Background: The ParA superfamily of polymerizing ATPases mediates DNA segregation ubiquitously in bacteria.
Results: Hyperactive ATPase variants of the ParA homolog, ParF, are defective in polymerization and segregation.
Conclusion: Conserved residues situated neither in canonical ATP motifs nor at interfaces implicated in ParF polymerization are crucial for protein function.
Significance: The architecture of the ParA nucleotide-binding pocket is key to precise DNA segregation.
Keywords: ATPases, Bacteria, Escherichia coli, Microbiology, Molecular Biology, Protein Self-assembly, ParA, ParF, Partition, Plasmid
Abstract
DNA segregation in bacteria is mediated most frequently by proteins of the ParA superfamily that transport DNA molecules attached via the segrosome nucleoprotein complex. Segregation is governed by a cycle of ATP-induced polymerization and subsequent depolymerization of the ParA factor. Here, we establish that hyperactive ATPase variants of the ParA homolog ParF display altered segrosome dynamics that block accurate DNA segregation. An arginine finger-like motif in the ParG centromere-binding factor augments ParF ATPase activity but is ineffective in stimulating nucleotide hydrolysis by the hyperactive proteins. Moreover, whereas polymerization of wild-type ParF is accelerated by ATP and inhibited by ADP, filamentation of the mutated proteins is blocked indiscriminately by nucleotides. The mutations affect a triplet of conserved residues that are situated neither in canonical nucleotide binding and hydrolysis motifs in the ParF tertiary structure nor at interfaces implicated in ParF polymerization. Instead the residues are involved in shaping the contours of the binding pocket so that nucleotide binding locks the mutant proteins into a configuration that is refractory to polymerization. Thus, the architecture of the pocket not only is crucial for optimal ATPase kinetics but also plays a key role in the polymerization dynamics of ParA proteins that drive DNA segregation ubiquitously in procaryotes.
Introduction
Accurate segregation of genetic material during cytokinesis is a key biological process in all organisms (1). However, only a fragmented understanding has been attained of the molecular mechanisms that drive genome partitioning in bacteria. Bacterial homologs of eucaryotic actin and tubulin that mediate segregation of certain plasmids have provided important insights into the partition process (2–6). In contrast, the mechanisms by which the ParA superfamily of P-loop ATPases mediate segregation are less well defined. The ubiquitous ParA proteins are involved most widely in bacterial genome segregation, including partitioning of low copy number antibiotic resistance and virulence plasmids, as well as of chromosomes (7–14). Current knowledge suggests that ATP-induced polymerization of ParA proteins either may propel tethered chromosomes or plasmids away from the central division zone or retraction of ParA filaments by polymer disassembly may draw attached genomes toward the cell poles in a process that simulates the action of the eucaryote mitotic spindle (15–24). The nucleoid may function as a substratum on which plasmid partitioning occurs (25, 26).
Multiresistance plasmid TP228 originates from Salmonella enterica serotype Newport and replicates stably at low copy number in Escherichia coli. The TP228 segrosome comprises the parH centromere that is coated cooperatively by the ParG centromere-binding factor. ParG in turn recruits the ParA homolog, ParF (27, 28). ParF displays low level, intrinsic autopolymerization in the absence of nucleotide in vitro. Polymerization is promoted dramatically by ATP binding, which induces the formation of extensive multistranded filaments (15). An extended N-terminal flexible tail in ParG remodels these polymers in both the absence and the presence of ATP (19). Centromere-binding proteins with diverse primary sequences elicit equivalent polymerization responses with their cognate ParA homologs, indicating a conserved mechanism of polymerization enhancement within the ParA superfamily (16, 20–22). In contrast with ATP, ADP represses ParF polymerization, suggesting that subcellular plasmid movement is driven by a cycle of ParF polymer assembly and disassembly that reflects the nucleotide-bound state of the protein (15, 29).
Determination of ParF structures has shown recently that the protein bound to ADP is monomeric. In contrast, ParF bound to the nonhydrolyzable ATP analog AMPPCP7 forms dimers that pack into dimer of dimer building blocks that assemble into polymers (29). ParG possesses an arginine finger-like motif in its flexible N-terminal tail that stimulates ATP hydrolysis by ParF. This stimulation is crucial for accurate segregation by tuning the kinetics of ATP hydrolysis within ParF filaments (19). Despite these observations, unraveling how nucleotide hydrolysis, polymerization, and depolymerization in the pervasive ParA superfamily integrate during segregation and how these activities are modulated by centromere-binding factors remain major challenges.
We previously highlighted a triplet of residues that are situated outside of canonical ATP motifs but that are highly conserved among short ParA homologs (∼200 amino acids), including ParF (30) (see Fig. 1A). Here, we demonstrate that all three amino acids are crucial for maintaining normal ATP hydrolysis kinetics of ParF: remarkably, mutation of any of the residues elicited strong ATPase hyperactivity. Two of the amino acids are key to maintaining the integrity of the nucleotide-binding pocket, whereas the third residue buttresses the position of the Walker A motif. The hyperactive ParF mutants no longer respond to stimulation by the arginine finger-like motif in ParG and display perturbations in both autopolymerization and nucleotide-induced filamentation, leading to marked segregation defects in vivo. We speculate that nucleotide binding fails to transduce the conformational change that elicits dimerization and fixes the mutant proteins into a configuration that is recalcitrant to polymer assembly. The data establish that maintaining the architecture of the ParF nucleotide-binding pocket is critical for optimal ATPase kinetics but also is fundamental to the polymerization dynamics that dictate the DNA segregation process.
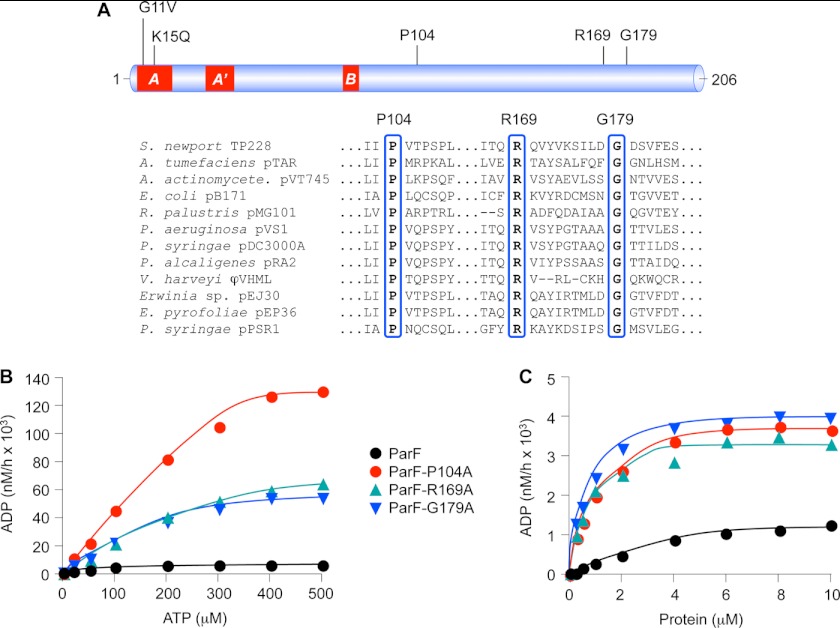
FIGURE 1.
Mutations in a triplet of conserved residues induce hyperactive ATPase profiles in ParF. A, linear representation of ParF with Walker A, A′, and B motifs highlighted in red. Mutation of Gly-11 and Lys-K15 residues in the A box ablates ATP hydrolysis (15). Positions of Pro-104, Arg-169, and Gly-179 residues and their conservation in selected ParF relatives (30) are shown. B, ATPase assays of wild-type ParF and derivatives bearing P104A, R169A, or G179A mutations. ATP hydrolysis is plotted with proteins (4 μm) at 0–500 μm ATP concentrations. C, ATPase assays of wild-type ParF and derivatives bearing P104A, R169A, or G179A mutations. ATP hydrolysis is plotted as a function of protein concentration with ATP at 5 μm. The data shown in A and B are typical results of experiments performed at least in duplicate.
EXPERIMENTAL PROCEDURES
Strains, Plasmids, and Recombinant DNA Techniques
E. coli DH5α (31) was used for plasmid propagation and cloning. Plasmids pFH450 (32) and pFH547 (30) are a segregation probe vector and the same plasmid harboring the parFGH cassette, respectively. Segregation assays were performed in E. coli BR825 polA (33) or KS3001 pcnB (34). A bacterial two-hybrid system (35) that was employed previously to monitor ParF-ParF and ParF-ParG associations (19, 27, 36) was used to assess the impact of ParF mutations on these interactions. The ParF and ParG proteins were produced from recombinant pET22b plasmids as described previously (27). Site-directed mutations of parF were constructed in pFH547, in two-hybrid vectors, and in expression plasmids by oligonucleotide swapping or by overlap extension PCR. Mutations were verified by sequencing.
Protein Purification and Biochemical Techniques
ParG and wild-type and mutated ParF were purified following overproduction as described previously (27). ATPase assays were performed with [α-35S]ATP, analyzed by thin layer chromatography, and quantified as detailed elsewhere (15). Assessment of two-hybrid interactions in vivo was determined by β-galactosidase assays. Polymerization of ParF and its derivatives was monitored by dynamic light scattering and sedimentation assays as outlined previously (15). Supershifting of ParG-parH complexes by ParF in gel retardation assays was examined as described recently using a biotinylated PCR product (28). Biotin end-labeled DNA was detected using chemiluminescent nucleic acid detection reagents (Thermo Scientific Pierce) (27). Fluorescence anisotropy measurements were performed in a Jovin-Yvon Horiba Fluoromax-3 spectrofluorimeter in a quartz microcuvette in a total volume of 150 μl. ATP binding activity of wild-type and mutated ParF proteins was assessed by anisotropy measurements of the fluorescent ATP analog MANT-ATP in 20 mm HEPES, 150 mm NaCl, 1 mm MgCl2, pH 7.0. The excitation wavelength (λex) and emission wavelengths (λem) were 356 and 442 nm, respectively. The MANT-ATP concentration was kept constant (0.9 μm), and the ParF concentration was increased from 0.25 to ∼5 μm. Ten measurements of fluorescence anisotropy were taken for each point, and the average value was plotted against ParF concentration. Fluorescence anisotropy of aromatic residues in ParF was also measured (λex = 280 nm; λem = 340 nm). Because of potential hydrolysis of MANT-ATP by ParF and mutants during measurements, a second set of ligand binding assays were done with the nonhydrolyzable MANT-ATPγS analog using the same conditions as for MANT-ATP. The data were analyzed by direct fitting to a single-site binding model using SigmaPlot.
RESULTS
Perturbation of the ParF Nucleotide-binding Pocket Induces Hyperactive ATP Hydrolysis
Residues Pro-104, Arg-169, and Gly-179 are highly conserved in ParF and related members of the ParA superfamily but are located distantly in the primary sequence from previously identified ATP binding and hydrolysis motifs (30) (Fig. 1A). The residues were changed individually to alanine, which did not appreciably disrupt the gross structure of the purified proteins based on circular dichroism analysis (supplemental Fig. S1).
ATPase kinetics of the purified proteins were examined. As demonstrated previously, wild-type ParF is a weak ATPase with a K0.5 value of ≈100 μm (15) (Fig. 1B). In contrast, the ParF-P104A, -R169A, and -G179A proteins displayed hyperactive ATPase profiles accompanied by modestly elevated K0.5 (≈150 μm). Notably, ParF-P104A hydrolyzed ATP more effectively at high substrate concentrations than either ParF-R169A or ParF-G179A proteins, suggesting a particularly fast catalytic rate for this mutant (Fig. 1C). Accordingly, the kcat values for mutant proteins were 10 to 20 times higher than wild-type ParF (Table 1). Analogously, when titrated with a fixed ATP concentration, the mutated proteins showed comparable profiles, depleting the substrate markedly faster than wild-type protein (Fig. 1B). The hyperactive mutants hereafter are collectively denoted ParFH for clarity.
TABLE 1.
Kinetic properties of wild-type and mutant ParF proteins
| Parameter | ParF | ParF-P104A | ParF-R169A | ParF-G179A |
|---|---|---|---|---|
| K0.5 | ∼100 μm | ∼150 μm | ∼150 μm | ∼120 μm |
| kcata | 1.6 | 32 | 15 | 14 |
| Minutes to hydrolyze one molecule of ATP | 37.5 | 1.875 | 4 | 4.285 |
a Molecules of ADP produced/molecule of protein in 60 min.
ParA ATPases possess three conserved motifs implicated in nucleotide binding and hydrolysis: deviant Walker A (P-loop), Walker A′, and Walker B motifs (37, 38) (Fig. 1A). Structures of monomeric ParF bound to ADP recently revealed that the protein adopts an α-β-α layered structure with a central seven-stranded β-sheet surrounded by several α-helices. The adenine nucleotide is situated on the C-terminal side of the parallel β-sheets and is recognized by nucleotide-binding motifs including the Walker A box (29). By contrast with ParF-ADP, ParF forms nucleotide sandwich dimers when bound to AMPPCP. These sandwich dimers pack into higher order, dimer of dimer units to produce a distinctive linear filament that reflects ParF polymer assembly (29).
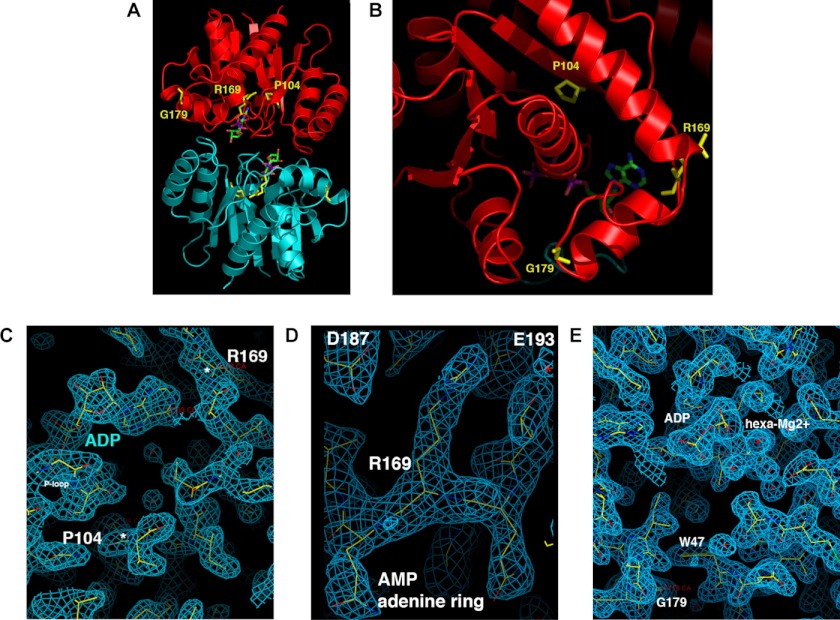
Conserved residue Pro-104 is part of a proline-rich patch in ParF. This patch inserts into a niche close to the adenine nucleotide-binding pocket of the adjacent subunit in ParF sandwich dimers (29). Considering its location near the Walker A motif in the ParF structure, the P104A change is expected to exert short range impacts on binding pocket conformation (Fig. 2, A–C). Arg-169 forms the “top” of the pocket with the amide nitrogen contacting the adenine N1 group (Fig. 2C). The arginine side chain is wedged between residues Asp-187 and Glu-193, which pins the short turn on which it is located into place. Thus, this side chain is vital in shaping the binding site (Fig. 2D). Although Gly-179 appears to be distant from the nucleotide-binding pocket (Fig. 2, A and B), altering the side chain at this location would clash with residue Trp-47. Trp-47 stacks against amino acids in the binding site and near the P-loop motif; changing the Gly-179 side chain is expected to induce a shift in the position of this motif (Fig. 2E). Thus, Pro-104, Arg-169, and Gly-179 are crucial for maintaining the architecture of the nucleotide-binding site. Changes at these positions are likely to modify nucleotide access and stability in the binding pocket, causing the hyperactivity of ParFH variants.
FIGURE 2.
Locations of residues Pro-104, Arg-169, and Gly-179 in the ParF crystal structure. A, ParF sandwich dimer bound to the nonhydrolyzable ATP analog AMPPCP (29). Monomers are shown in red and blue with locations of Pro-104, Arg-169, and Gly-179 highlighted on the former. B, close-up view of the Pro-104–Arg-169–Gly-179 triplet in monomeric ParF bound to ADP. C–E, wireframe representations in monomeric ParF bound to ADP of Pro-104 and Arg-169 with locations of the nucleotide and P-loop included, of Arg-169 and nearby residues Asp-187 and Glu-193, and of Gly-179 and Trp-47. The blue mesh represents the composite omit 2Fo-Fc map for the structure, contoured at 1σ.
Attenuated Nucleotide Binding by ParFH
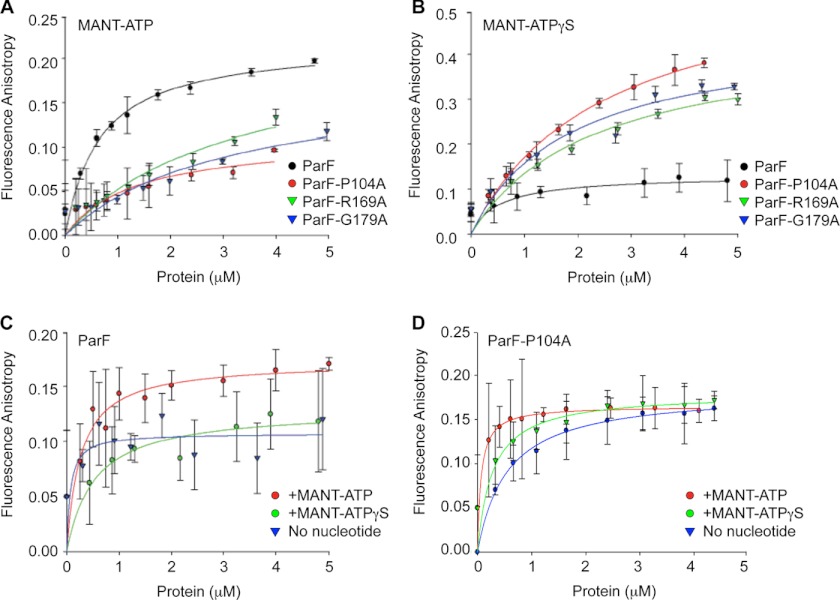
Nucleotide binding dynamics of ParF and ParFH were monitored by anisotropy of the fluorescent ATP analog MANT-ATP titrated with the four proteins (Fig. 3A). At low ParF concentrations, MANT-ATP anisotropy elevated sharply followed by a more gradual increase with an apparent Kd of ∼0.5 μm. By contrast, anisotropy values increased linearly with all three ParFH derivatives. Although saturation could not be reached with mutant proteins, anisotropy changes at the highest testable protein concentrations were ∼50% of wild-type protein (Fig. 3A).
FIGURE 3.
Fluorescence anisotropy studies of nucleotide binding by ParF and mutant proteins. A and B, anisotropy changes when MANT-ATP (0.9 μm) (A) or MANT-ATPγS (0.9 μm) (B) was titrated with increasing concentrations of wild-type ParF and ParFH proteins. C, intrinsic tryptophan fluorescence measurements of increasing concentrations of ParF without exogenous nucleotide or in the presence of MANT-ATP or MANT-ATPγS (0.9 μm each). D, intrinsic tryptophan fluorescence measurements of increasing concentrations of ParF-P104A without exogenous nucleotide or in the presence of MANT-ATP or MANT-ATPγS (0.9 μm each). The average fluorescence anisotropy values for 10 measurements for each point are shown.
In view of their dissimilar ATPase kinetics, differences in MANT-ATP anisotropy may reflect disparate rates of nucleotide binding or hydrolysis by wild-type and mutant proteins. Fluorescence anisotropy studies were performed with the nonhydrolyzable analog MANT-ATPγS to discriminate between these effects (Fig. 3B). Because there essentially is zero hydrolysis within the assay time scale, MANT-ATPγS anisotropy traces represent the true ligand binding curves. Wild-type ParF and ParFH each elicited a fast initial increase in MANT-ATPγS anisotropy. Values with ParF leveled off at concentrations >1 μm, yielding an apparent Kd of ∼0.5 μm, similar to that determined for MANT-ATP. In contrast, anisotropy in the presence of ParFH continued to increase with higher protein concentrations to give final values approximately twice that observed with wild-type ParF (Fig. 3B). The higher anisotropy with mutant proteins compared with wild-type ParF may indicate that the residency time of the fluorophore is longer or that its conformation is altered in the binding pockets of mutant proteins. Despite the higher MANT-ATPγS anisotropy, apparent Kd values for ParFH were >1 μm compared with ∼0.5 μm for wild-type protein. Thus, affinities of ParFH proteins for MANT-ATPγS are moderately reduced compared with wild-type ParF, revealing that the hyperactive ATPase profiles of mutant proteins arise from increased rates of hydrolysis rather than improved nucleotide binding.
Hyperactive ParF ATPase Function Disrupts DNA Segregation
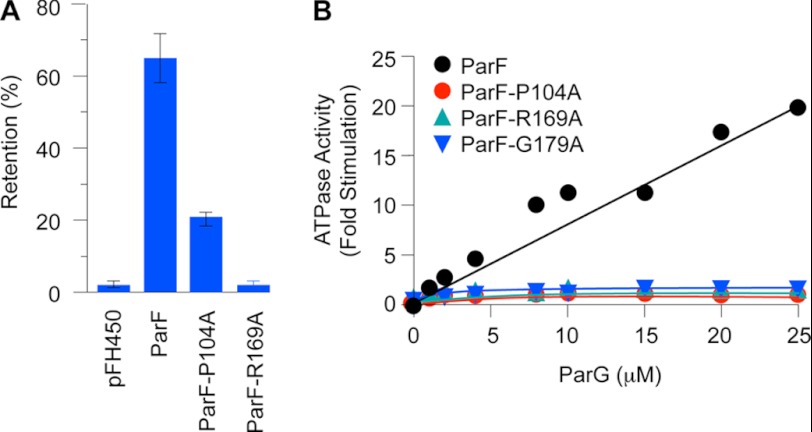
Partition assays with a low copy number vector were performed to assess the impact of ParFH mutations on DNA segregation. The vector is segregationally unstable in a polA host, showing <5% retention after ∼25 generations of nonselective growth (32). Insertion of the parFGH cassette improves plasmid retention levels to ∼70% (30) (Fig. 4A). Replacement of wild-type parF with the variant producing ParF-P104A reduced plasmid segregational stability markedly. The ParF-R169A protein exerted an even stronger effect, decreasing plasmid retention to the level of the empty vector (Fig. 4A).
FIGURE 4.
ATPase stimulation and segregation defects of hyperactive ParF ATPase mutants. A, partition assays of the segregation probe vector pFH450 (32) and the vector possessing either the wild-type parFGH cassette (pFH547) (30) or the same cassette bearing mutations that produce ParF-P104A and ParF-R169A. B, ATPase activities of ParFH mutants are not stimulated by ParG. Levels of ATP hydrolysis driven by ParF and mutated proteins as a function of ParG concentration are shown. ParF proteins were used at 0.5 μm. The data are expressed as fold stimulation of ATPase activity compared with basal activity without added ParG. The results in both panels are averages of at least three replicates.
A plasmid expressing parF-G179A could be propagated without any apparent detrimental effects at moderate copy number but failed repeatedly to produce transformant colonies at low copy number in the polA host. The ParF-G179A protein may exert a toxic effect so that the plasmid cannot be maintained at low copy number under selective conditions. The copy number of ColE1-like replicons is reduced by ∼90% in strains with pcnB mutations (39). ColE1-based plasmids either bearing the wild-type parFGH module or producing ParF-G179A were introduced equally efficiently into a pcnB strain. Partition tests showed that the plasmid possessing the intact parFGH cassette was retained in ∼75% of the population after ∼25 generations of nonselective growth. In contrast, the plasmid expressing parF-G179A was maintained in ∼35% of cells, a value similar to the empty vector. Thus, ParF-G179A confers a pronounced segregation defect that is detectable in a pcnB strain. In summary, residues Pro-104, Arg-169, and Gly-179 of ParF fulfill crucial roles in DNA segregation, with the G179A mutation additionally exerting a toxic effect on host cells under certain conditions.
Disruption of the ParF Nucleotide-binding Pocket Ablates ATPase Stimulation by the Arginine Finger-like Motif in ParG
The arginine finger-like loop in the ParG N-terminal flexible tail stimulates ATP hydrolysis by ParF ∼30-fold, which is critical for accurate DNA segregation (15, 19). Insertion of the motif in the ParF nucleotide-binding pocket is predicted to stabilize the transition state by neutralization of the negative charge that develops during phosphoryl transfer, akin to other arginine fingers. In view of the structural alterations induced in the nucleotide-binding pocket of ParFH mutants, the effects of these changes on enhancement of ATP hydrolysis by ParG were assessed. All three mutations entirely abolished the response to ParG (Fig. 4B). Thus, disrupting the integrity of the ParF nucleotide-binding pocket not only alters intrinsic ATPase kinetics (Fig. 1) but also interferes with correct alignment of the arginine finger-like motif supplied in trans by the ParG partner protein.
The interaction surfaces of ParG with ParF comprise both the N-terminal flexible tail bearing the arginine finger-like loop and the dimeric ribbon-helix-helix DNA-binding fold, from which a pair of these tails project (36, 40). ParF-ParG interactions can be monitored by a two-hybrid assay, either qualitatively by colony color on indicator growth media (19, 27, 36) or semiquantitatively by β-galactosidase assays. The ParF-ParG interaction is detectable as ∼150 β-galactosidase units compared with ∼20 units when ParG is absent (supplemental Fig. S2A). The ParF-P104A and -G179A proteins both elicited modest reductions in β-galactosidase levels compared with wild-type ParF, which may reflect less stable interactions with the ParG mobile tail. In contrast, interaction between ParF-R169A and ParG was significantly stronger than interaction of wild-type ParF with ParG (supplemental Fig. S2A). The ParG flexible tail may be tethered more rigidly in the nucleotide-binding pocket in ParF-R169A. Overall, the data show that binding pocket perturbations in ParF can elicit interactions with ParG that are moderately weaker or stronger but that in both cases lead to ablation of ATPase stimulation by the partner protein.
Polymerization of ParFH Mutants Is Inhibited Indiscriminately by Nucleotides
Although residues Pro-104, Arg-169, and Gly-179 are distant from ParF polymerization interfaces (29), the altered ATPase patterns provoked by changes at these positions may disrupt ParF polymerization dynamics. Two-hybrid analysis was employed initially to compare dimerization of wild-type and mutated proteins (27). Mutations were introduced into parF genes cloned in both bait and prey vectors and interactions assessed by β-galactosidase assays. ParF self-association produced ∼700 β-galactosidase units (supplemental Fig. S2B). Similar values were obtained both for self-association of ParF-G179A and for heterodimerization with wild-type ParF, revealing that the G179A change does not impair ParF subunit interactions in vivo (supplemental Fig. S2B). In contrast, self-association of ParF-R169A and interaction with wild-type protein both were detectably stronger than self-association of wild-type ParF, producing ∼1000–1200 β-galactosidase units. Conversely, ParF-P104A was unaffected in heterodimerization with wild-type ParF but did self-associate more weakly.
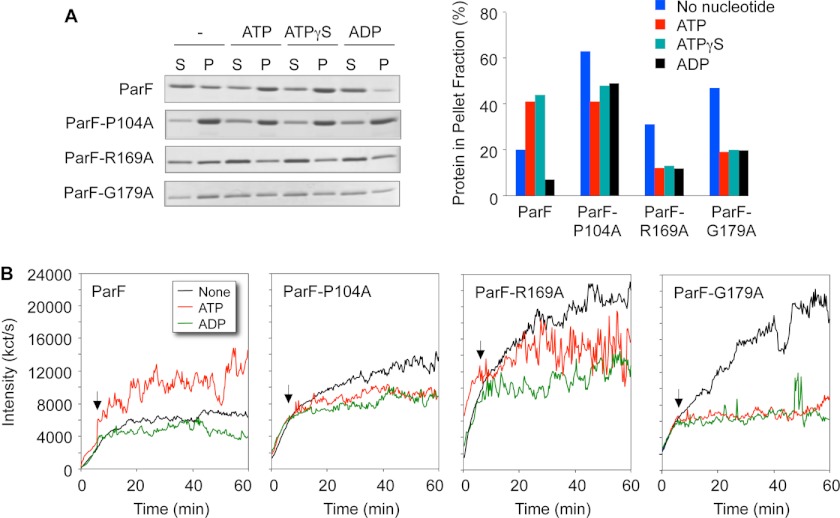
Polymerization of ParF is stimulated by ATP and the nonhydrolyzable analog ATPγS, whereas low level, intrinsic autopolymerization is blocked by ADP (15). Sedimentation assays demonstrated that ParFH proteins have increased autopolymerization tendencies compared with wild-type protein: without added nucleotide, higher proportions of mutated proteins than wild-type ParF were detected in pellet fractions (compare blue bars in Fig. 5A). Similarly, whereas autopolymerization of ParF reached a plateau after approximately 20 min in dynamic light scattering experiments, ParFH proteins continued to polymerize throughout the 60-min duration of the tests (Fig. 5B).
FIGURE 5.
Polymerization kinetics of wild-type ParF and ParFH. A, sedimentation assays in which proteins (4–8 μm) were incubated in the absence (−) or presence (2 mm) of nucleotides for 10 min at 30 °C, and the reactions were then centrifuged. In all, 100 and 33%, respectively, of the pellet (P) and supernatant (S) fractions were resolved on a 12% SDS gel and stained with Coomassie Blue. The percentages of ParF proteins detected in the pellet fractions are shown. B, polymerization of wild-type and mutated ParF proteins monitored by dynamic light scattering. Proteins (2 μm) were preincubated at 30 °C for 5 min, at which time ATP or ADP (500 μm) and MgCl2 (5 mm) were added (arrows). The reactions were followed for a further 55 min. The data in both panels are representative examples of experiments performed at least in duplicate with standard deviations ± 10%.
As noted in previous sedimentation assays (15, 19, 20), the mass of wild-type polymeric ParF entering the pellet fraction increased when incubated with ATP (red bars) or ATPγS (green bars), but autopolymerization was inhibited in the presence of ADP (black bars in Fig. 5A). Significantly, all three nucleotides blocked polymerization of ParFH. Each of the three proteins was inhibited similarly by ADP, ATP, and ATPγS, although the relative decreases in filamentation varied between the three mutated proteins. For example, the mass of polymeric ParF-G179A in the pellet fraction with each of the nucleotides was approximately half that observed without added nucleotide. Analogous patterns were observed in dynamic light scattering experiments. Thus, the addition of ATP entirely prevented polymerization of ParF-G179A as effectively as the addition of ADP (Fig. 5B). The data reveal that the nucleotide-binding pocket alterations in ParFH proteins provoke enhanced autopolymerization compared with wild-type ParF. However, the surge in ParF polymerization upon addition of ATP is severely dampened in ParFH proteins.
The unique tryptophan (Trp-47) in ParF buttresses residues in the nucleotide-binding site close to the P-loop motif (Fig. 2E). Intrinsic fluorescence of Trp-47 was used to report alterations that occur in the conformation of the surrounding environment upon nucleotide binding by wild-type protein and hyperactive ATPase variants. A significant increase in tryptophan fluorescence anisotropy occurred with higher wild-type ParF concentrations in the presence of MANT-ATP or MANT-ATPγS compared with reactions without nucleotide (Fig. 3C). Thus, both hydrolyzable and nonhydrolyzable ATP analogs induce conformational changes in ParF that correlate with nucleotide-induced polymerization of the protein (15). The data for ParF-P104A typify similar results obtained with the three hyperactive ATPase variants (Fig. 3D). In contrast with wild-type protein, a hyperbolic pattern of intrinsic tryptophan fluorescence anisotropy was evident with increasing ParF-P104A concentrations without added nucleotide. Because fluorescence anisotropy increases proportionally to the correlation time or molecular weight of the macromolecule, the anisotropy changes observed with increasing ParF-P104A concentration reflect the polymerization state of the protein. Thus, it is noteworthy that ParF-P104A autopolymerizes in the absence of nucleotide, whereas the wild-type ParF protein does not. However, the fluorescence anisotropy changes noted for ParF-P104A without nucleotide were attenuated when the protein was incubated with MANT-ATP or MANT-ATPγS (Fig. 3D). These data accord with the negative effect that nucleotides exert on polymerization (Fig. 5). Nucleotide binding may lock the mutant protein into a configuration that is not proficient for polymerization, thereby diminishing the intrinsic tryptophan fluorescence increases observed without nucleotide.
ParG Stabilizes Filaments Formed by ParFH
ParG potentiates ParF polymerization both in the absence and presence of exogenous ATP (15, 19). The interaction of the proteins during polymerization is evident in sedimentation assays: more ParF polymers are bundled in the presence of ParG than in its absence, either in reactions with or without added ATP. In parallel, ParG alone remains in the supernatant fraction, but a significant proportion enters the pellet when coincubated with ParF (supplemental Fig. S3A) (15). Because ParG failed to stimulate ATP hydrolysis by ParFH proteins (Fig. 4B) but sustained interactions with the proteins in two-hybrid analysis (supplemental Fig. S2A), the effect of ParG on polymerization was investigated in sedimentation assays. Experiments were performed with and without ATPγS. Without nucleotide, the levels of ParFH proteins entering the pellets when coincubated with ParG were at least equivalent to amounts found in the absence of the partner protein. For instance, approximately equal quantities of ParF-G179A pelleted when the protein was coincubated with (supplemental Fig. S3A) or without (Fig. 5A) ParG. Thus, ParG does not grossly affect the autopolymerization properties of ParFH.
As described above, ATPγS markedly reduced the concentrations of ParFH that pelleted in sedimentation assays compared with reactions without nucleotide (Fig. 5A). Strikingly, this effect was counteracted when both ParG and ATPγS were included in reactions: the levels of ParFH in pellet fractions were very similar to quantities observed for equivalent reactions without nucleotide (supplemental Fig. S3A). Thus, although ATPγS suppresses autopolymerization of ParFH, ParG overrides this effect. These data further reinforce the separable roles that ParG and ATP play in ParF filamentation dynamics.
Defective Segrosome Assembly by Hyperactive ATPase Mutants of ParF
Binding of ParG to the parH centromere yields a single nucleoprotein complex at saturating protein concentrations in gel retardation assays. ParF alone does not bind the site, but the ParG-parH complex is supershifted into a more slowly migrating species when ParF is added to reactions (28). Substitution of ParFH for wild-type ParF in binding assays diminished the quantities of supershifted complexes (supplemental Fig. S3B). Notably, very little segrosome formation was apparent in the case of ParF-G179A.
DISCUSSION
Genome segregation in bacteria is mediated most commonly by filamenting proteins of the ParA superfamily. ATP binding promotes ParA polymer extension from the segrosome assembled at the centromere site. A cycle of polymerization and depolymerization of ParA filaments is thought to move attached DNA molecules to opposite sides of the septal plane prior to cytokinesis. The nucleoid may function as a substratum during plasmid segregation. However, major gaps exist in understanding the kinetics of ATP binding and hydrolysis by ParA proteins, the role that centromere-binding factors play in stimulating nucleotide hydrolysis and controlling ParA polymerization, and the mechanism by which these processes are integrated to guarantee accurate segregation. Among P-loop ATPases, including the ParA superfamily, the Walker A motif forms a flexible segment that interacts with and positions the phosphate groups of the nucleotide, thereby making it susceptible to hydrolysis. A conserved aspartate residue in the B box is necessary for coordination of the essential Mg2+ ion co-factor, which in turn orients the nucleotide β- and γ-phosphates. Positioning of the two phosphate-binding motifs is ensured by hydrogen bonding between threonine/serine and aspartate residues of the A and B motifs, respectively (41, 42). ATP binding orchestrated by these motifs promotes polymerization of the ParF protein, which is crucial for DNA segregation (15, 29). Nucleotide binding is predicted to induce conformational changes in the ParF tertiary structure that prime the protein for polymerization (29). Here, we have established not only that the integrity of the nucleotide-binding site in ParF relies on the canonical ATP motifs but also that a triplet of conserved residues is key to maintaining the architecture of the pocket. These residues are dispersed around the catalytic niche: Pro-104 is part of a proline-rich patch near to the Walker A motif, Arg-169 forms the upper surface of the pocket, and Gly-179 sustains residue Trp-47 that in turn buttresses amino acids within the site (Fig. 2). Mutations at these positions attenuated ATP binding but, remarkably, converted ParF to a hyperactive ATPase (see summary in supplemental Table S1). We speculate that the mutations “relax” the binding pocket and encourage more promiscuous substrate access. Easier ingress of nucleotide to the pocket is predicted to offset the reduced ATP binding affinity and account for the elevated rates of hydrolysis.
The participation of an arginine residue in phosphoryl transfer during nucleotide hydrolysis is considered paradigmatic. In the case of the ParF ATPase, the ParG mobile N-terminal tail provides the requisite arginine in trans (19). Modulation of the nucleotide-binding pocket configuration by mutation of the Pro-104–Arg-169–Gly-179 triplet explains why the arginine finger-like motif in ParG no longer is proficient in stimulating ATP hydrolysis by ParF: reconfiguration of the binding pocket conformation prevents the precise alignment of the arginine finger-like motif supplied by ParG. Elucidation of the tertiary structures of apo- and nucleotide-bound ParF variants will provide intriguing insights into the molecular basis underpinning conversion of an intrinsically weak ATPase to a hyperactive version. It is feasible that this switch mimics an alternative nucleotide hydrolysis chemistry not involving an arginine finger that has been described recently (43).
Although ParFH proteins are hyperactive ATPases, they do not display identical properties. This is expected in view of the diverse contributions of the conserved residues to nucleotide-binding pocket architecture. First, the kinetics of ATP hydrolysis by ParF-P104A are noticeably different compared with those of other mutant proteins (Fig. 1C), confirming that the conserved triplet influences the integrity of the pocket differently. Second, segrosome assembly in vitro is impaired most severely by the G179A alteration (supplemental Fig. S3B). Because ParF assembles into the complex by interactions with ParG (28), the defect in ParF-G179A may reflect the modest reduction in interaction with ParG observed in two-hybrid assays (supplemental Fig. S2). Conversely, the ParF-R169A interaction with ParG is detectably stronger in two-hybrid analysis, but nevertheless the protein also is impaired in segrosome assembly. The strength of the ParF-ParG interaction may be fine-tuned for optimal complex formation: either weakening or enhancing this interaction may disrupt stable segrosome assembly. Third, whereas the P104A and R179A mutations induce segregation defects that are detectable in a standard partition assay, the low copy number vector bearing the segregation cassette that produces ParF-G179A is not transformable in host cells. The protein may be toxic in this context. Although the basis for ParF-G179A toxicity remains to be defined, it may stem from ParG-mediated stabilization of filaments formed by the protein (supplemental Fig. S3A). These polymers could sufficiently perturb plasmid replication at low copy number that selection of the plasmid following transformation is prevented. Alternatively, ParF-G179A polymers may interact with and inhibit a critical, unknown cellular target.
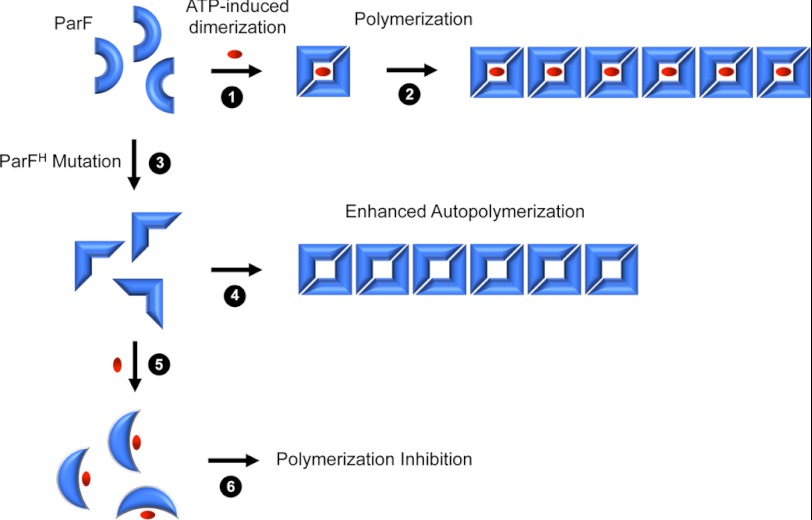
ATP binding is necessary and sufficient to induce polymerization of ParF above the intrinsic levels of polymerization that occur in the absence of nucleotide. Instead, ADP inhibits autopolymerization (15). Disruption of the nucleotide-binding pocket resulting in increased ATP hydrolysis exerted intriguing effects on ParF filamentation (Fig. 6 and supplemental Table S1). First, the structural changes enhanced autopolymerization of the protein. The altered configuration of the binding pocket may partially mimic the conformation of the site in the ATP-bound state thereby predisposing ParF to higher levels of inherent polymerization. ParF complexed with a nonhydrolyzable ATP analog forms dimers that assemble into the building blocks for polymerization (29). Interestingly, the R169A change also modestly enhanced ParF self-association in a two-hybrid assay (supplemental Fig. S2B). Second, whereas ATP binding stimulates polymerization of wild-type ParF, it suppressed filamentation when the structure of the binding pocket was modified. Here, the faster rate of ATP hydrolysis may produce a pool of ADP-bound protein that very effectively inhibits polymerization. However, nonhydrolyzable ATPγS blocked polymerization of the mutant proteins as efficiently as both ADP and ATP: nucleotide binding alone may trap the pocket in a configuration that is ineffective in relaying the conformational changes in the ParF tertiary structure that prime the protein for polymerization (Fig. 6).
FIGURE 6.
Nucleotide binding elicits different polymerization responses in wild-type ParF and ParFH. Steps 1 and 2, ATP (red oval) induces ParF dimerization (step 1 and Ref. 29) that primes the protein for polymerization (step 2 and Ref. 15). Step 3, ParFH mutations mimic the effect of ATP binding (step 3), so that the protein is more prone to autopolymerization (step 4). Steps 5 and 6, in contrast, nucleotide binding by ParFH causes a conformational change (step 5) that locks the protein into a configuration that is refractory to polymerization (step 6).
The stimulatory effects of ATP and ParG on polymerization of wild-type ParF are independent but additive. Thus, ParF bearing Walker A box mutations that ablate nucleotide binding or hydrolysis with consequent perturbations in filamentation remain sensitive to ParG-mediated stimulation of polymerization (15). These observations are reinforced by findings here that, despite the suppression of nucleotide-mediated polymerization in ParFH proteins, ParG promoted their polymerization as effectively as it promoted filamentation of wild-type ParF (supplemental Fig. S3A). Thus, although nucleotide binding may lock the mutant proteins into a configuration that is unproductive for ATP-induced polymerization (Fig. 6), ParG overrides this defect. The ParF-ParG co-structure has yet to be determined. However, in addition to the interaction of the arginine-finger like motif with the ParF nucleotide-binding pocket, the ParG N-terminal flexible tail makes contacts with ParF that independently promote polymerization (19). These contacts apparently are maintained in ParFH mutants. In conclusion, the conserved Pro-104–Arg-169–Gly-179 triplet is vital for maintaining the correct architecture of the nucleotide-binding pocket in ParF. Modifying the configuration of the site profoundly impacts ATPase kinetics, the capacity of the arginine finger-like motif to stimulate hydrolysis in trans, and ParF polymerization dynamics that drive the segregation process.
Supplementary Material
Acknowledgment
We thank Younghoon Lee (Korea Advanced Institute of Science and Technology) for supplying strain KS3001.
This work was supported, in whole or in part, by Biotechnology and Biological Sciences Research Council Grant G003114 (to F. H.). This work was also supported by Medical Research Council Grant G0801162 (to D. B.) and by National Institutes of Health Grant GM074815, an M. D. Anderson Trust Fellowship, and a Burroughs Wellcome Career Development Award (to M. A. S.).

This article contains supplemental Table S1 and Figs. S1–S3.
- AMPPCP
- adenosine 5′-(β,γ-methylene)-triphosphate
- MANT
- 2′(3′)-O-(N-methylanthraniloyl)
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
REFERENCES
- 1. Bloom K., Joglekar A. (2010) Towards building a chromosome segregation machine. Nature 463, 446–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Garner E. C., Campbell C. S., Weibel D. B., Mullins R. D. (2007) Reconstitution of DNA segregation driven by assembly of a prokaryotic actin homolog. Science 315, 1270–1274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Larsen R. A., Cusumano C., Fujioka A., Lim-Fong G., Patterson P., Pogliano J. (2007) Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 21, 1340–1352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Anand S. P., Akhtar P., Tinsley E., Watkins S. C., Khan S. A. (2008) GTP-dependent polymerization of the tubulin-like RepX replication protein encoded by the pXO1 plasmid of Bacillus anthracis. Mol. Microbiol. 67, 881–890 [DOI] [PubMed] [Google Scholar]
- 5. Ni L., Xu W., Kumaraswami M., Schumacher M. A. (2010) Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc. Natl. Acad. Sci. U.S.A. 107, 11763–11768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Drew K. R., Pogliano J. (2011) Dynamic instability-driven centering/segregating mechanism in bacteria. Proc. Natl. Acad. Sci. U.S.A. 108, 11075–11080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hayes F., Barillà D. (2006) The bacterial segrosome. A dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 4, 133–143 [DOI] [PubMed] [Google Scholar]
- 8. Hayes F., Barillà D. (2006) Assembling the bacterial segrosome. Trends Biochem. Sci. 31, 247–250 [DOI] [PubMed] [Google Scholar]
- 9. Schumacher M. A. (2007) Structural biology of plasmid segregation proteins. Curr. Opin. Struct. Biol. 17, 103–109 [DOI] [PubMed] [Google Scholar]
- 10. Schumacher M. A. (2008) Structural biology of plasmid partition. Uncovering the molecular mechanisms of DNA segregation. Biochem. J. 412, 1–18 [DOI] [PubMed] [Google Scholar]
- 11. Hayes F., Barillà D. (2010) Extrachromosomal components of the nucleoid. Recent developments in deciphering the molecular basis of plasmid segregation, in Bacterial Chromatin, (Dorman C. J., Dame R. T., eds.) pp. 49–70, Springer Publishing, Dordrecht, The Netherlands [Google Scholar]
- 12. Gerdes K., Howard M., Szardenings F. (2010) Pushing and pulling in prokaryotic DNA segregation. Cell 141, 927–942 [DOI] [PubMed] [Google Scholar]
- 13. Schumacher M. A. (2012) Bacterial plasmid partition machinery. A minimalist approach to survival. Curr. Opin. Struct. Biol. 22, 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Possoz C., Junier I., Espeli O. (2012) Bacterial chromosome segregation. Front. Biosci. 17, 1020–1034 [DOI] [PubMed] [Google Scholar]
- 15. Barillà D., Rosenberg M. F., Nobbmann U., Hayes F. (2005) Bacterial DNA segregation dynamics mediated by the polymerizing protein ParF. EMBO J. 24, 1453–1464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Lim G. E., Derman A. I., Pogliano J. (2005) Bacterial DNA segregation by dynamic SopA polymers. Proc. Natl. Acad. Sci. U.S.A. 102, 17658–17663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Adachi S., Hori K., Hiraga S. (2006) Subcellular positioning of F plasmid mediated by dynamic localization of SopA and SopB. J. Mol. Biol. 356, 850–863 [DOI] [PubMed] [Google Scholar]
- 18. Fogel M. A., Waldor M. K. (2006) A dynamic, mitotic-like mechanism for bacterial chromosome segregation. Genes Dev. 20, 3269–3282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Barillà D., Carmelo E., Hayes F. (2007) The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. Proc. Natl. Acad. Sci. U.S.A. 104, 1811–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Machón C., Fothergill T. J., Barillà D., Hayes F. (2007) Promiscuous stimulation of ParF protein polymerization by heterogeneous centromere binding factors. J. Mol. Biol. 374, 1–8 [DOI] [PubMed] [Google Scholar]
- 21. Bouet J. Y., Ah-Seng Y., Benmeradi N., Lane D. (2007) Polymerization of SopA partition ATPase. Regulation by DNA binding and SopB. Mol. Microbiol. 63, 468–481 [DOI] [PubMed] [Google Scholar]
- 22. Pratto F., Cicek A., Weihofen W. A., Lurz R., Saenger W., Alonso J. C. (2008) Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation. Nucleic Acids Res. 36, 3676–3689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ringgaard S., van Zon J., Howard M., Gerdes K. (2009) Movement and equipositioning of plasmids by ParA filament disassembly. Proc. Natl. Acad. Sci. U.S.A. 106, 19369–19374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ptacin J. L., Lee S. F., Garner E. C., Toro E., Eckart M., Comolli L. R., Moerner W. E., Shapiro L. (2010) A spindle-like apparatus guides bacterial chromosome segregation. Nat. Cell Biol. 12, 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castaing J. P., Bouet J. Y., Lane D. (2008) F plasmid partition depends on interaction of SopA with non-specific DNA. Mol. Microbiol. 70, 1000–1011 [DOI] [PubMed] [Google Scholar]
- 26. Havey J. C., Vecchiarelli A. G., Funnell B. E. (2012) ATP-regulated interactions between P1 ParA, ParB and non-specific DNA that are stabilized by the plasmid partition site, parS. Nucleic Acids Res. 40, 801–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barillà D., Hayes F. (2003) Architecture of the ParF-ParG protein complex involved in procaryotic DNA segregation. Mol. Microbiol. 49, 487–499 [DOI] [PubMed] [Google Scholar]
- 28. Wu M., Zampini M., Bussiek M., Hoischen C., Diekmann S., Hayes F. (2011) Segrosome assembly at the pliable parH centromere. Nucleic Acids Res. 39, 5082–5097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Schumacher M. A., Ye Q., Barge M. T., Zampini M., Barillà D., Hayes F. (2012) Structural mechanism of ATP induced polymerization of the partition factor ParF. Implications for DNA segregation. J. Biol. Chem. 287, 26146–26154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hayes F. (2000) The partition system of multidrug resistance plasmid TP228 includes a novel protein that epitomizes an evolutionarily-distinct subgroup of the ParA superfamily. Mol. Microbiol. 37, 528–541 [DOI] [PubMed] [Google Scholar]
- 31. Woodcock D. M., Crowther P. J., Doherty J., Jefferson S., DeCruz E., Noyer-Weidner M., Smith S. S., Michael M. Z., Graham M. W. (1989) Quantitative evaluation of Escherichia coli host strains for tolerance to cytosine methylation in plasmid and phage recombinants. Nucleic Acids Res. 17, 3469–3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hayes F. (1998) A family of stability determinants in pathogenic bacteria. J. Bacteriol. 180, 6415–6418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ludtke D. N., Eichorn B. G., Austin S. J. (1989) Plasmid-partition functions of the P7 prophage. J. Mol. Biol. 209, 393–406 [DOI] [PubMed] [Google Scholar]
- 34. Kim K. S., Sim S., Ko J. H., Lee Y. (2005) Processing of m1 RNA at the 3′ end protects its primary transcript from degradation. J. Biol. Chem. 280, 34667–34674 [DOI] [PubMed] [Google Scholar]
- 35. Karimova G., Pidoux J., Ullmann A., Ladant D. (1998) A bacterial two-hybrid system based on a reconstituted signal transduction pathway. Proc. Natl. Acad. Sci. U.S.A. 95, 5752–5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Carmelo E., Barillà D., Golovanov A. P., Lian L. Y., Derome A., Hayes F. (2005) The unstructured N-terminal tail of ParG modulates assembly of a quaternary nucleoprotein complex in transcription repression. J. Biol. Chem. 280, 28683–28691 [DOI] [PubMed] [Google Scholar]
- 37. Motallebi-Veshareh M., Rouch D. A., Thomas C. M. (1990) A family of ATPases involved in active partitioning of diverse bacterial plasmids. Mol. Microbiol. 4, 1455–1463 [DOI] [PubMed] [Google Scholar]
- 38. Koonin E. V. (1993) A superfamily of ATPases with diverse functions containing either classical or deviant ATP-binding motif. J. Mol. Biol. 229, 1165–1174 [DOI] [PubMed] [Google Scholar]
- 39. He L., Söderbom F., Wagner E. G., Binnie U., Binns N., Masters M. (1993) PcnB is required for the rapid degradation of RNAI, the antisense RNA that controls the copy number of ColE1-related plasmids. Mol. Microbiol. 9, 1131–1142 [DOI] [PubMed] [Google Scholar]
- 40. Golovanov A. P., Barillà D., Golovanova M., Hayes F., Lian L. Y. (2003) ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol. Microbiol. 50, 1141–1153 [DOI] [PubMed] [Google Scholar]
- 41. Walker J. E., Saraste M., Runswick M. J., Gay N. J. (1982) Distantly related sequences in the α- and β-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Saraste M., Sibbald P. R., Wittinghofer A. (1990) The P-loop. A common motif in ATP- and GTP-binding proteins. Trends Biochem. Sci. 15, 430–434 [DOI] [PubMed] [Google Scholar]
- 43. Scrima A., Thomas C., Deaconescu D., Wittinghofer A. (2008) The Rap-RapGAP complex. GTP hydrolysis without catalytic glutamine and arginine residues. EMBO J. 27, 1145–1153 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.