Abstract
The wall gives a Saccharomyces cerevisiae cell its osmotic integrity; defines cell shape during budding growth, mating, sporulation, and pseudohypha formation; and presents adhesive glycoproteins to other yeast cells. The wall consists of β1,3- and β1,6-glucans, a small amount of chitin, and many different proteins that may bear N- and O-linked glycans and a glycolipid anchor. These components become cross-linked in various ways to form higher-order complexes. Wall composition and degree of cross-linking vary during growth and development and change in response to cell wall stress. This article reviews wall biogenesis in vegetative cells, covering the structure of wall components and how they are cross-linked; the biosynthesis of N- and O-linked glycans, glycosylphosphatidylinositol membrane anchors, β1,3- and β1,6-linked glucans, and chitin; the reactions that cross-link wall components; and the possible functions of enzymatic and nonenzymatic cell wall proteins.
Keywords: adhesion, β-glucan, cell wall, cell wall protein, chitin, glycoprotein, glycosylphosphatidylinositol, mannan, septum
THE wall gives Saccharomyces cerevisiae its morphologies during budding growth, pseudohypha formation, mating, and sporulation; it preserves the cell’s osmotic integrity; and it provides a scaffold to present agglutinins and flocculins to other yeast cells. The wall consists of mannoproteins, β-glucans, and a small amount of chitin, which become cross-linked in various ways. Wall composition and organization vary during growth and development. During the budding cycle, deposition of chitin is tightly controlled, and expression of certain hydrolases involved in cell separation is daughter cell-specific. The wall can be weakened, and the cell consequently stressed, by treatment with polysaccharide binding agents such as Calcofluor White (CFW), Congo Red, sodium dodecyl sulfate (SDS), aminoglycoside antibiotics, and β-glucanase preparations or by mutational loss of capacity to make a wall component. Such stresses commonly activate the cell wall integrity (CWI) pathway (Levin 2011) and result in compensatory synthesis of wall material.
Up to a quarter of the genes in S. cerevisiae have some role in maintenance of a normal wall. From the results of a survey of deletion strains for cell wall phenotypes, De Groot et al. (2001) estimated that ∼1200 genes, not counting essential ones, impact the wall. Most of the effects, however, are indirect, and the number of genes that encode enzymes directly involved in biosynthesis or remodeling of the wall, or nonenzymatic wall proteins, is now ∼180 (see Supporting Information, Table S1). This review covers these proteins, with emphasis on the wall of vegetative cells during the budding cycle and in response to stress. Wall synthetic activities will be covered in the context of their cellular localization, starting with precursors in the cytoplasm, proceeding along the secretory pathway from the endoplasmic reticulum (ER) to the plasma membrane, and culminating with the events outside the plasma membrane that generate covalent cross-links between wall components. Additional information about individual proteins and the phenotypes of strains lacking them is presented in File S1, File S2, File S3, File S4, File S5, File S6, File S7, File S8, and File S9. Earlier work on the yeast cell wall has been reviewed by Ballou (1982), Fleet (1991), Orlean (1997), Kapteyn et al. (1999a), Cabib et al. (2001), Klis et al. (2002, 2006), and Lesage and Bussey (2006).
Wall Composition and Architecture
The wall accounts for 15–30% of the dry weight of a vegetative S. cerevisiae cell (Aguilar-Uscanga and François 2003; Yin et al. 2007). It is 110–200 nm wide, as estimated from transmission electron micrographs and by using an atomic force microscope to detect surface accessibility of “molecular rulers” consisting of versions of the plasma membrane sensor Wsc1 with different lengths (Dupres et al. 2010; Yamaguchi et al. 2011). The wall’s major components are β1,3- and β1,6-linked glucans, mannoproteins, and chitin, which can be covalently joined to form higher-order complexes. The β1,6-glucan, although quantitatively a minor component of the wall, has a central role in cross-linking wall components (Kollar et al. 1997). Some mannoproteins have or are predicted to have enzymatic activity as hydrolases or cross-linkers; others may have structural roles or mediate “social activity” by serving as mating agglutinins or flocculins. Among the latter, Flo1 and Flo11 promote formation of extensive mats of cells, or biofilms (Reynolds and Fink 2001; Beauvais et al. 2009; Bojsen et al. 2012).
Electron micrographs of thin sections through the wall of vegetative cells reveal two layers. The outer one is electron-dense, has a brush-like surface (Osumi et al. 1998) (Baba et al. 1989; Osumi et al. 1998); Kapteyn et al. 1999a; Hagen et al. 2004; Yamaguchi et al. 2011), and can be removed by proteolysis (Kopecka et al. 1974; Zlotnik et al. 1984); it therefore consists mostly of mannoproteins. The inner layer, more electron transparent, is microfibrillar and β-glucanase-digestible, indicating that its major components are glucans. The relative thicknesses of the two layers and their apparent organization can be altered in cell wall mutants.
Relative amounts and localization of individual wall components vary depending on cell cycle or developmental stage, growth phase, nutritional conditions, and wall stresses imposed by hypo-osmolarity, mutational loss of wall biosynthetic activities or wall proteins, or drug treatment. Variations in wall composition and organization impact the extent to which the wall is a barrier to export of soluble, secreted proteins to the medium. Some proteins can be retained by the wall outside the plasma membrane in the periplasmic space; in the case of Suc2, this is due to the ability of the protein to form large multimers (Orlean 1987). The barrier function of the wall is dependent on growth phase and cultural conditions, with the walls of growing cells being more porous (De Nobel and Barnett 1991). Native glycoproteins such as Cts1, as well as many heterologously expressed soluble glycoproteins with masses up to 400 kDa, can pass through the wall of logarithmically growing cells to the medium, whereas walls of stationary-phase cells are less porous (De Nobel et al. 1990; Kuranda and Robbins 1991). The relatively high porosity of walls of logarithmic-phase cells could reflect a lower degree of cross-linking, but the dissolution of septal material that occurs when dividing cells separate could also release wall proteins to the medium (see Order of incorporation of components into the cell wall). Perspectives on wall organization are provided by Kapteyn et al. (1999a), Klis et al. (2002, 2006), Latgé (2007), Pitarch et al. (2008), and Gonzalez et al. (2010a). The major wall components and strategies for isolating them are as follows.
Polysaccharides
Wall polysaccharides are typically separated into three fractions defined on the basis of their solubility in alkali and acid (Fleet 1991). These fractions contain differing relative amounts of β1,3- and β1,6-linked glucans and mannan (Magnelli et al. 2002) and also differ in whether and to what extent the glucans are cross-linked to chitin, which determines their solubility in alkali. Determination of Man-to-Glc ratios in total acid hydrolysates of walls has been useful in assessing the impact of mutations on wall composition (Ram et al. 1994; Dallies et al. 1998). Digestion of isolated walls and wall fractions with linkage-specific glycosidases has been used to quantify wall components and determine the fine structure of β1,6-glucan (Boone et al. 1990; Magnelli et al. 2002; Aimanianda et al. 2009), as well as to generate oligosaccharides for structural analysis and characterization of linkages between polymers (Kollar et al. 1995, 1997).
Chitin:
This polymer of β1,4-linked GlcNAc contributes only 1–2% of the dry weight of the wall of unstressed wild-type cells. Chitin is normally deposited in a ring in the neck between a mother cell and its emerging bud, in the primary division septum, and in the lateral walls of newly separated daughter cells. Chitin can be visualized in situ by staining with CFW, which reveals that most of it is present in division septa and bud scars. Chitin in lateral walls and in division septa can also be detected by immunoelectron microscopy (Shaw et al. 1991). Chitin levels are typically determined after extraction of walls with acid and alkali or hot SDS, followed by acid or enzymatic hydrolysis and quantification of GlcNAc (Kang et al. 1984; Orlean et al. 1985; Dallies et al. 1998; Magnelli et al. 2002). The average length of chitin in β-glucanase-digested septa is ∼110 GlcNAc residues (Kang et al. 1984). However, chitin occurs in three different and polydisperse forms in the wall: in addition to free chitin, some is bound to β1,3-glucan and present mainly in the neck between mother and daughter cell, whereas a lesser amount, found in lateral walls, is bound to β1,6-glucan, which is in turn linked to mannan and β1,3-glucan (Cabib and Duran 2005; Cabib 2009). Chitin levels increase in response to mating pheromones (Schekman and Brawley 1979; Orlean et al. 1985; see Sugar nucleotides) and delocalized chitin in lateral walls can increase to as much as 20% of the wall in S. cerevisiae mutants mounting the cell wall stress response (Kapteyn et al. 1997, 1999a; Popolo et al. 1997; Dallies et al. 1998; Ram et al. 1998; Osmond et al. 1999; Valdivieso et al. 2000; Magnelli et al. 2002; see Chitin synthesis in response to cell wall stress).
β-Glucans:
β-linked glucans compose 30–60% of the dry weight of the wall and can be separated into three fractions that contain both β1,3 and β1,6 linkages. The major fraction, which makes up ∼35% of the dry weight of the wall, is an acid- and alkali-insoluble β1,3-glucan with a degree of polymerization of ∼1500 and β1,3-linked glucan side chains initiated at branching β1,6-linked glucoses that represent ∼3% of the whole polymer (Fleet 1991). The nonreducing ends of β1,3-glucan chains in this fraction can be linked to chitin, rendering the β-glucan insoluble (Kollar et al. 1995; see below). A second β-glucan fraction, representing ∼20% of the dry weight of the wall, is similar in size and composition to the alkali-insoluble β1,3-glucan, but soluble in alkali because it is not cross-linked to chitin (Hartland et al. 1994). A third fraction, making up ∼5% of the dry weight of the wall, can be released from alkali-insoluble glucan by extraction with acid or digestion with endo-β1,3-glucanase (Manners et al. 1973; Boone et al. 1990). This fraction is a β1,6-glucan with a degree of polymerization of 140, in which 14% of the β1,6-linked residues bear a side-branching β1,3 Glc (Manners et al. 1973). A procedure involving serial digestion with purified hydrolases has also been used to separate and quantify β1,3- and β1,6-glucan, mannan, and chitin (Magnelli et al. 2002). The β1,6-glucan was released from the high-molecular-weight material remaining after treatment of walls with a mixture of β1,3-glucanase and chitinase by digestion with recombinant endo-β1,6-glucanase. The β1,6-glucan was therefore recovered as a mixture of oligosaccharides whose major component was Glcβ1,6Glc, and which also contained Glcβ1,6Glcβ1,6Glc and smaller amounts of Glcβ1,3Glcβ1,6Glc and Glcβ1,6Glcβ1,6Glc with a β1,3-Glc branching from its middle Glc (Magnelli et al. 2002). The degree of β1,3 branching inferred from the oligosaccharide profile was similar to that reported by Manners et al. (1973). This β1,6-glucan analysis would also include the β1,6-glucan present in the alkali-soluble cell wall fraction, which is not included in procedures involving alkali extraction. In another approach, β1,6-glucan was isolated following extraction of intact cells with hot SDS and mercaptoethanol, treatment with hot alkali under reducing conditions, and β1,3-glucanase digestion of the alkali-insoluble material (Aimanianda et al. 2009). The β1,3-glucanase releasable material was a β1,6-glucan of 190–200 glucoses with, on average, a β1,3-Glc or a β1,3-Glcβ1,3-Glc side branch on every fifth β1,6-linked glucose (Aimanianda et al. 2009).
Cross-links between polysaccharides:
Three types of linkages between wall polysaccharides have been described (Figure 1). The first is a β1,4-linkage between the reducing end of a chitin chain and the nonreducing end of a β1,3-linked glucan (Kollar et al. 1995), and up to half of the chitin chains in the wall may be linked to β-glucan in this way. Because there is about one chitin-β-glucan linkage per 8000 hexoses, these rare cross-links have a major impact on the solubility of β-glucan (Kollar et al. 1995). The second linkage is between the reducing end of chitin and the nonreducing end of a β1,3-Glc that branches off β1,6-glucan (Kollar et al. 1997; see Remodeling and Cross-Linking Activities at the Cell Surface). The configuration of this linkage is either β1,2- or β1,4-. The two types of chitin-β-glucan linkage are found in different parts of the wall. In the third linkage, the reducing ends of β1,6-glucan chains can be attached to β1,3-glucan, but the configuration is unknown (Kollar et al. 1997).
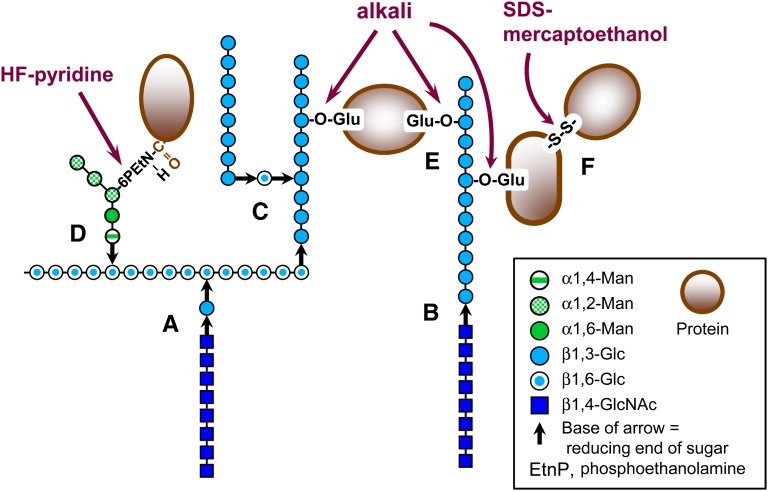
Figure 1 .
Wall components and cross-links between them. (A) Reducing end of chitin linked to a side-branching β1,3-Glc on β1,6-glucan. (B) Reducing end of chitin linked to a nonreducing end of β1,3-glucan. (C) Reducing end of β1,3-glucan chain linked to a side-branching β1,6-Glc on β1,3-glucan. (D) Reducing end of GPI glycan (possibly the α1,4-Man) to internal Glc in β1,6-glucan (linkage to nonreducing end of β1,6-glucan is also possible). (E) Ester linkages between β1,3-Glc and γ-carboxyl groups of glutamates in PIR protein internal repeats. (F) Disulfide link between CWP. Chemical treatments used to release CWP are indicated.
Cell wall mannoproteins
Yeast cell wall proteins can bear asparagine- (N-)linked glycans, O-linked manno-oligosaccharides, and often a glycosylphosphatidylinositol (GPI) as well. The N-linked glycans can be extended with an outer chain of 50 or more α1,6-linked Man that is extensively decorated with short α1,2-Man side branches terminated in α1,3-Man. Phosphodiester-linked mannoses can also be attached to α1,2-linked residues. Many glycoproteins also bear O-mannosyl glycans, which are often present in Ser/Thr-rich stretches.
Proteins relevant to the wall can be placed into one of three groups. The first contains those with the potential to participate in wall construction as hydrolases or transglycosidases. The second contains nonenzymatic agglutinins, flocculins, or β1,3-glucan cross-connectors (Klis et al. 2006, 2010; Dranginis et al. 2007; Goossens and Willaert 2010). Most, if not all the proteins in these two groups are glycosylated. Proteins that are covalently attached to cell wall glycan are referred to as CWP (Yin et al. 2005) and fall into the subgroups below. The third group consists of single-pass plasma membrane proteins with short C-terminal cytoplasmic domains and long Ser/Thr-rich extracellular regions. These include Wsc1, Wsc2, and Wsc3, which also have N-terminal cysteine-rich domains, as well as Mtl1 and Mid2. These are mechanosensors that detect cell wall stress and activate the CWI pathway (Rodicio and Heinisch 2010; Levin 2011). CWP and cell wall-active enzymes are discussed in Cell Wall-Active and Nonenzymatic Surface Proteins and Their Functions.
GPI proteins:
These receive a GPI that initially anchors them in the outer face of the plasma membrane, but many then become cross-linked to β1,6-glucan via a remnant of the GPI (Gonzalez et al. 2009). Results to date suggest that the GPI is cleaved between its GlcN residue and Man, whereupon the mannose’s reducing end is glycosidically linked to a nonreducing end of β1,6-glucan or to a Glc in a β1,6-Glc chain (Kollar et al. 1997; Fujii et al. 1999). The β1,6-glucan to which the GPI-CWP is attached is in turn linked to β1,3-glucan and chitin (Kapteyn et al. 1996; Van der Vaart et al. 1996; Kollar et al. 1997; Fujii et al. 1999; Figure 1). Some wall-bound GPI proteins may retain enzymatic activity, whereas others may have a structural role (Yin et al. 2005). GPI-CWP are released by treatment with hydrogen fluoride (HF)/pyridine, which cleaves the phosphodiester of the GPI that links Man and the phosphoethanolamine (Etn-P) moiety that is linked to protein (Yin et al. 2005). Proteins released in this way have a C-terminal GPI signal-anchor sequence, and this, and signals for wall anchorage of GPI-CWP, are discussed in Lumenal steps in GPI assembly and in Incorporation of GPI proteins into the cell wall. At least one GPI-CWP, Cwp1, can additionally be linked to the wall via an alkali-labile linkage (Kapteyn et al. 2001).
Mild alkali-releasable proteins:
These include four proteins with internal repeats (PIR proteins), which have multiple copies of the internal repeat sequence SQ[I/V][S/T/G]DGQ[I/V]Q[A][S/T/A] (Toh-E et al. 1993) [simplified to DGQ[hydrophobic amino acid]Q by Klis et al. (2010)] and are released by mild alkali or β1,3-glucanase (Mrša et al. 1997). PIR proteins have no GPI attachment sequence and are not linked to β1,6-glucan; rather, they are ester-linked to β1,3-glucan via side chains of amino acids in the repeat sequences (Ecker et al. 2006; see Incorporation of PIR proteins into the cell wall). Because PIR proteins can form several linkages to β1,3-glucan, they could interconnect glucans. Single PIR repeats are also present in certain GPI-CWP (see Incorporation of PIR proteins into the cell wall), and additional proteins lacking PIR sequences can be also extracted with alkali or β1,3-glucanase (Yin et al. 2005; see Cell Wall-Active and Nonenzymatic Surface Proteins and Their Functions).
Disulfide-linked proteins:
Various proteins can be released from the walls of living cells with sulfhydryl reagents, indicating that they are directly attached via disulfides or retained behind a network of disulfide-linked proteins (Orlean et al. 1986; Cappellaro et al. 1998; Moukadiri et al. 1999; Moukadiri and Zueco 2001; Insenser et al. 2010). Disulfide-linked mannoproteins create a barrier that protects wall polysaccharides from externally added glycosylhydrolases, making mercaptoethanol and protease pretreatment necessary for spheroplasting with lytic enzymes (Zlotnik et al. 1984). Furthermore, the ability of the cysteine-rich domain of Wsc1 to form disulfide cross-links is important for this mechanosensor in forming clusters and in functioning in CWI signaling (Heinisch et al. 2010; Dupres et al. 2011).
Strategies to identify CWP
Biochemistry and bioinformatics have been used to identify CWP. Because proteins can be associated with the wall in different ways, different treatments are necessary to release them. Separation and identification of individual CWP can be complicated by their heavy and heterogeneous glycosylation. CWP can be released from the wall by treatment with β1,3- and β1,6-glucanases (Van der Vaart et al. 1995; Mrša et al. 1997; Shimoi et al. 1998). In one approach, labeling of intact cells with a membrane-impermeable biotinylation reagent, followed successively by SDS and mercaptoethanol extraction and then mild alkali or β1,3-glucanase treatment, led to identification of nine “soluble cell wall” (Scw) and 11 “covalently linked cell wall” (Ccw) proteins (Mrša et al. 1997). In another approach, isolated walls, extracted with SDS, mercaptoethanol, NaCl, and EDTA, were then treated with HF/pyridine or mild alkali, and the CWP released were identified by mass spectrometry. Additional CWP were identified following proteolytic digestion of walls, the two procedures yielding 19 CWP, including GPI and PIR proteins and alkali-releasable proteins without PIR sequences (Yin et al. 2005, 2007). These studies led to the estimate that a dividing haploid cell contains ∼2 × 106 covalently attached CWPs and the suggestion that CWP form a densely packed surface layer (Yin et al. 2007). A strategy that also permitted identification of noncovalently associated surface proteins used treatment of intact cells with dithiothreitol followed by two-dimensional electrophoretic separation, or direct proteolytic digestion and isolation of peptides, and then mass spectrometric protein fingerprinting (Insenser et al. 2010). The 99 proteins so identified included CWP and glycosylhydrolases, as well as proteins associated with intracellular functions. The presence in the wall of proteins considered cytosolic raises the possibility that they reach the wall via a nonconventional export pathway (Nombela et al. 2006; Insenser et al. 2010). However, mercaptoethanol can make the plasma membrane permeable to cytosolic proteins (Klis et al. 2007).
Bioinformatics has been used identify proteins likely to receive a GPI anchor; hence, members of the major class of CWP. In silico surveys for GPI attachment sequences reveal that the S. cerevisiae proteome contains 60–70 potential GPI proteins, which often contain Ser/Thr-rich stretches (Caro et al. 1997; Hamada et al. 1998a; De Groot et al. 2003; Eisenhaber et al. 2004).
Cell wall phenotypes
Cell wall phenotypes that are typically scored are sensitivity to hypo-osmotic stress, which can be tested on half-strength yeast extract peptone medium (Valdivia and Schekman 2003); sensitivity or resistance to CFW and Congo Red; sensitivity to aminoglycosides, β1,3-glucan synthase inhibitors, caffeine, SDS, and K1 killer toxin; and sensitivity to β1,3-glucanase preparations (Ram et al. 1994; Hampsey 1997; Lussier et al. 1997b; De Groot et al. 2001).
Precursors and Carrier Lipids
Sugar nucleotides
Glycosyltransferases involved in wall biogenesis use UDP-Glc, UDP-GlcNAc, and GDP-Man or dolichol phosphate (Dol-P) Man or Dol-P-Glc as donors. UDP-Glc is formed from UTP and Glc-1-P by the essential UDPGlc pyrophosphorylase Ugp1 (Daran et al. 1995). Impairment of UDP-Glc synthesis ultimately impacts formation of cell wall β-glucans, although cells with no more than 5% of the activities of the phosphoglucomutases and Ugp1 that generate UDP-Glc are unaffected in growth and viability (Daran et al. 1997). GDP-Man is formed from Fru-6-P by the successive actions of phosphomannose isomerase (Pmi40), phosphomannomutase (Sec53), and GDP-Man pyrophosphorylase (Psa1/Srb1/Vig9), which are all encoded by essential genes, and loss of any of these enzyme activities leads to severe glycosylation and secretion defects (Hashimoto et al. 1997; Orlean 1997; Yoda et al. 2000). Elevated expression of GDP-Man pyrophosphorylase, which presumably increases GDP-Man levels, corrects the N-glycosylation defects in alg1 and alg2 mutants and the mannosylation and GPI synthetic defects in dpm1 cells (Janik et al. 2003). GDP-Man transport into the Golgi lumen is discussed in Sugar nucleotide transport.
The pathway for UDP-GlcNAc formation (Milewski et al. 2006) involves conversion of Fru-6-P to GlcN-6-P by glutamine:Fru-6-P amidotransferase Gfa1 (Watzele and Tanner 1989), N-acetylation of GlcN-6-P by Gna1 (Mio et al. 1999), conversion of GlcNAc-6-P to GlcNAc-1-P by the GlcNAc phosphate mutase Agm1/Pcm1 (Hofmann et al. 1994), and formation of UDP-GlcNAc by the pyrophosphorylase Uap1/Qri1 (Mio et al. 1998). Deficiencies in these enzymes lead to formation of short chains of undivided cells, swelling, and eventual lysis, a phenomenon known as glucosamineless death (Ballou et al. 1977; Mio et al. 1998, 1999). Glucosamine supply is highly regulated and impacts chitin levels, which increase in response to mating pheromones and cell wall stress (File S1).
Dolichol and dolichol phosphate sugars
Dolichol phosphate synthesis:
Yeast dolichols contain 14–18 isoprene units (Jung and Tanner 1973). Biosynthesis of dolichol (Schenk et al. 2001a; Grabinska and Palamarczyk 2002) starts with extension of trans farnesyl-PP by successive addition of cis-isoprene units by the homologous cis-prenyltransferases Rer2 and Srt1 (Sato et al. 1999; Schenk et al. 2001b). Rer2 is dominant and makes dolichols with 10–14 isoprene units, whereas dolichols made by Srt1 in cells lacking Rer2 contain 19–22 isoprenes. rer2Δ strains have severe defects in growth and in N- and O-glycosylation (Sato et al. 1999). The next two steps are likely the removal of the two phosphates from dehydrodolichyl diphosphate by unknown enzymes. The α-isoprene unit of the polyprenol is then reduced, and Dfg10 is responsible for much of this activity (Cantagrel et al. 2010; File S1). Dolichol is likely next phosphorylated by the CTP-dependent Dol kinase Sec59 (Heller et al. 1992).
Dol-PP generated on the lumenal side of the ER membrane after transfer of the N-linked oligosaccharide to protein is dephosphorylated to Dol-P and Pi on that side of the membrane by the phosphatase Cwh8/Cax4 (Van Berkel et al. 1999; Fernandez et al. 2001). CWH8-disruptants have an N-glycosylation defect and a growth defect that is partially suppressed by high-level expression of RER2, SEC59, and the lipid phosphatase gene LPP1. Cwh8 likely has a role in recycling of Dol-PP for use in new rounds of N-glycosylation on the cytoplasmic face of the ER membrane.
Dol-P-Man and Dol-P-Glc synthesis:
Dol-P-Man and Dol-P-Glc are the donors in the lumenal glycosyltransfers that occur in protein O-mannosylation and the assembly pathways for the Dol-PP-linked precursor in N-glycosylation and the GPI anchor precursor glycolipid. Dol-P-Man is formed upon transfer of Man from GDP-Man to Dol-P by the Dol-P-Man synthase Dpm1 (Orlean et al. 1988; Orlean 1990). Temperature-sensitive dpm1 mutants have cell wall defects, consistent with a general block of glycosylation and GPI anchoring, and these phenotypes are suppressed by high-level expression of RER2, which presumably elevates Dol-P levels (Orlowski et al. 2007).
Dol-P-Glc is formed from UDP-Glc and Dol-P. Deletion of the synthase gene, ALG5, is not lethal, and the disruptants show no obvious growth defects (Te Heesen et al. 1994). Because Dol-P-Man and Dol-P-Glc are used in lumenal reactions, and because spontaneous transmembrane translocation of these glycolipids is not favored energetically, their translocation may be protein-mediated. Assays for Dol-P-Man flipping have been reported (Haselbeck and Tanner 1982; Sanyal and Menon 2010), but a protein involved has yet to be identified. One possibility is that the Dol-P-Man and Dol-P-Glc-utilizing transferases are their own flippases (Burda and Aebi 1999).
Biosynthesis of Wall Components Along the Secretory Pathway
Cell surface proteins can be modified with N-glycans, O-linked manno-oligosaccharides, and a GPI anchor as they transit the secretory pathway. Initial attachment of these structures occurs in the ER lumen, and the glycans are modified in the Golgi before the glycoproteins are deposited in the plasma membrane or secreted from the cell, whereupon many become cross-linked to wall polysaccharides.
N-Glycosylation
N-glycosylation involves preassembly of a branched 14-sugar oligosaccharide on the carrier Dol-PP in the ER membrane and then transfer of the oligosaccharide to selected asparagines in the ER lumen (Burda and Aebi 1999; Helenius and Aebi 2004; Lehle et al. 2006; Larkin and Imperiali 2011). The first 7 sugars are transferred from sugar nucleotides on the cytosolic side of the ER membrane, and the remainder from Dol-P on the lumenal side (Figure 2).
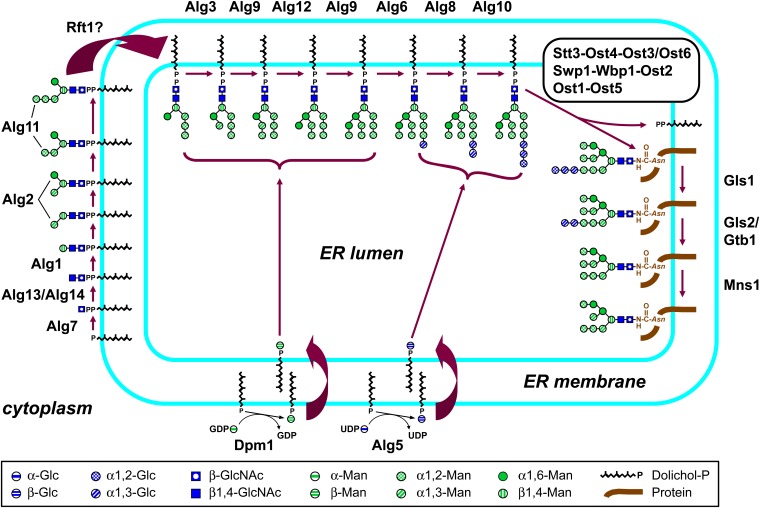
Figure 2 .
Assembly of the Dol-PP-linked precursor oligosaccharide in N-glycosylation, its transfer to protein, and subsequent glycan processing. Residues added at the cytoplasmic face of the ER membrane originate from sugar nucleotides, whereas Dol-P sugars generated at the cytoplasmic face of the membrane are the donors in lumenal transfers. Symbols are adaptations of those used by the Consortium of Glycobiology Editors in Essentials in Glycobiology (Varki and Sharon 2009).
Assembly and transfer of the Dol-PP-linked precursor oligosaccharide:
Steps on the cytoplasmic face of the ER membrane:
These steps are (i) transfer of GlcNAc-1-P from UDP-GlcNAc to Dol-P by Alg7, the target of the N-glycosylation inhibitor tunicamycin (Barnes et al. 1984), (ii) transfer of β1,4-GlcNAc from UDP-GlcNAc by heterodimeric Alg13/Alg14 (Bickel et al. 2005; Chantret et al. 2005; Gao et al. 2005), (iii) transfer of a β1,4-linked Man by Alg1 (Couto et al. 1984), (iv) successive transfer of an α1,3 and an α1,6 Man by Alg2 (O’Reilly et al. 2006; Kämpf et al. 2009), and (v) transfer of two α1,2-linked Man by Alg11 (Cipollo et al. 2001; O’Reilly et al. 2006; Absmanner et al. 2010). These proteins act in higher-order complexes (Gao et al. 2004; Noffz et al. 2009; File S2).
Transmembrane translocation of Dol-PP-oligosaccharides:
Dol-PP-GlcNAc2Man5 formed on the cytoplasmic face of the ER membrane is somehow translocated into the lumen (Burda and Aebi 1999; Helenius and Aebi 2002), and Rft1 is a candidate for the flippase (Helenius et al. 2002). Strains deficient in Rft1 accumulate Dol-PP-GlcNAc2Man5, but retain Alg3 Man-T activity and are unaffected in O-mannosylation or in GPI assembly, ruling out deficiences in Dol-P-Man supply to the lumen. Furthermore, high level expression of RFT1 partially suppresses the growth defect of alg11Δ and leads to increased levels of lumenal Dol-PP-GlcNAc2Man6-7 and an increase in glycosylation of the reporter carboxypeptidase Y, consistent with enhanced flipping of the suboptimal substrate Dol-PP-GlcNAc2Man3 (Helenius et al. 2002). However, although the above evidence is consistent with Rft1 being the flippase, depletion of Rft1 did not lead to loss of flipping activity measured in vitro (Frank et al. 2008; Rush et al. 2009; File S2).
Lumenal steps in Dol-PP-oligosaccharide assembly:
Dol-PP-GlcNAc2Man5 is extended by four Man and three Glc on the lumenal side of the ER membrane using Dol-P-Man and Dol-P-Glc as donors. Alg3 adds the sixth, α1,3-Man to the α1,6 Man of Dol-PP-GlcNAc2Man5 (Aebi et al. 1996; Sharma et al. 2001), Alg9 then transfers an α1,2-linked Man to the Man added by Alg3 (Burda et al. 1999; Cipollo and Trimble 2002), and Alg12 next adds the eighth, α1,6-Man to the Man added by Alg9 (Burda et al. 1999). Alg9 acts again to add the ninth Man, α1,2-linked Man to the Man added by Alg12 (Frank and Aebi 2005). Two α1,3-linked Glc are successively added by Alg6 and Alg8 to extend the arm of the heptasaccharide ending in the α1,2-linked Man transferred by Alg11, and finally, Alg10 adds an α1,2-Glc (Stagljar et al. 1994; Reiss et al. 1996; Burda and Aebi 1998). The six Dol-P-sugar-utilizing transferases are members of a family of multispanning membrane proteins that includes Man-T involved in GPI biosynthesis (Oriol et al. 2002).
Oligosaccharide transfer to protein:
GlcNAc2Man9Glc3 is transferred from Dol-PP to asparagines by the oligosaccharyltransferase complex (OST) (Knauer and Lehle 1999a; Yan and Lennarz 2005a; Kelleher and Gilmore 2006; Lehle et al. 2006; Weerapana and Imperiali 2006; Lennarz 2007; Larkin and Imperiali 2011). Acceptor asparagines occur in the sequon Asn-X-Ser/Thr, where X can be any amino acid except Pro. Mass spectrometric analyses of wall-derived peptides revealed that 85% of sequons were completely occupied, with preferential usage Asn-X-Thr over Asn-X-Ser sites (Schulz and Aebi 2009). Analyses of protein-linked N-glycans in mutants defective in the elaboration of the Dol-PP-linked precursor indicate that structures smaller than GlcNAc2Man9Glc3 can be transferred in vivo.
Yeast OST consists of Stt3, Ost1, Ost2, Wbp1, Swp1, Ost4, Ost5, and either of the paralogues Ost3 or Ost6. The first five are encoded by essential genes. Two OST complexes can be formed, containing either Ost3 or Ost6 (Schwarz et al. 2005; Spirig et al. 2005; Yan and Lennarz 2005b). The Ost3-containing complex is about four times as abundant as the Ost6-containing one (Spirig et al. 2005). Genetic interaction studies and coimmunoprecipitation and chemical cross-linking experiments suggest the existence of three OST subcomplexes: (i) Swp1-Wbp1-Ost2, (ii) Stt3-Ost4-Ost3, and (iii) Ost1-Ost5 (Karaoglu et al. 1997; Reiss et al. 1997; Spirig et al. 1997; Knauer and Lehle 1999b; Kim et al. 2003; Li et al. 2003; Kelleher and Gilmore 2006; File S2). OST complexes themselves may function as dimers (Chavan et al. 2006).
Stt3 is the catalytic subunit of OST. It can be chemically cross-linked to peptides derivatized with photoactivatable groups (Yan and Lennarz 2002; Nilsson et al. 2003), and its bacterial and protist homologs transfer glycans to protein substrates (Wacker et al. 2002; Kelleher and Gilmore 2006; Kelleher et al. 2007; Nasab et al. 2008; Hese et al. 2009). Ost3 and Ost6 have a lumenal thioreductase fold with a CXXC motif common to proteins involved in disulfide bond shuffling during oxidative protein folding (Kelleher and Gilmore 2006; Schulz et al. 2009), and the proteins likely form transient disulfide bonds with nascent proteins and promote efficient glycosylation of Asn-X-Ser/Thr sites by delaying oxidative protein folding (Schulz and Aebi 2009; Schulz et al. 2009). The Swp1p, Wbp1p, and Ost2p subcomplex may confer the preference of OST for GlcNAc2Man9Glc3 (Pathak et al. 1995; Kelleher and Gilmore 2006), Ost4 is required for recruitment of Ost3 and Ost6 to OST and also interacts with Stt3 (Karaoglu et al. 1997; Spirig et al. 1997; Knauer and Lehle 1999b; Kim et al. 2000, 2003; Spirig et al. 2005), and Ost1 may funnel nascent polypeptides to Stt3 (Lennarz 2007). OST may be subject to regulation by the CWI pathway via an interaction between Pkc1 or components of the PKC pathway with Stt3 (Park and Lennarz 2000; Chavan et al. 2003a; File S2).
N-glycan processing in the ER and glycoprotein quality control:
Protein-linked GlcNAc2Man9Glc3 is processed to glycans that are recognized by mechanisms that monitor correct protein folding and permit export from the ER or ensure degradation if the protein misfolds (Herscovics 1999; Aebi et al. 2010). Processing proceeds by removal of the α1,2-linked Glc by glucosidase I, Gls1/Cwh41 (Romero et al. 1997), and then of the two α1,3-linked Glc by soluble glucosidase II, a heterodimer of catalytic Gls2/Rot2 and Gtb1 (Trombetta et al. 1996; Wilkinson et al. 2006; Quinn et al. 2009; Figure 2). ER mannosidase I, Mns1, removes an α1,2 Man to generate GlcNAc2Man8 (Jakob et al. 1998; Herscovics 1999), and, if correctly folded, proteins bearing this glycan are exported from the ER. Un- or misfolded proteins are bound by protein disulfide isomerase Pdi1, some of which is in complex with Mns1 homolog Htm1, which trims the glycan to a GlcNAc2Man7 (Clerc et al. 2009; Gauss et al. 2011; File S2). Misfolded proteins with GlcNAc2Man7 are targeted to the cytosol for destruction by the ER-associated protein degradation (ERAD) system (Helenius and Aebi 2004). They are bound by the lectin Yos9 (Buschhorn et al. 2004; Bhamidipati et al. 2005; Kim et al. 2005; Szathmary et al. 2005) and in turn directed to the HRD-ubiquitin ligase complex of Hrd1 and Hrd3 for retrotranslocation to the cytoplasm (Bays et al. 2001; Deak and Wolf 2001; Gauss et al. 2006), where they are deglycosylated by peptide N-glycanase Png1 (Suzuki et al. 2000; Hirayama et al. 2010).
In mammals and Schizosaccharomyces pombe, following glucosidase II action, UDP-Glc:glycoprotein glucosyltransferase (UGGT) adds back an α1,3-Glc, allowing the monoglucosylated N-glycans to interact with the lumenal lectin domains of calnexin or calreticulin (Parodi 1999; Caramelo and Parodi 2007; Aebi et al. 2010). This interaction retains partially folded or misfolded proteins in the ER and buys them time to fold properly and be deglucosylated. Properly folded proteins are no longer recognized by UGGT and exported to the Golgi, whereas persistent misfolders are removed by ERAD. In S. cerevisiae, however, this quality control mechanism does not operate because UGGT activity is not detectable, and although the S. cerevisiae ER protein Kre5 is a sequence homolog of S. pombe UGGT, expression of the S. pombe UGGT cannot rescue the growth defect of kre5 mutants. However, kre5, as well as glucosidase I and II mutants and mutants in the calnexin homolog Cne1, are defective in β1,6-glucan synthesis, indicating roles for S. cerevisiae homologs of players in the UGGT/calnexin quality control system in β1,6-glucan synthesis (Jiang et al. 1996; Shahinian et al. 1998; Simons et al. 1998; see β1,6-Glucan).
Mannan elaboration in the Golgi:
N-linked glycans on proteins are extended with a Man10-14 core-type structure or with mannan outer chains containing up to 150–200 Man. Both structures can be modified with mannose phosphate (Figure 3) (Ballou 1990; Orlean 1997; Jigami 2008). The mannoses all originate from GDP-Man and are transferred by members of several families of redundant Golgi Man-T.
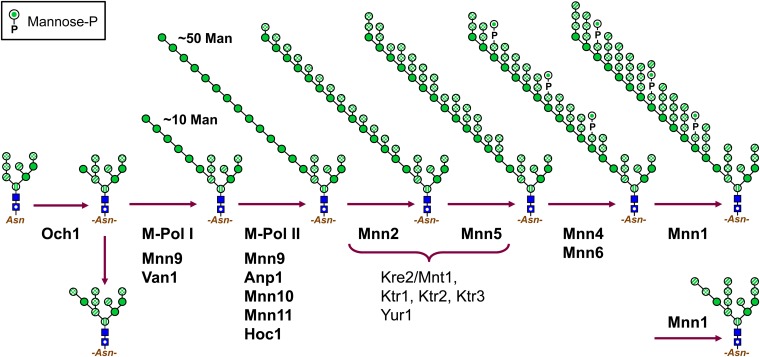
Figure 3 .
Formation of mannan outer chains and core-type N-glycans in the Golgi. Protein-bound Man8-GlcNAc2 structures are first acted on by the Och1 α1,6-Man-T in the cis-Golgi. The initiating α1,6-Man is then elongated with ∼10 α1,6-linked Man by mannan polymerase (M-Pol)-I, and this chain is then extended with up to ∼50 α1,6-linked Man by M-Pol-II. Kre2/Mnt1, Ktr1, Ktr2, Ktr3, and Yur1 collectively add α1,2-linked mannoses. Core-type glycans are formed when an α1,2-linked Man is added to the Och1-derived α1,6-Man. Symbols are as used in Figure 2.
Formation of core-type N-glycan and mannan outer chains:
Formation of core structures and mannan is initiated in the cis-Golgi by Och1, which transfers an α1,6-Man to the α1,3-Man of the N-glycan that had been added by Alg2 (Nakayama et al. 1997). OCH1 deletion is lethal in some strain backgrounds, and och1Δ strains have severe growth defects, highlighting the importance of mannan.
Synthesis of the poly-α1,6-mannan backbone is carried out in the cis-Golgi by two protein complexes: Man-Pol I, see containing homologs Mnn9 and Van1, and Man-Pol II, containing Mnn9, Anp1, Hoc1, and related Mnn10 and Mnn11 (Hashimoto and Yoda 1997; Jungmann and Munro 1998; Jungmann et al. 1999; File S2). M-Pol I acts first, with its Mnn9 subunit adding the first α1,6-Man to the Och1-derived Man, upon which 10–15 α1,6-Man are added in Van1-requiring reactions (Stolz and Munro 2002; Rodionov et al. 2009). This α1,6 backbone is further elongated with 40–60 α1,6-Man by M-Pol II, whose Mnn10 and Mnn11 subunits are responsible for the majority of the α1,6-Man-T activity (Jungmann et al. 1999). Hoc1’s role is unclear.
Core-type N-glycans are formed when an α1,2-Man is added to the Och1-derived Man, blocking elongation of an α1,6 mannan chain. The protein(s) involved have not been identified, but presumably either they, or M-Pol I, can tell from the context of an N-glycan which type of structure it is to bear (Lewis and Ballou 1991; Stolz and Munro 2002; Rodionov et al. 2009). Core-type structures are completed when that α1,2-Man, as well as the two other terminal α1,2-Man on the Man8GlcNAc2 structure, receives α1,3 mannoses from Mnn1.
Mannan side branching and mannose phosphate addition:
Branching of the α1,6 mannan backbone is initiated by the Mnn2 α1,2-Man-T, and Mnn5 adds a second α1,2-Man (Rayner and Munro 1998). Mnn2 and Mnn5 make up one of two Mnn1 subfamilies (Lussier et al. 1999). Five members of the Ktr1 protein subfamily, Kre2/Mnt1, Yur1, Ktr1, Ktr2, and Ktr3, also contribute to N-linked outer chain synthesis, acting collectively in the addition of the second and subsequent α1,2-mannoses to mannan side branches (Lussier et al. 1996, 1997a, 1999).
Core-type glycans and mannan can be modified with Man-P on α1,2-linked mannoses. Mnn6/Ktr6, a Ktr1 subfamily member, is mostly responsible for transferring Man-1-P from GDP-Man, generating GMP (Wang et al. 1997; Jigami and Odani 1999; File S2). Mnn4 is also involved in Man-P addition but does not resemble glycosyltransferases and may be regulatory (Odani et al. 1996). Levels of mannan phosphorylation are highest in the late log and stationary phases, when MNN4 expression is elevated (Odani et al. 1997). Terminal α1,2 mannoses and Man-1-Ps can be capped with α1,3-Man, added by Mnn1 (Ballou 1990; Yip et al. 1994).
O-Mannosylation
Many yeast proteins are modified on extracytoplasmic Ser or Thr residues with linear manno-oligosaccharides. The first Man is attached in α-linkage in the lumen of the ER, and up to four further Man are added by Man-T that act mostly in the Golgi.
Protein O-mannosyltransferases in the ER:
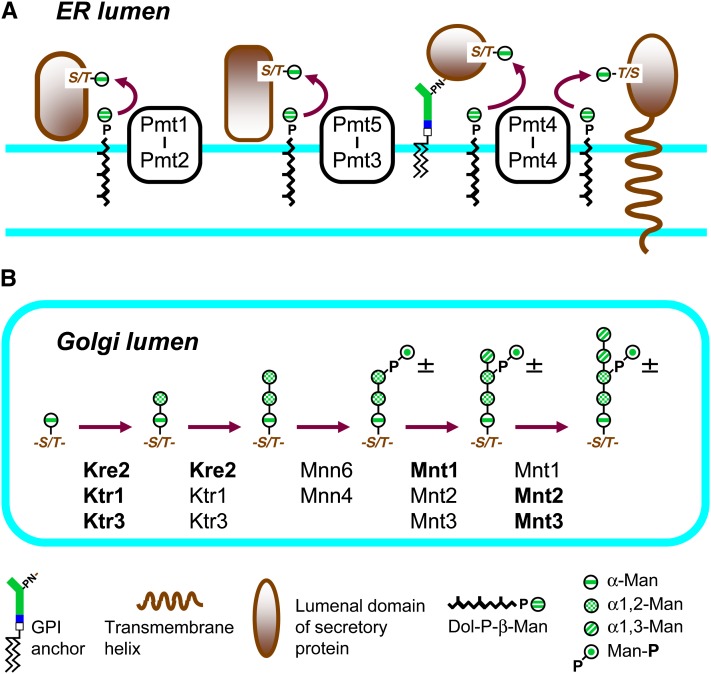
The first Man is transferred from Dol-P-Man (Strahl-Bolsinger et al. 1999; Lehle et al. 2006; Lommel and Strahl 2009). Consistent with the requirement for Dol-P-Man, O-mannosylation of the model protein Cts1 is blocked in a dpm1-Ts mutant (Orlean 1990). There are six protein O-mannosyltransferases (PMTs) in yeast. Prototypical Pmt1 is an ER protein with seven membrane-spanning domains with conserved residues important for catalysis and for interactions with acceptor peptides located in the first lumenal loop (Strahl-Bolsinger and Scheinost 1999; Girrbach et al. 2000; Lommel et al. 2011).
Pmts function as hetero- or homodimers, and the pairs that are formed are determined by membership of a subunit in one of three Pmt subfamilies. Pmt1 family members Pmt1 and Pmt5 can form heterodimers with members of the Pmt2 family (which also contains Pmt3 and Pmt6), for example, Pmt1-Pmt2 and Pmt5-Pmt3 dimers, which are the most prevalent complexes (Girrbach and Strahl 2003). Pmt4, the lone representative of the third family, forms homodimers.
Analyses of O-mannosylation of individual proteins in pmtΔ strains reveal that the different Pmt complexes have specificity for different protein substrates (File S3). Substrates for Pmt4 need to be attached to the membrane by a transmembrane domain or a GPI anchor and have an adjacent, lumenal Ser/Thr-rich domain, whereas Pmt1/Pmt2 substrates can be soluble or membrane-associated (Hutzler et al. 2007).
Because PMTs modify Ser and Thr, N-linked glycosylation sites are also potential targets, and this is the case with Cwp5. This protein contains a single sequon, NAT, that is normally O-mannosylated by Pmt4, but which receives an N-linked glycan in pmt4Δ cells (Ecker et al. 2003). O-mannosylation, therefore, normally precedes the action of OST on Cwp5 and may control N-glycosylation of this protein, and perhaps others as well.
Extension and phosphorylation of O-linked manno-oligosaccharide chains:
The Ser- or Thr-linked Man is extended with up to four α-linked Man by GDP-Man-dependent Man-T of the Ktr1 and Mnn1 families (Lussier et al. 1999; Figure 4; File S3). Transfer of the first two α1,2-Man is carried out by the Ktr1 subfamily members Ktr1, Ktr3, and Kre2 and extension of the trisaccharide chain with one or two α1,3-linked Man by Mnn1 family members Mnn1, Mnt2, and Mnt3 (Lussier et al. 1997a; Romero et al. 1999). The second α1,2-Man of an O-linked glycan can be modified with Man-1-P by Mnn6 with the involvement of the regulator Mnn4 (Nakayama et al. 1998).
Figure 4 .
Biosynthesis of O-linked glycans. (A) Addition of α-Man by protein O-mannosyltransferases in the ER lumen. Pmt4 homodimers act on membrane proteins or GPI proteins. Representative Pmt heterodimers are shown. (B) Extension of O-linked manno-oligosaccharides in the Golgi. Ktr1 family members have a collective role in adding α1,2-linked mannoses, and Mnt1 family members add α1,3-linked mannoses. The dominant Man-T active at each step are shown in boldface type. Man-P may be added to saccharides with two α1,2-linked Man.
Importance and functions of O-mannosyl glycans:
No individual PMT deletion is lethal, but strains lacking certain combinations of three Pmts, such as pmt1Δ pmt2Δ pmt4Δ or pmt2Δ pmt3Δ pmt4Δ, are inviable, even with osmotic support, indicating that yeast must carry out some minimum level of O-mannosylation to be viable or that one or more essential proteins need to be O-mannosylated (Gentzsch and Tanner 1996; Lommel et al. 2004). Moreover, strains lacking other combinations of Pmts, such as the pmt2Δ pmt3Δ and pmt2Δ pmt4Δ double nulls or the pmt1Δ pmt2Δ pmt3Δ triple null, are osmotically fragile, indicating impaired wall assembly (Gentzsch and Tanner 1996). Analyses of pmt mutants show that O-mannosylation can be important for function of individual O-mannosylated proteins (File S3).
The phenotypes of pmt mutants are mimicked by treatment with the rhodanine-3-acetic acid derivative OGT1458, which inhibits PMT activity in vitro (Orchard et al. 2004; Arroyo et al. 2011). OGT1458 was used to analyze genome-wide transcriptional changes in response to inhibition of O-mannosylation. Consistent with the importance of O-mannosylation in wall construction and protein stability, consequences of defective O-mannosylation were activation of the CWI pathway and the unfolded protein response (Arroyo et al. 2011). Furthermore, certain genes involved in N-linked mannan outer chain assembly were upregulated. This, together with the finding that PMT gene transcription is elevated when N-glycosylation is inhibited by tunicamycin (Travers et al. 2000), suggests that the N- and O-linked glycans of cell wall mannoproteins can compensate for one another to some extent (Arroyo et al. 2011).
GPI anchoring
GPI structure and proteins that receive GPIs:
GPI structure:
S. cerevisiae GPI anchors have the core structure protein CO-NH2-CH2-CH2-PO4-6-Manα1,2Manα1,6Manα1,4GlcNα1,6-myoinositol phospholipid. In addition, the third, α1,2-Man, bears a fourth α1,2-Man that is added during precursor assembly, and this Man may receive another α1,2- or α1,3-linked Man in the Golgi (Fankhauser et al. 1993). The α1,4- and α1,6-linked Man are also modified with Etn-P at their 2′- and 6′-OHs, respectively, and the 2-OH of inositol is transiently modified with palmitate (Orlean and Menon 2007; Pittet and Conzelmann 2007) (Figure 5). The lipid moiety, initially diacylglycerol, is remodeled to a diacylglycerol with C26, acyl chains, or, in many cases, to a ceramide (Conzelmann et al. 1992; Fankhauser et al. 1993).
Figure 5 .
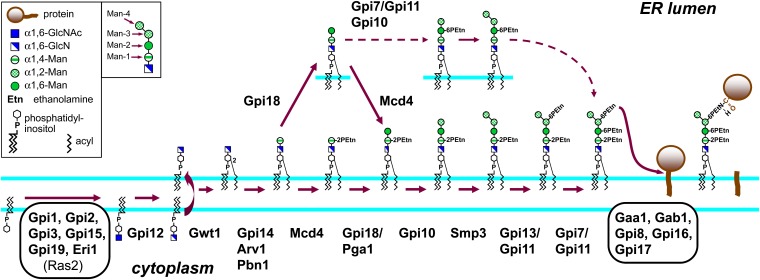
Biosynthesis of the GPI precursor and its transfer to protein in the ER membrane. GlcNAc addition to PI and de-N-acetylation of GlcNAc-PI to GlcN-PI occur at the cytoplasmic face of the ER membrane, and further additions to the GPI occur on the lumenal side of the ER membrane. Gpi18 and Mcd4 need not act in a defined order. Man3- and Man4-GPIs either bearing Etn-P on Man-2 but not Man-1 or without any Etn-Ps (not shown) have also been detected in radiolabeling experiments with certain late-stage GPI assembly mutants.
Identification of GPI proteins:
Biochemical demonstrations of a GPI on a yeast protein are rare, and the criterion of release of a protein by treatment with Ptd-Ins-specific phospholipase C (PI-PLC) is unreliable because although protein-bound GPIs are mostly sensitive to PI-PLC, this treatment does not always render the protein aqueous soluble in the commonly used Triton X-114 fractionation procedure (Conzelmann et al. 1990). Many GPI proteins become covalently linked to wall polysaccharide, and release from walls by treatment with HF/pyridine is a clue that the protein had received a GPI (see GPI proteins; Yin et al. 2005). The presence of a GPI is usually inferred from the results of in silico analyses of a protein’s sequence.
Features of a likely GPI protein are a hydrophobic N-terminal secretion signal and a C-terminal GPI signal-anchor sequence that includes the amino acid residue, ω, to which the GPI will be amide-linked. Amino acids N-terminal to ω are designated ω(−), and those C-terminal, are designated ω(+). Proceeding from the C-terminal amino acid of the unprocessed protein, the signal anchor signal consists of (i) a variable stretch of hydrophobic amino acids capable of spanning the membrane; (ii) a spacer region of moderately polar amino acids in positions ω+3 to ω+9 or more; (iii) the ω+2 residue, restricted mostly to G, A, or S; (iv) the ω residue itself, generally G, A, S, N, D, or C; and (v) a stretch of some 10 amino acids that may form a flexible linker region and whose relative polarity may influence plasma membrane or wall localization of the protein (Nuoffer et al. 1991, 1993; see Incorporation of GPI proteins into the cell wall). Some C-terminal sequences may contain alternative candidates for the ω and ω+2 amino acids. Evidence that a predicted GPI attachment sequence is functional can be obtained by fusing the sequence to the C terminus of a reporter protein and testing whether the reporter becomes expressed at the plasma membrane or in the wall (Hamada et al. 1998a).
Assembly of the GPI precursor and its attachment to protein in the ER:
At least 21 proteins are involved in GPI precursor synthesis and attachment to protein (Figure 5). Eighteen are encoded by essential genes, and mutants lacking any of the other noncatalytic proteins or GPI side-branching enzymes have severe growth defects. Additional information about GPI synthetic proteins and phenotypes associated with deficiencies in them is given in File S4.
Steps on the cytoplasmic face of ER membrane:
GPI assembly starts with transfer of GlcNAc from UDP-GlcNAc to PI. A complex of at least six proteins (GPI-GnT) is involved, of which Gpi3 is catalytic because it can be labeled with a photoactivatable UDP-GlcNAc analog (Kostova et al. 2000). GlcNAc transfer occurs at the cytoplasmic face of the ER membrane (Vidugiriene and Menon 1993; Watanabe et al. 1996; Tiede et al. 2000). Essential Gpi2, Gpi15, and Gpi19 (Leidich et al. 1995; Yan et al. 2001; Newman et al. 2005), and nonessential Gpi1 and Eri1 (Leidich and Orlean 1996; Sobering et al. 2004), are also required for GlcNAc-PI synthesis. ERI1 and GPI1 null mutants are temperature-sensitive. The mammalian orthologs of these proteins form a complex (Watanabe et al. 1998; Tiede et al. 2000; Eisenhaber et al. 2003; Murakami et al. 2005), and the yeast proteins likely also do, for Eri1 and Gpi19 associate with Gpi2 (Sobering et al. 2004; Newman et al. 2005). Roles of the noncatalytic subunits are unclear.
Ras2, in its GTP-bound form, can also join GPI-GnT (Sobering et al. 2004). Membranes from ras2Δ cells have 8- to 10-fold higher in vitro GPI-GnT activity than wild-type membranes, whereas membranes from cells expressing constitutively active Ras2-Val19 have almost undetectable activity. These findings indicate that Ras2-GTP is a negative regulator of GPI-GnT, and, depending on the degree to which the GTPase is activated, this could permit about a 200-fold range of GlcNAc-PI synthetic activity.
Once formed, GlcNAc-PI is de-N-acetylated at the cytoplasmic face of the ER membrane by Gpi12 (Vidugiriene and Menon 1993; Watanabe et al. 1999). GlcN-PI is the precursor likely to be translocated to the lumenal side of the ER membrane. Its flipping has been reconstituted in rat liver microsomes, but the protein involved is unknown (Vishwakarma and Menon 2005).
Lumenal steps in GPI assembly:
The inositol ring in GlcN-PI is next acylated on its 2-OH, making the glycolipid resistant to cleavage by PI-specific phospholipase C. The reaction uses acyl CoA as donor (Costello and Orlean 1992), and the acyl chain transferred in vivo is likely palmitate. Gwt1, the acyltransferase, was identified in a screen for resistance to 1-[4-butylbenzyl] isoquinoline, which inhibits surface expression of GPI proteins (Tsukahara et al. 2003; Umemura et al. 2003). Disruption of GWT1 is lethal or leads to slow growth and temperature sensitivity, depending on the strain background (Tsukahara et al. 2003). The inositol acyl chain may prevent GPIs from being translocated back to the cytoplasmic side of the ER membrane (Sagane et al. 2011), be important for later steps in GPI assembly or transfer to protein, or block the action of PI-specific phospholipases.
GlcN-(acyl)PI is next extended with four Man by GPI-Man-T I-IV, and the first three Man are concurrently modified with Etn-P by Etn-P-T I, II, and III. Dol-P-Man donates the mannoses because the dpm1 mutant accumulates GlcN-(acyl)PI (Orlean 1990). The first, α1,4-linked Man (Man-1, Figure 5) is added by Gpi14 (Maeda et al. 2001), and two additional proteins are involved at this step. One, Arv1, was originally implicated in ceramide and sterol metabolism. ARV1 disruptants are impaired in ER-to-Golgi transport of GPI proteins and accumulate GlcN-(acyl)PI in vitro (but not in vivo), although they are not defective in in vitro GPI-Man-T-I or Dpm1 activity or in N-glycosylation, and it was proposed that Arv1 has a role in delivering GlcN-(acyl)PI to Gpi14 (Kajiwara et al. 2008). The second protein, Pbn1, was implicated at the GPI-Man-T-I step because expression of both GPI14 and PBN1 is necessary to complement mammalian cell lines defective in Pbn1’s mammalian homolog Pig-X, and co-expression of PIG-X and the gene for Gpi14’s mammalian homolog, PIG-M, partially rescues the lethality of gpi14Δ (Ashida et al. 2005; Kim et al. 2007). Furthermore, Pbn1 depletion leads to accumulation of some of the ER form of the GPI protein Gas1, a phenotype of GPI precursor assembly mutants (Subramanian et al. 2006; File S4).
Addition of α1,6-linked Man-2 requires catalytic Gpi18 (Fabre et al. 2005; Kang et al. 2005) and Pga1 (Sato et al. 2007), which form a complex (Sato et al. 2007). Gpi18-deficient cells accumulate both a Man1-GPI with Etn-P esterified to its Man and an unmodified Man1-GPI, suggesting that GPI-Man-T-II can use either as acceptor (Fabre et al. 2005; Scarcelli et al. 2012).
Gpi10 and Smp3 successively add α1,2-linked Man-3 and Man-4 (Canivenc-Gansel et al. 1998; Sütterlin et al. 1998; Grimme et al. 2001). Smp3-dependent addition of Man-4 is essential because addition of this residue precedes addition of the Etn-P that subsequently becomes linked to protein (Grimme et al. 2001).
As the GPI glycan is extended, Etn-P moieties are added to the 2-OH of Man-1 and to the 6-OH of Man-2 and Man-3 (Orlean 2009). The Etn-Ps likely originate from Ptd-Etn (Menon and Stevens 1992; Imhof et al. 2000; File S4). The Etn-P-T-I, II, and III transferases are Mcd4, Gpi7, and Gpi13, respectively, which are 830- to 1100-amino-acid proteins predicted to have 10–14 transmembrane domains and a large lumenal loop containing sequences characteristic of the alkaline phosphatase superfamily that are important for function (Benachour et al. 1999; Gaynor et al. 1999; Galperin and Jedrzejas 2001; File S4). GPI-Etn-P-T-II and III also require small, hydrophobic Gpi11 for activity. mcd4 mutants accumulate unmodified Man1 and Man2-GPI (Wiedman et al. 2007; Scarcelli et al. 2012), suggesting that both structures can serve as Etn-P acceptors. From this, and because Gpi18-depleted cells accumulate Etn-P-modified Man1-GPI (Fabre et al. 2005), it seems that both Mcd4 and Gpi18 can use Man1-GPI as acceptor and then modify the GPI that the other has acted on (Figure 5; File S4). Etn-P transfer to Man-1 and GPI-dependent processing of Gas1 are inhibited by the terpenoid lactone YW3548 (Sütterlin et al. 1997, 1998). The Etn-P on Man-1 may enhance the ability of Gpi10 to add Man-3, promote export of GPI proteins from the ER, and be necessary for remodeling of the lipid moiety to ceramide (Zhu et al. 2006).
Gpi7 is the catalytic subunit of GPI-Etn-P-T-II, and GPI7 nulls, which are viable but temperature-sensitive, accumulate a Man4-GPI with Etn-P on Man-1 and Man-3 (Benachour et al. 1999). Essential Gpi11 was implicated at this step because Gpi11-deficient cells have similar GPI precursor accumulation profiles to gpi7Δ (Taron et al. 2000). The Etn-P on Man-2 enhances transfer of GPIs to protein, ER-to-Golgi transport of GPI proteins, GPI lipid remodeling to ceramide, transfer of GPI proteins to the wall, and targeting of certain GPI-anchored proteins in daughter cells (Benachour et al. 1999; Toh-E and Oguchi 1999; Richard et al. 2002; Fujita et al. 2004).
Gpi13 is the catalytic subunit of GPI-Etn-P-T-III. The major GPI accumulated upon Gpi13 depletion is a Man4-GPI with a single Etn-P on Man-1 (Flury et al. 2000; Taron et al. 2000). Gpi11 is likely involved in the GPI-Etn-P-T-III reaction because a gpi11-Ts mutant also accumulates a Man4-GPI with its Etn-P on Man-1 (K. Willis and P. Orlean, unpublished results), and human Gpi11 interacts with and stabilizes human Gpi13 (Hong et al. 2000).
GPI transfer to protein:
Man4-GPIs bearing three Etn-Ps are transferred to proteins with a C-terminal GPI signal-anchor sequence in a transamidation reaction in which the amino group of the Etn-P on Man-3 acts as nucleophile. Five essential membrane proteins are involved: Gaa1, Gab1, Gpi8, Gpi16, and Gpi17 (Hamburger et al. 1995; Benghezal et al. 1996; Fraering et al. 2001; Ohishi et al. 2000, 2001; Hong et al. 2003; Grimme et al. 2004). Gpi18 is catalytic because it resembles cysteine proteases and mutation of predicted active site residues eliminates its function (Meyer et al. 2000). The five transamidase subunits form a complex itself consisting of two subcomplexes: one containing Gaa1, Gpi8, and Gpi16, and the other, Gab1 and Gpi17 (Fraering et al. 2001; Grimme et al. 2004; Zhu et al. 2005). Roles for the noncatalytic subunits include recognition of the peptide and glycolipid substrates (Signorell and Menon 2009), and, in the case of Gab1 and Gpi8, possible interactions with the actin cytoskeleton (Grimme et al. 2004; File S4)
Remodeling of protein-bound GPIs:
Following GPI transfer to protein, both the anchor’s lipid and glycan remodeled (Figure 6; Fujita and Kinoshita 2010). The earliest event, which occurs in the ER, is removal of the inositol acyl moiety by lipase-related Bst1 (Tanaka et al. 2004; Fujita et al. 2006a). Next, the sn-2 acyl chain of the diacylglycerol is removed by the ER membrane protein Per1 to generate a lyso-GPI (Fujita et al. 2006b), whereupon a C26:0 acyl chain is transferred to the sn-2 position by Gup1 in the ER membrane (Bosson et al. 2006). Modifications of the GPI lipid by Bst1, Per1, and Gup1 are necessary for efficient transport of GPI proteins from the ER to the Golgi (File S4).
Figure 6 .
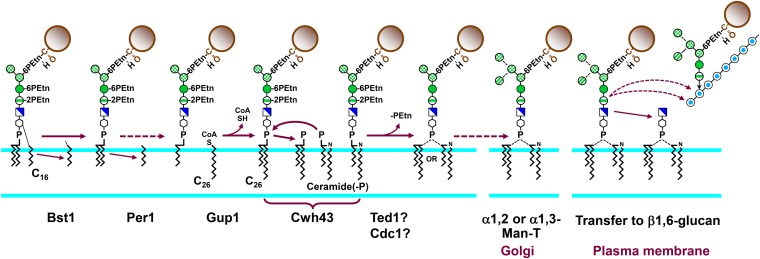
Remodeling of protein-bound GPIs. The inositol palmitoyl group and the sn-2 acyl chain are removed by Bst1 and Per1, respectively, and Gup1 transfers a C26:0 acyl chain to the sn-2 position. Cwh43 can replace diphosphatidic acid with ceramide phosphate (shown here) or diacylglycerol with ceramide. Etn-P on Man-1 and Man-2 may be removed by Ted1 and Cdc1. Steps through Etn-P removal occur in the ER. An α1,2- or an α1,3-linked Man is added to Man-4 in the Golgi by as yet unknown Man-T. At the plasma membrane, the GPI can be cleaved, possibly between GlcN and Man, and the reducing end of the GPI remnant transferred to β1,6-glucan. Symbols are as used in Figure 1 and Figure 5.
Many GPIs are next remodeled by replacement of their diacylglycerol with ceramide by Cwh43 (Martin-Yken et al. 2001; Ghugtyal et al. 2007; Umemura et al. 2007). Ceramide remodeling requires prior action of Bst1, and, because per1Δ and gup1Δ strains show defects in remodeling, the exchange reaction likely takes place after the first three lipid modification steps. The mechanism could involve a phospholipase-like reaction that replaces diphosphatidic acid with ceramide phosphate or diacylglycerol with ceramide (Ghugtyal et al. 2007; Fujita and Kinoshita 2010). Ceramide remodeling is not obligatory because certain GPI proteins, such as Gas1, reach the plasma membrane with a diacylglycerol-based anchor (Fankhauser et al. 1993). Moreover, ceramide remodeling does not seem to be required for incorporation of GPI proteins into the wall (Ghugtyal et al. 2007).
Further GPI processing events may be the removal of the Etn-P moieties from Man-2 and Man-1. This is inferred from the fact that mammalian PGAP5, which removes the side-branching Etn-P from Man-2 (Fujita et al. 2009), has two homologs in yeast: ER-localized Ted1 and Cdc1. Export of Gas1 is retarded in ted1Δ cells, and genetic interactions connect TED1 and CDC1 with processing and export of GPI proteins (Haass et al. 2007). Because Etn-P side chains are important for ceramide remodeling, they are likely removed after Cwh43 has acted.
Finally, a fifth, α1,2- or α1,3-linked Man can be added to Man-4 of protein-bound GPIs (Fankhauser et al. 1993). This modification is made to 20–30% of GPI proteins and occurs in the Golgi, but none of the many Golgi Man-T seems to be involved (Sipos et al. 1995; Pittet and Conzelmann 2007). On reaching the plasma membrane, the GPIs on many proteins become cross-linked to β1,6-glucan (see Incorporation of GPI proteins into the cell wall), and these GPI-CWP play structural or enzymatic roles in the wall (see Cell Wall-Active and Nonenzymatic Surface Proteins and Their Functions).
No individual GPI protein is essential in unstressed wild-type cells, so the lethality of mutations blocking GPI anchoring may be due to the collective effects of retarding ER exit and plasma membrane or wall anchorage of multiple proteins. Consistent with this, temperature-sensitive GPI anchoring mutants grown at semipermissive temperature have aberrant morphologies and shed wall proteins into the medium (Leidich and Orlean 1996; Vossen et al. 1997).
Sugar nucleotide transport
GDP-Man transport:
Cytoplasmically generated GDP-Man used by Golgi Man-T is transported into the Golgi lumen by Vrg4/Vig4. GMP, generated from GDP formed in Man-T reactions by GDPase activity, serves as antiporter. Vrg4/Vig4 is essential, and vrg4 mutants are defective in mannosylation of N- and O-linked glycans and mannosyl inositolphosphoceramides (Dean et al. 1997; Abe et al. 1999).
Two homologous Golgi proteins, Gda1 and Ynd1, have GDP-hydrolyzing activity. Gda1 has the highest activity toward GDP (Abeijon et al. 1989), and, consistent with GMP’s role as antiporter, rates of in vitro GDP-Man import into Golgi vesicles from gdaΔ cells are fivefold lower than those of vesicles from wild-type cells (Berninsone et al. 1994). Ynd1 is a broader specificity apyrase (Gao et al. 1999) that has a partially overlapping function with Gda1, and both Ynd1 and Gda1 are necessary for full elongation of N- and O-linked glycans (Gao et al. 1999; File S5).
Other sugar nucleotide transport activities:
Transport activities for UDP-Glc, UDP-GlcNAc, and UDP-Gal also occur in S. cerevisiae (Roy et al. 1998, 2000; Castro et al. 1999), and there are eight more candidate transporters (Dean et al. 1997; Esther et al. 2008) whose functions are unclear. UDP-Glc transport activity is present in the ER (Castro et al. 1999), and one possible need for it might be for a glucosylation reaction at an early stage of β1,6-glucan assembly (see β1,6-Glucan). Yea4 is an ER-localized UDP-GlcNAc transporter whose deletion impacts chitin synthesis (Roy et al. 2000; File S6). Hut1 is a candidate UDP-Gal transporter (Kainuma et al. 2001), although galactose has not been detected on S. cerevisiae glycans. Both Hut1 and Yea4 may have broader specificity and transport UDP-Glc (Esther et al. 2008).
Biosynthesis of Wall Components at the Plasma Membrane
Chitin
S. cerevisiae has three chitin synthase activities—CS I, CS II, and CS III—which require the catalytic proteins Chs1, Chs2, and Chs3, respectively. The Chs proteins are active in the plasma membrane although they originate from the rough ER. The pathways for trafficking and activation of Chs2 and Chs3 involve different sets of auxiliary proteins that ensure the correct spatial and temporal localization of chitin synthesis during septation.
Septum formation:
Factors determining the site at which a bud will be formed, and the proteins that recruit and organize the participants in septum formation, including septins and an actin–myosin contractile ring, are reviewed by Cabib et al. (2001), Cabib (2004), Roncero and Sanchez (2010), and Bi and Park (2012). Two chitin-containing structures are made during bud emergence and septum formation (Figure 7). The first is a ring deposited in the wall around the base of the emerging bud. This chitin is formed by Chs3 (Shaw et al. 1991), and, after cell separation, remains on the mother cell as a component of the bud scar. Upon completion of mitosis, the primary septum is formed by centripetal synthesis of chitin by Chs2 in the neck region between mother cell and bud (Shaw et al. 1991). Upon closure, the septum separates the plasma membranes of the two cells, accomplishing cytokinesis. In budding wild-type cells, the primary septum is thickened on both sides by deposition of a secondary septum that normally contains chitin, β1,3-glucan, β1,6-glucan, and covalently cross-linked mannoprotein (Rolli et al. 2009), resulting in a three-layered structure (Shaw et al. 1991).
Figure 7 .
Roles of chitin synthases II and III in chitin deposition during budding growth. (A) Chitin synthase III synthesizes a chitin ring (blue) around the base of the emerging bud. (B) The plasma membrane invaginates and chitin synthase II synthesizes the primary septum (red). No chitin is made in the lateral walls of the bud. (C) Secondary septa (green) are laid down on the mother- and daughter-cell sides of the primary septum, and chitin synthase III starts synthesizing lateral wall chitin in the bud (blue). (D) After cell separation, the bud scar (which is formed from the chitin ring made by Chs3), most of the primary septum made by Chs2, as well as secondary septal material deposited on the mother cell side, remain on the mother cell. The birth scar on the daughter cell contains residual chitin from the primary septum as well as secondary septal material. (E and F) Chitinase digestion of the primary septum from the daughter-cell side facilitates cell separation, and lateral wall chitin synthesis continues as the daughter cell grows. Figure is adapted from Cabib and Duran (2005).
Chs2 and Chs3 have important roles in septation and cytokinesis although in the absence of Chs2 or Chs3, or indeed of all three chitin synthases, cytokinesis can still take place. In chs2Δ mutants, the primary septum is missing, and a thick, amorphous septum is formed that contains chitin made by Chs3 (Shaw et al. 1991; Cabib and Schmidt 2003). chs3Δ mutants form a three-layered septum, but the neck region between mother cell and bud is elongated (Shaw et al. 1991). chs2Δ chs3Δ and chs1Δ chs2Δ chs3Δ strains grow very slowly on osmotically supported medium (Sanz et al. 2004; Schmidt 2004; File S6). The triple mutants, however, acquired a suppressor mutation that eliminated the need for osmotic support and conferred a growth rate as fast as that of a chs2Δ mutant although over a third of suppressed and unsuppressed cells in a culture were dead (Schmidt 2004).
For mother and daughter cells to separate, septal material must be degraded, a process that results from secretion of chitinase Cts1 (Kuranda and Robbins 1991), endo-β1,3-glucanases Eng1/Dse4 and Scw11 (Cappellaro et al. 1998; Colman-Lerner et al. 2001; Baladron et al. 2002; see Known and predicted enzymes), and possibly additional activities from the daughter cell’s side of the septum. Daughter cell-specific expression of these enzymes is under the control of the transcription factor Ace2 (Colman-Lerner et al. 2001).
Chitin synthase biochemistry:
Chs1, Chs2, and Chs3 use UDP-GlcNAc as donor and are members of GT family 2 of processive inverting glycosyltransferases, which includes hyaluronate and cellulose synthases. Yeast’s chitin synthases are predicted to have three to five transmembrane helices toward their C termini, and Chs3 likely has two more transmembrane domains nearer its N terminus (Jimenez et al. 2010; Merzendorfer 2011). Amino acid residues important for catalysis lie in a large cytoplasmic domain containing the signature sequences QXXEY, EDRXL, and QXRRW (Nagahashi et al. 1995; Saxena et al. 1995; Cos et al. 1998; Yabe et al. 1998; Ruiz-Herrera et al. 2002; Merzendorfer 2011). An additional motif, (S/T)WG(X)T(R/K), predicted to be extracellularly oriented (Merzendorfer 2011), lies near the protein’s C terminus (Cos et al. 1998; Merzendorfer 2011).
The molecular mechanism of chitin synthesis is not yet clear. By analogy with bacterial NodC, which synthesizes chito-oligosaccharides, and with nonfungal chitin synthases, chain extension would be at the nonreducing end (Kamst et al. 1999; Imai et al. 2003). This topic, and the issue of how the synthases overcome the steric challenge that each sugar in a β1,4-linked polymer is rotated by ∼180° relative to its neighbor, are discussed further in File S6.
Chitin made in vitro by CS I or CS III contains, on average, 115–170 GlcNAc residues (Kang et al. 1984; Orlean 1987). Chitin synthases presumably make chitin chains with a range of lengths, and the range would be predicted to shift to shorter chains as UDP-GlcNAc concentration drops below Km, resulting in lowered rates of chain extension. Indeed, purified Chs1 and membranes from cells overexpressing Chs2 make chito-oligosaccharides at low substrate concentrations (Kang et al. 1984; Yabe et al. 1998). Chitin made in vivo is polydisperse (Cabib and Duran 2005), and increased chitin chain lengths are seen in fks1Δ and gas1Δ mutants and CFW-treated cells, which mount the chitin stress response, whereas shorter chains were made in a strain expressing a Chs4 variant with lower in vitro CS III activity (Grabinska et al. 2007). However, GlcN treatment, which stimulates chitin synthesis in vivo (Bulik et al. 2003; see Sugar nucleotides), had little effect on polymer chain length (Grabinska et al. 2007).
S. cerevisiae’s three chitin synthases are all stimulated up to a few fold in vitro by high concentrations of GlcNAc (Sburlati and Cabib 1986; Orlean 1987). Possible explanations are that GlcNAc serves as a primer or allosteric activator in the chitin synthase reaction (see File S6).
S. cerevisiae’s chitin synthases and auxiliary proteins:
Chitin synthase I:
Most, if not all, Chs1 activity is detectable in vitro only after pretreatment of membranes or extensively purified Chs1 with trypsin (Duran and Cabib 1978; Kang et al. 1984; Orlean 1987). Proteolytically activated Chs1 has the highest in vitro activity of the chitin synthases assayed in membranes from wild-type cells (Sburlati and Cabib 1986; Orlean 1987), although Chs1 does not contribute measurably to chitin synthesis in vivo, even in the absence of Chs2 and Chs3 (Shaw et al. 1991). Although trypsin activation may mimic the effect of an endogenous activating protease, neither such an activator, nor an active, processed form of Chs1, have been identified.
Levels of protease-elicited Chs1 activity are the same in membranes from logarithmically growing and stationary-phase cells (Orlean 1987), and levels of Chs1 show little change during the cell division cycle (Ziman et al. 1996). CHS1 transcription and in vitro CS I activity increase in response to mating factors, but elevated in vitro activity is detectable only after trypsin activation (Schekman and Brawley 1979; Orlean 1987; Appeltauer and Achstetter 1989). However, Chs1 does not contribute to pheromone-induced chitin synthesis (Orlean 1987).
chs1Δ cultures contain the occasional lysed bud, a phenotype more pronounced in acidic medium but partially alleviated when Cts1 chitinase is also deleted (Cabib et al. 1989). Two explanations, which are not mutually exclusive, are that Chs1 may repair wall damage due to overdigestion of chitin by Cts1 or that Chs1 participates in septum synthesis and makes chitin during growth in acidic medium (Cabib et al. 1989; Bulawa 1993). Chs1 promotes wall association of at least one protein because small amounts of the GPI protein Gas1 are released into the medium from chs1Δ cells (Rolli et al. 2009).
Although the contribution of Chs1 to chitin synthesis is small, a wider role for the protein emerged from an analysis of the networks of genes that interact synthetically with CHS1 and CHS3 (Lesage et al. 2005). Most of the 57 genes in the CHS1 interaction network fell into two sets. One set contained genes that, when mutated, impact cell integrity or that themselves interact with genes involved in β1,3-glucan synthesis, indicating a role for Chs1 in buffering the wall against changes impacting its robustness. The other set contained genes involved in budding and in endocytic protein recycling, which in turn may impact Chs2 function, suggesting that Chs1 also buffers against deficiencies in Chs2. The CHS1-interacting genes were mostly distinct from the genes in the network that impacts Chs3 function, and, moreover, mutations in CHS1 itself or in the genes in the CHS1 interaction set do not trigger the Chs3-dependent chitin stress response. Chs1 and Chs3 therefore have distinct functions and one does not buffer against defects in the other (Lesage et al. 2005).
Chitin synthase II and proteins impacting its localization and activity:
Chs2 makes no more than 5% of the chitin in budding cells. Activity of endogenous Chs2 is detectable only in membranes from growing cells and can be stimulated by treatment with trypsin (Sburlati and Cabib 1986) although, in some studies, membrane preparations as well as partially purified Chs2 have significant in vitro activity without prior trypsin treatment, raising the possibility that full-size Chs2 makes chitin (Uchida et al. 1996; Oh et al. 2012). A soluble fraction from growing yeast cells, which stimulates Chs2 activity two- to fourfold but which must itself be pretreated with trypsin, has been described (Martínez-Rucobo et al. 2009). An endogenously activated, processed form of Chs2 has not been identified (File S6).
Levels of CHS2 expression and localization of the protein are coordinated with synthesis of the primary septum (Figure 8). CHS2 message levels peak just prior to primary septum formation at the G2/M phase (Pammer et al. 1992; Cho et al. 1998; Spellman et al. 1998), and levels of Chs2 and CS II activity then peak as the primary septum is made (Pammer et al. 1992; Choi et al. 1994a; Chuang and Schekman 1996). Upon completion of cytokinesis, levels of Chs2 and its message drop, indicating that both turn over rapidly.
Figure 8 .
Trafficking and regulation of Chs2. Cell cycle-regulated expression of CHS2 peaks at the G2-M phase transition, and Chs2 is synthesized at the ER. Phosphorylation of Chs2 by Cdk1 retains Chs2 in the ER. Upon chromosomal separation, Cdc14-dependent dephosphorylation of Chs2 allows release of the protein from the ER and its transit to the mother cell–bud junction. Inn1 and Cyk3, localized at the division site, are involved in Chs2 activation. After primary septum formation is complete, Chs2 is endocytosed and degraded. Localization, function, and subsequent removal of Chs2 when the primary septum is complete depend on phosphorylation by Dbf2. Figure is adapted from Lesage and Bussey (2006).
Temporal and spatial localization of Chs2 is impacted at at least two stages by protein kinases. Chs2 is synthesized in the ER during metaphase, but its release from the ER is coordinated with exit of the cell from mitosis and triggered upon inactivation of mitotic kinase by Sic1 (Zhang et al. 2006). The mitotic kinase Cdk1 likely acts directly on Chs2, which contains four CDK1 phosphorylation sites near its N terminus, because mutation of the target Ser residues to Glu leads to retention of Chs2 in the ER, whereas changing the serines to Ala leads to constitutive release of the mutant Chs2 even in the presence of high Cdk1 activity (Teh et al. 2009). Timed release of Chs2 from the ER after chromosome separation and exit of the cells from mitosis is triggered by dephosphorylation of the Cdk1 sites by the Cdc14 phosphatase, the terminal component of the mitotic exit network (MEN) cascade (Chin et al. 2012).
Exit of Chs2 from the ER and its delivery to the plasma membrane at the mother cell–bud junction is effected by COPII vesicles (Chuang and Schekman 1996; VerPlank and Li 2005; Zhang et al. 2006). Localization of Chs2 at the bud neck, correct formation of the primary septum, and removal of Chs2 at the end of cytokinesis depend on phosphorylation of Chs2 by the mitotic exit kinase Dbf2, also a component of MEN (Oh et al. 2012). Inn1 and Cyk3, whose localization to the division site is also regulated by MEN, are also involved in activation of Chs2 for primary septum formation (Nishihama et al. 2009; Meitinger et al. 2010; Oh et al. 2012). Overexpression of CYK3 leads to increased deposition of chitin at the division site in chs1Δ chs3Δ cells, where Chs2 is the sole chitin synthase (Oh et al. 2012). Cyk3 has a transglutaminase-like domain (Nishihama et al. 2009), but the nature of Cyk3’s effect on Chs2 is unclear and Inn1’s role in Chs2 activation is unknown. Additional phosphorylation sites are present in Chs2’s N-terminal domain (Martínez-Rucobo et al. 2009), but their roles are unclear.
Chs2 resides at the site of primary septum formation for only 7–8 min (Roh et al. 2002a; Zhang et al. 2006). The protein is degraded upon endocytosis and delivery to the vacuole (Chuang and Schekman 1996; Schmidt et al. 2002; VerPlank and Li 2005), and optimal endocytic turnover of Chs2 requires components of the endosomal sorting complexes required for transport (ESCRT) pathway (McMurray et al. 2011).
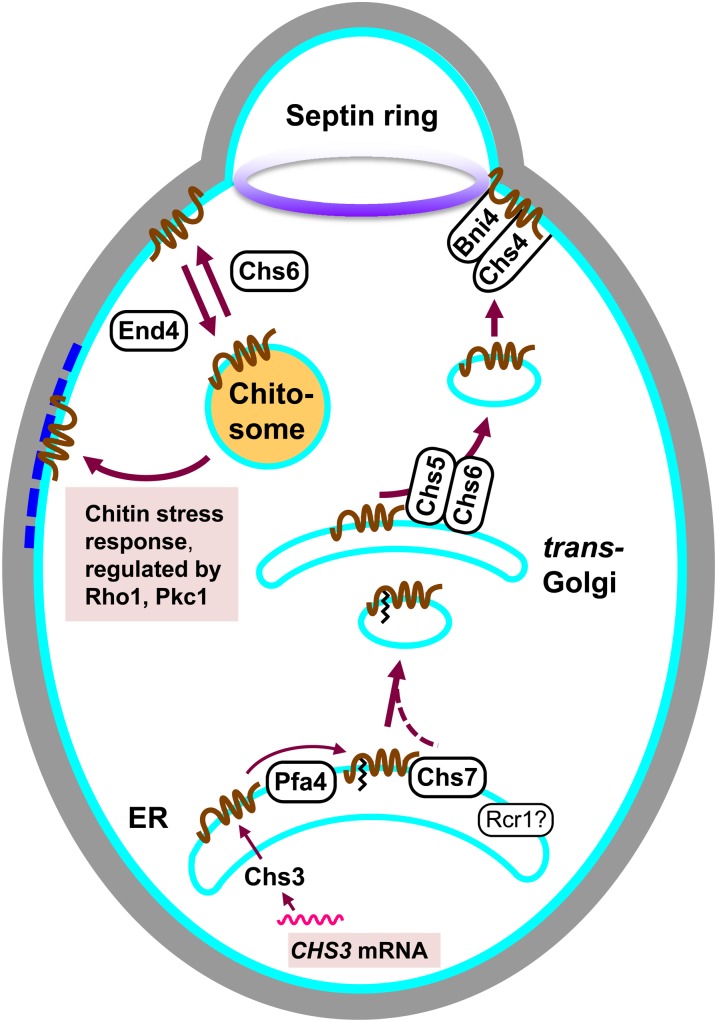
Chitin synthase III and proteins impacting its localization and activity:
Chitin synthase III is responsible for the synthesis of >90% of the chitin in unstressed vegetative cells, for the additional chitin made in the chitin stress response and in response to mating pheromones, and for the synthesis of the chitin that is de-N-acetylated to chitosan during ascospore wall formation. Cells deficient in CS III activity are resistant to CFW (Roncero et al. 1988). Chs3 is the transferase, but its function depends on its regulated transport from the ER to the plasma membrane, its removal from the plasma membrane and sequestration in intracellular vesicles called chitosomes, and its remobilization from chitosomes to the plasma membrane. A number of proteins are required for regulated Chs3 trafficking and for enzyme activity (Bulawa 1993; Trilla et al. 1999; Roncero 2002).
CS III [referred to as chitin synthase II by Orlean (1987)] is the major, if not only, activity detected in membrane fractions from logarithmically growing wild-type cells without prior treatment with trypsin and is trypsin sensitive (Orlean 1987). CS III activity determined in this way is presumably either due to constitutively active Chs3 or to an endogenously activated form of the protein. A pool of trypsin-activatable CS III was detected in detergent-treated membranes from chs1Δ chs2Δ cells or from cells lacking Chs4, an activator of CS III (Choi et al. 1994b; Trilla et al. 1997). The latter finding, together with the observation that overexpression of Chs4 lowers the extent to which trypsin activates CS III, suggested that trypsin treatment might mimic Chs4-dependent processing of Chs3. However, because no endogenously processed forms of Chs3 have been detected (Santos and Snyder 1997; Cos et al. 1998), and because Chs4 does not resemble any protease, the apparent zymogenicity of Chs3 in chs4Δ may be an artifact (Reyes et al. 2007).
Levels of CHS3 mRNA and Chs3 vary little during the budding cycle (Choi et al. 1994a; Chuang and Schekman 1996; Cos et al. 1998), indicating that CS III is regulated at the post-translational level. Sequences involved include the C-terminal extracellular region containing the motif (S/T)WG(X)T(R/K), which is required for in vitro CS III activity and chitin synthesis in vivo (Cos et al. 1998).
A number of proteins interact with Chs3 as it transits the secretory pathway (Figure 9). Chs3 is palmitoylated in the ER by Pfa4 (Lam et al. 2006; Montoro et al. 2011). pfa4Δ mutants are CFW-resistant and accumulate Chs3 in the ER, indicating a role for palmitoylation in Chs3 export (Lam et al. 2006). Chs3 has two palmitoylation sites in a cytoplasmic domain N-terminal to the proposed catalytic residues (Meissner et al. 2010). Exit of Chs3 from the ER also requires Chs7, an ER chaperone with six or seven transmembrane domains (Trilla et al. 1999) that interacts with Chs3. The effects of CHS7 deletion on chitin levels and CSIII activity are almost as severe as those of CHS3 deletion (Trilla et al. 1999). Chs3 aggregates in the ER in chs7Δ cells (Lam et al. 2006), and Chs7 is a limiting factor in export of Chs3 because simultaneous overexpression of CHS3 and CHS7 leads to elevated CSIII activity, whereas overexpression of CHS3 alone does not. Neither Pfa4 nor Chs7 is required for exit of Chs1 and Chs2 from the ER (Trilla et al. 1999; Lam et al. 2006). ER-membrane proteins Rcr1 and Yea4 also impact Chs3-dependent chitin synthesis in ways that are unclear (File S6).
Figure 9 .
Overview of Chs3 trafficking. Chs3, synthesized in the ER, requires palmitoylation by Pfa4 and association with Chs7 to exit the ER. In the trans-Golgi, Chs3 association with exomer components Chs5 and Chs6 facilitates incorporation of Chs3 into secretory vesicles for delivery to the plasma membrane at the site of chitin ring formation. Localization and activation of Chs3 depends on association with Chs4, whose association with the septin ring is mediated in turn via an interaction with Bni4. In cells with medium-sized buds, Chs3 is retrieved from the plasma membrane and sequestered in chitosomes in an endocytic process depending on End4 and later recruited back to the neck region in a Chs6-dependent manner. During the cell wall stress response, Rho1 and Pkc1 trigger mobilization of Chs3 from chitosomes to the plasma membrane for synthesis of extra chitin in the lateral wall. Figure is adapted from Lesage and Bussey (2006).
Transport of new Chs3 from the trans-Golgi to the plasma membrane, as well as Chs3 cycling from chitosomes to the plasma membrane, requires the peripheral Golgi membrane proteins Chs5 and Chs6 (Santos and Snyder 1997; Santos et al. 1997; Ziman et al. 1998). Chs6 and its homologs Bch1, Bch2, and Bud7, referred to as Chs5-Arf1-binding proteins, join with Chs5 to form exomer complexes that transiently bind Chs3 to promote its incorporation into secretory vesicles (Sanchatjate and Schekman 2006; Trautwein et al. 2006; Wang et al. 2006). Although Chs5 and Chs6 act in a complex, the two have different impacts on Chs3 activity and transport. chs5Δ and chs6Δ mutants make 25 and 10% of wild-type amounts of chitin, respectively, but whereas chs5Δ membranes lack in vitro CS III activity, this activity is normal in chs6Δ membranes (Bulawa et al. 1993; Santos et al. 1997). This may be because, in chs5Δ cells, Chs3 accumulates in late Golgi vesicles (Santos and Snyder 1997), whereas, in chs6Δ mutants, it collects in chitosomes, where it may encounter a chitosomal activator (Ziman et al. 1998). Exomer has a role in the transport of the chitin-β1,3-glucan cross-linker Crh2 to the cell surface. Cotransport of Chs3 and Crh2 would ensure colocalization of these proteins for efficient cross-linking of chitin to β1,3-glucan.
At the plasma membrane, Chs4 (Csd4/Skt5) interacts with Chs3 (DeMarini et al. 1997; Ono et al. 2000; Meissner et al. 2010) and has two roles apparently specific to Chs3. chs4Δ mutants lack in vitro CS III activity and make very little chitin (Bulawa 1993; Trilla et al. 1997). Overexpression of CHS4, but not CHS3, raises in vitro CS III activity (Bulawa 1993; Trilla et al. 1997; Ono et al. 2000) as well as levels of Chs3 in the plasma membrane (Reyes et al. 2007), suggesting that Chs4 is an activator of CS III. Stimulation of CS III by Chs4 requires a region of Chs4 to bind Chs3 because the ability of truncated forms of Chs4 to elicit CS III activity correlates with the ability of Chs4 fragments to interact with Chs3 in a two-hybrid analysis (Ono et al. 2000; Meissner et al. 2010). Chs4 has a C-terminal farnesylation site (Bulawa et al. 1993; Trilla et al. 1997; Grabinska et al. 2007) whose roles are discussed in File S6.
Chs4 not only activates Chs3, but also mediates Chs3 localization on the mother cell’s plasma membrane at the site of formation of the chitin ring prior to bud emergence. There, it interacts with the scaffold protein Bni4, which in turn associates with the septins (DeMarini et al. 1997; Kozubowski et al. 2003; Sanz et al. 2004). Absence of Bni4 leads to mislocalized deposition of chitin (DeMarini et al. 1997; Kozubowski et al. 2003; Sanz et al. 2004), and Chs4 is absent from the base of buds in small-budded cells (Sanz et al. 2004). In contrast to chs4Δ mutants, chitin synthesis and CS III activity are not dramatically affected in bni4Δ cells, suggesting that Bni4 is not required for CS III activity per se (Sanz et al. 2004).
Chs3 and Chs4 are associated with the plasma membrane just before formation of the chitin ring at the site of bud emergence and reside there in a ring at the base of the bud in many cells with small buds, then become scarcely detectable in cells with medium-sized buds, only to reappear, in a Bni4-independent manner, at both sides of the neck in cells with large buds prior to cytokinesis (Chuang and Schekman 1996; DeMarini et al. 1997; Santos and Snyder 1997; Kozubowski et al. 2003; Sanz et al. 2004). In between, Chs3 is retrieved from the membrane to chitosomes in an endocytic process dependent on End4/Sla2 (Chuang and Schekman 1996; Ziman et al. 1996, 1998), but is recruited back to the plasma membrane in a Chs6-dependent manner (Ziman et al. 1998; Wang et al. 2006). Chs3’s itinerary is consistent with the overall order of events in yeast cytokinesis.
Chitin synthesis in response to cell wall stress:
Cells with mutations affecting the formation of β-glucan, mannan, O-linked glycans, and GPI anchors respond by depositing additional chitin—as much as 10 times more than in wild-type cells—in their lateral walls in apparent compensation for compromised cell integrity (Gentzsch and Tanner 1996; Kapteyn et al. 1997, 1999a; Popolo et al. 1997; Dallies et al. 1998; Osmond et al. 1999; Garcia-Rodriguez et al. 2000; Valdivieso et al. 2000; Carotti et al. 2002; Lagorce et al. 2002; Magnelli et al. 2002; Sobering et al. 2004; Lesage et al. 2005). This chitin stress response, which is accompanied by increased precursor supply (see Precursors and Carrier Lipids), requires Chs3 and is dependent on Chs4, -5, -6, and -7 in gas1Δ cells (Valdivieso et al. 2000; Carotti et al. 2002). The response does not involve upregulation of the CHS genes, but, rather, an altered distribution of Chs3, which was seen in the plasma membrane of buds of gas1Δ and fks1Δ cells, and Chs4 was also delocalized (Garcia-Rodriguez et al. 2000; Valdivieso et al. 2000; Carotti et al. 2002; Valdivia and Schekman 2003). Interestingly, gas1Δ suppressed the lysed bud phenotype of chs1Δ, suggesting that the chitin stress response also repaired weakened bud walls (Valdivieso et al. 2000). The Chs3 making the stress response originates from chitosomes, and its translocation to the plasma membrane is regulated by Rho1 and Pkc1, which act early in the CWI pathway that triggers the chitin stress response (Valdivia and Schekman 2003).
Chitin synthase III in mating and ascospore wall formation:
Chitin synthase III is responsible for the extra chitin made in response to mating pheromones and for formation of the chitosan of ascospore walls. MATa cells treated with α-factor show a three- to fourfold increase in chitin, which is laid down diffusely in the shmoo (Schekman and Brawley 1979). Chs3 is necessary because no extra chitin is made in pheromone-treated chs3Δ cells, and the response is either abolished or much smaller in chs5Δ, chs6Δ, and chs4Δ cells, indicating that the machinery for trafficking and activation of Chs3 is required (Orlean 1987; Roncero et al. 1988; Bulawa 1993; Santos and Snyder 1997; Bulik et al. 2003). Consistent with its role in chitin deposition, Chs3 is localized at the periphery of the mating projection, and it remains there because it is not subject to endocytic turnover as it is in budding cells (Santos and Snyder 1997; Sacristan et al. 2012). Although the extra chitin synthesis in response to α-factor is presumably driven by the increased amount of UDP-GlcNAc made during the pheromone response (Orlean et al. 1985; Bulik et al. 2003), the mechanism behind pheromone-stimulated chitin synthesis by Chs3 is unclear. Levels of Chs3 increase sixfold upon α-factor treatment (Cos et al. 1998), but neither CHS3 transcription nor levels of chitin synthase III activity are elevated (Orlean 1987; Choi et al. 1994a). Factors that might limit total Chs3 activity might include prevention of the mobilization of the protein to the plasma membrane in shmoos or interference with interactions between Chs3 and regulatory proteins (Choi et al. 1994a).
The chitosan of the ascospore wall is initially synthesized as chitin by Chs3 (Pammer et al. 1992) and is then de-N-acetylated by chitin deacetylases Cda1 and Cda2, of which Cda2 has the dominant role (Mishra et al. 1997; Christodoulidou et al. 1999). From the sporulation defects in mutants in proteins involved in Chs3 trafficking in vegetative cells, Chs6 and Chs7, but not Chs5, have as-yet-undefined roles in ascospore maturation (Santos et al. 1997; Trilla et al. 1999). A Chs4 homolog, Shc1, has a regulatory role in chitosan synthesis (Sanz et al. 2002; File S6). Ascospore wall structure and assembly are reviewed by Neiman (2011).
β1,3-Glucan
De novo β1,3-glucan synthetic activity is associated with members of the Fks family, of which Fks1 and Fks2 require the soluble Rho1 GTPase as a regulatory subunit. In vitro activity is membrane-associated, uses UDP-Glc as donor, is stimulated by GTP via Rho1, and yields a product with a chain length of 60–80 glucoses (Shematek et al. 1980; Kang and Cabib 1986; Drgonová et al. 1996; Mazur and Baginsky 1996; Qadota et al. 1996). In vitro β1,3-glucan synthase activity is inhibited by acylated cyclic hexapeptides of the echinocandin group and by papulocandins, acylated derivatives of β1,4-galactosylglucose (Debono and Gordee 1994; Georgopapadakou and Tkacz 1995).
Fks family of β1,3-glucan synthases:
Fks1 (Cwh53/Etg1/Gsc1/Pbr1), Fks2, and Fks3 are in GT family 48, which also contains proteins implicated in callose sythesis in plants (Verma and Hong 2001). Fks1 has an N-terminal cytoplasmic domain that is followed by 6 transmembrane helices, a large cytoplasmic domain, and then 10 transmembrane helices (Inoue et al. 1995; Mazur et al. 1995; Qadota et al. 1996; Dijkgraaf et al. 2002; Okada et al. 2010). Three functional domains, mutations in which separately affect in vivo and in vitro β1,3 glucan synthetic activity, as well as cell polarity and endocytosis, have been distinguished (Okada et al. 2010; File S7). The phenotypes of fks1 mutants may in part reflect the involvement of the protein in processes other than β1,3-glucan biosynthesis. For example, mutations in both FKS1 and FKS2 result in lowered β1,6-glucan synthesis (Dijkgraaf et al. 2002). Fks1 is localized to the plasma membrane at sites of polarized growth and cell wall remodeling throughout the cell cycle, and this localization coincides with that of actin patches (Qadota et al. 1996; Dijkgraaf et al. 2002; Utsugi et al. 2002). Fks1 transits the secretory pathway because it accumulates intracellularly in vesicular transport mutants and its activity is sensitive to phytosphingosine levels in the ER (El-Sherbeini and Clemas 1995; Abe et al. 2001; File S7).
Roles of the Fks proteins in β1,3-glucan synthesis:
The Fks proteins show a degree of specialization. Deletion of FKS1 leads to slow growth, a 75% reduction in β1,3-glucan, and low in vitro β-1,3-glucan synthase activity, whereas the in vitro activity of fks2Δ membranes is nearly that of wild-type membranes and the disruptants have no defect in vegetative growth (Inoue et al. 1995; Mazur et al. 1995). Although this suggests that Fks1 is the major contributor to β1,3-glucan synthesis in budding cells, fks1Δ fks2Δ null mutants are inviable, indicating that Fks1 and Fks2 have overlapping functions (Inoue et al. 1995; Mazur et al. 1995). Consistent with this, overexpression of either FKS1 or FKS2 can partially correct the defects caused by deleting the other of the two genes (Mazur et al. 1995; Dijkgraaf et al. 2002), and, furthermore, the two proteins colocalize in sites of polarized growth in budding cells, although Fks1 is the most abundant (Dijkgraaf et al. 2002).
FKS1 and FKS2 show different expression patterns. FKS1 is expressed during budding growth and transcript levels peak in the late G1 and early S phases (Mazur et al. 1995; Ram et al. 1995; Lesage and Bussey 2006). FKS2 mRNA, in contrast, cannot be detected in budding cultures grown in glucose, but appears when glucose becomes depleted; when cells are grown on acetate, glycerol, or galactose; when cells are treated with α-factor or Ca2+; in fks1 mutants and mutants defective in the synthesis of other wall polymers; and when cells are stressed by shift to high temperature (Mazur et al. 1995; Zhao et al. 1998; Lesage and Bussey 2006). Induction of FKS2 is mediated via the PKC CWI and calcineurin pathways (Mazur et al. 1995; Ram et al. 1995; Zhao et al. 1998; Lagorce et al. 2003).
Fks2 is important in sporulation because fks2Δ fks2Δ diploids have a severe defect in this process (Mazur et al. 1995; Huang et al. 2005; Ishihara et al. 2007). Homozygous fks3Δ fks3Δ diploids also form abnormal spores, indicating a role for Fks3 in ascopore wall formation, although Fks3’s role in sporulation does not overlap with Fks2’s. It was proposed that Fks2 is primarily responsible for synthesis of β1,3-glucan in the ascospore wall and that Fks3, rather than functioning as a synthase, modulates glucan synthesis during ascospore wall formation (Ishihara et al. 2007; File S7).
After their export through the plasma membrane, β1,3-glucan chains can be cross-linked to chitin by Crh1 and Crh2 (Cabib 2009), and the polymer can be extended through the action of Gas1 family β1,3-glucanosyltransferases (Mouyna et al. 2000), and side-branching β1,6-linked glucoses as well as PIR proteins may be attached (Ecker et al. 2006; see Incorporation of PIR proteins into the cell wall and Exg1, Exg2, and Ssg/Spr1 exo-β1,3-glucanases).
Deficiencies in Fks1 are compensated for by Chs3-dependent chitin synthesis (Garcia-Rodriguez et al. 2000; Valdivieso et al. 2000; Carotti et al. 2002), and fks1Δ shows synthetic interactions with chs3Δ, chs4Δ, chs5Δ, chs6Δ, and chs7Δ (Osmond et al. 1999; Lesage et al. 2004), but correct synthesis of other wall constituents is also necessary when β1,3-glucan synthesis is compromised (Lesage et al. 2004). Analyses of the genome-wide responses to FKS1 deletion revealed upregulation of a “cell wall compensatory cluster” of 79 coregulated genes whose products include a range of proteins involved in wall synthesis and remodeling (Terashima et al. 2000; Lagorce et al. 2003). An overlapping set of genes, whose products function in the biosynthesis of chitin, β1,6-glucan, and mannan, as well as in the function of the secretory pathway and in maintenance of cell polarity, was identified in an analysis of the synthetic genetic interactions of fks1Δ (Lesage et al. 2004). This study showed that FKS2 made interactions only with FKS1 and that FKS3 made no interactions, consistent with differential expression of FKS2 and FKS3 (Lesage et al. 2004).
Rho1 GTPase, a regulatory subunit of β1,3-glucan synthase:
The essential Rho1 GTPase, which activates Pkc1 in the CWI pathway and is required for cell cycle progression and polarization of growth (Drgonová et al. 1999; Levin 2011), has a distinct role as a regulatory subunit of β1,3-glucan synthetic complexes containing Fks proteins. Evidence for this is that (i) Fks1 and Rho1 colocalize and coimmunoprecipitate, (ii) membranes from a temperature-sensitive rho1 mutant have a thermolabile β1,3-glucan synthase activity that can be corrected by adding back purified Rho1, (iii) membranes from cells expressing a consitutively active rho1 allele have GTP-independent β1,3-glucan synthase activity, and (iv) inactivation of Rho1 by ADP ribosylation eliminates the in vitro β1,3-glucan synthase activity of membranes from fks1Δ and fks2Δ strains (Drgonová et al. 1996; Mazur and Baginsky 1996; Qadota et al. 1996). Moreover, there are rho1 mutations that affect regulation of β1,3-glucan synthesis, but not other Rho1 functions, and the amino acids affected are different from those whose mutation causes cell cycle and polarization defects (Saka et al. 2001; Roh et al. 2002b). The amino acid changes in the β1,3-glucan synthesis-specific rho1 mutants might impact binding to Fks proteins, but the interacting domains on the regulatory and catalytic subunits have not been defined. The Rho1-Fks interaction at the cytoplasmic face of the plasma membrane, as well as activation of β1,3-glucan synthesis, requires Rho1 to be geranylgeranylated at its C terminus (Inoue et al. 1999).
β1,6-Glucan
Mutations in genes with products localized along the secretory pathway impact formation of β1,6-glucan (Shahinian and Bussey 2000; Lesage and Bussey 2006), but the biochemistry of β1,6-glucan synthesis is unclear. In vitro synthesis of β1,6-glucan is hard to detect, and no fungal enzyme has yet been shown to catalyze formation of a β1,6-glucosidic linkage using UDP-Glc as donor, although the linkage can be generated by the Bgl2 protein in a transglycosylation reaction (Goldman et al. 1995). Synthesis of β1,6-glucan is normal in alg5Δ mutants, indicating that Dol-P-Glc is not involved in formation of this polymer (Shahinian et al. 1998; Aimanianda et al. 2009).
In vitro synthesis of β1,6-glucan
Because β1,6-glucan is a linear polymer with side branches on average every fifth Glc (see β-glucans), it could be generated by a processive, UDP-Glc-dependent β1,6-glucan synthase and then branched or by assembly of shorter repeat units, whose glucoses originate from UDP-Glc. Detection of UDP-Glc-dependent formation of β1,6-glucan is complicated by the fact that UDP-Glc is also the donor in the synthesis of β1,3-glucan, glycogen, and glucolipids.
Two assays of the formation of β1,6-glucan using UDP-Glc as donor have been described. In the first, formation of β1,6-glucanase-sensitive polymer by membranes was detected by dot-blot assay using an anti-β1,6 glucan antibody (Vink et al. 2004). The reaction was distinguished from β1,3-glucan synthase because membranes from kre5 mutants, which make little β1,6-glucan but have normal β1,3-glucan synthetic capability, made little β1,6-glucan in vitro but had nearly wild-type β1,3-glucan synthase activities. Comparisons of the activities of wild-type and β1,6-glucan synthesis-defective strains revealed that levels of β1,6-glucan formed de novo correlated with the reduction in β1,6-glucan synthesis in vivo. It was proposed that the dot-blot assay measured β1,6-glucan chain extension and that higher rates of Glc transfer reflected the presence of more acceptor (Vink et al. 2004). The reaction was stimulated by GTP and higher β1,6-glucan synthetic activity was detected in membranes from cells overexpressing Rho1 GTPase, suggesting that β1,6-glucan synthase, like β1,3-glucan synthase, is Rho1-dependent (Vink et al. 2004).
In the second approach, formation of β1,6-glucan was measured in cells permeablized by osmotic shock and incubated with radiolabeled UDP-Glc (Aimanianda et al. 2009). The insoluble, radiolabeled β1,6-glucan formed was chemically identical to the branched β1,6-linked glucan isolated from cell walls, and radioactivity was distributed throughout the in situ product, indicating that de novo polymerization of β1,6-glucan had occurred (Aimanianda et al. 2009). Consistent with their severe in vivo defects in β1,6-glucan synthesis, permeabilized kre5 and kre9 mutants showed no in situ β1,6-glucan synthetic activity, but made β1,3-glucan. The β1,6-glucan synthetic activity in permeabilized cells was not stimulated by GTP. However, because β1,3-glucan synthesis mutants make less β1,6-glucan, and vice versa, formation of the two polymers may be coordinated in another way (Dijkgraaf et al. 2002; Aimanianda et al. 2009; see The Fks family of β1,3-glucan synthases).
Proteins involved in β1,6-glucan assembly
Mutants defective in β1,6-glucan synthesis were identified in screens for resistance to K1 killer toxin, which uses β1,6-glucan as its receptor (Hutchins and Bussey 1983), and in screens for CFW sensitivity (Ram et al. 1994; Lussier et al. 1997b; Orlean 1997; Shahinian and Bussey 2000; Pagé et al. 2003). In these mutants, levels of alkali-insoluble β1,6-glucan were lowered to different extents, and the proportions of β1,6- and β1,3-linked Glc residues in the alkali-insoluble glucan fraction were often altered. The finding that the proteins implicated in β1,6-glucan assembly were localized in the ER, Golgi, or plasma membrane, together with demonstrations of epistasis relationships and genetic interactions, led to the notion of a secretory pathway-based pathway for β1,6-glucan elaboration (Boone et al. 1990; reviewed by Orlean 1997 and Shahinian and Bussey 2000). However, β1,6-glucan is not detectable intracellularly (Montijn et al. 1999), and the roles of most of the proteins so far implicated are indirect. Proteins affecting the formation of β1,6-glucan will be discussed in the order of their location along the secretory pathway.
ER proteins:
Homologs of the UGGT/calnexin protein quality control machinery:
Four homologs of proteins involved in the UGGT/calnexin protein quality control system (see N-glycan processing in the ER and glycoprotein quality control) are required for formation of normal amounts of β1,6-glucan (Jiang et al. 1996; Abeijon and Chen 1998; Shahinian et al. 1998; Simons et al. 1998). These are diverged UGGT homologs Kre5, Gls1/Cwh41, Gls2/Rot2, and Cne1, of which Kre5 has the most important role because kre5 mutants make no more than 5% of normal amounts of β1,6-glucan (Meaden et al. 1990; Montijn et al. 1999; Levinson et al. 2002; Aimanianda et al. 2009). The contributions of the glucosidases and calnexin are likely indirect ones in maintaining normal levels of unknown components of the β1,6-glucan assembly machinery in the secretory pathway (Shahinian et al. 1998; Lesage and Bussey 2006). The essential function of Kre5 is other than as a UGGT in protein quality control because kre5Δ remained lethal in an alg8Δ gls2Δ background in which all N-glycans stayed monoglucosylated, thereby bypassing the need for UGGT activity (Shahinian et al. 1998). Kre5 could be a glucosyltransferase with a specialized role in quality control of β1,6-glucan assembly proteins (Levinson et al. 2002; Herrero et al. 2004; Lesage and Bussey 2006), or it could glucosylate the GPI glycan of future GPI-CWPs to generate a signal or attachment point for subsequent transfer to β1,6-glucan (Shahinian and Bussey 2000). S. cerevisiae has the necessary ER UDP-Glc transport activity to supply the donor (Castro et al. 1999).
N-glycosylation is important for wild-type levels of β1,6-glucan to be made. For example, stt3 mutants have a severe defect in β1,6-glucan synthesis and are synthetically lethal with kre5 and kre9 (Chavan et al. 2003b). This may reflect a requirement for N-glycosylation of one or more β1,6-glucan synthetic proteins or for an N-glycan to serve as acceptor for initiation of a β1,6-glucan chain (Lesage and Bussey 2006). Interestingly, mutations such as och1 and mnn9, which affect synthesis of the α1,6-mannan backbone, and mnn2, which blocks addition of the first, α1,2 side-branching Man, show elevated levels of β1,6-glucan (Magnelli et al. 2002; Pagé et al. 2003), suggesting that a balance is normally maintained between these two polymers.
Fungus-specific ER chaperones required for β1,6-glucan synthesis:
Mutations in genes encoding the ER-localized, fungus-specific membrane proteins Rot1, Big1, and Keg1 all cause a β1,6-glucan synthetic defect. ROT1 and BIG1 null mutants grow only with osmotic support, and even then very slowly, and in this and in the severity of their β1,6-glucan synthetic defect—a 95% reduction—they resemble kre5Δ strains (Bickle et al. 1998; Azuma et al. 2002; Machi et al. 2004). Levels of β1,3-glucan and chitin are elevated in rot1Δ and big1Δ. The β1,6-glucan defect in keg1Δ cells is similar to that in kre6Δ—about a 50% reduction (Nakamata et al. 2007).
Rot1, Big1, and Keg1 are small proteins that show no similarity to one another or to carbohydrate-active enzymes (Lesage and Bussey 2006). They seem to function as ER chaperones with varying degrees of importance for the stability of proteins involved in β1,6-glucan synthesis and may in some cases cooperate. Observations supporting this notion and indicating a relationship to Kre5 are discussed in File S8.
More widely distributed secretory pathway proteins:
Kre6 and Skn1:
Kre6 and Skn1 are homologous type 2 membrane proteins in GH family 16 of β-1,6/β-1,3-glucanases (Henrissat and Davies 1997; Montijn et al. 1999). kre6Δ cells make half normal amounts of β1,6-glucan, whereas skn1Δ cells make β1,6-glucan normally and have no growth defect. Expressed at high copy, Skn1 restores almost normal levels of β1,6-glucan to kre6Δ cells, and kreΔ skn1Δ double mutants are inviable or very slow growing, depending on the strain background, and make no more than 10% of normal amounts of β1,6-glucan (Roemer et al. 1993). From this, Kre6 and Skn1 seem to be functional homologs, with Kre6 normally having the dominant role in β1,6-glucan synthesis. As hydrolases or transglycosylases, Kre6 and Skn1 could act on a structure that serves as a precursor or acceptor in elaboration of β1,6-glucan or on a glycoprotein involved in β1,6-glucan synthesis (Lesage and Bussey 2006), but enzyme activity has yet to be demonstrated for these proteins. Much of Kre6 is ER-localized, where it interacts with Keg1, but the protein is also detectable in the Golgi, in secretory vesicles, and at the plasma membrane at sites of polarized growth (Li et al. 2002; Nakamata et al. 2007; Kurita et al. 2011; File S8). Localization of Skn1 has not been explored in detail. Skn1 also has a role in the formation of mannosyl diinositolphosphoryl ceramide [M(IP)2C], because skn1Δ, but not kre6Δ strains, is defective in M(IP)2C (Thevissen et al. 2005; File S8).
Kre6 has been implicated in the mode of action of a pyridobenzimidazole derivative identified in a screen for inhibitors of cell wall incorporation of a reporter GPI-CWP (Kitamura et al. 2009). Because cells treated with this compound showed lowered incorporation of radiolabeled Glc into a β1,6-glucan fraction, and because a resistant mutant had an amino acid substitution in Kre6, it was proposed that the compound is an inhibitor of β1,6-glucan synthesis and that Kre6 is its likely target (Kitamura et al. 2009).
Kre9 and Knh1:
Kre9 and Knh1 are 30 kDa, soluble, fungus-specific, O-mannosylated proteins that are secreted into the medium when overproduced (Brown and Bussey 1993; Dijkgraaf et al. 1996). The two are functional homologs, with Kre9 having the dominant role. kre9 nulls are slow growing and show an 80% reduction in β1,6-glucan, and the residual β1,6-glucan in them has about half the molecular mass as the wild-type polymer and is altered in its proportion of β1,6 and β1,6 linkages (Brown and Bussey 1993). The size and structure of β1,6-glucan made in knh1Δ cells is normal. Overexpression of KNH1 corrects the growth and β1,6-glucan defects of kre9Δ, but the kre9skn1 combination is synthetically lethal (Dijkgraaf et al. 1996). kre9Δ, but not knh1Δ, is also synthetically lethal with kre1Δ (see below) and kre6Δ, but not with skn1Δ. KRE9’s genetic interactions indicate that its product has a pleiotropic impact on β1,6-glucan formation, although its effects must be exerted after Kre5’s because the kre5Δ kre9Δ double null has the same phenotype as kre5Δ (Dijkgraaf et al. 1996; Shahinian and Bussey 2000). Neither Kre9 nor Knh1 shows similarity to proteins of known function. If they are not enzymes, Kre9 and Knh1 may serve to anchor β1,6-glucan in the cell wall (Lesage and Bussey 2006), but this must be reconciled with the finding that kre9 mutants have no UDP-Glc-dependent β1,6-glucan synthetic activity (Aimanianda et al. 2009).
Plasma membrane protein Kre1:
Kre1, a GPI protein, functions at the plasma membrane or in the wall. kre1Δ cells make 40% of wild-type levels of β1,6-glucan, but this glucan is smaller and its β1,3 side branches are not extended (Boone et al. 1990; Roemer and Bussey 1995). GPI attachment is necessary for Kre1’s function and cell surface localization (Breinig et al. 2004). The hydrophilic portion of Kre1 shows no similarity to known enzymes. Kre1 has a structural role and becomes cross-linked to other wall proteins (Breinig et al. 2004), and it also serves as a receptor for K1 killer toxin (File S8).
How might β1,6-glucan be made?:
Obstacles to identifying the β1,6-glucan synthase gene might be an inherent difficulty in obtaining hypomorphic alleles of an essential synthase gene or the existence of multiple redundant synthase genes whose individual mutation gives no phenotype (Lesage and Bussey (2006). Furthermore, there are no precedents in other organisms that could be exploited in bioinformatics-based approaches to β1,6-glucan synthesis. Although β1,6-glucan is widely distributed in the Fungi (Lesage and Bussey 2006), it is very rare elsewhere. The bacterium Actinobacillus suis makes a lipopolysaccharide containing a β1,6-glucan homopolymer (Monteiro et al. 2000), but the proteins involved in its formation are unknown. Some bacteria have GT2 family transferases that make polymers of β1,6-linked GlcNAc (Gerke et al. 1998; Itoh et al. 2008), but these enzymes resemble the S. cerevisiae Chs proteins. If β1,6-glucan is indeed formed directly from UDP-Glc, the β1,6-glucosyltransferase would represent a new GT family. Further possibilities are that a known yeast GT may also form β1,6-glucosidic linkages using UDP-Glc as donor or that β1,6-glucan is generated solely by transglycosylation.
Remodeling and Cross-Linking Activities at the Cell Surface
Order of incorporation of components into the cell wall
CWPs delivered by the secretory pathway meet up with chitin and β-glucans at the outer face of the plasma membrane and undergo cross-linking reactions that incorporate them into the wall. The order in which wall components are assembled has been inferred from analyses of the material formed when spheroplasts regenerate their walls and from the wall compositions of mutants unable to make a particular component (Kreger and Kopecká 1976; Roh et al. 2002b). The starting component is β1,3-glucan, which is necessary for incorporation of both β1,6-glucan and mannoproteins. Because β1,6-glucan was still attached to β1,3-glucan when GPI anchoring was inhibited (Roh et al. 2002b), and because incorporation of GPI-CWPs is lowered in β1,6-glucan synthesis mutants (Lu et al. 1995; Kapteyn et al. 1997), GPI-CWPs are likely incorporated after β1,6-glucan. Because chitin became detectable in the walls of daughter cells only after cytokinesis (Shaw et al. 1991), it was concluded that chitin is the last component to be incorporated into the wall (Roh et al. 2002b). The sequence β1,3-glucan→β1,6-glucan→mannoprotein must be able to accommodate changes in expression or assembly of individual components dictated by the cell cycle, cell wall stress, mating, or sporulation, as well as remodeling of individual polysaccharides. For example, a compensatory incorporation of PIR protein directly attached to β1,3-glucan is seen in β1,6-glucan synthesis mutants (Kapteyn et al. 1999b).
The model for the order of incorporation of wall components needs to be reconciled with the model for a bilayered wall, during whose formation CWPs are propelled to the cell surface, leaving polysaccharides nearer the plasma membrane. Furthermore, surface CWP may not be retained at the surface of wild-type cells. Thus, wild-type diploids expressing a Sag1-GFP fusion released a significant basal level of that glycoprotein into the medium (Gonzales et al. 2010). CWP may therefore routinely be shed during vegetative growth, perhaps upon digestion of the wall between mother cell and bud, along with secreted proteins such as chitinase and invertase (Kuranda and Robbins 1991). Cross-linking and remodeling reactions will be described next, and hydrolases of known or unknown functions, as well as nonenzymatic wall proteins are discussed in Cell Wall-Active and Nonenzymatic Surface Proteins and Their Functions.
Incorporation of GPI proteins into the wall
The ω(−) region of a GPI protein (see Identification of GPI proteins) influences whether the protein will be retained in the plasma membrane in lipid-anchored form or whether it can be transferred into the wall (Caro et al. 1997; Hamada et al. 1998a, 1999; De Sampaïo et al. 1999; Frieman and Cormack 2004). If this region includes two basic amino acids, the protein will be mostly retained in the plasma membrane (Caro et al. 1997; Frieman and Cormack 2003), but if basic residues are absent or replaced with hydrophobic ones, the predominant location is the wall (Hamada et al. 1998b, 1999; Frieman and Cormack 2003). However, having two basic amino acids in the ω(−) region does not guarantee membrane localization because the additional presence of a longer stretch of amino acids rich in Ser and Thr will override the dibasic motif and shift the protein to the wall (Frieman and Cormack 2004). Furthermore, not all wall-anchored GPI proteins have the amino acids suggested to promote incorporation into the wall (De Groot et al. 2003). In general, GPI proteins are partitioned between the membrane and wall to varying extents, and none may be restricted to only one location (Gonzales et al. 2009).
The nature of a GPI-protein’s mode of cell surface attachment can be critical. Ecm33, which is required for growth at elevated temperature (see Sps2 family), occurs mainly as a plasma membrane-anchored GPI protein, and this localization is required for in vivo function. Replacement of ω(−) amino acids ω-1-13 of Ecm33 with the corresponding amino acid sequences from wall-localized proteins resulted in increased cross-linking of Ecm33 to the wall, but also in loss of the protein’s ability to support growth at high temperature (Terashima et al. 2003).
The lipid-to-wall transfer reaction could be a one-step transglycosylation in which the GPI glycan is cleaved and its reducing end transferred to β1,6-glucan, or it could involve separate GPI cleavage and transglycosylation steps. Candidates for cross-linkers are Dfg5 and Dcw1, an essential, redundant pair of homologous GPI proteins that resemble an α1,6-endomannanase and are in GH Family 76 (Kitagaki et al. 2002). Single dfg5Δ and dcw1Δ mutants are viable, although dcw1Δ is hypersensitive to Zymolyase, but the combination of dfg5Δ and dcw1Δ is lethal (Kitagaki et al. 2002). Depletion of Dfg5 or Dcw1 by repressing their expression in the double-null background led to cell enlargement, delocalized chitin deposition, and secretion of a GPI-CWP protein into the growth medium (Kitagaki et al. 2002). dcw1Δ was also recovered in a screen for impaired cross-linking of GPI proteins (Gonzalez et al. 2010). The defects caused by loss of Dfg5 and Dcw1, together with the proteins’ resemblance to an α-endomannanase, are consistent with their having a role in GPI cleavage and/or transglycosylation. Homozygous DFG5 nulls are defective in filamentous growth (Mosch and Fink 1997).
GPI-CWP can be used to display heterologous proteins on the yeast cell surface (Schreuder et al. 1993; Van der Vaart et al. 1997; Gai and Wittrup 2007; Shibasaki et al. 2009). In one such system, heterologous proteins are fused to the Aga2 subunit of the α-mating agglutinin (see Flocculins and agglutinins), which is disulfide-linked to its partner, the GPI-CWP Aga1 (Boder and Wittrup 1997).
Incorporation of PIR proteins into the wall
The internal repeat (PIR) sequences of PIR proteins are required for the alkali-labile linkage that joins these proteins to β1,3-glucan (see Wall Composition and Architecture). Deletion of all PIR sequences from Pir1 and Pir4 leads to release of these proteins from the cells (Castillo et al. 2003; Sumita et al. 2005), indicating that the repeats are necessary for wall association. The more repeats, the stronger the binding: deletion of increasing numbers of Pir1’s repeats led to release of increasing amounts of Pir1 into the medium (Sumita et al. 2005).
Studies of Pir4/Ccw5, which has one PIR sequence and needs it for cell wall anchorage, revealed that the alkali-labile linkage was an ester between the γ-carboxyl group of glutamate and the hydroxyl groups of β1,3-glucan. The linkage was generated in a transglutaminase reaction with Q74 in the PIR sequence SQIGDGQ74[V/I]QAT[T/S] (Ecker et al. 2006). In addition to substitutions of Q74, individual mutations of Q69, D72, and Q76 also resulted in loss of wall anchorage of the protein, indicating that these residues have roles in the reaction. No transglutaminase has yet been identified, but Ecker et al. (2006) point out that, because the free energy of hydrolysis of the amide is high enough to drive formation of the ester linkage, the PIR proteins could catalyze their own attachment to β1,3-glucan.
The glucan attachment sequence of PIR repeats is also found in the GPI-CWP Tip1, Tir1, Cwp1, and Cwp2 (Van der Vaart et al. 1995). In the case of Cwp1, the PIR repeat may be used as an additional wall anchorage point because the protein is attached to the wall by both an alkali-labile and a GPI-dependent linkage (Kapteyn et al. 2001). Like GPI-CWP, PIR proteins can be used as carriers to direct surface expression of heterologous proteins fused to them (Andrés et al. 2005; Shimma et al. 2006).
Cross-linkage of chitin to β1,6- and β1,3-glucan
Related Crh1 and Crh2 generate cross-linkages between the reducing ends of chitin chains and both the nonreducing end of β1,3-glucose side branches on β1,6-glucan and the nonreducing ends of β1,3-glucan chains (Cabib et al. 2007; Cabib 2009), and their homolog Crr1 likely does so during ascospore wall assembly. These proteins are in GH family 16, and Crh2 and Crr1 also have a chitin-binding module (Rodriguez-Pena et al. 2000; Cabib et al. 2008). Crh1 and Crh2 are GPI proteins (Caro et al. 1997; Hamada et al. 1998a) whose localization matches that of Chs3. Crh1-GFP fusions are detectable at the site of bud emergence and later in the neck region between mother cell and bud, and Crh2-GFP is seen in the neck region throughout the budding cycle, as well as in the lateral wall (Rodriguez-Pena et al. 2000, 2002). Single crh1 and crh2 null mutants show Calcofluor White and Congo Red sensitivity, phenotypes enhanced in the double null, suggesting that Crh1 and Crh2 have a common wall-related function. Single crh mutants have a higher ratio of alkali soluble- to alkali-insoluble glucan, and this ratio is higher still in crh1Δ crh2Δ, indicating a role for Crh1 and Crh2 in linking β-glucan and chitin (Rodriguez-Pena et al. 2000).
Evidence that Crh1 and Crh2 are transglycosylases came from elegant studies by Cabib and coworkers, who used fluorescent, sulforhodamine-conjugated β1,3-gluco-oligosaccharides as acceptors and showed that they became cross-linked to chitin in bud scars and the lateral walls of live cells (Cabib et al. 2008). Fluorescent labeling was very weak in crh1Δ crh2Δ cells or in cells lacking Chs3, which makes the chitin normally bound to β1,3- and β1,6-glucan (Cabib and Duran 2005). The entire process of chitin polymerization and cross-linking could be reconstituted in detergent permeabilized, protease-treated cells. Cross-linking of fluorescent β1,3-gluco-oligosaccharides depended on the addition of UDP-GlcNAc (Cabib et al. 2008). Interestingly, the nascent chitin was generated in situ by Chs1, which is highly active in permeabilized cells, rather than by Chs3.
CRR1 shows sporulation-specific expression. Crr1-GFP fusions are localized on the surface of ascospores, and homozygous crr1Δ diploids have ascospore wall abnormalities, with irregular deposition of the outer dityrosine and chitosan layers over the inner β-glucan layer (Gómez-Esquer et al. 2004). Ascopores from Crr1-deficient diploids show increased sensitivity to heat shock and lytic enzymes, and these defects are exacerbated when the chitin deacetylases Cda1 and Cda2 are also absent. These findings suggest a role for Crr1 in generating cross-links between the β-glucan and chitosan or chitin during ascospore wall maturation (Gómez-Esquer et al. 2004).
Cell Wall-Active and Nonenzymatic Surface Proteins and Their Functions
Secreted, membrane, or wall proteins with known or conjectured roles in wall biogenesis, adhesion, and nutrition are surveyed here. The primary division is according to whether proteins have or are likely to have enzymatic activity or whether they are nonenzymatic, structural proteins. Both groups contain GPI proteins. Cell wall proteins have been reviewed by Klis et al. (2002, 2006), De Groot et al. (2005), Lesage and Bussey (2006), and Gonzalez et al. (2009), glycosylhydrolases by Adams (2004), agglutinins by Dranginis et al. (2007), and flocculins by Goossens and Willaert (2010). Additional information about these proteins is presented in File S9.
Known and predicted enzymes
Chitinases:
S. cerevisiae has two chitinases, Cts1 and Cts2. Cts1, an endochitinase, has an N-terminal catalytic domain, followed by a heavily O-mannosylated Ser/Thr-rich region, and lastly, a C-terminal chitin-binding domain (Kuranda and Robbins 1991). Cts1 is periplasmic, but much of it is secreted into the medium of cells grown in rich medium (Correa et al. 1982; Kuranda and Robbins 1991). Cts1 has a key role in cell separation because cts1Δ strains form aggregates of cells that remain joined at their chitin-containing septa, a phenotype mimicked when cells are treated with the chitinase inhibitor dimethyallosamidin (Kuranda and Robbins 1991). Cts1’s chitin-binding domain contributes to the enzyme’s localization in the septal region because Cts1 truncations lacking it only partially complement the cts1Δ separation defect (Kuranda and Robbins 1991). Cts2 may have a role in sporulation (Dünkler et al. 2008).
β1,3-glucanases:
Exg1, Exg2, and Ssg/Spr1 exo-β1,3-glucanases:
Exg1 is disulfide-linked (Cappellaro et al. 1998) whereas Exg2 is a surface-anchored GPI protein (Larriba et al. 1995; Caro et al. 1997). Single- or double-null mutants in EXG1 and EXG2 have no overt defects, although exg1Δ cells have slightly elevated levels of β1,6-glucan, and EXG1 overexpressers lower amounts of that polymer, suggesting roles for Exg1 and Exg2 in β-glucan remodeling (Jiang et al. 1995; Lesage and Bussey 2006). Ssg1/Spr1 is a sporulation-specific protein (File S9).
Bgl2, Scw4, Scw10, and Scw11 endo-β1,3-glucanases:
These are GPI-less secretory proteins. Bgl2 has endo-β1,3-glucanase activity in vitro (Mrša et al. 1993), but it can also create a β1,6 linkage between the reducing end that it generates by cleaving a β1,3-gluco-oligosaccharide and the nonreducing end of another β1,3-glucan chain (Goldman et al. 1995), and so could function as a β1,3-glucan branching enzyme. No enzymatic activity has been shown for Scw4, Scw10, or Scw11, although mutation of predicted catalytic residues in Scw10 abolished in vivo function (Sestak et al. 2004). Scw4, Scw10, and Bgl2 are wall-associated via disulfides (Cappellaro et al. 1998), but some Scw4 and Scw10 can also be linked to β1,3-glucan (Yin et al. 2005).
These proteins have roles in maintaining normal walls. bgl2Δ, scw4Δ, and scw10Δ strains grow like wild-type cells, but show CFW sensitivity and slightly increased chitin levels (Klebl and Tanner 1989; Cappellaro et al. 1998; Kalebina et al. 2003; Sestak et al. 2004), and bgl2Δ walls have elevated levels of alkali-soluble glucan (Sestak et al. 2004). Strains lacking both Scw4 and Scw10 are CFW-hypersensitive and morphologically abnormal, have doubled chitin content and increased alkali-soluble glucan, and show alterations in β1,3-glucan structure and in cross-linking of mannoproteins to the wall (Cappellaro et al. 1998; Sestak et al. 2004). The growth and morphological defects of scw4Δ scw10Δ are exacerbated by deletion of CHS3 or FKS2 (Sestak et al. 2004). From the phenotypes of strains expressing different relative amounts of Bgl2 and Scw10, it was proposed that levels of Bgl2 and Scw10 need to be balanced to ensure wall stability (Klebl and Tanner 1989; Sestak et al. 2004; File S9). Cells lacking Scw11 have a separation defect, and, consistent with this, Scw11 is a daughter cell-specific protein (Cappellaro et al. 1998; Colman-Lerner et al. 2001).
Eng1/Dse4 and Eng2/Acf2 endo-β1,3-glucanases:
These related proteins have endo-β1,3-glucanase activity in vitro, but different localizations. Eng1 is a GPI protein (Baladron et al. 2002; De Groot et al. 2003), whereas Eng2 is likely intracellular. Mutants lacking one or both proteins make normal walls, but eng1Δ cells have a separation defect, consistent with Eng1’s localization to the daughter side of the septum (Colman-Lerner et al. 2001; Baladron et al. 2002). ENG2 expression increases during sporulation, although eng2Δ diploids are not defective in that process (Baladron et al. 2002). Surprisingly, loss of multiple exo- and endo-β1,3-glucanases is not catastrophic because cells lacking Exg1, Exg2, Eng1, Eng2, and Bgl2 grow well and show only the eng1Δ separation defect (Cabib et al. 2008).
Gas1 family β1,3-glucanosyltransferases:
This family has five members, all of which have GPI attachment sites (Fankhauser et al. 1993; Caro et al. 1997; Popolo and Vai 1999; De Groot et al. 2003). Gas1, Gas3, and Gas5 can also be covalently linked to the wall (De Sampaïo et al. 1999; Yin et al. 2005). Gas proteins have β1,3-glucanosyltransfer activity: they cleave β1,3-glucosidic linkages within β1,3-glucan chains and then transfer the newly generated reducing end of the cleaved glycan to the nonreducing end of another β1,3-glucan molecule, thereby extending the recipient β1,3-glucan chain (Mouyna et al. 2000; Carotti et al. 2004; Ragni et al. 2007b; Mazan et al. 2011; File S9).
Gas1 has a major role in vegetative wall biogenesis. gas1Δ mutants are CFW-hypersensitive (Ram et al. 1994) and have less β1,3-glucan but more chitin and mannan in their walls (Ram et al. 1995; Popolo et al. 1997; Valdivieso et al. 2000). gas1Δ cells release β1,3-glucan to the medium (Ram et al. 1998), and Gas1’s β1,3-glucan elongase activity may therefore be necessary for incorporation of β1,3-glucan into the wall. In addition, analyses of the synthetic interaction network of gas1Δ revealed that survival in the absence of Gas1 requires correct assembly of β1,6-glucan (Tomishige et al. 2003; Lesage et al. 2004). Gas1 is detectable in the lateral wall, in the chitin ring in small-budded cells, and near the primary septum and remains in the bud scar after cell separation, and its localization is dependent on the presence of its GPI-attachment sequence (Rolli et al. 2009).
Gas3 and Gas5 likely have wall-related functions in vegetative cells (File S9). GAS2 and GAS4 are expressed only during sporulation, and diploids lacking both Gas2 and Gas4 have a severe sporulation defect. The inner glucan layer of the wall of double homozygous gas2gas4 null spores was disorganized and detached from chitosan, suggesting that the β1,3-glucanosyltransferase activity of Gas2 and Gas4 generates β1,3-glucan chains that associate optimally with chitosan (Ragni et al. 2007a).
Yapsin aspartyl proteases:
GPI-anchored aspartyl proteases of the yapsin family have roles in the turnover of CWPs and wall-localized enzymes. Yps1, Yps2/Mkc7, Yps3, and Yps6 are mostly plasma membrane-associated, whereas Yps7 is predicted to be wall anchored (Krysan et al. 2005; Gagnon-Arsenault et al. 2006). Yapsins cleave their substrates C-terminally to Lys or Arg or pairs of these residues (Olsen et al. 1998; Komano et al. 1999) and themselves undergo proteolytic processing to generate active enzyme (File S9).
Individual yapsin null mutants are sensitive to various wall-disrupting agents, and loss of multiple YPS genes leads to osmotically remedial, temperature-sensitive lysis defects, findings that indicate that the yapsins are involved in wall maintenance (Krysan et al. 2005). Walls from yps1Δ yps2Δ and yps1Δ yps2Δ yps3Δ yps6Δ yps7Δ null mutants showed lowered β1,3- and β1,6-glucan and elevated chitin levels, whereas mannan levels were unchanged, with the wall alterations being most pronounced in the quintuple mutant (Krysan et al. 2005). The β-glucan defects were due to decreased incorporation of these polymers into the wall, because synthesis of the two β-glucans was normal in the deletion strains. These findings suggest that yapsins act on wall hydrolases and transglycosidases, thereby regulating activity of the latter, and hence, incorporation of glucans into the wall (Krysan et al. 2005). Support for this came from identification of Gas1, Pir4, and Msb2 as Yps1 substrates (Gagnon-Arsenault et al. 2008; Vadaie et al. 2008). In addition to degrading or shedding proteins during wall remodeling, yapsins also have roles in mediating release of aberrantly folded or overexpressed GPI proteins that induce ER stress (Miller et al. 2010).
Nonenzymatic CWPs
Structural GPI proteins:
There are three families of GPI-CWP and several individual GPI-CWP that do not resemble known enzymes. Strains lacking one or more of these GPI-CWP have wall defects, and expression of some of these proteins can vary with cell cycle stage or be induced during mating or sporulation in response to cell wall stress or when oxygen levels are low. In general, GPI-CWPs have a collective role in maintaining cell wall stability (Lesage and Bussey 2006; Ragni et al. 2007c).
Sps2 family:
This group comprises Ecm33, Pst1, Sps2, and Sps22 (Caro et al. 1997). Of these, Ecm33 has an important role in vegetative walls. ecm33Δ cells are temperature-sensitive and sensitive to various wall-perturbing agents, have a disorganized wall with a thin or absent mannoprotein layer, and shed β1,6-glucan-linked mannoproteins and Pir2 that is possibly linked to β1,3-glucan more than normal in the medium (Lussier et al. 1997b; Pardo et al. 2004). pst1Δ cells have no obvious phenotype, but ecm33Δ pst1Δ double nulls show exacerbated sensitivity to various wall stresses. Ecm33’s and Pst1’s functions partially overlap, but the proteins are not fully redundant because overexpression of PST1 only weakly suppresses the ecm33Δ defects (Pardo et al. 2004). Sps2 and Sps22 are a redundant pair required for normal ascospore wall formation. Diploids lacking them form spores with abnormal β-glucan, chitosan, and dityrosine layers (Coluccio et al. 2004). Sps2 and Sps22 likely act at a similar stage in ascospore wall formation as Gas2, Gas4, and Crr1 in the formation of the β-glucan layer.
Tip1 family:
Tip1, Cwp1, Cwp2, Tir1, Tir2, Tir3, Tir4, and Dan1/Ccw13 are mostly small, Ser- and Ala-rich GPI-CWP that show differential expression during the cell cycle and during aerobic or anaerobic growth and can be localized differently on the cell surface. Cwp2 also contains a PIR repeat and so could be linked to β1,3-glucan (Klis et al. 2010).
Cwp1, Cwp2, Tip1, and Tir1 have roles in the vegetative wall. Deletion of their genes individually leads to CFW hypersensitivity (Van der Vaart et al. 1995), and cwp1Δ cwp2Δ double mutants show increased permeability to DNA-binding agents relative to the single nulls (Zhang et al. 2008). In addition, the walls of cwp2Δ and cwp1Δ cwp2Δ cells are thinner than those of the wild type (Van der Vaart et al. 1995; Zhang et al. 2008). Localization of these proteins correlates with their expression. Tip1 is expressed in G1 and found in mother cells only, whereas Cwp1, Cwp2, and Tir1 are expressed during the S-to-G2 transition, Cwp2 being found in small-to-medium-sized buds (Caro et al. 1998; Smits et al. 2006). Localization of Cwp2 and Tip1 is determined by the timing of their expression in the cell cycle (Smits et al. 2006; File S9). Tip1 and Tip2 are also heat- and cold-shock-inducible, and Tir1 and Tir4 are induced by cold shock (Kowalski et al. 1995; Abramova et al. 2001).
CWP1 and CWP2 are downregulated upon shift to anaerobic conditions, whereas Tip1, Tir1, Tir2, Tir3, Tir4, Dan1/Ccw13, and Dan4 are induced (Abramova et al. 2001). Of these, the Dan proteins are strongly repressed by oxygen. Strains lacking Tir1, Tir3, or Tir4 do not grow under anaerobic conditions. Shift to anaerobiosis therefore leads to remodeling of the wall (Abramova et al. 2001), although it is not clear how the anaerobically induced CWPs permit anaerobic growth.
Sed1 and Spi1:
These are two related, Ser/Thr-rich GPI-CWP whose expression is induced by nutrient limitation and stress. Sed1 is releasable from walls by treatment with β-glucanases or proteases (Shimoi et al. 1998). Association of Sed1 with the wall is dependent on Kre6 (Bowen and Wheals 2004), consistent with anchorage involving β1,6-glucan. SED1 expression is induced in the stationary phase, a time when the wall becomes thicker and more resistant to lytic enzymes (De Nobel et al. 1990). Consistent with a protective role in stationary-phase walls, sed1Δ cells become more sensitive to Zymolyase in that growth phase (Shimoi et al. 1998). Elevated Sed1 expression is also part of the compensatory response made by cells lacking multiple GPI-CWP (Hagen et al. 2004; File S9). Expression of SPI1 is induced by weak organic acids, and Sps1 is a major contributor to the β1,3-glucanase resistance that arises in response to this stress (Simoes et al. 2003). Low external pH also leads to formation of new alkali-labile linkages between GPI-CWPs and β1,3-glucan (Kapteyn et al. 2001).
Ccw12:
Ccw12 is a small, heavily glycosylated, Ser/Thr-rich GPI-CWP with two C-terminal repeats of an amino acid sequence critical for its function (File S9). Ccw12 is releasable by β1,3-glucanases (Mrša et al. 1999), but also has the potential to make disulfide cross-links because it has a three-Cys motif found in several S. cerevisiae flocculins (see Flocculins and agglutinins) and in wall proteins of other yeasts (Klis et al. 2010). Ccw12 is likely abundant because its gene has a very high codon adaptation index (Klis et al. 2010). Ccw12 has a major role in the wall, because cells lacking it are hypersensitive to CFW and other wall stressing agents and rounder than wild type, with thick, disorganized walls, lysis-prone buds, and elevated levels of chitin (Mrša et al. 1999; Hagen et al. 2004; Ragni et al. 2007c; Shankarnarayan et al. 2008). Man-to-Glc ratios in ccw12Δ cells are unchanged, but levels of alkali-soluble relative to alkali-insoluble glucan are higher, indicating altered organization and cross-linkage of wall components (Ragni et al. 2007c). Ccw12 localizes to sites of active wall synthesis, including the future bud site, the septum, the lateral walls of enlarging daughter cells, as well as the tips of mating projections, but then turns over, suggesting that it may stabilize walls as daughter cells and that mating projections are being formed (Ragni et al. 2011). Loss of Ccw12 alone activates the CWI pathway-mediated chitin stress response (Ragni et al. 2007c, 2011; see Chitin synthesis in response to cell wall stress), but deletion of additional GPI-CWP genes forces cells over a threshold that leads to triggering of a new compensatory response to loss of multiple GPI-CWP that depends on Sed1 and the non-GPI-CWP Srl1 (see File S7).
Other nonenzymatic GPI proteins:
Ccw14 (Ssr1/Icwp) is a β1,3-glucanase-extractable, Ser-rich GPI-CWP that has been localized to the inner cell wall (Moukadiri et al. 1997; Mrša et al. 1999; File S9). The protein has an eight-Cys-containing CFEM domain found in various fungal surface proteins (Kulkarni et al. 2003; De Groot et al. 2005) and, hence, a potential for disulfide formation. CCW14/SSR1 null mutants have no obvious growth defects, but show increased sensitivity to CFW, Congo Red, and Zymolyase. Overexpression of CCW14/SSR1 also leads to increased CFW and Congo Red sensitivity, although not Zymolyase sensitivity, suggesting that levels of Ccw14/Ssr1 relative to one or more other wall components need to be balanced (Moukadiri et al. 1997).
Dse2 and Egt2, which are unrelated to one another, are daughter cell-specific proteins with roles in cell separation. In haploids, Dse2 is concentrated in regions connecting mother and daughter cells (Colman-Lerner et al. 2001; Doolin et al. 2001), and Egt2 is localized to the septum (Fujita et al. 2004). Of these two GPI proteins (Hamada et al. 1998a; Terashima et al. 2002; De Groot et al. 2003), Egt2’s localization also depends on Gpi7 (Fujita et al. 2004). dse2Δ haploids show no defects, but homozygous DSE2 nulls show unipolar budding and form chains of cells. egt2Δ cells have separation defects similar to those of eng1Δ cells, a phenotype exacerbated in the double null, indicating that the two proteins act in parallel pathways involved in cell separation (Kovacech et al. 1996; Baladron et al. 2002).
The related Ser/Thr-rich GPI proteins Fit1, Fit2, and Fit3 (Hamada et al. 1999) have a nutritional role. Their expression is induced by iron limitation, and the proteins normally retain iron bound to ferrichrome because Zymolyase treatment of FIT-deletion mutants releases less iron from cells (Protchenko et al. 2001). Fit1 localizes to the wall, where it, Fit2, and Fit3 concentrate siderophore iron and facilitate subsequent uptake of the metal, highlighting a role of the wall in nutrient acquisition (Protchenko et al. 2001).
Flocculins and agglutinins:
GPI-CWP involved in cell–cell adhesion are the related Flo1, Flo5, Flo9, Flo10, and Flo11/Muc1 flocculins, the Aga1 and Fig2 pair, and the α-agglutinin Sag1 (Roy et al. 1991; Cappellaro et al. 1994; Chen et al. 1995; Caro et al. 1997; Erdman et al. 1998; Guo et al. 2000; Shen et al. 2001; Dranginis et al. 2007; Van Mulders et al. 2009; Goossens and Willaert 2010).
Flo1, Flo5, Flo9, and Flo10 are modular proteins composed of 1100–1500 amino acids. Major features are N-terminal PA14 domains that bind α-mannosides and mediate adhesion to adjacent cells, a central Ser/Thr-rich domain that is organized in repeat sequences and heavily glycosylated, and two or three conserved three-Cys repeats toward their C termini (Verstrepen and Klis 2006; Goossens and Willaert 2010; Klis et al. 2010; Veelders et al. 2010; Goossens et al. 2011). In addition, the Ser/Thr-rich domains of Flo1 and Flo11/Muc1 have short sequences enriched in Ile, Thr, and Val that are predicted to form intramolecular β-sheet-like interactions or amyloids, and both a soluble, GPI-less portion of Flo11/Muc1 and a Flo1-derived form fibrillar β-aggregates in vitro (Ramsook et al. 2010). Amyloid formation correlates with flocculation in vivo, for cells expressing Flo1 and Flo11/Muc1 that had been induced to flocculate in the presence of Ca2+ stained more brightly with an amyloid-binding dye, and amyloid formation may be part of the mechanism by which these proteins promote cell aggregation (Ramsook et al. 2010). FLO1, FLO5, FLO9, and FLO10 are not expressed in laboratory strains such as S288C because of a mutation in the transcriptional activator Flo8. However, activation of individual FLO genes confers the ability to flocculate (Guo et al. 2000; Van Mulders et al. 2009). Flo11/Muc1, a diverged Flo protein (Lambrechts et al. 1996; Lo and Dranginis 1996), is not involved in flocculation, but is required for pseudohypha formation by diploids, invasion of agar by haploids, and biofilm development (Lo and Dranginis 1998; Guo et al. 2000; Reynolds and Fink 2001; Dranginis et al. 2007; Bojsen et al. 2012).
Related Aga1 and Fig2 function in mating and localize to the mating projection (Erdman et al. 1998; Guo et al. 2000; Jue and Lipke 2002). Aga1 is a component of a-agglutinin that displays the Aga2 subunit, which is disulfide linked to it, and which confers binding specificity to α-agglutinin Sag1 (Orlean et al. 1986; Roy et al. 1991; Cappellaro et al. 1994; Shen et al. 2001). Fig2, which like Aga1 is expressed in both mating types, is required for formation of mating projections and maintenance of wall integrity during mating (Erdman et al. 1998; Guo et al. 2000; Zhang et al. 2002; File S9).
Sag1 has a long Ser/Thr-rich region in its C-terminal half that may hold up the N-terminal, Aga2-binding portion of the protein at the cell surface. Sag1’s N-terminal region contains three sequential domains that resemble variable immunoglobulin-like folds (Chen et al. 1995; Shen et al. 2001), the most C-terminal of which contains amino acids necessary for Aga2 binding (Wojciechowicz et al. 1993; Cappellaro et al. 1994; De Nobel et al. 1996).
Non-GPI-CWP:
PIR proteins:
Expression and localization of Pir1 (Ccw6), Pir2 (Ccw7/Hsp150), Pir3 (Ccw8), and Pir4 (Ccw5/Cis3) is regulated during cell cycle progression and in response to stress. PIR1, PIR2, and PIR3 show peaks of expression in early G1, whereas PIR4 expression is highest in G2 (Spellman et al. 1998). PIR2 is also induced by heat shock, treatment with CFW or Zymolyase, and nitrogen limitation (Russo et al. 1993; Toh-e et al. 1993; Yun et al. 1997; Boorsma et al. 2004). Consistent with their upregulation upon wall stress, all four PIR genes show elevated expression in an mpk1 mutant that constitutively activates the protein kinase C-dependent CWI pathway, an effect eliminated in mutants lacking the PKC pathway’s target transcription factor, Rlm1 (Jung and Levin 1999).
PIR proteins localize to different parts of the surface of budding cells (Sumita et al. 2005; File S9). Pir1 and Pir2 are found at bud scars of both haploids and diploids, Pir1 being localized inside the chitin ring. Some Pir1 and Pir2 and most Pir3 are also present in lateral walls (Yun et al. 1997). Pir4 has been reported be uniformly distributed at the cell surface or restricted to growing buds (Moukadiri et al. 1999; Sumita et al. 2005).
Strains lacking individual PIR proteins have subtle growth defects, but as more PIR genes are deleted, disruptants show a progressive increase in sensitivity to CFW, Congo Red, and heat shock, and cells become larger and irregularly shaped (Toh-e et al. 1993; Mrša and Tanner 1999). pir1Δ pir2Δ pir3Δ pir4Δ mutants show a loss of viability that is suppressed in osmotically supported medium (Teparic et al. 2004). These findings suggest a collective role for PIR proteins in maintenance of a normal wall. How these proteins contribute is unclear, because the carbohydrate composition of the quadruple PIR disruptant’s wall is unaltered, and the relative amounts of alkali-soluble and -insoluble glucan and chitin show modest changes (Teparic et al. 2004; Mazan et al. 2008). PIR proteins, however, impact permeability of the wall because the pir1Δ pir2Δ pir3Δ mutant is hypersensitive to membrane-active tobacco osmotin, whereas overexpression of PIR1, PIR2, or PIR3 confers osmotin resistance on walled cells but not spheroplasts (Yun et al. 1997). The effects of PIR protein levels on wall permeability are consistent with the role of these proteins in cross-linking β1,3-glucans (see Mild alkali-releasable proteins).
Scw3 (Sun4):
Haploids lacking this soluble cell wall protein (Cappellaro et al. 1998) are larger than wild-type cells and have a separation defect and thickened septa (Mouassite et al. 2000). Scw3/Sun4 is a member of the SUN family of proteins, of which Sim1 and Uth1 are also released from cell walls by dithiothreitol treatment (Velours et al. 2002). Uth1 and Scw3/Sun4 additionally localize to mitochondria (Velours et al. 2002), but the significance of this distribution of the SUN proteins is unclear. The biochemical function of the SUN proteins is unknown as they show no similarity to known enzymes (File S9).
Srl1:
This small Ser/Thr-rich protein is involved in the compensatory response to loss of multiple GPI-CWP (Hagen et al. 2004; File S9). It rescues the lysis defects of strains defective in the function of the “regulation of Ace2 and polarized morphogenesis” (RAM) signaling network when overexpressed (Kurischko et al. 2005), and some of it is tightly associated with the wall and released by β1,3-glucanase (Terashima et al. 2002). Slr1 localizes to the periphery of small buds (Shepard et al. 2003). srl1Δ mutants have no obvious morphological defects and show modest Calcofluor White sensitivity at 22°, but are hypersensitive to this agent at 37° (Kurischko et al. 2005). Mutants defective in RAM function are also suppressed by overexpression of Sim1 (see above) and Ccw12 (Kurischko et al. 2005), and slr1Δ and ccw12Δ show a strong genetic interaction in the RAM-defective background. The srl1Δ ccw12Δ strain is CFW hypersensitive at both 22° and 37°, and at 22°, but not at 37°, resembles mating pheromone-treated wild-type cells (Kurischko et al. 2005). Srl1 and Ccw12 have been proposed to have parallel functions in activation of a CWI pathway that operates when RAM signaling is defective (Kurischko et al. 2005).
What Is Next?
The biosynthesis of most individual yeast wall components is now understood in much detail and involves conserved pathways such as N-glycosylation and GPI anchoring and enzymes represented in other organisms, such as chitin and β1,3-glucan synthases. In contrast, β1,6-glucan formation and cross-linkage to GPI proteins and cross-linking of chitin to β-glucans are clearly restricted to certain yeasts and filamentous fungi, and the enzymes implicated in the latter processes, as well as certain CWP, are signatures of fungal cell walls, whose evolution of has been reviewed by Ruiz-Herrera and Ortiz-Castellanos (2010) and Xie and Lipke (2010).
Much of the work necessary to take both the conserved and the yeast-specific aspects of wall biogenesis to the next level must be biochemical and analytical. These efforts will involve charting new biochemical territory, such as determining how β1,6-glucan is made and defining the functions of wall-active proteins such as Ccw12, Ecm33, Kre1, and Kre9, which have key roles in wall biogenesis, but show no resemblance to proteins of known function and may not be enzymes. Other biochemical challenges are the mechanism and activation of chitin and β1,3-glucan synthases, the mechanism and conjectured flippase activities of the multispanning glycosyltransferases of the dolichol, O-mannosylation, and GPI pathways, the functions of the Etn-P side branches on GPIs, and the biochemical activities of predicted enzymes such as the Dcw1/Dfg5 pair, Kre5, Kre6, Scw4, and Scw10. The latter efforts require application of high-resolution techniques to analyze the fine structure and linkages of cell wall glycans (Magnelli et al. 2002; Aimanianda et al. 2009), which should highlight the reactions for which biochemists need to develop assays and screen mutants.
With the identification of so many proteins involved in cell wall biogenesis, and with ever-improving knowledge of wall composition, we can look forward to deepening our understanding of the complexities of yeast cell wall biogenesis.
Supplementary Material
Acknowledgments
I thank my students for their many contributions to GPI and wall biosynthesis. I also acknowledge the contributions of the late Yoshifumi Jigami to our field. I am grateful to three reviewers for their helpful comments. Work in my laboratory has been supported by grant GM-46220 from the National Institutes of Health and by a Burroughs Wellcome Scholar Award in Molecular Pathogenic Mycology.
Footnotes
Communicating editor: J. Thorner
Literature Cited
- Abe M., Hashimoto H., Yoda K., 1999. Molecular characterization of Vig4/Vrg4 GDP-mannose transporter of the yeast Saccharomyces cerevisiae. FEBS Lett. 458: 309–312. [DOI] [PubMed] [Google Scholar]
- Abe M., Nishida I., Minemura M., Qadota H., Seyama Y., et al. , 2001. Yeast 1,3-β-glucan synthase activity is inhibited by phytosphingosine localized to the endoplasmic reticulum. J. Biol. Chem. 276: 26923–26930. [DOI] [PubMed] [Google Scholar]
- Abeijon C., Chen L. Y., 1998. The role of glucosidase I (Cwh41p) in the biosynthesis of cell wall β-1,6-glucan is indirect. Mol. Biol. Cell 9: 2729–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abeijon C., Orlean P., Robbins P. W., Hirschberg C. B., 1989. Topography of glycosylation in yeast: characterization of GDP-mannose transport and lumenal guanosine diphosphatase activities in Golgi-like vesicles. Proc. Natl. Acad. Sci. USA 86: 6935–6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramova N., Sertil O., Mehta S., Lowry C. V., 2001. Reciprocal regulation of anaerobic and aerobic cell wall mannoprotein gene expression in Saccharomyces cerevisiae. J. Bacteriol. 183: 2881–2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Absmanner B., Schmeiser V., Kämpf M., Lehle L., 2010. Biochemical characterization, membrane association and identification of amino acids essential for the function of Alg11 from Saccharomyces cerevisiae, an α1,2-mannosyltransferase catalysing two sequential glycosylation steps in the formation of the lipid-linked core oligosaccharide. Biochem. J. 426: 205–217. [DOI] [PubMed] [Google Scholar]
- Adams D. J., 2004. Fungal cell wall chitinases and glucanases. Microbiology 150: 2029–2035. [DOI] [PubMed] [Google Scholar]
- Aebi M., Gassenhuber J., Domdey H., te Heesen S., 1996. Cloning and characterization of the ALG3 gene of Saccharomyces cerevisiae. Glycobiology 6: 439–444. [DOI] [PubMed] [Google Scholar]
- Aebi M., Bernasconi R., Clerc S., Molinari M., 2010. N-glycan structures: recognition and processing in the ER. Trends Biochem. Sci. 35: 74–82. [DOI] [PubMed] [Google Scholar]
- Aguilar-Uscanga B., François J. M., 2003. A study of the yeast cell wall composition and structure in response to growth conditions and mode of cultivation. Lett. Appl. Microbiol. 37: 268–274. [DOI] [PubMed] [Google Scholar]
- Aimanianda V., Clavaud C., Simenel C., Fontaine T., Delepierre M., et al. , 2009. Cell wall β-(1,6)-glucan of Saccharomyces cerevisiae: structural characterization and in situ synthesis. J. Biol. Chem. 284: 13401–13412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrés I., Gallardo O., Parascandola P., Javier Pastor F. I., Zueco J., 2005. Use of the cell wall protein Pir4 as a fusion partner for the expression of Bacillus sp. BP-7 xylanase A in Saccharomyces cerevisiae. Biotechnol. Bioeng. 89: 690–697. [DOI] [PubMed] [Google Scholar]
- Appeltauer U., Achstetter T., 1989. Hormone-induced expression of the CHS1 gene from Saccharomyces cerevisiae. Eur. J. Biochem. 181: 243–247. [DOI] [PubMed] [Google Scholar]
- Arroyo J., Hutzler J., Bermejo C., Ragni E., García-Cantalejo J., et al. , 2011. Functional and genomic analyses of blocked protein O-mannosylation in baker’s yeast. Mol. Microbiol. 79: 1529–1546. [DOI] [PubMed] [Google Scholar]
- Ashida H., Hong Y., Murakami Y., Shishioh N., Sugimoto N., et al. , 2005. Mammalian PIG-X and yeast Pbn1p are the essential components of glycosylphosphatidylinositol-mannosyltransferase I. Mol. Biol. Cell 16: 1439–1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma M., Levinson J. N., Pagé N., Bussey H., 2002. Saccharomyces cerevisiae Big1p, a putative endoplasmic reticulum membrane protein required for normal levels of cell wall β-1,6-glucan. Yeast 19: 783–793. [DOI] [PubMed] [Google Scholar]
- Baba M., Baba N., Ohsumi Y., Kanaya K., Osumi M., 1989. Three-dimensional analysis of morphogenesis induced by mating pheromone α factor in Saccharomyces cerevisiae. J. Cell Sci. 94: 207–216. [DOI] [PubMed] [Google Scholar]
- Baladrón V., Ufano S., Dueñas E., Martín-Cuadrado A. B., del Rey F., et al. , 2002. Eng1p, an endo-1,3-β-glucanase localized at the daughter side of the septum, is involved in cell separation in Saccharomyces cerevisiae. Eukaryot. Cell 1: 774–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballou C. E., 1982. Yeast cell wall and surface, pp. 335–360 in The Molecular Biology of the Yeast Saccharomyces: Metabolism and Gene Expression, edited by Strathern J. N., Jones E. W., Broach J. R. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Ballou C. E., 1990. Isolation, characterization, and properties of Saccharomyces cerevisiae mnn mutants with nonconditional protein glycosylation defects. Methods Enzymol. 185: 440–470. [DOI] [PubMed] [Google Scholar]
- Ballou C. E., Maitra S. K., Walker J. W., Whelan W. L., 1977. Developmental defects associated with glucosamine auxotrophy in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 74: 4351–4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes G., Hansen W. J., Holcomb C. L., Rine J., 1984. Asparagine-linked glycosylation in Saccharomyces cerevisiae: genetic analysis of an early step. Mol. Cell. Biol. 4: 2381–2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bays N. W., Gardner R. G., Seelig L. P., Joazeiro C. A., Hampton R. Y., 2001. Hrd1p/Der3p is a membrane-anchored ubiquitin ligase required for ER-associated degradation. Nat. Cell Biol. 3: 24–29. [DOI] [PubMed] [Google Scholar]
- Beauvais A., Loussert C., Prevost M. C., Verstrepen K., Latgé J. P., 2009. Characterization of a biofilm-like extracellular matrix in FLO1-expressing Saccharomyces cerevisiae cells. FEMS Yeast Res. 9: 411–419. [DOI] [PubMed] [Google Scholar]
- Benachour A., Sipos G., Flury I., Reggiori F., Canivenc-Gansel E., et al. , 1999. Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 274: 15251–15261. [DOI] [PubMed] [Google Scholar]
- Benghezal M., Benachour A., Rusconi S., Aebi M., Conzelmann A., 1996. Yeast Gpi8p is essential for GPI anchor attachment onto proteins. EMBO J. 15: 6575–6583. [PMC free article] [PubMed] [Google Scholar]
- Berninsone P., Miret J. J., Hirschberg C. B., 1994. The Golgi guanosine diphosphatase is required for transport of GDP-mannose into the lumen of Saccharomyces cerevisiae Golgi vesicles. J. Biol. Chem. 269: 207–211. [PubMed] [Google Scholar]
- Bhamidipati A., Denic V., Quan E. M., Weissman J. S., 2005. Exploration of the topological requirements of ERAD identifies Yos9p as a lectin sensor of misfolded glycoproteins in the ER lumen. Mol. Cell 19: 741–751. [DOI] [PubMed] [Google Scholar]
- Bi E., Park H.-O., 2012. Cell polarization and cytokinesis in budding yeast. Genetics 191: 347–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bickel T., Lehle L., Schwarz M., Aebi M., Jakob C. A., 2005. Biosynthesis of lipid-linked oligosaccharides in Saccharomyces cerevisiae: Alg13p and Alg14p form a complex required for the formation of GlcNAc2-PP-dolichol. J. Biol. Chem. 280: 34500–34506. [DOI] [PubMed] [Google Scholar]
- Bickle M., Delley P.-A., Schmidt A., Hall M. N., 1998. Cell wall integrity modulates RHO1 activity via the exchange factor ROM2. EMBO J. 17: 2235–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boder E. T., Wittrup K. D., 1997. Yeast surface display for screening combinatorial polypeptide libraries. Nat. Biotechnol. 15: 553–557. [DOI] [PubMed] [Google Scholar]
- Bojsen R. K., Andersen K. S., Regenberg B., 2012. Saccharomyces cerevisiae: a model to uncover molecular mechanisms for yeast biofilm biology. FEMS Immunol. Med. Microbiol. 65: 169–182. [DOI] [PubMed] [Google Scholar]
- Boone C., Sommer S. S., Hensel A., Bussey H., 1990. Yeast KRE genes provide evidence for a pathway of cell wall β-glucan assembly. J. Cell Biol. 110: 1833–1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boorsma A., de Nobel H., ter Riet B., Bargmann B., Brul S., et al. , 2004. Characterization of the transcriptional response to cell wall stress in Saccharomyces cerevisiae. Yeast 21: 413–427. [DOI] [PubMed] [Google Scholar]
- Bosson R., Jaquenoud M., Conzelmann A., 2006. GUP1 of Saccharomyces cerevisiae encodes an O-acyltransferase involved in remodeling of the GPI anchor. Mol. Biol. Cell 17: 2636–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen S., Wheals A. E., 2004. Incorporation of Sed1p into the cell wall of Saccharomyces cerevisiae involves KRE6. FEMS Yeast Res. 4: 731–735. [DOI] [PubMed] [Google Scholar]
- Breinig F., Schleinkofer K., Schmitt M. J., 2004. Yeast Kre1p is GPI-anchored and involved in both cell wall assembly and architecture. Microbiology 150: 3209–3218. [DOI] [PubMed] [Google Scholar]
- Brown J. L., Bussey H., 1993. The yeast KRE9 gene encodes an O glycoprotein involved in cell surface β-glucan assembly. Mol. Cell. Biol. 13: 6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E., 1993. Genetics and molecular biology of chitin synthesis in fungi. Annu. Rev. Microbiol. 47: 505–534. [DOI] [PubMed] [Google Scholar]
- Bulik D. A., Olczak M., Lucero H. A., Osmond B. C., Robbins P. W., et al. , 2003. Chitin synthesis in Saccharomyces cerevisiae in response to supplementation of growth medium with glucosamine and cell wall stress. Eukaryot. Cell 2: 886–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burda P., Aebi M., 1998. The ALG10 locus of Saccharomyces cerevisiae encodes the α-1,2 glucosyltransferase of the endoplasmic reticulum: the terminal glucose of the lipid-linked oligosaccharide is required for efficient N-linked glycosylation. Glycobiology 8: 455–462. [DOI] [PubMed] [Google Scholar]
- Burda P., Aebi M., 1999. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta 1426: 239–257. [DOI] [PubMed] [Google Scholar]
- Burda P., Jakob C. A., Beinhauer J., Hegemann J. H., Aebi M., 1999. Ordered assembly of the asymmetrically branched lipid-linked oligosaccharide in the endoplasmic reticulum is ensured by the substrate specificity of the individual glycosyltransferases. Glycobiology 9: 617–625. [DOI] [PubMed] [Google Scholar]
- Buschhorn B. A., Kostova Z., Medicherla B., Wolf D. H., 2004. A genome-wide screen identifies Yos9p as essential for ER-associated degradation of glycoproteins. FEBS Lett. 577: 422–426. [DOI] [PubMed] [Google Scholar]
- Cabib E., 2004. The septation apparatus, a chitin-requiring machine in budding yeast. Arch. Biochem. Biophys. 426: 201–207. [DOI] [PubMed] [Google Scholar]
- Cabib E., 2009. Two novel techniques for determination of polysaccharide cross-links show that Crh1p and Crh2p attach chitin to both β(1–6)- and β(1–3)glucan in the Saccharomyces cerevisiae cell wall. Eukaryot. Cell 8: 1626–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E., Duran A., 2005. Synthase III-dependent chitin is bound to different acceptors depending on location on the cell wall of budding yeast. J. Biol. Chem. 280: 9170–9179. [DOI] [PubMed] [Google Scholar]
- Cabib E., Schmidt M., 2003. Chitin synthase III activity, but not the chitin ring, is required for remedial septa formation in budding yeast. FEMS Microbiol. Lett. 224: 299–305. [DOI] [PubMed] [Google Scholar]
- Cabib E., Sburlati A., Bowers B., Silverman S. J., 1989. Chitin synthase 1, an auxiliary enzyme for chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 108: 1665–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabib E., Roh D. H., Schmidt M., Crotti L. B., Varma A., 2001. The yeast cell wall and septum as paradigms of cell growth and morphogenesis. J. Biol. Chem. 276: 19679–19682. [DOI] [PubMed] [Google Scholar]
- Cabib E., Blanco N., Grau C., Rodríguez-Peña J. M., Arroyo J., 2007. Crh1p and Crh2p are required for the cross-linking of chitin to β(1–6)glucan in the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 63: 921–935. [DOI] [PubMed] [Google Scholar]
- Cabib E., Farkas V., Kosík O., Blanco N., Arroyo J., et al. , 2008. Assembly of the yeast cell wall. Crh1p and Crh2p act as transglycosylases in vivo and in vitro. J. Biol. Chem. 283: 29859–29872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canivenc-Gansel E., Imhof I., Reggiori F., Burda P., Conzelmann A., et al. , 1998. GPI anchor biosynthesis in yeast: phosphoethanolamine is attached to the α1,4-linked mannose of the complete precursor glycophospholipid. Glycobiology 8: 761–770. [DOI] [PubMed] [Google Scholar]
- Cantagrel V., Lefeber D. J., Ng B. G., Guan Z., Silhavy J. L., et al. , 2010. SRD5A3 is required for converting polyprenol to dolichol and is mutated in a congenital glycosylation disorder. Cell 42: 203–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellaro C., Baldermann C., Rachel R., Tanner W., 1994. Mating type-specific cell-cell recognition of Saccharomyces cerevisiae: cell wall attachment and active sites of a- and α-agglutinin. EMBO J. 13: 4737–4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cappellaro C., Mrša V., Tanner W., 1998. New potential cell wall glucanases of Saccharomyces cerevisiae and their involvement in mating. J. Bacteriol. 180: 5030–5037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caramelo J. J., Parodi A. J., 2007. How sugars convey information on protein conformation in the endoplasmic reticulum. Semin. Cell Dev. Biol. 18: 732–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caro L. H. P., Tettelin H., Vossen J. H., Ram A. F. J., Van den Ende H., et al. , 1997. In silico identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13: 1477–1489. [DOI] [PubMed] [Google Scholar]
- Caro L. H., Smits G. J., van Egmond P., Chapman J. W., Klis F. M., 1998. Transcription of multiple cell wall protein-encoding genes in Saccharomyces cerevisiae is differentially regulated during the cell cycle. FEMS Microbiol. Lett. 161: 345–349. [DOI] [PubMed] [Google Scholar]
- Carotti C., Ferrario L., Roncero C., Valdivieso M. H., Duran A., et al. , 2002. Maintenance of cell integrity in the gas1 mutant of Saccharomyces cerevisiae requires the Chs3p-targeting and activation pathway and involves an unusual Chs3p localization. Yeast 19: 1113–1124. [DOI] [PubMed] [Google Scholar]
- Carotti C., Ragni E., Palomares O., Fontaine T., Tedeschi G., et al. , 2004. Characterization of recombinant forms of the yeast Gas1 protein and identification of residues essential for glucanosyltransferase activity and folding. Eur. J. Biochem. 271: 3635–3645. [DOI] [PubMed] [Google Scholar]
- Castillo L., Martinez A. I., Garcerá A., Elorza M. V., Valentín E., et al. , 2003. Functional analysis of the cysteine residues and the repetitive sequence of Saccharomyces cerevisiae Pir4/Cis3: the repetitive sequence is needed for binding to the cell wall β-1,3-glucan. Yeast 20: 973–983. [DOI] [PubMed] [Google Scholar]
- Castro O., Chen L. Y., Parodi A. J., Abeijón C., 1999. Uridine diphosphate-glucose transport into the endoplasmic reticulum of Saccharomyces cerevisiae: in vivo and in vitro evidence. Mol. Biol. Cell 10: 1019–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantret I., Dancourt J., Barbat A., Moore S. E. H., 2005. Two proteins homologous to the N- and C-terminal domains of the bacterial glycosyltransferase MurG are required for the second step of dolichyl linked oligosaccharide synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 280: 18551–18552. [DOI] [PubMed] [Google Scholar]
- Chavan M., Rekowicz M., Lennarz W., 2003a Insight into functional aspects of Stt3p, a subunit of the oligosaccharyl transferase. Evidence for interaction of the N-terminal domain of Stt3p with the protein kinase C cascade. J. Biol. Chem. 278: 51441–51447. [DOI] [PubMed] [Google Scholar]
- Chavan M., Suzuki T., Rekowicz M., Lennarz W., 2003b Genetic, biochemical, and morphological evidence for the involvement of N-glycosylation in biosynthesis of the cell wall β1,6-glucan of Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100: 15381–15386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chavan M., Chen Z., Li G., Schindelin H., Lennarz W. J., et al. , 2006. Dimeric organization of the yeast oligosaccharyl transferase complex. Proc. Natl. Acad. Sci. USA 103: 8947–8952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M.-H., Shen Z.-M., Bobin S., Kahn P. C., Lipke P. N., 1995. Structure of Saccharomyces cerevisiae α-agglutinin. J. Biol. Chem. 270: 26168–26177. [DOI] [PubMed] [Google Scholar]
- Chin C. F., Bennett A. M., Ma W. K., Hall M. C., Yeong F. M., 2012. Dependence of Chs2 ER export on dephosphorylation by cytoplasmic Cdc14 ensures that septum formation follows mitosis. Mol. Biol. Cell 23: 45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho R. J., Campbell M. J., Winzeler E. A., Steinmetz L., Conway A., et al. , 1998. A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2: 65–73. [DOI] [PubMed] [Google Scholar]
- Choi W. J., Santos B., Duran A., Cabib E., 1994a Are yeast chitin synthases regulated at the transcriptional or the posttranslational level? Mol. Cell. Biol. 14: 7685–7694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi W. J., Sburlati A., Cabib E., 1994b Chitin synthase 3 from yeast has zymogenic properties that depend on both the CAL1 and the CAL3 genes. Proc. Natl. Acad. Sci. USA 91: 4727–4730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christodoulidou A., Briza P., Ellinger A., Bouriotis V., 1999. Yeast ascospore wall assembly requires two chitin deacetylase isozymes. FEBS Lett. 460: 275–279. [DOI] [PubMed] [Google Scholar]
- Chuang J. S., Schekman R. W., 1996. Differential trafficking and timed localization of two chitin synthase proteins, Chs2p and Chs3p. J. Cell Biol. 135: 597–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cipollo J. F., Trimble R. B., 2002. The Saccharomyces cerevisiae alg12Δ mutant reveals a role for the middle-arm α1,2Man- and upper-arm α1,2Manα1,6Man- residues of Glc3Man9GlcNAc2-PP-Dol in regulating glycoprotein glycan processing in the endoplasmic reticulum and Golgi apparatus. Glycobiology 12: 749–762. [DOI] [PubMed] [Google Scholar]
- Cipollo J. F., Trimble R. B., Chi J. H., Yan Q., Dean N., 2001. The yeast ALG11 gene specifies addition of the terminal α1,2-Man to the Man5GlcNAc2-PP-dolichol N-glycosylation intermediate formed on the cytosolic side of the endoplasmic reticulum. J. Biol. Chem. 276: 21828–21840. [DOI] [PubMed] [Google Scholar]
- Clerc S., Hirsch C., Oggier D. M., Deprez P., Jakob C., et al. , 2009. Htm1 protein generates the N-glycan signal for glycoprotein degradation in the endoplasmic reticulum. J. Cell Biol. 184: 159–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman-Lerner A., Chin T. E., Brent R., 2001. Yeast Cbk1 and Mob2 activate daughter-specific genetic programs to induce asymmetric cell fates. Cell 107: 739–750. [DOI] [PubMed] [Google Scholar]
- Coluccio A., Bogengruber E., Conrad M. N., Dresser M. E., Briza P., et al. , 2004. Morphogenetic pathway of spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 3: 1464–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann A., Fankhauser C., Desponds C., 1990. Myoinositol gets incorporated into numerous membrane glycoproteins of Saccharomyces cerevisiae: incorporation is dependent on phosphomannomutase (SEC53). EMBO J. 9: 653–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conzelmann A., Puoti A., Lester R. L., Desponds C., 1992. Two different types of lipid moieties are present in glycophosphoinositol-anchored membrane proteins of Saccharomyces cerevisiae. EMBO J. 11: 457–466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correa J. U., Elango N., Polacheck I., Cabib E., 1982. Endochitinase, a mannan-associated enzyme from Saccharomyces cerevisiae. J. Biol. Chem. 257: 1392–1397. [PubMed] [Google Scholar]
- Cos T., Ford R. A., Trilla J. A., Duran A., Cabib E., et al. , 1998. Molecular analysis of Chs3p participation in chitin synthase III activity. Eur. J. Biochem. 256: 419–426. [DOI] [PubMed] [Google Scholar]
- Costello L. C., Orlean P., 1992. Inositol acylation of a potential glycosyl phosphoinositol anchor precursor from yeast requires acyl coenzyme A. J. Biol. Chem. 267: 8599–8603. [PubMed] [Google Scholar]
- Couto J. R., Huffaker T. C., Robbins P. W., 1984. Cloning and expression in Escherichia coli of a yeast mannosyltransferase from the asparagine-linked glycosylation pathway. J. Biol. Chem. 259: 378–382. [PubMed] [Google Scholar]
- Dallies N., Francois J., Paquet V., 1998. A new method for quantitative determination of polysaccharides in the yeast cell wall. Application to the cell wall defective mutants of Saccharomyces cerevisiae. Yeast 14: 1297–1306. [DOI] [PubMed] [Google Scholar]
- Daran J. M., Dallies N., Thines-Sempoux D., Paquet V., François J., 1995. Genetic and biochemical characterization of the UGP1 gene encoding the UDP-glucose pyrophosphorylase from Saccharomyces cerevisiae. Eur. J. Biochem. 233: 520–530. [DOI] [PubMed] [Google Scholar]
- Daran J. M., Bell W., François J., 1997. Physiological and morphological effects of genetic alterations leading to a reduced synthesis of UDP-glucose in Saccharomyces cerevisiae. FEMS Microbiol. Lett. 153: 89–96. [DOI] [PubMed] [Google Scholar]
- Deak P. M., Wolf D. H., 2001. Membrane topology and function of Der3/Hrd1p as a ubiquitin-protein ligase (E3) involved in endoplasmic reticulum degradation. J. Biol. Chem. 276: 10663–10669. [DOI] [PubMed] [Google Scholar]
- Dean N., 1999. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta 1426: 309–322. [DOI] [PubMed] [Google Scholar]
- Dean N., Zhang Y. B., Poster J. B., 1997. The VRG4 gene is required for GDP-mannose transport into the lumen of the Golgi in the yeast, Saccharomyces cerevisiae. J. Biol. Chem. 272: 31908–31914. [DOI] [PubMed] [Google Scholar]
- Debono M., Gordee R. S., 1994. Antibiotics that inhibit fungal cell wall development. Annu. Rev. Microbiol. 48: 471–497. [DOI] [PubMed] [Google Scholar]
- De Groot P. W., Ruiz C., Vázquez de Aldana C. R., Dueñas E., J Cid V., et al. , 2001. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp. Funct. Genomics 2: 124–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Groot P. W., Hellingwerf K. J., Klis F. M., 2003. Genome-wide identification of fungal GPI proteins. Yeast 20: 781–796. [DOI] [PubMed] [Google Scholar]
- De Groot P. W., Ram A. F., Klis F. M., 2005. Features and functions of covalently linked proteins in fungal cell walls. Fungal Genet. Biol. 42: 657–675. [DOI] [PubMed] [Google Scholar]
- DeMarini D. J., Adams A. E., Fares H., De Virgilio C., Valle G., et al. , 1997. A septin-based hierarchy of proteins required for localized deposition of chitin in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 139: 75–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nobel J. G., Klis F. M., Priem J., Munnik T., van den Ende H., 1990. The glucanase-soluble mannoproteins limit cell wall porosity in Saccharomyces cerevisiae. Yeast 6: 491–499. [DOI] [PubMed] [Google Scholar]
- De Nobel J. G., Barnett J. A., 1991. Passage of molecules through yeast cell walls: a brief essay-review. Yeast 7: 313–323. [DOI] [PubMed] [Google Scholar]
- De Nobel H., Lipke P. N., Kurjan J., 1996. Identification of a ligand binding site in Saccharomyces cerevisiae α-agglutinin. Mol. Biol. Cell 7: 143–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Sampaïo G., Bourdineaud J.-P., Laquin J.-M., 1999. A constitutive role for GPI anchors in Saccharomyces cerevisiae: cell wall targeting. Mol. Microbiol. 34: 247–256. [DOI] [PubMed] [Google Scholar]
- Dijkgraaf G. J., Brown J. L., Bussey H., 1996. The KNH1 gene of Saccharomyces cerevisiae is a functional homolog of KRE9. Yeast 12: 683–692. [DOI] [PubMed] [Google Scholar]
- Dijkgraaf G. J., Abe M., Ohya Y., Bussey H., 2002. Mutations in Fks1p affect the cell wall content of β-1,3- and β-1,6-glucan in Saccharomyces cerevisiae. Yeast 19: 671–690. [DOI] [PubMed] [Google Scholar]
- Doolin M. T., Johnson A. L., Johnston L. H., Butler G., 2001. Overlapping and distinct roles of the duplicated yeast transcription factors Ace2p and Swi5p. Mol. Microbiol. 40: 422–432. [DOI] [PubMed] [Google Scholar]
- Dranginis A. M., Rauceo J. M., Coronado J. E., Lipke P. N., 2007. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 71: 282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drgonová J., Drgon T., Tanaka K., Kollar R., Chen G. C., et al. , 1996. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science 272: 277–279. [DOI] [PubMed] [Google Scholar]
- Drgonova J., Drgon T., Roh D. H., Cabib E., 1999. The GTP-binding protein Rho1 is required for cell cycle progression and polarization of the yeast cell. J. Cell Biol. 146: 373–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dünkler A., Jorde S., Wendland J., 2008. An Ashbya gossypii cts2 mutant deficient in a sporulation-specific chitinase can be complemented by Candida albicans CHT4. Microbiol. Res. 163: 701–710. [DOI] [PubMed] [Google Scholar]
- Dupres V., Dufrêne Y. F., Heinisch J. J., 2010. Measuring cell wall thickness in living yeast cells using single molecular rulers. ACS Nano 4: 5498–5504. [DOI] [PubMed] [Google Scholar]
- Dupres V., Heinisch J. J., Dufrêne Y. F., 2011. Atomic force microscopy demonstrates that disulfide bridges are required for clustering of the yeast cell wall integrity sensor Wsc1. Langmuir 27: 15129–15134. [DOI] [PubMed] [Google Scholar]
- Duran A., Cabib E., 1978. Solubilization and partial purification of yeast chitin synthetase. Confirmation of the zymogenic nature of the enzyme. J. Biol. Chem. 253: 4419–4425. [PubMed] [Google Scholar]
- Ecker M., Mrša V., Hagen I., Deutzmann R., Strahl S., et al. , 2003. O-mannosylation precedes and potentially controls the N-glycosylation of a yeast cell wall glycoprotein. EMBO Rep. 4: 628–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecker M., Deutzmann R., Lehle L., Mrša V., Tanner W., 2006. Pir proteins of Saccharomyces cerevisiae are attached to β-1,3-glucan by a new protein-carbohydrate linkage. J. Biol. Chem. 281: 11523–11529. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Maurer-Stroh S., Novatchkova M., Schneider G., Eisenhaber F., 2003. Enzymes and auxiliary factors for GPI lipid anchor biosynthesis and post-translational transfer to proteins. Bioessays 25: 367–385. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B., Schneider G., Wildpaner M., Eisenhaber F., 2004. A sensitive predictor for potential GPI lipid modification sites in fungal protein sequences and its application to genome-wide studies for Aspergillus nidulans, Candida albicans, Neurospora crassa, Saccharomyces cerevisiae and Schizosaccharomyces pombe. J. Mol. Biol. 337: 243–253. [DOI] [PubMed] [Google Scholar]
- El-Sherbeini M., Clemas J. A., 1995. Cloning and characterization of GNS1: a Saccharomyces cerevisiae gene involved in synthesis of 1,3-β-glucan in vitro. J. Bacteriol. 177: 3227–3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman S. E., Lin L., Malczynski M., Snyder M., 1998. Pheromone-regulated genes required for yeast mating differentiation. J. Cell Biol. 140: 461–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esther C. R., Jr, Sesma J. I., Dohlman H. G., Ault A. D., Clas M. L., et al. , 2008. Similarities between UDP-glucose and adenine nucleotide release in yeast: involvement of the secretory pathway. Biochemistry 47: 9269–9278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre A.-L., Orlean P., Taron C. H., 2005. Saccharomyces cerevisiae Ybr004c and its human homologue are required for addition of the second mannose during glycosylphosphatidylinositol precursor assembly. FEBS J. 272: 1160–1168. [DOI] [PubMed] [Google Scholar]
- Fankhauser C., Homans S. W., Thomas-Oates J. E., McConville M. J., Desponds C., et al. , 1993. Structures of glycosylphosphatidylinositol membrane anchors from Saccharomyces cerevisiae. J. Biol. Chem. 268: 26365–26374. [PubMed] [Google Scholar]
- Fernandez F., Rush J. S., Toke D. A., Han G. S., Quinn J. E., et al. , 2001. The CWH8 gene encodes a dolichyl pyrophosphate phosphatase with a luminally oriented active site in the endoplasmic reticulum of Saccharomyces cerevisiae. J. Biol. Chem. 276: 41455–41464. [DOI] [PubMed] [Google Scholar]
- Fleet G. H., 1991. Cell walls, pp. 199–277 in The Yeasts, Vol. 4, Ed. 2, edited by Rose A. H., Harrison J. S. Academic Press, New York. [Google Scholar]
- Flury I., Benachour A., Conzelmann A., 2000. YLL031c belongs to a novel family of membrane proteins involved in the transfer of ethanolaminephosphate onto the core structure of glycosylphosphatidylinositol anchors in yeast. J. Biol. Chem. 275: 24458–24465. [DOI] [PubMed] [Google Scholar]
- Fraering P., Imhof I., Meyer U., Strub J. M., van Dorsselaer A., et al. , 2001. The GPI transamidase complex of Saccharomyces cerevisiae contains Gaa1p, Gpi8p, and Gpi16p. Mol. Biol. Cell 12: 3295–3306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank C. G., Aebi M., 2005. Alg9 mannosyltransferase is involved in two different steps of lipid-linked oligosaccharide biosynthesis. Glycobiology 15: 1156–1163. [DOI] [PubMed] [Google Scholar]
- Frank C. G., Sanyal S., Rush J. S., Waechter C. J., Menon A. K., 2008. Does Rft1 flip an N-glycan lipid precursor? Nature 454: E3–E4. [DOI] [PubMed] [Google Scholar]
- Frieman M. B., Cormack B. P., 2003. The ω-site sequence of glycosylphosphatidyl-inositol-anchored proteins in Saccharomyces cerevisiae can determine distribution between the membrane and the cell wall. Mol. Microbiol. 50: 883–896. [DOI] [PubMed] [Google Scholar]
- Frieman M. B., Cormack B. P., 2004. Multiple sequence signals determine the distribution of glycosylphosphatidylinositol proteins between the plasma membrane and cell wall in Saccharomyces cerevisiae. Microbiology 150: 3105–3114. [DOI] [PubMed] [Google Scholar]
- Fujii T., Shimoi H., Iimura Y., 1999. Structure of the glucan-binding sugar chain of Tip1p, a cell wall protein of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1427: 133–144. [DOI] [PubMed] [Google Scholar]
- Fujita M., Kinoshita T., 2010. Structural remodeling of GPI anchors during biosynthesis and after attachment to proteins. FEBS Lett. 584: 1670–1677. [DOI] [PubMed] [Google Scholar]
- Fujita M., Yoko-o T., Okamoto M., Jigami Y., 2004. GPI7 involved in glycosylphosphatidylinositol biosynthesis is essential for yeast cell separation. J. Biol. Chem. 279: 51869–51879. [DOI] [PubMed] [Google Scholar]
- Fujita M., Yoko-o T., Jigami Y., 2006a Inositol deacylation by Bst1p is required for the quality control of glycosylphosphatidylinositol-anchored proteins. Mol. Biol. Cell 17: 834–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Umemura M., Yoko-o T., Jigami Y., 2006b PER1 is required for GPI-phospholipase A2 activity and involved in lipid remodeling of GPI-anchored proteins. Mol. Biol. Cell 17: 5253–5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M., Maeda Y., Ra M., Yamaguchi Y., Taguchi R., et al. , 2009. GPI glycan remodeling by PGAP5 regulates transport of GPI-anchored proteins from the ER to the Golgi. Cell 139: 352–365. [DOI] [PubMed] [Google Scholar]
- Gagnon-Arsenault I., Tremblay J., Bourbonnais Y., 2006. Fungal yapsins and cell wall: a unique family of aspartic peptidases for a distinctive cellular function. FEMS Yeast Res. 6: 966–978. [DOI] [PubMed] [Google Scholar]
- Gagnon-Arsenault I., Parise L., Tremblay J., Bourbonnais Y., 2008. Activation mechanism, functional role, and shedding of glycosylphosphatidylinositol-anchored Yps1p at the Saccharomyces cerevisiae cell surface. Mol. Microbiol. 69: 982–983. [DOI] [PubMed] [Google Scholar]
- Gai S. A., Wittrup K. D., 2007. Yeast surface display for protein engineering and characterization. Curr. Opin. Struct. Biol. 17: 467–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galperin M. Y., Jedrzejas M. J., 2001. Conserved core structure and active site residues in alkaline phosphatase superfamily enzymes. Proteins 45: 318–324. [DOI] [PubMed] [Google Scholar]
- Gao X. D., Kaigorodov V., Jigami Y., 1999. YND1, a homologue of GDA1, encodes membrane-bound apyrase required for Golgi N- and O-glycosylation in Saccharomyces cerevisiae. J. Biol. Chem. 274: 21450–21456. [DOI] [PubMed] [Google Scholar]
- Gao X. D., Nishikawa A., Dean N., 2004. Physical interactions between the Alg1, Alg2, and Alg11 mannosyltransferases of the endoplasmic reticulum. Glycobiology 14: 559–570. [DOI] [PubMed] [Google Scholar]
- Gao X. D., Tachikawa H., Sato T., Jigami Y., Dean N., 2005. Alg14 recruits Alg13 to the cytoplasmic face of the endoplasmic reticulum to form a novel bipartite UDP-N-acetylglucosamine transferase required for the second step of N-linked glycosylation. J. Biol. Chem. 280: 36254–36262. [DOI] [PubMed] [Google Scholar]
- Garcia-Rodriguez L. J., Trilla J. A., Castro C., Valdivieso M. H., Duran A., et al. , 2000. Characterization of the chitin biosynthesis process as a compensatory mechanism in the fks1 mutant of Saccharomyces cerevisiae. FEBS Lett. 478: 84–88. [DOI] [PubMed] [Google Scholar]
- Gauss R., Jarosch E., Sommer T., Hirsch C., 2006. A complex of Yos9p and the HRD ligase integrates endoplasmic reticulum quality control into the degradation machinery. Nat. Cell Biol. 8: 849–854. [DOI] [PubMed] [Google Scholar]
- Gauss R., Kanehara K., Carvalho P., Ng D. T., Aebi M., 2011. A complex of Pdi1p and the mannosidase Htm1p initiates clearance of unfolded glycoproteins from the endoplasmic reticulum. Mol. Cell 42: 782–793. [DOI] [PubMed] [Google Scholar]
- Gaynor E. C., Mondesert G., Grimme S. J., Reed S. I., Orlean P., et al. , 1999. MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol. Biol. Cell 10: 627–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentzsch M., Tanner W., 1996. The PMT gene family: protein O-glycosylation in Saccharomyces cerevisiae is vital. EMBO J. 15: 5752–5759. [PMC free article] [PubMed] [Google Scholar]
- Georgopapadakou N. H., Tkacz J. S., 1995. The fungal cell wall as a drug target. Trends Microbiol. 3: 98–104. [DOI] [PubMed] [Google Scholar]
- Gerke C., Kraft A., Süssmuth R., Schweitzer O., Götz F., 1998. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J. Biol. Chem. 273: 18586–18593. [DOI] [PubMed] [Google Scholar]
- Ghugtyal V., Vionnet C., Roubaty C., Conzelmann A., 2007. CWH43 is required for the introduction of ceramides into GPI anchors in Saccharomyces cerevisiae. Mol. Microbiol. 65: 1493–1502. [DOI] [PubMed] [Google Scholar]
- Girrbach V., Strahl S., 2003. Members of the evolutionarily conserved PMT family of protein O-mannosyltransferases form distinct protein complexes among themselves. J. Biol. Chem. 278: 12554–12562. [DOI] [PubMed] [Google Scholar]
- Girrbach V., Zeller T., Priesmeier M., Strahl-Bolsinger S., 2000. Structure-function analysis of the dolichyl phosphate-mannose: protein O-mannosyltransferase ScPmt1p. J. Biol. Chem. 275: 19288–19296. [DOI] [PubMed] [Google Scholar]
- Goldman R. C., Sullivan P. A., Zakula D., Capobianco J. O., 1995. Kinetics of β-1,3 glucan interaction at the donor and acceptor sites of the fungal glucosyltransferase encoded by the BGL2 gene. Eur. J. Biochem. 227: 372–378. [DOI] [PubMed] [Google Scholar]
- Gómez-Esquer F., Rodríguez-Peña J. M., Díaz G., Rodríguez E., Briza P., et al. , 2004. CRR1, a gene encoding a putative transglycosidase, is required for proper spore wall assembly in Saccharomyces cerevisiae. Microbiology 150: 3269–3280. [DOI] [PubMed] [Google Scholar]
- Gonzalez M., Lipke P. N., Ovalle R., 2009 GPI proteins in biogenesis and structure of yeast cell walls, pp. 321–356 in The Enzymes, Vol. XXVI, Glycosylphosphatidylinositol (GPI) Anchoring of Proteins, edited by A. K. Menon, T. Kinoshita, P. Orlean, and F. Tamanoi. Academic Press/Elsevier, New York. [Google Scholar]
- Gonzalez M., De Groot P. W. J., Klis F. M., Lipke P. N., 2010a Glycoconjugate structure and function in fungal cell walls, pp. 169–183 in Microbial Glycobiology. Structures, Relevance and Applications, edited by Moran A. P., Holst O., Brennan P. J., von Itzstein M. Academic Press/Elsevier, New York. [Google Scholar]
- Gonzalez M., Goddard N., Hicks C., Ovalle R., Rauceo J. M., et al. , 2010b A screen for deficiencies in GPI-anchorage of wall glycoproteins in yeast. Yeast 27: 583–596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens K., Willaert R., 2010. Flocculation protein structure and cell-cell adhesion mechanism in Saccharomyces cerevisiae. Biotechnol. Lett. 32: 1571–1585. [DOI] [PubMed] [Google Scholar]
- Goossens K. V., Stassen C., Stals I., Donohue D. S., Devreese B., et al. , 2011. The N-terminal domain of the Flo1 flocculation protein from Saccharomyces cerevisiae binds specifically to mannose carbohydrates. Eukaryot. Cell 10: 110–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabinska K., Palamarczyk G., 2002. Dolichol biosynthesis in the yeast Saccharomyces cerevisiae: an insight into the regulatory role of farnesyl diphosphate synthase. FEMS Yeast Res. 2: 259–265. [DOI] [PubMed] [Google Scholar]
- Grabinska K. A., Magnelli P., Robbins P. W., 2007. Prenylation of S. cerevisiae Chs4p affects chitin synthase III activity and chitin chain length. Eukaryot. Cell 6: 328–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimme S. J., Westfall B. A., Wiedman J. M., Taron C. H., Orlean P., 2001. The essential Smp3 protein is required for addition of the side-branching fourth mannose during assembly of yeast glycosylphosphatidylinositols. J. Biol. Chem. 276: 27731–27739. [DOI] [PubMed] [Google Scholar]
- Grimme S. J., Gao X.-D., Martin P. S., Tu K., Tcheperegine S. E., et al. , 2004. Deficiencies in the endoplasmic reticulum (ER)-membrane protein Gab1p perturb transfer of glycosylphosphatidylinositol to proteins and cause perinuclear ER-associated actin bar formation. Mol. Biol. Cell 15: 2758–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B., Styles C. A., Feng Q., Fink G., 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. USA 97: 12158–12163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haass F. A., Jonikas M., Walter P., Weissman J. S., Jan Y. N., et al. , 2007. Identification of yeast proteins necessary for cell-surface function of a potassium channel. Proc. Natl. Acad. Sci. USA 104: 18079–18084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen I., Ecker M., Lagorce A., Francois J. M., Sestak S., et al. , 2004. Sed1p and Srl1p are required to compensate for cell wall instability in Saccharomyces cerevisiae mutants defective in multiple GPI-anchored mannoproteins. Mol. Microbiol. 52: 1413–1425. [DOI] [PubMed] [Google Scholar]
- Hamada K., Fukuchi S., Arisawa M., Baba M., Kitada K., 1998a Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol. Gen. Genet. 258: 53–59. [DOI] [PubMed] [Google Scholar]
- Hamada K., Terashima H., Arisawa M., Kitada K., 1998b Amino acid sequence requirement for efficient incorporation of glycosylphosphatidylinositol-associated proteins into the cell wall of Saccharomyces cerevisiae. J. Biol. Chem. 273: 26946–26953. [DOI] [PubMed] [Google Scholar]
- Hamada K., Terashima H., Arisawa M., Yabuki N., Kitada K., 1999. Amino acid residues in the ω-minus region participate in cellular localization of yeast glycosylphosphatidylinositol-attached proteins. J. Bacteriol. 181: 3886–3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamburger D., Egerton M., Riezman H., 1995. Yeast Gaa1p is required for attachment of a completed GPI anchor onto proteins. J. Cell Biol. 129: 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampsey M., 1997. A review of phenotypes in Saccharomyces cerevisiae. Yeast 13: 1099–1133. [DOI] [PubMed] [Google Scholar]
- Hartland R. P., Vermeulen C. A., Klis F. M., Sietsma J. H., Wessels J. G., 1994. The linkage of (1–3)-β-glucan to chitin during cell wall assembly in Saccharomyces cerevisiae. Yeast 10: 1591–1599. [DOI] [PubMed] [Google Scholar]
- Haselbeck A., Tanner W., 1982. Dolichyl phosphate-mediated mannosyl transfer through liposomal membranes. Proc. Natl. Acad. Sci. USA 79: 1520–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto H., Yoda K., 1997. Novel membrane protein complexes for protein glycosylation in the yeast Golgi apparatus. Biochem. Biophys. Res. Commun. 241: 682–686. [DOI] [PubMed] [Google Scholar]
- Hashimoto H., Sakakibara A., Yamasaki M., Yoda K., 1997. Saccharomyces cerevisiae VIG9 encodes GDP-mannose pyrophosphorylase, which is essential for protein glycosylation. J. Biol. Chem. 272: 16308–16314. [DOI] [PubMed] [Google Scholar]
- Heinisch J. J., Dupres V., Wilk S., Jendretzki A., Dufrêne Y. F., 2010. Single-molecule atomic force microscopy reveals clustering of the yeast plasma-membrane sensor Wsc1. PLoS ONE 5: e11104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius J., Aebi M., 2002. Transmembrane movement of dolichol linked carbohydrates during N-glycoprotein biosynthesis in the endoplasmic reticulum. Semin. Cell Dev. Biol. 13: 171–178. [DOI] [PubMed] [Google Scholar]
- Helenius A., Aebi M., 2004. Roles of N-linked glycans in the endoplasmic reticulum. Annu. Rev. Biochem. 73: 1019–1049. [DOI] [PubMed] [Google Scholar]
- Helenius J., Ng D. T. W., Marolda C. L., Walter P., Valvano M. A., et al. , 2002. Translocation of lipid-linked oligosaccharides across the ER membrane requires Rft1 protein. Nature 415: 447–450. [DOI] [PubMed] [Google Scholar]
- Heller L., Orlean P., Adair W. L., 1992. Saccharomyces cerevisiae sec59 cells are deficient in dolichol kinase activity. Proc. Natl. Acad. Sci. USA 89: 7013–7016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henrissat B., Davies G. J., 1997. Structural and sequence-based classification of glycoside hydrolases. Curr. Opin. Struct. Biol. 7: 637–644. [DOI] [PubMed] [Google Scholar]
- Herrero A. B., Magnelli P., Mansour M. K., Levitz S. M., Bussey H., et al. , 2004. KRE5 gene null mutant strains of Candida albicans are avirulent and have altered cell wall composition and hypha formation properties. Eukaryot. Cell 3: 1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herscovics A., 1999. Processing glycosidases of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1426: 275–285. [DOI] [PubMed] [Google Scholar]
- Hese K., Otto C., Routier F. H., Lehle L., 2009. The yeast oligosaccharyltransferase complex can be replaced by STT3 from Leishmania major. Glycobiology 19: 160–171. [DOI] [PubMed] [Google Scholar]
- Hirayama H., Seino J., Kitajima T., Jigami Y., Suzuki T., 2010. Free oligosaccharides to monitor glycoprotein endoplasmic reticulum-associated degradation in Saccharomyces cerevisiae. J. Biol. Chem. 285: 12390–12404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann M., Boles E., Zimmermann F. K., 1994. Characterization of the essential yeast gene encoding N-acetylglucosamine-phosphate mutase. Eur. J. Biochem. 221: 741–747. [DOI] [PubMed] [Google Scholar]
- Hong Y., Maeda Y., Watanabe R., Inoue N., Ohishi K., et al. , 2000. Requirement of PIG-F and PIG-O for transferring phosphoethanolamine to the third mannose in glycosylphosphatidylinositol. J. Biol. Chem. 275: 20911–20919. [DOI] [PubMed] [Google Scholar]
- Hong Y., Ohishi K., Kang J. Y., Tanaka S., Inoue N., et al. , 2003. Human PIG-U and yeast Cdc91p are the fifth subunit of GPI transamidase that attaches GPI-anchors to proteins. Mol. Biol. Cell 14: 1780–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. S., Doherty H. K., Herskowitz I., 2005. The Smk1p MAP kinase negatively regulates Gsc2p, a 1,3-β-glucan synthase, during spore wall morphogenesis in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 102: 12431–12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchins K., Bussey H., 1983. Cell wall receptor for yeast killer toxin: involvement of (1→6)-β-D-glucan. J. Bacteriol. 154: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutzler J., Schmid M., Bernard T., Henrissat B., Strahl S., 2007. Membrane association is a determinant for substrate recognition by PMT4 protein O-mannosyltransferases. Proc. Natl. Acad. Sci. USA 104: 7827–7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai T., Watanabe T., Yui T., Sugiyama J., 2003. The directionality of chitin biosynthesis: a revisit. Biochem. J. 374: 755–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof I., Canivenc-Gansel E., Meyer U., Conzelmann A., 2000. Phosphatidylethanolamine is the donor of the phosphorylethanolamine linked to the α1,4-linked mannose of yeast GPI structures. Glycobiology 10: 1271–1275. [DOI] [PubMed] [Google Scholar]
- Inoue S. B., Takewaki N., Takasuka T., Mio T., Adachi M., et al. , 1995. Characterization and gene cloning of 1,3-β-D-glucan synthase from Saccharomyces cerevisiae. Eur. J. Biochem. 231: 845–854. [DOI] [PubMed] [Google Scholar]
- Inoue S. B., Qadota H., Arisawa M., Watanabe T., Ohya Y., 1999. Prenylation of Rho1p is required for activation of yeast 1,3-β-glucan synthase. J. Biol. Chem. 274: 38119–38124. [DOI] [PubMed] [Google Scholar]
- Insenser M. R., Hernáez M. L., Nombela C., Molina M., Molero G., et al. , 2010. Gel and gel-free proteomics to identify Saccharomyces cerevisiae cell surface proteins. J. Proteomics 73: 1183–1195. [DOI] [PubMed] [Google Scholar]
- Ishihara S., Hirata A., Nogami S., Beauvais A., Latge J. P., et al. , 2007. Homologous subunits of 1,3-β-glucan synthase are important for spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 6: 143–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh Y., Rice J. D., Goller C., Pannuri A., Taylor J., et al. , 2008. Roles of pgaABCD genes in synthesis, modification, and export of the Escherichia coli biofilm adhesin poly-β-1,6-N-acetyl-D-glucosamine. J. Bacteriol. 190: 3670–3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob C. A., Burda P., Roth J., Aebi M., 1998. Degradation of misfolded endoplasmic reticulum glycoproteins in Saccharomyces cerevisiae is determined by a specific oligosaccharide structure. J. Cell Biol. 142: 1223–1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janik A., Sosnowska M., Kruszewska J., Krotkiewski H., Lehle L., et al. , 2003. Overexpression of GDP-mannose pyrophosphorylase in Saccharomyces cerevisiae corrects defects in dolichol-linked saccharide formation and protein glycosylation. Biochim. Biophys. Acta 1621: 22–30. [DOI] [PubMed] [Google Scholar]
- Jiang B., Ram A. F., Sheraton J., Klis F. M., Bussey H., 1995. Regulation of cell wall β-glucan assembly: PTC1 negatively affects PBS2 action in a pathway that includes modulation of EXG1 transcription. Mol. Gen. Genet. 248: 260–269. [DOI] [PubMed] [Google Scholar]
- Jiang B., Sheraton J., Ram A. F., Dijkgraaf G. J. P., Klis F. M., et al. , 1996. CWH41 encodes a novel endoplasmic reticulum membrane N-glycoprotein involved in β1,6-glucan assembly. J. Bacteriol. 178: 1162–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jigami Y., 2008. Yeast glycobiology and its application. Biosci. Biotechnol. Biochem. 72: 637–648. [DOI] [PubMed] [Google Scholar]
- Jigami Y., Odani T., 1999. Mannosylphosphate transfer to yeast mannan. Biochim. Biophys. Acta 1426: 335–345. [DOI] [PubMed] [Google Scholar]
- Jimenez C., Sacristan C., Roncero M. I., Roncero C., 2010. Amino acid divergence between the CHS domain contributes to the different intracellular behaviour of family II fungal chitin synthases in Saccharomyces cerevisiae. Fungal Genet. Biol. 47: 1034–1043. [DOI] [PubMed] [Google Scholar]
- Jue C. K., Lipke P. N., 2002. Role of Fig2p in agglutination in Saccharomyces cerevisiae. Eukaryot. Cell 1: 843–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung P., Tanner W., 1973. Identification of the lipid intermediate in yeast mannan biosynthesis. Eur. J. Biochem. 37: 1–6. [DOI] [PubMed] [Google Scholar]
- Jung U. S., Levin D. E., 1999. Genome-wide analysis of gene expression regulated by the yeast cell wall integrity signalling pathway. Mol. Microbiol. 34: 1049–1057. [DOI] [PubMed] [Google Scholar]
- Jungmann J., Munro S., 1998. Multi-protein complexes in the cis Golgi of Saccharomyces cerevisiae with α-1,6-mannosyltransferase activity. EMBO J. 17: 423–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungmann J., Rayner J. C., Munro S., 1999. The Saccharomyces cerevisiae protein Mnn10p/Bed1p is a subunit of a Golgi mannosyltransferase complex. J. Biol. Chem. 274: 6579–6585. [DOI] [PubMed] [Google Scholar]
- Kainuma M., Chiba Y., Takeuchi M., Jigami Y., 2001. Overexpression of HUT1 gene stimulates in vivo galactosylation by enhancing UDP-galactose transport activity in Saccharomyces cerevisiae. Yeast 18: 533–541. [DOI] [PubMed] [Google Scholar]
- Kajiwara K., Watanabe R., Pichler H., Ihara K., Murakami S., et al. , 2008. Yeast ARV1 is required for efficient delivery of an early GPI intermediate to the first mannosyltransferase during GPI assembly and controls lipid flow from the endoplasmic reticulum. Mol. Biol. Cell 19: 2069–2082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalebina T. S., Farkas V., Laurinavichiute D. K., Gorlovoy P. M., Fominov G. V., et al. , 2003. Deletion of BGL2 results in an increased chitin level in the cell wall of Saccharomyces cerevisiae. Antonie van Leeuwenhoek 84: 179–184. [DOI] [PubMed] [Google Scholar]
- Kämpf M., Absmanner B., Schwarz M., Lehle L., 2009. Biochemical characterization and membrane topology of Alg2 from Saccharomyces cerevisiae as a bifunctional α1,3- and 1,6-mannosyltransferase involved in lipid-linked oligosaccharide biosynthesis. J. Biol. Chem. 284: 11900–11912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamst E., Bakkers J., Quaedvlieg N. E. M., Pilling J., Kijne J. W., et al. , 1999. Chitin oligosaccharide synthesis by Rhizobia and Zebrafish embryos starts by glycosyl transfer to O4 of the reducing-terminal residue. Biochemistry 38: 4045–4052. [DOI] [PubMed] [Google Scholar]
- Kang J. Y., Hong Y., Ashida H., Shishioh N., Murakami Y., et al. , 2005. PIG-V involved in transferring the second mannose in glycosylphosphatidylinositol. J. Biol. Chem. 280: 9489–9497. [DOI] [PubMed] [Google Scholar]
- Kang M. S., Cabib E., 1986. Regulation of fungal cell wall growth: a guanine nucleotide-binding, proteinaceous component required for activity of (1→3)-β-D-glucan synthase. Proc. Natl. Acad. Sci. USA 83: 5808–5812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang M. S., Elango N., Mattia E., Au-Young J., Robbins P. W., et al. , 1984. Isolation of chitin synthetase from Saccharomyces cerevisiae. Purification of an enzyme by entrapment in the reaction product. J. Biol. Chem. 259: 14966–14972. [PubMed] [Google Scholar]
- Kapteyn J. C., Montijn R. C., Vink E., de la Cruz J., Llobell A., et al. , 1996. Retention of Saccharomyces cerevisiae cell wall proteins through a phosphodiester-linked β-1,3-/β-1,6-glucan heteropolymer. Glycobiology 6: 337–345. [DOI] [PubMed] [Google Scholar]
- Kapteyn J. C., Ram A. F., Groos E. M., Kollar R., Montijn R. C., et al. , 1997. Altered extent of cross-linking of β1,6-glucosylated mannoproteins to chitin in Saccharomyces cerevisiae mutants with reduced cell wall β1,3-glucan content. J. Bacteriol. 179: 6279–6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapteyn J. C., Van Den Ende H., Klis F. M., 1999a The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta 1426: 373–383. [DOI] [PubMed] [Google Scholar]
- Kapteyn J. C., Van Egmond P., Sievi E., Van Den Ende H., Makarow M., et al. , 1999b The contribution of the O-glycosylated protein Pir2p/Hsp150 to the construction of the yeast cell wall in wild-type cells and β1,6-glucan-deficient mutants. Mol. Microbiol. 31: 1835–1844. [DOI] [PubMed] [Google Scholar]
- Kapteyn J. C., ter Riet B., Vink E., Blad S., De Nobel H., et al. , 2001. Low external pH induces HOG1-dependent changes in the organization of the Saccharomyces cerevisiae cell wall. Mol. Microbiol. 39: 469–479. [DOI] [PubMed] [Google Scholar]
- Karaoglu D., Kelleher D. J., Gilmore R., 1997. The highly conserved Stt3 protein is a subunit of the yeast oligosaccharyltransferase and forms a subcomplex with Ost3p and Ost4p. J. Biol. Chem. 272: 32513–32520. [DOI] [PubMed] [Google Scholar]
- Karaoglu D., Kelleher D. J., Gilmore R., 2001. Allosteric regulation provides a molecular mechanism for preferential utilization of the fully assembled dolichol-linked oligosaccharide by the yeast oligosaccharyltransferase. Biochemistry 40: 12193–12206. [DOI] [PubMed] [Google Scholar]
- Kelleher D. J., Gilmore R., 2006. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16: 47R–62R. [DOI] [PubMed] [Google Scholar]
- Kelleher D. J., Banerjee S., Cura A. J., Samuelson J., Gilmore R., 2007. Dolichol-linked oligosaccharide selection by the oligosaccharyltransferase in protist and fungal organisms. J. Cell Biol. 177: 29–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Park H., Montalvo L., Lennarz W. J., 2000. Studies on the role of the hydrophobic domain of Ost4p in interactions with other subunits of yeast oligosaccharyl transferase. Proc. Natl. Acad. Sci. USA 97: 1516–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Yan Q., Von Heijne G., Caputo G. A., Lennarz W. J., 2003. Determination of the membrane topology of Ost4p and its subunit interactions in the oligosaccharyltransferase complex in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 100: 7460–7464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W., Spear E. D., Ng D. T., 2005. Yos9p detects and targets misfolded glycoproteins for ER-associated degradation. Mol. Cell 19: 753–764. [DOI] [PubMed] [Google Scholar]
- Kim Y. U., Ashida H., Mori K., Maeda Y., Hong Y., et al. , 2007. Both mammalian PIG-M and PIG-X are required for growth of GPI14-disrupted yeast. J. Biochem. 142: 123–129. [DOI] [PubMed] [Google Scholar]
- Kitagaki H., Wu H., Shimoi H., Ito K., 2002. Two homologous genes, DCW1 (YKL046c) and DFG5, are essential for cell growth and encode glycosylphosphatidylinositol (GPI)-anchored membrane proteins required for cell wall biogenesis in Saccharomyces cerevisiae. Mol. Microbiol. 46: 1011–1022. [DOI] [PubMed] [Google Scholar]
- Kitamura A., Someya K., Hata M., Nakajima R., Takemura M., 2009. Discovery of a small-molecule inhibitor of β-1,6-glucan synthesis. Antimicrob. Agents Chemother. 53: 670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klebl F., Tanner W., 1989. Molecular cloning of a cell wall exo-β-1,3-glucanase from Saccharomyces cerevisiae. J. Bacteriol. 171: 6259–6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klis F. M., Mol P., Hellingwerf K., Brul S., 2002. Dynamics of cell wall structure in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 26: 239–256. [DOI] [PubMed] [Google Scholar]
- Klis F. M., Boorsma A., De Groot P. W., 2006. Cell wall construction in Saccharomyces cerevisiae. Yeast 23: 185–202. [DOI] [PubMed] [Google Scholar]
- Klis F. M., de Jong M., Brul S., de Groot P. W., 2007. Extraction of cell surface-associated proteins from living yeast cells. Yeast 24: 253–258. [DOI] [PubMed] [Google Scholar]
- Klis F. M., Brul S., De Groot P. W., 2010. Covalently linked wall proteins in ascomycetous fungi. Yeast 27: 489–493. [DOI] [PubMed] [Google Scholar]
- Knauer R., Lehle L., 1999a The oligosaccharyltransferase complex from yeast. Biochim. Biophys. Acta 1426: 259–273. [DOI] [PubMed] [Google Scholar]
- Knauer R., Lehle L., 1999b The oligosaccharyltransferase complex from Saccharomyces cerevisiae. Isolation of the OST6 gene, its synthetic interaction with OST3, and analysis of the native complex. J. Biol. Chem. 274: 17249–17256. [DOI] [PubMed] [Google Scholar]
- Kollár R., Petráková E., Ashwell G., Robbins P. W., Cabib E., 1995. Architecture of the yeast cell wall. The linkage between chitin and beta(1→3)-glucan. J. Biol. Chem. 270: 1170–1178. [DOI] [PubMed] [Google Scholar]
- Kollár R., Reinhold B. B., Petráková E., Yeh H. J., Ashwell G., et al. , 1997. Architecture of the yeast cell wall. β(1→6)-glucan interconnects mannoprotein, β(1→3)-glucan, and chitin. J. Biol. Chem. 272: 17762–17775. [DOI] [PubMed] [Google Scholar]
- Komano H., Rockwell N. C., Wang G. T., Krafft G. A., Fuller R. S., 1999. Purification and characterization of the yeast glycosylphosphatidylinositol-anchored, mono-basic specific aspartyl protease yapsin 2 (Mkc7). J. Biol. Chem. 274: 24431–24437. [DOI] [PubMed] [Google Scholar]
- Kopecká M., Phaff H. J., Fleet G. H., 1974. Demonstration of a fibrillar component in the cell wall of the yeast Saccharomyces cerevisiae and its chemical nature. J. Cell Biol. 62: 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kostova Z., Rancour D. M., Menon A. K., Orlean P., 2000. Photoaffinity labelling with P3-(4-azidoanilido)uridine 5′-triphosphate identifies Gpi3p as the UDP-GlcNAc-binding subunit of the enzyme that catalyses formation of GlcNAc-phosphatidylinositol, the first glycolipid intermediate in glycosylphosphatidylinositol synthesis. Biochem. J. 350: 815–822. [PMC free article] [PubMed] [Google Scholar]
- Kovacech B., Nasmyth K., Schuster T., 1996. EGT2 gene transcription is induced predominantly by Swi5 in early G1. Mol. Cell. Biol. 16: 3264–3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowalski L. R., Kondo K., Inouye M., 1995. Cold-shock induction of a family of TIP1-related proteins associated with the membrane in Saccharomyces cerevisiae. Mol. Microbiol. 15: 341–353. [DOI] [PubMed] [Google Scholar]
- Kozubowski L., Panek H., Rosenthal A., Bloecher A., DeMarini D. J., et al. , 2003. A Bni4-Glc7 phosphatase complex that recruits chitin synthase to the site of bud emergence. Mol. Biol. Cell 14: 26–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kreger D. R., Kopecká M., 1976. On the nature and formation of the fibrillar nets produced by protoplasts of Saccharomyces cerevisiae in liquid media: an electronmicroscopic, x-ray diffraction and chemical study. J. Gen. Microbiol. 92: 207–220. [DOI] [PubMed] [Google Scholar]
- Krysan D. J., Ting E. L., Abeijon C., Kroos L., Fuller R. S., 2005. Yapsins are a family of aspartyl proteases required for cell wall integrity in Saccharomyces cerevisiae. Eukaryot. Cell 4: 1364–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni R. D., Kelkar H. S., Dean R. A., 2003. An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28: 118–121. [DOI] [PubMed] [Google Scholar]
- Kuranda M. J.Robbins P. W., 1991. Chitinase is required for cell separation during growth of Saccharomyces cerevisiae. J. Biol. Chem. 266: 19758–19767. [PubMed] [Google Scholar]
- Kurischko C., Weiss G., Ottey M., Luca F. C., 2005. A role for the Saccharomyces cerevisiae regulation of Ace2 and polarized morphogenesis signaling network in cell integrity. Genetics 171: 443–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurita T., Noda Y., Takagi T., Osumi M., Yoda K., 2011. Kre6 protein essential for yeast cell wall β-1,6-glucan synthesis accumulates at sites of polarized growth. J. Biol. Chem. 286: 7429–7438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagorce A., Le Berre-Anton V., Aguilar-Uscanga B., Martin-Yken H., Dagkessamanskaia A., et al. , 2002. Involvement of GFA1, which encodes glutamine-fructose-6-phosphate amidotransferase, in the activation of the chitin synthesis pathway in response to cell-wall defects in Saccharomyces cerevisiae. Eur. J. Biochem. 269: 1697–1707. [DOI] [PubMed] [Google Scholar]
- Lagorce A., Hauser N. C., Labourdette D., Rodriguez C., Martin-Yken H., et al. , 2003. Genome-wide analysis of the response to cell wall mutations in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 278: 20345–20357. [DOI] [PubMed] [Google Scholar]
- Lam K. K., Davey M., Sun B., Roth A. F., Davis N. G., et al. , 2006. Palmitoylation by the DHHC protein Pfa4 regulates the ER exit of Chs3. J. Cell Biol. 174: 19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambrechts M. G., Bauer F. F., Marmur J., Pretorius I. S., 1996. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. USA 93: 8419–8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin A., Imperiali B., 2011. The expanding horizons of asparagine-linked glycosylation. Biochemistry 50: 4411–4426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larriba G., Andaluz E., Cueva R., Basco R. D., 1995. Molecular biology of yeast exoglucanases. FEMS Microbiol. Lett. 125: 121–126. [DOI] [PubMed] [Google Scholar]
- Latgé J.-P., 2007. The cell wall: a carbohydrate armour for the fungal cell. Mol. Microbiol. 66: 279–290. [DOI] [PubMed] [Google Scholar]
- Lehle L., Strahl S., Tanner W., 2006. Protein glycosylation, conserved from yeast to man: a model organism helps elucidate congenital human diseases. Angew. Chem. Int. Ed. Engl. 45: 6802–6818. [DOI] [PubMed] [Google Scholar]
- Leidich S. D., Orlean P., 1996. Gpi1, a Saccharomyces cerevisiae protein that participates in the first step in glycosylphosphatidylinositol anchor synthesis. J. Biol. Chem. 271: 27829–27837. [DOI] [PubMed] [Google Scholar]
- Leidich S. D., Kostova Z., Latek R. R., Costello L. C., Drapp D. A., et al. , 1995. Temperature-sensitive yeast GPI anchoring mutants gpi2 and gpi3 are defective in the synthesis of N-acetylglucosaminyl phosphatidylinositol. Cloning of the GPI2 gene. J. Biol. Chem. 270: 13029–13035. [DOI] [PubMed] [Google Scholar]
- Lennarz W. J., 2007. Studies on oligosaccharyl transferase in yeast. Acta Biochim. Pol. 54: 673–677. [PubMed] [Google Scholar]
- Lesage G., Bussey H., 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70: 317–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Sdicu A. M., Menard P., Shapiro J., Hussein S., et al. , 2004. Analysis of β-1,3 glucan assembly in Saccharomyces cerevisiae using a synthetic interaction network and altered sensitivity to caspofungin. Genetics 167: 35–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage G., Shapiro J., Specht C. A., Sdicu A. M., Menard P., et al. , 2005. An interactional network of genes involved in chitin synthesis in Saccharomyces cerevisiae. BMC Genet. 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin D. E., 2011. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: the cell wall integrity signaling pathway. Genetics 189: 1145–1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levinson J. N., Shahinian S., Sdicu A. M., Tessier D. C., Bussey H., 2002. Functional, comparative and cell biological analysis of Saccharomyces cerevisiae Kre5p. Yeast 19: 1243–1259. [DOI] [PubMed] [Google Scholar]
- Lewis M. S., Ballou C. E., 1991. Separation and characterization of two α1,2-mannosyltransferase activities from Saccharomyces cerevisiae. J. Biol. Chem. 266: 8255–8261. [PubMed] [Google Scholar]
- Li G., Yan Q., Oen H. O., Lennarz W. J., 2003. A specific segment of the transmembrane domain of Wbp1p is essential for its incorporation into the oligosaccharyl transferase complex. Biochemistry 42: 11032–11039. [DOI] [PubMed] [Google Scholar]
- Li H., Pagé N., Bussey H., 2002. Actin patch assembly proteins Las17p and Sla1p restrict cell wall growth to daughter cells and interact with cis-Golgi protein Kre6p. Yeast 19: 1097–1112. [DOI] [PubMed] [Google Scholar]
- Lo W. S., Dranginis A. M., 1996. FLO11, a yeast gene related to the STA genes, encodes a novel cell surface flocculin. J. Bacteriol. 178: 7144–7151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo W. S., Dranginis A. M., 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by S. cerevisiae. Mol. Biol. Cell 9: 161–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel M., Strahl S., 2009. Protein O-mannosylation: conserved from bacteria to humans. Glycobiology 19: 816–828. [DOI] [PubMed] [Google Scholar]
- Lommel M., Bagnat M., Strahl S., 2004. Aberrant processing of the WSC family and Mid2p cell surface sensors results in cell death of Saccharomyces cerevisiae O-mannosylation mutants. Mol. Cell. Biol. 24: 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lommel M., Schott A., Jank T., Hofmann V., Strahl S., 2011. A conserved acidic motif is crucial for enzymatic activity of protein O-mannosyltransferases. J. Biol. Chem. 286: 39768–39775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu C. F., Montijn R. C., Brown J. L., Klis F., Kurjan J., et al. , 1995. Glycosyl phosphatidylinositol-dependent cross-linking of α-agglutinin and β1,6-glucan in the Saccharomyces cerevisiae cell wall. J. Cell Biol. 128: 333–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M., Sdicu A. M., Camirand A., Bussey H., 1996. Functional characterization of the YUR1, KTR1, and KTR2 genes as members of the yeast KRE2/MNT1 mannosyltransferase gene family. J. Biol. Chem. 271: 11001–11008. [DOI] [PubMed] [Google Scholar]
- Lussier M., Sdicu A. M., Bussereau F., Jacquet M., Bussey H., 1997a The Ktr1p, Ktr3p, and Kre2p/Mnt1p mannosyltransferases participate in the elaboration of yeast O- and N-linked carbohydrate chains. J. Biol. Chem. 272: 15527–15531. [DOI] [PubMed] [Google Scholar]
- Lussier M., White A. M., Sheraton J., di Paolo T., Treadwell J., et al. , 1997b Large scale identification of genes involved in cell surface biosynthesis and architecture in Saccharomyces cerevisiae. Genetics 147: 435–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lussier M., Sdicu A. M., Bussey H., 1999. The KTR and MNN1 mannosyltransferase families of Saccharomyces cerevisiae. Biochim. Biophys. Acta 1426: 323–334. [DOI] [PubMed] [Google Scholar]
- Machi K., Azuma M., Igarashi K., Matsumoto T., Fukuda H., et al. , 2004. Rot1p of Saccharomyces cerevisiae is a putative membrane protein required for normal levels of the cell wall 1,6-β-glucan. Microbiology 150: 3163–3173. [DOI] [PubMed] [Google Scholar]
- Maeda Y., Watanabe R., Harris C. L., Hong Y., Ohishi K., et al. , 2001. PIG-M transfers the first mannose to glycosylphosphatidylinositol on the lumenal side of the ER. EMBO J. 20: 250–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magnelli P., Cipollo J. F., Abeijon C., 2002. A refined method for the determination of Saccharomyces cerevisiae cell wall composition and β-1,6-glucan fine structure. Anal. Biochem. 301: 136–150. [DOI] [PubMed] [Google Scholar]
- Manners D. J., Masson A. J., Patterson J. A., Bjorndal H., Lindberg B., 1973. The structure of a β-1,6-glucan from yeast cell walls. Biochem. J. 135: 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Rucobo F. W., Eckhardt-Strelau L., Terwisscha van Scheltinga A. C., 2009. Yeast chitin synthase 2 activity is modulated by proteolysis and phosphorylation. Biochem. J. 417: 547–554. [DOI] [PubMed] [Google Scholar]
- Martin-Yken H., Dagkessamanskaia A., De Groot P., Ram A., Klis F., et al. , 2001. Saccharomyces cerevisiae YCRO17c/CWH43 encodes a putative sensor/transporter protein upstream of the BCK2 branch of the PKC1-dependent cell wall integrity pathway. Yeast 18: 827–840. [DOI] [PubMed] [Google Scholar]
- Mazan M., Mazánová K., Farkas V., 2008. Phenotype analysis of Saccharomyces cerevisiae mutants with deletions in Pir cell wall glycoproteins. Antonie van Leeuwenhoek 94: 335–342. [DOI] [PubMed] [Google Scholar]
- Mazan M., Ragni E., Popolo L., Farkas V., 2011. Catalytic properties of the Gas family β1,3-glucanosyltransferases active in fungal cell wall biogenesis as determined by a novel fluorescent assay. Biochem. J. 438: 275–282. [DOI] [PubMed] [Google Scholar]
- Mazur P., Baginsky W., 1996. In vitro activity of 1,3-β-D-glucan synthase requires the GTP-binding protein Rho1. J. Biol. Chem. 271: 14604–14609. [DOI] [PubMed] [Google Scholar]
- Mazur P., Morin N., Baginsky W., el Sherbeini M., Clemas J. A., et al. , 1995. Differential expression and function of two homologous subunits of yeast 1,3-β-D-glucan synthase. Mol. Cell. Biol. 15: 5671–5681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurray M. A., Stefan C. J., Wemmer M., Odorizzi G., Emr S. D., et al. , 2011. Genetic interactions with mutations affecting septin assembly reveal ESCRT functions in budding yeast cytokinesis. Biol. Chem. 392: 699–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meaden P., Hill K., Wagner J., Slipetz D., Sommer S. S., et al. , 1990. The yeast KRE5 gene encodes a probable endoplasmic reticulum protein required for (1–6)-β-D-glucan synthesis and normal cell growth. Mol. Cell. Biol. 10: 3013–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner D., Odman-Naresh J., Vogelpohl I., Merzendorfer H., 2010. A novel role of the yeast CaaX protease Ste24 in chitin synthesis. Mol. Biol. Cell 21: 2425–2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meitinger F., Petrova B., Lombardi I. M., Bertazzi D. T., Hub B., et al. , 2010. Targeted localization of Inn1, Cyk3 and Chs2 by the mitotic-exit network regulates cytokinesis in budding yeast. J. Cell Sci. 123: 1851–1861. [DOI] [PubMed] [Google Scholar]
- Menon A. K., Stevens V. L., 1992. Phosphatidylethanolamine is the donor of the ethanolamine residue linking a glycosylphosphatidylinositol anchor to protein. J. Biol. Chem. 267: 15277–15280. [PubMed] [Google Scholar]
- Merzendorfer H., 2011. The cellular basis of chitin synthesis in fungi and insects: common principles and differences. Eur. J. Cell Biol. 90: 759–769. [DOI] [PubMed] [Google Scholar]
- Meyer U., Benghezal M., Imhof I., Conzelmann A., 2000. Active site determination of Gpi8p, a caspase-related enzyme required for glycosylphosphatidylinositol anchor addition to proteins. Biochemistry 39: 3461–3471. [DOI] [PubMed] [Google Scholar]
- Milewski S., Gabriel I., Olchowy J., 2006. Enzymes of UDP-GlcNAc biosynthesis in yeast. Yeast 23: 1–14. [DOI] [PubMed] [Google Scholar]
- Miller K. A., DiDone L., Krysan D. J., 2010. Extracellular secretion of overexpressed glycosylphosphatidylinositol-linked cell wall protein Utr2/Crh2p as a novel protein quality control mechanism in Saccharomyces cerevisiae. Eukaryot. Cell 11: 1669–1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mio T., Yabe T., Arisawa M., Yamada-Okabe H., 1998. The eukaryotic UDP-N-acetylglucosamine pyrophosphorylases. Gene cloning, protein expression, and catalytic mechanism. J. Biol. Chem. 273: 14392–14397. [DOI] [PubMed] [Google Scholar]
- Mio T., Yamada-Okabe T., Arisawa M., Yamada-Okabe H., 1999. Saccharomyces cerevisiae GNA1, an essential gene encoding a novel acetyltransferase involved in UDP-N-acetylglucosamine synthesis. J. Biol. Chem. 274: 424–429. [DOI] [PubMed] [Google Scholar]
- Mishra C., Semino C. E., McCreath K. J., de la Vega H., Jones B. J., et al. , 1997. Cloning and expression of two chitin deacetylase genes of Saccharomyces cerevisiae. Yeast 13: 327–336. [DOI] [PubMed] [Google Scholar]
- Monteiro M. A., Slavic D., St. Michael F. J., Brisson R., MacInnes J. I., et al. , 2000. The first description of a (1→6)-β-D-glucan in prokaryotes: (1→6)-β-D-glucan is a common component of Actinobacillus suis and is the basis for a serotyping system. Carbohydr. Res. 329: 121–130. [DOI] [PubMed] [Google Scholar]
- Montijn R. C., Vink E., Müller W. H., Verkleij A. J., Van Den Ende H., et al. , 1999. Localization of synthesis of β1,6-glucan in Saccharomyces cerevisiae. J. Bacteriol. 181: 7414–7420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro A. G., Ramirez S. C., Quiroga R., Taubas J. V., 2011. Specificity of transmembrane protein palmitoylation in yeast. PLoS ONE 6: e16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosch H. U., Fink G. R., 1997. Dissection of filamentous growth by transposon mutagenesis in Saccharomyces cerevisiae. Genetics 145: 671–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouassite M., Camougrand N., Schwob E., Demaison G., Laclau M., et al. , 2000. The ‘SUN’ family: yeast SUN4/SCW3 is involved in cell septation. Yeast 16: 905–919. [DOI] [PubMed] [Google Scholar]
- Moukadiri I., Zueco J., 2001. Evidence for the attachment of Hsp150/Pir2 to the cell wall of Saccharomyces cerevisiae through disulfide bridges. FEMS Yeast Res. 1: 241–245. [DOI] [PubMed] [Google Scholar]
- Moukadiri I., Armero J., Abad A., Sentandreu R., Zueco J., 1997. Identification of a mannoprotein present in the inner layer of the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 179: 2154–2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moukadiri I., Jaafar L., Zueco J., 1999. Identification of two mannoproteins released from cell walls of a Saccharomyces cerevisiae mnn1 mnn9 double mutant by reducing agents. J. Bacteriol. 181: 4741–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouyna I., Fontaine T., Vai M., Monod M., Fonzi W. A., et al. , 2000. Glycosylphosphatidylinositol-anchored glucanosyltransferases play an active role in the biosynthesis of the fungal cell wall. J. Biol. Chem. 275: 14882–14889. [DOI] [PubMed] [Google Scholar]
- Mrša V., Tanner W., 1999. Role of NaOH-extractable cell wall proteins Ccw5p, Ccw6p, Ccw7p and Ccw8p (members of the Pir protein family) in stability of the Saccharomyces cerevisiae cell wall. Yeast 15: 813–820. [DOI] [PubMed] [Google Scholar]
- Mrša V., Klebl F., Tanner W., 1993. Purification and characterization of the Saccharomyces cerevisiae BGL2 gene product, a cell wall endo-β-1,3-glucanase. J. Bacteriol. 175: 2102–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mrša V., Seidl T., Gentzsch M., Tanner W., 1997. Specific labelling of cell wall proteins by biotinylation. Identification of four covalently linked O-mannosylated proteins of Saccharomyces cerevisiae. Yeast 13: 1145–1154. [DOI] [PubMed] [Google Scholar]
- Mrša V., Ecker M., Strahl-Bolsinger S., Nimtz M., Lehle L., et al. , 1999. Deletion of new covalently linked cell wall glycoproteins alters the electrophoretic mobility of phosphorylated wall components of Saccharomyces cerevisiae. J. Bacteriol. 181: 3076–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami Y., Siripanyaphinyo U., Hong Y., Tashima Y., Maeda Y., et al. , 2005. The initial enzyme for glycosylphosphatidylinositol biosynthesis requires PIG-Y, a seventh component. Mol. Biol. Cell 16: 5236–5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagahashi S., Sudoh M., Ono N., Sawada R., Yamaguchi E., et al. , 1995. Characterization of chitin synthase 2 of Saccharomyces cerevisiae. Implication of two highly conserved domains as possible catalytic sites. J. Biol. Chem. 270: 13961–13967. [DOI] [PubMed] [Google Scholar]
- Nakamata K., Kurita T., Bhuiyan M. S., Sato K., Noda Y., et al. , 2007. KEG1/YFR042w encodes a novel Kre6-binding endoplasmic reticulum membrane protein responsible for β-1,6-glucan synthesis in Saccharomyces cerevisiae. J. Biol. Chem. 282: 34315–34324. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Nakanishi-Shindo Y., Tanaka A., Haga-Toda Y., Jigami Y., 1997. Substrate specificity of α-1,6-mannosyltransferase that initiates N-linked mannose outer chain elongation in Saccharomyces cerevisiae. FEBS Lett. 412: 547–550. [DOI] [PubMed] [Google Scholar]
- Nakayama K., Feng Y., Tanaka A., Jigami Y., 1998. The involvement of mnn4 and mnn6 mutations in mannosylphosphorylation of O-linked oligosaccharide in yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1425: 255–262. [DOI] [PubMed] [Google Scholar]
- Nasab F. P., Schulz B. L., Gamarro F., Parodi A. J., Aebi M., 2008. All in one: Leishmania major STT3 proteins substitute for the whole oligosaccharyltransferase complex in Saccharomyces cerevisiae. Mol. Biol. Cell 19: 3758–3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neiman A. M., 2011. Sporulation in the budding yeast Saccharomyces cerevisiae. Genetics 189: 737–765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman H. A., Romeo M. J., Lewis S. E., Yan B. C., Orlean P., et al. , 2005. Gpi19, the Saccharomyces cerevisiae homologue of mammalian PIG-P, is a subunit of the initial enzyme for glycosylphosphatidylinositol anchor biosynthesis. Eukaryot. Cell 4: 1801–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson I., Kelleher D. J., Miao Y., Shao Y., Kreibich G., et al. , 2003. Photocross-linking of nascent chains to the STT3 subunit of the oligosaccharyltransferase complex. J. Cell Biol. 161: 715–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R., Schreiter J. H., Onishi M., Vallen E. A., Hanna J., et al. , 2009. Role of Inn1 and its interactions with Hof1 and Cyk3 in promoting cleavage furrow and septum formation in S. cerevisiae. J. Cell Biol. 185: 995–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noffz C., Keppler-Ross S., Dean N., 2009. Hetero-oligomeric interactions between early glycosyltransferases of the dolichol cycle. Glycobiology 19: 472–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nombela C., Gil C., Chaffin W. L., 2006. Non-conventional protein secretion in yeast. Trends Microbiol. 14: 15–21. [DOI] [PubMed] [Google Scholar]
- Nuoffer C., Jeno P., Conzelmann A., Riezman H., 1991. Determinants for glycophospholipid anchoring of the Saccharomyces cerevisiae GAS1 protein to the plasma membrane. Mol. Cell. Biol. 11: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuoffer C., Horvath A., Riezman H., 1993. Analysis of the sequence requirements for glycosylphosphatidylinositol anchoring of Saccharomyces cerevisiae Gas1 protein. J. Biol. Chem. 268: 10558–10563. [PubMed] [Google Scholar]
- Odani T., Shimma Y., Tanaka A., Jigami Y., 1996. Cloning and analysis of the MNN4 gene required for phosphorylation of N-linked oligosaccharides in Saccharomyces cerevisiae. Glycobiology 6: 805–810. [DOI] [PubMed] [Google Scholar]
- Odani T., Shimma Y., Wang X. H., Jigami Y., 1997. Mannosylphosphate transfer to cell wall mannan is regulated by the transcriptional level of the MNN4 gene in Saccharomyces cerevisiae. FEBS Lett. 420: 186–190. [DOI] [PubMed] [Google Scholar]
- Oh Y., Chang K.-J., Orlean P., Wloka C., Deshaies R., et al. , 2012. Mitotic exit kinase Dbf2 directly phosphorylates chitin synthase Chs2 to regulate cytokinesis in budding yeast. Mol. Biol. Cell 23: 2445–2456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi K., Inoue N., Maeda Y., Takeda J., Riezman H., et al. , 2000. Gaa1p and Gpi8p are components of a glycosylphosphatidylinositol (GPI) transamidase that mediates attachment of GPI to proteins. Mol. Biol. Cell 11: 1523–1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohishi K., Inoue N., Kinoshita T., 2001. PIG-S and PIG-T, essential for GPI anchor attachment to proteins, form a complex with GAA1 and GPI8. EMBO J. 20: 4088–4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H., Abe M., Asakawa-Minemura M., Hirata A., Qadota H., et al. , 2010. Multiple functional domains of the yeast 1,3-β-glucan synthase subunit Fks1p revealed by quantitative phenotypic analysis of temperature-sensitive mutants. Genetics 184: 1013–1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen V., Guruprasad K., Cawley N. X., Chen H.-C., Blundell T. L., et al. , 1998. Cleavage efficiency of the novel aspartic protease yapsin 1 (Yap3p) enhanced for substrates with arginine residues flanking the P1 site: correlation with electronegative active-site pockets predicted by molecular modeling. Biochemistry 37: 2768–2777. [DOI] [PubMed] [Google Scholar]
- Ono N., Yabe T., Sudoh M., Nakajima T., Yamada-Okabe T., et al. , 2000. The yeast Chs4 protein stimulates the trypsin-sensitive activity of chitin synthase 3 through an apparent protein-protein interaction. Microbiology 146: 385–391. [DOI] [PubMed] [Google Scholar]
- Orchard M. G., Neuss J. C., Galley C. M., Carr A., Porter D. W., et al. , 2004. Rhodanine-3-acetic acid derivatives as inhibitors of fungal protein mannosyl transferase 1 (PMT1). Bioorg. Med. Chem. Lett. 14: 3975–3978. [DOI] [PubMed] [Google Scholar]
- O’Reilly M. K., Zhang G., Imperiali B., 2006. In vitro evidence for the dual function of Alg2 and Alg11: essential mannosyltransferases in N-linked glycoprotein biosynthesis. Biochemistry 45: 9593–9603. [DOI] [PubMed] [Google Scholar]
- Oriol R., Martinez-Duncker I., Chantret I., Mollicone R., Codogno P., 2002. Common origin and evolution of glycosyltransferases using Dol-P-monosaccharides as donor substrate. Mol. Biol. Evol. 19: 1451–1463. [DOI] [PubMed] [Google Scholar]
- Orlean P., 1987. Two chitin synthases in Saccharomyces cerevisiae. J. Biol. Chem. 262: 5732–5739. [PubMed] [Google Scholar]
- Orlean P., 1990. Dolichol phosphate mannose synthase is required in vivo for glycosyl phosphatidyl-inositol membrane anchoring, O mannosylation, and N glycosylation of protein in Saccharomyces cerevisiae. Mol. Cell. Biol. 10: 5796–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P., 1997. Biogenesis of yeast wall and surface components, pp. 229–362 in The Molecular and Cellular Biology of the Yeast Saccharomyces. Cell Cycle and Cell Biology, Vol. 3, edited by Pringle J. R., Broach J. R., Jones E. W. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Orlean P., 2009 Phosphoethanolamine addition to glycosylphosphatidylinositols, pp. 117–132 in The Enzymes, Vol. XXVI, Glycosylphosphatidylinositol (GPI) Anchoring of Proteins, edited by A. K. Menon, T. Kinoshita, P. Orlean, and F. Tamanoi, Academic Press/Elsevier, New York. [Google Scholar]
- Orlean P., Menon A. K., 2007. GPI anchoring of protein in yeast and mammalian cells or: how we learned to stop worrying and love glycophospholipids. J. Lipid Res. 48: 993–1011. [DOI] [PubMed] [Google Scholar]
- Orlean P., Arnold E., Tanner W., 1985. Apparent inhibition of glycoprotein synthesis by S. cerevisiae mating pheromones. FEBS Lett. 184: 313–317. [DOI] [PubMed] [Google Scholar]
- Orlean P., Ammer H., Watzele M., Tanner W., 1986. Synthesis of an O-glycosylated cell surface protein induced in yeast by α-factor. Proc. Natl. Acad. Sci. USA 83: 6263–6266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlean P., Albright C., Robbins P. W., 1988. Cloning and sequencing of the yeast gene for dolichol phosphate mannose synthase, an essential protein. J. Biol. Chem. 263: 17499–17507. [PubMed] [Google Scholar]
- Orlowski J., Machula K., Janik A., Zdebska E., Palamarczyk G., 2007. Dissecting the role of dolichol in cell wall assembly in the yeast mutants impaired in early glycosylation reactions. Yeast 24: 239–252. [DOI] [PubMed] [Google Scholar]
- Osmond B. C., Specht C. A., Robbins P. W., 1999. Chitin synthase III: synthetic lethal mutants and “stress related” chitin synthesis that bypasses the CSD3/CHS6 localization pathway. Proc. Natl. Acad. Sci. USA 96: 11206–11210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osumi M., 1998. The ultrastructure of yeast: cell wall structure and formation. Micron 29: 207–233. [DOI] [PubMed] [Google Scholar]
- Pagé N., Gerard-Vincent M., Menard P., Beaulieu M., Azuma M., et al. , 2003. A Saccharomyces cerevisiae genome-wide mutant screen for altered sensitivity to K1 killer toxin. Genetics 163: 875–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pammer M., Briza P., Ellinger A., Schuster T., Stucka R., et al. , 1992. DIT101 (CSD2, CAL1), a cell cycle-regulated yeast gene required for synthesis of chitin in cell walls and chitosan in spore walls. Yeast 8: 1089–1099. [DOI] [PubMed] [Google Scholar]
- Pardo M., Monteoliva L., Vázquez P., Martínez R., Molero G., et al. , 2004. PST1 and ECM33 encode two yeast cell surface GPI proteins important for cell wall integrity. Microbiology 150: 4157–4170. [DOI] [PubMed] [Google Scholar]
- Park H., Lennarz W. J., 2000. Evidence for interaction of yeast protein kinase C with several subunits of oligosaccharyl transferase. Glycobiology 10: 737–744. [DOI] [PubMed] [Google Scholar]
- Parodi A. J., 1999. Reglucosylation of glycoproteins and quality control of glycoprotein folding in the endoplasmic reticulum of yeast cells. Biochim. Biophys. Acta 1426: 287–295. [DOI] [PubMed] [Google Scholar]
- Pathak R., Hendrickson T. L., Imperiali B., 1995. Sulfhydryl modification of the yeast Wbp1p inhibits oligosaccharyl transferase activity. Biochemistry 34: 4179–4185. [DOI] [PubMed] [Google Scholar]
- Pitarch A., Nombela C., Gil C., 2008. Cell wall fractionation for yeast and fungal proteomics. Methods Mol. Biol. 425: 217–239. [DOI] [PubMed] [Google Scholar]
- Pittet M., Conzelmann A., 2007. Biosynthesis and function of GPI proteins in the yeast Saccharomyces cerevisiae. Biochim. Biophys. Acta 1771: 405–420. [DOI] [PubMed] [Google Scholar]
- Popolo L., Vai M., 1999. The Gas1 glycoprotein, a putative wall polymer cross-linker. Biochim. Biophys. Acta 1426: 385–400. [DOI] [PubMed] [Google Scholar]
- Popolo L., Gilardelli D., Bonfante P., Vai M., 1997. Increase in chitin as an essential response to defects in assembly of cell wall polymers in the ggp1Δ mutant of Saccharomyces cerevisiae. J. Bacteriol. 179: 463–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Protchenko O., Ferea T., Rashford J., Tiedeman J., Brown P. O., et al. , 2001. Three cell wall mannoproteins facilitate the uptake of iron in Saccharomyces cerevisiae. J. Biol. Chem. 276: 49244–49250. [DOI] [PubMed] [Google Scholar]
- Qadota H., Python C. P., Inoue S. B., Arisawa M., Anraku Y., et al. , 1996. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-β-glucan synthase. Science 272: 279–281. [DOI] [PubMed] [Google Scholar]
- Quinn R. P., Mahoney S. J., Wilkinson B. M., Thornton D. J., Stirling C. J., 2009. A novel role for Gtb1p in glucose trimming of N-linked glycans. Glycobiology 19: 1408–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni E., Coluccio A., Rolli E., Rodriguez-Peña J. M., Colasante G., et al. , 2007a GAS2 and GAS4, a pair of developmentally regulated genes required for spore wall assembly in Saccharomyces cerevisiae. Eukaryot. Cell 6: 302–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ragni E., Fontaine T., Gissi C., Latgé J. P., Popolo L., 2007b The Gas family of proteins of Saccharomyces cerevisiae: characterization and evolutionary analysis. Yeast 24: 297–308. [DOI] [PubMed] [Google Scholar]
- Ragni E., Sipiczki M., Strahl S., 2007c Characterization of Ccw12p, a major key player in cell wall stability of Saccharomyces cerevisiae. Yeast 24: 309–319. [DOI] [PubMed] [Google Scholar]
- Ragni E., Piberger H., Neupert C., García-Cantalejo J., Popolo L., et al. , 2011. The genetic interaction network of CCW12, a Saccharomyces cerevisiae gene required for cell wall integrity during budding and formation of mating projections. BMC Genomics 12: 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram A. F., Wolters A., Ten Hoopen R., Klis F. M., 1994. A new approach for isolating cell wall mutants in Saccharomyces cerevisiae by screening for hypersensitivity to calcofluor white. Yeast 10: 1019–1030. [DOI] [PubMed] [Google Scholar]
- Ram A. F., Brekelmans S. S., Oehlen L. J., Klis F. M., 1995. Identification of two cell cycle regulated genes affecting the β1,3-glucan content of cell walls in Saccharomyces cerevisiae. FEBS Lett. 358: 165–170. [DOI] [PubMed] [Google Scholar]
- Ram A. F., Kapteyn J. C., Montijn R. C., Caro L. H., Douwes J. E., et al. , 1998. Loss of the plasma membrane-bound protein Gas1p in Saccharomyces cerevisiae results in the release of β1,3-glucan into the medium and induces a compensation mechanism to ensure cell wall integrity. J. Bacteriol. 180: 1418–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsook C. B., Tan C., Garcia M. C., Fung R., Soybelman G., et al. , 2010. Yeast cell adhesion molecules have functional amyloid-forming sequences. Eukaryot. Cell 9: 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner J. C., Munro S., 1998. Identification of the MNN2 and MNN5 mannosyltransferases required for forming and extending the mannose branches of the outer chain mannans of Saccharomyces cerevisiae. J. Biol. Chem. 273: 26836–26843. [DOI] [PubMed] [Google Scholar]
- Reiss G., te Heesen S., Zimmerman J., Robbins P. W., Aebi M., 1996. Isolation of the ALG6 locus of Saccharomyces cerevisiae required for glucosylation in the N-linked glycosylation pathway. Glycobiology 6: 493–498. [DOI] [PubMed] [Google Scholar]
- Reiss G., te Heesen S., Gilmore R., Zufferey R., Aebi M., 1997. A specific screen for oligosaccharyltransferase mutations identifies the 9 kDa OST5 protein required for optimal activity in vivo and in vitro. EMBO J. 16: 1164–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes A., Sanz M., Duran A., Roncero C., 2007. Chitin synthase III requires Chs4p-dependent translocation of Chs3p into the plasma membrane. J. Cell Sci. 120: 1998–2009. [DOI] [PubMed] [Google Scholar]
- Reynolds T. B., Fink G. R., 2001. Bakers’ yeast, a model for fungal biofilm formation. Science 291: 878–881. [DOI] [PubMed] [Google Scholar]
- Richard M., De Groot P., Courtin O., Poulain D., Klis F., et al. , 2002. GPI7 affects cell-wall protein anchorage in Saccharomyces cerevisiae and Candida albicans. Microbiology 148: 2125–2133. [DOI] [PubMed] [Google Scholar]
- Rodicio R., Heinisch J. J., 2010. Together we are strong: cell wall integrity sensors in yeasts. Yeast 27: 531–540. [DOI] [PubMed] [Google Scholar]
- Rodionov D., Romero P. A., Berghuis A. M., Herscovics A., 2009. Expression and purification of recombinant M-Pol I from Saccharomyces cerevisiae with α-1,6 mannosylpolymerase activity. Protein Expr. Purif. 66: 1–6. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Peña J. M., Cid V. J., Arroyo J., Nombela C., 2000. A novel family of cell wall-related proteins regulated differently during the yeast life cycle. Mol. Cell. Biol. 20: 3245–3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Peña J. M., Rodriguez C., Alvarez A., Nombela C., Arroyo J., 2002. Mechanisms for targeting of the Saccharomyces cerevisiae GPI-anchored cell wall protein Crh2p to polarised growth sites. J. Cell Sci. 115: 2549–2558. [DOI] [PubMed] [Google Scholar]
- Roemer T., Bussey H., 1995. Yeast Kre1p is a cell surface O-glycoprotein. Mol. Gen. Genet. 249: 209–216. [DOI] [PubMed] [Google Scholar]
- Roemer T., Delaney S., Bussey H., 1993. SKN1 and KRE6 define a pair of functional homologs encoding putative membrane proteins involved in β-glucan synthesis. Mol. Cell. Biol. 13: 4039–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh D. H., Bowers B., Schmidt M., Cabib E., 2002a The septation apparatus, an autonomous system in budding yeast. Mol. Biol. Cell 13: 2747–2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh D. H., Bowers B., Riezman H., Cabib E., 2002b Rho1p mutations specific for regulation of β(1→3)glucan synthesis and the order of assembly of the yeast cell wall. Mol. Microbiol. 44: 1167–1183. [DOI] [PubMed] [Google Scholar]
- Rolli E., Ragni E., Calderon J., Porello S., Fascio U., et al. , 2009. Immobilization of the glycosylphosphatidylinositol-anchored Gas1 protein into the chitin ring and septum is required for proper morphogenesis in yeast. Mol. Biol. Cell 20: 4856–4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero P. A., Dijkgraaf G. J., Shahinian S., Herscovics A., Bussey H., 1997. The yeast CWH41 gene encodes glucosidase I. Glycobiology 7: 997–1004. [DOI] [PubMed] [Google Scholar]
- Romero P. A., Lussier M., Véronneau S., Sdicu A. M., Herscovics A., et al. , 1999. Mnt2p and Mnt3p of Saccharomyces cerevisiae are members of the Mnn1p family of α-1,3-mannosyltransferases responsible for adding the terminal mannose residues of O-linked oligosaccharides. Glycobiology 9: 1045–1051. [DOI] [PubMed] [Google Scholar]
- Roncero C., 2002. The genetic complexity of chitin synthesis in fungi. Curr. Genet. 41: 367–378. [DOI] [PubMed] [Google Scholar]
- Roncero C., Sánchez Y., 2010. Cell separation and the maintenance of cell integrity during cytokinesis in yeast: the assembly of a septum. Yeast 27: 521–530. [DOI] [PubMed] [Google Scholar]
- Roncero C., Valdivieso M., Ribas J. C., Duran A., 1988. Isolation and characterization of Saccharomyces cerevisiae mutants resistant to calcofluor white. J. Bacteriol. 170: 1950–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A., Lu C. F., Marykwas D. L., Lipke P. N., Kurjan J., 1991. The AGA1 product is involved in cell surface attachment of the Saccharomyces cerevisiae cell adhesion glycoprotein a-agglutinin. Mol. Cell. Biol. 11: 4196–4206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S. K., Yoko-o T., Ikenaga H., Jigami Y., 1998. Functional evidence for UDP-galactose transporter in Saccharomyces cerevisiae through the in vivo galactosylation and in vitro transport assay. J. Biol. Chem. 273: 2583–2590. [DOI] [PubMed] [Google Scholar]
- Roy S. K., Chiba Y., Takeuchi M., Jigami Y., 2000. Characterization of yeast Yea4p, a uridine diphosphate-N-acetylglucosamine transporter localized in the endoplasmic reticulum and required for chitin synthesis. J. Biol. Chem. 275: 13580–13587. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J., Ortiz-Castellanos L., 2010. Analysis of the phylogenetic relationships and evolution of the cell walls from yeasts and fungi. FEMS Yeast Res. 10: 225–243. [DOI] [PubMed] [Google Scholar]
- Ruiz-Herrera J., Gonzalez-Prieto J. M., Ruiz-Medrano R., 2002. Evolution and phylogenetic relationships of chitin synthases from yeasts and fungi. FEMS Yeast Res. 1: 247–256. [DOI] [PubMed] [Google Scholar]
- Rush J. S., Gao N., Lehrman M. A., Matveev S., Waechter C. J., 2009. Suppression of Rft1 expression does not impair the transbilayer movement of Man5GlcNAc2-P-P-dolichol in sealed microsomes from yeast. J. Biol. Chem. 284: 19835–19842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo P., Simonen M., Uimari A., Teesalu T., Makarow M., 1993. Dual regulation by heat and nutrient stress of the yeast HSP150 gene encoding a secretory glycoprotein. Mol. Gen. Genet. 239: 273–280. [DOI] [PubMed] [Google Scholar]
- Sacristan C., Reyes A., Roncero C., 2012. Neck compartmentalization as the molecular basis for the different endocytic behaviour of Chs3 during budding or hyperpolarized growth in yeast cells. Mol. Microbiol. 83: 1124–1135. [DOI] [PubMed] [Google Scholar]
- Sagane K., Umemura M., Ogawa-Mitsuhashi K., Tsukahara K., Yoko-o T., et al. , 2011. Analysis of membrane topology and identification of essential residues for the yeast endoplasmic reticulum inositol acyltransferase Gwt1p. J. Biol. Chem. 286: 14649–14658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saka A., Abe M., Okano H., Minemura M., Qadota H., et al. , 2001. Complementing yeast rho1 mutation groups with distinct functional defects. J. Biol. Chem. 276: 46165–46171. [DOI] [PubMed] [Google Scholar]
- Sanchatjate S., Schekman R., 2006. Chs5/6 complex: a multiprotein complex that interacts with and conveys chitin synthase III from the trans-Golgi network to the cell surface. Mol. Biol. Cell 17: 4157–4166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B., Snyder M., 1997. Targeting of chitin synthase 3 to polarized growth sites in yeast requires Chs5p and Myo2p. J. Cell Biol. 136: 95–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos B., Duran A., Valdivieso M. H., 1997. CHS5, a gene involved in chitin synthesis and mating in Saccharomyces cerevisiae. Mol. Cell. Biol. 17: 2485–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanyal S., Menon A. K., 2010. Stereoselective transbilayer translocation of mannosyl phosphoryl dolichol by an endoplasmic reticulum flippase. Proc. Natl. Acad. Sci. USA 107: 11289–11294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz M., Trilla J. A., Duran A., Roncero C., 2002. Control of chitin synthesis through Shc1p, a functional homologue of Chs4p specifically induced during sporulation. Mol. Microbiol. 43: 1183–1195. [DOI] [PubMed] [Google Scholar]
- Sanz M., Castrejon F., Duran A., Roncero C., 2004. Saccharomyces cerevisiae Bni4p directs the formation of the chitin ring and also participates in the correct assembly of the septum structure. Microbiology 150: 3229–3241. [DOI] [PubMed] [Google Scholar]
- Sato K., Noda Y., Yoda K., 2007. Pga1 is an essential component of glycosylphosphatidylinositol-mannosyltransferase II of Saccharomyces cerevisiae. Mol. Biol. Cell 18: 3472–3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato M., Sato K., Nishikawa S., Hirata A., Kato J., et al. , 1999. The yeast RER2 gene, identified by endoplasmic reticulum protein localization mutations, encodes cis-prenyltransferase, a key enzyme in dolichol synthesis. Mol. Cell. Biol. 19: 471–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena I. M., Brown R. M., Jr, Fevre M., Geremia R. A., Henrissat B., 1995. Multidomain architecture of β-glycosyl transferases: implications for mechanism of action. J. Bacteriol. 177: 1419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sburlati A., Cabib E., 1986. Chitin synthetase 2, a presumptive participant in septum formation in Saccharomyces cerevisiae. J. Biol. Chem. 261: 15147–15152. [PubMed] [Google Scholar]
- Scarcelli J. J., Colussi P. A., Fabre A.-L., Boles E., Orlean P., et al. , 2012. Uptake of radiolabeled GlcNAc into Saccharomyces cerevisiae via native hexose transporters and its in vivo incorporation into GPI precursors in cells expressing heterologous GlcNAc kinase. FEMS Yeast Res. 12: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schekman R., Brawley V., 1979. Localized deposition of chitin on the yeast cell surface in response to mating pheromone. Proc. Natl. Acad. Sci. USA 76: 645–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk B., Fernandez F., Waechter C. J., 2001a The ins(ide) and out(side) of dolichyl phosphate biosynthesis and recycling in the endoplasmic reticulum. Glycobiology 11: 61R–70R. [DOI] [PubMed] [Google Scholar]
- Schenk B., Rush J. S., Waechter C. J., Aebi M., 2001b An alternative cis-isoprenyltransferase activity in yeast that produces polyisoprenols with chain lengths similar to mammalian dolichols. Glycobiology 11: 89–98. [DOI] [PubMed] [Google Scholar]
- Schmidt M., 2004. Survival and cytokinesis of Saccharomyces cerevisiae in the absence of chitin. Microbiology 150: 3253–3260. [DOI] [PubMed] [Google Scholar]
- Schmidt M., Bowers B., Varma A., Roh D.-H., Cabib E., 2002. In budding yeast, contraction of the actomyosin ring and formation of the primary septum depend on each other. J. Cell Sci. 115: 293–302. [DOI] [PubMed] [Google Scholar]
- Schreuder M. P., Brekelmans S., van den Ende H., Klis F. M., 1993. Targeting of a heterologous protein to the cell wall of Saccharomyces cerevisiae. Yeast 9: 399–409. [DOI] [PubMed] [Google Scholar]
- Schulz B. L., Aebi M., 2009. Analysis of glycosylation site occupancy reveals a role for Ost3p and Ost6p in site-specific N-glycosylation efficiency. Mol. Cell. Proteomics 8: 357–364. [DOI] [PubMed] [Google Scholar]
- Schulz B. L., Stirnimann C. U., Grimshaw J. P., Brozzo M. S., Fritsch F., et al. , 2009. Oxidoreductase activity of oligosaccharyltransferase subunits Ost3p and Ost6p defines site-specific glycosylation efficiency. Proc. Natl. Acad. Sci. USA 106: 11061–11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M., Knauer R., Lehle L., 2005. Yeast oligosaccharyltransferase consists of two functionally distinct sub-complexes, specified by either the Ost3p or Ost6p subunit. FEBS Lett. 579: 6564–6568. [DOI] [PubMed] [Google Scholar]
- Sestak S., Hagen I., Tanner W., Strahl S., 2004. Scw10p, a cell-wall glucanase/transglucosidase important for cell-wall stability in Saccharomyces cerevisiae. Microbiology 150: 3197–3208. [DOI] [PubMed] [Google Scholar]
- Shahinian S., Bussey H., 2000. β-1,6-Glucan synthesis in Saccharomyces cerevisiae. Mol. Microbiol. 35: 477–489. [DOI] [PubMed] [Google Scholar]
- Shahinian S., Dijkgraaf G. J., Sdicu A. M., Thomas D. Y., Jakob C. A., et al. , 1998. Involvement of protein N-glycosyl chain glucosylation and processing in the biosynthesis of cell wall β-1,6-glucan of Saccharomyces cerevisiae. Genetics 149: 843–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shankarnarayan S., Malone C. L., Deschenes R. J., Fassler J. S., 2008. Modulation of yeast Sln1 kinase activity by the Ccw12 cell wall protein. J. Biol. Chem. 283: 1962–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma C. B., Knauer R., Lehle L., 2001. Biosynthesis of lipid-linked oligosaccharides in yeast: the ALG3 gene encodes the Dol-P-Man:Man5GlcNAc2-PP-Dol mannosyltransferase. Biol. Chem. 382: 321–328. [DOI] [PubMed] [Google Scholar]
- Shaw J. A., Mol P. C., Bowers B., Silverman S. J., Valdivieso M. H., et al. , 1991. The function of chitin synthases 2 and 3 in the Saccharomyces cerevisiae cell cycle. J. Cell Biol. 114: 111–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shematek E. M., Braatz J. A., Cabib E., 1980. Biosynthesis of the yeast cell wall. I. Preparation and properties of β-(1→3)glucan synthetase. J. Biol. Chem. 255: 888–894. [PubMed] [Google Scholar]
- Shen Z. M., Wang L., Pike J., Jue C. K., Zhao H., et al. , 2001. Delineation of functional regions within the subunits of the Saccharomyces cerevisiae cell adhesion molecule a-agglutinin. J. Biol. Chem. 276: 15768–15775. [DOI] [PubMed] [Google Scholar]
- Shepard K. A., Gerber A. P., Jambhekar A., Takizawa P. A., Brown P. O., et al. , 2003. Widespread cytoplasmic mRNA transport in yeast: identification of 22 bud-localized transcripts using DNA microarray analysis. Proc. Natl. Acad. Sci. USA 100: 11429–11434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibasaki S., Maeda H., Ueda M., 2009. Molecular display technology using yeast-arming technology. Anal. Sci. 25: 41–49. [DOI] [PubMed] [Google Scholar]
- Shimma Y., Saito F., Oosawa F., Jigami Y., 2006. Construction of a library of human glycosyltransferases immobilized in the cell wall of Saccharomyces cerevisiae. Appl. Environ. Microbiol. 72: 7003–7012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoi H., Kitagaki H., Ohmori H., Iimura Y., Ito K., 1998. Sed1p is a major cell wall protein of Saccharomyces cerevisiae in the stationary phase and is involved in lytic enzyme resistance. J. Bacteriol. 180: 3381–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signorell A., Menon A. K., 2009 Attachment of a GPI anchor to protein, pp. 133–149 in The Enzymes, Vol. XXVI, Glycosylphosphatidylinositol (GPI) Anchoring of Proteins, edited by A. K. Menon, T. Kinoshita, P. Orlean, and F. Tamanoi. Academic Press/Elsevier, New York. [Google Scholar]
- Simoes T., Teixeira M. C., Fernandes A. R., Sa-Correia I., 2003. Adaptation of Saccharomyces cerevisiae to the herbicide 2,4-dichlorophenoxyacetic acid, mediated by Msn2p- and Msn4p-regulated genes: important role of SPI1. Appl. Environ. Microbiol. 69: 4019–4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons J. F., Ebersold M., Helenius A., 1998. Cell wall 1,6-β-glucan synthesis in Saccharomyces cerevisiae depends on ER glucosidases I and II, and the molecular chaperone BiP/Kar2p. EMBO J. 17: 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sipos G., Puoti A., Conzelmann A., 1995. Biosynthesis of the side chain of yeast glycosylphosphatidylinositol anchors is operated by novel mannosyltransferases located in the endoplasmic reticulum and the Golgi apparatus. J. Biol. Chem. 270: 19709–19715. [DOI] [PubMed] [Google Scholar]
- Smits G. J., Schenkman L. R., Brul S., Pringle J. R., Klis F. M., 2006. Role of cell cycle-regulated expression in the localized incorporation of cell wall proteins in yeast. Mol. Biol. Cell 17: 3267–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobering A. K., Watanabe R., Romeo M. J., Yan B. C., Specht C. A., et al. , 2004. Yeast Ras regulates the complex that catalyzes the first step in GPI-anchor biosynthesis at the ER. Cell 117: 637–648. [DOI] [PubMed] [Google Scholar]
- Spellman P. T., Sherlock G., Zhang M. Q., Iyer V. R., Anders K., et al. , 1998. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol. Biol. Cell 9: 3273–3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirig U., Glavas M., Bodmer D., Reiss G., Burda P., et al. , 1997. The STT3 protein is a component of the yeast oligosaccharyltransferase complex. Mol. Gen. Genet. 256: 628–637. [DOI] [PubMed] [Google Scholar]
- Spirig U., Bodmer D., Wacker M., Burda P., Aebi M., 2005. The 3.4-kDa Ost4 protein is required for the assembly of two distinct oligosaccharyltransferase complexes in yeast. Glycobiology 15: 1396–1406. [DOI] [PubMed] [Google Scholar]
- Stagljar I., te Heesen S., Aebi M., 1994. New phenotype of mutations deficient in glucosylation of the lipid-linked oligosaccharide: cloning of the ALG8 locus. Proc. Natl. Acad. Sci. USA 91: 5977–5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz J., Munro S., 2002. The components of the Saccharomyces cerevisiae mannosyltransferase complex M-Pol I have distinct functions in mannan synthesis. J. Biol. Chem. 277: 44801–44808. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Scheinost A., 1999. Transmembrane topology of Pmt1p, a member of an evolutionarily conserved family of protein O-mannosyltransferases. J. Biol. Chem. 274: 9068–9075. [DOI] [PubMed] [Google Scholar]
- Strahl-Bolsinger S., Gentzsch M., Tanner W., 1999. Protein O-mannosylation. Biochim. Biophys. Acta 1426: 297–307. [DOI] [PubMed] [Google Scholar]
- Subramanian S., Woolford C. A., Drill E., Lu M., Jones E. W., 2006. Pbn1p: an essential endoplasmic reticulum membrane protein required for protein processing in the endoplasmic reticulum of budding yeast. Proc. Natl. Acad. Sci. USA 103: 939–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumita T., Yoko-o T., Shimma Y., Jigami Y., 2005. Comparison of cell wall localization among Pir family proteins and functional dissection of the region required for cell wall binding and bud scar recruitment of Pir1p. Eukaryot. Cell 4: 1872–1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sütterlin C., Escribano M. V., Gerold P., Maeda Y., Mazon M. J., et al. , 1998. Saccharomyces cerevisiae GPI10, the functional homologue of human PIG-B, is required for glycosylphosphatidylinositol-anchor synthesis. Biochem. J. 332: 153–159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutterlin C., Horvath A., Gerold P., Schwarz R. T., Wang Y., et al. , 1997. Identification of a species-specific inhibitor of glycosylphosphatidylinositol synthesis. EMBO J. 16: 6374–6383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Park H., Hollingsworth N. M., Sternglanz R., Lennarz W. J., 2000. PNG1, a yeast gene encoding a highly conserved peptide: N-glycanase. J. Cell Biol. 149: 1039–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szathmary R., Bielmann R., Nita-Lazar M., Burda P., Jakob C. A., 2005. Yos9 protein is essential for degradation of misfolded glycoproteins and may function as lectin in ERAD. Mol. Cell 19: 765–775. [DOI] [PubMed] [Google Scholar]
- Tanaka S., Maeda Y., Tashima Y., Kinoshita T., 2004. Inositol deacylation of glycosylphosphatidylinositol-anchored proteins is mediated by mammalian PGAP1 and yeast Bst1p. J. Biol. Chem. 279: 14256–14263. [DOI] [PubMed] [Google Scholar]
- Taron C. H., Wiedman J. M., Grimme S. J., Orlean P., 2000. Glycosylphosphatidylinositol biosynthesis defects in Gpi11p- and Gpi13p-deficient yeast suggest a branched pathway and implicate Gpi13p in phosphoethanolamine transfer to the third mannose. Mol. Biol. Cell 11: 1611–1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh E. M., Chai C. C., Yeong F. M., 2009. Retention of Chs2p in the ER requires N-terminal CDK1-phosphorylation sites. Cell Cycle 8: 2965–2976. [PubMed] [Google Scholar]
- Te Heesen S., Lehle L., Weissmann A., Aebi M., 1994. Isolation of the ALG5 locus encoding the UDP-glucose:dolichyl-phosphate glucosyltransferase from Saccharomyces cerevisiae. Eur. J. Biochem. 224: 71–79. [DOI] [PubMed] [Google Scholar]
- Teparic R., Stuparevic I., Mrša V., 2004. Increased mortality of Saccharomyces cerevisiae cell wall protein mutants. Microbiology 150: 3145–3150. [DOI] [PubMed] [Google Scholar]
- Terashima H., Yabuki N., Arisawa M., Hamada K., Kitada K., 2000. Up-regulation of genes encoding glycosylphosphatidylinositol (GPI)-attached proteins in response to cell wall damage caused by disruption of FKS1 in Saccharomyces cerevisiae. Mol. Gen. Genet. 264: 64–74. [DOI] [PubMed] [Google Scholar]
- Terashima H., Fukuchi S., Nakai K., Arisawa M., Hamada K., et al. , 2002. Sequence-based approach for identification of cell wall proteins in Saccharomyces cerevisiae. Curr. Genet. 40: 311–316. [DOI] [PubMed] [Google Scholar]
- Terashima H., Hamada K., Kitada K., 2003. The localization change of Ybr078w/Ecm33, a yeast GPI-associated protein, from the plasma membrane to the cell wall, affecting the cellular function. FEMS Microbiol. Lett. 218: 175–180. [DOI] [PubMed] [Google Scholar]
- Thevissen K., Idkowiak-Baldys J., Im Y. J., Takemoto J., François I. E., et al. , 2005. SKN1, a novel plant defensin-sensitivity gene in Saccharomyces cerevisiae, is implicated in sphingolipid biosynthesis. FEBS Lett. 579: 1973–1977. [DOI] [PubMed] [Google Scholar]
- Tiede A., Nischan C., Schubert J., Schmidt R. E., 2000. Characterisation of the enzymatic complex for the first step in glycosylphosphatidylinositol biosynthesis. Int. J. Biochem. Cell Biol. 32: 339–350. [DOI] [PubMed] [Google Scholar]
- Toh-e A., Oguchi T., 1999. Las21 participates in extracellular/cell surface phenomena in Saccharomyces cerevisiae. Genes Genet. Syst. 74: 241–256. [DOI] [PubMed] [Google Scholar]
- Toh-e A., Yasunaga S., Nisogi H., Tanaka K., Oguchi T., et al. , 1993. Three yeast genes, PIR1, PIR2 and PIR3, containing internal tandem repeats, are related to each other, and PIR1 and PIR2 are required for tolerance to heat shock. Yeast 9: 481–494. [DOI] [PubMed] [Google Scholar]
- Tomishige N., Noda Y., Adachi H., Shimoi H., Takatsuki A., et al. , 2003. Mutations that are synthetically lethal with a gas1Δ allele cause defects in the cell wall of Saccharomyces cerevisiae. Mol. Genet. Genomics 269: 562–573. [DOI] [PubMed] [Google Scholar]
- Trautwein M., Schindler C., Gauss R., Dengjel J., Hartmann E., et al. , 2006. Arf1p, Chs5p and the ChAPs are required for export of specialized cargo from the Golgi. EMBO J. 25: 943–954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travers K. J., Patil C. K., Wodicka L., Lockhart D. J., Weissman J. S., et al. , 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101: 249–258. [DOI] [PubMed] [Google Scholar]
- Trilla J. A., Cos T., Duran A., Roncero C., 1997. Characterization of CHS4 (CAL2), a gene of Saccharomyces cerevisiae involved in chitin biosynthesis and allelic to SKT5 and CSD4. Yeast 13: 795–807. [DOI] [PubMed] [Google Scholar]
- Trilla J. A., Duran A., Roncero C., 1999. Chs7p, a new protein involved in the control of protein export from the endoplasmic reticulum that is specifically engaged in the regulation of chitin synthesis in Saccharomyces cerevisiae. J. Cell Biol. 145: 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta E. S., Simons J. F., Helenius A., 1996. Endoplasmic reticulum glucosidase II is composed of a catalytic subunit, conserved from yeast to mammals, and a tightly bound noncatalytic HDEL-containing subunit. J. Biol. Chem. 271: 27509–27516. [DOI] [PubMed] [Google Scholar]
- Tsukahara K., Hata K., Sagane K., Watanabe N., Kuromitsu J., et al. , 2003. Medicinal genetics approach towards identifying the molecular target of a novel inhibitor of fungal cell wall assembly. Mol. Microbiol. 48: 1029–1042. [DOI] [PubMed] [Google Scholar]
- Uchida Y., Shimmi O., Sudoh M., Arisawa M., Yamada-Okabe H., 1996. Characterization of chitin synthase 2 of Saccharomyces cerevisiae. II: both full size and processed enzymes are active for chitin synthesis. J. Biochem. 119: 659–666. [DOI] [PubMed] [Google Scholar]
- Umemura M., Okamoto M., Nakayama K., Sagane K., Tsukahara K., et al. , 2003. GWT1 gene is required for inositol acylation of glycosylphosphatidylinositol anchors in yeast. J. Biol. Chem. 278: 23639–23647. [DOI] [PubMed] [Google Scholar]
- Umemura M., Fujita M., Yoko-o T., Fukamizu A., Jigami Y., 2007. Saccharomyces cerevisiae CWH43 is involved in the remodeling of the lipid moiety of GPI anchors to ceramides. Mol. Biol. Cell 18: 4304–4316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsugi T., Minemura M., Hirata A., Abe M., Watanabe D., et al. , 2002. Movement of yeast 1,3-β-glucan synthase is essential for uniform cell wall synthesis. Genes Cells 7: 1–9. [DOI] [PubMed] [Google Scholar]
- Vadaie N., Dionne H., Akajagbor D. S., Nickerson S. R., Krysan D. J., et al. , 2008. Cleavage of the signaling mucin Msb2 by the aspartyl protease Yps1 is required for MAPK activation in yeast. J. Cell Biol. 181: 1073–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivia R. H., Schekman R., 2003. The yeasts Rho1p and Pkc1p regulate the transport of chitin synthase III (Chs3p) from internal stores to the plasma membrane. Proc. Natl. Acad. Sci. USA 100: 10287–10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdivieso M. H., Ferrario L., Vai M., Duran A., Popolo L., 2000. Chitin synthesis in a gas1 mutant of Saccharomyces cerevisiae. J. Bacteriol. 182: 4752–4757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Berkel M. A., Rieger M., te Heesen S., Ram A. F., van den Ende H., et al. , 1999. The Saccharomyces cerevisiae CWH8 gene is required for full levels of dolichol-linked oligosaccharides in the endoplasmic reticulum and for efficient N-glycosylation. Glycobiology 9: 243–253. [DOI] [PubMed] [Google Scholar]
- Van der Vaart J. M., Caro L. H. P., Chapman J. W., Klis F. M., Verrips C. T., 1995. Identification of three mannoproteins in the cell wall of Saccharomyces cerevisiae. J. Bacteriol. 177: 3104–3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Vaart J. M., te Biesebeke R., Chapman J. W., Klis F. M., Verrips C. T., 1996. The β-1,6-glucan containing side-chain of cell wall proteins of Saccharomyces cerevisiae is bound to the glycan core of the GPI moiety. FEMS Microbiol. Lett. 145: 401–407. [DOI] [PubMed] [Google Scholar]
- Van der Vaart J. M., te Biesebeke R., Chapman J. W., Toschka H. Y., Klis F. M., et al. , 1997. Comparison of cell wall proteins of Saccharomyces cerevisiae as anchors for cell surface expression of heterologous proteins. Appl. Environ. Microbiol. 63: 615–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Mulders S. E., Christianen E., Saerens S. M., Daenen L., Verbelen P. J., et al. , 2009. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 9: 178–190. [DOI] [PubMed] [Google Scholar]
- Varki A., Sharon N., 2009. Chapter 1 Historical Background and Overview, in Essentials of Glycobiology, Ed. 2, edited by A. Varki, R. D. Cummings, J. D. Esko, H. H. Freeze, P. Stanley, et al. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- Veelders M., Brückner S., Ott D., Unverzagt C., Mösch H. U., et al. , 2010. Structural basis of flocculin-mediated social behavior in yeast. Proc. Natl. Acad. Sci. USA 107: 22511–22516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velours G., Boucheron C., Manon S., Camougrand N., 2002. Dual cell wall/mitochondria localization of the ‘SUN’ family proteins. FEMS Microbiol. Lett. 207: 165–172. [DOI] [PubMed] [Google Scholar]
- Verma D. P., Hong Z., 2001. Plant callose synthase complexes. Plant Mol. Biol. 47: 693–701. [DOI] [PubMed] [Google Scholar]
- VerPlank L., Li R., 2005. Cell cycle-regulated trafficking of Chs2 controls actomyosin ring stability during cytokinesis. Mol. Biol. Cell 16: 2529–2543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstrepen K. J., Klis F. M., 2006. Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60: 5–15. [DOI] [PubMed] [Google Scholar]
- Vidugiriene J., Menon A. K., 1993. Early lipid intermediates in glycosyl-phosphatidylinositol anchor assembly are synthesized in the ER and located in the cytoplasmic leaflet of the ER membrane bilayer. J. Cell Biol. 121: 987–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink E., Rodriguez-Suarez R. J., Gérard-Vincent M., Ribas J. C., de Nobel H., et al. , 2004. An in vitro assay for (1→6)-β-D-glucan synthesis in Saccharomyces cerevisiae. Yeast 21: 1121–1131. [DOI] [PubMed] [Google Scholar]
- Vishwakarma R. A., Menon A. K., 2005. Flip-flop of glycosylphosphatidylinositols (GPI’s) across the ER. Chem. Commun. (Camb.) 2005: 453–455. [DOI] [PubMed] [Google Scholar]
- Vossen J. H., Müller W. H., Lipke P. N., Klis F. M., 1997. Restrictive glycosylphosphatidylinositol anchor synthesis in cwh6/gpi3 yeast cells causes aberrant biogenesis of cell wall proteins. J. Bacteriol. 179: 2202–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker M., Linton D., Hitchen P. G., Nita-Lazar M., Haslam S. M., et al. , 2002. N-linked glycosylation in C. jejuni and its functional transfer to E. coli. Science 298: 1790–1793. [DOI] [PubMed] [Google Scholar]
- Wang C. W., Hamamoto S., Orci L., Schekman R., 2006. Exomer: a coat complex for transport of select membrane proteins from the trans-Golgi network to the plasma membrane in yeast. J. Cell Biol. 174: 973–983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.-H., Nakayama K.-I., Shimma Y.-i., Tanaka A., Jigami Y., 1997. MNN6, a member of the KRE2/MNT1 family, is the gene for mannosylphosphate transfer in Saccharomyces cerevisiae. J. Biol. Chem. 272: 18117–18124. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Kinoshita T., Masaki R., Yamamoto A., Takeda J., et al. , 1996. PIG-A and PIG-H, which participate in glycosylphosphatidylinositol anchor biosynthesis, form a protein complex in the endoplasmic reticulum. J. Biol. Chem. 271: 26868–26875. [DOI] [PubMed] [Google Scholar]
- Watanabe R., Inoue N., Westfall B., Taron C. H., Orlean P., et al. , 1998. The first step of glycosylphosphatidylinositol biosynthesis is mediated by a complex of PIG-A, PIG-H, PIG-C and GPI1. EMBO J. 17: 877–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe R., Ohishi K., Maeda Y., Nakamura N., Kinoshita T., 1999. Mammalian PIG-L and its yeast homologue Gpi12p are N-acetylglucosaminylphosphatidylinositol de-N-acetylases essential in glycosylphosphatidylinositol biosynthesis. Biochem. J. 339: 185–192. [PMC free article] [PubMed] [Google Scholar]
- Watzele G., Tanner W., 1989. Cloning of the glutamine:fructose-6-phosphate amidotransferase gene from yeast. Pheromonal regulation of its transcription. J. Biol. Chem. 264: 8753–8758. [PubMed] [Google Scholar]
- Weerapana E., Imperiali B., 2006. Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology 16: 91R–101R. [DOI] [PubMed] [Google Scholar]
- Wiedman J. M., Fabre A.-L., Taron B. W., Taron C. H., Orlean P., 2007. In vivo characterization of the GPI assembly defect in yeast mcd4-174 mutants and bypass of the Mcd4p-dependent step in mcd4 null mutants. FEMS Yeast Res. 7: 78–83. [DOI] [PubMed] [Google Scholar]
- Wilkinson B. M., Purswani J., Stirling C. J., 2006. Yeast GTB1 encodes a subunit of glucosidase II required for glycoprotein processing in the endoplasmic reticulum. J. Biol. Chem. 281: 6325–6333. [DOI] [PubMed] [Google Scholar]
- Wojciechowicz D., Lu C.-F., Kurjan J., Lipke P. N., 1993. Cell surface anchorage and ligand-binding domains of the Saccharomyces cerevisiae cell adhesion protein α-agglutinin, a member of the immunoglobulin superfamily. Mol. Cell. Biol. 13: 2554–2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Lipke P. N., 2010. On the evolution of fungal and yeast cell walls. Yeast 27: 479–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yabe T., Yamada-Okabe T., Nakajima T., Sudoh M., Arisawa M., et al. , 1998. Mutational analysis of chitin synthase 2 of Saccharomyces cerevisiae. Identification of additional amino acid residues involved in its catalytic activity. Eur. J. Biochem. 258: 941–947. [DOI] [PubMed] [Google Scholar]
- Yamaguchi M., Namiki Y., Okada H., Mori Y., Furukawa H., et al. , 2011. Structome of Saccharomyces cerevisiae determined by freeze-substitution and serial ultrathin-sectioning electron microscopy. J. Electron Microsc. (Tokyo) 60: 321–335. [DOI] [PubMed] [Google Scholar]
- Yan A., Lennarz W. J., 2005a Unraveling the mechanism of protein N-glycosylation. J. Biol. Chem. 280: 3121–3124. [DOI] [PubMed] [Google Scholar]
- Yan A., Lennarz W. J., 2005b Two oligosaccharyl transferase complexes exist in yeast and associate with two different translocons. Glycobiology 15: 1407–1415. [DOI] [PubMed] [Google Scholar]
- Yan B. C., Westfall B. A., Orlean P., 2001. Ynl038wp (Gpi15p) is the Sacccharomyces cerevisiae homologue of human Pig-Hp and participates in the first step in glycosylphosphatidylinositol assembly. Yeast 18: 1383–1389. [DOI] [PubMed] [Google Scholar]
- Yan Q., Lennarz W. J., 2002. Studies on the function of the oligosaccharyltransferase subunits: Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 277: 47692–47700. [DOI] [PubMed] [Google Scholar]
- Yin Q. Y., de Groot P. W., Dekker H. L., de Jong L., Klis F. M., et al. , 2005. Comprehensive proteomic analysis of Saccharomyces cerevisiae cell walls: identification of proteins covalently attached via glycosylphosphatidylinositol remnants or mild alkali-sensitive linkages. J. Biol. Chem. 280: 20894–20901. [DOI] [PubMed] [Google Scholar]
- Yin Q. Y., de Groot P. W., de Jong L., Klis F. M., De Koster C. G., 2007. Mass spectrometric quantitation of covalently bound cell wall proteins in Saccharomyces cerevisiae. FEMS Yeast Res. 7: 887–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip C. L., Welch S. K., Klebl F., Gilbert T., Seidel P., et al. , 1994. Cloning and analysis of the Saccharomyces cerevisiae MNN9 and MNN1 genes required for complex glycosylation of secreted proteins. Proc. Natl. Acad. Sci. USA 91: 2723–2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoda K., Kawada T., Kaibara C., Fujie A., Abe M., et al. , 2000. Defect in cell wall integrity of the yeast Saccharomyces cerevisiae caused by a mutation of the GDP-mannose pyrophosphorylase gene VIG9. Biosci. Biotechnol. Biochem. 64: 1937–1941. [DOI] [PubMed] [Google Scholar]
- Yun D. J., Zhao Y., Pardo J. M., Narasimhan M. L., Damsz B., et al. , 1997. Stress proteins on the yeast cell surface determine resistance to osmotin, a plant antifungal protein. Proc. Natl. Acad. Sci. USA 94: 7082–7087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Kashimshetty R., Ng K. E., Tan H. B., Yeong F. M., 2006. Exit from mitosis triggers Chs2p transport from the endoplasmic reticulum to mother-daughter neck via the secretory pathway in budding yeast. J. Cell Biol. 174: 207–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Bennett D., Erdman S. E., 2002. Maintenance of mating cell integrity requires the adhesin Fig2p. Eukaryot. Cell 1: 811–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Liang Y., Zhang X., Xu Y., Dai H., et al. , 2008. Deletion of yeast CWP genes enhances cell permeability to genotoxic agents. Toxicol. Sci. 103: 68–76. [DOI] [PubMed] [Google Scholar]
- Zhao C., Jung U. S., Garrett-Engele P., Roe T., Cyert M. S., et al. , 1998. Temperature-induced expression of yeast FKS2 is under the dual control of protein kinase C and calcineurin. Mol. Cell. Biol. 18: 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Fraering P., Vionnet C., Conzelmann A., 2005. Gpi17p does not stably interact with other subunits of glycosylphosphatidylinositol transamidase in Saccharomyces cerevisiae. Biochim. Biophys. Acta 1735: 79–88. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Vionnet C., Conzelmann A., 2006. Ethanolaminephosphate side chain added to GPI anchor by Mcd4p is required for ceramide remodeling and forward transport of GPI proteins from ER to Golgi. J. Biol. Chem. 281: 19830–19839. [DOI] [PubMed] [Google Scholar]
- Ziman M., Chuang J. S., Schekman R. W., 1996. Chs1p and Chs3p, two proteins involved in chitin synthesis, populate a compartment of the Saccharomyces cerevisiae endocytic pathway. Mol. Biol. Cell 7: 1909–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziman M., Chuang J. S., Tsung M., Hamamoto S., Schekman R., 1998. Chs6p-dependent anterograde transport of Chs3p from the chitosome to the plasma membrane in Saccharomyces cerevisiae. Mol. Biol. Cell 9: 1565–1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zlotnik H., Fernandez M. P., Bowers B., Cabib E., 1984. Saccharomyces cerevisiae mannoproteins form an external cell wall layer that determines wall porosity. J. Bacteriol. 159: 1018–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.