Abstract
Isolation by distance and landscape connectivity are fundamental factors underlying speciation and evolution. To understand how landscapes affect gene flow and shape population structures, island species provide intrinsic study objects. We investigated the effects of landscapes on the population structure of the endangered frog species, Odorrana ishikawae and O. splendida, which each inhabit an island in southwest Japan. This was done by examining population structure, gene flow and demographic history of each species by analyzing 12 microsatellite loci and exploring causal environmental factors through ecological niche modeling (ENM) and the cost-distance approach. Our results revealed that the limited gene flow and multiple-population structure in O. splendida and the single-population structure in O. ishikawae were maintained after divergence of the species through ancient vicariance between islands. We found that genetic distance correlated with geographic distance between populations of both species. Our landscape genetic analysis revealed that the connectivity of suitable habitats influences gene flow and leads to the formation of specific population structures. In particular, different degrees of topographical complexity between islands are the major determining factor for shaping contrasting population structures of two species. In conclusion, our results illustrate the diversification mechanism of organisms through the interaction with space and environment. Our results also present an ENM approach for identifying the key factors affecting demographic history and population structures of target species, especially endangered species.
Keywords: population genetics, ecological niche modeling, cost-path analysis, gene flow, microsatellite, endangered species
Introduction
One of the fundamental aspects for understanding speciation and genetic variation across spatial scales is isolation by distance (IBD), which was introduced by Wright (1943). IBD expresses genetic variation and gene flow as a function of distance and advocates greater genetic affinity between geographically close populations through genetic drift. On the other hand, the environment of an organism's local ecosystem is spatially heterogeneous. According to Wallace's classical biogeographical region theory (Wallace, 1876), the distribution of animals is spatially divided by environmental barriers, a fundamental principle underlying the dispersal of individuals. Even neighboring populations can be isolated and genetically differentiated when unfavorable landscapes prevent migration between populations. Therefore, identifying how landscapes affect the gene flow and population structure is an intrinsic aspect in population and conservation biology (Manel et al., 2003; Coulon et al., 2004; Spear et al., 2005; Stevens et al., 2006).
Recent advances in geospacial information technology and highly polymorphic genetic markers have provided the opportunity to approach this point. As a result, studies in the field of landscape genetics are conducted by integrating landscape ecology and population genetics (Manel et al., 2003; Storfer et al., 2007). One of the successful schemes for revealing how landscapes affect population structure is examining the correlation between genetic distance, estimated from molecular markers, and cost distance, estimated from hypothetical cost surfaces, to establish the effect of multiple environmental and landscape factors using geographic information systems (GIS) (Coulon et al., 2004; Spear et al., 2005; Stevens et al., 2006; Spear and Storfer, 2008; Wang et al., 2009). In this scheme, ‘costs' assignments are the most important and should be generated according to the mobility of target species (Storfer et al., 2007). However, direct observation of movements of individuals is often impossible in field studies (Wang et al., 2009). Therefore, ‘costs' are predetermined by authors by incorporating ecological and behavioral evidence (Coulon et al., 2004; Spear et al., 2005; Stevens et al., 2006; Spear and Storfer, 2008) or by enumerating every combination of cost assignment (Wang et al., 2009). However, the ecology and behavior of some species, including endangered species, is not always clear. In addition, even if some information is known, it can be difficult to incorporate observation information based on environmental and landscape factors onto the cost-matrix layer. One of the promising alternatives to predetermination is integration of ecological niche modeling (ENM) for cost assignment (Graham et al., 2004; Wang et al., 2008; Richards-Zawacki, 2009).
ENM predicts geographical distribution of a species by mathematically representing its known environmental and geographic distribution with habitat suitability values on a map while integrating complex environmental variables (Elith and Leathwick, 2009). Results can then be used to generate the cost surface by inverting the suitability to cost value of each cell. This approach is useful for species that have insufficient ecological information and can be influenced by unanticipated environmental factors. In addition, ENM can be a powerful tool for studying endangered species because it estimates the extent of suitable and potentially suitable habitats, including areas where populations are extinct.
Island species, particularly archipelago species, provide intrinsic study objects for illustrating how landscapes can affect and shape population structures through comparison of islands with other types of landscapes. Typically, archipelago populations of terrestrial animals, including amphibians, diverge following continuous isolation due to fragmentation of land bridges by tectonic deformation and/or strait formation. Subsequently, different environments and landscapes will shape local population structures on each island like a natural ‘experiment' (MacArthur and Wilson, 1967). Unfortunately, most island species are endangered and populations are in decline (Frankham et al., 2002).
Ishikawa's frogs, Odorrana ishikawae and O. splendida, are examples of such endangered island species and are endemic to the Amami and Okinawa Islands, respectively, in southwestern Japan. These are sister species of family Ranidae, and O. splendida was recently split from O. ishikawae based on morphological differences, genetic differences and reproductive isolation due to hybrid inviability and abnormal spermatogenesis in hybrids (Kuramoto et al., 2011). Recent habitat loss due to deforestation and development has caused serious population declines in both species, which are particularly sensitive to habitat modification and loss because of their intrinsically small habitats in mountain torrents surrounded by forests. Therefore, O. ishikawae has been listed as a class B1 endangered species in the IUCN Red List of Threatened Species (Kaneko and Matsui, 2011) and designated as a natural asset in both Okinawa and Kagoshima prefectures. No O. ishikawae have been observed in the Nago and Motobu areas in the middle of Okinawa Island since 1993, and populations in these areas are believed to be extinct (Utsunomiya, 1995). Despite the current knowledge, the ecology of both species is not yet fully understood. With regards to the breeding ecology of O. ishikawae, eggs are typically deposited in water in underground holes located along the slopes of rocky upper streams in forest regions (Katsuren et al., 1977; Utsunomiya et al., 1979). However, the ecology of adults of both species in the non-breeding season has not been revealed due to infrequent observation of individuals (Utsunomiya, 1995). The study of the population structures of these two Ishikawa's frogs provides a unique opportunity to identify how environments affect gene flow and conservation through habitat connectivity and natural demography.
In this study, we examine the gene flow, population structure and demographic history of Ishikawa's frogs. Causal factors for shaping population structure are estimated by examining the correlation between genetic distance and least cost distance based on suitability values of ENM using six environmental variables. Finally, we discuss how each environmental variable influences gene flow and leads to the formation of contrasting population structures.
Materials and methods
Sample collection
Sampling was conducted annually during spring from 2004 to 2010 in mountain torrents of Amami (O. splendida) and Okinawa (O. ishikawae) islands across the current distribution range of each species. We used a sampling scheme that enabled us to clarify genetic divergence and test the effects of environmental structures on genetic structure. Therefore, we treated adjacent localities belonging to the same river system as single sites. In Santaro (SAN) and Mt Nangou (NAN), we sampled adult O. splendida individuals that emerged onto a paved road near the mountain torrents.
In total, we collected 127 individuals of O. splendida from 12 sites and 33 individuals of O. ishikawae from six sites across their current distribution areas (Figure 1; Table 1). Toes of adults and tails of tadpoles were clipped once collected. To avoid collecting descendants of only a few parents, we tried to collect multiple adults and tadpoles from multiple tributaries in each locality during the breeding season. However, multiple specimens were sampled in some populations, but only tadpoles were sampled at Shinokawa (SHI) and Mt Kanmuri (KAN) because no adults were observed during the non-breeding season and rarely or not repeatedly even during the breeding season at some sites, in addition to the endangered status of both species. Furthermore, no individuals were observed at some sites of O. splendida after 2010 because landslides unfortunately caused by the record torrential rain inflicted destructive damage on some sites in Amami Isl. (O. splendida) in October 2010.
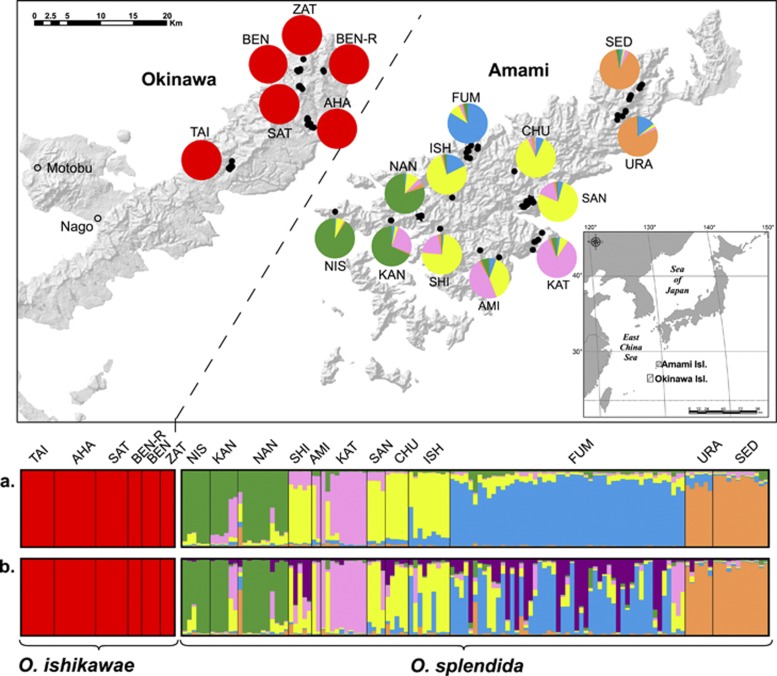
Figure 1.
Sampling localities of O. ishikawae (Okinawa Island) and O. splendida (Amami Island) and assigned genetic clusters from STRUCTURE. Map: sampling sites of individuals are indicated with solid circles on the shaded leaf maps of each island. Abbreviations are listed in Table 1. Colored pie charts show the total ratios of assigned genetic clusters of individuals in each population from STRUCTURE 2.3.3 (Hubisz et al., 2009) using the admixture model with the LOCPRIOR option. Color scheme: Graphical output of assigned genetic cluster from STRUCTURE 2.3.3. Each vertical bar represents an individual, and bars are divided into proportions of assigned genetic clusters based on (a) the admixture model with the LOCPRIOR option and (b) the admixture model without any priors.
Table 1. Genotypic data of 12 microsatellite loci in populations of O. ishikawae and O. splendid.
| Species | Population (abbreviations) | N | NA | HO | HE | FIS |
|---|---|---|---|---|---|---|
| O. ishikawae | Zatsun (ZAT) | 3 | 2.58 | 0.389 | 0.356 | −0.124 |
| Benoki (BEN) | 4 | 2.33 | 0.333 | 0.299 | −0.066 | |
| Benoki-River (BEN-R) | 3 | 2.67 | 0.444 | 0.380 | −0.161 | |
| Sate (SAT) | 7 | 3.50 | 0.393 | 0.347 | −0.138 | |
| Aha (AHA) | 9 | 4.33 | 0.370 | 0.397 | 0.087 | |
| Taiho (TAI) | 7 | 3.33 | 0.440 | 0.370 | −0.183 | |
| Mean | 5.5 | 3.12 | 0.395 | 0.358 | −0.0438 | |
| O. splendida | Sedome (SED) | 12 | 3.17 | 0.410 | 0.433 | 0.012 |
| Urakami (URA) | 6 | 2.67 | 0.375 | 0.363 | −0.023 | |
| Fukumoto (FUM) | 51 | 7.42 | 0.414 | 0.538 | 0.184 | |
| Ishira (ISH) | 9 | 5.33 | 0.500 | 0.499 | −0.044 | |
| Chuosen (CHU) | 5 | 3.50 | 0.467 | 0.447 | −0.098 | |
| Santaro (SAN) | 4 | 3.42 | 0.438 | 0.443 | −0.026 | |
| Katoku (KAT) | 10 | 4.25 | 0.424 | 0.458 | 0.030 | |
| Aminoko (AMI) | 2 | 2.08 | 0.292 | 0.344 | 0.162 | |
| Shinokawa (SHI) | 5 | 3.33 | 0.517 | 0.487 | −0.096 | |
| Mt Nangou (NAN) | 11 | 4.25 | 0.427 | 0.461 | 0.019 | |
| Mt Kanmuri (KAN) | 6 | 3.00 | 0.340 | 0.380 | 0.090 | |
| Nishikomi (NIS) | 6 | 3.25 | 0.361 | 0.418 | 0.168 | |
| Mean | 10.6 | 3.81 | 0.414 | 0.439 | 0.0315 |
Abbreviations: FIS, fixation index; HE, expected heterozygosity; HO, observed heterozygosity; N, number of individuals; nA, number of alleles.
Genotyping
DNA was extracted using the DNeasy Tissue and Blood Kit (Qiagen, Hilden, Germany), and 12 microsatellite loci were examined (Igawa et al., 2011). Loci were amplified using KOD FX (TOYOBO, Osaka, Japan) or ExTaq (TAKARA BIO, Otsu, Japan) and the method was described in Igawa et al. (2011). Polymerase chain reaction was conducted using a Veriti 200 thermal cycler (Applied Biosystems, Life Technologies, Foster City, CA, USA) and a GeneAmp 9700 system (Applied Biosystems). Amplified fragments were electrophoresed on an ABI3130xl analyzer (Applied Biosystems), and allele sizes were determined using GeneScan LIZ 500 (Applied Biosystems) as an internal size standard and genotyped using GeneMapper 4.0 (Applied Biosystems). We used MICRO-CHECKER V2.2.3 (Van Oosterhout et al., 2004) to check for the presence of null alleles. To test for repeatability in microsatellite scoring, we repeated all steps from amplification through scoring on a set of 32 samples (20% of total sample size). We explored the departure from HWE (Hardy–Weinberg equilibrium) and LD (linkage disequilibrium) of our loci using GENEPOP 4.0 (Rousset, 2008) and Genalex (Peakall and Smouse, 2006). The tests for HWE and LD were assessed using the Markov chain algorithm (Guo and Thompson, 1992).
Population structure and gene flow analysis
To explore population structure, we used the individual-based approach (which does not assume population) and the population-based approach. In the individual-based approach, we used Bayesian clustering implemented in STRUCTURE 2.3.3 (Hubisz et al., 2009), which assigns genetic population units of each individual. To explore the population structure of each species, we conducted separate analyses and varied the number of clusters (K) from 1 to 10. We used admixture models for MCMC (Markov Chain Monte Carlo) inference with and without prior information on the locality of samples (LOCPRIOR). We ran 1 000 000 MCMC repetitions after discarding the first 100 000 iterations as burnin data. Ten simulations were completed for each estimated K. To estimate a realistic K value, we analyzed our results according to the method of Evanno et al. (2005), in which log-likelihood values and their variance from ten repetitive runs for each K were used to calculate ΔK. This parameter is an ad hoc statistic used in simulations to accurately identify the true number of genetic clusters at the uppermost level of hierarchical structure. It also provides an objective alternative to selecting K instead of simply choosing a K value with the highest log probability, which tends to overestimate K (Evanno et al., 2005).
For the population-based approach, AMOVA (analysis of molecular variance), pairwise FST, and the exact test of population differentiation were performed using Arlequin ver 3.5.1.2 (Excoffier and Lischer, 2010). As mentioned above, some populations had small sample sizes. Therefore, we carefully checked the results from the population-based approach for any biased data due to a small sample size. Pairwise genetic distance and a phylogenetic tree based on the DA distance of Nei et al. (1983) with 1000 bootstrap iterations were performed using POPTREE2 (Takezaki et al., 2010).
To infer demographic history, we focused on the migration rate between populations of each species. To estimate gene flow between populations, we used the following two methods: a genetic assignment method implemented in BAYESASS+ v1.3 (Wilson and Rannala, 2003); and a coalescent method implemented in MIGRATE v3.1.6 (Beerli and Palczewski, 2010). Although both methods estimate asymmetrical dispersal rates, genetic assignment methods typically measure recent dispersal rates, while coalescent methods estimate long-term averages (Pearse and Crandall, 2004). BAYESASS+ uses fully Bayesian MCMC resampling to estimate migration rate (m), allele frequency (P) and inbreeding value (F) as variables. We ran a total of 3 000 000 MCMC iterations and sampled the chain every 2000 iterations, discarding the first 1 000 000 iterations as burnin data. The chains were compared for convergence by performing multiple runs with different initial random seeds. We checked that accepted numbers of proposed changes were between 40 and 60% in accordance with BAYESASS+ 1.3 documentation.
MIGRATE v3.1.6 uses coalescent simulations to estimate pairwise migration rates (M=m/μ, where m=dispersal rate per generation and μ=mutation rate per generation) and effective population size (θ=4Neμ) for each population (Beerli and Palczewski, 2010). We averaged the results from three independent runs using the program's option to combine the results of the last chains of each run (Beerli and Palczewski, 2010). Each run used 10 short chains with 50 000 generations, sampled every 20 generations, and eight long chains with 5000 generations, sampled every 20 generations. We used the infinite alleles model (IAM) of microsatellite mutation with default heating to swap between chains and Gelman's R statistic to assess convergence.
We first conducted analyses among all samples without separating either species. However, the estimated m values between populations of different species using BAYESASS+v1.3 (Wilson and Rannala, 2003) were not larger than the values with uninformative data. In addition, although the results from MIGRATE v3.1.6 showed partial slight migration between populations of different species, values varied between independent runs. Therefore, we focused on the gene flow among populations in each species.
Bottleneck detection
To detect molecular signatures of historical bottlenecks, we used two methods implemented in BOTTLENECK V1.2.02 (Cornuet and Luikart, 1996; Piry et al., 1999) and M-RATIO (Garza and Williamson, 2001). BOTTLENECK investigates deviations from expected heterozygote excess relative to allelic diversity. During population bottlenecks, rare alleles are lost due to drift at a faster rate than loss of heterozygosity (Nei et al., 1975), and BOTTLENECK utilizes this disparity to detect past bottlenecks. We performed analyses under all three available microsatellite mutational models: IAM, SMM (stepwise mutational model), and TPM (two-phase mutational model) with 80% single-step mutations and 20% multiple-step mutations. M-RATIO calculates the statistical significance of the M-statistic in each population compared with that of a simulated population (Garza and Williamson, 2001). In this test, M is the ratio between the number of alleles at a given locus and the total range of allele size. Again, because rare alleles are lost more regularly during a population bottleneck, M will be reduced in populations that have undergone a significant population size reduction, unless all rare alleles are in the tails of the allele size distribution. We parameterized the TPM used in M-RATIO by assigning an 80% rate for single-step mutations, with mean values of 2.8 repeats for the size change of multiple-step mutations, as found in the literature (Garza and Williamson, 2001). We performed calculations under three θ (4 Neμ) values (0.05, 0.1 and 0.5). Because the sample size was less than that of the mean (6.5), we excluded ZAT, BEN and BEN-R of O. ishikawae and URA, CHU, SAN, AMI, SHI, KAN and NIS of O. splendida from the analyses.
ENM and landscape genetics
To clarify the environmental factors that shape population structures, we employed least cost-path analyses (LCPA) and ENM. Least cost path (LCP) is a function implemented in GIS for determining the shortest path from source to destination on the cost surface. With appropriate cost assignment for movement of target species, LCP shows likely dispersal corridors (Storfer et al., 2007; Wang et al., 2009). To generate cost layers for LCPA, we translated suitability values from ENM into cost by assuming that habitat suitability should have a similar cost to movements on a larger scale (Hirzel et al., 2006; Wang et al., 2008; Richards-Zawacki, 2009; Wang and Summers, 2010). For selecting appropriate models, we constructed cost layers accounting for 63 estimated models based on combinations of the six environmental variables and compared the cost layers with those predicted by our genetic analysis (Supplementary Table S4). Analysis was conducted as follows: (1) inference of expected habitat suitability from combinations of six environmental variables by ENM; (2) estimation of LCP, where cost is the inverse of habitat suitability [inferred in (1)] and (3) comparative analysis of correlations between LCP distance [estimated in (2)] and genetic distance estimated by genetic analysis.
We first performed ENM analysis using MAXENT V3.3.1 (Phillips et al., 2006), which estimates habitat suitability based on environmental variables via maximum entropy modeling. To conduct ENM and landscape genetics, we selected the following six geographical and ecological GIS layers that potentially influence habitat suitability and movement of frogs: elevation, slope angle, watershed, terrain wetness index (TWI), soil and vegetation. Elevation was prepared using a 10-m grid DEM (digital elevation map) provided by The Geospatial Information Authority of Japan. Slope angle was prepared using a spatial analyst extension implemented in ArcGIS 9.3 (ESRI, Redlands, CA, USA). TWI was prepared in the same manner by using the following equation (Wilson and Gallant, 2000):
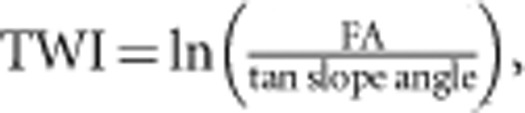 where FA is flow accumulation calculated by estimating water coverage on the DEM using ArcGIS. FA was also used to estimate watershed after extraction of various threshold FA values (10, 100 and 1000) to determine ecologically meaningful data for estimation. We calculated watershed based on >100 FA values in the following analyses because of the high ROC (receiver operating characteristic) values. Vegetation was prepared using a 1:50 000 (equivalent to a 500-m grid) vegetation map (Natural Environmental Information GIS, 6th and 7th ed., Biodiversity Center, Nature Conservation Bureau, Ministry of the Environment). Soil was prepared using a 1 200 000 (equivalent to a 2-Km grid) soil classification map (Basic Land Classification Survey, National Land Survey Division, Land and Water Bureau, Ministry of Land, Infrastructure, Transport and Tourism). Because MAXENT requires the same resolution layers, we prepared various resized layers (10, 50 and 250 m grid scales) and confirmed almost the same results on the 10- and 50-m grid scale using all six environmental variables. We then used the 50-m grid scale for following ENM and LCPA. We compiled lists of 26 and 35 occurrence localities of O. ishikawae and O. splendida based on our own sample collection points. Because our goal was to find the causal environmental factors explaining population structures, we used all combinations of six variables for each species. This resulted in a total of 63 suitability models (Supplementary Table S5). Values in each model range from 0 to 1, and higher scores indicated a more suitable habitat. To verify the accuracy of estimation, we performed ROC analysis to test how well the distribution model discriminates between the presence and absence by randomly selecting 25% of occurrence records and 10 000 cells as background points (Phillips et al., 2006).
where FA is flow accumulation calculated by estimating water coverage on the DEM using ArcGIS. FA was also used to estimate watershed after extraction of various threshold FA values (10, 100 and 1000) to determine ecologically meaningful data for estimation. We calculated watershed based on >100 FA values in the following analyses because of the high ROC (receiver operating characteristic) values. Vegetation was prepared using a 1:50 000 (equivalent to a 500-m grid) vegetation map (Natural Environmental Information GIS, 6th and 7th ed., Biodiversity Center, Nature Conservation Bureau, Ministry of the Environment). Soil was prepared using a 1 200 000 (equivalent to a 2-Km grid) soil classification map (Basic Land Classification Survey, National Land Survey Division, Land and Water Bureau, Ministry of Land, Infrastructure, Transport and Tourism). Because MAXENT requires the same resolution layers, we prepared various resized layers (10, 50 and 250 m grid scales) and confirmed almost the same results on the 10- and 50-m grid scale using all six environmental variables. We then used the 50-m grid scale for following ENM and LCPA. We compiled lists of 26 and 35 occurrence localities of O. ishikawae and O. splendida based on our own sample collection points. Because our goal was to find the causal environmental factors explaining population structures, we used all combinations of six variables for each species. This resulted in a total of 63 suitability models (Supplementary Table S5). Values in each model range from 0 to 1, and higher scores indicated a more suitable habitat. To verify the accuracy of estimation, we performed ROC analysis to test how well the distribution model discriminates between the presence and absence by randomly selecting 25% of occurrence records and 10 000 cells as background points (Phillips et al., 2006).
We then used the inverse of these estimated values for LCP analysis (because lower suitability should have a higher cost) to assign cost to each cell on the cost layer (Hirzel et al., 2006; Wang et al., 2008; Richards-Zawacki, 2009; Wang and Summers, 2010). We conducted LCP analysis on these cost layers for each species by using the Spatial Analyst extension in ArcGIS. Pairwise LCP distances were calculated between six populations of O. ishikawae and 11 populations of O. splendida (excluding AMI due to the small sample size and scattered collecting sites). The center point of each collecting site in each population was input into the GIS.
Correlation of LCP with the Euclidean distance matrix and the genetic distance matrix was investigated using Mantel's test (Mantel, 1967). We used linearized FST values (Rousset, 1997) for genetic distance because r values correlate better with Euclidean distance and LCP distances than with other indexes. The tests were conducted using Fstat version 2.9.3 (Goudet, 2001). Correlation significance was determined through 2000 random matrix permutations. Then, to identify the most appropriate habitat suitability model for explaining population structures, we used AIC (Akaike, 1973; Burnham and Anderson, 2002; Spear et al., 2005) after selecting the top 10 models by r value.
Results
Genotypic data
For the 12 microsatellite markers examined in this study, the total number of alleles ranged from 2 to 48 with a mean of 15.917 per locus. The number of alleles varied from 1 to 23 and from 1 to 35, with mean values of 6.167 and 11.167 per locus in O. ishikawae and O. splendida, respectively (Appendix I). Of these, the mean number of private alleles was 4.750 (77.02%) and 9.975 (87.30%) in O. ishikawae and O. splendida, respectively. In particular, two alleles of OishiP-6 were completely segregated between the species, but were fixed within each population. Although the observed heterozygosity was similar between both species (0.394 and 0.419 in O. ishikawae and O. splendida, respectively), the fixation index (F) was largely different (0.003 and 0.175 in O. ishikawae and O. splendida, respectively) (Appendix I). Microchecker (Van Oosterhout et al., 2004) indicated null alleles in six loci (OishiM-12, OishiM-18, OishiM-20, OishiP-7, OishiP-9 and OishiP-12) in some populations. We conducted 141 HWE tests representing every polymorphic locus/population combination (α=0.000355 after Bonferroni correction) and detected significant deviation from HWE in five tests of four loci (OishiM-18, OishiM-20, OishiP-7 and OishiP-9). Significant LD was detected only in the Fukumoto population (FUM) at the combination of Oishi-M20/OishiP-9 (α=0.000123 after Bonferroni correction). Because none of these loci (OishiM-12, OishiM-18, OishiM-20, OishiP-7, OishiP-9 and OishiP-12) showed significant LD in most populations, we did not exclude them from analysis. We scored each individual twice to confirm consistent results between runs. Although we tried twice to amplify some loci of specific individuals, we could not unambiguously score 1% of genotypes, which were subsequently classified as missing data. In each population, there were only minor differences in allelic diversity (nA), observed heterozygosity and expected heterozygosity (Ho and He) and fixation index (FIS) (Table 1). The greater number of alleles in FUM can be explained by the larger sample size (n=51). We subsequently conducted analyses based on two data sets: with and without the loci that deviated from the HWE and the samples with null alleles. Then, we confirmed that the results were qualitatively similar in both cases.
Population structure and gene flow estimation
Pairwise FST and Nei's DA distances showed great genetic divergence between populations of different species (mean FST=0.517±0.001 and mean DA=0.934±0.001) (Supplementary Table S2). AMOVA in O. ishikawae indicated no population structure, showing molecular variation of −0.82% among populations (P=0.80) and 100.82% within populations. On the other hand, AMOVA in O. splendida revealed significant population structure, showing molecular variation of 9.12% among populations (P<0.01) and 90.88 within populations. Pairwise FST and Nei's DA distances showed similar results (Supplementary Table S2). Within O. ishikawae, almost all pairwise FST values were negative (FST=−0.081–0.021) with a mean FST of −0.015±0.001 and pairwise DA distances of <0.2 (DA=0.080–0.174) with a mean DA of 0.139±0.001. Whereas, within O. splendida, pairwise FST and DA were higher and varied more widely (FST=0.004–0.231 and DA=0.116–0.449) than those of O. ishikawae with a mean FST of 0.111±0.003 and DA of 0.295±0.005. A neighbor-joining tree based on the DA distance showed clusters consisting of geographically neighboring populations of O. splendida (Supplementary Figure S2). However, the relationships between clusters of O. splendida and between populations of O. ishikawae were ambiguous, except for the paraphyly of ZAT compared with other populations.
The results of Bayesian clustering analysis implemented in STRUCTURE v2.3.3 (Hubisz et al., 2009) are summarized in Supplementary Figure S1. In O. ishikawae, mean posterior probability values did not differ for each K value, and ΔK from K=2 to K=9 was ambiguous in both admixture models with and without LOCPRIOR, which implies no population structure in O. ishikawae (K=1). Whereas in O. splendida, although the highest mean posterior probability value (L(K)) was K=6 in the admixture model and K=9 in the admixture model with LOCPRIOR, ΔK supported K=6 in the admixture model and K=5 in the admixture model with LOCPRIOR, which are most likely true genetic clusters (Supplementary Figure S1). At K=5 in the admixture model with LOCPRIOR in O. splendida, there was a cluster group in the northern population of SED and URA, a second group in the western population FUM, a third group in the central population of ISH, CHU, SAN and SHI, a fourth group in the south eastern population KAT, and a fifth group in the south western (Sotsukou peninsula) population of NAN, KAN and NIS. The two individuals of AMI presented different estimated ancestries of south eastern (KAT) and central (ISH, CHU, SAN and SHI) populations, respectively. At K=6 in the admixture model in O. splendida, although the general framework of clustering did not differ from the admixture model with LOCPRIOR, FUM and SHI were included as two major estimated ancestries.
BAYESASS+ detected low migration rates between TAI and the other O. ishikawae populations (m=0.140–0.242), but migration rates were not significant between remaining populations from zero (mean m=0.021). This trend was not seen in the results of MIGRATE, which showed low variation in migration rate between all populations pairs (Supplementary Table S3). For O. splendida, low migration rates were detected between the following neighboring populations: from SED to URA (m=0.128), from FUM to ISH, CHU and SAN (m=0.165, 0.097 and 0.050), and from KAT to SAN, AMI and SHI (m=0.067, 0.049 and 0.088). Unexpectedly higher migration rates were also detected between the following geographically distant populations: from SED to NAN, KAN and NIS (m=0.195, 0.117 and 0.129) (Supplementary Table S4).
Bottleneck detection
A Wilcoxon test implemented in BOTTLENECK (Piry et al., 1999) revealed a significant excess of heterozygosity compared with the expected equilibrium (P<0.05) in the following populations of O. splendida: FUM (P=0.019 in the IAM model) and NAN (P=0.016 in the SMM model) (Table 2). M-RATIO (Graza and Wiliamson, 2001) found a significant population bottleneck signal in one population of O. ishikawae, AHA (under all θ values), and in four population of O. splendida, SED (under all θ values), FUM (under θ=0.05 and 0.1), KAT (under all θ values) and NAN (under all θ values). There were no significant bottleneck signals in other cases (Table 2).
Table 2. Summary of bottleneck tests implemented in BOTTLENECK and M-RATIO.
| Species | Pop. | BOTTLENECK | M-RATIO | ||||
|---|---|---|---|---|---|---|---|
| |
|
IAM |
TPM |
SMM |
θ=0.05 |
θ=0.1 |
θ=0.5 |
| O. ishikawae | SAT | 0.139 | 0.095 | 0.119 | 0.069 | 0.083 | 0.204 |
| AHA | 0.106 | 0.103 | 0.066 | <10–3 | <10–3 | 0.001 | |
| TAI | 0.491 | 0.496 | 0.068 | 0.080 | 0.089 | 0.070 | |
| O. splendida | SED | 0.254 | 0.518 | 0.483 | <10–3 | <10–3 | <10–3 |
| FUM | 0.019 | 0.255 | 0.270 | 0.027 | 0.031 | 0.071 | |
| KAT | 0.448 | 0.074 | 0.140 | <10–3 | <10–3 | 0.036 | |
| NAN | 0.510 | 0.083 | 0.016 | 0.017 | 0.002 | 0.010 | |
For BOTTLENECK, three mutation models were used: infinity allele model (IAM), two-phase mutation model (TPM) and stepwise mutation model (SMM). For M-RATIO, three θ values were used: 0.05, 0.1 and 0.5. Values in bold type indicate statistical significance (P<0.05). ZAT, BEN and BEN-R of O. ishikawae and URA, CHU, SAN, AMI, SHI, KAN and NIS of O. splendida were excluded due to small sample sizes (n<7).
ENM and least cost-path analysis
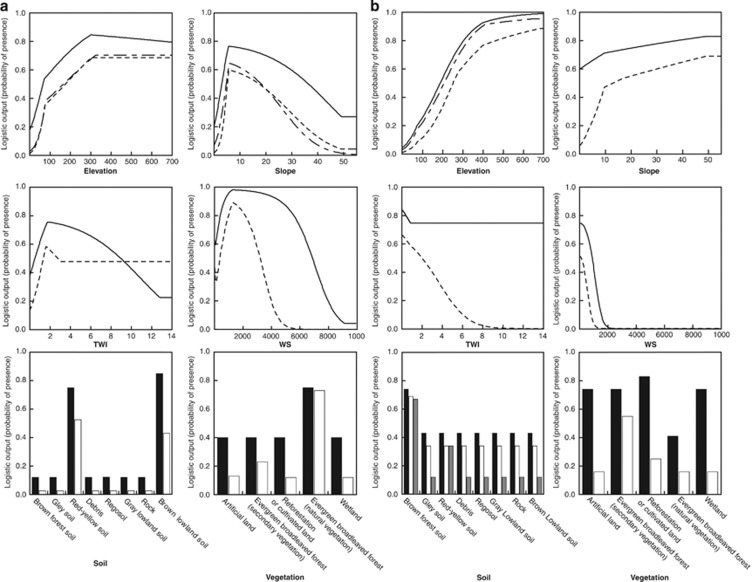
The 63 ecological niche models for each species are summarized in Supplementary Table S5. The area under the curve (AUC) values for ROC varied from 0.656 to 0.951 (mean 0.869) in O. ishikawae and from 0.670 to 0.905 (mean 0.825) in O. splendida (Supplementary Table S5). The AUC values in full models using all variables were 0.951 and 0.903 in O. ishikawae and O. splendida, respectively, indicating good fitting to the species distribution model (Phillips et al., 2006). The estimated suitability map was congruent with the known distribution areas of each species (Utsunomiya, 1995) (Supplementary Figure S3). For O. ishikawae, higher suitability values were also well distributed in the Nago and Motobu areas, where populations had been previously observed, but not since 1993 (Utsunomiya et al., 1979). The assigned probabilities of each variable were also consistent with the current knowledge of O. ishikawae and O. splendida. According to the full model of O. ishikawae, the probability of each variable showed that high elevation, low slope angle, moderate TWI and watershed, moisture rich soils (red-yellow and brown lowland soils) and evergreen broad-leaved forest were assigned as high presence probabilities of O. ishikawae (Figure 2a). While according to the full model of O. splendida, high elevation, small watershed, brown-forest soil, and reforestation or land cultivation were assigned differently as high presence probabilities (Figure 2b). O. ishikawae dwells in upper streams in the mountain area covered by natural evergreen forest, and their spawning is observed in holes around the river source area (Katsuren et al., 1977; Utsunomiya et al., 1979), while O. splendida similarly dwells in river bed water in mountain areas (Oumi, 2006), covered mainly by secondary evergreen forest (Miyawaki, 1989), and is also found on the border of the forest and on cultivated land (Koba, 1955). However, some models based on single variables showed lower AUC values (Supplementary Table S5), indicating different probability assignments of each variable compared with full models (TWI and watershed in O. ishikawae and slope and TWI in O. splendida) (Figure 2). Although these results seemed to be poor for predicting species distribution, we used them for constructing cost surfaces because we could not negate the possibility that they affected population structures. By inverting the probability of the 63 ecological niche models, we obtained cost-surface layers consisting of cells with assigned cost values varying from 1 to 100.
Figure 2.
Assigned probabilities of the presence for each environmental variable. The graphs show assigned probabilities for six environmental variables [elevation, slope, TWI, watershed (WS), soil and vegetation] in MAXENT V3.3.1 (Phillips et al., 2006) for (a) O. ishikawae and (b) O. splendida. Solid, dashed and dash-dot lines indicate the probabilities in full models using all six variables, models using only a single variable, and best-fitting models explaining population structures, respectively. Black, white and gray rectangles in bar graphs also indicate the probabilities in full models, models with a single variable and best-fitting models, respectively.
Landscape genetics
We found linear correlation between linearized FST and Euclidean distance (Supplementary Figure S4A), suggesting an IBD pattern in both species. Regression lines drawn through the scatterplot of these two variables showed a higher slope within O. splendida than within O. ishikawae. Mantel's test on these distance matrixes indicated a high Mantel's r value (r=0.657) and significant correlation in O. splendida (P=0.001; Table 3), but despite the high Mantel's r value (r=0.597), there was no significant correlation in O. ishikawae (P=0.064). A series of Mantel's tests examining the correlation between LCP and genetic distance indicated generally positive correlations. For O. ishikawae, Mantel's r value varied from 0.592 to 0.611 depending on the LCP distance model (Supplementary Table S5), and the top 10 models with values of >0.607 were selected as candidates of best-fitting models (Table 3). However, these models showed only marginal correlation with genetic distance (P>0.05), except for LCP distance by the elevation+slope angle and elevation+vegetation models, which showed a significant correlation (P=0.043 and P=0.046, respectively). Because the AICc values of the elevation+slope angle model (−47.187) were smaller than those of the elevation+vegetation model (−44.913), the former model was selected as the best-fitting model for explaining genetic structure (Table 3; Figure 3). For O. splendida, Mantel's r values varied from 0.648 to 0.699 (Supplementary Table S5) and all LCP distance models showed a significant correlation with genetic distances (P=0.001). Within the top 10 models with values of >0.674, LCP distance by the elevation+soil+vegetation and elevation+soil models had the highest Mantel's r value (r=0.699), and the latter was selected as the best-fitting model with the smallest AICc value (−138.033) (Table 3; Figure 3).
Table 3. Summary of resultant values from ecological niche modeling (AUC and variable contribution), Mantel's tests (Mantel's r value, P-value) and AICc.
| Candidate model | AUC | Variable contribution (%) | Mantel's r | P-value | AICc | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| |
|
Elevation |
Slope |
TWI |
Watershed |
Soil |
Vegetation |
|
|
|
| O. ishikawae | ||||||||||
| Elevation+soil+slope+TWI | 0.923 | 28.1 | 13.6 | 16.4 | – | 41.9 | – | 0.611 | 0.073 | −40.209 |
| Elevation+soil+slope | 0.884 | 32.2 | 19.8 | – | – | 47 | – | 0.611 | 0.054 | −42.630 |
| Elevation | 0.770 | 100.0 | – | – | – | – | – | 0.610 | 0.075 | −47.187 |
| Elevation+slope | 0.826 | 69.5 | 30.5 | – | – | – | – | 0.610 | 0.043 | −44.945 |
| Elevation+soil | 0.853 | 44.4 | – | – | – | 55.6 | – | 0.609 | 0.058 | −44.937 |
| Elevation+soil+TWI | 0.902 | 38.7 | – | 13.1 | – | 48.2 | – | 0.608 | 0.066 | −42.596 |
| Elevation+soil+vegetation | 0.917 | 17.4 | – | – | – | 29.8 | 52.8 | 0.608 | 0.067 | −42.594 |
| Elevation+soil+slope+watershed | 0.933 | 26.7 | 15.8 | – | 23.1 | 34.4 | – | 0.608 | 0.068 | −40.063 |
| Elevation+vegetation | 0.889 | 29.0 | – | – | – | – | 71.0 | 0.607 | 0.046 | −44.913 |
| Elevation+soil+slope+TWI+watershed | 0.940 | 27.1 | 9.9 | 5.1 | – | 33.5 | – | 0.607 | 0.063 | −37.632 |
| Full model | 0.951 | 17.1 | 5.6 | 2.7 | 12.2 | 23.5 | 38.9 | 0.604 | 0.053 | – |
| Euclidian | – | – | – | – | – | – | – | 0.597 | 0.064 | – |
| O. splendida | ||||||||||
| Elevation+soil+vegetation | 0.900 | 75.4 | – | – | – | 18.2 | 6.5 | 0.699 | 0.001 | −135.740 |
| Elevation+soil | 0.891 | 80.9 | – | – | – | 19.1 | – | 0.699 | 0.001 | −138.033 |
| Elevation+soil+slope | 0.900 | 80.5 | 1.3 | – | – | 18.2 | – | 0.698 | 0.001 | −135.687 |
| Elevation+soil+slope+TWI | 0.898 | 79.9 | 0.8 | 0.2 | – | 19.1 | – | 0.698 | 0.001 | −133.218 |
| Elevation+soil+slope+vegetation | 0.905 | 76.4 | 1.0 | – | – | 15.9 | 6.7 | 0.697 | 0.001 | −133.148 |
| Elevation+soil+TWI | 0.893 | 80.6 | – | 0.4 | – | 19.0 | – | 0.697 | 0.001 | −135.635 |
| Elevation+soil+slope+TWI+vegetation | 0.901 | 74.6 | 1.6 | 0.2 | – | 16.2 | 7.4 | 0.696 | 0.001 | −130.598 |
| Elevation+soil+TWI+vegetation | 0.900 | 75.6 | – | 0.4 | – | 16.9 | 7.1 | 0.695 | 0.001 | −133.051 |
| Elevation+soil+slope+watershed | 0.896 | 77.2 | 0.7 | – | – | 20.7 | 1.3 | 0.676 | 0.001 | −131.857 |
| Elevation+soil+slope+vegetation+watershed | 0.902 | 74.4 | 0.8 | – | 1.4 | 16.3 | 7.1 | 0.674 | 0.001 | −131.731 |
| Full model | 0.903 | 73.3 | 1.2 | 0.2 | 2.3 | 15.8 | 7.2 | 0.669 | 0.001 | – |
| Euclidian | – | – | – | – | – | – | – | 0.657 | 0.001 | – |
Abbreviations: AUC, area under the curve; TWI, terrain wetness index.
Values in bold type indicate statistical significance (P<0.05) and the lowest AICc values within the models are statistically significant in Mantel's test.
Figure 3.
Cost-matrix layers and inferred LCP among populations in best-fitting models for each species. The maps shows cost-matrix layers for O. ishikawae (Okinawa Isl.: left) and O. splendida (Amami Isl.: right). Warmer colors indicate lower costs of movements. Open circles indicate center points of populations, and lines indicate estimated LCP among the points.
Discussion
Population structure and gene flow
Our genotypic data revealed a clear divergence between O. splendida and O. ishikawae. The percentage of private alleles in each species, FST estimates, and the phylogenetic tree all indicated a fairly high level of divergence between species. Especially in OishiP-6, different fixed alleles between species were also observed (Supplementary Table S1). These results were supported by the previous mitochondrial (16S rRNA) geneology, which showed monophyletic clades of each species and higher nucleotide divergence between species (1.44–2.16%) than those within species (0–0.18% within O. ishikawae and 0–0.54% within O. splendida) (Kuramoto et al., 2011). Matsui et al. (2005) estimated the time of this split during late Pliocene to early Pleistocene (3.2–1.5 Ma) using molecular dating based on mitochondrial genes (12S and 16S rRNA). They also presumed that the ancestor of O. ishikawae and O. splendida had already reached the northernmost range of O. splendida by the middle to late Miocene (18.0–7.9 Ma) via an ancient land bridge connecting the Eurasian continent with the present area of the Ryukyu Archipelago, and the ancestor subsequently diverged into two species following the isolation of the Ryukyu Archipelago during the late Miocene to the middle Pliocene (Kimura, 2000). Taken together, our results and mitochondrial data show that both species likely diverged through ancient vicariance to two islands and became completely isolated without gene flow between them.
Furthermore, our genetic cluster assignment revealed that the contrasting population structures were maintained within each species: all populations of O. ishikawae were grouped into a single genetic cluster, while populations of O. splendida were divided into multiple genetic clusters (Figure 1). This is obvious from AMOVA and the fixation index within species (Appendix I), which detected a significant population structure in O. splendida and notably higher F values for O. splendida than for O. ishikawae. Assuming complete isolation of the species after vicariance and no recent bottlenecks in either species, higher migration rates between populations of O. ishikawae than those of O. splendida were expected. However, estimates of gene flow between populations are globally low for both species. For O. ishikawae, only recent directional migration from Taiho (TAI) to other populations was detected. The TAI population is located on the southern end of the present distribution of O. ishikawae across Mt Yonaha, which is the highest mountain in Okinawa. Thus, the population structure of O. ishikawae should be maintained through recent directional migration from the south to the north, suggesting source-sink dynamics (Pulliam, 1988). Gene flow between O. splendida populations is limited to only between neighboring populations, suggesting that each genetic group as assigned by structure is highly isolated. Especially for the southwest populations in Sotsukou Peninsula (NAN, KAN and NIS) Kuramoto et al. (2011) reported larger body size of individuals from these populations (large type) than that from the other O. splendida populations (common type). Our genetic assignment and phylogenetic analyses grouped corresponding populations of these two morphotypes into distinct genetic clusters and monophyletic clades (Figure 1; Supplementary Figure S2), which suggests that these morphotypes are genetically different and that their morphological differences are possibly derived from genetic differences. However, mitochondrial geneology by the same authors showed paraphyly and small nucleotide divergences between haplotypes in these populations. Although there is no record of observation or ecological evidence, this disparity might be explained by the detected gene flow from SED (common type) to the southwest population (large type) (Supplementary Table S4). Alternatively, this might be due to mitochondrial introgression and/or incomplete lineage sorting during the demographic and evolutionary history of O. splendida.
Bottleneck effects
We found evidence of population bottlenecks in AHA in O. ishikawae and all populations under almost all conditions in M-RATIO, but only two populations of O. splendida under the fraction of conditions in BOTTLENECK (Table 2). Because BOTTLENECK and M-RATIO measure different properties of bottlenecks, our results do not necessarily contradict each other. Simulations have shown that BOTTLENECK is more effective for identifying populations that have recently experienced a severe reduction in population size (Piry et al., 1999), while M-RATIO is likely to identify a bottleneck that occurred in the distant past, when the reduction in population size was fairly severe, and also when the effective population size of ancestral population is large (Williamson-Natesan, 2005). Thus, our results indicate the genetic signature of a bottleneck in the distant past, specifically in O. splendida, but there is little signature in the recent past in either species.
Regarding the conservation of Ishikawa's frogs, there are multiple ongoing issues for the declining population sizes: habitat loss due to deforestation and development, over exploitation for the pet trade (Kaneko and Matsui, 2011), and predation by the invasive small Indian mongoose, Herpestes javanicus (Watari et al., 2008). However, the influence of these issues causing population declines was not reflected in our results from a genetic aspect. Even before these issues became prominent, Ishikawa's frog populations were smaller than those of other frog species on the two islands (Okada, 1931; Koba, 1955). Therefore, our results suggest that the present populations of O. splendida have not yet fully recovered from bottlenecks in the distant past and have maintained small sizes due to the limited habitat and distribution range of the small islands. On the other hand, the influence of bottlenecks in O. ishikawae was probably limited, even in the distant past.
Landscape genetics
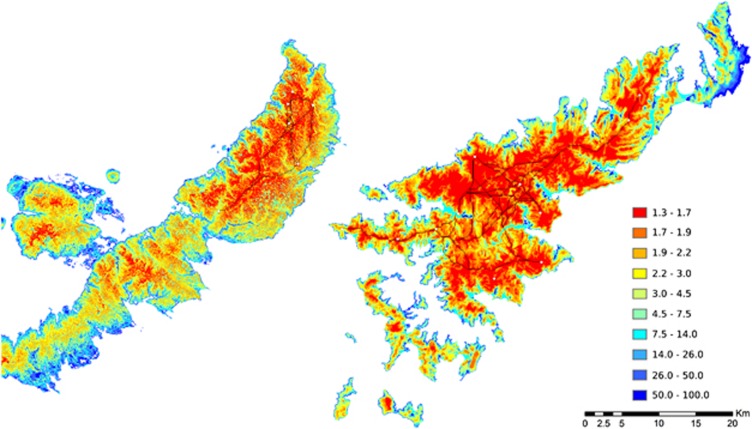
Our results showed a correlation between genetic and Euclidean geographic distances in both species, showing that the genetic diversity of Ishikawa's frogs could be explained by IBD (Supplementary Figure S4A). However, the patterns of population structures in each species are different as described in previous section (Figure 1), suggesting the existence of environmental and landscape features that limit migration and gene flow between populations in O. splendida. This is also apparent because correlation with genetic distance was improved when habitat suitability was incorporated into the distance between populations as the costs for movement (Table 3). Enumerated top 10 models in both species included elevation as a major factor contributing to population structure. This strongly suggests that topographical effects, particularly connectivity of high-elevation mountain areas, are a major factor for shaping population structure of both species, which is in agreement with topographical features on the islands in the present study: a single mountain mass expanding north–south in the northern area of Okinawa Island and multiple mountain masses divided by lowlands and rivers on Amami Island (Supplementary Figure S5). The effect of different environments and landscapes on the two islands is also obvious from the slope of regression lines when plotting genetic distance and LCP distance, instead of Euclidian distance, on best-fitting models (Supplementary Figure S4B), suggesting that the models successfully correct topological difference between the two islands by incorporating the effects of landscapes and environments on the rate of gene flow. Funk et al. (2005) proposed the ‘valley-mountain' model, based on data from Columbia spotted frogs, Rana luteiventris. This model determines the population structure of amphibians that inhabit mountain ridges by measuring the high level of gene flow between lowland populations and restricted gene flow between high-elevation populations and between low- and high-elevation populations. Although our results partially follow this model, the population structures of the two frog species in our study are the result of more extreme isolation of populations in different mountain ridges with a high level of gene flow between the same mountain ridges and no gene flow between lowlands. This difference can be accounted for by the different ecological history and suitable niche of Ishikawa's frogs inhabiting only high elevations and that of Columbia spotted frogs inhabiting a variety of habitats ranging from low to high elevation.
The best-fitting models for each species differed in detail. For O. splendida, soil type was selected as a secondary factor, and brown-forest soil was assigned as higher suitability and low cost for movement, in the best-fitting model (Table 3; Figure 2b). Because of its rich wetness and organic compound for sustaining broad-leaf forests, this assignment is also reasonable, considering the moisture-requiring physiology of amphibians. In the southwest islands of Japan, red-yellow soil, which has low humidity and few organic compounds, is zonal soil which is predominantly distributed in hilly regions near coasts and lowlands, while brown-forest soil is limited to mountain areas (Matsui and Isotani, 1990). Thus, the distribution of soil type should also affect migration between disjunctive mountain ridges leading to isolated populations of O. splendida. Whereas for O. ishikawae, although soil type was not included in the best-fitting model, it was included in the other selected models as a major contributing factor, and red-yellow soil or brown lowland soil was assigned as higher suitability in the single and full models (Table 3; Figure 2a). This difference in soil between the two species could be attributed to the different distribution of soil between the mountain areas (>100 m in elevation) of each island: 43% of brown-forest soil and 54% of red-yellow soil in Amami Isl., and 85% of red-yellow soil and 14% of brown lowland soil in Okinawa Isl. In addition, the present distribution area of O. ishikawae called ‘Yambaru' is an area on a single mountain mass that is globally covered with natural broad-leaf evergreen forests that has high rainfall and humidity, in contrast to the mountain area of Amami Isl., which is covered with secondary forest. Thus, red-yellow soil in this area did not serve as a strong barrier against migration, leading to population isolation in O. ishikawae.
On the other hand, slope angle was selected as a secondary factor in O. ishikawae, while lower slope angle was assigned as higher habitat suitability and low costs for movements in the best-fitting model (Table 3; Figure 2a), suggesting that a steep slope is a factor that consistently inhibits gene flow in amphibians (Funk et al., 2005; Spear et al., 2005; Lowe et al., 2006; Giordano et al., 2007). Meanwhile for O. splendida, steeper angle was oppositely assigned as slightly higher habitat suitability in the full and single models (Figure 2b). This difference in relationship between the species is also attributable to different distribution of slope angles between the mountain areas (>100 m in elevation) of the two islands: attenuation distribution from lower to higher angle (mean 11.88 °±7.27 °) in Okinawa Isl., and near normal distribution in Amami Isl. (mean 19.49 °±8.56 °). Because of steeper slope in the mountain area of Amami Isl., O. splendida might have adapted steeper slopes, and slope angle might not have been important for habitat selection or migration of O. splendida, showing a lower contribution ratio of slope angle in the full and the selected models (Table 3).
From a conservation perspective, fragmented populations have a smaller population size and carry a higher extinction risk than a single, large unfragmented population due to the significant deleterious effects on inbreeding and loss of genetic diversity (Frankham et al., 2002). As mentioned above, each population of O. splendida has been highly isolated, and the area inhabited by each population should be smaller than that of O. ishikawae. Thus, the extinction risk of each genetic cluster of O. splendida should potentially be high and is likely to be affected by extinction through deforestation. Therefore, further ecological observation studies to confirm our dispersal model and continuous monitoring activity of Ishikawa's frogs are needed.
The present study revealed that two sister species of Ishikawa's frog were diverged by vicariance to form contrasting population structures on separate islands. Together, these population structures of both species were shaped by the IBD model, but the differences between the two species were caused by the different patterns of suitable habitat connectivity between islands. Particularly, because of the important effects of elevation on habitat suitability, habitats fragmented by complicated topography of multiple mountain mosses divided by lowlands on Amami Island have shaped multiple genetic clusters of O. splendida, while the continuous habitat consisting of simple topography of a single mountain mass on Okinawa Island has maintained a single genetic cluster of O. ishikawae. In addition, soil type and slope angle have had a different effect on each species and have thus contributed to shaping contrasting population structures. Regarding conservation implications, the bottleneck tests show that severe population size reduction occurred in O. splendida in the distant past, but not in the recent past. To conclude the impact of the population decline of Ishikawa's frogs, further comprehensive studies integrating surveys from other aspects, such as population estimates, are needed. Furthermore, our ENM and landscape genetic analysis approach suggested that the extinct populations of O. ishikawae should be genetically different populations from present populations, which potentially have an extinction risk due to small suitable habitats. Finally, by integrating, genetic data, ENM, and LCPA, we were able to elucidate the detailed scenario, leading to contrasting population structures in the island frogs O. ishikawae and O. splendida. The ENM approach will be a strong tool for planning future comprehensive managements of target species together with surrounding ecosystems, especially for endangered species facing population fragmentation.
Data archiving
Our genotype data were deposited in Dryad.
Acknowledgments
We thank the Board of Education of Okinawa and Kagoshima Prefectures for permission to collect the materials used in this study. We are also grateful to Professor H Ota, Institute of Natural and Environmental Sciences, University of Hyogo, Japan, for his kindness in providing samples of O. ishikawae from Okinawa, Japan. We also thank Professor T Fujii, Faculty of Human Culture and Science, Hiroshima Prefectural University, Japan, and Mr N Sato, Institute for Amphibian Biology, Graduate School of Science, Hiroshima University, Japan for sample collection in the field and Dr Y Watari, Japan Forest Technology Association for useful suggestion for ENM analyses. This work was supported by a Grant-in-Aid for Scientific Research (B and C) (No. 24310173 and No. 20510216) to MS and a Grant-in-Aid for Young Scientists (B) (No. 23710282) to TI from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on Heredity website (http://www.nature.com/hdy)
Supplementary Material
References
- Akaike H.1973Information theory and an extension of the maximum likelihood principleIn: Petrov BN, Csaki F, (eds)Second International Symposium on Information Theory Akademiai Kiado: Budapest; 267–281. [Google Scholar]
- Beerli P, Palczewski M. Unified framework to evaluate panmixia and migration direction among multiple sampling locations. Genetics. 2010;185:313–326. doi: 10.1534/genetics.109.112532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR.2002Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach2nd edn., Springer: New York [Google Scholar]
- Cornuet JM, Luikart G. Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics. 1996;144:2001–2014. doi: 10.1093/genetics/144.4.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon A, Cosson JF, Angibault JM, Cargnelutti B, Galan M, Morellet N, et al. Landscape connectivity influences gene flow in a roe deer population inhabiting a fragmented landscape: an individual-based approach. Mol Ecol. 2004;13:2841–2850. doi: 10.1111/j.1365-294X.2004.02253.x. [DOI] [PubMed] [Google Scholar]
- Elith J, Leathwick JR. Species distribution models: ecological explanation and prediction across space and time. Annu Rev Ecol Evol Syst. 2009;40:677–697. [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Lischer HEL. Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour. 2010;10:564–567. doi: 10.1111/j.1755-0998.2010.02847.x. [DOI] [PubMed] [Google Scholar]
- Frankham R, Ballou JD, Briscoe DA. Introduction to Conservation Genetics. Cambridge University Press: New York; 2002. [Google Scholar]
- Funk WC, Blouin MS, Corn PS, Maxell BA, Pilliod DS, Amish S, et al. Population structure of Columbia spotted frogs (Rana luteiventris) is strongly affected by the landscape. Mol Ecol. 2005;14:483–496. doi: 10.1111/j.1365-294X.2005.02426.x. [DOI] [PubMed] [Google Scholar]
- Garza JC, Williamson EG. Detection of reduction in population size using data from microsatellite loci. Mol Ecol. 2001;10:305–318. doi: 10.1046/j.1365-294x.2001.01190.x. [DOI] [PubMed] [Google Scholar]
- Giordano AR, Ridenhour BJ, Storfer A. The influence of altitude and topography on genetic structure in the long-toed salamander (Ambystoma macrodactulym) Mol Ecol. 2007;16:1625–1637. doi: 10.1111/j.1365-294X.2006.03223.x. [DOI] [PubMed] [Google Scholar]
- Goudet J.2001FSTAT, a program to estimate and test gene diversities and fixation indices (version 2.9.3). http://www2.unil.ch/izea/softwares/fstat.html .
- Graham CH, Ron SR, Santos JC, Schneider CJ, Moritz C. Integrating phylogenetics and environmental niche models to explore speciation mechanisms in dendrobatid frogs. Evolution. 2004;58:1781–1793. doi: 10.1111/j.0014-3820.2004.tb00461.x. [DOI] [PubMed] [Google Scholar]
- Guo SW, Thompson EA. Performing the exact test of Hardy-Weinberg proportion for multiple alleles. Biometrics. 1992;48:361–372. [PubMed] [Google Scholar]
- Hirzel AH, Lelay G, Helfer V, Randin C, Guisan A. Evaluating the ability of habitat suitability models to predict species presences. Ecol Modell. 2006;199:142–152. [Google Scholar]
- Hubisz MJ, Falush D, Stephens M, Pritchard JK. Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour. 2009;9:1322–1332. doi: 10.1111/j.1755-0998.2009.02591.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igawa T, Okuda M, Oumi S, Katsuren S, Kurabayashi A, Umino T, et al. Isolation and characterization of twelve microsatellite loci of endangered Ishikawa's frog (Odorrana ishikawae) Conserv Genet Resour. 2011;3:421–424. [Google Scholar]
- Kaneko Y, Matsui M.2011Odorrana ishikawae. In: IUCN (ed) IUCN 2011. IUCN Red List of Threatened Species. (Version 2011. 1) . www.iucnredlist.org .
- Katsuren S, Tanaka S, Ikehara S.1977A brief observation on the breeding site and eggs of a frog, Rana ishikawae (Stejneger) in Okinawa IslandIn: Ikehara S, (ed)Ecological Studies of Nature Conservation of the Ryukyu Islands—(III). Naha Okinawa, University of Ryukyus: Okinawa 49–54.
- Kimura M. Palaeogeography of the Ryukyu Islands. Tropics. 2000;10:5–24. [Google Scholar]
- Koba K.1955Herpetological Fauna of Amamioshima, Loo Choo Islands Mem Fac Edu Kumamoto Univ 3145–162.in Japanese. [Google Scholar]
- Kuramoto M, Satou N, Oumi S, Kurabayashi A, Sumida M. Inter- and intra- island divergence in Odorrana ishikawae (Anura, Ranidae) of the Ryukyu Archipelago of Japan, with description of a new species. Zootaxa. 2011;2767:25–40. [Google Scholar]
- Lowe WH, Likens GE, McPeek MA, Buso DC. Linking direct and indirect data on dispersal: isolation by slope in a headwater stream salamander. Ecology. 2006;87:334–339. doi: 10.1890/05-0232. [DOI] [PubMed] [Google Scholar]
- MacArthur RH, Wilson EO. The Theory of Island Biogeography. Princeton University Press: New Jersey; 1967. [Google Scholar]
- Manel S, Schwartz MK, Luikart G, Taberlet P. Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol. 2003;18:189–197. [Google Scholar]
- Mantel N. The detection of disease clustering and a generalized regression approach. Cancer Res. 1967;27:209–220. [PubMed] [Google Scholar]
- Matsui K, Isotani T.1990Different nature of the hills across north and southIn: Matsui K, Takeuchi K, Tamura T, (eds)Natural Environment in Hills Kokon shoin: Tokyo; 5–10.in Japanese. [Google Scholar]
- Matsui M, Shimada T, Ota H. Multiple invasions of the Ryukyu Archipelago by Oriental frogs of the subgenus Odorrana with phylogenetic reassessment of the related subgenera of the genus. Rana Mol Phylogenet Evol. 2005;37:733–742. doi: 10.1016/j.ympev.2005.04.030. [DOI] [PubMed] [Google Scholar]
- Miyawaki A.1989Gebietsmonographie und räumliche Ordnung der VegetationIn: Miyawaki A, (ed)Vegetation of Japan Okinawa & Ogasawara Shibundo: Tokyo; 465–539.in Japanese. [Google Scholar]
- Nei M, Maruyama T, Chakraborty R. The bottleneck effect and genetic variability in populations. Evolution. 1975;29:1–10. doi: 10.1111/j.1558-5646.1975.tb00807.x. [DOI] [PubMed] [Google Scholar]
- Nei M, Tajima F, Tateno Y. Accuracy of estimated phylogenetic trees from molecular data. II. Gene frequency data. J Mol Evol. 1983;19:153–170. doi: 10.1007/BF02300753. [DOI] [PubMed] [Google Scholar]
- Okada Y. The Tailless Batrachians of the Japanese Empire. Imperial Agricultural experiment station: Tokyo; 1931. [Google Scholar]
- Oumi S.2006Reproduction of Ishikawa's frog on Amami Oshima Island Bulletin of The Herpetological Society of Japan 2006104–108.in Japanese. [Google Scholar]
- Peakall R, Smouse PE. genalex 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes. 2006;6:288–295. doi: 10.1093/bioinformatics/bts460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearse DE, Crandall KA. Beyond FST: analysis of population genetic data for conservation. Conserv Genet. 2004;5:585–602. [Google Scholar]
- Phillips S, Anderson R, Schapire R. Maximum entropy modeling of species geographic distributions. Ecol Modell. 2006;190:231–259. [Google Scholar]
- Piry S, Luikart G, Cornuet JM. BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered. 1999;90:502–503. [Google Scholar]
- Pulliam HR. Sources, sinks and population regulation. Am Nat. 1988;132:652–661. [Google Scholar]
- Richards-Zawacki CL. Effects of slope and riparian habitat connectivity on gene flow in an endangered Panamanian frog, Atelopus varius. Divers Distrib. 2009;15:796–806. [Google Scholar]
- Rousset F. Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics. 1997;145:1219. doi: 10.1093/genetics/145.4.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousset F. Genepop'007: a complete reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour. 2008;8:103–106. doi: 10.1111/j.1471-8286.2007.01931.x. [DOI] [PubMed] [Google Scholar]
- Spear SF, Peterson CR, Matocq MD, Storfer A. Landscape genetics of the blotched tiger salamander (Ambystoma tigrinum melanostictum) Mol Ecol. 2005;14:2553–2564. doi: 10.1111/j.1365-294X.2005.02573.x. [DOI] [PubMed] [Google Scholar]
- Spear SF, Storfer A. Landscape genetic structure of coastal tailed frogs (Ascaphus truei) in protected vs. managed forests. Mol Ecol. 2008;17:4642–4656. doi: 10.1111/j.1365-294X.2008.03952.x. [DOI] [PubMed] [Google Scholar]
- Stevens VM, Verkenne C, Vandewoestijne S, Wesselingh RA, Baguette M. Gene flow and functional connectivity in the natterjack toad. Mol Ecol. 2006;15:2333–2344. doi: 10.1111/j.1365-294X.2006.02936.x. [DOI] [PubMed] [Google Scholar]
- Storfer A, Murphy MA, Evans JS, Goldberg CS, Robinson S, Spear SF, et al. Putting the “landscape” in landscape genetics. Heredity. 2007;98:128–142. doi: 10.1038/sj.hdy.6800917. [DOI] [PubMed] [Google Scholar]
- Takezaki N, Nei M, Tamura K. POPTREE2: software for constructing population trees from allele frequency data and computing other population statistics with windows interface. Mol Biol Evol. 2010;27:747–752. doi: 10.1093/molbev/msp312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsunomiya T.1995. Rana ishikawae. In: Ministry of Agriculture,Forestry and Fisheries (ed) Basic Data for Rare Wild Aquatic Organisms (II) Japan Fisheries Resource Conservation Association: Tokyo; 429–475.in Japanese. [Google Scholar]
- Utsunomiya Y, Utsunomiya T, Katsuren S. some ecological observations of Rana ishikawae, a rare frog endemic to Ryukyu Islands. Proc Jpn Acad Ser B Phys Biol Sci. 1979;55:233–237. [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. Micro-Checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes. 2004;4:535–538. [Google Scholar]
- Wallace AR. The Geographical Distribution of Animals: With a Study of the Relations of Living and Extinct Faunas as Elucidating the Past Chances of the Earth's Surface. Vol 1. Harper & Brothers: New York; 1876. [Google Scholar]
- Wang IJ. Fine-scale population structure in a desert amphibian: landscape genetics of the black toad (Bufo exsul) Mol Ecol. 2009;18:3847–3856. doi: 10.1111/j.1365-294X.2009.04338.x. [DOI] [PubMed] [Google Scholar]
- Wang IJ, Savage WK, Shaffer HB. Landscape genetics and least-cost path analysis reveal unexpected dispersal routes in the California tiger salamander (Ambystoma californiense) Mol Ecol. 2009;18:1365–1374. doi: 10.1111/j.1365-294X.2009.04122.x. [DOI] [PubMed] [Google Scholar]
- Wang IJ, Summers K. Genetic structure is correlated with phenotypic divergence rather than geographic isolation in the highly polymorphic strawberry poison-dart frog. Mol Ecol. 2010;19:447–458. doi: 10.1111/j.1365-294X.2009.04465.x. [DOI] [PubMed] [Google Scholar]
- Wang Y-H, Yang K-C, Bridgman CL, Lin L-K. Habitat suitability modelling to correlate gene flow with landscape connectivity. Landsc Ecol. 2008;23:989–1000. [Google Scholar]
- Watari Y, Takatsuki S, Miyashita T. Effects of exotic mongoose (Herpestes javanicus) on the native fauna of Amami-Oshima Island, southern Japan, estimated by distribution patterns along the historical gradient of mongoose invasion. Biol Invasions. 2008;10:7–17. [Google Scholar]
- Williamson-Natesan EG. Comparison of methods for detecting bottlenecks from microsatellite loci. Conserv Genet. 2005;6:551–562. [Google Scholar]
- Wilson GA, Rannala B. Bayesian inference of recent migration rates using multilocus genotypes. Genetics. 2003;163:1177–1191. doi: 10.1093/genetics/163.3.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson JP, Gallant JC. Terrain Analysis: Principles and Applications. John Wiley & Sons: New Jersey; 2000. [Google Scholar]
- Wright S. Isolation by distance. Genetics. 1943;28:114–318. doi: 10.1093/genetics/28.2.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.