (and not the more general allelic spectrum) that even if α, β are known, we can obtain information only about
Thus in fitting admixture graphs to f-statistics, we can, without loss of generality, fit all the genetic drift specific to the admixed population on the lineage directly ancestral to the admixed population (the lineage leading from C to G in Figure 1C).
The outgroup case
Care though is needed in interpretation. Consider Figure 1E. Here a similar calculation to the one just given shows (again assuming orthogonality of drift on each edge) that
| (3) |
Note that Y has little to do with the admixture into C and we obtain the same F3 value for any population Y that splits off from A more anciently than X.
We call this case, where we have apparent admixture between A and Y, the outgroup case, and it needs to be carefully considered when recovering population relationships.
Estimates of mixing proportions
We want to estimate, or at least bound, the mixing proportions that have resulted in the ancestral population of C. With further strong assumptions on the phylogeny we can get quite precise estimates even without accurate surrogates for the ancestral populations (see Reich et al. 2009 and the F4-ratio estimation that we describe below, for examples). Also if we have data from populations that are accurate surrogates for the ancestral admixing population (and we can ignore the drift post admixture), the problem is much easier. For instance, in Patterson et al. (2010) we give an estimator that works well even when the sample sizes of the relevant populations are small, and we have multiple admixing populations whose deep phylogenetic relationships we may not understand. Here we show a method that obtains useful bounds, without requiring full knowledge of the phylogeny, although the bounds are not very precise. Note that although our three-population test remains valid even if the populations A, B are admixed, the mixing proportions we calculate are not meaningful unless the assumed phylogeny is at least roughly correct. Indeed even discussing mixing from an ancestral population of A hardly makes sense if A is admixed itself subsequent to the admixing event in C. This is discussed further when we present data from Human Genome Diversity Panel (HGDP) populations.
In much of the work in this article, we analyze some populations A, B, C and need an outgroup, which split off from the ancestral population of A, B, C before the population split of A, B. For example, in Figure 1E, Y is such an outgroup. Usually, when studying a group of populations within a species, a plausible outgroup can be proposed. The outgroup assumption can then be checked using the methods of this article, by adding an individual from a more distantly related population, which can be treated as a second outgroup. For instance, with human populations from Eurasia, Yoruba or San Bushmen from sub-Saharan Africa will often be plausible outgroups.1 Our second outgroup here is simply being used to check a phylogenetic assumption in our primary analysis, and we do not require polymorphism at the root for this narrow purpose. Chimpanzee is always a good second outgroup for studies of humans.
Consider the phylogeny of Figure 1F. Here α, β are mixing parameters (α + β = 1) and we show drift distances along the graph edges. Note that here we use a, b,…, as branch lengths (F2 distances), not sample or population allele frequencies as we do elsewhere in this article. Thus, for example, F2(O, X) = u. Now we can obtain estimates of
We also have estimates of
Set Yi = Zi − Z0, i = 0…5, which eliminates u. This shows that any population O which is a true outgroup should (up to statistical noise) give similar estimates for Yi (Figure 1F). We have three inequalities:
Using αa = Y1, βb = Y2 we can rewrite these as
giving lower and upper bounds on α, which we write as αL,αU in the tables of results that follow. These bounds can be computed by a program qpBound in the ADMIXTOOLS software package that we make available with this article.
Although these bounds will be nearly invariant to choices of the outgroup O, choices for the source populations A, B may make a substantial difference. We give an example in a discussion of the relationship of Siberian populations to Europeans. In principle we can give standard errors for the bounds, but these are not easily interpretable, and we think that in most cases systematic errors (for instance, that our phylogeny is not exactly correct) are likely to dominate.
We observe that in some cases the lower bound exceeds the upper, even when the Z-score for admixture of population C is highly significant. We interpret this as suggesting that our simple model for the relationships of the three populations is wrong. A negative Z-score indeed implies that C has a complex history, but if A or B also has a complex history, then a recovered mixing coefficient α has no real meaning.
Estimation and normalization
With all our f-statistics it is critical that we can compute unbiased estimates of the population F-parameter for a single SNP, with finite sample sizes. Without that, our estimates will be biased, even if we average over many unlinked SNPs. The explicit formulae for f2, f3, f4 that we present in Appendix A (previously given in Reich et al. 2009, Supplementary Material) are in fact minimum variance unbiased estimates of the corresponding F-parameters, at least for a single marker.
The expected (absolute) values of an f-statistic, such as f3, strongly depends on the distribution of the derived allele frequencies of the SNPs examined; for example, if many SNPs are present that have a low average allele frequency across the populations being examined, then the magnitude of f3 will be reduced. To see this, suppose that we are computing f3(C; A, B), and as before a′, b′, c′ are population frequencies of an allele in A, B, C. If the allele frequencies are small, then it is obvious that the expected value of f3(C; A, B) will be small in absolute magnitude as well. Importantly, however, the sign of an f-statistic is not dependent on the absolute magnitudes of the allele frequencies (all that it depends on is the relative magnitudes across the populations being compared). Thus, a significant deviation of an f-statistic from 0 can serve as a statistically valid test for admixture, regardless of the ascertainment of the SNPs that are analyzed. However, to reduce the dependence of the value of the f3-statistic on allele frequencies for some of our practical computations, in all of the empirical analyses we report below, we normalize using an estimate for each SNP of the heterozygosity of the target population C. Specifically, for each SNP i, we compute unbiased estimates , of both
Now we normalize our f3-statistic computing
This greatly reduces the numerical dependence of f3 on the allelic spectrum of the SNPs examined, without making much difference to statistical significance measures such as a Z-score. We note that we use f3 and interchangeably in many places in this article. Both of these statistics give qualitatively similar results and thus if the goal is only to test if f3 has negative expected value then the inference should be unaffected.
D-statistics
The D-statistic test was first introduced in Green et al. (2010) where it was used to evaluate formally whether modern humans have some Neandertal ancestry. Further theory and applications of D-statistics can be found in Reich et al. (2010) and Durand et al. (2011). A very similar statistic f4 was used to provide evidence of admixture in India (Reich et al. 2009), where we called it a four-population test. The D-statistic was also recently used as a convenient statistic for studying locus-specific introgression of genetic material controlling coloration in Heliconius butterflies (Dasmahapatra et al. 2012).
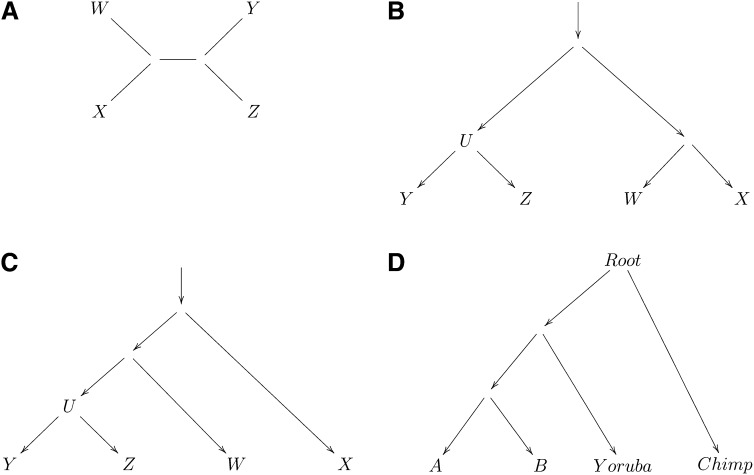
Let W, X, Y, Z be four populations, with a phylogeny that corresponds to the unrooted tree of Figure 3A. For SNP i suppose variant population allele frequencies are w′, x′, y′, z′, respectively. Choose an allele at random from each of the four populations. Then we define a “BABA” event to mean that the W and Y alleles agree, and the X and Z alleles agree, while the W and X alleles are distinct. We define an “ABBA” event similarly, now with the W and Z alleles in agreement. Let Numi and Deni be the numerator and denominator of the statistic:
For SNP data these values can be computed using either population or sample allele frequencies. Durand et al. (2011) showed that replacing population allele frequencies (w′, y′, etc.) by the sample allele frequencies yields unbiased estimates of Numi, Deni. Thus if w, x, y, z are sample allele frequencies we define
and, in a similar spirit to our normalized f3-statistic we define the D-statistic D(W, X; Y, Z) as
summing both the numerator and denominator over many SNPs and only then taking the ratio. If we ascertain in an outgroup, then if (W, X) and (Y, Z) are clades in the population tree, it is easy to see that E[Numi] = 0. We can compute a standard error for D using the weighted block jackknife (Busing et al. 1999). The number of standard errors that this quantity is from zero forms a Z-score, which is approximately normally distributed and thus yields a formal test for whether (W, X) indeed forms a clade.
Figure 3 .
D-statistics provide formal tests for whether an unrooted phylogenetic tree applies to the data, assuming that the analyzed SNPs are ascertained as polymorphic in a population that is an outgroup to both populations (Y, Z) that make up one of the clades. (A) A simple unrooted phylogeny, (B) phylogenies in which (Y, Z) and (W, X) are clades that diverge from a common root, (C) phylogenies in which (Y, Z) are a clade and W and X are increasingly distant outgroups, and (D) a phylogeny to test if human Eurasian populations (A, B) form a clade with respect to sub-Saharan Africans (Yoruba).
More generally, if the relationship of the analyzed populations is as shown in Figure 3B or Figure 3C and we ascertain in an outgroup or in {W, X} then D should be zero up to statistical noise. The reason is that if U is the ancestral population to Y, Z and u′, y′, z′ are population allele frequencies in U, Y, Z, then E[y′ − z′|u′] = E[y′|u′] − E[z′|u′] = 0. Here there is no need to assume polymorphism at the root of the tree, as for a SNP to make a nonzero contribution to D we must have polymorphism at both {Y, Z} and {W, X}. If the tree assumption is correct, drift between Y, Z and between W, X are independent so that E[Numi] = 0. Thus testing whether D is consistent with zero constitutes a test for whether (W, X) and (Y, Z) are clades in the population tree.
As mentioned earlier, D-statistics are very similar to the four-population test statistics introduced in Reich et al. (2009). The primary difference is in the computation of the denominator of D. For statistical estimation, and testing for “treeness,” the D-statistics are preferable, as the denominator of D, the total number of ABBA and BABA events, is uninformative for whether a tree phylogeny is supported by the data, while D has a natural interpretation: the extent of the deviation on a normalized scale from −1 to 1.
As an example, let us assume that two human Eurasian populations A, B are a clade with respect to West Africans (Yoruba). Assume the phylogeny shown in Figure 3D and that we ascertain in an outgroup to A, B. Then
F4-ratio estimation
F4-ratio estimation, previously referred to as f4-ancestry estimation in Reich et al. (2009), is a method for estimating ancestry proportions in an admixed population, under the assumption that we have a correct historical model.
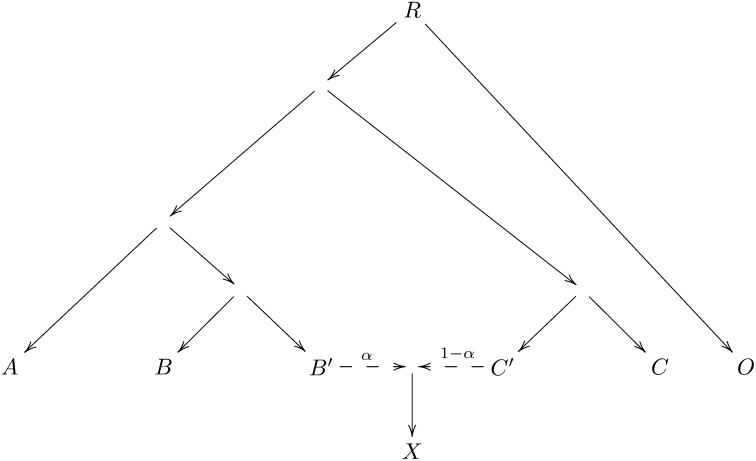
Consider the phylogeny of Figure 4. The population X is an admixture of populations B′ and C′ (possibly with subsequent drift). We have genetic data from populations A, B, X, C, O.
Figure 4 .
A phylogeny explaining f4-ratio estimation.
Since F4(A, O; C′, C) = 0 it follows that
| (4) |
Thus an estimate of α is obtained as
| (5) |
where the estimates in both numerator and denominator are obtained by summing over many SNPs.
As we can obtain unbiased F4-statistics by sampling a single allele from each population, we can apply this test to sequence data, where we pick a single allele, from a high-quality read, for all relevant populations at each polymorphic site. In practice this must be done with care as both sequencing error that is correlated between samples and systematic misalignment of reads to a reference sequence can distort the statistics.
Examples of F4-ratio estimation
Reich et al. (2009) provide evidence that most human South Asian populations can be modeled as a mixture of Ancestral North Indians (ANI) and Ancestral South Indians (ASI) and that if we set, using the labeling above,
Label Population
A Adygei
B CEU (HapMap European Americans)
X Indian (many populations)
C Onge (indigenous Andamanese)
O Papuan (Dai and HapMap YRI West Africans also work)
we get estimates of the mixing coefficients that are robust, have quite small standard errors, and are in conformity with other estimation methods. See Reich et al. (2009, Supplementary S5) for further details.
As another example, in Reich et al. (2010) and Green et al. (2010) evidence was given that there was gene flow (introgression) from Neandertals into non-Africans. Further, a sister group to Neandertals, “Denisovans” represented by a fossil from Denisova cave, Siberia, shows no evidence of having contributed genes to present-day humans in mainland Eurasia (Reich et al. 2010, 2011). The phylogeny is that of Figure 4 if we set:
Label Population
A Denisova
B Neandertal
X French (or almost any population from Eurasia)
C Yoruba
O Chimpanzee
Here B′ is the population of Neandertals that admixed, which forms a clade with the Neandertals from Vindija that were sequenced by Green et al. (2010). So for this example, we obtain an estimate of α, the proportion of Neandertal gene flow into French as 0.022 ± 0.007 (see Reich et al. 2010, SI8, for more detail).
Simulations to test the accuracy of f- and D-statistic-based historical inferences
We carried out coalescent simulations of five populations related according to Figure 4, using ms (Hudson 2002). Detailed information about the simulations is given in Appendix D.
Table 1 shows that using the three-population test, D-statistics, and F4-ratio estimation, we reliably detect mixture events and obtain accurate estimates of mixture proportions, even for widely varied demographic histories and strategies for discovering polymorphisms.
Table 1 . Behavior of f- and D-statistics for a simulated scenarios of admixture.
| Scenario | Fst(C, B) | Fst(O, B) | D(A, B; C, O) | D(A, X; C, O) | f3(B; A, C) | f3(X; A, C) | f4-ratio |
|---|---|---|---|---|---|---|---|
| Baseline | 0.10 | 0.14 | 0.00 | −0.08 | 0.002 | −0.005 | 0.47 |
| Vary sample size | |||||||
| n = 2 from each population | 0.10 | 0.14 | 0.00 | −0.08 | 0.002 | −0.005 | 0.47 |
| Vary SNP ascertainment | |||||||
| Use all sites (full sequencing data) | 0.10 | 0.13 | 0.00 | −0.11 | 0.001 | −0.002 | 0.47 |
| Polymorphic in a single B individual | 0.10 | 0.16 | −0.01 | −0.06 | 0.003 | −0.006 | 0.47 |
| Polymorphic in a single C individual | 0.10 | 0.16 | 0.00 | −0.13 | 0.003 | −0.007 | 0.46 |
| Polymorphic in a single X individual | 0.11 | 0.16 | 0.00 | −0.11 | 0.003 | −0.007 | 0.49 |
| Polymorphic in two individuals: B and O | 0.10 | 0.16 | −0.01 | −0.08 | 0.002 | −0.005 | 0.46 |
| Vary demography | |||||||
| NA = 2,000 (vs. 50,000) pop A bottleneck | 0.10 | 0.14 | 0.00 | −0.08 | 0.002 | −0.005 | 0.48 |
| NB = 2,000 (vs. 12,000) pop B bottleneck | 0.14 | 0.17 | 0.00 | −0.08 | 0.011 | −0.004 | 0.48 |
| NC = 1,000 (vs. 25,000) pop C bottleneck | 0.16 | 0.14 | 0.00 | −0.08 | 0.002 | −0.005 | 0.46 |
| NX = 500 (vs. 10,000) pop X bottleneck | 0.10 | 0.14 | 0.00 | −0.08 | 0.002 | 0.004 | 0.47 |
| NABB′ = 3,000 (vs. 7,000) ABB′ bottleneck | 0.14 | 0.17 | 0.00 | −0.09 | 0.002 | −0.007 | 0.47 |
We carried out simulations for populations related according to Figure 4 using ms (Hudson 2002) with the command: ./ms 110 1000000 -t 1 -I 5 22 22 22 22 22 -n 1 8.0 -n 2 2.5 -n 3 5.0 -n 4 1.2 -n 5 1.0 -es 0.001 5 0.47 -en 0.001001 6 1.0 -ej 0.0060 5 4 -ej 0.007 6 2 -en 0.007001 2 0.33 -ej 0.01 4 3 -en 0.01001 3 0.7 -ej 0.03 3 2 -en 0.030001 2 0.25 -ej 0.06 2 1 -en 0.060001 1 1.0. We chose parameters to produce pairwise FST similar to that for A = Adygei, B = French, X = Uygur, C = Han and O = Yoruba. The baseline simulations correspond to n = 20 samples from each population; SNPs ascertained as heterozygous in a single individual from the outgroup O; and a mixture proportion of α = 0.47. Times are in generations with the subscript indicating the populations derived from the split: tadmix = 40, tBB′ = 240, tABB′ = 400, tCC′ = 280, tABB′ = 400, tABB′CC′ =1,200, tO = 2,400. The diploid population sizes are indicated by a subscript corresponding to the population to which they are ancestral in Figure 4 and are: NA = 50,000, NB = 12,000, NB′ = 10,000, NBB′ = 12,000, NC′ = 25,000, NX = NC′= 10,000, NCC′ = 3,300, NO = 80,000, NABB′ = 7,000, NABB′CC′ = 2,500, NABB′CC′O = 10,000. All simulations involved 106 replicates except for the run involving 2 samples (a single heterozygous individual) from each population, where we increased this to 107 replicates to accommodate the noisier results.
The simulations also document important features of our methods. As mentioned earlier, the only case where the f3-statistic for a population that is truly admixed fails to be negative is when the population has experienced a high degree of population-specific genetic drift after the admixture occurred. Further, the D-statistics show a substantial deviation from 0 only when an admixture event occurred in the history of the four populations contributing to the statistic. Finally, the estimates of admixture proportions using F4-ratio estimation are accurate for all ascertainment strategies and demographies.
Effect of ascertainment process on f- and D-statistics
So far, we have assumed that we have sequence data from all populations and ascertainment is not an issue. However, the ascertainment of polymorphisms (for example, enriching the set of analyzed SNPs for ancestry informative markers) can modulate the magnitudes of F3, F4, and D. Empirically, we observe that in commercial SNP arrays developed for genome-wide association studies (like Affymetrix 6.0 and Illumina 610-Quad), ascertainment does indeed affect the observed magnitudes of these statistics, but importantly, does not cause them to be biased away from zero if this is their expected value in the absence of complex ascertainment (e.g., for complete genome sequencing data). This is key to the robustness of our tests for admixture: since our tests are largely based on evaluating whether particular f- or D-statistics are consistent with zero, and SNP ascertainment almost never causes a deviation from zero, the ascertainment process does not appear to be contributing to spuriously significant signals of admixture. We have verified this through two lines of analysis. First, we carried out simulations showing that tests of admixture (as well as F4-ratio estimation) perfomed using these methods are robust to very different SNP ascertainment strategies (Table 1). Second, we report analyses of data from a new SNP array with known ascertainment that we designed specifically for studies of population history. Even when we use radically different ascertainment schemes, and even when we use widely used commercial SNP arrays, inferences about history are indistinguishable (Table 9).
Table 9 . Z-scores produce consistent inferences whatever outgroup we use.
| Outgroup (O) | Yoruba | San | Chimpanzee | Gorilla | Orangutan | Macaque |
|---|---|---|---|---|---|---|
| D(O, Karitiana; Sardinian, French) | 10.5 | 8.9 | 7.3 | 7.0 | 6.9 | 6.7 |
| D(O, San; Sardinian, Han) | N/A | N/A | −1.1 | −0.8 | −0.5 | −0.5 |
Admixture graph fitting:
We next describe qpGraph, our tool for building a model of population relationships from f-statistics. We first remark that given n populations P1, P2, …, Pn, then
-
1.
the f-statistics (f2, f3 and f4) span a linear space VF of dimension ,
-
2.
all f-statistics can be found as linear sums of statistics , and
-
3.
fix a population (say P1). Then all f-statistics can be found as linear sums of statistics .
These statements are true, both for the theoretical F-values, and for our f-statistics, at least when we have no missing data, so that for all populations our f-statistics are computed on the same set of markers.
Requirements 2 and 3, above, describe bases for the vector space VF. We usually find the basis of 3 to be the most convenient computationally. More detail can be found in Reich et al. (2009, Supplement paragraph 2.3).
Thus choose a basis. From genotype data we can calculate as follows:
-
1.
f-statistics on the basis. Call the resulting long vector f.
-
2.
An estimated error covariance Q of f using the weighted block jackknife (Busing et al. 1999).
Now, given a graph topology, as well as graph parameters (edge values and admixture weights), we can calculate g, the expected value of f.
A natural score function is
| (6) |
an approximate log-likelihood. Note that nonindependence of the SNPs is taken into account by the jackknife. A technical problem is that for n large our estimate Q of the error covariance is not stable. In particular, the smallest eigenvalue of Q may be unreasonably small. This is a common issue in multivariate statistics. Our program qpGraph allows a least-squares option with a score function
| (7) |
where λ is a small constant introduced to avoid numerical problems. The score S2 is not basis independent, but in practice seems robust.
Maximizing or is straightforward, at least if n is moderate, which is the only case in which we recommend using qpGraph. We note that given the admixture weights, both score functions , are quadratic in the edge lengths, and thus can be maximized using linear algebra. This reduces the maximization to the choice of admixture weights. We use the commercial routine nag_opt_simplex from the Numerical Algorithms Group (http://www.nag.com/numeric/cl/manual/pdf/e04/e04ccc.pdf), which has an efficient implementation of least squares. Users of qpGraph will need to have access to nag, or substitute an equivalent subroutine.
Interpretation and limitations of qpGraph
-
1.
A major use of qpGraph is to show that a hypothesized phylogeny must be incorrect. This generalizes our D-statistic test, which is testing a simple tree on four populations.
-
2.
After fitting parameters, study of which f-statistics fit poorly can lead to insights as to how the model must be wrong.
-
3.
Overfitting can be a problem, especially if we hypothesize many admixing events, but only have data for a few populations.
Simulations validate the performance of qpGraph
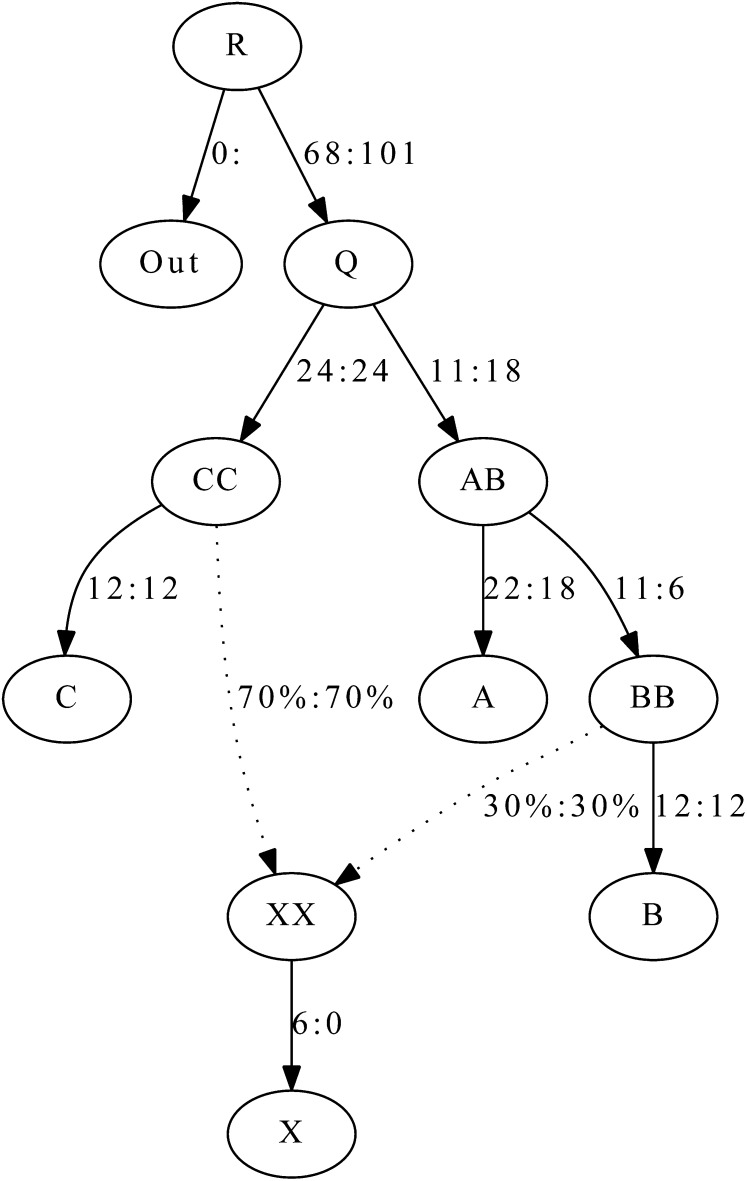
We show in Figure 5 an example in which we simulated a demography with five observed populations Out, A, B, C, and X and one admixture event. We simulated 50,000 unlinked SNPs, ascertained as heterozygous in a single diploid individual from the outgroup Out. Sample sizes were 50 in all populations and the historical population sizes were all taken to be 10,000. We show that we can accurately recover the drift lengths and admixture proportions using qpGraph.
Figure 5 .
Admixture graph fitting: We show an admixture graph fitted by qpGraph for simulated data. We simulated 50,000 unlinked SNPs ascertained as heterozygous in a single diploid individual from the outgroup Out. Sample sizes were 50 in all populations and the historical population sizes were all taken to be 10,000. The true values of parameters are before the colon and the estimated values afterward. Mixture proportions are given as percentages, and branch lengths are given in units of Fst (before the colon) and f2 values (after). F2 and Fst are multiplied by 1000. The fitted admixture weights are exact, up to the resolution shown, while the match of branch lengths to the truth is rather approximate.
Rolloff:
Our fifth technique, rolloff, studies the decay of admixture linkage disequilibrium with distance to infer the date of admixture. Importantly, we do not consider multimarker haplotypes, but instead study the joint allelic distribution at pairs of markers, where the markers are stratified into bins by genetic distance. This method was first introduced in Moorjani et al. (2011) where it was used to infer the date of sub-Saharan African gene flow into southern Europeans, Levantines, and Jews.
Suppose we have an admixed population and for simplicity assume that the population is homogeneous (which usually implies that the admixture is not very recent).
Let us also assume that admixture occurred over a very short time span (pulse admixture model), and since then our admixed (target) population has not experienced further large-scale immigration from the source populations. Call the two admixing (ancestral) populations A, B. Consider two alleles on a chromosome in an admixed individual at loci that are a distance d apart. Then n generations after admixture, with probability e−nd the two alleles belonged, at the admixing time, to a single chromosome.
Suppose we have a weight function w at each SNP that is positive when the variant allele has a higher frequency in population A than in B and negative in the reverse situation. For each SNP s, let w(s) be the weight for SNP s. For every pair of SNPs s1, s2, we compute an LD-based score z(s1, s2) which is positive if the two variant alleles are in linkage disequilibrium; that is, they appear on the same chromosome more often than would be expected assuming independence. For diploid unphased data, which is what we have here, we simply let v1, v2 be the vectors of genotype counts of the variant allele, dropping any samples with missing data. Let m be the number of samples in which neither s1 or s2 has missing data. Let ρ be the Pearson correlation between v1, v2. We apply a small refinement, insisting that m ≥ 4 and clipping ρ to the interval [−0.9, 0.9]. Then we use Fisher’s z-transformation,
which is known to improve the tail behavior of z. In practice this refinement makes little difference to our results.
Now we form a correlation between our z-scores and the weight function. Explicitly, for a bin-width x, define the “bin” , d = x, 2x, 3x,… by the set of SNP pairs (s1, s2), where
where ui is the genetic position of SNP si.
Then we define A(d) to be the correlation coefficient
| (8) |
Here in both numerator and denominator we sum over pairs of SNPs approximately d units apart (counting SNP pairs into discrete bins). In this study, we set a bin size of 0.1 cM in all our examples. In practice, different choices of bin sizes only qualitatively affect the results (Moorjani et al. 2011).
Having computed A(d) over a suitable distance range, we fit
| (9) |
by least squares and interpret n as an admixture date in generations. Equation 9 follows because a recombination event on a chromosome since admixture decorrelates the alleles at the two SNPs being considered, and e−nd is the probability that no such event occurred. (Implicitly, we assume here that the number of recombinations over a genetic interval of d in n generations is Poisson distributed with mean nd. Because of crossover interference, this is not exact, but it is an excellent approximation for the d and n relevant here.)
By fitting a single exponential distribution to the output, we have assumed a single pulse model of admixture. However, in the case of continuous migration we can expect the recovered date to lie within the time period spanned by the start and end of the admixture events. We further discuss rolloff date estimates in the context of continuous migration in applications to real data (below). We estimate standard errors using a weighted block jackknife (Busing et al. 1999) where we drop one chromosome in each run.
Choice of weight function
In many applications, we have access to two modern populations A, B, which we can regard as surrogates for the true admixing populations, and in this context we can simply use the difference of empirical frequencies of the variant allele as our weight. For example, to study the admixture in African Americans, very good surrogates for the ancestral populations are Yoruba and North Europeans. However, a strength of rolloff is that it provides unbiased dates even without access to accurate surrogates for the ancestral populations. That is, rolloff is robust to use of highly divergent populations as surrogates. In cases when the ancestrals are no longer extant or data from the ancestrals are not available, but we have access to multiple admixed populations with differing admixture proportions (as for instance happens in India (Reich et al. 2009), we can use the “SNP loadings” generated from principal component analysis (PCA) as appropriate weights. This also gives unbiased dates for the admixture events.
Simulations to test rolloff
We ran three sets of simulations. The goals of these simulations were
-
1.
to access the accuracy of the estimated dates, in cases for which data from accurate ancestral populations are not available,
-
2.
to investigate the bias seen in Moorjani et al. (2011),
-
3.
to test the effect of genetic drift that occurred after admixture.
We describe the results of each of these investigations in turn.
-
1.
First, we report simulation results that test the robustness of inferences of dates of admixture when data from accurate ancestral populations are not available. We simulated data for 20 individuals using phased data from HapMap European Americans (CEU) and HapMap West Africans (YRI), where the mixture date was set to 100 generations before present and the proportion of European ancestry was 20%. We ran rolloff using pairs of reference populations that were increasingly divergent from the true ancestral populations used in the simulation. The results are shown in Table 2 and are better than those of the rather similar simulations in Moorjani et al. (2011). Here we use more SNPs (378K instead of 83K) and 20 admixed individuals rather than 10. The improved results likely reflect the fact that we are analyzing larger numbers of admixed individuals and SNPs in these simulations, which improves the accuracy of rolloff inferences by reducing sampling noise in the calculation of the Z-score. In analyzing real data, we have found that the accuracy of rolloff results improves rapidly with sample size; this feature of rolloff contrasts markedly with allele frequency correlation statistics like f-statistics where the accuracy of estimation increases only marginally as sample sizes increase above five individuals per population.
-
2.
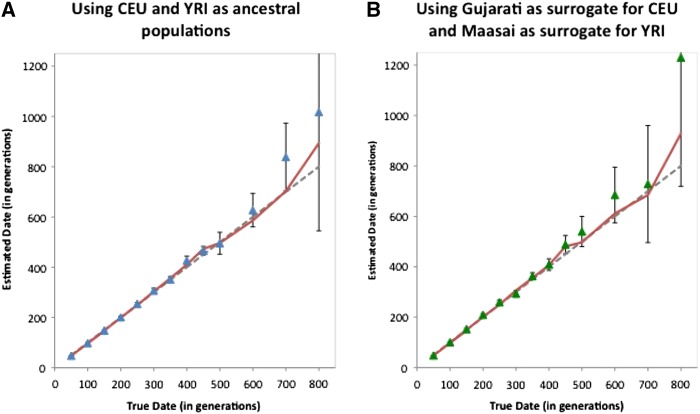
Second, we report simulation results investigating the bias seen in Moorjani et al. (2011). Moorjani et al. (2011) showed that low sample size and admixture proportion can cause a bias in the estimated dates. In our new simulations, we generated haplotypes for 100 individuals using phased data from HapMap CEU and HapMap YRI, where the mixture date was between 50 and 800 generations ago (Figure 6) and the proportion of European ancestry was 20%. We ran rolloff with two sets of reference populations: (1) the true ancestral populations (CEU and YRI) and (2) the divergent populations Gujarati (Fst(CEU, Gujarati) = 0.03 and Maasai (Fst(YRI, Maasai) = 0.03). We show the results for one run and the mean date from each group of 10 runs in Figures 6, A and B. These results show no important bias, and the date estimates, even in the more difficult case where we used Gujarati and Maasai as assumed ancestrals, are tightly clustered near the “truth” up to 500 generations (around 15,000 years). This shows that the bias is removed with larger sample sizes.
-
3.
The simulations reported above sample haplotypes without replacement, effectively removing the impact of genetic drift after admixture. To study the effect of drift post-dating admixture, we performed simulations using the MaCS coalescent simulator (Chen et al. 2009). We simulated data for one chromosome (100 Mb) for three populations (say, A, B, and C). We set the effective population size (Ne) for all populations to 12,500, the mutation rate to 2 × 10−8/bp/generation, and the recombination rate to 1.0 × 10−8/bp/generation. Consider the phylogeny in Figure 1C. G is an admixed population that has 80%/20% ancestry from E and F, with an admixture time (t) set to be 30, 100, or 200 generations before the present. Populations A, B, C are formed by drift from E, F, G, respectively. Fst(A, B) = 0.16 (similar to that of Fst(YRI, CEU)). We performed rolloff analysis with C as the target (n = 30) and A and B as the reference populations. We estimated the standard error using a weighted block jackknife where the block size was set to 10 cM. The estimated dates of admixture were 28 ± 4, 97 ± 10, and 212 ± 19 corresponding the true admixture dates of 30, 100, and 200 generations, respectively. This shows that the estimated dates are not measurably affected by genetic drift post-dating the admixture event.
Table 2 . Performance of rolloff.
| Reference populations | Fst(1) | Fst(2) | Estimated date ± SE |
|---|---|---|---|
| CEU, YRI | 0.000 | 0.000 | 107 ± 4 |
| Basque, Mandenka | 0.009 | 0.009 | 106 ± 4 |
| Druze, LWK(HapMap) | 0.017 | 0.008 | 105 ± 4 |
| Gujarati(HapMap), Maasai | 0.034 | 0.026 | 107 ± 4 |
We simulated data for 20 admixed individuals with 20%/80% CEU and YRI admixture that occurred 100 generations ago. We ran rolloff using “reference populations” shown above that were increasing divergent from CEU (Fst(1)) and YRI (Fst(2)). Estimated dates are shown in generations.
Figure 6 .
rolloff simulation results: We simulated data for 100 individuals of 20% European and 80% African ancestry, where the mixture occurred between 50 and 800 generations ago. Phased data from HapMap3 CEU and YRI populations was used for the simulations. We performed rolloff analysis using CEU and YRI (A) and using Gujarati and Maasai (B) as reference populations. We plot the true date of mixture (dotted line) against the estimated date computed by rolloff (points in blue A and green B). Standard errors were calculated using the weighted block jackknife. To test the bias in the estimated dates, we repeated each simulation 10 times. The estimated date based on the 10 simulations is shown in red.
A SNP array designed for population genetics
We conclude our presentation of our methods by describing a new experimental resource and publicly available data set that we have generated for facilitating studies of human population history and that we use in many of the applications that follow.
For studies that aim to fit models of human history to genetic data, it is highly desirable to have an exact record of how polymorphisms were chosen. Unfortunately, conventional SNP arrays developed for medical genetics have a complex ascertainment process that is nearly impossible to reconstruct and model (but see Wollstein et al. 2010). While the methods reported in our study are robust in theory and also in simulation to a range of strategies for how polymorphisms were ascertained (Table 1), we nevertheless wished to empirically validate our findings on a data set without such uncertainties.
Here, we report on a novel SNP array that we developed that is now released as the Affymetrix Human Origins array. This includes 13 panels of SNPs, each ascertained in a rigorously documented way that is described in File S1, allowing users to choose the one most useful for a particular analysis. The first 12 are based on a strategy used in Keinan et al. (2007), discovering SNPs as heterozygotes in a single individual of known ancestry for whom sequence data are available (from Green et al. 2010; Reich et al. 2010) and then confirming the site as heterozygous with a different assay. After the validation steps described in File S1 (which serves as technical documentation for the new SNP array), we had the following number of SNPs from each panel: San, 163,313; Yoruba, 124,115; French, 111,970; Han, 78,253; Papuan (two panels), 48,531 and 12,117; Cambodian, 16,987; Bougainville, 14,988; Sardinian, 12,922; Mbuti, 12,162; Mongolian, 10,757; Karitiana, 2,634. The 13th ascertainment consisted of 151,435 SNPs where a randomly chosen San allele was derived (different from the reference Chimpanzee allele) and a randomly chosen Denisova allele (Reich et al. 2010) was ancestral (same as chimpanzee). The array was designed so that all sites from panels 1–13 had data from chimpanzee as well as from Vindija Neandertals and Denisova, but the values of the Neandertal and Denisova alleles were not used for ascertainment (except for the 13th ascertainment).
Throughout the design process, we avoided sources of bias that could cause inferences to be affected by genetic data from human samples other than the discovery individual. Our identification of candidate SNPs was carried out entirely using sequencing reads mapped to the chimpanzee genome (PanTro2), so that we were not biased by the ancestry of the human reference sequence. In addition, we designed assays blinded to prior information on the positions of polymorphisms and did not take advantage of prior work that Affymetrix had done to optimize assays for SNPs already reported in databases. After initial testing of 1,353,671 SNPs on two screening arrays, we filtered to a final set of 542,399 SNPs that passed all quality-control criteria. We also added a set of 84,044 “compatibility SNPs” that were chosen to have a high overlap with SNPs previously included on standard Affymetrix and Illumina arrays, to facilitate coanalysis with data collected on other SNP arrays. The final array contains 629,443 unique and validated SNPs, and its technical details are described in File S1.
We successfully genotyped the array in 934 samples from the HGDP and made the data publicly available on August 12, 2011, at ftp://ftp.cephb.fr/hgdp_supp10/. The present study analyzes a curated version of this data set in which we have used principal component analysis (Patterson et al. 2006) to remove samples that are outliers relative to others from their same populations; 828 samples remained after this procedure. This curated data set is available for download from the Reich laboratory website (http://genetics.med.harvard.edu/reich/Reich_Lab/Datasets.html).
Results and Discussion
Initial application to data: South African Xhosa
The Xhosa are a South African population whose ancestors are mostly Bantu speakers from the Nguni group, although they also have some Bushman ancestors (Patterson et al. 2010). We first ran our three-population test with San (HGDP) (Cann et al. 2002) and Yoruba (HapMap) (International Hapmap 3 Consortium 2010) as source populations and 20 samples of Xhosa as the target population, a sample set already described in Patterson et al. (2010). We obtain an f3-statistic of −0.009 with a Z-score of −33.5, as computed with the weighted block jackknife (Busing et al. 1999).
Note that the admixing Bantu-speaking population is known to have been Nguni and certainly was not Nigerian Yoruba. However, as explained earlier, this is not crucial, if the actual admixing population is related genetically (Bantu speakers have an ancient origin in West Africa). If α is the admixing proportion of San here, we obtain using our bounding technique with Han Chinese as an outgroup,
Although this interval is wide, it does show that the Bushmen have made a major contribution to Xhosa genomes.
Xhosa: rolloff
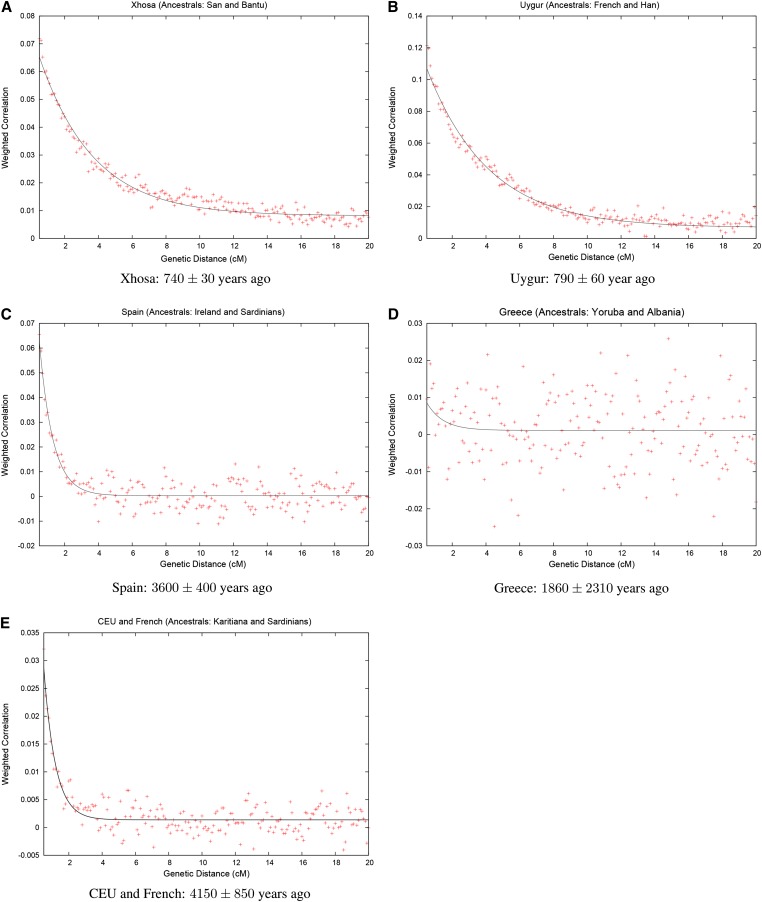
We then applied our rolloff technique, using San and Yoruba as the reference populations, obtaining a very clear exponential admixture LD curve (Figure 7A). We estimate a date of 25.3 ± 1.1 generations, yielding a date of about 740 ± 30 years before present (YBP) assuming 29 years per generation (we also assume this generation time in the analyses that follow) (Fenner 2005).
Figure 7 .
rolloff analysis of real data: We applied rolloff to compute admixture LD between all pairs of markers in each admixed population. We plot the correlation as a function of genetic distance for (A) Xhosa, (B) Uygur, (C) Spain, (D) Greece, and (E) CEU and French. The title of each includes information about the reference populations that were used for the analysis. We fit an exponential distribution to the output of rolloff to estimate the date of the mixture (estimated dates ±SE shown in years). We do not show inter-SNP intervals of <0.5 cM as we have found that at this distance admixture LD begins to be confounded by background LD.
Archeological and linguistic evidence show that the Nguni are a population that migrated south from the Great Lakes area of East Africa. For the dating of the migration we quote:
From an archaeological perspective, the first appearance of Nguni speakers can be recognized by a break in ceramic style; the Nguni style is quite different from the Early Iron Age sequence in the area. This break is dated to about AD 1200 (Huffman 2010).
More detail on Nguni migrations and archeology can be found in Huffman (2004).
Our date is slightly more recent than the dates obtained from the archeology, but very reasonable, since gene flow from the Bushmen into the Nguni plausibly continued after initial contact.
Admixture of the Uygur
The Uygur are known to be historically admixed, but we wanted to try our methods on them. We analyzed a small sample (nine individuals from HGDP) (Cann et al. 2002). Our three-population test using French and Japanese as sources and Uygur as target gives a Z-score of −76.1, a remarkably significant value. Exploring this a little further, we get the results shown in Table 3.
Table 3 . f3(Uygur; A, B).
| Source populations | f3 | Z |
|---|---|---|
| French, Japanese | −0.0255 | −76.109 |
| French, Han | −0.0254 | −77.185 |
| Russian, Japanese | −0.0216 | −68.232 |
| Russian, Han | −0.0217 | −68.486 |
Using Han instead of Japanese is historically more plausible and statistically not significantly different. Our bounding methods suggest that the West Eurasian admixture α is in the range
We used French and Han for the source populations here. Russian as a source is significantly weaker than French. We believe that the likely reason is that our Russian samples have more gene flow from East Asia than the French samples, and this weakens the signal. We confirm this by finding that D(Yoruba, Han; French, Russian) = 0.192, Z = 26.3. The fact that we obtain very similar statistics when we substitute a very different sub-Saharan African population (HGDP San) for Yoruba (D = 0.189, Z = 23.9) indicates that the gene flow does not involve an African population, and instead the findings reflect gene flow between relatives of the Han and Russians.
Uygur: rolloff
Applying rolloff we again get a very clear decay curve (Figure 7B). We estimate a date of 790 ± 60 YBP.
Uygur genetics has been analyzed in two articles by Xu, Jin, and colleagues (Xu et al. 2008; Xu and Jin 2008), using several sets of samples, one of which is the same set of HGDP samples we analyze here. Xu and Jin, primarily using ancestry informative markers (AIMs), estimate West Eurasian admixture proportions of ∼50%, in agreement with our analysis, but also an admixture date estimate using STRUCTURE 2.0 (Falush et al. 2003) of more than 100 generations that is substantially older than ours.
Why are the admixture dates that we obtain so much more recent than those suggested by Xu and Jin? We suspect that STRUCTURE 2.0 systematically overestimates the admixture date, when the reference populations (source populations for the admixture) are not close to the true populations, so that the assumed distribution of haplotypes is in error. It has been suggested (Mackerras 1972) that the West Eurasian component was Tocharian, an ancient Indo-European-speaking population, whose genetics are essentially unknown. Xu and Jin used 60 European American (HapMap CEU) samples to model the European component in the Uygur, and if the admixture is indeed related to the Tocharians it is plausible that they were substantially genetically drifted relative to the CEU, providing a potential explanation for the discrepancy.
Our date of ∼800 years before present is not in conformity with Mackerras(1972), who places the admixture in the eighth century of the common era. Our date though is rather precisely in accordance with the rise of the Mongols under Genghis Khan (1206–1368), a turbulent time in the region that the Uygur inhabit. Could there be multiple admixture events and we are primarily dating the most recent?
Northern European gene flow into Spain
While investigating the genetic history of Spain, we discovered an interesting signal of admixture involving Sardinia and northern Europe. We made a data set by merging genotypes from samples from the population reference sample (POPRES) (Nelson et al. 2008), HGDP (Li et al. 2008), and HapMap Phase 3 (International Hapmap 3 Consortium 2010). We ran our three-population test on triples of populations using Spain as a target (admixed population). We had 137 Spanish individuals in our sample. With Sardinian fixed as a source, we find a clear signal using almost any population from northern Europe. Table 4 gives the top f3-statistics with corresponding Z-scores. The high score for the Russian and Adygei is likely to be partially confounded with the effect discussed in the section on flow from Asia into Europe (below).
Table 4 . Three-population test results showing northern European gene flow into Spain.
| X (data set) | Sample size | f3(Sardinian, X; Spain) | Z-score |
|---|---|---|---|
| Russian (H) | 25 | −0.0025 | −22.90 |
| Norway | 3 | −0.0021 | −9.49 |
| Ireland | 62 | −0.0020 | −24.31 |
| Poland | 22 | −0.0019 | −18.88 |
| Sweden | 11 | −0.0018 | −13.21 |
| Orcadian (H) | 15 | −0.0018 | −14.59 |
| Scotland | 5 | −0.0017 | −10.01 |
| Russia | 6 | −0.0016 | −9.82 |
| UK | 388 | −0.0015 | −28.21 |
| CEU (HapMap) | 113 | −0.0015 | −21.79 |
| Netherlands | 17 | −0.0014 | −12.45 |
| Germany | 75 | −0.0013 | −19.36 |
| Czech | 11 | −0.0012 | −9.33 |
| Hungary | 19 | −0.0012 | −11.98 |
| Belgium | 43 | −0.0010 | −13.76 |
| Adygei (H) | 17 | −0.0010 | −7.44 |
| Austria | 14 | −0.0009 | −7.89 |
| Bosnia | 9 | −0.0008 | −5.68 |
| Croatia | 8 | −0.0007 | −5.33 |
| Swiss-German | 84 | −0.0007 | −11.67 |
| French (H) | 28 | −0.0005 | −6.33 |
| Swiss-French | 760 | −0.0005 | −11.77 |
| Switzerland | 168 | −0.0005 | −9.60 |
| France | 92 | −0.0004 | −8.07 |
| Romania | 14 | −0.0004 | −3.62 |
| Serbia | 3 | −0.0004 | −1.75 |
| Basque (H) | 24 | −0.0001 | −1.08 |
| Portugal | 134 | 0.0001 | 2.15 |
| Macedonia | 4 | 0.0003 | 1.60 |
| Swiss-Italian | 13 | 0.0004 | 3.11 |
| Albania | 3 | 0.0004 | 1.75 |
| Greece | 7 | 0.0006 | 4.27 |
| Tuscan (H) | 8 | 0.0009 | 5.88 |
| Italian (H) | 12 | 0.0009 | 7.86 |
| Italy | 225 | 0.0009 | 16.58 |
| Cyprus | 4 | 0.0014 | 6.56 |
Here the CEU are from HapMap3, and the HGDP populations are indicated by (H) in parentheses.
A geographical structure is clear, with the largest magnitude f3-statistics seen for source populations that are northern European or Slavic. The Z-score is unsurprisingly more significant for populations with a larger sample size. (Note that positive Z-scores are not meaningful here.) We were concerned that the Slavic scores might be confounded by a central Asian component and therefore decided to concentrate our attention on Ireland as a surrogate for the ancestral population as they have a substantial sample size (n = 62).
Spain: rolloff
We applied rolloff to Spain using Ireland and Sardinians as the reference populations. In Figure 7C we show a rolloff curve. The rolloff of signed LD out to about 2 cM is clear and gives an admixture age of 3600 ± 400 YBP (the standard error was computed using a block jackknife with a block size of 5 cM).
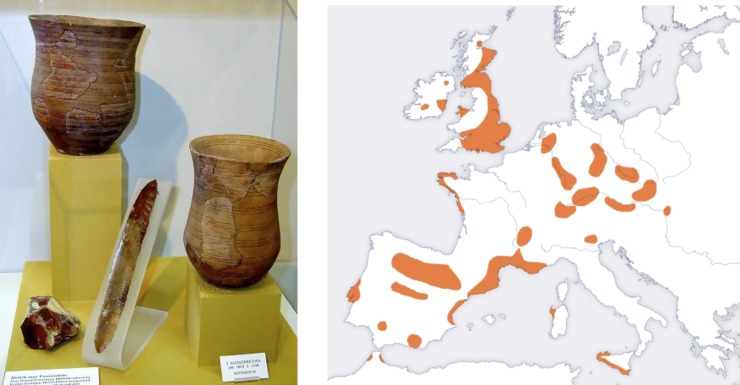
We have detected here a signal of gene flow from populations related to present-day northern Europeans into Spain around 2000 B.C. We discuss a likely interpretation. At this time there was a characteristic pottery termed “bell-beakers” believed to correspond to a population spread across Iberia and northern Europe. We hypothesize that we are seeing here a genetic signal of the “Bell-Beaker culture” (Harrison 1980). Initial cultural flow of the Bell-Beakers appears to have been from South to North, but the full story may be complex. Indeed one hypothesis is that after an initial expansion from Iberia there was a reverse flow back to Iberia (Czebreszuk 2003); this “reflux” model is broadly concordant with our genetic results, and if this is the correct explanation it suggests that this reverse flow may have been accompanied by substantial population movement. (See Figures 8, 9, and 10.)
Figure 8 .
Bell-Beaker culture. On the left we show some Beaker culture objects (from Bruchsal City Museum). On the right we show a map of Bell-Beaker attested sites. We are grateful to Thomas Ihle for the Bruchsal Museum photograph. It is licensed under the Creative Commons Attribution-Share Alike 3.0 Unported license, and a GNU Free documentation license. The map is public domain, licensed under a creative commons license, and adapted from a map in Harrison (1980).
Figure 9 .
Northeast Asian-related admixture in northern Europe. A proposed model of population relationships that can explain some features observed in our genetic data.
Figure C1 .
(A) Appendix C, Theorem 1. (B) Appendix C, Theorem 2.
It is important to point out that we are not detecting gene flow from Germanic peoples (Suevi, Vandals, Visigoths) into Spain even though it is known that they migrated into Iberia around 500 A.D. We believe such migration must have occurred, based on the historical record (and perhaps is biasing our admixture date to be too recent), but any accompanying gene flow must have occurred at a lower level than the much earlier flow we discuss.
An example of the outgroup case
Populations closely related geographically often mix genetically, which leaves a clear signal in PCA plots. An example is that isolation-by-distance effects dominate much of the genetic patterning of Europe (Lao et al. 2008; Novembre et al. 2008). This can lead to significant f3-statistics and is related to the outgroup case we have already discussed. Here is an example. We find
[YRI are HapMap Yoruba Nigerians (International Hapmap 3 Consortium 2010).] Sub-Saharan populations (including HGDP San) all give a Z < −4.0 when paired with Albania, and even f3(Greece; Albania, Papuan) = −0.0033 (Z = −3.5). There may be a low level of sub-Saharan ancestry in our Greek samples, contributing to our signal, but the consistent pattern of highly significant f3-statistics suggests that we are primarily seeing an outgroup case.
We attempted to date Albanian-related gene flow into Greece using rolloff [with HapMap Yoruba and Albanian as the source populations (Figure 7D)]. However, the technique evidently fails here. Formally we get a date of 62 ± 77 generations, which is not significantly different from zero. It is possible that the admixture is very old (>500 generations) or the gene flow was continuous at a low level, and our basic rolloff model does not work well here.
Admixture events detected in Human Genome Diversity Panel populations
We ran our f3-statistic on all possible triples of populations from the HGDP, genotyped on an Illumina 650Y array (Table 5) (Rosenberg 2006; Li et al. 2008).
Table 5 . Three-population test in HGDP.
| Source1 | Source2 | Target | f3 | Z-score | αL | αU | ZSan | ZHan | αL > αR |
|---|---|---|---|---|---|---|---|---|---|
| Japanese | Italian | Uygur | −0.0259 | −74.79 | 0.484 | 0.573 | −46.08 | −42.31 | |
| Japanese | Italian | Hazara | −0.0230 | −74.05 | 0.46 | 0.615 | −45.19 | −42.22 | |
| Yoruba | Sardinian | Mozabite | −0.0211 | −56.95 | 0.288 | 0.304 | −40.65 | −31.16 | |
| Mozabite | Surui | Maya | −0.0149 | −19.67 | 0.165 | 0.408 | −11.51 | −9.40 | |
| Yoruba | San | Bantu-SA | −0.0107 | −31.39 | 0.677 | 0.839 | −24.67 | −16.70 | |
| Yoruba | Sardinian | Palestinian | −0.0107 | −36.70 | 0.07 | 0.157 | −25.64 | −18.35 | |
| Yoruba | Sardinian | Bedouin | −0.0104 | −33.73 | 0.07 | 0.185 | −23.37 | −14.24 | |
| Druze | Yi | Burusho | −0.0090 | −27.62 | 0.558 | 0.731 | −15.94 | −13.59 | |
| Sardinian | Karitiana | Russian | −0.0086 | −20.68 | 0.694 | 0.923 | −10.07 | −10.98 | |
| Druze | Karitiana | Pathan | −0.0084 | −22.25 | 0.547 | 0.922 | −10.68 | −9.37 | |
| Han | Orcadian | Tu | −0.0076 | −20.64 | 0.875 | 0.926 | −12.38 | −8.98 | |
| Mbuti | Orcadian | Makrani | −0.0076 | −19.56 | 0.038 | 0.151 | −11.87 | −6.61 | |
| Han | Orcadian | Mongola | −0.0075 | −19.21 | 0.879 | 0.916 | −12.63 | −8.16 | |
| Han | French | Xibo | −0.0069 | −16.92 | 0.888 | 0.922 | −9.52 | −8.19 | |
| Druze | Dai | Sindhi | −0.0067 | −21.99 | 0.467 | 0.877 | −12.25 | −8.40 | |
| Sardinian | Karitiana | French | −0.0060 | −18.36 | 0.816 | 0.964 | −9.55 | −9.33 | |
| Dai | Italian | Cambodian | −0.0060 | −13.16 | 0.846 | 0.928 | −6.78 | −6.43 | |
| Sardinian | Karitiana | Adygei | −0.0057 | −13.03 | 0.635 | 0.956 | −5.60 | −5.59 | |
| Biaka | Sardinian | Bantu-Kenya | −0.0054 | −13.42 | 0.405 | 0.834 | −9.65 | −7.15 | |
| Sardinian | Karitiana | Tuscan | −0.0052 | −11.26 | 0.803 | 0.962 | −5.12 | −4.76 | |
| Sardinian | Pima | Italian | −0.0045 | −12.48 | 0.84 | 0.97 | −7.48 | −5.66 | |
| Druze | Karitiana | Balochi | −0.0044 | −11.58 | 0.483 | 0.96 | −6.96 | −6.30 | |
| Daur | Dai | Han | −0.0026 | −13.20 | 0.664 | 0.26 | −7.89 | −6.31 | * |
| Han | Orcadian | Han-NChina | −0.0025 | −7.09 | 0.958 | 0.97 | −4.16 | −2.74 | |
| Han | Yakut | Daur | −0.0025 | −9.05 | 0.6 | 0.588 | −6.91 | −5.78 | * |
| Druze | Karitiana | Brahui | −0.0025 | −6.43 | 0.47 | 0.964 | −2.23 | −2.41 | |
| Hezhen | Dai | Tujia | −0.0021 | −6.97 | 0.452 | 0.39 | −4.36 | −3.94 | * |
| Sardinian | Karitiana | Orcadian | −0.0019 | −4.31 | 0.803 | 0.952 | −2.18 | −3.24 | |
| She | Yakut | Oroqen | −0.0017 | −5.13 | 0.422 | 0.296 | −4.99 | −2.44 | * |
This table only lists the most significantly negative f3-statistics observed in HGDP samples. For each target population, we loop over all possible pairs of source populations, and report the pair that produces the most negative f3-statistic. Here we only print results for target populations for which the most negative f3-statistic is significant after correcting for multiple hypothesis testing; that is, the Z-score is more than 4 standard errors below zero. For the line with Bantu-SA as target, we used HGDP Han as an outgroup. In four cases indicated by an asterisk in the last column, the lower bound on the admixture proportion αL is greater than the upper bound αR, suggesting that our proposed three-population phylogeny is not feasible. We suspect that here the admixing (source) populations are themselves admixed. The 2 Z-score columns are with San and Han het ascertainment respectively.
Here we show for each HGDP target population (column 3) the two-source populations with the most negative (most significant) f3-statistic. We compute Z using the block jackknife as we did earlier and just show entries with Z < −4. We bound α, the mixing coefficient involving the first source population, as
where αL, αU are computed with HGDP San as outgroup using the methodology of estimating mixing proportions that we have already discussed.
In four cases indicated by an asterisk in the last column, αL > αR, suggesting that our three-population phylogeny is not feasible. We suspect (and in some cases the table itself proves) that here the admixing (source) populations are themselves admixed.
It is likely that there are other lines in our table where our source populations are admixed, but that this has not been detected by our rather coarse admixing bounds. In such situations our bounds may be misleading.
Many entries are easily interpretable, for instance the admixture of Uygur (Xu et al. 2008; Xu and Jin 2008) (which we have already discussed), Hazara, Mozabite (Corander and Marttinen 2006; Li et al. 2008), and Maya (Mao et al. 2007) are historically attested. The entry for Bantu-SouthAfrica is likely detecting the same phenomenon that we already discussed in connection with the Xhosa.
However, there is much of additional interest here. Note, for example, the entry for Tu, a people with a complex history and clearly with both East Asian and West Eurasian ancestry. It is important to realize that the finding here by no means implies that the target population is admixed from the two given source populations. For example, in the second line, we do not believe that Japanese, or modern Italians, have contributed genes to the Hazara. Instead one should interpret this line as meaning that an East Asian population related genetically to a population ancestral to the Japanese has admixed with a West Eurasian population. As another example, the most negative f3-statistic for the Maya arises when we use as source populations Mozabite (north African) and Surui (an indigenous population of South America in whom we have detected no post-Colombian gene flow). The Mozabites are themselves admixed, with sub-Saharan and West Eurasian gene flow. We think that the Maya samples have three-way admixture (European, West African, and Native American) and the incorrect two-way admixture model is simply doing the best it can (Table 5).
Insensitivity to the ascertainment of polymorphisms
In the Materials and Methods section we described a novel SNP array with known ascertainment that we developed specifically for population genetics (now available as the Affymetrix Human Origins array). The array contains SNPs ascertained in 13 different ways, 12 of which involved ascertaining a heterozygote in a single individual of known ancestry from the HGDP. We genotyped 934 unrelated individuals from the HGDP (Cann et al. 2002) and here report the value of f3-statistics on either SNPs ascertained as a heterozygote in a single HGDP San individual, or at SNPs ascertained in a single Han Chinese (Table 6). We show Z-statistics for these two ascertainments in the last two columns. The number of SNPs used is reduced relative to the 644,247 analyzed in Li et al. (2008); we had 124,440 SNPs for the first ascertainment and 59,251 for the second ascertainment, after removing SNPs at hypermutable CpG dinucleotides. Thus, we expect standard errors on f3 to be larger and the Z-scores to be smaller, as we observe. The correlation coefficient between the Z-scores for the 2008 data (Z2008) and our newly ascertained data are in each case ∼0.99. We were concerned that this correlation coefficient might be inflated by the very large Z-statistics for some populations, such as the Hazara and Uygur, but the correlation coefficients remain very large if we divide the table into two halves and analyze separately the most significant and least significant entries.
Table 6 . Correlation of Z-scores with distinct ascertainments.
| Selected Z | Correlation Z2008, ZSan | Correlation Z2008, ZHan |
|---|---|---|
| Most negative Z | 0.981 | 0.995 |
| Least negative Z | 0.875 | 0.944 |
| Overall | 0.987 | 0.991 |
Ascertainment on a San heterozygote or a Han heterozygote are very different phylogenetically, and the San are unlikely to have been used in the construction of the 2008 SNP panel, so the consistency of findings for these distinct ascertainment processes provides empirical evidence, confirming our expectations from theory and findings from simulation (Table 1) that the SNP ascertainment process does not have a substantial effect on inferences of admixture from the f3-statistics (Table 6).
Evidence for Northeast Asian-related genetic material in Europe
We single out from Table 5 the score for French arising as an admixture of Karitiana, an indigenous population from Brazil, and Sardinians. The Z-score of −18.4 is unambiguously statistically significant. We do not of course think that there has been substantial gene flow back into Europe from Amazonia.
The only plausible explanation we can see for our signal of admixture into the French is that an ancient northern Eurasian population contributed genetic material to both the ancestral population of the Americas and the ancestral population of northern Europe. This was quite surprising to us, and in the remainder of the article this is the effect we discuss.
We are not dealing here with the outgroup case, where the effect is simply caused by Sardinian-related gene flow into the French. If that were the case, then we would expect to see that (French, Sardinian) are approximately a clade with respect to sub-Saharan Africa and Native Americans. There is some modest level of sub-Saharan (probably West African related) gene flow from Africa into Sardinia as is shown by analyses in Moorjani et al. (2011), but no evidence for gene flow from the San (Bushmen), which is indeed historically most unlikely. But if we compute D(San, Karitiana, French, Sardinian) we obtain a value of −0.0178 and a Z-score of −18.1. Thus we have here gene flow “related” to South America into mainland Europe to a greater extent than into Sardinia.
Further confirmation
We merged two SNP array data sets that included data from Europeans and other relevant populations: POPRES (Nelson et al. 2008) and HGDP (Li et al. 2008). We considered only populations with a sample size of at least 10.
We considered European populations with Sardinian and Karitiana as sources and computed the statistic f3(X; Karitian, Sardinian), where X is various European populations. We also added Druze, as a representative population of the Middle East (Table 7). The effect is pervasive across Europe, with nearly all populations showing a highly significant effect. Orcadians and Cyprus are island populations with known island-specific founder events that could plausibly mask admixture signals produced by the three-population test, so the absence of the signal in these populations does not provide compelling evidence that they are not admixed. Our Cypriot samples are also likely to have some proportion of Levantine ancestry (like the Druze) that does not seem to be affected by whatever historical events are driving our negative f3-statistic. We can use any Central American or South American population to demonstrate this effect, in place of the Karitiana.
Table 7 . f3(X; Karitiana, Sardinian/Basque).
| Sardinian | Basque | |||
|---|---|---|---|---|
| X | f3 | Z | f3 | Z |
| Russian | −0.0084 | −15.78 | −0.0074 | −15.04 |
| Romania | −0.0070 | −13.86 | −0.0036 | −7.05 |
| Hungary | −0.0069 | −14.65 | −0.0045 | −9.44 |
| English | −0.0068 | −9.20 | −0.0047 | −6.54 |
| Croatia | −0.0065 | −10.09 | −0.0036 | −5.32 |
| Turkey | −0.0064 | −7.81 | −0.0021 | −2.51 |
| Russia | −0.0063 | −8.56 | −0.0044 | −6.01 |
| Macedonia | −0.0062 | −6.70 | −0.0019 | −2.06 |
| Scotland | −0.0061 | −7.53 | −0.0045 | −5.52 |
| Yugoslavia | −0.0058 | −14.66 | −0.0020 | −4.68 |
| Portugal | −0.0058 | −16.84 | −0.0021 | −5.93 |
| French | −0.0057 | −13.81 | −0.0030 | −7.14 |
| Austria | −0.0057 | −11.32 | −0.0029 | −5.38 |
| Sweden | −0.0057 | −9.44 | −0.0042 | −7.49 |
| Spain | −0.0056 | −16.43 | −0.0024 | −7.24 |
| France | −0.0056 | −15.67 | −0.0028 | −7.66 |
| Australia | −0.0056 | −13.88 | −0.0034 | −8.89 |
| Switzerland | −0.0055 | −15.08 | −0.0025 | −6.98 |
| Swiss-French | −0.0055 | −15.48 | −0.0025 | −7.37 |
| Czech | −0.0054 | −9.39 | −0.0034 | −6.07 |
| Belgium | −0.0054 | −12.55 | −0.0029 | −6.98 |
| Adygei | −0.0053 | −9.27 | −0.0020 | −3.35 |
| Bosnia | −0.0051 | −8.35 | −0.0019 | −3.07 |
| Swiss-German | −0.0050 | −12.75 | −0.0022 | −5.99 |
| Germany | −0.0049 | −12.09 | −0.0027 | −7.03 |
| UK | −0.0048 | −12.40 | −0.0031 | −8.63 |
| Swiss-Italian | −0.0048 | −9.31 | −0.0009 | −1.76 |
| TSI | −0.0047 | −13.46 | −0.0001 | −0.39 |
| CEU | −0.0047 | −11.72 | −0.0029 | −7.79 |
| Greece | −0.0046 | −7.11 | 0.0002 | > 0 |
| Netherlands | −0.0043 | −8.09 | −0.0023 | −4.51 |
| Tuscan | −0.0043 | −6.94 | 0.0001 | > 0 |
| Italian | −0.0043 | −8.37 | 0.0002 | > 0 |
| Poland | −0.0040 | −7.94 | −0.0023 | −4.69 |
| Ireland | −0.0038 | −8.10 | −0.0025 | −6.28 |
| Cyprus | −0.0024 | −2.53 | 0.0036 | > 0 |
| Orcadian | −0.0018 | −3.11 | −0.0002 | −0.32 |
| Druze | 0.0040 | > 0 | 0.009763 | > 0 |
If we replace the Sardinian population by Basque as a source, the effect is systematically smaller, but still enormously statistically significant for most of the populations of Europe (Table 7). We note that in our three populations from mainland Italy [TSI (Hapmap Tuscans), Tuscan, and Italian] the effect essentially disappears when using Basque as a source, although it is quite clear and significant with Sardinian. This is not explored further here, but suggests that further investigation of the genetic relationships of Basque, Sardinian, and other populations of Europe might be fruitful.
Replication using a novel SNP array
The signal above is overwhelmingly statistically significant but we found the effect quite surprising, especially as on common-sense grounds one would expect substantial recent gene flow from the general Spanish and French populations into the Basque, and from mainland Italy into Sardinia, which would weaken the observed effect. We wanted to exclude the possibility that what we are seeing here is an effect of how SNPs were chosen for the medical genetics array used for genotyping. Could the ascertainment be producing false-positive signals of admixture? If, for example, SNPs were chosen specifically so that the population frequencies were very different in Sardinia and northern Europe, an artifactual signal would be expected to arise. This seemed implausible but we had no way to exclude it.
We therefore returned to analysis of data from the Affymetrix Human Origins SNP array with known ascertainment. We show statistics for f3(French; Karitiana, Saridinian) for all 13 ascertainments and compare them to the statistics for the genotype data from the Illumina 650Y array developed for medical genetics (Li et al. 2008) (Table 8).
Table 8 . Three-population test with 14 ascertainments shows the robustness of the signal of Northeast Asian-related admixture in northern Europeans.
| f3(French; Karitiana, Sardinian) | Z | N | Ascertainment |
|---|---|---|---|
| −0.006 | −18.36 | 586414 | Li et al. (2008) |
| −0.007 | −11.49 | 107525 | French |
| −0.006 | −9.06 | 69626 | Han |
| −0.006 | −8.19 | 40725 | Papuan |
| −0.005 | −9.43 | 92566 | San |
| −0.006 | −9.92 | 82416 | Yoruba |
| −0.006 | −5.27 | 7193 | MbutiPygmy |
| −0.003 | −1.91 | 2396 | Karitiana |
| −0.004 | −4.33 | 12400 | Sardinian |
| −0.006 | −5.84 | 12963 | Melanesian |
| −0.006 | −5.91 | 15171 | Cambodian |
| −0.006 | −5.48 | 9655 | Mongola |
| −0.007 | −6.55 | 10166 | Papuan |
| −0.006 | −11.55 | 83385 | Denisova/San |
Two different Papuan samples were used for ascertainment. The last column indicates the ascertainment used, while the column headed N is the number of SNPs contributing to f3, so that SNPs monomorphic in all samples of (Karitiana, Sardinian, French) are not counted.
All our Z-scores are highly significant with a very wide range of ascertainments, except for the ascertainment consisting of finding a heterozygote in a Karitiana sample, where the number of SNPs involved is small (thus reducing power). We can safely conclude that the effect is real and that the French have a complex history.
There is evidence that the effect here is substantially stronger in northern than in southern Europe. We confirm this using the statistic D(San, Karitiana; French, Italian), which has a Z-score of −6.4 on the Illumina 650Y SNP array panel and −3.5 on our population genetics panel ascertained with a San heterozygote. These results show that the Karitiana are significantly more closely related to the French than to the Italians. The Italian samples here are from Bergamo, northern Italy. A likely explanation for these findings is discussed below where we apply rolloff to date this admixture event.
As an aside we have repeatedly assumed that backmutations (or recurrent mutations) are not importantly affecting our results. As evidence that this assumption is reasonable, in Table 9 we compute two of our most important D-statistic-based tests for treeness using a variety of increasingly distant outgroups ranging from modern human outgroups to chimpanzee, gorilla, orangutan, and macaque. Results are entirely consistent across this enormous range of genetic divergence. For example, for the crucial statistic D(Outgroup, Karitiana; Sardinian, French), which demonstrates the signal of Northeast Asian-related admixture in northern Europeans, we find that Z-scores are consistently positive with high significance whichever outgroup is used. As a second example, when we test if the San are consistent with being an outgroup to two Eurasian populations through the statistic D(Outgroup, San; Sardinian, Han) we detect no significant deviation from zero whichever outgroup is used.
Siberian populations
We obtained Illumina SNP array data from Hancock et al. (2011) from the Naukan and Chukchi, Siberian peoples who live in extreme northeastern Siberia. After merging with the 2008 Illumina 650Y SNP array data on HGDP samples (Li et al. 2008) we obtain the f3-statistics in Table 10.
Table 10 . The signal of admixture in the French is robust to the Northeast Asian-related population that is used as the surrogate for the ancestral admixing population.
| Sources; Target | f3 | Z | αL | αU | N |
|---|---|---|---|---|---|
| Karitiana, Sardinian; French | −0.006 | −18.36 | 0.036 | 0.184 | 586406 |
| Naukan, Sardinian; French | −0.005 | −16.73 | 0.051 | 0.176 | 393216 |
| Chukchi, Sardinian; French | −0.005 | −15.92 | 0.056 | 0.174 | 393466 |
We can assume here that we have a common admixture event to explain. Although the statistics for Chukchi are (slightly) weaker than those in the Native Americans, we obtain better bounds on the mixing coefficient α of between 5% and 18%. We caution that if the Sardinians are themselves admixed with Asian ancestry although less so than other Europeans (a scenario we think is historically plausible), then we will have underestimated the Asian-related mixture proportion in Europeans.
We wanted to test if (French, Sardinian) form a clade relative to (Karitiana, Chukchi), which would, for example, be the case if the admixing population to northern Europe had a common ancestor with an ancestor of Karitiana and Chukchi. In our data set,
while this hypothesis predicted D = 0. Thus, we can rule out this alternative hypothesis.
One possible explanation for these findings is that the ancestral Karitiana were closer genetically to the northern Eurasian population that contributed genes to northern Europeans than are the Chukchi. The original migration into the Americas occurred at least 15,000 YBP, so there is ample time for some population inflow into the Chukchi peninsula since then. However, the Chukchi and Naukan samples show no evidence of recent west Eurasian admixture, and we specifically tested for ethnic Russian admixture, finding nothing.
We carried out a rolloff analysis in which we attempted to learn about the date of the admixture events in the history of northern Europeans. We pooled samples from CEU, a population of largely northern European origin (International Hapmap 3 Consortium 2010) with HGDP French to form our target admixed population, wishing to maximize the sample size. The surrogate ancestral populations for this analysis are Karitiana and Sardinian.
The admixture date we are analyzing here is old, and to improve the performance of rolloff here and in the analysis of northern European gene flow into Spain reported above, we filtered out two regions of the genome that have substantial structural variation that is not accurately modeled by rolloff, which assumes Poisson-distributed recombination events between two alleles (Mills et al. 2011). The two regions we filtered out were HLA on chromosome 6 and the p-telomeric region on chromosome 8, which we found in practice contributed to anomalous rolloff signals in some of our analyses. Our signals should be robust to removal of small genomic regions.
In Figure 7E we show the rolloff results. The signal is clear enough, although noisy. We estimate an admixture date of 4150 ± 850 YBP. Our standard errors computed using a block jackknife (block size of 5 cM) are uncomfortably large here.
However, this date must be treated with great caution. We obtained a data set from the Illumina iControl database (http://www.illumina.com/science/icontroldb.ilmn) of “Caucasians” and after curation have 1232 samples of European ancestry genotyped on an Illumina SNP array panel. We merged the data with the HGDP Illumina 650Y genotype data obtaining a data set with 561,268 SNPs. Applying rolloff to this sample with HGDP Karitiana and Sardinians as sources, we get a much more recent date of 2200 ± 762 YBP. We think that this is not a technical problem with rolloff, but rather, it is an issue of interpretation that is a challenge for all methods for estimating dates of admixture events.
Our admixture signal is stronger in northern Europe as we showed above in the context of discussing the statistic D(San, Karitiana; French, Italian). It seems plausible that the initial admixture might have been exclusively in northern Europe, but since this ancient event, there has been extensive gene flow within Europe, as shown, for example, in Lao et al. (2008) and Novembre et al. (2008). But if northern and southern Europe have differing amounts of “Asian” admixture, this intra-European flow is confounding to our analysis. The more recent gene flow between northern and southern Europe will contribute to our inferring too recent a date. Admixture into one section of a population, followed by slow mixing within the population, may be quite common in human history and will substantially complicate the dating for any genetic method.
Interpretation in light of ancient DNA
Ancient DNA studies have documented a clean break between the genetic structure of the Mesolithic hunter–gatherers of Europe and the Neolithic first farmers who followed them. Mitochondrial analyses have shown that the first farmers in central Europe, belonging to the linear pottery culture (LBK), were genetically strongly differentiated from European hunter–gatherers (Bramanti et al. 2009), with an affinity to present-day Near Eastern and Anatolian populations (Haak et al. 2010). More recently, new insight has come from analysis of ancient nuclear DNA from three hunter–gatherers and one Neolithic farmer who lived roughly contemporaneously at about 5000 YBP in what is now Sweden (Skoglund et al. 2012). The farmer’s DNA shows a signal of genetic relatedness to Sardinians that is not present in the hunter–gatherers who have much more relatedness to present-day northern Europeans. These findings suggest that the arrival of agriculture in Europe involved massive movements of genes (not just culture) from the Near East to Europe and that people descending from the Near Eastern migrants initially reached as far north as Sweden with little mixing with the hunter–gatherers they encountered. However, the fact that today, northern Europeans have a strong signal of admixture of these two groups, as proven by this study and consistent with the findings of (Skoglund et al. 2012), indicates that these two ancestral groups subsequently mixed.
Combining the ancient DNA evidence with our results, we hypothesize that agriculturalists with genetic ancestry close to modern Sardinians immigrated into all parts of Europe along with the spread of agriculture. In Sardinia, the Basque country, and perhaps other parts of southern Europe they largely replaced the indigenous Mesolithic populations, explaining why we observe no signal of admixture in Sardinians today to the limits of our resolution. In contrast, the migrants did not replace the indigenous populations in northern Europe and instead lived side-by-side with them, admixing over time (perhaps over thousands of years). Such a scenario would explain why northern European populations today are admixed and also have a rolloff admixture date that is substantially more recent than the initial arrival of agriculture in northern Europe.
An alternative history that could produce the signal of Asian-related admixture in northern Europeans is admixture from steppe herders speaking Indo-European languages, who after domesticating the horse would have had a military and technological advantage over agriculturalists (Anthony 2007). However, this hypothesis cannot explain the ancient DNA result that northern Europeans today appear admixed between populations related to Neolithic and Mesolithic Europeans (Skoglund et al. 2012), and so even if the steppe hypothesis has some truth, it can explain only part of the data.
We show an admixture graph that corresponds to our hypothesis in Figure 9.
To test the predictions of our hypothesized historical scenario, we downloaded the recently published DNA sequence of the Tyrolean “Iceman” (Keller et al. 2012). The Iceman lived (and died) in the Tyrolean Alps close to the border of modern Austria and Italy. From isotopic analysis (Muller et al. 2003) he was probably born within 60 miles of the site at which he was found. To analyze the Iceman data, we applied similar filtering steps as those applied in the analysis of the Neandertal genome (Green et al. 2010). After filtering on map quality and sequence quality of a base as described in that study, we chose a random read covering each base of the Affymetrix Human Origins array. This produced nearly 590,000 sites for analysis.
Our D-statistic analysis suggests that the Iceman and the HGDP Sardinians are consistent with being a clade, providing formal support for the findings of Keller et al. (2012) who reported that the Iceman is close genetically to modern Sardinians based on PCA. Concretely, our test for whether they are a clade is
| (10) |
This D-statistic shows no significant deviation from zero, in contrast with the highly significant evidence that the Iceman and French are not a clade:
Our failure to detect a signal of admixture using the D-statistic is not due to reduced power on account of having only one sample, since when we recompute the statistic of (10) using each of the 26 French individuals in turn in place of Iceman, the Z-scores are all significant, ranging from −3.1 to −8.5. These results imply that Iceman has less northeast Asian-related ancestry than a typical modern North European, but the data are consistent with Iceman having the same amount of northeast Asian-related ancestry as Sardinians. Further confirmation for this interpretation comes from the very similar magnitude f3-statistics that we observe when using either Sardinians or Iceman as a source for the admixture:
The Z-score for Iceman is of smaller magnitude than that for the Sardinian samples, because with a single individual we have much more sampling noise. However, the important quantity in this context is the magnitude of the f3-statistic. Thus the Iceman harbors less northeast Asian-related genetic material than modern French, and the northeast Asian-related genetic material is not detectably different in Iceman and the HGDP Sardinians, to the limits of our resolution.
A caveat to these analyses is that the relatively poor quality and highly fragmented DNA sequence fragments from Iceman may occasionally align incorrectly to the reference human genome sequence (and in particular, may do so at a rate higher than that of the comparison data from present-day humans), which could in theory bias the D-statistics. However, our point here is simply that to the limits of the analyses we have been able to carry out, Iceman and modern Sardinians are consistent with forming a clade, supporting the hypothesis we sketched out above.
Although the Iceman lived near where he was found, it cannot be logically excluded that his genetic ancestry was unusual for the region. For instance, his parents might have been migrants from ancient Sardinia. However, the Iceman does not carry the signal of northeast Asian ancestry that we have detected in northern Europeans, and lived at least 2000 years after the arrival of farming in Europe. If his genome was typical of the region in which he lived, the northeast Asian-related genetic material that is currently widespread in northern Italy and southern Austria must be due to admixture events and/or migrations that occurred well after the advent of agriculture in the region, supporting the hypothesis, presented above, that Neolithic farmers of near eastern origin initially largely replaced the indigenous Mesolithic population of southern Europe and that only well afterward did they develop the signal of major admixture that they harbor today.
Summary of inferences about European history from our methods
Our methods for analyzing genetic data have led to several novel inferences about history, showing the power of the approaches. In particular, we have presented evidence suggesting that the genetic history of Europe from around 5000 B.C. includes:
-
1.
the arrival of Neolithic farmers probably from the Middle East,
-
2.
nearly complete replacement of the indigenous Mesolithic southern European populations by Neolithic migrants and admixture between the Neolithic farmers and the indigenous Europeans in the north,
-
3.
substantial population movement into Spain occurring around the same time as the archeologically attested Bell-Beaker phenomenon (Harrison 1980),
-
4.
subsequent mating between peoples of neighboring regions, resulting in isolation-by-distance (Lao et al. 2008; Novembre et al. 2008). This tended to smooth out population structure that existed 4000 years ago.
Further, the populations of Sardinia and the Basque country today have been substantially less influenced by these events.
Software
We release a software package, ADMIXTOOLS, that implements five methods: the three-population test, D-statistics, F4-ratio estimation, admixture graph fitting, and rolloff. In addition, it computes lower and upper bounds on admixture proportions based on f3-statistics. ADMIXTOOLS can be downloaded from the following URL: http://genetics.med.harvard.edu/reich/Reich_Lab/Software.html.
Data sets used
The following data sets are used:
HapMap Phase 3 (International Hapmap 3 Consortium 2010), HGDP genotyped on the Illumina 650K array (Li et al. 2008), HGDP genotyped on the Affymetrix Human Origins array, POPRES (Nelson et al. 2008), Siberian data (Hancock et al. 2011), and Xhosa data (Patterson et al. 2010).
Supplementary Material
Acknowledgments
We are grateful to Mark Achtman, David Anthony, Vanessa Hayes, and Mike McCormick for instructive and helpful conversations, Mark Daly for a useful technical suggestion, and Thomas Huffman for references on the history of the Nguni. Joe Felsenstein made us aware of some references we would otherwise have missed. Wolfgang Haak corrected some of our misinterpretations of the Bell-Beaker culture and shared some valuable references. We thank Anna Di Rienzo for early access to the data of (Hancock et al. 2011) from peoples of Siberia. We thank Graham Coop, Rasmus Nielsen, and several anonymous referees whose reading of the manuscript allowed us to make numerous improvements and clarifications. This work was supported by U.S. National Science Foundation HOMINID grant 1032255, and by National Institutes of Health grant GM100233.
Appendix A: Unbiased Estimates of f-Statistics
Fix a marker (SNP) for now. We have populations A, B, C, D in which the variant allele frequencies are a′, b′, c′, d′, respectively. Sample counts of the variant and reference alleles are nA, n′A, etc. Set
so that sA is the total number of alleles observed in population A. Define a = nA/sA, the sample allele frequency in A, with b, c, d defined similarly. Thus a′, b′, c′, d′ are population frequencies and a, b, c, d are allele frequencies in a finite sample. We first define
so that 2hA is the heterozygosity of population A. Set
| . |
Then is an unbiased estimator of hA. We now can show that
are unbiased estimates of F2(A, B), F3(C; A, B), and F4(A, B; C, D), respectively. For completeness we give estimates in the same spirit for Fst(A, B). We define
which we note differs from the definition of Cavalli-Sforza in his magisterial book Cavalli-Sforza et al. (1994), and (at least in the case of unequal sample sizes) the definition in Weir and Cockerham (1984).
Write N, D for the numerator and denominator of the above expression. Then N = F2(A, B), and we have already given an unbiased estimator. We can write D = N + hA + hB and so an unbiased estimator for D is
This definition and these estimators were used in Reich et al. (2009) and are implemented in our widely used program smartpca Patterson et al. (2006). An article in preparation explores Fst in much greater detail.
Appendix B: Visual Interpretation of f-Statistics
The expected value of f-statistics can be computed in a visually interpretable way by writing down all the possible genetic drift paths through the admixture graph relating the populations involved in the f-statistic. For each of the statistics we compute
F2(A, C): Overlap between the genetic drift paths A → C, A → C
F3(C; A, B): Overlap between the genetic drift paths C → A, C → B
F4(A, E; D, C): Overlap between the genetic drift paths A → E, D → C
If there is no admixture, then the expected value of an f-statistic can be computed from the overlap of the two drift paths in the single phylogenetic tree relating the populations. If admixture occurred, the drift can take alternative paths, and we need to write down trees corresponding to each of the possible paths and weight their contribution by the probability that the drifts take that path.
There is a loose analogy here to the Feynman diagrams (Kotikov 1991a,b), used by particle physicists to perform computations about the strength of the interaction among fundamental particles such as quarks and photons. The Feynman diagrams correspond exactly to the terms of a mathematical equation (a path integral) and provide a way to compute its value. Each corresponds to a different path by which particles can interact. By writing down all possible Feynman diagrams relating two particles (all possible ways that they can interact through intermediate particles), computing the contribution to the integral from each Feynman diagram, and combining the results, one can compute the strength of the interaction.
Figure 2 shows how this strategy can be used to obtain expected values for f2-, f3-, and f4-statistics. The material below is meant to be read in conjunction with that figure:
The expected value of f2(C, A) can be computed by the overlaps of the genetic drifts C → A, C → A over all four possible paths in the tree with weights α2, αβ, βα, and β2. The expected values can be counterintuitive. For example, Neandertal gene flow into non-Africans has most probably reduced rather than increased allelic frequency differentiation between Africans and non-Africans. If A is Yoruba, C is French, and B is Neandertal, and we set a = 0.026, c = 0.036, d = 0.068, e + f + g = 0.33, α = 0.975 (reasonable parameter values based on previous work), then we compute the expected value of f2(C, A) to be 0.127. Using the same equation but α = 1 (no Neandertal admixture), we get f2 = 0.130:
| . |
If population C is admixed, there is a negative term in the expected value of f3(C; A, B), which arises because the genetic drift paths C → A and C → B can take opposite directions through the deepest part of the tree. The observation of a negative value provides unambiguous evidence of population mixture in the history of population C:
The expected value of f4(A, E; D, C) can be computed from the overlap of drifts A → E and D → C. Here there are two possible paths for D → C, with weights α and β, resulting in two graphs whose expected contribution to f4 are 0 and −αg so that E[f4] = −αg. Thus, by taking the ratio of the f4-statistics for a population that is admixed and one where α is equal to 1, we have an estimate of α.
Appendix C: Mathematical Analysis of F3
In the article we use a′ for population allele frequencies in a population A and a for sample frequencies. Here we switch notation and write a, b, c, …, for population frequencies in A, B, C, ….
We consider three populations A, B, C with a root population R, and consider F3 = E[(c − a)(c − b)] under various ascertainment schemes.
Theorem 1. Assuming that genetic drift is neutral, no backmutation, and no recurrent mutations and that A, B, C have a simple phylogeny, with no mixing events, then under the following ascertainments,
-
1.
no ascertainment, such as in sequence data,
-
2.
ascertainment in an outgroup, which split from R more remotely than A, B, C,
-
3.
ascertainment by finding a heterozygote in a single individual of {A, B, C}, where we also assume the population of R is in mutation-drift equilibrium so that the probability that a polymorphic derived allele with population frequency r ∝ 1/r Ewens (1963).
Proof. The first two cases are clear, since drift on edges of the tree rooted at R are orthogonal. This is the situation discussed at length in the main article. The case where we ascertain that a heterozygote is more complicated and our discussion involves some substantial algebra, which we carried out with Maple.
First consider the tree shown in Figure C1A. Here we show drift distances on the diffusion scale for R → X, X → A, X → C. So, for example, the probability that two random alleles of A have a most recent common ancestor (MRCA) more ancient than X is . We let allele frequencies in A, B, C, X, R be a, b, c, x, r, respectively. If we ascertain in C, then E[r − a] = E[r − b] = 0, and E[(r − a)(r − b)] = E[(r − x)2] ≥ 0. The case of ascertainment in A is more complex: Write E0 for the expectation simply assuming R is polymorphic and in mutation-drift equilibrium. Then E[(c − a)(c − b)] under ascertainment of a heterozygote in A is given by
| (A1) |
Thus it is necessary and sufficient to show E0[(c − a)(c − b)a(1 − a)] ≥ 0:
So it is enough to prove E[(r − a)(r − b)] ≥ 0. But
Let K(p, q; τ) be the transition function of the Wright–Fisher diffusion so that for 0 < p, q < 1
where X(τ) is the allele frequency at time τ on the diffusion time scale.
We make extensive use of Kimura’s theorem giving an explicit representation of K.
Theorem 2 (Kimura 1955).
| (A2) |
where Ji are explicit polynomials (Jacobi or Gegenbauer polynomials) orthogonal on the unit interval with respect to the function w(x) = x(1 − x). Numi are normalization constants with
and λ(i) is given by
| (A3) |
We need to show that
We deal with polynomials in . To simplify the notation set,
Using Kimura’s theorem and the orthogonality of Jacobi polynomials, this integral can be expressed in closed form.
We consider ascertainment of a heterozygote in A. Now calculation shows that
where Q = 5 + 3v2 + u(5 + 3v2) − 2v2(u2 + u3 + u4).
Noting that 0 ≤ v, u ≤ 1,
Next consider the tree shown in Figure C1B. First suppose we ascertain a heterozygote in A,
and so we want to show
A similar calculation to that above shows that
as required. Next suppose we ascertain a heterozygote in C. We now want to show
We find
where
We need to show Q ≥ 0. Expanding Q into monomials with coefficients ±1 there are six negative terms, each of which can be paired with a positive term of lower degree.
This completes the proof.
Summarizing, our three-population test is rigorous if there is ascertainment in an outgroup only (or no ascertainment as in sequence data). It also is rigorous with a variety of other simple ascertainments. Further in practice, on commercial SNP arrays, highly significant false positives do not seem to arise as we show in Table 5.
Appendix D: Simulations to Test f-Statistic Methodology
To test the robustness of our f-statistic methodology, we carried out coalescent simulations of five populations related according to Figure 4, using ms (Hudson 2002).
Our simulations involved specifying six dates:
-
1.
tadmix: Date of admixture between populations B′ and C′.
-
2.
tBB′: Date of divergence of populations B and B′.
-
3.
tCC′: Date of divergence of populations C and C′.
-
4.
tABB′: Date of divergence of population A from the B, B′ clade.
-
5.
tABB′CC′: Date of divergence of the A, B, B′ and C, C′ clades.
-
6.
tO: Date of divergence of the A, B, B′, C, C′ clade and the outgroup O.
We assumed that all populations were constant in size in the periods between when they split, with the following diploid sizes:
-
1.
Nx: Size in the ancestry of population X.
-
2.
NB′: Size in the ancestry of population B′’.
-
3.
NB: Size in the ancestry of population B.
-
4.
NC′: Size in the ancestry of population C′.
-
5.
NC: Size in the ancestry of population C.
-
6.
NO: Size in the recent ancestry of the outgroup O.
-
7.
NBB′: Size in the common ancestry of B and B′.
-
8.
NCC′: Size in the common ancestry of C and C′.
-
9.
NABB′: Size in the common ancestry of A, B, and B′.
-
10.
NABB′CC′: Size in the common ancestry of A, B, B′, C, and C′.
-
11.
NABB′CC′O: Size in the common ancestry of all populations.
We picked population sizes, times, and Fst to approximately match empirical data for
A: Adygei, West Eurasian
B: French, West Eurasian
C: Han, East Asian
X: Uygur, Admixed
Y: Yoruba, Outgroup
Thus, our baseline simulations correspond to a roughly plausible scenario for some of the genetic history of Eurasia, with Yoruba serving as an outgroup. We then varied parameters, as well as ascertainments of SNPs, and explored how this affected the observed values from simulation.
In Table 1 we show baseline demographic parameters, as well as several alternatives that each involved varying a single parameter compared with the baseline. Each alternate parameter set was separately assessed by simulation (including different SNP ascertainments).
Table 1 shows the results. We find that:
Fst-statistics change as expected depending on SNP ascertainment and demographic history.
The consistency of D-statistics with 0 in the absence of admixture is robust to SNP ascertainment. Substantially nonzero values are observed only when the test population is admixed (X) and not when it is unadmixed (B).
f3-statistics are negative when the test population is admixed (X) except for high population-specific drift, which masks the signal as expected. Statistics are always positive when the test population is unadmixed (B), regardless of ascertainment.
Thus, these simulations show that inferences about history based on the f-statistics are robust to ascertainment process as we argued in the main text on theoretical grounds.
Footnotes
Communicating editor: R. Nielsen
There is no completely satisfactory term for the ‘Khoisan’ peoples of southern Africa; see Barnard (1992, introduction) for a sensitive discussion. We prefer ‘Bushmen’ following Barnard. However, the standard name for the HGDP Bushmen sample is ‘San’ in the genetic literature [for example Cann et al. (2002)], and we use this specifically to refer to these samples.
Literature Cited
- Alexander D. H., Novembre J., Lange K., 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19: 1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthony D. W., 2007. The Horse, the Wheel, and Language: How Bronze-Age Riders from the Eurasian Steppes Shaped the Modern World, Princeton University Press, Princeton, NJ. [Google Scholar]
- Barnard A., 1992. Hunters and Herders of Southern Africa. A comparative ethnography of the Khoisan peoples, Cambridge University Press, Cambridge, UK. [Google Scholar]
- Beerli P., Felsenstein J., 2001. Maximum likelihood estimation of a migration matrix and effective population sizes in n subpopulations by using a coalescent approach. Proc. Natl. Acad. Sci. USA 98: 4563–4568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bramanti B., Thomas M. G., Haak W., Unterlaender M., Jores P., et al. 2009. Genetic discontinuity between local hunter-gatherers and central Europe’s first farmers. Science 326: 137–140. [DOI] [PubMed] [Google Scholar]
- Brisbin A., 2010. Linkage analysis for categorical traits and ancestry assignment in admixed individuals, Cornell University, Ithaca. [Google Scholar]
- Busing F., Meijer E., van der Leeden R., 1999. Delete-m jackknife for unequal m. Stat. Comput. 9: 3–8. [Google Scholar]
- Cann H., de Toma C., Cazes L., Legrand M., Morel V., et al. 2002. A human genome diversity cell line panel. Science 296: 261–262. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L. L., Edwards A. W., 1967. Phylogenetic analysis. Models and estimation procedures. Am. J. Hum. Genet. 19: 233–257. [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza L., Menozzi P., Piazza A., 1994. The History and Geography of Human Genes, Princeton University Press, Princeton, NJ. [Google Scholar]
- Chen G., Marjoram P., Wall J., 2009. Fast and flexible simulation of DNA sequence data. Genome Res. 19: 136–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corander J., Marttinen P., 2006. Bayesian identification of admixture events using multilocus molecular markers. Mol. Ecol. 15: 2833–2843. [DOI] [PubMed] [Google Scholar]
- Czebreszuk J., 2003 Bell beakers from west to east, pp. 476–485, Vol. 8000. Ancient Europe 8000 B.C. - A.D 1000: An encyclopedia of the barbarian world, Vol. 2, edited by P. I. Bogucki and P. J. Crabtree. Charles Scribner’s Sons, New York. [Google Scholar]
- Dasmahapatra K. K., Walters J. R., Briscoe A. D., Davey J. W., Whibley A., et al. 2012. Butterfly genome reveals promiscuous exchange of mimicry adaptations among species. Nature 487: 94–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand E. Y., Patterson N., Reich D., Slatkin M., 2011. Testing for ancient admixture between closely related populations. Mol. Biol. Evol. 28: 2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens W., 1963. The diffusion equation and a pseudo-distribution in genetics. J. R. Stat. Soc., B 25: 405–412. [Google Scholar]
- Falush D., Stephens M., Pritchard J., 2003. Inference of population structure using multilocus genotype data: Linked loci, and correlated allele frequencies. Genetics 164: 1567–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenner J. N., 2005. Cross-cultural estimation of the human generation interval for use in genetics-based population divergence studies. Am. J. Phys. Anthropol. 128: 415–423. [DOI] [PubMed] [Google Scholar]
- Green R. E., Krause J., Briggs A. W., Maricic T., Stenzel U., et al. 2010. A draft sequence of the Neandertal genome. Science 328: 710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W., Balanovsky O., Sanchez J. J., Koshel S., Zaporozhchenko V., et al. 2010. Ancient DNA from European early neolithic farmers reveals their near eastern affinities. PLoS Biol. 8: e1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock A. M., Witonsky D. B., Alkorta-Aranburu G., Beall C. M., Gebremedhin A., et al. 2011. Adaptations to climate-mediated selective pressures in humans. PLoS Genet. 7: e1001375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison R. J., 1980. The Beaker Folk. Thames & Hudson, London. [Google Scholar]
- Hudson R. R., 2002. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics 18: 337–338. [DOI] [PubMed] [Google Scholar]
- Huffman, T., 2010 Prehistory of the Durban area. http://www.sahistory.org.za/durban/prehistory-durban-area.
- Huffman T. N., 2004. The archaeology of the Nguni past. Southern African Humanities 16: 79–111. [Google Scholar]
- International Hapmap 3 Consortium, 2010. Integrating common and rare genetic variation in diverse human populations. Nature 467: 52–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keinan A., Mullikin J., Patterson N., Reich D., 2007. Measurement of the human allele frequency spectrum demonstrates greater genetic drift in East Asians than in Europeans. Nat. Genet. 39: 1251–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A., Graefen A., Ball M., Matzas M., Boisguerin V., et al. 2012. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nature Communications 3: 698. [DOI] [PubMed] [Google Scholar]
- Kimura M., 1955. Solution of a process of random genetic drift with a continuous model. Proc. Natl. Acad. Sci. USA 41: 144–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotikov A., 1991a Differential equation method. the calculation of N-point Feynman diagrams. Phys. Lett. B 267: 123–127. [Google Scholar]
- Kotikov A., 1991b Differential equations method: the calculation of vertex-type Feynman diagrams. Phys. Lett. B 259: 314–322. [Google Scholar]
- Lao O., Lu T., Nothnagel M., Junge O., Freitag-Wolf S., et al. 2008. Correlation between genetic and geographic structure in Europe. Curr. Biol. 18: 1241–1248. [DOI] [PubMed] [Google Scholar]
- Lathrop G. M., 1982. Evolutionary trees and admixture: phylogenetic inference when some populations are hybridized. Ann. Hum. Genet. 46: 245–255. [DOI] [PubMed] [Google Scholar]
- Li J., Absher D., Tang H., Southwick A., Casto A., et al. 2008. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319: 1100–1104. [DOI] [PubMed] [Google Scholar]
- Mackerras C., 1972. The Uighur Empire According to the Tang Dynastic Histories, Australian National University Press, Canberra. [Google Scholar]
- Mao X., Bigham A. W., Mei R., Gutierrez G., Weiss K. M., et al. 2007. A genomewide admixture mapping panel for Hispanic/Latino populations. Am. J. Hum. Genet. 80: 1171–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills R. E., Walter K., Stewart C., Handsaker R. E., Chen K., et al. 2011. Mapping copy number variation by population-scale genome sequencing. Nature 470: 59–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moorjani P., Patterson N., Hirschhorn J. N., Keinan A., Hao L., et al. 2011. The history of African gene flow into Southern Europeans, Levantines, and Jews. PLoS Genet. 7: e1001373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller W., Fricke H., Halliday A. N., McCulloch M. T., Wartho J. A., 2003. Origin and migration of the Alpine Iceman. Science 302: 862–866. [DOI] [PubMed] [Google Scholar]
- Nei M., 1987. Molecular evolutionary genetics, Columbia University Press, New York. [Google Scholar]
- Nelson M. R., Bryc K., King K. S., Indap A., Boyko A. R., et al. 2008. The Population Reference Sample, POPRES: a resource for population, disease, and pharmacological genetics research. Am. J. Hum. Genet. 83: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novembre J., Johnson T., Bryc K., Kutalik Z., Boyko A., et al. 2008. Genes mirror geography within Europe. Nature 456: 98–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Price A., Reich D., 2006. Population Structure and Eigenanalysis. PLoS Genet. 2: e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N., Petersen D. C., van der Ross R. E., Sudoyo H., Glashoff R. H., et al. 2010. Genetic structure of a unique admixed population: implications for medical research. Hum. Mol. Genet. 19: 411–419. [DOI] [PubMed] [Google Scholar]
- Pickrell J., Pritchard J., 2012. Inference of population splits and mixtures from genome-wide allele frequency data PLoS Genetics (in press). [Google Scholar]
- Pool J. E., Nielsen R., 2009. Inference of historical changes in migration rate from the lengths of migrant tracts. Genetics 181: 711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price A. L., Tandon A., Patterson N., Barnes K. C., Rafaels N., et al. 2009. Sensitive detection of chromosomal segments of distinct ancestry in admixed populations. PLoS Genet. 5: e1000519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J., Stephens M., Donnelly P., 2000. Inference of population structure using multilocus genotype data. Genetics 155: 945–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D., Thangaraj K., Patterson N., Price A. L., Singh L., 2009. Reconstructing Indian population history. Nature 461: 489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D., Green R. E., Kircher M., Krause J., Patterson N., et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature 468: 1053–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D., Patterson N., Kircher M., Delfin F., Nandineni M. R., et al. 2011. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am. J. Hum. Genet. 89: 516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg N., 2006. Standardized subsets of the HGDP-CEPH Human Genome Diversity Cell Line Panel, accounting for atypical and duplicated samples and pairs of close relatives. Ann. Hum. Genet. 70: 841–847. [DOI] [PubMed] [Google Scholar]
- Sankararaman S., Sridhar S., Kimmel G., Halperin E., 2008. Estimating local ancestry in admixed populations. Am. J. Hum. Genet. 82: 290–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skoglund P., Malmstrom H., Raghavan M., Stora J., Hall P., et al. 2012. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science 336: 466–469. [DOI] [PubMed] [Google Scholar]
- Thompson E., 1975. Human Evolutionary trees, Cambridge University Press, Cambridge, UK. [Google Scholar]
- Waddell P., Penny D., 1996 Evolutionary trees of apes and humans from DNA sequences pp. 53–74 in Handbook of Human Symbolic Evolution, edited by A. Lock and C. Peter. Wiley-Blackwell, New York. [Google Scholar]
- Weir B., Cockerham C. C., 1984. Estimating f-statistics for the analysis of population structure. Evolution 38: 1358–1370. [DOI] [PubMed] [Google Scholar]
- Wollstein A., Lao O., Becker C., Brauer S., Trent R. J., et al. 2010. Demographic history of Oceania inferred from genome-wide data. Curr. Biol. 20: 1983–1992. [DOI] [PubMed] [Google Scholar]
- Xu S., Jin L., 2008. A genome-wide analysis of admixture in Uyghurs and a high-density admixture map for disease-gene discovery. Am. J. Hum. Genet. 83: 322–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Huang W., Qian J., Jin L., 2008. Analysis of genomic admixture in Uyghur and its implication in mapping strategy. Am. J. Hum. Genet. 82: 883–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.