Abstract
During the development of female mammals, one of the two X chromosomes is inactivated, serving as a dosage-compensation mechanism to equalize the expression of X-linked genes in females and males. While the choice of which X chromosome to inactivate is normally random, X chromosome inactivation can be skewed in F1 hybrid mice, as determined by alleles at the X chromosome controlling element (Xce), a locus defined genetically by Cattanach over 40 years ago. Four Xce alleles have been defined in inbred mice in order of the tendency of the X chromosome to remain active: Xcea < Xceb < Xcec < Xced. While the identity of the Xce locus remains unknown, previous efforts to map sequences responsible for the Xce effect in hybrid mice have localized the Xce to candidate regions that overlap the X chromosome inactivation center (Xic), which includes the Xist and Tsix genes. Here, we have intercrossed 129S1/SvImJ, which carries the Xcea allele, and Mus musculus castaneus EiJ, which carries the Xcec allele, to generate recombinant lines with single or double recombinant breakpoints near or within the Xce candidate region. In female progeny of 129S1/SvImJ females mated to recombinant males, we have measured the X chromosome inactivation ratio using allele-specific expression assays of genes on the X chromosome. We have identified regions, both proximal and distal to Xist/Tsix, that contribute to the choice of which X chromosome to inactivate, indicating that multiple elements on the X chromosome contribute to the Xce.
Keywords: mouse genetics, X inactivation, quantitative trait loci, Xce
IN female mammals, either one of the two X chromosomes becomes inactivated during development of the embryo. This random form of X chromosome inactivation (XCI) was first proposed by Lyon (1961) to explain the mosaic pattern of X-linked phenotypes observed in coats of various mammals. XCI serves as a dosage-compensation mechanism to equalize the expression of most X-linked genes in females and males. The steps to random XCI during development of the embryonic lineage are thought to include counting of the number of X chromosomes and the choice of which will be active or inactive, followed by initiation, spreading and finally maintenance of the inactive state throughout development (Heard et al. 1997; Wutz 2011). While choice of which X to inactivate is known to be a primary event occurring early in development, when one X chromosome carries a detrimental mutation, preferential inactivation of the X chromosome with the mutation is typically observed (Morey and Avner 2010). This form of skewed XCI is exemplified in human cells and is most likely due to a secondary cell survival effect in choice (Puck and Willard 1998; Amos-Landgraf et al. 2006). In mice, random XCI is observed in homozygous females carrying X chromosomes from the same genetic background, whereas skewed XCI can be observed when females are heterozygous for X chromosomes from different backgrounds. In contrast to the situation observed in many human females, the process of this skewed XCI in mice is considered to be a primary event in the choice of which X chromosome will remain active (Rastan 1982; Morey and Avner 2010).
Early studies on various structural anomalies of the X chromosome, including X autosome translocations (t(X;A)) in both human and mouse cells, led to the genetic identification of the X inactivation center (XIC/Xic) region (reviewed by Heard et al. 1997). The XIC/Xic was defined as the region on the X chromosome that contains the elements required for XCI. Within the XIC/Xic, the X-inactivation-specific transcript locus (XIST/Xist) was cloned—first in human (Brown et al. 1991) and then in mice (Borsani et al. 1991; Brockdorff et al. 1991). XIST/Xist encodes a long noncoding RNA that is exclusively expressed on the inactive X chromosome. Upon XCI, Xist expression is induced on the future inactive X chromosome, where Xist RNA coats the X chromosome and facilitates spreading of inactivation of genes in cis. On the future active X chromosome, Xist is silenced during XCI. In mice, Lee and colleagues identified an antisense regulator of Xist, Tsix, whose product is also a noncoding RNA (Lee et al. 1999). Tsix expression represses Xist in cis and was shown to be involved in the choice process (Lee and Lu 1999). Numerous targeting and mutation studies of Xist and Tsix have shown the requirement for Xist and Tsix expression in regulating XCI (Payer and Lee 2008). Notably, however, single-copy transgenes spanning Xist/Tsix and integrated at autosomal loci in male ES cells did not initiate XCI upon differentiation (Heard et al. 1999), suggesting that Xist and Tsix alone do not define all of the cis elements of the Xic required for XCI. Furthermore, despite the apparent requirement for Tsix in choice, the relationship between Xist/Tsix and skewing of X inactivation is not well understood.
To explain the skewed XCI detected in mice heterozygous for X chromosomes of divergent backgrounds, Cattanach proposed the presence of the X-chromosome-controlling element (Xce) (Cattanach 1970). The Xce is defined as the cis element influencing choice in XCI in mice. Thus far, four variants of the Xce locus have been described: Xcea, Xceb, Xcec, and Xced (Cattanach and Rasberry 1991). The alleles are ordered in their tendency to remain active: Xcea < Xceb < Xcec < Xced (Cattanach and Williams 1972; West and Chapman 1978; Johnston and Cattanach 1981). In heterozygous Xcea/Xcec mice, for example, the X inactivation ratio is approximately 0.25, reflecting that ∼25% of cells will have an active X chromosome with the Xcea allele and ∼75% of the cells will have an active X chromosome with the Xcec allele (Plenge et al. 2000). In contrast, in mice homozygous for Xcea/Xcea or Xcec/Xcec, where XCI is random, the X inactivation ratio is ∼0.50, reflecting that ∼50% of cells will have one X chromosome active and the other ∼50% of cells will have the other X chromosome active. It has been proposed that a hypothetical blocking factor, originally proposed to function in counting of X chromosomes, may interact at the Xce locus on the future active X where it blocks X chromosome from inactivation and thereby contributes to choice during XCI (Lyon 1971; Brown and Chandra 1973; Russell and Cacheiro 1978; Rastan 1983; Avner and Heard 2001; Percec et al. 2003). One interpretation of this model is that the Xce is defined by a discrete locus to which a trans-acting blocking factor binds and thereby blocks XCI and that allelic differences in binding affinity explain the differing activities of the Xce alleles (Percec et al. 2003). It is therefore of great interest to define the X chromosome region responsible for the Xce and the nature of the alleles that determine the Xce effect.
Mapping of Xce was initially performed in mice with an X chromosome recombinant for the Xcea and Xceb alleles. Original studies placed the Xce between the tabby (Ta) and the phosphoglycerate kinase (Pgk-1) genes (Cattanach et al. 1970, 1982, 1989; Cattanach and Papworth 1981). The Xce region was subsequently narrowed to the sequence between Ta and blotchy (Moblo) (Cattanach and Papworth 1981; Cattanach et al. 1989; Simmler et al. 1993). Further fine mapping of recombinant alleles with new microsatellite markers suggested that Xist and Xce were distinct elements (Simmler et al. 1993). Chadwick et al. (2006) refined Xce to a region <2 Mb that included the Xist/Tsix genes. The simplest interpretation of these two studies is that the Xce is a single locus within the X chromosome sequence common to both candidate Xce regions. Within the Xic, several protein-coding and noncoding genes and genetic elements have been identified and shown to affect XCI (Clerc and Avner 2003; Lee 2011; Morey and Avner 2011; Pontier and Gribnau 2011); however, none of these has been shown to contribute to the Xce effect.
Here we further analyze and map the Xce in mice carrying the 129S1/SvImJ (129S1) Xcea allele and the Mus musculus castaneus (Cast) Xcec allele (Courtier et al. 1995; Plenge et al. 2000). We generated male mice recombinant for portions of the 129S1 (Xcea) and Cast (Xcec) X chromosomes. We progeny tested these males by mating them to 129S1 females and determining the X chromosome inactivation ratio by measuring allele-specific expression of X-linked genes in female progeny. By comparing X inactivation ratios in females inheriting the recombinant alleles and control females that are heterozygous or homozygous for Xce, we identify multiple X chromosome loci that contribute to the Xce effect. We show that both sequences proximal and distal to and spanning Xist/Tsix affect skewing/choice of XCI. Our results, therefore, indicate that for the 129S1 (Xcea) and Cast (Xcec) alleles, “Xce” may be defined by multiple X chromosome strain-specific differences including differences within the Xist/Tsix region.
Materials and Methods
Mice
New X chromosome recombinant lines were generated to map Xce (Figure 1). Male and female 129S1/SvImJ (129S1) mice, with an Xcea allele, and Mus musculus castaneus EiJ (Cast) males, with an Xcec allele, were purchased from JAX. Because Cast breeding pairs are difficult to maintain, we generated mice with a Cast X chromosome (CastX) on an otherwise mixed 129S1/Cast background. 129S1 females were first mated to Cast males and then F1 female progeny were mated to Cast males. N2 progeny with a Cast X chromosome were identified. The N2 CastX females were then mated to 129S1 males, to isolate additional CastX males. These CastX males (CastXm) were mated to N2 CastX females (CastXf) to maintain the CastX mice. We also saved and progeny tested N2 progeny with a recombinant breakpoint at the very distal ends of the X chromosome (recombinant males 246m and 88m; Figure 2 and Supporting Information, Table S1). Recombinant male 246m was Cast at the proximal end of the X chromosome and otherwise 129S1. In female progeny of 246m mated to 129S1 females, Xce should be homozygous. These female progeny were then bred with 129S1 males to progeny test inheritance of recombinant allele from the mother (246f; Figure 2 and Table S1). For mapping of the Xce, we used the 129S1, Cast, and CastX mice to establish, maintain, and progeny test new recombinant lines (see below). We designated the mating schemes to generate recombinant X chromosomes as RX1 and RX2. All animal work was conducted in accordance with the Institutional Animal Care and Use Committee.
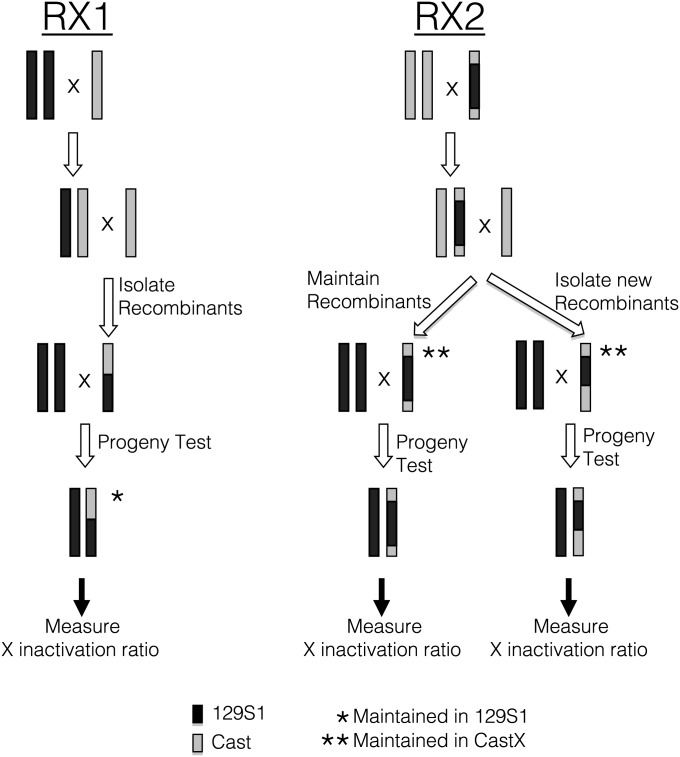
Figure 1.
RX1 and RX2 breeding schemes. In the RX1 breeding scheme, 129S1 females were first mated to Cast males and then F1 female progeny were mated to Cast or CastX males. Male progeny with a recombinant X chromosome were mated to 129S1 females for progeny testing. Tail tips were collected from 2- to 3-week-old females to measure the X inactivation ratios. Mice were bred to 129S1 mice to maintain the existing recombinant X chromosome. In the RX2 breeding scheme, CastX females were mated to a male with a double recombinant X chromosome that was Cast at distal ends and 129S1 in the middle region. Female progeny with a CastX and the double recombinant X chromosome were mated with CastX males to maintain existing and to generate new recombinant alleles. Male progeny with a recombinant X chromosome were mated with 129S1 females and toe and ear samples were collected from female progeny to measure the X inactivation ratio. To maintain existing or to generate new recombinant X chromosomes, males with the recombinant X chromosome were crossed to CastX females and then female progeny were mated with CastX males. 129S1 and Cast DNA are designated by solid and shaded bars, respectively.
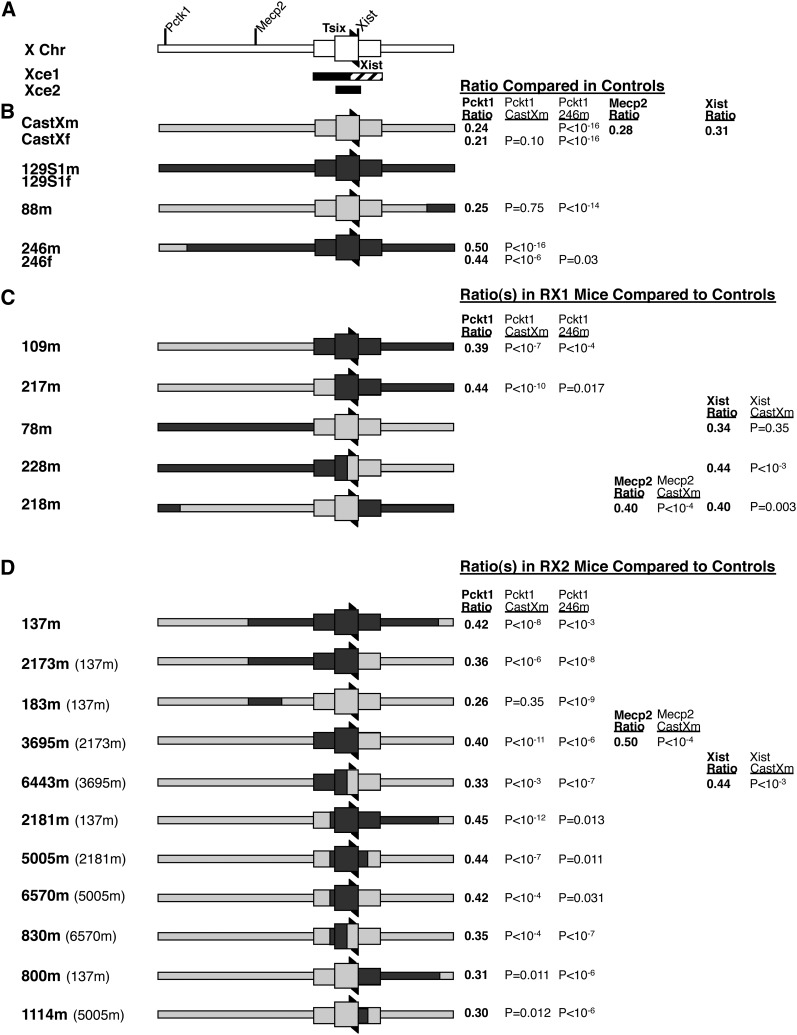
Figure 2.
Measuring the X inactivation ratios in progeny of mice with control vs. RX1 and RX2 chromosomes suggests that multiple regions comprise the Xce. (A) Schematic of X chromosome and previously defined candidate Xce regions designated as Xce1 (solid and cross-hatched bar beneath X chromosome; Simmler et al. 1993) and Xce2 (solid bar beneath X chromosome; Chadwick et al. 2006). For Xce1, the solid and cross-hatched bar refers to the initially reported candidate Xce region and the solid bar refers to the candidate Xce region based on the genotype analysis of mice phenotyped that contained Xcea and Xceb alleles (Cattanach and Williams 1972; Simmler et al. 1993). Chromosomal boundaries of Xce1, Xce2, and recombinant chromosomes analyzed below are in Table S1. Right and left arrowheads indicate orientation and location of Tsix and Xist, respectively. Genes that were used to measure the X inactivation ratio are also indicated. (B) Schematic of control chromosomes that were progeny tested. Here and below, light gray indicates Cast DNA and dark gray indicates 129S1 DNA. “m” and “f” noted after a number indicates a male or female was progeny tested, respectively. Recombinant alleles 246m and 88m were isolated during the establishment of CastX mice and 246f was derived from mating the 246m male to a 129S1 female. Female offspring from the CastXm control cross are heterozygous at the Xce whereas female progeny from the 246m control cross are homozogous at the Xce. Note that offspring from both crosses are heterozygous at Pctk1, which is used for the phenotyping (the X inactivation ratio) of Xce. Males (CastXm, 88m and 246m) were progeny tested by mating with 129S1 females. Females (CastXf and 246f) were progeny tested by mating with 129S1 males. The mean X inactivation ratio in female progeny using designated assay is reported to the right (Pctk1 and Mecp2 ratios are percentage expression from 129S1 allele; Xist ratios are percentage expression from Cast allele). The ratios were compared to ratios in progeny of control males (CastXm and 246m) and P-values (P) of the ratio comparison are also shown to the right. (C) RX1- and (D) RX2-derived recombinant X chromosomes. All males were progeny tested by mating with 129S1 females. As described in B, the mean X inactivation ratio(s) and significance (P) are reported to the right. When the ratio differs (P < 0.05) from the CastXm progeny ratio, this indicates that Xce of the RX1/RX2 recombinant allele is no longer fully heterozygous (i.e., there is 129S1 sequence in the Xce region on the RX1/RX2 recombinant chromosome). When the X inactivation ratio differs from the 246m progeny ratio (homozygous Xce), this indicates that Xce regions are within the Cast sequence of the RX1 or RX2 derived alleles (i.e., Xce is at least partially heterozygous). In the allele designation to the left, the labels in parentheses refer to the grandparental allele.
RX1 mating scheme
129S1 females were mated to Cast males. F1 females were mated to Cast or CastX males to isolate males with recombinant X. These males were progeny tested by breeding with 129S1 females and measuring the X inactivation ratio in female progeny. The RX1 female progeny were also bred to 129S1 males to maintain the RX1 allele. The breeding scheme is illustrated in Figure 1. The RX1 recombinant X chromosomes are summarized in Table S1. Each name corresponds to the mouse in which the recombinant chromosome was initially identified.
RX2 mating scheme
The founding double recombinant male was the offspring of a mating between a CastX male and a mixed background female heterozygous for Cast and 129S1. This recombinant male was mated to a Cast X female. Subsequent female progeny were then mated to CastX males and the original X chromosome recombinant was maintained, or new recombinants were identified that mapped near the Xce boundaries identified by Chadwick et al. (2006). Recombinant males generated by the RX2 breeding scheme were progeny tested by mating to 129S1 females. Mice with new and existing recombinant X chromosomes were established or maintained by crossing to CastX mice. The breeding scheme is illustrated in Figure 1. The RX2 recombinant X chromosomes are summarized in Table S1. Each name corresponds to the mouse in which the recombinant chromosome was initially identified.
Progeny test breeding
RX1- and RX2-derived male mice were mated to 129S1 females. Tissues from 2- to 3-week old female progeny were collected for measuring the X inactivation ratio, which we previously referred to as the X inactivation pattern (Plenge et al. 2000; Percec et al. 2003). The X inactivation ratio was calculated as the fraction of RNA expressed from the 129S1 (or Cast) allele relative to the total level of RNA expressed from the 129S1 and Cast alleles for the designated X-linked genes. From progeny of RX1-derived males, tail tips were collected for RNA analysis and ear clips for DNA analysis. From killed progeny of RX2-derived males, toe and ear samples were collected for RNA analysis and toe samples for DNA analysis, as previously described (Percec et al. 2003).
RNA isolation
From 2- to 3-week old mice, toe and ear samples or tail samples were collected and stored at −80° for RNA isolation. Tissues were first pulverized with a pestle in an Eppendorf tube on dry ice/ethanol bath and then RNA was isolated using the High Pure RNA tissue kit (Roche) as previously described (Percec et al. 2003). Toe and ear (TE) RNAs were eluted in 90 μl of kit elution buffer (EB) and tail (t) RNAs were eluted in 50 μl of EB. RNA concentrations were measured on the NanoDrop (Thermo Scientific). cDNA was synthesized from 500 ng RNA using M-MLV RT (Invitrogen) in a 20-μl reaction as previously described (Percec et al. 2003).
Allele-specific expression assays
The Pctk1 (renamed Cdk16 for cyclin-dependent kinase 16) expression assay was conducted as previously described (Percec et al. 2002 and Figure S1). For all other assays, 0.5 μM of each primer and 1 μl of cDNA were added to RubyTaq PCR master mix per manufacturer’s guidelines (Affymetrix). Allele-specific expression assays for Mecp2, Xist, Hprt, Abc7 (current name Abcb7), and Jarid1c (current name Kdm5c) were designed. Table S2 provides the sequence and location of primers, SNPs assayed, annealing conditions for PCR reaction, and enzyme used to detect the SNP within PCR product. Digested PCR products were resolved on a 12% polyacrylamide gel.
Mecp2 and Xist were used for allele-specific expression in addition to genotyping of recombinant alleles (Figure S1). The Hprt, Abc7, and Jarid1c assays were used for genotyping of recombinant alleles.
DNA isolation and genotyping
DNA was extracted from tail, toe, or ear samples of 2- to 3-week old mice, as previously described (Percec et al. 2002). Supernatant was stored at –20°. Genotyping was carried out using X chromosome microsatellite markers and SNPs listed in Table S1 and Table S2. (All chromosomal locations are in accordance with the UCSC genome browser July 2007 (NCBI37/mm9) assembly.) We initially genotyped the X chromosome with DXMit53, DXMit62, DXMit18, DXMit64, and DXMit249 map pairs and then fine mapped recombinant alleles as shown in Table S1. Table S2 lists SNP and restriction fragment length polymorphism (RFLP) assays. SNPs used were originally identified by Perlegen (presently curated at http://www.ncbi.nlm.nih.gov/projects/SNP/) or identified by sequencing of genomic Cast and 129S1 amplified DNA. For genotyping PCR, 1 μl of supernatant and 0.5 μM of each primer were added to GoTaq Green PCR master mix according to manufacturer’s recommendations (Promega). After an initial denaturing step at 94° for 2 min, amplification was performed for 35 cycles at 94° for 15 sec, 57° for 15 sec, and 72° for 20 sec. For SNP analysis, PCR products were either sequenced or digested with appropriate restriction enzyme for RFLP analysis. The PCR products were resolved on a 12% polyacrylamide gel.
Statistical analysis
Mean and standard deviation of X inactivation ratios were determined and graphically illustrated using Microsoft Excel data analysis tools. Assuming the null hypothesis, the difference of two means was determined using a two-tail t-test assuming unequal variances.
Preserving CastX, RX1, and RX2 lines
We maintained the CastX mice for subsequent studies, but it was not feasible to maintain the RX1 and RX2 mice. We did, however, cryopreserve sperm isolated from many of the RX1 and RX2 mice. Using these sperm, mice with the recombinant RX1 X chromosome can most readily be rederived by intracytoplasmic sperm injection (ICSI) of 129S1 oocytes and mice with the recombinant RX2 X chromosome can most readily be rederived by ICSI of CastX oocytes. In addition, we have isolated early passage female mouse embryonic fibroblasts (MEFs) from 12.5 to 14.5 days post coitum embryos from 129S1 females crossed to CastX, RX1, or RX2 males. These MEFs could be used to derive induced pluripotent stem cell (iPSC) lines that may be used to study initial steps of random XCI, provided iPSC clones become fully reprogrammed and are capable of undergoing random XCI upon differentiation.
Results
Strategy for defining Xce
To define more precisely the genetic location of Xce, we generated new mouse strains with recombinant X chromosomes. Previous studies have shown that Xce is linked to Xist/Tsix sequences within or near the Xic (Simmler et al. 1993; Chadwick et al. 2006). Figure 2A and Table S1 show previously defined Xce candidate regions. However, one study has suggested that X chromosome sequences proximal to the Xist/Tsix region may contribute to the Xce effect (Simmler et al. 1993). These studies typically employed mice with a single recombination along the X chromosome to progeny test and define boundaries of the Xce candidate region by QTL analysis (Simmler et al. 1993). Here, we have used mice with Cast (Xcec allele) and 129S1 (Xcea allele) X chromosomes to generate X chromosome alleles with single or double recombination breakpoints. We mapped the breakpoint(s) using microsatellite markers and sequencing. The breakpoint(s) of several of the recombinant X chromosomes coincided with the previously proposed proximal or distal Xce candidate region boundaries (Cattanach and Papworth 1981; Cattanach et al. 1991; Simmler et al. 1993; Chadwick et al. 2006). We progeny tested males with the recombinant X chromosomes by mating them to 129S1 females and assessing the X inactivation ratio in female progeny, measuring allele-specific expression analysis of at least one gene on the X chromosome that undergoes XCI. Furthermore, we compared these ratios to X inactivation ratios in progeny of control mice in which Xce is either heterozygous (ratio ∼0.25) or homozygous (ratio ∼0.50) (Plenge et al. 2000).
In summary, we progeny tested numerous males with different recombinant X chromosomes that are described in Figure 2 and testing of several of the recombinants indicate that the Xce is a dispersed element. The most direct evidence that regions proximal to Xist/Tsix contribute to the Xce effect is concluded from the analysis of the 217m, 228m, 6443m, 830m recombinant males. The most direct evidence that regions distal to Xist/Tsix contribute to the Xce effect is concluded from the analysis of the 218m, 800m, and 1114m recombinant males.
Establishing mouse lines to map Xce using mice with Xcea and Xcec alleles
Using 129S1 and Cast mice, we generated control and recombinant lines to identify discrete regions along the X chromosome that could define the location of sequences that contribute to the Xce QTL. Specifically, we generated males with single and double recombinant X chromosomes using two mating schemes designated as recombinant X chromosome 1 and 2 (RX1 and RX2) (Figure 1; see Materials and Methods). During the generation of the CastX mice, we also identified mice with breakpoints near the proximal and distal ends of the X chromosome, 246m and 88m, respectively (Figure 2B and Table S1). As described below, the 246m mice were especially useful for control progeny testing, by measuring the X inactivation ratio when Xce is homozygous by descent.
Analysis of control mice
Multiple factors may affect the X inactivation ratio measurement in addition to the Xce alleles. These include the direction of mating, strain background, tissue chosen for the gene expression measurements, as well as variations detected for a specific gene expression assay (Plenge et al. 2000; Percec et al. 2002; Chadwick and Willard 2005). We performed a series of control experiments to determine how these factors may affect the outcome of our progeny testing of RX1- and RX2-derived X chromosome recombinant mice. For these experiments we used the previously established Pctk1 assay to measure the X inactivation ratio ((Percec et al. 2002) and Figure S1A). This assay requires that the very proximal end of the X chromosomes is heterozygous for Cast and 129S1, in a region not likely to affect the X inactivation ratio (Figure 2A), as previously reported (Plenge et al. 2000).
For control experiments, we isolated both males (m) and females (f) with designated X chromosomes for progeny testing. Mice with the CastXm, CastXf, 88m, 246m, and 246f chromosomes were isolated from 129S1 and Cast breeding (Figure 2B). The male 199m with a CastX chromosome (Table S1) was generated from the RX1 breeding scheme. The female 2175f with CastX chromosomes (Table S1) was the offspring of a first-generation recombinant female mated to CastX male in the RX2 breeding scheme. For reasons explained below, 199m and 2175f mice were separately progeny tested from CastXm and CastXf, respectively.
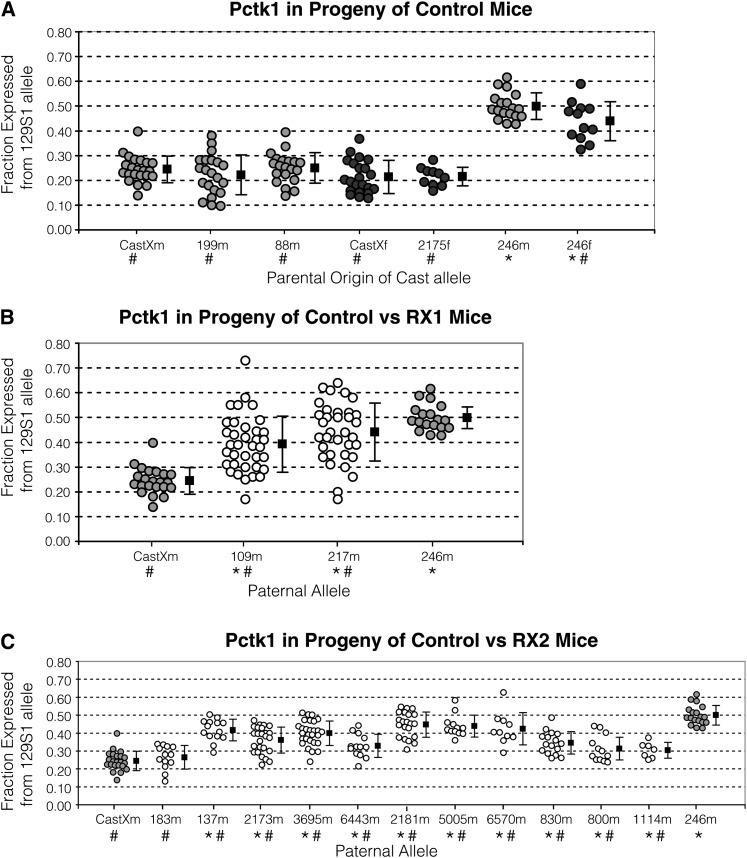
For control progeny testing experiments, we mated males with the Cast Pctk1 allele to 129S1 females, mated females with a Cast Pctk1 allele to 129S1 males, and assayed the X inactivation ratio in female progeny from each mating. First we assayed progeny in which Xce is heterozygous (progeny of CastXm, CastXf, 88m, 199m, and 2175f; Figures 2B and 3A and Table S1). The mean X inactivation ratio in progeny inheriting the CastX chromosome from the father (CastXm, ratio = 0.24, SD = 0.054) did not significantly differ from the ratio in progeny inheriting the CastX chromosome from the mother (CastXf, ratio = 0.21, SD = 0.066, P-value, P, = 0.10), suggesting that direction of mating did not affect the X inactivation ratio. The mean ratios in progeny inheriting the CastXm vs. the 88m (ratio = 0.25, SD = 0.061, P = 0.75) paternal X chromosomes did not differ, indicating that the distal 129S1 sequence on the 88m X chromosome was not contributing to the Xce QTL (Figures 2B and 3A and Table S1). The ratios in progeny inheriting the maternal X chromosome from different mating schemes did not differ (Figure 3A; compare CastXf vs. 2175f [ratio = 0.21, SD = 0.038, P = 0.93]). Using toe and ear RNA (CastXm progeny) vs. tail RNA (199m progeny [ratio = 0.22, SD = 0.081, P = 0.30]) for the analysis did not affect the X inactivation ratio. The latter two control experiments indicated that strain background differences and tissues chosen for the analysis did not affect the outcome. However, there was a greater variance in gene expression measurements when tail RNA was used for the analysis instead of toe and ear RNA (Figure 3A; compare CastXm vs. 199m). We therefore used toe and ear RNA for gene expression measurements in RX2 progeny.
Figure 3.
The X inactivation ratio as measured by allelic Pctk1 expression. The x-axis lists progeny tested mice with the designated allele described in Figure 2. Mice were mated with 129S1 mice for progeny testing. The ratio was measured in individual female progeny that are represented by circles. Shading of the circle describes the parent of progeny tested animal: control male (light gray circles); control female (dark gray circles); RX1 and RX2 derived recombinant lines (open circles). RNA isolated from tissues of 2- to 3-week mice was analyzed. The y-axis provides the ratio, measured as the fraction of total RNA that is expressed from the 129S1 allele (Xcea). To the right of each group of the ratio measurements, the corresponding mean (solid square) and standard deviation (black lines) are provided. Using a t-test of two samples assuming unequal variance, the ratios were compared to control animal with the CastXm or 246m X chromosome. Below each allele on the x-axis, ★ indicates the ratio differed from CastXm ratio and # indicates the ratio differed from the 246m ratio, according to P-value from two-tailed t-test (P-values are noted below). (A) Pctk1 expression in progeny of control mice. The mean X inactivation ratio of CastXm did not differ from 199m (P = 0.30), 88m (P = 0.75), CastXf (P = 0.10), and 2175f (P = 0.09), but it did differ from the ratio of 246m (P < 10−16) and 246f (P < 10−6). The ratio of 246m (P < 10−13) and 246f (P < 10−6) both significantly differed from ratio of all other controls. The ratio of 246m significantly differed from 246f (P = 0.033). (B) Pctk1 in progeny of control vs. RX1 mice. The ratio in CastXm progeny significantly differed from the ratio in 109m (P < 10−7) and 217m (P < 10−10) progeny. The ratio in 246m progeny also significantly differed from ratio in 109m (P < 10−4) and 217m (P = 0.017) progeny. (C) Pctk1 in progeny of control vs. RX2 mice. The ratio in CastXm and 183m progeny did not differ (P = 0.35). The ratio in CastXm significantly differed from the ratio in 137m (P < 10−8), 2173m (P < 10−6), 3695m (P < 10−11), 6443m (P < 10−3), 2181m (P < 10−12), 5005m (P < 10−7), 6570m (P < 10−4), 830m (P < 10−4), 800m (P = 0.011), and 1114m (P = 0.012) progeny. The ratio in 246m progeny significantly differed from the ratio in 137m (P < 10−3), 2173m (P < 10−8), 183m(P < 10−9), 3695m (P < 10−6), 6443m (P < 10−7), 2181m (P = 0.013), 5005m (P = 0.011), 6570m (P = 0.031), 830m (P < 10−7), 800m (P < 10−6), and 1114m (P < 10−6) progeny.
We also performed control experiments in progeny in which the Xce is homozygous (progeny of 246m and 246f; Figures 2B and 3A and Table S1), to further test if directionality of cross affects the X inactivation ratio. Although the assays to determine the Xce QTL were quite different, previous studies have found that the direction of mating either did not affect the ratio (Johnston and Cattanach 1981) or did influence the X inactivation ratios in heterozygous females (Chadwick and Willard 2005). As noted above, the direction of mating did not affect such measurements in our study of Xce heterozygous progeny. In contrast, the ratios significantly differed in Xce homozygous progeny of 246m (ratio = 0.50, SD = 0.054) and 246f (ratio = 0.44, SD = 0.079, P = 0.033) mice (Figures 2B and 3A), suggesting that the direction of the mating can affect the ratios.
The observation that the ratio is higher when the Cast Pctk1 allele is inherited from the father (246m) than when inherited from the mother (246f) suggests that there still remains some memory of the paternal mark for the imprinted XCI during random XCI (see Lee 2011 for review of imprinted and random XCI). Hence, we progeny tested all of the RX1- and RX2-derived mice in one direction: recombinant males crossed to 129S1 females. For all analyses we compared X inactivation ratios to the heterozygous Xce (CastXm) and, when possible, to Xce homozygous (246m) offspring.
Progeny testing of recombinant males derived from RX1 and RX2 schemes indicates that sequences proximal and distal to Xist/Tsix affect the X inactivation ratio
The RX1 breeding scheme produced male mice with five different recombinant X chromosome breakpoints (Figures 1 and 2C and Table S1). Genotyping revealed that four lines were the result of a single recombination and one line (218m) was the consequence of a double recombination event between the Cast and 129S1 chromosomes. To progeny test RX1-derived mice, recombinant males were mated to 129S1 females and the X inactivation ratio was determined in female progeny. For each of these five recombinants several mice (from the same or multiple generations) were generally progeny tested. We did not observe differences in X inactivation ratios in progeny from males from different generations with the same X chromosome (data not shown). The same was true for RX2 mice (below). Therefore, we combined all of the measurements for mice with the same recombinant X chromosome.
Two of the RX1-derived chromosomes (109m and 217m) were Cast at Pctk1, thus enabling us to use Pctk1 to measure X inactivation ratios in female progeny. The recombination site of the 109m chromosome coincides with the proximal boundary for the Xce candidate region proposed by Simmler et al. (1993) (Xce1 in Figure 2A and Table S1). The recombination site of the 217m allele coincides with the Xce candidate region boundary determined by Chadwick et al. (2006) (Xce2 in Figure 2A and Table S1). The mean X inactivation ratios measured in both 109m (ratio = 0.39; SD = 0.113) and 217m (ratio = 0.44; SD = 0.117) progeny were significantly greater than the X inactivation ratio measured in the offspring from the CastXm control mating (ratio = 0.24; SD = 0.054; P < 10−7) (Figure 2C and 3B). The loss of skewing in XCI indicated that Xce was homozygous in 109m and 217m progeny. However, the X inactivation ratios in the 109m and 217m offspring were significantly less than in the progeny of 246m control mice (ratio = 0.50; SD = 0.054; P < 0.02) (Figure 2C and 3B), indicating that the Xce was not entirely homozygous in both the 109m or 217m progeny. This was surprising since the proposed Xce candidate regions Xce1 and Xce2 (Figure 2A and Table S1) were homozygous for 129S1 in the 109m and 217m progeny, respectively.
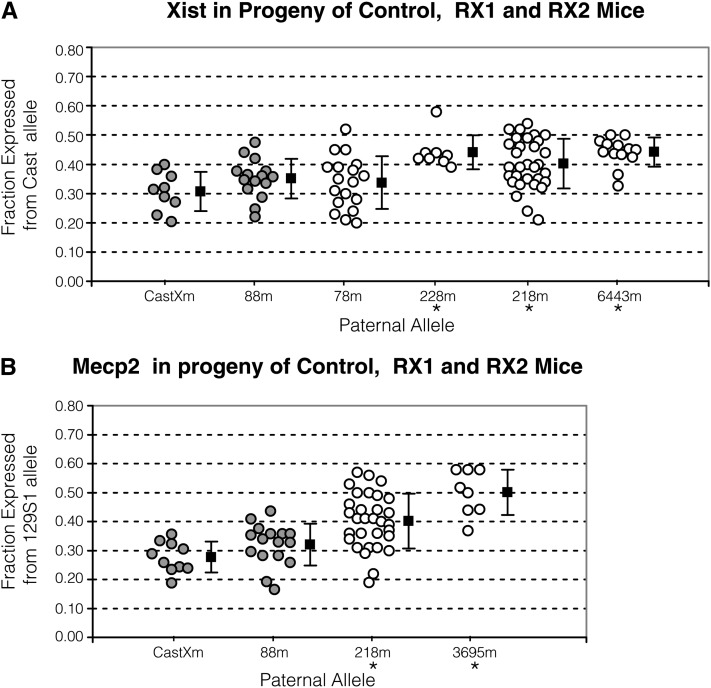
The other RX1-derived recombinant males were also of great interest because the 78m and 218m X chromosome breakpoints coincided with an Xce1 and Xce2 boundary, respectively, and the 228m breakpoint was within the Xce2 candidate interval, just proximal to Xist/Tsix (Figure 2C and Table S1). Because mice with these X chromosomes were 129S1 at Pctk1, we could not measure the X inactivation ratio by the Pctk1 assay. We therefore established allele-specific expression assays at Mecp2 and Xist (Figure S1, B and C). The mean X inactivation ratios as measured by Xist in the progeny of mice with the 78m (Xist ratio = 0.34; SD = 0.090) and the CastXm (Xist ratio = 0.31; SD = 0.067; P = 0.35) X chromosomes were not different (Figures 2C and 4A). Therefore, sequences proximal to the 78m X chromosome breakpoint did not contribute to the Xce effect. In contrast, the mean X inactivation ratio as measured by Mecp2 and/or Xist in progeny of mice with the 218m (Xist ratio = 0.40 and SD = 0.084; Mecp2 ratio = 0.40 and SD = 0.095) or 228m (Xist ratio = 0.44; SD = 0.058) X chromosomes did differ significantly from the ratios measured in CastXm (Mecp2 ratio = 0.277; SD = 0.053; P < 0.003) control progeny (Figures 2C and 4). These results indicate that sequences proximal to the 228m X chromosome breakpoint and distal to the 218m X chromosome breakpoint are contributing to the Xce effect.
Figure 4.
The X inactivation ratio as measured by allelic Xist and Mecp2 expression. The x-axis lists progeny tested mice with the designated allele described in Figure 2. Mice were mated with 129S1 mice for progeny testing. See Figure 3 legend for detail. (A) Xist in progeny of control, RX1 and RX2 progeny. The y-axis provides the ratio, measured as the fraction of total RNA that is expressed from the Cast allele. The mean X inactivation ratio in CastXm and 78m progeny did not differ (P = 0.35). The ratio in CastXm progeny significantly differed from the ratio in 228m (P < 10−3), 218m (P = 0.003), and 6443m (P < 10−3) progeny. (B) Mecp2 in progeny of control, RX1 and RX2 progeny. The y-axis provides the ratio, measured as the fraction of total RNA that is expressed from the 129S1 allele. The ratio in CastXm progeny differed from the ratio in 218m (P < 10−4) and 3695m (P < 10−4) progeny.
Progeny testing of recombinant males derived from the RX2 scheme confirms that sequences proximal and distal to Xist/Tsix affect the X inactivation ratio. The RX2 breeding scheme was used to establish and progeny test mice with 11 different recombinant X chromosomes (Figures 1 and 2D and Table S1). Many of these lines had recombination sites that coincided with or were within the Xce1 and Xce2 candidate intervals (Figure 2 and Table S1). RX2 males were mated with 129S1 females and the X inactivation ratio was measured in female progeny, typically scoring offspring from more than one mouse. All recombinant X chromosomes in progeny-tested mice were Cast at Pctk1, and therefore the X inactivation ratios of progeny were compared to that of Xce heterozygous control (CastXm) progeny and Xce homozygous control (246m) progeny. Only in progeny inheriting the 183m (ratio = 0.26; SD = 0.066) X chromosome was the X inactivation ratio similar to that measured in CastXm progeny (P = 0.35), suggesting therefore that Xce is heterozygous in such mice (Figures 2D and 3C). Thus our analysis of the 183m allele supported the analysis of RX1-derived 78m offspring showing that sequences proximal to the Xce1 boundary were not affecting the Xce QTL (Figure 2).
The X inactivation ratio as measured by Pctk1 in the progeny of 10 of the RX2 lines (mean ratios ranges from 0.30 to 0.45) was significantly greater (i.e., less skewed) than that measured in CastXm control progeny (P-values ranged from 0.012 to 10−12), which are heterozygous for Xce (Figures 2D, 3C, and 4). This indicated that in the progeny of these RX2-derived mice, the Xce was at least partly homozygous. Because Xist/Tsix sequences were heterozygous in many of the progeny (6443m [ratio = 0.33; SD = 0.064], 830m [ratio = 0.35; SD = 0.063], 800m [ratio = 0.31; SD = 0.074] and 1114m [ratio = 0.30; SD = 0.044] in Figure 2D and Table S1), these results support the analysis of the RX1-derived 228m and 218m chromosomes, suggesting that sequences both proximal and distal to Xist/Tsix contribute to the Xce effect. The mean X inactivation ratios in progeny tended to be higher when sequences spanning Xist/Tsix were homozygous (137m [ratio = 0.42; SD = 0.061]), 2173m [ratio = 0.36; S.D.=0.073], 3695m [ratio = 0.40; S.D.=0.068], 2181m [ratio = 0.45; SD = 0.070], 5005m [ratio = 0.44; SD = 0.061], 6570m [ratio = 0.42; SD = 0.090]), indicating that Xist/Tsix sequences did, however, contribute to the Xce effect.
The X inactivation ratio in progeny of all 11 RX2-derived lines was also significantly smaller than the ratios of the 246m control progeny (P-value ranged from 0.031 to 10−9), which are homozygous for Xce (Figures 2D and 3C). These results indicate that sequences that remain heterozygous in the progeny are contributing to the Xce QTL. This is difficult to explain in the context of the 137m progeny testing results and, as discussed above, the RX1 109m progeny testing results. Previous mapping studies of the Xce (Simmler et al. 1993; Chadwick et al. 2006; and Figure 2A) indicate that 137m and 109m progeny should be entirely homozygous for Xce and that XCI should, therefore, be random (∼50:50). Our results suggest that elements outside of the originally mapped Xce candidate interval are somehow contributing to skewed XCI in mice with 129S1 and Cast X chromosomes.
Discussion
The Xce has been defined as an X chromosome locus that influences the randomness of XCI in female mice (Cattanach and Williams 1972; Cattanach 1975), scored originally as a QTL by vibrissae counts and coat color variegation and more recently by direct measurements of X-linked allele-specific gene expression. We set out to map the Xce in mice heterozygous for the 129S1 Xcea allele and the Cast Xcec allele. Two breeding schemes (RX1 and RX2; see Figure 1) produced mice with single and double recombinant X chromosomes, which were subsequently used in test crosses to map the Xce QTL. We compared the X inactivation ratio to control progeny in which the entire X was either nearly completely heterozygous for the 129S1 and Cast X chromosomes (thus heterozygous for Xcea and Xcec; skewed ratio ∼0.25) or nearly completely homozygous for the 129S1 X chromosome (homozygous for Xcea; ratio ∼0.50). To our surprise, we identified multiple regions on the X chromosome that influence the randomness of XCI in female mice, including a region proximal to and another distal to Xist/Tsix (6443m, 830m, 800m, and 1114m in Figures 2D and 3C and Figure 5). Our data, however, indicate that sequences spanning Xist/Tsix also contribute to the Xce effect because X inactivation ratios tended to be higher (that is, XCI showed less skewing) when sequences including Xist/Tsix were homozygous for 129S1 (Figures 2D and 3C). As we found multiple regions that affected the X inactivation ratio, we conclude that no single discrete region defines the full Xce QTL.
Figure 5.
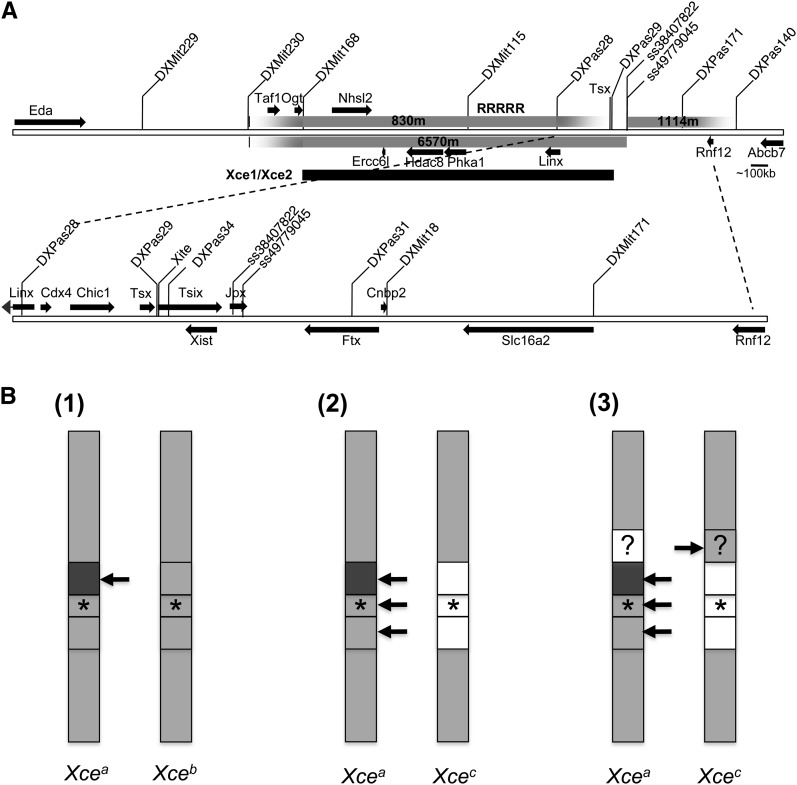
Xce regions and models. (A) Map of breakpoints for RX2 derived 830m, 6570m, and 1114m X chromosomes. Below is the Xce candidate region (Xce1/Xce2) that overlaps the region mapped by Simmler et al. (1993) and Chadwick et al. (2006). (See RX2 chromosomes and Xce1 and Xce2 in Table S1.) Relative location of genes (arrows) and genetic markers is extrapolated from UCSC Genome Browser on Mouse July 2007 (NCBI37/mm9) Assembly. On the upper extended view, only genes >10 kb are indicated within the 830m X chromosome region; RRRRR designates the location of a highly repetitive sequence. Expanded view of X chromosome region between DXPas28 and Rnf12 is shown below. (B) Regions along the X chromosome that may be responsible for the Xce effect are demarcated in females heterozygous for Xcea and Xceb (B(1)) and heterozygous for Xcea and Xcec (B(2) and (3)) chromosomes. Regions are shaded differently where sequence is contributing to the Xce effect; the darker the shade the more likely the chromosome is chosen to be inactivated. The arrows point to the X most likely chosen to be inactive when the corresponding region is heterozygous. The Xist/Tsix locus is designated by ★. The proximal boundary of Xist/Tsix is between DXPas28 and DXPas29 and the distal boundary is between Xist and DXPas31. Boundaries of demarcated regions proximal and distal to ★ are inferred from Simmler et al. (1993) in (B(1)) and our data in (B(2)) and (B(3)). (B(1)) Based on Simmler et al., the Xce effect in females with an Xcea and Xceb chromosome is due to differences in sequence proximal to DXPas29 and within sequence spanning DXPas28 and Eda. Based on the overlapping Xce candidate region Chadwick et al. (2006) identified, this candidate Xce region may further be reduced to sequences within DXMit168 and DXPas29 (B(2)). Our data suggest that at least three discrete loci on the X chromosome that may contribute to the Xce effect in females with an Xcea and Xcec chromosome. Our analysis indicates that Xcec Xist/Tsix spanning sequence (★) contributes to the Xce effect. The minimal Xce region proximal to Xist/Tsix is defined by the RX2 830m X chromosome 129S1 sequence spanning DXMit168 and DXpas28. The minimal Xce region distal to ★ is defined by the RX2 1114m chromosome with 129S1 sequence spanning ss49779045 and DXMit171. (B(3)) As depicted by the corresponding regions (open) on the Xcea chromosome and (shaded) on Xcec chromosome, if multiple X chromosome Xce loci contribute to the Xce effect, then perhaps one locus promotes preferential inactivation the Xcec chromosome.
At first glance, our conclusions appear to contradict the original definition and earlier mapping studies of Xce. Review of the Xce and Xic literature, however, indicates otherwise. Evidence of nonrandom XCI and definition of the Xcea and Xceb alleles was first reported by Cattanach and Williams (1972). In this careful genetic study, different strains of inbred mice were mated with tester mice either carrying an X-linked Tabby (Eda) or Vbr (Atp7A) mutation. The degree of nonrandomness in XCI was reflected in the scoring of secondary vibrissae number in Ta progeny and measuring coat color variegation in Vbr progeny. Findings from this study led to the conclusion that, in Xcea/Xceb heterozygotes, the X chromosome with the Xcea allele is more likely to be the inactive X. Subsequently, in matings between a wild-derived M. M. musculus mouse with Xcec and inbred Xcea or Xceb strains of mice, using the polymorphism within Pgk-1 to detect allele-specific protein expression, West and Chapman (1978) demonstrated that the X chromosome with the Xceb allele is more likely to be inactivated in Xceb/Xcec heterozygotes. While X;autosome translocations [T(X;A)] and X chromosome deletions in mice have defined the Xic (Rastan 1983; Rastan and Robertson 1985; Cattanach et al. 1991; Heard et al. 1997), these studies did not necessarily define the location of Xce, as this depends on heterozygosity for the sequences responsible for the Xce effect. Nevertheless, analysis of Xce phenotype in T(X;A) mice and X-linked Tabby (Eda), Mottled (Atp7A), and Pgk-1 phenotypes led to the conclusion that the Xce is between Ta (Eda) and Pgk-1 (Cattanach and Papworth 1981; Simmler et al. 1993). If Xce is absolutely required for choice (that is, deciding which X chromosome to inactivate) then the Xic may define the Xce. However, if Xce merely influences choice (that is, skewing XCI depending on the two Xce alleles present), then approaches used to define the Xic may not define the Xce.
Simmler et al. (1993) further mapped Xce within the candidate interval spanning Ta (Eda) and Mottled (Atp7A), which includes Xist/Tsix, by identifying three new microsatellite markers: DXPas28 and DXPas29 downstream of Xist and DXPas31 upstream of Xist. With these markers, they genotyped the inbred mice that were used by Cattanach and Williams (1972) to characterize nonrandom XCI in mice with Xcea and Xceb alleles. To their surprise, they observed that one inbred strain phenotyped as Xceb (JU/Ct) exhibited an Xceb genotype at DXPas28 but Xcea at DXpas29 and DXpas31. This led them to conclude that Xce was distinct from Xist.
Some of our recombinant lines (RX1 228m and RX2 6443m and 830m; Figure 2) also suggested that Xce lies proximal to Xist and distal to Eda (Table S1 and Figure 5A). However, using allele-specific expression as a measure of the X inactivation ratio, we were also able to detect gradations indicating that (1) the Xce QTL is not an all or none effect and (2) sequences spanning Xist/Tsix seem to contribute to the Xce effect (e.g., ratios in 3695m tended to be higher than those in 6443m progeny and higher in 6570m than those in 830m progeny; Figures 2D and 3C). Our data also indicate that sequences distal to Xist can affect the X inactivation ratio and therefore contribute to the Xce effect. As we discuss below, there are multiple candidates within this region that may contribute to the Xce QTL (Figure 5A).
Is there a discrepancy between the Simmler et al. (1993) study and our mapping data? Not necessarily. The difference in results might be explained by the different Xce alleles that were used for each mapping study (Cattanach and Williams 1972; Simmler et al. 1993; Chadwick et al. 2006; and Figure 5B.) All the mapping studies culminating with the Simmler et al. (1993) analysis had been performed using mice with Xcea and Xceb alleles that are far more related to each other than to mice with Xcec allele. While Xce mapping by Chadwick et al. (2006) used Xcea and Xcec recombinant lines and Xceb and Xcec recombinant lines, in these cases, mapping of the Xce candidate interval was always associated with Xist, which is in agreement with our observation that Xist/Tsix sequences also contributes to the Xce effect. This Xce candidate interval was, however, contiguous with part of the interval mapped by Simmler et al. (1993) and therefore mostly validates the region proximal to Xist/Tsix as including the Xce (Figure 5A). Thus the present study is first to show that more than one X chromosome region may contribute to the Xce QTL.
With the identification of Xist, subsequent generation of targeted deletions at the Xist locus and the analysis of Xist spanning trangenes have uncovered essential elements of the Xic (Payer and Lee 2008; Morey and Avner 2011). The Xce, however, has remained elusive. In female cells inheriting an Xist deletion allele, skewed XCI of the wild-type allele occurs (Marahrens et al. 1998). By contrast, in female cells inheriting a deletion allele of Tsix, the antisense repressor of Xist, skewed XCI of the deletion allele occurs (Lee and Lu 1999). Furthermore, deletion of Xite, which is required for full expression of Tsix, also results in skewed XCI of the deletion allele. The latter study led Lee and colleagues (Ogawa and Lee 2003) to speculate that Xite is a candidate for the Xce. The CG-rich DXPas34 sequence, however, despite being a major regulator of Tsix transcription, was less attractive as a candidate for Xce, because it does not carry allele-specific DNA methylation marks at the time in development when choice is made (Prissette et al. 2001; Vigneau et al. 2006). Nevertheless, Tsix is a major determinant of choice, and Xce alleles that are distinct from the Xist/Tsix locus may exert their role in XCI by modifying the function/expression of Tsix in cis.
We have demonstrated that sequences proximal and distal to Xist/Tsix contribute to the Xce effect. Recently identified elements within the broad Xic candidate region have been shown to affect XCI by affecting choice. Distal to Xist, the noncoding genes Jpx and Ftx and the protein-coding gene Rnf12 have been shown to influence XCI by affecting Xist expression (Jonkers et al. 2009; Tian et al. 2010; Chureau et al. 2011; Pontier and Gribnau 2011) (Figure 5A). SNPs in any of these genes may contribute to the loss of skewing observed in progeny of the RX2-derived male 1114m. Proximal to Xist, there are multiple genes within the overlapping candidate Xce region identified by Simmler et al. (1993) and Chadwick et al. (2006) and our RX2-derived 830m X chromosome (Figure 5A). The distal recombination breakpoint for RX2-derived male 830m (and 3695m) lies between DXPas28 and Exon 4 of Tsx (Table S1 and Table S2); therefore, Xite can be excluded but Tsx may still be part of this Xce candidate region. This is interesting because Tsx is reported to affect Tsix expression (Anguera et al. 2011). In addition, the expression and transgenic analysis of the noncoding gene Linx, which is within the 830m X chromosome distal recombination breakpoint, suggests that Linx expression affects XCI choice and therefore Linx may also be a candidate for the Xce QTL (Figure 5A) (Nora et al. 2012).
We observed an unanticipated trend when comparing the X inactivation ratios in the progeny from RX1-derived males 109m (ratio = 0.39) and 217m (ratio = 0.44) and the X inactivation ratios in the progeny of RX2-derived males 2173m (ratio = 0.36) and 3695m (ratio = 0.40) (Figure 2 and 3). In the progeny of 109m and 2173m males, a longer region was homozygous for 129S1 proximal to Xist than in the progeny of 217m and 3695m males. We therefore expected to measure higher X inactivation rations in progeny of 109m and 2173m relative to ratios measured in progeny of 217m and 3695m males, respectively. We observed the opposite trend. Although not anticipated, this is consistent with Xce being a QTL defined by multiple X-linked loci. Our results could be explained by an Xce locus that promotes preferential XCI of the Cast allele rather than the 129S1 allele [Figure 5B(3)]. Closely linked loci may have opposite effects as was reported for QTL on chromosome 2 that affect body weight (Mollah and Ishikawa 2011). We cannot rule out that this trend is due to background effects in the RX1-and RX2-derived mice but because we observed this trend in both RX1- and RX2-derived mice, which were generated and maintained in different backgrounds (Figure 1), an X-linked locus may best explain our observation. Thus, to define and characterize X-linked sequences that define the Xce QTL, each candidate locus needs to be tested independently.
Finally there are numerous models to consider to explain how and when Xce alleles function. Binding of a blocking factor to a unique entity on the future active X chromosome has long been proposed to contribute to choice in XCI (Lyon 1971; Brown and Chandra 1973; Russell and Cacheiro 1978; Rastan 1983). Under this model, skewed XCI would result from the preferential binding of the blocking factor to one or the other Xce allele (Percec et al. 2003). Hence, “strong” Xce alleles are preferentially associated with the future active X chromosome and “weak” Xce alleles are preferentially associated with the future inactive X chromosome. A simple model for skewed XCI in which a blocking factor binds to a unique Xce element, however, is not supported by our Xce mapping study of Xcea 129S1 and Xcec Cast X chromosomes. This model may need to be broadened to consider multiple binding sites that could act additively or synergistically to influence the choice of one or the other X for X inactivation. Another possibility is that Xce alleles contribute to the stochastic process Monkhorst et al. (2008) proposed to regulate counting and choice during XCI. The stochastic model predicts that SNPs found within cis-acting activators or repressors of XCI that are external to the Xist/Tsix locus can affect choice during XCI and could be within Xce candidate regions we have mapped. Moreover, there may be numerous times during development when Xce alleles function: before X-inactivation is triggered as alluded X chromosome analyses in ES cell (Mlynarczyk-Evans et al. 2006; Monkhorst et al. 2008), when X-chromosome pairing that occurs at the beginning stages of XCI (Bacher et al. 2006; Xu et al. 2006), or even during the short time after XCI is initiated when XCI is reversible (Wutz and Jaenisch 2000).
In conclusion, we demonstrate that X chromosome regions proximal to, including and distal to Xist/Tsix, contribute to the choice in XCI in mice with Xcea 129S1 and Xcec Cast X chromosomes, consistent with Xce being a QTL. In these mice, XCI is highly skewed with preferential inactivation of the Xcea 129S1 X chromosome. In contrast, Xce mapping using mice with relatively closer Xcea and Xceb X chromosomes, in which XCI is less skewed with preferential inactivation of the Xcea, led to mapping of Xce to a region proximal Xist/Tsix (Cattanach and Williams 1972; Simmler et al. 1993). This is consistent with the proposal that “only one locus is involved” in Xce if Xce acts upon the XCI process vs. cell selection (Cattanach and Williams 1972). With the recent identification of numerous X-linked genes and genetic elements that contribute to the XCI process and uncovering the stochastic nature of this process (Monkhorst et al. 2008; Jonkers et al. 2009; Tian et al. 2010; Chureau et al. 2011; Pontier and Gribnau 2011; Nora et al. 2012); however, the number and diversity of loci that define Xce may be more complex than originally envisioned and vary with the relatedness of the X chromosomes being evaluated.
Supplementary Material
Acknowledgments
We thank Jesse Mager for his contributions on experimental design during the early stages of this work. We are grateful to Sebastien Vigneau and Nora Engel for critical reading of the manuscript. This work was funded in part by grant GM74768 (to M.S.B.) from the National Institutes of Health.
Footnotes
Communicating editor: T. C.-t. Wu
Literature Cited
- Amos-Landgraf J. M., Cottle A., Plenge R. M., Friez M., Schwartz C. E., et al. , 2006. X chromosome-inactivation patterns of 1,005 phenotypically unaffected females. Am. J. Hum. Genet. 79: 493–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anguera M. C., Ma W., Clift D., Namekawa S., Kelleher R. J., 3rd, et al. , 2011. Tsx produces a long noncoding RNA and has general functions in the germline, stem cells, and brain. PLoS Genet. 7: e1002248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avner P., Heard E., 2001. X-chromosome inactivation: counting, choice and initiation. Nat. Rev. Genet. 2: 59–67. [DOI] [PubMed] [Google Scholar]
- Bacher C. P., Guggiari M., Brors B., Augui S., Clerc P., et al. , 2006. Transient colocalization of X-inactivation centres accompanies the initiation of X inactivation. Nat. Cell Biol. 8: 293–299. [DOI] [PubMed] [Google Scholar]
- Borsani G., Tonlorenzi R., Simmler M. C., Dandolo L., Arnaud D., et al. , 1991. Characterization of a murine gene expressed from the inactive X chromosome. Nature 351: 325–329. [DOI] [PubMed] [Google Scholar]
- Brockdorff N., Ashworth A., Kay G. F., Cooper P., Smith S., et al. , 1991. Conservation of position and exclusive expression of mouse Xist from the inactive X chromosome. Nature 351: 329–331. [DOI] [PubMed] [Google Scholar]
- Brown C. J., Ballabio A., Rupert J. L., Lafreniere R. G., Grompe M., et al. , 1991. A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44. [DOI] [PubMed] [Google Scholar]
- Brown S. W., Chandra H. S., 1973. Inactivation system of the mammalian X chromosome. Proc. Natl. Acad. Sci. USA 70: 195–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattanach B. M., 1970. Controlling elements in the mouse X-chromosome. 3: influence upon both parts of an X divided by rearrangement. Genet. Res. 16: 293–301. [DOI] [PubMed] [Google Scholar]
- Cattanach B. M., 1975. Control of chromosome inactivation. Annu. Rev. Genet. 9: 1–18. [DOI] [PubMed] [Google Scholar]
- Cattanach B. M., Papworth D., 1981. Controlling elements in the mouse. V. Linkage tests with X-linked genes. Genet. Res. 38: 57–70. [DOI] [PubMed] [Google Scholar]
- Cattanach B. M., Rasberry C., 1991. Identification of the Mus spretus Xce allele. Mouse Genome 89: 565–566. [Google Scholar]
- Cattanach B. M., Williams C. E., 1972. Evidence of non-random X chromosome activity in the mouse. Genet. Res. 19: 229–240. [DOI] [PubMed] [Google Scholar]
- Cattanach B. M., Perez J. N., Pollard C. E., 1970. Controlling elements in the mouse X-chromosome. II. Location in the linkage map. Genet. Res. 15: 183–195. [DOI] [PubMed] [Google Scholar]
- Cattanach B. M., Bücher T., Andrews S. J., 1982. Location of Xce in the mouse X chromosome and effects of Pgk-1 expression. Genet. Res. 40: 103–104. [Google Scholar]
- Cattanach B. M., Rasberry C., Evans E. P., Burtenshaw M. D., 1989. Further Xce linkage data. Mouse News Let. 83: 165. [Google Scholar]
- Cattanach B. M., Rasberry C., Evans E. P., Dandolo L., Simmler M. C., et al. , 1991. Genetic and molecular evidence of an X-chromosome deletion spanning the tabby (Ta) and testicular feminization (Tfm) loci in the mouse. Cytogenet. Cell Genet. 56: 137–143. [DOI] [PubMed] [Google Scholar]
- Chadwick L. H., Willard H. F., 2005. Genetic and parent-of-origin influences on X chromosome choice in Xce heterozygous mice. Mamm. Genome 16: 691–699. [DOI] [PubMed] [Google Scholar]
- Chadwick L. H., Pertz L. M., Broman K. W., Bartolomei M. S., Willard H. F., 2006. Genetic control of X chromosome inactivation in mice: definition of the Xce candidate interval. Genetics 173: 2103–2110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chureau C., Chantalat S., Romito A., Galvani A., Duret L., et al. , 2011. Ftx is a noncoding RNA which affects Xist expression and chromatin structure within the X-inactivation center region. Hum. Mol. Genet. 20: 705–718. [DOI] [PubMed] [Google Scholar]
- Clerc P., Avner P., 2003. Multiple elements within the Xic regulate random X inactivation in mice. Semin. Cell Dev. Biol. 14: 85–92. [DOI] [PubMed] [Google Scholar]
- Courtier B., Heard E., Avner P., 1995. Xce haplotypes show modified methylation in a region of the active X chromosome lying 3′ to Xist. Proc. Natl. Acad. Sci. USA 92: 3531–3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heard E., Clerc P., Avner P., 1997. X-chromosome inactivation in mammals. Annu. Rev. Genet. 31: 571–610. [DOI] [PubMed] [Google Scholar]
- Heard E., Mongelard F., Arnaud D., Avner P., 1999. Xist yeast artificial chromosome transgenes function as X-inactivation centers only in multicopy arrays and not as single copies. Mol. Cell. Biol. 19: 3156–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston P. G., Cattanach B. M., 1981. Controlling elements in the mouse. IV. Evidence of non-random X-inactivation. Genet. Res. 37: 151–160. [DOI] [PubMed] [Google Scholar]
- Jonkers I., Barakat T. S., Achame E. M., Monkhorst K., Kenter A., et al. , 2009. RNF12 is an X-Encoded dose-dependent activator of X chromosome inactivation. Cell 139: 999–1011. [DOI] [PubMed] [Google Scholar]
- Lee J. T., 2011. Gracefully ageing at 50, X-chromosome inactivation becomes a paradigm for RNA and chromatin control. Nat. Rev. Mol. Cell Biol. 12: 815–826. [DOI] [PubMed] [Google Scholar]
- Lee J. T., Lu N., 1999. Targeted mutagenesis of Tsix leads to nonrandom X inactivation. Cell 99: 47–57. [DOI] [PubMed] [Google Scholar]
- Lee J. T., Davidow L. S., Warshawsky D., 1999. Tsix, a gene antisense to Xist at the X-inactivation centre. Nat. Genet. 21: 400–404. [DOI] [PubMed] [Google Scholar]
- Lyon M. F., 1961. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 190: 372–373. [DOI] [PubMed] [Google Scholar]
- Lyon M. F., 1971. Possible mechanisms of X chromosome inactivation. Nat. New Biol. 232: 229–232. [DOI] [PubMed] [Google Scholar]
- Marahrens Y., Loring J., Jaenisch R., 1998. Role of the Xist gene in X chromosome choosing. Cell 92: 657–664. [DOI] [PubMed] [Google Scholar]
- Mlynarczyk-Evans S., Royce-Tolland M., Alexander M. K., Andersen A. A., Kalantry S., et al. , 2006. X chromosomes alternate between two states prior to random X-inactivation. PLoS Biol. 4: e159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollah M. B., Ishikawa A., 2011. Intersubspecific subcongenic mouse strain analysis reveals closely linked QTLs with opposite effects on body weight. Mamm. Genome 22: 282–289. [DOI] [PubMed] [Google Scholar]
- Monkhorst K., Jonkers I., Rentmeester E., Grosveld F., Gribnau J., 2008. X inactivation counting and choice is a stochastic process: evidence for involvement of an X-linked activator. Cell 132: 410–421. [DOI] [PubMed] [Google Scholar]
- Morey C., Avner P., 2010. Genetics and epigenetics of the X chromosome. Ann. N. Y. Acad. Sci. 1214: E18–E33. [DOI] [PubMed] [Google Scholar]
- Morey C., Avner P., 2011. The demoiselle of X-inactivation: 50 years old and as trendy and mesmerising as ever. PLoS Genet. 7: e1002212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora E. P., Lajoie B. R., Schulz E. G., Giorgetti L., Okamoto I., et al. , 2012. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485: 381–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa Y., Lee J. T., 2003. Xite, X-inactivation intergenic transcription elements that regulate the probability of choice. Mol. Cell 11: 731–743. [DOI] [PubMed] [Google Scholar]
- Payer B., Lee J. T., 2008. X chromosome dosage compensation: how mammals keep the balance. Annu. Rev. Genet. 42: 733–772. [DOI] [PubMed] [Google Scholar]
- Percec I., Plenge R. M., Nadeau J. H., Bartolomei M. S., Willard H. F., 2002. Autosomal dominant mutations affecting X inactivation choice in the mouse. Science 296: 1136–1139. [DOI] [PubMed] [Google Scholar]
- Percec I., Thorvaldsen J. L., Plenge R. M., Krapp C. J., Nadeau J. H., et al. , 2003. An N-ethyl-N-nitrosourea mutagenesis screen for epigenetic mutations in the mouse. Genetics 164: 1481–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plenge R. M., Percec I., Nadeau J. H., Willard H. F., 2000. Expression-based assay of an X-linked gene to examine effects of the X-controlling element (Xce) locus. Mamm. Genome 11: 405–408. [DOI] [PubMed] [Google Scholar]
- Pontier D. B., Gribnau J., 2011. Xist regulation and function explored. Hum. Genet. 130: 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prissette M., El-Maarri O., Arnaud D., Walter J., Avner P., 2001. Methylation profiles of DXPas34 during the onset of X-inactivation. Hum. Mol. Genet. 10: 31–38. [DOI] [PubMed] [Google Scholar]
- Puck J. M., Willard H. F., 1998. X inactivation in females with X-linked disease. N. Engl. J. Med. 338: 325–328. [DOI] [PubMed] [Google Scholar]
- Rastan S., 1982. Timing of X-chromosome inactivation in postimplantation mouse embryos. J. Embryol. Exp. Morphol. 71: 11–24. [PubMed] [Google Scholar]
- Rastan S., 1983. Non-random X-chromosome inactivation in mouse X-autosome translocation embryos: location of the inactivation centre. J. Embryol. Exp. Morphol. 78: 1–22. [PubMed] [Google Scholar]
- Rastan S., Robertson E. J., 1985. X-chromosome deletions in embryo-derived (EK) cell lines associated with lack of X-chromosome inactivation. J. Embryol. Exp. Morphol. 90: 379–388. [PubMed] [Google Scholar]
- Russell L. B., Cacheiro N. L., 1978. The use of mouse X-autosome translocations in the study of X-inactivation pathways and nonrandomness. Basic Life Sci. 12: 393–416. [DOI] [PubMed] [Google Scholar]
- Simmler M. C., Cattanach B. M., Rasberry C., Rougeulle C., Avner P., 1993. Mapping the murine Xce locus with (CA)n repeats. Mamm. Genome 4: 523–530. [DOI] [PubMed] [Google Scholar]
- Tian D., Sun S., Lee J. T., 2010. The long noncoding RNA, Jpx, is a molecular switch for X chromosome inactivation. Cell 143: 390–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigneau S., Augui S., Navarro P., Avner P., Clerc P., 2006. An essential role for the DXPas34 tandem repeat and Tsix transcription in the counting process of X chromosome inactivation. Proc. Natl. Acad. Sci. USA 103: 7390–7395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West J. D., Chapman V. M., 1978. Variation for X chromosome expression in mice detected by electrophoresis of phosphoglycerate kinase. Genet. Res. 32: 91–102. [DOI] [PubMed] [Google Scholar]
- Wutz A., 2011. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat. Rev. Genet. 12: 542–553. [DOI] [PubMed] [Google Scholar]
- Wutz A., Jaenisch R., 2000. A shift from reversible to irreversible X inactivation is triggered during ES cell differentiation. Mol. Cell 5: 695–705. [DOI] [PubMed] [Google Scholar]
- Xu N., Tsai C. L., Lee J. T., 2006. Transient homologous chromosome pairing marks the onset of X inactivation. Science 311: 1149–1152. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.